Abstract
Billions of people worldwide have experienced irreversible kidney injuries, which is mainly attributed to the complexity of drug-induced nephrotoxicity. Consequently, there is an urgent need for uncovering the mechanisms of nephrotoxicity caused by compounds. In the present study, a network-based methodology was applied to explore the mechanisms of nephrotoxicity induced by specific compounds. Initially, a total of 42 nephrotoxic compounds and 60 kinds of syndromes associated with nephrotoxicity were collected from public resources. Afterward, network localization and separation algorithms were used to map the targets of compounds and diseases into the human interactome. By doing so, 199 statistically significant nephrotoxic networks displaying the interaction between compound targets and disease genes were obtained, which played pivotal roles in compounds-induced nephrotoxicity. Subsequently, enrichment analysis pinpointed core Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathways that highlight commonalities in nephrotoxicity induced by nephrotoxic compounds. It was found that nephrotoxic compounds primarily induce nephrotoxicity by mediating the advanced glycosylation end products-receptor for advanced glycosylation end products signaling pathway in diabetic complications, human cytomegalovirus infection, lipid and atherosclerosis, Kaposi sarcoma–associated herpesvirus infection, apoptosis, and the phosphatidylinositol 3-kinase-Akt pathways. These results provide valuable insights for preventing drug-induced nephrotoxicity. Furthermore, the approaches we used are also helpful in conducting research on other kinds of toxicities.
Keywords: MT: Bioinformatics, nephrotoxicity, network localization, network separation, disease, compound
Graphical abstract

Drug-induced nephrotoxicity is one of the biggest threats to human health. Xi and colleagues constructed nephrotoxic networks by using network analysis algorithms and identified key genes that are closely associated with nephrotoxicity. Meanwhile, the mechanisms of nephrotoxic compounds-induced nephrotoxicity are unveiled. The results will provide clues for the prevention of drug-induced nephrotoxicity.
Introduction
The kidneys, as the main metabolic and excretory organ, can excrete hazardous substances from our body. Drugs and their metabolites pose a particular risk of kidney damage during the process of glomerular filtration and tubular reabsorption, given the rich blood flow and high oxygen consumption of the kidneys.1 Nephrotoxicity (renal toxicity), as one of the primary adverse effects of drugs, refers to the toxic side effects of drugs on renal function and the induction of toxic reactions in the kidney.2 According to data jointly released by the American Society of Nephrology, the International Society of Nephrology, and the European Renal Association, over 850 million people worldwide have experienced irreversible damage to their renal function.3 More than 40% of acute kidney injuries (AKIs) in patients were attributed to drug-induced nephrotoxicity.4 In addition, drug-induced kidney injury is a major cause of the termination of candidate drugs in clinical trials and withdrawal from markets. To date, the research on drug nephrotoxicity has primarily focused on, for example, animal models, 2-dimensional cell models, organoids, organ chips, network pharmacology, and metabolomics. Gu et al.5 evaluated the nephrotoxicity of esculentoside A in Phytolacca acinosa Roxb. by using renal organoids derived from human induced pluripotent stem cells. It was found that esculentoside A–induced nephrotoxicity was mainly involved in the epithelial-mesenchymal transition via the stimulator of interferon genes signaling pathway. Cohen et al.6 integrated tissue-embedded microsensors into a vascularized proximal tubular spherical chip, which not only clarified nephrotoxic mechanisms that could not be uncovered by animal experiments but also confirmed that their method has an advantage in elucidating the mechanisms for drug-induced nephrotoxicity. He et al.7 identified potential biomarkers and therapeutic targets by integrating network pharmacology and metabolomics techniques. Subsequently, the mechanism of cantharidin-induced nephrotoxicity in HK-2 cells was revealed. This result provides a theoretical basis for reducing the toxic side effects of cantharidin in clinical applications.
Conventionally experimental methods are costly, labor-intensive, and time-consuming, whereas computational approaches have great advantages in enhancing the efficiency and diminishing the costs of pharmaceutical analysis.8 Network medicine, with the booming development of systems biology, has the ability to not only integrate multiomics data but also gain deeper insights into biological associations between complex phenotypes and disease states at the molecular level.9,10 Although the current human interactome is incomplete and unexplored,11 adequate coverage of the protein–protein interaction (PPI) network is sufficient to effectively address most cutting-edge issues in network medicine.12,13 Dai et al.14 used network-based approaches to construct the overlapping network of hepatotoxic disease genes and three hexabromocyclododecane (HBCD) diastereoisomer targets on the basis of the human interactome. Subsequently, gene enrichment analysis was performed to investigate the mechanisms of HBCD-induced hepatotoxicity according to the shortest path length between compound targets and disease genes. Network-based drug repurposing developed dramatically during the coronavirus disease 2019 (COVID-19) pandemic. Wang et al.15 built the COVID-19 disease module based on the human interactome and identified the core targets playing critical roles in the pathogenic progress of COVID-19, which were expected to be significant guides in the repositioning of the prioritization analysis of registered drugs and repurposed drugs. Morselli Gysi et al.13 used experimental PPIs recorded in public databases to construct the human interactome. Subsequently, network diffusion and network proximity algorithms were implemented to identify drugs able to be repurposed against severe acute respiratory syndrome coronavirus-2, which was conducive to reduce research costs and stimulate the development of innovative drugs. The accomplishments of the aforementioned studies collectively demonstrate the efficacy of network-based methodologies in drug and disease research.
At present, the prediction and mechanism analysis of drug nephrotoxicity remain significant challenges for the scientific community. Moreover, critical pathways and mechanisms for most nephrotoxic compounds have not been comprehensively elucidated. In the present work, to explore the molecular mechanisms of nephrotoxicity in more detail, we classified compounds-induced nephrotoxicity into heterogeneous pathological changes and clinical manifestations. The categories of the collected nephrotoxic compounds were also investigated based on their chemical structures. Network-based localization and network-based separation algorithms16 were used to construct nephrotoxic compound targets-nephrotoxic disease genes networks. Subsequently, enrichment analysis for core genes and intersection calculation were performed with the aim of screening out core Gene Ontology (GO) terms and core Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways. Ultimately, the mechanisms of compounds-induced nephrotoxicity were probed to reflect the commonality of nephrotoxicity caused by nephrotoxic compounds. The workflow for our research is depicted in Figure 1.
Figure 1.
Workflow for the analysis of nephrotoxic compounds-induced nephrotoxicity
Results
Nephrotoxic compounds and syndromes classification
A total of 42 nephrotoxic compounds, 60 pathological changes, and clinical manifestations were assembled from PubMed and (China National Knowledge Infrastructure (CNKI, https://www.cnki.net/) (Table S1). Synonymous terms of 60 pathological changes and clinical manifestations gathered by Medical Subject Headings (MeSH, https://meshb.nlm.nih.gov/search), Unified Medical Language System (UMLS, https://uts.nlm.nih.gov/uts/umls/home), Human Phenotype Ontology (HPO, https://hpo.jax.org/app/), and the literature are listed in Table S1A. Moreover, a total of 5,299 compound targets and 58,288 disease genes were gathered from 15 biomedical databases after removing redundancy (Table S2). In addition, we explored the categories of the collected 42 nephrotoxic compounds (Table S1B) based on their chemical structures. Figure S1 illustrates that among the 42 nephrotoxic compounds, 13 were classified as alkaloid, making it the category with the highest number of nephrotoxic compounds. Following alkaloids, the categories with significant representation were terpenoid and phenol. This result suggests that researchers should be vigilant about these three categories of nephrotoxic compounds because compounds belonging to these categories may possess nephrotoxicity.
Significantly distributed modules in the human interactome
Based on the two criteria of network-based localization algorithm (see supplemental methods), we constructed 10 nephrotoxic compound modules and 22 nephrotoxic disease modules that were significantly distributed in the human interactome (Tables S3A and S3B). The molecular formula, PubChem Compound Identification (CID), and molecular weight (g/mol) for the 10 nephrotoxic compounds are presented in Table 1. These 32 modules obeyed the following requirements: (1) more than 25 compound targets or disease genes, (2) the largest connected component (lcc) size S was significantly larger than the random expectation lcc[rand], and (3) p < 0.05. Using an intersection-based approach, we conducted an analysis to determine whether there are common targets or genes shared between the 10 nephrotoxic compounds and 22 nephrotoxic syndromes, respectively. It was found that among the 22 nephrotoxic syndromes, only 21 shared 1 gene, indicating that these syndromes may operate through distinct molecular mechanisms, as evidenced by the lack of significant gene overlap among them (Table S3C). Meanwhile, among the 10 nephrotoxic compounds, only 9 share a common target (Table S3D), suggesting that they may induce different kinds of nephrotoxic syndromes. This result highlights potential variations in their underlying mechanisms and resulting clinical effects.
Table 1.
