Summary
KcsA is a potassium channel with a plethora of structural and functional information, but its activity in the KcsA-producing actinomycete membranes remains elusive. To determine lipid species involved in channel-modulation, a surface plasmon resonance (SPR)-based methodology, characterized by immobilization of membrane proteins under a membrane environment, was applied. Dianionic cardiolipin (CL) showed extremely higher affinity for KcsA than monoanionic lipids. The SPR experiments further demonstrated that CL bound not only to the N-terminal M0 helix, a lipid-sensor domain, but to the M0 helix-deleted mutant. In contrast, monoanionic lipids interacted primarily with the M0 helix. This indicates the presence of an alternative CL-binding site, plausibly in the transmembrane domain. Single-channel recordings demonstrated that CL enhanced channel opening in an M0-independent manner. Taken together, the action of monoanionic lipids is exclusively mediated by the M0 helix, while CL binds both the M0 helix and its specific site, further enhancing the channel activity.
Subject areas: Biological sciences, Biochemistry, Membrane architecture
Graphical abstract
Highlights
-
•
Cardiolipin has a stronger interaction with KcsA than monoanionic lipids
-
•
Cardiolipin binds to both the lipid-sensing M0 helix and the transmembrane site
-
•
The action of monoanionic lipids on KcsA is exclusively mediated by the M0 helix
-
•
Cardiolipin can enhance KcsA channel opening without binding to the M0 helix
Biological sciences; Biochemistry; Membrane architecture
Introduction
The structural and functional integrity of membrane proteins (MPs) is determined by the membrane environment.1,2,3,4,5,6,7,8,9,10,11,12,13,14 The effects of membranes on MP structure and function can be explained by direct interactions between these MPs and the membrane lipids,2,3,4,5,6,7,8,9,10,11,12 or physicochemical properties, such as thickness, tension, and lateral pressure.13,14,15,16,17,18,19 In particular, recent crystallographic studies of MPs have revealed the presence of specifically binding lipid molecules,20,21,22,23 suggesting the prevalence of direct MP–lipid interactions. However, systematic studies on this topic are still incipient, mainly because of the lack of a practical experimental methodology. Therefore, understanding whether specific lipids affect the structural and functional features of individual MP systems, and the related mechanisms involved, remains a challenge.24
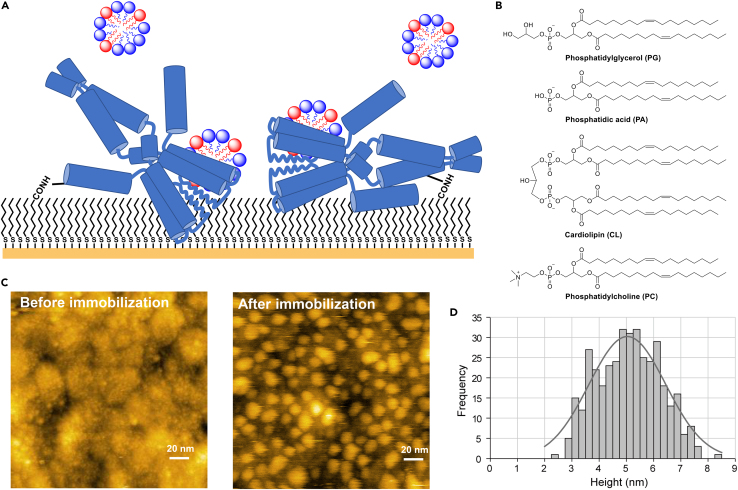
To address this issue, we recently developed a surface plasmon resonance (SPR)-based method for quantitative analysis of the interactions between MPs and lipids (Figure 1A),25 which we called the self-assembled monolayer-assisted MP-lipid interaction analysis (SAMPLIA). In this method, the SPR sensor chip surface was modified with a self-assembled monolayer (SAM), which allowed for more efficient immobilization of MPs and provided a partial membranous environment to these immobilized MPs. The affinity of various types of lipids to MPs was sensitively and quantitatively evaluated by the addition of a solubilized lipid solution to the immobilized MPs. Examination of halobacterial MP bacteriorhodopsin (bR) using the SAMPLIA method not only quantified the interaction with various lipids, but also identified a native glycolipid, S-TGA-1, as the specifically-interacting lipid.25 Further analysis revealed that S-TGA-1 promoted trimer formation of bR and enhanced its light-driven proton pump activity.26 Although bR is one of the most extensively studied MPs, the effect of native lipids on bR was not fully elucidated until our study. These findings demonstrate the general applicability of the SAMPLIA method to various MPs, which could facilitate the investigation of the role of lipids in MP activity.
Figure 1.
SAMPLIA method for KcsA–lipid interaction
(A) Schematic of KcsA–lipid interaction analysis. KcsA molecules are covalently immobilized onto a sensor chip coated with SAM composed of mercaptoalkylcarboxylic acid. Solubilized lipids are added to detect the interaction with KcsA.25
(B) Chemical structures of lipids.
(C) AFM images of SAM-coated SRP sensor chip before and after fl-KcsA immobilization. To facilitate observation, KcsA was immobilized at a lower concentration than in the SPR measurements.
(D) Height distribution pattern of fl-KcsA on the SAM-coated SPR sensor chip. The gray line indicates the result of Gaussian fitting. The height was 5.0 ± 1.4 nm (n = 385).
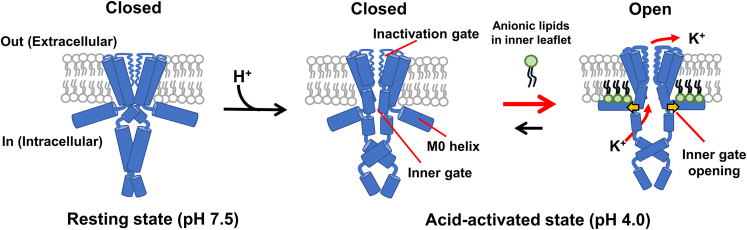
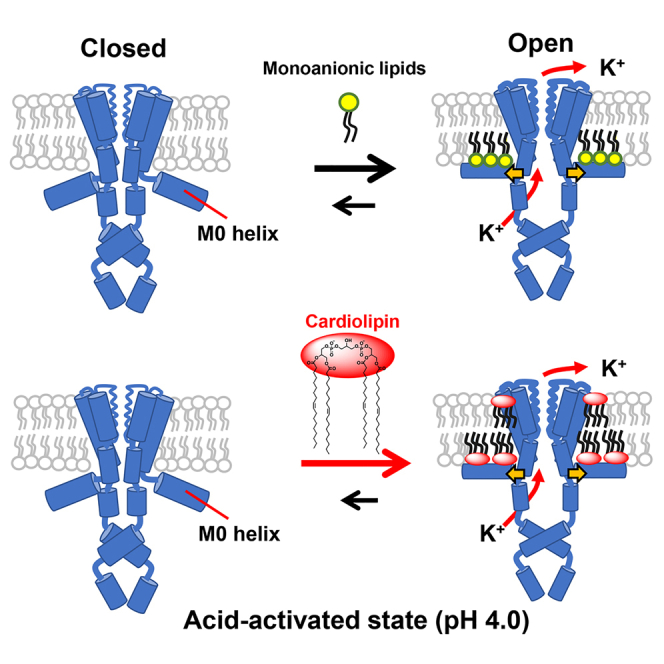
KcsA from Streptomyces lividans, is a homotetrameric potassium channel with high structural similarity to eukaryotic congeners.27,28,29 KcsA is activated by an intracellular acidic pH,30,31,32,33 and negatively charged phospholipids are believed to modulate channel activity (Figure 2).34,35,36,37,38,39 Since the co-crystal structure showed that phosphatidylglycerol (PG) is bound at the inter-subunit groove on the extracellular side of KcsA,29,35 a number of studies have postulated that the lipid binding to the groove is related to the gating properties of the channel.39 Some studies hypothesized that the binding stabilizes the conductive conformation of the selectivity filter and inhibits the inactivation process,40,41,42,43 whereas others contradictorily assumed that the binding prevents channel cluster formation and consequently modulates channel conductance.44,45
Figure 2.
