Summary
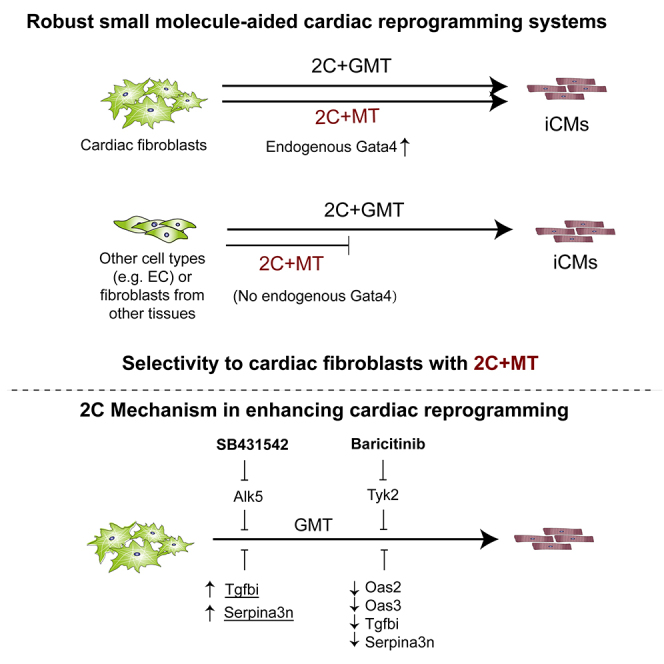
Direct cardiac reprogramming to induce cardiomyocyte-like cells, e.g., by GMT (Gata4, Mef2c and Tbx5), is a promising route for regenerating damaged heart in vivo and disease modeling in vitro. Supplementation with additional factors and chemical agents can enhance efficiency but raises concerns regarding selectivity to cardiac fibroblasts and complicates delivery for in situ cardiac reprogramming. Here, we screened 2000 chemicals with known biological activities and found that a combination of 2C (SB431542 and Baricitinib) significantly enhances cardiac reprogramming by GMT. Without Gata4, MT (Mef2c and Tbx5) plus 2C could selectively reprogram cardiac fibroblasts with enhanced efficiency, kinetics, and cardiomyocyte function. Moreover, 2C significantly enhanced cardiac reprogramming in human cardiac fibroblasts. 2C synergistically enhances cardiac reprogramming by inhibiting Alk5, Tyk2 and downregulating Oas2, Oas3, Serpina3n and Tgfbi. 2C enables selective and robust cardiac reprogramming that can greatly facilitate disease modeling in vitro and advance clinical therapeutic heart regeneration in vivo.
Subject areas: Cardiovascular medicine, Cell biology
Graphical abstract
Highlights
-
•
Combining ALK5 and JAKs/TYK2 inhibitors (2C) greatly improves cardiac reprogramming
-
•
Cardiomyocyte-like cells induced with 2C possess enhanced functional properties
-
•
Gata4 is dispensable for high-efficiency reprogramming in the presence of 2C
-
•
Cardiac fibroblast selectivity is achieved by removing Gata4 in the presence of 2C
Cardiovascular medicine; Cell biology
Introduction
Due to the limited regenerative capacity of the adult mammalian heart,1,2 direct cardiac reprogramming has emerged as a promising therapeutic strategy for replenishing injured heart tissues by converting scar (fibroblasts) into functional heart cells (cardiomyocytes).3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18 However, low efficiency in the induction of functional cardiomyocytes (CM) has hampered its broad application. To address this issue, several groups have developed cocktails that stimulate cardiomyocyte differentiation by adding one or more transcription factors to GMT, such as Hand2,6 Hand2+Nkx2.5,19 Akt1+Hand2,15 or PHF7.18 Although effective in murine cells, cardiac reprogramming in human cells requires a greater number of additional reprogramming factors,4,8,9,10,12 which increases the difficulty of in vivo delivery.
Compounding the complexity of this issue, different combinations of GMT-based cardiac reprogramming factors have been used to successfully reprogram a broad range of fibroblasts in mice, such as those derived from embryos19,20,21,22 or tail-tips,3,13,15,16,22,23,24,25 raising the concern that direct delivery of GMT in vivo may result in severe off-target effects, especially reprogramming cells in other organs into beating cardiomyocytes. Although supplementing GMT with additional transcription factors is a reasonable strategy for enhancing the efficiency of cardiac reprogramming, the increased difficulty of in vivo delivery and substantial risks of reprogramming cells other than cardiac fibroblasts (CF) suggests the need for a robust and CF-specific reprogramming system to advance the therapeutic application of this technology in vivo. The expression of cardiogenic genes (i.e., Gata4, Mef2c, Tbx20) in CF has been shown to contribute to heart development and repair,26 suggesting that the close examination of the cardiogenic transcriptomic signature in CF can facilitate the assembly of a well-tailored combination of factors for the selective reprogramming of CF, but not fibroblasts from other tissues.
In addition to transcription factors, small molecules have also been widely applied to manipulate cell fate or identity,27 such as in chemically induced pluripotent stem cells (CiPSCs),28,29,30 chemically induced neurons (CiNs),31,32 extended pluripotent stem cells (EPSCs),33 human chemically induced pluripotent stem cells (hCiPS),34 and in the long-term maintenance of primary human hepatocytes in vitro.35 Moreover, several small molecules have been found to enhance cardiac reprogramming efficiency, including the ALK5 inhibitor, SB43154220; ALK4/5/7 inhibitor, A83-0122; ROCK1/2 inhibitor, Y-2763222; Mll1 H3K4 methyltransferase inhibitor, MM40836; the WNT inhibitor, XAV9394; the Notch inhibitor, DAPT17; and IMAP, comprising IGF-1 (insulin-like growth factor-1), MM589 (an Mll1 inhibitor), A83–01 (a transforming growth factor β inhibitor), and PTC-209 (a Bmi1 inhibitor).37 Despite the development of these chemicals or chemical combinations, overall reprogramming efficiency remains low, suggesting that further advances are needed in identifying signaling pathways or signaling pathway networks that can be modulated to promote cardiac reprogramming.
In this study, we screen a panel of chemical inhibitors or combinations of inhibitors for activity in signaling pathways required for the epigenetic regulation of cell identity, and subsequently developed a high-efficiency cardiac reprogramming cocktail, 2C+MT, that selectively targets CF for reprogramming, but not fibroblasts from skin, tail-tip, brain, lung, or liver. These findings open the possibility of inducing direct reprogramming in situ without harming off-target cell types.
Results
The 2C combination boosts cardiac reprogramming
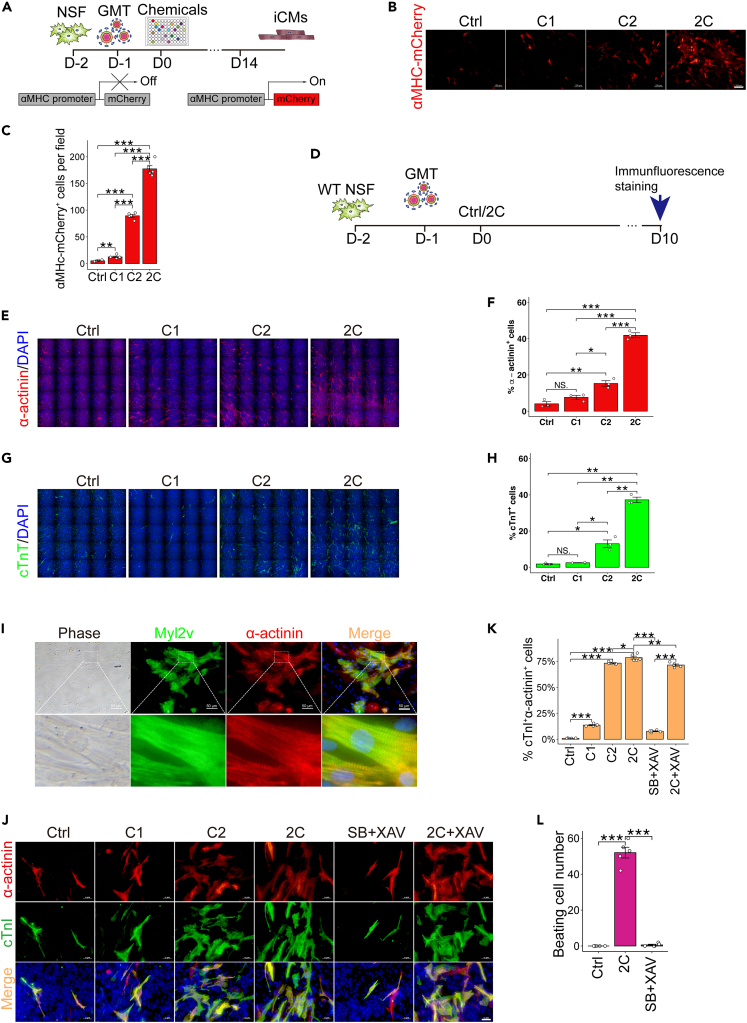
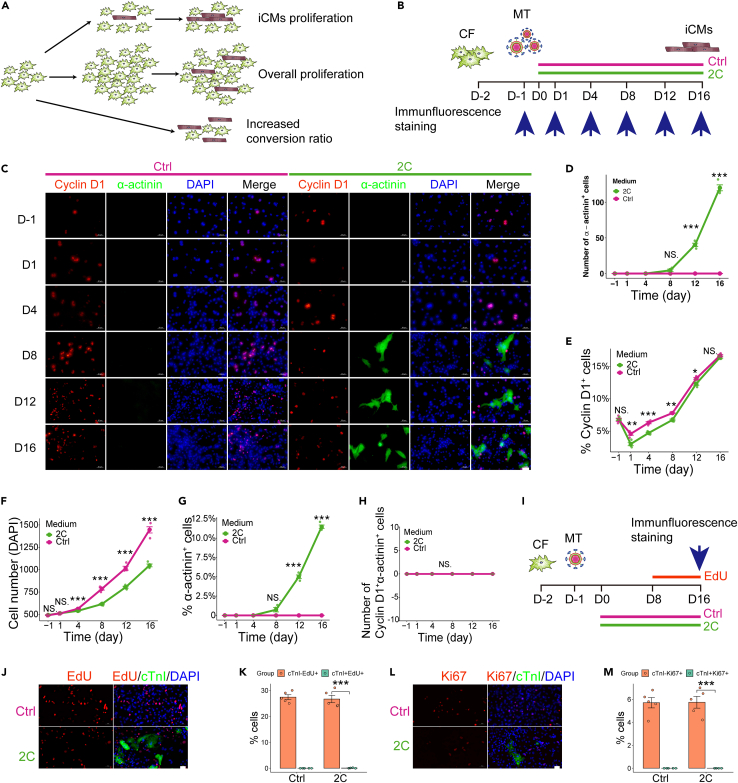
To identify chemicals that can enhance cardiac reprogramming, we screened neonatal skin fibroblasts38 (NSF) isolated from transgenic mice expressing mCherry driven by the cardiomyocyte-specific promoter, αMHC.3,39 In total, ∼2000 compounds from both commercially available and proprietary in-house libraries were tested for their effects in NSF cells infected with GMT (Gata4, Mef2c, and Tbx5)-expressing lentivirus. The potentially induced cardiomyocyte-like cells (iCMs) were detected by the activation of αMHC-mCherry at two weeks post treatment (Figure 1A). Reprogramming efficiency was quantified by counting mCherry-positive cells or immunostaining instead of flow cytometric analysis, since in situ counting cell numbers can significantly get rid of false positive cells by checking cardiomyocyte-like morphology (rod-shaped with increased size than Ctrl or unconverted fibroblasts or even with well-organized sarcomere), and by avoiding the bias of trypsin that selectively dissociates fibroblasts than iCMs before flow cytometric sorting analysis (Video S1). This screen identified two compounds that appeared to synergistically enhance cardiac reprograming. In the DMSO-treated control group, approximate quantification by fluorescence microscopy revealed an average of 5 ɑMHC-mCherry+ cells in each field of view, while an average of 13.7 αMHC-mCherry+ cells were detected in the C1 (SB431542) group, and an average of 87.3 induced cells per field of view were observed in the C2 (Baricitinib) group. By contrast, 178.3 αMHC-mCherry+ cells per field of view were observed in the 2C (SB431542 and Baricitinib) combination group (Figures 1B and 1C). In wild-type NSF (Figure 1D), we performed immunofluorescence staining for α-actinin (sarcomeric alpha actinin, depicted in Figure 1E and quantified in 1F). Our findings indicate that 2C+GMT induced 41.9% α-actinin+ cells, whereas the NSF control, DMSO control, C1, and C2 treatments resulted in substantially lower proportions of α-actinin+ cells at 0%, 4.1%, 7.6%, and 15.3%, respectively. In Figures 1G and 1H, 2C+GMT induced a 37.2% increase in cTnT+ (cardiac troponin T) cells, whereas the NSF control, DMSO control, C1, and C2 treatments resulted in lower percentages of cTnT+ cells at 0%, 1.8%, 2.5%, and 13%, respectively. Furthermore, the large majority of iCMs induced by 2C+GMT co-expressed Myl2v (myosin light chain 2) and α-actinin and had a well-defined sarcomere structure (Figure 1I).
