Abstract
Canonical heterotrimeric G-proteins (G-proteins) are comprised of Gα, Gβ, and Gγ subunits. G-proteins regulate multiple crucial plant growth and development processes, incorporating environmental responses. Besides Gα, Gβ and Gγ, the discovery of atypical Gα subunits termed as extra-large G-proteins or extra-large GTP-binding proteins (XLGs) makes G-protein signaling unique in plants. The C-terminus of XLG shares similarities with the canonical Gα subunits; the N-terminus harbors a nuclear localization signal (NLS) and is rich in cysteine. The earlier explorations suggest XLG's role in flowering, the development of embryos and seedlings, root morphogenesis, stamen development, cytokinin-induced development, stomatal opening and regulation of rice grain filling. The XLGs are also known to initiate signaling cascades that prime plants against a variety of abiotic and biotic stresses. They are also engaged in controlling several agronomic parameters such as rice panicle length, grain filling, grain size, and biomass, highlighting their potential contribution to crop improvement. The present review explores the remarkable properties of non-canonical Gα subunits (XLGs) and reflects on the various developmental, abiotic and biotic stress signaling pathways controlled by them. Moreover, the bottleneck dilemma of how a tiny handful of XLGs control a multiplicity of stress-responsive activities is partially resolved in this review by addressing the interaction of XLGs with different interacting proteins. XLG proteins presented in this review can be exploited to gain access to highly productive and stress-tolerant plants.
Keywords: G-proteins, Extra-large GTP-binding proteins (XLGs), Agronomic parameter, Abiotic stress, Biotic stress
Introduction
G-proteins, or guanine nucleotide-binding proteins, operate as molecular switches in the cell. They facilitate the propagation of signals from several exterior stimuli to the inside of a cell (Trusov and Botella 2016). G-proteins are categorized into two types: monomeric G-proteins (small GTPases) and heterotrimeric G-proteins (Bhardwaj et al. 2020; Pandey 2020; Ganotra et al. 2023). Developmental, abiotic, and biotic stress responses are only a handful of the numerous activities that both types of G-proteins are known to modulate in plants (Tuteja and Sopory 2008; Trusov and Botella 2016; Bhardwaj et al. 2020; Pandey 2020; Ganotra et al. 2023). The heterotrimeric G-proteins (hereafter G-proteins) comprise three structurally and functionally different subunits: Gα, Gβ and Gγ (Botella 2012; Pandey 2020). The Gα subunit is capable of both GTP-binding and GTP hydrolysis; the Gβ subunit interacts with a myriad of proteins owing to its seven WD40 repeats; the Gγ subunit remains tethered to the Gβ subunit, forming a Gβγ dimer (Trusov et al. 2007; Maruta et al. 2021a). The three core G-protein subunits are in a trimeric complex in the inactive state, with GDP linked to Gα. Conventionally, in mammalian and yeast systems, the activation of G-protein signaling occurs when a ligand binds to a serpentine transmembrane receptor, namely the G-protein-coupled receptor (GPCR) (Chakraborty and Raghuram 2022). The GDP is exchanged for GTP on Gα during activation, which is triggered by signal perception by GPCR, leading to the separation of GTP-bound Gα and the Gβγ dimer (McIntire 2009; Pandey 2020). The downstream signaling is then subsequently controlled by the segregated Gα subunit and Gβγ complex, which interact with a number of downstream effectors (McIntire 2009; Ganotra et al. 2023). The signal is stopped when the Gα subunit triggers the hydrolysis of GTP to GDP, resulting in Gα-GDP being released from its effector and re-associated with Gβγ complex (McIntire 2009). The GTPase activating protein (GAP) activity of the regulator of G-protein signaling (RGS) accelerates the intrinsic GTP hydrolysis on the Gα subunit (Ganotra et al. 2023). According to some studies, GPCRs interact with Gα proteins in plants, but nevertheless, it has not been established that they can activate Gα via promoting GDP to GTP exchange (Trusov and Botella 2016). According to Hackenberg et al. (2017), numerous plants lack an RGS protein homolog; therefore, it is uncertain if RGS-mediated deactivation is the main mechanism regulating the G-protein cycle.
The Gα subunits of plants split off from a shared eukaryotic ancestor many years ago and have subsequently pursued distinct evolutionary trajectories ever since (Anantharaman et al. 2011). The evolutionary findings indicate that many primitive and predominantly unicellular eukaryotes have lost their whole G-protein complex, whereas multicellular eukaryotes contain several Gα subunits (Anantharaman et al. 2011). Plants contain two different kinds of Gα subunits: a canonical Gα subunit and a non-canonical Gα subunit known as the extra-large GTP-binding protein, or XLG (Maruta et al. 2021a). The canonical subunits, or subunits structurally related to those found in animals, were formerly thought to be the only G-protein subunits existing in plants. The repertoire of G-protein subunits was later broadened to incorporate non-canonical proteins such as XLGs that are specific to plants (Table 1) (Ding et al. 2008). The components that constitute the XLGβγ trimer are present across the whole land plant lineage, while their occurrence in algae is erratic (Mohanasundaram et al. 2022). According to a study, the XLG subunits underwent substantial gene duplication and gene fusion during the evolution of the charophycean algae (Urano et al. 2016; Mohanasundaram et al. 2022). Charophyte algae like Coleochaeta orbicularis and Klebsormidium flaccidiium show the presence of XLG genes (CoXLG: GBSL01023349.1; KlXLG: kfl00304_0070) (Hackenberg et al. 2016). The genome of Marchantia polymorpha contains only a single copy of the XLG gene (Mapoly0129s0046.1) (Bowman et al. 2017). According to a report, Physcomitrium patens (moss) lacks the canonical Gα protein and contains only one XLG gene (Pp1s147_153V6.1), indicating that the XLG subunit might have replaced the functions of Gα in the moss (Hackenberg et al. 2016; Maruta et al. 2021b). The XLG homologs have also been identified in Sphagnum fallax and the lycophyte Selaginella moellendorffii (Hackenberg et al. 2016).
Table 1.
List of XLGs present in different plants
| Plants | Family | XLGs | Number of XLGs | References |
|---|---|---|---|---|
| Arabidopsis thaliana | Brassicaceae |
AtXLG1 (At2g23460), AtXLG2 (At4g34390), AtXLG3 (At1g31930) |
3 | Assmann (2005); Chakravorty et al. (2011) |
| Oryza sativa | Poaceae |
OsXLG1 (Os12g40190), OsXLG3a (Os11g10050), OsXLG3b (Os06g02130), OsXLG4 (Os10g02814) |
4 | Cantos et al. (2023) |
| Zea mays | Poaceae | ZmXLG1 (GRMZM2G127739), ZmXLG3a (GRMZM2G016858), ZmXLG3b (GRMZM2G429113) | 3 | Wu et al. (2018) |
| Brassica nigra | Brassicaceae |
One XLG1 (BniXLG1‐B1: BniB027874) Three XLG2 (BniXLG2‐B1: BniB032952; BniXLG2‐B2: BniB002451 and BniXLG2‐B3: BniB048389) Two XLG3 (BniXLG3‐B1: BniB016337; BniXLG3‐B2: BniB042917) |
6 | Tiwari et al. (2021) |
| Brassica rapa | Brassicaceae |
One XLG1 (BraXLG1‐A1: Bra032166), Three XLG2 (BraXLG2‐A1: Bra011526; BraXLG2‐A2: Bra017647 and BraXLG2‐A3: Bra034623) Two XLG3 (BraXLG3‐A1: Bra23220; BraXLG3‐A2: Bra033865) |
6 | Tiwari et al. (2021) |
| Brassica juncea | Brassicaceae | Two BjuXLG1, five BjuXLG2, three BjuXLG3 | 10 | Tiwari et al. (2021) |
| Nicotiana benthamiana | Solanaceae | NbXLG1 (Niben101Scf00372g05021), NbXLG2 (Niben101Scf04286g01030), NbXLG3 (Niben101Scf01202g02006), NbXLG4 (Niben101Scf05674g05014), NbXLG5 (Niben101Scf06100g02001), NbXLG6 (Niben101Scf04383g01013), NbXLG7 (Niben101Scf01249g03025) | 7 | Li et al. (2022) |
| Solanum tuberosum | Solanaceae | StXLG1 (XP_006361258), StXLG2 (XP_006352927), StXLG3 (XP_015159201), StXLG4 (XP_006346509), StXLG5 (XP_006338247) | 5 | Li et al. (2022) |
| Solanum lycopersicum | Solanaceae |
SlXLG1 (Sl02g090160), SlXLG2 (Sl01g109110), SlXLG3 (Sl08g005310), SlXLG4 (Sl08g076160), SLXLG5 (Sl03g097980) |
5 | Li et al. (2022) |
| Fagopyrum tataricum | Polygonaceae |
FtXLG1 (FtPinG0009603600.01), FtXLG2 (FtPinG0004968100.01), FtXLG3 (FtPinG0004619900.01), FtXLG4 (FtPinG0004387400.01), FtXLG5 (FtPinG0000898500.01), FtXLG6 (FtPinG0005205100.01) |
6 | Liu et al. (2021) |
| Ananas comosus | Bromeliaceae | AcXLG1, AcXLG2, AcXLG3, AcXLG4 | 4 | Li et al. (2022) |
| Fragaria ananassa | Rosaceae | FaXLG1, FaXLG2, FaXLG3, FaXLG4 | 4 | Li et al. (2022) |
| Malus domestica | Rosaceae | MdXLG1, MdXLG2, MdXLG3, MdXLG4, MdXLG5, MdXLG6, MdXLG7 | 7 | Li et al. (2022) |
| Coleochaeta orbicularis | Coleochaetaceae | Present | Number not reported | Hackenberg et al. (2016) |
| Klebsormidium flaccidiium | Klebsormidiaceae | Present | Number not reported | Hackenberg et al. (2016) |
| Marchantia polymorpha | Marchantiaceae |
MpXLG (Mapoly0129s0046.1) |
1 | Bowman et al. (2017); Wu et al. (2022) |
| Physcomitrium patens | Funariaceae |
PpXLG (Pp1s147_153V6.1) |
1 | Hackenberg et al. (2016); Maruta et al. (2021b) |
XLGs were ignored for a long time considering that, aside from homology, there was no convincing evidence that they were associated with plant G-protein signaling (Ghusinga et al. 2022). However, XLGs resurfaced when it was shown that the elimination of all XLGs in conjunction with the conventional Gα subunit produced phenotypes similar to Gβγ mutants (Ghusinga et al. 2022). Moreover, myriad of studies implicate that the spectrum of G-protein heterotrimer combinations is expanded by XLG-Gβγ heterotrimers, which offer alternative signaling paradigms for fine-tuning plant G-protein responses. Furthermore, the possibility of signal partition and competition between Gα and XLGs opens up a new frontier of cell signaling in plants (Ghusinga et al. 2022). The existence of these peculiar G-proteins distinguishes and paradoxically characterizes plant G-protein signaling (Urano et al. 2016). Notably, various agronomically significant plant architecture and resilience to abiotic and biotic stresses are controlled by XLGs in both redundant and specific manner (Cantos et al. 2023). Consequently, considering the several roles of XLGs throughout plant growth and stress responses, they serve as additional crucial nodes in plant G-protein signaling (Ding et al. 2008; Maruta et al. 2015; Urano et al. 2016; Tiwari et al. 2021). Further studies of important crop species may assist in identifying novel physiological and architectural traits as well as stress responses linked to XLGs involving their mechanism of action, which may be useful for crop improvement. This review offers insights into the structural details and interactomes of XLGs in plants. The review also discusses the functions of XLGs in plant growth, abiotic and biotic stress responses, and proposes various pathways involving XLGs to regulate these biological processes in plants.
