Abstract
The innate immune system of mammals is formed by a complex web of interacting proteins, which together constitute the first barrier of entry for infectious pathogens. Genes from the E3-ubiquitin ligase tripartite motif (TRIM) family have been shown to play an important role in the innate immune system by restricting the activity of different retrovirus species. For example, TRIM5 and TRIM22 have both been associated with HIV restriction and are regarded as crucial parts of the antiretroviral machinery of mammals. Our analyses of positive selection corroborate the great significance of these genes for some groups of mammals. However, we also show that many species lack TRIM5 and TRIM22 altogether. By analyzing a large number of mammalian genomes, here we provide the first comprehensive view of the evolution of these genes in eutherians, showcasing that the pattern of accumulation of TRIM genes has been dissimilar across mammalian orders. Our data suggest that these differences are caused by the evolutionary plasticity of the immune system of eutherians, which have adapted to use different strategies to combat retrovirus infections. Altogether, our results provide insights into the dissimilar evolution of a representative family of restriction factors, highlighting an example of adaptive and idiosyncratic evolution in the innate immune system.
Keywords: TRIM, TRIM5, immunogenetics, retrovirus evolution, positive selection, restriction factors
Significance.
Some members of the TRIM family of genes have been previously identified as restriction factors that inhibit the replication of retroviruses such as HIV. Although previous studies have described the evolution of TRIM genes in different species, this study is the first comparative analysis of the TRIM6/34/5/22 gene cluster to examine the evolution of these genes across a large number of mammals. Our findings reveal significant dissimilarities in the evolution of TRIM genes across mammalian lineages. For instance, some lineages have repeatedly acquired copies of TRIM5 or TRIM22, whereas others have lost these genes completely. Due to TRIM genes’ role in our immune system, understanding how they have evolved might prove invaluable knowledge to combat retroviral diseases in the future.
Introduction
The E3-ubiquitin ligase tripartite motif (TRIM) proteins comprise a large family of proteins associated with diverse cellular functions of the immune system (Ozato et al. 2008; van Gent et al. 2018; Yang et al. 2020). These proteins canonically consist of amino-terminal RING, B-box, and coiled-coil domains, which can be associated with different carboxy-terminal domains in each protein (Ozato et al. 2008). Some closely related paralogous genes from this family can be found grouped together in the genome of most eutherian mammals, forming a gene cluster commonly referred to as TRIM6/34/5/22. Genes from this cluster have previously been shown to be important to the innate immunity of mammals. TRIM5 is a potent restriction factor of retroviruses, which was first identified as a repressor of HIV-1 replication in Old World monkey cells (Hatziioannou et al. 2004; Keckesova et al. 2004; Stremlau et al. 2004; Yap et al. 2004). Since then, TRIM5 was shown to also be able to restrict other virus species in a capsid-specific manner, such as the murine leukemia virus of primates, the visna/maedi virus of ovines, the feline immunodeficiency virus of cats, and even the endogenous rabbit endogenous lentivirus type K (RELIK) virus of lagomorphs (Yap et al. 2004; Saenz et al. 2005; Fletcher et al. 2010; Jáuregui et al. 2012; Yap and Stoye 2013). Subsequent studies showed that the TRIM22 protein is also capable of species-specific restriction, which was linked with suppression of activity of different viruses (Gao et al. 2009; Di Pietro et al. 2013; Jing et al. 2019). TRIM5’s and TRIM22's specificity have been mapped to their carboxy-terminal PRYSPRY domain, where the strong signs of positive selection have been detected in multiple lineages of mammals (Sawyer et al. 2005; Tareen et al. 2009; Águeda-Pinto et al. 2019; Fernandes et al. 2022). In primates, a high number of polymorphisms observed for TRIM5 was also attributed to balancing selection (Newman et al. 2006). Whereas TRIM5’s and TRIM22's functional properties are well described in the literature, TRIM6’s and TRIM34's functions in the innate system remain somewhat elusive. It is known, however, that these proteins are also associated with the innate immune system. For example, TRIM6 was shown to upregulate type-1 interferon–stimulated gene activity, whereas TRIM34 was shown to be able to restrict HIV in a TRIM5-dependent manner (Rajsbaum et al. 2014; Ohainle et al. 2020).
As expected for genes that interact with the innate immune system in very different ways, great dissimilarities have been observed in the patterns of evolution of the TRIM genes. TRIM5 and TRIM22 duplications have been shown to be abundant in some eutherian orders (Si et al. 2006; Tareen et al. 2009; Boso et al. 2019; Fernandes et al. 2022), likely because maintaining multiple copies of these genes allows them to independently evolve to restrict different viruses simultaneously (Sawyer et al. 2007; Daugherty and Malik 2012). Alternatively, it is possible that some of these new TRIM copies are undergoing neofunctionalization and are not necessarily involved in the same processes of immune response as the ancestral gene. In contrast to TRIM5 and TRIM22, TRIM6 and TRIM34 are usually more evolutionarily conserved and are found as a single copy in the genome of most species (Sawyer et al. 2007; Boso et al. 2019; Fernandes et al. 2022). However, because no large-scale comparative study of these genes had yet been performed, it remained hard to tell whether TRIM genes are part of the core antiviral immune defense system of all eutherian mammals or instead their function could be fulfilled by other families of restriction factors of retroviruses, allowing for the degeneration of the TRIM6/34/5/22 gene cluster in some lineages.
Recently, a large number of restriction factors of retroviruses evolving under positive selection has been described in mammals (Duggal and Emerman 2012; Colomer-Lluch et al. 2018). Some of these genes exhibit patterns of adaptive evolution and have been subject to successive events of paralogous expansion or, in some cases, have degenerated and pseudogenized (Sawyer et al. 2007; Ito et al. 2020; Yap et al. 2020). Together, they integrate a large arsenal of antiretroviral weapons that have engaged retroviruses in evolutionary arms race for millions of years. As time passed, however, lineages of mammals have started to rely on these proteins in different ways. Primates, for example, were shown to accumulate a large number of apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3 (APOBEC3) genes compared with other eutherians (Ito et al. 2020) but have not acquired even a single TRIM6/34/5/22 duplication (Sawyer et al. 2007). In contrast, rodents are known to maintain a large number of functional TRIM5 paralogs but display a low number of APOBEC3 paralogs compared with other mammalian orders (Boso et al. 2019; Ito et al. 2020). To provide a comprehensive view of how TRIM genes have evolved in eutherians, in this study, we compared the evolutionary history of the TRIM6/34/5/22 gene cluster in 53 mammal species, showcasing the extraordinary evolutionary plasticity of this representative family of restriction factors in mammals.
