Abstract
Both homeologous exchanges and homeologous expression bias are generally found in most allopolyploid species. Whether homeologous exchanges and homeologous expression bias differ between repeated allopolyploid speciation events from the same progenitor species remains unknown. Here, we detected a third independent and recent allotetraploid origin for the model grass Brachypodium hybridum. Our homeologous exchange with replacement analyses indicated the absence of significant homeologous exchanges in any of the three types of wild allotetraploids, supporting the integrity of their progenitor subgenomes and the immediate creation of the amphidiploids. Further homeologous expression bias tests did not uncover significant subgenomic dominance in different tissues and conditions of the allotetraploids. This suggests a balanced expression of homeologs under similar or dissimilar ecological conditions in their natural habitats. We observed that the density of transposons around genes was not associated with the initial establishment of subgenome dominance; rather, this feature is inherited from the progenitor genome. We found that drought response genes were highly induced in the two subgenomes, likely contributing to the local adaptation of this species to arid habitats in the third allotetraploid event. These findings provide evidence for the consistency of subgenomic stability of parental genomes across multiple allopolyploidization events that led to the same species at different periods. Our study emphasizes the importance of selecting closely related progenitor species genomes to accurately assess homeologous exchange with replacement in allopolyploids, thereby avoiding the detection of false homeologous exchanges when using less related progenitor species genomes.
Keywords: allopolyploids, comparative genomics, drought response, ecological adaptation, homeologous exchanges, homeologous expression bias, subgenome dominance, recurrent origins
Introduction
Allopolyploidy plays a significant role in driving plant speciation and has had a profound impact on the genomes and phenomes of most angiosperms (Soltis et al. 2009; Van de Peer et al. 2017). All modern diploid flowering plants are considered downsized paleopolyploids, as they have returned to the diploid state by eliminating redundant sequences (Doyle et al. 2008; Jiao et al. 2011; Soltis et al. 2016). Some small genome diploids, like Brachypodium, have experienced several rounds of polyploidization and diploidization throughout their history (Salse et al. 2008). Recent meso- and neopolyploids in Brachypodium and other grasses have emerged through new hybridizations and whole genome duplication (WGD) events (Stebbins 1985; Soltis and Soltis 2021; Sancho et al. 2022), a process which is still ongoing today. Allopolyploid speciation appears to occur rapidly due to the effective reproductive isolation of the allopolyploid from its progenitor species (Te Beest et al. 2012; Oxelman et al. 2017). In fact, biological competition, niche competition, or better adaptation to new environments could lead to the rapid displacement of parental populations by successful hybrid allopolyploids (Leitch and Leitch 2008). Polyploidy is widely recognized as a significant source of genomic and evolutionary novelty, contributing to adaptive speciation (Soltis et al. 2014; Van de Peer et al. 2017; Baduel et al. 2018; Nieto Feliner et al. 2020). However, to truly understand the impact of polyploidy on evolutionary innovation and the formation of new species, it is essential to analyze it within a comprehensive pangenomic framework that takes into account both the progenitor species and the resulting allopolyploids. This analysis assists in revealing the genomic diversity of allopolyploid species, which is a combination of the diversities inherited from their progenitor genomes and those acquired through hybridization, polyploidization, and diploidization with evolutionary novelty (Gordon et al. 2020; Scarlett et al. 2022).
One of the immediate purported consequences attributed to allopolyploidy is the need for genomic and regulatory changes to accommodate the combination of different genomes within a single nucleus (Edger et al. 2019). These alterations aim to stabilize the mixed nuclear environment, and as a result, homeologous exchanges (HEs), which involve chromosomal recombinations between homeologs, have been observed in both recently synthesized and natural plant allopolyploids (e.g. Tragopogon, Gossypium, Brassica, Fragaria, Cucumis) (Chester et al. 2012; Chalhoub et al. 2014; Guo et al. 2014; Edger et al. 2019; Yu et al. 2021; Deb et al. 2023). HEs are believed to be responsible for generating evolutionary novelty, phenotypes, and contributing to speciation (Mason and Wendel 2020; Deb et al. 2023). However, other synthetic or natural allopolyploids, including Arabidopsis, Capsella, Trifolium, and Eragrostis, do not exhibit evidence of homeologous recombinations (Douglas et al. 2015; Griffiths et al. 2019; VanBuren et al. 2020; Burns et al. 2021). Another consequence often associated with allopolyploidy is subgenome dominance or homeolog expression bias (HEB), which is considered a mechanism that facilitates genome stabilization by resolving (epi)genetic conflicts in the allopolyploid (Edger et al. 2019). Subgenome dominance correlates with HEs when HE involves replacement (deletion/duplication) (Deb et al. 2023). When the dominant subgenome contains a greater number of gene copies and highly expressed homeologs compared to the submissive subgenome, it also leads to subgenome dominance (Alger and Edger 2020). Subgenome dominance can be established immediately after WGD or gradually over evolutionary time (Flagel and Wendel 2010; Edger et al. 2017). It is believed that subgenome dominance has contributed to the adaptive success of the diploidized allopolyploid, with a number of evidences from different angiosperm groups (Alger and Edger 2020; Deb et al. 2023). However, some allopolyploids do not exhibit significant HEB (Scarlett et al. 2022), which contradicts the expected pattern. The recently proposed “genome chimera” model offers a new perspective to reconcile these conflicting patterns within a comprehensive framework. It suggests that subgenome dominance, particularly HEB, simply reflects the differences in genomic background in terms of gene expression and regulation (e.g. transcription efficiency) between the two progenitor genomes, rather than being an inherent evolutionary trait of neo-allotetraploids (Zhang et al. 2023).
Meiosis-regulating genes play a crucial role in the cytogenetic mechanisms that influence plant response to hybridization and WGD, as they are responsible for ensuring homologous pairing and stabilizing nascent allopolyploids (Jenczewski and Alix 2004). In Triticum and Brassica allopolyploids, candidate meiotic genes that prevent meiotic crossovers between homeologs have been identified, thereby avoiding mis-segregation, aneuploidy, and gamete infertility (Martín et al. 2018; Higgins et al. 2021). Notably, Ph1 and Ph2-type genes have shown major qualitative effects, particularly in allopolyploid wheat (Martín et al. 2021; Serra et al. 2021). The establishment of meiotic stability after allopolyploidization has occurred gradually over generations in certain angiosperm groups, possibly due to selection for particular allele combinations that confer higher fertility (Cifuentes et al. 2010; Mason and Wendel 2020), while in other recent polyploids meiotic stability might have been acquired immediately (Jenczewski et al. 2002). The rapid stability of allopolyploids may depend on the divergence of progenitors (Ramsey and Schemske 2002), progenitor genotypes (Steige and Slotte 2016), and the epigenetic effects of transposable elements (TEs; Vicient and Casacuberta 2017; Wendel et al. 2018). Although the structural and functional impact of subgenomes on the adaptive success of new allopolyploid phenotypes in specific ecological niches has been investigated in several plant polyploids (van de Peer et al. 2021), it has not been fully assessed within a multiple-origin evolutionary scenario of the same polyploid species.
Brachypodium hybridum has emerged as a model system for stable allopolyploidy in grasses, specifically immediate amphidiploidy after WGD (Catalán et al. 2012; Scholthof et al. 2018; Gordon et al. 2020; Hasterok et al. 2022; Scarlett et al. 2022). Previous phylogenetic and cytogenetic studies have demonstrated that B. hybridum (2n = 4x = 30, x = 10 + 5), an allotetraploid grass, originated recurrently and bidirectionally from two extant diploid progenitor species B. stacei (2n = 2x = 20, x = 10) and B. distachyon (2n = 2x = 10, x = 5) (Catalán et al. 2012; Lopez-Alvarez et al. 2012; Scholthof et al. 2018). The native Mediterranean niche of B. hybridum overlaps with those of its progenitor species, with higher overlap observed with the more mesic B. distachyon niche in the northern part of the Mediterranean range and with the more aridic B. stacei niche in the southern part of the range (López-Alvarez et al. 2015). Genome-wide analyses of B. hybridum and its progenitor species have dated the recurrent origins of the allotetraploid to two different geological times in the Western Mediterranean region, approximately 1.4 million years ago (Ma) for the ancestral D-plastotypes (D-anc) (maternal parent B. distachyon) and 0.14 Ma for the recent S-plastotypes (Western-Med) (maternal parent B. stacei) (Gordon et al. 2020; Hasterok et al. 2022; Scarlett et al. 2022). B. hybridum also served as control for testing the accuracy of phylogenetic subgenomic detection algorithms, as it is the only polyploid species of the genus with known extant diploid progenitor genomes while all the others have one or more “ghost” subgenomes (Sancho et al. 2022). Both allopolyploid hybrids have displayed high subgenomic integrity with respect to the progenitor species genomes, although the D-anc has shown evidence of post-WGD structural change and slight but significant biased subgenomic gene loss (Scarlett et al. 2022). However, it is important to note that these results were based on comparative genomics and phylogenomics using incomplete Illumina-based reference genomes of B. hybridum S-recent plastotype (West-Med), B. stacei, and B. distachyon, a more complete PacBio-based reference genome of B. hybridum D-anc plastotype, and low resequenced genomes of several other B. hybridum and B. distachyon accessions (Gordon et al. 2020; Scarlett et al. 2022). Additionally, analyzing the B. hybridum D-anc genome presents a challenge due to the considerable divergence of its BhS and BhD subgenomes compared to their respective B. stacei and B. distachyon progenitor lineages, as the two subgenomic lineages split well before the recent radiations of the current B. stacei and B. distachyon clades (Gordon et al. 2020).
Here, we conducted a comprehensive analysis of this allopolyploid model system by employing comparative genomics and evolutionary analyses. We extensively studied a wide range of B. hybridum accessions from the relatively unexplored Eastern Mediterranean region, as well as other Mediterranean localities. Our research involved the generation of a complete PacBio-based reference genome for an accession from the East-Med region, along with several population-level resequenced genomes. To ensure accurate assessments, we selected the most closely related progenitor genomes from the B. stacei (Gordon et al. 2020; Mu et al. 2023) and B. distachyon (Gordon et al. 2017) pangenomes. Our analysis revealed a novel origin for a B. hybridum S-plastotype in the Eastern Mediterranean (East-Med) region, specifically in Evolution Canyon I (ECI), and other localities of Israel. These populations emerged from distinct local ancestors and are distributed within a more arid latitudinal belt compared to the previously known ancestral and recent B. hybridum populations in the west (supplementary fig. S1 and supplementary table S1, Supplementary Material online). ECI provides an ideal geographic setting for investigating microscale evolutionary models, as it encompasses two contrasting slopes: the xeric African south-facing slope with ecotypes adapted to drought and heat stresses and the forested European north-facing slope with ecotypes adapted to more mesic habitats and less stressful conditions (Nevo 2012). Utilizing the B. hybridum grass complex as a model system, we examined the extent of HEs, HEB, and the meiotic regulation mechanisms that contribute to the rapid stability of the allotetraploids within a time-course evolutionary scenario involving three recurrent origins of B. hybridum in different geographical and ecological contexts. Furthermore, we uncovered the functional contributions of the diploid progenitor subgenomes to the adaptive success of the newly discovered B. hybridum allotetraploid plants to the arid conditions of the African slope (AS) in ECI, Israel.
Results
Genomic Landscape of Brachypodium hybridum ECI
Combining PacBio HiFi and Hi-C sequencing technologies, we generated the de novo assembled Brachypodium hybridum ECI (Bhyb-ECI) reference genome (supplementary fig. S2 and supplementary table S2, Supplementary Material online). The final genome assembly captured 527.87 Mb base pairs with 15 pseudo-chromosomes (supplementary fig. S2B and supplementary tables S2A and B, Supplementary Material online). The genome size of Bhyb-ECI is similar to those of the reference B. hybridum S-plastotype (Bhyb-ABR113) genome (530.5 Mb) and the reference B. hybridum D-plastotype (Bhyb-26) genome (528.49 Mb) (Joint Genome Institute [JGI]; Phytozome, https://genome.jgi.doe.gov/). The de novo assembly of the Bhyb-ECI reference genome showed a high contiguity, the contig N50 was 18.70 Mb, and most chromosomes were composed of fewer than five contigs and with few gaps (supplementary fig. S2C and supplementary table S2D, Supplementary Material online). The base call accuracy (QualityValue [QV]) and assembly completeness were 38.79% and 99.38% with >99% and >98.5% mapping and coverage rates according to Illumina paired-end read mapping, respectively (supplementary table S2E, Supplementary Material online). Almost complete sets of Benchmarking sets of Universal Single-Copy Orthologs (BUSCO) genes (98.8%) were detected in the Bhyb-ECI assembly (supplementary fig. S2D, Supplementary Material online). In addition, the LTR Assembly Index (LAI) indicated a high long-terminal repeat (LTR) retrotransposon completeness (supplementary fig. S2C, Supplementary Material online), showing values higher than those reported for Bhyb-ABR113 (v1.1), but similar to those of Bhyb-26. Together, these metrics demonstrate high consistency and completeness of the new genome assembly, as well as greatly improved contiguity and repetitive sequence completeness.