Structure of XLGs
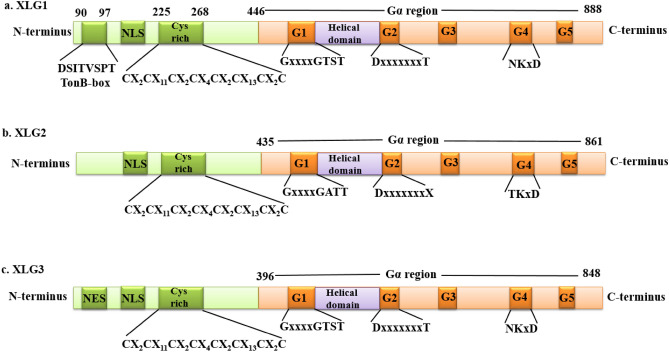
The model plant A. thaliana contains three XLGs: AtXLG1, AtXLG2 and AtXLG3 (Assmann 2005; Chakravorty et al. 2011). XLG1 comprises 888 amino acids and has a molecular mass of 99 kDa, twice that of the conventional Gα (Lee and Assmann 1999). The C-terminal region of AtXLG1 is about 405 amino acids long and shares 26% identity and 50% similarity with Saccharomyces cerevisiae GPA1 (Gα protein) and 32% identity and 54% similarity with Arabidopsis GPα1 (Lee and Assmann 1999). The Gα subunit, which pertains to the GTPase superfamily, has a GTPase domain that is remarkably conserved (McIntire 2009). Guanine nucleotide binding and hydrolysis are associated with five regions of the GTPase domain, numbered G-1 to G-5 (Temple and Jones 2007). The G-1 to G-5 portions of the GTPase domain are mostly but not entirely conserved in the AtXLG1 protein (Temple and Jones 2007). With the exception of a lysine residue being absent, the sequence in AtXLG1 between amino acid residues 485 and 498 resembles the consensus sequence of the G-1 region (Lee and Assmann 1999). AtXLG1 has a threonine residue in lieu of a lysine residue (Ding et al. 2008; Urano et al. 2016). While AtXLG1 maintains the same sequence as the G-2 consensus sequence from amino acids 661 to 669, a conserved arginine residue found in Gα-proteins is swapped for glutamate in this protein (Ding et al. 2008). The least conserved region in AtXLG1 is the G-3 portion (Lee and Assmann 1999). All known Gα-proteins contain three amino acid residues aspartate, glycine, and glutamine in their G-3 region, but AtXLG1 lacks them. Between amino acid residues 770 and 777, AtXLG1 shares the same sequence as G-4 (Ding et al. 2008). The G-5 domain cannot be specifically identified due to the limited information. In addition to sharing an identical aspartate towards the end of G-5, AtXLG1 possesses a conserved serine substitution at the first threonine residue of G-5 (Lee and Assmann 1999). Furthermore, between the G-1 and G-2 portions, XLG1 features a helical domain, and between the G-3 and G-4 regions, XLG1 has an aspartate/glutamate-rich loop (Ding et al. 2008). XLG1 has a peculiar N-terminal region that has about 400 amino acids (Ding et al. 2010). AtXLG1 has a TonB-box at its N-terminus. The TonB-box is localized in the transport proteins of the outer membrane of bacteria and in a few proteins of eukaryotes (Postle and Kadner 2003). The consensus sequence identified as (90DSITVSPT97) characterizes the TonB-box. The identification of a TonB-box in AtXLG1 opens up enticing prospects for the involvement of this unique plant protein in energy transduction and signaling processes. As a possibility, the TonB-box function could be to bestow specificity on AtXLG1's interactions with upstream or downstream proteins (Ding et al. 2008). Another intriguing aspect of the N-terminus of AtXLG1 is the presence of a cysteine rich region exhibiting CX2CX11CX2CX4CX2CX13CX2C (where X represents the number of amino acid residues between two cysteine residues); this region extends from 225 to 268 amino acid residues (Urano et al. 2013). Despite not matching any known zinc finger, the regularly spaced cysteine region is evocative of zinc finger domains that are engaged in the interactions between protein and DNA (Ding et al. 2008).
Two additional homologs of XLG1, namely, XLG2 and XLG3 have also been reported in the genome of A. thaliana (Ding et al. 2008). XLG1446−888, XLG2435−861, and XLG3396−848 each feature a C-terminal Gα domain that shares 26.1, 23.2, and 28.5% identity, respectively, with the canonical Gα protein (Liang et al. 2017). Although XLG2 and XLG3 share the majority of the Gα domain characteristics, there are some differences in the amino acid sequence. In the G-1 region (characterized as GxxxxGKST), XLG1 and XLG3 contain GxxxxGTST, while XLG2 contains GxxxxGATT. In the G-2 region (characterized as DxxxxxxxT), XLG1 and XLG3 have the conserved amino acids D and T, while XLG2 lacks the conserved T amino acid in the G-2 region (Ding et al. 2008). However, it possesses the conserved D in the G-2 region. The three AtXLGs lack the conserved D, G and Q in the G-3 region, which is featured by DxxGQ (Fig. 1) (Ding et al. 2008). In the G-4 region, which is characterized by NKxD, XLG1 and XLG3 contain all the conserved residues, but in XLG2, N is substituted by T (Ding et al. 2008). The XLG2 and XLG3 also possess a similar cysteine-rich amino pattern as is present in the XLG1. However, XLG2 and XLG3 lack a TonB box consensus sequence, in contrast to XLG1 (Lee and Assmann 1999; Ding et al. 2008). According to a study, each of the three XLGs has a putative nuclear localization signal (NLS), and XLG3 is responsible for encoding the nuclear export signal (NES) (Urano et al. 2013; Liang et al. 2017). The amino-termini of the coding regions of XLG were linked upstream of a GFP reporter and then transformed in Vicia faba in order to evaluate the assumption that the N-termini of XLG proteins possess an NLS. As a result, the nucleus was found to be the location of fluorescence for all three fused XLG proteins (Ding et al. 2008). According to a study, the varied cofactor requirements of XLG1, XLG2 and XLG3 are presumably caused by changes in the amino acid sequences of these proteins (Heo et al. 2012).
Fig. 1.
Comparison of the structures of XLG1, XLG2 and XLG3 of A. thaliana. All three XLGs possess an N-terminus and a C-terminus region. The C-terminus of all the XLGs is similar to the canonical Gα subunit and harbors five G-boxes: G-1, G-2, G-3, G-4 and G-5. A helical domain is present between the G-1 and G-2 region. The N-terminus of XLGs has a nuclear localization signal (NLS) and cysteine-rich regions marked as CX2CX11CX2CX4CX2CX13CX2C (where X represents the number of amino acid residues between two cysteine residues). The C-termini of XLG1, XLG2 and XLG3 range from 446–888, 435–861 and 396–848 amino acids respectively. The G-1, G-2 and G-4 regions of XLG1 and XLG3 are similar; however, these regions in XLG2 differ in the amino acid sequence, as shown in the figure. Furthermore, XLG1 has a TonB-box, which is missing in other XLGs. Nuclear export signal (NES) is present in XLG3, although it is absent in XLG1 and XLG2
The AtXLG1 gene shows the presence of 7 exons and 6 introns (Lee and Assmann 1999). OsXLG1 and OsXLG2 both have 9 exons and 8 introns, whereas OsXLG3 has 8 exons and 9 introns (Biswal et al. 2022). Additionally, the XLG genes in the Brassica lineage have independently expanded and have 6–8 exons. In contrast, the introns in XLG genes have substantially diverse sizes and sequences, indicating independent evolution (Tiwari et al. 2021). In addition to the XLG genes, the genome of A. thaliana has four genes that are referred to as the “XLG-related proteins” (Ding et al. 2008). These XLG-related proteins do not include any Gα domains or the cysteine-rich region that is present in XLGs. Nonetheless, the C-terminus of XLG-related proteins exhibits a slight resemblance to some portions of the N-termini of XLG proteins (Ding et al. 2008).
XLGs lack key residues for GTP-binding
It is uncertain whether XLGs bind guanine nucleotides or not, but the available evidence indicates that, if they do, their mechanism of binding differs from the classical Gα subunit (Liang et al. 2017). Since XLG2 shows poor binding with GTP in vitro, it is not envisaged that XLG2 would be nucleotide bound in plant cells at the expected GTP concentrations. Notably, it has been reported that the XLG2 interacts with the RGS, Gβγ, and defense-related receptor-like kinases (RLKs) with an affinity similar to that of classical Gα subunits, independent of GTP-binding (Liang et al. 2016; Lou et al. 2020; Maruta et al. 2021a, b). A study by Heo et al. (2012) reported that XLG2 demonstrated GTP hydrolysis and binding by utilizing Ca2+ as a cofactor rather than Mg2+. In the presence of Mg2+, GPA1 has been observed to bind GTPγS quickly (Sprang 1997; Lou et al. 2020). However, XLG2C (C-terminus of XLG2) failed to bind GTPγS when Mg2+ was present (Heo et al. 2012). For the GTPase activities, XLG2 proteins favour Ca2+ rather than Mg2+ as a cofactor (Heo et al. 2012; Chakravorty et al. 2015). It has been demonstrated that Ca2+ stimulates GTPγS binding with XLG2C, but in the absence of any divalent ion, GTPγS does not exhibit binding with XLG2C (Heo et al. 2012). Contrarily, GPA1 demonstrates reduced GTPγS binding when Ca2+ is present compared to Mg2+ (Heo et al. 2012). XLG1 and XLG3 function similar to XLG2C in that they also bind GTPγS when Ca2+ is present (Heo et al. 2012; Trusov and Botella 2016). The G-1 to G-3 domains of Gα proteins serve as vital regions for the binding of Mg2+ with the α, β and γ phosphates of guanine nucleotides (Sprang 2016). For the guanine ring to bind, the G-4 and G-5 domains are critical. The G-1, G-2, and G-4 portions of the XLG2 protein display the presence of conserved amino acid residues (Lee and Assmann 1999). However, the G-3 and G-5 regions' conserved motifs are absent from XLG2 proteins (Lou et al. 2020). Despite having a conserved threonine amino acid residue present in the G-2 region, which is essential for binding to Mg2+, XLG2 employs Ca2+ but not Mg2+ as a cofactor (Lee and Assmann 1999; Urano et al. 2013; Heo et al. 2012). Unexpectedly, the Physcomitrium patens XLG protein, unlike other G-like proteins, was active when Mg2+ was present instead of Ca2+ (Hackenberg et al. 2016).
