Abstract
Background and Purpose: Addressing the need for accurate dose calculation in MRI-only radiotherapy, the generation of synthetic Computed Tomography (sCT) from MRI has emerged. Deep learning (DL) techniques, have shown promising results in achieving high sCT accuracies. However, existing sCT synthesis methods are often center-specific, posing a challenge to their generalizability. To overcome this limitation, recent studies have proposed approaches, such as multicenter training . Material and methods: The purpose of this work was to propose a multicenter sCT synthesis by DL, using a 2D cycle-GAN on 128 prostate cancer patients, from four different centers. Four cases were compared: monocenter cases, monocenter training and test on another center, multicenter trainings and a test on a center not included in the training and multicenter trainings with an included center in the test. Trainings were performed using 20 patients. sCT accuracy evaluation was performed using Mean Absolute Error, Mean Error and Peak-Signal-to-Noise-Ratio. Dose accuracy was assessed with gamma index and Dose Volume Histogram comparison. Results: Qualitative, quantitative and dose results show that the accuracy of sCTs for monocenter trainings and multicenter trainings using a seen center in the test did not differ significantly. However, when the test involved an unseen center, the sCT quality was inferior. Conclusions: The aim of this work was to propose generalizable multicenter training for MR-to-CT synthesis. It was shown that only a few data from one center included in the training cohort allows sCT accuracy equivalent to a monocenter study.
Keywords: Radiotherapy treatment planning, Synthetic-CT, Magnetic resonance imaging, cycle-GAN, Multicenter
1. Introduction
In the standard workflow of radiation therapy (RT), CT scan is the gold standard for dose calculation. However, MRI provides better soft-tissue contrast than CT [1]. MRI allows for a more accurate delineation of the prostate gland [2], translating into a reduction in doses delivered to proximity organs at risk [3], [4]. MRI-guidance also recently demonstrated a decrease in both genitourinary and gastrointestinal acute toxicity after prostate radiotherapy [5], [6]. However, combining MRI and CT requires a registration step which introduces uncertainties and registration errors especially in the pelvic region [7], [8], up to 2 mm for the prostate [9]. An MR-only RT workflow allows to skip the registration step, and has a growing interest with the rising implementation of MR-linac devices (MRI combined with a linear accelerator) [9]. The main drawback of MRI is the absence of electron density information which is essential in dose calculation [10]. To address this issue, the generation of synthetic CTs (sCTs) from MRI by deep learning methods has previously been proposed [11], [12]. The aim of these methods is to find a correspondence between CT and MRI. However, DL methods depend on the training cohort: they are center-dependent, acquisition device, the anatomical localization, the MR sequence and the acquisition parameters. Due to the differences in intensity distribution of MRI and CT across various centers, the occurrence of artifacts in images, and varying fields of view, generalizable training cannot be achieved using monocentrer training, as highlighted in several studies [13], [14], [15]. Multicenter training has been proposed as an alternative approach, to allow for a better robustness of the model [14], [16], [17]. Only a very few studies were performed in the pelvic area [14], [18] as it is a challenge to obtain a large number of centers to build multicenter cohorts. To achieve a reliable generation in a supervised context, a crucial aspect is the implementation of a series of preprocessing steps to standardize the database and allow uniformity and consistency in the data. One of the widely used architectures is the generative adversarial networks (GANs) [11], [19] and its variants such as cycle-GAN [20]. In this study, a 2D cycle-GAN approach for multicenter MR-to-CT synthesis was used in the pelvic area to perform dose calculation directly from MR images. The purpose of this work was to obtain accurate sCTs from any center that could be integrated in an MRI-only radiotherapy workflow.
2. Material and Methods
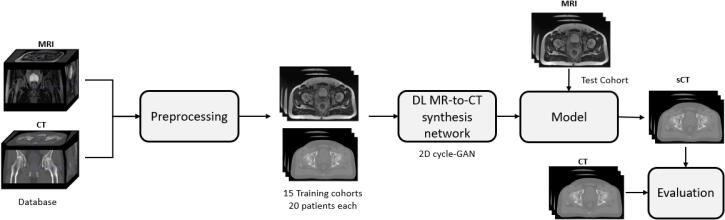
The Fig. 1 presents the workflow of this study on MR-to-CT synthesis. First, images are preprocessed, then they are divided into training cohorts of paired and registered CT/MRI to train the model. Afterwards, the model is used to synthesise a CT. Finally, the sCT is compared to the CT.
Fig. 1.
Workflow of the sCT generation from multicenter cohort.
