Abstract
c-Met has been an attractive target of prognostic and therapeutic studies in various cancers. TPX-0022 is a macrocyclic inhibitor of c-Met, c-Src and CSF1R kinases and is currently in phase I/II clinical trials in patients with advanced solid tumors harboring MET gene alterations. In this study, we determined the co-crystal structures of the c-Met/TPX-0022 and c-Src/TPX-0022 complexes to help elucidate the binding mechanism. TPX-0022 binds to the ATP pocket of c-Met and c-Src in a local minimum energy conformation and is stabilized by hydrophobic and hydrogen bond interactions. In addition, TPX-0022 exhibited potent activity against the resistance-relevant c-Met L1195F mutant and moderate activity against the c-Met G1163R, F1200I and Y1230H mutants but weak activity against the c-Met D1228N and Y1230C mutants. Overall, our study reveals the structural mechanism underlying the potency and selectivity of TPX-0022 and the ability to overcome acquire resistance mutations and provides insight into the development of selective c-Met macrocyclic inhibitors.
Keywords: C-Met, TPX-0022, Crystal structure, Resistance-relevant mutations, C-Src
Graphical Abstract
1. Introduction
c-Met, also known as hepatocyte growth factor receptor (HGFR), is a receptor tyrosine kinase encoded by the proto-oncogene MET, and HGF is the only known ligand[1]. The c-Met signaling pathway plays a significant role in many physiological processes, such as embryonic development, organ regeneration, and tissue damage repair[2]. Dysregulation of c-Met activity, including MET amplification, overexpression, mutation, and rearrangement, is widely observed in human cancers such as non-small cell lung cancer (NSCLC) [1], [3], gastric cancer[4], and hepatocellular carcinoma[5]. MET alterations are associated with cancer development, metastatic progression and poor prognosis[6], [7], [8]. Additionally, MET alterations (mainly gene amplification) constitute the most frequent cause of bypass pathway activation as an acquired resistance mechanism to EGFR tyrosine kinase inhibitors (TKIs) [9]. Therefore, the development of c-Met-targeted drugs has been an effective and promising strategy for the treatment of cancers harboring MET alterations [10], [11], [12], [13].
c-Met protein is composed of semaphorin domain, plexin-semaphorin-integrin (PSI) domain, immunoglobulin-plexin-transcription (IPT) domain, transmembrane domain (TM), juxtamembrane domain (JM), kinase domain, and docking sites[14]. Monoclonal antibodies targeting the semaphorin domain and kinase inhibitors targeting the kinase domain are effective strategies for the development of c-Met-targeted drugs. Small molecule inhibitors targeting the c-Met kinase domain have made inspiring progress in recent years. Six inhibitors have been approved by the FDA or NMPA for clinical treatment of cancer (Fig. S1) [15], [16], [17], [18], [19]. The indications for these approved drugs are mainly for NSCLC harboring MET gene alterations [11]. Meanwhile, some c-Met inhibitors are under clinical trials for gastric cancer and hepatocellular carcinoma [10]. TPX-0022 (also known as elzovantinib), developed by Turning Point Therapeutics, is an orally bioavailable c-Met inhibitor that also targets CSF1R and c-Src kinases [20], [21]. Unlike most linear c-Met inhibitors, TPX-0022 possesses a compact multicyclic scaffold, which could offer unique drug-like profiles such as desirable conformational rigidity, improved oral bioavailability, enhanced metabolic stability and cell permeability (Fig. 1A) [22]. TPX-0022 was granted fast track approval and orphan drug designation by the FDA for the treatment of gastric cancer in 2021. Preclinical data indicated that TPX-0022 blocked both HGF- and CSF1-mediated signaling and showed activity in multiple MET- and CSF1R-dependent cell lines and xenograft models[21]. An ongoing phase I/II study (NCT03993873) will evaluate the safety, tolerability, pharmacokinetics and efficacy of TPX-0022 for patients with advanced NSCLC and gastric cancer harboring MET genetic alterations. The initial results are encouraging[23].
Fig. 1.
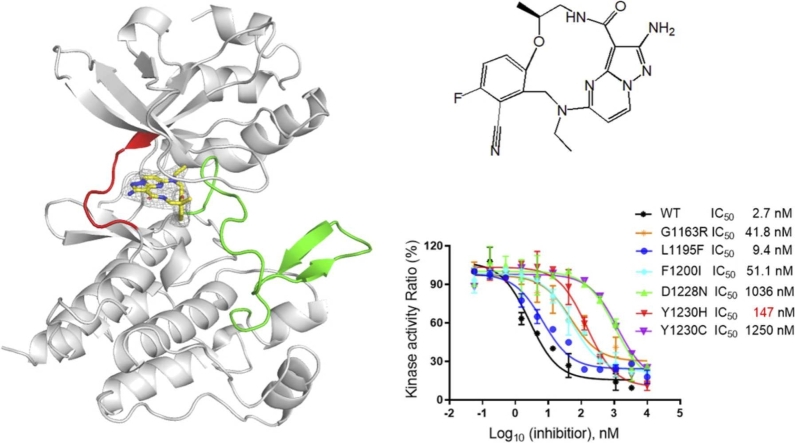
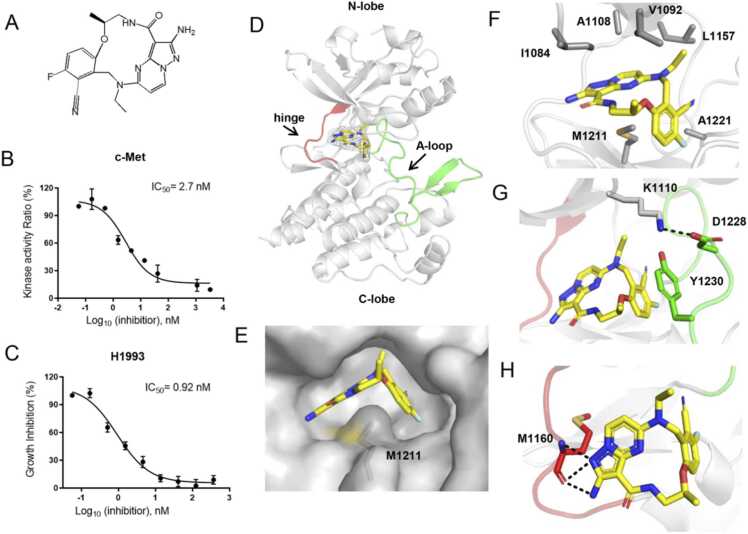
Structural basis of the TPX-0022/c-Met interaction. (A) Chemical structure of TPX-0022. (B) The potency of TPX-0022 against c-Met was analysed using an in vitro kinase assay kit. (C) Cellular activity determination of TPX-0022 using NCI-H1993 cells. Data in (B/C) are represented as the mean ± SD of n = 3 independent experiments. (D) The overall structure of the c-Met/TPX-0022 complex. c-Met kinase is colored gray. TPX-0022 is colored yellow with the electron density map (2mFo-DFc) at 1σ showing the density. The hinge loop (red) and A-loop (green) are highlighted. (E) Surface presentation of c-Met shows the binding pocket of TPX-0022. (F) Hydrophobic interaction between c-Met and TPX-0022. (G) Role of residues Y1230, K1110 and D1228 in the interaction with TPX-0022. (H) Hydrogen bond interactions between c-Met and TPX-0022. Salt bridges and hydrogen bonds are indicated as black dotted lines.