The 10 nephrotoxic compounds studied in this article
| Nephrotoxic compound | Molecular formula | PubChem CID | Molecular weight (g/mol) |
|---|---|---|---|
| Tetrandrine | C38H42N2O6 | 73078 | 622.7 |
| Aloe-emodin | C15H10O5 | 10207 | 270.24 |
| Arecoline | C8H13NO2 | 2230 | 155.19 |
| Evodiamine | C19H17N3O | 442088 | 303.4 |
| Matrine | C15H24N2O | 91466 | 248.36 |
| Cinnamaldehyde | C9H8O | 637511 | 132.16 |
| Rhein | C15H8O6 | 10168 | 284.22 |
| Geniposide | C17H24O10 | 107848 | 388.4 |
| Podophyllotoxin (Podofilox) | C22H22O8 | 10607 | 414.4 |
| Gossypol | C30H30O8 | 3503 | 518.6 |
The shortest path length between compound targets and disease genes
The lower shortest path length between compound targets and disease genes denotes the higher degree of aggregation. In 10 compound modules and 22 disease modules, the shortest path length (=0, 1, 2, >2, N/A) between compound targets and disease genes for each two modules was calculated through a network-based separation algorithm (see the dis_ab column of Tables S4, S5, S6, S7, S8, S9, S10, S11, S12, and S13). dis_ab = 0 means that the disease gene and compound target are overlapped in the network, whereas dis_ab = 1 indicates that the disease gene or compound target is one step away from the compound target or disease gene, and so forth. dis_ab = N/A indicates that there is no path or connection between the disease gene or compound target and the compound target or disease gene. The distribution of the shortest path length between most compound targets and disease genes was observed to be ≤3, indicating that each paired module tended to cluster together in the network. Taking the AKI module (lcc size S = 1,417, Z score of observed lcc ≈ 3.00/3.11, p < 0.05; see Table S3A and supplemental methods) and the tetrandrine module (lcc size S = 70, Z score of observed lcc ≈ 62.56/61.39, p < 0.001; see Table S3B) as examples, the shortest path length between all 108 compound targets and 1,359 of 1,649 disease genes was ≤3 (including common nodes, see Table S13). Consequently, for the subsequent analysis, only nodes with dis_ab ≤ 1 were considered for further analysis.
Nephrotoxic compound targets-disease genes networks
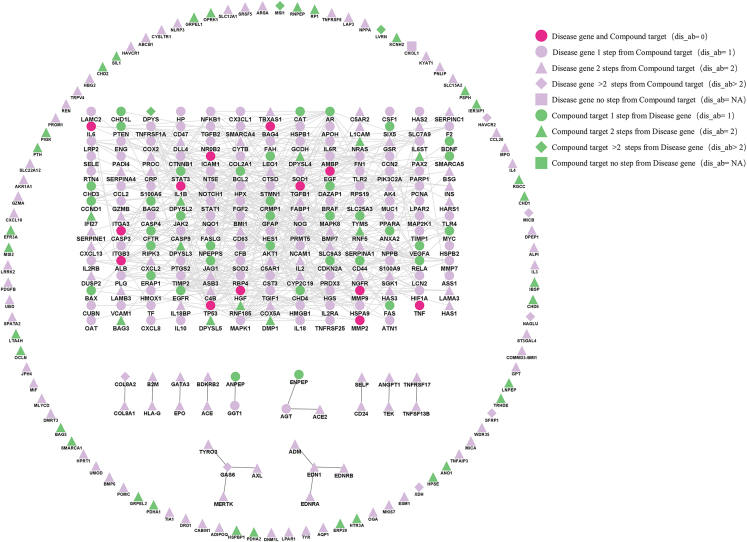
Based on 32 significantly distributed modules, 10 nephrotoxic compound modules were sequentially matched with 22 nephrotoxic disease modules, which formed 220 compounds targets-diseases genes networks. The 199 significantly distributed compound targets-disease genes networks were obtained by network-based localization algorithm (see Tables S4, S5, S6, S7, S8, S9, S10, S11, S12, and S13), and the 21 compound targets-disease genes networks that were not significantly distributed in the human interactome are listed in Table S14. With matrine targets-tubular necrosis genes network as an example, the lcc size S = 193 (Z score of observed lcc ≈ 13.78/14.25, p < 0.001) was significantly larger than the random expectation lcc[rand] = 5.438/5.222 (see Table S10). Figure 2 displays various sets formed by nodes with different degrees of aggregation in the matrine targets-tubular necrosis genes network. The outermost circle of the network listed discrete genes that had no interactions. The rectangle in the inner parts refers to the lcc interconnected by 193 core genes (including 13 common nodes, 47 compound targets, and 133 disease genes), whereas the remaining 11 are partially connected genes. The annotations for parameters of network-based localization are sufficiently described in Table S10. According to the statistics from Table S10, the shortest length path for 304 nodes out of all 315 nodes was ≤2 in the network. We also found that the common nodes for matrine and tubular necrosis were located in the core gene set. The other 9 representative nephrotoxic compounds targets-disease genes networks are provided in Figure S2. Except for aloe emodin targets-bicarbonaturia genes network, most common nodes shared by disease genes and compound targets for the remaining 8 networks were located in the core gene set.
Figure 2.
Matrine targets-tubular necrosis genes network
Distinct colors have distinct meanings. Rose: common nodes between disease genes and compound targets. Purple: disease genes. Green: compound targets. Different shapes have different implications. Rose circle: disease gene and compound target overlapped. Purple circle: disease gene was 1 step away from compound target. Green circle: compound target was 1 step away from disease gene. Triangle: disease gene or compound target was 2 steps away from compound target or disease gene. Diamond: disease gene or compound target was >2 steps away from compound target or disease gene. Square: disease gene or compound target was 0 steps away from compound target or disease gene.
Enrichment results for the nephrotoxic networks
We performed GO annotations and KEGG functional enrichment analysis on core genes with dis_ab ≤1 for 199 significantly distributed nephrotoxic compound targets-disease genes networks (see enrichment analysis column of Tables S4, S5, S6, S7, S8, S9, S10, S11, S12, and S13). The results of GO and KEGG enrichment analysis are listed in Tables S15, S16, S17, S18, S19, S20, S21, S22, S23, and S24. Taking the results of the matrine targets-tubular necrosis genes network (see Table S21) as an example, the chord diagrams in Figure 3 displayed the top 10 GO terms, the top 20 KEGG terms and the enriched core genes. Figure 3A indicates that the core genes were mainly involved in gland development, cellular response to chemical stress, gliogenesis, response to lipopolysaccharide, regulation of apoptotic signaling pathway, cellular response to oxidative stress, response to molecule of bacterial origin, response to oxidative stress, epithelial cell proliferation, and response to reactive oxygen species (ROS). Among them, cellular response to chemical stress, regulation of apoptotic signaling pathway, cellular response to oxidative stress, epithelial cell proliferation, and response to ROS may act as important roles in the process of matrine-induced tubular necrosis. Figures 3B and 3C display the top 10 molecular function (MF) terms and top 10 cellular component (CC) terms. The most significant MF and CC terms were cytokine receptor binding and vesicle lumen, respectively. The top 20 KEGG terms (Figure 3D) implied that the core genes in matrine targets-tubular necrosis genes network may be engaged in the advanced glycosylation end products-receptor for advanced glycosylation end products (AGE-RAGE) signaling pathway in diabetic complications, hepatitis B, tumor necrosis factor (TNF) signaling pathway, proteoglycans in cancer, pancreatic cancer, lipid and atherosclerosis, human cytomegalovirus infection, Kaposi sarcoma–associated herpesvirus (KSHV) infection, malaria, fluid shear stress and atherosclerosis, hepatitis C, phosphatidylinositol 3-kinase (PI3K)-Akt signaling pathway, toxoplasmosis, colorectal cancer, Chagas disease, bladder cancer, influenza A, measles, epidermal growth factor receptor tyrosine kinase inhibitor resistance, and tuberculosis. Notably, the most significantly enriched KEGG pathway was the AGE-RAGE signaling pathway in diabetic complications (padj = 1.04E−20, pAdjustMethod = Bonferroni). The KEGG pathway with the highest number of enriched genes was the PI3K-Akt signaling pathway. Therefore, it was speculated that matrine-induced tubular necrosis was closely related to the AGE-RAGE signaling pathway in diabetic complications and the PI3K-Akt signaling pathway.
Figure 3.
Results of enrichment analysis for matrine targets-tubular necrosis genes network
(A) Top 10 biological process (BP) terms. (B) Top 10 MF terms. (C) Top 10 CC terms. (D) Top 20 KEGG terms. The left half of the chord diagrams represents enriched core genes with dis_ab ≤1. The right half of the chord diagrams is the range of padj.