Gating states of the KcsA channel
The gate does not open at a neutral pH. At an acidic pH on the intracellular side, the channel becomes acid-activated; the pH sensor in the cytoplasmic domain changes conformation, and the inner gate is ready to open via a global twisting motion. Negatively charged lipids in the inner leaflet make the channel fully open via binding to the N-terminal amphipathic M0 helix.34
In contrast, our functional study using asymmetric membranes revealed that monoanionic lipids in the inner leaflet stabilized the open conformation via electrostatic interactions with the N-terminal amphipathic M0 helix, which is a lipid sensor lying at the membrane interface (Figure 2).34,46 Thus, the study clearly demonstrated that negatively charged lipids are required for the channel to be fully open.34,46
In this study, the SAMPLIA method was first applied to quantitatively analyze the interactions between KcsA and anionic lipids (Figure 1B). Although PG has been frequently used as a specific lipid for KcsA activation, it is not present in KcsA-producing actinomycete membranes,47,48,49 where phosphatidic acid (PA) and cardiolipin (CL) are the main anionic lipid constituents.47 Therefore, the binding of PA and CL to KcsA was examined using PG as a positive control. In parallel, single-channel recordings using the contact bubble bilayer (CBB) method50,51,52 were performed to examine the functional contribution of these lipids to the gating of the KcsA channel. To distinguish M0 helix-dependent and -independent actions, full-length KcsA (fl-KcsA) and an M0-deleted mutant (ΔM0-KcsA) were used. The combined approach of the SPR-based interaction analysis and the electrophysiological method revealed the presence of M0 helix-independent action of CL on the KcsA channel.
Results
CL interacts more strongly than PG and PA with KcsA
KcsA was immobilized on SAM-modified sensor chips at ∼3000 resonance units, which enabled us to clearly observe the interactions between KcsA and lipid molecules.25 According to the crystal structure of KcsA,29 the length of the SAM (approximately 1 nm) was insufficiently thick to completely cover and accommodate the transmembrane domain (TMD) of KcsA; thus, a considerable part of the protein should be outside of the SAM surface (Figure 1A).
To confirm the immobilization of KcsA and gain insight into the orientation of immobilized KcsA, we performed atomic force microscope (AFM) observations of the SAM-modified SPR sensor chip with and without fl-KcsA immobilization (Figure 1C). The histogram is shown for the height of KcsA channel from the SAM surface (Figure 1D), which ranges from 2.5 to 8 nm. Given that the width and longitudinal length of fl-KcsA channel are ca. 3.5 and 10 nm, respectively,53,54,55 and that the SAM length is 1 nm, the histogram range is consistent with the dimension of the channel when slightly immersed into the SAM. Fitting with the Gaussian distribution of the height histogram gave the mean value of 5.0 ± 1.4 nm. These data suggest that the channel is somewhat immersed into the SAM and inclined at various angles from horizontal to vertical centering around mid-angles, thus taking random orientation. This random orientation would allow for unrestricted contact between the immobilized KcsA and extraneously added lipids. We further evaluated the particle size of lipid-detergent mixtures and confirmed that the lipids were solubilized in SPR running buffer containing the detergent Tween 20 (Table S1).
Prior to analyzing the interaction between fl-KcsA and anionic lipids, we examined the binding of immobilized fl-KcsA to tetrabutylammonium (TBA), a specific blocker of KcsA (Figure S1).56 KcsA that was co-crystallized with TBA revealed a binding configuration of TBA, with its four butyl chains extending to the subunits.57 The binding affinity of TBA to KcsA deduced from the present SPR study (Figure S1) strongly suggests that KcsA retains its tetrameric form on the chip (Figure 1A).
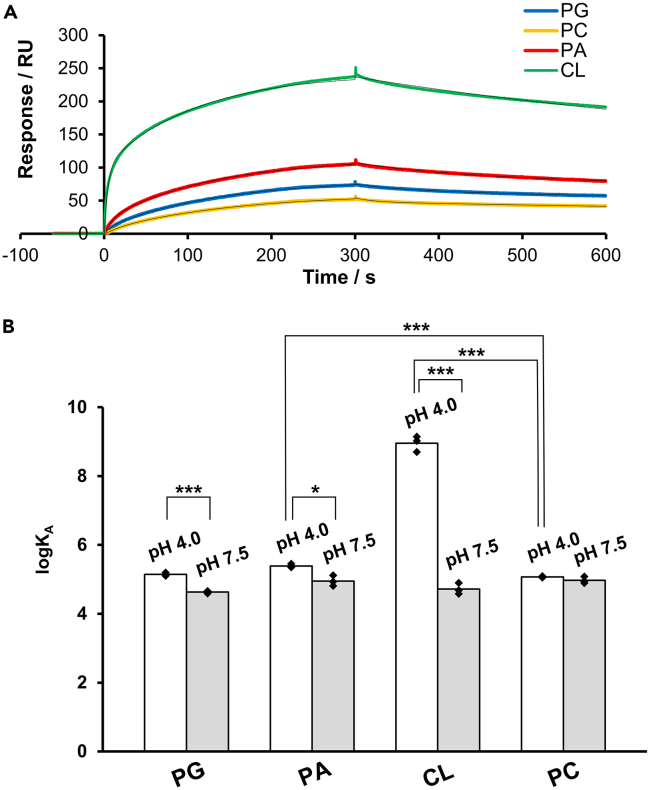
Next, we evaluated the interaction between PG and fl-KcsA to confirm the validity of the SAMPLIA method.34,35,36,37 Since KcsA adopts an acid-activated state at acidic pH and a resting state at neutral pH (Figure 2), both acidic (pH 4.0) and neutral (pH 7.5) buffers were used for the analysis. The sensorgrams were recorded upon starting of perfusion containing lipids, and were fitted to a 1:2 heterogeneous ligand binding model that assumed that the lipid binds to KcsA at two different sites (Table S2).58,59 The smaller association constants (KA) were considered to be due to non-specific or less relevant binding, because their maximum binding values (Rmax) were much smaller than those of the larger KAs (Table S2), indicating little contribution to binding. Therefore, we focused on larger KA values in the subsequent experiments. Further verification of this treatment for the two KA values is given in the supplementary note including Figure S3 and Table S6. The SAMPLIA method revealed that PG binds fl-KcsA more strongly at pH 4.0 than at pH 7.5 (Figures 3; S2; and Table S2). On the other hand, the interaction between fl-KcsA and PC was not sensitive to the pH change (Figure 3B; and Table S2). This result is consistent with those of previous studies showing that the KcsA channel opens in the presence of PG at acidic pH,34,36,37 verifying that the SAMPLIA method can be used to properly evaluate KcsA–lipid interactions.
Figure 3.
Analysis of the interaction of PG, PA, CL, and PC with fl-KcsA
(A) SPR sensorgrams showing the interactions of PG, PC, PA, and CL with fl-KcsA immobilized on the C6-SAM modified sensor chip; 100 μM of the lipids dissolved in acidic buffer (pH 4.0) were injected at a flow rate of 30 μL min−1. Colored lines indicate experimentally obtained sensorgrams, while black lines indicate fitted curves.
(B) the affinity of PG, PA, CL, and PC toward fl-KcsA in acidic (pH 4.0) and neutral (pH 7.5) buffers, calculated from the sensorgrams by fitting to a 1:2 heterogeneous ligand binding model, as listed in Table S2, and higher association constants were adopted (see text). Asterisks denote statistical significance (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). Data presented as mean ± SD (n = 3) are given in Table S2. See also Figure S2.
We subsequently evaluated the interaction of fl-KcsA with PA and CL, which are main components of Streptomyces membranes. PA also showed significantly higher interaction with fl-KcsA at pH 4.0 than at 7.5, and when compared at pH 4.0, PA had a significantly higher affinity for fl-KcsA than PC (Figures 3; S2; and Table S2). Surprisingly, CL showed a drastically different affinity for KcsA between acidic and neutral conditions, and the affinity constant KA between CL and KcsA reached almost 109 at pH 4.0 (Figure 3; and Table S2), showing that CL has extremely high affinity for acid-activated KcsA. At pH 7.5, CL and other anionic lipids showed affinity comparable to that of PC (Figure 3B), suggesting that the interaction of these lipids is less specific for resting-state KcsA. The extremely high affinity of CL for KcsA at pH 4 indicates that a specific binding site appears for CL in acid-activated KcsA. This was further supported by the kinetic data of the interaction (Table S3), which shows that CL has a markedly smaller koff at pH 4.0 than other monoanionic lipids, suggesting its slow dissociation from the specific binding site.
In this SAMPLIA experiment, we used lipid concentration of 100 μM. Considering that CL is reported to account for more than 10% of total phospholipid in S. lividans,60 the lipid concentrations adopted in the SAMPLIA would not be far from physiological conditions in terms of contact probability between KcsA and CL.
