Abstract
Background
Glioblastoma (GBM) stem-like cells (GSCs) are crucial drivers of treatment resistance and tumor recurrence. While the concept of “migrating” cancer stem cells was proposed a decade ago, the roles and underlying mechanisms of the heterogeneous populations of GSCs remain poorly defined.
Methods
Cell migration using GBM cell lines and patient-derived GSCs was examined using Transwell inserts and the scratch assay. Single-cell RNA sequencing data analysis were used to map GSC drivers to specific GBM cell populations. Xenografted mice were used to model the role of brain-type fatty acid-binding protein 7 (FABP7) in GBM infiltration and expansion. The mechanism by which FABP7 and its fatty acid ligands promote GSC migration was examined by gel shift and luciferase gene reporter assays.
Results
A subpopulation of FABP7-expressing migratory GSCs was identified, with FABP7 upregulating SOX2, a key modulator for GBM stemness and plasticity, and ZEB1, a prominent factor in GBM epithelial-mesenchymal transition and invasiveness. Our data indicate that GSC migration is driven by nuclear FABP7 through activation of RXRα, a nuclear receptor activated by polyunsaturated fatty acids (PUFAs).
Conclusion
Infiltrative progression in GBM is driven by migratory GSCs through activation of a PUFA-FABP7-RXRα neurogenic pathway.
Keywords: epithelial-to-mesenchymal transition, fatty acid-binding protein 7, glioblastoma, migratory cancer stem cells, retinoid-X-receptor alpha
Graphical Abstract
Graphical Abstract.
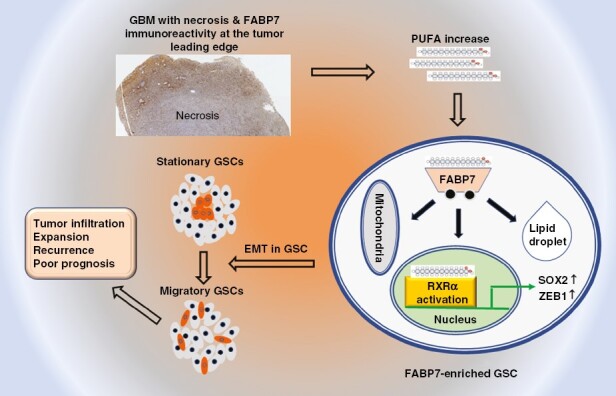
Key Points.
We identify a subset of migratory glioblastoma stem-like cells that express the polyunsaturated fatty acid-binding protein, fatty acid-binding protein 7 (FABP7).
Stationary-to-migratory transition is driven by a PUFA-FABP7-RXRα pathway.
PUFA-FABP7-RXRα orchestrates stemness, epithelial-to-mesenchymal transition, and invasion in glioblastoma cells.
Importance of the Study.
We identify and characterize a migratory stem-like cell population in glioblastoma. We show that stationary-to-migratory transition in glioblastoma stem-like cells is driven by a fatty acid-binding protein 7 (FABP7)-RXRα neurogenic pathway that is triggered by polyunsaturated fatty acids. Our findings suggest that inhibition of the FABP7-RXRα pathway may target the cancer stem cell population responsible for glioblastoma infiltration and expansion.
GBM is the most aggressive and lethal type of brain tumor, with a median survival of 12–15 months.1 Clinical management of GBM is mainly palliative because of its highly infiltrative nature and poor response to treatment. Glioblastoma stem-like cells (GSCs) promote invasive tumor growth, recurrence, and resistance to treatment.2 Molecular biomarkers and pathways that dictate stemness and infiltrative properties are needed to inform future strategies for the treatment of GBM.
Both stationary and migratory cancer stem cells (CSCs) have been described in colorectal cancer.3 Stationary CSCs are primarily restricted to the differentiated regions of cancer, whereas migrating CSCs are disseminated outside the main tumor mass leading to invasive growth. Migrating CSCs have been postulated to be derived from stationary CSCs that have undergone epithelial-to-mesenchymal transition (EMT).3,4 As such, migrating CSCs combine two critical traits that drive tumor progression: Stemness and motility.3 Although CSC heterogeneity in GBM has been documented, little is known about the roles of the different GSC populations.
Fatty acid-binding protein 7 (FABP7) is a member of the FABP family that preferentially binds to polyunsaturated fatty acids (PUFAs) and is predominantly expressed in the radial glial cells (neural progenitor/stem cells) of the developing brain.5 FABP7 has recently been linked to the uptake, transport, and metabolism of lipids in slow-cycling GBM cells that display enhanced invasion and chemoresistance.6 In addition, GSCs accumulate higher levels of PUFAs compared to non-stem cells,7 and we have shown that FABP7 promotes PUFA uptake in GSCs.8
Here, we identify a subpopulation of FABP7-expressing GSCs with increased migratory and infiltrative properties, which is driven by FABP7-mediated-induction of stem cell factor SOX2 and EMT factor ZEB1. We provide evidence that FABP7 functions through nuclear factor RXRα to activate a PUFA-mediated signaling pathway that promotes GBM aggressiveness.
Materials and Methods
Patient-Derived Cells
Primary GBM tumor tissues (>95% of which are expected to be IDH-wild-type9) from consented patients were collected under the Health Research Ethics Board of Alberta –HREBA.CC-14-0070. GSCs were generated and cultured as previously described8 (A4 and ED series). To generate patient-derived adherent GBM cells, cells from A4-004 patients were also directly cultured in DMEM supplemented with 10% FCS.
GBM Xenograft and Xenograft-Derived Cells
U87 cells stably transfected with empty episomal expression vector (pREP4) or pREP4-FABP7 were selected and cultured in hygromycin.10 Cells (3 × 106) were injected into the right flank of 6–8 weeks nude mice (CD-1, nu/nu, Charles River Laboratory, 6/group). Tumor size was measured with calipers twice a week. At the endpoint, mice were euthanized, and tumors were removed from both the primary injection site as well as metastatic intestinal site detected upon dissection. Primary and metastatic tumor tissues were processed for tissue culture and immunostaining.8,11 For our orthotopic xenograft models, U87 pREP4 and pREP4-FABP7 cells, or patient-derived A4-007 cells, were injected intracranially (105 cells in 5 µL) at 2 mm depth into the right frontal cortex of immunodeficient mice (3 mice/group) (see Supplementary Materials and Methods for additional details). All animal experiments were in accordance with the approved guidelines of the Cross Cancer Institute Animal Care Committee—protocol AC06120.
Tissue Microarray (TMA) Analysis
A GBM TMA12 was immunostained using an in-house-developed anti-FABP7 antibody (1:500).13 Cytoplasmic and nuclear FABP7 immuno-reactivity was scored as previously described12 (see Supplementary Materials and Methods for more details).
Cell Migration Assays
A4-004 cells were cultured on laminin-coated coverslips in neurosphere (serum-free) medium until they reached ~80% confluence. A top-to-bottom scratch was introduced in the middle of the coverslip with a P20 pipet tip. Cells were allowed to migrate into the gap over a period of 24 hours. Cells accumulating in the closing gap (CG) were defined as fast-migrating cells, whereas cells that stayed in the confluent region (CR) were defined as stationary or slow-migrating cells. Cells were fixed in 4% paraformaldehyde, and coimmunostained with antibodies to FABP7 (in-house antibody; 1:400 dilution) and SOX2 (1:400, Cell Signaling, #3579), Nestin (1:400, Abcam, ab22035), or ZEB1 (1:400, Invitrogen, #14-9741-82). Anti-rabbit or anti-mouse Alexa-488 or Alexa-555 were used as secondary antibodies (Invitrogen, 1:400). The percentage of co-immunostained cells was quantified using the Multi-Wavelength Cell Scoring Module of the MetaXpress 6 Software (Molecular Devices, USA).