Figure 1.
The 2C combination boosts cardiac reprogramming
(A) Schematic representation of the screening strategy for chemicals that enhance cardiac reprogramming.
(B and C) Representative images (B) and quantification (C) for αMHc-mCherry+ cells 2 weeks after transduction with GMT (Gata4, Mef2c and Tbx5), in combination with Ctrl (DMSO), C1 (SB431542), C2 (Baricitinib), or 2C (SB431542+Baricitinib) treatment of neonatal mouse skin fibroblasts (NSF). n = 5 independent experiments. Scale bars, 100 μm.
(D) Schematic representation of iCMs reprogramming in wild type mouse skin fibroblasts.
(E and F) Representative immunocytochemistry images (E) and quantification (F) of α-actinin+ cells from NSF 10 days after transduction with GMT, in combination with Ctrl (DMSO), C1, C2, and 2C treatment. n = 3 independent experiments.
(G and H) Representative immunocytochemistry images (G) and quantification (H) of cTnT+ cells from NSF 10 days after transduction with GMT, in combination with Ctrl (DMSO), C1, C2, and 2C treatment. n = 3 independent experiments.
(I) Representative immunocytochemistry images of Myl2v+ and α-actinin+ cells 2 weeks after transduction with GMT, in combination with 2C treatment of NSF. Scale bars, 50 μm.
(J and K) Representative immunocytochemistry images (J) and quantification (K) of cTnI+α-actinin+ cells from NSF 14 days after transduction with GMT, in combination with Ctrl (DMSO), C1, C2, 2C, SB+XAV (SB431542+XAV939), and 2C+XAV (2C+XAV939) treatment. n = 5 independent experiments.
(L) Quantification of the number of spontaneous beating cells 2 weeks after transduction with GMT, in combination with Ctrl, 2C, or SB+XAV treatment in NSF. n = 5 independent experiments. All data are presented as the means ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus the relevant control. NS., not significant.
Further optimization showed that 2 μM concentrations of C1 and C2 were the most effective dosage for inducing the highest proportion of cTnT+ cells (Figures S1A–S1D), while the highest efficiency was obtained through continuous treatment during induction, indicated by the high percentage of α-actinin+ cells and the number of spontaneous beating cells (a reliable readout of iCMs functionality) (Figure S1E). Based on our evidence of consistent co-expression of cTnI (cardiac troponin I), cTnT, and α-actinin in iCMs (Figures S2A and S2B), we used these markers interchangeably throughout our study due to the low availability of each antibody.
It was previously reported that the SB431542+XAV939 (SB+XAV) combination could significantly increase cardiac reprogramming efficiency.4 In comparison with 2C, SB+XAV showed moderate improvement on the percentage of cTnI+ and α-actinin+ cells (Figure 1J, quantified in 1K). Regarding the number of spontaneous beating cells, 2C was much more efficient (Figure 1L, Videos S2, S3, and S4). Taken together, these results showed that the 2C cocktail could boost the efficiency of mouse cardiomyocyte reprogramming over that of vehicle control.
2C+MT selectively reprograms cardiac fibroblasts
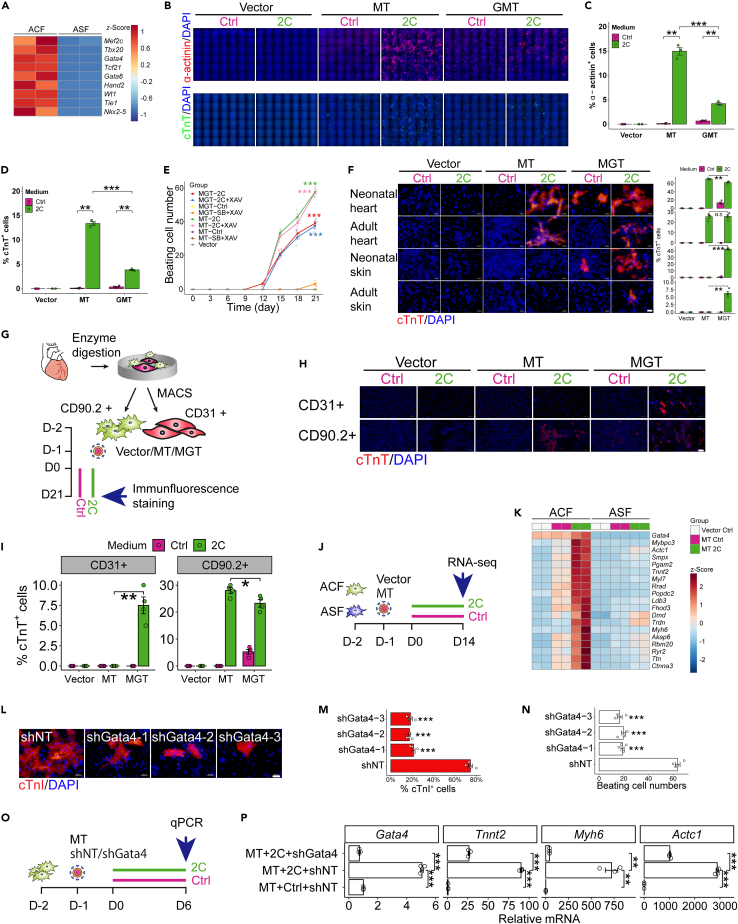
Besides the above-mentioned improvement of cardiac reprogramming in skin fibroblasts, we also tested the effect of 2C in cardiac fibroblasts. Adult mouse cardiac fibroblasts and neonatal mouse cardiac fibroblasts were isolated according to a previously described protocol40 and confirmed by immunofluorescence staining of cTnT and α-actinin to exclude any contamination with cardiomyocytes (Figure S3). Consistent with previous report,26 our RNA-seq results showed that several cardiogenic genes (i.e., Gata4, Mef2c, Tbx20, Hand2, Nkx2-5) were expressed in adult mouse cardiac fibroblasts (ACF) compared to adult mouse skin fibroblasts (ASF) (Figure 2A). Inspired by this result, 2C was tested with each individual transcription factor and each combination of two factors. Using the cTnT marker, we found that 2C could also enhance cardiac reprogramming in cells infected with only MT (Mef2c and Tbx5)-expressing lentivirus, but not other combinations (Figure S4A, quantified in S4B). The combination of MT with 2C resulted in the induction of 15% α-actinin+ cells and 13.4% cTnT+ cells (shown in Figure 2B and quantified in Figures 2C and 2D). In contrast, treatments with GMT alone yielded only 0.6% α-actinin+ cells and 0.4% cTnT+ cells. To further fine-tune the stoichiometry of Mef2c (M) and Tbx5 (T), we expressed the M and T transcription factors in distinct ratios (Figures S5A and S5B) using a 2A “self-cleaving” peptide system. 2A peptides are short (18–22 amino acids) viral oligopeptides that mediate cleavage of polypeptides during translation in eukaryotic cells.41,42,43 We found that the bicistronic M-T2A-T combination resulted in higher reprogramming efficiency than T-P2A-M, M-P2A-T or M-E2A-T, based on the increased percentage of α-actinin+ cells and the greater number of spontaneous beating cells (Figures S5C–S5E). Further comparison of the 2C+MT (2C+M-T2A-T) treatment with the tri-cistronic MGT construct,24 in various combinations with 2C and/or SB+XAV, showed that the 2C+M-T2A-T combination induced significantly more spontaneous beating cells than any other combination, including 2C+MGT (Figure 2E, Videos S5 and S6).
Figure 2.
2C+MT selectively reprograms cardiac fibroblasts
(A) Cardiogenic genes expression in adult mouse cardiac fibroblasts (ACF) and adult mouse skin fibroblasts (ASF), determined by RNA-seq. n = 2 independent experiments.
(B–D) Representative immunocytochemistry images (B) and quantification of α-actinin+ cells (C), cTnT+ cells (D) 10 days after transduction with vector, MT (M+T), or GMT (G+M+T), in combination with Ctrl (DMSO) or 2C treatment of neonatal mouse cardiac fibroblasts. n = 3 independent experiments.
(E) Quantification of the number of spontaneous beating cardiomyocytes over time for groups transduced with vector control, MT (M-T2A-T), or MGT (M-P2A-G-T2A-T) in various combinations with Ctrl, 2C, SB+XAV, or 2C+XAV treatment. n = 5 independent experiments.
(F) Representative immunocytochemistry images and quantification of cTnT+ cells 3 weeks post transduction with MT (M-T2A-T) or MGT, in combinations with Ctrl or 2C treatment of various fibroblasts derived from indicated organs/tissues. n = 5 independent experiments. Scale bars, 50 μm.
(G) Schematic representation of CD90.2+ cardiac fibroblasts, CD31+ endothelial cells sorting by MACS (magnetic-activated cell sorting) and cardiac reprogramming.
(H and I) Representative immunocytochemistry images (H) and quantification (I) of cTnT+ cells 3 weeks post transduction with MT or MGT, in combinations with Ctrl or 2C treatment in CD90.2+ fibroblasts or CD31+ endothelial cells. n = 4 independent experiments. Scale bars, 50 μm.
(J) Schematic representation of cardiac reprogramming and in ACF and ASF.
(K) Heatmap of the relative expression of a set of cardiomyocyte-specific genes in vector+Ctrl, MT+Ctrl, or MT+2C treated cells derived from ACF and ASF, 2 weeks post transduction. n = 2 independent experiments.
(L–N) Representative immunocytochemistry images (L) and quantification (M) of cTnI+ cells and the number of spontaneous beating cells (N) 3 weeks after transduction with MT and shNT (Non-target control) or shGata4 in neonatal cardiac fibroblasts. All groups were treated with 2C. n = 4 independent experiments. Scale bars, 50 μm.
(O) Schematic representation of cardiac reprogramming in neonatal mouse cardiac fibroblasts treated with shNT or shGata4.
(P) Relative mRNA expression level of Gata4, Tnnt2, Myh6, and Actc1 in neonatal cardiac fibroblasts treated with shNT or shGata4 determined by qPCR. n = 3 independent experiments. All data are presented as the means ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus the relevant control. NS., not significant.
Testing a range of cell types showed that 2C+MT (2C+M-T2A-T) could selectively reprogram cTnT+ iCMs from mouse neonatal cardiac fibroblasts and adult cardiac fibroblasts at 70.5% and 25.5% respectively (Figure 2F). However, either negligible or no reprogramming effects of 2C+MT were observed in mouse neonatal skin fibroblasts or adult skin fibroblasts. By contrast, 2C+MGT (tri-cistronic MGT) treatment led to efficient reprogramming in all these fibroblast types. We tested various fibroblasts derived from different tissues/organs (skin, heart, brain, lung, liver) with consistent results (Figure S6). To further determine if 2C+MT selectively reprogram cardiac fibroblasts, we sorted CD90.2+ cardiac fibroblasts and CD31+ endothelial cells by MACS (magnetic-activated cell sorting) and performed iCMs reprogramming (Figure 2G). Our result showed that 2C+MT efficiently induced iCMs from CD90.2+ cardiac fibroblasts, while no cTnT+ iCMs could be detected in CD31+ endothelial cells (Figure 2H, quantified in 2I). These results indicated that 2C+MGT might be widely applicable for in vitro cell reprogramming in different fibroblasts, especially skin fibroblasts, which are easily accessible. However, the markedly stronger selectivity of 2C+MT suggested its potential in vivo therapeutic application for expanding beating cardiomyocytes while avoiding fibroblasts conversion in other tissues, such as skin.