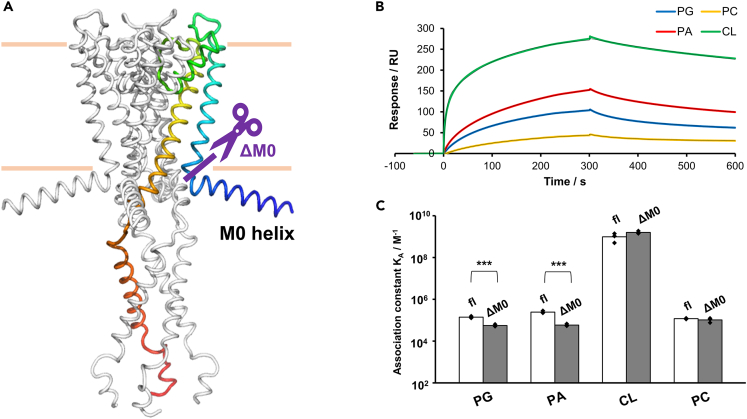
CL binds to an alternative site other than the M0 helix
Previous studies have shown that the M0 helix enables the sensing of monoanionic phospholipids, such as PG, in the membrane and shifts the equilibrium toward the open conformation of KcsA.34 To understand the contribution of the M0 helix to the high-affinity binding of dianionic CL, an M0 helix-deleted mutant of KcsA, ΔM0-KcsA (Figure 4A), was used. CL bound to ΔM0-KcsA tightly, and the KA value was indistinguishable from that to the fl-KcsA channel (Figures 4B and 4C). In contrast, binding of PA and PG was significantly weaker to the ΔM0-KcsA channel than to fl-KcsA (p < 0.001) (Figures 4B and 4C). These results indicate that CL has an additional binding site other than the M0 helix, while the M0 helix is the main binding site for PA and PG. On the other hand, PC binding did not differ between fl-KcsA and ΔM0-KcsA (Figures 4B and 4C), suggesting that PC binds less-specifically to KcsA without specifically recognizing the M0 helix.
Figure 4.
Effect of truncation of the M0 helix on the affinity of lipids toward KcsA
(A): Location of deletion on KcsA. ΔM0-KcsA is the mutant in which the N-terminal M0-helix domain was truncated (purple line).
(B): SPR sensorgrams showing the interactions of PG, PA, CL, and PC with ΔM0-KcsA immobilized on the C6-SAM modified sensor chip; 100 μM of the lipids dissolved in acidic buffer (pH 4.0) were injected at a flow rate of 30 μL min−1. Colored lines indicate experimentally obtained sensorgrams, while black lines indicate fitted curves.
(C) Affinity of PG, PA, CL, and PC toward fl-KcsA and ΔM0-KcsA in acidic buffer (pH 4.0). Each affinity was calculated from the sensorgrams by fitting to a 1:2 heterogeneous ligand binding model, as listed in Table S4. Asterisks denote statistical significance (∗∗∗p < 0.001). Data presented as mean ± SD (n = 3) are given in Table S4.
Then, the involvement of the M0 helix in CL binding was examined using an M0 helix-mimicking peptide (Figure 5A). In this experiment, liposomes containing anionic lipids were immobilized on the SPR sensor chip, whose surface was modified with an alkyl chain (Figure 5B),61 and the interaction with the peptide was evaluated. Although some methods are known for immobilizing lipid molecules on the sensor chip, such as using biotin-tagged lipids, the liposome immobilization adopted in this study seems more appropriate because the M0 domain is thought to interact with lipid molecules present in the bilayers, as schematically depicted in Figure 1A. Figure 5C shows the resultant sensorgrams, which oddly began to decrease during the association process (up to 300 s), and thus could not be fitted by theoretical curves. This is probably because the M0 peptide had some effect on the immobilized liposomes and stripped them from the sensor chip surface. Nevertheless, the data confirmed that the M0 peptide strongly associated with CL to a comparable extent as the monoanionic lipids, PG and PA (Figure 5C). These results clearly show that CL interacts with acid-activated KcsA at multiple sites, including the M0 helix, whereas PA and PG bind preferentially to the M0 helix.
Figure 5.
Analysis of the interactions of M0 peptide with liposomes immobilized on the SPR sensor chip
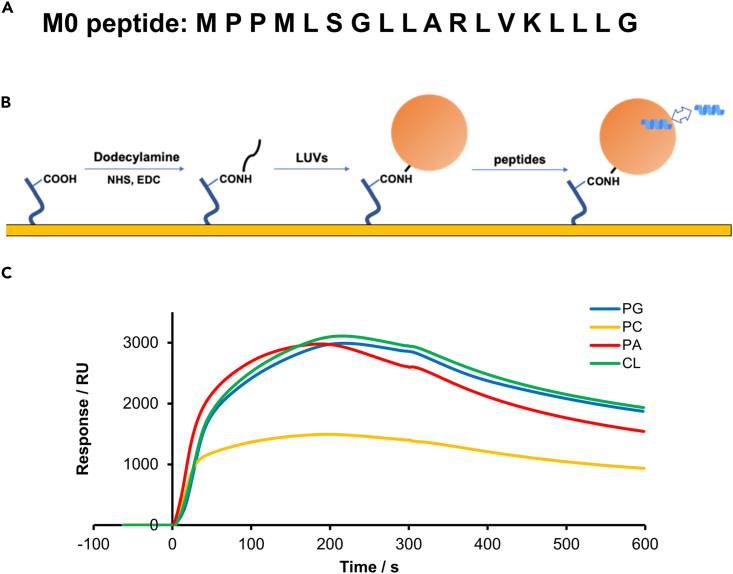
(A) Amino acid sequence of wild type M0 peptide.
(B) Procedure for analyzing the interaction.61 Dodecylamine was coupled with COOH group on the CM5 sensor chip via a conventional amine coupling method. Next, 100 nm LUV solution was injected to be immobilized onto the chip surface. Then the peptide aqueous solution was added to the immobilized liposomes and their interactions were analyzed.
(C) Sensorgrams indicating the interaction of M0 peptide to pure PC, PG/PC (1:9), PA/PC (1:9), and CL/PC (1:9) membranes in acidic buffer (pH 4.0).
CL enhances opening of ΔM0-KcsA channel
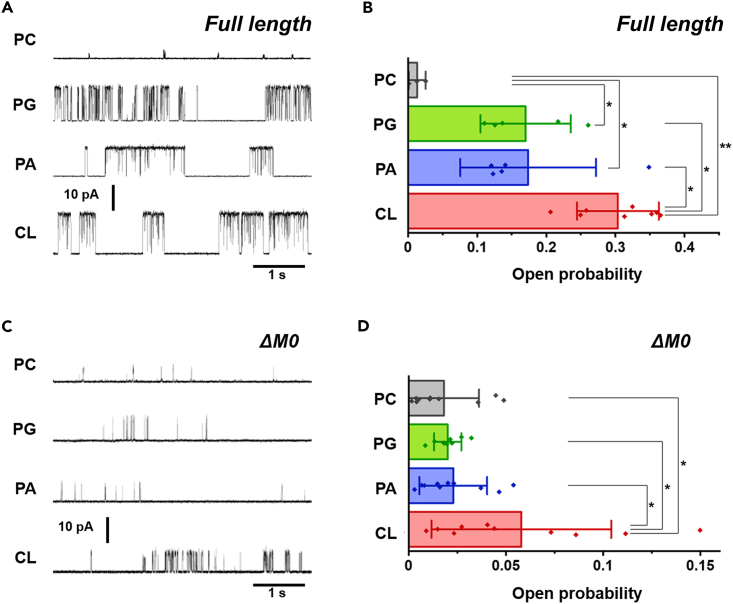
To gain further insight into the functional outcomes of CL binding, we performed single-channel current recordings of acid-activated KcsA (Figure 2) using the contact bubble bilayer (CBB) method.50,51,52 As we previously reported, the open probability (Popen) of fl-KcsA in the pure 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) bilayer was approximately 1% (Figures 6A, 6B; and Table S5),34 and this value was used as a control in the present study. In the presence of anionic lipids (25 wt %), the fl-KcsA channel exhibited substantially high Popen values. For example, Popen was 17% for the PG-containing membrane (Figure 6A, 6B; Table S5), which is consistent with our previous report using bilayers containing 100% anionic lipids.34 In the case of CL (Figure 6A, and 6B), the Popen value (30%) was significantly higher than those of other monoanionic lipids (p < 0.05), which is relevant to the higher CL binding affinity revealed by the SPR experiments (Figure 3).
Figure 6.
Single-channel recordings of KcsA in various lipid membranes
The KcsA channel is activated by intracellular acidic pH.
Therefore, by making the pH conditions asymmetric (pH4/pH7.5) across the membrane, the orientation of the “active” channel in the membrane can be controlled so that only channel molecule with its cytoplasmic domain facing acidic side contributes to the single-channel current.
