Abstract
Background
Bone scans are often used to identify bone metastases, but their low specificity may necessitate further studies. Deep learning models may improve diagnostic accuracy but require both medical and programming expertise. Therefore, we investigated the feasibility of constructing a deep learning model employing ChatGPT for the diagnosis of bone metastasis in bone scans and to evaluate its diagnostic performance.
Method
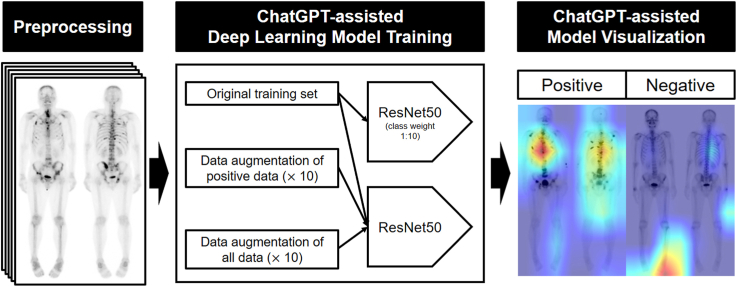
We examined 4626 consecutive cancer patients (age, 65.1 ± 11.3 years; 2334 female) who had bone scans for metastasis assessment. A nuclear medicine physician developed a deep learning model using ChatGPT 3.5 (OpenAI). We employed ResNet50 as the backbone network and compared the diagnostic performance of four strategies (original training set, original training set with 1:10 class weight, 10-fold data augmentation for positive images only, and 10-fold data augmentation for all images) to address the class imbalance. We used a class activation map algorithm for visualization.
Results
Among the four strategies, the deep learning model with 10-fold data augmentation for positive cases only, using a batch size of 16 and an epoch size of 150, achieved the area under curve of 0.8156, the sensitivity of 56.0 %, and specificity of 88.7 %. The class activation map indicated that the model focused on disseminated bone metastases within the spine but might confuse them with benign spinal lesions or intense urinary activity.
Conclusions
Our study illustrates that a clinical physician with rudimentary programming skills can develop a deep learning model for medical image analysis, such as diagnosing bone metastasis in bone scans using ChatGPT. Model visualization may offer guidance in enhancing deep learning model development, including preprocessing, and potentially support clinical decision-making processes.
Keywords: Convolutional neural network, Deep learning, ChatGPT, Bone scan, Bone metastasis
Graphical abstract
1. Introduction
Cancer staging is vital for determining treatment strategies and prognostic outcomes. Typically, metastatic cases receive systemic treatment instead of surgery, resulting in less favorable prognoses compared with nonmetastatic cases. Common solid organ sites for metastases include the lungs, liver, and bone. Approximately 5 % of cancer patients present bone metastasis at the time of diagnosis [[1], [2], [3]], highlighting the importance of selecting the optimal imaging technique for detecting bone metastasis.
Different imaging modalities are available for diagnosing bone metastases, including CT, MR, PET, and bone scans [4]. Among them, bone scans are the most commonly conducted imaging modality due to their high sensitivity and the convenience of scanning the entire skeletal system at once [5,6]. Nonetheless, the relatively low specificity of bone scans can lead to false-positive results, which may lead to inappropriate treatment and unnecessary adverse effects while necessitating additional CT and MR scans to rule out benign lesions [7].
In recent years, deep learning algorithms, such as convolutional neural networks (CNNs), have made significant advances in various image processing tasks, including classification, regression, and image generation, and they have shown great potential in nuclear medicine and molecular imaging fields [[8], [9], [10]]. As the most commonly performed planar imaging technique in nuclear medicine, bone scans are suitable for 2D-CNN modeling, requiring relatively fewer training parameters compared with labeled data [11,12]. Several recent studies have attempted to develop deep learning models for diagnosing bone metastasis in bone scans [[13], [14], [15], [16], [17], [18], [19]]. However, creating a deep learning model using medical data requires a combination of medical and programming expertise, which poses a challenge for researchers without a well-organized team.
Recently, an open-source artificial intelligence (AI) chatbot known as ChatGPT has been made available, and it generates programming codes even for beginners [20]. As part of the generative pretrained transformer language model family, ChatGPT enables users to receive responses in the Python programming language by asking questions casually and conversationally. However, to our knowledge, no study has yet been published on developing a medical image data deep learning model using an open-source AI platform. In this study, we aimed to investigate the feasibility of developing a deep learning model for detecting bone metastasis in bone scans using a chatbot.