Results
Origin and Phylogeny of the TRIM6/34/5/22 Genes
To ask about how the TRIM6/34/5/22 gene cluster has evolved broadly across mammalian evolutionary history, we curated a large data set of TRIM sequences (see Materials and Methods). Our final data set comprises sequences from a total of 53 species from 12 orders of Eutheria: Primates, Rodentia, Chiroptera, Artiodactyla, Carnivora, Lagomorpha, Perissodactyla, Pilosa, Macroscelidea, Sirenia, Eulipotyphla, and Pholidota. The sequences were obtained through Protein Basic Local Alignment Search Tool (BLASTP) searches performed in mammalian infraclasses Marsupialia and Eutheria (the latter of which is further subdivided into three lineages: Boreoeutheria, Afrotheria, and Xenarthra). Although all genes from the TRIM6/34/5/22 gene cluster were detected in both boreoeutherians and afrotherians, neither TRIM34 nor TRIM22 was found in the only xenarthran species in the data set: Choloepus didactylus (two-toed sloth). Further, we did not identify any sequence hits in marsupial species, suggesting the whole gene cluster is missing in this group. Under the hypothesis of either an Afrotheria or Atlantogenata (Xenarthra + Afrotheria) rooting for eutherian mammals (Morgan et al. 2013; Upham et al. 2019; Álvarez-Carretero et al. 2022; Foley et al. 2023), these results suggest that the TRIM6/34/5/22 gene cluster originated sometime between the divergence of eutherians and marsupials (166–123 Ma) (Upham et al. 2019; Álvarez-Carretero et al. 2022) and the radiation of the crown group Eutheria (ca. 97–78 Ma; Upham et al. 2019; Álvarez-Carretero et al. 2022; Foley et al. 2023). The origin of this gene cluster is remarkably recent when compared with other known elements of the innate immune system, many of which are shared among all animals or even with prokaryotes (Wein and Sorek 2022; Iwama and Moran 2023). To rule out that the TRIM6/34/5/22 gene cluster could be missing in Marsupialia but present in more distant relatives of Eutheria, we performed additional BLASTP searches on all available noneutherian NCBI genomes and found no matches for any of the genes (closest matches were TRIM39 sequences).
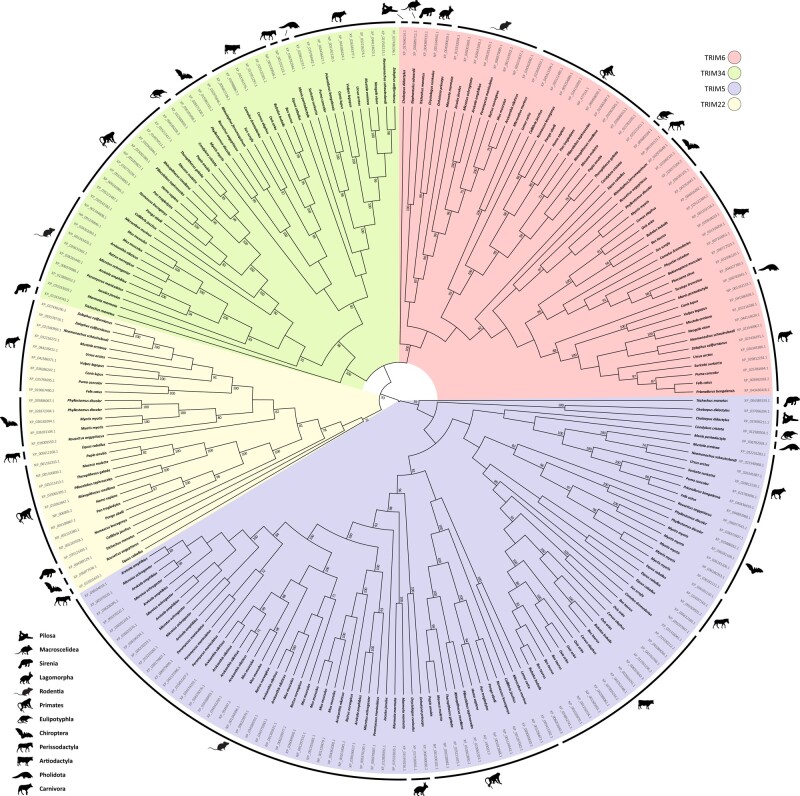
Next, we analyzed the phylogeny of TRIM6, TRIM34, TRIM5, and TRIM22 orthologs to better understand the evolutionary history of these genes in mammals. A maximum-likelihood (ML) tree built with the sequences of all 53 species in our data set (fig. 1) displays the most likely topology for the genes in the TRIM6/34/5/22 cluster. The bootstrap values calculated for the nodes that represent splits between different TRIM genes on the ML tree were very high overall (>93%), with the exception of the node representing the split between TRIM6 and TRIM34, for which the bootstrap was 62%. The phylogeny of TRIM genes has been inconsistent across different studies in the past. Our tree, although unrooted, is topologically consistent with Williams et al. (2019), Carthagena et al. (2009), and Sardiello et al. (2008) but inconsistent with Sawyer et al. (2007), favoring the hypothesis that TRIM6 and TRIM34 form sister taxa in the topology (fig. 1).
Fig. 1.
Phylogenetic analysis of the TRIM6/34/5/22 genes of eutherians. An ML tree of TRIM6, TRIM34, TRIM5, and TRIM22 genes extracted from eutherian genomes was built with IQ-Tree software (Nguyen et al. 2015) implemented in the W-IQ-TREE web server (Trifinopoulos et al. 2016). A total of 1,000 bootstrap repetitions were performed to ensure the robustness of data. Only bootstrap values >70% are shown.
Gene Copy Number Has Diverged in the TRIM6/34/5/22 Cluster
We examined our phylogeny to ask if there are differences in the pattern of duplication and deletion events of TRIM genes across mammalian evolution. TRIM5 has been relatively well studied in the past due to its role in HIV-1 restriction (Stremlau et al. 2004), and a large number of duplications on this locus have been described by previous studies (Si et al. 2006; Tareen et al. 2009; Boso et al. 2019; Fernandes et al. 2022). We similarly find that duplications of the TRIM5 locus have been very common in many of the mammalian groups included in our data set (fig. 2). For example, clades such as Rodentia, Yangochiroptera, Ruminantia as well as the Equus caballus species (horse) have accumulated up to seven copies of the TRIM5 gene in their genome (fig. 2A). Extending these findings, we detected a novel duplication of TRIM5 in the xenarthran species C. didactylus, indicating that events of paralogous expansion of TRIM5 can also occur outside of the Boreoeutheria lineage (figs. 1 and 2A). The two TRIM5 sequences found in this species are quite similar (90% identity after pairwise alignment), suggesting that they arose from a relatively new duplication. Although TRIM5 is often duplicated in mammalian genomes, it is also missing from the genome of some species, such as all cetacean species included in our data set, some members of Yinpterochiroptera and Carnivora, and Elephantulus edwardii (the Cape elephant shrew).
