Summary
Background
The real-world impact of bivalent vaccines for wild type (WA.1) and Omicron variant (BA.5) is largely unknown in immunocompromised patients with Multiple Myeloma (MM). We characterize the humoral and cellular immune responses in patients with MM before and after receiving the bivalent booster, including neutralizing assays to identify patterns associated with continuing vulnerability to current variants (XBB1.16, EG5) in the current post-pandemic era.
Methods
We studied the humoral and cellular immune responses before and after bivalent booster immunization in 48 MM patients. Spike binding IgG antibody levels were measured by SARS-CoV-2 spike binding ELISA and neutralization capacity was assessed by a SARS-CoV-2 multi-cycle microneutralization assays to assess inhibition of live virus. We measured spike specific T-cell function using the QuantiFERON SARS-CoV-2 (Qiagen) assay as well as flow-cytometry based T-cell. In a subset of 38 patients, high-dimensional flow cytometry was performed to identify immune cell subsets associated with lack of humoral antibodies.
Findings
We find that bivalent vaccination provides significant boost in protection to the omicron variant in our MM patients, in a treatment specific manner. MM patients remain vulnerable to newer variants with mutations in the spike portion. Anti-CD38 and anti-BCMA therapies affect the immune machinery needed to produce antibodies.
Interpretation
Our study highlights varying immune responses observed in MM patients after receiving bivalent COVID-19 vaccination. Specifically, a subgroup of MM patients undergoing anti-CD38 and anti-BCMA therapy experience impairment in immune cells such DCs, B cells, NK cells and TFH cells, leading to an inability to generate adequate humoral and cellular responses to vaccination.
Funding
National Cancer Institute (National Institutes of Health), National Institute of Allergy and Infectious Diseases (National Institutes of Health), NCI Serological Sciences Network for COVID-19 (SeroNet) and The Icahn School of Medicine at Mount Sinai.
Keywords: COVID-19, Bivalent vaccine, Multiple Myeloma, SARS-CoV-2, Omicron, Hematological malignancy
Research in context.
Evidence before this study
Our group has previously reported that Multiple Myeloma (MM) patients have sub-optimal antibody and T cell immune responses after mRNA COVID-19 vaccination, however the efficacy of bivalent vaccination against wild type and BA.5 strains is not known in this immunocompromised population. We searched PubMed from inception of database to July 23, 2023 for articles published for real-world COVID-19 bivalent booster effectiveness using the terms "Bivalent SARS-CoV-2 vaccine" OR "Bivalent COVID-19 vaccine" OR "Bivalent booster vaccine" OR "Bivalent omicron" OR "mRNA bivalent booster" OR "mRNA bivalent booster" AND "Hematology" or "Multiple Myeloma". Although effectiveness of bivalent booster vaccination is highly reported in healthy immunocompetent individuals, the protective capacity is unknown in immunocompromised patients such as those with MM. To date there have been no studies that combines an integrative view of humoral and cellular immunity in response for the bivalent vaccination.
Added value of this study
To our knowledge our study in the first to combine a multifaceted approach to assessing the effect on humoral and cellular immunity of the bivalent vaccination on patients with Multiple Myeloma. We provide insight into neutralization capacity of variants of interest to highlight continued susceptibility to fatal COVID-19 infections in MM patients post bivalent vaccination. We implement a rapidly deployable method to measuring T cell activity to gain insight into the protective capacity after bivalent vaccination. Our data provides insights into underlying immune machinery that is defective in MM patients with sub-optimal SARS-CoV-2 bivalent vaccination responses which can serve as an immune signature to identify patients at risk for severe COVID-19 manifestations.
Implications of all the available evidence
Multiple Myeloma patients are more prone to fatal infections due to a compromised immune system. Understanding the effect of bivalent vaccination is important to guide the public health recommendations. Our study identifies beneficial protective effects conferred by bivalent omicron vaccination but ongoing susceptibilities with emerging new variants. We report an underlying immune cellular phenotype that is associated with increased vulnerability to infection which can be helpful to inform management of patients.
Introduction
Multiple Myeloma (MM) is a cancer of plasma cells that results in defective humoral immunity. MM patients are prone to infections due to the immunosuppression that is brought on by the disease and treatment.1, 2, 3, 4 In 2019, the severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) emerged on the world stage causing viral pneumonia, acute lung injury, acute respiratory distress syndrome, and death.5 SARS-CoV-2 vaccines have been proven to be highly effective at preventing severe disease and mortality in healthy individuals; however, immunocompromised individuals with hematologic malignancies remain at increased risk for severe COVID-19 manifestations.6,7 We and others have shown that the immune responses elicited by current COVID-19 mRNA vaccines are often sub-optimal in immunocompromised patients.8 Our previous data showed that immunocompromised myeloma patients are at increased risk of morbidity and mortality from COVID-19 and both antibody responses as well as COVID-19-specific T cell responses after complete mRNA vaccination (two doses) were lower in MM patients as compared to the healthy population.9,10 We noted that patients undergoing anti-CD38 and novel anti-B cell maturation antigen (BCMA) targeted therapies have a greater likelihood of producing sub-optimal SARS-CoV-2 anti-S IgG antibody levels thus leading to decreased humoral and cellular protection against severe COVID-19 manifestations. This difference remained even after receiving a monovalent booster vaccination (e.g., third vaccine dose) and was associated with lower serum neutralization of the Omicron BA.1 variant in half of the Myeloma patients studied.8 As ancestral variants were completely replaced by antigenically highly diverse Omicron lineages, updated Bivalent (Ancestral and Omicron BA.4/BA.5) mRNA booster vaccines were deployed starting in fall of 2022 to provide continued immunity against the antigenically diverse Omicron variants.11 The real-world impact of the bivalent mRNA vaccine is largely unknown in patients with MM, as is the underlying cause for the suboptimal vaccine immune responses mounted by a subset of MM patients undergoing treatment. Here, we describe the humoral and cellular effects of the bivalent mRNA vaccination in a cohort of MM patients treated at the Icahn school of Medicine at Mount Sinai. We note that the majority of MM patients greatly benefited from the bivalent booster vaccination both at the humoral as well as at the cellular level. Moreover, we identified a deficiency in neutralizing capacity for the recently emerged XBB.1.5, XBB.1.16 and EG variants and identified an immune phenotype predictive of sub-optimal antibody responses after bivalent vaccination.
Methods
Study design
Biospecimen and data were obtained from the observational longitudinal clinical sample collection from patients with emerging viral infections research study (approved by the Program for the Protection of Human Subjects at the Icahn School of Medicine at Mount Sinai Institutional Review Board, IRB-17-00791/STUDY-16-01215). Participants provided informed consent prior to sample and data collection. We used biospecimen collected from 48 patients with Multiple Myeloma (MM) before and after receiving a Pfizer or Moderna bivalent vaccine boosters containing ancestral as well as BA.5 spike mRNA.
Specimen collection
Biospecimens used in this study. Peripheral blood was collected in heparin green tops (Cat#362761), BD Vacutainer CPT (Cat#367985), and BD SST™ Serum Separation Tubes (Cat#0268396) via venipuncture. Peripheral blood mononuclear cells (PBMC) were isolated within a few hours of collection as per the CPT manufacturer’s instructions and cryopreserved in liquid nitrogen. Samples were collected 31 ± 63 (0–260) days before and 16 ± 8 (6–31) days after bivalent booster vaccination. Detailed demographic characteristics and vaccination information for each study participant and group are summarized in Table 1.
Table 1.
Clinical characteristics of patients with Multiple Myeloma receiving Bivalent SARS-CoV-2 Vaccination.
| Variable | MM cohort |
|
|---|---|---|
| (N = 48) | ||
| Age (y) | 71 | [50–84] |
| Male gender | 54% | 26 |
| Vaccine Type Initial Dose | ||
| Pfizer-BioNTech | 69% | 33 |
| Moderna | 27% | 13 |
| Unknown | 4% | 2 |
| Bivalent as 3rd dose | 4% | 2 |
| Bivalent as 4th dose | 54% | 26 |
| Bivalent as 5th dose | 40% | 19 |
| Bivalent as 6th dose | 2% | 1 |
| Timing of after bivalent dose (d) | 47.5 | [6–183] |
| Had documented COVID-19 | 33% | 16 |
| Disease isotype | ||
| IgG | 50% | 24 |
| IgA | 21% | 10 |
| LC | 21% | 10 |
| Other | 2% | 1 |
| SMM | 6% | 3 |
| ≥ 3 previous lines of treatment | 28% | 7 |
| >5 previous lines of treatment | 16% | 10 |
| Disease response status | ||
| CR or sCR | 48% | 23 |
| VGPR | 21% | 10 |
| PR or MR | 13% | 6 |
| SD or PD | 6% | 3 |
| Unable to assess | 13% | 6 |
| Treatment regimen at bivalent vaccination contains | ||
| Immunomodulatory drug | 44% | 21 |
| Proteasome inhibitor | 17% | 8 |
| Anti-CD38 mAb | 50% | 24 |
| Anti-SLAMF7 mAb | 2% | 1 |
| BCMA-targeted therapy | 13% | 6 |
| BCMA-targeted bispecific | 8% | 4 |
| CAR T cell therapy | 4% | 2 |
| Other bispecific (non-BCMA) | 6% | 3 |
| Other therapy (incl. venetoclax, selinexor, alkylators) | 6% | 3 |
| Previous ASCT | 42% | 20 |
| No active treatment | 15% | 7 |
Values are presented as percentage (n) or median [range]. Disease response status and treatment regimen were registered at the date of administration of the first dose of mRNA vaccine.