The majority of c-Met inhibitors currently approved or under development are type I or type II inhibitors. Type I inhibitors occupy the ATP-binding pocket in the active conformation, while type II inhibitors bind to the pocket in the inactive conformation and extend to a nearby allosteric pocket[11], [24]. For all TKIs, it is difficult to avoid the development of acquired drug resistance in long-term targeted drug therapy. The resistance mechanisms related to MET inhibitors include on-target resistance and off-target resistance. The most common type of on-target resistance is secondary site mutations in the kinase domain of MET, such as the G1163, D1228 and Y1230 mutations for type I inhibitors and the L1195 and F1200 mutations for type II inhibitors [25].
TPX-0022 was reported to be a type I inhibitor, whereas the detailed structural mechanism of TPX-0022 acting on its targets remains unclear. Whether TPX-0022 can overcome c-Met resistance-associated mutations is also unknown. Here, we determined the structural basis of TPX-0022 binding to c-Met and the inhibitory activity of TPX-0022 against common resistance-related c-Met mutations. In addition, we determined the structure of the c-Src/TPX-0022 complex and compared in detail the binding properties of TPX-0022 with other macrocyclic kinase inhibitors. These structural and biochemical data help elucidate the binding mechanism of TPX-0022 and facilitate the design and optimization of prospective macrocyclic kinase inhibitors.
2. Materials and methods
2.1. Plasmids
The kinase domain of human wild-type c-Met (residues 1038–1346) with an N-terminal His-SUMO tag followed by an Ulp1 protease cleavage site was cloned into a modified pET28a vector. Chicken Src (residues 251–533) was cloned into a modified pET28a vector with an N-terminal 6 ×His tag followed by a PreScission protease cleavage site. The c-Met mutations (G1163R, L1195F, F1200I, D1228N, Y1230H, Y1230C) were introduced by PCR using primers with the desired mutations.
2.2. Protein expression and purification
Plasmids were transformed into E. coli BL21(DE3) cells together with YopH to obtain nonphosphorylated c-Met or c-Src protein. Cells were grown in LB medium at 37 °C until the OD600 reached 0.8 and then induced with 0.5 mM IPTG for 16 h at 12 °C or 18 °C. Cells were harvested by centrifugation for the next purification.
All steps of purification were performed at 4 °C. Cells were resuspended in lysis buffer containing 20 mM Tris-HCl pH 8.5, 500 mM NaCl, 10% (v/v) glycerol and 0.5% (v/v) Triton X-100. Then, the cells were lysed by ultrasonic for a total of 30 min with the program of 100w power, 2 s of operation and 4 s of interval. The lysate was clarified by centrifugation at 18000g for 30 min. The proteins were first purified by Ni-NTA affinity chromatography. The purified His-tagged recombinant proteins were pooled and digested overnight with Ulp1 or PreScission protease and dialyzed simultaneously. Then, the reaction mixtures were reloaded onto the His-Trap column, and the target proteins were collected in the flow through. Anion exchange chromatography (Mono Q) and size exclusion chromatography (SEC) were exploited for further purification. The purified c-Met proteins were stored in buffer containing 20 mM Tris-HCl pH 8.5, 150 mM NaCl, 10% (v/v) glycerol and 1 mM TCEP, while the c-Src proteins were stored in buffer containing 20 mM Tris-HCl pH 8.0, 150 mM NaCl, 5% (v/v) glycerol, and 1 mM TCEP. The proteins were concentrated to approximately 5–10 mg/mL and flash frozen for storage at − 80 °C for subsequent studies.
2.3. Kinase inhibition assay
The ADP-Glo™ Kinase Assay Kit (Part No #V9101, Promega) was used to perform kinase inhibition assays. The experiments were performed according to the manufacturer’s instructions. All kinase inhibition assays were performed with optimized kinase assay buffer consisting of 40 mM Tris-HCl 8.0, 20 mM NaCl, 0.1 mg/mL BSA, 20 mM MgCl2, 1 mM TCEP, and 4% DMSO.[26] Briefly, TPX-0022 in triple dilutions (ranging from 0.01 nM to 25 μM) was incubated with kinases (0.1 μM) in white opaque 384-well plates for 30 min at room temperature. Second, 5 ×ATP plus poly (4:1 Glu, Tyr) peptides were added to start the kinase reactions. The final reaction consisted of 0.04 μM kinase, 50 μM Tyr4 peptide and 10 μM ATP in kinase buffer. Then, the reactions were terminated by the addition of stop buffer ADP-Glo after half an hour of incubation. The produced ADP was completely converted to ATP after 40 min of incubation. Finally, fluorescence was measured on a multimode plate reader (Perkin-Elmer) after the addition of detection reagent for 30–60 min. A three-parameter log [inhibitor] versus response model (Equation: Y=Bottom+(Top-Bottom)/(1 +10^((X-LogIC50)))) in GraphPad Prism software was used to determine the IC50 values.