Commonality of nephrotoxic compounds-induced nephrotoxicity
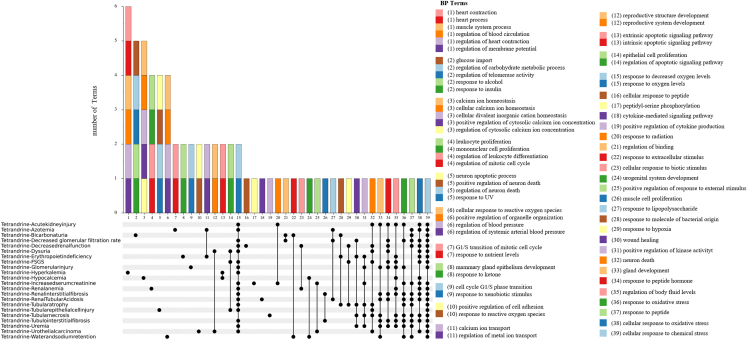
After 10 nephrotoxic compounds were paired with multiple nephrotoxic diseases to combine into the “pairs,” GO intersection terms and KEGG intersection terms were calculated based on the enrichment results of 199 compound targets-disease genes networks (see Table S25). For instance, the nephrotoxic compound tetrandrine was matched with 22 nephrotoxic diseases, forming a total of 22 pairs (Table S25). Figure 4 displays the intersection terms of GO-biological process between each pair. It is evident from the UpSet plot that the GO-biological process intersection term with the highest number of pairs is located in column 39. Twenty pairs in column 39 form an intersection term together. This core GO-biological process was the cellular response to chemical stress. Figure 5 illustrates that the KEGG intersection terms involving the highest number of pairs were distributed in the first column of the UpSet plot. These core KEGG pathways were the AGE-RAGE signaling pathway in diabetic complications, chronic myeloid leukemia, hepatitis B, human cytomegalovirus infection, and prostate cancer, respectively. Intersection terms of GO-MF and GO-CC for tetrandrine-induced nephrotoxicity are provided in Figures S3 and S4, which involve the highest number of pairs were ubiquitin protein ligase binding as well as ubiquitin-like protein ligase binding, membrane microdomain, and membrane raft, respectively. In general, the core KEGG terms for 10 nephrotoxic compounds-induced nephrotoxicity were chronic myeloid leukemia, AGE-RAGE signaling pathway in diabetic complications, prostate cancer, human cytomegalovirus infection, hepatitis B, lipid and atherosclerosis, KSHV infection, PI3K-Akt signaling pathway, and apoptosis (Figure 6). In addition, the core GO-MF and GO-CC for 10 nephrotoxic compounds-induced nephrotoxicity are shown in Figure S5.
Figure 4.
Intersection terms of GO- BP for tetrandrine-induced nephrotoxicity
Nephrotoxic compound tetrandrine was matched with 22 nephrotoxic diseases to form 22 pairs. Intersection calculation was performed on the GO-BP results of each pair. The columns in the bar charts represent BP intersection terms shared by the pairs of the UpSet parts. BP intersection terms are listed at right; each number and each color corresponds with those of the bar charts.
Figure 5.
Intersection terms of KEGG for tetrandrine-induced nephrotoxicity
Nephrotoxic compound tetrandrine was matched with 22 nephrotoxic diseases to form 22 pairs. Intersection calculation was performed on the KEGG results of each pair. The columns in the bar charts represent KEGG intersection terms shared by the pairs of the UpSet parts. KEGG intersection terms are listed at right; each number and each color corresponds with those of the bar charts.
Figure 6.
Core KEGG and GO-BP for 10 nephrotoxic compounds-induced nephrotoxicity
The intersection terms of KEGG and BP with the highest number of pairs involved are defined as core KEGG and core GO-BP.
Discussion
In this study, we constructed nephrotoxic networks using network-based algorithms based on the human interactome. We then performed calculations to identify core GO and KEGG terms to investigate the mechanisms of compounds-induced nephrotoxicity. To conduct a comprehensive analysis, we initially divided compounds-induced nephrotoxicity into 60 pathological changes and clinical manifestations and classified nephrotoxic compounds into 13 categories according to chemical structures. Then, a network-based localization algorithm was used for the acquisition of significantly distributed modules of 10 nephrotoxic compounds and 22 nephrotoxic diseases. In addition, a network-based separation algorithm was used to calculate the shortest path length between compound targets and disease genes for the further filtration of nodes with dis_ab ≤1. Subsequently, a network-based localization algorithm was used to construct the 199 significantly distributed compound targets-disease genes networks. Finally, we performed GO and KEGG enrichment analysis on core genes with dis_ab ≤1 in the 199 nephrotoxic networks. Intersection terms of GO and KEGG were calculated to identify core GO and core KEGG terms that may act pivotal roles in the process of nephrotoxic compounds-induced nephrotoxicity.
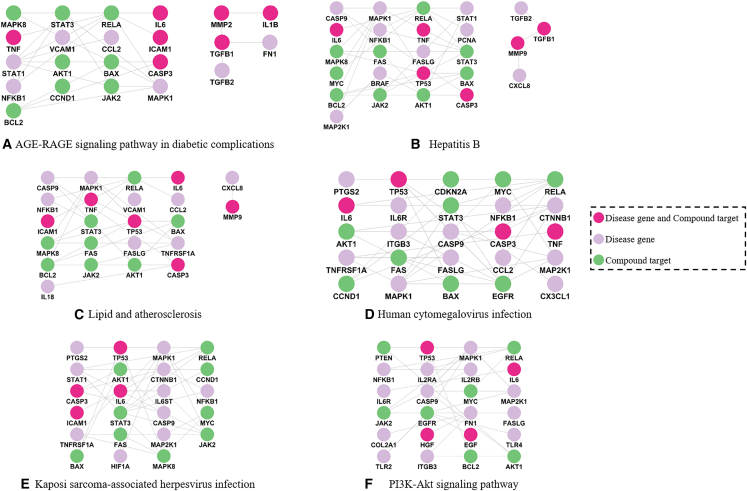
Take matrine-induced tubular necrosis as an example, six core KEGG pathways were identified (Figure 3D). The genes and targets enriched in these pathways were used for the construction of 6 networks (Figure 7), which display the relationships between them. To investigate whether these six predicted pathways play significant roles in the process of matrine-induced tubular necrosis, their biological mechanisms were analyzed. With regard to the AGE-RAGE signaling pathway in diabetic complications, it is intimately linked to the inflammation and fibrosis of the kidneys.17,18 AGEs accumulation has been implicated in ROS generation, apoptosis, and injury of renal tubular cells.19 A pictorial representation of the AGE-RAGE signaling pathway in diabetic complications involved in matrine-induced tubular necrosis is provided in Figure S6. Matrine, when acting on the AGE-RAGE signaling pathway, may lead to the accumulation of ROS, thereby exacerbating oxidative stress. Activation of NFKB1 (nuclear factor κB) in this context can promote the expression of cytokines such as TNF, interleukin-1β (IL-1β), and interleukin-6 (IL-6). In addition, the induction of apoptosis can occur through the upregulation of caspase 3 (CASP3) expression. Hepatitis B cirrhosis, triggered by the prolonged infection of hepatitis B virus, is related to AKI in cirrhotic patients.20,21 For lipid and atherosclerosis, atherosclerosis has a relationship with chronic kidney disease.22 Lipid accumulation may subsequently develop into lipotoxicity, which further induces pathological injury in proximal tubular cells.23 Most of the patients who experience human cytomegalovirus (HCMV) disease have HCMV DNA in their renal tubular epithelial cells.24 Human cytomegalovirus can promote the formation of renal fibrosis by activation of TGF-β1 after the infection of renal tubular epithelial cells. It may also play a role in renal tubular injury and interstitial nephritis.25,26 KSHV prevalence gradually becomes higher in end-stage renal disease (ESRD) patients.27 Patients with ESRD were more likely provoked by acute tubular necrosis.28 The PI3K-Akt signaling pathway is a pivotal signal transduction pathway that regulates cell proliferation, apoptosis, and inflammatory activity.29,30 Numerous studies have shown that the PI3K-Akt signaling pathway plays a critical role in the pathogenesis of drug-induced nephrotoxicity.31,32,33,34,35,36 Renal tubular cell function can also be regulated by the PI3K-Akt pathway in diabetes mellitus patients.37 Obviously, these 6 core KEGG pathways were directly or indirectly correlated with renal toxicity. To the best of our knowledge, chronic myeloid leukemia and prostate cancer were less often documented in related research, especially for the associations with nephrotoxicity. These two core KEGG pathways deserve further investigation and analysis. It is apparent that 5 common nodes—AKT serine/threonine kinase 1 (AKT1), IL-6, mitogen-activated protein kinase 1 (MAPK1), nuclear factor kappa B subunit 1 (NFKB1), and RELA ptoto-oncogene (RELA)—fully appeared in 6 networks. Inhibiting the expression of mitochondrial AKT1 in renal proximal tubules will aggravate the development of renal tubular injuries.38 When renal tubular epithelial cells undergo necrosis, they will release proinflammatory cytokines such as IL-6, followed by the initiation of signal transducer and activator of transcription 3 signaling, which may increase the infiltration of inflammatory cells in the kidney.39 MAPK1 participates in the activation of the NF-κB pathway, which is associated with cell apoptosis and inflammatory response. The depletion of MAPK1 alleviates renal injury and inflammation.40 The activation of NF-κB has correlation with ROS generation and cellular apoptosis. NF-κB signaling pathway controls the expression of several inflammatory cytokines that take part in modulating kidney inflammation.41 RELA, a key component of the NF-κB signaling pathway, plays a pivotal role in the progression of inflammation.42,43 Numerous studies have previously demonstrated that drug-induced nephrotoxicity is associated with inflammation, oxidative stress, immune response, apoptosis, and necrosis.4,44,45,46,47,48 Thus, it can be concluded that the above 6 core KEGG pathways and 5 common genes play significant roles in the progression of matrine-induced tubular necrosis, and nephrotoxic compounds-induced nephrotoxicity is regulated by multitargets and multipathways.
Figure 7.