We predicted 72,685 high-confidence protein coding genes (supplementary table S3A, Supplementary Material online). The average coding sequence region length, exon length, and exon number are similar to those of other representative genomes of the B. hybridum, B. stacei, and B. distachyon species (supplementary fig. S3A, Supplementary Material online). More than 96% of the predicted genes of the Bhyb-ECI genome had homologs in public functional databases (supplementary table S3B, Supplementary Material online). BUSCO analysis in the Bhyb-ECI genome indicated that 99.8% complete genes were present in our gene model predictions (supplementary fig. S2D, Supplementary Material online), indicating the high completeness of the gene model annotation. Most BUSCO genes (98.8%) were retained in duplicate, corresponding to homeologs present in both subgenomes of the allotetraploid. A total of 3,764 genes in the Bhyb-ECI genome were classified as putative transcription factors, belonging to 67 gene families and representing 5.1% of the total predicted genes (supplementary table S3C, Supplementary Material online). Moreover, 219.62 Mb of repetitive element sequences accounted for 41.60% of the total Bhyb-ECI genome (supplementary table S3D, Supplementary Material online); these figures were similar to those detected in previous B. hybridum assemblies (Bhyb-ABR113, 38.13%, Gordon et al. 2020; Bhyb-26, 41.51%, Scarlett et al. 2022).
We partitioned the Bhyb-ECI genome into two subgenomes based on the collinearity of its sequence to the genomes of the respective progenitor species (Fig. 1). The B. distachyon-like subgenome (BhD) consisted of 5 chromosomes and 38,330 genes (275.83 Mb), while the B. stacei-like subgenome (BhS) comprised 10 chromosomes and 33,766 genes (252.04 Mb) (Fig. 1A and supplementary tables S2D and S3A, Supplementary Material online). The sizes of the subgenome and the numbers of genes in Bhyb-ECI BhD and BhS were consistent with those of the respective progenitor species genomes (B. stacei Bsta-ECI and Bsta-ABR114, and B. distachyon Bdis-Bd21 and Bdis-Bd1-1, which were approximately 250 Mb and 275 Mb, respectively). The high-quality assembly of our Bhyb-ECI reference genome at the chromosome level, along with its comprehensive annotation (Fig. 1A), establishes a solid foundation for investigating the evolutionary mechanisms that underlie the recurrent speciation events of allotetraploid B. hybridum in diverse spatio-temporal scenarios and ecological contexts.
Fig. 1.
Genome organization and evolutionary landscape of the newly sequenced Brachypodium hybridum Bhyb-ECI reference genome. A) Overview of B. hybridum Bhyb-ECI genome assembly/synteny with progenitor species genomes B. stacei Bsta-ECI and B. distachyon Bdis-Bd1-1. The tracks indicate: (a) chromosomes, (b) GC contents, (c) TE densities, (d) gene models' densities, and (e) collinearity of syntenic genes. B) Conserved synteny between the two progenitor species' genomes and the respective B. hybridum (Bhyb-ECI, Bhyb-ABR113, and Bhyb-26) BhD and BhS subgenomes showing highly structural divergence of chromosomes from the two subgenomes, consistent with the high divergence between chromosomes of the two corresponding progenitors' genomes.
Evolutionary Trajectories of Ancestral and Recent B. hybridum Allotetraploids Across Multiple Genomic Dimensions
To analyze recurrent allopolyploidizations of B. hybridum within an evolutionary framework, we enlarged our search to the BhD and BhS subgenomes of the other available B. hybridum allotetraploids (Bhyb-ABR113 and Bhyb-26), and to other B. distachyon (Bdis-Bd21 and Bdis-Bd1-1) and B. stacei (Bsta-ABR114 and Bsta-ECI) progenitor species genomes. Genome collinearity analysis detected high synteny between the genomic profiles of the Bhyb-ECI BhD and BhS subgenomes and their corresponding Brachypodium progenitor species' genomes and other allotetraploids' subgenomes (Fig. 1B). Next, we performed ortholog-clustering analysis using all genes of the studied Brachypodium genomes and the O. sativa genome (supplementary table S4, Supplementary Material online). This pangenome-level search rendered a total of 15,042 core ortholog gene clusters (OGCs) present in all genomes, 17,301 in all Brachypodium genomes, 19,745 in all S (B. stacei-type) (sub-)genomes and 19,420 in all D (B. distachyon-type) (sub-)genomes (supplementary fig. S3B, Supplementary Material online). In contrast, all ten Brachypodium (sub-)genomes had very few (sub-)genomes-specific dispensable OGCs (supplementary fig. S3B, Supplementary Material online) and genes compared with common OGCs (supplementary fig. S3C, Supplementary Material online). These results supported the conservative genome landscape at gene level among B. hybridum, B. distachyon, and B. stacei. Furthermore, a total of 12,481 single-copy ortho-homeologous genes (SCOGs) identified in Brachypodium and Oryza (supplementary fig. S3C, Supplementary Material online) were used to investigate the origins of eastern Mediterranean Bhyb-ECI and the other B. hybridum lineages through time-measured phylogenomic analysis.
A dated Bayesian chronogram (Fig. 2A) based on the 12,481 SCOGs and a secondary calibration for the Brachypodium/Oryza split inferred different divergence ages between the B. distachyon and B. stacei progenitor species lineages and their respective Bhd and BhS child lineages for each allotetraploid. The splits for the more ancestral D-anc Bhyb-26 and the more recently evolved West-Med Bhyb-ABR113 subgenomic lineages were estimated to have occurred 1.87–1.76 Ma and 1.21(0.69)−0.48 Ma, respectively, while the inferences for the East-Med Bhyb-ECI subgenomic lineages indicated relatively recent splits of 0.48–0.30 Ma (Fig. 2A). These age estimates were slightly older than those based on the distributions of non-zero synonymous substitutions per synonymous site (Ks) (Gaut et al. 1996) of ortho-homeologous genes in intergenomic syntenic blocks (Bhyb-ECID vs Bdis-Bd1-1, Kspeak ∼ 0.0035, t = 0.27 Ma; Bhyb-ECIS vs Bsta-ECI, Kspeak ∼ 0.0031, t = 0.24 Ma, μ = 6.5 × 10−9) (supplementary fig. S4, Supplementary Material online). We also performed pairwise-count of gene pairs that had no synonymous substitutions constructing two quadruple ortho-homeologous gene chains of D-(sub)genomes (Bhyb-ECID, Bhyb-ABR113D, Bdis-Bd1-1, Bdis-Bd21) and S-(sub)genomes (Bhyb-ECIS, Bhyb-ABR113S, Bsta-EC, Bsta-ABR114) based on ortho-homeologous genes in each genome set to test the closeness of genomic/subgenomic relationships. The D-genome-type contained 24,531 gene chains and S-genome-type 25,010; the pairwise-count tests indicated that Bhyb-ECID and Bhyb-ECIS have significantly more zero ks gene pairs with, respectively, Bdis-Bd1-1 and Bsta-ECI than with Bdis-Bd21 and Bsta-ABR114 (supplementary fig. S4, Supplementary Material online). These results supported the closeness of the Bhyb-ECI BhS and BhD subgenomes to the local East-Med Bsta-ECI and Bdis-Bd1-1 genomes, a finding consistent with the phylogenetic evidence (Fig. 2A).
Fig. 2.
Phylogenomics, cross-bracing, and evolutionary model analyses. Phylogenomic analysis based on 12,481 nuclear SCOGs of Brachypodium stacei (Bsta-ECI and Bsta-ABR114), B. distachyon (Bdis-Bd1-1 and Bdis-Bd21), and B. hybridum (nuclear BhD and BhS subgenomes) of the eastern Mediterranean Bhyb-ECI and western Mediterranean Bhyb-ABR113 and Bhyb-26 allotetraploids, and O. sativa (see supplementary table S3, Supplementary Material online for more information). A) Dated Bayesian tree of all representative Brachypodium species genomes and subgenomes. Estimated nodal ages are indicated in black; O. sativa was used to root the tree. B) Inferred cross-bracing ages for the respective formations of the ancient (D-anc Bhyb-26) and recent Western Mediterranean (West-Med Bhyb-ABR113) and Eastern Mediterranean (East-Med Bhyb-ECI) B. hybridum clades (see coalescent-based SNP and AFLP package for phylogenetic analysis (SNAPP) tree computed from 30,000 filtered SCOG SNPs and used as baseline for the cross-bracing estimates, supplementary fig. S5, Supplementary Material online). All branches showed strong posterior probability support (1.0) in both topologies. C) Posterior probabilities of the four compared evolutionary scenarios on the alternative origins of Brachypodium hybridum allotetraploids estimated separately for the D and the S genomes/subgenomes through approximated Bayesian computation and RF methods. Scenario 1: three independent origins of ancestral (Bhyb-26), recent West-Mediterranean (Bhyb-ABR113), and very recent East-Mediterranean (Bhyb-ECI) allotetraploids; scenario 2: two independent origins of ancestral (Bhyb-26) and recent West-Mediterranean (Bhyb-ABR113) allotetraploids, followed by introgression of Bhyb-ABR113 with East-Mediterranean progenitors; scenario 3: single ancestral origins for the three allotetraploids; scenario 4: single recent origin for the three allotetraploids. Four thousand data sets per scenario were simulated for the training set and a forest of 2,000 random trees was generated for model choice. Posterior probabilities are indicated within parentheses for each scenario. Scenario 1 showed the highest posterior probability for both D and S genomes/subgenomes, supporting the three independent origins of B. hybridum allotetraploids, each from different progenitors.
A coalescence-based dating analysis performed with 30,000 filtered single nucleotide polymorphism (SNP) data from the SCOGs data set inferred similar divergence times for the splits of the ancestral Bhyb-26 subgenomic lineages (2.33 and 1.98 Ma) but more recent ages for those of Bhyb-ABR113 (0.89 and 0.55 Ma) and especially Bhyb-ECI (0.14 and 0.13 Ma) (supplementary fig. S5, Supplementary Material online). As the B. hybridum BhS and BhD splits corresponding to the same hybridization event showed nearly the same age for the ancestral event and close ages for the recent events, we used the cross-bracing approach that forces the split times of each parental genome to be contemporaneous. We obtained single-age estimates for the respective origins of D-anc Bhyb-26, which formed 2.16 Ma (95% Highest Posterior Density [HPD] 2.54–1.78), and of recent West-Med Bhyb-ABR113 (0.72 Ma; 95% HPD 0.88–0.56), that are relatively consistent though older than previous estimates (Gordon et al. 2020), and a very recent origin for the newly studied East-Med Bhyb-ECI (0.13 Ma; 95% HPD 0.24–0.02) (Fig. 2B). To further assess the maternal ancestor of the newly studied East-Med B. hybridum lineage we performed phylogenomic analysis based on whole plastome sequences of several B. hybridum, B. distachyon and B. stacei accessions. The plastome topology revealed that Bhyb-ECI as well as other individuals of this population and Israel showed a strongly supported B. stacei-type maternally inherited plastome, representing thus a new East-Med S-plastotype lineage that originated independently from the West-Med S-plastotype lineage (supplementary fig. S6A, Supplementary Material online). The plastome tree also suggested that the B. hybridum ECI plastomes were not inherited from local B. stacei ECI ancestors but from other eastern or western B. stacei sources, although the results were inconclusive due to low support of intraclade branches (supplementary fig. S6A, Supplementary Material online). Plastome lengths were longer and less variable in S-plastotypes than in D-plastotypes (136,289–136,333 bp for B. stacei and S-plastotype allotetraploids, and 134,991–135,423 bp for B. distachyon and D-plastotype allotetraploids) (supplementary fig. S6B, Supplementary Material online), and fell within the expected length ranges indicated previously (Sancho et al. 2018; Gordon et al. 2020).
To rule out possible introgressions of recent eastward-dispersed West-Med B. hybridum with eastern Mediterranean progenitor species that could falsify the putative third independent origin of the very recent East-Med B. hybridum, we tested alternative evolutionary scenarios for different origins of B. hybridum using filtered SCOG SNP data and coalescence approximated Bayesian computation and supervised machine learning methods. To test biologically meaningful scenarios, we split the full dataset into the D (sub)-genome and the S (sub)-genome subsets, considering that B. hybridum could backcross with one or the other progenitor species but not with both at the same time. We compared the same four alternative scenarios separately for the D and the S lineages with DIYABC-random forest (RF) (Fig. 2C and supplementary fig. S7 and supplementary table S5, Supplementary Material online). Scenario 1 (three independent origins of ancestral [Bhyb-26], recent West-Med [Bhyb-ABR113], and very recent East-Med [Bhyb-ECI] allotetraploids) was selected as the model with the highest number of votes and highest posterior probability over scenario 2 (two independent origins of ancestral and recent West-Med allotetraploids, followed by introgression of recent West-Med B. hybridum with eastern Mediterranean B. distachyon or B. stacei progenitors) and other less supported scenarios (scenario 3: single ancestral origins for the three allotetraploids; scenario 4: single recent origin for the three allotetraploids) (Fig. 2C and supplementary fig. S7 and supplementary table S5, Supplementary Material online).