(A) Typical single-channel current traces of the fl-KcsA channel in PC (control) and anionic-lipid-containing PC membranes at 100 mV. Single-channel conductance differs depending on lipid membrane, owing to the surface potential of negatively charged lipids.34
(B) Open probability (Popen) in the membranes with different compositions for fl-KcsA. Error bars represent mean ± SD (n = 3–8). Asterisks denote statistical significance (∗p < 0.05, ∗∗p < 0.01). Popen values are listed in Table S5.
(C) Typical single-channel current traces of the ΔM0 mutant at 100 mV.
(D) Popen of the ΔM0 mutant in the membranes with different compositions for ΔM0-KcsA. Error bars represent mean ± SD (n = 9–10). Asterisks denote statistical significance (p < 0.05). Popen values are listed in Table S5.
We then performed single-channel recording experiments for ΔM0-KcsA (Figures 6C and 6D). In the absence of anionic lipids (control), Popen (2%) was comparable to the fl-KcsA channel (Table S5), indicating that the M0 helix itself does not modify channel gating. In the presence of PA and PG, the Popen values did not increase and were indistinguishable from that of the control (Figure 6D). Thus, the effects of PA and PG were exclusively mediated by the M0 helix. In contrast, CL enhanced gating even without the M0 helix (Popen 6%, p < 0.05). Thus, binding of CL to a site different from the M0 helix rendered the channel open. This is consistent with the SPR data showing that ΔM0-KcsA maintained CL binding but weakened the binding to PG or PA (Figure 4C). These results indicate that the CL binding site of the ΔM0-KcsA channel, plausibly in the TMD, is exclusive to CL and not interchangeable with monoanionic lipids. We should also mention that Popen of ΔM0-KcsA in the presence of CL remained lower than that of fl-KcsA. Therefore, CL binding to both M0-helix and TMD contributes to higher Popen of fl-KcsA.
Discussion
The important roles of lipids in MP stability, structure, and function have been reported in various studies.9,62,63 The mechanisms of such effects have generally been explained from two perspectives. One aspect is the effect of lipids on MP function through alternating membrane physicochemical properties, such as membrane fluidity, curvature, lateral pressure, membrane tension, and hydrophobic match. The other aspect is the direct binding of lipids to MPs, which may stabilize specific conformations, change clustering features, and/or affect their functions. KcsA is a good target for in-depth study of lipid effects, because the channel is affected by both aspects; KcsA senses not only membrane tension17,18 but also anionic phospholipids. In terms of the direct effect of anionic lipids on KcsA, two different modes of interaction have been proposed: electrostatic interactions between anionic lipids and the N-terminal M0 helix,34 and interactions between anionic lipids and the inter-subunit groove on the extracellular surface of KcsA.39,40,41,42,43,44,45 Thus, the groove present in the TMD is the candidate for an alternative binding site for CL. Given the pH-dependent binding of CL to KcsA (Figure 3), this groove would dramatically enhance its affinity for CL in the acid-activated state.
Due to the inherent disorder of the M0 helix, either its conformation or its specific binding to lipids has not been revealed.28,45,64 In this study, we demonstrated that the M0 helix is the main binding site for anionic lipids; in particular, monovalent PG and PA bind preferentially to the M0 helix (Figure 4). On the other hand, the lipid-bound crystal structures showed that PG binds to the extracellular inter-subunit groove of KcsA,29,35 and NMR indicated that PA binds to the groove.65 Considering that PA and PG have KA of 105 order for ΔM0-KcsA (Figure 4), they can bind to the groove to some extent, and might be observed in the crystal and NMR structures. Single-channel recordings demonstrated that deletion of the M0 helix completely abolished the effects of monovalent PG and PA (Figure 6). These affinity and functional data collectively confirmed that monoanionic PG and PA may bind to the groove in a less specific manner, but their main binding site is the M0 helix and their action on KcsA is exclusively mediated by their binding to the M0 helix. This is consistent with our previous report demonstrating that PG in the outer membrane did not influence the channel function.34
Importantly, we demonstrated that CL opened the ΔM0-KcsA channel by binding to an alternative site, presumably the inter-subunit groove. There are three Arg residues (Arg 52, 64, 89) around the groove.35,43 The dianionic nature of CL likely allows for its tight binding to these residues, which cannot be attained by monoanionic lipids, consequently modulating the gating activity. Thus, the groove may discriminate the charge number of anionic lipids via multiple electrostatic interactions, distinguishing dianionic CL from monoanionic lipids. The detailed binding site and mode of interaction of CL are currently being studied, and will be reported in due course.
With two distinct binding sites, CL showed a higher open probability for fl-KcsA than the other monoanionic lipids (Figure 6B). Although CL is known to promote the channel opening of KcsA,28,35,37,42,66,67,68 its contribution to the M0 helix has not been addressed so far. The simultaneous binding of CL to both the M0-helix and the groove site may have a synergistic effect on KcsA channel activity, resulting in a significantly higher open probability.
Here, we refer to the SAMPLIA method, which is characteristic of utilizing the SAM.25 Since the SAM is not long enough to fully accommodate the hydrophobic regions of KcsA, its hydrophobic regions are partially exposed to the aqueous phase, making it possible to interact efficiently with extraneous lipid molecules (Figure 1A). The length of the SAM is considered important for the interaction analysis because a long SAM alkyl chain could bury the channel and hamper the contact of the protein with the lipid molecules. Furthermore, the hydrophobic nature of the SAM-modified sensor chip is expected to enable a high level of KcsA immobilization and provide a partially membranous environment for the immobilized KcsA molecules. The affinity data obtained from the SAMPLIA agree with those of the functional experiments, thus demonstrating the applicability of this method to KcsA.
In conclusion, the high-affinity binding of CL to ΔM0-KcsA indicates that CL has an additional binding site, which is dianion-selective (Figures 4 and 6C, 6D). The binding site is most likely the groove site where dianionic CL binds to the multiple positive charges in adjacent subunits. On the other hand, the M0 helix site is interchangeable for mono- and di-anionic lipids. These mechanistic views will be examined in future. Historically, the KcsA channel has been used as a prototypical channel due to the extensive structural and functional information available, whereas the physiologically relevant function of the KcsA channel in KcsA-producing actinomycete membrane has not been addressed. Since CL is a main component of the native membrane,47 the finding that CL binds more strongly to KcsA, and thus renders the channel more active by a mechanism distinct from other monoanionic lipids, provides invaluable insight into the physiological function of the KcsA channel in situ in the life cycle of Streptomyces. CL has recently attracted significant attention as signaling lipid via its binding to proteins,69 and this study provides an additional insight into its effect on MPs via specific binding motifs. In addition, the procedure established in this study, namely identification of channel-binding lipids by SAMPLIA and subsequent physiological analysis, will pave the way for a comprehensive investigation of the biological actions of diverse membrane lipids on channels.