Relationships of enriched targets and genes in six core KEGG pathways
Several drawbacks were embodied in our study. First, the incompleteness of the human interactome will result in the lack of exploration for the mechanisms of other nephrotoxic compounds and diseases. Second, a network-based separation algorithm can also be used to measure pathobiological similarities between two disease modules.16 Therefore, we will further compare the pathobiological and clinical similarities for 60 pathological changes and clinical manifestations of nephrotoxicity. Third, in some cases, nephrotoxic compounds-induced nephrotoxicity is dose dependent. In future research, sound attention should be paid to the doses of nephrotoxic compounds.
Conclusions
The results of the compound classification highlight the importance of exercising caution when dealing with substances classified as alkaloid, terpenoid, and phenol, due to the potential risks associated with nephrotoxicity. By using network analysis, core nephrotoxic compound targets and disease genes having significant implications in nephrotoxicity were identified. The core GO terms and KEGG pathways that play crucial roles in the progression of nephrotoxic compounds-induced nephrotoxicity were comprehensively analyzed. Although our study has several drawbacks, it will undeniably lay the foundation for the safety assessment and clinically reasonable application of nephrotoxic drugs. What is more, the methods used in this study have the potential to extend their utility to other investigations within the realm of toxicity research.
Materials and methods
Nephrotoxic compounds and diseases collection
To explore the diversity of compounds-induced nephrotoxicity, we conducted a literature search using the key words “nephrotoxicity” and “renal toxicity” to identify literature-recorded nephrotoxic compounds and nephrotoxic diseases. The majority of nephrotoxic compounds were successfully mapped with their PubChem CID through the PubChem database,49 whereas the remaining compounds, which had no PubChem CID, were also considered in this research. To investigate the main categories of nephrotoxic compounds, classification of the collected nephrotoxic compounds was conducted based on chemical structures through the PubChem database. Mulliner et al. classified drug-induced liver injury (also known as hepatotoxicity) into 96 types based on clinical chemistry findings and morphological findings.50 For the construction of the hepatotoxicity lexicon and the filtration of hepatotoxicity term candidates in the Literature Mining for Toxicology database, 2 selection criteria formulated by Cañada et al.51 were as follows: (1) a trigger of hepatotoxicity (e.g., transaminitis, steatosis, hepatotoxic) was included in one noun phrase of the terms, and (2) one trigger standing for the hepatobiliary system (e.g., hepatocyte, liver) together with toxicity or adverse event–associated words (e.g., injury, necrosis) were both contained in the same noun phrase. Inspired by the above methods of hepatotoxicity classification, nephrotoxic compounds-induced nephrotoxicity was divided into heterogeneous pathological changes and clinical manifestations (nephrotoxic diseases), which were assembled from PubMed and CNKI. Various synonymous terms of the pathological changes and clinical manifestations were gathered by MeSH-Entry Term(s), UMLS-Atom(s), and HPO-Synonym(s).52 The rest of the nephrotoxic diseases that were not documented in the above three public resources were also taken into account in this research.
Nephrotoxic compound targets and disease genes collection
To acquire comprehensive and relevant data, a total of 15 biomedical repositories, including 7 compound targets databases and 8 disease genes databases, were adopted. Human targets of nephrotoxic compounds were collected from An Intelligent Network Pharmacology Platform Unique for Traditional Chinese Medicine (INPUT 2.0),53 Herbal Ingredients’ Targets Platform (HIT 2.0),54 an integrative database of traditional Chinese medicine enhanced by symptom mapping (SymMap version 2),55 a high-throughput experiment- and reference-guided database of traditional Chinese medicine (HERB),56 an encyclopedia of traditional Chinese medicine (ETCM),57 the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP),58 and the Search Tool for Interactions of Chemicals (STITCH).59 All synonymous terms of nephrotoxic diseases were used as search terms for the collection of human genes of diseases. Disease genes were manually curated from the NCBI-Gene (https://www.ncbi.nlm.nih.gov/gene), the Universal Protein Knowledgebase (Uniprot),60 Online Mendelian Inheritance in Man (OMIM),61 GeneCards,62 the Therapeutic Target Database (TTD),63 DisGeNET,64 the Comparative Toxicogenomics Database (CTD),65 and Centers for Disease Control and Prevention Public Health Genomics and Precision Health Knowledge Base (PHGKB)-Phenopedia.66 Redundant compound targets and disease genes were removed.
Selection of significantly distributed modules
The module was established after importing compound targets or disease genes into the human interactome constructed by Menche et al.16 Components of the human interactome are listed in the supplemental methods. A network-based localization algorithm was used to obtain modules that were significantly distributed in the human interactome. The gene symbol of the collected compound targets and disease genes was transferred into uniform ENTREZ gene ID by the conversion tool of The Database for Annotation, Visualization and Integrated Discovery (DAVID, https://david.ncifcrf.gov/home.jsp).67 Subsequently, the network-based localization algorithm was used to individually import compound targets and disease genes into the human interactome for the construction of compound modules and disease modules. The lcc is generated by the highest number of targets and/or genes that are directly connected to one another in the human interactome. To assess whether the modules generated by compound targets or disease genes were significantly distributed in the human interactome, we performed random simulations 1,000 times. During each simulation, we randomly selected sets containing the same number of targets or genes as each module,14 generating the random expectation lcc[rand]. Retention and abandonment of the module were determined based on two criteria: (1) the number of compound targets or disease genes, and (2) the value of lcc[rand] and p value in the random expectation. According to the percolation theory,16 only compounds or diseases with more than 25 compound targets or disease genes could have an observable module in the human interactome. The lcc size S is used to describe the number of targets and/or genes that are interconnected to form lcc. If the lcc size S is significantly larger than the random expectation lcc[rand], the observed module is not randomly agglomerated by compound targets or disease genes.16 Therefore, only modules that have p < 0.05 and meet the above requirements will be retained; these are called significantly distributed modules. For the reliability of the results, network-based localization for each module was performed twice. After ignoring targets or genes that were not in the human interactome, significantly distributed compound modules and disease modules were obtained.
Calculation of the shortest path length
Each significantly distributed nephrotoxic compound module was paired with each significantly distributed nephrotoxic disease module. To find nodes (compound targets or/and disease genes) with shorter distances to one another, the network-based separation algorithm was used to calculate the shortest path length (dis_ab)14,68 between compound targets and disease genes for each paired module,
| (Equation 1) |
where a and b are nodes of the nephrotoxic disease module and nephrotoxic compound module, respectively. A represents the set of nephrotoxic disease genes, and B is the set of nephrotoxic compound targets.
Construction of nephrotoxic compound targets-disease genes networks
Each significantly distributed compound module was matched with multiple significantly distributed disease modules. Hence, the network-based localization algorithm was exploited to construct nephrotoxic compound targets-disease genes networks. According to the above-mentioned criteria for retention and abandonment of modules, only significantly distributed nephrotoxic compound targets-disease genes networks were retained. For each nephrotoxic compound targets-disease genes network, network-based localization was performed twice using the same methodology. To screen out core nodes, which may play important roles in nephrotoxic compounds-induced nephrotoxicity, according to Equations 2 and 3, the nodes in a network were classified into diverse sets (i.e., discrete gene set, partially connected gene set, core gene set) based on the extent of agglomeration in the network. If there was no interaction between nephrotoxic compound targets and disease genes in the human interactome, then nodes were defined as discrete genes. Nodes in the lcc belonged to the core gene set. The remaining nodes were taken as a partially connected gene.
| (Equation 2) |
Nd and Ng are nodes in a discrete gene set and gene set obtained by importing nephrotoxic compound targets and disease genes into the human interactome, respectively. Ni stands for nodes in a gene–gene interaction network formed by compound targets and disease genes that have interactions in the human interactome.
| (Equation 3) |
Np means nodes in the set of partially connected genes, and Nc stands for nodes in the core gene set. Nodes in the core gene set are equivalent to nodes in the lcc.
Then, ENTREZ gene ID of compound targets and disease genes was converted into a uniform gene symbol using the biological DataBase network (bioDBnet, https://biodbnet-abcc.ncifcrf.gov/). The visualization of the compound targets-disease genes networks was accomplished by Cytoscape version 3.9.1.69
Enrichment analysis for core genes with the shortest path length ≤−1
In the human interactome, a higher extent of node agglomeration indicates greater functional and biological similarities.16 Thus, we performed GO annotations and KEGG functional enrichment analysis (pAdjustMethod = Bonferroni, pvalueCutoff = 0.05) on core genes (the nodes of lcc) with dis_ab ≤1 by using clusterProfiler version 4.4.470 in significantly distributed compound targets-disease genes networks. Only the top 10 GO terms and the top 20 KEGG terms were reserved in this research. The enrichment results were visualized by Sangerbox3.0 (http://vip.sangerbox.com/home.html).71
Core GO terms and core KEGG pathways
For the above-mentioned significantly distributed compound targets-disease genes networks, each nephrotoxic compound was systematically paired with multiple nephrotoxic diseases, forming what we refer to as pairs. Afterward, GO and KEGG intersection terms were calculated using Hiplot (ORG) (https://hiplot.cn/)72 based on enrichment results of the networks. UpSet diagrams plotted by ChiPlot (https://www.chiplot.online/) were applied to seek out GO and KEGG pathways that could reflect the commonality of compounds-induced nephrotoxicity. We defined intersection terms of GO and KEGG with the highest number of pairs as core GO and core KEGG. In addition, ChiPlot was applied to plot Sankey diagrams for the core GO and core KEGG.