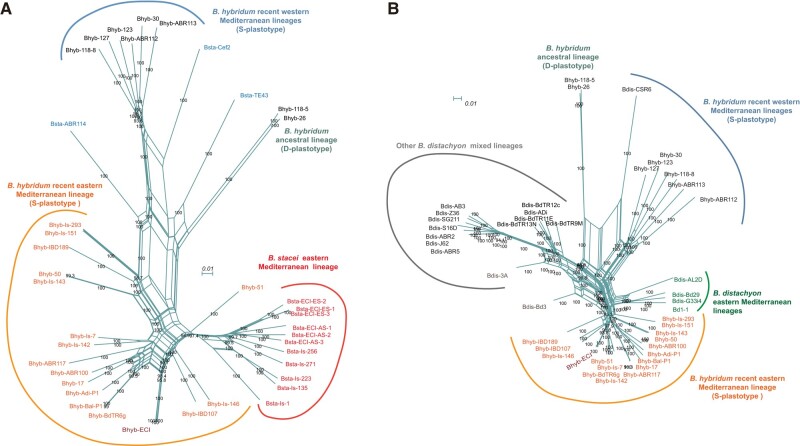
To improve our understanding of the origins of the B. hybridum ECI population and other populations in Israel, we performed a population-level genomic study of their individuals and of available samples of other B. hybridum populations and of progenitor species B. distachyon and B. stacei populations from the eastern, central and western parts of the Mediterranean region (supplementary fig. S1, Supplementary Material online). Independent S-genome (B. stacei and B. hybridum BhS) and D-genome (B. distachyon and B. hybridum BhD) neighbor-networks constructed from syntenic SNP data showed the close relationships of the B. hybridum ECI and Israel lineages to B. stacei and B. distachyon lineages from Israel and the East-Med region, respectively, and their divergences from B. hybridum and progenitor species lineages from the central and western Mediterranean region (Fig. 3 and supplementary fig. S8, Supplementary Material online). The S-genome network retrieved close links between B. hybridum ECI and other populations from Israel to the B. stacei populations from the same settings (Fig. 3A and supplementary table S1, Supplementary Material online). The D-genome network recovered close ties of the B. hybridum Israel populations to B. distachyon East-Mediterranean populations from near Israel (Fig. 3B and supplementary table S1, Supplementary Material online). S-genomic relationships from multivariate PCA also discriminated the B. hybridum S-plastotype (plus B. stacei) eastern and western Mediterranean populations along PCA1 while the B. hybridum D-plastotype populations separated along PCA2 (supplementary fig. S8A, Supplementary Material online). Within the very closely related and recent S genomes/subgenomes population structure analysis detected three optimal genomic groups (best K = 3) differentiating the eastern B. hybridum populations from eastern B. stacei populations and western B. stacei + B. hybridum populations (supplementary fig. S8B, Supplementary Material online). Some individuals from eastern B. hybridum populations showed signatures of admixture with the other two genomic groups. The genetic diversity (θπ) value of East-Med B. hybridum (5.21e−4) population group was lower than that of the West-Med B. hybridum (6.01e−4) group (supplementary fig. S8C, Supplementary Material online), a result consistent with the more recent allotetraploidy of the former group (Fig. 2A). The pair-wised fixation index (FST) between the three S-genome population groups showed significantly higher genomic divergence between the East-Med vs West-Med B. hybridum groups (0.528 and 0.547) than between East-Med B. stacei and East-Med B. hybridum (0.218) (supplementary fig. S8C, Supplementary Material online). These population scale results strongly support the in situ origin of the recently evolved East-Med B. hybridum lineage (S-plastotype) from local ancestors in this area (Fig. 3 and supplementary fig. S8, Supplementary Material online).
Fig. 3.
Population-level evolutionary analysis based on nuclear syntenic SNP data of B. hybridum (BhS subgenome) and its progenitor species B. stacei and of B. hybridum (BhD subgenome) and its progenitor species distachyon. Phylogenetic neighbor-networks of S genomes (B. stacei and B. hybridum BhS) (A) and of D genomes (B. distachyon and B. hybridum BhD) (B) computed with SplitsTree showing the respective divergence of B. hybridum and B. stacei and of B. hybridum and B. distachyon eastern Mediterranean populations from populations of these species from the western Mediterranean region. Only nodes with bootstrap >90 are shown.
Conservative Genomic Traits Between B. hybridum Subgenomes and Progenitor Genomes
To assess the genomic consequences after polyploidy, we performed comparative genomic analysis between Bhyb-ECI and its closest progenitor's genomes (Bsta-ECI and Bdis-Bd1-1). The GC contents of Bhyb-ECID and Bhyb-ECIS were 46.43% and 45.52%, respectively, which were similar than the GC content of the two progenitor species and other corresponding (sub-)genomes (46.42% and 45.43%, respectively) (supplementary fig. S9A, Supplementary Material online). Further analysis indicated that repetitive element content was greater in the two subgenomes of Bhyb-ECI than in the corresponding progenitor species' genomes, especially in regions near encoding genes (supplementary figs. S9B and C and supplementary table S6, Supplementary Material online), except for recent LTR Copia and Gypsy insertion burst detected in both subgenomes and in the corresponding progenitors' genomes (supplementary fig. S9D, Supplementary Material online). By comparing the whole Bhyb-ECI genome to the concatenated progenitor species genomes (Bsta-ECI and Bdis-Bd1-1), we found 486.58 Mb of collinear regions and 38.15 Mb of rearranged regions in the Bhyb-ECI subgenomes (supplementary fig. S9 and supplementary table S7, Supplementary Material online), consistent with the high syntenic conservation of the Bhyb-ABR113 genome detected previously (Gordon et al. 2020). These rearrangements included 12.82 Mb (n = 147) of inversions, 6.18 Mb (n = 1,539) of translocations, and 19.15 Mb (n = 8,444) of polymorphic duplications (supplementary fig. S10 and supplementary table S7, Supplementary Material online). At gene level, we found 27,171 orthologous gene pairs in Bhyb-ECID and Bdis-Bd1-1, 14,408 of which had no synonymous substitutions (Ks = 0, 53.02%), and 10,314 (37.95%) had identical sequences, whereas we detected 30,206, 17,780 (58.86%), and 13,372 (44.26%) gene pairs in similar Bhyb-ECIS and Bsta-ECI comparisons (supplementary table S8, Supplementary Material online). When we scrutinized the other allotetraploids, we found 27,873, 14,649 (52.55%), and 10,550 (37.85%) orthologous genes pairs in Bhyb-ABR113S and Bsta-ABR114, 30,371, 9,952 (32.7%), and 5,744 (18.91%) in Bhyb-ABR113D and Bdis-Bd21, 21,633, 1,279 (5.91%), and 393 (1.81%) in Bhyb-26S and Bsta-ABR114, and 22,741, 1,550 (6.81%), and 451 (1.98%) in Bhyb-26D and Bdis-Bd21 comparisons (supplementary table S8, Supplementary Material online).
Low Occurrence of HEs and Similar Expression of Meiotic Genes in BhD and BhS Subgenomes of B. hybridum Allotetraploids
To encompass various spatio-temporal scenarios involving multiple origins of B. hybridum, we constructed two types of progenitor genomes: Ref-East (concatenated Bsta-ECI and Bdis-Bd1-1) and Ref-West (concatenated Bsta-ABR114 and Bdis-Bd21) (see supplementary fig. S11 and supplementary table S9, Supplementary Material online). When aligning B. hybridum samples from the eastern Mediterranean to the merged progenitor genomes (Ref-East and Ref-West), we observed a significantly lower number of outlier blocks with the Ref-East genome (supplementary fig. S11A and supplementary table S9D, Supplementary Material online). The two ancestral D-plastotype B. hybridum samples exhibited a similar and substantial number of putative deletion and duplication blocks when mapped to either the Ref-East or Ref-West merged genomes (supplementary fig. S11B and supplementary table S9D, Supplementary Material online). These findings highlight the crucial influence of selecting an appropriate reference progenitor genome for accurately identifying HEs (supplementary figs. S11A and C, Supplementary Material online). Consequently, we performed our analysis on an additional 21 East-Med B. hybridum samples using Ref-East genomes and 6 West-Med B. hybridum samples using Ref-West genomes to ensure reliable results. Only five reliable “HE with replacement” swaps were identified in the eastern Mediterranean B. hybridum samples when mapped to their closest Ref-East genomes (supplementary table S9A, Supplementary Material online); one of these swaps occurred in Bhyb-ECI and other 11 samples (supplementary fig. S11A and supplementary table S9A, Supplementary Material online). In contrast, four “HE with replacement” swaps were identified in western Mediterranean B. hybridum samples and two ancestral (D-anc) B. hybridum samples when using Ref-W as reference genome (supplementary tables S9B and C, Supplementary Material online), likely due to under-sampling of close progenitor genomes. Notably, no “reciprocal HE” events were found in any of the three B. hybridum lineages. Our analysis revealed that HE events were infrequent in Bhyb-ECI and the other studied allotetraploids (supplementary fig. S11 and supplementary table S9, Supplementary Material online), suggesting that the two duplicated subgenomes maintain stable bivalent chromosome pairing during meiosis.
Interestingly, we observed geographical variations in the depth patterns of outlier regions between the BhS subgenomes of western and eastern Mediterranean B. hybridum accessions and their respective B. stacei progenitor genomes. Specifically, the Bhyb-ABR113S subgenome exhibited a similar pattern of deletion regions with the Bsta-ABR114 genome when using Ref-East as the reference genome. However, this pattern differed (with the absence of deletions) when comparing the same chromosomal region in the Bsta-ECIS subgenome to its Bsta-ECI progenitor genome using the same reference genome (supplementary fig. S11D, Supplementary Material online). These results confirm the highly conserved genomic structure between the progenitor genomes and their respective descendant BhS subgenomes, further supporting the distinct evolutionary origins of the recent East-Med and West-Med S-plastotypes B. hybridum lineages.
The comparative analysis of the functional expression of meiotic Ph1 and Ph2 orthologs in the subgenomes of the studied B. hybridum allotetraploids revealed a balanced expression of Ph2-type genes from the BhD and BhS subgenomes in the leaf tissues of the D-anc Bhyb-26 allotetraploid (52.511 vs 44.134) and similarly low but equivalent values in the roots (2.7035 vs 2.6857) (supplementary table S10, Supplementary Material online). On the other hand, the expression of Ph1-type genes from both subgenomes exhibited only residual but similar values in these tissues. In the recent West-Med Bhyb-ABR113 allotetraploid the expression levels in BhD and BhS were low and equivalent for Ph1-type (1.8761 vs 1.4200) and Ph2-type (1.9465 vs 1.2292) genes in spikelet tissues and nearby absent in leaves. In the recent East-Med Bhyb-ECI allotetraploid, the overall expressions were generally low but balanced between the two subgenomes (Ph2-type genes in roots: 0.6940 vs 0.3946; Ph1-type genes in leaves: 0.5016 vs 0.1214, and roots: 0.2961 vs 0.5315) (supplementary table S10, Supplementary Material online).
Lack of HEB and Evidence That Gene Expression Changes Contributed to Ecological Adaptations of B. hybridum Allotetraploids
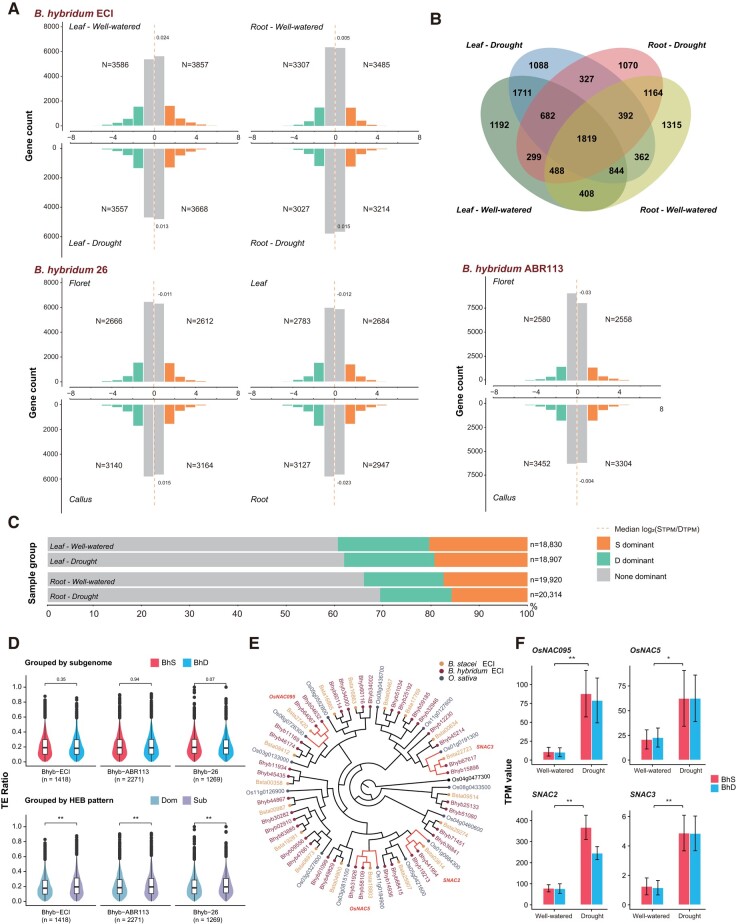
To investigate subgenome dominance patterns in Bhyb-ECI of B. hybridum and to compare it with its other allotetraploids (Bhyb-ABR113, Bhyb-26), we analyzed gene expression in leaf and root tissues of Bhyb-ECI samples under well-watered (control) and drought conditions (Fig. 4 and supplementary table S11, Supplementary Material online). We also examined gene expression in callus and floret tissues of Bhyb-ABR113 and in callus, floret, leaf and root tissues of Bhyb-26 under control conditions (Gordon et al. 2020; Scarlett et al. 2022). In contrast to many allotetraploid species that exhibit subgenome dominance, our pairwise comparisons showed that Log2(BhSTPM/BhDTPM) values were close to zero in all groups analyzed (Fig. 4A and supplementary table S11, Supplementary Material online). These results suggest that there was no apparent HEB between BhD and BhS homeologs in different tissues across the three ancestral and recent B. hybridum allopolyploid lineages (Fig. 4A), nor under any growth condition in Bhyb-ECI (Figs. 4B and C). At the gene level, we conducted a more comprehensive analysis of Bhyb-ECI, focusing on 1,819 out of 23,313 syntenic homeologous gene pairs between BhD and BhS that exhibited HEB in all groups studied (leaf-well watered, leaf-drought, root-well watered, and root-drought) (Fig. 4B). Among them, 1,418 gene pairs maintained consistent expression bias patterns across all four case study groups and did not show any subgenome dominance (supplementary table S12, Supplementary Material online).