2.1. Image data
In this study, 128 patients with prostate cancer from four datasets (D1, D2, D3 and D4) had CT and MR scans (standard MRI (for D1, D2, and D3) and MRI-Linac (for D4)) in the treatment position. The dataset D1 is composed by 39 patients from one care center, CT scans were acquired with a GE LightSpeedRT large-bore scanner or a Toshiba Aquilion. For MR images, 3D T2-weighted SPACE sequences were acquired on a 3T Siemens Skyra MRI scanner [21]. For the second dataset (D2), the 30 CT scans were acquired on a Philips BigBore and the T2 MRIs on an 1.5T Siemens Skyra MRI scanner. Bladders are injected on the CTs with a contrast agent. The third dataset (D3) is the public GoldAtlas [22] composed of 19 patients from 3 different centers with 1.5T and 3T MRI. Finally, the fourth dataset (D4) is composed of 40 patients, CT were acquired on a GE Light-SpeedRT16 and T2 MRI were acquired on a 0.35T MRIdian (ViewRay) MRI-Linac. For all CT and MR images, an expert delineated the target volume (prostate) and organs at risk (OARs, i.e. bladder, rectum and bones) except for the GoldAtlas base where the bones were not delineated. The studies involving human participants were reviewed and approved by Eugene Marquis Center (CLRCC) Ethics Committee. The patients/participants provided their written informed consent to participate in this study.
2.2. Image preprocessing
To eliminate outlier values, a thresholding ([-1000;1600] HU for the CTs and [0;500] for MRIs) was performed. To correct MRI non-uniformity, images were preprocessed by: (1) N4 bias field correction [23]; (2) histogram equalization (levels: 1024, match points: 7, threshold at mean intensity); and (3) filtering by gradient anisotropic diffusion [24]. Each CT was registered to its corresponding MRI with a symmetric rigid registration performed by the treatment planning system (TPS), followed by a hybrid (contours and intensities based) non-rigid registration using demons algorithm [25]. The registered CT was considered as the ground truth. Then, each MRI and CT were cropped to maintain a common field of view (FOV) of maximum 8 cm above and under the center of the prostate to improve the coherence. However, this FOV cannot be obtained on all images due to small FOV during acquisition, especially for MRI from D2. For these cases, the intersection between the 8 cm FOV and the acquisition FOV was kept. Afterwards, a 99.5% percentile contrast stretching was used to reduce the dynamic range of the histogram. To achieve dimensions of 256*256*128, B-spline resampling was used. The dataset D3 received the highest degree of resampling as the number of slices along the axial axis was comparatively low. Finally, 3D images were divided along the axial plan into 128 2D slices. After, images from the four centers were divided into training cohorts of 20 patients (18 for D3) and testing cohorts with the rest of the database. Table 1 presents the composition of the 15 different combinations of the four datasets. Ten patients from each center were used to evaluate the accuracy of each training.
Table 1.
Composition methodology of the training cohorts. Rows represent the centers included in the corresponding dataset and the column represent the number of patient of each dataset for each training.
| Number of center in the training | Training dataset | D1 | D2 | D3 | D4 |
|---|---|---|---|---|---|
| 1 | D1 | 20 | - | - | - |
| D2 | - | 20 | - | - | |
| D3 | - | - | 18 | - | |
| D4 | - | - | - | 20 | |
| 2 | D1 & D2 | 10 | 10 | - | - |
| D1 & D3 | 10 | - | 10 | - | |
| D1 & D4 | 10 | - | - | 10 | |
| D2 & D3 | - | 10 | 10 | - | |
| D2 & D4 | - | 10 | - | 10 | |
| D3 & D4 | - | - | 10 | 10 | |
| 3 | D1 & D2 & D3 | 7 | 7 | 7 | - |
| D1 & D2 & D4 | 7 | - | 7 | 7 | |
| D1 & D3 & D4 | 7 | - | 7 | 7 | |
| D2 & D3 & D4 | - | 7 | 7 | 7 | |
| 4 | D1 & D2 & D3 & D4 | 5 | 5 | 5 | 5 |
2.3. Experimental design
2.3.1. Deep learning architecture
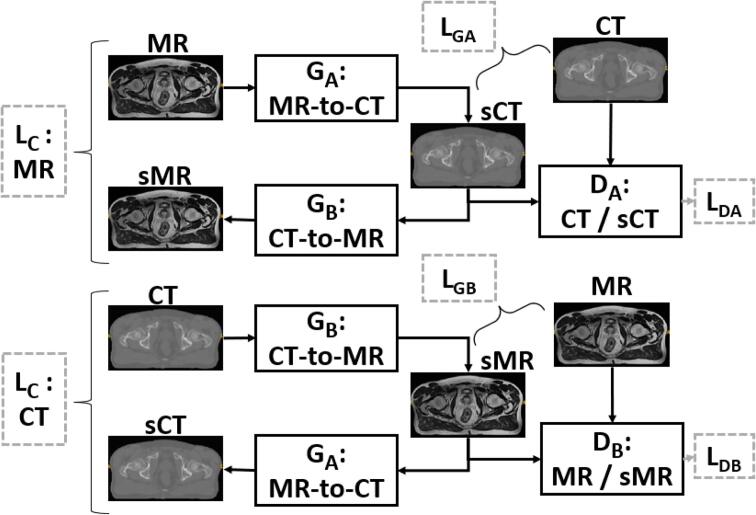
The Generative Adversarial Network (GAN) is composed of two adversarial networks: the generator and the discriminator [19]. A generator (), a 9 block ResNet, was used to convert MRIs into synthetic CTs (sCTs). The discriminator () that is a 70*70 PatchGAN network, classifies the sCT and gives the probability that it is a “real-CT” or “fake” (fig. 1). With loss functions associated with the two adversarial networks, the generator adjusts its generation parameters. The number of epochs was empirically adjusted to allow a stabilization of these parameters.
