Abstract
The gntT gene of Escherichia coli is specifically induced by gluconate and repressed via catabolite repression. Thus, gluconate is both an inducer and a repressor of gntT expression since gluconate is a catabolite-repressing sugar. In a gntR deletion mutant, the expression of a chromosomal gntT::lacZ fusion is both high and constitutive, confirming that GntR is the negative regulator of gntT. Indeed, GntR binds to two consensus gnt operator sites; one overlaps the −10 region of the gntT promoter, and the other is centered at +120 with respect to the transcriptional start site. The binding of GntR to these sites was proven in vitro by gel redardation assays and in vivo by site-directed mutagenesis of the binding sites. Binding of GntR to the operators is eliminated by gluconate and also by 6-phosphogluconate at a 10-fold-higher concentration. Interestingly, when gntR deletion strains are grown in the presence of gluconate, there is a twofold decrease in gntT expression which is independent of catabolite repression and binding of GntR to the operator sites. This novel response of gntR mutants to the inducer is termed ultrarepression. Transcription of gntT is activated by binding of the cyclic AMP (cAMP)-cAMP receptor protein (CRP) complex to a CRP binding site positioned at −71 upstream of the gntT transcription start site.
Escherichia coli metabolizes gluconate via the Entner-Doudoroff pathway (EDP) (6, 12, 13), which is, in addition to the gluconate transport and gluconate kinase activities, specifically induced by gluconate (15). Recent experiments suggested that some of the genes involved in gluconate metabolism are required for E. coli to colonize the mouse large intestine (43). It was shown that at least the eda gene, which encodes the 2-keto-3-deoxy-6-phosphogluconate aldolase of the EDP, and the gntP gene, which encodes one of the four gluconate transporters of E. coli (33), play a crucial role during the colonization (22, 43).
Two sets of genes are involved in transport and phosporylation of gluconate (1, 20, 51). The main system, GntI, contains gntT and gntU, encoding high- and low-affinity gluconate transporters (approximate Kms of 6 and 212 μM), respectively, and gntK, a thermoresistant gluconokinase (8). The GntII system, which was discovered in a GntI deletion mutant, contains gntW and gntV, encoding another high-affinity gluconate transporter and a thermosensitive gluconokinase (20, 45). Thus, there are four known gluconate transporters, including GntP (22). Together with three other E. coli proteins of unknown function (Dsdx, ORFo454, and ORFf388), these seven proteins comprise a novel transporter family (33, 48). There is controversy as to why E. coli possesses so many gluconate transporters and which are expressed during growth on gluconate. Recent results show that gntT and gntU are expressed during growth on gluconate (34, 35, 45). In contrast to gntU, gntT shows a pronounced peak of expression very early in the logarithmic phase and is expressed at lower levels in late logarithmic phase (35). It was also found that gntT is maximally induced by 0.5 mM gluconate, whereas gntU shows the highest expression in medium with 10 to 100 mM gluconate (unpublished data). Thus, it appears that GntT is important for growth on low gluconate concentrations, for entry into logarithmic phase, and for cometabolism of gluconate and glucose (35).
Previous genetic studies indicated that the GntI system is, together with the EDP genes edd and eda, negatively regulated by the gntR gene product (10, 45, 11). GntR belongs to the GalR-LacI family of regulators and possesses an N-terminal helix-turn-helix DNA-binding motif, suggesting that GntR fulfills a regulatory role similar to that of LacI for the lac operon (45, 46, 47). Recently we postulated a consensus sequence for a gnt operator which was found in all gluconate-inducible genes (35). Furthermore, the genes of the GntI system are subject to catabolite repression. Interestingly, gluconate is itself a catabolite-repressing sugar (36). In this study, we show for the first time that the operator sequence is indeed the binding site for the negative regulator GntR and that gluconate is the true inducer of gntT, by inactivation of GntR binding to the operator. The results in this report also demonstrate that gntT is regulated by the cyclic AMP (cAMP)-cAMP receptor protein (CRP) complex, which was found be essential for full induction.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The E. coli strains, plasmids, and phages used in this study are listed in Table 1. All chromosomal single gntT::lacZ fusion were constructed in P90C; DH5α was used for constructing and propagating plasmids. E. coli strains were routinely grown at 37°C in Luria broth (LB) (28) with or without added carbohydrate (0.4%), and growth was monitored by measuring the turbidity (optical density [OD]) with a Spectronic 601 (Milton Roy Co.) spectrophotometer.
TABLE 1.
Bacterial strains, plasmids, and phage used in this study
| Strain, plasmid, or phage | Relevant genotype | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH5α | lacZΔM15 recA | Bethesda Research Laboratories |
| P90C | araΔ(pro-lac) thi | 41 |
| DPB271 | recD::Tn10 F− λ− | B. Bender |
| HT28 | W3110 Δcya::Kanr | 19 |
| RH74 | Δcya-851 ilv::Tn10 | 26 |
| RH77 | Δcya-851 Δcrp zhd-731::Tn10 | 26 |
| HT216 | RH74 gntR::Kanr | This study |
| M15 | recA+ uvr+ F−mtl gal ara lac (pREP4) | 50 |
| NP100 | P90C (λRS88 gntT::lacZ) | 35 |
| PR201 | NP100 gntR::Kanr | This study |
| Plasmids | ||
| pQE-30 | Expression vector; His6 affinity tag | 42 |
| pRS551 | bla′ lacZ+ (operator fusion vector) | 41 |
| pRS552 | bla′ lacZ+ (protein fusion vector) | 41 |
| pTC229 | pTC221 Kanr in StuI | This study |
| pNP-41 | gntR in pQE30 (gntR overexpression) | This study |
| pNP-52 | crp in pQE30 (crp overexpression) | This study |
| Phage λRS88 | bla′ lacZ imm434 ind | 41 |
Construction of single-copy gntT::lacZ fusions.
gntT::lacZ operon and protein fusions, present in the chromosome as lambda lysogens in single copy, were constructed by using the system of Simons et al. (41). DNA fragments of the promoter region of gntT were amplified by PCR with primers carrying either an EcoRI or a BamHI site, one at each end. These fragments were cloned into the vector pRS551 (operon fusion vector). Single-copy fusions were made by homologous recombination with the lambda phage λRS88 and integration of the lysogenic phage into the chromosome (41). The copy number of the fusion in the chromosome was checked by the method of Powell et al. (37).
Site-directed mutagenesis.