2.4. Crystallization
Crystals were obtained by the hanging drop vapour diffusion method. Protein-inhibitor complexes were prepared by mixing protein (5–10 mg/mL) with inhibitor at a 1:1.5 molar ratio and incubated on ice for 30 min. c-Met/TPX-0022 crystals were grown for 7 days at 18 °C using a well solution containing 0.1 M HEPES (pH 7.8) and 15–30% (v/v) PEG 8000, while crystals of c-Src/TPX-0022 were grown in 3 days at 18 °C using a well solution containing 0.1 M MES (pH 6.4), 2% glycerol, 8% PEG 4000, 50 mM sodium acetate, and 10 mM MgCl2. Prior to diffraction experiments, crystals were cryoprotected by supplementing the mother liquor with 20% glycerol and then cooled in liquid nitrogen.
2.5. Data collection and structure determination
Crystal datasets were collected at the BL02U [27] and BL19U1 [28] beamlines of the Shanghai Synchrotron Radiation Facility (SSRF). The diffraction data were processed using XDS [29]. Phases were determined by molecular replacement using Phenix. Phaser[30] and the apo c-Met structure (PDB ID: 6SD9)[31]. The inhibitors were drawn by ChemDraw software, and the models were built by CCP4 software. Phenix.ligandfit was run to place the ligand, and the correlation coefficient (CC) value was above 0.7. Then, structures were refined with phenix.refine, and model building was performed using WinCoot[32]. Structural graphics were presented by PyMOL [33]. Data collection and refinement statistics are presented in Table S1.
2.6. Molecular docking
Computational docking was performed to predict the binding of TPX-0022 to CSF1R, c-Src, c-Met WT and c-Met mutations. PDB 3LCD, 4U5J, 8GVJ and 5HLW with all waters and ligands removed as the protein models for CSF1R, c-Src, c-Met WT and c-Met Y1230H mutant. Structures of c-Met G1163R, L1195F, F1200I, D1228N were acquired by mutating the residues on the basis of the c-Met WT structure. The c-Met Y1230C structure was acquired by mutating H1230 to Cys on the basis of the c-Met Y1230H structure. Polar hydrogens, Gasteiger charges and rotatable bonds to the inhibitors were assigned by the AutoDock Tools program[34]. The docking simulations were carried out using proteins with flexible procket and a flexible ligand. A docking grid with dimensions of 40 * 40 * 40 points in the x-, y-, and z-axis directions was built, which encompassed the entire ligand-binding clefts.
2.7. Metadynamic studies
Schrödinger Maestro software was used to perform the torsional angle scan analysis. We applied metadynamics to sample the conformation of TPX-0022 using the dihedral angle of the CH2N(C2H3) linker as the collective variable. The OPLS3 force field was used, and the SPC solvent model was used to solvate the compound. The system was relaxed using default settings and simulated in the NPT ensemble with the temperature at 300 K and the pressure set at 1 atm. The Gaussian height was set to 0.03 kcal/mol, and the bias potential was added every 0.09 ps. Metadynamic analysis was used for the simulations.
2.8. Prediction of ADME/drug-likeness properties
Absorption parameters, distribution parameters, metabolism parameters and drug- likeness properties for the macrocyclic inhibitors were predicted using the ADMET program SwissADME, which is developed by the Swiss Institute of Bioinformatics and freely available at www.swissadme.ch[35].
3. Results
3.1. Structural basis of TPX-0022 binding to c-Met
We first performed a kinase assay to verify the binding activity of TPX-0022 to c-Met. The results indicated that TPX-0022 potently inhibited c-Met with an IC50 of 2.7 nM (Fig. 1B). In the NSCLC NCI-H1993 cell line harboring c-Met alteration, TPX-0022 effectively inhibited cell proliferation with an IC50 of 0.92 nM (Fig. 1C). The intracellular inhibitory activity of TPX-0022 was stronger than that of the kinase assay, possibly due to differences in experimental methods or the inhibition of multiple kinases by the inhibitor.
To further analyse the molecular interactions between TPX-0022 and c-Met, we determined the co-crystal structure of TPX-0022 with c-Met at 2.7 Å. The statistics of data collection and model refinement are listed in Table S1. As shown in Fig. 1D, c-Met exhibits a typical protein tyrosine kinase structure, consisting of a bilobed architecture (N-lobe and C-lobe). TPX-0022 binds to the ATP-binding cavity of c-Met in a type-I inhibitor binding mode (Fig. 1D, Fig. S2A). TPX-0022 bent into a “U-shaped” binding conformation and wrapped around Met1211 (Fig. 1E). It establishes hydrophobic contacts with residues I1084, V1092, A1108, L1157, M1211 and A1221 in the ATP pocket (Fig. 1F). In addition, the benzonitrile group of TPX-0022 forms a π-stacking interaction with the aromatic side chain of Y1230 of the A-loop, and the orientation of the A-loop is stabilized by a salt bridge between D1228 and K1110 (Fig. 1G). The pyrazolo[1,5-а]pyrimidin-2-ylaminne group of TPX-0022 forms two hydrogen bonds with the main chain of Met1160 in the hinge loop of c-Met (Fig. 1H). These van der Waals interactions help stabilize the c-Met/TPX-0022 complex.