To investigate the migration of FABP7-expressing cells in real-time, U87 cells were transfected with pcDNA3.1-GFP or pcDNA3.1-GFP-FABP7 using polyethylenimine.14 At 80% cell confluency, a scratch was introduced and real-time cell migration was analyzed with a Incucyte Live-Cell Analysis System (Sartorius, Germany).
Cell migration was also analyzed using Transwell inserts (8 µM pore size) as previously described.11 To address the correlation between FABP7 expression and cell migration in patient-derived GSCs, we used populations of A4-004 cells which were ~70% positive for FABP7. Thirty thousand cells were added to the top chambers of paired duplicate Transwells. Cells were allowed to migrate from the top chamber containing DMEM/F12 medium towards the bottom chamber containing DMEM/F12 medium supplemented with B27, EGF, and FGF (neurosphere medium). After 30 hours, nonmigratory cells (top chamber of Transwell #1; cells removed from the bottom chamber with a Q-tip) and migratory cells that had migrated across the membrane (bottom chamber of Transwell #2; cells removed from the top chamber with a Q-tip) were processed for DAPI nuclear staining and FABP7 immunostaining, and membranes released from the top and bottom chambers as previously described.15 FABP7 positive cells and total (DAPI-stained) cells were counted using MetaXpress 6 Software Multi-Wavelength Cell Scoring Module (Molecular Devices, USA). Four independent sets of duplicate wells were analyzed.
Neurosphere Formation Assay
A4-007 cells were seeded in ultra-low attachment 24-well plates (1000 cells/well) and cultured in serum-free neurosphere medium. Cells were treated with 0, 10, and 20 µM FABP7 inhibitor (SBFI-26, AOBIOUS, INC) after 24 hours. Neurosphere numbers were counted and photographed after 6 days.
GBM Single-Cell RNA Sequencing (scRNA-seq) Data Analysis
scRNA-seq data from 8 GBM (IDH-wild type) patients16 were downloaded from the Broad Institute database. The data were analyzed with R using the scRNA-seq Seurat algorithm (https://satijalab.org/seurat/articles/pbmc3k_tutorial.html). See Supplementary Materials and Methods for more details.
Gel Shift Assays
Gel shifts were carried out using a peroxisome proliferator-activated receptor response element (PPRE or DR1, 5’-TCCTGTTTCCAGT-3’)17 probe as previously described.13 Mutated DR1 (5’-TTCTGTTTCAACC -3’) was used as a control probe. Nuclear extracts (1 µg) from U87 transfectants cultured under specified conditions were preincubated with 1 µg poly(dIdC), followed by addition of labeled probe. For super-shift assays, 1 µL of anti-PPARs, anti-RARs, or anti-RXRs antibodies (see Supplementary Table 1 for antibody information) was added to the binding reaction 10 minutes after addition of the labeled probe and incubated at room temperature for 30 minutes. DNA-protein complexes were resolved in a 5% native polyacrylamide gel and exposed to X-Ray film.
Luciferase Reporter Assay
Xenograft-derived cells with siRNA depletion of FABP7 or RXRα were seeded in 24-well plates (30 000 cells/well) and transfected with the luciferase reporter construct (PPRE 3-TK-Luc, Addgene) using polyethyleneimine (PEI). Luciferase activity was measured and analyzed as previously described.11
Quantitative RT-PCR and Western Blot Analysis
Total RNA from GBM cell lines was prepared using the Trizol reagent (Invitrogen). First-strand cDNA was synthesized using SuperScript reverse transcriptase II (Invitrogen). RT-qPCR was performed using gene-specific TaqMan probes (FABP7, Hs00361426_A1; SOX2, Hs00162669_m1; NES, Hs04187831_g1; GAPDH, Hs03929097_g1) on QuantStudio 6 Flex (ThermoFisher). Whole-cell lysates (40 µg/lane) were separated by SDS-PAGE and transferred to nitrocellulose membranes. Membranes were immunoblotted with primary and secondary antibodies (listed in Supplementary Table 1). Cytosolic and nuclear extracts were prepared using the NE-PER Nuclear and Cytoplasmic Reagent kit (Thermo Fisher).
Statistical Analysis
A gene expression dataset of 453 GBM tissues (TCGA HG-U133A, 92.5% IDH-wild type) was used for correlation analysis of FABP7 and either neural stemness or EMT markers. For TMA analysis, the log-rank test using MedCalc version 14.12.0 (MedCalc Software) was employed to compare Kaplan–Meier survival probabilities between patient populations stratified based on FABP7 immuno-reactivity. One-way ANOVA (when comparing more than 2 groups) or two-sided student t-test (when comparing 2 groups) were employed to compute the significance of the difference between experimental treatments. Two-way ANOVA was used to examine the significance of variance caused by each factor and covariance caused by the interaction of 2 experimental factors.
Results
Identification of Migrating GSCs
We found that FABP7 expression correlates with neural stem cell markers SOX2 and nestin in established GBM cell lines (Supplementary Figure 1A) and patient-derived GSC cultures based on RT-qPCR (Supplementary Figures 1B–D). As patient-derived GSC cultures show heterogeneity in their expression of FABP7, we carried out the scratch assay on A4-004, a patient-derived GSC culture characterized by moderate FABP7 levels, followed by immunostaining, to see whether FABP7 might be preferentially expressed in migratory cells. We observed significantly increased FABP7-immunoreactivity in the CG of the scratch compared to the CR (63.4% vs. 18.7%) (Figures 1A-C). We also observed 76.7% coimmunostaining of FABP7 with SOX2 and 87.4% coimmunostaining of FABP7 with nestin in fast-migrating cells compared to 24.7% containing with SOX2 and 69.1% costaining with nestin in stationary cells located in the CRs (Figures 1D-G). Immunofluorescence staining of cells from the top or bottom side of the Transwell membrane showed a significantly higher percentage of FABP7-positive cells in the migrated (bottom) compared to nonmigrated (top) cell populations (90% vs. 60%; Supplementary Figure 2).
Figure 1.