PCR was used to generate mutations in the operator sequences of the gntT gene. Mutations within the internal operator sequence were produced by PCR using a set of mismatched 3′ primers carrying substitutions and insertions within the internal operator site (P102 to P106) and a 5′ primer with the wild-type sequence (P231) (16). These DNA fragments were cloned into the operon fusion vector pRS551. Mutations within the external gnt operator sequence and the CRP binding site were generated by a three-step recombinant PCR method first described by Higuchi et al. (16). In a separate reaction, two overlapping primary PCR products were synthesized by using outside primers (P230 and P231) of the gntT fragment and overlapping, mismatched inside primers (P206B to P221B) containing the same mutation. The two PCR products were purified by electroelution from an agarose gel and subjected to a second PCR to form a heteroduplex product. This secondary PCR product, containing the mutation in both strands, was amplified with the two outside primers. Double mutations were constructed by the same method, by using DNA containing the first mutation as a template for the three-step recombinant PCR method. The PCR products were cloned into the operator fusion vector pRS551 and then recombined into λRS88 for construction of single-copy fusions as described above. The following primers were used. Wild-type primers P230 (5′-GCGGATCCCCGATAGCAACAATGACTAATG-3′) and P231 (5′-CGGAATTCTGAAAGGTGTGCGCGATCTCAC-3′) were used to amplify the entire promoter region of gntT; wild-type primer P101 (5′-GCGGATCCCATTTGTTATGGGTAACGTCAATTT-3′) and primers P102 (5′-GCGGATCCCATTTGTTATGCAGGTAACGTCAATTT-3′), P103 (5′-GCGGATCCCATTTGTTATGAGGTAACGTCAATTT-3′), P104 (5′-GCGGATCCCATTTGTTATGGGCGACGTCAATTT-3′), P105 (5′ GCGGATCCTTTCATTTGCGCTGGGTAACGTCAA-3′), and P106 (5′-GCGGATCCATTTGTTCTGGGTAACGTCAATTT-3′) were used to create mutations within the internal gnt operator site; primers P206 (5′-TGAATGATACGGTCGACATCTGGCGTTT-3′), P206B (5′ AAACGCCAGATGTCGACCGTATCATTCA-3′), P208 (5′-ATGATACGGGTACCCCATGGCGTTTGAGAA-3′), P208B (5′-TTCTCAAACGCCATGGGGTACCCGTATCAT-3′), P211 (5′-CATGTGAATGACCCGGGTAACATCTGGC-3′), P211B (5′-GCCAGATGTTACCCGGGTCATTCACATG-3′), P212 (5′-CCAGATGTTACCATGGTATCATTCACA-3′), and P212B (5′-TGTGAATGATACCATGGTAACATCTGG-3′) were used to create mutations within the external gnt operator site; primer P220s (5′-TGAGAGGTTGGTCGACTTATCGCGGGGA-3′), P220B (5′ TCCCCGCGATAAGTCGACCAACCTCTCA-3′), P221 (5′-TTTAAATTATCGATGGTTGGTCATAT-3′), and P221B (5′-ATATGACCAACCATCGATAATTTAAA-3′) were used to create mutations within the cAMP-CRP binding site. All DNA fragments carrying PCR-generated mutations were verified by DNA sequence analysis.
DNA band migration retardation.
DNA band migration retardation analyses were performed as described by Carey (5). DNA fragments used in the assays were labeled with [α-32P]dATP during PCR. The binding reactions contained, in a 20-μl final volume, the labeled DNA fragment, 2 μg of sonicated herring sperm DNA, 4 mM Tris-HCl (pH 7.0), 5 mM sodium chloride, 2 mM magnesium chloride, 7.5% glycerol, 2 mM dithiothreitol, and 0.1 mM phenylmethylsulfonyl fluoride. For some experiments, the labeled DNA was first digested with a restriction enzyme and then purified by Sephadex-G50 chromatography. Protein, sugars, and cAMP were added as indicated. The samples were incubated for 10 min at room temperature and loaded onto a 5 or 6% polyacrylamide gel (20 cm by 16 cm by 1 mm thick; 29:1 acrylamide/bisacrylamide). The gel was equilibrated overnight at 4°C in 1× Tris-borate-EDTA buffer (pH 8.3) and prerun for 2 h at 200 V prior to sample loading. For the loading of the samples, the voltage was increased to 300 V. As soon as the last sample had entered the gel, the voltage was reduced to 200 V. After electrophoresis for 1 to 2 h, the gel was dried on paper at 80°C and autoradiographed on Kodak BIOMAX MS film.
Isolation of GntR and CRP.
GntR and CRP were isolated by using the His tag modification system from Qiagen (Hilden, Germany) (17, 42). A 1,130-bp DNA fragment containing the coding region of the gntR gene was synthesized by PCR using primers P701 (5′-GCGGATCCATGAAAAAGAAAAGACCCGTAC-3′) and P702 (5′-GGGGTACCGTGCCCCCACAATACAAGAA-3′), and a 634-bp DNA fragment containing the coding region of the crp gene was synthesized by using primers PCRP1 (5′-GCGGATCCATGGTGCTTGGCAAACCGCAAA-3′) and PCRP3 (5′-GGGGTACCACGGGATTAACGAGTGCCGTAA-3′). After being digested with the endonucleases BamHI and KpnI, the fragments were cloned into the vector pQE30, which contains an isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible T5 promoter (4). The resulting plasmids, pNT70 and pNT82, containing the gntR and crp genes, respectively, fused to six histidine codons at the 5′ end, were transformed into E. coli M15, which overexpresses the lacI gene from pREP4 (50). For the isolation of the recombinant proteins, the resulting E. coli strains M15(pNT70) and M15(pNT82) were grown in LB medium to an OD at 600 nm (OD600) of 0.8 or 1.5, respectively. Expression of the proteins was induced by the addition of 1 to 2 mM IPTG for 2 to 4 h. The cells were then suspended in cold buffer (50 mM sodium phosphate [pH 8.0], 300 mM NaCl, 10 mM imidazole) and disrupted by sonication. Cell debris was removed by centrifugation, and the supernatant was mixed with nickel-nitrilotriacetate matrix and incubated for 2 h on ice. The matrix was washed three times with buffer (50 mM sodium phosphate [pH 6.0], 300 mM NaCl, 10% glycerol, 20 mM imidazole). The proteins were eluted with the latter buffer, containing 250 mM imidazole. The proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), carried out as described by Laemmli (25); the gel was stained with Coomassie brilliant blue as described by Sambrook et al. (40).
β-Galactosidase assays.
β-Galactosidase activity was determined in permeabilized cells as described by Miller (30) and is expressed in Miller units. Each value is the average of at least three independent measurements.
Genetic methods.
Transduction was performed as described by Lennox (27), using bacteriophage P1. A gntR (kanamycin-resistant [Kanr]) insertion mutant was constructed as follows. A 1,020-bp Kanr gene block (Pharmacia, Piscataway, N.J.) was ligated into the StuI site of plasmid pTC221, carrying the gntR gene on a 3.4-kb DNA fragment. The resulting plasmid, pTC29, was digested with PvuII, purified by electroelution from an agarose gel, and transformed by electroporation into E. coli DPB271. Kanr transformants were analyzed by PCR.
Chemicals and enzymes.
[α-32P]dATP was purchased from Amersham International (Buckinghamshire, United Kingdom). Biochemicals and endonucleases were from Boehringer (Mannheim, Germany), Merck (Darmstadt, Germany), or Sigma (St. Louis, Mo.).
RESULTS
Effect of the gntR mutation on gntT expression.