For this study, the 2D Cycle-GAN architecture proposed by Zhu et al. [20] was used in a supervised context. Fig. 2 details the Cycle-GAN architecture. This architecture combines two GANs in opposition. For each ResNet generator (), a PatchGAN discriminator () was associated. First, MR images are input to the first generator which is trained to convert them into a sCT. The second generator converts CTs into synthetic MR images (sMR). The architecture works following the steps detailed in Fig. 1: (1) After half a cycle: a similarity measure is computed between sCT and reference CT by a loss function () between sMR and reference MR by the same loss function (). (2) The sCT is sent to the input of generator and the sMR to the input of generator . (3) At the end of a cycle, we obtain an sCT generated from a sMR and a sMR generated from a sCT. They are evaluated with cycle losses (). For each synthetic image, the discriminator determines the probability of having a real image, the result is evaluated by the associated discriminator loss ( and ).
Fig. 2.
Cycle-GAN model adapted from [20]. Generator converts MR images into sCT or sMR into sCT, does the opposite. The perceptual loss as and is computed, it compares the sCT (resp sMR) with the reference CT (resp MR). At each iteration, the cycle loss () compares the real CT (respectively MR) with the sCT (resp. sMR) obtained with the sMR (resp. the sCT). Finally, the BCE loss function (resp. the ) is computed and determines the probability of having a real CT (resp. real MR).
2.3.2. Loss functions
In this work, the VGG-based perceptual loss [26] was used for the generators as and . This loss allows to separate content and style of each image. In this study, only four layers for the content were used (relu1_2, 2_2, 3_3, 4_3) and the style term was not used as in most studies of the literature [26]. Feature maps of MR and sCT are compared with the mean square error (MSE). This loss was chosen for the MR-to-CT synthesis because it allows a great accuracy of the sCTs [27]. For the discriminators, the Binary Cross Entropy (BCE) was used. The L1-norm was used for the cycle loss .
2.4. Postprocessing
To avoid inconsistencies outside the body that could affect dosimetric analysis, a value of −1000 HU was applied outside the MRI body contour.
2.5. Implementation
The cycle-GAN was implemented in Python3.8 using Pytorch 1.12 with CUDA 11.7. The model was trained and tested on an NVIDIA RTX A6000 with 48 GigaBytes of VRAM. Models were trained for 200 epochs and the learning rate set to 0.0002. Model parameters were chosen according to the last epoch. To assess the robustness of these models, a 4-fold cross validation was performed for all the trainings.
2.6. Evaluation
To evaluate the accuracy of each sCT, different voxel-wise metrics (or full-reference metrics) were used. They evaluate the error between the sCT and the CT. They are described as follows: the mean absolute error (MAE) in HU
| (1) |
the mean error (ME) in HU,
| (2) |
and the peak signal to noise ratio (PSNR) in dB.
| (3) |
with MSE as the mean square error (L2 norm between sCT and reference CT), and Q the amplitude. These metrics were computed on five different volumes: the body contour (whole pelvis), the prostate, the bladder, the rectum and the bones (except for D3).
The results were analysed using four distinct cases: case A) monocenter study, which uses data from the same dataset for both training and testing; case B) monocenter training, test done on a different dataset; case C) multicenter study using unseen data in the testing phase, where training is done on at least two centers and testing is carried out on a separate dataset; and case D) multicenter study using seen data, where training is done on data from at least two datasets and testing is done on data from a dataset included in the training.