Data and code availability
The analyzed data are provided in the supplemental information. The pipeline for our toxicological network analysis is available at https://github.com/MarvinXi/Nephrotoxicity.
Acknowledgments
This work was supported by the Natural Science Foundation of Sichuan (no. 2023NSFSC0683), the Innovation Team and Talents Cultivation Program of National Administration of Traditional Chinese Medicine (no. ZYYCXTD-D-202209), the Foundation of Education Department of Liaoning Province (grant no. LJKZ0280), and the Natural Science Foundation of Liaoning Province (grant no. 2023-MS-288).
Author contributions
Conceptualization, K.X. and W.C. Data curation, K.X. and M.Z. Visualization, K.X., M.Z., and M.L. Methodology, Q.T. Writing – original draft, K.X. Writing – review & editing, Q.Z. and W.C.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2023.102075.
Contributor Information
Qi Zhao, Email: zhaoqi@lnu.edu.cn.
Wei Chen, Email: greatchen@ncst.edu.cn.
Supplemental information
Synonymous terms for 50 nephrotoxic diseases were assembled from Medical Subject Headings, Unified Medical Language System, and Human Phenotype Ontology. Ten nephrotoxic diseases which were not documented in above three biomedical thesauruses were annotated with literature DOI. The collected 42 nephrotoxic compounds were classified into 13 categories based on chemical structures. Five nephrotoxic compounds which had no PubChem CID were annotated with literature DOI.
The module was established after importing compound targets or disease genes into the human interactome. Significantly distributed disease modules (A) and compound modules (B) were labelled as “√”, modules which were not significantly distributed in the human interactome were marked as “× ”. Common genes for the 21 statistically significant nephrotoxic diseases (C) and common targets for the 9 statistically significant nephrotoxic compounds (D).
lcc size S: The number of nodes which are interconnected to form lcc. diameter d_s: The average value of the shortest distance from one node to the next closest node for one module in the human interactome, which also called the mean shortest distance. lcc[rand]: The random expectation lcc[rand]. z-score: Description for the size of observed lcc. dis ab: The shortest path length between compound targets and disease genes. Enrichment Analysis (dis ab ≤ 1) stood for nodes in core gene set with dis ab ≤ 1 were chosen for GO and KEGG enrichment analysis. Distinct colors had distinct meanings, Rose: Common node between disease gene and compound target. Purple: Disease gene. Green: Compound target.
lcc size S: The number of nodes which are interconnected to form lcc. diameter d_s: The average value of the shortest distance from one node to the next closest node for one module in the human interactome, which also called the mean shortest distance. lcc[rand]: The random expectation lcc[rand]. z-score: Description for the size of observed lcc. dis ab: The shortest path length between compound targets and disease genes. Enrichment Analysis (dis ab ≤ 1) stood for nodes in core gene set with dis ab ≤ 1 were chosen for GO and KEGG enrichment analysis. Distinct colors had distinct meanings, Rose: Common node between disease gene and compound target. Purple: Disease gene. Green: Compound target.
lcc size S: The number of nodes which are interconnected to form lcc. diameter d_s: The average value of the shortest distance from one node to the next closest node for one module in the human interactome, which also called the mean shortest distance. lcc[rand]: The random expectation lcc[rand]. z-score: Description for the size of observed lcc. dis ab: The shortest path length between compound targets and disease genes. Enrichment Analysis (dis ab ≤ 1) stood for nodes in core gene set with dis ab ≤ 1 were chosen for GO and KEGG enrichment analysis. Distinct colors had distinct meanings, Rose: Common node between disease gene and compound target. Purple: Disease gene. Green: Compound target.
lcc size S: The number of nodes which are interconnected to form lcc. diameter d_s: The average value of the shortest distance from one node to the next closest node for one module in the human interactome, which also called the mean shortest distance. lcc[rand]: The random expectation lcc[rand]. z-score: Description for the size of observed lcc. dis ab: The shortest path length between compound targets and disease genes. Enrichment Analysis (dis ab ≤ 1) stood for nodes in core gene set with dis ab ≤ 1 were chosen for GO and KEGG enrichment analysis. Distinct colors had distinct meanings, Rose: Common node between disease gene and compound target. Purple: Disease gene. Green: Compound target.
lcc size S: The number of nodes which are interconnected to form lcc. diameter d_s: The average value of the shortest distance from one node to the next closest node for one module in the human interactome, which also called the mean shortest distance. lcc[rand]: The random expectation lcc[rand]. z-score: Description for the size of observed lcc. dis ab: The shortest path length between compound targets and disease genes. Enrichment Analysis (dis ab ≤ 1) stood for nodes in core gene set with dis ab ≤ 1 were chosen for GO and KEGG enrichment analysis. Distinct colors had distinct meanings, Rose: Common node between disease gene and compound target. Purple: Disease gene. Green: Compound target.
lcc size S: The number of nodes which are interconnected to form lcc. diameter d_s: The average value of the shortest distance from one node to the next closest node for one module in the human interactome, which also called the mean shortest distance. lcc[rand]: The random expectation lcc[rand]. z-score: Description for the size of observed lcc. dis ab: The shortest path length between compound targets and disease genes. Enrichment Analysis (dis ab ≤ 1) stood for nodes in core gene set with dis ab ≤ 1 were chosen for GO and KEGG enrichment analysis. Distinct colors had distinct meanings, Rose: Common node between disease gene and compound target. Purple: Disease gene. Green: Compound target.
lcc size S: The number of nodes which are interconnected to form lcc. diameter d_s: The average value of the shortest distance from one node to the next closest node for one module in the human interactome, which also called the mean shortest distance. lcc[rand]: The random expectation lcc[rand]. z-score: Description for the size of observed lcc. dis ab: The shortest path length between compound targets and disease genes. Enrichment Analysis (dis ab ≤ 1) stood for nodes in core gene set with dis ab ≤ 1 were chosen for GO and KEGG enrichment analysis. Distinct colors had distinct meanings, Rose: Common node between disease gene and compound target. Purple: Disease gene. Green: Compound target.
lcc size S: The number of nodes which are interconnected to form lcc. diameter d_s: The average value of the shortest distance from one node to the next closest node for one module in the human interactome, which also called the mean shortest distance. lcc[rand]: The random expectation lcc[rand]. z-score: Description for the size of observed lcc. dis ab: The shortest path length between compound targets and disease genes. Enrichment Analysis (dis ab ≤ 1) stood for nodes in core gene set with dis ab ≤ 1 were chosen for GO and KEGG enrichment analysis. Distinct colors had distinct meanings, Rose: Common node between disease gene and compound target. Purple: Disease gene. Green: Compound target.
lcc size S: The number of nodes which are interconnected to form lcc. diameter d_s: The average value of the shortest distance from one node to the next closest node for one module in the human interactome, which also called the mean shortest distance. lcc[rand]: The random expectation lcc[rand]. z-score: Description for the size of observed lcc. dis ab: The shortest path length between compound targets and disease genes. Enrichment Analysis (dis ab ≤ 1) stood for nodes in core gene set with dis ab ≤ 1 were chosen for GO and KEGG enrichment analysis. Distinct colors had distinct meanings, Rose: Common node between disease gene and compound target. Purple: Disease gene. Green: Compound target.
lcc size S: The number of nodes which are interconnected to form lcc. diameter d_s: The average value of the shortest distance from one node to the next closest node for one module in the human interactome, which also called the mean shortest distance. lcc[rand]: The random expectation lcc[rand]. z-score: Description for the size of observed lcc. dis ab: The shortest path length between compound targets and disease genes. Enrichment Analysis (dis ab ≤ 1) stood for nodes in core gene set with dis ab ≤ 1 were chosen for GO and KEGG enrichment analysis. Distinct colors had distinct meanings, Rose: Common node between disease gene and compound target. Purple: Disease gene. Green: Compound target.
These networks were marked as “×”.
Intersection calculation was performed on the results of GO and KEGG enrichment analysis for each pair.
