Abstract
The Escherichia coli glycine cleavage repressor protein (GcvR) negatively regulates expression of the glycine cleavage operon (gcv). In this study, the gcvR translational start site was determined by N-terminal amino acid sequence analysis of a GcvR-LacZ fusion protein. Primer extension analysis of the gcvR promoter region identified a primary transcription start site 27 bp upstream of the UUG translation start site and a minor transcription start site approximately 100 bp upstream of the translation start codon. The -10 and -35 promoter regions upstream of the primary transcription start site were defined by mutational analysis. Expression of a gcvR-lacZ fusion was unaltered in the presence of glycine or inosine, molecules known to induce or repress expression of gcv, respectively. In addition, it was shown that gcvR-lacZ expression is neither regulated by the glycine cleavage activator protein (GcvA) nor autogenously regulated by GcvR. From DNA sequence analysis, it was predicted that the translation start codon of the downstream bcp gene overlaps the gcvR stop codon, suggesting that these genes may form an operon. However, a down mutation in the -10 promoter region of gcvR had no effect on the expression of a downstream bcp-lacZ fusion, and primer extension analysis of the bcp promoter region demonstrated that bcp has its own promoter within the gcvR coding sequence. These results show that gcvR and bcp do not form an operon. Furthermore, the deletion of bcp from the chromosome had no effect on gcv-lacZ expression.
The Escherichia coli glycine cleavage enzyme system catalyzes the cleavage of glycine into carbon dioxide, ammonia, and 5,10-methylenetetrahydrofolate (9). Glycine is required for both protein and purine biosynthesis, while 5,10-methylenetetrahydrofolate serves as a one-carbon donor in the biosynthesis of purines, methionine, thymine, and numerous methylated products (15). Three components of the glycine cleavage enzyme system, the GcvT, GcvH, and GcvP proteins, are encoded by the gcv operon (17). Induced by glycine (13, 17, 28) and repressed by purines (8, 27), it appears that expression of the gcv operon is regulated in order to balance cellular requirements for glycine and one-carbon units.
Currently, four proteins, the leucine-responsive regulatory protein (Lrp), the purine repressor protein (PurR), the glycine cleavage activator protein (GcvA), and the glycine cleavage repressor protein (GcvR), have been shown to be involved in regulating expression of the gcv operon. Lrp is a global regulatory protein involved in the control of transcription of numerous genes involved in amino acid metabolism (6) and is required for normal induction of gcv (11, 21). Lrp binds to multiple sites upstream of the gcv promoter, suggesting a direct role for Lrp in gcv expression (21). Whether Lrp interacts with RNA polymerase, one of the other regulatory proteins, or plays a structural role by bending DNA is unknown.
PurR is a negative regulator of many genes involved in nucleotide metabolism (30), including gcv (27). PurR mediates a twofold decrease in gcv transcription in response to purine supplementation and has been shown to bind to the gcv control region, overlapping the gcv transcription start site (20, 27).
GcvA and GcvR work in concert to further regulate gcv expression. In glucose minimal medium, GcvR negatively regulates gcv expression, resulting in a low, basal level of gcv expression (8). In glycine-supplemented cultures, GcvR repression is relieved, and GcvA activates gcv expression (8, 28). In purine-supplemented cultures, both GcvA and GcvR are required to mediate a PurR-independent repression of gcv (8, 27). GcvA has been shown to bind to three sites upstream of the gcv promoter, and mutations have verified that all three sites are required for normal GcvA-mediated activation and repression of gcv (29).
It has been shown that gcv expression can be altered by changing the ratio of GcvA to GcvR (8). Overexpression of GcvA leads to constitutive activation of gcv, even in the absence of glycine, while overexpression of GcvR causes superrepression of gcv, even in the absence of purines. Therefore, it is important to understand how the expression of gcvA and gcvR is regulated. Expression of gcvA is negatively autoregulated and is unaffected by glycine or purine supplementation (26). It has also been shown that GcvR has no effect on gcvA expression (8). In this study, we characterized the promoter region of gcvR and investigated transcriptional regulation of gcvR. In addition, we show that bcp, a gene located immediately downstream of gcvR, is not in an operon with gcvR and plays no role in the regulation of gcv expression.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The E. coli K-12 strains and plasmids used in this study are described in Table 1.