To assess the sCT accuracy, a dose evaluation was conducted. A treatment of 60 Gy (20 fractions) was planned in VMAT on the reference CT using RaySearch RayStation v.12A TPS. The beam parameters were then applied to the sCTs of the corresponding patient. Dose endpoints were Dose-Volume Histogram (DVH) absolute differences between dose calculated on CTs and on sCTs, and 3D gamma analysis (criteria: local analysis, 1%/1 mm, with dose threshold = 10%) between dose distributions on CTs and sCTs. For dose calculation, 1 set of test sCTs was chosen for each case and dataset. For dataset 1, chosen sets were trained on D1 for case A, D3 for case B, D2&D3&D4 for case C and D1&D2&D3&D4 for case D. For dataset 2, chosen case were respectively D2, D4, D1&D3&D4 and D1&D2&D3&D4. For D3, D3, D1, D1&D2&D3 and D1&D2&D3&D4 and finally for D4: D4, D2, D1&D2&D3 and D1&D2&D3&D4.
Wilcoxon tests were performed to determine whether the results were significantly different from the monocenter training for each dataset. P-values that were smaller than 0.05 were considered as significant.
3. Results
On average, it took 8 hours to complete a training session with 20 paired CT/MRI for 200 epochs. The generation of a single sCT took approximately 10 s.
3.1. Qualitative results
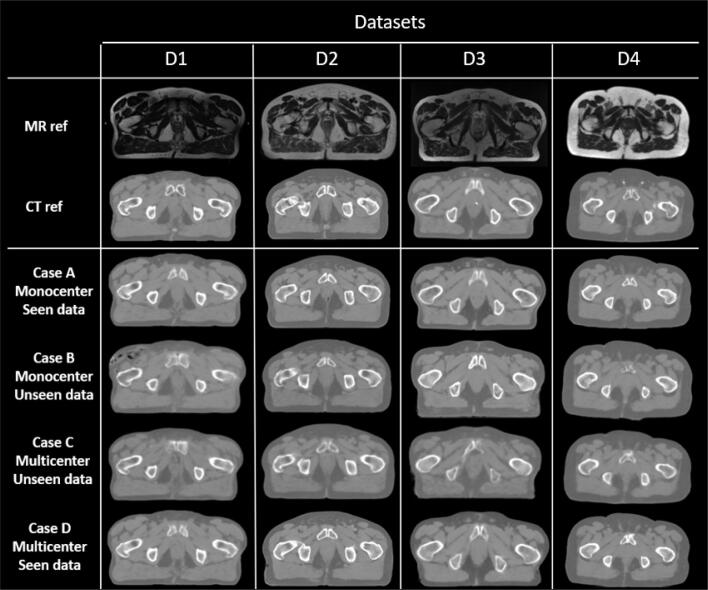
The Fig. 3 shows the qualitative results of the sCT synthesis. It can be observed that monocenter synthesis (case A) and multicenter synthesis using seen data (case D) are similar, whereas multicenter synthesis using unseen data (case C) and monocentric trainings and tests done on another dataset (case B) are realistic but the image quality is significantly inferior.
Fig. 3.
Preprocessed CT and MRI and image and sCT results according to the test dataset and the case: case A) monocenter study, case B) monocenter training using unseen dataset in the test, case C) multicenter training using unseen data in the test, and case D) multicenter training using seen data in the test.
3.2. Quantitative results
Table 2 presents the MAE results in the body contour for all centers for the different trainings. The lowest MAE results were obtained for dataset D1 with 30.0 HU for the monocentric training with seen data. Multicenter results using seen data in training were not significantly different compared with monocentric cases for the four datasets. For all datasets, MAE results were lower when data from the dataset were seen in the training than when they were not. For example, for D1 as test dataset, MAE results were between 31.3 HU and 34.4 HU for seen data in multicenter training, between 45.8 HU and 54.5 HU for unseen data in multicenter training and between 48.6 HU and 65.4 HU for a training on another dataset.
Table 2.