To further validate the selectivity of 2C+MT in cardiac fibroblast, we performed RNA-seq in ACF and ASF (Figure 2J). 2C+MT treatment led to the up-regulation of a panel of cardiac genes (i.e., Gata4, Actc1, Tnnt2, Myh6, Ryr2, and Ttn) in cardiac fibroblasts, while no significant up-regulation of these genes was detected in skin fibroblasts (Figure 2K). To validate the hypothesis that 2C+MT enables selective cardiac reprogramming in cardiac fibroblasts dependent on the expression of endogenous cardiogenic genes, we induced Gata4 knockdown in neonatal mouse cardiac fibroblasts by shRNA, which resulted in significantly fewer cTnI+ (Figure 2L, quantified in 2M) and spontaneous beating cells (Figure 2N) induced by 2C+MT compared to that in the non-target shRNA controls (shNT). Moreover, we performed qPCR 6 days post induction to determine the expression of cardiac genes (Figure 2O). 2C+MT significantly up-regulated the expression of Gata4, Tnnt2, Myh6, and Actc1, all of which were down-regulated by the knockdown of Gata4 (Figure 2P). These results indicated that endogenous Gata4 expression in cardiac fibroblasts can be further enhanced/activated by 2C to enable selective cardiac reprogramming.
To determine if both C1 and C2 are essential to substitute Gata4 in the GMT treatment, we treated mouse neonatal cardiac fibroblasts infected with MT-expressing lentivirus with C1, C2, 2C, SB+XAV, 2C+XAV (2C+XAV939), or DMSO (Ctrl). Quantification of cTnT+ cells and spontaneous beating cells revealed that MT in combination with either C1, C2, or SB+XAV could induce cTnT+ cells at low efficiency (Figures S7A and S7B), but spontaneous beating iCMs could only be detected in the presence of 2C, and that XAV did not further enhance the effects of 2C (Figure S7C). These results thus indicated that 2C synergistically boosts the efficiency and kinetics of MT-mediated cardiac reprogramming, selectively in cardiac fibroblasts, and is essential for inducing spontaneous beating cells.
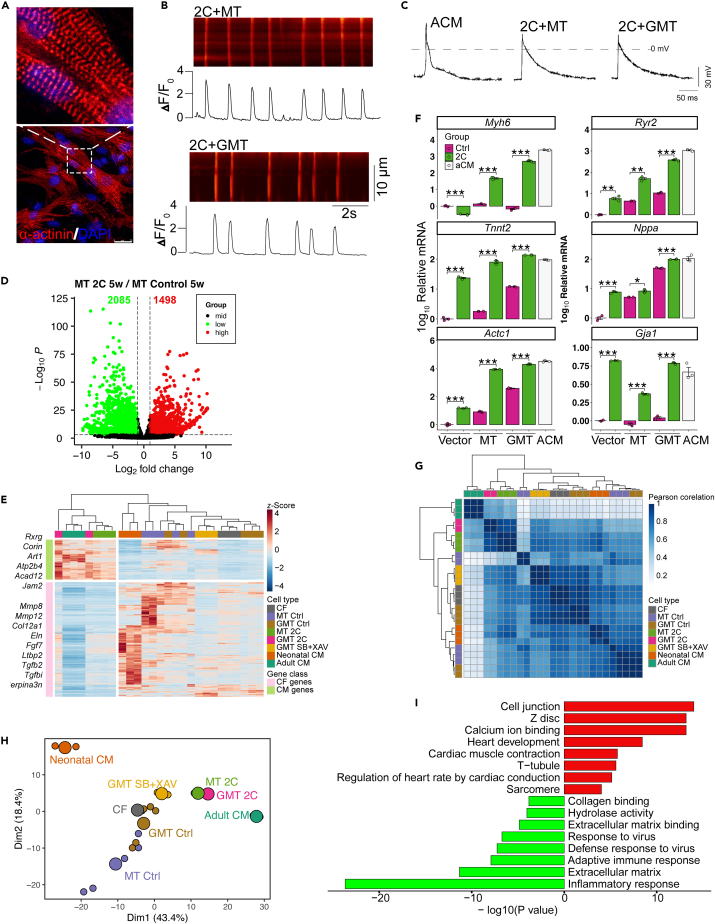
2C+MT-reprogrammed induced cardiomyocyte-like cells functionally and transcriptomically resemble adult cardiomyocytes
Morphological observations confirmed that sarcomere structure was well-organized in iCMs induced by 2C+MT (Figure 3A, Video S7), while 2C+MT or 2C+GMT induced iCMs with spontaneous calcium transients (Figure 3B, Video S8) and action potential (Figure 3C). To better understand how 2C affects cardiac fibroblasts reprogramming, we used RNA-seq for the transcriptomic profiling of iCMs induced by 2C or other combinations. In total, 1498 genes were differentially up-regulated in 2C treatments while 2085 genes were differentially down-regulated at 5 weeks post induction (Figure 3D). Transcriptomic profiling showed that 2C treatment led to significantly higher cardiomyocyte-specific gene expression than that in control groups (i.e., control or SB+XAV) (Figure 3E), while fibroblast-related genes were significantly down-regulated compared with their expression in SB+XAV treatments. Subsequent qPCR analysis of cardiomyocyte identity and functional markers also confirmed that iCMs derived from mouse neonatal cardiac fibroblasts expressing MT, GMT, or the empty vector control, exposure to 2C resulted in significant up-regulation of the cardiomyocyte-specific sarcomere genes (Myh6, Tnnt2 and Actc1) and the functional markers (Ryr2, Nppa and Gja1) (Figure 3F).
Figure 3.
2C+MT-reprogrammed iCMs Functionally and Transcriptomically Resemble Adult Cardiomyocytes
(A) Representative immunocytochemistry images of 2C+MT-induced iCMs cultured for 5 weeks display well-organized sarcomere structure. Scale bars, 25 μm.
(B) 2C+MT and 2C+GMT-induced iCMs cultured for 5 weeks show spontaneous Ca2+ oscillation. n = 3–5 independent experiments.
(C) 2C+MT- or 2C+GMT-induced iCMs cultured for 5 weeks display action potentials resembling those of adult mouse ventricular cardiomyocytes. n = 3–5 independent experiments.
(D) Volcano map of differentially expressed genes (DEGs) between 2C and control (Ctrl) treatment on MT-transduced cardiac fibroblasts 5 weeks post induction. n = 2–5 independent experiments.
(E) Relative expression of a set of cardiomyocytes (CM) and cardiac fibroblast-related (CF) genes determined by RNA-seq. n = 2–5 independent experiments.
(F) Relative mRNA level of cardiomyocyte-specific gene expression in cells transduced with vector control, MT, or GMT in combination with Ctrl (DMSO) or 2C for 5 weeks, determined by qPCR. ACM represents adult mouse ventricular cardiomyocyte. n = 3 independent experiments.
(G) Pearson correlation analysis of DEGs between 2C and control (Ctrl) treatment on 5-week cultured MT-transduced cardiac fibroblasts. n = 2–5 independent experiments.
(H) Principal component analysis (PCA) of DEGs between 2C and control (Ctrl) treatment on MT-transduced cardiac fibroblasts show that 2C+MT- or 2C+GMT-induced iCMs resemble adult cardiomyocyte. n = 2–5 independent experiments.
(I) Enriched gene ontology (GO) terms for differentially regulated genes in iCMs treated with 2C+MT and Ctrl+MT for 5 weeks. All data are presented as the means ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus the relevant control. NS., not significant. 4.
According to the Pearson corelation analysis (Figure 3G) and principal component analysis (PCA) (Figure 3H), 2C promoted fibroblast conversion toward an adult cardiomyocyte gene program compared with GMT/MT alone or GMT/MT+SB431542+XAV939. Gene ontology (GO) term enrichment analyses suggested that the up-regulated genes were associated with cell junction, Z-disc, calcium ion binding, heart development, cardiac muscle contraction, T-tubule, and sarcomere, while the downregulated genes were associated with inflammatory response, extracellular matrix, and response to virus (Figure 3I). Furthermore, 2C enhanced cardiac reprogramming progressively (Figures S8A–S8E). These cumulative results implied that 2C treatment could enhance functionality in reprogrammed iCMs, most likely through inducing an adult cardiomyocyte signature, resulting in calcium transients and action potential.
2C enhance cardiac reprogramming in human cardiac fibroblasts
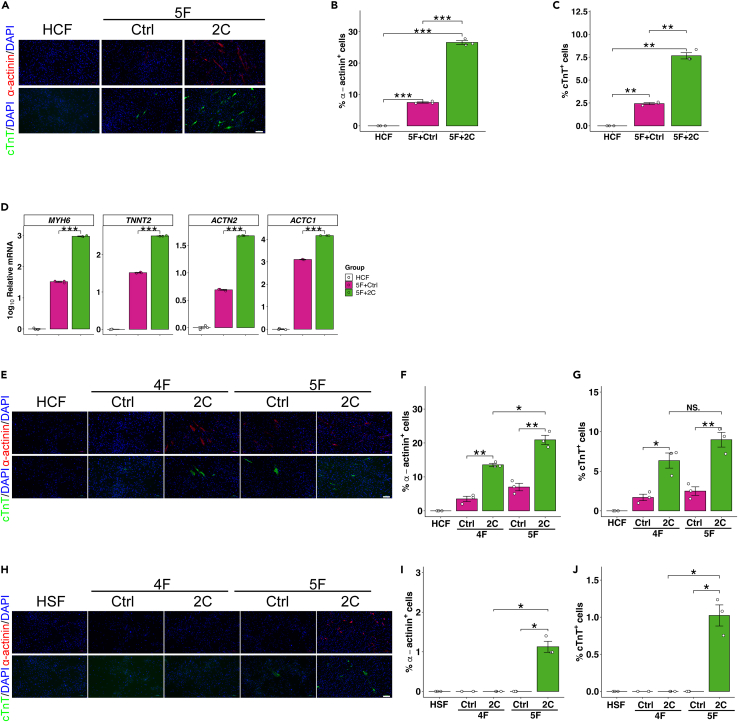
Direct cardiac reprogramming in human cells is substantially more complicated than in mouse fibroblasts, requiring more reprogramming factors and longer induction time, while resulting in lower reprogramming efficiency and less functional state.4,8,9,10,12 Since 2C could increase the efficiency, kinetics, and selectivity of cardiac reprogramming in mouse cells while reducing the number of requisite transgenes and improving functionality in iCMs, we then evaluated the effects of 2C in reprogramming human cardiac fibroblasts. To this end, we transduced human cardiac fibroblasts with GATA4, MEF2C, TBX5, MESP1 and MYOCD (5F), as described by Wada and colleagues,10 and treated the cells with solvent control (Ctrl) or 2C. Human cardiac fibroblasts we used were confirmed by the expression of Periostin and without the expression of ɑ-actinin, cTnT or cMHC (cardiac myosin heavy chain) in Figure S9. Immunofluorescence staining for α-actinin (depicted in Figure 4A and quantified in 4B) revealed that 2C+5F induced 26.6% of α-actinin+ cells, whereas the DMSO control resulted in only 7.4% α-actinin+ cells 2 weeks post-induction. Similarly, immunofluorescence staining for cTnT (depicted in Figure 4A and quantified in 4C) demonstrated that 2C+5F induced 7.7% cTnT+ cells, whereas the DMSO control resulted in only 2.4% cTnT+ cells. Additionally, qPCR analysis indicated a significant upregulation of cardiac genes (i.e., MYH6, TNNT2, ACTN2, and ACTC1) in the 2C+5F-induced cardiomyocytes (Figure 4D). These findings collectively demonstrate the efficacy of 2C in enhancing the reprogramming of human cardiac fibroblasts.
Figure 4.
2C enhance cardiac reprogramming in human cardiac fibroblasts
(A–C) Representative immunocytochemistry images (A) and quantification of α-actinin+ cells (B), cTnT+ cells (C) 2 weeks after 5F (GATA4, MEF2C, TBX5, MESP1 and MYOCD) transfection, in combination with Ctrl (DMSO) or 2C treatment of human cardiac fibroblasts. n = 3 independent experiments. Scale bars, 100 μm.
(D) Relative mRNA level of cardiomyocyte-specific genes (MYH6, TNNT2, ACTN2 and ACTC1) determined by qPCR, related to A and B. n = 3 independent experiments.
(E–G) Representative immunocytochemistry images (E) and quantification of α-actinin+ cells (F), cTnT+ cells (G) 2 weeks after 5F or 4F (MEF2C, TBX5, MESP1 and MYOCD) transfection, in combination with Ctrl (DMSO) or 2C treatment of human cardiac fibroblasts. n = 3 independent experiments. Scale bars, 100 μm.
(H–J) Representative immunocytochemistry images (H) and quantification of ɑ-actinin+ cells (I), cTnT+ cells (J) 2 weeks after 5F or 4F (MEF2C, TBX5, MESP1 and MYOCD) transfection, in combination with Ctrl (DMSO) or 2C treatment of human skin fibroblasts. n = 3 independent experiments. Scale bars, 100 μm. All data are presented as the means ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus the relevant control. NS., not significant.