3.2. Potency of TPX-0022 against c-Met resistance-relevant mutations
On-target secondary mutation is one of the main mechanisms driving acquired resistance to c-Met inhibitors. We used a kinase assay to investigate the inhibitory potency of TPX-0022 against six common mutations of c-Met, including G1163R, D1228N, Y1230H and Y1230C, which are frequently induced by type I inhibitors, as well as L1195F and F1200I, which are observed for type II inhibitors.
For mutants L1195F and F1200I, TPX-0022 still exhibited inhibition activities of 9.4 nM and 51.1 nM, respectively (Fig. 2A). Residues L1195 and F1200 are not involved in direct interactions with TPX-0022 but play a role in hydrophobic interactions with F1223 to stabilize the A-loop (Fig. 2B). Mutation-induced changes in the hydrophobic environment may result in a 3- to 18-fold reduction in potency.
Fig. 2.
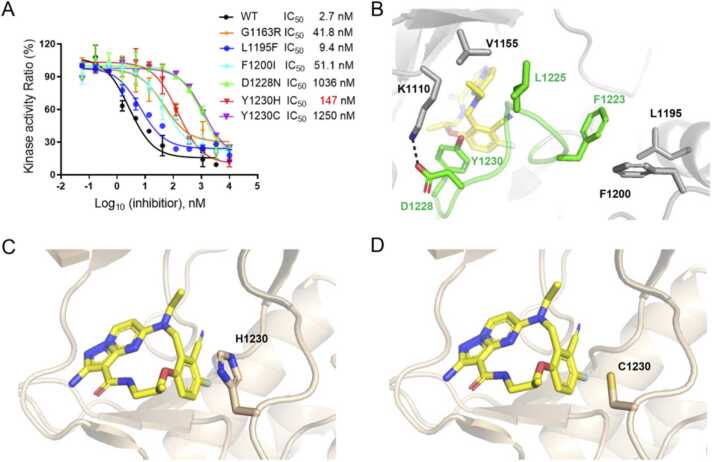
Potency of TPX-0022 against c-Met resistance-relevant mutations. (A) Potency of TPX-0022 against c-Met resistance-relevant mutations using kinase assays. (B) The positions of resistance-relevant mutations in the c-Met/TPX-0022 structure. Salt bridges are indicated as black dotted lines. (C) The modelled c-Met Y1230H/TPX-0022 complex structure was acquired by superposition of the c-Met Y1230H/compound 14 (PDB ID: 5HLW) structure with the c-Met/TPX-0022 structure. (D) The modelled c-Met Y1230C/TPX-0022 complex. The c-Met Y1230C structure was acquired by mutating H1230 to Cys on the basis of the c-Met Y1230H structure. TPX-0022 is colored yellow. c-Met WT is colored gray. c-Met Y1230H and c-Met Y1230C are colored wheat.
For mutant G1163R, TPX-0022 exhibited inhibition activity of 41.8 nM (Fig. 2A). Residue G1163 is located at the edge of pocket and forms van der Waals interactions with TPX-0022 (Fig. S3), and G1163R mutation resulted in a 15-fold reduction in potency. The inhibitory potency of TPX-0022 against D1228N was reduced by approximately 383-fold with an IC50 value of 1036 nM (Fig. 2A). D1228 is engaged in a salt bridge with K1110, which helps stabilize the A-loop of c-Met (Fig. 2B). Mutation D1228N could result in breakage of the salt bridge and consequently affect the interaction between TPX-0022 and the A-loop. Meanwhile, Y1230 forms a critical π-stacking interaction with TPX-0022. Y1230C resulted in a 460-fold decrease in potency, probably owing to destruction of the π-stacking interaction. Surprisingly, TPX-0022 still exhibited moderate inhibitory potency against Y1230H with an IC50 value of 147 nM (Fig. 2A). A modelled structure of TPX-0022 with the Y1230H mutant indicated that the imidazole group of Y1230H could also form a π-stacking interaction with the pyrazolo[1,5-а]pyrimidin group of TPX-0022 (Fig. 2C), while the Y1230C mutant could not (Fig. 2D). In addition, the predicted binding energy of TPX-0022 to c-Met WT, Y1230H, Y1230C was − 9.7, − 8.8, − 8.3 kcal•mol-1, and Ki values were 78.9, 307.1 and 737.8 nM, respectively (Table S2), indicating that the Y1230C mutant resulted in a more severe impact to c-Met/TPX-0022 interaction than the Y1230H mutant.
3.3. Structural basis of the binding of TPX-0022 to c-Src
In addition to c-Met, TPX-0022 also shows potent inhibitory ability to c-Src and CSF1R, which is thought to have the potential to improve clinical efficacy[21]. Kinase assays indicated that TPX-0022 had similar inhibitory potency against c-Met and c-Src, with IC50 values of 2.7 nM and 3.7 nM, respectively (Figs. 1B, 3A). To help elucidate the binding mechanism, we determined the structure of TPX-0022 in complex with c-Src at a resolution of 2.8 Å (Table S1).
Fig. 3.