MAE results (in HU) comparing the reference CT with the synthetic-CTs in the body for the different trainings. Rows represent the datasets used for training and the colomns represent the dataset used for test. Values are the MAEs between CT and sCT for all test patients associated to the standard deviation. Results in grey represent case A (monocentric case with seen data), results in italic represent case B (monocentric training with unseen data), results not in bold nor italic nor grey represent case C (multicentric training with unseen data in the test), and results in bold represent case D (multicenter case with seen data). Not statistically different trainings (column by column) are recognized by an asterisk.
 |
Detailed results of ME and PSNR are presented in supplementary materials. The first table presents ME results, it can be observed that most of the time, values the closest to 0 correspond to cases A and D: when the same datasets are used in the training and the test. The second table presents the PSNR results: for cases A and D PSNR is higher. For instance, for D1, the PSNR ranges between 31.1 dB and 31.7 dB for cased A and D and between 26.4 dB and 28.9 dB for cases B and C.
3.3. Dose evaluation results
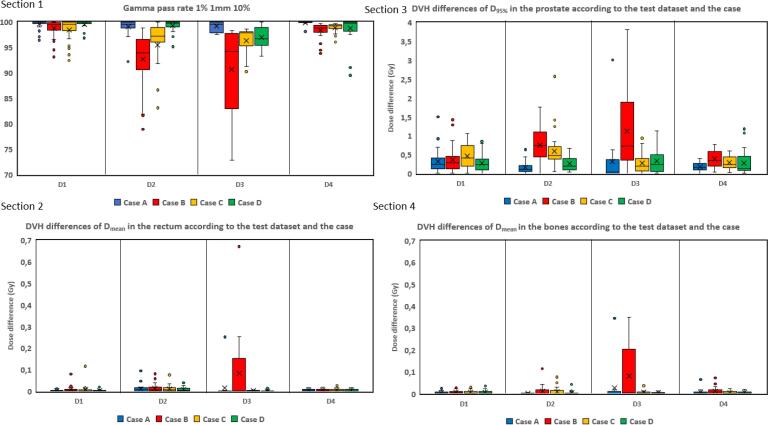
The Fig. 4 shows gamma pass rate results. Wilcoxon tests showed that for cases A and D of datasets D1, D2 and D3 gamma pass rate were not significantly different. However, for dataset D4, p-value was 0.01 between cases A and D. Section 2 presents absolute differences between the D95 of the reference CT and the D95 of each case sCT in the prostate according to the test dataset and the case. On average, the dose difference is inferior at 1 Gy on the target (prostate) for cases A and D and between 1 and 3 Gy for cases B and C. Section 3, 4 present the absolute difference between mean doses of rectum and bones respectively. For the rectum, the dose difference is always under 0.7 Gy even for the worse case and under 0.04 Gy for cases A and D. For the bones, the dose difference is under 0.35 Gy for all patients for the four cases. Due to strong artifacts on case B sCT, dose calculation was not possible on one patient from the GoldAtlas dataset. This patient was entirely removed from the dose study.
Fig. 4.
Results of sCT dose evaluation. Section 1 shows the gamma pass rate results according to the test dataset (D1-D4) and the case (A,B,C or D). Sections 2, 3, 4 presents absolute dose differences. Section 2, presents the results of the absolute differences between the of the dose on reference CT and the of calculated dose on sCT in the prostate according to the test dataset and the case. Section 3 presents the results of the absolute differences between the calculated on the reference CT and the calculated on sCT in the rectum according to the test dataset and the case. Section 4 presents the same results as Section 3 but in the bones.
4. Discussion
With this study, it was confirmed that a deep learning model trained on one set of data cannot be generalized or presents a decrease of performances on other datasets. Indeed, the worst results in terms of image evaluation and dose evaluation were obtained when the test was performed on a dataset not included in the training cohort (cases B and C) (Table 2, Fig. 4). It underlines that when there is a significant difference between training and test data, the DL model struggles to accurately synthesize sCT. However, the results between monocentric studies with seen data (case A) and multicenter studies with a seen dataset (case D) were equivalent (Fig. 3, Fig. 4, Table 2). Results highlight two points: firstly, a training cohort containing data from all centers (case D) is stable for all of them. Secondly, the model only requires five pairs of CT/MR images of each center to allow equivalent performances as the monocentric scenario (case A), whereas a training cohort with a size of five data does not converge. Tests were performed using a training cohort composed of five data, the DL models were not able to synthesize sCT.
Furthermore, as shown in the Fig. 4, the bones and rectum show the highest MAE, which can be attributed to specific factors. With bones, the high intensity of the structures means that a significant error in HU may not be visually apparent. On the other hand, the variability in rectum MAE can be explained by the presence of gas pockets on CT but not on MRI (often the case for the dataset D1) which are not always reconstructed by DL algorithms [28].