XLGs with an array of interacting partners regulate physiological functions in plants
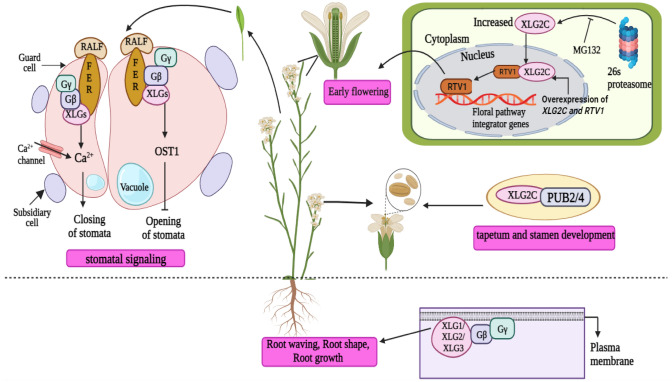
XLGs are known to play various functions in the developmental processes of plants (Urano et al. 2016). A crucial step in the effective reproduction of flowering plants is the transition from the vegetative to the reproductive stages. According to studies, XLG2 aids in the early flowering of A. thaliana (Heo et al. 2012). Related to Vernalization 1 (RTV1) acts as an interacting partner of XLG2, and the C-terminus of GTP-bound XLG2 has been shown to physically interact with RTV1 in vitro as well as in planta (Heo et al. 2012; Trusov and Botella 2016). The activity of RTV1 has been found to be enhanced by the interaction between XLG2 and RTV1 (Trusov and Botella 2016). The mutant xlg2c that is unable to bind to GTP did not show interaction with RTV1 (Fig. 2) (Heo et al. 2012). It has been further reported that floral pathway integrators, namely, FLOWERING LOCUS T (FT), LEAFY (LFY), and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1), exhibited increased expression when RTV1 and XLG2 were overexpressed simultaneously (Liang et al. 2017). These plants displayed noticeably earlier flowering than those in which only RTV1 was overexpressed (Maruta et al. 2015). In this way, the regulation of blooming time in A. thaliana is accomplished by the interplay between RTV1 and XLG2. Additionally, it has been shown that XLGs negatively affect root length in A. thaliana (Ding et al. 2010). The xlg1-1 xlg2-1 xlg3-1 triple mutants have been shown to have substantially longer primary roots than wild-type (WT) plants when grown in the dark; this phenomenon was not present in the xlg single mutants (Ding et al. 2008; Urano et al. 2013). XLG3 participates in the modulation of root responses. The root-waving and root-skewing phenomena have been revealed to be positively affected by XLG3 and AGB1 (Pandey et al. 2008; Ding et al. 2010). These proteins modulate the hormonal variables that govern root-waving and root-skewing in plants (Pandey et al. 2008).
Fig. 2.
The physiological roles of XLGs in A. thaliana. The XLGs are known to influence the early flowering in A. thaliana by interacting with RTV1. The overexpression of RTV1 and XLG2 boosts the expression of floral pathway integrators. The C-terminal region of GTP-bound XLG2C physically interacts with RTV1 and activates it, thereby triggering the binding of RTV1 to the promoter regions of floral pathway integrator genes. This stimulates the early flowering of the plants. The XLGs regulate the development of the tapetum and stamen by interacting with PUB2/4. XLGs, particularly XLG3, influence root parameters like root shape, root waving, root skewing and root growth by interacting with the Gβ subunit. XLGs are also implicated in the stomatal signaling cascades. XLGs (XLG1, XLG2 and XLG3) and Gβ subunits are involved in RALF1-FERONIA signaling which may raise the levels of cytosolic calcium and promote stomatal closure. When OST1 is involved in this interaction, it inhibits the stomatal opening
One of the key elements affecting the yield per plant of rice and other cereal crops is the grain weight. Numerous studies have found that XLGs are involved in controlling various agronomic parameters, including rice panicle length, grain filling, grain size and biomass (Cui et al. 2020; Biswal et al. 2022; Zhao et al. 2022). Numerous xlg mutants have been developed to explore the functions of XLGs in regulating various plant agronomic characteristics (Table 2) (Wu et al. 2018; Cui et al. 2020; Biswal et al. 2022; Zhao et al. 2022). For instance, three genes, ZmXLG1, ZmXLG3a, and ZmXLG3b, were knocked out to determine the role of ZmXLGs in maize development; all Zmxlg triple mutants showed a significant delay in development due to lethality at the seedling stage (Wu et al. 2018). Similar abnormalities were also exhibited by the triple xlg mutants Osxlg1/Osxlg2/Osxlg4, suggesting that XLGs are critical for the survival of rice (Biswal et al. 2022). Moreover, a knockout mutant of Gβ also show lethality at the seedling stage in rice as well as maize (Gao et al. 2019; Cantos et al. 2023), thereby implying that XLGs and Gβ(γ) interact to promote the development of plants.
Table 2.
List of observed phenotypes conferred by mutations in XLG gene(s) in various plants
| XLG mutants | Plant | XLG mutant phenotype | References |
|---|---|---|---|
| xlg2c | Arabidopsis thaliana | Unable to promote early flowering | Heo et al. (2012) |
| xlg 1/2/3 triple mutant | Arabidopsis thaliana | Longer roots than the wild-type | Ding et al. (2008) |
| xlg1/2/3 triple mutant | Arabidopsis thaliana | Numerous stomata | Roy Choudhury et al. (2020) |
| xlg1/2/3 triple mutant | Arabidopsis thaliana | Improper development of stamen and tapetum | Wang et al. (2017) |
| Osxlg1 (pxlg1) | Oryza sativa | More panicles | Cui et al. (2020) |
| Osxlg2-1 | Oryza sativa | Longer grains | Biswal et al. (2022) |
| Osxlg4-1 | Oryza sativa | Longer grains | Biswal et al. (2022) |
| Osxlg1-1 | Oryza sativa | Shorter grains | Biswal et al. (2022) |
| Osxlg1-2, 4–2; Osxlg2-5, 4–2 double mutants | Oryza sativa | Reduction in grain size | Biswal et al. (2022) |
| Osxlg1,2,4–3; Osxlg1,2,4–5; Osxlg1,2,4–6 triple mutants | Oryza sativa | Reduction in grain size | Biswal et al. (2022) |
| Osxlg1-1, Osxlg2-1, Osxlg4-1 single mutants | Oryza sativa | Improved growth, improved aerial biomass, more tillers | Biswal et al. (2022) |
| Osxlg1 single mutants | Oryza sativa | Marginally increased plant height, longer grains and panicles, increased seed weight | Zhao et al. (2022) |
| Osxlg3 single mutants | Oryza sativa | Shorter plants, shorter panicles and smaller grains | Zhao et al. (2022) |
| ZmXLG1, ZmXLG3a, ZmXLG3b triple mutants | Zea mays | Delay in development, lethality at seedling stage | Wu et al. (2018) |
| PpXLG mutant | Physcomitrium patens | Unable to mature into sporophytes | Hackenberg et al. (2016) |
| xlg3 | Arabidopsis thaliana | Hypersensitivity to ethylene | Ding et al. (2008) |
| xlg1/2/3 triple mutant | Arabidopsis thaliana | Hypersensitivity to cadmium | Urano et al. (2016) |
| xlg2-1, xlg3-1 single mutants | Arabidopsis thaliana | Hypersensitivity to tunicamycin | Chakravorty et al. (2015) |
| Osxlg1-1, Osxlg2-1, Osxlg4-1 single mutants | Oryza sativa | Shoot and root lengths were equivalent to WT under basal and salt conditions | Biswal et al. (2022) |
| Osxlg4 | Oryza sativa | Exhibited increased tolerance to cold and drought stress | Cantos et al. (2023) |
| Atxlg2 | Arabidopsis thaliana | More susceptibility to P. syringae | Zhu et al. (2009) |
| xlg1 xlg2 xlg3 triple mutants | Arabidopsis thaliana | Impaired activation of MAPK cascades, more susceptible to pathogen | Wang et al. (2023) |
| Nbxlg3,5 and Nbxlg4 | Nicotiana benthamiana | Less ROS production and reduced expression of PTI5 and ACRE31 when infected by P. syringae or S.sclerotiorum or P. parasitica | Li et al. (2022) |
| BjuXLG-RNAi lines | Brassica juncea | Progression of disease and deposition of fungal mass, reduced production of glucosinolates when infected by S. sclerotiorum | Tiwari et al. (2021) |
|
Knock down of BjuXLGs (BjuXLG1, BjuXLG2 and BjuXLG3) |
Brassica juncea | Decreased expression of defense marker genes | Tiwari et al. (2021) |
| Osxlg1 | Oryza sativa | Susceptible to the bacterial pathogen X. oryzae | Zhao et al. (2022) |
| Osxlg2, Osxlg3 | Oryza sativa | Impaired resistance to M. oryzae | Zhao et al. (2022) |
In Zea mays, XLGs have been reported to perform redundant functions, some of which are independent of Gα subunits while others work in conjunction with Gα subunits (Wu et al. 2018; Cantos et al. 2023). Dwarfism is a common phenotype of all double Zmxlg mutants, and it is enhanced when the Gα subunit COMPACT PLANT2 (CT2) is also mutated in maize (Wu et al. 2018). Although the shoot apical meristem (SAM) of the Zmxlg triple mutant is normal, the decreased SAM size of the ct2 mutant is amplified when paired with any xlg double mutant (Wu et al. 2018; Cantos et al. 2023). These findings suggest that CT2 and XLGs perform overlapping functions in regulating height and the apical meristem of maize, but CT2 cannot compensate for the XLGs during the early stages of maize development, where they are necessary for survival after the germination stage of maize. Additionally, when ZmXLGs were knocked out using CRISPR/Cas9, it did not result in ear fasciation or improve this trait in the ct2 mutants, indicating that CT2 plays an independent role in the development of the inflorescence meristem in maize (Wu et al. 2018; Cantos et al. 2023). However, in Arabidopsis, the XLGs and Gα subunits play similar roles in regulating stomatal numbers. The number of stomata in gpa1 mutants is fewer than that in WT plants; however, it is higher in agb1 mutants. Along with the agb1 mutants, the xlg1/2/3 mutants have been demonstrated to also have increased stomatal densities than the WT plants (Roy Choudhury et al. 2020). Surprisingly, the phenotype produced by xlg1/2/3.gpa1 mutants was identical to the gpa1 mutant, indicating that the XLGs and GPA1 subunits share a similar mechanism for this response (Roy Choudhury et al. 2020). It also implies that GPA1 acts downstream of the XLG subunits. This indicates a very intricate relationship between G-proteins in which each protein may affect stomatal development independently in addition to being genetically and physiologically related in parallel pathways (Roy Choudhury et al. 2020). Furthermore, a G-protein-dependent process is used by the Rapid Alkalinization Factor (RALF) to control stomatal apertures (Yu et al. 2018). According to a study, GPA1 and the XLG proteins may compete or divide in several G-protein plant signaling facets (Yu et al. 2018). The fact that AGB1 and the XLGs (XLG1, XLG2 and XLG3), but not canonical Gα (GPA1), are essential for RALF1-mediated stomatal opening and closing in Arabidopdis lends credence to the idea that GPA1 and the XLGs play unique roles in guard cell responses orchestrated by RALF1 (Fig. 2) (Yu et al. 2018; Wang and Botella 2022). Using bimolecular fluorescence complementation, it has been demonstrated that AGB1 and the RLK, namely FERONIA (FER), show interaction. FER-RALF and XLG (XLG1, XLG2 and XLG3) proteins are involved in stomatal closure by raising intracellular calcium. OPEN STOMATA 1 (OST1) inhibits the stomatal opening by cross-talking with FER-RALF and XLGs (Qu et al. 2019).
Ubiquitin-mediated proteasomal degradation is a crucial regulatory pathway to control protein activity in cells. In a study, the A. thaliana XLGs were found to interact with the PUB2 and PUB4 plant U-box E3 ligases (Wang et al. 2017). The pub4 single mutant, the pub2/4 double mutant, and the xlg1/2/3 triple mutant were reported to have comparable phenotypic abnormalities and were found to be impaired in the cytokinin response. Moreover, they lacked a proper tapetum and stamen, indicating the involvement of XLGs in stamen development (Fig. 2) (Wang et al. 2017). The abnormalities of pub4 and the xlg triple mutant were found to be only partially rescued by overexpression of Arabidopsis Response Regulator (ARR10), which is a positive regulator of cytokinin signaling (Wang et al. 2017). The proteasomal-mediated degradation mechanism controls the amount of XLG2 protein in the cells (Zhu et al. 2009). A proteasome inhibitor, benzyloxycarbonyl-L-leucyl-L-leucyl-L-norvaline 4-methyl-coumaryl-7-amide (MG132), was infused into leaves by Zhu et al. (2009) to impede the activity of the proteasomal pathway. It was shown that MG132 treatment dramatically elevated the amount of XLG2 that accumulated in the lines that overexpressed XLG2 (Fig. 2) (Zhu et al. 2009).