References
- 1.Soo J.Y.C., Jansen J., Masereeuw R., Little M.H. Advances in predictive in vitro models of drug-induced nephrotoxicity. Nat. Rev. Nephrol. 2018;14:378–393. doi: 10.1038/s41581-018-0003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perazella M.A. Renal vulnerability to drug toxicity. Clin. J. Am. Soc. Nephrol. 2009;4:1275–1283. doi: 10.2215/CJN.02050309. [DOI] [PubMed] [Google Scholar]
- 3.Jager K.J., Kovesdy C., Langham R., Rosenberg M., Jha V., Zoccali C. A single number for advocacy and communication-worldwide more than 850 million individuals have kidney diseases. Kidney Int. 2019;96:1048–1050. doi: 10.1016/j.kint.2019.07.012. [DOI] [PubMed] [Google Scholar]
- 4.Yang B., Xie Y., Guo M., Rosner M.H., Yang H., Ronco C. Nephrotoxicity and Chinese Herbal Medicine. Clin. J. Am. Soc. Nephrol. 2018;13:1605–1611. doi: 10.2215/CJN.11571017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu S., Wu G., Lu D., Wang Y., Tang L., Zhang W. Human kidney organoids model of Esculentoside A nephrotoxicity to investigate the role of epithelial-mesenchymal transition via STING signaling. Toxicol. Lett. 2023;373:172–183. doi: 10.1016/j.toxlet.2022.11.019. [DOI] [PubMed] [Google Scholar]
- 6.Cohen A., Ioannidis K., Ehrlich A., Regenbaum S., Cohen M., Ayyash M., Tikva S.S., Nahmias Y. Mechanism and reversal of drug-induced nephrotoxicity on a chip. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abd6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He T., Liu J., Wang X., Duan C., Li X., Zhang J. Analysis of cantharidin-induced nephrotoxicity in HK-2 cells using untargeted metabolomics and an integrative network pharmacology analysis. Food Chem. Toxicol. 2020;146 doi: 10.1016/j.fct.2020.111845. [DOI] [PubMed] [Google Scholar]
- 8.Chen S., Li Z., Zhang S., Zhou Y., Xiao X., Cui P., Xu B., Zhao Q., Kong S., Dai Y. Emerging biotechnology applications in natural product and synthetic pharmaceutical analyses. Acta Pharm. Sin. B. 2022;12:4075–4097. doi: 10.1016/j.apsb.2022.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pandey A.K., Loscalzo J. Network medicine: an approach to complex kidney disease phenotypes. Nat. Rev. Nephrol. 2023;19:463–475. doi: 10.1038/s41581-023-00705-0. [DOI] [PubMed] [Google Scholar]
- 10.Sadegh S., Skelton J., Anastasi E., Bernett J., Blumenthal D.B., Galindez G., Salgado-Albarrán M., Lazareva O., Flanagan K., Cockell S., et al. Network medicine for disease module identification and drug repurposing with the NeDRex platform. Nat. Commun. 2021;12:6848. doi: 10.1038/s41467-021-27138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X.W., Madeddu L., Spirohn K., Martini L., Fazzone A., Becchetti L., Wytock T.P., Kovács I.A., Balogh O.M., Benczik B., et al. Assessment of community efforts to advance network-based prediction of protein-protein interactions. Nat. Commun. 2023;14:1582. doi: 10.1038/s41467-023-37079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luck K., Kim D.K., Lambourne L., Spirohn K., Begg B.E., Bian W., Brignall R., Cafarelli T., Campos-Laborie F.J., Charloteaux B., et al. A reference map of the human binary protein interactome. Nature. 2020;580:402–408. doi: 10.1038/s41586-020-2188-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morselli Gysi D., do Valle Í., Zitnik M., Ameli A., Gan X., Varol O., Ghiassian S.D., Patten J.J., Davey R.A., Loscalzo J., Barabási A.L. Network medicine framework for identifying drug-repurposing opportunities for COVID-19. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2025581118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai W., Tang T., Dai Z., Shi D., Mo L., Zhang Y. Probing the Mechanism of Hepatotoxicity of Hexabromocyclododecanes through Toxicological Network Analysis. Environ. Sci. Technol. 2020;54:15235–15245. doi: 10.1021/acs.est.0c03998. [DOI] [PubMed] [Google Scholar]
- 15.Wang H., Zhang J., Lu Z., Dai W., Ma C., Xiang Y., Zhang Y. Identification of potential therapeutic targets and mechanisms of COVID-19 through network analysis and screening of chemicals and herbal ingredients. Briefings Bioinf. 2022;23:bbab373. doi: 10.1093/bib/bbab373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Menche J., Sharma A., Kitsak M., Ghiassian S.D., Vidal M., Loscalzo J., Barabási A.L. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science. 2015;347 doi: 10.1126/science.1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y., Liang C., Liu X., Liao B., Pan X., Ren Y., Fan M., Li M., He Z., Wu J., Wu Z. AGEs increased migration and inflammatory responses of adventitial fibroblasts via RAGE, MAPK and NF-kappaB pathways. Atherosclerosis. 2010;208:34–42. doi: 10.1016/j.atherosclerosis.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 18.Cai H.D., Su S.L., Qian D.W., Guo S., Tao W.W., Cong X.D., Tang R., Duan J.A. Renal protective effect and action mechanism of Huangkui capsule and its main five flavonoids. J. Ethnopharmacol. 2017;206:152–159. doi: 10.1016/j.jep.2017.02.046. [DOI] [PubMed] [Google Scholar]
- 19.Ishibashi Y., Yamagishi S.i., Matsui T., Ohta K., Tanoue R., Takeuchi M., Ueda S., Nakamura K.i., Okuda S. Pravastatin inhibits advanced glycation end products (AGEs)-induced proximal tubular cell apoptosis and injury by reducing receptor for AGEs (RAGE) level. Metabolism. 2012;61:1067–1072. doi: 10.1016/j.metabol.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Cullaro G., Kanduri S.R., Velez J.C.Q. Acute Kidney Injury in Patients with Liver Disease. Clin. J. Am. Soc. Nephrol. 2022;17:1674–1684. doi: 10.2215/CJN.03040322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y., Yuan W., Fang M., Guo H., Zhang X., Mei X., Zhang Y., Ji L., Gao Y., Wang J., et al. Determination of HMGB1 in hepatitis B virus-related acute-on-chronic liver failure patients with acute kidney injury: Early prediction and prognostic implications. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.1031790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Go A.S., Chertow G.M., Fan D., McCulloch C.E., Hsu C.Y. Chronic kidney disease and the risks of death, cardiovascular events, and hospitalization. N. Engl. J. Med. 2004;351:1296–1305. doi: 10.1056/NEJMoa041031. [DOI] [PubMed] [Google Scholar]
- 23.Liu L., Ning X., Wei L., Zhou Y., Zhao L., Ma F., Bai M., Yang X., Wang D., Sun S. Twist1 downregulation of PGC-1α decreases fatty acid oxidation in tubular epithelial cells, leading to kidney fibrosis. Theranostics. 2022;12:3758–3775. doi: 10.7150/thno.71722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y.T., Emery V.C., Surah S., Jarmulowicz M., Sweny P., Kidd I.M., Griffiths P.D., Clark D.A. Extensive human cytomegalovirus (HCMV) genomic DNA in the renal tubular epithelium early after renal transplantation: Relationship with HCMV DNAemia and long-term graft function. J. Med. Virol. 2010;82:85–93. doi: 10.1002/jmv.21662. [DOI] [PubMed] [Google Scholar]
- 25.Shimamura M., Murphy-Ullrich J.E., Britt W.J. Human cytomegalovirus induces TGF-β1 activation in renal tubular epithelial cells after epithelial-to-mesenchymal transition. PLoS Pathog. 2010;6 doi: 10.1371/journal.ppat.1001170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vichot A.A., Formica R.N., Jr., Moeckel G.W. Cytomegalovirus glomerulopathy and cytomegalovirus interstitial nephritis on sequential transplant kidney biopsies. Am. J. Kidney Dis. 2014;63:536–539. doi: 10.1053/j.ajkd.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang Q., Wang X., Liu Z., Zhu M., Ding M., Minhas V., Wood C., Zhang T. Seroprevalence of human herpesvirus 8 and its impact on the hemoglobin level in patients of end stage of renal diseases. J. Med. Virol. 2018;90:338–343. doi: 10.1002/jmv.24937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foley R.N., Sexton D.J., Reule S., Solid C., Chen S.C., Collins A.J. End-stage renal disease attributed to acute tubular necrosis in the United States, 2001-2010. Am. J. Nephrol. 2015;41:1–6. doi: 10.1159/000369832. [DOI] [PubMed] [Google Scholar]
- 29.Shi H.H., Chen L.P., Wang C.C., Zhao Y.C., Wang Y.M., Xue C.H., Zhang T.T. Docosahexaenoic acid-acylated curcumin diester alleviates cisplatin-induced acute kidney injury by regulating the effect of gut microbiota on the lipopolysaccharide- and trimethylamine-N-oxide-mediated PI3K/Akt/NF-κB signaling pathway in mice. Food Funct. 2022;13:6103–6117. doi: 10.1039/d1fo04178a. [DOI] [PubMed] [Google Scholar]
- 30.Fang L., Zhang Y., Wang Q., Zang Y., Li Z., Duan Z., Ren J., Xu Z. A polysaccharide from Huaier ameliorates cisplatin nephrotoxicity by decreasing oxidative stress and apoptosis via PI3K/AKT signaling. Int. J. Biol. Macromol. 2019;139:932–943. doi: 10.1016/j.ijbiomac.2019.07.219. [DOI] [PubMed] [Google Scholar]
- 31.Zhang W., Hou J., Yan X., Leng J., Li R., Zhang J., Xing J., Chen C., Wang Z., Li W. Platycodon grandiflorum Saponins Ameliorate Cisplatin-Induced Acute Nephrotoxicity through the NF-κB-Mediated Inflammation and PI3K/Akt/Apoptosis Signaling Pathways. Nutrients. 2018;10:1328. doi: 10.3390/nu10091328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuwana H., Terada Y., Kobayashi T., Okado T., Penninger J.M., Irie-Sasaki J., Sasaki T., Sasaki S. The phosphoinositide-3 kinase gamma-Akt pathway mediates renal tubular injury in cisplatin nephrotoxicity. Kidney Int. 2008;73:430–445. doi: 10.1038/sj.ki.5002702. [DOI] [PubMed] [Google Scholar]
- 33.Sherif I.O., Al-Shaalan N.H., Sabry D. Ginkgo Biloba Extract Alleviates Methotrexate-Induced Renal Injury: New Impact on PI3K/Akt/mTOR Signaling and MALAT1 Expression. Biomolecules. 2019;9:691. doi: 10.3390/biom9110691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu P., Xue Y., Zheng B., Liang Y., Zhang J., Shi J., Chu X., Han X., Chu L. Crocetin attenuates the oxidative stress, inflammation and apoptosisin arsenic trioxide-induced nephrotoxic rats: Implication of PI3K/AKT pathway. Int. Immunopharm. 2020;88 doi: 10.1016/j.intimp.2020.106959. [DOI] [PubMed] [Google Scholar]