Fig. 4.
Unbiased homeologous gene expression in Brachypodium hybridum. A) Histograms showing the distribution of HEB of genes in the BhD and BhS subgenomes of B. hybridum Bhyb-ECI (leaf and root tissues of samples grown under well-watered vs drought conditions), B. hybridum Bhyb-ABR113 (callus and floret tissues of plants grown under control conditions), and B. hybridum Bhyb-26 (callus, floret, leaf, and root tissues of plants grown under control conditions). Log2 fold-changing (BhSTPM/BhDTPM) indicates the degree of HEB. Homeologous gene pairs were classified as biased toward BhS or BhD dominance. B) B. hybridum Bhyb-ECI: overlap of homeologous gene pairs that exhibit HEB between different tissue and treatment groups (leaf-well watered; leaf-drought; root-well watered; root-drought). C) Numbers and percentages of HEB genes from the four sample groups across the B. hybridum Bhyb-ECI gene expression atlas. D) Comparisons of TE density in genes in the homologous gene pair showed the same expression bias patterns in all case study groups in Bhyb-ECI, Bhyb-ABR113, and Bhyb-26, respectively. Grouped by subgenome (BhD and BhS) and grouped by expression bias type (Dom: dominant, Sub: submissive), **P-value < 0.01 (paired Wilcoxon test). E) Phylogenetic relationships and orthology prediction of abiotic stress-related NAC genes in Bhyb-ECI and Bsta-ECI using O. sativa as an outgroup. F) Significant differential gene expression of BhD and BhS homeologs of four representative NAC transcription factors in individuals grown under well-watered vs drought conditions. Values shown are TPM; *P-value < 0.05, **P-value < 0.01 (Wilcoxon test).
This unbiased baseline provided an opportunity to investigate the cause of HEB, which is indicative of subgenome dominance and diploidization, as well as a potential constraint for stronger purifying selection (Cheng et al. 2018). Initially, we divided these consistently biased gene pairs into two subgenomic groups, BhD and BhS, and assessed the selection pressure by comparing the non-synonymous and synonymous substitution rates (Ka, Ks) of ortho-homeologous gene pairs with those of the corresponding progenitor species genomes. We found no evidence of significant differences (P = 0.53) between genes from the BhD and BhS subgenomes (supplementary fig. S12A, Supplementary Material online). Likewise, when we divided them based on expression levels, there were no significant differences (P = 0.44) in Ka/Ks between dominant genes (DGs) and their corresponding submissive genes (SGs) (supplementary fig. S12B, Supplementary Material online), suggesting that selection pressure is not a direct cause of HEB. Consistent with previous reports in B. distachyon (Gordon et al. 2017) and other plants (Hollister and Gaut 2009), gene expression levels in Bhyb-ECI showed a negative correlation with the density of nearby TEs in the whole genome background (rPearson = −0.30, P = 2.2e−16) (supplementary fig. S13, Supplementary Material online). Investigation of the TE density near these 1,418 stable gene pairs with biased expression in Bhyb-ECI did not detect a significant difference (P = 0.35) between the two subgenomes (Fig. 4D). However, there was a significant difference (P = 3.341−7) between DGs and their corresponding SGs (Fig. 4D).
Interestingly, when we examined the density of TEs near the orthologous genes of these gene pairs within the respective progenitor genomes (Bdis-Bd1-1 and Bsta-ECI), we discovered significant differences not only when grouping them by genomes but also when grouping them based on expression bias type (supplementary fig. S14, Supplementary Material online). A similar pattern of stable expression bias was also observed in Bhyb-ABR113 (ngenepair = 2,541) and Bhyb-26 (ngenepair = 1,478) (Fig. 4D and supplementary fig. S14, Supplementary Material online), indicating that it is a common pattern following recurrent allotetraploidy in B. hybridum. Furthermore, we observed that the difference values were predominantly centered around 0 in both recent B. hybridum lineages when comparing TE density near orthologous gene pairs between the progenitor genomes and subgenomes (supplementary fig. S13, Supplementary Material online). The conservation of both sequence and nearby TE density in the genes of stable biased gene pairs suggests that this HEB pattern may have been inherited from the progenitor genomes rather than emerging solely as a result of the allotetraploidy event. These findings are in line with the recent “nuclear chimera” model (Zhang et al. 2023), which posits that the two subgenomes of an allotetraploid maintain their gene expression patterns and regulatory networks until chromosomal rearrangement events disrupt the balance.
To validate this hypothesis, we investigated whether HEB maintained or altered the expression patterns of genes under drought conditions. Surprisingly, we observed a higher number of unbiased gene pairs in the leaf and root tissues of plants subjected to drought compared to those grown under well-watered conditions (Fig. 4C). This unexpected result contradicts previous findings in allotetraploid Eragrostis (VanBuren et al. 2020). Only 162 (1.69%) and 310 (3.38%) gene pairs exhibited converse dominance patterns in leaves and roots, respectively, between plants in well-watered and drought conditions (supplementary table S13, Supplementary Material online). In leaves, 4,894 genes (50.92%) with HEB maintained the same dominance pattern under both conditions, and in roots, this was observed for 3,553 genes (38.65%). Our findings suggest that approximately half of the homeologous gene pairs underwent changes in dominance patterns when plants were exposed to drought stress (supplementary table S13, Supplementary Material online), indicating independent gene regulation dynamics of the two subgenomes in response to drought.
We conducted an analysis of stress response of transcription factors belonging to the NAC gene family (You et al. 2015) in Bhyb-ECI. In B. hybridum, we identified two homeologous copies of most NAC genes, whereas B. stacei and Oryza sativa had only single copies (Fig. 4E). Interestingly, the majority of these genes were expressed in both leaf and root tissues of plants grown under both well-watered and drought conditions (supplementary fig. S15, Supplementary Material online), indicating constitutive expression regardless of the growth conditions. Additionally, we examined the expression of four well-known drought response genes (ONAC095, OsNAC5, SNAC2, and SNAC3) in Bhyb-ECI. We found that all of these genes were strongly induced in response to drought stress in both BhD and BhS homeologs (Fig. 4F).
Discussion
Genomic Stability and Subtle Evolutionary Novelties of the Recurrently Originated B. hybridum Allopolyploids: Most Traits Were Probably Inherited From Parents
Our study has made significant progress in understanding the genomic evolutionary history of the recurrently originated B. hybridum allopolyploid model system, building upon and reconciling previous findings (Gordon et al. 2020; Hasterok et al. 2022; Scarlett et al. 2022). Through comparative genomic and phylogenomic analyses using more comprehensive reference genomes of the three allotetraploid hybrids and their closely related progenitor species, we have confirmed their independent origins (Figs. 2 and 3). This suggests that post-WGD diploidization likely occurred immediately in these three allotetraploids, as there is no evidence of significant genomic rearrangements or subgenome dominance. The survival of a “successful” allopolyploid involves overcoming genomic dis-homeostasis, such as changes in chromosome numbers, unstable meiotic pairing, potential reconfiguration of gene expression regulatory networks (Comai 2005; Nieto Feliner et al. 2020; Deb et al. 2023) and initial competition with progenitor species (Te Beest et al. 2012). Our analyses have uncovered a third spontaneous origin of B. hybridum in the eastern Mediterranean region (East-Med lineage; Figs. 2 and 3 and supplementary figs. S1 and S5, Supplementary Material online), expanding the evolutionary timeline for the recurrent formation of this allotetraploid (Gordon et al. 2020; Scarlett et al. 2022). Moreover, backcrossing the allotetraploid with any of its diploid progenitors is highly challenging due to differences in ploidy levels, chromosome numbers, and postzygotic barriers (Catalan et al. 2016; Sancho et al. 2022). Our coalescence-RF analysis unequivocally rules out potential introgressions and other less reliable single origins, thus confirming the three separate origins (Fig. 2C). These results align with previous and current observations that have not detected intermediate chromosome numbers between those of B. hybridum and its diploid progenitors (Catalan et al. 2016), and with the different karyotype structure of the B. stacei and B. distachyon chromosomes (Gordon et al. 2020). Consequently, the potential occurrence of crossovers between “homeologous” S and D chromosomes within the B. hybridum nucleus is highly unlikely.
Our tests also provide support for the immediate amphidiploidy of the inter-specific annual hybrids, which suggests that they could reproduce within the same year and establish themselves as new allotetraploids (Sancho et al. 2022). The geographically distant locations of the two relatively recent West-Med (0.72 Ma) and very recent East-Med (0.13 Ma) B. hybridum S-plastotype allopolyploidization events (B. stacei-type maternal parents; Fig. 2 and supplementary figs. S5 and S6, Supplementary Material online), along with their proximity to current western and eastern Mediterranean populations of B. stacei and B. distachyon progenitors (Fig. 3 and supplementary fig. S8, Supplementary Material online) support parallel in situ speciation events. These hybrids likely originated from different local ancestors but resulted in the same allotetraploid species. The extensive distributions of these lineages across the circum-Mediterranean region (supplementary fig. S1, Supplementary Material online) suggest a rapid and successful adaptation to their original niches (Te Beest et al. 2012) and quick colonization of new niches after long-distance-dispersals (Scholthof et al. 2018). In contrast, the ancestral D-anc B. hybridum lineage is restricted to a small area in Southern Spain (2.16 Ma, Fig. 2 and supplementary fig. S5, Supplementary Material online), which overlaps with the distribution of the West-Med lineage (supplementary fig. S1, Supplementary Material online) and is likely its place of origin. However, the ancestral lineage's formation from a B. distachyon-type maternal parent (D-plastotype) and its old age, estimated to have preceded the divergence of the current progenitor species lineages (e.g. B. distachyon, ∼1 Ma, Sancho et al. 2018; Gordon et al. 2020; B. stacei, ∼0.22 Ma; unpublished data), make it an “orphan” allotetraploid with potentially extinct progenitor parents. Nonetheless, it exhibits similar phenotypic features to the recent S-plastotype allotetraploids. The different age estimates obtained for the D-anc and West-Med B. hybridum lineages in Gordon et al. (2020) (1.4 and 0.14 Ma, respectively) and in the current study (2.16 and 0.72 Ma; Fig. 2 and supplementary fig. S5, Supplementary Material online) may be affected by differences in sampling size and the use of more conserved SNP data set in this study. However, the age estimate of the very recent East-Med lineage (0.13 Ma; Fig. 2 and supplementary fig. S5, Supplementary Material online) is a consequence of selecting appropriate close progenitor D (Bdis-Bd1-1) and S (Bsta-EC) genomes (Fig. 3 and supplementary fig. S8, Supplementary Material online), which are unknown for the D-anc lineage and not available for the West-Med B. hybridum lineage (Figs. 2 and 3 and supplementary figs. S5 and S8, Supplementary Material online).
Despite being considered an important aspect of the evolution and establishment of many nascent plant allopolyploids (Mason and Wendel 2020; Deb et al. 2023), our results highlight the lack of significant HEs in the formation of the three B. hybridum lineages. We identified very few reliable cases of “HEs with replacement” (supplementary fig. S11 and supplementary table S9, Supplementary Material online; Scarlett et al. 2022), and no “reciprocal HE” events in all of these lineages. The absence of “reciprocal HEs” is consistent with the fact that these wild allotetraploids are not recent early-generation allotetraploids but rather late-generation allotetraploids (Figs. 2A and B and supplementary fig. S5, Supplementary Material online). In this context, the original “reciprocal HEs” would have undergone mosaicization and transformed into “HEs with replacement” over their evolutionary history (Deb et al. 2023). In contrast to the findings of Scarlett et al. (2022), which detected more genome rearrangements in the ancestral Bhyb-26 genome than in the more recent West-Med Bhyb-ABR113 genome, our genome-wide HE analyses indicate the overall lack of HE events in any of the three B. hybridum lineages (supplementary fig. S11 and supplementary table S9, Supplementary Material online). Our HE data do not support the hypothesis of gradual polyploid genome evolution in B. hybridum (Scarlett et al. 2022), but rather suggest immediate amplidiploidization of each allotetraploid in each temporal scenario, possibly due to the distinct structural karyotypes of two parental subgenomes (Gordon et al. 2020). Meiotic Ph1 and Ph2 ortholog expression analyses also support these findings (supplementary table S10, Supplementary Material online), indicating stable bivalent formation in all types of hybrids. Collectively, our assessments indicate that no major structural diploidization rearrangements occurred after the three recurrent allopolyploidization events (Fig. 1B and supplementary figs. S9 and S10 and supplementary tables S7, S9, and S10, Supplementary Material online), while some TE mobilization around genes in the BhD subgenome (supplementary fig. S9, Supplementary Material online) indicates that Bhyb-ECI, like the other B. hybridum lineages (Bhyb-ABR113; Bhyb-26; supplementary fig. S9, Supplementary Material online; Gordon et al. 2020; Scarlett et al. 2022), may have experienced transposon proliferations following allotetraploidization.