Our study also shows differences between monocentric with seen data performances: the best MAE results are obtained with dataset D1 (Table 2). This can be explained by several reasons: 1) the registration between MRI and CT was better for this center (quantified with the Dice metric). 2) For center D2, the acquisition FOV was shorter: the DL model learns less information about the area under the prostate so it can be less performant. 3) Dataset D3 is composed of three different centers: the variability between these centers was important [22]. Moreover, due to the small number of slices that make up the 8 cm FOV in this center, the resampling process has a deeper impact, resulting in a significant loss of information. To face these issues, the registration for datasets D2 and D4 needs to be improved.
To improve the robustness of MR to CT synthesis, additional centers could be added in the training cohort including a higher diversity in acquisition devices, intensity ranges, acquisition parameters. But, an acquisition protocol needs to be standardized for all centers to obtain the right FOV and resolution. Moreover, a further point to improve is the correction of 2D artifacts by a 3D training [29]. However, the main drawback of these architectures is their high computing complexity and the large number of data required for the training. These problems could be solved by high-performance computing stations and data-augmentation. Furthermore, this study underlines that dose calculation on sCT is accurate for clinical integration. Gamma analysis shows that for the multicenter model (case D) the gamma pass rate was above 92% for all patients. Moreover, the DVH analysis shows that the target volume still receives the prescribed dose with less than 1.5% of DVH difference and the OARs are also as preserved as on the reference planning CT with a DVH difference inferior of 50 cGy for cases A and D. What can be observed with the results, is that the worst dose results were obtained on the dataset D3 whereas worse MAEs were obtained for the dataset D2. It shows that there is not a correlation between MAE and dose. Furthermore, it was observed that the results of the GoldAtlas dataset were better for the patients from the seven patients from the first center of this dataset and the worse for the six patients of the third center. In the literature, Bird et al. [17], used a dataset comprising 90 anorectal patients drawn from two medical centers and applied a 2D conditional GAN. The average results for MAE stood at 35 HU, and the deviation in dose-volume histogram (DVH) was less than 0.7%. In contrast, when considering the D1 dataset for comparison, our findings indicated an average of 32 HU for image assessment and an average of 1,0% for dose evaluation. Then, despite the differences in MRI acquisition systems between the centers (high and low magnetic fields: from 0.35T to 3T), equivalent results were obtained for monocentric cases (cases A) and multicenter cases when the training contains data from the test center (cases D), even with few data of each center. Since MRI Linac is used for both planning and daily images, the same DL models can be applied to both. Thus, the proposed multicenter sCT generation can also be applied to daily MRI for dose monitoring. The idea is to estimate the delivered dose by calculating the ”dose of the day” and then the accumulated dose. Additionally, patients with unique characteristics such as higher body mass, rectum ablation, the presence of prostheses and so one [30] may be considered outliers within the training dataset. This study specifically observed a few patients with significantly higher body mass, which posed challenges for the cycle-GAN in reconstructing larger adipose volumes. To address these issues, it is crucial to create an expanded training database that includes individuals with diverse attributes and a wider range of imaging devices. This step is essential for improving the accuracy of the model for a larger patient population. Moreover, while this model can be evaluated using a real ground truth (the reference CT), in clinical practice, the ground truth is not always available. In such cases, sCTs can be visually assessed by clinicians, or alternative evaluation methods without references are being developed [31].
This evidence from this work should lead to further improvements in the clinical implementation of MRI in radiation oncology.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
The present work was funded by CominLabs with the CEMMTAUR project 2022. This research was partially supported by a PhD scholarship Grant from University of Rennes (France).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.phro.2023.100511.
Contributor Information
Blanche Texier, Email: blanche.texier@univ-rennes1.fr.
Jean-Claude Nunes, Email: jean-claude.nunes@uni-rennes.fr.