TABLE 1.
E. coli K-12 strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| Strainsa | ||
| GS162 | Wild type | This lab |
| GS597 | metJ97 | This lab |
| GS1029 | Δ(gcvA):ΣaadA | 26 |
| GS1053 | gcvR::Tn10 | 8 |
| GS1066 | recD::Tn10 | This lab |
| GS1111 | Δ(gcvR bcp)Σneo | This study |
| Plasmids | ||
| pGS311 | Single-copy cloning vector | 8 |
| pGS334 | Multicopy gcvR+ plasmid | 8 |
| pGS338 | Single-copy gcvR+ plasmid | 8 |
| pGS343 | gcvR-lacZ fusion plasmid | This study |
| pGS393 | pGS343 with -13A gcvR promoter mutation | This study |
| pGS394 | pGS343 with -36A gcvR promoter mutation | This study |
| pGS429 | pGS334 with -13A gcvR promoter mutation | This study |
| pGS430 | bcp-lacZ fusion plasmid | This study |
| pGS431 | pGS430 with -13A gcvR promoter mutation | This study |
| pGS433 | pBR322 with SspI site replaced by SmaI site | This study |
| pGS434 | Multicopy gcvR+ bcp+ plasmid | This study |
| pGS437 | Δ(gcvR bcp)Σneo in pGS434 | This study |
| pGS451 | pGS437 with extra 2 kb of homologous DNA upstream of the Δ(gcvR bcp)Σneo | This study |
All also carry thi, pheA905, ΔlacU169, araD129, and rpsL150 mutants.
Media and growth conditions.
The complex medium used was Luria-Bertani (LB) broth (14). The glucose minimal (GM) medium used was minimal salts (25) supplemented with 0.4% glucose, phenylalanine (50 μg/ml), and thiamine (1 μg/ml). Additional supplements were added, where indicated, at the following concentrations: inosine, 50 μg/ml; glycine, 300 μg/ml; ampicillin, 150 μg/ml; and kanamycin, 30 μg/ml. λ lysogens carry the cI857 mutation resulting in a temperature-sensitive repressor and were grown at 30°C. All other strains were grown at 37°C.
Nucleotide sequencing.
Dideoxynucleotide DNA sequencing was performed by using a Sequenase version 2.0 sequencing kit (United States Biochemical, Cleveland, Ohio).
Construction of gcvR-lacZ and bcp-lacZ translational fusions.
The gcvR-lacZ fusion plasmid pGS343 was constructed by cloning the EcoRI-SspI fragment carrying the control region and initial coding sequence of gcvR from plasmid pGS334 (Fig. 1) into the EcoRI-SmaI sites of the lac fusion plasmid pMC1403 (7), forming an in-frame fusion of gcvR to the lacZYA genes. Plasmids bearing mutations in the -10 (-13A) and -35 (-36A) regions of the gcvR-lacZ fusion promoter (pGS393 and pGS394, respectively) were created by the PCR megaprimer mutagenesis procedure (18), using primers GcvR7 (5′-GCATACATCAATCAGAACGG-3′) and GcvR8 (5′-GCATGTTTTTTTTATGCATTCCTTAAG-3′) (mutations are underlined; see Fig. 2). Plasmid pGS343 was used as a template.
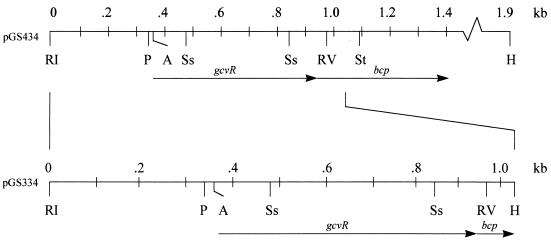
FIG. 1.
Diagram of the EcoRI-HindIII fragments of plasmids pGS334 and pGS434. The locations and directions of transcription of gcvR and bcp are indicated by arrows. The EcoRI sites of both fragments as well as the HindIII site of the pGS334 fragment were PCR generated. A, AflII; H, HindIII; RI, EcoRI; RV, EcoRV; P, Ppu10I; Ss, SspI; St, StuI.
FIG. 2.