Limitations of the study
This study has revealed the specific interaction between of CL and KcsA, shedding light on its consequential effects on channel gating. While a putative CL binding pocket is discussed in the text, the precise binding site remains undetermined. Moreover, this investigation has raised new questions about which of the two KcsA gates CL affects, along with the mechanism by which CL facilitates channel opening. In pursuit of resolving these inquiries, the ongoing research involves the mutagenesis of KcsA, followed by comprehensive binding and channel assay experiments.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| E. coli BL21(DE3)pLysS | Promega | L1195 |
| Chemicals, peptides, and recombinant proteins | ||
| Cardiolipin | Avanti Polar Lipids | Cat#710335P, CAS: 115404-77-8 |
| Phosphatidylglycerol (DOPG) | Avanti Polar Lipids | Cat#840475P, CAS: 67254-28-8 |
| Phosphatic acid (DOPA) | Avanti Polar Lipids | Cat#840875P, CAS: 108392-02-5 |
| Phosphatidylcholine (DOPC) | Avanti Polar Lipids | Cat#850375P, CAS: 4235-95-4 |
| 6-mercaptohexanoic acid | Sigma-Aldrich | Cat#674974, CAS: 17689-17-7 |
| 6-mercaptohexanol | Sigma-Aldrich | Cat#725226, CAS: 1633-78-9 |
| Succinic acid | Fujifilm-Wako | Cat#194-04335, CAS: 110-15-6 |
| HEPES | Fujifilm-Wako | Cat#346-01373, CAS: 7365-45-9 |
| KCl | Fujifilm-Wako | Cat#163-03545, CAS: 7447-40-7 |
| EDTA | Fujifilm-Wako | Cat#345-01865, CAS: 6381-92-6 |
| Tween 20 | TCI | Cat#T2530, CAS: 9005-64-5 |
| Dodecylamine | Sigma-Aldrich | Cat#325163-5ML, CAS: 124-22-1 |
| 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide | TCI | Cat#D1601, CAS: 25952-53-8 |
| N-hydrosuccinimide | TCI | Cat#H0623, CAS: 6066-82-6 |
| HBS-EP buffer | Cytiva | Cat#BR100188 |
| Tetrabutylammonium chloride | TCI | Cat#T0055, CAAS 1113-67-0 |
| Phosphatidylcholine (POPC) | Avanti Polar Lipids | Cat#850457, CAS: 26853-31-6 |
| HBS-N buffer | Cytiva | Cat#BR100828 |
| DMSO | Fujifilm-Wako | Cat#043-07216, CAS: 67-68-5 |
| Ethanolamine-HCl | TCI | Cat#A0298, CAS:2002-24-6 |
| NaOH | Fujifilm-Wako | Cat#194-18865, CAS: 1310-73-2 |
| 2-Propanol | Fujifilm-Wako | Cat#161-09163, CAS: 67-63-0 |
| SDS | Fujifilm-Wako | Cat#196-08675, CAS: 151-21-3 |
| fl-KcsA | Iwamoto et al.56 | N/A |
| ΔM0-KcsA | Iwamoto and Oiki34 | N/A |
| M0 peptide | GenScript | N/A |
| n-dodecyl-β-D-maltoside | Dojindo | Cat#D316, CAS: 69227-93-6 |
| Succinic acid | Nacalai | Cat#32402-05, CAS: 110-15-6 |
| Isopropyl β-d-thiogalactopyranoside | Nacalai | Cat#19742-36, CAS:367-93-1 |
| Potassium phosphate buffer | Promega | V446A-C |
| 2-mercaptoethanol | Nacalai | Cat#21417-52, CAS:60-24-2 |
| Imidazole | Nacalai | Cat#19004-35, CAS:288-32-4 |
| Critical commercial assays | ||
| CM5 sensor chip | Cytiva | Cat#BR100530 |
| SIA Kit Au | Cytiva | Cat#BR100405 |
| Recombinant DNA | ||
| pET29c(+) | EMD Biosciences | N/A |
| Software and algorithms | ||
| BiaEvaluation software | Cytiva | N/A |
| AFMViewer version 220418 software | This paper | N/A |
| pCLAMP software | Molecular Devices | Ver. 10.7 |
| BiaEvaluation software | Cytiva | N/A |
| Other | ||
| Biacore T100 | Cytiva | N/A |
| AFM system | Yamamoto70 | N/A |
| Cantilever | Olympus | BL-AC10DS-A2 |
| Contact bubble bilayer system | Iwamoto and Oiki50 | N/A |
| Patch-clamp amplifier | Molecular Devices | Axopatch 200B |
| Analog-to-digital converter | Molecular Devices | Digidata 1550A |
| Plasma cleaner | Harrick Plasma | PDC-32G |
| Scanning electron microscope | Hitachi High-Tech | S-4100 |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Nobuaki Matsumori (matsmori@chem.kyushu-univ.jp).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and study participant details
This study did not include in vivo experiments. Our experimental model was constituted by recombinant proteins expressed in E. coli.
Method details
Materials
All lipids used in this study were purchased from Avanti Polar Lipids (Alabaster, AL, USA). The sensor chips for the SPR measurements were purchased from Cytiva (Little Chalfont, Buckinghamshire, UK). Any other reagents and materials were all purchased from FUJIFILM Wako Pure Chemical Corp. (Osaka, Japan), Tokyo Chemical Inc. (Tokyo, Japan), or Sigma-Aldrich (St. Louis, MO, USA). M0 mimicking peptide was purchased from custom peptide synthesis service of GenScript (Piscataway, NJ, USA).
Expression and purification of fl- and ΔM0-KcsA
Expression and purification of the fl- and ΔM0-KcsA channels were carried out as previously reported.34,55 In brief, the gene for the fl- or ΔM0-KcsA channel with the hexa-His-tag at the C-terminal end was inserted in the pET29c(+) vector and expressed in E. coli BL21(DE3)pLysS cells. Protein expression was induced by addition of 0.5 mM isopropyl β-d-thiogalactopyranoside (IPTG). Two hours after induction with IPTG, E. coli cells expressing KcsA channels were harvested and disrupted by sonication. The membrane fractions were collected by ultracentrifugation and solubilized in buffer (20 mM potassium phosphate, 200 mM KCl, 20 mM 2-mercaptoethanol, 50 mM imidazole, pH 7.4) containing 1% n-dodecyl β-d-maltoside. Histidine-tagged channels were purified with a Co2+-based metalchelate chromatography resin. Purified channels were eluted by 400 mM imidazole at a protein concentration of 0.5-3 mg mL−1.
Preparation of SPR sensor chip and immobilization of KcsA
The SAM-modified sensor chip was prepared using bare Au sensor chip (SIA Kit Au, Cytiva) and a mixture of 6-mercaptohexanoic acid and 6-mercaptohexanol (8:2), as previously reported.25 fl-KcsA or ΔM0-KcsA solubilized with 0.06% n-dodecyl-β-D-maltoside (DDM) was immobilized onto SAM-modified sensor chip, according to a standard amino coupling method. HBS-EP buffer (10 mM HEPES [pH 7.4], 150 mM NaCl, 3 mM EDTA, 0.05 % (v/v) Tween 20) was used as the running buffer. Briefly, COOH groups on the surface were activated by injecting a mixture of 390 mM 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide (EDC) and 100 mM N-hydrosuccinimide (NHS) for 7 min, followed by the immobilization of KcsA by injecting the solubilized KcsA solution (100 μg mL−1) for 20 min. Unreacted carboxyl groups were deactivated by injecting the blocking solution (1 M ethanolamine-HCl, pH 8.5) for 7 min, at a flow rate of 5 μL min−1.
KcsA-lipid interaction analysis
SPR analysis were carried out at 25.0°C using Biacore T100 system (GE Healthcare, Chicago, IL, USA). In all protocols, acidic buffer (10 mM succinic acid [pH 4.0], 200 mM KCl, 3 mM EDTA, 0.5% (v/v) Tween 20) and neutral buffer (10 mM HEPES [pH 7.5], 200 mM KCl, 3 mM EDTA, 0.5% (v/v) Tween 20) were used as running buffers. Evaluation of KcsA-lipid interactions was made within 2 days after KcsA immobilization. Analyte samples were prepared by dissolving dried lipids into the buffer at the concentration of 100 μM, and the samples were sonicated prior to use. All procedures were automated, using repetitive cycles of sample injection and regeneration. The binding assays were performed after three cycles of start-up injections to normalize the two flow cells, and just before the injection of lipid solutions, the buffer alone was injected in the first two cycles in order to obtain baseline value. Afterward, lipid solutions were injected for 180 s at a flow rate of 30 μL min−1, followed by 180 s of dissociation at the same flow rate. The surface of the chip was regenerated by three sequential injection of regeneration solution (10 mM NaOH, 0.5% (v/v) Tween 20) for 30 s at a flow rate of 30 μL min−1. Association and dissociation of lipid molecules were expressed as sensorgrams, representing the time-dependent changes. To remove the contribution from non-specific binding between SAM and lipids, a blank channel without KcsA immobilization was used as a reference, and the response of the reference channel (SAM-lipid interactions) was subtracted from that of the sample channel. The evaluation of interactions was performed using BiaEvaluation software (Cytiva), and kinetic parameters were extracted by a local fit of the corrected sensorgrams using a 1:2 heterogeneous ligand binding model.58,59 For more description on the heterogeneous binding model, see Table S2 caption. The correlations of the fitting were evaluated using χ2 analyses and residual plots.
AFM observation of immobilized KcsA channel
The SAM-coated sensor chip was cut into 1-2 mm squares by a diamond cutter and ultrasonically cleaned in EtOH for 3 min and then in Milli-Q water for 3 min. The cleaned sensor chip was fixed to a 2 mm diameter cylindrical glass with a double-sided tape and was imaged using a laboratory-built AFM.70 The AFM imaging was performed in MilliQ-water. The cantilevers used were 9 μm long micro-cantilevers with nominal spring constant of 0.1 N/m, and resonance frequency of ∼500 kHz in water (BL-AC10DS-A2; Olympus, Tokyo, Japan). The probe tips were fabricated on each cantilever by electron beam deposition method using the scanning electron microscope S-4100 (Hitachi High-Tech, Tokyo, Japan). The probe tips were sharpened in argon plasma using the plasma cleaner PDC-32G (Harrick Plasma, Ithaca, NY, USA). AFM measurements were taken by amplitude modulation mode. The free oscillation amplitude of the cantilever A0 was 1nm. The set-point amplitude was set as 0.9 A0 during the imaging. The scanning rate was 20 lines/s.