Interestingly, our analyses have revealed the potential for inaccurate detection of HEs referred to as “false positive outlier blocks” when aligning the B. hybridum genomes to inappropriate reference progenitor genomes (supplementary fig. S11A, Supplementary Material online). This issue becomes more pronounced in the “orphan” ancestral B. hybridum allotetraploid, which exhibits the highest number of questionable duplications and deletions when mapped to either the Ref-West or Ref-East merged progenitor genomes (supplementary fig. S11B, Supplementary Material online). Hence, it is plausible that the significant gene losses and TE proliferations previously detected in the ancestral D-anc Bhyb-26 compared to the recent West-Med Bhyb-ABR113 (Scarlett et al. 2022) could be attributed not only to gradual polyploid evolution but also to the lack of appropriate ancestral progenitor genomes for comparative genomics. Similarly, ambiguous evidence of HEs may have been misidentified in other plant allopolyploids (Edger et al. 2020) when close progenitor genomes are unknown (Liston et al. 2020) and comprehensive pangenomic sampling of progenitor genomes and hybrids is absent (Sancho et al. 2022). In contrast, the observed geographical variation in HE patterns between the western and eastern Mediterranean B. hybridum BhS subgenomes and their respective B. stacei progenitor genomes (supplementary fig. S11 and supplementary table S9, Supplementary Material online) confirms a highly conserved genomic structure in the progenitor genomes and their descendant B. hybridum subgenomes, suggesting that most of their genomic characteristics were probably inherited from the parents. Only subtle evolutionary novelties, such as TE turnovers, appear to have been acquired in each lineage (supplementary fig. S11 and supplementary table S9, Supplementary Material online).
Subgenomic Background Rather Than HEB Drives the Adaptive Success of B. hybridum
In line with previous studies conducted on B. hybridum Bhyb-ABR113 (Gordon et al. 2020) and B. hybridum Bhyb-26 (Scarlett et al. 2022), the Bhyb-ECI genome does not exhibit any signs of subgenome dominance in both leaves and roots (Figs. 4A–C). However, it is worth noting that subgenome dominance does occur rapidly after the formation of allopolyploids, even after multiple generations (Yu et al. 2021). Contrary to the expectation that relaxed purifying selection may drive the diploidization process of the allopolyploid in a few generations or over time (Douglas et al. 2015; Deb et al. 2023), our observations indicate that selection pressure is not a direct cause of biased expression in a small group of genes (Fig. 4B). This holds true for both the comparison between BhS and BhD subgenomes and the comparison between dominant and submissive genes (supplementary fig. S12, Supplementary Material online). Furthermore, although our data indicate a difference in TE density surrounding these genes, which could be related to gene expression regulation and methylation (Lippman et al. 2004) and potentially lead to initial biased expression (Fig. 4D), further analysis within a progenitor-allotetraploid framework suggests that this difference between DGs and SGs may be inherited from the progenitors' genomes (supplementary fig. S14, Supplementary Material online) in Bhyb-ECI. This finding has also been confirmed in Bhyb-ABR113 and Bhyb-26 (Fig. 4D and supplementary fig. S13, Supplementary Material online) and may potentially be present in other plant genomes as well (Mason and Wendel 2020; Scarlett et al. 2022; Deb et al. 2023). By integrating our findings on the absence of biased expression in all three B. hybridum lineages and our analysis of stably biased expression gene pairs in an unbiased genomic background, we provide additional support for the “nuclear chimera” model (Zhang et al. 2023) from a different perspective.
The adaptive advantages of allopolyploids have been widely reported (Van de Peer et al. 2017). Previous studies have indicated that B. hybridum occupies a similar ecological niche to its two progenitor species within its native circum-Mediterranean range and has successfully established populations in other regions worldwide (Lopez-Alvarez et al. 2015; Catalan et al. 2016). However, while the distribution of B. hybridum overlaps with that of its progenitor B. distachyon species in higher latitudes and less arid regions of the western Mediterranean, it only coincides with the range of the warm-adapted B. stacei in lower latitudes and extremely arid conditions found in most of Israel (supplementary fig. S1, Supplementary Material online; Lopez-Alvarez et al. 2015; Penner et al. 2020). Interestingly, we discovered that drought stress can significantly alter the biased expression patterns in Bhyb-ECI (supplementary table S13, Supplementary Material online). Furthermore, the conserved subgenomic gene expression patterns of Bhyb-ECI suggest subtle gene loss, sub- or neo-functionalization, which would have maintained separate gene expression regulatory networks in the two subgenomes (Woodhouse et al. 2014). Our findings also suggest that there is an increased capacity for gene expression or regulation networks in response to drought stress (Fig. 4B), which could contribute to the environment adaptability of allotetraploid B. hybridum. Notably, the overexpression of drought-responsive NAC family genes in Bhyb-ECI under drought conditions, similar to those found in the arid habitat ECI-AS of Israel where B. distachyon is currently absent (Lopez-Alvarez et al. 2015; Mu et al. 2023), corresponds to BhD homeologs rather than to BhS homeologs (Fig. 4F and supplementary fig. S15, Supplementary Material online). These pieces of evidences suggest that a plausible drought tolerant B. distachyon ecotype, similar to those found in the East-Mediterranean region (Decena et al. 2021), may have been the potential parent of the current Bhyb-ECI and that its inherited genomic background, rather than biased expression, contributed to its adaptive success. The presence of multiple homeologous gene copies, generated through allotetraploidy and maintained throughout evolutionary history, may have enhanced the stress tolerance of B. hybridum in ECI, particularly in the dry AS environment and similar habitats throughout Israel and the Mediterranean region.
Materials and Methods
Brachypodium hybridum ECI Reference Genome
A new reference genome for B. hybridum from a sample collected at ECI (Bhyb-ECI; Lower Nahal Oren, Mount Carmel, Israel, 32°43′N; 34°58′E) was sequenced using a PacBio Sequel2 and Illumina HiSeq X Ten platform. In total 28.97 Gb of circular consensus sequencing reads and 29.95 Gb of clean pair-end were generated for Bhyb-ECI. For Hi-C libraries, DNA was extracted from fresh leaves, chromatin was fixed with formaldehyde in the nucleus, and cross-linked DNA was digested with DpnII. Approximately 166.76 Gb of Hi-C raw reads were yielded using an Illumina HiSeq X (supplementary table S2A, Supplementary Material online).
Genome sizes were estimated using K-mer methods. K-mers counting program (KMC; v3) (Kokot et al. 2017) and FindGSE (v1.94) R package (Sun et al. 2018) were employed to generate K-mer distributions and calculate genome size (supplementary fig. S2A, Supplementary Material online). HiFi reads of the Bhyb-ECI genome were assembled using NextDenovo (v2.5.0) (Hu et al. 2023). Low-quality Hi-C reads were filtered using Fastp (v0.20.0) (Chen et al. 2018). Juicer (v1.9.9) (Durand et al. 2016) and 3D-DNA (v201008) (Dudchenko et al. 2017) were employed to anchor contigs to the 15 chromosomes of Bhyb-ECI (supplementary table S2C, Supplementary Material online). Genome assembly quality and completeness was assessed using Merqury (v1.3) (Rhie et al. 2020) and BUSCO (v3.0.2) (Simão et al. 2015) with the embryophya_odb10 dataset. LAI values were generated with LTR-retriever (v2.9.0) (Ou and Jiang 2018). In addition, SAMtools (v1.1) (Li et al. 2009) and Bamdst (https://github.com/shiquan/bamdst, v1.0.9) were applied for calculating mapping depth and coverage. Contig NG (Bradnam et al. 2013) was estimated using customized scripts.
The repetitive DNA sequences (tandem repeats and TEs) of the Bhyb-ECI genome were identified with TandemRepeatsFinder (TRF; v4.09b) (Benson 1999), RepeatModeler (v2.0) (Price et al. 2005), LTR-retriever (v2.9.0) (Ou and Jiang 2018), RepeatMasker (v4.0.7) (Chen 2004), and RepeatProteinMasker (included in the RepeatMasker package) based on the published Repbase (v20181026) (Jurka et al. 2005). All results were merged to non-redundancy repeat sequence annotation, and genome sequence in repeat region masked using Bedtools (v2.29.1) (Quinlan and Hall 2010). Insertion times of the most abundant Gypsy and Copia retrotransposon families of the B. distachyon (Bdis-Bd21 and Bdis-Bd1-1) and B. stacei (Bsta-ABR114 and Bsta-ECI) genomes, and B. hybridum (Bhyb-ECI, Bhyb-ABR113, and Bhyb-26) BhD and BhS subgenomes were calculated using LTR-retriever with automatic default options and the rice's mutation rate (1.3e−8).
The gene prediction strategy involved the integration of multiple methods (ab initio, homology-based, and transcriptomic). Augustus (v3.2.3) (Stanke et al. 2006) was utilized for ab initio gene prediction. For homology-based prediction, we collected genome data from representative grass species, including B. distachyon (Bdis-Bd21), B. stacei (Bsta-ABR114, Bsta-ECI) B. hybridum (Bhyb-ABR113, Bhyb-26) and Oryza sativa (GCF_001433935.1), and employed GeMoMa (v1.6.4) (Keilwagen et al. 2019) to search for orthologous gene structures by mapping protein sequences of these species to Bhyb-ECI. Both alignment-based and de novo-based methods were employed for transcriptomic-based gene predictions. For the alignment-based approach, HISAT2 (v2.2.1) (Kim et al. 2019) was applied to map RNA-Seq data to the sequenced genome. StringTie (v2.1.6) (Pertea et al. 2015) and Transdecoder (https://github.com/TransDecoder/TransDecoder) were employed to generate the gene predictions based on information from the alignment. In the de novo-based method, we used Trinity (v2.9.1) (Haas et al. 2013) and the Program to Assemble Spliced Alignments (PASA)-pipeline (v2.4.1) (Haas et al. 2003) to assemble transcripts and obtain gene coding regions of genomes. All the gene models predicted through these approaches were integrated with EvidenceModeler (v1.1.1) (Haas et al. 2008) and this step generated a final gene feature file for functional annotation and downstream analysis.
BUSCO (v3.0.2) (Simão et al. 2015) with the embryophya_odb10 dataset was used as a measure of gene model completeness. The functional information of gene model predictions was annotated using InterproScan (v5.36) (Jones et al. 2014). Blast (diamond blastp) program was utilized to search for homeologous candidates with three public databases, Swiss-Prot (Bairoch and Apweiler 2000), RefSeq non-redundant data base (NR) (Pruitt et al. 2007), and TrEMBL. Gene Ontology (GO) term annotations integrated results from InterProScan, the EggONG (v3.5) (Huerta-Cepas et al. 2019) and Blast2GO pipeline (v6.0) (Conesa et al. 2005). Transcription factors within the gene models were identified using iTAK (web version) (Zheng et al. 2016). The subgenomes of allotetraploid B. hybridum Bhyb-ECI were identified merging the two progenitor species genomes (B. stacei Bsta-ECI and B. distachyon Bdis-Bd1-1) and performing genome synteny analysis using whole-genome duplication integrated analysis (WGDi; v0.6.1) (Sun et al. 2022). The genome collinearity between all Brachypodium genomes (Bhyb-ABR113, Bhyb-26, Bhyb-ECI, Bsta-ABR114, Bsta-ECI, Bdis-Bd21, Bdis-Bd1-1) was performed with JCVI library (Tang et al. 2008).
Phylogenomics, Cross-bracing Analysis and Testing of Alternative Origin Scenarios
For a more accurate inference of the evolutionary trajectory of the newly sequenced B. hybridum Bhyb-ECI genome, a phylogenetic framework was built containing: (i) the B. stacei reference genome Bsta-ABR114 and the new B. stacei Bsta-ECI genome generated from an individual collected in ECI (Mu et al. 2023); (ii) the B. distachyon reference genome Bdis-Bd21 and the potential closest B. distachyon genomes available within the pangenome data set of this second progenitor species (Bdis-Bd1-1); (iii) the two other reference genomes of B. hybridum (ancestral Bhyb-26 D-plastotype, recent Bhyb-ABR113 S-plastotype) (Gordon et al. 2020; Scarlett et al. 2022); and (iv) O. sativa used as out group. The phylogenetic scenario was constructed using their coding gene sequences. We obtained a high-quality, single-copy gene data set using OrthoMCL (v2.0.9) (Li et al. 2003), aligning the protein sequence of single-copy genes into a concatenated multiple sequence alignment (MSA) using MAFFT (v7.505) (Katoh and Toh 2008) and constructed a phylogenetic tree based on this MSA using Bayesian Evolutionary Analysis Sampling Trees (BEAST; v2.4.7) (General Time Reversible [GTR], gamma+ site heterogeneity, invariant sites, relaxed molecular clock, and Yule tree prior models, 10 million Mountain Chain Monte Carlo (MCMC) chain length, logging parameters every 500) (Bouckaert et al. 2014).
The homeologous gene chains (e.g. Bdis-Bd1-1-gene1, Bdis-Bd21-gene1, Bhyb-ECID-gene1, Bhyb-ABR113D-gene1, and Bhyb-26D-gene1) and their synonymous substitutions per synonymous site (Ks) were analyzed with WGDi (v0.6.1) (Sun et al. 2022) using the YN00 model. The divergence times between the two subgenomes of B. hybridum and those of its progenitor species for the three recurrent origins of the allotetraploid were estimated using the equation T = Ks/2r, where r stands for a divergence rate of 6.5 × 10−9 (Gaut et al. 1996).