- 35.Li X., Zou Y., Xing J., Fu Y.Y., Wang K.Y., Wan P.Z., Zhai X.Y. Pretreatment with Roxadustat (FG-4592) Attenuates Folic Acid-Induced Kidney Injury through Antiferroptosis via Akt/GSK-3β/Nrf2 Pathway. Oxid. Med. Cell. Longev. 2020;2020 doi: 10.1155/2020/6286984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng H.L., Zhang H.Y., Zhu C.L., Li H.Y., Cui S., Jin J., Piao S.G., Jiang Y.J., Xuan M.Y., Jin J.Z., et al. L-Carnitine protects against tacrolimus-induced renal injury by attenuating programmed cell death via PI3K/AKT/PTEN signaling. Acta Pharmacol. Sin. 2021;42:77–87. doi: 10.1038/s41401-020-0449-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu Z., Jia K., Wang H., Gao F., Zhao S., Li F., Hao J. METTL14-regulated PI3K/Akt signaling pathway via PTEN affects HDAC5-mediated epithelial-mesenchymal transition of renal tubular cells in diabetic kidney disease. Cell Death Dis. 2021;12:32. doi: 10.1038/s41419-020-03312-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin H.Y.H., Chen Y., Chen Y.H., Ta A.P., Lee H.C., MacGregor G.R., Vaziri N.D., Wang P.H. Tubular mitochondrial AKT1 is activated during ischemia reperfusion injury and has a critical role in predisposition to chronic kidney disease. Kidney Int. 2021;99:870–884. doi: 10.1016/j.kint.2020.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J., Xiong M., Fan Y., Liu C., Wang Q., Yang D., Yuan Y., Huang Y., Wang S., Zhang Y., et al. Mecp2 protects kidney from ischemia-reperfusion injury through transcriptional repressing IL-6/STAT3 signaling. Theranostics. 2022;12:3896–3910. doi: 10.7150/thno.72515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang H., Mou H., Xu X., Liu C., Zhou G., Gao B. LncRNA KCNQ1OT1 (potassium voltage-gated channel subfamily Q member 1 opposite strand/antisense transcript 1) aggravates acute kidney injury by activating p38/NF-κB pathway via miR-212-3p/MAPK1 (mitogen-activated protein kinase 1) axis in sepsis. Bioengineered. 2021;12:11353–11368. doi: 10.1080/21655979.2021.2005987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu X., Meng X., Xu M., Zhang X., Zhang Y., Ding G., Huang S., Zhang A., Jia Z. Celastrol ameliorates cisplatin nephrotoxicity by inhibiting NF-κB and improving mitochondrial function. EBioMedicine. 2018;36:266–280. doi: 10.1016/j.ebiom.2018.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Y., Xiong H., Liu D., Hill C., Ertay A., Li J., Zou Y., Miller P., White E., Downward J., et al. Autophagy inhibition specifically promotes epithelial-mesenchymal transition and invasion in RAS-mutated cancer cells. Autophagy. 2019;15:886–899. doi: 10.1080/15548627.2019.1569912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang T.T., Wang B., Li Z.L., Wen Y., Feng S.T., Wu M., Liu D., Cao J.Y., Yin Q., Yin D., et al. Kim-1 Targeted Extracellular Vesicles: A New Therapeutic Platform for RNAi to Treat AKI. J. Am. Soc. Nephrol. 2021;32:2467–2483. doi: 10.1681/ASN.2020111561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sahu B.D., Kuncha M., Sindhura G.J., Sistla R. Hesperidin attenuates cisplatin-induced acute renal injury by decreasing oxidative stress, inflammation and DNA damage. Phytomedicine. 2013;20:453–460. doi: 10.1016/j.phymed.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 45.Amarasiri S.S., Attanayake A.P., Arawwawala L.D.A.M., Mudduwa L.K.B., Jayatilaka K.A.P.W. Barleria prionitis L. extracts ameliorate doxorubicin-induced acute kidney injury via modulation of oxidative stress, inflammation, and apoptosis. J. Tradit. Complement. Med. 2023;13:500–510. doi: 10.1016/j.jtcme.2023.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kong J., Kui H., Tian Y., Kong X., He T., Li Q., Gu C., Guo J., Liu C. Nephrotoxicity assessment of podophyllotoxin-induced rats by regulating PI3K/Akt/mTOR-Nrf2/HO1 pathway in view of toxicological evidence chain (TEC) concept. Ecotoxicol. Environ. Saf. 2023;264 doi: 10.1016/j.ecoenv.2023.115392. [DOI] [PubMed] [Google Scholar]
- 47.Wang M., Yang N., Wu X., Zou T., Zheng J., Zhu H., Zhao C., Wang J. Insight into Nephrotoxicity and Processing Mechanism of Arisaema erubescens (Wall.) Schott by Metabolomics and Network Analysis. Drug Des. Dev. Ther. 2023;17:1831–1846. doi: 10.2147/DDDT.S406551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Abd-Ellatif R.N., Nasef N.A., El-Horany H.E.S., Emam M.N., Younis R.L., El Gheit R.E.A., Elseady W., Radwan D.A., Hafez Y.M., Eissa A., et al. Adrenomedullin Mitigates Doxorubicin-Induced Nephrotoxicity in Rats: Role of Oxidative Stress, Inflammation, Apoptosis, and Pyroptosis. Int. J. Mol. Sci. 2022;23 doi: 10.3390/ijms232314570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B., et al. PubChem 2023 update. Nucleic Acids Res. 2023;51:D1373–D1380. doi: 10.1093/nar/gkac956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mulliner D., Schmidt F., Stolte M., Spirkl H.P., Czich A., Amberg A. Computational Models for Human and Animal Hepatotoxicity with a Global Application Scope. Chem. Res. Toxicol. 2016;29:757–767. doi: 10.1021/acs.chemrestox.5b00465. [DOI] [PubMed] [Google Scholar]
- 51.Cañada A., Capella-Gutierrez S., Rabal O., Oyarzabal J., Valencia A., Krallinger M. LimTox: a web tool for applied text mining of adverse event and toxicity associations of compounds, drugs and genes. Nucleic Acids Res. 2017;45:W484–W489. doi: 10.1093/nar/gkx462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Köhler S., Gargano M., Matentzoglu N., Carmody L.C., Lewis-Smith D., Vasilevsky N.A., Danis D., Balagura G., Baynam G., Brower A.M., et al. The Human Phenotype Ontology in 2021. Nucleic Acids Res. 2021;49:D1207–D1217. doi: 10.1093/nar/gkaa1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li X., Tang Q., Meng F., Du P., Chen W. INPUT: An intelligent network pharmacology platform unique for traditional Chinese medicine. Comput. Struct. Biotechnol. J. 2022;20:1345–1351. doi: 10.1016/j.csbj.2022.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan D., Zheng G., Wang C., Chen Z., Mao T., Gao J., Yan Y., Chen X., Ji X., Yu J., et al. HIT 2.0: an enhanced platform for Herbal Ingredients' Targets. Nucleic Acids Res. 2022;50:D1238–D1243. doi: 10.1093/nar/gkab1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu Y., Zhang F., Yang K., Fang S., Bu D., Li H., Sun L., Hu H., Gao K., Wang W., et al. SymMap: an integrative database of traditional Chinese medicine enhanced by symptom mapping. Nucleic Acids Res. 2019;47:D1110–D1117. doi: 10.1093/nar/gky1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fang S., Dong L., Liu L., Guo J., Zhao L., Zhang J., Bu D., Liu X., Huo P., Cao W., et al. HERB: a high-throughput experiment- and reference-guided database of traditional Chinese medicine. Nucleic Acids Res. 2021;49:D1197–D1206. doi: 10.1093/nar/gkaa1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Xu H.Y., Zhang Y.Q., Liu Z.M., Chen T., Lv C.Y., Tang S.H., Zhang X.B., Zhang W., Li Z.Y., Zhou R.R., et al. ETCM: an encyclopaedia of traditional Chinese medicine. Nucleic Acids Res. 2019;47:D976–D982. doi: 10.1093/nar/gky987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ru J., Li P., Wang J., Zhou W., Li B., Huang C., Li P., Guo Z., Tao W., Yang Y., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J. Cheminf. 2014;6:13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Szklarczyk D., Santos A., von Mering C., Jensen L.J., Bork P., Kuhn M. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016;44:D380–D384. doi: 10.1093/nar/gkv1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Amberger J.S., Bocchini C.A., Scott A.F., Hamosh A. OMIM.org: leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 2019;47:D1038–D1043. doi: 10.1093/nar/gky1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stelzer G., Rosen N., Plaschkes I., Zimmerman S., Twik M., Fishilevich S., Stein T.I., Nudel R., Lieder I., Mazor Y., et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinformatics. 2016;54:1.30.1–1.30.33. doi: 10.1002/cpbi.5. [DOI] [PubMed] [Google Scholar]
- 63.Zhou Y., Zhang Y., Lian X., Li F., Wang C., Zhu F., Qiu Y., Chen Y. Therapeutic target database update 2022: facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res. 2022;50:D1398–D1407. doi: 10.1093/nar/gkab953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Piñero J., Ramírez-Anguita J.M., Saüch-Pitarch J., Ronzano F., Centeno E., Sanz F., Furlong L.I. The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res. 2020;48:D845–D855. doi: 10.1093/nar/gkz1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Davis A.P., Grondin C.J., Johnson R.J., Sciaky D., Wiegers J., Wiegers T.C., Mattingly C.J. Comparative Toxicogenomics Database (CTD): update 2021. Nucleic Acids Res. 2021;49:D1138–D1143. doi: 10.1093/nar/gkaa891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yu W., Clyne M., Khoury M.J., Gwinn M. Phenopedia and Genopedia: disease-centered and gene-centered views of the evolving knowledge of human genetic associations. Bioinformatics. 2010;26:145–146. doi: 10.1093/bioinformatics/btp618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sherman B.T., Hao M., Qiu J., Jiao X., Baseler M.W., Lane H.C., Imamichi T., Chang W. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucleic Acids Res. 2022;50:W216–W221. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Guney E., Menche J., Vidal M., Barábasi A.L. Network-based in silico drug efficacy screening. Nat. Commun. 2016;7 doi: 10.1038/ncomms10331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wu T., Hu E., Xu S., Chen M., Guo P., Dai Z., Feng T., Zhou L., Tang W., Zhan L., et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation. 2021;2 doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shen W., Song Z., Zhong X., Huang M., Shen D., Gao P., Qian X., Wang M., He X., Wang T., et al. Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. iMeta. 2022;1:e36. doi: 10.1002/imt2.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li J., Miao B., Wang S., Dong W., Xu H., Si C., Wang W., Duan S., Lou J., Bao Z., et al. Hiplot: a comprehensive and easy-to-use web service for boosting publication-ready biomedical data visualization. Briefings Bioinf. 2022;23:bbac261. doi: 10.1093/bib/bbac261. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Synonymous terms for 50 nephrotoxic diseases were assembled from Medical Subject Headings, Unified Medical Language System, and Human Phenotype Ontology. Ten nephrotoxic diseases which were not documented in above three biomedical thesauruses were annotated with literature DOI. The collected 42 nephrotoxic compounds were classified into 13 categories based on chemical structures. Five nephrotoxic compounds which had no PubChem CID were annotated with literature DOI.