Furthermore, when employing 2C+4F (comprising MEF2C, TBX5, MESP1, and MYOCD), we observed an induction of 13.6% α-actinin+ cells (depicted in Figure 4E and quantified in 4F) and 6.3% cTnT+ cells (depicted in Figure 4E and quantified in 4G) in human cardiac fibroblasts (HCF). In contrast, negligible or no reprogramming effects (Figures 4H–4J) of 2C+4F were observed in human skin fibroblasts (HSF). Taken together, our results indicate that 2C+5F significantly enhance cardiac reprogramming in both human cardiac and skin fibroblasts, whereas 2C+4F selectively reprogram human cardiac fibroblasts.
2C increases the proportion of induced cardiomyocyte-like cells and inhibits cell proliferation
Considering our above data showing that 2C can increase the efficiency and functionality of cardiac reprogramming, we then investigated whether and how the effects of 2C might be related to cell proliferation by testing three models (Figure 5A). First, we hypothesized that 2C enhanced the proliferation of converted iCMs. Second, we speculated that 2C could promote proliferation in the total cell population. Our third hypothesis was that 2C directly increased the conversion ratio. To test these hypotheses, we conducted immunofluorescence staining to simultaneously quantify proliferating cells (indicated by Cyclin D1+ cells), iCMs (indicated by α-actinin+ cells), and total cell numbers (DAPI+ cells) at various time points over 16 days (Figure 5B). These assays showed that 2C enhanced cardiac reprogramming efficiency in a time-dependent manner by increasing the absolute number of α-actinin+ iCMs (Figures 5C and 5D), inhibiting cell proliferation (Figure 5E), decreasing the absolute number of total cells (Figure 5F) and thus resulting in the higher ratio of iCMs to total cells (Figure 5G). No Cyclin D1 and α-actinin double-positive cells could be detected throughout the induction (Figure 5H).
Figure 5.
2C increases the proportion of iCMs and inhibits cell proliferation
(A) Schematic representation of three models of the enhancement of 2C on cardiac reprogramming and cell proliferation.
(B) Schematic of reprogramming system and detection of cell proliferation.
(C) Representative immunocytochemistry images for Cyclin D1, α-actinin and DAPI at the indicated time points. Scale bars, 50 μm.
(D) Quantification of the absolute number of α-actinin+ cells over time for groups treated with Ctrl (DMSO) or 2C. n = 4 independent experiments.
(E) Quantification of the percentage of Cyclin D1+ cells over time for groups treated with Ctrl (DMSO) or 2C. n = 4 independent experiments.
(F) Quantification of the number of cells (indicated by DAPI+ cells) over time for groups treated with Ctrl (DMSO) or 2C. n = 4 independent experiments.
(G) Quantification of the percentage of α-actinin+ cells over time for groups treated with Ctrl (DMSO) or 2C. n = 4 independent experiments.
(H) Quantification of the number of Cyclin D1+ α-actinin+ cells over time for groups treated with Ctrl (DMSO) or 2C. n = 4 independent experiments.
(I) Schematic of EdU incorporation and Ki67 staining assay.
(J and K) Representative immunocytochemistry images (J) and quantification (K) of cTnI-EdU+ and cTnI+EdU+ cells. n = 5 independent experiments. Scale bars, 20 μm.
(L and M) Representative immunocytochemistry images (L) and quantification (M) of cTnI-Ki67+ and cTnI+Ki67+ cells. n = 5 independent experiments. Scale bars, 20 μm. All data are presented as the means ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus the relevant control. NS., not significant.
We also performed EdU incorporation (Figures 5I–5K) and Ki67 staining (Figures 5L and 5M) to further confirm that 2C+MT induced iCMs without inducing iCMs proliferation. These cumulative data indicated that 2C enhances cardiac reprogramming by increasing the absolute number of iCMs while inhibiting cell proliferation and thus increasing the conversion ratio.
2C+MT-reprogrammed induced cardiomyocyte-like cells maintain a stable cardiomyocyte fate
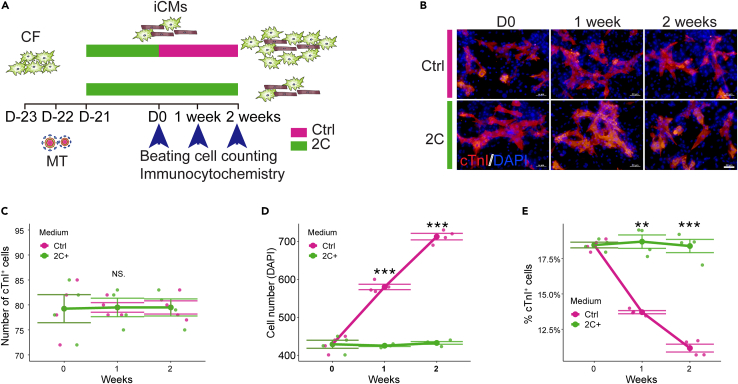
In addition to optimizing the 2C treatment duration (Figure S1E), we also asked whether the iCMs retained their cardiomyocyte-like phenotype in the absence of 2C. To answer this question, mouse cardiac fibroblasts were infected with MT-expressing lentivirus and treated with 2C for 3 weeks, after which they were cultured for another 2 weeks with or without 2C (Figure 6A). Immunofluorescent staining revealed that the reprogrammed iCMs indeed maintained their cardiomyocyte-like fate, indicated by the stable number of cTnI+ cells detected in cultures without 2C (Figures 6B and 6C). By contrast, the number of total cells rapidly increased in the absence of 2C (Figure 6D), resulting in a decreased proportion of cTnI+ cells (Figure 6E). These data indicated that the fibroblasts which did not undergo reprogramming began to rapidly proliferate following withdrawal of 2C treatment. Further, the iCMs induced by 2C+MT were stably reprogrammed and did not require 2C to maintain a persistent cardiomyocyte phenotype.
Figure 6.
2C+MT induced iCMs maintain a stable cardiomyocyte fate
(A) Schematic of reprogramming system to test the stability of MT+2C-induced iCMs.
(B) Representative immunocytochemistry images for cTnI and DAPI at the indicated time points. Scale bars, 50 μm.
(C) Quantification of the number of cTnI+ cells over time for groups treated with Ctrl (DMSO) or 2C. n = 4 independent experiments.
(D) Quantification of the number of cells (indicated by DAPI+ cells) over time for groups treated with Ctrl (DMSO) or 2C. n = 4 independent experiments.
(E) Quantification of the percentage of cTnI+ cells over time for groups treated with Ctrl (DMSO) or 2C. n = 4 independent experiments. All data are presented as the means ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus the relevant control. NS., not significant.
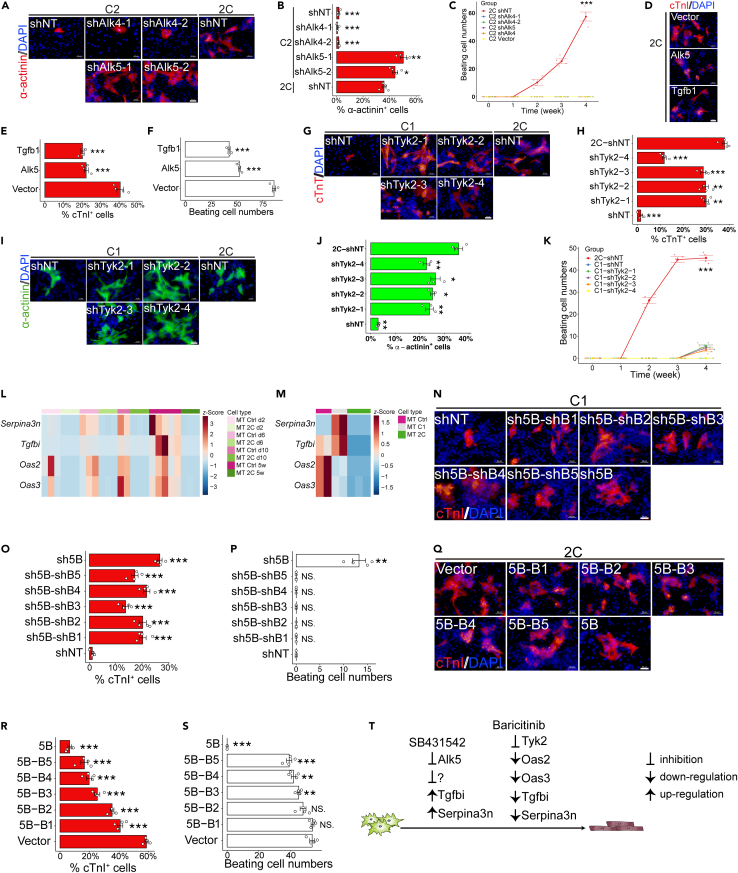
C2 synergizes cardiomyocyte induction via the suppression of C1-activated molecular barriers
Alk5 (Transforming Growth Factor β Receptor 1, also known as Tgfbr1) stands as a well-established target for C143,44, while C2 has been associated with targeting Jak1, Jak2, Jak3, and Tyk2.45 To discern the impact of 2C on the reprogramming process through these canonical targets, we conducted tests involving various chemicals known to share these regulatory targets with C1 and C2, respectively. Our findings revealed that all the chemicals tested, with the same targets as C1, significantly enhanced cardiac reprogramming efficiency, as indicated by the proportion of cTnT+ cells (Figure S10A). These results align with previous reports emphasizing the role of inhibiting the TGF-beta signaling pathway in cardiac reprogramming.4,20,22
Furthermore, we aimed to mimic the effects of C1 by conducting knockdown experiments targeting the receptors Alk4 and Alk5. Alk4 knockdown did not exhibit any enhancement of cardiac reprogramming. However, Alk5 knockdown yielded a substantial increase in the percentage of α-actinin+ cells, indicating a clear enhancement of cardiac reprogramming (Figures 7A and 7B). Notably, under Alk5 knockdown conditions, no spontaneous beating cells were detected (Figure 7C), suggesting that C1 may promote cardiac reprogramming through additional targets. Conversely, overexpression of Alk5 or Tgfb1 (transforming growth factor β 1) resulted in significantly lower reprogramming efficiency, evidenced by a reduced proportion of cTnI+ cells and a decrease in the number of beating cells (Figures 7D–7F). These observations imply that the inhibition of Alk5 is crucial for cardiac reprogramming but does not entirely account for the enhanced effects elicited by C1.
Figure 7.
C2 synergizes cardiomyocyte induction via the suppression of C1-activated molecular barriers
(A and B) Representative immunocytochemistry images (A) and quantification (B) for α-actinin+ cells 3 weeks after MT transduction, in combination with shNT, shAlk4, or shAlk5 and cultured in C2 or 2C medium. n = 4 independent experiments. Scale bars, 50 μm.
(C) Quantification of spontaneous beating cells over time, related to A and B.
(D–F) Representative immunocytochemistry images (D), quantification of cTnI+ cells (E), and quantification of beating cells 3 weeks after MT transduction, in combination with 2C medium and over-expression of vector (vector control), Alk5 or Tgfb1. n = 4 independent experiments. Scale bars, 50 μm.
(G and H) Representative immunocytochemistry images (G) and quantification of cTnT+ cells (H) 3 weeks after MT transduction, in combination with C1 or 2C medium and knock-down of NT (non-target control) or Tyk2. n = 4 independent experiments. Scale bars, 50 μm.
(I and J) Representative immunocytochemistry images (I) and quantification of α-actinin+ cells (J) 3 weeks after MT transduction, in combination with C1 or 2C medium and knock-down of NT (non-target control) or Tyk2. n = 4 independent experiments. Scale bars, 50 μm.
(K) Quantification of spontaneous beating cells over time, related to G-J. n = 4 independent experiments.
(L) Relative expression of Oas2, Oas3, Serpina3n, and Tgfbi in cardiac fibroblasts transduced with MT, in combination with Ctrl (DMSO) or 2C medium over time. n = 2–3 independent experiments.
(M) Relative expression of Oas2, Oas3, Serpina3n, and Tgfbi in cardiac fibroblasts 3 weeks after transduction with MT, in combination with Ctrl (DMSO), C1 or 2C medium. n = 2–3 independent experiments.