2. Material and methods
2.1. Subjects
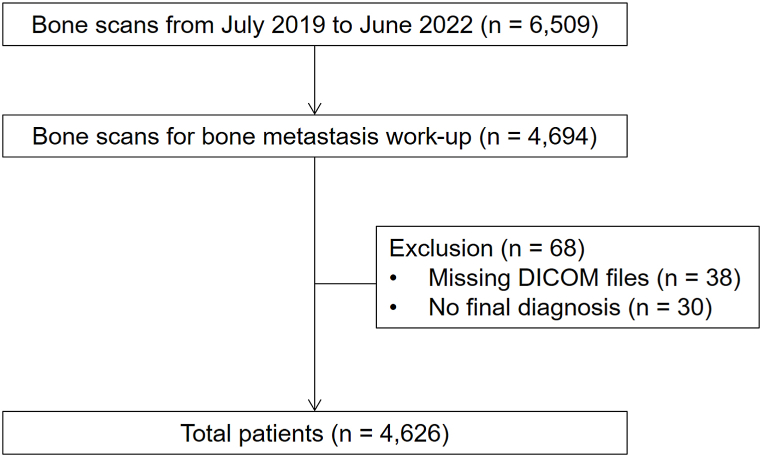
In this study, we included 4626 consecutive cancer patients who underwent bone scans for bone metastasis evaluation between July 2019 and June 2022 at our institution (Fig. 1). Patients who underwent bone scans for reasons other than the detection of bone metastases (e.g., trauma or infection) were excluded from the study. The Institutional Review Board of our institution approved this study (IRB no. 2023-02-008), and informed consent was waived due to its retrospective nature.
Fig. 1.
Flow diagram of patient enrollment.
2.2. Bone scan acquisition, preprocessing, and classification
Whole-body bone scans were conducted 2–4 h after injecting a median dose of 740 MBq (20 mCi) 99mTc-hydroxymethylene diphosphonate intravenously. A dual-head gamma camera (NM830, GE Healthcare) with low-energy, high-resolution, and sensitivity parallel-hole collimators that scans at a speed of 22 cm/min was utilized. A blend ratio of 60 % was used with Clarity 2D processing.
We converted the original DICOM files of whole-body bone scans (256 × 1024 pixels anterior and posterior images) to png files in the [0, 255] range. The intensity was standardized by setting the level as the total accumulated counts of the anterior and posterior images divided by 100,000, and the width as twice the level. Images were then concatenated into a single 512 × 1024 pixel-sized image.
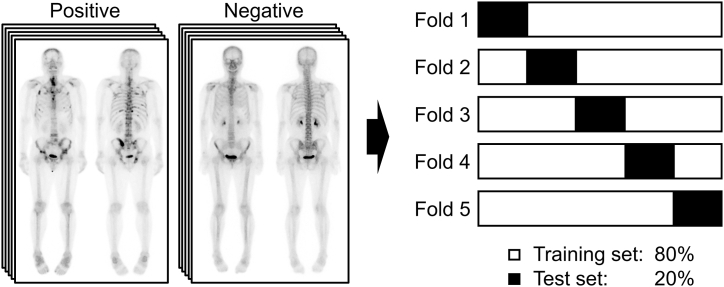
A nuclear medicine board-certified physician (S.H.L.) with 12 years of experience in bone scan interpretation produced clinical reports for all bone scans, grading them from 1 to 5 according to the level of certainty [21]. We classified grades 1 and 2 as “negative” and grades 4 and 5 as “positive,” and grade 3 images were re-evaluated and classified based on correlative imaging and follow-up. We excluded images that could not be accurately classified. Any healed bone metastases that were not visible on the bone scan image were considered negative. The bone scan images were allocated as follows: 80 % for training sets and 20 % for testing sets; five lots were configured to guarantee no overlap between the test sets (Fig. 2). All test sets were independent of the training sets.
Fig. 2.
Dataset composition. Five different 8:2 training sets and test sets were constructed not to overlap the test sets.
2.3. Deep learning model
The deep learning model was developed by a nuclear medicine board-certified physician (S.H.L.) using ChatGPT 3.5 (OpenAI). He possessed rudimentary knowledge of Python and underwent a deep learning fundamentals lecture courses for approximately 10 days but did not have significant programming experience. An initial deep learning model without errors was obtained through multiple questions on ChatGPT (Supplementary Material 1).
We used an NVIDIA GeForce RTX 3070 Ti laptop GPU (VRAM: 8 GB) for the execution, with Python as the programming language and Pytorch for model programming. The initial deep learning model employed ResNet50, a type of deep learning neural network model that exhibits powerful performance with relatively fewer parameters than previous models (e.g., AlexNet and VGG) in medical image analysis [22], as a backbone network, utilizing batch sizes of 32, 16, and 8, 200 epochs, an Adam optimizer, and a learning rate of 0.001.
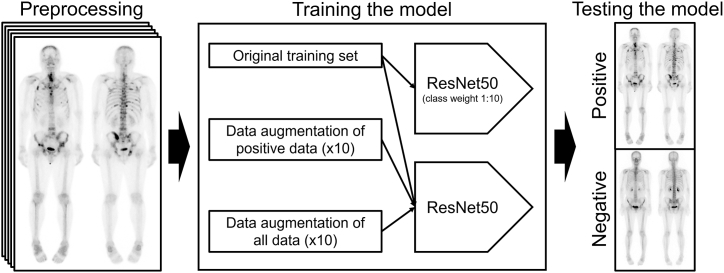
To improve diagnostic performance, we asked further questions and adopted some of the answers to identify optimal strategies (Supplementary material 2). We compared the results from four strategies to address the class imbalance of our data (positive = 8.6 %, negative = 91.4 %): 1) Strategy 1: original training set, 2) Strategy 2: original training set with 1:10 class weight, 3) Strategy 3: 10-fold data augmentation for positive images only, and 4) Strategy 4: 10-fold data augmentation for all images. Data augmentation involved random shifting (up and down within 5 pixels, left and right within 2 pixels), flipping, and rotation (within 2°). We did not augment the test sets. These strategies used ResNet50 as a backbone network, a batch size of 16, 200 epochs, an Adam optimizer, and a learning rate of 0.001. Fig. 3 presents a summary of the deep learning model setup process.