The module was established after importing compound targets or disease genes into the human interactome. Significantly distributed disease modules (A) and compound modules (B) were labelled as “√”, modules which were not significantly distributed in the human interactome were marked as “× ”. Common genes for the 21 statistically significant nephrotoxic diseases (C) and common targets for the 9 statistically significant nephrotoxic compounds (D).
lcc size S: The number of nodes which are interconnected to form lcc. diameter d_s: The average value of the shortest distance from one node to the next closest node for one module in the human interactome, which also called the mean shortest distance. lcc[rand]: The random expectation lcc[rand]. z-score: Description for the size of observed lcc. dis ab: The shortest path length between compound targets and disease genes. Enrichment Analysis (dis ab ≤ 1) stood for nodes in core gene set with dis ab ≤ 1 were chosen for GO and KEGG enrichment analysis. Distinct colors had distinct meanings, Rose: Common node between disease gene and compound target. Purple: Disease gene. Green: Compound target.
lcc size S: The number of nodes which are interconnected to form lcc. diameter d_s: The average value of the shortest distance from one node to the next closest node for one module in the human interactome, which also called the mean shortest distance. lcc[rand]: The random expectation lcc[rand]. z-score: Description for the size of observed lcc. dis ab: The shortest path length between compound targets and disease genes. Enrichment Analysis (dis ab ≤ 1) stood for nodes in core gene set with dis ab ≤ 1 were chosen for GO and KEGG enrichment analysis. Distinct colors had distinct meanings, Rose: Common node between disease gene and compound target. Purple: Disease gene. Green: Compound target.
lcc size S: The number of nodes which are interconnected to form lcc. diameter d_s: The average value of the shortest distance from one node to the next closest node for one module in the human interactome, which also called the mean shortest distance. lcc[rand]: The random expectation lcc[rand]. z-score: Description for the size of observed lcc. dis ab: The shortest path length between compound targets and disease genes. Enrichment Analysis (dis ab ≤ 1) stood for nodes in core gene set with dis ab ≤ 1 were chosen for GO and KEGG enrichment analysis. Distinct colors had distinct meanings, Rose: Common node between disease gene and compound target. Purple: Disease gene. Green: Compound target.
lcc size S: The number of nodes which are interconnected to form lcc. diameter d_s: The average value of the shortest distance from one node to the next closest node for one module in the human interactome, which also called the mean shortest distance. lcc[rand]: The random expectation lcc[rand]. z-score: Description for the size of observed lcc. dis ab: The shortest path length between compound targets and disease genes. Enrichment Analysis (dis ab ≤ 1) stood for nodes in core gene set with dis ab ≤ 1 were chosen for GO and KEGG enrichment analysis. Distinct colors had distinct meanings, Rose: Common node between disease gene and compound target. Purple: Disease gene. Green: Compound target.
lcc size S: The number of nodes which are interconnected to form lcc. diameter d_s: The average value of the shortest distance from one node to the next closest node for one module in the human interactome, which also called the mean shortest distance. lcc[rand]: The random expectation lcc[rand]. z-score: Description for the size of observed lcc. dis ab: The shortest path length between compound targets and disease genes. Enrichment Analysis (dis ab ≤ 1) stood for nodes in core gene set with dis ab ≤ 1 were chosen for GO and KEGG enrichment analysis. Distinct colors had distinct meanings, Rose: Common node between disease gene and compound target. Purple: Disease gene. Green: Compound target.
lcc size S: The number of nodes which are interconnected to form lcc. diameter d_s: The average value of the shortest distance from one node to the next closest node for one module in the human interactome, which also called the mean shortest distance. lcc[rand]: The random expectation lcc[rand]. z-score: Description for the size of observed lcc. dis ab: The shortest path length between compound targets and disease genes. Enrichment Analysis (dis ab ≤ 1) stood for nodes in core gene set with dis ab ≤ 1 were chosen for GO and KEGG enrichment analysis. Distinct colors had distinct meanings, Rose: Common node between disease gene and compound target. Purple: Disease gene. Green: Compound target.
lcc size S: The number of nodes which are interconnected to form lcc. diameter d_s: The average value of the shortest distance from one node to the next closest node for one module in the human interactome, which also called the mean shortest distance. lcc[rand]: The random expectation lcc[rand]. z-score: Description for the size of observed lcc. dis ab: The shortest path length between compound targets and disease genes. Enrichment Analysis (dis ab ≤ 1) stood for nodes in core gene set with dis ab ≤ 1 were chosen for GO and KEGG enrichment analysis. Distinct colors had distinct meanings, Rose: Common node between disease gene and compound target. Purple: Disease gene. Green: Compound target.
lcc size S: The number of nodes which are interconnected to form lcc. diameter d_s: The average value of the shortest distance from one node to the next closest node for one module in the human interactome, which also called the mean shortest distance. lcc[rand]: The random expectation lcc[rand]. z-score: Description for the size of observed lcc. dis ab: The shortest path length between compound targets and disease genes. Enrichment Analysis (dis ab ≤ 1) stood for nodes in core gene set with dis ab ≤ 1 were chosen for GO and KEGG enrichment analysis. Distinct colors had distinct meanings, Rose: Common node between disease gene and compound target. Purple: Disease gene. Green: Compound target.
lcc size S: The number of nodes which are interconnected to form lcc. diameter d_s: The average value of the shortest distance from one node to the next closest node for one module in the human interactome, which also called the mean shortest distance. lcc[rand]: The random expectation lcc[rand]. z-score: Description for the size of observed lcc. dis ab: The shortest path length between compound targets and disease genes. Enrichment Analysis (dis ab ≤ 1) stood for nodes in core gene set with dis ab ≤ 1 were chosen for GO and KEGG enrichment analysis. Distinct colors had distinct meanings, Rose: Common node between disease gene and compound target. Purple: Disease gene. Green: Compound target.
lcc size S: The number of nodes which are interconnected to form lcc. diameter d_s: The average value of the shortest distance from one node to the next closest node for one module in the human interactome, which also called the mean shortest distance. lcc[rand]: The random expectation lcc[rand]. z-score: Description for the size of observed lcc. dis ab: The shortest path length between compound targets and disease genes. Enrichment Analysis (dis ab ≤ 1) stood for nodes in core gene set with dis ab ≤ 1 were chosen for GO and KEGG enrichment analysis. Distinct colors had distinct meanings, Rose: Common node between disease gene and compound target. Purple: Disease gene. Green: Compound target.
These networks were marked as “×”.
Intersection calculation was performed on the results of GO and KEGG enrichment analysis for each pair.
Data Availability Statement
The analyzed data are provided in the supplemental information. The pipeline for our toxicological network analysis is available at https://github.com/MarvinXi/Nephrotoxicity.