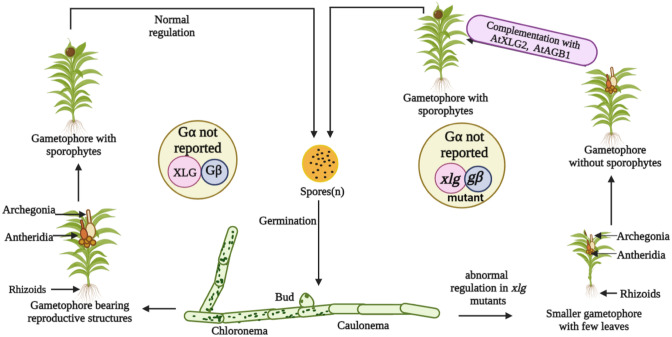
Throughout the evolution of plants, a bryophyte, namely, Physcomitrium patens, has held a special place (Rensing et al. 2020). The fully sequenced genome of this moss does not encode for the classical Gα protein, although it possesses genes encoding Gβ and Gγ proteins. The genome of P. patens also contains a single gene for the XLG protein (Hackenberg et al. 2016; Pandey et al. 2022). This makes it an interesting case from the perspective of G-protein signaling. Research has shown that the conventional Gα protein in P. patens is biochemically and physiologically superseded by the XLG protein, which regulates its development by acting in a genetic process similar to one of the Gβ proteins (Hackenberg et al. 2016). Besides that, the deletion of the chromosomal loci of DPpXLG and DPpGβ2 (deletion mutants) produced gametophores that were smaller, had a slower growth rate, and possessed fewer leaves. These gametophores produce characteristic reproductive structures, but strikingly, they could not mature into sporophytes, indicating that in P. patens, PpXLG (Pp1s147_153V6.1) and PpGβ2 are critical for sporophyte development (Fig. 3) (Hackenberg et al. 2016; Pandey et al. 2022). The homologous genes from A. thaliana, AtXLG2 and AtAGB1, have been reported to complement the mutant phenotypes of PpXLG and PpGβ2, indicating that their function has remained mostly unaltered throughout the evolutionary history of plants (Hackenberg et al. 2016).
Fig. 3.
The importance of XLGs in the life cycle of Physcomitrium patens. The moss, Physcomitrium patens, exhibits an alteration of generation between a long gametophytic phase and a small sporophytic phase. The genome of this moss lacks the Gα subunit, although it has Gβ and Gγ subunits, which makes it a fascinating case study for G-proteins. When XLG, along with the Gβ subunit, is present, moss completes its life cycle. However, if XLG and Gβ subunits are lacking, the moss is unable to produce sporophytes. Moreover, it also shows slower growth and fewer leaves. If the XLG2 and Gβ of A. thaliana are introduced into xlg and gβ mutants of Physcomitrium patens, it becomes capable of producing sporophyte and completing its life cycle
Abiotic stress responses by XLGs and their interacting partners
The plants being sessile are persistently exposed to changing environmental conditions that impact their growth and development, causing significant crop yield losses across the globe (Mantri et al. 2012). However, the plants have adapted a number of mechanisms to handle the challenging conditions (Mantri et al. 2012). The XLG proteins were initially identified in A. thaliana, and it was found that they are necessary for the plants to respond to abscisic acid (ABA), ethylene (ET), auxin, and various other abiotic stresses (Pandey et al. 2008). In A. thaliana, the xlg3 or agb1 mutants have been demonstrated to show hypersensitivity to ET (Ding et al. 2008; Roy Choudhury et al. 2020). According to a study, XLGs respond to cadmium stress, as the xlg1/2/3 triple mutant and agb1-2 mutant show cadmium hypersensitivity (Urano et al. 2016). In A. thaliana, the xlg mutants also exhibit hypersensitivity to tunicamycin (Oliveira et al. 2022). When compared to the WT, the xlg single mutants, primarily xlg2-1 and xlg3-1, displayed hypersensitivity to tunicamycin and a notable increase in stunted seedlings (Chakravorty et al. 2015; Oliveira et al. 2022). Moreover, it has been demonstrated that the xlg triple mutant replicates the hypersensitive phenotype of agb1-2, thereby indicating that XLGs may participate with AGB1/AGG1 and AGB1/AGG2 in a heterotrimeric complex (Chakravorty et al. 2015; Maruta et al. 2021a, b). Wu et al. (2022) found that G-protein drives the transition from metabolic and transcriptional homeostasis to a stress-ready condition in A. thaliana and Marchantia polymorpha by comparing the metabolomic, phenotypic, and transcriptome profiles of Gα, Gβ and xlg null mutants under basal and salt-stress conditions. This stress preparedness strengthens the ABA responses and phenylpropanoid pathway to safeguard the plant from further challenges (Ferrero-Serrano et al. 2022; Wu et al. 2022). The study also highlights that the networks controlling transcription and metabolism have remained unchanged throughout the history of land plants, while the function of plant-specific XLGs has shown divergence (Wu et al. 2022). The roles of XLGs in abiotic stress tolerance have also been examined in Nicotiana benthamiana. It has been found that NbXLG3 and NbXLG5 negatively affect the plant's response to abiotic challenges, including polyethylene glycol (PEG), high salt and mannitol (Li et al. 2022). A study in rice has shown that all single Osxlg mutants exhibited shoot and root lengths that were equivalent to WT under basal and salt conditions, but the Osxlg1,4 double mutant displayed noticeably longer root lengths when compared to WT. The study further indicates that mutations in Osxlg1,4 initiate a signaling cascade inside the plant, which retards shoot growth (Biswal et al. 2022). However, when examined in Osxlg2 and Osxlg4 mutants, it was observed that XLG2 both alone and in conjunction with XLG4 enhances salt tolerance in rice (Cui et al. 2020; Biswal et al. 2022). Recently, a study reported that the mutants of Osxlg4 show increased tolerance to cold stress and drought stress in comparison to the other Osxlg mutants (Cantos et al. 2023). In summary, these findings suggest that the XLG proteins are involved in numerous abiotic stress responses in plants.
Asparagine-rich protein 1 (AtNRP1) and AtNRP2 have been identified as potential interactomes of AtXLG2 and AtXLG3 (Liang et al. 2017; Camargos et al. 2019). In A. thaliana, early response to dehydration (ERD15), which plays a role in stomatal opening and during ABA responses, has also been proposed to show interaction with XLG3 (Aalto et al. 2012; Camargos et al. 2019). The homolog of AtERD15 identified from Glycine max shows binding to the promoter regions of NRP/DCD genes (development and cell death domain-containing asparagine-rich protein) and initiates the signaling cascades that lead to cell death during stress (Aalto et al. 2012; Camargos et al. 2019). Therefore, it can be strongly implicated that the interaction between XLGs and NRP/DCD may contribute to developmental and stress responses (Camargos et al. 2019).
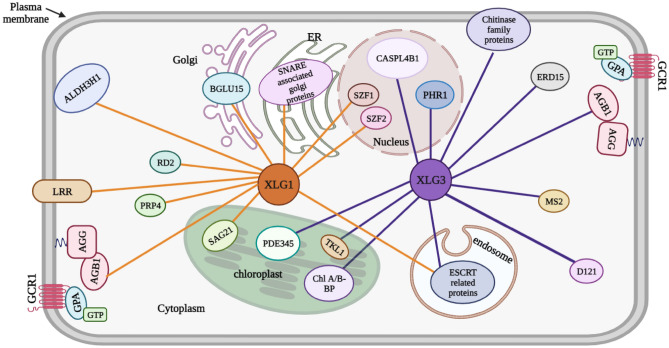
Interacting partners of XLG regulate the localization of XLG proteins inside various organelles (Liang et al. 2017). The endosomal sorting complex required for transport (ESCRT)-related protein interacts with XLG1 and XLG3 in the endosome (Liang et al. 2017). In A. thaliana, some of the XLG protein partners seem to be confined to the nucleus. For instance, the two transcription factors salt-inducible zinc finger 1 (SZF1) and SZF2 show interaction with XLG1 and XLG3 inside the nucleus (Fig. 4, Table 3) (Sun et al. 2007; Liang et al. 2017; Wu et al. 2018). SZF1 and SZF2 gene expression is controlled by the interaction of Gβ subunits with XLGs and other regulators (Liang et al. 2017). Plants under salinity stress develop more quickly when SZF1 and SZF2 are expressed (Sun et al. 2007). Thus, it can be speculated that XLGs may contribute to salinity stress by interacting with SZF1 and SZF2. Therefore, further research to comprehend the mechanisms involved in the signaling cascades modulated by XLGs along with their interacting proteins will eventually pave the way to overcome worldwide crop losses marked by abiotic stress.
Fig. 4.
The network of interacting partners of XLG1 and XLG3 in A. thaliana. XLG1 interacts with various proteins in different cellular compartments like nucleus, endoplasmic reticulum, cytosol, chloroplast, etc. XLG1 interacts with SZF1 and SZF2 (transcription factors) inside the nucleus. It possibly interacts with SAG21, BGLU15 and SNARE-associated Golgi proteins inside the chloroplast, Golgi and endoplasmic reticulum, respectively. LRR, ALDH3H1 and AGB1 show interaction with XLG1 at the plasma membrane. RD2 and PRP4 have been predicted to interact with XLG1 in the cytosol. XLG3 may interact with CASPL4B1 and PHR1 inside the nucleus. ERD15, MS2 and DI21 have been predicted to interact with XLG3 in the cytosol. Other proteins like Chl A/B binding proteins, PDE345 and TKL1 interact with XLG3 inside the chloroplast (Liang et al. 2017; https://string-db.org; https://www.arabidopsis.org). The possible functions of these interactions have been listed in Table 3. Senescence-associated gene 21 (SAG21); Beta-glucosidase 15 (BGLU 15); Leucine-rich repeats (LRRs); Aldehyde dehydrogenase 3H1 (ALDH3H1); Responsive to desiccation 2 (RD2); Proline-rich protein (PRP4); Casp-like protein 4B1 (CASPL4B1); Phosphate-starvation response 1 (PHR1); Early response to dehydration 15 (ERD15); Methionine synthase 2 (MS2); Drought-induced 21 (DI21); Pigment defective 345 (PDE345); Transketolase 1 (TKL1)
Table 3.