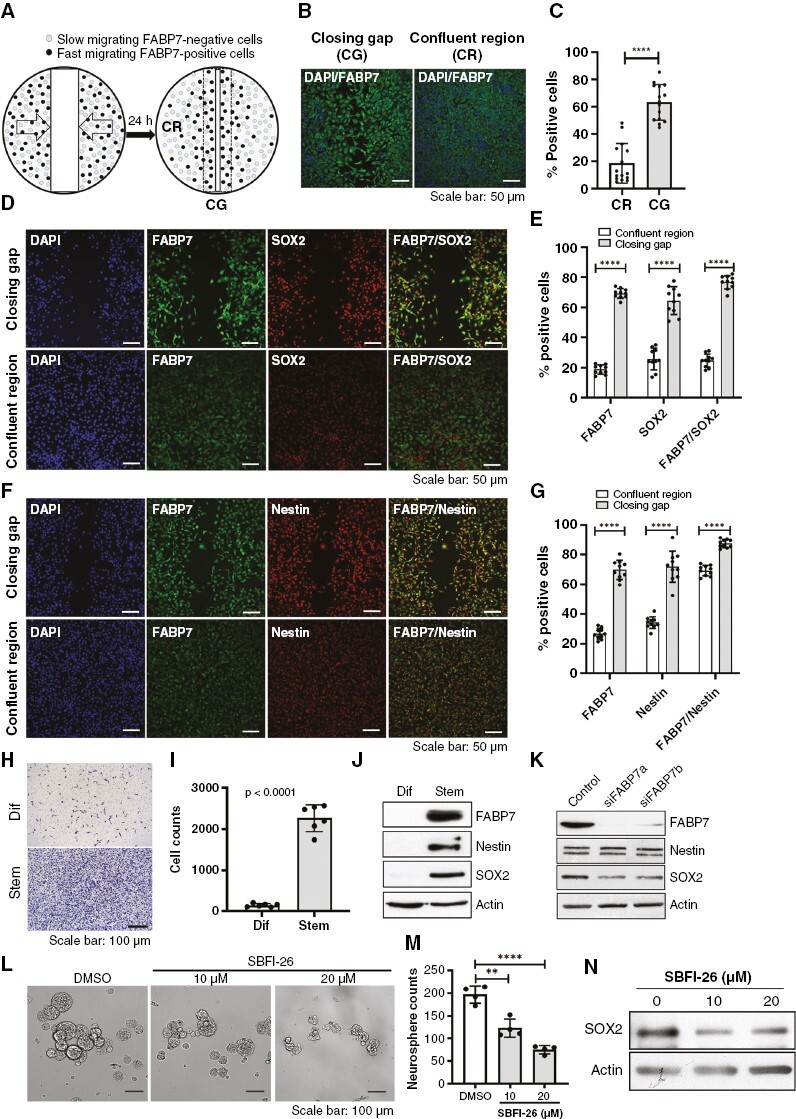
Correlation of fatty acid-binding protein 7 (FABP7) immunoreactivity with neural stem cell markers in migrating GSCs. (A) Schematic illustration of experimental design that combines the scratch assay and fluorescence immunostaining. confluent region (CR), closing gap (CG). (B, C) Representative images (B) and quantitative analysis (C) of FABP7 immunoreactivity in GSCs (A4-004 cells) in the CG and CR 24 hours post scratch formation. (D-G) Coimmunostaining of FABP7 and SOX2 (D, E) or FABP7 and nestin (F, G) in A4-004 cells migrating into the CG and in the CR 24 hours after introduction of the scratch. (H, I) Representative images (H) and statistical analysis (I) of the Transwell assay comparing the migration of A4-004 cells cultured in serum-supplemented DMEM (Dif) (adherent conditions; associated with a more differentiated phenotype) to that of cells cultured in serum-free neurosphere medium (Stem). (J) Western blot showing differential expression of FABP7, SOX2 and nestin in A4-004 cells cultured under adherent (Dif) and neurosphere (Stem) conditions. (K) Western blot showing a reduction in SOX2 protein levels but not nestin upon FABP7 knockdown in A4-004 cells cultured under neurosphere conditions. (L-M) Representative images (L) and histogram (M) showing neurosphere formation in A4-007 cells cultured in the absence (DMSO) or presenceof FABP7 inhibitor SBFI-26. (N) Western blot showing reduced levels of SOX2 in A4-007 cells treated with SBFI-26. N = 2 to 4, in triplicate. **** denotes P < .0001.
Using Transwell inserts, we observed a >10-fold increase in the migration of A4-004 cells cultured under neurosphere conditions compared to regular (FCS-containing) differentiation conditions (Figures 1H,I). We further observed abundant expression of FABP7, along with SOX2 and nestin, in A4-004 cells cultured under neurosphere conditions compared to regular conditions (Figure 1J). Expression of SOX2 was reduced when FABP7 was depleted in A4-004 cells using siRNAs. Nestin expression was not affected (Figure 1K). These data suggest that FABP7 is an upstream regulator of SOX2.
To further pursue the importance of FABP7 in GBM self-renewal capacity, we examined the effect of FABP7 inhibition on neurosphere formation in patient-derived A4-007 GSCs. We observed a significant reduction in neurosphere numbers when cells were treated with the FABP7 inhibitor SBFI-26 (Figures 1L,M), along with decreased SOX2 levels (Figure 1N).
Single-cell RNA sequencing analysis showed that FABP7 is specifically expressed in cell populations with typical GBM/mesenchymal stem cell signatures (Supplementary Figures 3A–G). A strong positive correlation between FABP7 mRNA levels and SOX2 (r = 0.537, P < .001) or NES; r = 0.676, P < .001) was observed in a GBM patient cohort dataset (Supplementary Figure 3H). These combined results suggest that FABP7 may be a biomarker for migratory GSCs.
FABP7 Regulates EMT in GSCs
EMT is believed to be a key process that allows CSCs to transition from stationary to migratory states.3 By analyzing a single-cell RNAseq dataset, we found that ZEB1, a zinc finger and homeodomain transcription factor that controls EMT and tumor progression in various cancers including GBM,18 is present in the same clusters as FABP7 (Figure 2A). The percentage of cells expressing ZEB1 in FABP7-positive cells was significantly higher than in FABP7-negative cells (15% vs. 5%, P = 1.38e-27; Figure 2B). Similar results in FABP7-positive versus FABP7-negative cells were observed for CDH2 encoding N-cadherin (35% vs. 6%, P = 1.96e-123) (Figures 2C,D). N-cadherin is a transmembrane adhesion protein fundamental to the EMT process.19 Analysis of a GBM patient cohort (TGCA HG U133A) revealed a significant positive correlation between FABP7 and either ZEB1 (r = 0.53, P < .001) or CDH2 (r = 0.69, P < .001), but a negative correlation between FABP7 and CDH1 (encoding E-cadherin, normally lost in cells that have undergone EMT, r = −0.49, P < .001) (Figure 2E).
Figure 2.
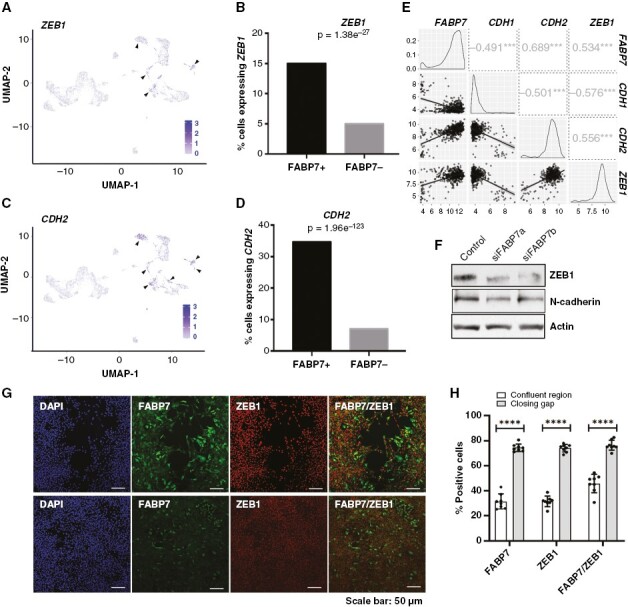
Expression of epithelial-to-mesenchymal transition markers is correlated with and regulated by fatty acid-binding protein 7 (FABP7) in GBM cells. (A, C) UMAP depicting ZEB1 (A) and CDH2 (C) expressing cells in GBM cell clusters generated through scRNA-seq analysis. (B, D) Analysis of the scRNA-seq data showing significantly higher percentages of cells expressing ZEB1 (B) and CDH2 (encoding N-cadherin) (D) in FABP7-positive cells compared to FABP7-negative cells. (E) Correlation of FABP7 RNA levels with that of ZEB1 and CDH2 in a TCGA dataset (HG U-133A) comprised of 453 GBM patients. Numbers in squares denote correlation coefficients, ***, P < .001. (F) Western blot showing decreased expression of ZEB1 and N-cadherin resulting from FABP7 knockdown in A4-004 cells cultured in neurosphere medium. (G) Representative images showing coimmunostaining of FABP7 and ZEB1 (G) in A4-004 GSCs in the closing gap and confluent regions 24 hours after introduction of the scratch. (H) Histogram showing quantitation and significance test of the data from the FABP7, ZEB1 and combined FABP7/ZEB1 fluorescence immunostaining data shown in (G). N = 2, in triplicate. **** denotes P < .0001.