Fig. 3.
Overview of the deep learning model to classify bone metastasis positive and negative of bone scans. After unifying the intensities of the bone scan images in the preprocessing stage, the diagnostic performance in the test set is compared after training with four different strategies.
2.4. Visualization
We utilized a class activation map algorithm to generate visual representations of the specific regions of interest that the deep learning model prioritized when evaluating bone scan images for the presence or absence of bone metastasis. The heatmap visualization code was created by a nuclear medicine physician (S.H.L.) with the assistance of ChatGPT 3.5 and ChatGPT 4.0 (OpenAI, Supplementary Material 3). The resulting heatmaps were evaluated by two nuclear medicine physicians (H.J.S. and S.H.L.) to ascertain the distinguishing features used by the trained model to classify the bone scan images as positive or negative.
2.5. Statistical analysis
We used the scikit-learn library to compare the diagnostic performances of deep learning models by calculating the area under the curve (AUC), sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), accuracy, and crossentropy loss of test sets. Parameters for test datasets were calculated every 50 epochs. S.H.L. generated the code for calculations with guidance from ChatGPT 3.5 (Supplementary Material 4).
3. Results
3.1. Clinical characteristics of the patients
A total of 4626 patients with various malignancies were included in this study (Fig. 1). The most common underlying malignancy was breast cancer (38.3 %), followed by prostate cancer (27.2 %) and lung cancer (11.3 %), all of which are representative tumors that can lead to osteoblastic bone metastasis (76.8 %). Bone metastasis was present in 400 patients (8.6 %). The clinical characteristics of the patients are summarized in Table 1.
Table 1.
Patients’ characteristics.
| Characteristics | n = 4626 |
|---|---|
| Age, year, mean ± SD | 65.1 ± 11.3 |
| Sex | |
| Female, n (%) | 2334 (50.5 %) |
| Male, n (%) | 2292 (49.5 %) |
| Underlying malignancy | |
| Breast cancer, n (%) | 1774 (38.3 %) |
| Prostate cancer, n (%) | 1260 (27.2 %) |
| Lung cancer, n (%) | 522 (11.3 %) |
| Renal cell carcinoma, n (%) | 413 (8.9 %) |
| Bladder tumor, n (%) | 336 (7.3 %) |
| Urothelial cancer, n (%) | 78 (1.7 %) |
| Colorectal cancer, n (%) | 36 (0.8 %) |
| Esophageal cancer, n (%) | 34 (0.7 %) |
| Head and neck cancer, n (%) | 30 (0.6 %) |
| Hepatocellular carcinoma, n (%) | 20 (0.4 %) |
| Soft tissue tumor, n (%) | 16 (0.3 %) |
| Neuroendocrine tumor, n (%) | 13 (0.2 %) |
| Malignancy of unknown origin, n (%) | 11 (0.2 %) |
| Stomach cancer, n (%) | 11 (0.2 %) |
| Thymus tumor, n (%) | 11 (0.2 %) |
| Cervical cancer, n (%) | 8 (0.2 %) |
| Testis tumor, n (%) | 8 (0.2 %) |
| Pancreas cancer, n (%) | 7 (0.2 %) |
| Primary bone tumor, n (%) | 7 (0.2 %) |
| Cholangiocarcinoma, n (%) | 6 (0.1 %) |
| Ovarian cancer, n (%) | 6 (0.1 %) |
| Thyroid cancer, n (%) | 4 (0.1 %) |
| Lymphoma, n (%) | 3 (0.1 %) |
| Multiple myeloma, n (%) | 3 (0.1 %) |
| Uterine cancer, n (%) | 2 (0.0 %) |
| Gallbladder cancer, n (%) | 2 (0.0 %) |
| Brain tumor, n (%) | 1 (0.0 %) |
| Penile cancer, n (%) | 1 (0.0 %) |
| Skin cancer, n (%) | 1 (0.0 %) |
| Vaginal cancer, n (%) | 1 (0.0 %) |
| Wilm's tumor, n (%) | 1 (0.0 %) |
| Presence of bone metastasis | |
| Negative, n (%) | 4226 (91.4 %) |
| Positive, n (%) | 400 (8.6 %) |
SD, standard deviation.