Fig. 2.
Summary of the number of copies of each TRIM paralog in the genome of eutherian species. Each amino acid sequence was categorized as TRIM6, TRIM34, TRIM5, or TRIM22 according to the phylogeny available in figure 1. (A) The number of paralog sequences are displayed for different species of eutherians. In yellow are sequences that had to be removed from the alignment from which the tree was derived, and their gene annotation was instead extracted directly from the NCBI database. The cladogram displayed in the table was derived from the work of Upham et al. (2019). (B) Number of species in which each TRIM gene is found as a single copy, duplicated, or absent in our data set.
TRIM22 duplications could also be detected in our data set, but in fewer groups compared with TRIM5 (fig. 2B). We previously described a large number of independent duplications of the TRIM22 gene that have affected the Chiroptera order (Fernandes et al. 2022), but our results now reveal that TRIM22 duplications can be observed in at least two other species: Zalophus californianus (California sea lion) and Eq. caballus (fig. 2A). The first of these duplication events seems to have occurred rather recently in the context of Carnivora evolution, as it has affected Z. californianus but not the closely related Neomonachus schauinslandi (separation time 24–14 Ma [Upham et al. 2019; Foley et al. 2023]). The TRIM22 sequences obtained from the genome of Z. californianus are also very similar (98.8% identical after pairwise alignment), which corroborates the hypothesis that they have originated recently. In contrast, the duplication that has affected Eq. caballus seems to be particularly old. The TRIM22 genes of Eq. caballus differ significantly in their amino acid composition, only displaying 58.1% identity after pairwise alignment, which was the lowest value in all pairs of TRIM22 paralogs we analyzed. If more TRIM22 sequences from species related to Eq. caballus were identified, we would be able to more definitely date the origin of this duplication. However, complete genomes from the order Perissodactyla are unfortunately rare. The only other sequenced TRIM6/34/5/22 cluster available (from Ceratotherium simum) was determined unfit for our final data set because it was split into multiple genomic scaffolds. Our initial BLAST searches, however, did find multiple TRIM22 copies for Ce. simum, hinting that TRIM22 duplications in Perissodactyla could be common. In contrast to the duplication events in Chiroptera and Perissodactyla, TRIM22 has also undergone independent events of deletion in many mammalian lineages (fig. 2). In our data set, TRIM22 is missing in Rodentia, Artiodactyla, Lagomorpha, Strepsirrhini, C. didactylus, E. edwardii, Condylura cristata, Manis pentadactyla, Suricata suricatta, and Prionailurus bengalensis, suggesting that at least nine independent events of deletion of the TRIM22 locus have taken place in Eutheria (fig. 2A).
Contrasting to TRIM5 and TRIM22, the evolution of TRIM34 in Eutheria has been much less dynamic. Only a single duplication could be identified in this locus, in the Mus musculus (house mouse) species. The two paralogous TRIM34 proteins are identical, suggesting that the duplication is extremely recent. We confirmed that each of the two sequences represents a different locus by analyzing their introns, which display differences in their nucleotide composition. As far as we know, this duplication is a unique feature of M. musculus and is not present in other rodent species (Tareen et al. 2009; Boso et al. 2019). As with TRIM5 and TRIM22, TRIM34 is also absent in some groups of mammals, such as Cetacea, Lagomorpha, C. didactylus, E. Edwardii, and Felis catus. Overall, the pattern of evolution of TRIM34 seems to be significantly different from that of TRIM5 and TRIM22. Duplications of the latter two are common, and many copies of the genes can accumulate in a single species’ genome. In contrast, TRIM34 duplications seem to be very rare, implying different selective pressures on the maintenance of a single copy of this TRIM gene in mammalian genomes.
In sum, we find the evolutionary patterns of genes from the TRIM6/34/5/22 gene cluster to be extremely diverse. Dissimilar profiles of duplications can be observed not only between the different TRIM genes (fig. 2B) but also among lineages of Eutheria (fig. 2A), suggesting the evolution of this gene cluster is very plastic.
Synteny of TRIM Genes in the TRIM6/34/5/22 Gene Cluster Varies between Mammalian Lineages
To better understand how duplications of TRIM genes may have affected the organization of the TRIM6/34/5/22 gene cluster in different species, we created a synteny map of the flanking genes of this region of the genome in select species (fig. 3). The synteny analysis reveals that a large number of duplications and inversions have shaped the TRIM6/34/5/22 gene cluster in most species, resulting in a poor conservation in the organization of TRIM genes across mammal genomes. This is especially evident in the most species-rich orders such as Chiroptera and Rodentia, which display radically different arrangements of TRIM6/34/5/22 from species to species (compare Myotis myotis with Rousettus aegyptiacus in fig. 3). Nevertheless, the upstream genomic portion of the gene cluster seems to be well preserved in most groups, usually consisting of an olfactory receptor family 52 subfamily b member 6 (OR52B6) gene followed by a single TRIM6 gene. After that, the gene cluster becomes less predictable, as the remaining TRIM genes of each species are intercalated by a variable number of olfactory receptor and ubiquitin-like genes, such as OR52H1, OR52B4, and UBQLN3 (fig. 3).
Fig. 3.
Synteny map of the genomic organization of the TRIM6/34/5/22 gene cluster. The genomic organization of the TRIM6/34/5/22 gene cluster of different species of mammals was analyzed using NCBI's Genome Data Viewer. The direction of transcription is indicated by arrows. Half black arrows mark pseudogenes.
In some mammalian species, the TRIM6/34/5/22 gene cluster has been radically reshaped. For example, TRIM6 is not preceded by OR52B6 in cetacean genomes and is instead located far from any genes recognizable in the synteny of other members of Artiodactyla (fig. 3). A BLAST search performed with a transcript of the TRIM6-neighboring FAM160A2 gene (XM_019948115.2) of Tursiops truncatus (common bottlenose dolphin) found a homolog sequence for this locus in ruminant species such as Ovis aries, Bos taurus, and Cervus elaphus but located approximately one million nucleotides upstream of their TRIM6/34/5/22 gene cluster. This suggests that TRIM6 may be the only gene that survived a series of duplications, recombinations, and deletions that have completely reshaped the TRIM6/34/5/22 gene cluster in Cetacea, thus explaining why there are no signs of closeby TRIM34, TRIM5, or TRIM22 genes in any genomes of this lineage (figs. 2A and 3). Another interesting genomic organization is that of B. taurus, which is the only species in which the TRIM6/34/5/22 gene cluster was split across different chromosomes (fig. 3). A TRIM5 gene copy was translocated from chromosome 15 of this species and instead is found in chromosome 9. The translocated gene has not degenerated and aligns well with other TRIM5 copies from this species, suggesting that it remains functional. Overall, we find that the synteny of genes in the TRIM6/34/5/22 is very poorly conserved, with the exception of the upstream-most portion of the cluster where TRIM6 is placed.