Abbreviations: y, years; mo, months; COVID-19, coronavirus disease 2019; Ig, immunoglobulin; MM, multiple myeloma; SMM, smoldering multiple myeloma; HD, healthy donor; CR, complete response; sCR, stringent complete response; VGPR, very good partial response; PR, partial response; MR, minimal response; SD, stable disease; PD, progressive disease; ASCT, autologous stem cell transplant; mAb, monoclonal antibody; BCMA, B-cell maturation antigen; CAR, chimeric antigen receptor.
SARS-CoV-2 antibody ELISA
Spike binding IgG antibodies to SARS-CoV-2 were measured using an established, quantitative two-step ELISA termed Mount Sinai Antibody test/Kantaro which received FDA emergency use authorization and is described in detail in the referenced manuscripts.12,13 The assay shows a performance of 100% specificity and 95% sensitivity in an in-house evaluation and is performed by the Pathology laboratory of the Mount Sinai Health System. Results are expressed in artificial units/mL (AU/mL).
Flow cytometry assay to detect immune cell populations
An in-house antibody panel (Panel A) developed to immunophenotype the myeloid and lymphoid cells in peripheral blood. Thawed PBMC were initially stained with Live/Dead Fixable Blue Dead Cell Stain Kit (L23105, Thermofisher Scientific) for 15 min at room temperature. Viability dye stained PBMC were further stained with Panel A in multiple staining steps at different temperatures. PBMC were stained at room temperature for 15 min with a cocktail of 14 antibodies, washed and further stained with a cocktail of remaining antibodies in Panel A for 30 min on ice. Antibodies in Panel A stained at room temperature include CCR6-BUV496 (clone 11Ag), CD45RA-BUV563 (clone HI100), CD28-BUV737 (clone 28.2) (all from BD Biosciences), TCR gamma-delta-PerCP-eFluor710 (clone B1.1, Thermofisher), CCR7-BV421 (clone G043H7), CXCR3-BV510 (clone G025H7), CD27-BV570 (clone O323), CXCR5-BV605 (clone J25D4), CRTH2-BV711 (clone BM16), PD-1-BV750 (clone EH12.1H7), CD25-PE (clone M-A251), CD66b-PE-Dazzzle 594 (clone QA17A51), CCR4-PE-FIRE 810 (clone L291H4), CD11c-Alexa700 (clone Bu15) (all from BioLegend). Antibodies in Panel A stained on ice include CD4-BUV395 (clone SK3), CD56-BUV615 (clone NCAM16.2), HLA-DR-BUV661 (clone G46-6), CD3-BUV805 (clone UCHT1), CD20-BV480 (clone 2H7) (all from BD Biosciences), CD1c-SuperBright 436 (clone L161), CD123-eFluor450 (clone 6H6), CD8-NFB555 (clone OKT8), CD19-NFB610-70S (clone HIB19), CD14-NFB660-40S (clone MEM-15), CD127-PE-Cy5.5 (clone eBioRDR5), CD16-NFR685 (clone 3G8) (all from Thermofisher), IgM-BV650 (clone MHM-88), IgD-BV785 (IA6-2), CD11b-PerCP (clone M1/70), CD57-PerCP-Cy5.5 (clone HNK-1), CD24-PE-Cy5 (clone ML5), IgG Fc-PE-Cy7 (clone M1310G05) CD38-APC-FIRE810 (clone HIT2) (all from BioLegend), IgA-APC-VIO770 (clone IS11-8E10, Miltenyi Biotec). Each antibody was used at a dilution of 1:25. All antibody cocktail preparations included True-Stain Monocyte buffer (Biolegend), CellBlox Monocyte and Macrophage blocking buffer (Thermofisher) and Super Bright Complete Staining buffer (Thermofisher) at a dilution of 1:20 to avoid nonspecific dye-dye and dyes to cell interaction. Detailed panel information is listed in Supplemental Table S1. Cells were acquired on Cytek Aurora Flow Cytometer (Cytek Biosciences). Flow data was compensated on Cytek Aurora acquisition software SpectroFlo and compensated.fcs files were exported to Flowjo software (BD Biosciences) for analysis. Supervised hierarchal gating was employed to delineate major cell types in PBMC. Total cells were initially gated to remove dead cells, doublets and CD66b+ cells. From the live CD66b negative cell gate monocytes were identified based on expression of markers CD16 and CD14 (CD16hi/-CD14−/+). Monocyte-negative cells were sequentially gated for lineage cell types. B Cells were identified from monocyte negative gate by expression of CD19 or CD20. T cells were extracted from B cell negative by the presence of CD3 and HLA-DR. Dendritic Cells (DCs) were identified from the T cell negative gate by the expression of CD123+ plasmacytoid dendritic cells (pDC, CD123+CD1c−) and CD1c+ conventional dendritic cells (cDC, CD123-CD1c+). CD4 and CD8 T cells were identified from the T cell gate. TFH cells were identified as CD4 T Cells that express CXCR5. Finally, DC cell-negative cells were plotted as CD56 vs CD16 to identify NK cells (CD56hiCD16- and CD56dimCD16+ NK cells). Full gating strategy is depicted in Supplemental Figure S1A.
Intracellular cytokine staining flow cytometry (ICS-flow) T cell assay
We used combined activation induced marker (AIM) and intracellular cytokine staining (ICS) assays to detect SARS CoV-2 specific T cell responses. T cell assays were carried out in RPMI supplemented with 10% Human Ab serum (R&D Systems), 1x Glutamax (Lonza), 1x Penicillin-Streptomycin. PBMC were stimulated for 6 h with a pool of spike peptides (15-mer sequences with 11 amino acids overlap spanning the entire spike protein of either BA.5, or wild-type, Miltenyi Biotec) at concentrations recommended by the manufacturer or with water as control along with co-stimulators for CD28 and CD49d (i.e., anti-CD28 clone CD28.1 and anti-CD49d clone 9F109, both from Biolegend). Culture conditions also included antibodies to detect CD4 activation marker CD154 (CD154-PE, clone 24-31 Biolegend) and Monensin (BioLegend). Stimulations with Staphylococcal enterotoxin B (SEB) were used as positive control when samples were available. Post stimulation, cells were washed and stained with Live/Dead Fixable Blue Dead Cell Stain Kit for 15 min at room temperature followed by surface staining with a cocktail of antibodies comprising of CD3-BUV805 (clone UCHT1), CD4-BUV395 (clone SK3), CD8-BUV496 (clone RPA-T8), CD45RA-BUV563 (clone HI100), PD-1-BUV615 (clone EH12.1), HLA-DR-BUV661 (clone G46-6) (all from BD Biosciences), CCR7-BV510 (clone G043H7), CD27-BV570 (clone O323), CD69-BV605 (clone FN50), CD200-BV711 (clone OX-104), CXCR5-BV785 (clone J252D4), ICOS-PE-Dazzle594 (clone QA17A51), OX40-PE-Cy5 (clone Ber-ACT35), 4-1bb-APC-Fire750 (clone 4B4-1) (all from BioLegend) and CD19-NFB610-70S (cloneHIB19). After surface marker staining, PBMCs were fixed with 4% paraformaldehyde (PFA) and permeabilized with BD perm buffer (BD Biosciences) and stained with a cocktail of antibodies to cytokines IL-4 (IL-4-BUV737, clone MP4-25D2), IL-17 (IL-17-BV650, clone N49-653) (both from BD Biosciences), IFN-g (IFN-g-BV421, clone B27), TNF-a (TNF-a-PE-Cy7, clone MaB11), IL-2 (IL-2-APC, clone MQ1-17H12) and GM-CSF (GM-CSF-PerCP-Cy5.5) (all from BioLegend). Detailed panel information is listed in Supplemental Table S2. The cells were acquired on Cytek Aurora Flow Cytometer. Flow data was compensated on Cytek Aurora acquisition software SpectroFlo and compensated.fcs files were exported to Flowjo software (BD Biosciences) for analysis. Data was gated to exclude dead cells and doublets and then further gated on forward scatter (FSC-A) vs side scatter (SSC-A) plot to identify lymphocytes. CD3 vs CD19 plots on lymphocytes were used to identify total CD3+ T cells. Total T cells were gated to identify CD4+ and CD8+ T cells. Activated CD4+ T cell population were identified by the expression of activation markers CD154 or CD69 as described in our previous report.9 Full gating strategy is depicted in Supplemental Figure S1B. Total cytokine responses in CD4+ T cells were quantified by performing Boolean gating for each cytokine on activated CD4+ T cells. Events from each cytokine combination were pooled and divided by total CD4 T cells events to calculate the frequency of total cytokine positive CD4+ T cells. Finally, SARS-CoV-2 spike-specific CD4+ T cell response was calculated by subtracting water control total cytokine frequencies from SARS-CoV-2 spike peptide-stimulated conditions. Negative values were designated as zero. PBMC from healthy donors prior to any SARS-CoV-2 vaccination or SARS-CoV-2 exposure were stimulated similarly and were used as a control group.