We carried out the scratch assay using A4-004 cells followed by coimmunostaining with antibodies to FABP7 and ZEB1. ZEB1, like FABP7, was detected in a significantly higher proportion of fast-migrating GSCs in the CG compared to stationary GSCs in the CR remote from the gap. A significantly higher proportion of cells showed FABP7 and ZEB1 coimmunoreactivity in the CG compared to the cells in the CR (76% vs. 45%, P < .0001; Figures 2G,H). Finally, we observed marked reduction of ZEB1 and N-cadherin in A4-004 cells upon FABP7 depletion (Figure 2F). These combined results show a clear link between FABP7 expression, EMT status, and GSC migration, with FABP7 residing upstream of key EMT markers.
FABP7 Promotes Invasive GBM Tumor Growth In Vivo
Immunostaining of human GBM tumor tissue showed preferential expression of FABP7 and SOX2 in the leading (infiltrative) edge of the tumor (Supplementary Figure 4). To further address the role of FABP7 in GBM cells, we used the FABP7-negative U87 GBM cell line transiently transfected with pEGFP-N1 or pEGFP-N1-FABP7 expression constructs, followed by the scratch assay. For this experiment, cells migrating into the CG were measured in real-time for 48 hours using the Incucyte live-cell analysis system. Both the density of cells and GFP intensity were higher in the CG for GFP-FABP7-expressing cells compared to control cells (Figures 3A,B; Supplementary Figure 5A), further supporting a role for FABP7 in GBM cell migration.
Figure 3.
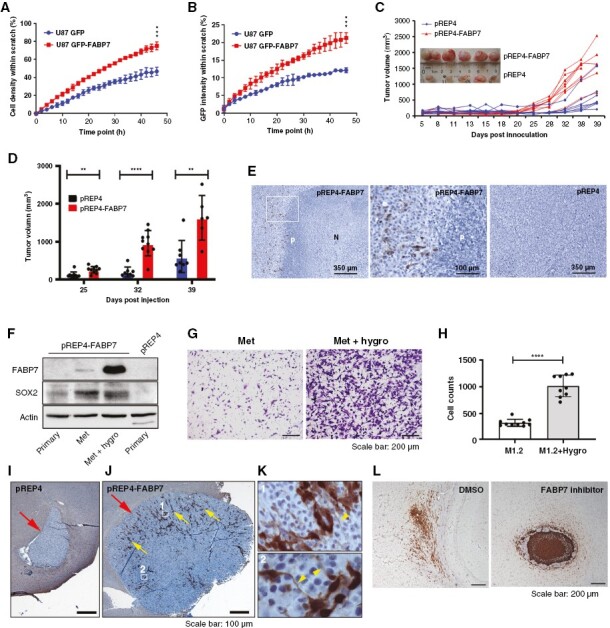
Fatty acid-binding protein 7 (FABP7) drives xenograft GBM growth and metastasis. (A) Real-time dynamic changes incell density in the scratch as a function of migration start time (Incucyte). (B) Real-time dynamic changes in the percentage of GFP intensity within the closing gap (Incucyte). (C, D) Tumor volume as a function of days post injection of U87-pREP4 and U87-pREP4-FABP7 cells in the flank of nude mice (C) and statistical analysis of tumor volume (D) at specific time points (inset shows photograms of tumors). (E) Representative images showing FABP7 immunostaining in xenograft tumors generated from FABP7-expressing (pREP4-FABP7; left, middle – magnification of the squared area of the left panel) and control (pREP4) U87 cells (right panel). (F) Western blot showing expression of FABP7 and SOX2 in primary cultures of xenograft tumor tissues, as well as in intestinal metastasis cultured in the absence and presence of hygromycin. (G) Representative images of Transwell assay comparing migration of the intestinal metastasis-derived cells without (Met) or with hygromycin selection (Met + hygro). (H) Histogram showing quantification and statistical significance of the results obtained using the Transwell assay. (I-K) Representative images of tumors formed from U87 pREP4 (I) and U87-pREP4-FABP7 (J) cells orthotopically injected into the brains of NSG mice. Magnified images (K) show the morphology of FABP7-positive cells from the indicated areas in M with arrowheads indicating extended cell processes. Large arrows in I and J point to tumors. Small arrows in J point to FABP7 immunoreactive cells. (L) Representative images showing the distribution of A4-007 FABP7-positive cells orthotopically injected into the brains of NSG mice (left panel), and the effect of intraperitoneal injection of the FABP7 inhibitor SBFI-26 on tumor formation (right panel). The flank xenograft experiments were performed twice with 6 mice in each group per experiment and the two orthotopic xenograft experiments were each carried out once with 3 mice in each group. P denotes pseudopalisade. N, necrosis. **, P < .01. ***, P < .001. ****, P < .0001.
We then generated stable cell lines by transfecting U87 cells with either pREP4 or pREP4-FABP7 followed by hygromycin selection (Supplementary Figure 5B). Neurosphere formation was significantly increased in FABP7-expressing U87 cells compared to control cells (Supplementary Figures 5C,D), indicating that FABP7 confers stemness features to GBM cells. Next, we examined how FABP7 might affect GBM tumor growth in vivo using a flank model. Tumors generated from stable FABP7-expressing U87 cells grew significantly faster starting from 25 days postinjection compared to control cells (Figures 3C, D). Immuno-histochemical analysis of FABP7 in the xenograft tumor tissues revealed widespread necrosis and pseudo-palisading in tumors from pREP4-FABP7 cells, but not in the tumors derived from control cells (Figure 3E). FABP7 immuno-reactivity was mainly observed surrounding the pseudo-palisading cell layers (Figure 3E) and in the leading edge of the tumor (Supplementary Figures 6A,C), where SOX2 was also detected (Supplementary Figures 6B,D). We also observed colocalization of nuclear FABP7 with β-Catenin, a critical signaling molecule with its nuclear translocation implicated in GBM stemness and tumor progression (Supplementary Figure 7).
Upon dissection of mice injected with U87-FABP7 cells, we observed metastatic intestinal tumors in addition to tumors in the flank. We cultured cells from both the primary and metastatic tumors in the absence or presence of hygromycin. Western blot analysis showed no (primary site) or weak (intestinal tumors) expression of FABP7 in cells cultured in the absence of hygromycin. There was a dramatic increase in FABP7 levels in intestinal tumor cells when cultured in the presence of hygromycin. Notably, SOX2 was preferentially expressed in metastatic tumors (Figure 3F). The hygromycin-resistant metastatic cells with high FABP7 expression showed considerably higher migratory ability compared to the weakly expressing non-hygromycin-selected cells (Figures 3G,H). These results suggest a direct correlation between FABP7 expression and aggressive GBM growth.
U87 pREP4 and U87 pREP4-FABP7 cells were also orthotopically injected into the brains of SCID mice. 9.4T MRI scans showed a trend towards faster growth for U87 pREP4-FABP7 compared to U87 pREP4 cells (Supplementary Figure 8). Immunostaining again showed a strong FABP7 signal in the expanding front of tumors generated from U87 pREP4-FABP7 cells, which had extended cell protrusions/processes, features of stemness, invasion, and treatment resistance in GBM20,21 (Figures 3I-K). We also examined the effect of FABP7 inhibition on the orthotopic growth of patient-derived A4-007 GSCs. We observed highly infiltrative tumor growth in all three mice intraperitoneally injected with the solvent control DMSO (Figure 3L and Supplementary Figure 9). In contrast, there was marked suppression of GBM cell infiltration in the brains of two of the three mice intraperitoneally injected with the FABP7 inhibitor SBFI-26, with tumors showing encapsulation with minimal infiltration (Figure 3L and Supplementary Figure 9). The third mouse showed a partial response to the FABP7 inhibitor, forming a partially encapsulated tumor with dense growth (Supplementary Figure 9).