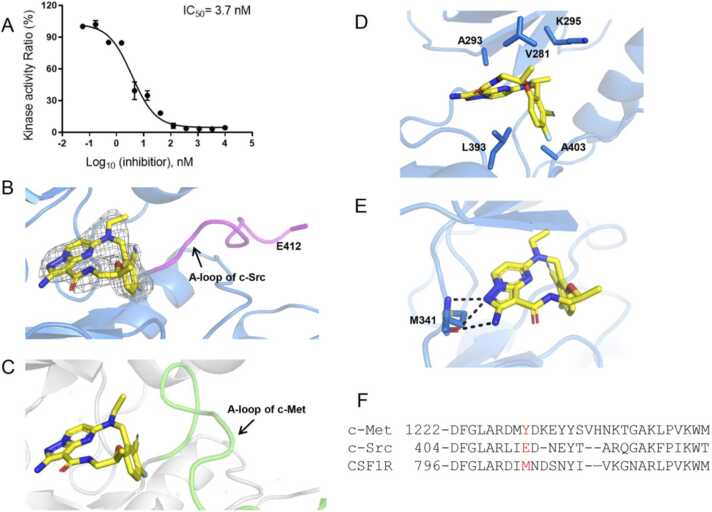
Structural basis of the c-Src/TPX-0022 interaction. (A) Potency of TPX-0022 against c-Src was determined by kinase inhibition assay. (B) Binding mode of TPX-0022 to c-Src. The A-loop is highlighted in magenta. The electron density map (2mFo-DFc) at 1σ shows the density of TPX-0022. (C) Binding mode of TPX-0022 to c-Met. The A-loop is highlighted in green. (D) The hydrophobic interaction between c-Src and TPX-0022. (E) The hydrogen bonds between c-Src and TPX-0022. (F) Sequence alignment of the A-loop residues of kinase proteins. Hydrogen bonds are shown as black dotted lines. c-Src is colored marine. c-Met is colored gray. TPX-0022 is colored yellow.
The overall structure of the c-Src/TPX-0022 complex is similar to the c-Met/TPX-0022 structure, with an RMSD of 0.8 Å (Fig. 3B-C, S4). TPX-0022 binds to the ATP pocket of c-Src in a similar “U-shaped” conformation and forms hydrophobic contacts with residues V281, A293, K295, L393 and A403 in the ATP pocket (Fig. 3D). In the hinge loop of c-Src, the pyrazolo[1,5-а]pyrimidin group of TPX-0022 also forms two hydrogen bonds with the backbone of M341 (homologous to M1160 in c-Met) (Fig. 3E). A major difference between the two kinases is the orientation of the A-loop (Fig. 3B-C). The A-loop of c-Src is more flexible and moves away from the hinge, where some residues (amino acids 413–423) have no density. In addition, the π-stacking interaction between TPX-0022 and Tyr1230 of the c-Met A-loop is not observed in the c-Src/TPX-0022 complex. The corresponding residue E412 in c-Src is oriented towards the solvent (Figs. 3B, 3F).
Although the benzonitrile group of TPX-0022 does not form a π-stacking interaction with the A-loop of c-Src, it exhibits the same conformation as in the c-Met/TPX-0022 complex, which is perpendicular to the pyrazolo[1,5-а]pyrimidin group. To explore the effect of this unique conformation of TPX-0022, we performed torsional angle scan analysis to calculate the free energy of TPX-0022 in different conformations. TPX-0022 has two local minimum energy conformations under aqueous conditions, including “unfolded” and “folded” conformations, with dihedral angles of approximately − 126° – − 111° and 56–71° (Fig. S5). When bound to c-Met or c-Src, TPX-0022 adopts a conformation with a dihedral angle of 63° or 61°. If docked to CSF1R, c-Met or c-Src, TPX-0022 also displays a similar conformation (Fig. S6). Therefore, TPX-0022 seems to be fixed in a favourable “folded” conformation to reduce entropy loss upon its binding and thus improve affinity against kinases [36].
3.4. Comparison with other macrocyclic compounds of kinases
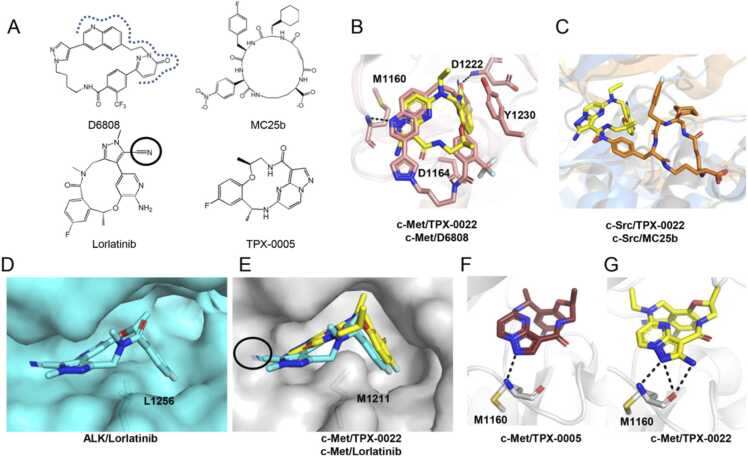
To understand the binding properties of macrocyclic inhibitors to c-Met, we compared TPX-0022 with the c-Met-specific macrocyclic inhibitor D6808 reported in our previous study (Fig. 4A) [37]. The c-Met structure is similar in the c-Met/D6808 complex (PDB ID: 8GVJ) and c-Met/TPX-0022 complex, with an RMSD of 0.528 Å (Fig. 4B). D6808 also binds to the ATP-binding pocket in a type I binding mode. In comparison with TPX-0022, D6808 has a larger cyclic scaffold and a larger interaction surface with c-Met. In addition to two hydrogen bonds with the backbone atoms of M1160 and D1222, it forms a hydrogen bond with the side chain of D1164 (Fig. 4B, S3B, S7A). Moreover, the trifluoromethyl group of D6808 undergoes multipolar interactions with the side-chain amide residues of N1167 and the positively charged guanidinium side chains of R1208 and R1166 (Fig. S7B)[38]. These residues are not conserved in c-Src, which may be a reason why D6808 has a low inhibitory affinity against c-Src (Fig. S7C) [37]. Although D6808 and TPX-0022 behaved similarly in vitro, D6808 had relatively poor oral PK profiles with a low maximum concentration and low exposure, whereas TPX-0022 performed well in clinical trials[23]. ADME/drug-likeness prediction suggests that D6808 has some properties that are inferior to those of TPX-0022 (Table S3).
Fig. 4.