Cells
African green monkey Vero.E6 cells expressing transmembrane protease serine 2 (TMPRSS2) were cultured at 37 °C with 5% CO2 in Dulbecco’s modified Eagle medium (DMEM, Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (FBS), 1× nonessential amino acids (NEAA), 1% penicillin-streptomycin, 3 μg/mL puromycin (InvivoGen) and 100 μg/mL Normocin (InvivoGen).
Primary SARS-CoV-2 isolates
The SARS-CoV-2 isolate USA-WA1/2020 was used as ancestral reference (BEI Resources, NR-52281). The Mount Sinai Pathogen Surveillance Program provided the following sequence verified primary SARS-CoV-2 isolates: USA/NY-MSHSPSP-PV67190/2022 (Omicron BA.5), USA/NY-MSHSPSP-PV87883/2023 (XBB.1.16), USA/NY-MSHSPSP-PV88029/2023 (EG.1), and USA/NY-MSHSPSP-PV87839/2023 (XBB.1.5.10 with F456L).
SARS-CoV-2 multi-cycle microneutralization assays
Serum samples collected before and after bivalent booster vaccination from study participants were used to determine the neutralization of ancestral (WA.1), Omicron BA.5 and three currently circulating Omicron lineages. All procedures were performed in a biosafety level 3 (BSL-3) facility at the Icahn School of Medicine at Mount Sinai following standard safety guidelines. The day before infection, Vero-E6-TMPRSS2 cells were seeded in 96-well high binding cell culture plates (Costar, #07620009) at a density of 20,000 cells/well in complete Dulbecco’s modified Eagle medium (cDMEM). All sera were heat inactivated at 56 °C for 1 h.
Virus neutralization assays
We determined serum mediated neutralization of SARS-CoV-2 using a multi-cycle replication assay format and authentic replication competent primary viral isolates. For the two viruses (ancestral and BA.5) included in the bivalent booster formulation, we serially diluted sera over 7 points starting at 1:10 to open data suitable to calculate ID50 values. Briefly, Sera were serially diluted (3-fold) in minimum essential media (MEM; Gibco, #11430-030) supplemented with 2 mM l-glutamine (Gibco, #25030081), 0.1% sodium bicarbonate (w/v, HyClone), 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 100/mL penicillin, 100 μg/mL streptomycin (Gibco) and 0.2% bovine serum albumin (BSA, MP Biomedicals, Cat#810063) starting at 1:10. In addition, sera was tested at a 1:10 dilution in duplicate plates for all five viruses (ancestral, BA.5, XBB.1.16, EG.1 and XBB.1.5.10) to provide percent neutralization at maximum serum input.
For each virus, serially diluted Remdesivir (Medkoo Bioscience Inc.) was included as an internal control and to determine assay variation. Diluted sera were incubated with 10,000 TCID50 of WT USA-WA1/2020, USA/NY-MSHSPSP-PV67190/2022 (BA.5.2.1), USA/NY-MSHSPSP-PV87883/2023 (XBB.1.16), USA/NY-MSHSPSP-PV88029/2023 (EG.1), or USA/NY-MSHSPSP-PV87839/2023 (XBB.1.5.10 + F456L) for 1 h at RT, followed by the transfer of 120 μl of the virus-sera mix to Vero-E6-TMPRSS2 plates. Infections were left to proceed for 1 h at 37 °C.
For the SARS-CoV-2 multi-cycle neutralization assay with serially diluted sera the inoculum was removed after 1 h incubation at 37 °C. 100 μl/well of the corresponding sera serial dilutions plus 100 μl/well of infection media supplemented with 2% fetal bovine serum (FBS; Gibco, #10082-147) were added to the cells. For the SARS-CoV-2 multi-cycle neutralization assay with only one 1:10 dilution, 80 μl/well of infection media supplemented with 2% fetal bovine serum (FBS; Gibco, #10082-147) was added to the cells with virus/sera.
Plates were incubated for 48 h at 37 °C followed by fixation overnight at 4 °C in 200 μl/well of a 10% formaldehyde solution. For staining of the nucleoprotein, formaldehyde solution was removed, and cells were washed with PBS (pH 7.4) (Gibco) and permeabilized by adding 150 μl/well of 0.1% Triton X-100 (Fisher Bioreagents) in PBS for 15 min at room temperature (RT). Permeabilization solution was removed, plates were washed with 200 μl/well of PBS (Gibco) twice, and blocked with 3% BSA in PBS for 1 h at RT. During this time the primary antibody was biotinylated according to manufacturer protocol (Thermo Scientific EZ-Link NHS-PEG4-Biotin). Blocking solution was removed and 100 μl/well of biotinylated mAb 1C7C7, a mouse anti-SARS nucleoprotein monoclonal antibody generated at the Center for Therapeutic Antibody Development at The Icahn School of Medicine at Mount Sinai ISMMS (Millipore Sigma), at a concentration of 1 μg/mL in 1% BSA in PBS was added for 1 h at RT. Cells were washed with 200 μl/well of PBS twice and 100 μl/well of HRP-conjugated streptavidin (Thermo Fisher Scientific) diluted in 1% BSA in PBS were added at a 1:2000 dilution for 1 h at RT. Cells were washed twice with PBS, and 100 μl/well of o-phenylenediamine dihydrochloride (Sigmafast OPD; Sigma-Aldrich) were added for 10 min at RT, followed by addition of 50 μl/well of a 3 M HCl solution (Thermo Fisher Scientific). Optical density (OD) was measured (490 nm) using a microplate reader (Synergy H1; Biotek).
After subtraction of background and calculation of the percentage of neutralization with respect to the “virus only” control, a nonlinear regression curve fit analysis was performed to calculate the 50% inhibitory dilution (ID50), with top and bottom constraints set to 100% and 0%, respectively for the serially diluted sera. The percentage of inhibition (range 0–100%) was calculated for the samples tested in duplicates at a single dilution of 1:10. Values under 30% were set to 0. All samples were analyzed in a blinded manner. Additional methods are described in more detail in our previous publication.14
QuantiFERON SARS-CoV-2 RUO test
SARS-CoV-2 specific T cell responses were measured using the QuantiFERON SARS-CoV-2 RUO assay (Qiagen cat. no. 626215, cat. no. 626015), which is an interferon gamma release capture assay. Whole blood was collected via venipuncture and aliquoted to 3 tubes containing SARS-CoV-2 peptides, a positive Mitogen control tube, and a negative Nil control tube. The three antigen containing tubes have different propriety blends of peptides and are designated as Ag1 (containing CD4+ epitopes derived from the S1 subunit RBD of the Spike protein), Ag2 (containing CD4+ and CD8+ epitopes from the S1 and S2 subunits of the Spike protein) and Ag3 (containing CD4+ and CD8+ epitopes from S1 and S2, plus immunodominant CD8+ epitopes derived from whole genome). Stimulated whole blood was incubated for 24 h at 37 degrees. After incubation plasma? From the stimulated samples was isolated and used for the detection of IFN-γ using an enzyme-linked immunosorbent assay (ELISA)-based platform. Specimens were processed as per the manufacturer’s guidelines. IFN-γ concentration in IU/mL from negative control samples were subtracted from SARS-CoV-2 peptide stimulated samples to obtain absolute IFN-γ concentration in IU/mL.
Statistical analysis
Normal distribution was tested using a Shapiro–Wilk test. The Mann–Whitney U test was used to determine significance for all continuous variables that were not normally distributed. Differences between paired samples was tested using Wilcoxon signed-rank test. Chi-square test was used to determine significance in outcome measures. Spearman’s correlation analysis was used to find significance between nonparametric distributed variables. A two-sided alpha <0.05 was considered statistically significant. Differences between continuous variables and contingency variables were done using R (v4.0.2). All statistical tests were run with R (v4.0.2).
Role of the funders
The funders had no role in study design, data collection, data analysis, data interpretation, or writing of this manuscript.
Results
Anti-SARS-CoV-2 spike (S) IgG levels were measured a median of 22 days prior to the bivalent vaccination and were compared to post-dose levels at least one week after the administration (median 47 days) in forty-eight MM patients. Summarized demographic characteristics of the study participants are shown in Table 1. Three patients had asymptomatic MM but most patients were on treatment, with a median of two lines of therapy at the time of vaccination (range 0–16). One third (n = 16) of our MM patients had reported having had COVID-19 at least once in the past three years of the pandemic.