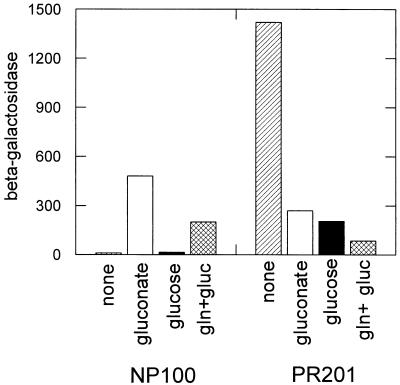
To investigate the role of GntR in repression of the gntT gene, the expression of a single-copy chromosomal gntT::lacZ fusion was measured in a gntR mutant. This gntR null mutant was constructed by insertion of a Kanr cassette into the StuI site of the gntR gene, followed by linear transformation and specific recombination in a recD strain and then transduction of the gntR mutation into the gntT::lacZ fusion strain E. coli NP100 to create E. coli PR201. The gntT::lacZ fusion was fully derepressed in E. coli PR201 and showed a threefold higher β-galactosidase activity in the absence of gluconate compared to the fully induced level in the wild type grown on gluconate (Fig. 1). This result confirms that GntR is the negative regulator of gntT.
FIG. 1.
Expression of the gntT::lacZ operon fusion in the wild type (NP100) and in a gntR mutant strain (PR201). The strains were growing in LB plus 0.4% of the indicated carbon sources. β-Galactosidase activities were measured in late log growth phase.
Catabolite repression of gntT.
Expression of the gntT::lacZ fusion in the gntR mutant (PR201) remained subject to catabolite repression. Growth of E. coli PR201 in the presence of gluconate, glucose, or the mixture of glucose plus gluconate resulted in 5-, 7-, or 18-fold repression, respectively (Fig. 1). As expected, this catabolite repression order follows the relative cAMP concentrations measured for cells grown on these sugars (14). Thus, gluconate serves not only as an inducer of gntT but also as a repressor, most likely via cAMP-dependent catabolite repression.
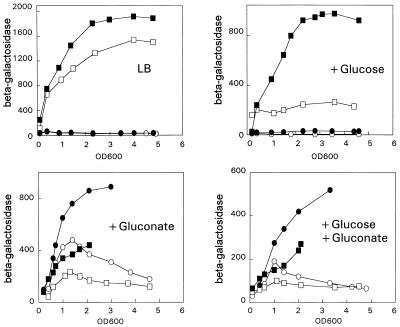
Next, cAMP-dependent catabolite repression was investigated in detail. The effect of exogenous cAMP on expression of the gntT::lacZ fusion was measured in the wild-type and gntR mutant strains during growth on LB and on LB containing glucose, gluconate, or a mixture of both sugars at different time points in batch cultures (Fig. 2). In the wild-type strain, there was no induction of β-galactosidase activity in the absence of gluconate, regardless of the presence of cAMP, which again indicates that the repression by GntR is cAMP independent. In wild-type cells grown in the presence of gluconate, the addition of cAMP caused a significant increase in the β-galactosidase activity, confirming that catabolite repression of the gntT gene is cAMP dependent (Fig. 2). Addition of cAMP to the wild type growing under catabolite-repressing conditions was in no case able to increase expression of the gntT::lacZ fusion to the fully derepressed level of the gntR mutant. Also, cAMP addition to the gntR mutant strain growing on glucose caused a fivefold increase in β-galactosidase activity, but again to a level lower than that of E. coli PR201 grown on LB without added sugar (Fig. 2). Thus, addition of cAMP was not sufficient to fully relieve catabolite repression. The recent finding that expression of the crp gene is modulated by catabolite-repressing sugars, including gluconate, may at least partially explain why addition of cAMP does not fully overcome catabolite repression (18).
FIG. 2.
Expression of the gntT::lacZ fusion in wild-type and gntR mutant strains in the presence and absence of cAMP. The cells were grown in LB medium with carbon sources as indicated. Symbols: circles, wild type (NP100); squares, gntR mutant (PR201); open and filled symbols, in the absence and presence, respectively, of 5 mM cAMP. β-Galactosidase was measured in the cultures at the indicated OD600 points.
To investigate the role of the CRP and adenylate cyclase in catabolite repression of the gntT gene by glucose and gluconate, crp and cya null mutations were transduced into the strain carrying the chromosomal gntT::lacZ fusion. In the presence of gluconate, both the crp and the cya mutants showed fourfold-lower β-galactosidase activity (Table 2). However, this low level of expression was still gluconate inducible. As expected, addition of cAMP to the cya mutant, but not to the crp mutant, relieved the catabolite repression to a level equivalent to that of the wild type. In summary, these results demonstrate that gntT is positively regulated by CRP.
TABLE 2.
Effects of the cya and crp mutations on gntT::lacZ expression
| E. coli strain | Relevant genotype | β-Galactosidase activity (U) with indicated carbon source
|
|||||
|---|---|---|---|---|---|---|---|
| None
|
Gluconate
|
Glucose
|
|||||
| −cAMP | +cAMPa | −cAMP | +cAMP | −cAMP | +cAMP | ||
| NP100 | Wild type | 10 | 10 | 460 | 680 | 0 | 0 |
| RH112 | crp | 20 | 15 | 105 | 70 | 0 | 0 |
| HT110 | cya | 30 | 25 | 115 | 620 | 0 | 0 |
| RH114 | crp cya | 0 | 10 | 90 | 60 | 0 | 0 |
| RH216 | crp gntR | 50 | 60 | 45 | 35 | 50 | 55 |
| HT216 | cya gntR | 75 | 585 | 55 | 380 | 60 | 290 |
Added to a final concentration of 5 mM.
Site-directed mutagenesis of the putative operator sites of the gntT promoter.
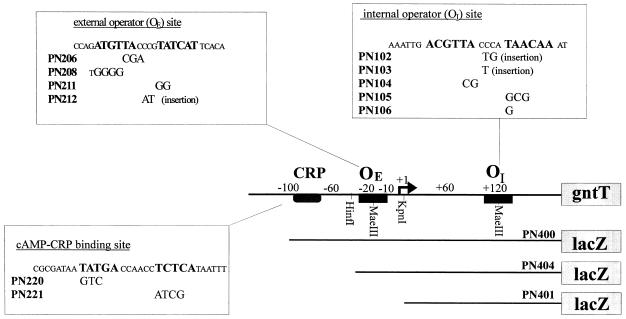
Two putative gnt operators were identified on the basis of similarity to sequences found in all other gluconate-inducible promoters (35). To check whether the postulated gnt operator sites within the gntT promoter region are in fact binding sites for the GntR protein, the effects of mutations within these sequences were measured. Mutations of the internal operator of the gntT::lacZ fusion were constructed by a recombinant PCR method using mismatched primers (Fig. 3). Mutations of the external operator site were generated by using a three-step PCR (Fig. 3). The activities of these fusions were determined from single-copy fusions generated by homologous recombination with λRS88 followed by integration into the chromosome.
FIG. 3.
Schematic representation of the gntT promoter region showing the site-directed mutations within the two operator sites and the cAMP-CRP binding site. The binding sites are shown in boldface; the specific mutations are indicated below the wild-type binding sequences. The deletion subclones (PN400, PN401, and PN404) of the gntT promoter region are shown under the line map.
Replacement of highly conserved bases within the internal operator site in strains PN104 and PN105 caused constitutive expression of β-galactosidase in cells grown in medium without gluconate (Table 3). Even the replacement of a single conserved base (PN106) caused a significant derepression of gntT::lacZ fusion activity. Insertion of two base pairs (TG) into the spacer sequence of the operator site (strain PN102) resulted in a similar constitutive expression of the gntT::lacZ fusion (Table 3), suggesting that the length of the spacer is crucial for binding of GntR. Even a single-base-pair insertion in mutant NP103 had a significant effect on the regulation of the gntT::lacZ fusion. None of the internal operator mutations were affected with respect to catabolite repression by glucose or gluconate. These results demonstrate that the internal operator site is indeed an important regulatory sequence. Thus, it appears that any change to the highly conserved operator site is sufficient to affect GntR binding.