3.2. Deep learning models
We first explored the optimal batch size and epoch size for training the models based on the mean test crossentropy loss and AUC of the 5-fold datasets (Table 2). In batch sizes of 32 and 16, test loss increased at the epoch size of 200 compared with the epoch sizes of 50, 100, and 150, and test loss increased at batch size 8 at epoch sizes of 100, 150, and 200 compared with an epoch size of 50. For batch sizes 32 and 16, the AUC was also lower at an epoch size of 200 compared with epoch sizes of 50, 100, and 150. Based on the results of the initial deep learning model, we chose a batch size of 16 and an epoch size of 150 as optimal due to the relatively low test loss (0.2894) and high AUC (0.7952) compared with other combinations. A batch size of 16 was chosen because it provided a balance between computational efficiency and model performance, while an epoch size of 150 allowed for sufficient model training without overfitting or underfitting.
Table 2.
Mean test loss and area under the curve of 5-fold datasets according to the batch size and epoch size.
| Epoch size | Batch size = 32 | Batch size = 16 | Batch size = 8 | Overall | |
|---|---|---|---|---|---|
| 50 | 0.2649 | 0.2858 | 0.2741 | 0.2749 | |
| 100 | 0.2692 | 0.2875 | 0.3120 | 0.2896 | |
| Loss | 150 | 0.2835 | 0.2894 | 0.3116 | 0.2948 |
| 200 | 0.3235 | 0.3249 | 0.3162 | 0.3216 | |
| Overall | 0.2853 | 0.2969 | 0.3035 | 0.2952 | |
| 50 | 0.3300 | 0.3400 | 0.2850 | 0.3183 | |
| 100 | 0.2900 | 0.2775 | 0.3450 | 0.3042 | |
| Sensitivity | 150 | 0.3325 | 0.2875 | 0.2925 | 0.3042 |
| 200 | 0.2600 | 0.2825 | 0.3975 | 0.3133 | |
| Overall | 0.3031 | 0.2969 | 0.3300 | 0.3100 | |
| 50 | 0.9647 | 0.9546 | 0.9768 | 0.9654 | |
| 100 | 0.9780 | 0.9737 | 0.9584 | 0.9700 | |
| Specificity | 150 | 0.9624 | 0.9785 | 0.9768 | 0.9726 |
| 200 | 0.9735 | 0.9678 | 0.9501 | 0.9638 | |
| Overall | 0.9697 | 0.9686 | 0.9655 | 0.9679 | |
| 50 | 0.7963 | 0.7941 | 0.7965 | 0.7956 | |
| 100 | 0.7927 | 0.7951 | 0.7932 | 0.7937 | |
| AUC | 150 | 0.7923 | 0.7952 | 0.7952 | 0.7942 |
| 200 | 0.7878 | 0.7889 | 0.7987 | 0.7918 | |
| Overall | 0.7923 | 0.7933 | 0.7959 | 0.7938 |
AUC, area under the curve.
To overcome the relatively low sensitivity of the initial deep learning model (overall sensitivity: 31 %), we compared the diagnostic performances of four different model training strategies using the selected batch size of 16 (Table 3). In Strategies 1, 2, and 4, the loss of test datasets increased with an epoch size of 200 compared with an epoch size of 150. The AUC of the test datasets decreased with an epoch size of 200 compared with an epoch size of 150 in all strategies. We determined that an epoch size of 150 was optimal for four different strategies when changing the batch size.
Table 3.
Mean test loss and area under the curve of 5-fold datasets according to the training set modification and epoch size (batch size = 16).
| Epoch size | Strategy 1 Original |
Strategy 2 Class weight (1:0) |
Strategy 3 × 10 data (positive only) |
Strategy 4 × 10 data (all) |
Overall | |
|---|---|---|---|---|---|---|
| 50 | 0.2858 | 0.4646 | 0.3427 | 0.2978 | 0.3477 | |
| 100 | 0.2875 | 0.3940 | 0.3536 | 0.3485 | 0.3459 | |
| Loss | 150 | 0.2894 | 0.3542 | 0.3646 | 0.3471 | 0.3388 |
| 200 | 0.3249 | 0.4323 | 0.3113 | 0.3540 | 0.3556 | |
| Overall | 0.2969 | 0.4113 | 0.3431 | 0.3369 | 0.3470 | |
| 50 | 0.3400 | 0.4400 | 0.5200 | 0.3050 | 0.4013 | |
| 100 | 0.2775 | 0.4975 | 0.4825 | 0.2925 | 0.3875 | |
| Sensitivity | 150 | 0.2875 | 0.3900 | 0.5600 | 0.2400 | 0.3694 |
| 200 | 0.2825 | 0.5425 | 0.4425 | 0.3300 | 0.3994 | |
| Overall | 0.2969 | 0.4675 | 0.5013 | 0.2919 | 0.3894 | |
| 50 | 0.9546 | 0.8537 | 0.8933 | 0.9669 | 0.9171 | |
| 100 | 0.9737 | 0.8765 | 0.8926 | 0.9796 | 0.9306 | |
| Specificity | 150 | 0.9785 | 0.9257 | 0.8873 | 0.9860 | 0.9444 |
| 200 | 0.9678 | 0.8540 | 0.9344 | 0.9619 | 0.9295 | |
| Overall | 0.9686 | 0.8775 | 0.9019 | 0.9736 | 0.9304 | |
| 50 | 0.7941 | 0.7877 | 0.8129 | 0.8209 | 0.8039 | |
| 100 | 0.7951 | 0.7959 | 0.8162 | 0.8064 | 0.8034 | |
| AUC | 150 | 0.7952 | 0.7945 | 0.8156 | 0.8053 | 0.8027 |
| 200 | 0.7889 | 0.7927 | 0.8114 | 0.7998 | 0.7982 | |
| Overall | 0.7933 | 0.7927 | 0.8140 | 0.8081 | 0.8020 |
AUC, area under the curve.