Positive Selection Has Shaped TRIM Genes Differently in Mammals
Antiviral proteins are often rapidly evolving in many lineages of mammalian phylogenies due to the selective pressure imposed by pathogens (Daugherty and Malik 2012). Next, we asked if we can observe evidence of rapid evolution of the TRIM6/34/5/22 gene cluster in mammals. The rate of selection (ω) of a protein is a measure of how selective pressures acting on the phenotype of organisms have affected the underlying DNA sequence of the protein (or of a particular amino acid or domain of the protein; Eyre-Walker 2006). To determine how selection has affected the TRIM6/34/5/22 gene cluster during its evolution, we split our data into subsections corresponding to the different TRIM genes and ran separate adaptive branch-site random effects likelihood (aBSREL) analyses for each of them (fig. 4 and table 1; Smith et al. 2015). The model of selection implemented by aBSREL adjusts amino acids with a variable number of rate of selection classes along each branch of the protein phylogeny, allowing the strength and extent of positive selection to be distinguished for each group.
Fig. 4.
Positive selection acting on TRIM loci throughout the eutherian phylogeny. We performed independent aBSREL (Smith et al. 2015) analyses for each gene of the TRIM6/34/5/22 gene cluster to evaluate how positive selection has shaped these proteins during the evolutionary history of Eutheria. Each branch is partitioned according to the proportion of sites that belong to different ω classes (red, for positive selection; light gray, for neutral selection; and black for purifying selection). Thick branches are those in which the evidence of positive selection was deemed significant (P < 0.05). “Branch IDs” are indicated for relevant branches and serve as entry keys for table 1. The sections of the TRIM34 tree where no signs of positive selection were found have been collapsed.
Table 1.
Detailed ω Ratios and Proportion of Codons Affected by Positive Selection in TRIM Loci
| Phylogeny | Branch ID | ω | Amino Acids Affected (%) |
|---|---|---|---|
| Br1 | >8.0 | 11.0 | |
| Br2 | >8.0 | 2.4 | |
| Br3 | >8.0 | 3.6 | |
| Br4 | >8.0 | 4.2 | |
| Br5 | >8.0 | 6.8 | |
| Br6 | >8.0 | 3.0 | |
| Br7 | >8.0 | 5.2 | |
| Br8 | >8.0 | 9.8 | |
| Br9 | >8.0 | 18.5 | |
| Br10 | >8.0 | 13.0 | |
| Br11 | >8.0 | 13.8 | |
| Br12 | >8.0 | 5.4 | |
| Br13 | >8.0 | 2.4 | |
| Br14 | >8.0 | 1.7 | |
| TRIM5 | NP_001355682.1 | >8.0 | 16.0 |
| XP_004590030.2 | 3.7 | 31.8 | |
| XP_012580504.1 | 5.0 | 25.2 | |
| XP_013210282.1 | >8.0 | 1.4 | |
| XP_015333035.1 | 4.7 | 26.5 | |
| XP_015853207.2 | >8.0 | 11.8 | |
| XP_017196941.1 | >8.0 | 9.4 | |
| XP_021540968.1 | >8.0 | 6.6 | |
| XP_027835244.2 | >8.0 | 5.0 | |
| XP_028372907.1 | >8.0 | 7.0 | |
| XP_034342250.1 | >8.0 | 19.1 | |
| XP_036009560.1 | >8.0 | 8.9 | |
| XP_036181100.1 | >8.0 | 3.1 | |
| XP_042089056.1 | >8.0 | 10.1 | |
| XP_045413292.1 | >8.0 | 3.9 | |
| Br15 | >8.0 | 13.4 | |
| XP_023501633.1 | >8.0 | 16.5 | |
| TRIM22 | XP_035886067.1 | >8.0 | 2.2 |
| XP_036077536.1 | >8.0 | 6.7 | |
| XP_036181104.1 | >8.0 | 3.3 | |
| TRIM34 | XP_025784462.1 | >8.0 | 1.1 |
Note.—Entries in this table are identified by branch IDs indicated in figure 4.
We find that both TRIM5 and TRIM22 have evolved under positive selection in the past, although they were affected in different ways. For TRIM5, the positive selection signal is lineage pervasive, affecting the ancestors of living mammals throughout the entire phylogeny (fig. 4). In fact, the signals of positive selection in this gene date back to the radiation of the two major Boreoeutheria superorders: Laurasiatheria and Euarchontoglires (respectively branches Br2 and Br3 in fig. 4). This means that TRIM5 has been evolving under positive selection for at least 77 Myr, as that is the approximate radiation time for Euarchontoglires, the oldest of the two groups (Upham et al. 2019). Positive selection could also be observed in the most recent branches of the TRIM5 phylogeny, indicating diversification of this gene is still ongoing in many lineages. For example, the recent expansions of TRIM5 in My. myotis, Phyllostomus discolor, O. aries, M. musculus, Arvicanthis niloticus, and Peromyscus maniculatus were all targeted by strong (ω > 8) positive selection (fig. 4). We also show that positive selection has affected groups that have a single copy of TRIM5, such as Lagomorpha, Condylura cristata, Marmota marmota, and Lemur catta.
In contrast to TRIM5, positive selection in the TRIM22 locus seems to be a less common occurrence, affecting only five branches in the phylogeny. Of note, only lineages that display more than one copy of TRIM22 were targeted by diversifying selection in this gene (namely Chiroptera and Eq. caballus). This suggests that selection in TRIM22 may be dependent or otherwise correlated with events of duplication, something that we did not observe for TRIM5. Selection was strongly positive (ω > 8) in all the branches it was detected, although the number of amino acids affected varied greatly from branch to branch (2.2–16.5%, see table 1).
Overall, we did not observe significant signals of rapid evolution in TRIM6 or TRIM34. This is in agreement with previous work showing that these genes are usually highly conserved (Sawyer et al. 2007). However, we did identify an exception: a strong signature of positive selection in the TRIM34 protein of Puma concolor. The TRIM34 ortholog of this species displays a highly variable section of around 23 nucleotides in length between its coiled-coil and SPRY domains (supplementary fig. S3, Supplementary Material online), which was likely considered the evidence of positive selection by aBSREL. To our knowledge, this is the first report of signals of positive selection on TRIM34. However, at present, we cannot rule out the possibility that this unconserved region results from a sequencing error, so these data should be interpreted with caution.
Altogether, these patterns of selection highlight the divergent evolutionary histories of TRIM genes. Although TRIM6 and TRIM34 tend to be more conserved in mammals, TRIM5 and TRIM22 are often subject to periods of intense positive selection and genomic relocation.