TABLE 3.
Effects of mutations in the regulatory region of gntT
| E. coli strain | β-Galactosidase activity (U) with indicated carbon source(s)
|
|||
|---|---|---|---|---|
| None | Gluconate | Glucose | Gluconate + glucose | |
| NP100 (wild type) | 10 | 510 | 0 | 280 |
| NP102 | 780 | 520 | 100 | 320 |
| NP103 | 280 | 490 | 60 | 260 |
| NP104 | 840 | 490 | 100 | 330 |
| NP105 | 920 | 500 | 90 | 310 |
| NP106 | 110 | 610 | 40 | 300 |
| PN206 | 1,320 | 560 | 190 | 190 |
| PN211 | 60 | 80 | 40 | 40 |
| PN208 | 1,260 | 460 | 170 | 210 |
| PN212 | 1,180 | 480 | 160 | 190 |
| PN220 | 0 | 130 | 60 | 60 |
| PN221 | 0 | 110 | 50 | 70 |
| PN1256 | 1,460 | 640 | 190 | 280 |
| PN400 | 0 | 590 | 0 | 290 |
| PN401 | 10 | 10 | 0 | 0 |
| PN404 | 0 | 140 | 40 | 60 |
Strains NP206, NP208, and NP212 carry different substitutions or insertions within the external operator site (Fig. 3), which overlaps the −10 region of the promoter. Each of these mutations showed a level of β-galactosidase activity in the absence of gluconate which was twice as high as that of the internal operator mutants and similar to the level measured in the gntR mutant grown in the absence of sugar. External operator site mutants grown in medium with gluconate or the mixture of gluconate and glucose showed a level of catabolite repression equivalent to that of the wild type. Cells grown with glucose exhibited a derepressed level of β-galactosidase which was almost twofold higher than that of the internal operator mutants. The mutation in strain PN211, with a two-base substitution in the external operator which also replaces the most highly conserved region of the −10 hexamer, TATCAT, with GGTCAT, resulted in only a low constitutive synthesis of β-galactosidase under all growth conditions. The double mutant NP1256, which contains mutations in both operator sites, was constructed by using one of the mutant gntT::lacZ fusion as the template for a second site-directed mutagenesis. This strain showed a level of β-galactosidase equivalent to the fully derepressed level of the gntR mutant. In summary, the results clearly demonstrate that both operators are required for repression of gntT by GntR. Of the two operators, it appears that the external operator is perhaps the more important, since these mutants are derepressed to a greater extent than the internal operator mutants.
Mutations of the cAMP-CRP binding site.
Two potential binding sites for the cAMP-CRP complex within the promoter region of gntT were identified on the basis of similarity to the consensus binding site, positioned at −71 (TATGAccaaccTCTCA) and −13 (TGTTAcccgtaTCATT) with respect to the transcriptional start site (Fig. 3). The latter CRP binding site overlaps the external operator site and could be a hybrid site for both proteins. However, the effects of base pair substitutions in the left half of this site (NP206 and NP208) that should have eliminated cAMP-CRP binding did not affect the positive regulation of gntT. On the other hand, replacement of three or four base pairs within the left or right half of the putative cAMP-CRP binding site at −71 bp upstream of the transcriptional start site resulted in a strong decrease in β-galactosidase activity in strains PN220 and PN221 (Table 3), respectively, a phenotype similar to that observed in the crp and cya mutants. These results clearly demonstrate that the cAMP-CRP binding site at −71 is indeed involved in the catabolite repression and the positive regulation of gntT. PN220 and PN221 remained gluconate inducible, again indicating that induction and catabolite repression are independent.
Deletion mutants.
To check whether additional regulatory elements are present within the gntT promoter region, deletions of the gntT::lacZ fusion were constructed. Deletion of the entire region upstream of the cAMP-CRP binding site beyond position −91, in mutant PN400 (Fig. 3), resulted in no significant difference in regulation by comparison to the wild type (Table 3). Mutant strain PN404, which was constructed by removing the distal EcoRI-HinfI fragment containing the cAMP-CRP binding site, showed very low but still gluconate-inducible β-galactosidase activity, confirming results described above. During the course of previous work, primer extension analysis had revealed two ends for the gntT mRNA (35), but the existence of a second promoter is not confirmed by these data. Since no expression was measured in the mutant carrying a deletion of the entire promoter region (PN401), it appears that there is only a single promoter for gntT, and it is suggested that the second primer extension signal is the result of transcript processing.
Additional regulatory effect of GntR.
During the course of this work, it became apparent that GntR might affect gntT expression by a mechanism that is independent of negative control by operator binding as well as activation by cAMP-CRP. Unexpectedly, expression of the gntT::lacZ fusion in the gntR mutant grown in the presence of gluconate was approximately one-half of that of the wild type grown under the same conditions (a finding called the 50% effect) (Fig. 1 and 2). This result was duplicated in the presence of cAMP (Fig. 2). Furthermore, the crp gntR double mutant expressed the gntT::lacZ fusion in the presence of gluconate at one-half of the level of the crp mutant in a gntR+ background, regardless of whether cAMP was added (Table 2). Similarly, the cya gntR double mutant expressed the gntT::lacZ fusion in the presence of gluconate at one-half of the level of the cya mutant in a gntR+ background. In this case, addition of cAMP increased expression, but the gntR mutant retained the 50% effect. Thus, it appears that the twofold decrease in expression of the gntT::lacZ fusion in the gntR mutant when grown in the presence of gluconate is independent of catabolite repression. The fully derepressed double-operator-site mutant, PN1256, expressed the gntT::lacZ fusion when grown on gluconate at a level similar to that of the induced level of the wild type grown under the same conditions (Table 3), indicating that the 50% effect is independent of GntR binding and, hence, the negative regulation of the gntT gene; that is, that the gntR mutation is required for the observed 50% effect during growth on gluconate.
Since a pronounced peak of gntT expression was observed during early log phase (35), a pattern of expression similar to that of the Fis protein (3), it was possible that Fis is the regulator of gntT. Although a sequence (GTTTGAGAATCACCA, at −42) with similarity to the consensus binding sequence of Fis (GNN[C/T][A/G]NN[T/A]NN[T/C][G/A]NNC) (3) could be identified within the gntT promoter region, a fis null mutation had no effect on expression of the gntT::lacZ fusion.
Purification of GntR and CRP.