We then assessed the diagnostic performances of the models trained with 80 % of the total dataset using the chosen batch size of 16 and epoch size of 150 (Table 4). The overall AUC values for Strategies 1, 2, 3, and 4 were 0.7952, 0.7945, 0.8156, and 0.8053, respectively, confirming that Strategy 3 had the highest AUC, followed by Strategy 4. The sensitivity of Strategy 3 (0.5600) was the highest, although the specificity of Strategy 3 (0.8873) was lower than those of Strategies 1 and 4. Based on our investigation, a deep learning model with 10-fold data augmentation for positive cases only (Strategy 3) with a batch size of 16 and an epoch size of 150 can effectively detect bone metastasis on bone scan images in patients with various malignancies.
Table 4.
Diagnostic performances of strategies trained with 80 % of the total dataset (epoch size = 150).
| Strategy | Fold 1 (n = 925) | Fold 2 (n = 925) | Fold 3 (n = 925) | Fold 4 (n = 925) | Fold 5 (n = 926) | Overall (n = 4626) | |
|---|---|---|---|---|---|---|---|
| AUC | 1. Original | 0.7248 | 0.8123 | 0.7982 | 0.8595 | 0.7814 | 0.7952 |
| 2. Class weight (1:0) | 0.7236 | 0.8071 | 0.8045 | 0.8478 | 0.7897 | 0.7945 | |
| 3. × 10 data (positive only) | 0.7687 | 0.8228 | 0.8458 | 0.8568 | 0.7838 | 0.8156 | |
| 4. × 10 data (all) | 0.7479 | 0.8135 | 0.8446 | 0.8677 | 0.7528 | 0.8053 | |
| Sensitivity | 1. Original | 0.1750 | 0.4000 | 0.2500 | 0.3250 | 0.2875 | 0.2875 |
| 2. Class weight (1:0) | 0.4250 | 0.3750 | 0.6000 | 0.3125 | 0.2375 | 0.3900 | |
| 3. × 10 data (positive only) | 0.5125 | 0.6000 | 0.6750 | 0.6500 | 0.3625 | 0.5600 | |
| 4. × 10 data (all) | 0.1625 | 0.3125 | 0.2625 | 0.2500 | 0.2125 | 0.2400 | |
| Specificity | 1. Original | 0.9882 | 0.9751 | 0.9645 | 0.9870 | 0.9775 | 0.9785 |
| 2. Class weight (1:0) | 0.8533 | 0.9503 | 0.8568 | 0.9834 | 0.9846 | 0.9257 | |
| 3. × 10 data (positive only) | 0.8686 | 0.8911 | 0.8249 | 0.8911 | 0.9610 | 0.8873 | |
| 4. × 10 data (all) | 0.9834 | 0.9846 | 0.9751 | 0.9976 | 0.9894 | 0.9860 | |
| PPV | 1. Original | 0.5833 | 0.6038 | 0.4000 | 0.7027 | 0.5476 | 0.5675 |
| 2. Class weight (1:0) | 0.2152 | 0.4167 | 0.2840 | 0.6410 | 0.5938 | 0.4301 | |
| 3. × 10 data (positive only) | 0.2697 | 0.3429 | 0.2673 | 0.3611 | 0.4677 | 0.3418 | |
| 4. × 10 data (all) | 0.4815 | 0.6579 | 0.5000 | 0.9091 | 0.6538 | 0.6405 | |
| NPV | 1. Original | 0.9267 | 0.9450 | 0.9314 | 0.9392 | 0.9355 | 0.9356 |
| 2. Class weight (1:0) | 0.9400 | 0.9414 | 0.9577 | 0.9379 | 0.9318 | 0.9418 | |
| 3. × 10 data (positive only) | 0.9495 | 0.9592 | 0.9640 | 0.9641 | 0.9410 | 0.9556 | |
| 4. × 10 data (all) | 0.9254 | 0.9380 | 0.9332 | 0.9336 | 0.9300 | 0.9320 | |
| Accuracy | 1. Original | 0.9178 | 0.9254 | 0.9027 | 0.9297 | 0.9179 | 0.9187 |
| 2. Class weight (1:0) | 0.8162 | 0.9005 | 0.8346 | 0.9254 | 0.9201 | 0.8794 | |
| 3. × 10 data (positive only) | 0.8378 | 0.8659 | 0.8119 | 0.8703 | 0.9093 | 0.8590 | |
| 4. × 10 data (all) | 0.9124 | 0.9265 | 0.9135 | 0.9330 | 0.9222 | 0.9215 |
AUC, area under the curve; PPV, positive predictive value; NPV, negative predictive value.