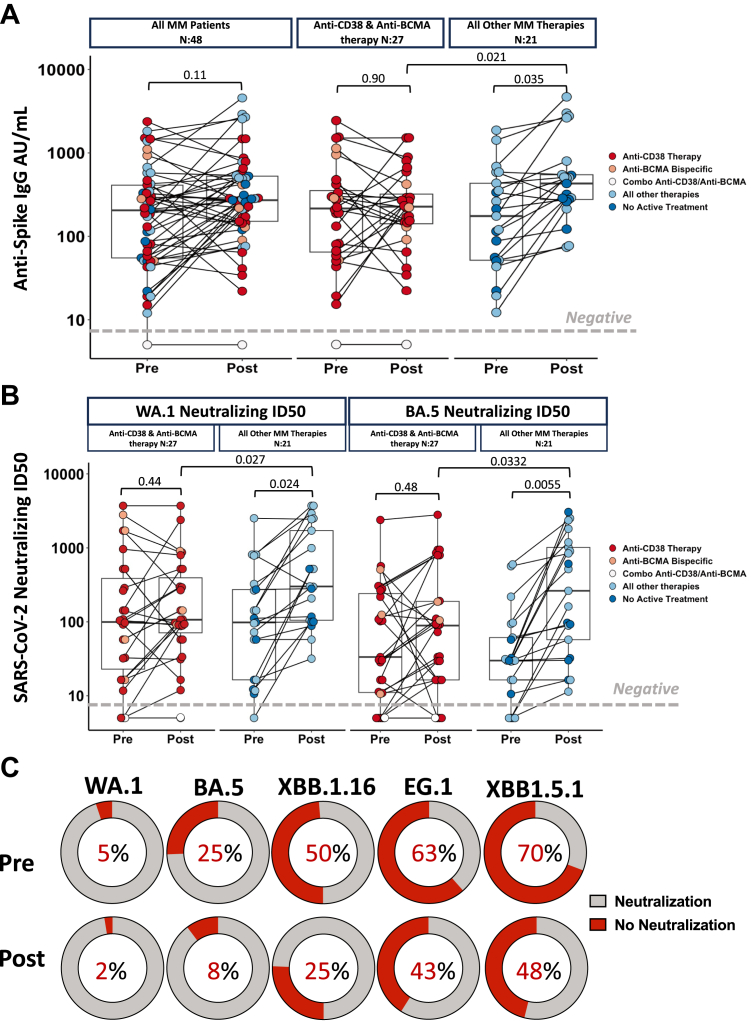
The serological effect of the bivalent vaccination in MM patients is nuanced, as illustrated in Fig. 1A. Consistent with our prior results showing decreased Anti-S IgG production in patients undergoing anti-CD38 and anti-BCMA antibody therapy following vaccination, the overall cohort did not demonstrate a significant increase post bivalent vaccination (median 196 AU/mL prior to bivalent to median 276 AU/mL post bivalent, Wilcoxon signed-rank test p = 0.11). This lack of response was driven by patients undergoing anti-CD38 and anti-BCMA antibody therapy (median 205 AU/mL prior to bivalent to median 248 AU/mL post bivalent, p = 0.9) as compared to MM patients undergoing other therapies who demonstrated a significant increase (median 190.5 AU/mL prior to bivalent to median 455 AU/mL post bivalent, Wilcoxon signed-rank test p = 0.035, Fig. 1A) in anti-S antibody titers after booster vaccination. Of note, the only patient from our cohort that remained seronegative after bivalent vaccination were undergoing a combination of both anti-CD38 and anti-BCMA therapy.
Fig. 1.
Humoral responses to Bivalent Vaccination in patients with Multiple Myeloma. (A) Left panel: anti-SARS-CoV-2 spike IgG antibody levels before and after SARS-CoV-2 bivalent vaccination in MM patients. Right panel: anti-SARS-CoV-2 spike IgG antibody levels before and after SARS-CoV-2 bivalent vaccination by major treatment groups. Antibody concentrations measured in artificial units per mL (AU/mL) and are depicted on a log-10 scale. The gray horizontal dotted line indicates the lower limit of detection (5 AU/mL). (B) Neutralizing antibody ID50 to WA.1 wild-type SARS-CoV-2 strain for MM subject groups split according to major treatment groups. Neutralizing antibody ID50 to Omicron SARS-CoV-2 strain for MM subject groups split according to major treatment groups. The gray horizontal dotted line indicates the lower limit of detection. Dots are colored to indicate treatment regimen at the time of vaccination. p-values represent comparison using the paired Wilcoxon signed-rank test. (C) Percent of MM patients that are unable to neutralize ancestral strain (WA.1) and variants of interest BA.5, XBB1.16, EG.1 and XBB1.5.1 pre and post bivalent vaccination. Threshold for neutralization was at least 30% inhibition of variant. Gray denotes patients able to neutralize while red denotes those not able to neutralize.
An important outstanding question is whether the bivalent mRNA vaccine induced immune response offers increased protection against the current variants.15 We next investigated how the serum neutralizing capacity for ancestral SARS-CoV-2 (WA.1) and Omicron BA.5 viruses changed in response to bivalent vaccination in our MM patients. Bivalent vaccination significantly increases neutralizing capacity in subsets of MM patients in a treatment-specific manner. Patients on anti-CD38 and anti-BCMA antibody therapy did not significantly increase neutralizing capacity to WA.1 (median ID50 = 104.2 prior to bivalent to median ID50 = 107.6 post bivalent, Wilcoxon signed-rank test p = 0.42) or BA.5 (median ID50 = 30.6 prior to bivalent to median ID50 = 72.8 post bivalent, Wilcoxon signed-rank test p = 0.48), while MM patients on other therapies benefitted significantly as illustrated by the increased neutralizing capacity of WA.1 (median ID50 = 104.8 prior to bivalent to median ID50 = 409.5 post bivalent, Wilcoxon signed-rank test p = 0.024) and BA.5 (median ID50 = 30 prior to bivalent to median ID50 = 212.6 post bivalent, Wilcoxon signed-rank test p = 0.0055). Of note, the increase in neutralizing capacity after the bivalent vaccination was more prominent in the neutralization of the BA.5 viral variant with a three-fold increase post vaccination compared to 1.7 fold increase for the ancestral WA.1 strain (Fig. 1B). Serum neutralizing activity was in line with the anti-S binding IgG titers as patients undergoing therapy with anti-CD38 and anti-BCMA antibody therapy had lower neutralization both WA.1 (patients undergoing anti-CD38 and anti-BCMA antibody median ID50 = 107.5, all other therapies ID50 = 409.5, p = 0.0332) and BA.5 (patients undergoing anti-CD38 and anti-BCMA antibody median ID50 = 72.8, all other therapies ID50 = 212.6, Mann–Whitney U test, p = 0.027) compared to patients receiving other therapies (Fig. 1B). Anti-Spike IgG levels strongly correlated with neutralizing ID50 values of WA.1 pre (Spearman (95% CIs), R = 0.9 (0.83, 0.94), p < 0.0001, Supplementary Figure S2A) and post bivalent vaccination (Spearman (95% CIs), R = 0.87 (0.77, 0.92), p < 0.0001, Supplementary Figure S2A) as well as and BA.5 pre (Spearman (95% CIs), R = 0.81 (0.68, 0.89), p < 0.0001, Supplementary Figure S2B) and bivalent post vaccination (Spearman (95% CIs), R = 0.69 (0.51, 0.81), p < 0.0001, Supplementary Figure S2B).
For a subset of 40 MM patients we further assessed to what extent the bivalent vaccine improved on cross-neutralization of newly emerging variants of interest which were not included in the vaccine preparation. The bivalent booster dose significantly reduced the number of sera that failed to neutralize ancestral and BA.5 viruses and improved neutralization of XBB.16, EG.1 and XBB.1.5.1 (with an additional 456 L in spike) (Fig. 1C, Supplementary Figure S3A). Patients undergoing anti-BCMA bispecific and anti-CD38 targeting therapy did not have a significant increase post vaccination that was present in patients on other therapies (Supplementary Figures S3A–H). Regardless of therapy, MM patients remained highly vulnerable to variants EG.1 and XBB1.5.1 as sera collected post bivalent booster failed to neutralize in over 40% of patients. Of note, the spike region of XBB1.5.1 isolate has the same set of mutations as EG.5 highlighting the potential of this recently spreading Omicron variant to cause morbidity in patients with MM.