The interacting partners of XLGs and their predicted functions in A. thaliana
| XLGs | Interacting partners | Predicted functions | Interaction localization |
|---|---|---|---|
| XLG1 | SZF1 | Salt-stress responses | Nucleus |
| XLG1 | SZF2 | Salt-stress responses | Nucleus |
| XLG1 | SAG21 | Involved in leaf senescence | Chloroplast |
| XLG1 | BGLU 15 | Beta-glucosidase activity | Golgi |
| XLG1 | SNARE-associated golgi protein | Vesicle fusion | Endoplasmic reticulum |
| XLG1 | LRR | Works as a receptor | Plasma membrane |
| XLG1 | ALDH3H1 | Water-deprivation responses | Plasma membrane |
| XLG1 | RD2 | Desiccation responses | Cytosol |
| XLG1 | PRP4 | Localization on the cell wall | Cytosol |
| XLG1 | ESCRT-related protein | Involved in trafficking | Endosome |
| XLG1 | AGB1 | Developmental responses | Plasma membrane |
| XLG3 | CASPL4B1 | Unknown | Nucleus |
| XLG3 | PHR1 | Phosphate starvation responses | Nucleus |
| XLG3 | ERD15 | Water-deprivation responses | Cytosol |
| XLG3 | MS2 | Cadmium stress responses | Cytosol |
| XLG3 | DI21 | Drought stress responses | Cytosol |
| XLG3 | Chitinase family protein | Chitinase activity during fungal attack | Plasma membrane |
| XLG3 | AGB1 | Developmental responses | Plasma membrane |
| XLG3 | Chl A/B-Binding Protein | Unknown | Chloroplast |
| XLG3 | PDE345 | Fructose-bisphosphate aldolase activity | Chloroplast |
| XLG3 | TKL1 | Cadmium stress responses | Chloroplast |
| XLG3 | ESCRT-related protein | Involved in trafficking | Endosome |
Biotic stress responses by XLGs
Plants, unlike animals, lack an adaptive immune system. Usually, plants are outfitted with the systems needed to identify encroaching pathogens and send out systemic signals at the location of pathogen invasion (Kaur et al. 2022; Zhang et al. 2022). The G-proteins are recognized to be important in regulating plant immunity to biotic stress (Nitta et al. 2015). In A. thaliana Gα has a protective role against the bacterium Pseudomonas syringae, possibly via modulating stomatal function and thereby limiting bacterial access inside the leaf (Zeng and He 2010). The Gα subunit in Brassica sp. is involved in controlling agronomic parameters like seed germination, silique dimensions and seed weight (Kumar et al. 2014). Gβ and Gγ subunits are implicated in stress responses in Brassica sp. (Kumar et al. 2014). Numerous studies have also revealed the importance of XLG proteins in plant defensive responses (Chakravorty et al. 2015; Maruta et al. 2015; Liang et al. 2018; Tiwari et al. 2021). When various pathogens, such as P. syringae (a bacterium), Alternaria brassicicola (a necrotrophic fungus), and Fusarium oxysporum (a hemibiotrophic fungus), infect xlg and agb1 mutants of A. thaliana, their defense responses are significantly impaired in the same manner and to the same extent (Maruta et al. 2015). XLG3 has been shown to provide resistance against F. oxysporum; however, XLG1 has not been shown to contribute to plant immunity (Maruta et al. 2015). The study additionally shows that the Gβγ dimer interacts physically with XLGs located at the plasma membrane to regulate plant responses against pathogens (Chakravorty et al. 2015; Maruta et al. 2021a). When pathogen-associated elicitors like flagellin 22 (flg22) and elf18 are present, XLG2 and XLG3 participate redundantly in ROS production (Maruta et al. 2015). In A. thaliana, the infection caused by P. syringae was observed to quickly raise the expression levels of XLG2 and XLG3, and the loss-of-function mutation in XLG2 makes the plants more vulnerable to the pathogen in addition to compromising the synthesis of pathogen-responsive genes (Zhu et al. 2009; Maruta et al. 2021a). Using co-immunoprecipitation tests, it was discovered that XLG2 shows interaction with the A. thaliana Gβ subunit, which has earlier been reported to trigger tolerance against various pathogens (Ishikawa 2009; Liu et al. 2013; Nitta et al. 2015). Generally, XLG2 protein is found in lower concentrations in the cells of uninfected leaves; however, within 30 min of Pst avrRpm1 inoculation, the XLG2 transcript begins to accumulate considerably, reaching its peak 3 h later (Zhu et al. 2009). Moreover, the continuous overexpression of XLG2 causes numerous defense-related genes such as AtMPK3, RbohC, and PAD3 to produce aberrant transcripts that are smaller in size, emphasizing the possibility of XLG2 involvement in the transcriptional and/or post-transcriptional regulation of genes involved in defense responses (Zhu et al. 2009). Seedling growth arrest and consequent lethality affect the Zmxlg triple mutant (Wu et al. 2018). Cell death traits and high expression of pathogenesis-related genes, namely PR1 and PR5, suggest that lethality is caused by a hyperactive immune response or that increased PR expression is triggered in response to cell death (Wu et al. 2018; Cantos et al. 2023). Considering that ZmGβ knockout also causes seedling lethality and that ZmXLGs physically interact with ZmGβγ, it is possible that XLG/Gβγ signaling plays a significant role in immunity or cell-death responses, independent of CT2 (Wu et al. 2020).
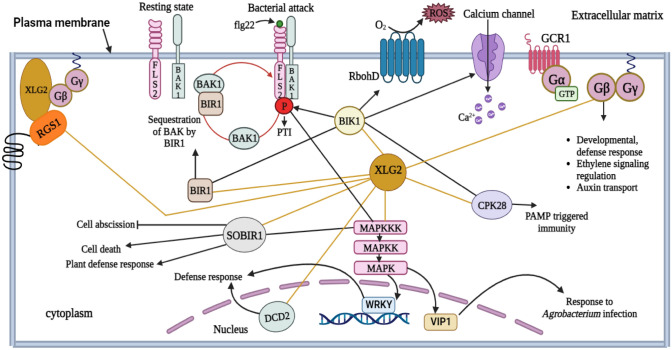
Flagellin-Sensitive 2 (FLS2) and Chitin Elicitor Receptor Kinase 1 (CERK1) are the members of the RLK family (Heese et al. 2007; Petutschnig et al. 2022) Chitin and other components of the fungus cell wall show association with CERK1 (Petutschnig et al. 2022). FLS2, a pattern recognition receptor (PRR) in the plasma membrane of plants, is a useful paradigm for comprehending how the innate immune signaling cascade works in plants (Yuan et al. 2021). FLS2 teams up with Brassinosteroid insensitive 1-associated kinase 1 (BAK1), a co-receptor, to detect the bacterial telltale protein, flg22 (Chinchilla et al. 2007; Sun et al. 2013; Yuan et al. 2021). It subsequently transmits this signal to a receptor-like cytoplasmic kinase (RLCK), Botrytis-induced kinase 1 (BIK1), which then stimulates a number of plant defense mechanisms (Fig. 5) (Lu et al. 2010; Petutschnig et al. 2022). ROS are produced as a result of NADPH oxidase, with RbohD being phosphorylated by BIK1 (Wang and Botella 2022). According to several findings, XLG2, Gβ, and Gγ1/2 act downstream of PRRs to mediate the defense responses in A. thaliana (Ishikawa 2009; Liang et al. 2016). As BIK1 is prone to proteasomal degradation, the G-proteins (XLG2, Gβ, and Gγ1/2) suppress the proteasomal degradation of BIK1 prior to being activated by flg22, maintaining optimal signaling proficiency (Liang et al. 2016; Yu et al. 2022). Upon flg22 elicitation, XLG2 detaches from the Gβ subunit (Stateczny et al. 2016). It results in the phosphorylation of the N-terminus of XLG2 at Ser530 by BIK1 (Liang et al. 2016). This phosphorylation results in the generation of ROS by RbohD, possibly through the interaction of XLG2 with RbohD. The generated ROS provide tolerance against P. syringae pv tomato (Pst) (Liang et al. 2016). Interestingly, the association between XLG2 and RbohD was observed even when flg22 was absent, suggesting that XLG2 interacts with RbohD on a constitutive basis (Oliveira et al. 2022). In this way, FLS2-mediated immunity is regulated by direct interactions between XLG2, FLS2 and BIK1 (Fig. 6). Furthermore, it has also been shown that XLG2 mediates the activation of ROS generation through a BIK1 independent mechanism (Zhong et al. 2019). According to a study, the stability of G-proteins in the FLS2 receptor complex during the resting state has been shown to be significantly influenced by RGS (Stateczny et al. 2016; Liang et al. 2018; Zhong et al. 2019). RGS1 speeds up the GTP hydrolysis in XLG2 in order to stabilize the XLG2-Gβγ trimer in the FLS2 complex (Liang et al. 2018; Zhong et al. 2019). When pathogen associated molecular patterns (PAMPs) are recognized by RLKs, BAK1 phosphorylates and activates BIK1, which then phosphorylates RGS1 at numerous locations in the C-terminus, with Ser431 serving as the main site (Liang et al. 2018; Erickson et al. 2022; Oliveira et al. 2022). As a result, RGS1 segregates from FLS2 and the Gα subunit (Liang et al. 2018). Furthermore, flg22-induced RGS1 internalization occurs with the aid of clathrin-mediated endocytosis in a β-arrestin-like mechanism mediated by the vacuolar sorting protein 26 (VPS26) (Oliveira et al. 2022). Since GPA1 and XLG2 are self-activating Gα proteins, they rapidly convert GDP to GTP when RGS1 is absent. The activated Gα protein (GPA1 or XLG2) separates from Gβγ and interacts with the appropriate effectors to start the downstream signaling process (Tunc-Ozdemir et al. 2017; Xu et al. 2019). XLG2 and GPA1 are known to be involved in different facets of immunological responses (Zhong et al. 2019). While XLG2 increases PAMP-triggered ROS generation and the transcription of genes related to defense, GPA1 regulates stomatal closure (Zeng and He 2010; Liang et al. 2016, 2018; Zhong et al. 2019). It suggests that RGS uses different downstream G-protein subunits and different defensive mechanisms to contribute to the immunological responses of plants (Zhong et al. 2019). A recent study in A. thaliana reveals that all three XLGs show interaction with the MAPK proteins, MAPKKK3/5-MKK4/5-MPK3/6, to further strengthen the plant's resistance against pathogens, as the xlg1 xlg2 xlg3 triple mutants lack MAPK activation and are more susceptible to the pathogen (Wang et al. 2023). Thus, the crosstalk between XLGs and various cell surface receptors initiates signaling cascades involving several downstream effectors that eventually help plants withstand biotic stress.
Fig. 5.
The network of interacting partners of XLG2 in A. thaliana. Among the three XLGs, XLG2 plays a major role during biotic stress. XLG2 interacts with BIK1, which phosphorylates the FLS2 and BAK1 complex and activates them during pathogen attack to trigger PTI and initiate the MAPK cascade to activate the defense-related genes. BIK1 also leads to RbohD-mediated ROS production and the opening of the calcium channel. XLG2 interacts with BIR1 in the cytosol; BIR1 acts as a negative regulator of defense responses by sequestering BAK1. However, FLS2 outcompetes BIR1 and forms a complex with BAK1 following the pathogen attack. XLG2 interacts with (Suppressor of BIR1) SOBIR1 and Calcium-dependent protein kinase 28 (CPK28) in the cytosol to promote plant defense responses. It also shows interaction with DCD2 inside the nucleus. RGS1 and AGB1 show interaction with XLG2 at the plasma membrane (Liang et al. 2017, 2018; Xu et al. 2019; https://string-db.org; https://www.arabidopsis.org)
Fig. 6.