FABP7 Activates RXRα, a Nuclear Receptor Induced by Polyunsaturated Fatty Acids in Brain
Docosahexaenoic acid (DHA), the preferred ligand of FABP7, activates RXR in rodent brain22 and enhances nuclear translocation of FABP7.23 This gives rise to the possibility that nuclear FABP7 may be transferring DHA to DHA-responsive transcription factor(s) such as PPARs and RXRs. We, therefore, cultured U87 pREP4 control and U87 pREP4-FABP7 cells in the absence or presence of DHA and carried out gel shift assays using nuclear extracts prepared from these cultures. While there was little difference in the binding of U87 control and U87 FABP7 nuclear extracts to32P-labeled DR1 (PPAR/RXR binding element), the signal was markedly enhanced when FABP7-expressing cells were cultured in the presence of DHA (Figure 4A). Addition of 100X excess wild-type cold competitor resulted in complete loss of the specific DR1-protein interactions (Figure 4B). When DR1 was mutated at nucleotides 2/10/11/13 of the DR1 consensus sequence, the intensity of the shifted bands was significantly reduced (Figure 4B).
Figure 4.
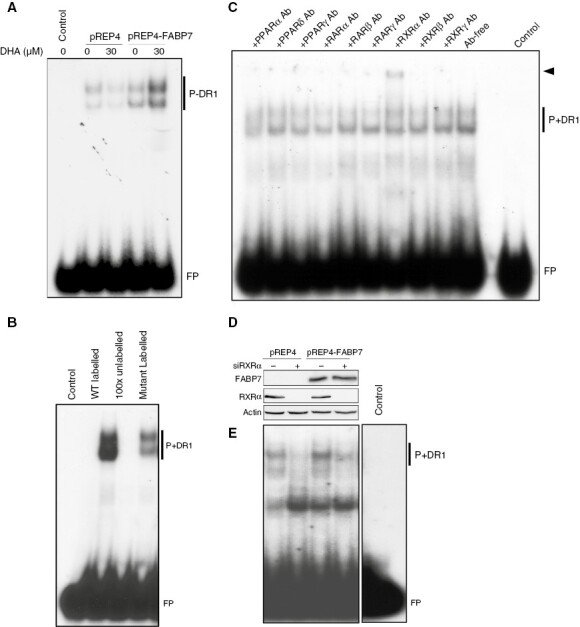
Fatty acid-binding protein 7 (FABP7) facilitates PUFA-induced activation of RXRα. (A) Gel shift showing that DHA treatment enhances formation of nuclear protein-DR1 complexes (P+DR1) in U87 pREP4-FABP7, but not in control cells (pREP4). (B) Inclusion of excess unlabeled PPRE probe (100X) in gel shifts effectively competed with the radio-labeled probe, resulting in undetectable protein-DR1 complexes (P+DR1). Gel shifts with a radio-labeled mutant DR1 probe considerably reduced protein-DR1 complex formation. (C) Supershift assay with antibodies against nine nuclear receptors shows a supershifted band (arrowhead) exclusively in the presence of anti-RXRα antibody. (D) Western blot showing knockdown of RXRα in stable U87 transfectants. (E) RXRα depletion resulted in reduced protein-DR1 complex formation in both U87 pREP4 and U87 pREP4-FABP7 transfectants. Each gene shift experiment was carried out 2 to 4 times. FP denotes free probe. Ab, antibody.
We carried out super-shift assays to identify the nuclear receptor(s) that bind to DR1 in GBM cell nuclear extracts. We observed a clear super-shifted band in reaction mixtures incubated with RXRα antibody (Figure 4C; arrow). Although RXRs usually function as heterodimers with PPAR or RARs, no super-shifted bands were observed with anti-PPAR or anti-RAR antibodies. The DR1-protein complex was reduced in both U87 control and FABP7-expressing cell lines after RXRα depletion (Figures 4D,E). These results demonstrate that FABP7 enhances the binding of RXRα to its recognition element DR1, suggesting a role for FABP7 in facilitating PUFA-mediated activation of RXRα.
RXRα Mediates the Promigratory/Prostemness Roles of FABP7 in GBM Cells
Identification of RXRα as a possible mediator of nuclear FABP7 function suggests a role for RXRα in the induction of the FABP7-dependent GSC migratory state. To address this potential role, we knocked down RXRα in control and FABP7-depleted A4-004 GSCs. We found that the number of migrating cells was significantly reduced (~50%) upon either FABP7 or RXRα depletion. Depletion of both FABP7 and RXRα further reduced cell migration (by ~75%) (Figures 5A,B), suggesting an interrelationship between RXRα and FABP7-induced GSC migration.
Figure 5.
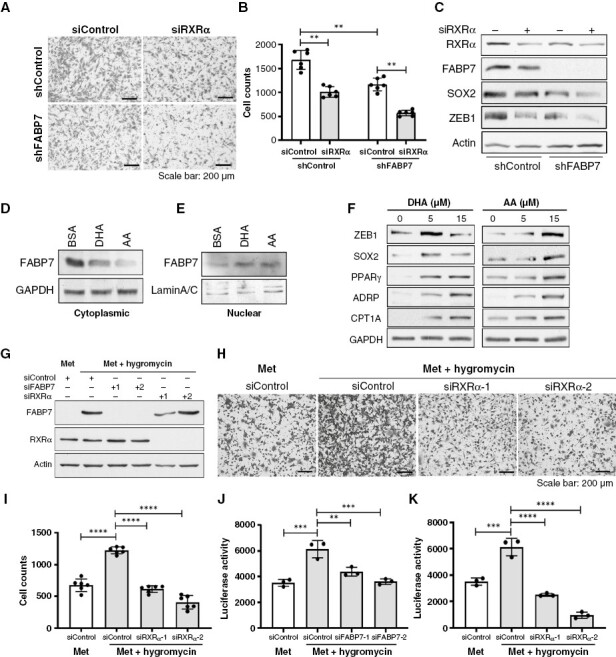
Fatty acid-binding protein 7 (FABP7) functions through RXRα in GSCs. (A, B) RXRα depletion in FABP7-depleted A4-004 cells (GSCs) further reduces cell migration using the Transwell assay compared to cells depleted of either FABP7 or RXRα alone. Representative images (A) and cell counts with statistical analysis (B) are shown. (C) Western blots showing expression of stem cell marker SOX2 and epithelial-to-mesenchymal transition (EMT) marker ZEB1 upon depletion of FABP7 and/or RXRα in A4-004. (D, E) Western blots showing changes in FABP7 levels in the cytoplasm (D) and nucleus (E) of A4-004 cells growing in neurosphere medium supplemented with BSA or 15 μM of either DHA or AA for 24 hours. (F) Western blots showing PUFA regulation of key proteins involved in cell stemness (SOX2), EMT (ZEB1) and fatty acid metabolism (PPARγ, ADRP, CPT1A) in A4-004 cells. Cells were supplemented with the indicated concentrations of DHA or AA for 24 hours in neurosphere medium. (G) Western blots showing expression of FABP7 and RXRα in cells derived from a xenograft intestinal metastasis (Met) with or without hygromycin selection, after FABP7 and/or RXRα depletion using two different siRNAs each (labeled 1 and 2). (H, I) Representative images (H) and bar chart (I) of the Transwell migration assay showing the effect of hygromycin selection and RARα depletion on GBM xenograft-derived metastatic cells. (J, K) Bar charts of luciferase reporter assay showing the effect of FABP7 (J) or RXRα (K) knockdown on PPRE/DR1-driven transcription activity in GBM xenograft-derived metastatic cells. Migration and luciferase assays were carried out 2 to 3 times, in triplicate. Westerns were repeated 2–4 times. **, P < .01; ***, P < .001; ****, P < .0001.
Next, we examined whether RXRα might be directly implicated in the upregulation of SOX2 and ZEB1 expression. Our western blot analysis showed that either FABP7 or RXRα depletion led to reduced levels of SOX2 and ZEB1 (Figure 5C), suggesting a role for both these proteins in the regulation of SOX2 and ZEB1. Codepletion of FABP7 and RXRα resulted in a further decrease in SOX2 and ZEB1 expression (Figure 5C).