Next, COOH groups on the surface were treated with 20 μL of a mixture of 195 mM EDC and 50 mM NHS for 10 min, washed with 10 mM acetic acid (20 μL, 5 times), and then reacted with solubilized KcsA solution (3.2 μg mL−1 in 10 mM acetic acid pH 4.0) for 10 min. The surface was then washed with the SPR running buffer (10 mM succinic acid [pH 4.0], 200 mM KCl, 3 mM EDTA, 0.5% (v/v) Tween 20) (20 μL, twice) and with Milli-Q water (20 μL, twice), and AFM observation was made in the same manner as above.
The AFM images were processed using a laboratory-built software, AFMViewer (version 220418). A Gaussian filter was applied to the AFM images to reduce the noise and analyze the height of KcsA. To analyze the height of KcsA, the histogram was fitted by the integral from x to x + Δx of single Gaussian distribution, h(x), given by:
where N is the number of particles, μ is the mean of the distribution, σ is the standard deviation of the distribution, Δx is the bin width of histogram, and erf(x) is the error function, given by:
Preparation of dodecylamine-modified sensor chip
The CM5 sensor chip surface was modified with dodecylamine in the same manner as the previous report.61 The modification was performed at 25.0°C on a Biacore T100 system, using HBS-N (10 mM HEPES [pH 7.4], 150 mM NaCl) as a running buffer. Before modification the chip surface was washed with 50 mM NaOH/2-propanol [3/2 (v/v)] for 3 times at a flow rate of 20 μL min−1. In the modification of the chip surface, carboxyl groups on the surface were activated by injecting a mixture of 390 mM EDC and 100 mM NHS for 7 min, followed by the immobilization of dodecylamine by injecting the dodecylamine (1.0 mg mL−1, 5.4 mM) in 10 mM NaOAc solution (pH 5.0, 10% (v/v) DMSO) for 30 min, and the deactivation of unreacted carboxyl group by injecting 1 M ethanolamine-HCl (pH 8.5) for 7 min, at a flow rate of 5 μL min−1.
M0 peptide-membrane interaction analysis
Liposome immobilization on the dodecylamine-modified sensor chip was performed as previously reported.61 DOPC (5.0 mg, 6.4 μmol) and 0.71 μmol of DOPG, DOPA, or tetraC18:1 CL were mixed in MeOH/CHCl3, dried under N2 gas flow, and suspended in 1 mL of acidic buffer (10 mM succinic acid [pH 4.0], 200 mM KCl, 3 mM EDTA), to prepare multilamellar vesicles (MLVs). The MLV suspension was extruded through 200 nm, and then 100 nm polycarbonate membranes (LiposoFast Liposome Factory, Sigma-Aldrich, St. Louis, MO, USA) (21 passes for each type of membrane), to prepare 100 nm large unilamellar vesicles (LUVs). The LUV solution was then diluted with the buffer used, to prepare 1 mM LUV solution. The following measurements were performed at 25.0°C on a Biacore T100 system. LUV solution was injected for 40 min over flowcell on the dodecylamine-modified sensor chip at 2 μL min−1 to immobilize LUV onto the chip surface, and washed with 50 mM NaOH solution for 3 times at a flow rate of 20 μL min−1 to remove immobilized LUV. The amount of immobilized LUV was recorded after 10 min of signal stabilization. Afterward, the peptide solution (10 μg mL−1) was injected for 300 s at a flow rate of 10 μL min−1, followed by 300 s of dissociation at the same flow rate. The surface of the chip was regenerated by two sequential injections of 0.5% (w/v) SDS for 120 s and 50 mM NaOH/2-propanol [3/2 (v/v)] at a flow rate of 20 μL min−1. To remove the contribution from non-specific binding of the peptide to the chip surface, a blank channel without liposome was used as a reference, and the response of the reference channel was subtracted from that of the sample channel. The kinetic parameters were extracted by a local fit of the corrected sensorgrams using a 1:1 interaction (Langmuir interaction), performed by BiaEvaluation software (Cytiva).
Single-channel recordings
Prior to the single-channel recordings, purified fl-KcsA or ΔM0-KcsA channel was reconstituted into liposomes by dilution as follows. First, pre-prepared POPC or POPC/anionic lipid (3/1, w/w) liposomes were suspended in 200 mM KCl solution, yielding a 2 mg mL−1 liposome solution. Then, an aliquot of purified KcsA solution containing 0.06% DDM was diluted 50 times with the liposome solution, obtaining the KcsA-reconstituted proteoliposomes with a lipid/protein ratio of 2000 (w/w). Just before the single-channel recording, a small amount of concentrated buffer (succinic acid for pH 4 or HEPES for pH 7.5) was added to the proteoliposome solution to adjust the pH (final buffer concentration was 10 mM). The contact bubble bilayer (CBB) method was applied for the single-channel recordings, as described in the previous report.47 Briefly, the proteoliposome solutions of different pHs were filled into separate bubble-forming pipettes (tip diameter, approximately 50 μm). The tip of the pipette was immersed in hexadecane in the chamber while observing with an inverted microscope (IX73; Olympus). A small water bubble (diameter, approximately 100 μm) was inflated at the tip of the pipette by pushing the proteoliposome solution out of the pipette into the hexadecane. A lipid monolayer forms spontaneously at the water-hexadecane interface of the bubble. Finally, a lipid bilayer (i.e., CBB) was prepared by contacting two water bubbles lined with a lipid monolayer. The KcsA channel spontaneously transfers from the liposomal membrane to the CBB. The channel insertion into the CBB was detected as the appearance of the single-channel current with the membrane potential applied. Since the KcsA channel is activated when the pH of the intracellular side becomes acidic, only the channel molecule whose cytoplasmic domain faces the acidic bubble contributes to the single-channel under asymmetric pH conditions (pH 4/pH 7.5). Thus, the acidic and neutral bubbles can be defined as the intracellular (in) and extracellular (out) sides, respectively, in the CBB experiments. The single-channel current was recorded using a patch-clamp amplifier (Axopatch 200B, Molecular Devices, San Jose, CA). The data was passed through a low-pass filter (1 kHz cutoff frequency) and stored in PC using an analog-to-digital converter (5 kHz sampling; Digidata 1550A; Molecular Devices) and pCLAMP software (Molecular Devices).
Quantification and statistical analysis
All results were expressed as mean ± SD, and scatterplots of all values are superimposed. One-way analysis of variance (ANOVA) with Tukey's test was performed to evaluate the statistical significance.
Additional resources
There are no additional resources to be reported.
Acknowledgments
We acknowledge Dr. Misuzu Ueki for preparation of channel proteins, Ms. Masako Takashima and Ms. Masami Miyagoshi for technical assistance in single-channel recordings. This work was supported by the Japan Society for the Promotion of Science KAKENHI (JP17K07360, JP20H03219, and JP21K19212 to M.I.; JP16H00759, JP16K15179, JP17H04017, and JP20H00497 to S.O.; JP21H05040 and JP23H04963 to D.Y.; JP15H03121, JP16H00773, JP20H00405, JP22K19115, and JP23H04881 to N.M.), and JST ERATO (Lipid Active Structure Project).
Author contributions
M.M. carried out SPR analysis. I.M. and O.S. prepared KcsA and its mutants, and performed single-channel recordings. Y.N. and D.Y. performed sample preparation and observation of AFM. M.N. and I.M. supervised the whole study. All authors wrote the manuscript.
Declaration of interests
The authors declare no competing interest.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: November 14, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108471.
Contributor Information
Masayuki Iwamoto, Email: iwamoto@u-fukui.ac.jp.
Nobuaki Matsumori, Email: matsmori@chem.kyushu-univ.jp.