Discussion
Plasticity of TRIM Evolution across Mammals
A striking feature of the evolution of immune-related proteins is the plasticity associated with their genomic organization, overall abundance, and innovation at the amino acid level. This effect is well described for the adaptive immune system, whose ability to combine different genes in order to create a pathogen-specific response has led to incredible diversification among vertebrate lineages (Sun et al. 2013). More recently, many genes involved in innate immunity were also shown to have evolved under positive selection or to have undergone events of expansion and deletion that have created significant genomic diversity across relatively closely related species (Boso and Kozak 2020; Ito et al. 2020; Yap et al. 2020; Ahmad et al. 2022). For example, independent events of TRIM5 duplication have been described in many groups of mammals, including rodents, bats, and cows (Sawyer et al. 2007; Tareen et al. 2009; Fernandes et al. 2022). Similarly, we have previously shown that TRIM22 has undergone multiple independent events of duplication in the Chiroptera order (Fernandes et al. 2022). In this study, we expand on these findings, showing that TRIM copy number, organization, and sequence conservation have been remarkably dynamic across mammalian evolution.
Various models could explain the divergent accumulation of antiviral TRIM genes across genomes. Specific to TRIM proteins’ antiretroviral activities, one hypothesis is that duplications of TRIM loci were selected in the mammalian lineages that have been more affected by retroviruses in the past. Maintaining multiple copies of TRIM genes might have been advantageous for those lineages because each gene copy could independently evolve to restrict a different virus species (Sawyer et al. 2007; Daugherty and Malik 2012). A relation between gene expansion events and history of interaction with retroviruses was previously described in mammals for a different family of restriction factors, APOBEC3. For this family of genes, a mild correlation between the number of accumulated gene copies and the percentage of the genome of a species that corresponded to endogenous retroviruses (ERVs) could be observed (Ito et al. 2020). Our data suggest that the same correlation can be observed for TRIM genes, although we note that the variables are even more weakly associated (supplementary fig. S2, Supplementary Material online). Both the total number of TRIM paralogs and the number of TRIM5 copies increase as a factor of the genomic prevalence of ERVs, with respective coefficients of 0.07 and 0.1 (P < 0.05 in both cases). However, this correlation is clearly not sufficient to explain why TRIM accumulation has been so diverse among mammal groups, and therefore, we must also explore the influence of other elements in this process.
One of the factors we believe to be largely influential in the genomic organization and abundance of TRIM genes is the plasticity associated with the evolution of immune-related proteins. It is possible that the main factor driving diversity in the TRIM6/34/5/22 gene cluster of mammals is simply that each lineage has evolved different tools to deal with retrovirus activity. These tools may range from rapid accumulation of TRIM5 and TRIM22 copies to delegation of function to other protein families or perhaps even loss of specific antiretroviral restriction activity altogether in some cases. There may also be ecological components in the direction of evolution that each lineage takes (such as increased risk of retroviral infection), although our data showcase that orders with similar organizations of TRIM6/34/5/22 share few ecological traits, suggesting that this process is largely stochastic. As a group starts to rely more and more on a specific mechanism to counteract retroviral infection, it tends to progressively become dependent on it. Similarly, evolutionary pathways that have been closed long ago can hardly be reactivated, as degeneration has likely already ruined most of the functional properties of the related proteins. This results in patterns of evolution that are mostly consistent between closely related lineages, such as within each order or suborder, but differ greatly among higher groups, something that can indeed be observed in our data (fig. 2).
Intriguingly, we recently described the potential role of heterologous TRIM complexes formed from more than one TRIM paralog in retroviral defense and the expansion of apparent antiviral specificities mediated by two TRIM proteins acting together in retroviral restriction (Ohainle et al. 2020; Twentyman et al. 2023). The dynamic nature of the evolution of this clade of TRIM genes in mammals may suggest that such interdependence of TRIMs and novel antiviral specificities may be a larger, unappreciated aspect of antiviral TRIM biology. More work in this area will define how broadly this might apply as a feature of TRIM evolution.
Specific Antiretroviral Activity in TRIM5-Deficient Species
TRIM5 plays an important role in the innate immune defenses against retroviral invaders due to its ability to engage and restrict the capsid of different virus species (Stremlau et al. 2004; Yap et al. 2004; Saenz et al. 2005; Fletcher et al. 2010; Jáuregui et al. 2012; Yap and Stoye 2013; Ganser-Pornillos and Pornillos 2019). Because TRIM5-mediated restriction is capsid specific, it is thought that positive selection on this gene is reflective of the need for the TRIM5 protein to keep up with the fast-paced mutation rate characteristic of viruses. To attest to the importance of genes from the TRIM6/34/5/22 gene cluster as restriction factors of retroviruses, we performed gene-wide analyses of positive selection using aBSREL (Smith et al. 2015). These analyses have confirmed that TRIM5 and TRIM22 have both been subject to strong positive selection in different groups of mammals in the past. In this context, positive selection might be driven by evolutionary arms race between TRIM genes and virus species. As viruses continuously evolve to escape TRIM restriction, TRIM proteins also experience a selective pressure to change in order to maintain function, resulting in high rates of positive selection. Some have also suggested that balancing selection might play a bigger role in the evolution of TRIM genes than previously appreciated by ensuring high allele diversity within populations. We find that a scenario in which both balancing and positive selection contribute to drive molecular diversification in TRIM proteins in different times and lineages is the most likely. For example, lineages that maintain a single copy of TRIM5 might be subject to strong balancing selection on this locus in order to keep high sequence diversity within populations, whereas recent TRIM expansions are likely to experience positive selection as the duplicated genes adapt to better restrict different viruses.
The finding of broad signals of positive selection in TRIM5 signifies the importance of TRIM5 in the immune capabilities of mammalian species throughout their evolution. However, the TRIM5 locus is also notably missing in some mammal species. For example, cats express a truncated and nonfunctional TRIM5, whereas in dogs, this gene is completely absent (Sawyer et al. 2007; McEwan et al. 2009). Here, we show that many more mammalian groups are missing TRIM5 and, in some cases, also most other genes in the TRIM6/34/5/22 gene cluster (fig. 2A). To account for the lack of TRIM5-mediated antiretroviral activity in TRIM5-deficient species, it has been questioned whether other TRIM genes such as TRIM22 could be fulfilling TRIM5's role as targeted antiviral effectors (Sawyer et al. 2007). In this study, we show that none of the TRIM genes that are closely related to TRIM5 were affected by positive selection in the lineages where TRIM5 is missing, suggesting that they are not acting as TRIM5 substitutes in the species we analyzed. The hypothesis that TRIM22 fulfills the role of TRIM5 in TRIM5-deficient species also does not explain how lineages that lack both genes handle retroviral infection.