Long-chain PUFAs such as DHA and arachidonic acid (AA) are preferred ligands of FABP75,24 and have recently been shown to promote GSC maintenance25 and brain tumor metastasis26 by modulating cell signaling via the nucleus.25–28 Western blots showed a significant decrease in cytoplasmic FABP7 in A4-004 cells cultured in either 15 μM DHA or AA, with the latter showing the strongest effect (Figure 5D). In contrast, there was an increase in nuclear FABP7 levels in cells treated with either AA or DHA (Figure 5E). These results support PUFA-driven nuclear translocation of FABP7, an essential step in nuclear PUFA-mediated signaling. We further observed marked induction of SOX2 and ZEB1 by DHA at 5 μM with attenuated induction at 15 μM. Induction of SOX2 and ZEB1 was also observed when cells were cultured in 15 μM AA but not 5 µM AA. Importantly, three proteins associated with lipid accumulation and utilization were also induced by DHA and AA: PPARγ, an RXR heterodimerizing partner implicated in lipogenesis; ADRP, a typical lipid droplet marker; and CPT1A, a rate-limiting enzyme governing beta-oxidation of fatty acids in the mitochondria (Figure 5F). Next, we tested the effect of DHA- and AA-supplementation on A4-007 GSCs. Increases in nuclear FABP7 and SOX2, along with cytoplasmic CPT1A were also observed in these cells (Supplementary Figure 10). These results suggest that environmental PUFAs may enhance nuclear receptor (eg, RXR and PPAR) activity, increase lipid accumulation and promote PUFA-derived energy production in FABP7-expressing GSCs.
To further investigate the functional relationship between FABP7 and RXRα in promoting GBM invasiveness, we used the xenograft metastatic cells described earlier. While cell migration was increased 2-fold upon hygromycin selection, RXRα knockdown completely eliminated this effect (Figures 5G–I), with similar results observed upon FABP7 knockdown (Supplementary Figure 11). DR1-driven luciferase reporter assay showed that RXRα transactivation (indicated by luciferase activity) was significantly enhanced in hygromycin-selected cells (by ~2-fold), an effect that was abrogated by depletion of either FABP7 (Figures 5G,J) or RXRα (Figures 5G,K). Overexpression of RXRα in FABP7-depleted cells significantly restored DR1-driven luciferase activity. Cell migration was rescued by RXRα in siFABP7-1 depleted cells, with a trend towards rescued observed in siFABP7-2-depleted (Supplementary Figures 12A–D). These results further support a role for RXRα in mediating the proneoplastic function of FABP7 in GBM.
Nuclear FABP7 is Associated With Poor Prognosis of GBM Patients
To evaluate the expression and clinical implication of FABP7 in GBM, we immunostained a TMA prepared with GBM tissues from 116 patients.12 The median 5-year overall survival time for this GBM cohort was 12.7 months. Clear immunoreactivity scoring data were obtained for 99 patients, of which 83% showed cytoplasmic FABP7 positivity and 77.4% showed nuclear positivity (Figures 6A,B). We further found that high levels of nuclear, but not cytoplasmic, FABP7 were significantly associated with a worse prognosis, with a hazard ratio of 2.02 (P = .023) (Figures 6C,D), which is in agreement with previous reports.29,30 Interestingly, the prognostic significance of nuclear FABP7 was magnified in a subpopulation of younger patients below the median age (<55 years) but disappeared in the older subpopulation (≥55 years) (Figures 6E,F).
Figure 6.
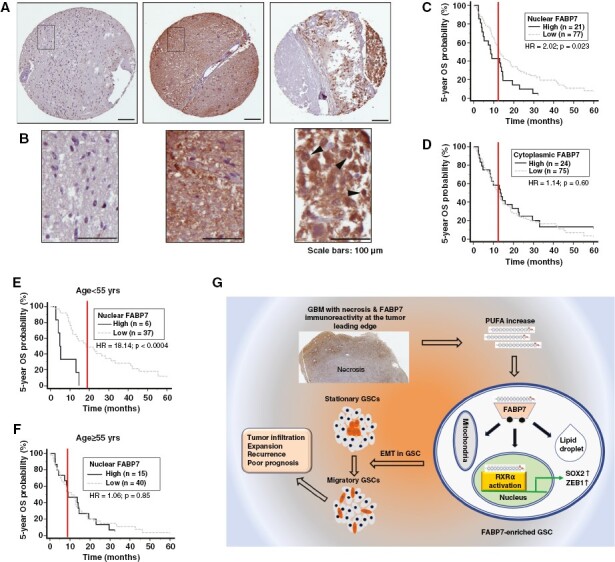
Immunohistochemical analysis of a GBM TMA. (A) Representative immunostaining images showing negative (left panel), uniform strong cytoplasmic and nuclear (middle panel), and heterogenous intratumoral and subcellular distribution of Fatty acid-binding protein 7 (FABP7) (right panel). (B) Magnified images from the squared regions shown in (A). Arrowheads point to FABP7 immunoreactivity in cell nucleus. (C, D) Patient overall survival (OS) curves generated based on FABP7 nuclear (C) and cytoplasmic immunoreactivity (D). (E, F) Patient overall survival curves generated based on nuclear FABP7 immunoreactivity in a patient population under (E) or above (F) the median age of 55. Three to six cores from each tumor on TMA slides were immunostained and scored. HR denotes hazard ratio. Scale bars: 100 μm. (G) Graphical summary of the role of the FABP7-RXRα neurogenic pathway in determining stationary to migratory transition in GSC and tumor progression.
Discussion
CSCs, including GSCs, represent a small fraction of tumor cells that drive tumor invasion, progression, and treatment resistance.31 These cells show significant heterogeneity as well as plasticity.32,33,34 CSCs have been classified as stationary (mostly found in the more differentiated aspects of the tumors) or mobile/migratory (predominately located at the tumor-host interface where they drive tumor infiltration and metastasis).3 The determining process underlying the migratory state of CSCs is believed to be the acquisition of the EMT phenotype.3 Recent studies have shown dynamic changes in the epithelial-and-mesenchymal states of CSCs during tumor progression, with the more mesenchymal CSCs located at the invasive edge of primary tumors.35,36 Diversity of neural stem cells in the subventricular zone of adult normal brain has also been reported, with FABP7 expression being indicative of activated neural stem cells.37 FABP7 shows inter-tumoral expression heterogeneity in GBM, with highest levels found in the classical molecular subtype. FABP7 is also found at much higher levels in IDH-wild-type GBM tumors compared to IDH mutant tumors that are no longer categorized as GBM (Supplementary Figure 13).
Using patient- and xenograft-derived cell line models, we found that FABP7 expression marks a subpopulation of GSCs with enhanced cell migration. Our data show that FABP7 serves as a driver of migration in GSCs by upregulating EMT. In keeping with immunostaining data showing elevated FABP7 in the invading regions of human GBM tumors,10 FABP7-immunoreactive cells were predominantly observed in the migratory tumor regions located adjacent to pseudo-palisading cell layers, as well as the expanding (U87 FABP7)/infiltrative (patient-derived A4-007) fronts of xenograft brain tumors (Figures 3E, I-L; Supplementary Figures 4A,C and 6A,C). Furthermore, infiltrative tumor growth in brain was directly associated with FABP7 as intraperitoneal injections of the FABP7 inhibitor SBFI-26 resulted in greatly reduced infiltration (Figures 3L). SBFI-26 has been used as a potent inhibitor of FABP7,6 although it can also inhibit FABP5 activity. Our findings are in keeping with FABP7 serving as a critical biomarker for activated migratory GSCs driving GBM infiltration.