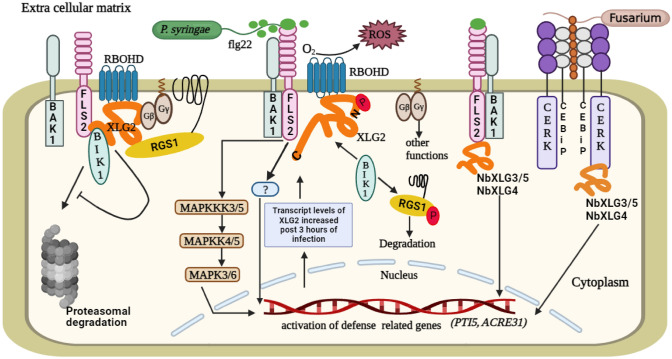
The roles of XLGs during biotic stress in plants. The interaction between Flagellin-Sensitive 2 (FLS2), XLG2 and Botrytis-induced kinase 1 (BIK1) plays important roles during defense responses in plants. FLS2, located in the plasma membrane, acts as a receptor for the flagellin 22 (flg22) protein present in the bacterial flagella. Brassinosteroid insensitive 1-associated kinase 1 (BAK1) works as a co-receptor with FLS2. Since BIK1 is prone to proteasomal degradation, XLG2, Gβ and Gγ1/2 suppress its degradation prior to the pathogen attack in order to maintain signal proficiency. When the pathogen (P. syringae) attacks the plant cell, flg22 present in its flagella binds to FLS2 and activates it. This results in the dimerization of FLS2 and BAK1, which initiate the defense signaling cascades. BIK1, XLG2, Gβ and Gγ1/2 work downstream of the FLS2. Upon activation, XLG2 detaches from the Gβ subunit and BIK1 phosphorylates XLG2 at the N-terminus. This results in the RbohD-dependent production of reactive oxygen species (ROS) through the interaction of XLG2 and RbohD. The ROS provides resistance against the pathogen. Furthermore, this receptor activation also leads to MAPK cascades that activate the defense related genes in the nucleus to strengthen the plant’s immunity. RGS1 stabilises the FLS2 receptor complex in its resting state by hydrolyzing the GTP of XLG2. But when this receptor complex is in an active state, BIK1 phosphorylates RGS1, which may lead to the degradation of RGS1. Another receptor complex, chitin elicitor receptor kinase (CERK)-chitin elicitor binding protein (CEBiP), acts as a receptor for the chitin molecules present in the fungus to mediate resistance against fungal pathogens. XLG3/5 and XLG4 in Nicotiana benthamiana interact with the CERK-CEBiP and FLS2-BAK1 receptor complex to activate the defense related genes that ultimately provide tolerance against the fungus
XLGs, particularly NbXLG3/5 and NbXLG4, have been proven to impart resistance to Nicotiana benthamiana against a variety of pathogens, including P. syringae pv. tomato DC3000 (a bacterial pathogen), Sclerotinia sclerotiorum (a fungal pathogen), and a multitude of oomycetes pathogens in the genus Phytophthora such as P. parasitica, P. infestans, and P. capsici (Li et al. 2022). When the mutant lines Nbxlg3, 5 and Nbxlg4 of N. benthamiana were subjected to bacterial flg22 and fungal chitin, they produced noticeably less ROS and exhibited reduced expression of pathogenesis-related genes transcriptional activator, PTI5 and ACRE3,1 in comparison to the WT plants, indicating that NbXLG3/5 and NbXLG4 help in the production of ROS during the biotic stress (Li et al. 2022). Furthermore, NbXLGs have been demonstrated to interact with the Gβγ to form heterotrimers (Li et al. 2022). NbXLG3, NbXLG5, and NbXLG4 also show coupling with the immune receptors FLS2 and CERK1 to protect the plants against pathogens (Li et al. 2022). Another study in N. benthamiana has found that XLG2 rapidly translocates inside the nucleus in response to flg22 elicitation and this translocation requires NLS (Ma et al. 2022). Furthermore, flg22-induced phosphorylation of XLG2 at Serines 141, 148, 150, and 151 is critical for its nuclear localization. MUT9-like kinases (MLKs) that negatively affect plant immunity are suppressed by XLG2 inside the nucleus (Ma et al. 2022). By suppressing the kinase activity of MLKs, XLG2 stimulates the expression of defense genes, thereby enhancing plant defense mechanisms (Ma et al. 2022). A study indicates that XLGs also play a role during biotic stress in Oryza sativa. OsXLG1 enhances the tolerance of rice plants to Xanthomonas oryzae, as mutants lacking OsXLG1 were extremely susceptible to the bacterial pathogen (Zhao et al. 2022). OsXLG2 and OsXLG3 provide tolerance to the fungus Magnaporthe oryzae, as the mutants Osxlg2 and Osxlg3 showed impaired tolerance to the fungal pathogen (Zhao et al. 2022). These findings suggest that XLGs are important components of plant defense mechanisms against various pathogens. Further research is necessary to understand the molecular mechanisms underlying the role of XLGs in biotic stress responses in different plant species.
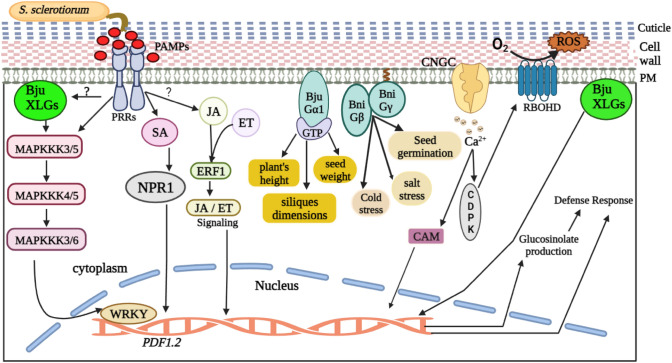
In recent decades, the productivity of Brassica species has been severely constrained because of their innate sensitivity to pathogens. S. sclerotiorum, the causative agent of Sclerotinia stem rot, significantly lowers the production of B. juncea and has been challenging to control because of inadequate host resistance (Sharma et al. 2018; Singh et al. 2022). This pathogenic fungus interferes with plant defense by altering a myriad of signaling pathways, defense hormones in plants and stress-related metabolites (Wang et al. 2019). Research found that in plants, XLGs modulate the defense mechanisms and stress-related compounds in response to S. sclerotiorum, as the BjuXLG-RNAi lines result in the progression of disease and deposition of fungal mass (Tiwari et al. 2021). All three BjuXLG genes (BjuXLG1, BjuXLG2 and BjuXLG3) in B. juncea are known to impart resistance during the preliminary stages of S. sclerotiorum infection; however, BjuXLG3 appears to play a more prominent role as the infection progresses (Tiwari et al. 2021). Moreover, the knockdown of BjuXLGs genes leads to a decreased expression pattern of defense marker genes PDF1.2 and WRKY33, thereby affecting the host’s resistance against S. sclerotiorum (Fig. 7) (Tiwari et al. 2021). Since the phytoalexins produced by A. thaliana under the control of the pathogen-inducible transcription factor WRKY33 offer resistance in response to S. sclerotiorum, it can be inferred that the decreased expression of WRKY33 in BjuXLG-RNAi lines may hinder the accumulation of phytoalexins specific to Brassica, which in turn may contribute to the increased sensitivity of Brassica juncea to the fungus (Mao et al. 2011; Stotz et al. 2011; Tiwari et al. 2021). The glucosinolates present in the plants of the family Brassicaceae, coupled with the by-products of their hydrolysis, are crucial for guarding plants from infections and pests (Sotelo et al. 2015; Chen et al. 2020). A favorable correlation exists between the presence of glucosinolates and B. napus tolerance to S. sclerotiorum (Abuyusuf et al. 2018). Through the modulation of glucosinolate synthesis, XLGs in Brassica juncea have been found to elicit immunological responses (Tiwari et al. 2021). The study further highlights that XLGs and glucosinolate pathways may interact with one another in B. juncea to elicit immunological responses, as the BjuXLG-RNAi lines displayed diminished amounts of glucosinolates in the leaves on S. sclerotiorum infection, in which the levels of aliphatic glucosinolates were severely impacted (Tiwari et al. 2021). Therefore, future research can shed light on many additional metabolites that XLGs may trigger to strengthen the defense system of plants during stress conditions.
Fig. 7.
XLGs provide tolerance to Brassica sp. against a variety of stress. When S. sclerotiorum attacks Brassica, PAMPs are produced, which are recognised by pattern recognition receptors (PRRs). This initiates cross-talk in the cell, which sets off defense responses. PRRs, by an unknown mechanism, activate BjuXLGs, which in turn activate mitogen-activated protein kinase (MAPK) cascades that trigger the activation of defense-related genes. PRRs mediate defense responses through phytohormones. BjuXLGs also aid in the synthesis of glucosinolates during biotic stress responses. The Gα subunit of B. juncea in GTP-bound form mediates agronomic traits like plants’s height, silique dimensions, and seed weight. However, the Gβ and Gγ subunits of B. nigra are implicated in cold and salt stress responses, along with other agronomic trait regulation. A cyclic-nucleotide gated channel (CNGC) helps in the influx of calcium (Ca2+) that also triggers defense responses by producing RbohD-dependent ROS (Ding et al. 2021)
Perspectives and future directions
Identification of the majority, if not all, of the elements of plant G-proteins, marks the beginning of finding solutions for real-world agronomic problems. In plants, a subclass of G-proteins known as XLG is involved in multiple processes, including flowering, root architecture, stomatal signaling, growth and abiotic and biotic stress. It is essential to ascertain how the XLGs integrate into the conventional G-protein signaling cascade in anticipation of analysing the mechanism of signal transmission across the plant cell. A thorough comprehension of the XLG proteins involved in the G-protein signaling cascade may help identify the potential breeding targets for the numerous agronomically significant traits linked to the G-proteins, such as biomass production, flowering time, abiotic stress and disease resistance. In this review, we have incorporated emerging insights about the involvement of XLG proteins in regulating pathways that facilitate plant growth and environmental stress management. The information on XLG proteins included in this review can be augmented by implementing techniques like CRISPR/Cas9-mediated gene editing, overexpression, and RNAi approaches to engineer economically significant crops. As a result, farmers and agriculturists would gain access to highly productive, stress-tolerant plants in a future marked by global warming and climate change. Future-focused research should therefore be capable of mapping the agronomic potential of XLG proteins.
Acknowledgements
Authors acknowledge the University Grant Commission, New Delhi, India for funding this research.
Author contributions
NT, DB and SG conceptualised the idea. BS, JG, KS and DB have written the paper. BS and BB designed the figures. NT, DB and SG edited the paper.
Funding
No funding was received to assist with the preparation of this manuscript.
Data availability
Not applicable.
Declarations
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Deepak Bhardwaj, Email: dbhardwajpmb@gmail.com.
Narendra Tuteja, Email: narendratuteja@gmail.com.