Alternatively, it has been proposed that TRIM5-deficient mammals may have not been significantly affected by retrovirus during their evolution, thus allowing for the degeneration of TRIM5 (Sawyer et al. 2007; McEwan et al. 2009). If that is the case, it is possible that some species have not developed replacing mechanisms of specific antiretroviral activity following the loss of TRIM5. Of note, cat cells have been extensively used in in vitro HIV studies precisely because they are permissive to retroviral infection (McEwan et al. 2009). Supporting this hypothesis, a previous genomic analysis of the TRIM5-deficient Canis lupus reported that both the absolute and relative prevalences of ERVs are lower in the genome of this species when compared with that of humans or mice (Lindblad-Toh et al. 2005). In cetaceans, in which the TRIM6/34/5/22 gene cluster has been dismantled (figs. 2 and 3), a similarly low number of ERVs has been reported (Hayward et al. 2015; Zheng et al. 2021). Altogether, this could indicate that retrovirus epidemics may have been less common among TRIM5-deficient lineages when compared with other groups. That said, counting ERVs is complicated and depends greatly on the quality of the analyzed genome (Zheng et al. 2021), meaning that comparisons made with this parameter must be taken with caution. It is also not clear how exactly the history of retrovirus epidemics relates to the deletion of restriction factors in a lineage. For example, it is possible that lineages that maintain a highly effective antiretroviral arsenal in their genome can prevent widespread contamination in the early stages of an epidemic, which could lead to a low ERV genomic prevalence.
Another possibility to explain how TRIM5-deficient species handle retroviral activity is that there are proteins from other gene families that substitute TRIM5's function. In recent years, a myriad of new restriction factors of retroviruses has been characterized in humans (Duggal and Emerman 2012; Colomer-Lluch et al. 2018; Boso and Kozak 2020), constituting what is now known to be a complex web of interactions between the host immune system and invading retroviruses. For example, the APOBEC3 gene has evolved dynamically during mammalian evolution both in terms of copy number and variation in protein sequence, which has led to divergent functions. One APOBEC3 paralog, APOBEC3H, has also been shown to have undergone positive selection in cats, possibly due to an evolutionary arms race against FIV (Zhang et al. 2018). Therefore, it is reasonable to assume that TRIM5-deficient carnivorans are still capable of some level of target-specific retrovirus restriction, although they seem to have stopped relying on TRIM5 for this purpose. Of note, only one carnivoran species in our data set (N. schauinslandi) displays signals of positive selection on TRIM5, suggesting that carnivorans might favor other proteins as specific antiviral effectors.
Conclusion
This study provides a comprehensive view of the evolution of the TRIM6/34/5/22 gene cluster in mammals. By analyzing the topology of a phylogenetic tree built with TRIM sequences obtained from 12 major eutherian orders, we conclude that this gene cluster has most likely originated between the divergence of eutherians and marsupials and the radiation of eutherian superorders. We also show that the pattern of evolution of these genes is dissimilar among eutherian lineages, as both duplications and deletions of TRIM loci have created great diversity in terms of genomic organization. These dissimilarities, our data suggest, highlight the evolutionary plasticity of the innate immune system of mammals. For example, different mammalian lineages have evolved in different ways to handle retroviral infections, either heavily relying on TRIM proteins for this purpose or instead entirely discarding them, possibly in favor of other proteins with similar functional activity. Altogether, our results provide an in-depth view of the evolution of TRIM6/34/5/22 genes, showcasing the incredibly diverse evolutionary histories of this representative family of restriction factors across Eutheria.
Materials and Methods
Phylogenetic Analyses
To retrieve sequence data, we sequentially performed nondiscriminative BLASTP searches (Altschul et al. 1997) in all annotated mammal genomes available in NCBI (https://www.ncbi.nlm.nih.gov/genome/annotation_euk/all/) using the human TRIM6, TRIM34, TRIM5, and TRIM22 proteins as queries. These searches yielded 2,198 protein sequences, which were then aligned with Clustal Omega (Sievers et al. 2011). From the resulting alignment, a phylogenetic tree was created in FastTree (supplementary data S1, Supplementary Material online; Price et al. 2010) and used to separate 1,281 proteins identified as either TRIM6, TRIM34, TRIM5, or TRIM22 (which are the most recent TRIM paralogs and group together in the tree) from the remaining sequences. For the purpose of simplicity, the TRIM12 and TRIM30 genes of rodents were labeled as TRIM5, which they derive from Tareen et al. (2009). These 1,281 TRIM sequences, belonging to 170 different mammal species, were then used to evaluate the integrity and continuity of the TRIM6/34/5/22 gene cluster in each species’ genome. To ensure good quality of data, the initial 170 species were narrowed down to 53 species in which this gene cluster either had its chromosomal location annotated or was unplaced in the genome but available as a single, unfragmented genomic scaffold. This was done to reduce the odds that genes are miscounted in species, which could happen for species that have a TRIM6/34/5/22 gene cluster that we could not account for due to gaps in the assembly. Even though we selected high completeness and well-annotated genomes, this can still be a concern, and it is possible that a few gene copies have been missed.
From previous work regarding the same gene cluster in bats (Fernandes et al. 2022), we knew that TRIM proteins can sometimes be incorrectly annotated on NCBI databases. To investigate the existence of such misidentified sequences in our data set, we generated a new, more accurate phylogenetic tree (supplementary data S2, Supplementary Material online) using IQ-Tree (Nguyen et al. 2015) and looked for mislabeled sequences in the phylogeny. Twenty misidentified sequences were found (see supplementary table S1, Supplementary Material online), mostly cases of sequences annotated as TRIM5, but that unambiguously group with TRIM34 proteins in the tree. These sequences were then correctly labeled and reintroduced in our database. The phylogenetic tree also allowed us to identify two TRIM6–TRIM34 tandem hybrid loci that were incorrectly annotated as TRIM6 genes (LOC114499369 and LOC107135587). It is unclear, however, whether a TRIM6–TRIM34 hybrid protein is truly expressed from these genes, so, for now, we included TRIM6 and TRIM34 proteins from both loci in the data set separately. To rule out the possibility that gene conversion was the cause of the inconsistencies between gene annotation and our phylogeny, we ran exploratory recombination detection analyses on the alignment with recombination detection program (RDP) (using the RDP, MAXCHI, and GENECONV methods; Martin et al. 2021). No signal of recombination was detected.
Finally, we chose a single protein sequence to represent each locus in the final data set (except for the hybrid TRIM6–TRIM34 loci, as mentioned above). The protein chosen for each locus was the longest one that did not exceed 600 amino acids in length and that was not annotated as a “low-quality protein” on NCBI. Unfortunately, the length criteria meant that four genes (LOC123642340, LOC122913544, LOC117029703, and LOC117029704) were left unrepresented in the data set and could not be included in subsequent analyses. The final sequence data set of a single protein per locus was then used to build a new amino acid alignment using the command-line implementation of Clustal Omega (Sievers et al. 2011) in two iterations with default parameters (supplementary data S3, Supplementary Material online). From this alignment, the ML tree available in figure 1 was generated in the W-IQ-TREE web server (Trifinopoulos et al. 2016) using the JTT substitution model with five gamma categories. This model was selected as the best fit for our data according to the corrected Akaike information criterion (AICc) estimator in MEGA-X (Kumar et al. 2018).