(N–P) Representative immunocytochemistry images (N) and quantification (O) of cTnI+ cells and spontaneous beating cells (P) 3 weeks after transduction with MT and knock-down of NT (Non-target control), 5B (Tyk2 (B1), Oas2 (B2), Oas3 (B3), Serpina3n (B4) and Tgfbi (B5)) and each combination of 4 factors. n = 4 independent experiments. All groups were cultured in C1 medium. Scale bars, 50 μm.
(Q–S) Representative immunocytochemistry images (Q) and quantification (R) of cTnI+ cells and spontaneous beating cells (S) 3 weeks after transduction with MT and over-expression of vector, 5B, and each combination of 4 factors. n = 4 independent experiments. All groups were cultured in 2C medium. Scale bars, 50 μm.
(T) Schematic of the downstream of C1 and C2. All data are presented as the means ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus the relevant control. NS., not significant.
However, an investigation into the role of C2, coupled with JAK inhibitors, demonstrated that treatment with 19 out of 56 Jak inhibitors led to an increased percentage of cTnT+ cells, with 9 of them also significantly increasing the number of beating cells (Figures S10B and S10C). To genetically validate whether C2 enhances cardiac reprogramming through the inhibition of Jak1, Jak2, Jak3, or Tyk2, we conducted knockdown experiments using two independent shRNAs for each gene. Knockdown of Jak1, Jak2, and Jak3 did not exert significant effects on the percentage of ⍺-actinin+ cells, cTnT+ cells, or the number of spontaneous beating cells (Figures S11A–S11E). In contrast, Tyk2 knockdown resulted in an increased ratio of α-actinin+ cells, and cTnT+ cells (Figures 7G–7J). Notably, even after an extended induction period spontaneous beating cells were barely detectable (Figure 7K). Conversely, Tyk2 overexpression did not negate the effects of C2, even when used at various concentrations of C2 (Figures S11F–S11H).
Stat3 serves as a primary downstream target of Jak signaling, activated via phosphorylation of tyrosine residue at 705 (Y705)44,45,46,47,48 or serine residue at 727 (S727).49,50,51,52,53 To examine the role of Stat3, we introduced mutations in the Y705 and S727 residues to simulate the phosphorylated and dephosphorylated states of Stat3. Surprisingly, the percentage of α-actinin+ cells and the number of beating cells remained unaffected in both WT and mutated Stat3 (Figure S11I).
Subsequent RNA-seq analysis conducted on 2C+MT, C1+MT, or MT control fibroblasts over a five-week time course identified four genes - Oas2, Oas3, Serpina3n, and Tgfbi (transforming growth factor β induced) - that were significantly downregulated in the presence of 2C throughout the entire experimental period (Figure 7L). Intriguingly, Serpina3n and Tgfbi were upregulated in C1+MT cells but downregulated by 2C (Figure 7M). Immunofluorescence staining further demonstrated that the combined knockdown of these five genes, referred to as the "5B" set (Tyk2, Oas2, Oas3, Serpina3n, and Tgfbi), significantly increased the percentage of cTnI+ cells and the number of beating cells, underscoring their role as inhibitors of cardiac reprogramming (Figures 7N–7P). In contrast, the overexpression of the 5B gene set effectively countered the effects of 2C+MT, resulting in a reduced percentage of cTnI+ cells and beating cells (Figures 7Q–7S). Moreover, in line with these findings in mouse cells, TGFBI, SERPINA3, and OAS3 were also significantly downregulated by 2C, while OAS2 did not exhibit significant downregulation (Figure S12). These data suggest that 2C may function similarly, although not identically, in human cardiac fibroblast (hiCM) reprogramming. Collectively, these results indicate that C1 enhances cardiac reprogramming through the inhibition of the TGF-beta signal pathway, consistent with previous reports.4,20,22,54 C1 also exhibits some adverse effects that can inhibit cardiac reprogramming, but these effects are mitigated by C2, explaining their synergistic effects (Figures 1B–1H). These cumulative findings demonstrate that the components of 2C function synergistically to enhance cardiac reprogramming by modulating multiple downstream effectors (Figure 7T).
Discussion
In this study, we found that 2C significantly enhances the efficiency of GMT-mediated reprogramming. Testing each individual transcription factor and each combination of two factors revealed that 2C+MT can selectively reprogram cardiac fibroblasts with significantly higher efficiency and kinetics than GMT. 2C+4F (MEF2C, TBX5, MESP1, and MYOCD) also selectively convert human cardiac fibroblasts into iCMs. Several cardiogenic genes, such as Gata4, are known to be expressed in cardiac fibroblasts,26 potentially accounting for the high selectivity of this cocktail. Moreover, 2C further upregulates endogenous Gata4 expression in cardiac fibroblasts, but does not induce any detectable Gata4 expression in fibroblasts from other organs. In addition to 2C, Bmi1 knock-down16 or PHF7 over-expression18 can both complement the loss of Gata4 in reprogramming mouse cardiac fibroblasts, although further investigation is required to determine whether those factor combinations are also selective to cardiac fibroblasts. More recently, Ascl1+Mef2c was also reported to induce iCMs from cardiac fibroblasts, highlighting the cross-lineage potential of Ascl1, but shows poor selectivity toward cardiac fibroblasts, since MEF can also be reprogrammed.55
Our findings strengthen the basis for in vivo study or applications of direct cardiac reprogramming. Increased selectivity with a reduced number of requisite genetic factors may also facilitate the therapeutic application of in situ direct cardiomyocyte induction from cardiac fibroblasts. Additionally, although evaluating the effects of 2C in vivo falls beyond the scope of the current study, we found that 2C significantly down-regulates inflammatory signaling activity, which is activated in damaged heart tissue56,57 These findings further support that 2C may be potentially applied in cardiac reprogramming in vivo to overcome inflammation-associated molecular barriers, aligning well with another study that showed cardiac reprogramming efficiency is enhanced via ZNF281-mediated suppression of inflammation.58
Although we found that 2C is required for the highest efficiency reprogramming in the first 3 weeks of cardiomyocyte induction, (Figure S1E), iCMs induced by 2C+MT are stably reprogrammed and do not require 2C to maintain a persistent cardiomyocyte phenotype (Figure 6). Thus, the short-term use of 2C might decrease potential toxicity and off-target effects, which can be leveraged to develop a feasible and safe therapeutic approach to repair damaged heart tissue in vivo.
C1, an Alk5 inhibitor,59 is now widely recognized for its capacity to induce a variety of cell types, including iPSCs,60 iNs,32 cardiac progenitor cells,61 cardiac Purkinje cells,62 and iCMs.4,14,20,54,63 Other studies have demonstrated that inhibiting pro-fibrotic signaling, a major barrier to cardiac lineage reprogramming, can substantially increase reprogramming efficiency.22 Although Alk5 inhibition is essential for cardiac reprogramming, this function alone cannot fully account for the effects of C1 on reprogramming In addition to Alk5 inhibition, we found that the pan-Jak inhibitor, C2, also conferred positive effects on reprogramming, which aligns well with other studies that showed two Jak inhibitors (Erlotinib and Ruxolitinib) could improve cardiac reprogramming, indicated by the elevated expression of cardiomyocyte markers.13 Similarly, ZNF281 has been shown to benefit cardiac reprogramming by modulating inflammatory gene expression.58 These reports, in conjunction with our own findings in this study, highlight the role of Jak-Stat signaling as a molecular barrier to cardiac reprogramming. However, relatively few pan-Jak inhibitors that share structural similarity with C2 can also efficiently induce spontaneous beating iCMs, indicating further investigation are required to elucidate the mechanism underpin C2 enhancement of iCMs. Moreover, we found that C2 promotes cardiac reprogramming via the suppression of C1-activated molecular barriers - Tgbfi and Serpina3n.
In summary, we established a robust cardiac reprogramming system to convert mouse or human cardiac fibroblasts into cardiomyocytes selectively and efficiently, opening new potential avenues for the clinical translation of targeted reprogramming by optimizing reprogramming cocktails and reducing the number of genetic factors.
Limitations of the study
While we have reported the effectiveness of the cardiac fibroblast-selective reprogramming factor combination, 2C+MT, further investigations are warranted to ascertain the selectivity and efficiency of 2C+MT in reprogramming cardiac fibroblasts into cardiomyocyte-like cells in mouse models of myocardium infarction in vivo. Additionally, due to limitations related to the availability of human cardiac fibroblasts, this study did not determine the minimal factor combination required for cardiac reprogramming in the presence of 2C. Although we have identified several factors as downstream effectors of 2C, their contributions to cardiac reprogramming require further elucidation in future studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Cardiac Troponin T Monoclonal Antibody (13-11) | Thermo Fisher | Cat#MA5-12960; RRID:AB_11000742 |
| Anti-Cardiac Troponin I | abcam | Cat#ab56357; RRID:AB_880622 |
| Anti-Sarcomeric Alpha Actinin | abcam | Cat#ab9465; RRID:AB_307264 |
| Anti-Cyclin D1 | abcam | Cat#ab16663; RRID:AB_443423 |
| Anti-Myosin Light Chain 2 | abcam | Cat#ab48003; RRID:AB_2282315 |
| Anti-Ki67 | abcam | Cat#ab15580; RRID:AB_443209 |
| Anti-Periostin | abcam | Cat#ab14041; RRID:AB_2299859 |
| Anti-cMHC | abcam | Cat#ab207926; RRID:AB_3075445 |
| Donkey anti-Mouse Alexa Fluor 488 | Thermo Fisher | Cat#A-21202; RRID:AB_141607 |
| Donkey anti-Mouse Alexa Fluor 555 | Thermo Fisher | Cat#A-31570; RRID:AB_2536180 |
| Donkey anti-Rabbit Alexa Fluor 488 | Thermo Fisher | Cat#A-21206; RRID:AB_2535792 |
| Donkey anti-Rabbit Alexa Fluor 555 | Thermo Fisher | Cat#A-31572; RRID:AB_162543 |
| Donkey anti-Goat Alexa Fluor 488 | Thermo Fisher | Cat#A-11055; RRID:AB_2534102 |
| Donkey anti-Goat Alexa Fluor 555 | Thermo Fisher | Cat#A-21432; RRID:AB_2535853 |
| CD90.2 MicroBeads | Miltenyi | Cat#130-121-278; RRID:AB_3073748 |
| Chemicals, peptides, and recombinant proteins | ||
| SB431542 | Selleck | Cat#S1067 |
| Baricitinib | Selleck | Cat#S2851 |
| Doxycycline hyclate | Sigma-Aldrich | Cat#D9891 |
| XAV939 | Selleck | Cat#S1180 |
| Critical commercial assays | ||
| pEASY®-Uni Seamless Cloning and Assembly Kit | TransGen | Cat#CU101 |
| RNeasy Plus Mini Kit | QIAGEN | Cat#74134 |
| QIAGEN Plasmid Mega Kit | QIAGEN | Cat#12183 |
| Fluo4-AM | Thermo Fisher | Cat#F14201 |
| TransScript® One-Step gDNA Removal and cDNA Synthesis SuperMix | TransGen | Cat#AT311 |
| ChamQ SYBR qPCR Master Mix(Without ROX) | Vazyme | Cat# Q321-02 |
| Deposited data | ||
| RNA-seq | GEO Database | GSE213021 |
| Experimental models: Cell lines | ||
| NHCF | Lonza | Cat#CC2904 |
| BJ | ATCC | Cat#CRL-2522 |
| Experimental models: Organisms/strains | ||
| Mouse: B6; D2-Tg(Myh6∗-mCherry)2Mik/J | The Jackson Laboratory | Mouse strain #: 021577 |