3.3. Visualization
To generate visual representations of the specific regions of interest that the deep learning model prioritized when evaluating bone scan images for the presence or absence of bone metastasis, employing a class activation map algorithm indicated that the deep learning model focused on identifying disseminated bone metastases within the spine.
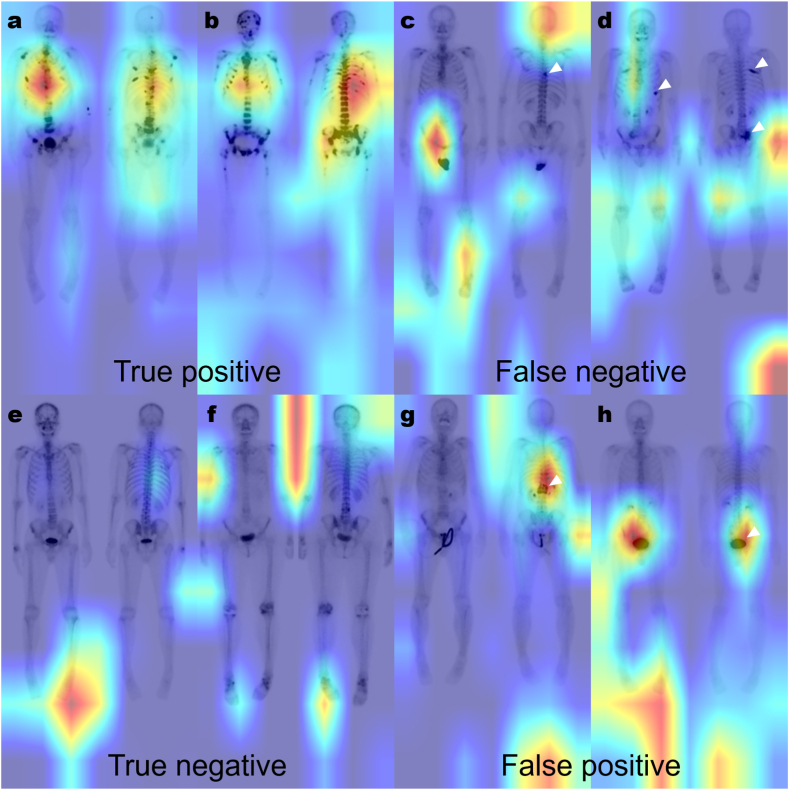
We trained all 4626 datasets with 10-fold data augmentation for positive cases only (Strategy 3) with a batch size of 16 and an epoch size of 150. Our final model classified 368 out of 400 positive cases as true positives (sensitivity: 0.9200) and 3671 out of 4226 negative cases as true negatives (specificity: 0.8687). Representative cases of our model are shown in Fig. 4. In true-positive cases, the model seemed to be weighted on disseminated bone metastases located in the spine, especially the T-spine (Fig. 4a and b). However, in the case of a single lesion (Fig. 4c) or bone metastases located outside the spine (Fig. 4d), there were cases classified as false negatives. In true-negative cases, no significant lesions were seen in the entire skeleton (Fig. 4e and f). However, benign spine lesions (such as traumatic compression fracture, Fig. 4g) or large intense bladder activity (Fig. 4h) could be classified as false positives. Additionally, there were instances where the trained model prioritized the region outside the body as the area of interest. (Fig. 4d, f, g, and h).
Fig. 4.
Representative cases of the final model. True-positive cases show disseminated bone metastases weighted in the spine, especially the T-spine (a, b). However, patients with a single lesion in the T-spine (c, white arrowhead) and bone metastases outside the spine (d, white arrowhead) are classified as false negatives. There is no significant region of interest in the trained model in the entire skeleton in true-negative cases (e, f). However, a traumatic compression fracture (g, white arrowhead) and large intense urinary bladder activity (h, white arrowhead) can be classified as false positives. The region outside the body can be the area of interest for the trained model (d, f, g, h).
4. Discussion
This study demonstrates the feasibility of developing a deep learning model to diagnose bone metastasis in bone scans with the aid of the open-source AI chatbot ChatGPT. The primary challenge in developing deep learning models for medical imaging is the need for proficiency and expertise in both medical knowledge and programming. However, in this study, a nuclear medicine specialist with basic knowledge of Python and deep learning concepts developed a deep learning model using ChatGPT, proving the feasibility of utilizing AI chatbots such as ChatGPT for medical image deep learning model development. Our model achieved promising performance in detecting bone metastases in bone scans of patients with various malignancies, with an AUC of 0.8156 for Strategy 3, which utilized data augmentation exclusively for positive cases.