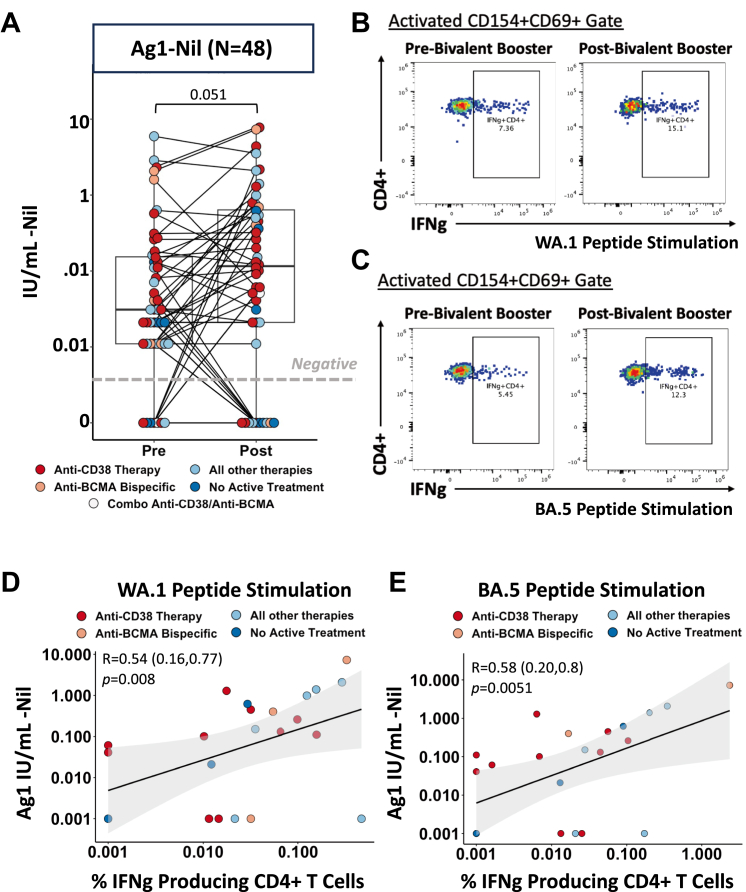
In addition to humoral responses, we profiled T cell responses by implementing a QuantiFERON SARS-CoV-2 RUO assay (Qiagen) in our MM population. This assay has been shown to be a rapidly deployable clinical test to measure cellular immunity in vaccinated healthcare workers to ascertain cellular protection.16 We tested three conditions composed of different SARS-CoV-2 peptide pools pre and post bivalent vaccination. Ag1 which contains CD4+ epitopes from the S1 subunit of the receptor binding domain of the spike protein (Fig. 2A), Ag2 peptide pool which contains CD4+ and CD8+ epitopes from the S1 and S2 subunits of the spike protein (Supplementary Figure S4A), and Ag3 which encompasses CD4+ and CD8+ epitopes as well as immunodominant CD8+ epitopes derived from the whole genome (Supplementary Figure S4B) on our MM patients.17 Bivalent vaccination resulted in a general increase of T cell responses in all peptide stimulated conditions tests albeit not reaching statistically significance (p = 0.057) regardless of therapy. Using our published flow cytometry assay9 we tested IFNg production in MM patient peripheral blood nuclear cells (PBMCs) to both ancestral strand (Fig. 2B) and BA.5 variant (Fig. 2C) pre and post bivalent vaccination. As we have published prior, vaccine responses are primarily CD4+ T cell dominant, therefore our FACS based activation induced marker (AIM) assay is maximized to capture the cytokine production of CD4+ T Cells. We observed a significant correlation between the INF-ɣ values induced by Ag1 peptide pool by CD4+ T cells and the IFN-ɣ produced by CD4+ T cells measured by FACS-based T Cell assays to both WA.1 stimulation (Spearman (95% CIs), R = 0.54 (0.16, 0.77), p = 0.008, Fig. 2D) and BA.5 variant stimulation (Spearman (95% CIs), R = 0.58 (0.20, 0.8), p = 0.0051, Fig. 2D). There was also a significant correlation between Ag2 T cells responses and flow-based T cell to both WA.1 (Spearman (95% CIs), R = 0.60 (0.24, 0.8), p = 0.0027, Supplementary Figure S4B) and BA.5 (Spearman (95% CIs), R = 0.54 (0.15, 0.78), p = 0.01, Supplementary Figure S4C) but not Ag3 in either WA.1 (Spearman (95% CIs), R = 0.29 (−0.14, 0.62), p = 0.19, Supplementary Figure S4E) or BA.5 (Spearman (95% CIs), R = 0.39 (−0.04, 0.7), p = 0.077, Supplementary Figure S4F) post vaccination which may be in part due to our FACS assay being optimized to quantifying CD4+ T cell responses. SARS-CoV-2 specific T cell responses significantly correlated with anti-Spike IgG levels in MM patients (Spearman (95% CIs), R = 0.49 (0.24, 0.68), p = 0.00041, Supplementary Figure S5A), WA.1 neutralizing ID50 (Spearman (95% CIs), R = 0.6 (0.37, 0.76), p < 0.0001, Supplementary Figure S5B) and BA.5 neutralizing ID50 (Spearman (95% CIs), R = 0.54 (0.29, 0.71), p = 0.00012, Supplementary Figure S5C) post bivalent vaccination reinforcing the fact that T cell activity is not a dominant form of compensative protection in sub-optimal MM patients with diminished humoral responses.9
Fig. 2.
T Cell responses to Bivalent Vaccination in patients with Multiple Myeloma. (A) SARS-CoV-2 specific CD4+ T cell responses in MM patients measured by Qiagen QuantiFERON assay. Results show IFN gamma in IU/mL produced by AG1 peptide pool stimulation containing CD4+ epitopes from the S1 subunit of the ancestral Spike protein minus the IFN gamma produced from negative control condition. Dots are colored to indicate treatment regimen at the time of vaccination. p-values represent comparison using the paired Wilcoxon signed-rank test. (B and C) Representative FACS dot plots of IFN gamma expressing CD4+ T cells in a MM patient, pre and post bivalent vaccination, after 6 h stimulation with (B) ancestral SARS-CoV-2 CD4 peptide pools and (C) Omicron BA.5 SARS-CoV-2 CD4 peptide pools. Cytokine expressing CD4+ T cells were identified within activation gates defined by expression of CD4+ T cell activation markers CD154 and CD69. (D) Spearman’s rank correlation between SARS-CoV-2 specific T cell response from Qiagen QuantiFERON kit and IFN gamma producing CD4+ T cells stimulated with ancestral peptide strains. Shaded gray indicates the 95% confidence bands for the regression line and values in parenthesis indicate 95% confidence intervals for correlation coefficient. IFN gamma-expressing CD4+ T cells were calculated by subtracting water control frequencies from the CD4+ T cell response for each subject. (E) Spearman’s rank correlation between SARS-CoV-2 specific T cell response from Qiagen QuantiFERON kit and IFN gamma producing CD4+ T cells stimulated with Omicron BA.5 peptide strains. Shaded gray indicates the 95% confidence bands for the regression line and values in parenthesis indicate 95% confidence intervals for correlation coefficient. IFN gamma-expressing CD4+ T cells were calculated by subtracting water control frequencies from the CD4+ T cell response for each subject. Colors of dots indicate treatment regimen at the time of vaccination.
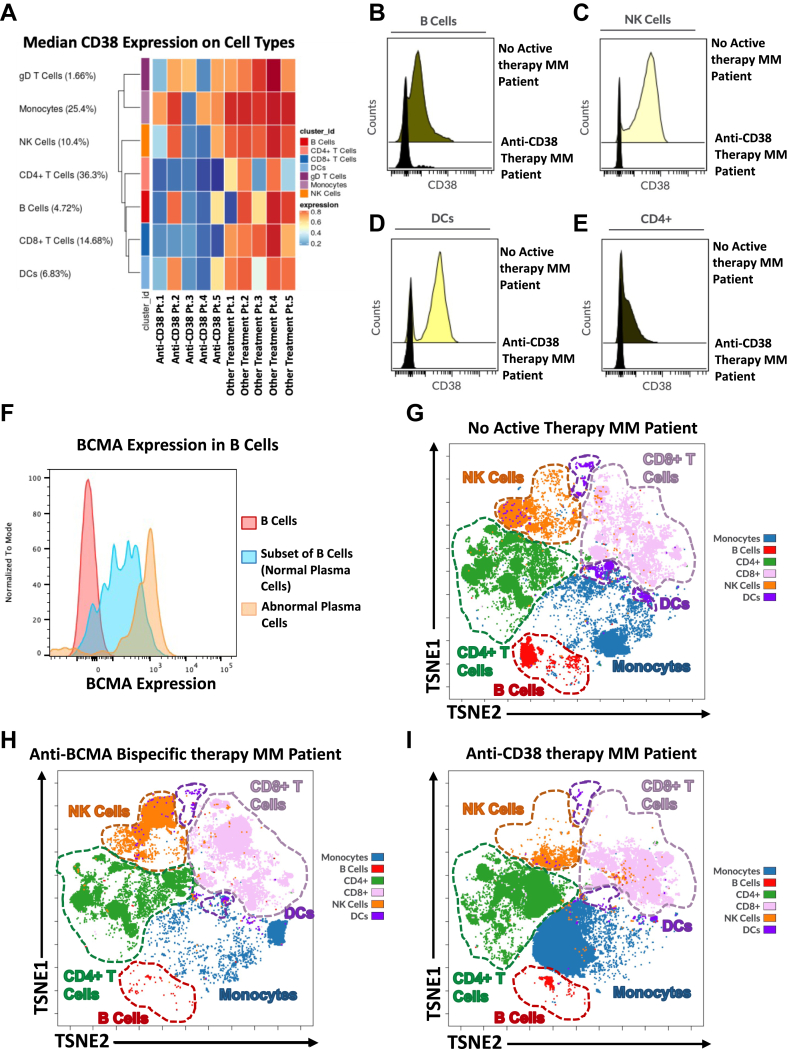
To characterize the cellular microenvironment factors involved in the sub-optimal antibody production upon vaccination seen in the subset of MM patients on anti-CD38 and anti-BCMA treatments, we performed high dimensional immunophenotyping using spectral flow cytometry on a subset of 38 patients. Patients with lowest antibody titers post bivalent booster vaccination exhibit the most compromised SARS-CoV-2 humoral and cellular responses. We separated the lowest quantile (patients with Anti-S IgG <156 AU/mL) (hereafter referred to as “sub-optimal cohort”) and compared their cellular immune phenotypes to the remaining MM patients (hereafter referred to as “responding cohort”).