Complete nucleotide and deduced amino acid sequences of the E. coli gcvR gene and partial nucleotide and deduced amino acid sequences of the E. coli bcp gene. The gcvR and bcp transcription start sites (+1), -10 and -35 promoter regions, and ribosome binding sites (RBS) are underlined and in boldface. A potential secondary gcvR transcription start site and its promoter are underlined. Mutations in the gcvR promoter constructed in vitro are shown below the sequence. The gcvR amino acid sequence in boldface was verified by N-terminal amino acid sequencing.
The bcp-lacZ fusion plasmid pGS430 was constructed by cloning the EcoRI-EcoRV fragment carrying gcvR and the initial coding sequence of bcp from pGS334 (Fig. 1) into the EcoRI-SmaI sites of pMC1403. Plasmid pGS431, carrying a gcvR promoter mutation upstream of a bcp-lacZ fusion, was constructed in two steps. First, the EcoRI-AflII fragment carrying the wild-type gcvR promoter was deleted from plasmid pGS334 (Fig. 1) and replaced with the same fragment carrying the -13A mutation from pGS393. This plasmid was designated pGS429. Then, a bcp-lacZ fusion was constructed downstream of this gcvR promoter mutation by cloning the EcoRI-EcoRV fragment from pGS429 into the EcoRI-SmaI sites of pMC1403.
All fusions were cloned into bacteriophage λgt2 (16) as previously described (23), and the resultant phage were used to lysogenize the appropriate strains. Lysogens were tested for the presence of a single copy of the λ phage by infection with phage λcI90c17 (19).
Purification and N-terminal amino acid sequencing of a GcvR-LacZ fusion protein.
Wild-type strain GS162 carrying the gcvR-lacZ fusion plasmid pGS343 was grown in LB-ampicillin, and the GcvR-LacZ fusion protein was extracted and partially purified by affinity chromatography (22). To further purify the fusion protein, approximately 5 μg of fusion protein was electrophoresed on a sodium dodecyl sulfate–10% polyacrylamide (PA) gel and electroblotted onto a polyvinylidene difluoride membrane. The sequence of the first 14 amino acids was determined by automated N-terminal amino acid sequencing at the University of Iowa Protein Facility.
Primer extension analysis.
To determine the location of the gcvR promoter, primer extension mapping was performed with a Promega (Madison, Wis.) primer extension kit. Wild-type strain GS162 carrying the gcvR+ plasmid pGS334 was grown in LB-ampicillin, and total RNA was isolated as previously described (3). Primer extension reactions were carried out by the Promega protocol, using the 32P-labeled primer GcvR4 (5′-GCAACTACTGACATGACGGGTG-3′), which hybridizes 71 bp downstream of the gcvR translation start site. Reaction products were electrophoresed on a 5% PA gel next to DNA sequencing reactions also generated with primer GcvR4.
To determine the location of the bcp promoter, primer extension mapping was performed as described below, using SuperScript II RNase H− reverse transcriptase (RT) (Gibco BRL, Gaithersburg, Md.). Total RNA was isolated from strain GS597 as previously described (3). Approximately 5 μg of RNA and either 100 fmol (reaction 1) or 1 pmol (reaction 2) of 32P-labeled primer Bcp2 (5′-CTCCGTCTTGATCCGGCAAGC-3′) were mixed in 1× RT buffer (50 mM Tris-HCl [pH 8.3], 75mM KCl, 3 mM MgCl2), heated to 60°C for 10 min, and then allowed to anneal at 25°C for 10 min. Extension reactions were carried out in 1× RT buffer, 400 nM deoxynucleoside triphosphates 100 μg of bovine serum albumin per ml, and 1 mM dithiothreitol in a total volume of 10 μl at 42°C for 30 min, using 10 (reaction 1) or 100 (reaction 2) U of RT. Reactions were stopped by adding 8 μl of sequencing stop mix. The products of each reaction were electrophoresed on a 5% PA gel next to DNA sequencing reactions also generated with primer Bcp2.
β-Gal assays.
β-Galactosidase (β-Gal) enzyme assays were performed as described by Miller (14). All results are the average of two or more assays, with each sample being determined in triplicate. All standard deviations are ≤12% of the mean.
Construction of a gcvR-bcp chromosomal deletion.