| Mouse: CD-1(ICR) | Beijing Vital River LaboratoryAnimal Technology Co., Ltd | Mouse strain #: 201 |
| Oligonucleotides | ||
| Separated attachement, Table S1 | ||
| Recombinant DNA | ||
| FU-tet-o-Puro-Vector | This paper | N/A |
| FU-tet-o-Puro-EGFP | This paper | N/A |
| FU-tet-o-Puro-Gata4 | This paper | N/A |
| FU-tet-o-Puro-Mef2c | This paper | N/A |
| FU-tet-o-Puro-Tbx5 | This paper | N/A |
| FU-tet-o-Puro-MGT | This paper | N/A |
| FU-tet-o-Puro-T-P2A-M | This paper | N/A |
| FU-tet-o-Puro-M-P2A-T | This paper | N/A |
| FU-tet-o-Puro-M-T2A-T | This paper | N/A |
| FU-tet-o-Puro-M-E2A-T | This paper | N/A |
| tetO-hGATA4 | addgene | Cat#46030 |
| tetO-hMEF2C | addgene | Cat#46031 |
| tetO-hTBX5 | addgene | Cat#46032 |
| FU-tet-o-hMESP1 | This paper | N/A |
| FU-tet-o-hMYOCD | This paper | N/A |
| FU-Ubc-rtTA-Neo | This paper | N/A |
| pLenti-BleoR-Vector | This paper | N/A |
| pLenti-BleoR-EGFP | This paper | N/A |
| pLenti-BleoR-Stat3 | This paper | N/A |
| pLenti-BleoR-Stat3-S727A | This paper | N/A |
| pLenti-BleoR-Stat3-S727D | This paper | N/A |
| pLenti-BleoR-Stat3-Y705E | This paper | N/A |
| pLenti-BleoR-Stat3-Y705E S727D | This paper | N/A |
| pLenti-BleoR-Stat3-Y705F | This paper | N/A |
| pLenti-BleoR-Stat3-Y705F S727A | This paper | N/A |
| pLKO.1-NT | Sigma-Aldrich | SHC016H |
| pLKO.1-Jak1-1 | Sigma-Aldrich | TRCN0000023290 |
| pLKO.1-Jak1-2 | Sigma-Aldrich | TRCN0000023292 |
| pLKO.1-Jak2-1 | Sigma-Aldrich | TRCN0000023650 |
| pLKO.1-Jak2-2 | Sigma-Aldrich | TRCN0000023652 |
| pLKO.1-Jak3-1 | Sigma-Aldrich | TRCN0000023475 |
| pLKO.1-Jak3-2 | Sigma-Aldrich | TRCN0000023478 |
| pLKO.1-Tyk2-1 | Sigma-Aldrich | TRCN0000025886 |
| pLKO.1-Tyk2-2 | Sigma-Aldrich | TRCN0000025942 |
| pLKO.1-Tyk2-3 | Sigma-Aldrich | TRCN0000025964 |
| pLKO.1-Tyk2-4 | Sigma-Aldrich | TRCN0000025903 |
| FU-tet-o-BSD-Tyk2 | This paper | N/A |
| pLKO.1-Alk4-1 | Sigma-Aldrich | TRCN0000022574 |
| pLKO.1-Alk4-2 | Sigma-Aldrich | TRCN0000022575 |
| pLKO.1-Alk5-1 | Sigma-Aldrich | TRCN0000322043 |
| pLKO.1-Alk5-2 | Sigma-Aldrich | TRCN0000322044 |
| FU-tet-o-BSD-Vector | This paper | N/A |
| FU-tet-o-BSD-Alk5 | This paper | N/A |
| FU-tet-o-BSD-Tgfb1 | This paper | N/A |
| pLKO.1-Oas2-1 | Sigma-Aldrich | TRCN0000075813 |
| pLKO.1-Oas2-2 | Sigma-Aldrich | TRCN0000075814 |
| pLKO.1-Oas3-1 | Sigma-Aldrich | TRCN0000075823 |
| pLKO.1-Oas3-2 | Sigma-Aldrich | TRCN0000075824 |
| pLKO.1-Tgfbi-1 | Sigma-Aldrich | TRCN0000054809 |
| pLKO.1-Tgfbi-2 | Sigma-Aldrich | TRCN0000054810 |
| pLKO.1-Serpina3n-1 | Sigma-Aldrich | TRCN0000080455 |
| pLKO.1-Serpina3n-2 | Sigma-Aldrich | TRCN0000080456 |
| FU-tet-o-BSD-Oas2 | This paper | N/A |
| FU-tet-o-BSD-Oas3 | This paper | N/A |
| FU-tet-o-BSD-Tgfbi | This paper | N/A |
| FU-tet-o-BSD-Serpina3n | This paper | N/A |
| pLKO.1-Gata4-1 | Sigma-Aldrich | TRCN0000095214 |
| pLKO.1-Gata4-2 | Sigma-Aldrich | TRCN0000095215 |
| pLKO.1-Gata4-3 | Sigma-Aldrich | TRCN0000095216 |
| Software and algorithms | ||
| R version 4.2.1 | R Project | https://www.r-project.org |
| RStudio Desktop 2022.07.1+554 | Rstudio | https://www.rstudio.com |
| DESeq2 1.36.0 | DESeq2 | https://bioconductor.org/packages/release/bioc/html/DESeq2.html |
| tidyverse | tidyverse | https://www.tidyverse.org |
| EnhancedVolcano 1.14.0 | EnhancedVolcano | https://github.com/kevinblighe/EnhancedVolcano |
| ImageJ-Fiji | National Institutes of Health, USA | https://fiji.sc/ |
| Other | ||
| DMEM,high glucose | HyClone | Cat#SH30022.01 |
| MEDIUM 199 | Thermo Fisher | Cat#11150-059 |
| KnockOut Serum Replacement (KSR) | Thermo Fisher | Cat#A3181502 |
| Fetal Bovine Serum (FBS) | VISTECH | Cat#SE100-011 |
| GlutaMAX | Thermo Fisher | Cat#35050–061 |
| MEM Non-Essential Amino Acids | Thermo Fisher | Cat#11140-050 |
| Penicillin-streptomycin | Thermo Fisher | Cat#15140-122 |
| DAPI | Sigma-Aldrich | Cat#D9542 |
| Polybrene | Sigma-Aldrich | Cat#H9268-10G |
| PBS | Corning | Cat#21-040-CVR |
| Trypsin-EDTA | Thermo Fisher | Cat#25200-056 |
| Puromycin | InvivoGen | Cat#ant-pr-1 |
| Geneticin | InvivoGen | Cat#ant-gn-1 |
| Blasticidin | InvivoGen | Cat#ant-bl-05 |
| EdU | Abbkine | Cat#KTA2031-CN |
Resource availability
Lead contact
-
•
Further information and requests for resources and reagents shouldbe directed to and will be fulfilled by the lead contact, Yang Zhao (yangzhao@pku.edu.cn).
Materials availability
-
•
This study did not generate new unique reagents.
Data and code availability
-
•
This paper does not report original code.
-
•
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
Experimental model and subject details
Plasmid construction
Coding sequences of Gata4, Mef2c, and Tbx5 were amplified by PCR from pMXs-MGT (addgene, Cat#111810, a gift from the Qian lab). The FU-tet-o-hOct4 (addgene, Cat#19778) was cut by EcoR I. Inserts and linearized vector were assembled using a pEASY-Uni Seamless Cloning and Assembly Kit (TransGen, Cat#CU101). tetO-hGATA4 (addgene, #46030), tetO-hMEF2C (addgene, #46031), and tetO-hTBX5 (addgene, #46032) were purchased from addgene. FU-tet-o-hMESP1 and t FU-tet-o-hMYOCD were constructed in this work. Detailed list of plasmids is available at key resources table. For each vector used in this article, we validated proper gene expression at desired levels (overexpression or knockdown, data not shown here).
Method details
Viral packaging and transduction
Lentiviral vector (15 μg) together with pMDLg/pRRE, RSV/Rev, and VSV-G (5 μg for each), were co-transfected into 293T cells with the Ca3(PO4)2-method in 10-cm dishes and incubated 12-16 hours. On the following day, the medium was changed. 2 days after transfection, viral supernatant was collected and filtered through 0.45 μm filters (Millipore, Cat#SLHP033RB) and then placed onto target fibroblasts supplemented with 8 μg/mL polybrene (Sigma-Aldrich, Cat#H9268-10G). Lentiviruses were used freshly or frozen at -80°C for future use.
Animals and surgery
All animal experiments were performed according to the Animal Protection Guidelines of Peking University and all procedures conformed to the guidelines from the NIH Guide for the Care and Use of Laboratory Animals. Transgenic mice expressing mCherry driven by the cardiomyocyte-specific promoter, ɑMHC (B6;D2-Tg(Myh6∗-mCherry)2Mik/J, The Jackson Laboratory, #021577), were a gift from Dr. Kotlikoff.39 MI was performed in 5-week-old male ICR mice of the B6;129S4 genetic background (Jackson Laboratory #008214) induced by permanent ligation of the left anterior descending coronary artery (LAD) with a 7-0 prolene suture as described previously.64 Euthanasia was performed by deep (4%) isoflurane anesthesia followed by exsanguination, consistent with American Veterinary Medical Association guidelines.
Primary cell cultures
Isolated neonatal (1.5-day-old CD-1 mice) hearts were minced into small pieces less than 1 mm3 in size and digested with collagenase type II solution. After 4-6 days, cells were separated by magnetic cell sorting (MACS) to obtain CD90.2+ neonatal fibroblasts according to a previously described protocol.40 Adult mouse (5 weeks) cardiac fibroblasts and fibroblasts from neonatal mouse lung, neonatal mouse liver, neonatal mouse brain, adult mouse skin and adult mouse lung were isolated using a similar protocol without MACS. MICF (cardiac fibroblasts isolated from adult mouse after myocardial infarction) were isolated from infarct and border regions of adult heart, 4-5 days after surgery. All fibroblasts used were confirmed by immunofluorescence staining to exclude any contamination with cardiomyocytes. Neonatal mouse cardiomyocytes were isolated according to a previously described protocol.65 Adult mouse cardiomyocytes were isolated according to a previously described protocol.66 Human cardiac fibroblasts were purchased from Lonza (Cat#CC2904). Human skin fibroblasts (BJ) ware purchased from ATCC (Cat#CRL-2522).
iCMs reprogramming
P1 neonatal mouse cardiac fibroblasts were plated into a 24-well plate at a density of 1 x 105cells per well in FB medium (10% FBS in DMEM supplemented with 1% penicillin-streptomycin). On the following day, fibroblasts were transduced with MT (FU-tet-o-MT, FUdeltaGW-rtTA, MOI = 10) lentivirus in transfection medium (10% FBS in DMEM supplemented with 8 μg/mL polybrene). On the following day, the medium was changed to 2C medium (DMEM/M199 (4:1) supplemented with 10% FBS,10% KnockOut Serum Replacement, 1% MEM Non-Essential Amino Acids, 1% GlutaMAX, 1% penicillin-streptomycin, 2 μg/mL Doxycycline hyclate, 2 μM SB431542, 2 μM Baricitinib). Three days after infection, reprogramming medium was supplemented with 1 μg/mL Puromycin and 200 μg/mL Geneticin. The medium was changed every 3-4 days. For human cells, P6-P8 cells were plated into a 24-well plate at a density of 2 x 105cells per well for reprogramming.
Chemical screening
Neonatal mouse skin fibroblasts (NSF) were isolated from transgenic mice expressing mCherry driven by the cardiomyocyte-specific promoter, αMHC. NSF were plated into a 24-well plate at a density of 1 x 105 cells per well in FB medium. The following day, NSF were infected with GMT-expressing lentivirus (FU-tet-o-Gata4, FU-tet-o-Mef2c, FU-tet-o-Tbx5 and FUdeltaGW-rtTA, ∼100 μL unconcentrated virus, MOI = 10) in transfection medium. After 1 day, the medium was changed to reprogramming medium with Ctrl (DMSO) or 2 μM indicated chemicals. The medium was changed every 2-3 days. Two weeks after induction, ɑMHC-mCherry expression was quantified by fluorescence microscopy. In total, we screened ∼2,000 compounds from both commercially available and proprietary in-house libraries.
Immunocytochemistry
After fixation with 4% paraformaldehyde (DingGuo, Cat#AR-0211) at room temperature for 30 min, cells were permeabilized with PBS-0.3% Triton X-100 (Sigma-Aldrich, Cat#T8787) and blocked with blocking buffer (PBS supplemented with 2.5% donkey serum (Jackson Immuno Research, Cat#017-000-121)) at 37°C for 1 hour. Primary antibody (Cardiac Troponin T, Thermo Fisher, Cat#MA5-12960; Cardiac Troponin I, abcam, Cat#ab56357; Sarcomeric Alpha Actinin, abcam, Cat#ab9465; Cyclin D1, abcam, Cat#ab16663; Myosin Light Chain 2, abcam, Cat#ab48003; Ki67, abcam, Cat#ab15580; Periostin, abcam, Cat#ab14041; cMHC, abcam, Cat#ab207926) incubation with appropriate dilutions were performed at 4°C overnight in blocking buffer. The following day, cells were washed with PBS three times and probed with secondary antibodies at 37°C for 1 hour in blocking buffer. Cells were then washed with PBS three times and DNA was stained with DAPI solution (Sigma-Aldrich, Cat#D9542). For the quantification, 3-5 independent experiments were used for scanning of whole wells and analysed in Image J software, or 5 fields were randomly selected in a blinded manner and the indicated cells were counted manually in each experiment.
qPCR
Total RNA was isolated with RNeasy Plus Mini Kit (QIAGEN, Cat#74134) and reverse-transcribed into cDNA using TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix (TransGen, Cat#AT311). qRT-PCR was performed using the q225 Real-Time PCR system (Kubo Tech, Cat#q225) and ChamQ SYBR qPCR Master Mix (Vazyme, Cat#Q321-02) according to the manufacturer’s instructions. The primer sequences used for qRT-PCR are listed in the key resources table. mRNA levels were normalized by comparison to Gapdh mRNA.