Most MM patients (90%) in our sub-optimal cohort were undergoing anti-CD38 and anti-BCMA antibody therapy compared to 40% in the responding patient group (p = 0.0089). We investigated the therapeutic targets of these treatments and confirmed their presence in the cell types that are required for functional and healthy humoral protection. CD38 is a multifunctional protein that is widely expressed on various cell types which is upregulated by a large variety of inflammatory signals and is frequently used as a marker of cell activation.18 In untreated MM patients we observed widespread expression of CD38 in T cells, monocytes, natural killer (NK) cells, B cells and dendritic cells (DCs) (Fig. 3A). In MM patients receiving anti-CD38 therapy we detected a depletion of CD38 expressing cell populations (Fig. 3B–E). BCMA expression is seen in healthy differentiated B cells, plasma cells, and MM tumor cells which are clonal plasma cells in the bone marrow (Fig. 3F). Healthy plasma cells are a critical component in the production of antibodies in functional humoral immunity. We compared the phenotypic composition of the immune cell milieu in patients that were untreated (Fig. 3G) to those patients that were receiving anti-BCMA (Fig. 3H) and anti-CD38 targeting therapy (Fig. 3I) and found a stark decrease in the major cell types that express the therapeutic targets.
Fig. 3.
CD38 and BCMA expression in MM patient Samples. (A) Median CD38 expression in immune cell types in 5 MM patients receiving anti-CD38 targeting therapy and 5 MM patients receiving other treatment regimens not including an anti-CD38 targeting therapy in peripheral blood patients. (B–E) Representative histogram of CD38 expression by flow cytometry in the peripheral blood of patients receiving anti-CD38 therapy compared to patients not undergoing any active therapy. (B) CD38 expression on B cells (C) CD38 expression on NK Cells (D) CD38 expression on Dendritic Cells (DCs) (E) CD38 expression on CD4+ T cells. (F) Histogram of BCMA expression in the plasma cell B cell subset in the bone marrow sample of an MM patient. (G–I) Representative TSNE illustrating the composition of the immune cell milieu in (G) Reference patient currently not on any active treatment (H) patients receiving anti-BCMA bispecific therapy and (I) patients on anti-CD38 therapy.
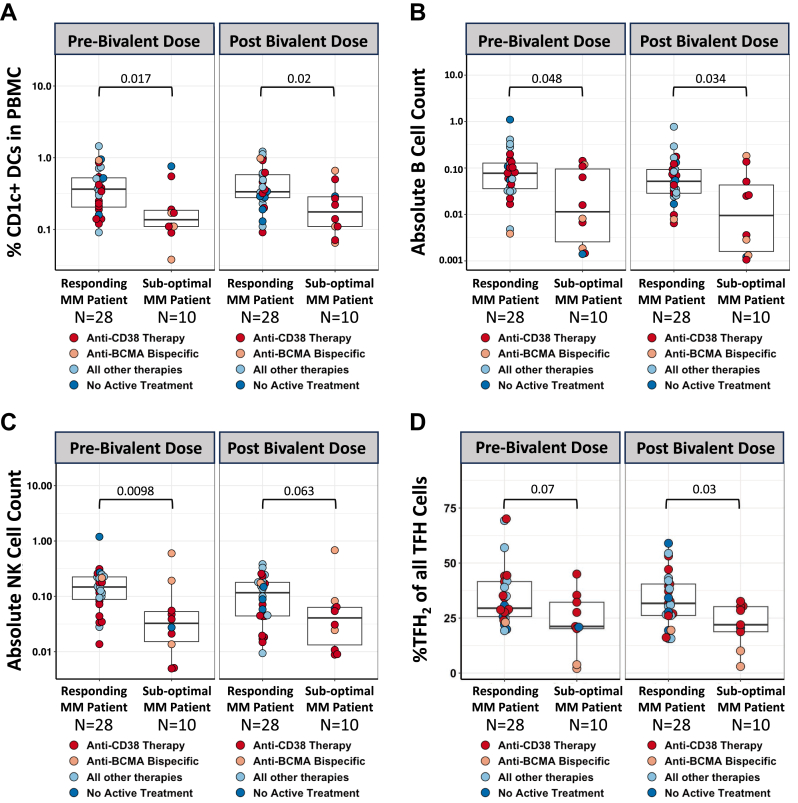
Immune populations involved in robust antibody production, such as DCs, B cells, NK cells and T follicular helper cells (TFH), were compared between sub-optimal MM patients and responding MM patients pre and post bivalent booster vaccination. Patients with sub-optimal antibody production consistently displayed lower levels of antigen presenting cells such as a subset of DCs (Fig. 4A) and B cells expressing either CD19 or CD20 (Fig. 4B, Supplementary Figure S6A). Further investigation into the B cell population revealed differences in the frequency of B cells expressing the classical chemokine receptor CXCR5 (Supplementary Figure S6B). CXCR5 helps direct B cells into the follicles of secondary lymphoid organs.19 Sub-optimal MM patients consistently had diminished amount of CXCR5+ B cells compared to responding MM patients (Supplementary Figure S6C). Sub-optimal patients also displayed lower counts of NK cells compared to responding patients pre and post bivalent vaccination (Fig. 4C). Additionally, responding patients had a NK phenotype that was predominantly mature cytolytic NK phenotype (CD56dim CD16high NK Cells Supplementary Figure S7A) which was severely decreased in sub-optimal patients pre and post bivalent vaccine (Supplementary Figure S7B). CD4+ CXCR5+ cells (TFH cells) are a class of T cells that orchestrate germinal center B cell maturation which is critical in the context of infection and cellular environment.20 Patients with sub-optimal antibody responses had lower presence of TFH2 cells characterized as CXCR3-CCR6-CCR4+ TFH cells known to induce differentiation of B cells, antibody secretion, and control immunoglobulin (Ig) isotype switching (Fig. 4D).21, 22, 23
Fig. 4.
Phenotypic differences between responding and sub-optimal Multiple Myeloma patients during bivalent vaccination. Suboptimal MM patients are denoted as the lowest quartile of anti-SARS-CoV-2 spike IgG antibody levels antibody production compared to responding MM patients. (A) Frequencies of CD1c+ dendritic cells in peripheral blood mononuclear cells pre and post bivalent vaccination. (B) Absolute NK Cell counts in peripheral blood pre and post bivalent vaccination in MM patients. (C) Absolute B Cell counts in peripheral blood pre and post bivalent vaccination in MM patients. (D) Frequency of CXCR5+ CD4+ (TFH cells) expressing a Th2 phenotype (CXCR3-CCR6-CCR4+) in peripheral blood peripheral blood pre and post bivalent vaccination in MM patients. Dots are colored to indicate treatment regimen at the time of vaccination. p-values represent comparison using Mann–Whitney U test.
Our findings of a phenotype associated with sub-optimal SARS-CoV-2 protection was retrospectively validated in our cohort of patients post the third dose of SARS-CoV-2 vaccination. We compared patients that were seronegative post third dose and in the lowest quartile of antibody production (sub-optimal MM patients) against the remaining three quartiles which were deemed as responding patients. Full demographics of the third dose cohort are reported in Supplementary Table S3. We found the same deficiencies in the machinery that was present in our cohort of MM undergoing bivalent SARS-CoV-2 vaccination. The most vulnerable sub-optimal MM patients pre and post the third dose had deficiencies in a subset of DCs (Supplementary Figure S8A), NK cells (Supplementary Figure S8B), B cells (Supplementary Figure S8C), and TFH2 T Cells (Supplementary Figure S8D). The subtypes of mature cytolytic NK cells (Supplementary Figure S7C) and CXCR5+ B cells (Supplementary Figure S6D) were also reduced in the MM patients that failed to produce robust antibody responses.