An approximately 1.9-kb fragment carrying gcvR and bcp was cloned by PCR amplification of Kohara λ phage 424 (10), using primers GcvR1 (5′-GAATTCGCAATTACCGGAATGCGCC-3′) and Bcp1 (5′-GCGCATGGGCATTCACCGGACGGC-3′). This fragment was then digested with EcoRI and HindIII, and the resulting fragment was cloned into the EcoRI-HindIII sites of plasmid pGS433, which is a pBR322 (5) derivative in which the SspI site has been replaced with a SmaI site. This plasmid was designated pGS434 (Fig. 1). A deletion from codon 35 of gcvR through codon 49 of bcp (Fig. 1) was made by digesting pGS434 with SspI and StuI. The deleted fragment was then replaced with a blunt-ended, SalI-HindIII fragment carrying the kanamycin resistance gene (neo) from Tn5 (4). This plasmid was designated pGS437.
Initial attempts to integrate this deletion into the chromosome by linear transformation were unsuccessful, most likely due to a limiting amount of homologous DNA flanking the neo gene. Therefore, an additional 2 kb of flanking DNA was introduced upstream of the neo gene by cloning an EcoRI-Ppu10I fragment from Kohara λ phage 424 carrying approximately 2.3 kb of DNA upstream of the gcvR coding sequence into the EcoRI-Ppu10I sites of pGS437, creating plasmid pGS451. This plasmid was digested with EcoRI and HindIII and used to perform a linear transformation of strain GS1066 (recD::Tn10). Transformants were selected on LB-kanamycin and checked for ampicillin sensitivity by spotting on LB-ampicillin. One kanamycin-resistant, ampicillin-sensitive transformant was designated GS1111.
Nucleotide sequence accession number.
The nucleotide sequence data reported here are in the GenBank database under accession no. AFO23337.
RESULTS AND DISCUSSION
Nucleotide sequence of gcvR.
Plasmid pGS334 carries the E. coli gcvR gene and a portion of the downstream bcp gene on an approximately 1-kb EcoRI-HindIII fragment (Fig. 1). In this study, the nucleotide sequence of most of this PCR-generated fragment was determined (Fig. 2). This sequence differs from the original sequence of this region as determined by S. C. Andrews et al. (GenBank accession no. M37689) but is identical to the sequence of this region as determined by F. R. Blattner et al. and Y. Yamamoto et al. (GenBank accession no. ECAE000335 and D90877, respectively).
gcvR translation start site.
To determine where translation of gcvR begins, a gcvR-lacZ fusion plasmid was constructed, the GcvR-LacZ fusion protein was purified, and the sequence of the first 14 N-terminal amino acids was determined (Fig. 2; see Materials and Methods). This sequence indicates that translation initiates at a UUG codon located 9 bp downstream of a potential ribosome binding site (Fig. 2). The formylmethionine encoded by the UUG translation initiation codon is apparently removed from the nascent polypeptide. These data suggest that GcvR is a 189-amino-acid protein with a molecular mass of 20.6 kDa. A similarity search performed with the BLAST (1) program failed to identify any proteins with significant similarity to the deduced amino acid sequence of GcvR.
Only 1% of all sequenced E. coli genes use a UUG codon to initiate translation (91% use AUG; 8% use GUG) (12), and in such cases, translational efficiency is reduced three- to fivefold (24). Since overproduction of the GcvR protein results in repression of the gcv operon (8), the use of this codon as the gcvR translation start site may serve to reduce translation of gcvR, preventing superrepression of gcv.
gcvR promoter mapping.
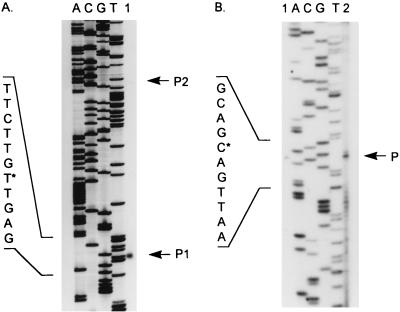
To determine the location of the gcvR promoter, primer extension mapping was performed on RNA isolated from the wild-type strain GS162 bearing the multicopy gcvR+ plasmid pGS334. As shown in Fig. 3A, two primer extension products were synthesized. The major primer extension product (P1) migrated in alignment with an A residue located 27 bp upstream of the gcvR translation start site (Fig. 2). This A residue was designated +1. The minor primer extension product (P2) indicates that a second transcription start site may also exist approximately 100 bp upstream of the gcvR translation start codon. A potential ς70 promoter sequence is present upstream of both start sites (Fig. 2).