Appendix A. Supplementary materials
The following are the Supplementary data to this article:
References
- 1.Dirix P., Haustermans K., Vandecaveye V. The Value of Magnetic Resonance Imaging for Radiotherapy Planning. Semin Radiat Oncol. 2014;24:151–159. doi: 10.1016/j.semradonc.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Gao Z., Wilkins D., Eapen L., Morash C., Wassef Y. Gerig L.A study of prostate delineation referenced against a gold standard created from the visible human data. Radiother oncol. 2007;85:239–246. doi: 10.1016/j.radonc.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Steenbakkers R.J., Deurloo K.E., Nowak P.J., Lebesque J.V., van Herk M., Rasch C.N. Reduction of dose delivered to the rectum and bulb of the penis using mri delineation for radiotherapy of the prostate. Int J Radiat Oncol Biol Phys. 2003;57:1269–1279. doi: 10.1016/S0360-3016(03)01446-9. [DOI] [PubMed] [Google Scholar]
- 4.Pathmanathan A.U., McNair H.A., Schmidt M.A., Brand D.H., Delacroix L., Eccles C.L., et al. Comparison of prostate delineation on multimodality imaging for mr-guided radiotherapy. Br J Radiol. 2019;92:20180948. doi: 10.1259/bjr.20180948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kishan A.U., Lamb J., Casado M., Wang X., Ma T.M., Low D., et al. Magnetic resonance imaging-guided versus computed tomography-guided stereotactic body radiotherapy for prostate cancer (mirage): Interim analysis of a phase iii randomized trial. JAMA Oncol. 2022;9:365–373. doi: 10.1001/jamaoncol.2022.6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruynzeel A.M., Tetar S.U., Oei S.S., Senan S., Haasbeek C.J., Spoelstra F.O., et al. A prospective single-arm phase 2 study of stereotactic magnetic resonance guided adaptive radiation therapy for prostate cancer: early toxicity results. Int J Radiat Oncol Biol Phys. 2019;105:1086–1094. doi: 10.1016/j.ijrobp.2019.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Ulin K., Urie M.M., Cherlow J.M. Results of a multi-institutional benchmark test for cranial ct/mr image registration. Int J Radiat Oncol Biol Phys. 2010;77:1584–1589. doi: 10.1016/j.ijrobp.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Florkow MC, Zijlstra F, Kerkmeijer LG, Maspero M, van den Berg CA, van Stralen M et al. The impact of mri-ct registration errors on deep learning-based synthetic ct generation. In: Medical Imaging 2019: Image Processing volume 10949 p. 831–7; 2019.
- 9.Nyholm T., Nyberg M., Karlsson M.G., Karlsson M. Systematisation of spatial uncertainties for comparison between a mr and a ct-based radiotherapy workflow for prostate treatments. Radiat Oncol. 2009;4:1–9. doi: 10.1186/1748-717X-4-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seco J., Evans P.M. Assessing the effect of electron density in photon dose calculations. Med Phys. 2006;33:540–552. doi: 10.1118/1.2161407. [DOI] [PubMed] [Google Scholar]
- 11.Boulanger M., Nunes J.-C., Chourak H., Largent A., Tahri S., Acosta O., et al. Deep learning methods to generate synthetic CT from MRI in radiotherapy: A literature review. Phys Med. 2021;89:265–281. doi: 10.1016/j.ejmp.2021.07.027. [DOI] [PubMed] [Google Scholar]
- 12.Spadea M.F., Maspero M., Zaffino P., Seco J. Deep learning based synthetic-ct generation in radiotherapy and pet: a review. Med Phys. 2021;48:6537–6566. doi: 10.1002/mp.15150. [DOI] [PubMed] [Google Scholar]
- 13.Perone C.S., Ballester P., Barros R.C., Cohen-Adad J. Unsupervised domain adaptation for medical imaging segmentation with self-ensembling. NeuroImage. 2019;194:1–11. doi: 10.1016/j.neuroimage.2019.03.026. [DOI] [PubMed] [Google Scholar]
- 14.Brou Boni K.N.D., Klein J., Vanquin L., Wagner A., Lacornerie T., Pasquier D., et al. Mr to ct synthesis with multicenter data in the pelvic area using a conditional generative adversarial network. Phys Med Biol. 2020;65 doi: 10.1088/1361-6560/ab7633. [DOI] [PubMed] [Google Scholar]
- 15.Karani N, Chaitanya K, Baumgartner C, Konukoglu E.A lifelong learning approach to brain mr segmentation across scanners and protocols. In: Medical Image Computing and Computer Assisted Intervention–MICCAI 2018: 21st International Conference, Granada, Spain, September 16–20, 2018, Proceedings, Part I p. 476–84; 2018.