Several deep learning models have been investigated for detecting bone metastases in bone scans. Our ChatGPT assisted model exhibited a diagnostic performance of 56.0 % sensitivity and 88.7 % specificity, which are slightly lower than the values reported in prior studies (sensitivity range: 59.9%–94.0 %, specificity range: 85.5%–99.3 %, Table 5) [[13], [14], [15], [16], [17], [18], [19]]. These discrepancies could be attributed to differences in preprocessing, bone metastasis prevalence, underlying malignancies, sample size, and deep learning models between our study and earlier investigations. Most previous studies utilized distinct preprocessing techniques, such as extracting only valid body regions from bone scan images [15,17,18], eliminating artifacts such as urine contamination or injection sites [13,14], the segmentation of urinary bladders [19], or the left-to-right flipping of posterior images and their subsequent merging with anterior images [17]. However, we did not perform such preprocessing in our study, except for the uniform setting of the intensity, which may have impeded effective learning due to artifacts or areas outside the body. Upon examining the heatmaps in our study, we observed that areas outside the body, urine contamination, and the urinary bladder might have confounded the model's training. We anticipate that implementing appropriate preprocessing will improve diagnostic performance.
Table 5.
Diagnostic performance of detecting bone metastases in bone scans of previous studies.
| Authors | Type of CNN | Primary tumors | n | Prv (%) | Acc (%) | Sen (%) | Spe (%) | Prc (%) | Rec (%) |
|---|---|---|---|---|---|---|---|---|---|
| Papandrianos et al. [13] | 4-layer CNN | Prostate | 778 | 41.9 | 91.6 | 92.7 | 96.0 | ||
| Papandrianos et al. [14] | 3-layer CNN | Breast | 408 | 54.1 | 92.5 | 94.0 | 92.0 | 93.4 | 93.8 |
| Pi et al. [15] | Inception-V3 | Lung (31 %) Breast (24 %) Prostate (10 %) Other (12 %) Benign (22 %) |
15,474 | 37.5 | 95.0 | 93.2 | 96.1 | ||
| Hsieh et al. [16] | ResNet50V2 | Breast (59 %) H&N (12 %) Prostate (7 %) Lung (5 %) Liver (3 %) Other (14 %) |
37,427 | 8.1 | 96.1 | 59.9 | 99.3 | 87.8 | |
| Guo et al. [17] | 26-layer CNN | Lung | 945 | 64.8 | 83.1 | 87.0 | 87.0 | ||
| Han et al. [18] | GLUE 2D-CNN | Prostate | 9133 | 32.7 | 90.0 | 82.8 | 93.5 | ||
| Liu et al. [19] | ResNet 34 | Prostate Lung Breast Gastrointestinal |
621 | 43.9 | 88.6 | 92.6 | 85.5 |
Acc, accuracy; CNN, convolutional neural network; GLUE, global–local unified emphasis; H&N, head and neck; Prc, precision, Prv, prevalence; Rec, recall; Sen, sensitivity; Spe, specificity.
In our investigation, the bone metastasis positivity rate was low at 8.6 %, in contrast to the prevalence of 32.7%–64.8 % reported in earlier studies [[13], [14], [15],[17], [18], [19]]. Only Hsieh et al.'s study reported a similar positivity rate of 8.1 % out of 37,427 images [16]. Our outcomes are more representative of the actual prevalence, considering the general 5 % incidence of bone metastasis in cancer patients [[1], [2], [3]]. While some research has focused on a single tumor type [13,14,17,18], both the study by Hsieh et al. and our study involved patients with various cancer types, making our results more generalizable. Both investigations used ResNet50 as the backbone network, with Hsieh's model demonstrating 59.9 % sensitivity and 99.3 % specificity, indicating low sensitivity and high specificity, similar to our model. In the case of class imbalance, low-frequency data do not learn features as effectively as high-frequency data, potentially leading to reduced sensitivity with fewer positive cases [23]. To address the low prevalence, our Strategy 3 implemented 10-fold data augmentation for positive cases exclusively in the training set, increasing sensitivity from 28.8 % to 56.0 % compared with the strategy with the original data (Strategy 1).
Constructing deep learning models utilizing ChatGPT presented some obstacles. ChatGPT does not always offer the optimal answer to simple and straightforward questions [24]. When inquired about creating a deep learning model, only a basic model with several convolutional layers is provided. More comprehensive responses are provided when queries are more specific, such as backbone networks (Supplementary material 1). Moreover, ChatGPT does not ensure error-free code. In our case, it took approximately five questions for ChatGPT 3.5 to generate an initial error-free deep learning model using ResNet50. However, creating heatmap code proved more challenging even with dozens of inquiries, and we ultimately completed it with the aid of ChatGPT 4.0. As ChatGPT 4.0 can identify images as well as text [25], open-source AI is expected to be increasingly beneficial in developing appropriate models in the future.