To verify that the tree topology is correct and that no sequence has been misidentified in the phylogeny, we used the same alignment from which the ML was derived to build a tree using Bayesian Inference in MrBayes (Ronquist et al. 2012) with default parameters (supplementary data S4, Supplementary Material online). The Bayesian tree confirmed the phylogenetic relations between the TRIM6/34/5/22 genes and corroborated the previous classification of all TRIM sequences included in our data set.
Synteny Map
The synteny of the TRIM6/34/5/22 gene cluster was mapped for key species using data obtained from NCBI's Genome Data Viewer (fig. 3). We included representative species with distinct organizations of TRIM6/34/5/22 in the synteny map in order to provide a broad view of the diverse syntenies of this region in mammalian genomes. A version of the synteny map including more species is provided in the supplementary table S2, Supplementary Material online. During the examination of the genomic annotation of species we considered for the synteny map, we encountered three TRIM genes annotated as functional that were not retrieved by our initial BLAST searches (LOC116151278, LOC244183, and LOC106780875). All sequences derived from these genes are very short (<300 amino acids, compared with the usual length of around 500 amino acids of other TRIM proteins) and align very poorly with other TRIM transcripts (supplementary data S5, Supplementary Material online). It is unclear whether these sequences truly represent functional TRIM genes, and thus, they were not included in our data set.
Positive Selection Analyses
To determine the effects of selection on TRIM6, TRIM34, TRIM5, and TRIM22, we ran separate aBSREL (Smith et al. 2015) analyses for each gene. Alignments were created with Clustal Omega (Sievers et al. 2011) using the previously obtained amino acid sequences. Some sequences had to be removed from the alignments prior to the analyses because they contained either poorly conserved regions or early stop codons, both of which can be indicative of either pseudogenization or sequencing errors that could compromise the quality of the analyses. The accession number for all removed sequences is available in supplementary table S3, Supplementary Material online. Then, the phylogenetic tree previously built using all TRIM6/34/5/22 sequences was split into subsections corresponding to each gene and passed as input to the command-line implementation of aBSREL on HyPhy (Smith et al. 2015). All branches were considered foreground branches for the analyses. Because of the unlikely grouping of two sequences in the phylogenetic tree of TRIM22 (XP_036077536.1 from R. aegyptiacus and XP_023501633.1 from Eq. caballus), we included an additional iteration of aBSREL for this gene using a tree in which these branches were relocated as sister clades of their paralog pairs (supplementary fig. S1, Supplementary Material online). The remaining trees closely matched the expected phylogeny for the analyzed species (Upham et al. 2019).
Supplementary Material
Acknowledgments
We thank Prof. Harmit Malik for his helpful comments on an earlier draft of this manuscript. Work supported by National Funds through Fundação para a Ciência e a Tecnologia (FCT) in the scope of the project UIDP/50027/2020. FCT also supported the Investigator grant of PJE (CEECIND/CP1601/CT0005).
Contributor Information
Alexandre P Fernandes, CIBIO, Centro de Investigação em Biodiversidade e Recursos Genéticos, InBIO Laboratório Associado, Campus de Vairão, Universidade do Porto, Vairão, Portugal; Departamento de Biologia, Faculdade de Ciências, Universidade do Porto, Porto, Portugal; BIOPOLIS Program in Genomics, Biodiversity and Land Planning, CIBIO, Campus de Vairão, Vairão, Portugal.
Molly OhAinle, Division of Immunology and Molecular Medicine, Department of Molecular and Cell Biology, University of California Berkeley, Berkeley, California, USA.
Pedro J Esteves, CIBIO, Centro de Investigação em Biodiversidade e Recursos Genéticos, InBIO Laboratório Associado, Campus de Vairão, Universidade do Porto, Vairão, Portugal; Departamento de Biologia, Faculdade de Ciências, Universidade do Porto, Porto, Portugal; BIOPOLIS Program in Genomics, Biodiversity and Land Planning, CIBIO, Campus de Vairão, Vairão, Portugal.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Data Availability
All scripts used for our analyses are available at https://github.com/aleferna12/TRIM-Evolution. Data files including alignments and Newick phylogenetic trees are available in the Supplementary material online and in the linked repository.
Literature Cited
- Águeda-Pinto A, et al. 2019. Not so unique to primates: the independent adaptive evolution of TRIM5 in Lagomorpha lineage. PLoS One 14:e0226202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad HI, et al. 2022. Positive selection drives the adaptive evolution of mitochondrial antiviral signaling (MAVS) proteins-mediating innate immunity in mammals. Front Vet Sci. 8:814765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, et al. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Álvarez-Carretero S, et al. 2022. A species-level timeline of mammal evolution integrating phylogenomic data. Nature 602:263–267. [DOI] [PubMed] [Google Scholar]
- Boso G, Kozak CA. 2020. Retroviral restriction factors and their viral targets: restriction strategies and evolutionary adaptations. Microorganisms 8:1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boso G, et al. 2019. Evolution of the rodent Trim5 cluster is marked by divergent paralogous expansions and independent acquisitions of TrimCyp fusions. Sci Rep. 9:11263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthagena L, et al. 2009. Human TRIM gene expression in response to interferons. PLoS One 4:e4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomer-Lluch M, Ruiz A, Moris A, Prado JG. 2018. Restriction factors: from intrinsic viral restriction to shaping cellular immunity against HIV-1. Front Immunol. 9:2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daugherty MD, Malik HS. 2012. Rules of engagement: molecular insights from host-virus arms races. Annu Rev Genet. 46:677–700. [DOI] [PubMed] [Google Scholar]
- Di Pietro A, et al. 2013. TRIM22 inhibits influenza A virus infection by targeting the viral nucleoprotein for degradation. J Virol. 87:4523–4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggal NK, Emerman M. 2012. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat Rev Immunol. 12:687–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre-Walker A. 2006. The genomic rate of adaptive evolution. Trends Ecol Evol. 21:569–575. [DOI] [PubMed] [Google Scholar]
- Fernandes AP, Águeda-Pinto A, Pinheiro A, Rebelo H, Esteves PJ. 2022. Evolution of TRIM5 and TRIM22 in bats reveals a complex duplication process. Viruses 14:345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher AJ, Hué S, Schaller T, Pillay D, Towers GJ. 2010. Hare TRIM5α restricts divergent retroviruses and exhibits significant sequence variation from closely related lagomorpha TRIM5 genes. J Virol. 84:12463–12468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley NM, et al. 2023. A genomic timescale for placental mammal evolution. Science 380:eabl8189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganser-Pornillos BK, Pornillos O. 2019. Restriction of HIV-1 and other retroviruses by TRIM5. Nat Rev Microbiol. 17:546–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao B, Duan Z, Xu W, Xiong S. 2009. Tripartite motif-containing 22 inhibits the activity of hepatitis B virus core promoter, which is dependent on nuclear-located RING domain. Hepatology 50:424–433. [DOI] [PubMed] [Google Scholar]