Comparison of macrocyclic compounds of kinases. (A) The chemical structures of the compounds. The dashed line marks the contact surface of D6808 with c-Met similar to TPX-0022. (B) Superimposition of the structures of the c-Met/TPX-0022 complex and c-Met/D6808 complex (PDB ID: 8GVJ). (C) Superimposition of the structures of the c-Src/TPX-0022 complex and c-Src/MC25b complex (PDB ID: 5BMM). (D) Binding mode of lorlatinib to the ALK kinase domain (PDB ID: 4CLI). (E) The modelled structure of c-Met/lorlatinib complex. The model was acquired by superposition of the c-Met/TPX-0022 structure with the AKL/lorlatinib structure. The Black circle indicates the part where clashing may occur between lorlatinib and c-Met. (F) The hydrogen bond between c-Met and TPX-0005. The model was acquired by superposition of the c-Met/TPX-0022 structure with the TRK/TPX-0005 structure. (G) The hydrogen bonds between c-Met and TPX-0022. TPX-0022 is colored yellow. c-Met is colored in gray. c-Src is colored in marine. The c-Src/MC25b complex is colored orange. The ALK/lorlatinib complex is colored cyan. TPX-0005 is colored in ruby. Hydrogen bonds are indicated as black dotted lines.
Furthermore, we compared the binding patterns of TPX-0022 to several macrocyclic inhibitors of kinase, including Src-specific MC25b [39], the FDA-approved Lorlatinib[23], and TPX-0005, which is structurally similar to TPX-0022[40]. Lorlatinib and TPX-0005 are macrocyclic kinase inhibitors targeting AKL/ROS and ALK/ROS1/TRK, respectively. In the structure of the c-Src/MC25b complex (PDB ID: 5BMM), MC25b moves away from the hinge loop of c-Src and occupies the adenine binding pocket, a pocket underneath the b3-aC loop, and a pocket near the peptide binding patch (Fig. 4C). The binding site is totally different from that of TPX-0022 and other acyclic inhibitors of c-Src (Fig. S8). Compared with ALK, c-Met has a smaller pocket in the hinge region, which may be incompatible with Lorlatinib (Fig. 4D-E, S9A). In addition, M1211 in β7 of the c-Met C-lobe bulges towards the N-lobe, whereas ALK and most other tyrosine kinases are leucine with a shorter chain in the responding location (Fig. 4D-E, S9B). A superposition of the c-Met/TPX-0022 structure with the TRK/TPX-0005 structure (PDB ID: 7VKO) indicates that the pyrazolo[1,5-а]pyrimidin group of TPX-0005 is unable to form a hydrogen bond interaction with the hinge of c-Met due to the absence of an amino group, which may impact the binding of TPX-0005 to c-Met (Figs. 4F, 4G). Overall, these structural properties of c-Met may be helpful for designing highly selective c-Met macrocyclic inhibitors.
4. Discussion
Macrocyclic drugs represent attractive candidates in drug discovery due to their unique drug-like profiles[22]. Compared to the acyclic counterparts, macrocyclization can change the biological and physiochemical properties, such as good pharmacokinetic and pharmacodynamic parameters, increased oral bioavailability, and improved metabolic stability and cell permeability[41]. Fourteen macrocycle drugs have been approved by the FDA, and lorlatinib is the only kinase inhibitor [42]. In addition to lorlatinib, many macrocycle kinase inhibitors have been developed.
A portion of efforts have been made to develop macrocycle c-Met inhibitors, such as TPX-0022. Our structural analysis of TPX-0022 with the targets suggests some essentials for the design of c-Met-specific macrocyclic inhibitors. First, the A-loop of c-Met has an unusual conformation that forms a turn between the β-sheet and the α C-helix and offers an opportunity for the design of selective inhibitors [43]. A π-stacking interaction with Y1230, located in the A-loop of c-Met, is crucial for type I inhibitors and has been observed in TPX-0022, D6808, and most acyclic inhibitors [37]. Additionally, the hinge loop of c-Met plays an important role in the interaction with TPX-0022, and this region is not conserved among kinases (Fig. S9A). Consequently, Lorlatinib with a bulky group would clash with the hinge of c-Met, and TPX-0005, which lacks a hydrogen bond with the hinge loop, has low affinity for c-Met. Notably, the highly rigid conformation of macrocyclic compounds limits their binding conformation. When binding to c-Met, TPX-0022, as well as D6808, is wrapped around M1211 and forms a local minimum energy conformation with a small dihedral angle. Therefore, the conformation of macrocyclic inhibitors and possible clashes with M1211 should also be taken into account when designing macrocyclic inhibitors for c-Met.
Acquired resistance appears frequently after long-term cancer treatment with kinase inhibitors. Acquired resistance of MET-altered cancers can emerge either through mechanisms related to MET alterations (on-target) or through activating shunting mechanisms (off-target) [44]. The binding sites are different for different types of inhibitors, leading to inhibitor-specific MET alterations. The most frequent on-target mutation sites include G1163, D1228 and Y1230 for type I inhibitors and L1195 and F1200 for type II inhibitors. In general, type I inhibitors had potent activity against mutations of L1195 and F1200 but low affinity against mutations of D1228 and Y1230, such as capmatinib, tepotinib and savolitinib (Fig. S10)[45]. TPX-0022 could also not overcome D1228N and Y1230C but still had moderate inhibition activity against Y1230H. The rigid structure of TPX-0022 may play a role in maintaining close proximity to Y1230H to form π-stacking interactions. The location of G1163 at the edge of the pocket and the compact multicyclic scaffold of TPX-0022 may explain why TPX-0022 retains moderate inhibitory potency against G1163R. Notably, the potency of TPX-0022 and crizotinib was also decreased to 50–200 nM for the F1200I mutant. In contrast, type II inhibitors, such as cabozantinib, could overcome mutations of D1228 and Y1230. Strategies to overcome resistance to these "on-target"-dependent mutations are mainly the cross-use of type I and type II c-Met inhibitors. However, this cross-use strategy may not always be effective, and an accurate IC50 should be considered for optimal efficacy.