We compared those clock-based estimations with coalescent-based estimations analyzed with SNAPP and a cross-bracing approach implemented by Gordon et al. (2020) to calculate the ages of the different origins of B. hybridum. We filtered the syntenic polymorphic positions of the concatenated genes for a reduced data set of all subgenomes of the three studied B. hybridum samples and selected clade members of B. distachyon and B. stacei, and run the SNAPP search using BEAST (v.2.4.7) (Bouckaert et al. 2014) imposing a normal prior distribution for a secondary age constrain at the Brachypodium crown node (11.6 ± 1.0 Ma), a 1/x distribution for clock rate and lambda (Yule model), and a uniform distribution for theta. The adequacy of parameters was checked using TRACER v.1.7 (Rambaut et al. 2018); most parameters showed effective sample size > 200, and a maximum clade credibility tree was computed after discarding 10% of the saved trees as burn-in. We followed the cross-bracing approach of McCann et al. (2018) adapted to SNAPP data by Gordon et al. (2020), using a cross-bracing normal distribution prior of 0 ± 0.02 (which enforces very low probability on trees that differ in node height and makes the age distributions of the cross-braced nodes become nearly congruent with respect to mean and shape) and running separate SNAPP searches for each allopolyploidization event (ancestral, recent West-Mediterranean, and recent East Mediterranean).
To test alternative evolutionary scenarios for different origins of B. hybridum we employed approximate Bayesian computation and supervised machine learning methods implemented in DIYABC-RF v.1.1.1-beta (Collin et al. 2021) for the separate Brachypodium D and S(sub)genomic filtered SCOG SNP data sets (7,780 and 40,199 SNP loci for the more and less divergent D and S genomic data sets, respectively). The DIYABC-RF approach enabled efficient discrimination among scenarios and estimation of the posterior probabilities. We compared four alternative scenarios for three (ancestral, recent, and very recent), two (ancestral and recent + introgression), single ancestral, and single recent origins of B. hybridum with the D and S datasets (Fig. 2C). For all scenarios, training sets were generated using 4,000 simulations per model, and prior distributions were uniform and set to default values. We identified the most likely scenario of each set using the RF module of DIYABC-RF, computing 2,000 random trees per model, as recommended in the manual, and the RF algorithm for model choice based on linear discriminant analysis.
Population Genomics
The genomes of another 6 B. hybridum samples from the ECI population plus 6 additional samples from other localities from Israel were sequenced using the Illumina HiSeq X Ten platform and other representative circum-Mediterranean regions. B. distachyon (21 samples), B. stacei (13 samples) and B. hybridum (18 samples) accessions collected from NCBI database (supplementary supplementary fig. S1 and supplementary table S1, Supplementary Material online) were employed for population genomics analysis (Gordon et al. 2020; Scarlett et al. 2022; Mu et al. 2023).
The plastomes of B. hybridum Bhyb-ECI and Israel samples and several other accessions were assembled using Getorganelle (v1.7.1) (Jin et al. 2020). Whole plastome sequences (including large single copy region [LSC], SSC, IRa, and IRb regions) of B. hybridum and its progenitor species were aligned with multiple alignment using Fast Fourier Transform (MAFFT; v7.505) (Katoh and Toh 2008) and used to construct a maximum-likelihood tree with IQTREE (v1.6.12) (Nguyen et al. 2015) for inferring the maternal origins of the allotetrapoids.
The pair-end reads of each sample (from newly sequenced data and from NCBI data) were first filtered with Fastp (v0.20.0) to trim low-quality reads, and mapped to their corresponding genomes (Bsta-ECI for B. stacei, Bdis-Bd21 for B. distachyon and Bhyb-ECI for B. hybridum) using BWA-MEM2 (v2.2.1) (Vasimuddin et al. 2019) to generate unbiased sequence alignment files, and then the second alignment was filtered and sorted with SAMtools (v1.1) (Li et al. 2009). SNP from population samples of each species were called using “HaplotypeCaller” and “GenotypeGVCF” module and filtered using “VariantFiltration” (QualByDepth [QD] < 2.0 || FisherStrand (FS) > 200.0 || ReadPosRankSum < −20.0 and QD < 2.0 || FS > 60.0 || RMSMappingQuality (MQ) < 40.0 || MQRankSum < −12.5 || ReadPosRankSum < −8.0) module of Genome Analysis Toolkit (GATK; v3.8.1) (Depristo et al. 2011). Extra SNPs filtering steps were performed using custom Perl script with the following criteria: (i) labeling SNPs with non-information (./.) which had low (<1/3 of average chromosome) or high (>3-fold chromosome average) mapping depth; (ii) labeling SNPs with non-information (./.) showing quality scores ( Genotype Quality [GQ]) of genotypes < 10. Homeologous regions of the B. stacei S genomes and B. hybridum BhS subgenomes, and of B. distachyon D genomes and B. hybridum BhD subgenomes, were detected with Minimap2 (Li 2018) and then transformed to chain file using transanno (https://github.com/informationsea/transanno). The SNPs' coordinates of B. stacei samples were converted to those of the B. hybridum S subgenome, and those of B. distachyon samples to B. hybridum D subgenome using Crossmap (https://github.com/liguowang/CrossMap, v0.6.5). The VariantCallFormat (VCF) files were transformed to Fast-All (FASTA) format using customized Perl script, filtering sites with <80% information. Phylogenetic neighbor-networks were constructed separately for the S and D genomes/subgenomes using SplitsTree4 (Huson and Bryant 2006). The ADMIXTURE (100 bootstraps) (v1.3.0) (Alexander et al. 2009) and “smartpca” packages in EIGENSOFT (v7.2.1) (Patterson et al. 2006) were employed for population structure and Pirncipal Component Analysis (PCA) analysis. The Fixation statistic (FST) and the nucleotide genetic diversities (θπ) were estimated using pixy (v1.2.7) (Korunes and Samuk 2021) with a 10 kb window size.
HE Analysis
HE regions between the BhD and BhS subgenomes were identified following Chalhoub et al. (2014), and the HE regions were mainly detected based on mapping depth and sequence collinearity. We first detected whether different reference genomes affected the identification of HEs using two combined progenitor genome types, represented as Ref-East for the concatenated Bsta-ECI + Bdis-Bd1-1 genomes, and Ref-West for the concatenated Bsta-ABR114 + Bdis-Bd21 genomes. The syntenic collinearity blocks between progenitor genomes (Bsta-ECI vs Bdis-Bd1-1, and Bsta-ABR114 vs Bdis-Bd21) were generated by linking aligned regions with distances less than 20 kb identified with Minimap2 (v2.21) (Li 2018).
Using mapping depth approach, we performed analyses of the HEs pattern in the B. hybridum allopolyploids at the population scale intending to cover all spatio-temporal scenarios for the multiple origins of B. hybridum. The paired-end reads of each sample were mapped to these concatenated references genomes using BWA-MEM2 (v2.2.1), filtering the secondary alignments and sorting them using SAMTools (v1.1) to generate the Binary Alignment/Map (BAM) files. Then, the mapping depth was calculated with non-overlapping 10 kb window using Bamdst. The window had an outlier depth (within the 1.5–5 fold and 0–0.5 fold ranges of the corresponding chromosome average depth for each sample) which was used as threshold to detect the potential duplications and deletions windows. Adjacent windows were linked together if they had the same mark type. The regions spanning more than six windows (60 kb) and located in the two progenitor species genomes' syntenic collinearity blocks were considered as putative HE regions and transformed into coordinates in the B. hybridum genomes (Bhyb-ECI, Bhyb-ABR113, and Bhyb-26). The few detected HEs corresponded to “HE with replacement” events (reciprocal duplications/deletions in the respective homeologous chromosomes), while no “reciprocal HE” events (syntenic swapped regions between the respective homeologous chromosomes) (Mason and Wendel 2020; Deb et al. 2023) were detected in any of the three late-generation allotetraploids.
Comparative Genome Analysis
The GC content, and the repetitive element content of total genome and near encoding genes were calculated through custom Perl scripts. The LTR insert time were estimated by LTR-retriever (v2.9.0) (Ou and Jiang 2018). The homeologous gene pair between subgenomes and corresponding progenitor genomes, and between the two subgenomes in three B. hybridum genomes were analyzed with WGDi (v0.6.1), and Ka and Ks values were obtained using WGDi (v0.6.1) with the YN00 model (v0.6.1) (Sun et al. 2022). The structural variations between subgenomes and corresponding progenitor genomes were identified by Nucmer (v4.0.0beta2) (Marçais et al. 2018) and SyRI (v1.6) (Goel et al. 2019). The Ph1 (Martín et al. 2021) and Ph2 (Serra et al. 2021) orthologs in B. hybridum genomes were identified on Basic Local Alignment Search Tool (BLAST) (blastp) searching homologous proteins against hexaploid bread wheat (Triticum aestivum L., AABBDD 2n = 6x = 42) (Ph1 wheat homologs were located in chromosomes 3A, 3B, and 3D).
Drought Stress Experiment, Transcriptome Sequencing and Gene Expression Analysis
A drought stress experiment was performed with three biological replicates of a B. hybridum ECI-AS sample, aiming to elucidate if its BhS subgenome was dominant over its BhD subgenome under strong soil water deficit conditions, similar to those of its native arid ECI AS site, and searching for drought-tolerance genes. Seeds from the individual samples were germinated and grown for 14 h light/10 h dark cycles and a constant temperature of 23 ± 2 °C and then plants were assigned to two treatment groups, drought in which water was withheld for one week, and control in which plants were watered every two days for the duration of the experiment. Leaf and root samples from control and drought-treated plants were collected for transcriptomic analysis. Total RNA was extracted from each sample using the QIAGEN RNeasy plant mini kit, and sequenced using a DNBseq-T7 system following RNA library construction with oligo dT methods. The transcriptomic data of the ancestral D-anc Bhyb-26 (SRR20045864, SRR20045873, SRR20045874, and SRR20045875) and recent West-Med Bhyb-ABR113 (SRR4094443, SRR4094444, SRR4094445, SRR4094446, SRR11836559, and SRR11836560) were collected from National Center for Biotechnology Information (NCBI). Transcriptome analysis was conducted using the “HISAT2-Stringtie-DESeq” pipeline (Conesa et al. 2016). RNA-Seq reads were mapped to the corresponding genome with HISAT2 (v2.2.1) (Kim et al. 2019) and gene expression abundances (transcripts per kilobase per million mapped reads, TPM) were calculated using StringTie (v2.1.6) (Pertea et al. 2015).
HEB Analysis in B. hybridum Genomes
HEB was performed through syntenic gene pairs analysis (Yang et al. 2016) in the East-Med Bhyb-ECI genome (two treatment conditions and two tissues), the ancestral D-anc Bhyb-26 (one treatment condition and four tissues) (Scarlett et al. 2022) and recent West-Med Bhyb-ABR113 (one treatment condition and two tissues) (Gordon et al. 2020) genomes. Gene pairs between the BhS and BhD subgenomes of the three types of allotetraploids were identified with WGDi. After removing gene pairs with TPM values < 1 in all samples, differentially expressed gene pairs (with >2-fold TPM differences) were regarded as expression-biased gene pairs, exhibiting either BhS or BhD dominance. The gene in each pair with the higher expression level was classified as a DG, the other gene as a SG, and the two genes included in non-dominance gene pairs were regarded as neutral genes. All of analysis were performed with custom Perl scripts.
Statistical Analysis
GO enrichment analysis was performed with GOATools (v1.1.6) (Klopfenstein et al. 2018) with Benjamini-Hochberg procedure (BH) correction methods (Benjamini and Hochberg 1995). R (v4.0; https://www.r-project.org/) utilities were employed for statistic test in this study, and the R package ggplot2 (v3.3.5) (https://ggplot2.tidyverse.org/) was used to visualize the results.
Supplementary Material
Acknowledgments
We thank the handling editor and three anonymous reviewers for their valuable comments to an early version of the manuscript.
Contributor Information
Wenjie Mu, State Key Laboratory of Herbage Innovation and Grassland Agro-Ecosystem, College of Ecology, Lanzhou University, Lanzhou 730000, China; State Key Laboratory for Animal Disease Control and Prevention, College of Veterinary Medicine, Lanzhou University, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, China.
Kexin Li, State Key Laboratory of Herbage Innovation and Grassland Agro-Ecosystem, College of Ecology, Lanzhou University, Lanzhou 730000, China.
Yongzhi Yang, State Key Laboratory of Herbage Innovation and Grassland Agro-Ecosystem, College of Ecology, Lanzhou University, Lanzhou 730000, China.
Adina Breiman, Department of Evolutionary and Environmental Biology, University of Tel-Aviv, Tel-Aviv 6997801, Israel.
Jiao Yang, State Key Laboratory of Herbage Innovation and Grassland Agro-Ecosystem, College of Ecology, Lanzhou University, Lanzhou 730000, China.
Ying Wu, State Key Laboratory of Herbage Innovation and Grassland Agro-Ecosystem, College of Ecology, Lanzhou University, Lanzhou 730000, China.
Mingjia Zhu, State Key Laboratory of Herbage Innovation and Grassland Agro-Ecosystem, College of Ecology, Lanzhou University, Lanzhou 730000, China.
Shuai Wang, State Key Laboratory for Animal Disease Control and Prevention, College of Veterinary Medicine, Lanzhou University, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, China.
Pilar Catalan, Escuela Politecnica Superior de Huesca, Universidad de Zaragoza, Huesca 22071, Spain.
Eviatar Nevo, Institute of Evolution, University of Haifa, Haifa 3498838, Israel.
Jianquan Liu, State Key Laboratory of Herbage Innovation and Grassland Agro-Ecosystem, College of Ecology, Lanzhou University, Lanzhou 730000, China.
Supplementary Material
Supplementary material is available at Molecular Biology and Evolution online.