- Hatziioannou T, Perez-Caballero D, Yang A, Cowan S, Bieniasz PD. 2004. Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5alpha. Proc Natl Acad Sci U S A. 101:10774–10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward A, Cornwallis CK, Jern P. 2015. Pan-vertebrate comparative genomics unmasks retrovirus macroevolution. Proc Natl Acad Sci U S A. 112:464–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J, Gifford RJ, Sato K. 2020. Retroviruses drive the rapid evolution of mammalian genes. Proc Natl Acad Sci U S A. 117:610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwama RE, Moran Y. 2023. Origins and diversification of animal innate immune responses against viral infections. Nat Ecol Evol. 7:182–193. [DOI] [PubMed] [Google Scholar]
- Jáuregui P, et al. 2012. Ovine TRIM5α can restrict visna/maedi virus. J Virol. 86:9504–9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing H, et al. 2019. Nuclear localization signal in TRIM22 is essential for inhibition of type 2 porcine reproductive and respiratory syndrome virus replication in MARC-145 cells. Virus Genes. 55:660–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keckesova Z, Ylinen LMJ, Towers GJ. 2004. The human and African green monkey TRIM5alpha genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc Natl Acad Sci U S A. 101:10780–10785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35:1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, et al. 2005. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438:803–819. [DOI] [PubMed] [Google Scholar]
- Martin DP, et al. 2021. RDP5: a computer program for analyzing recombination in, and removing signals of recombination from, nucleotide sequence datasets. Virus Evol. 7:veaa087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan WA, et al. 2009. Truncation of TRIM5 in the Feliformia explains the absence of retroviral restriction in cells of the domestic cat. J Virol. 83:8270–8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan CC, et al. 2013. Heterogeneous models place the root of the placental mammal phylogeny. Mol Biol Evol. 30:2145–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman RM, et al. 2006. Balancing selection and the evolution of functional polymorphism in Old World monkey TRIM5alpha. Proc Natl Acad Sci U S A. 103:19134–19139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohainle M, et al. 2020. TRIM34 restricts HIV-1 and SIV capsids in a TRIM5α-dependent manner. PLoS Pathog. 16:e1008507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozato K, Shin D-M, Chang T-H, Morse HC III. 2008. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 8:849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajsbaum R, et al. 2014. Unanchored K48-linked polyubiquitin synthesized by the E3-ubiquitin ligase TRIM6 stimulates the interferon-IKKε kinase-mediated antiviral response. Immunity 40:880–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, et al. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61:539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saenz DT, Teo W, Olsen JC, Poeschla EM. 2005. Restriction of feline immunodeficiency virus by Ref1, Lv1, and primate TRIM5alpha proteins. J Virol. 79:15175–15188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, Cairo S, Fontanella B, Ballabio A, Meroni G. 2008. Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol Biol. 8:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Emerman M, Malik HS. 2007. Discordant evolution of the adjacent antiretroviral genes TRIM22 and TRIM5 in mammals. PLoS Pathog. 3:e197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Wu LI, Emerman M, Malik HS. 2005. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci U S A. 102:2832–2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si Z, et al. 2006. Evolution of a cytoplasmic tripartite motif (TRIM) protein in cows that restricts retroviral infection. Proc Natl Acad Sci U S A. 103:7454–7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, et al. 2015. Less is more: an adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol Biol Evol. 32:1342–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stremlau M, et al. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848–853. [DOI] [PubMed] [Google Scholar]
- Sun Y, Wei Z, Li N, Zhao Y. 2013. A comparative overview of immunoglobulin genes and the generation of their diversity in tetrapods. Dev Comp Immunol. 39:103–109. [DOI] [PubMed] [Google Scholar]
- Tareen SU, Sawyer SL, Malik HS, Emerman M. 2009. An expanded clade of rodent Trim5 genes. Virology 385:473–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifinopoulos J, Nguyen L-T, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44:W232–W235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twentyman J, Khalifeh A, Felton AL, Emerman M, Ohainle M. 2023. Primate TRIM34 is a broadly-acting, TRIM5-dependent lentiviral restriction factor. Retrovirology 20:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upham NS, Esselstyn JA, Jetz W. 2019. Inferring the mammal tree: species-level sets of phylogenies for questions in ecology, evolution, and conservation. PLoS Biol. 17:e3000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent M, Sparrer KMJ, Gack MU. 2018. TRIM proteins and their roles in antiviral host defenses. Annu Rev Virol. 5:385–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wein T, Sorek R. 2022. Bacterial origins of human cell-autonomous innate immune mechanisms. Nat Rev Immunol. 22:629–638. [DOI] [PubMed] [Google Scholar]
- Williams FP, Haubrich K, Perez-Borrajero C, Hennig J. 2019. Emerging RNA-binding roles in the TRIM family of ubiquitin ligases. Biol Chem. 400:1443–1464. [DOI] [PubMed] [Google Scholar]
- Yang W, Gu Z, Zhang H, Hu H. 2020. To TRIM the immunity: from innate to adaptive immunity. Front Immunol. 11:02157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap MW, Nisole S, Lynch C, Stoye JP. 2004. Trim5alpha protein restricts both HIV-1 and murine leukemia virus. Proc Natl Acad Sci U S A. 101:10786–10791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap MW, Stoye JP. 2013. Apparent effect of rabbit endogenous lentivirus type K acquisition on retrovirus restriction by lagomorph Trim5αs. Philos Trans R Soc Lond B Biol Sci. 368:20120498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap MW, Young GR, Varnaite R, Morand S, Stoye JP. 2020. Duplication and divergence of the retrovirus restriction gene Fv1 in Mus caroli allows protection from multiple retroviruses. PLoS Genet. 16:e1008471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, et al. 2018. Feline APOBEC3s, barriers to cross-species transmission of FIV? Viruses 10:186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J, Wang J, Gong Z, Han G-Z. 2021. Molecular fossils illuminate the evolution of retroviruses following a macroevolutionary transition from land to water. PLoS Pathog. 17:e1009730. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All scripts used for our analyses are available at https://github.com/aleferna12/TRIM-Evolution. Data files including alignments and Newick phylogenetic trees are available in the Supplementary material online and in the linked repository.