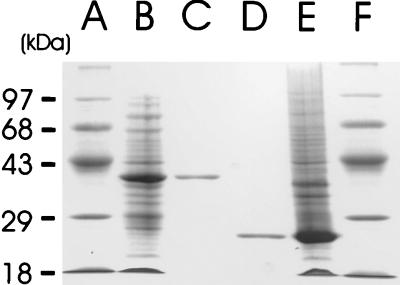
GntR and CRP were isolated by using a His tag modification system as described in Materials and Methods. The coding regions of both genes, including the original start and stop codons, were amplified by PCR and cloned into the expression vector pQE30. The resulting fusion genes carried six additional His codons at their 5′ ends. After overexpression of the His-tagged GntR and CRP in E. coli M15(pNP-41) and M15(pNP-52), respectively, the proteins were isolated in native form by high-affinity chromatography on nitrilotriacetate resin. The purified GntR and CRP had apparent molecular masses of 35 to 37 and 23 to 25 kDa, respectively, as determined by SDS-PAGE (Fig. 4), in agreement with the predicted values of 36.8 and 24.3 kDa, respectively. The procedure yielded proteins of greater than 95% purity.
FIG. 4.
Overexpression and purification of GntR and CRP, demonstrated by SDS-PAGE of the purified His-tagged proteins. Lanes A and F, molecular weight standards; lanes B and E, cell extracts of strains M15(pNP-41) and M15(pNP-52) after IPTG induction; lanes C and D, purified GntR and CRP, respectively.
Binding of GntR to the gntT operator sites.
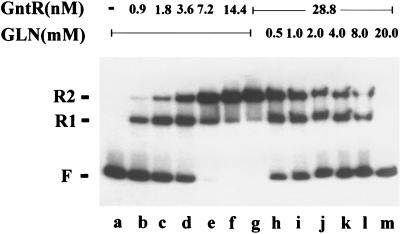
To test the binding of GntR to the operators of gntT and the effects of potential inducers on such binding, we studied GntR-operator complex formation with the purified GntR protein by gel electrophoretic mobility assays. A DNA fragment containing the proposed regulatory region of gntT including both operator sites was amplified and radioisotopically labeled by using PCR. The DNA was incubated with different amounts of purified GntR before electrophoresis. Two DNA bands with reduced mobility, corresponding to DNA with one or two GntR molecules bound, were formed (Fig. 5 and 7). The DNA-protein interaction was found not to be cooperative, since the formation of the DNA-protein complex was linearly proportional to the concentration of DNA and GntR protein (data not shown). If the binding of GntR to the operator sites of gntT is specific, unlabeled DNA carrying these binding sites should compete for binding, whereas nonspecific DNA should not. Addition of a 10-fold excess of unlabeled operator DNA completely abolished the formation of a labeled GntR-DNA complex, whereas a 100-fold molar excess of unlabeled sonicated herring sperm DNA was insufficient to compete for the binding with the labeled fragment (data not shown). These results indicated that GntR binding is specific for the operator. To test the effect of gluconate on the formation of the GntR-operator complex, the binding assay was carried out in the presence of different gluconate concentrations. As shown in Fig. 5, gluconate reduced the formation of the binary complex in proportion to its concentration, probably by reducing the binding affinity of GntR to the operator sites. Several other sugars and sugar acids were tested for this effect on formation of the repressor-DNA complex, and only 6-phosphogluconate was found to inhibit the protein-DNA interaction, but a concentration 10-fold higher than that of gluconate was required, suggesting that the phosphorylated product of gluconate can act as an weak inducer of the gntT gene (data not shown). The sugars glucuronate, 5-ketogluconate, glucose-6-phosphate, fructose, glycerol, and l-idonate had no effect on binding (data not shown).
FIG. 5.
Electrophoretic mobility of the GntR-operator complexes. Increasing amounts of purified GntR were added to a 456-bp gntT fragment containing the putative regulatory region of the gntT gene in the absence or presence of different concentrations of gluconate. In the presence of GntR, two GntR-operator complexes (R1 and R2) were formed. F, free DNA. All samples contained 8.1 nM gntT DNA and the designated concentrations of GntR and gluconate.
FIG. 7.
Effects of mutations in the internal and external operator sites on the formation of the repressor-operator complex. Addition of different concentrations of purified GntR to the 456-bp gntT fragment gave a mixture containing free DNA (F) and 1:1 (R1) and 2:1 (R2) repressor complexes, respectively. Samples a to i and j to n contained 6 and 2.2 nM gntT DNA, respectively. Samples a to c, d to f, g to i, and j to n contained gntT fragments from strains NP105, NP206, NP1256, and NP220, respectively.
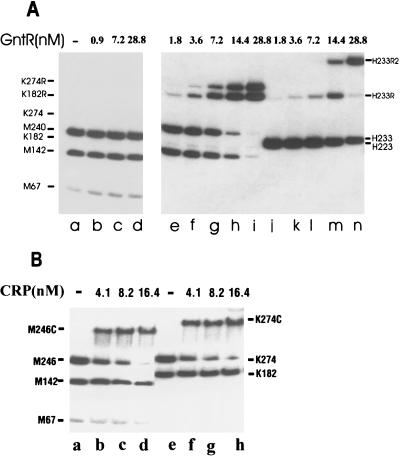
The presence of several restriction sites within the gntT regulatory region afforded the opportunity to test whether cleavage of the consensus sequence of the operators would eliminate binding and to confirm the relative locations of the operator sites. The restriction enzyme MaeIII specifically cleaves the gntT fragment within the left half-site of each operator. None of the three labeled MaeIII fragments showed binding to GntR in the gel retardation assay (Fig. 6A), indicating that the GntR binding sites were destroyed by digestion with MaeIII. Both the 182- and 274-bp KpnI fragments, each carrying one of the operator sites, were shifted in the presence of GntR, while only one of the two HinfI fragments produced DNA bands with lower mobility during electrophoresis (Fig. 6A). These data indicated that the two operator sites are separated by the KpnI site and are downstream of the HinfI site.
FIG. 6.
Formation of GntR- and CRP-DNA complexes with cleaved gntT DNA fragment. Digestion of the gntT fragment with different restriction enzymes resulted in following fragments: MaeIII (67, 142, and 240 bp), KpnI (182 and 274 bp), and HinfI (223 and 233 bp). The formation of complexes was tested with GntR (A) and CRP (B). Complexes of the DNA fragment with one GntR molecule, two GntR molecules, and Crp were given the suffixes R, R2, and C, respectively.
To prove that GntR binds to the postulated operator sequences, we constructed mutants carrying base pair substitutions and insertions which were predicted to affect the binding of GntR to these sites (Fig. 7 and unpublished data). The substitution of two, three, or four base pairs within the left or right half of the internal or the external operator site and the insertion of two base pairs into the spacer sequences prevented the binding of GntR in gel retardation assays. On the other hand, the mutants carrying a substitution of one base pair in the right half site and an insertion of a single base pair into the spacer sequence of the internal operator site (PN106 and PN103, respectively) could form complexes only in the presence of eight- and fivefold-higher GntR concentrations (data not shown), suggesting that these mutations caused a lower affinity of GntR to these sites. Likewise, the mutant carrying a two-base-pair substitution in the right half-site of the internal operator (PN211) could form the binary complex, but only with a 12-fold-higher GntR concentration (data not shown). Mutations of just one of the two operators eliminated formation of the binary complex, while no complex was observed with a mutation which eliminated both operators (Fig. 7).
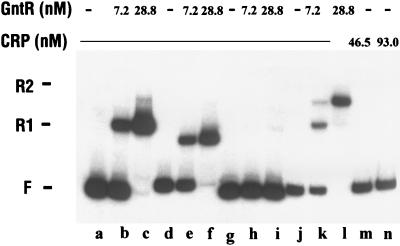
Binding of CRP to the gntT promoter.