Discussion
Our study highlights the nuances in immune responses and continuing vulnerability following bivalent COVID-19 immunizations in MM patients. Bivalent vaccination has been reported to reduce the rate of COVID-19 and increase neutralization capacity to omicron variants infection in patients with hematological neoplasms.24,25 We show that bivalent boosters critically improve neutralization of both ancestral (WA.1) as well as the BA.5 Omicron variant significantly in Myeloma patients that are not on anti-CD38 or anti-BCMA therapeutics, which correlates with both anti-Spike IgG titers and T cell responses. MM patients enhanced their ability to neutralize recently emerged antigenically diverse XBB variants after bivalent vaccination. Indeed, when only analyzing the lack of neutralization, the bivalent booster vaccination provided a clear benefit for neutralization of all five SARS-CoV-2 viruses tested. When comparing sera collected before and after the bivalent booster vaccination, the frequency of sera that failed to neutralize was reduced by 2-fold, not only for the viruses included in the bivalent booster (ancestral, BA.5) but also for XBB.1.16. This benefit was not observed for EG.1 and XBB.1.5.10 which display substantially greater neutralization resistance. These findings underscore the necessity to provide an updated booster vaccine dose containing XBB.1.5 spike.26
Safety profiles of anti-CD38 and anti-BCMA therapies report lymphopenia in the majority of patients with over 70% of patients having at least one infection throughout therapy.27,28 Immune composition has been reported to being altered with NK cell fratricide in anti-CD38 therapy and B cell reduction in anti-BCMA therapy.29,30 Our results demonstrate that patients on anti-CD38 or anti-BCMA therapeutics have increased vulnerability to COVID-19 despite bivalent vaccination as their immune machinery is unable to mount responses to these booster vaccines.
There is an essential interaction between DCs, NK cells, TFH and B cells for initiation, coordinating and sustaining the humoral antibody responses.31 Our high dimensional cellular analysis pinpoints the cellular defects likely responsible for generating sub-optimal antibody and T cell responses that renders them unable to respond effectively to bivalent vaccination. These defects in immune populations in response to anti-BCMA and anti-CD38 therapies have consequences for patient selection, MM management (especially duration of maintenance therapy in patients who are in remission), as well as vaccine development in the future. This is critical as we have documented ongoing susceptibility and potential for persistent SARS-CoV-2 replication in immunocompromised patients.32 Patients receiving combination anti-CD38 and anti-BCMA therapy may be most vulnerable as demonstrated by the lack of any anti-Spike IgG titers and neutralizing capacity even after repeated vaccination. Retrospectively, we see that the compromised cellular populations conferring sub-optimal responses to bivalent vaccination were consistently diminished in the sub-optimal cohort of patients most lacking humoral and cellular protection post third dose. Ongoing studies will determine whether the compromised immune machinery and the reduced T cell activity identified herein can further discriminate patients at high risk for non-COVID infectious complications such as CMV and other infections from novel anti-BCMA therapies.
Our data also suggest that the QuantiFERON SARS-CoV-2 RUO assay, especially the Ag2 peptide pool, could be deployed for clinical use, bridging the need for suitable laboratory tests to measure SARS-CoV-2 specific T cell responses. As we and others have reported the T cell responses are mainly driven by CD4 stimulation, however, convalescent patients have shown to produce robust CD8 T cell responses allowing for Ag2 to provide a more encompassing view of the immune response in MM patients.33 We compared QuantiFERON SARS-CoV-2 RUO assay to a “gold standard” spectral flow-cytometry based assay which, though comprehensive, is extremely time consuming and not available outside of a few specialized labs and found the best correlation with the Ag2 peptide pool stimulation. The QuantiFERON test could theoretically be deployed wherever similar kit-based tuberculosis testing is currently being carried out adding another parameter to consider in the care of these immunocompromised MM patients.
Our study suffers from several limitations. First, our sample size is limited. However, even with limited numbers we see a statistically significant differences in the effect of the bivalent vaccine on immune responses. The statistical significance reached with the sample sizes we have are indicative that the trends in our report are impactful and sound. Second, recall bias may play a role in the cumulative infection status that we have documented. Our annotation for positive COVID19 infection was done through chart review. Throughout the pandemic our patients would have to test negative to receive treatment and see physicians at out institute. However, as the COVID-19 restrictions have lessened, documentation of COVID-19 infection is reliant of patient reporting allowing for potential underreporting as some patients may be asymptotic and not testing during periods between therapies. Finally, our study highlights the effect of bivalent vaccination at one timepoint post vaccine not providing an insight into long-term effects of vaccination and impact of decay in protective capacities. As we continue monitoring our patients, we will provide further studies into the kinetics and continued magnitude of protection in relation to bivalent vaccination.
In conclusion, we have identified a phenotype deficient in key immune cell machinery that is associated with suboptimal vaccine immune responses in MM patients. Therapies may reduce tumor burden but also fundamentally change the immune microenvironment allowing for opportunistic infections to evolve and thrive. As SARS-CoV-2 becomes an endemic respiratory viral pathogen, MM patients and other immunocompromised populations may need specialized consideration for annual or booster vaccinations. Future studies are needed to explore if other immunizations are equally impacted by specific MM treatment lines.
Our neutralizing assay results demonstrate that MM patients benefited significantly from bivalent vaccination with a clear increase in neutralizing capacity to BA.5, however a subset of patients undergoing anti-CD38 and anti-BCMA bispecific therapy will remain susceptible to currently circulating, newly emerged viral strains such as EG.5. Patients should be encouraged to receive the booster vaccine against XBB1.5 that will be available this fall based on our results, and ongoing studies will inform on the protective nature of the that booster in terms of neutralization in the immunocompromised population.
Contributors
Viviana Simon, Ania Wajnberg, Miriam Merad, Samir Parekh and other members of the PVI/MM/Seronet Study Group contributed conceptualization, methodology, analysis, and resources for this work. Adolfo Aleman, Morgan van Kesteren, Kseniya Serebryakova, Katerina Kappes, Komal Srivastava, Neko Lyttle, Jessica R Nardulli, Charles R. Gleason and members of the PVI/MM/Seronet Study Group contributed to organizational aspects of the clinical studies, patient recruitment, data collection and analysis. Carlos Cordon-Cardo, Viviana Simon, and PVI/MM/Seronet Study Group contributed to design, data collection, analysis, visualization and interpretation of serological data. Adolfo Aleman, Ariel Kogan Zajdman, Bhaskar Upadhyaya and Lucia Y Chen contributed to design, execution, analysis, visualization and interpretation phenotypic assessment and T cell assays. Sundar Jagannath, Ania Wajnberg, Samir Parekh and Viviana Simon were involved in different aspects of patient care. Adolfo Aleman, Morgan van Kesteren, Sundar Jagannath, Viviana Simon and Samir Parekh contributed to interpretation of the data and conceptualization of the first manuscript draft. Morgan van Kesteren, Jacob Mischka, Jessica Nardulli, Harm van Bakel, Emilia Mia Sordillo, Charles R. Gleason, Komal Srivastava, Annika Oostenink, Gianna Y. Cai, Christian Cognigni, Viviana Simon and members of the PVI/MM/Seronet Study Group contributed to the neutralization assays and serological quantification. Adolfo Aleman, Viviana Simon and Samir Parekh have accessed and verified the underlying data. All co-authors provided critical edits to the initial manuscript draft. All authors have read and approved the final version of the published manuscripts.
Data sharing statement
Raw data was generated at the Icahn School of Medicine at Mount Sinai. De-identified datasets generated and analyzed during the current study are available from the corresponding author upon reasonable request.
Declaration of interests
The Icahn School of Medicine at Mount Sinai has filed patent applications relating to SARS-CoV-2 serological assays which list Viviana Simon and Carlos Cardon-Cordo as co-inventors. Mount Sinai has spun out a company, Kantaro, to market serological tests for SARS-CoV-2. Sundar Jagannath reports consulting fees for Bristol Myers Squibb (Celgene), Janssen, Karyopharm Therapeutics, Merck, Sanofi, and Takeda Pharmaceuticals. Samir Parekh reports consulting fees from Foundation Medicine and research funding from Bristol Myers Squibb (Celgene), Karyopharm, and Amgen. The other authors reported no relevant conflicts of interest.
Acknowledgements
We thank participants for their generosity and willingness to participate in longitudinal COVID-19 research studies. None of this work would be possible without their contributions. We acknowledge the clinical and research staff at the Center of Excellence for Multiple Myeloma at Mount Sinai. S.P. is supported by National Cancer Institute (NCI) R01 CA244899, CA252222 and receives research funding from Amgen, Celgene/BMS, and Karyopharm. This work and PVI/MM/Seronet Study Group was partially funded by the NIAID Collaborative Influenza Vaccine Innovation Centers (CIVIC) contract 75N93019C00051, NIAID Center of Excellence for Influenza Research and Surveillance (CEIRS, contracts HHSN272201400008C and HHSN272201400006C), and NIAID grants U01AI141990 and U01AI150747; by the generous support of the JPB Foundation and the Open Philanthropy Project (research grant 2020-215611 [5384]); and by anonymous donors. This effort was supported by the Serological Sciences Network (SeroNet) in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract 75N91019D00024, task order 75N91021F00001. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. We acknowledge support of the Center of Excellence for Multiple Myeloma Philanthropy.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104886.