References
- Aalto MK, Helenius E, Kariola T, Pennanen V, Heino P, Hõrak H, Puzõrjova I, Kollist H, Palva ET. ERD15—an attenuator of plant ABA responses and stomatal aperture. Physiol Mol Biol Plants. 2012;182:19–28. doi: 10.1016/j.plantsci.2011.08.009. [DOI] [PubMed] [Google Scholar]
- Abuyusuf M, Robin AHK, Lee JH, Jung HJ, Kim HT, Park JI, Nou IS. Glucosinolate profiling and expression analysis of glucosinolate biosynthesis genes differentiate white mold resistant and susceptible cabbage lines. Int J Mol Sci. 2018;19(12):4037. doi: 10.3390/ijms19124037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Abhiman S, de Souza RF, Aravind L. Comparative genomics uncovers novel structural and functional features of the heterotrimeric GTPase signaling system. Gene. 2011;475(2):63–78. doi: 10.1016/j.gene.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assmann SM. G proteins go green: a plant G protein signalling FAQ sheet. Science. 2005;310(5745):71–73. doi: 10.1126/science.1118580. [DOI] [PubMed] [Google Scholar]
- Bhardwaj D, Sahoo RK, Naqvi AR, Lakhanpaul S, Tuteja N. Pea Gβ subunit of G proteins has a role in nitric oxide-induced stomatal closure in response to heat and drought stress. Protoplasma. 2020;257:1639–1654. doi: 10.1007/s00709-020-01529-6. [DOI] [PubMed] [Google Scholar]
- Biswal AK, Wu TY, Urano D, Pelissier R, Morel JB, Jones AM, Biswal AK. Novel mutant alleles reveal a role of the extra-large G protein in rice grain filling, panicle architecture, plant growth, and disease resistance. Front Plant Sci. 2022;12:2821. doi: 10.3389/fpls.2021.782960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botella JR. Can heterotrimeric G proteins help to feed the world? Trends Plant Sci. 2012;17(10):563–568. doi: 10.1016/j.tplants.2012.06.002. [DOI] [PubMed] [Google Scholar]
- Bowman JL, Kohchi T, Yamato KT, Jenkins J, Shu S, Ishizaki K, Yamaoka S, Nishihama R, Nakamura Y, Berger F, Adam C. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell. 2017;171(2):287–304. doi: 10.1016/j.cell.2017.09.030. [DOI] [PubMed] [Google Scholar]
- Cantos CF, de Pamphilis CW, Assmann SM. Extra-large G proteins have extra-large effects on agronomic traits and stress tolerance in maize and rice. Trends Plant Sci. 2023 doi: 10.1016/j.tplants.2023.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty N, Raghuram N. Life, death and resurrection of plant GPCRs. Plant Mol Biol. 2022 doi: 10.1007/s11103-022-01323-3. [DOI] [PubMed] [Google Scholar]
- Chakravorty D, Trusov Y, Zhang W, Acharya BR, Sheahan MB, McCurdy DW, Assmann SM, Botella JR. An atypical heterotrimeric G-protein γ-subunit is involved in guard cell K+-channel regulation and morphological development in Arabidopsis thaliana. Plant. 2011;J67(5):840–851. doi: 10.1111/j.1365-313X.2011.04638.x. [DOI] [PubMed] [Google Scholar]
- Chakravorty D, Gookin TE, Milner MJ, Yu Y, Assmann SM (2015) Extra-large G proteins expand the repertoire of subunits in Arabidopsis heterotrimeric G protein signaling. J Plant Physiol 169(1):512–529. 10.1104/pp.15.00251 [DOI] [PMC free article] [PubMed]
- Chen J, Ullah C, Reichelt M, Beran F, Yang ZL, Gershenzon J, Hammerbacher A, Vassão DG. The phytopathogenic fungus Sclerotinia sclerotiorum detoxifies plant glucosinolate hydrolysis products via an isothiocyanate hydrolase. Nat Commun. 2020;11(1):3090. doi: 10.1038/s41467-020-16921-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nürnberger T, Jones JD, Felix G, Boller T. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defense. Nature. 2007;448(7152):497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- Cui Y, Jiang N, Xu Z, Xu Q. Heterotrimeric G proteins are involved in the regulation of multiple agronomic traits and stress tolerance in rice. BMC Plant Biol. 2020;20:1–13. doi: 10.1186/s12870-020-2289-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Camargos LF, Fraga OT, Oliveira CC, da Silva JCF, Fontes EPB, Reis PAB. Development and cell death domain-containing asparagine-rich protein (DCD/NRP): an essential protein in plant development and stress responses. Theor Exp Plant Physiol. 2019;31:59–70. doi: 10.1007/s40626-018-0128-z. [DOI] [Google Scholar]
- Ding L, Pandey S, Assmann SM. Arabidopsis extra-large G proteins (XLGs) regulate root morphogenesis. Plant J. 2008;53(2):248–263. doi: 10.1111/j.1365-313X.2007.03335.x. [DOI] [PubMed] [Google Scholar]
- Ding L, Gookin TE, Assmann SM. Unconventional GTP-binding proteins in plants. Integr G Proteins Signal Plants. 2010 doi: 10.1007/978-3-642-03524-1_14. [DOI] [Google Scholar]
- Ding LN, Li T, Guo XJ, Li M, Liu XY, Cao J, Tan XL. Sclerotinia stem rot resistance in rapeseed: recent progress and future prospects. J Agric Food Chem. 2021;69(10):2965–2978. doi: 10.1021/acs.jafc.0c07351. [DOI] [PubMed] [Google Scholar]
- Erickson J, Weckwerth P, Romeis T, Lee J. What’s new in protein kinase/phosphatase signaling in the control of plant immunity? Essays Biochem. 2022;66(5):621–634. doi: 10.1042/EBC20210088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrero-Serrano Á, Chakravorty D. Plants and heterotrimeric G proteins: expect the unexpected. Mol Plant. 2022;16(3):506–508. doi: 10.1016/j.molp.2022.12.017. [DOI] [PubMed] [Google Scholar]
- Ganotra J, Sharma B, Biswal B, Bhardwaj D, Tuteja N. Emerging role of small GTPases and their interactome in plants to combat abiotic and biotic stress. Protoplasma. 2023;260(4):1007–1029. doi: 10.1007/s00709-022-01830-6. [DOI] [PubMed] [Google Scholar]
- Gao Y, Gu H, Leburu M, Li X, Wang Y, Sheng J, Fang H, Gu M, Liang G. The heterotrimeric G protein β subunit RGB1 is required for seedling formation in rice. Rice. 2019;12(1):1–14. doi: 10.1186/s12284-019-0313-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghusinga KR, Elston TC, Jones AM. Towards resolution of a paradox in plant G-protein signaling. J Plant Physiol. 2022;188(2):807–815. doi: 10.1093/plphys/kiab534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg D, Perroud PF, Quatrano R, Pandey S. Sporophyte formation and life cycle completion in moss requires heterotrimeric G-proteins. J Plant Physiol. 2016;172(2):1154–1166. doi: 10.1104/pp.16.01088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackenberg D, McKain MR, Lee SG, Roy Choudhury S, McCann T, Schreier S, Harkess A, Pires JC, Wong GK, Jez JM, Kellogg EA. Gα and regulator of G-protein signaling (RGS) protein pairs maintain functional compatibility and conserved interaction interfaces throughout evolution despite frequent loss of RGS proteins in plants. New Phytol. 2017;216(2):562–575. doi: 10.1111/nph.14180. [DOI] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, Schroeder JI, Peck SC, Rathjen JP. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA. 2007;104(29):12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heo JB, Sung S, Assmann SM. Ca2+-dependent GTPase, extra-large G protein 2 (XLG2), promotes activation of DNA-binding protein related to vernalization 1 (RTV1), leading to activation of floral integrator genes and early flowering in Arabidopsis. J Biol Chem. 2012;287(11):8242–8253. doi: 10.1074/jbc.M111.317412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa A. The Arabidopsis G-protein β-subunit is required for defense response against Agrobacterium tumefaciens. Biosci. 2009;73(1):47–52. doi: 10.1271/bbb.80449. [DOI] [PubMed] [Google Scholar]
- Kaur S, Samota MK, Choudhary M, Choudhary M, Pandey AK, Sharma A, Thakur J. How do plants defend themselves against pathogens-Biochemical mechanisms and genetic interventions. Physiol Mol Biol Plants. 2022;28(2):485–504. doi: 10.1007/s12298-022-01146-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Arya GC, Bisht NC. Differential expression and interaction specificity of the heterotrimeric G-protein family in Brassica nigra reveal their developmental-and condition-specific roles. Plant Cell Physiol. 2014;55(11):1954–1968. doi: 10.1093/pcp/pcu126. [DOI] [PubMed] [Google Scholar]
- Lee YRJ, Assmann SM. Arabidopsis thaliana ‘extra-large GTP-binding protein’ (AtXLG1): a new class of G-protein. Plant Mol Biol. 1999;40:55–64. doi: 10.1023/A:1026483823176. [DOI] [PubMed] [Google Scholar]
- Li Y, Zhang Q, Gong L, Kong J, Wang X, Xu G, et al. Extra-large G proteins regulate disease resistance by directly coupling to immune receptors in Nicotiana benthamiana. Phytopathol Res. 2022;4(1):49. doi: 10.1186/s42483-022-00155-9. [DOI] [Google Scholar]
- Liang X, Ding P, Lian K, Wang J, Ma M, Li L, Li L, Li M, Zhang X, Chen S, Zhang Y. Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. Elife. 2016;5:e13568. doi: 10.7554/eLife.13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Gao Y, Jones AM. Extra-large G-protein interactome reveals multiple stress response functions and partner-dependent XLG subcellular localization. Front Plant Sci. 2017;8:1015. doi: 10.3389/fpls.2017.01015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Ma M, Zhou Z, Wang J, Yang X, Rao S, Bi G, Li L, Zhang X, Chai J, Chen S. Ligand-triggered de-repression of Arabidopsis heterotrimeric G proteins coupled to immune receptor kinases. Cell Res. 2018;28(5):529–543. doi: 10.1038/s41422-018-0027-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ding P, Sun T, Nitta Y, Dong O, Huang X, Yang W, Li X, Botella JR, Zhang Y. Heterotrimeric G proteins serve as a converging point in plant defense signaling activated by multiple receptor-like kinases. J Plant Physiol. 2013;161(4):2146–2158. doi: 10.1104/pp.112.212431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Ye X, Zou L, Xiang D, Wu Q, Wan Y, Wu X, Zhao G. Genome-wide identification of genes involved in heterotrimeric G-protein signaling in Tartary buckwheat (Fagopyrum tataricum) and their potential roles in regulating fruit development. Int J Biol Macromol. 2021;171:435–447. doi: 10.1016/j.ijbiomac.2021.01.016. [DOI] [PubMed] [Google Scholar]
- Lou F, Abramyan TM, Jia H, Tropsha A, Jones AM. An atypical heterotrimeric Gα protein has substantially reduced nucleotide binding but retains nucleotide-independent interactions with its cognate RGS protein and Gβγ dimer. J Biomol Struct Dyn. 2020;38(17):5204–5218. doi: 10.1080/07391102.2019.1704879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, He P. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA. 2010;107(1):496–501. doi: 10.1073/pnas.0909705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma M, Wang W, Fei Y, Cheng HY, Song B, Zhou Z, Zhao Y, Zhang X, Li L, Chen S, Wang J. A surface-receptor-coupled G protein regulates plant immunity through nuclear protein kinases. Cell Host Microbe. 2022;30(11):1602–1614. doi: 10.1016/j.chom.2022.09.012. [DOI] [PubMed] [Google Scholar]
- Mantri N, Patade V, Penna S, Ford R, Pang E (2012) Abiotic stress responses in plants: present and future. In: Abiotic Stress Responses in Plants: Metabolism, Productivity, and Sustainability, pp 1–19