Calcium imaging
Calcium imaging was performed according to the standard protocol. Briefly, cells were loaded with 10 μM fluo4-AM (invitrogen, Cat#F14201) for 5 min at 37°C in the dark. Line-scan images were performed on a confocal microscope (Carl Zeiss, Cat#LSM-710) and acquired at sampling rates of 3.78 ms/line and 0.03 μm/pixel.
Electrophysiological recording
Whole-cell patch clamping was applied for action potential recording by an Axon 200B patch-clamp amplifier (Axon Instruments, USA). The action potentials were recorded in current clamp mode. Pipette resistances were 1.5–3 MΩ. The external solution contained (in mmol/L): 140 NaCl, 5.4 KCl, 1.8 CaCl2,1 MgCl2, 10 Glucose, 10 HEPES (pH 7.4 adjusted with NaOH). The pipette solution contained 120 KCl, 1 MgCl2, 3 MgATP, 10 HEPES,10 EGTA at pH 7.2 adjusted with KOH.
RNA-Seq and transcriptome analysis
Total RNA was isolated using a RNeasy Plus Mini Kit (QIAGEN, Cat#74134). Total amounts and integrity of RNA were assessed using the RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, CA, USA). Total RNA was used as input material for RNA sample preparations. Differential expression analysis was performed using the DESeq2 R package67 (1.36.0) and visualized using the EnhancedVolcano R package (1.14.0). Genes with an adjusted P value < 10-4 and fold-change > 2 were assigned as differentially expressed. Gene ontolgy (GO) term enrichment analyses were performed using the DAVID 6.8 functional annotation tool.68 Terms that had a P-value <0.05 were defined as significantly enriched.
Quantification and statistical analysis
Values were presented as means ± SEM. The normality of the data was tested using Shapiro-Wilk’s method. The unpaired t-test was used to determine the significance of differences between two groups. Statistical analysis was performed using R 4.2.1. A value of P < 0.05 was considered statistically significant (∗), a P value of < 0.01 was considered highly significant (∗∗), a P value of < 0.001 was considered very highly significant (∗∗∗), and a P value of > 0.05 was labelled as n.s (not significant). The statistical details of experiments can be found in the figure legends. All data are representative of multiple independent experiments.
Acknowledgments
We thank Li Qian (University of North Carolina, Chapel Hill) for kindly providing the pMXs-MGT plasmid and detailed cardiac reprogramming protocol. We thank Yu Nie (Fuwai Hospital) for kindly providing the αMHC-mCherry mice. This work is supported by the National Key Research and Development Program of China (2018YFA0800504), the National Natural Science Foundation of China (31922020), and fundings provided by Plastech Pharmaceutical Technology Co., LTD.
Author contributions
Y.T. designed and performed experiments, analyzed data; Y.Y. assisted with some data collection about human cardiac reprogramming; Z.Y. assisted with chemical screening; L.W. and S.W. contribute to a part of the calcium imaging and all electrophysiological recording; Y.Z. conceived this work and supervised this study. Y.T. and Y.Z. wrote the article.
Declaration of interests
Y.Z. is a shareholder of Plastech, as a founder and CSO of Plastech. A patent on this work has been applied (PCT/CN2021/109183). All authors declare no competing financial interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: November 14, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108466.
Supplemental information
References
- 1.Laflamme M.A., Murry C.E. Heart regeneration. Nature. 2011;473:326–335. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Porrello E.R., Mahmoud A.I., Simpson E., Hill J.A., Richardson J.A., Olson E.N., Sadek H.A. Transient regenerative potential of the neonatal mouse heart. Science. 2011;331:1078–1080. doi: 10.1126/science.1200708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ieda M., Fu J.D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B.G., Srivastava D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell. 2010;142:375–386. doi: 10.1016/j.cell.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mohamed T.M.A., Stone N.R., Berry E.C., Radzinsky E., Huang Y., Pratt K., Ang Y.S., Yu P., Wang H., Tang S., et al. Chemical Enhancement of In Vitro and In Vivo Direct Cardiac Reprogramming. Circulation. 2017;135:978–995. doi: 10.1161/CIRCULATIONAHA.116.024692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qian L., Huang Y., Spencer C.I., Foley A., Vedantham V., Liu L., Conway S.J., Fu J.D., Srivastava D. In vivo reprogramming of murine cardiac fibroblasts into induced cardiomyocytes. Nature. 2012;485:593–598. doi: 10.1038/nature11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song K., Nam Y.J., Luo X., Qi X., Tan W., Huang G.N., Acharya A., Smith C.L., Tallquist M.D., Neilson E.G., et al. Heart repair by reprogramming non-myocytes with cardiac transcription factors. Nature. 2012;485:599–604. doi: 10.1038/nature11139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyamoto K., Akiyama M., Tamura F., Isomi M., Yamakawa H., Sadahiro T., Muraoka N., Kojima H., Haginiwa S., Kurotsu S., et al. Direct In Vivo Reprogramming with Sendai Virus Vectors Improves Cardiac Function after Myocardial Infarction. Cell Stem Cell. 2018;22:91–103.e5. doi: 10.1016/j.stem.2017.11.010. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Y., Liu Z., Welch J.D., Gao X., Wang L., Garbutt T., Keepers B., Ma H., Prins J.F., Shen W., et al. Single-Cell Transcriptomic Analyses of Cell Fate Transitions during Human Cardiac Reprogramming. Cell Stem Cell. 2019;25:149–164.e9. doi: 10.1016/j.stem.2019.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nam Y.J., Song K., Luo X., Daniel E., Lambeth K., West K., Hill J.A., DiMaio J.M., Baker L.A., Bassel-Duby R., Olson E.N. Reprogramming of human fibroblasts toward a cardiac fate. Proc. Natl. Acad. Sci. USA. 2013;110:5588–5593. doi: 10.1073/pnas.1301019110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wada R., Muraoka N., Inagawa K., Yamakawa H., Miyamoto K., Sadahiro T., Umei T., Kaneda R., Suzuki T., Kamiya K., et al. Induction of human cardiomyocyte-like cells from fibroblasts by defined factors. Proc. Natl. Acad. Sci. USA. 2013;110:12667–12672. doi: 10.1073/pnas.1304053110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao N., Huang Y., Zheng J., Spencer C.I., Zhang Y., Fu J.D., Nie B., Xie M., Zhang M., Wang H., et al. Conversion of human fibroblasts into functional cardiomyocytes by small molecules. Science. 2016;352:1216–1220. doi: 10.1126/science.aaf1502. [DOI] [PubMed] [Google Scholar]
- 12.Fu J.D., Stone N.R., Liu L., Spencer C.I., Qian L., Hayashi Y., Delgado-Olguin P., Ding S., Bruneau B.G., Srivastava D. Direct reprogramming of human fibroblasts toward a cardiomyocyte-like state. Stem Cell Rep. 2013;1:235–247. doi: 10.1016/j.stemcr.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hashimoto H., Wang Z., Garry G.A., Malladi V.S., Botten G.A., Ye W., Zhou H., Osterwalder M., Dickel D.E., Visel A., et al. Cardiac Reprogramming Factors Synergistically Activate Genome-wide Cardiogenic Stage-Specific Enhancers. Cell Stem Cell. 2019;25:69–86.e5. doi: 10.1016/j.stem.2019.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stone N.R., Gifford C.A., Thomas R., Pratt K.J.B., Samse-Knapp K., Mohamed T.M.A., Radzinsky E.M., Schricker A., Ye L., Yu P., et al. Context-Specific Transcription Factor Functions Regulate Epigenomic and Transcriptional Dynamics during Cardiac Reprogramming. Cell Stem Cell. 2019;25:87–102.e9. doi: 10.1016/j.stem.2019.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou H., Dickson M.E., Kim M.S., Bassel-Duby R., Olson E.N. Akt1/protein kinase B enhances transcriptional reprogramming of fibroblasts to functional cardiomyocytes. Proc. Natl. Acad. Sci. USA. 2015;112:11864–11869. doi: 10.1073/pnas.1516237112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Y., Wang L., Vaseghi H.R., Liu Z., Lu R., Alimohamadi S., Yin C., Fu J.D., Wang G.G., Liu J., Qian L. Bmi1 Is a Key Epigenetic Barrier to Direct Cardiac Reprogramming. Cell Stem Cell. 2016;18:382–395. doi: 10.1016/j.stem.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abad M., Hashimoto H., Zhou H., Morales M.G., Chen B., Bassel-Duby R., Olson E.N. Notch Inhibition Enhances Cardiac Reprogramming by Increasing MEF2C Transcriptional Activity. Stem Cell Rep. 2017;8:548–560. doi: 10.1016/j.stemcr.2017.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garry G.A., Bezprozvannaya S., Chen K., Zhou H., Hashimoto H., Morales M.G., Liu N., Bassel-Duby R., Olson E.N. The histone reader PHF7 cooperates with the SWI/SNF complex at cardiac super enhancers to promote direct reprogramming. Nat. Cell Biol. 2021;23:467–475. doi: 10.1038/s41556-021-00668-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Addis R.C., Ifkovits J.L., Pinto F., Kellam L.D., Esteso P., Rentschler S., Christoforou N., Epstein J.A., Gearhart J.D. Optimization of direct fibroblast reprogramming to cardiomyocytes using calcium activity as a functional measure of success. J. Mol. Cell. Cardiol. 2013;60:97–106. doi: 10.1016/j.yjmcc.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ifkovits J.L., Addis R.C., Epstein J.A., Gearhart J.D. Inhibition of TGFbeta signaling increases direct conversion of fibroblasts to induced cardiomyocytes. PLoS One. 2014;9 doi: 10.1371/journal.pone.0089678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christoforou N., Chellappan M., Adler A.F., Kirkton R.D., Wu T., Addis R.C., Bursac N., Leong K.W. Transcription factors MYOCD, SRF, Mesp1 and SMARCD3 enhance the cardio-inducing effect of GATA4, TBX5, and MEF2C during direct cellular reprogramming. PLoS One. 2013;8 doi: 10.1371/journal.pone.0063577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao Y., Londono P., Cao Y., Sharpe E.J., Proenza C., O'Rourke R., Jones K.L., Jeong M.Y., Walker L.A., Buttrick P.M., et al. High-efficiency reprogramming of fibroblasts into cardiomyocytes requires suppression of pro-fibrotic signalling. Nat. Commun. 2015;6:8243. doi: 10.1038/ncomms9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirai H., Katoku-Kikyo N., Keirstead S.A., Kikyo N. Accelerated direct reprogramming of fibroblasts into cardiomyocyte-like cells with the MyoD transactivation domain. Cardiovasc. Res. 2013;100:105–113. doi: 10.1093/cvr/cvt167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L., Liu Z., Yin C., Asfour H., Chen O., Li Y., Bursac N., Liu J., Qian L. Stoichiometry of Gata4, Mef2c, and Tbx5 influences the efficiency and quality of induced cardiac myocyte reprogramming. Circ. Res. 2015;116:237–244. doi: 10.1161/CIRCRESAHA.116.305547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L., Huang P., Near D., Ravi K., Xu Y., Liu J., Qian L. Isoform Specific Effects of Mef2C during Direct Cardiac Reprogramming. Cells. 2020;9 doi: 10.3390/cells9020268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Furtado M.B., Costa M.W., Pranoto E.A., Salimova E., Pinto A.R., Lam N.T., Park A., Snider P., Chandran A., Harvey R.P., et al. Cardiogenic genes expressed in cardiac fibroblasts contribute to heart development and repair. Circ. Res. 2014;114:1422–1434. doi: 10.1161/CIRCRESAHA.114.302530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vandana J.J., Lacko L.A., Chen S. Phenotypic technologies in stem cell biology. Cell Chem. Biol. 2021;28:257–270. doi: 10.1016/j.chembiol.2021.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou P., Li Y., Zhang X., Liu C., Guan J., Li H., Zhao T., Ye J., Yang W., Liu K., et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science. 2013;341:651–654. doi: 10.1126/science.1239278. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Y., Zhao T., Guan J., Zhang X., Fu Y., Ye J., Zhu J., Meng G., Ge J., Yang S., et al. A XEN-like State Bridges Somatic Cells to Pluripotency during Chemical Reprogramming. Cell. 2015;163:1678–1691. doi: 10.1016/j.cell.2015.11.017. [DOI] [PubMed] [Google Scholar]