FIG. 3.
Primer extension analysis of gcvR and bcp transcripts. Primer extension products were synthesized as described in Materials and Methods and are marked by arrows. DNA sequencing ladders were generated with the same primer as that used in the primer extension reactions. The nucleotide sequence of the complementary strand of each promoter region is indicated to the left. The nucleotide designated +1 for each promoter is indicated with an asterisk. (A) gcvR primer extension analysis. Lanes: A, C, G, and T, DNA sequencing ladders; 1, primer extension products (P1, primary transcription start site; P2, minor transcription start site). (B) bcp primer extension analysis. Lanes 1, primer extension reaction 1; A, C, G, and T, DNA sequencing ladders; 2, primer extension reaction 2.
To verify the -10 and -35 regions of the promoter sequence upstream of the primary start site, a mutation was introduced in each region, and the effect of each mutation on promoter function was determined. Wild-type strain GS162 was lysogenized with λ phage carrying either wild-type (λgcvR-lacZ) or mutant (λgcvR-113A-lacZ and λgcvR-36A-lacZ) fusions. The resulting lysogens were grown in GM medium and GM medium supplemented with either glycine or inosine (molecules known to induce or repress gcvT-lacZ expression, respectively) and assayed for β-Gal activity (Table 2). Expression of the wild-type gcvR-lacZ fusion is not affected by the addition of glycine or inosine, suggesting that these molecules do not indirectly affect gcv operon expression by altering gcvR expression. The -13A mutation resulted in a 15-fold decrease in gcvR-lacZ expression compared to the wild-type level, and the -36A mutation resulted in a 9-fold decrease in gcvR-lacZ expression compared to the wild-type level, regardless of supplementation. Thus, it appears that the primary transcription start site determined by primer extension mapping is correct.
TABLE 2.
Expression of a gcvR-lacZ fusion from wild-type and mutant gcvR promoters
| Strain | β-Gal activity (Miller units)a
|
||
|---|---|---|---|
| GM | Glycine | Inosine | |
| GS162λgcvR-lacZ | 73 | 78 | 69 |
| GS162λgcvR-13A-lacZ | 5 | 5 | 4 |
| GS162λgcvR-36A-lacZ | 7 | 8 | 7 |
Cells were grown in GM medium with supplements as indicated and assayed for β-Gal activity. Each value is the average of two separate determinations, each performed in triplicate. All standard deviations are ≤11% of the mean.
The importance of the minor promoter of gcvR is still unclear. If this promoter functions in vivo to control expression of gcvR, it appears to be relatively weak compared to the primary promoter, since the -13A and -36A mutations in the primary promoter eliminate more than 90% of gcvR-lacZ expression under all growth conditions (Table 2). These results are consistent with the primer extension results which show that nearly all of the gcvR mRNA transcript initiates from the primary promoter (Fig. 3A).
gcvR expression is not regulated by GcvA or GcvR.
Since it is known that GcvR-mediated regulation of gcv requires GcvA, it is possible that GcvA regulates gcvR expression. To test this hypothesis, lysogens GS162λgcvR-lacZ and GS1029λgcvR-lacZ (gcvA) were grown in GM medium and GM medium supplemented with either glycine or inosine and assayed for β-Gal activity. As shown in Table 3, GcvA has no effect on gcvR-lacZ expression. This was true for all growth conditions.
TABLE 3.
gcvR-lacZ expression is not regulated by GcvA or GcvR
| Strain | Relevant genotype | β-Gal activity (Miller units)a
|
||
|---|---|---|---|---|
| GM | Glycine | Inosine | ||
| GS162λgcvR-lacZ | Wild type | 91 | 91 | 88 |
| GS1029λgcvR-lacZ | gcvA | 94 | 94 | 90 |
| GS1053λgcvR-lacZ | gcvR | 101 | 94 | 97 |
Cells were grown in GM medium with supplements as indicated and assayed for β-Gal activity. Each value is the average of two separate determinations, each performed in triplicate. All standard deviations are ≤10% of the mean.