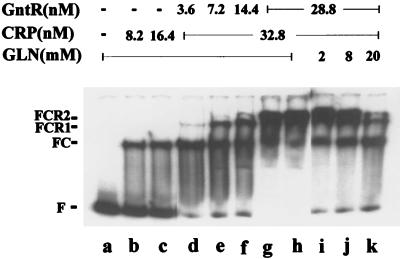
The purified CRP protein was tested for binding to the gntT promoter region by gel electrophoretic mobility assay (Fig. 6B). When the labeled gntT fragment was incubated with CRP, only one band with reduced mobility was formed, and as expected, this occurred only in the presence of cAMP. Since it could be argued that the His-tagged CRP might have different binding properties, the native enzyme was tested and found to give the same results (data not shown). A gntT DNA fragment carrying a mutation within the CRP binding site showed no formation of a complex with cAMP-CRP. Also, assays with restriction endonuclease cleaved gntT DNA showed that only the fragments carrying the CRP site at −71 were able to bind CRP. When the gntT promoter fragment was incubated with both GntR and CRP-cAMP, a ternary complex consisting of one GntR molecule and one cAMP-CRP complex bound or two GntR molecules and one cAMP-CRP complex bound was formed (Fig. 8). These results indicated that cAMP-CRP binds specifically to the site centered at −71 but not to the hypothetical site at −13. The binding of the cAMP-CRP complex is essential for full induction of gntT, and this binding is neither cooperative nor inhibited by GntR.
FIG. 8.
Formation of a ternary and a tertiary complex of GntR, CRP, and gntT operator shown by electrophoretic mobility assay. The gntT promoter fragment was incubated with different amounts of CRP and GntR in the presence of 5 mM cAMP. Besides the free DNA (F), three DNA bands with reduced mobility, FC, FCR1, and FCR2, corresponding to DNA bound to one CRP, one CRP and one GntR, and one CRP and two GntR molecules, respectively, were formed.
DISCUSSION
In E. coli, gntT is specifically induced in the presence of gluconate, fully repressed in the absence of gluconate, and partially induced on a mixture of glucose and gluconate. A strain in which the gntR gene is knocked out by the insertion of a Kanr cassette showed constitutive expression of a single-copy chromosomal gntT::lacZ operon fusion, even in the absence of an inducer (Fig. 1). Since this constitutive expression was threefold higher than the gluconate-induced expression in wild-type cells, it appears that gluconate can serve as both an inducer and a repressor for gntT expression.
It has long been known that gluconate is a very effective substrate for catabolite repression and acts by decreasing the cellular cAMP concentration to nearly the same extent as glucose. In fact, a mixture of glucose and gluconate caused one of the most catabolite repressing conditions measured (14, 31, 32). The results presented here demonstrate that a higher cellular cAMP concentration relieves catabolite repression of gntT by gluconate and also by glucose. The fact that gntT expression is very low in the absence of adenylate cyclase (cya) or catabolite receptor protein (crp), even in the presence of gluconate, shows that the positive regulation by cAMP-CRP is essential for full induction of gntT. However, addition of cAMP does not fully relieve the repressing effect of glucose and gluconate, which indicates that besides cAMP, another regulatory factor is involved in the catabolite repression. In the general model, the variable cAMP concentration alone is believed to be the mediator of catabolite repression. The intracellular cAMP pool is regulated by phosphorylated EIIAGlc, which stimulates adenylate cyclase to produce cAMP (36, 23, 39). More recently, Hogema et al. (18) showed that catabolite-repressing sugars lower not only the intracellular cAMP pool but also the intracellular concentration of CRP. Furthermore, EIIGlc seemed not be involved in the regulation of the CRP concentration by the non-phosphoenolpyruvate:sugar phosphotransferase system (PTS) substrate gluconate, since a crr mutation did not affect their results (18, 49). The reason why exogenous cAMP does not fully relieve the catabolite repression of gntT by gluconate could be the simultaneous, cAMP-independent reduction of the CRP concentration by gluconate (18). It should be pointed out that it is still not known how gluconate, a non-PTS carbon source, regulates the intracellular concentrations of cAMP and CRP. One possibility is that gluconate by itself, or a regulatory component of the gluconate regulon, interacts directly or indirectly with adenylate cyclase and the crp gene.
The other important question is why gluconate acts on gntT expression both as an inducer and as a repressor. Derepression of the gnt regulon by addition of cAMP during growth on gluconate or by deletion of gntR caused a growth inhibition, probably as a result of the accumulation of the toxic metabolite methylglyoxal as has been reported previously (2). The growth inhibition caused by cAMP and gluconate was accentuated in the gntR mutant (Fig. 2), indicating that the growth inhibition is likely the result of overexpression of the gluconate catabolic pathway. The accumulation of methylglyoxal is probably the result of an unregulated flux of carbon from gluconate to glyceraldehyde-3-phosphate, which bypasses the key allosteric control points, phosphofructokinase and pyruvate kinase. The formation of methylglyoxal from dihydroxyacetone phosphate is catalyzed by the methylglyoxal synthetase. Obviously, there is an important balance between induction and repression of gluconate catabolism.
A comparison of negative control of the gnt regulon to that of the gal and lac regulons, the two paradigms of negative control (7), highlights an interesting difference. The galR mutant, when grown in the presence of the inducer galactose, shows a twofold-higher level of induction by comparison to the fully induced wild type (44). The mechanism of ultrainduction in the gal regulon is now understood to be due to the presence of an additional repressor, galS (47). The lacI mutant is derepressed for lacZ expression to exactly the same extent as the fully induced wild type, and the inducer IPTG has no effect on lacZ expression in the lacI mutant (21). Paradoxically, the gntR mutation causes a 50% lower expression of the gntT::lacZ fusion in the presence of the inducer gluconate by comparison to the wild type (Fig. 1). Furthermore, the crp gntR and cya gntR double mutants expressed the gntT::lacZ fusion in the presence of gluconate at one-half of the level of the crp or cya mutant in a gntR+ background. Finally, mutation of both operators in the fully derepressed mutant, PN1256, did not cause a 50% lowering of gntT expression when cells were grown on gluconate. Thus, it appears that the twofold decrease in expression of the gntT::lacZ fusion in the gntR mutant when grown in the presence of gluconate is independent of catabolite repression, as well as GntR binding to the operators. The simplest interpretation of this phenotype, which we can call ultrarepression, by analogy to the ultrainduction of the gal operon in a galR mutant (44), is that another regulatory factor may be involved in the regulation of gntT. One possibility is that GntR is essential for the expression of an activator, which acts positively on the transcription of gntT in the presence of gluconate, and that this activation is lost in the gntR mutant. The data presented here indicate that Fis (3) is not involved. However, it is possible that the closely related YjgS protein, which is 46% identical to GntR, is involved in the positive regulation of gntT. This possibility is particularly intriguing since the regulatory region of the GntII genes contains two gnt operator consensus sequences (35).