There has been tremendous growth in the development of c-Met inhibitors in recent years, and four inhibitors have been approved for marketing in the last three years alone. In this study, we describe the structural basis of the marocyclic c-Met inhibitor TPX-0022 against c-Met and c-Src and its ability to overcome resistance mutations. The structural characteristics of the c-Met ATP pocket, mainly in the A-loop, hinge and β7 shite, are subtly different from those of other kinases, which may have potential for the development of selective c-Met marocyclic inhibitors.
Funding
This work was supported by the National Natural Science Foundation of China (grants 81570537, 81974074, 82273496, 82202920 and 31900880), Hunan Provincial Science and Technology Department (2018RS3026 and 2021RC4012), China Postdoctoral Science Foundation (2019M652805), Science and Technology Planning Project of Hunan Province (2018TP1017), Hunan Provincial Natural Science Foundation (2023JJ40954, 2023JJ20092) and Central South University Innovation-Driven Research Programme (2023CXQD076).
CRediT authorship contribution statement
L.Q. and H.L. performed the experiments; S.D., M.G., X.C., J.L., and H.Z. performed data collection and structure determination. L.Q., H.L., M.L., X.L., Z.C., H.W. and Y.C. analysed the data. L.Q., H.W. and Y.C. prepared the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank the staff from the BL02U/BL19U1 beamline of the National Facility for Protein Science in Shanghai (NFPS) at the Shanghai Synchrotron Radiation Facility for assistance during data collection.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.csbj.2023.11.028.
Contributor Information
Hudie Wei, Email: hudiewei18@163.com.
Yongheng Chen, Email: yonghenc@163.com.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
Supplementary material
.
Data availability
The coordinates and structure factors are deposited in the Protein Data Bank under accession codes 8K78 and 8K79.
References
- 1.Moiseenko F., et al. Management and treatment of non-small cell lung cancer with MET alteration and mechanisms of resistance. Curr Treat Options Oncol. 2022;23(12):1664–1698. doi: 10.1007/s11864-022-01019-2. [DOI] [PubMed] [Google Scholar]
- 2.Su P., Zhang M., Kang X. Targeting c-Met in the treatment of urologic neoplasms: current status and challenges. Front Oncol. 2023;13 doi: 10.3389/fonc.2023.1071030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang Q., et al. MET inhibitors for targeted therapy of EGFR TKI-resistant lung cancer. J Hematol Oncol. 2019;12(1) doi: 10.1186/s13045-019-0759-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawakami H., Okamoto I. MET-targeted therapy for gastric cancer: the importance of a biomarker-based strategy. Gastric Cancer. 2016;19(3):687–695. doi: 10.1007/s10120-015-0585-x. [DOI] [PubMed] [Google Scholar]
- 5.Bouattour M., et al. Recent developments of c-Met as a therapeutic target in hepatocellular carcinoma. Hepatology. 2018;67(3):1132–1149. doi: 10.1002/hep.29496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang X., Liao H.Y., Zhang H.H. Roles of MET in human cancer. Clin Chim Acta. 2022;525:69–83. doi: 10.1016/j.cca.2021.12.017. [DOI] [PubMed] [Google Scholar]
- 7.Guo R., et al. MET-dependent solid tumours - molecular diagnosis and targeted therapy. Nat Rev Clin Oncol. 2020;17(9):569–587. doi: 10.1038/s41571-020-0377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakamoto M., Patil T. MET alterations in advanced non-small cell lung cancer. Lung Cancer. 2023;178:254–268. doi: 10.1016/j.lungcan.2023.02.018. [DOI] [PubMed] [Google Scholar]
- 9.Leonetti A., et al. Resistance mechanisms to osimertinib in EGFR-mutated non-small cell lung cancer. Br J Cancer. 2019;121(9):725–737. doi: 10.1038/s41416-019-0573-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Herpe F., Van Cutsem E. The role of cMET in gastric cancer-a review of the literature. Cancers (Basel) 2023;15(7) doi: 10.3390/cancers15071976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang C., Lu X. Targeting MET: discovery of small molecule inhibitors as non-small cell lung cancer therapy. J Med Chem. 2023;66(12):7670–7697. doi: 10.1021/acs.jmedchem.3c00028. [DOI] [PubMed] [Google Scholar]
- 12.Centuori S.M., Bauman J.E. c-met signaling as a therapeutic target in head and neck cancer. Cancer J. 2022;28(5):346–353. doi: 10.1097/PPO.0000000000000619. [DOI] [PubMed] [Google Scholar]
- 13.Garon E.B., Brodrick P. Targeted therapy approaches for MET abnormalities in non-small cell lung cancer. Drugs. 2021;81(5):547–554. doi: 10.1007/s40265-021-01477-2. [DOI] [PubMed] [Google Scholar]
- 14.Van Der Steen N., et al. cMET exon 14 skipping: from the structure to the clinic. J Thorac Oncol. 2016;11(9):1423–1432. doi: 10.1016/j.jtho.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 15.Ai J., et al. Preclinical evaluation of SCC244 (Glumetinib), a novel, potent, and highly selective inhibitor of c-met in MET-dependent cancer models. Mol Cancer Ther. 2018;17(4):751–762. doi: 10.1158/1535-7163.MCT-17-0368. [DOI] [PubMed] [Google Scholar]
- 16.Puccini A., et al. Safety and tolerability of c-MET inhibitors in cancer. Drug Saf. 2019;42(2):211–233. doi: 10.1007/s40264-018-0780-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Markham A. Tepotinib: first approval. Drugs. 2020;80(8):829–833. doi: 10.1007/s40265-020-01317-9. [DOI] [PubMed] [Google Scholar]
- 18.Markham A. Savolitinib: first approval. Drugs. 2021;81(14):1665–1670. doi: 10.1007/s40265-021-01584-0. [DOI] [PubMed] [Google Scholar]
- 19.Dhillon S. Capmatinib: first approval. Drugs. 2020;80(11):1125–1131. doi: 10.1007/s40265-020-01347-3. [DOI] [PubMed] [Google Scholar]
- 20.Deng W., et al. Abstract 1325: TPX-0022, a polypharmacology inhibitor of MET/CSF1R/SRC inhibits tumor growth by promoting anti-tumor immune responses. Cancer Res. 2019;79(13_Supplement) 1325-1325. [Google Scholar]
- 21.Goel V.K., et al. Abstract 1444: TPX-0022, a potent MET/SRC/CSF1R inhibitor that modulates the tumor immune microenvironment in preclinical models. Cancer Res. 2021;81(13_Supplement) 1444-1444. [Google Scholar]
- 22.Sun D. Recent advances in macrocyclic drugs and microwave-assisted and/or solid-supported synthesis of macrocycles. Molecules. 2022;27(3) doi: 10.3390/molecules27031012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.〈https://www.clinicaltrials.gov/ct2/show/NCT03993873〉(accessed on August 20,2019).