- Mao G, Meng X, Liu Y, Zheng Z, Chen Z, Zhang S. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell. 2011;23(4):1639–1653. doi: 10.1105/tpc.111.084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta N, Trusov Y, Brenya E, Parekh U, Botella JR. Membrane-localized extra-large G proteins and Gβγ of the heterotrimeric G proteins form functional complexes engaged in plant immunity in Arabidopsis. Plant Physiol. 2015;167(3):1004–1016. doi: 10.1104/pp.114.255703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta N, Trusov Y, Jones AM, Botella JR. Heterotrimeric G proteins in plants: canonical and atypical Gα subunits. Int J Mol Sci. 2021;22(21):11841. doi: 10.3390/ijms222111841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruta N, Trusov Y, Urano D, Chakravorty D, Assmann SM, Jones AM, Botella JR. GTP binding by Arabidopsis extra-large G protein 2 is not essential for its functions. Plant Physiol. 2021;186(2):1240–1253. doi: 10.1093/plphys/kiab119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntire WE. Structural determinants involved in the formation and activation of G protein βγ dimers. Neurosignals. 2009;17(1):82–99. doi: 10.1159/000186692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanasundaram B, Dodds A, Kukshal V, Jez JM, Pandey S. Distribution and the evolutionary history of G-protein components in plant and algal lineages. Plant Physiol. 2022;189(3):1519–1535. doi: 10.1093/plphys/kiac153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta Y, Ding P, Zhang Y. Heterotrimeric G proteins in plant defense against pathogens and ABA signaling. Environ Exp Bot. 2015;114:153–158. doi: 10.1016/j.envexpbot.2014.06.011. [DOI] [Google Scholar]
- Oliveira CC, Jones AM, Fontes EPB, Reis PABD. G-Protein phosphorylation: aspects of binding specificity and function in the plant kingdom. Int J Mol Sci. 2022;23(12):6544. doi: 10.3390/ijms23126544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S. Plant receptor-like kinase signaling through heterotrimeric G-proteins. J Exp Bot. 2020;71(5):1742–1751. doi: 10.1093/jxb/eraa016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey S, Monshausen GB, Ding L, Assmann SM. Regulation of root-wave response by extra-large and conventional G proteins in Arabidopsis thaliana. Plant J. 2008;55(2):311–322. doi: 10.1111/j.1365-313X.2008.03506.x. [DOI] [PubMed] [Google Scholar]
- Pandey S, Roy Choudhury S, Ha CV, Mohanasundaram B, Li M, Dodds A. Evolutionarily conserved and non-conserved roles of heterotrimeric Gα proteins of plants. Plant Cell Physiol. 2022;63(6):817–828. doi: 10.1093/pcp/pcac045. [DOI] [PubMed] [Google Scholar]
- Petutschnig E, Anders J, Stolze M, Meusel C, Hacke R, Much L, Schwier M, Gippert AL, Kroll S, Fasshauer P, Wiermer M. EXTRA LARGE G-PROTEIN2 mediates cell death and hyperimmunity in the chitin elicitor receptor kinase 1–4 mutant. Plant Physiol. 2022;189(4):2413–2431. doi: 10.1093/plphys/kiac214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postle K, Kadner RJ. Touch and go: tying TonB to transport. Mol Microbiol. 2003;49(4):869–882. doi: 10.1046/j.1365-2958.2003.03629.x. [DOI] [PubMed] [Google Scholar]
- Qu X, Cao B, Kang J, Wang X, Han X, Jiang W, Shi X, Zhang L, Cui L, Hu Z, Zhang Y. Fine-tuning stomatal movement through small signaling peptides. Front Plant Sci. 2019;10:69. doi: 10.3389/fpls.2019.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, Goffinet B, Meyberg R, Wu SZ, Bezanilla M. The moss Physcomitrium (Physcomitrella) patens: a model organism for non-seed plants. Plant Cell. 2020;32(5):1361–1376. doi: 10.1105/tpc.19.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy Choudhury S, Li M, Lee V, Nandety RS, Mysore KS, Pandey S. Flexible functional interactions between G-protein subunits contribute to the specificity of plant responses. Plant J. 2020;102(2):207–221. doi: 10.1111/tpj.14714. [DOI] [PubMed] [Google Scholar]
- Sharma P, Samkumar A, Rao M, Singh VV, Prasad L, Mishra DC, Bhattacharya R, Gupta NC. Genetic diversity studies based on morphological variability, pathogenicity and molecular phylogeny of the Sclerotinia sclerotiorum population from Indian mustard (Brassica juncea) Front Microbiol. 2018;9:1169. doi: 10.3389/fmicb.2018.01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M, Avtar R, Lakra N, Hooda E, Singh VK, Bishnoi M, Kumari N, Punia R, Kumar N, Choudhary RR. Genetic and proteomic basis of sclerotinia stem rot resistance in Indian mustard [Brassica juncea (L.) Czern&Coss.] Genes. 2022;12(11):1784. doi: 10.3390/genes121117848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotelo T, Lema M, Soengas P, Cartea ME, Velasco P. In vitro activity of glucosinolates and their degradation products against brassica-pathogenic bacteria and fungi. Appl Environ Microbiol. 2015;81(1):432–440. doi: 10.1128/AEM.03142-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprang SR. G protein mechanisms: insights from structural analysis. Annu Rev Biochem. 1997;66(1):639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- Sprang SR. Invited review: activation of G proteins by GTP and the mechanism of Gα-catalyzed GTP hydrolysis. Biopolymers. 2016;105(8):449–462. doi: 10.1002/bip.22836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stateczny D, Oppenheimer J, Bommert P. G protein signaling in plants: minus times minus equals plus. Curr Opin Plant Biol. 2016;34:127–135. doi: 10.1016/j.pbi.2016.11.001. [DOI] [PubMed] [Google Scholar]
- Stotz HU, Sawada Y, Shimada Y, Hirai MY, Sasaki E, Krischke M, Brown PD, Saito K, Kamiya Y. Role of camalexin, indole glucosinolates, and side chain modification of glucosinolate-derived isothiocyanates in defense of Arabidopsis against Sclerotinia sclerotiorum. Plant J. 2011;67(1):81–93. doi: 10.1111/j.1365-313X.2011.04578.x. [DOI] [PubMed] [Google Scholar]
- Sun J, Jiang H, Xu Y, Li H, Wu X, Xie Q, Li C. The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol. 2007;48(8):1148–1158. doi: 10.1093/pcp/pcm088. [DOI] [PubMed] [Google Scholar]
- Sun Y, Li L, Macho AP, Han Z, Hu Z, Zipfel C, Zhou JM, Chai J. Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science. 2013;342(6158):624–628. doi: 10.1126/science.1243825. [DOI] [PubMed] [Google Scholar]
- Temple BR, Jones AM. The plant heterotrimeric G-protein complex. Annu Rev Plant Biol. 2007;58:249–266. doi: 10.1146/annurev.arplant.58.032806.103827. [DOI] [PubMed] [Google Scholar]
- Tiwari R, Kaur J, Bisht NC. Extra-large G-proteins influence plant response to Sclerotinia sclerotiorum by regulating glucosinolate metabolism in Brassica juncea. Physiol Mol Plant Pathol. 2021;22(10):1180–1194. doi: 10.1111/mpp.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusov Y, Botella JR. Plant G-proteins come of age: breaking the bond with animal models. Front Chem. 2016;4:24. doi: 10.3389/fchem.2016.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusov Y, Rookes JE, Tilbrook K, Chakravorty D, Mason MG, Anderson D, Chen JG, Jones AM, Botella JR. Heterotrimeric G protein γ subunits provide functional selectivity in Gβγ dimer signaling in Arabidopsis. Plant Cell. 2007;19(4):1235–1250. doi: 10.1105/tpc.107.050096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunc-Ozdemir M, Li B, Jaiswal DK, Urano D, Jones AM, Torres MP. Predicted functional implications of phosphorylation of regulator of G protein signaling protein in plants. Front Plant Sci. 2017;8:1456. doi: 10.3389/fpls.2017.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuteja N, Sopory SK. Plant signaling in stress: G-protein coupled receptors, heterotrimeric G-proteins and signal coupling via phospholipases. Plant Signal Behav. 2008;3(2):79–86. doi: 10.4161/psb.3.2.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Chen JG, Botella JR, Jones AM. Heterotrimeric G protein signalling in the plant kingdom. Open Biol. 2013;3(3):120186. doi: 10.1098/rsob.120186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urano D, Maruta N, Trusov Y, Stoian R, Wu Q, Liang Y, Jaiswal DK, Thung L, Jackson D, Botella JR, Jones AM. Saltational evolution of the heterotrimeric G protein signaling mechanisms in the plant kingdom. Sci Signal. 2016;9(446):ra93. doi: 10.1126/scisignal.aaf9558. [DOI] [PubMed] [Google Scholar]
- Wang Y, Botella JR. Heterotrimeric G protein signaling in abiotic stress. Plants. 2022;11(7):876. doi: 10.3390/plants11070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wu Y, Yu B, Yin Z, Xia Y. EXTRA-LARGE G PROTEINs interact with E3 ligases PUB4 and PUB2 and function in cytokinin and developmental processes. Plant Physiol. 2017;173(2):1235–1246. doi: 10.1104/pp.16.00816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Ma LY, Cao J, Li YL, Ding LN, Zhu KM, Yang YH, Tan XL. Recent advances in mechanisms of plant defense to Sclerotinia sclerotiorum. Front Plant Sci. 2019;10:1314. doi: 10.3389/fpls.2019.01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang H, Wang P, Zhong H, Liu W, Zhang S, Xiong L, Wu Y, Xia Y. Arabidopsis EXTRA-LARGE G PROTEIN 1 (XLG1) functions together with XLG2 and XLG3 in PAMP-triggered MAPK activation and immunity. J Integr Plant Biol. 2023;65(3):825–837. doi: 10.1104/pp.16.00816. [DOI] [PubMed] [Google Scholar]
- Wu Q, Regan M, Furukawa H, Jackson D. Role of heterotrimeric Gα proteins in maize development and enhancement of agronomic traits. PLoS Genet. 2018;14(4):e1007374. doi: 10.1371/journal.pgen.1007374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Xu F, Liu L, Char SN, Ding Y, Je BI, Schmelz E, Yang B, Jackson D. The maize heterotrimeric G protein β subunit controls shoot meristem development and immune responses. Proc Natl Acad Sci. 2020;117(3):1799–1805. doi: 10.1073/pnas.1917577116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TY, Krishnamoorthi S, Boonyaves K, Al-Darabsah I, Leong R, Jones AM, Ishizaki K, Liao KL, Urano D. G protein controls stress readiness by modulating transcriptional and metabolic homeostasis in Arabidopsis thaliana and Marchantia polymorpha. Mol Plant. 2022;15(12):1889–1907. doi: 10.1016/j.molp.2022.10.020. [DOI] [PubMed] [Google Scholar]
- Xu L, Yao X, Zhang N, Gong BQ, Li JF. Dynamic G protein alpha signaling in Arabidopsis innate immunity. Biochem. 2019;516(3):1039–1045. doi: 10.1016/j.bbrc.2017.07.040. [DOI] [PubMed] [Google Scholar]
- Yu Y, Chakravorty D, Assmann SM. The G protein β-subunit, AGB1, interacts with FERONIA in RALF1-regulated stomatal movement. Plant Physiol. 2018;176(3):2426–2440. doi: 10.1104/pp.17.01277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Derkacheva M, Rufian JS, Brillada C, Kowarschik K, Jiang S, Derbyshire P, Ma M, DeFalco TA, Morcillo RJ, Stransfeld L. The Arabidopsis E3 ubiquitin ligase PUB4 regulates BIK1 and is targeted by a bacterial type-III effector. Embo J. 2022;41(23):107257. doi: 10.15252/embj.2020107257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M, Jiang Z, Bi G, Nomura K, Liu M, Wang Y, Cai B, Zhou JM, He SY, Xin XF. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature. 2021;592(7852):105–109. doi: 10.1038/s41586-021-03316-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, He SY. A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis. Plant Physiol. 2010;153(3):1188–1198. doi: 10.1104/pp.110.157016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Li C, Si J, Han Z, Chen D. Action mechanisms of effectors in plant-pathogen interaction. Int J Mol Sci. 2022;23(12):6758. doi: 10.3390/ijms23126758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Shi Y, Jiang G, Wu Y, Ma M, Zhang X, Liang X, Zhou JM. Rice extra-large G proteins play pivotal roles in controlling disease resistance and yield-related traits. New Phytol. 2022;234(2):607–617. doi: 10.1111/nph.17997. [DOI] [PubMed] [Google Scholar]
- Zhong CL, Zhang C, Liu JZ. Heterotrimeric G protein signaling in plant immunity. J Exp Bot. 2019;70(4):1109–1118. doi: 10.1093/jxb/ery426. [DOI] [PubMed] [Google Scholar]
- Zhu H, Li GJ, Ding L, Cui X, Berg H, Assmann SM, Xia Y. Arabidopsis extra large G-protein 2 (XLG2) interacts with the Gβ subunit of heterotrimeric G protein and functions in disease resistance. Mol Plant. 2009;2(3):513–525. doi: 10.1093/mp/ssp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.