In the second part of this work, we used site-directed mutagenesis and DNA band migration retardation assays to prove that the gnt operators serve as binding sites for the negative regulator GntR and that the true inducers of the gnt regulon are gluconate and, to a lesser extent, 6-phosphogluconate. The putative site for binding of the cAMP-CRP complex was also tested. One or two copies of the highly conserved consensus sequence postulated to be the gnt operator, consisting of two inverted hexamers separated by a GC-rich spacer sequence of 4 bp (ATGTTA[N4; GC rich]TAACAT), are present in the regulatory regions of all gluconate-inducible genes (edd, gntKU, gntT, gntV, and yjgV) (35). Gel retardation assays showed that GntR binds to two different sites within the promoter region of gntT. Gluconate and also a 10-fold-higher concentration of 6-phosphogluconate were found to inhibit the formation of the GntR-DNA complexes (Fig. 5). These results confirm that regulation of the gntT gene by GntR is similar to other negatively regulated systems. In the absence of an inducer, the GntR repressor binds to the two operator sites, resulting in repression of the gntT gene. The inducer, gluconate (or 6-phosphogluconate), acts to eliminate binding by GntR, probably by reducing the DNA binding affinity of GntR to the operator. These results confirm earlier genetic studies indicating that gluconate, and 6-phosphogluconate in pgi gnd double mutants which can accumulate this intermediate, serves as an inducer of the gnt regulon (24, 38).
Proof that GntR binds to the postulated operator sequences was obtained by using mutants which were predicted to affect the binding of GntR to these sites. Substitutions within the left or right half of the operator sites and insertions in the spacer sequences resulted in a high, constitutive expression of the gntT::lacZ fusion in the absence of gluconate. Binding assays showed that these mutations specifically prevented the binding of GntR to these sites. Furthermore, when the operators were destroyed by digestion with MaeIII, no binding of GntR was detected. In summary, these results demonstrate that GntR binds to two operator sites at gntT and that these protein-DNA interactions are highly specific.
CRP was found to bind, under in vitro conditions in the presence of cAMP, to the binding site at −71 (Fig. 6B and 8) but not to the putative site at −13, which was postulated to be a hybrid of GntR and CRP binding sites. Mutations within the CRP binding site at −71 prevented the binding of the cAMP-CRP complex. Furthermore, these mutations caused a significant decrease of gntT::lacZ fusion expression under induced conditions (Table 2), as did deletion of the distal region of the gntT promoter, including the CRP binding site at −71. Together these results demonstrated that the cAMP-CRP complex activates the transcription of gntT by binding to the −71 binding site and that full activity of the gntT promoter is dependent on this activation.
Since mutation of either of the operator sites caused a derepressed, constitutive expression of the gntT gene, it appears that there may be some kind of interaction between these sites. We favor the idea that the repression of gntT is the result of DNA looping through interaction between the two GntR molecules as shown for many other operons in E. coli, such as the ara, gal, lac, and deo operons (29). A greater extent of derepression results from mutation of the external operator site by comparison to the internal operator site mutants, suggesting that binding of GntR to the external site results in a stronger repression of the gntT gene. This can be explained by the fact that the external operator site overlaps the −10 region of the promoter, perhaps causing competition between RNA polymerase and GntR for binding to the promoter.
ACKNOWLEDGMENTS
This work was funded by grants from the DOE, Division of Energy Biosciences (DE-FG02-95ER20178), and the NSF (MCB-9723593).
We thank Sankar Adhya for providing purified CRP.
REFERENCES
- 1.Bächi B, Kornberg H L. Genes involved in the uptake and catabolism of gluconate by Escherichia coli. J Gen Microbiol. 1975;90:321–335. doi: 10.1099/00221287-90-2-321. [DOI] [PubMed] [Google Scholar]
- 2.Bächi B, Kornberg H L. Utilization of gluconate by Escherichia coli. A role of adenosine 3′:5′-cyclic monophosphate in the induction of gluconate catabolism. Biochem J. 1975;150:23–128. doi: 10.1042/bj1500123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ball C A, Osuna R, Ferguson K C, Johnson R C. Dramatic changes in fis levels upon nutrient upshift in Escherichia coli. J Bacteriol. 1992;174:8043–8056. doi: 10.1128/jb.174.24.8043-8056.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bujard H, Gentz R, Lanzer M, Stueber D, Mueller M, Ibrahimi I, Haeuptle M T, Dobberstein B. A T5 promoter based transcription-translation system for the analysis of proteins in vivo and in vitro. Methods Enzymol. 1987;155:416–433. doi: 10.1016/0076-6879(87)55028-5. [DOI] [PubMed] [Google Scholar]
- 5.Carey J. Gel retardation. Methods Enzymol. 1991;208:103–117. doi: 10.1016/0076-6879(91)08010-f. [DOI] [PubMed] [Google Scholar]
- 6.Conway T. The Entner-Doudoroff pathway: history, physiology, and molecular biology. FEMS Microbiol Rev. 1992;103:1–27. doi: 10.1111/j.1574-6968.1992.tb05822.x. [DOI] [PubMed] [Google Scholar]
- 7.Choy H, Adhya S. Negative control. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1287–1299. [Google Scholar]
- 8.Cohen S S. Gluconokinase and the oxidative path of glucose-6-phosphate utilization. J Biol Chem. 1951;189:617–628. [PubMed] [Google Scholar]
- 9.Conway T. The Entner-Doudoroff pathway: history, physiology, and molecular biology. FEMS Microbiol Rev. 1992;103:1–27. doi: 10.1111/j.1574-6968.1992.tb05822.x. [DOI] [PubMed] [Google Scholar]
- 10.De Rekarte U D, Cortes M, Porco A, Nino G, Isturiz T. Mutations affecting gluconate catabolism in Escherichia coli. Genetic mapping of loci for the low affinity transport and the thermoresistant gluconokinase. J Basic Microbiol. 1994;34:363–370. doi: 10.1002/jobm.3620340602. [DOI] [PubMed] [Google Scholar]
- 11.Egan S, Fliege R, Tong S, Shibata A, Wolf R E, Jr, Conway T. Molecular characterization of the Entner-Doudoroff pathway in Escherichia coli: sequence analysis and localization of promoters for the edd-eda operon. J Bacteriol. 1992;174:4638–4646. doi: 10.1128/jb.174.14.4638-4646.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenberg R C, Dobrogosz W J. Gluconate metabolism in Escherichia coli. J Bacteriol. 1967;93:941–949. doi: 10.1128/jb.93.3.941-949.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Entner N, Doudoroff M. Glucose and gluconic acid oxidation of Pseudomonas saccharophila. J Biol Chem. 1952;196:853–862. [PubMed] [Google Scholar]
- 14.Epstein W, Rothman-Denes L B, Hesse J. Adenosine 3′:5′-cyclic monophosphate as mediator of catabolite repression in Escherichia coli. Proc Natl Acad Sci USA. 1975;72:2300–2304. doi: 10.1073/pnas.72.6.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fraenkel D G. Glycolysis, pentose phosphate pathway, and Entner-Doudoroff pathway. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 142–150. [Google Scholar]