- 24.Zhan Z., et al. Discovery of 10H-Benzo[b]pyrido[2,3-e][1,4]oxazine AXL Inhibitors via Structure-Based Drug Design Targeting c-Met Kinase. J Med Chem. 2023;66(1):220–234. doi: 10.1021/acs.jmedchem.2c00962. [DOI] [PubMed] [Google Scholar]
- 25.Fujino T., et al. Sensitivity and resistance of MET exon 14 mutations in lung cancer to Eight MET tyrosine kinase inhibitors in vitro. J Thorac Oncol. 2019;14(10):1753–1765. doi: 10.1016/j.jtho.2019.06.023. [DOI] [PubMed] [Google Scholar]
- 26.Wu D., et al. LY2874455 potently inhibits FGFR gatekeeper mutants and overcomes mutation-based resistance. Chem Commun (Camb) 2018;54(85):12089–12092. doi: 10.1039/c8cc07546h. [DOI] [PubMed] [Google Scholar]
- 27.Qi-Sheng, et al. Upgrade of macromolecular crystallography beamline BL17U1at SSRF. Nucl Sci Tech. 2018;v.29(05):3–9. [Google Scholar]
- 28.The protein complex crystallography beamline (BL19U1) at the Shanghai Synchrotron Radiation Facility. Nuclear Science and Techniques, 2019. 30(11): p. 1–11.
- 29.Juers D.H., et al. Acta crystallographica section D biological crystallography. Acta Crystallogr Sect D Biol Crystallogr. 2013:1139–1153. doi: 10.1107/S0907444907045040. [DOI] [PubMed] [Google Scholar]
- 30.Liebschner D., et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr D Struct Biol. 2019;75(Pt 10):861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collie G.W., et al. Structural and molecular insight into resistance mechanisms of first generation cMET inhibitors. ACS Med Chem Lett. 2019;10(9):1322–1327. doi: 10.1021/acsmedchemlett.9b00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P., et al. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66(Pt 4):486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schrodinger, L.L.C., The PyMOL Molecular Graphics System, Version 1.8. 2015.
- 34.Morris G.M., et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daina A., Michielin O., Zoete V. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci Rep. 2017;7(1) doi: 10.1038/srep42717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Engelhardt H., et al. Start selective and rigidify: the discovery path toward a next generation of EGFR tyrosine kinase inhibitors. J Med Chem. 2019;62(22):10272–10293. doi: 10.1021/acs.jmedchem.9b01169. [DOI] [PubMed] [Google Scholar]
- 37.Wang C., et al. Discovery of D6808, a highly selective and potent macrocyclic c-met inhibitor for gastric cancer harboring MET gene alteration treatment. J Med Chem. 2022;65(22):15140–15164. doi: 10.1021/acs.jmedchem.2c00981. [DOI] [PubMed] [Google Scholar]
- 38.Muller K., Faeh C., Diederich F. Fluorine in pharmaceuticals: looking beyond intuition. Science. 2007;317(5846):1881–1886. doi: 10.1126/science.1131943. [DOI] [PubMed] [Google Scholar]
- 39.Aleem S., et al. Structural and biochemical basis for intracellular kinase inhibition by Src-specific peptidic macrocycles. Cell Chem Biol. 2016;23(9):1103–1112. doi: 10.1016/j.chembiol.2016.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murray B.W., et al. Molecular characteristics of repotrectinib that enable potent inhibition of TRK fusion proteins and resistant mutations. Mol Cancer Ther. 2021;20(12):2446–2456. doi: 10.1158/1535-7163.MCT-21-0632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amrhein J.A., Knapp S., Hanke T. Synthetic opportunities and challenges for macrocyclic kinase inhibitors. J Med Chem. 2021;64(12):7991–8009. doi: 10.1021/acs.jmedchem.1c00217. [DOI] [PubMed] [Google Scholar]
- 42.Syed Y.Y. Lorlatinib: first global approval. Drugs. 2019;79(1):93–98. doi: 10.1007/s40265-018-1041-0. [DOI] [PubMed] [Google Scholar]
- 43.Cui J.J., et al. Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK) J Med Chem. 2011;54(18):6342–6363. doi: 10.1021/jm2007613. [DOI] [PubMed] [Google Scholar]
- 44.Recondo G., et al. Molecular mechanisms of acquired resistance to MET tyrosine kinase inhibitors in patients with MET Exon 14-Mutant NSCLC. Clin Cancer Res. 2020;26(11):2615–2625. doi: 10.1158/1078-0432.CCR-19-3608. [DOI] [PubMed] [Google Scholar]
- 45.Fujino T., et al. Foretinib can overcome common on-target resistance mutations after capmatinib/tepotinib treatment in NSCLCs with MET exon 14 skipping mutation. J Hematol Oncol. 2022;15(1):79. doi: 10.1186/s13045-022-01299-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Data Availability Statement
The coordinates and structure factors are deposited in the Protein Data Bank under accession codes 8K78 and 8K79.