- 30.Zhao T., Fu Y., Zhu J., Liu Y., Zhang Q., Yi Z., Chen S., Jiao Z., Xu X., Xu J., et al. Single-Cell RNA-Seq Reveals Dynamic Early Embryonic-like Programs during Chemical Reprogramming. Cell Stem Cell. 2018;23:31–45.e7. doi: 10.1016/j.stem.2018.05.025. [DOI] [PubMed] [Google Scholar]
- 31.Hu W., Qiu B., Guan W., Wang Q., Wang M., Li W., Gao L., Shen L., Huang Y., Xie G., et al. Direct Conversion of Normal and Alzheimer's Disease Human Fibroblasts into Neuronal Cells by Small Molecules. Cell Stem Cell. 2015;17:204–212. doi: 10.1016/j.stem.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 32.Li X., Zuo X., Jing J., Ma Y., Wang J., Liu D., Zhu J., Du X., Xiong L., Du Y., et al. Small-Molecule-Driven Direct Reprogramming of Mouse Fibroblasts into Functional Neurons. Cell Stem Cell. 2015;17:195–203. doi: 10.1016/j.stem.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 33.Yang Y., Liu B., Xu J., Wang J., Wu J., Shi C., Xu Y., Dong J., Wang C., Lai W., et al. Derivation of Pluripotent Stem Cells with In Vivo Embryonic and Extraembryonic Potency. Cell. 2017;169:243–257.e25. doi: 10.1016/j.cell.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guan J., Wang G., Wang J., Zhang Z., Fu Y., Cheng L., Meng G., Lyu Y., Zhu J., Li Y., et al. Chemical reprogramming of human somatic cells to pluripotent stem cells. Nature. 2022;605:325–331. doi: 10.1038/s41586-022-04593-5. [DOI] [PubMed] [Google Scholar]
- 35.Xiang C., Du Y., Meng G., Soon Yi L., Sun S., Song N., Zhang X., Xiao Y., Wang J., Yi Z., et al. Long-term functional maintenance of primary human hepatocytes in vitro. Science. 2019;364:399–402. doi: 10.1126/science.aau7307. [DOI] [PubMed] [Google Scholar]
- 36.Liu L., Lei I., Karatas H., Li Y., Wang L., Gnatovskiy L., Dou Y., Wang S., Qian L., Wang Z. Targeting Mll1 H3K4 methyltransferase activity to guide cardiac lineage specific reprogramming of fibroblasts. Cell Discov. 2016;2 doi: 10.1038/celldisc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo Y., Lei I., Tian S., Gao W., Hacer K., Li Y., Wang S., Liu L., Wang Z. Chemical suppression of specific C-C chemokine signaling pathways enhances cardiac reprogramming. J. Biol. Chem. 2019;294:9134–9146. doi: 10.1074/jbc.RA118.006000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lichti U., Anders J., Yuspa S.H. Isolation and short-term culture of primary keratinocytes, hair follicle populations and dermal cells from newborn mice and keratinocytes from adult mice for in vitro analysis and for grafting to immunodeficient mice. Nat. Protoc. 2008;3:799–810. doi: 10.1038/nprot.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jesty S.A., Steffey M.A., Lee F.K., Breitbach M., Hesse M., Reining S., Lee J.C., Doran R.M., Nikitin A.Y., Fleischmann B.K., Kotlikoff M.I. c-kit+ precursors support postinfarction myogenesis in the neonatal, but not adult, heart. Proc. Natl. Acad. Sci. USA. 2012;109:13380–13385. doi: 10.1073/pnas.1208114109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang L., Liu Z., Yin C., Zhou Y., Liu J., Qian L. Improved Generation of Induced Cardiomyocytes Using a Polycistronic Construct Expressing Optimal Ratio of Gata4, Mef2c and Tbx5. J. Vis. Exp. 2015:53426. doi: 10.3791/53426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu Z., Chen O., Wall J.B.J., Zheng M., Zhou Y., Wang L., Vaseghi H.R., Qian L., Liu J. Systematic comparison of 2A peptides for cloning multi-genes in a polycistronic vector. Sci. Rep. 2017;7:2193. doi: 10.1038/s41598-017-02460-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahier A., Jarriault S. Simultaneous expression of multiple proteins under a single promoter in Caenorhabditis elegans via a versatile 2A-based toolkit. Genetics. 2014;196:605–613. doi: 10.1534/genetics.113.160846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daniels R.W., Rossano A.J., Macleod G.T., Ganetzky B. Expression of multiple transgenes from a single construct using viral 2A peptides in Drosophila. PLoS One. 2014;9 doi: 10.1371/journal.pone.0100637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Veeriah S., Brennan C., Meng S., Singh B., Fagin J.A., Solit D.B., Paty P.B., Rohle D., Vivanco I., Chmielecki J., et al. The tyrosine phosphatase PTPRD is a tumor suppressor that is frequently inactivated and mutated in glioblastoma and other human cancers. Proc. Natl. Acad. Sci. USA. 2009;106:9435–9440. doi: 10.1073/pnas.0900571106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Peyser N.D., Du Y., Li H., Lui V., Xiao X., Chan T.A., Grandis J.R. Loss-of-Function PTPRD Mutations Lead to Increased STAT3 Activation and Sensitivity to STAT3 Inhibition in Head and Neck Cancer. PLoS One. 2015;10 doi: 10.1371/journal.pone.0135750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X., Guo A., Yu J., Possemato A., Chen Y., Zheng W., Polakiewicz R.D., Kinzler K.W., Vogelstein B., Velculescu V.E., Wang Z.J. Identification of STAT3 as a substrate of receptor protein tyrosine phosphatase T. Proc. Natl. Acad. Sci. USA. 2007;104:4060–4064. doi: 10.1073/pnas.0611665104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen Y.W., Guo T., Shen L., Wong K.Y., Tao Q., Choi W.W.L., Au-Yeung R.K.H., Chan Y.P., Wong M.L.Y., Tang J.C.O., et al. Receptor-type tyrosine-protein phosphatase kappa directly targets STAT3 activation for tumor suppression in nasal NK/T-cell lymphoma. Blood. 2015;125:1589–1600. doi: 10.1182/blood-2014-07-588970. [DOI] [PubMed] [Google Scholar]
- 48.Su F., Ren F., Rong Y., Wang Y., Geng Y., Wang Y., Feng M., Ju Y., Li Y., Zhao Z.J., et al. Protein tyrosine phosphatase Meg2 dephosphorylates signal transducer and activator of transcription 3 and suppresses tumor growth in breast cancer. Breast Cancer Res. 2012;14:R38. doi: 10.1186/bcr3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tkach M., Rosemblit C., Rivas M.A., Proietti C.J., Díaz Flaqué M.C., Mercogliano M.F., Beguelin W., Maronna E., Guzmán P., Gercovich F.G., et al. p42/p44 MAPK-mediated Stat3Ser727 phosphorylation is required for progestin-induced full activation of Stat3 and breast cancer growth. Endocr. Relat. Cancer. 2013;20:197–212. doi: 10.1530/ERC-12-0194. [DOI] [PubMed] [Google Scholar]
- 50.Tammineni P., Anugula C., Mohammed F., Anjaneyulu M., Larner A.C., Sepuri N.B.V. The import of the transcription factor STAT3 into mitochondria depends on GRIM-19, a component of the electron transport chain. J. Biol. Chem. 2013;288:4723–4732. doi: 10.1074/jbc.M112.378984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo X., Ribeiro M., Bray E.R., Lee D.H., Yungher B.J., Mehta S.T., Thakor K.A., Diaz F., Lee J.K., Moraes C.T., et al. Enhanced Transcriptional Activity and Mitochondrial Localization of STAT3 Co-induce Axon Regrowth in the Adult Central Nervous System. Cell Rep. 2016;15:398–410. doi: 10.1016/j.celrep.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gough D.J., Corlett A., Schlessinger K., Wegrzyn J., Larner A.C., Levy D.E. Mitochondrial STAT3 supports Ras-dependent oncogenic transformation. Science. 2009;324:1713–1716. doi: 10.1126/science.1171721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wegrzyn J., Potla R., Chwae Y.J., Sepuri N.B.V., Zhang Q., Koeck T., Derecka M., Szczepanek K., Szelag M., Gornicka A., et al. Function of mitochondrial Stat3 in cellular respiration. Science. 2009;323:793–797. doi: 10.1126/science.1164551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Singh V.P., Pinnamaneni J.P., Pugazenthi A., Sanagasetti D., Mathison M., Wang K., Yang J., Rosengart T.K. Enhanced Generation of Induced Cardiomyocytes Using a Small-Molecule Cocktail to Overcome Barriers to Cardiac Cellular Reprogramming. J. Am. Heart Assoc. 2020;9 doi: 10.1161/JAHA.119.015686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang H., Keepers B., Qian Y., Xie Y., Colon M., Liu J., Qian L. Cross-lineage potential of Ascl1 uncovered by comparing diverse reprogramming regulatomes. Cell Stem Cell. 2022;29:1491–1504.e9. doi: 10.1016/j.stem.2022.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frangogiannis N.G. Regulation of the inflammatory response in cardiac repair. Circ. Res. 2012;110:159–173. doi: 10.1161/CIRCRESAHA.111.243162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nahrendorf M., Pittet M.J., Swirski F.K. Monocytes: protagonists of infarct inflammation and repair after myocardial infarction. Circulation. 2010;121:2437–2445. doi: 10.1161/CIRCULATIONAHA.109.916346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou H., Morales M.G., Hashimoto H., Dickson M.E., Song K., Ye W., Kim M.S., Niederstrasser H., Wang Z., Chen B., et al. ZNF281 enhances cardiac reprogramming by modulating cardiac and inflammatory gene expression. Genes Dev. 2017;31:1770–1783. doi: 10.1101/gad.305482.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Callahan J.F., Burgess J.L., Fornwald J.A., Gaster L.M., Harling J.D., Harrington F.P., Heer J., Kwon C., Lehr R., Mathur A., et al. Identification of novel inhibitors of the transforming growth factor beta1 (TGF-beta1) type 1 receptor (ALK5) J. Med. Chem. 2002;45:999–1001. doi: 10.1021/jm010493y. [DOI] [PubMed] [Google Scholar]
- 60.Maherali N., Hochedlinger K. Tgfbeta signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr. Biol. 2009;19:1718–1723. doi: 10.1016/j.cub.2009.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu J.S.L., Palano G., Lim C., Moggio A., Drowley L., Plowright A.T., Bohlooly-Y M., Rosen B.S., Hansson E.M., Wang Q.D., Yusa K. CRISPR-Knockout Screen Identifies Dmap1 as a Regulator of Chemically Induced Reprogramming and Differentiation of Cardiac Progenitors. Stem Cell. 2019;37:958–972. doi: 10.1002/stem.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prodan N., Ershad F., Reyes-Alcaraz A., Li L., Mistretta B., Gonzalez L., Rao Z., Yu C., Gunaratne P.H., Li N., et al. Direct reprogramming of cardiomyocytes into cardiac Purkinje-like cells. iScience. 2022;25 doi: 10.1016/j.isci.2022.105402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang H., Cao N., Spencer C.I., Nie B., Ma T., Xu T., Zhang Y., Wang X., Srivastava D., Ding S. Small molecules enable cardiac reprogramming of mouse fibroblasts with a single factor, Oct4. Cell Rep. 2014;6:951–960. doi: 10.1016/j.celrep.2014.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gao E., Lei Y.H., Shang X., Huang Z.M., Zuo L., Boucher M., Fan Q., Chuprun J.K., Ma X.L., Koch W.J. A novel and efficient model of coronary artery ligation and myocardial infarction in the mouse. Circ. Res. 2010;107:1445–1453. doi: 10.1161/CIRCRESAHA.110.223925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ehler E., Moore-Morris T., Lange S. Isolation and culture of neonatal mouse cardiomyocytes. J. Vis. Exp. 2013;79 doi: 10.3791/50154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Judd J., Lovas J., Huang G.N. Isolation, Culture and Transduction of Adult Mouse Cardiomyocytes. J. Vis. Exp. 2016;2016 doi: 10.3791/54012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
This paper does not report original code.
-
•
RNA-seq data have been deposited at GEO and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.