Since many regulatory proteins are known to regulate their own expression, we tested the ability of GcvR to regulate gcvR-lacZ expression. Lysogens GS162λgcvR-lacZ and GS1053λgcvR-lacZ (gcvR) were grown in GM medium and GM medium supplemented with either glycine or inosine and assayed for β-Gal activity. As shown in Table 3, GcvR does not autoregulate its own expression. This is true for all growth conditions. Furthermore, when GS162λgcvR-lacZ was grown in rich (LB) medium, or when GM medium-grown cells were harvested from stationary phase rather than exponential phase, there was no significant change in β-Gal activity (data not shown), suggesting that gcvR expression does not respond to any other component in LB medium or to growth phase. Thus, whatever the role is for GcvR in the negative regulation of the gcv operon, the mechanism appears to be independent of changes in GcvR levels in the cell.
The bcp gene is not in an operon with gcvR.
The bcp gene, encoding the bacterioferritin comigratory protein, is located immediately downstream of gcvR. It is predicted that this open reading frame, first reported by Andrews et al. (2), encodes a 156-amino-acid protein of unknown function. Our sequence of gcvR suggests that gcvR and bcp may form an operon, as the UAA stop codon of gcvR overlaps the predicted AUG start codon of bcp (Fig. 2). To test this hypothesis, a translational bcp-lacZ fusion was constructed downstream of the wild-type gcvR gene as well as downstream of a mutant gcvR gene carrying the -13A promoter mutation described above. Both fusions were cloned into phage λgt2, and the phage was used to lysogenize the wild-type strain GS162. The resulting lysogens were grown in GM medium and assayed for β-Gal activity. The lysogen carrying the wild-type gcvR promoter had 325 U of activity, and the lysogen carrying the mutant gcvR promoter had 323 U of activity. Thus, a mutation in the -10 region of the primary gcvR promoter, which results in a 15-fold decrease in gcvR-lacZ expression, has no effect on the expression of a downstream bcp-lacZ fusion. These results suggest that gcvR and bcp do not form an operon.
To further demonstrate that gcvR and bcp do not form an operon, we tested whether bcp has its own promoter within the gcvR coding sequence. Primer extension mapping was performed on RNA isolated from strain GS597. As shown in Fig. 3B, transcription of bcp begins at a G nucleotide, 8 bp downstream of a potential ς70 promoter and 41 bp upstream of the putative bcp translation start site as predicted by Andrews et al. (2) (Fig. 2). The presence of multiple products in lane 2 compared to the presence of a single product in lane 1 is most likely due to nonspecific primer annealing and extension due to the 10-fold increase in the amount of primer and RNA polymerase used in this extension reaction.
The bcp gene has no effect on the regulation of gcv expression.
Even though gcvR and bcp are transcribed separately, their proximity still suggested that bcp might play a role in the regulation of gcv expression. To test whether bcp plays a role in gcv expression, we constructed strain GS1111, which carries a chromosomal deletion of both the gcvR and bcp genes (see Materials and Methods). Lysogens GS162λgcvT-lacZ, GS1053λgcvT-lacZ (gcvR), and GS1111λgcvT-lacZ (ΔgcvR bcp), each transformed with the single-copy vector pGS311, and lysogen GS1111λgcvT-lacZ, transformed with the single-copy gcvR+ plasmid pGS338, were grown in GM medium and GM medium supplemented with either glycine or inosine and assayed for β-Gal activity (Table 4). GS1053λgcvT-lacZ[pGS311] and GS1111λgcvT-lacZ[pGS311] had similar levels of gcvT-lacZ expression under all conditions. In addition, transformation of GS1111λgcvT-lacZ with pGS338 resulted in wild-type levels of gcvT-lacZ expression under all conditions, confirming that bcp is not involved in the regulation of gcv expression.
TABLE 4.