Author Contributions
J.L. led the work. J.L., E.N., and P.C. designed the study. K.L. and A.B. conducted the sampling. W.M. and Y.Y. performed the experiments and the analyses. J.Y., Y.W., M.Z., S.W., and P.C. contributed to the analyses. P.C., J.L., E.N., and W.M. wrote the manuscript. All authors read and commented on the manuscript.
Funding
J.L. (Jianquan Liu) was supported by the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (no. 2019QZKK0502) and Strategic Priority Research Program of Chinese Academy of Sciences (no. XDB31010300). P.C. (Pilar Catalan) was funded by the Spanish Ministry of Science and Innovation (grant no. PID2019-108195GB-I00), the Spanish Aragon Government (grant no. LMP82_21), and the Spanish Aragon Government-European Social Fund Bioflora (grant no. A01-23R). All the computation works were supported by the Big Data Computing Platform for Western Ecological Environment and Regional Development, and the Supercomputing Center of Lanzhou University.
Data Availability
Data for the empirical data analysis and scripts and data used to run the genomic and evolutionary analyses are deposited at public database: the sequencing data were deposited at NCBI database (https://www.ncbi.nlm.nih.gov/), and the project number is PRJNA791713. The genome assembly and main script used in analysis were uploaded at Github (https://github.com/Axolotl233/Brachypodium_hybridum-ECI-genome).
References
- Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009:19(9):1655–1664. 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alger EI, Edger PP. One subgenome to rule them all: underlying mechanisms of subgenome dominance. Curr Opin Plant Biol. 2020:54:108–113. 10.1016/j.pbi.2020.03.004. [DOI] [PubMed] [Google Scholar]
- Baduel P, Bray S, Vallejo-Marin M, Kolář F, Yant L. The “polyploid hop”: shifting challenges and opportunities over the evolutionary lifespan of genome duplications. Front Ecol Evol. 2018:6:1–19. 10.3389/fevo.2018.00117. [DOI] [Google Scholar]
- Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000:28(1):45–48. 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995:57:289–300. 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999:27(2):573–580. 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Hnert DK, Vaughan T, Wu C-H. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 2014:10(4):1003537. 10.1371/journal.pcbi.1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradnam KR, Fass JN, Alexandrov A, Baranay P, Bechner M, Birol I, Boisvert S, Chapman JA, Chapuis G, Chikhi R, et al. Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species. Gigascience. 2013:2(1):1–31. 10.1186/2047-217X-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns R, Mandáková T, Gunis J, Soto-Jiménez LM, Liu C, Lysak MA, Novikova PY, Nordborg M. Gradual evolution of allopolyploidy in Arabidopsis suecica. Nat Ecol Evol. 2021:5(10):1367–1381. 10.1038/s41559-021-01525-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catalan P, López-Álvarez D, Díaz-Pérez A, Sancho R, López-Herránz ML. Phylogeny and evolution of the genus Brachypodium. In: Vogel J, editor. Genetics and genomics of Brachypodium. Series plant genetics and genomics: crops models. New York: Springer; 2016. p. 9–38. [Google Scholar]
- Catalán P, Müller J, Hasterok R, Jenkins G, Mur LAJ, Langdon T, Betekhtin A, Siwinska D, Pimentel M, López-Alvarez D. Evolution and taxonomic split of the model grass Brachypodium distachyon. Ann Bot. 2012:109(2):385–405. 10.1093/aob/mcr294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalhoub B, Denoeud F, Liu S, Parkin IAP, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B, et al. Early allopolyploid evolution in the post-neolithic Brassica napus oilseed genome. Science. 2014:345(6199):950–953. 10.1126/science.1253435. [DOI] [PubMed] [Google Scholar]
- Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics Chapter. 2004:4:1–14. 10.1002/0471250953.bi0410s25. [DOI] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018:34(17):i884–i890. 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng F, Wu J, Cai X, Liang J, Freeling M, Wang X. Gene retention, fractionation and subgenome differences in polyploid plants. Nat Plants. 2018:4(5):258–268. 10.1038/s41477-018-0136-7. [DOI] [PubMed] [Google Scholar]
- Chester M, Gallagher JP, Symonds VV, Cruz da Silva AV, Mavrodiev EV, Leitch AR, Soltis PS, Soltis DE. Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). Proc Natl Acad Sci U S A. 2012:109(4):1176–1181. 10.1073/pnas.1112041109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes M, Grandont L, Moore G, Chèvre AM, Jenczewski E. Genetic regulation of meiosis in polyploid species: new insights into an old question. New Phytol. 2010:186(1):29–36. 10.1111/j.1469-8137.2009.03084.x. [DOI] [PubMed] [Google Scholar]
- Collin F, Durif G, Raynal L, Lombaert E, Gautier M, Vitalis R, Marin J, Estoup A. Extending approximate Bayesian computation with supervised machine learning to infer demographic history from genetic polymorphisms using DIYABC random forest. Mol Ecol Resour. 2021:21(8):2598–2613. 10.1111/1755-0998.13413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comai L. The advantages and disadvantages of being polyploid. Nat Rev Genet. 2005:6(11):836–846. 10.1038/nrg1711. [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005:21(18):3674–3676. 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, Szcześniak MW, Gaffney DJ, Elo LL, Zhang X, et al. A survey of best practices for RNA-Seq data analysis. Genome Biol. 2016:17:1–19. 10.1186/s13059-015-0866-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb SK, Edger PP, Pires JC, McKain MR. Patterns, mechanisms, and consequences of homoeologous exchange in allopolyploid angiosperms: a genomic and epigenomic perspective. New Phytol. 2023:238(6):2284–2304. 10.1111/nph.18927. [DOI] [PubMed] [Google Scholar]
- Decena MA, Sergio G, Agostini F, Sancho R, Contreras-moreira B, Des Marais DL, Hernandez P, Catalán P. Comparative genomics, evolution and drought-induced expressions of dehydrin genes in model Brachypodium grasses. Plants. 2021:10(12):2664. 10.3390/plants10122664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, Del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011:43(5):491–501. 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas GM, Gos G, Steige KA, Salcedo A, Holm K, Josephs EB, Arunkumar R, Ågren JA, Hazzouri KM, Wang W, et al. Hybrid origins and the earliest stages of diploidization in the highly successful recent polyploid Capsella bursa-pastoris. Proc Natl Acad Sci U S A. 2015:112(9):2806–2811. 10.1073/pnas.1412277112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Flagel LE, Paterson AH, Rapp RA, Soltis DE, Soltis PS, Wendel JF. Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet. 2008:42(1):443–461. 10.1146/annurev.genet.42.110807.091524. [DOI] [PubMed] [Google Scholar]
- Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, Shamim MS, Machol I, Lander ES, Aiden AP, et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017:356(6333):92–95. 10.1126/science.aal3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand NC, Shamim MS, Machol I, Rao SSP, Huntley MH, Lander ES, Aiden EL. Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst. 2016:3(1):95–98. 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, McKain MR, Yocca AE, Knapp SJ, Qiao Q, Zhang T. Reply to: revisiting the origin of octoploid strawberry. Nat Genet. 2020:52(1):5–7. 10.1038/s41588-019-0544-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, Poorten TJ, VanBuren R, Hardigan MA, Colle M, McKain MR, Smith RD, Teresi SJ, Nelson ADL, Wai CM, et al. Origin and evolution of the octoploid strawberry genome. Nat Genet. 2019:51(3):541–547. 10.1038/s41588-019-0356-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger PP, Smith R, McKain MR, Cooley AM, Vallejo-Marin M, Yuan Y, Bewick AJ, Ji L, Platts AE, Bowman MJ, et al. Subgenome dominance in an interspecific hybrid, synthetic allopolyploid, and a 140-year-old naturally established neo-allopolyploid monkeyflower. Plant Cell. 2017:29(9):2150–2167. 10.1105/tpc.17.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel LE, Wendel JF. Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytol. 2010:186(1):184–193. 10.1111/j.1469-8137.2009.03107.x. [DOI] [PubMed] [Google Scholar]
- Gaut BS, Morton BR, McCaig BC, Clegg MT. Substitution rate comparisons between grasses and palms: synonymous rate differences at the nuclear gene adh parallel rate differences at the plastid gene rbcL. Proc Natl Acad Sci U S A. 1996:93(19):10274–10279. 10.1073/pnas.93.19.10274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel M, Sun H, Jiao W-B, Schneeberger K. SyRI: finding genomic rearrangements and local sequence differences from whole-genome assemblies. Genome Biol. 2019:20(1):1–13. 10.1186/s13059-019-1911-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SP, Contreras-Moreira B, Levy JJ, Djamei A, Czedik-Eysenberg A, Tartaglio VS, Session A, Martin J, Cartwright A, Katz A, et al. Gradual polyploid genome evolution revealed by pan-genomic analysis of Brachypodium hybridum and its diploid progenitors. Nat Commun. 2020:11(1):1–16. 10.1038/s41467-019-13993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon SP, Contreras-Moreira B, Woods DP, Des Marais DL, Burgess D, Shu S, Stritt C, Roulin AC, Schackwitz W, Tyler L, et al. Extensive gene content variation in the Brachypodium distachyon pan-genome correlates with population structure. Nat Commun. 2017:8(1):2184. 10.1038/s41467-017-02292-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths AG, Moraga R, Tausen M, Gupta V, Bilton TP, Campbell MA, Ashby R, Nagy I, Khan A, Larking A, et al. Breaking free: the genomics of allopolyploidy-facilitated niche expansion in white clover. Plant Cell. 2019:31(7):1466–1487. 10.1105/tpc.18.00606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Wang X, Gundlach H, Mayer KFX, Peterson DG, Scheffler BE, Chee PW, Paterson AH. Extensive and biased intergenomic nonreciprocal DNA exchanges shaped a nascent polyploid genome, Gossypium (cotton). Genetics. 2014:197(4):1153–1163. 10.1534/genetics.114.166124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RKJr, Hannick LI. Improving the Arabidopsis Genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003:31(19):5654–5666. 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, et al. De Novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat Protoc. 2013:8:1494–1512. 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, White O, Robin CR, Wortman JR. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008:9(1):1–22. 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasterok R, Catalan P, Hazen SP, Roulin AC, Vogel JP, Wang K, Mur LAJ. Brachypodium: 20 years as a grass biology model system; the way forward? Trends Plant Sci. 2022:27(10):1002–1016. 10.1016/j.tplants.2022.04.008. [DOI] [PubMed] [Google Scholar]
- Higgins EE, Howell EC, Armstrong SJ, Parkin IAP. A major quantitative trait locus on chromosome A9, BnaPh1, controls homoeologous recombination in Brassica napus. New Phytol. 2021:229(6):3281–3293. 10.1111/nph.16986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollister JD, Gaut BS. Epigenetic silencing of transposable elements: a trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 2009:19(8):1419–1428. 10.1101/gr.091678.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Wang Z, Sun Z, Hu B, Ayoola AO, Liang F, Li J, Sandoval JR, Cooper DN, Ye K. An efficient error correction and accurate assembly tool for noisy long reads. bioRxiv. 10.1101/2023.03.09.531669, 12 March 2023, preprint: not peer reviewed. [DOI] [PMC free article] [PubMed]
- Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, et al. EggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019:47(D1):D309–D314. 10.1093/nar/gky1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006:23(2):254–267. 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Jenczewski E, Alix K. From diploids to allopolyploids: the emergence of efficient pairing control genes in plants. CRC Crit Rev Plant Sci. 2004:23(1):21–45. 10.1080/07352680490273239. [DOI] [Google Scholar]
- Jenczewski E, Eber F, Manzanares-Dauleux MJ, Chevre AM. A strict diploid-like pairing regime is associated with tetrasomic segregation in induced autotetraploids of kale. Plant Breed. 2002:121(2):177–179. 10.1046/j.1439-0523.2002.00672.x. [DOI] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS, et al. Ancestral polyploidy in seed plants and angiosperms. Nature. 2011:473(7345):97–100. 10.1038/nature09916. [DOI] [PubMed] [Google Scholar]
- Jin JJ, Bin YW, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020:21(1):241. 10.1186/s13059-020-02154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Binns D, Chang H-Y, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014:30(9):1236–1240. 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005:110(1–4):462–467. 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008:9(4):286–298. 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Keilwagen J, Hartung F, Grau J. Gemoma: homology-based gene prediction utilizing intron position conservation and RNA-seq data. Methods Mol Biol. 2019:1962:161–177. 10.1007/978-1-4939-9173-0_9. [DOI] [PubMed] [Google Scholar]
- Kim D, Paggi JM, Park C, Bennett C, Salzberg SL. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019:37(8):907–915. 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfenstein DV, Zhang L, Pedersen BS, Ramírez F, Vesztrocy AW, Naldi A, Mungall CJ, Yunes JM, Botvinnik O, Weigel M, et al. GOATOOLS: a python library for gene ontology analyses. Sci Rep. 2018:8(1):1–17. 10.1038/s41598-018-28948-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokot M, Dlugosz M, Deorowicz S. KMC 3: counting and manipulating k-mer statistics. Bioinformatics. 2017:33(17):2759–2761. 10.1093/bioinformatics/btx304. [DOI] [PubMed] [Google Scholar]