Our results reveal a crucial role for nuclear FABP7 through activation of RXRα, a nuclear receptor and transcription factor enriched in mammalian brain.22,38,39 DHA and AA are ligands of FABP724 and RXRα22,40 that have been implicated in the neurogenesis of both developing and adult brains.5,22,38,41 We show that FABP7-mediated activation of RXRα is enhanced in GBM cells cultured in DHA- or AA-supplemented medium. Importantly, supplementation of GSC culture medium with DHA or AA significantly induced expression of proteins involved in lipid storage, fatty acid beta-oxidation, and invasion. We further found that RXRα transactivation parallels FABP7 expression and mediates FABP7-induced cell migration in xenograft-derived metastatic GBM cells.
In GBM tumors, PUFAs are released from necrotic tumor tissue, resulting in increased levels of AA and DHA in the tumor microenvironment.42 Furthermore, upregulation of the PUFA synthesis enzyme ELOVL2 in GBM has been shown to promote de novo synthesis of DHA, resulting in enhanced EGFR signaling and GSC maintenance.25 Shakia et al. further showed that an increased PUFA content in GSCs, but not in non-GSCs, enhanced cell viability and self-renewal.7 Together, these studies indicate a correlation between endogenous PUFA content and GBM stemness/invasiveness. However, how PUFAs affect stemness and cell viability remains poorly understood. Based on our observations, we speculate that as GBM progresses, levels of endogenous PUFAs increase due to membrane breakdown of necrotic cells42 or de novo production of PUFAs.25 Free PUFAs in GSCs are bound by FABP7 and delivered to the nucleus where they activate the nuclear receptor RXRα. The resulting upregulation of EMT and stemness markers leads to the transition of FABP7-expressing GSCs from the quiescent/stationary to an activated/migratory state (Figure 6G).
The discovery of GSCs has led to the hypothesis that GBMs may arise from aberrant neural stem cells.43 The subventricular zone of the lateral ventricles (SVZ) is the major source of neural stem cells and remains FABP7 positive in adult brain.44,45 Loss of FABP7-positive stem/progenitor cells correlates with reduced neurogenesis in aging rodents and postnatal humans.37 In animal models, gliomas have been shown to develop from neural stem cells in the SVZ or from glial progenitor cells.46,47 Patients with GBM lesions adjacent to the SVZ show multifocal lesions at diagnosis, distant recurrence, and poor prognosis compared to GBMs that are distant from the SVZ.48,49 Interestingly, recent studies indicate that the cell types found in GBM recapitulate a normal neurodevelopmental hierarchy with glial progenitor-like cells at the apex, and that human adult GBM indeed harbor radial glial-like cells which share expression of radial glial and stem cell markers including FABP7.33,50 Based on evidence from the literature and the findings reported in this study, we propose that the FABP7-mediated neurogenic program in adult normal brain is appropriated by GBM thereby promoting infiltration of normal brain parenchyma.
In summary, we have identified a FABP7-expressing migratory subpopulation of GSCs that appear to underlie the infiltrative properties characteristic of GBM tumors. Our data indicate that FABP7 orchestrates stemness and EMT in GBM by facilitating PUFA-induced activation of RXRα, a nuclear receptor involved in brain neurogenesis. We propose that inhibiting FABP7 or RXRα may provide a targeted approach to prevent infiltration and recurrence in GBM.
Supplementary Material
Acknowledgments
We thank Drs. Xuejun Sun and Guobin Sun for their help with cell imaging data acquisition and analysis, Liz Monckton for technical assistance, and Drs. Hua Chen and Kenneth Petruk for their kind donation of the ED-series GBM cultures.
Contributor Information
Rong-Zong Liu, Department of Oncology, University of Alberta, Cross Cancer Institute, Edmonton, AB, Canada.
Won-Shik Choi, Department of Oncology, University of Alberta, Cross Cancer Institute, Edmonton, AB, Canada.
Saket Jain, Department of Oncology, University of Alberta, Cross Cancer Institute, Edmonton, AB, Canada.
Xia Xu, Department of Oncology, University of Alberta, Cross Cancer Institute, Edmonton, AB, Canada.
Darryl D Glubrecht, Department of Oncology, University of Alberta, Cross Cancer Institute, Edmonton, AB, Canada.
Anthony G Tessier, Department of Oncology, University of Alberta, Cross Cancer Institute, Edmonton, AB, Canada.
Jacob C Easaw, Department of Oncology, University of Alberta, Cross Cancer Institute, Edmonton, AB, Canada.
B Gino Fallone, Department of Oncology, University of Alberta, Cross Cancer Institute, Edmonton, AB, Canada.
Roseline Godbout, Department of Oncology, University of Alberta, Cross Cancer Institute, Edmonton, AB, Canada.
Funding
Canadian Institutes of Health Research (#130314 and #190313) and the Terry Fox Research Institute – Prairie Cancer Research Consortium
Conflict of interest statement
The authors declare no conflict of interest.
Authorship statement
Conceptualization: RG and RZL. Experimentation: RZL, WSC, SJ, XX, MEE, DDG, AGT and BGF. Tissue microarray: JCE. Manuscript - Drafting: RZL; Editing: RG and RZL; approved by all authors. Supervision: RG.
References
- 1. Buckner JC, Brown PD, O’Neill BP, et al. Central nervous system tumors. Mayo Clin Proc. 2007;82(10):1271–1286. [DOI] [PubMed] [Google Scholar]
- 2. Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN.. Cancer stem cells in glioblastoma. Genes Dev. 2015;29(12):1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T.. Opinion: Migrating cancer stem cells - an integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5(9):744–749. [DOI] [PubMed] [Google Scholar]
- 4. Thankamony AP, Saxena K, Murali R, Jolly MK, Nair R.. Cancer stem cell plasticity - A deadly deal. Front Mol Biosci. 2020;7(Apr 30):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu RZ, Mita R, Beaulieu M, Gao Z, Godbout R.. Fatty acid binding proteins in brain development and disease. Int J Dev Biol. 2010;54(8-9):1229–1239. [DOI] [PubMed] [Google Scholar]
- 6. Hoang-Minh LB, Siebzehnrubl FA, Yang C, et al. Infiltrative and drug-resistant slow-cycling cells support metabolic heterogeneity in glioblastoma. EMBO J. 2018;37(23):e98772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shakya S, Gromovsky AD, Hale JS, et al. Altered lipid metabolism marks glioblastoma stem and non-stem cells in separate tumor niches. Acta Neuropathol Commun. 2021;9(1):101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Choi WS, Xu X, Goruk S, et al. FABP7 facilitates uptake of docosahexaenoic acid in glioblastoma neural stem-like Cells. Nutrients. 2021;13(8):2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alzial G, Renoult O, Paris F, et al. Wild-type isocitrate dehydrogenase under the spotlight in glioblastoma. Oncogene. 2022;41(5):613–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mita R, Coles JE, Glubrecht DD, et al. B-FABP-expressing radial glial cells: The malignant glioma cell of origin? Neoplasia. 2007;9(9):734–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu RZ, Choi WS, Jain S, et al. The FABP12/PPARgamma pathway promotes metastatic transformation by inducing epithelial-to-mesenchymal transition and lipid-derived energy production in prostate cancer cells. Mol Oncol. 2020;14(12):3100–3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu RZ, Li S, Garcia E, et al. Association between cytoplasmic CRABP2, altered retinoic acid signaling, and poor prognosis in glioblastoma. Glia. 2016;64(6):963–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu RZ, Graham K, Glubrecht DD, et al. A fatty acid-binding protein 7/RXRbeta pathway enhances survival and proliferation in triple-negative breast cancer. J Pathol. 2012;228(3):310–321. [DOI] [PubMed] [Google Scholar]