Bcp is not involved in the regulation of gcvT-lacZ expression
| Strain | Relevant genotype | β-Gal activity (Miller units)a
|
||
|---|---|---|---|---|
| GM | Glycine | Inosine | ||
| GS162λgcvT-lacZ[pGS311] | Wild type | 159 | 1020 | 15 |
| GS1053λgcvT-lacZ[pGS311] | gcvR | 1,443 | 1,710 | 1,012 |
| GS1111λgcvT-lacZ[pGS311] | gcvR bcp | 1,400 | 1,559 | 909 |
| GS1111λgcvT-lacZ[pGS338] | gcvR bcp[gcvR+] | 205 | 1,150 | 16 |
Cells were grown in GM medium with supplements as indicated and assayed for β-Gal activity. Each value is the average of two separate determinations, each performed in triplicate. All standard deviations are ≤12% of the mean.
Possible roles for GcvA and GcvR in the regulation of gcv.
Expression of the gcv operon is induced in the presence of glycine and repressed in the presence of purines (8, 13, 17, 27, 28). Since overproduction of GcvA leads to activation of gcv, while overproduction of GcvR results in repression of gcv (8), it is possible that glycine and inosine alter gcv expression indirectly by altering the ratio of GcvA to GcvR. However, previous results (8, 26) as well as those presented here demonstrate that glycine and inosine have no effect on the expression of gcvA and gcvR. Both genes also appear to be transcribed constitutively with respect to medium richness and growth phase.
Since both GcvA and GcvR are required for normal regulation of gcv, it is also possible that they regulate gcv indirectly by regulating the expression of one another. However, it has been shown that no reciprocal regulation occurs. Only a twofold autoregulation by GcvA occurs.
Several models to explain how GcvA and GcvR interact to regulate gcv expression have been proposed. In one model, GcvA homocomplexes function as activators, while GcvA-GcvR heterocomplexes function as repressors. In this model, the coregulators determine the type of complex formed; glycine leads to the formation of activation complexes, while inosine leads to the formation of repression complexes. Increasing the amount of either GcvA or GcvR would force the formation of activation or repression complexes, respectively. In a second model, GcvR may synthesize the corepressor required for the repressor function of GcvA. In a gcvR mutant, insufficient corepressor would lead to constitutive gcv expression. If GcvR is overproduced, too much corepressor causes superrepression of gcv. In another model, GcvR may negatively regulate gcv by modifying the structure of GcvA, changing it from an activator to a repressor in response to the coregulators. Purification of GcvR protein and identification of the actual coregulators of gcv expression should help us to determine which of these models, if any, is correct.
ACKNOWLEDGMENT
This work was supported by Public Health Service grant GM26878 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Andrews S C, Harrison P M, Guest J R. A molecular analysis of the 53.3 minute region of the Escherichia coli linkage map. J Gen Microbiol. 1991;137:361–367. doi: 10.1099/00221287-137-2-361. [DOI] [PubMed] [Google Scholar]
- 3.Baker R F, Yanofsky C. The periodicity of RNA polymerase initiations: a new regulatory feature of transcription. Proc Natl Acad Sci USA. 1968;60:313–320. doi: 10.1073/pnas.60.1.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck E, Ludwig G, Auerswald E A, Reiss B, Schaller H. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982;19:327–336. doi: 10.1016/0378-1119(82)90023-3. [DOI] [PubMed] [Google Scholar]
- 5.Bolivar F, Rodriguez R L, Greene P J, Betlach M C, Heyneker H L, Boyer H W, Crosa J H, Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2:95–113. [PubMed] [Google Scholar]
- 6.Calvo J M, Matthews R G. The leucine-responsive regulatory protein, a global regulator of metabolism in Escherichia coli. Microbiol Rev. 1994;58:466–490. doi: 10.1128/mr.58.3.466-490.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casadaban M J, Chou J, Cohen S N. In vitro gene fusions that join an enzymatically active β-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980;143:971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghrist A C, Stauffer G V. Characterization of the Escherichia coli gcvR gene encoding a negative regulator of gcv expression. J Bacteriol. 1995;177:4980–4984. doi: 10.1128/jb.177.17.4980-4984.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kikuchi G. The glycine cleavage system: composition, reaction mechanism, and physiological significance. Mol Cell Biochem. 1973;1:169–187. doi: 10.1007/BF01659328. [DOI] [PubMed] [Google Scholar]