Supplemental information
References
- 1.Singer S.J., Nicolson G.L. The fluid mosaic model of the structure of cell membranes. Science. 1972;175:720–731. doi: 10.1126/science.175.4023.720. [DOI] [PubMed] [Google Scholar]
- 2.Hunte C., Richers S. Lipids and membrane protein structures. Curr. Opin. Struct. Biol. 2008;18:406–411. doi: 10.1016/j.sbi.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Yeagle P.L. Non-covalent binding of membrane lipids to membrane proteins. Biochim. Biophys. Acta. 2014;1838:1548–1559. doi: 10.1016/j.bbamem.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 4.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dowhan W. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu. Rev. Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- 6.Sanders C.R., Mittendorf K.F. Tolerance to changes in membrane lipid composition as a selected trait of membrane proteins. Biochemistry. 2011;50:7858–7867. doi: 10.1021/bi2011527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Contreras F.X., Ernst A.M., Wieland F., Brügger B. Specificity of intramembrane protein-lipid interactions. Cold Spring Harb. Perspect. Biol. 2011;3 doi: 10.1101/cshperspect.a004705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dawaliby R., Trubbia C., Delporte C., Masureel M., Van Antwerpen P., Kobilka B.K., Govaerts C. Allosteric regulation of G protein-coupled receptor activity by phospholipids. Nat. Chem. Biol. 2016;12:35–39. doi: 10.1038/nchembio.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laganowsky A., Reading E., Allison T.M., Ulmschneider M.B., Degiacomi M.T., Baldwin A.J., Robinson C.V. Membrane proteins bind lipids selectively to modulate their structure and function. Nature. 2014;510:172–175. doi: 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patrick J.W., Boone C.D., Liu W., Conover G.M., Liu Y., Cong X., Laganowsky A. Allostery revealed within lipid binding events to membrane proteins. Proc. Natl. Acad. Sci. USA. 2018;115:2976–2981. doi: 10.1073/pnas.1719813115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akutsu H. Strategies for elucidation of the structure and function of the large membrane protein complex, FoF1-ATP synthase, by nuclear magnetic resonance. Biophys. Chem. 2023;296 doi: 10.1016/j.bpc.2023.106988. [DOI] [PubMed] [Google Scholar]
- 12.Dürr U.H.N., Gildenberg M., Ramamoorthy A. The magic of bicelles lights up membrane protein structure. Chem. Rev. 2012;112:6054–6074. doi: 10.1021/cr300061w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janmey P.A., Kinnunen P.K.J. Biophysical properties of lipids and dynamic membranes. Trends Cell Biol. 2006;16:538–546. doi: 10.1016/j.tcb.2006.08.009. [DOI] [PubMed] [Google Scholar]
- 14.Phillips R., Ursell T., Wiggins P., Sens P. Emerging roles for lipids in shaping membrane-protein function. Nature. 2009;459:379–385. doi: 10.1038/nature08147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cantor R.S. The influence of membrane lateral pressures on simple geometric models of protein conformational equilibria. Chem. Phys. Lipids. 1999;101:45–56. doi: 10.1016/s0009-3084(99)00054-7. [DOI] [PubMed] [Google Scholar]
- 16.Levitan I., Fang Y., Rosenhouse-Dantsker A., Romanenko V. Cholesterol and ion channels. Subcell. Biochem. 2010;51:509–549. doi: 10.1007/978-90-481-8622-8_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwamoto M., Oiki S. Constitutive Boost of an Active KcsA Channel under the Intrinsic Lipid Bilayer Tension Attenuated by Cholesterol in the Membrane. Proc. Natl. Acad. Sci. USA. 2018;115:13117–13122. doi: 10.1073/pnas.1812282115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iwamoto M., Oiki S. Hysteresis of a Tension-Sensitive K+ Channel Revealed by Time-Lapse Tension Measurements. JACS Au. 2021;1:467–474. doi: 10.1021/jacsau.0c00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bozelli J.C., Jr., Aulakh S.S., Epand R.M. Membrane shape as determinant of protein properties. Biophys. Chem. 2021;273 doi: 10.1016/j.bpc.2021.106587. [DOI] [PubMed] [Google Scholar]
- 20.Lee A.G. Lipid-protein interactions in biological membranes: a structural perspective. Biochim. Biophys. Acta. 2003;1612:1–40. doi: 10.1016/s0005-2736(03)00056-7. [DOI] [PubMed] [Google Scholar]
- 21.Long S.B., Tao X., Campbell E.B., MacKinnon R. Atomic structure of a voltage-dependent K+ channel in a lipid membrane-like environment. Nature. 2007;450:376–382. doi: 10.1038/nature06265. [DOI] [PubMed] [Google Scholar]
- 22.Gimpl G. Interaction of G protein coupled receptors and cholesterol. Chem. Phys. Lipids. 2016;199:61–73. doi: 10.1016/j.chemphyslip.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 23.Corradi V., Sejdiu B.I., Mesa-Galloso H., Abdizadeh H., Noskov S.Y., Marrink S.J., Tieleman D.P. Emerging Diversity in Lipid–Protein Interactions. Chem. Rev. 2019;119:5775–5848. doi: 10.1021/acs.chemrev.8b00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agasid M.T., Robinson C.V. Probing membrane protein-lipid interactions. Curr. Opin. Struct. Biol. 2021;69:78–85. doi: 10.1016/j.sbi.2021.03.010. Epub 2021 Apr 27. PMID: 33930613. [DOI] [PubMed] [Google Scholar]
- 25.Inada M., Kinoshita M., Sumino A., Oiki S., Matsumori N. A concise method for quantitative analysis of interactions between lipids and membrane proteins. Anal. Chim. Acta. 2019;1059:103–112. doi: 10.1016/j.aca.2019.01.042. [DOI] [PubMed] [Google Scholar]
- 26.Inada M., Kinoshita M., Matsumori N. Archaeal Glycolipid S-TGA-1 Is Crucial for Trimer Formation and Photocycle Activity of Bacteriorhodopsin. ACS Chem. Biol. 2020;15:197–204. doi: 10.1021/acschembio.9b00756. [DOI] [PubMed] [Google Scholar]
- 27.Heginbotham L., Odessey E., Miller C. Tetrameric stoichiometry of a prokaryotic K+ channel. Biochemistry. 1997;36:10335–10342. doi: 10.1021/bi970988i. [DOI] [PubMed] [Google Scholar]
- 28.Doyle D.A., Morais Cabral J., Pfuetzner R.A., Kuo A., Gulbis J.M., Cohen S.L., Chait B.T., MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Y., Morais-Cabral J.H., Kaufman A., MacKinnon R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 A resolution. Nature. 2001;414:43–48. doi: 10.1038/35102009. [DOI] [PubMed] [Google Scholar]
- 30.Thompson A.N., Posson D.J., Parsa P.V., Nimigean C.M. Molecular mechanism of pH sensing in KcsA potassium channels. Proc. Natl. Acad. Sci. USA. 2008;105:6900–6905. doi: 10.1073/pnas.0800873105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeuchi K., Takahashi H., Kawano S., Shimada I. Identification and characterization of the slowly exchanging pH-dependent conformational rearrangement in KcsA. J. Biol. Chem. 2007;282:15179–15186. doi: 10.1074/jbc.M608264200. [DOI] [PubMed] [Google Scholar]
- 32.Perozo E., Cortes D.M., Cuello L.G. Structural rearrangements underlying K+-channel activation gating. Science. 1999;285:73–78. doi: 10.1126/science.285.5424.73. [DOI] [PubMed] [Google Scholar]
- 33.Chill J.H., Louis J.M., Delaglio F., Bax A. Local and global structure of the monomeric subunit of the potassium channel KcsA probed by NMR. Biochim. Biophys. Acta. 2007;1768:3260–3270. doi: 10.1016/j.bbamem.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iwamoto M., Oiki S. Amphipathic antenna of an inward rectifier K+ channel responds to changes in the inner membrane leaflet. Proc. Natl. Acad. Sci. USA. 2013;110:749–754. doi: 10.1073/pnas.1217323110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valiyaveetil F.I., Zhou Y., MacKinnon R. Lipids in the structure, folding, and function of the KcsA K+ channel. Biochemistry. 2002;41:10771–10777. doi: 10.1021/bi026215y. [DOI] [PubMed] [Google Scholar]