- 14. Liu RZ, Garcia E, Glubrecht DD, et al. CRABP1 is associated with a poor prognosis in breast cancer: Adding to the complexity of breast cancer cell response to retinoic acid. Mol Cancer. 2015;14(Jul 5):129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xu X, Wang Y, Choi WS, Sun X, Godbout R.. Super resolution microscopy reveals DHA-dependent alterations in glioblastoma membrane remodelling and cell migration. Nanoscale. 2021;13(21):9706–9722. [DOI] [PubMed] [Google Scholar]
- 16. Neftel C, Laffy J, Filbin MG, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178(4):835–849.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kast-Woelbern HR, Dana SL, Cesario RM, et al. Rosiglitazone induction of Insig-1 in white adipose tissue reveals a novel interplay of peroxisome proliferator-activated receptor gamma and sterol regulatory element-binding protein in the regulation of adipogenesis. J Biol Chem. 2004;279(23):23908–23915. [DOI] [PubMed] [Google Scholar]
- 18. Siebzehnrubl FA, Silver DJ, Tugertimur B, et al. The ZEB1 pathway links glioblastoma initiation, invasion and chemoresistance. EMBO Mol Med. 2013;5(8):1196–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Loh CY, Chai JY, Tang TF, et al. The E-cadherin and n-cadherin switch in epithelial-to-mesenchymal transition: Signaling, therapeutic implications, and challenges. Cells. 2019;8(10):1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Osswald M, Jung E, Sahm F, et al. Brain tumour cells interconnect to a functional and resistant network. Nature. 2015;528(7580):93–98. [DOI] [PubMed] [Google Scholar]
- 21. Wang X, Liang J, Sun H.. The network of tumor microtubes: An improperly reactivated neural cell network with stemness feature for resistance and recurrence in gliomas. Front Oncol. 2022;12(Jun 29):921975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. de Urquiza AM, Liu S, Sjoberg M, et al. Docosahexaenoic acid, a ligand for the retinoid X receptor in mouse brain. Science. 2000;290(5499):2140–2144. [DOI] [PubMed] [Google Scholar]
- 23. Mita T, Mayanagi T, Ichijo H, et al. Docosahexaenoic acid promotes axon outgrowth by translational regulation of tau and collapsin response mediator protein 2 expression. J Biol Chem. 2016;291(10):4955–4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu LZ, Sanchez R, Sali A, Heintz N.. Ligand specificity of brain lipid-binding protein. J Biol Chem. 1996;271(40):24711–24719. [DOI] [PubMed] [Google Scholar]
- 25. Gimple RC, Kidwell RL, Kim LJY, et al. Glioma stem cell-specific superenhancer promotes polyunsaturated fatty-acid synthesis to support EGFR signaling. Cancer Discov. 2019;9(9):1248–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zou Y, Watters A, Cheng N, et al. Polyunsaturated fatty acids from astrocytes activate PPARgamma signaling in cancer cells to promote brain metastasis. Cancer Discov. 2019;9(12):1720–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bensaad K, Favaro E, Lewis CA, et al. Fatty acid uptake and lipid storage induced by HIF-1alpha contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 2014;9(1):349–365. [DOI] [PubMed] [Google Scholar]
- 28. Wu X, Geng F, Cheng X, et al. Lipid droplets maintain energy homeostasis and glioblastoma growth via autophagic release of stored fatty acids. iScience. 2020;23(10):101569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Liang Y, Bollen AW, Aldape KD, Gupta N.. Nuclear FABP7 immunoreactivity is preferentially expressed in infiltrative glioma and is associated with poor prognosis in EGFR-overexpressing glioblastoma. BMC Cancer. 2006;6(Apr 19):97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kaloshi G, Mokhtari K, Carpentier C, et al. FABP7 expression in glioblastomas: Relation to prognosis, invasion and EGFR status. J Neurooncol. 2007;84(3):245–248. [DOI] [PubMed] [Google Scholar]
- 31. Vermeulen L, de Sousa e Melo F, Richel DJ, Medema JP.. The developing cancer stem-cell model: Clinical challenges and opportunities. Lancet Oncol. 2012;13(2):e83–e89. [DOI] [PubMed] [Google Scholar]
- 32. Chen R, Nishimura MC, Bumbaca SM, et al. A hierarchy of self-renewing tumor-initiating cell types in glioblastoma. Cancer Cell. 2010;17(4):362–375. [DOI] [PubMed] [Google Scholar]
- 33. Couturier CP, Ayyadhury S, Le PU, et al. Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy. Nat Commun. 2020;11(1):3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dirkse A, Golebiewska A, Buder T, et al. Stem cell-associated heterogeneity in glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment. Nat Commun. 2019;10(1):1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bocci F, Gearhart-Serna L, Boareto M, et al. Toward understanding cancer stem cell heterogeneity in the tumor microenvironment. Proc Natl Acad Sci U S A. 2019;116(1):148–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pastushenko I, Brisebarre A, Sifrim A, et al. Identification of the tumour transition states occurring during EMT. Nature. 2018;556(7702):463–468. [DOI] [PubMed] [Google Scholar]
- 37. Giachino C, Basak O, Lugert S, et al. Molecular diversity subdivides the adult forebrain neural stem cell population. Stem Cells. 2014;32(1):70–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cao H, Li MY, Li G, et al. Retinoid X receptor alpha regulates DHA-dependent spinogenesis and functional synapse formation in vivo. Cell Rep. 2020;31(7):107649. [DOI] [PubMed] [Google Scholar]
- 39. Simandi Z, Horvath A, Cuaranta-Monroy I, et al. RXR heterodimers orchestrate transcriptional control of neurogenesis and cell fate specification. Mol Cell Endocrinol. 2018;471(Aug 15):51–62. [DOI] [PubMed] [Google Scholar]
- 40. Lengqvist J, Mata De Urquiza A, Bergman AC, et al. Polyunsaturated fatty acids including docosahexaenoic and arachidonic acid bind to the retinoid X receptor alpha ligand-binding domain. Mol Cell Proteomics. 2004;3(7):692–703. [DOI] [PubMed] [Google Scholar]
- 41. He C, Qu X, Cui L, Wang J, Kang JX.. Improved spatial learning performance of fat-1 mice is associated with enhanced neurogenesis and neuritogenesis by docosahexaenoic acid. Proc Natl Acad Sci U S A. 2009;106(27):11370–11375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hirano H, Takeshima H, Niiro M, et al. The environment of increased concentration of docosahexaenoic acid in glioblastoma may suppress the anti-tumor effect of macrophages. Oncol Rep. 2005;13(6):1185–1191. [PubMed] [Google Scholar]
- 43. Achanta P, Sedora Roman NI, Quinones-Hinojosa A.. Gliomagenesis and the use of neural stem cells in brain tumor treatment. Anticancer Agents Med Chem. 2010;10(2):121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Matsumata M, Sakayori N, Maekawa M, et al. The effects of Fabp7 and Fabp5 on postnatal hippocampal neurogenesis in the mouse. Stem Cells. 2012;30(7):1532–1543. [DOI] [PubMed] [Google Scholar]
- 45. Sundholm-Peters NL, Yang HK, Goings GE, Walker AS, Szele FG.. Radial glia-like cells at the base of the lateral ventricles in adult mice. J Neurocytol. 2004;33(1):153–164. [DOI] [PubMed] [Google Scholar]
- 46. Alcantara Llaguno S, Chen J, Kwon CH, et al. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell. 2009;15(1):45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Assanah M, Lochhead R, Ogden A, et al. Glial progenitors in adult white matter are driven to form malignant gliomas by platelet-derived growth factor-expressing retroviruses. J Neurosci. 2006;26(25):6781–6790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chaichana KL, McGirt MJ, Frazier J, et al. Relationship of glioblastoma multiforme to the lateral ventricles predicts survival following tumor resection. J Neurooncol. 2008;89(2):219–224. [DOI] [PubMed] [Google Scholar]
- 49. Lim DA, Cha S, Mayo MC, et al. Relationship of glioblastoma multiforme to neural stem cell regions predicts invasive and multifocal tumor phenotype. Neuro Oncol. 2007;9(4):424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang R, Sharma R, Shen X, et al. Adult human glioblastomas harbor radial glia-like cells. Stem Cell Rep. 2020;15(1):275–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