- 16.Higuchi R, Krummel B, Saiki R K. A general method of in vitro preparation and specific mutagenesis of DNA fragments: study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hochuli E. Purification of recombinant proteins with metal chelate adsorbent. Genet Eng. 1990;12:87–98. doi: 10.1007/978-1-4613-0641-2_6. [DOI] [PubMed] [Google Scholar]
- 18.Hogema B M, Arents J C, Inada T, Aiba H, van Dam K, Postma P W. Catabolite repression by glucose 6-phosphate, gluconate and lactose in Escherichia coli. Mol Microbiol. 1997;24:857–867. doi: 10.1046/j.1365-2958.1997.3991761.x. [DOI] [PubMed] [Google Scholar]
- 19.Inada T, Takahashi H, Mizuno T, Aiba H. Down regulation of cAMP production by cAMP receptor protein in Escherichia coli: an assessment of the contributions of transcriptional and posttranscriptional control of adenylate cyclase. Mol Gen Genet. 1996;253(1–2):198–204. doi: 10.1007/s004380050313. [DOI] [PubMed] [Google Scholar]
- 20.Istúriz T, Palmero E, Vitelli-Flores J. Mutations affecting gluconate catabolism in Escherichia coli. Genetic mapping of the locus for the thermosensitive gluconokinase. J Gen Microbiol. 1986;132:3209–3212. doi: 10.1099/00221287-132-11-3209. [DOI] [PubMed] [Google Scholar]
- 21.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 22.Klemm P, Tong S, Nielsen H, Conway T. The gntP gene of Escherichia coli involved in gluconate uptake. J Bacteriol. 1996;178:61–67. doi: 10.1128/jb.178.1.61-67.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolb A, Busby S, Buc H, Garges S, Adhya S. Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- 24.Kornberg H L, Soutar A K. Utilization of gluconate by Escherichia coli. Biochem J. 1973;134:489–498. doi: 10.1042/bj1340489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 26.Lange R M, Hengge-Aronis R. Complex transcriptional control of the sigma S-dependent stationary-phase-induced and osmotically regulated osmY (csi-5) gene suggests novel roles for Lrp, cyclic AMP (cAMP) receptor protein-cAMP complex, and integration host factor in the stationary-phase response of Escherichia coli. J Bacteriol. 1993;175:7910–7917. doi: 10.1128/jb.175.24.7910-7917.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lennox E S. Transduction of linked characters of the host by bacteriophage P1. Virology. 1955;1:190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- 28.Luria S E, Delbrück M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matthews K S. DNA looping. Microbiol Rev. 1992;56:123–136. doi: 10.1128/mr.56.1.123-136.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 31.Okinaka R T, Dobrogosz W J. Enhanced catabolite repression in Escherichia coli by growth on combined substrates. J Bacteriol. 1966;92:526–527. doi: 10.1128/jb.92.2.526-527.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paigen K, Williams B. Catabolite repression and other control mechanisms in carbohydrate utilization. Adv Microb Physiol. 1970;4:251–324. [Google Scholar]
- 33.Peekhaus N, Tong S, Murray E, Reizer J R, Saier M, Conway T. Initial characterization of a novel transporter family that includes multiple Escherichia coli gluconate transporters and their homologs. FEMS Microbiol Lett. 1996;147:233–238. doi: 10.1111/j.1574-6968.1997.tb10247.x. [DOI] [PubMed] [Google Scholar]
- 34.Porco A, Isturiz T. Selection of lacZ operon fusions in genes of gluconate metabolism in E. coli. Characterization of a gntT::lacZ fusion. Acta Cient Venez. 1991;42:270–275. [PubMed] [Google Scholar]
- 35.Porco A, Peekhaus N, Bausch C, Tong S, Isturiz T. Molecular genetic characterization of the Escherichia coli gntT gene of GntI, the main system for gluconate metabolism. J Bacteriol. 1997;179:1584–1590. doi: 10.1128/jb.179.5.1584-1590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Postma P W, Lengeler J W, Jacobsen G R. Phosphoenolpyruvate:carbohydrate phosphotransferase systems of bacteria. Microbiol Rev. 1993;57:543–594. doi: 10.1128/mr.57.3.543-594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Powell B S, Court D L, Nakamura Y, Rivas M P, Turnbough C L. Rapid confirmation of single copy lambda prophage integration by PCR. Nucleic Acids Res. 1994;25:5765–5766. doi: 10.1093/nar/22.25.5765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robin A, Kepes A. Inducible gluconate permease in a gluconate kinase-deficient mutant of Escherichia coli. Biochim Biophys Acta. 1975;406:50–59. doi: 10.1016/0005-2736(75)90041-3. [DOI] [PubMed] [Google Scholar]
- 39.Saier M H, Ramseier T M, Reizer J. Regulation of carbon utilization. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1325–1343. [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Simons R W, Houman F, Kleckner N. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96. doi: 10.1016/0378-1119(87)90095-3. [DOI] [PubMed] [Google Scholar]
- 42.Stueber D, Bannwarth W, Meloen J R L, Matile H. New B cell epitopes in the Plasmodium falciparum malaria circumsporozoite protein. Eur J Immunol. 1990;20:819–824. doi: 10.1002/eji.1830200416. [DOI] [PubMed] [Google Scholar]
- 43.Sweeney N J, Laux D C, Cohen P S. Escherichia coli F-18 and E. coli K-12 eda mutants do not colonize the streptomycin-treated mouse large intestine. Infect Immun. 1996;64:3504–3511. doi: 10.1128/iai.64.9.3504-3511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tokeson J P E, Garges S, Adhya S. Further inducibility of a constitutive system: ultrainduction of the gal operon. J Bacteriol. 1991;173:2319–2327. doi: 10.1128/jb.173.7.2319-2327.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tong S, Porco A, Isturiz T, Conway T. Cloning and molecular characterization of the Escherichia coli gntR, gntK, and gntU genes of GntI, the main system for gluconate metabolism. J Bacteriol. 1996;178:3260–3269. doi: 10.1128/jb.178.11.3260-3269.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Valentin-Hansen P, Holst B, Sogaard-Andersen L, Martinussen J, Nesvera J, Douthwaite S R. Design of cAMP-CRP-activated promoters in Escherichia coli. Mol Microbiol. 1991;5:433–437. doi: 10.1111/j.1365-2958.1991.tb02126.x. [DOI] [PubMed] [Google Scholar]
- 47.Weickert M, Adhya S. A family of bacterial regulators homologous to Gal and Lac repressors. J Biol Chem. 1992;267:15869–15874. [PubMed] [Google Scholar]
- 48.Yamada M, Kawai T, Izu H. Analysis of Escherichia coli gntT and gntU genes and comparison of the products with their homologues. Biosci Biotechnol Biochem. 1996;60:1548–1550. doi: 10.1271/bbb.60.1548. [DOI] [PubMed] [Google Scholar]
- 49.Yang J K, Bloom R W, Epstein W. Catabolite and transient repression in Escherichia coli do not require enzyme I of the phosphotransferase system. J Bacteriol. 1979;38:275–279. doi: 10.1128/jb.138.1.275-279.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zamenhof P J, Villarejo M. Construction and properties of Escherichia coli strains exhibiting alpha-complementation of β-galactosidase fragments in vivo. J Bacteriol. 1972;124:171–178. doi: 10.1128/jb.110.1.171-178.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zwaig N, Nagel de Zwaig R, Isturiz T, Wecksler M. Regulatory mutation affecting the gluconate system in Escherichia coli. J Bacteriol. 1973;114:469–473. doi: 10.1128/jb.114.2.469-473.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]