Contributor Information
Viviana Simon, Email: viviana.simon@mssm.edu.
Samir Parekh, Email: samir.parekh@mssm.edu.
PVI/MM/Seronet Study Group:
Hala Alshammary, Dalles Andre, Radhika Banu, Katherine Beach, María Carolina Bermúdez-González, Ajai Chari, Yuexing Chen, Hearn Cho, Adolfo Firpo, Ana Silvia Gonzalez-Reiche, Eun Hye Kim, Giulio Kleiner, Florian Krammer, Jacob Mauldin, Rao Mendu, Brian Monahan, Shambavi Richard, Joshua Richter, Cesar Rodriguez, Adrianna Rossi, Ashley Salimbangon, Laryssa Sanchez, and Daniel Verina
Appendix A. Supplementary data
References
- 1.Giuliani N., Accardi F., Marchica V., et al. Novel targets for the treatment of relapsing multiple myeloma. Expert Rev Hematol. 2019;12(7):481–496. doi: 10.1080/17474086.2019.1624158. [DOI] [PubMed] [Google Scholar]
- 2.Shah U.A., Mailankody S. Emerging immunotherapies in multiple myeloma. BMJ. 2020;370 doi: 10.1136/bmj.m3176. [DOI] [PubMed] [Google Scholar]
- 3.Nucci M., Anaissie E. Infections in patients with multiple myeloma in the era of high-dose therapy and novel agents. Clin Infect Dis. 2009;49(8):1211–1225. doi: 10.1086/605664. [DOI] [PubMed] [Google Scholar]
- 4.Blimark C., Holmberg E., Mellqvist U.H., et al. Multiple myeloma and infections: a population-based study on 9253 multiple myeloma patients. Haematologica. 2015;100(1):107–113. doi: 10.3324/haematol.2014.107714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Kuraishy H.M., Al-Gareeb A.I., Mohammed A.A., Alexiou A., Papadakis M., Batiha G.E. The potential link between Covid-19 and multiple myeloma: a new saga. Immun Inflamm Dis. 2022;10(12) doi: 10.1002/iid3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bakouny Z., Hawley J.E., Choueiri T.K., et al. COVID-19 and cancer: current challenges and perspectives. Cancer Cell. 2020;38(5):629–646. doi: 10.1016/j.ccell.2020.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulligan M.J., Lyke K.E., Kitchin N., et al. Phase I/II study of COVID-19 RNA vaccine BNT162b1 in adults. Nature. 2020;586(7830):589–593. doi: 10.1038/s41586-020-2639-4. [DOI] [PubMed] [Google Scholar]
- 8.Aleman A., Van Oekelen O., Upadhyaya B., et al. Augmentation of humoral and cellular immune responses after third-dose SARS-CoV-2 vaccination and viral neutralization in myeloma patients. Cancer Cell. 2022;40(5):441–443. doi: 10.1016/j.ccell.2022.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aleman A., Upadhyaya B., Tuballes K., et al. Variable cellular responses to SARS-CoV-2 in fully vaccinated patients with multiple myeloma. Cancer Cell. 2021;39(11):1442–1444. doi: 10.1016/j.ccell.2021.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van Oekelen O., Gleason C.R., Agte S., et al. Highly variable SARS-CoV-2 spike antibody responses to two doses of COVID-19 RNA vaccination in patients with multiple myeloma. Cancer Cell. 2021;39(8):1028–1030. doi: 10.1016/j.ccell.2021.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chalkias S., Harper C., Vrbicky K., et al. A bivalent Omicron-containing booster vaccine against Covid-19. N Engl J Med. 2022;387(14):1279–1291. doi: 10.1056/NEJMoa2208343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stadlbauer D., Amanat F., Chromikova V., et al. SARS-CoV-2 seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr Protoc Microbiol. 2020;57(1) doi: 10.1002/cpmc.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stadlbauer D., Tan J., Jiang K., et al. Repeated cross-sectional sero-monitoring of SARS-CoV-2 in New York City. Nature. 2021;590(7844):146–150. doi: 10.1038/s41586-020-2912-6. [DOI] [PubMed] [Google Scholar]
- 14.Carreno J.M., Alshammary H., Tcheou J., et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602(7898):682–688. doi: 10.1038/s41586-022-04399-5. [DOI] [PubMed] [Google Scholar]
- 15.Carreno J.M., Singh G., Simon V., Krammer F., PVI Study Group Bivalent COVID-19 booster vaccines and the absence of BA.5-specific antibodies. Lancet Microbe. 2023;4(8) doi: 10.1016/S2666-5247(23)00118-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krüttgen A., Klingel H., Haase G., Haefner H., Imöhl M., Kleines M. Evaluation of the QuantiFERON SARS-CoV-2 interferon-ɣ release assay in mRNA-1273 vaccinated health care workers. J Virol Methods. 2021;298 doi: 10.1016/j.jviromet.2021.114295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaganathan S., Stieber F., Rao S.N., et al. Preliminary evaluation of QuantiFERON SARS-CoV-2 and QIAreach anti-SARS-CoV-2 total test in recently vaccinated individuals. Infect Dis Ther. 2021;10(4):2765–2776. doi: 10.1007/s40121-021-00521-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glaría E., Valledor A.F. Roles of CD38 in the immune response to infection. Cells. 2020;9(1):228. doi: 10.3390/cells9010228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henneken M., Dörner T., Burmester G.R., Berek C. Differential expression of chemokine receptors on peripheral blood B cells from patients with rheumatoid arthritis and systemic lupus erythematosus. Arthritis Res Ther. 2005;7(5):R1001–R1013. doi: 10.1186/ar1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavazzoni C.B., Hanson B.L., Podestà M.A., et al. Follicular T cells optimize the germinal center response to SARS-CoV-2 protein vaccination in mice. Cell Rep. 2022;38(8) doi: 10.1016/j.celrep.2022.110399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeh C.-H., Finney J., Okada T., Kurosaki T., Kelsoe G. Primary germinal center-resident T follicular helper cells are a physiologically distinct subset of CXCR5hiPD-1hi T follicular helper cells. Immunity. 2022;55(2):272–289.e7. doi: 10.1016/j.immuni.2021.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui D., Tang Y., Jiang Q., et al. Follicular helper T cells in the immunopathogenesis of SARS-CoV-2 infection. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.731100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu M., Jiang Y., Wang J., et al. Distribution of distinct subsets of circulating T follicular helper cells in Kawasaki disease. BMC Pediatr. 2019;19(1):43. doi: 10.1186/s12887-019-1412-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tadmor T., Melamed G., Alapi H., Patalon T., Rokach L. The effectiveness of bivalent mRNA Omicron containing booster vaccines among patients with hematological neoplasms. Anticancer Res. 2023;43(7):3129–3134. doi: 10.21873/anticanres.16485. [DOI] [PubMed] [Google Scholar]
- 25.Ehmsen S., Pedersen R.M., Bang L.L., et al. BQ.1.1, XBB.1, and XBB.1.5 neutralization after bivalent mRNA COVID-19 booster in patients with cancer. Cancer Cell. 2023;41(4):649–650. doi: 10.1016/j.ccell.2023.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zappa M., Verdecchia P., Angeli F. Severe acute respiratory syndrome coronavirus 2 evolution: how mutations affect XBB.1.5 variant. Eur J Intern Med. 2023;112:128–132. doi: 10.1016/j.ejim.2023.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moreau P., Garfall A.L., van de Donk N.W.C.J., et al. Teclistamab in relapsed or refractory multiple myeloma. N Engl J Med. 2022;387(6):495–505. doi: 10.1056/NEJMoa2203478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nahi H., Chrobok M., Gran C., et al. Infectious complications and NK cell depletion following daratumumab treatment of multiple myeloma. PLoS One. 2019;14(2) doi: 10.1371/journal.pone.0211927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geraldes C., Neves M., Chacim S., da Costa F.L. Practical considerations for the daratumumab management in Portuguese routine clinical practice: recommendations from an expert panel of hematologists. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.817762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang C. Teclistamab: first approval. Drugs. 2022;82(16):1613–1619. doi: 10.1007/s40265-022-01793-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Teijaro J.R., Farber D.L. COVID-19 vaccines: modes of immune activation and future challenges. Nat Rev Immunol. 2021;21(4):195–197. doi: 10.1038/s41577-021-00526-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzalez-Reiche A.S., Alshammary H., Schaefer S., et al. Sequential intrahost evolution and onward transmission of SARS-CoV-2 variants. Nat Commun. 2023;14(1):3235. doi: 10.1038/s41467-023-38867-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sekine T., Perez-Potti A., Rivera-Ballesteros O., et al. Robust T cell immunity in convalescent individuals with asymptomatic or mild COVID-19. Cell. 2020;183(1):158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.