- Korunes KL, Samuk K. Pixy: unbiased estimation of nucleotide diversity and divergence in the presence of missing data. Mol Ecol Resour. 2021:21(4):1359–1368. 10.1111/1755-0998.13326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitch AR, Leitch IJ. Genomic plasticity and the diversity of polyploid plants. Science. 2008:320(5875):481–483. 10.1126/science.1153585. [DOI] [PubMed] [Google Scholar]
- Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018:34(18):3094–3100. 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009:25(16):2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003:13(9):2178–2189. 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, Gendrel A-V, Black M, Vaughn MW, Dedhia N, Richard McCombie W, Lavine K, Mittal V, May B, Kasschau KD, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004:430(6998):471–476. 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- Liston A, Wei N, Tennessen JA, Li J, Dong M, Ashman T. Revisiting the origin of octoploid strawberry. Nat Genet. 2020:52(1):2–4. 10.1038/s41588-019-0543-3. [DOI] [PubMed] [Google Scholar]
- Lopez-Alvarez D, Lopez-Herranz ML, Betekhtin A, Catalan P. A DNA barcoding method to discriminate between the model plant Brachypodium distachyon and its close relatives B. stacei and B. hybridum (Poaceae). PLoS One. 2012:7(12):e51058. 10.1371/journal.pone.0051058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Alvarez D, Manzaneda AJ, Rey PJ, Giraldo P, Benavente E, Allainguillaume J, Mur L, Caicedo AL, Hazen SP, Breiman A, et al. Environmental niche variation and evolutionary diversification of the Brachypodium distachyon grass complex species in their native circum-Mediterranean range. Am J Bot. 2015:102(7):1073–1088. 10.3732/ajb.1500128. [DOI] [PubMed] [Google Scholar]
- Marçais G, Delcher AL, Phillippy AM, Coston R, Salzberg SL, Zimin A. MUMmer4: a fast and versatile genome alignment system. PLoS Comput Biol. 2018:14(1):e1005944. 10.1371/journal.pcbi.1005944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín AC, Alabdullah AK, Moore G. A separation-of-function ZIP4 wheat mutant allows crossover between related chromosomes and is meiotically stable. Sci Rep. 2021:11(1):1–13. 10.1038/s41598-021-01379-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín AC, Borrill P, Higgins J, Alabdullah A, Ramírez-González RH, Swarbreck D, Uauy C, Shaw P, Moore G. Genome-wide transcription during early wheat meiosis is independent of synapsis, ploidy level, and the Ph1 locus. Front Plant Sci. 2018:871:1–19. 10.3389/fpls.2018.01791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason AS, Wendel JF. Homoeologous exchanges, segmental allopolyploidy, and polyploid genome evolution. Front Genet. 2020:11:1–10. 10.3389/fgene.2020.01014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann J, Jang TS, Macas J, Schneeweiss GM, Matzke NJ, Novák P, Stuessy TF, Villaseñor JL, Weiss-Schneeweiss H. Dating the species network: allopolyploidy and repetitive DNA evolution in American daisies (Melampodium sect. Melampodium, Asteraceae). Syst Biol. 2018:67(6):1010–1024. 10.1093/sysbio/syy024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu W, Li K, Yang Y, Breiman A, Yang J, Wu Y, Wu S, Zhu M, Liu J, Nevo E, et al. Scattered differentiation of unlinked loci across the genome underlines ecological divergence of the selfing grass Brachypodium stacei. Proc Natl Acad Sci U S A. 2023:120(45):e2304848120. 10.1073/pnas.2304848120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevo E. “Evolution Canyon,” a potential microscale monitor of global warming across life. Proc Natl Acad Sci U S A. 2012:109(8):2960–2965. 10.1073/pnas.1120633109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015:32(1):268–274. 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto Feliner G, Casacuberta J, Wendel JF. Genomics of evolutionary novelty in hybrids and polyploids. Front Genet. 2020:11:792. 10.3389/fgene.2020.00792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou S, Jiang N. LTR_retriever: a highly accurate and sensitive program for identification of long terminal repeat retrotransposons. Plant Physiol. 2018:176(2):1410–1422. 10.1104/pp.17.01310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxelman B, Brysting AK, Jones GR, Marcussen T, Oberprieler C, Pfeil BE. Phylogenetics of allopolyploids. Annu Rev Ecol Evol Syst. 2017:48(1):543–557. 10.1146/annurev-ecolsys-110316-022729. [DOI] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and Eigen analysis. PLoS Genet. 2006:2(12):2074–2093. 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penner S, Dror B, Aviezer I, Bar-Lev Y, Salman-Minkov A, Mandakova T, Šmarda P, Mayrose I, Sapir Y. Phenology and polyploidy in annual Brachypodium species (Poaceae) along the aridity gradient in Israel. J Syst Evol. 2020:58(2):189–199. 10.1111/jse.12489. [DOI] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015:33(3):290–295. 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005:21(Suppl 1):351–358. 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007:35(Database):61–65. 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010:26(6):841–842. 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. Posterior summarization in Bayesian phylogenetics using tracer 1.7. Syst Biol. 2018:67(5):901–904. 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey J, Schemske DW. Neopolyploidy in flowering plants. Annu Rev Ecol Syst. 2002:33(1):589–639. 10.1146/annurev.ecolsys.33.010802.150437. [DOI] [Google Scholar]
- Rhie A, Walenz BP, Koren S, Phillippy AM. Merqury: reference-free quality, completeness, and phasing assessment for genome assemblies. Genome Biol. 2020:21(1):245. 10.1186/s13059-020-02134-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salse J, Bolot S, Throude M, Jouffe V, Piegu B, Quraishi UM, Calcagno T, Cooke R, Delseny M, Feuillet C. Identification and characterization of shared duplications between rice and wheat provide new insight into grass genome evolution. Plant Cell. 2008:20(1):11–24. 10.1105/tpc.107.056309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho R, Cantalapiedra CP, López-Alvarez D, Gordon SP, Vogel JP, Catalán P, Contreras-Moreira B. Comparative plastome genomics and phylogenomics of Brachypodium: flowering time signatures, introgression and recombination in recently diverged ecotypes. New Phytol. 2018:218(4):1631–1644. 10.1111/nph.14926. [DOI] [PubMed] [Google Scholar]
- Sancho R, Inda LA, Díaz-Pérez A, Des Marais DL, Gordon S, Vogel JP, Lusinska J, Hasterok R, Contreras-Moreira B, Catalán P. Tracking the ancestry of known and “ghost” homeologous subgenomes in model grass Brachypodium polyploids. Plant J. 2022:109(6):1535–1558. 10.1111/tpj.15650. [DOI] [PubMed] [Google Scholar]
- Scarlett VT, Lovell JT, Shao M, Phillips J, Shu S, Lusinska J, Goodstein DM, Jenkins J, Grimwood J, Barry K, et al. Multiple origins, one evolutionary trajectory: gradual evolution characterizes distinct lineages of allotetraploid Brachypodium. Genetics. 2022:223(2):iyac146. 10.1093/genetics/iyac146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholthof KBG, Irigoyen S, Catalan P, Mandadi KK. Brachypodium: a monocot grass model genus for plant biology. Plant Cell. 2018:30(8):1673–1694. 10.1105/tpc.18.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra H, Svačina R, Baumann U, Whitford R, Sutton T, Bartoš J, Sourdille P. Ph2 encodes the mismatch repair protein MSH7-3D that inhibits wheat homoeologous recombination. Nat Commun. 2021:12(1):1–10. 10.1038/s41467-021-21127-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015:31(19):3210–3212. 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Soltis DE, Albert VA, Leebens-Mack J, Bell CD, Paterson AH, Zheng C, Sankoff D, DePamphilis CW, Wall PK, Soltis PS. Polyploidy and angiosperm diversification. Am J Bot. 2009:96(1):336–348. 10.3732/ajb.0800079. [DOI] [PubMed] [Google Scholar]
- Soltis PS, Liu X, Marchant DB, Visger CJ, Soltis DE. Polyploidy and novelty: Gottlieb's legacy. Philos Trans R Soc B Biol Sci. 2014:369(1648):20130351. 10.1098/rstb.2013.0351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soltis PS, Soltis DE. Plant genomes: markers of evolutionary history and drivers of evolutionary change. Plants People Planet. 2021:3(1):74–82. 10.1002/ppp3.10159. [DOI] [Google Scholar]
- Soltis DE, Visger CJ, Blaine Marchant D, Soltis PS. Polyploidy: pitfalls and paths to a paradigm. Am J Bot. 2016:103(7):1146–1166. 10.3732/ajb.1500501. [DOI] [PubMed] [Google Scholar]
- Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B. AUGUSTUS: a b initio prediction of alternative transcripts. Nucleic Acids Res. 2006:34(Web Server):435–439. 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stebbins GL. Polyploidy, hybridization and the invasion of new habitats. Ann Missouri Bot Gard. 1985:72(4):824–832. 10.2307/2399224. [DOI] [Google Scholar]
- Steige KA, Slotte T. Genomic legacies of the progenitors and the evolutionary consequences of allopolyploidy. Curr Opin Plant Biol. 2016:30:88–93. 10.1016/j.pbi.2016.02.006. [DOI] [PubMed] [Google Scholar]
- Sun H, DIng J, Piednoël M, Schneeberger K. FindGSE: estimating genome size variation within human and Arabidopsis using k-mer frequencies. Bioinformatics. 2018:34(4):550–557. 10.1093/bioinformatics/btx637. [DOI] [PubMed] [Google Scholar]
- Sun P, Jiao B, Yang Y, Shan L, Li T, Li X, Xi Z, Wang X, Liu J. WGDI: a user-friendly toolkit for evolutionary analyses of whole-genome duplications and ancestral karyotypes. Mol Plant. 2022:15(12):1841–1851. 10.1016/j.molp.2022.10.018. [DOI] [PubMed] [Google Scholar]
- Tang H, Bowers JE, Wang X, Ming R, Alam M, Paterson AH. Synteny and collinearity in plant genomes. Science. 2008:320(5875):486–488. 10.1126/science.1153917. [DOI] [PubMed] [Google Scholar]
- Te Beest M, Le Roux JJ, Richardson DM, Brysting AK, Suda J, Kubešová M, Pyšek P. The more the better? The role of polyploidy in facilitating plant invasions. Ann Bot. 2012:109(1):19–45. 10.1093/aob/mcr277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuren R, Man Wai C, Wang X, Pardo J, Yocca AE, Wang H, Chaluvadi SR, Han G, Bryant D, Edger PP, et al. Exceptional subgenome stability and functional divergence in the allotetraploid Ethiopian cereal teff. Nat Commun. 2020:11(1):1–11. 10.1038/s41467-020-14724-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Peer Y, Ashman TL, Soltis PS, Soltis DE. Polyploidy: an evolutionary and ecological force in stressful times. Plant Cell. 2021:33(1):11–26. 10.1093/plcell/koaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Peer Y, Mizrachi E, Marchal K. The evolutionary significance of polyploidy. Nat Rev Genet. 2017:18(7):411–424. 10.1038/nrg.2017.26. [DOI] [PubMed] [Google Scholar]
- Vasimuddin M, Misra S, Li H, Aluru S. Efficient architecture-aware acceleration of BWA-MEM for multicore systems. 2019 IEEE 33rd international parallel & distributed processing symposium (IPDPS); 20–24 May, Rio de Janeiro, Brazil. 2019. p. 314–324.
- Vicient CM, Casacuberta JM. Impact of transposable elements on polyploid plant genomes. Ann Bot. 2017:120(2):195–207. 10.1093/aob/mcx078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF, Lisch D, Hu G, Mason AS. The long and short of doubling down: polyploidy, epigenetics, and the temporal dynamics of genome fractionation. Curr Opin Genet Dev. 2018:49:1–7. 10.1016/j.gde.2018.01.004. [DOI] [PubMed] [Google Scholar]
- Woodhouse MR, Cheng F, Pires JC, Lisch D, Freeling M, Wang X. Origin, inheritance, and gene regulatory consequences o genome dominance in polyploids. Proc Natl Acad Sci U S A. 2014:111(14):5283–5288. 10.1073/pnas.1402475111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Liu D, Wang X, Ji C, Cheng F, Liu B, Hu Z, Chen S, Pental D, Ju Y, et al. The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat Genet. 2016:48(10):1225–1232. 10.1038/ng.3657. [DOI] [PubMed] [Google Scholar]
- You J, Zhang L, Song B, Qi X, Chan Z. Systematic analysis and identification of stress-responsive genes of the NAC gene family in Brachypodium distachyon. PLoS One. 2015:10(3):1–20. 10.1371/journal.pone.0122027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Wang P, Li J, Zhao Q, Ji C, Zhu Z, Zhai Y, Qin X, Zhou J, Yu H, et al. Whole-genome sequence of synthesized allopolyploids in Cucumis reveals insights into the genome evolution of allopolyploidization. Adv Sci. 2021:8(9):1–15. 10.1002/advs.202004222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Lingkui Z, Cui Y, Yang Y, Wu J, Liang J, Li X, Zhang X, Zhang Y, Guo Z, et al. The lack of negative association between TE load and subgenome dominance in synthesized Brassica allotetraploids. Proc Natl Acad Sci U S A. 2023:120(42):e2305208120. 10.1073/pnas.2305208120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Jiao C, Sun H, Rosli HG, Pombo MA, Zhang P, Banf M, Dai X, Martin GB, Giovannoni JJ, et al. iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol Plant. 2016:9(12):1667–1670. 10.1016/j.molp.2016.09.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data for the empirical data analysis and scripts and data used to run the genomic and evolutionary analyses are deposited at public database: the sequencing data were deposited at NCBI database (https://www.ncbi.nlm.nih.gov/), and the project number is PRJNA791713. The genome assembly and main script used in analysis were uploaded at Github (https://github.com/Axolotl233/Brachypodium_hybridum-ECI-genome).