- 16.Lenkowicz J., Votta C., Nardini M., Quaranta F., Catucci F., Boldrini L., et al. A deep learning approach to generate synthetic ct in low field mr-guided radiotherapy for lung cases. Radiother oncol. 2022;176:31–38. doi: 10.1016/j.radonc.2022.08.028. [DOI] [PubMed] [Google Scholar]
- 17.Bird D., Nix M.G., McCallum H., Teo M., Gilbert A., Casanova N., et al. Multicentre, deep learning, synthetic-ct generation for ano-rectal mr-only radiotherapy treatment planning. Radiother Oncol. 2021;156:23–28. doi: 10.1016/j.radonc.2020.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cusumano D., Lenkowicz J., Votta C., Boldrini L., Placidi L., Catucci F., et al. A deep learning approach to generate synthetic ct in low field mr-guided adaptive radiotherapy for abdominal and pelvic cases. Radiother Oncol. 2020;153:205–212. doi: 10.1016/j.radonc.2020.10.018. [DOI] [PubMed] [Google Scholar]
- 19.Goodfellow I., Pouget-Abadie J., Mirza M., Xu B., Warde-Farley D., Ozair S., et al. Generative adversarial networks. Commun ACM. 2020;63:139–144. doi: 10.1145/3422622. [DOI] [Google Scholar]
- 20.Zhu J-Y, Park T, Isola P, Efros AA Unpaired image-to-image translation using cycle-consistent adversarial networks. In: 2017 IEEE International Conference on Computer Vision (ICCV) volume abs/1703.10593 2017 p. 2242–51. doi: 10.1109/ICCV.2017.244.
- 21.Dowling J.A., Sun J., Pichler P., Rivest-Hénault D., Ghose S., Richardson H., et al. Automatic Substitute Computed Tomography Generation and Contouring for Magnetic Resonance Imaging (MRI)-Alone External Beam Radiation Therapy From Standard MRI Sequences. Int J Radiat Oncol Biol Phys. 2015;93:1144–1153. doi: 10.1016/j.ijrobp.2015.08.045. [DOI] [PubMed] [Google Scholar]
- 22.Nyholm T., Svensson S., Andersson S., Jonsson J., Sohlin M., Gustafsson C., et al. Mr and ct data with multiobserver delineations of organs in the pelvic area–part of the gold atlas project. Med Phys. 2018;45:1295–1300. doi: 10.1002/mp.12748. [DOI] [PubMed] [Google Scholar]
- 23.Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA et al. N4itk: improved n3 bias correction. IEEE T Med Imaging 2010; 29: 1310–20. doi: 1.0.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed]
- 24.Perona P., Malik J. Scale-space and edge detection using anisotropic diffusion. IEEE T Pattern Anal. 1990;12:629–639. doi: 10.1109/34.56205. [DOI] [Google Scholar]
- 25.Rivest-Hénault D, Greer P, Fripp J, Dowling J. Structure-Guided Nonrigid Registration of CT-MR Pelvis Scans with Large Deformations in MR-Based Image Guided Radiation Therapy. In: Erdt Marius, Linguraru Marius George, Oyarzun Laura Cristina, Shekhar Raj, Wesarg Stefan, González Ballester Miguel Angel et al. (Eds.), Clinical Image-Based Procedures. Translational Research in Medical Imaging Springer International Publishing p. 65–73; 2014.
- 26.Johnson J, Alahi A, Fei-Fei L. Perceptual Losses for Real-Time Style Transfer and Super-Resolution in: Leibe Bastian, Matas Jiri, Sebe Nicu, Welling Max (Eds.), Computer Vision - ECCV 2016 Springer International Publishing p. 694–711; 2016.
- 27.Tahri S., Barateau A., Cadin C., Chourak H., Ribault S., Nozahic F., et al. A high-performance method of deep learning for prostate MR-only radiotherapy planning using an optimized Pix2Pix architecture. Phys Med. 2022;103:108–118. doi: 10.1016/j.ejmp.2022.10.003. [DOI] [PubMed] [Google Scholar]
- 28.Maspero M., Tyyger M.D., Tijssen R.H.N., Seevinck P.R., Intven M.P.W., van den Berg C.A.T. Feasibility of magnetic resonance imaging-only rectum radiotherapy with a commercial synthetic computed tomography generation solution. Phys Imag Radiat Oncol. 2018;7:58–64. doi: 10.1016/j.phro.2018.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu Y, Lei Y, Zhou J, Wang T, David SY, Beitler JJ et al. Synthetic ct-aided mri-ct image registration for head and neck radiotherapy. In: Medical Imaging 2020: Biomedical Applications in Molecular, Structural, and Functional Imaging volume 11317 p. 572–78; 2020.
- 30.Dowling J., O’Connor L., Acosta O., Raniga P., de Crevoisier R., Nunes J.-C., et al. Biomedical Image Synthesis and Simulation. Elsevier; 2022. Image synthesis for mri-only radiotherapy treatment planning; pp. 423–445. [Google Scholar]
- 31.Chourak H., Barateau A., Tahri S., Cadin C., Lafond C., Nunes J.-C., et al. Quality assurance for mri-only radiation therapy: A voxel-wise population-based methodology for image and dose assessment of synthetic ct generation methods. Front Oncol. 2022;12 doi: 10.3389/fonc.2022.968689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.