- 10.Kohara Y, Akiyama K, Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987;50:495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- 11.Lin R, D’Ari R, Newman E B. λ placMu insertions in genes of the leucine regulon: extension of the regulon to genes not regulated by leucine. J Bacteriol. 1992;174:1948–1955. doi: 10.1128/jb.174.6.1948-1955.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makrides S C. Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol Rev. 1996;60:512–538. doi: 10.1128/mr.60.3.512-538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meedel T H, Pizer L I. Regulation of one-carbon biosynthesis and utilization in Escherichia coli. J Bacteriol. 1974;118:905–910. doi: 10.1128/jb.118.3.905-910.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 15.Mudd S H, Cantoni G L. Biological transmethylation, methyl-group neogenesis and other “one-carbon” metabolic reactions dependent upon tetrahydrofolic acid. Compr Biochem. 1964;15:1–47. [Google Scholar]
- 16.Panasenko S M, Cameron J R, Davis R W, Lehman I R. Five hundred-fold overproduction of DNA ligase after induction of a hybrid lambda lysogen constructed in vitro. Science. 1977;196:188–189. doi: 10.1126/science.322281. [DOI] [PubMed] [Google Scholar]
- 17.Plamann M D, Rapp W D, Stauffer G V. Escherichia coli K12 mutants defective in the glycine cleavage enzyme system. Mol Gen Genet. 1983;192:15–20. doi: 10.1007/BF00327641. [DOI] [PubMed] [Google Scholar]
- 18.Sarkar G, Sommer S. The “megaprimer” method of site-directed mutagenesis. BioTechniques. 1990;8:404–407. [PubMed] [Google Scholar]
- 19.Shimada K, Weisberg R A, Gottesman M E. Prophage λ at unusual chromosomal locations. I. Location of the secondary attachment sites and properties of the lysogens. J Mol Biol. 1972;63:483–503. doi: 10.1016/0022-2836(72)90443-3. [DOI] [PubMed] [Google Scholar]
- 20.Stauffer L T, Fogarty S J, Stauffer G V. Characterization of the Escherichia coli gcv operon. Gene. 1994;142:17–22. doi: 10.1016/0378-1119(94)90349-2. [DOI] [PubMed] [Google Scholar]
- 21.Stauffer L T, Stauffer G V. Characterization of the gcv control region from Escherichia coli. J Bacteriol. 1994;176:6159–6164. doi: 10.1128/jb.176.20.6159-6164.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Steers E, Jr, Cuartrecasas P, Pollard H B. The purification of β-galactosidase from Escherichia coli by affinity chromatography. J Biol Chem. 1971;246:196–200. [PubMed] [Google Scholar]
- 23.Urbanowski M L, Stauffer G V. Autoregulation by tandem promoters of the Salmonella typhimurium LT2 metJ gene. J Bacteriol. 1986;165:740–745. doi: 10.1128/jb.165.3.740-745.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vellanoweth R L, Rabinowitz J C. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol Microbiol. 1992;6:1105–1114. doi: 10.1111/j.1365-2958.1992.tb01548.x. [DOI] [PubMed] [Google Scholar]
- 25.Vogel H J, Bonner D M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 26.Wilson R L, Stauffer G V. DNA sequence and characterization of GcvA, a LysR family regulatory protein for the Escherichia coli glycine cleavage enzyme system. J Bacteriol. 1994;176:2862–2868. doi: 10.1128/jb.176.10.2862-2868.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson R L, Stauffer L T, Stauffer G V. Roles of the GcvA and PurR proteins in negative regulation of the Escherichia coli glycine cleavage enzyme system. J Bacteriol. 1993;175:5129–5134. doi: 10.1128/jb.175.16.5129-5134.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson R L, Steiert P S, Stauffer G V. Positive regulation of the Escherichia coli glycine cleavage enzyme system. J Bacteriol. 1993;175:902–904. doi: 10.1128/jb.175.3.902-904.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson R L, Urbanowski M L, Stauffer G V. DNA binding sites of the LysR-type regulator GcvA in the gcv and gcvA control regions of Escherichia coli. J Bacteriol. 1995;177:4940–4946. doi: 10.1128/jb.177.17.4940-4946.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zalkin H, Nygaard P. Biosynthesis of purine nucleotides. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Maganasik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 561–579. [Google Scholar]