- 36.Deol S.S., Domene C., Bond P.J., Sansom M.S.P. Anionic phospholipid interactions with the potassium channel KcsA: simulation studies. Biophys. J. 2006;90:822–830. doi: 10.1529/biophysj.105.071407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marius P., Alvis S.J., East J.M., Lee A.G. The interfacial lipid binding site on the potassium channel KcsA is specific for anionic phospholipids. Biophys. J. 2005;89:4081–4089. doi: 10.1529/biophysj.105.070755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heginbotham L., Kolmakova-Partensky L., Miller C. Functional reconstitution of a prokaryotic K + channel. J. Gen. Physiol. 1998;111:741–749. doi: 10.1085/jgp.111.6.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Y., McDermott A.E. Inactivation in the potassium channel KcsA. J. Struct. Biol. X. 2019;3 doi: 10.1016/j.yjsbx.2019.100009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raja M. The potassium channel KcsA: a model protein in studying membrane protein oligomerization and stability of oligomeric assembly? Arch. Biochem. Biophys. 2011;510:1–10. doi: 10.1016/j.abb.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 41.Marius P., Zagnoni M., Sandison M.E., East J.M., Morgan H., Lee A.G. Binding of anionic lipids to at least three nonannular sites on the potassium channel KcsA is required for channel opening. Biophys. J. 2008;94:1689–1698. doi: 10.1529/biophysj.107.117507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van der Cruijsen E.A.W., Prokofyev A.V., Pongs O., Baldus M. Probing conformational changes during the gating cycle of a potassium channel in lipid bilayers. Biophys. J. 2017;112:99–108. doi: 10.1016/j.bpj.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weingarth M., Prokofyev A., van der Cruijsen E.A.W., Nand D., Bonvin A.M.J.J., Pongs O., Baldus M. Structural determinants of specific lipid binding to potassium channels. J. Am. Chem. Soc. 2013;135:3983–3988. doi: 10.1021/ja3119114. [DOI] [PubMed] [Google Scholar]
- 44.Poveda J.A., Marcela Giudici A., Lourdes Renart M., Morales A., González-Ros J.M. Towards understanding the molecular basis of ion channel modulation by lipids: mechanistic models and current paradigms. Biochim. Biophys. Acta Biomembr. 2017;1859:1507–1516. doi: 10.1016/j.bbamem.2017.04.003. [DOI] [PubMed] [Google Scholar]
- 45.Molina M.L., Giudici A.M., Poveda J.A., Fernández-Ballester G., Montoya E., Renart M.L., Fernández A.M., Encinar J.A., Riquelme G., Morales A., González-Ros J.M. Competing lipid-protein and protein-protein interactions determine clustering and gating patterns in the potassium channel from streptomyces lividans (KcsA) J. Biol. Chem. 2015;290:25745–25755. doi: 10.1074/jbc.M115.669598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cortes D.M., Cuello L.G., Perozo E. Molecular architecture of full-length KcsA: role of cytoplasmic domains in ion permeation and activation gating. J. Gen. Physiol. 2001;117:165–180. doi: 10.1085/jgp.117.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sandoval-Calderón M., Guan Z., Sohlenkamp C. Knowns and unknowns of membrane lipid synthesis in streptomycetes. Biochimie. 2017;141:21–29. doi: 10.1016/j.biochi.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schauner C., Dary A., Lebrihi A., Leblond P., Decaris B., Germain P. Modulation of lipid metabolism and spiramycin biosynthesis in Streptomyces ambofaciens unstable mutants. Appl. Environ. Microbiol. 1999;65:2730–2737. doi: 10.1128/aem.65.6.2730-2737.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoischen C., Gura K., Luge C., Gumpert J. Lipid and fatty acid composition of cytoplasmic membranes from Streptomyces hygroscopicus and its stable protoplast-type L form. J. Bacteriol. 1997;179:3430–3436. doi: 10.1128/jb.179.11.3430-3436.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iwamoto M., Oiki S. Contact bubble bilayers with flush drainage. Sci. Rep. 2015;5:9110. doi: 10.1038/srep09110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iwamoto M., Oiki S. Lipid bilayer experiments with contact bubble bilayers for patch-clampers. J. Vis. Exp. 2019;143 doi: 10.3791/58840. [DOI] [PubMed] [Google Scholar]
- 52.Oiki S., Iwamoto M. Channel-Membrane Interplay in Lipid Bilayer Membranes Manipulated through Monolayer Technologies. Biol. Pharm. Bull. 2018;41:303–311. doi: 10.1248/bpb.b17-00708. [DOI] [PubMed] [Google Scholar]
- 53.Shimizu H., Iwamoto M., Konno T., Nihei A., Sasaki Y.C., Oiki S. Global twisting motion of single molecular KcsA potassium channel upon gating. Cell. 2008;132:67–78. doi: 10.1016/j.cell.2007.11.040. [DOI] [PubMed] [Google Scholar]
- 54.Sumino A., Sumikama T., Iwamoto M., Dewa T., Oiki S. The open gate structure of the membrane-embedded KcsA potassium channel viewed from the cytoplasmic side. Sci. Rep. 2013;3:1063. doi: 10.1038/srep01063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sumino A., Uchihashi T., Oiki S. Oriented reconstitution of the full-length KcsA potassium channel in a lipid bilayer for AFM imaging. J. Phys. Chem. Lett. 2017;8:785–793. doi: 10.1021/acs.jpclett.6b03058. [DOI] [PubMed] [Google Scholar]
- 56.Iwamoto M., Shimizu H., Inoue F., Konno T., Sasaki Y.C., Oiki S. Surface structure and its dynamic rearrangements of the KcsA potassium channel upon gating and tetrabutylammonium blocking. J. Biol. Chem. 2006;281:28379–28386. doi: 10.1074/jbc.M602018200. [DOI] [PubMed] [Google Scholar]
- 57.Faraldo-Gómez J.D., Kutluay E., Jogini V., Zhao Y., Heginbotham L., Roux B. Mechanism of Intracellular Block of the KcsA K+ Channel by Tetrabutylammonium: Insights from X-ray Crystallography, Electrophysiology and Replica-Exchange Molecular Dynamics Simulations. J. Mol. Biol. 2007;365:649–662. doi: 10.1016/j.jmb.2006.09.069. [DOI] [PubMed] [Google Scholar]
- 58.Morton T.A., Myszka D.G., Chaiken I.M. Interpreting complex binding kinetics from optical biosensors: a comparison of analysis by linearization, the integrated rate equation, and numerical integration. Anal. Biochem. 1995;227:176–185. doi: 10.1006/abio.1995.1268. [DOI] [PubMed] [Google Scholar]
- 59.Khalifa M.B., Choulier L., Lortat-Jacob H., Altschuh D., Vernet T. BIACORE data processing: an evaluation of the global fitting procedure. Anal. Biochem. 2001;293:194–203. doi: 10.1006/abio.2001.5119. [DOI] [PubMed] [Google Scholar]
- 60.Lejeune C., Abreu S., Chaminade P., Dulermo T., David M., Werten S., Virolle M.J. Impact of Phosphate Availability on Membrane Lipid Content of the Model Strains, Streptomyces lividans and Streptomyces coelicolor. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.623919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mouri R., Konoki K., Matsumori N., Oishi T., Murata M. Complex formation of amphotericin B in sterol-containing membranes as evidenced by surface plasmon resonance. Biochemistry. 2008;47:7807–7815. doi: 10.1021/bi800334p. [DOI] [PubMed] [Google Scholar]
- 62.Lee A.G. Lipid-protein interactions. Biochem. Soc. Trans. 2011;39:761–766. doi: 10.1042/BST0390761. [DOI] [PubMed] [Google Scholar]
- 63.Williamson I.M., Alvis S.J., East J.M., Lee A.G. The potassium channel KcsA and its interaction with the lipid bilayer. Cell. Mol. Life Sci. 2003;60:1581–1590. doi: 10.1007/s00018-003-3172-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Uysal S., Vásquez V., Tereshko V., Esaki K., Fellouse F.A., Sidhu S.S., Koide S., Perozo E., Kossiakoff A. Crystal structure of full-length KcsA in its closed conformation. Proc. Natl. Acad. Sci. USA. 2009;106:6644–6649. doi: 10.1073/pnas.0810663106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Poveda J.A., Giudici A.M., Renart M.L., Millet O., Morales A., González-Ros J.M., Oakes V., Furini S., Domene C. Modulation of the potassium channel KcsA by anionic phospholipids: Role of arginines at the non-annular lipid binding sites. Biochim. Biophys. Acta Biomembr. 2019;1861 doi: 10.1016/j.bbamem.2019.183029. [DOI] [PubMed] [Google Scholar]
- 66.Raja M. The role of phosphatidic acid and cardiolipin in stability of the tetrameric assembly of potassium channel KcsA. J. Membr. Biol. 2010;234:235–240. doi: 10.1007/s00232-010-9251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim D.M., Dikiy I., Upadhyay V., Posson D.J., Eliezer D., Nimigean C.M. Conformational heterogeneity in closed and open states of the KcsA potassium channel in lipid bicelles. J. Gen. Physiol. 2016;148:119–132. doi: 10.1085/jgp.201611602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Alvis S.J., Williamson I.M., East J.M., Lee A.G. Interactions of anionic phospholipids and phosphatidylethanolamine with the potassium channel KcsA. Biophys. J. 2003;85:3828–3838. doi: 10.1016/S0006-3495(03)74797-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Maguire J.J., Tyurina Y.Y., Mohammadyani D., Kapralov A.A., Anthonymuthu T.S., Qu F., Amoscato A.A., Sparvero L.J., Tyurin V.A., Planas-Iglesias J., et al. Known unknowns of cardiolipin signaling: The best is yet to come. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2017;1862:8–24. doi: 10.1016/j.bbalip.2016.08.001. 1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sumino A., Yamamoto D., Iwamoto M., Dewa T., Oiki S. Gating-Associated Clustering–Dispersion Dynamics of the KcsA Potassium Channel in a Lipid Membrane. J. Phys. Chem. Lett. 2014;5:578–584. doi: 10.1021/jz402491t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.