Our study had a few limitations. First, the dataset used in our investigation was obtained from a single institution with relatively small sample size, potentially limiting the generalizability of the findings. Unfortunately, due to the difficulty in obtaining publicly available bone scan images, cross-validation using either publicly available bone scans or images from other institutions will be required in the future. Second, our deep learning model was constructed by a physician with basic knowledge of Python and deep learning concepts, which might limit the model's performance compared with models developed by experts in both medicine and programming. Nevertheless, our findings demonstrate the feasibility of employing ChatGPT for developing deep learning models in medical imaging.
5. Conclusions
Our study illustrates that even an individual with rudimentary programming skills can develop a deep learning model for diagnosing bone metastasis in bone scans using ChatGPT, an open-source AI chatbot. Data augmentation of low-prevalence classes can be a promising solution to address the class imbalance problem, as opposed to class weighting or data augmentation of the entire dataset. Examining the model's heatmaps might direct future model development to improve diagnostic performance.
Data availability statement
The data used in our study has not been deposited into a publicly available repository. The datasets used and/or analyzed during the current study will be made available from the corresponding author upon reasonable request.
Additional information
The underlying code for this study is included in the supplementary materials.
Ethics approval
The present study was approved by the institutional review board of our institution (IRB no. 2023-02-008) and with the 2013 Helsinki declaration and its later amendments or comparable ethical standards.
Consent to participate
The institutional review board at our institution approved this retrospective study, and the requirement to obtain informed consent was waived.
Consent for publication
The institutional review board at our institution approved this retrospective study, and the requirement to obtain informed consent was waived.
CRediT authorship contribution statement
Hye Joo Son: Writing – original draft, Funding acquisition, Formal analysis. Soo-Jong Kim: Validation, Methodology. Sehyun Pak: Project administration, Data curation. Suk Hyun Lee: Writing – review & editing, Visualization, Validation, Supervision, Resources, Project administration, Methodology, Investigation, Formal analysis, Conceptualization.
Declaration of generative AI and AI-assisted technologies in the writing process
During the preparation of this work the authors used ChatGPT 4.0 (OpenAI) to improve the readability and quality of the writing after the initial draft. After using this tool, the authors reviewed and edited the content as needed and takes full responsibility for the content of the publication.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the research fund of Dankook University in 2021.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e22409.
Appendix A. Supplementary data
The following is/are the supplementary data to this article.
References
- 1.Hernandez R.K., Wade S.W., Reich A., Pirolli M., Liede A., Lyman G.H. Incidence of bone metastases in patients with solid tumors: analysis of oncology electronic medical records in the United States. BMC Cancer. 2018;18:44. doi: 10.1186/s12885-017-3922-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang J.F., Shen J., Li X., et al. Incidence of patients with bone metastases at diagnosis of solid tumors in adults: a large population-based study. Ann. Transl. Med. 2020;8:482. doi: 10.21037/atm.2020.03.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jiang W., Rixiati Y., Zhao B., Li Y., Tang C., Liu J. Incidence, prevalence, and outcomes of systemic malignancy with bone metastases. J. Orthop. Surg. 2020;28 doi: 10.1177/2309499020915989. [DOI] [PubMed] [Google Scholar]
- 4.Yang H.L., Liu T., Wang X.M., Xu Y., Deng S.M. Diagnosis of bone metastases: a meta-analysis comparing 1⁸FDG PET, CT, MRI and bone scintigraphy. Eur. Radiol. 2011;21:2604–2617. doi: 10.1007/s00330-011-2221-4. [DOI] [PubMed] [Google Scholar]
- 5.Agrawal K., Marafi F., Gnanasegaran G., Van der Wall H., Fogelman I. Pitfalls and limitations of radionuclide planar and hybrid bone imaging. Semin. Nucl. Med. 2015;45:347–372. doi: 10.1053/j.semnuclmed.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 6.Lee W.W., Ryu J.S. KSNM 60 in general nuclear medicine: the old dream comes true. Nucl Med Mol Imaging. 2022;56:71–79. doi: 10.1007/s13139-021-00731-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rajarubendra N., Bolton D., Lawrentschuk N. Diagnosis of bone metastases in urological malignancies--an update. Urology. 2010;76:782–790. doi: 10.1016/j.urology.2009.12.050. [DOI] [PubMed] [Google Scholar]
- 8.Sadik M., López-Urdaneta J., Ulén J., et al. Artificial intelligence increases the agreement among physicians classifying focal skeleton/bone marrow uptake in hodgkin's lymphoma patients staged with [18F]FDG PET/CT—a retrospective study. Nucl Med Mol Imaging. 2023;57:110–116. doi: 10.1007/s13139-022-00765-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park J., Kang S.K., Hwang D., et al. Automatic lung cancer segmentation in [18F]FDG PET/CT using a two-stage deep learning approach. Nucl Med Mol Imaging. 2023;57:86–93. doi: 10.1007/s13139-022-00745-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia E.V., Piccinelli M. Preparing for the artificial intelligence revolution in nuclear cardiology. Nucl Med Mol Imaging. 2023;57:51–60. doi: 10.1007/s13139-021-00733-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu J., Yang Y., Singhrao K., et al. Deep learning approaches using 2D and 3D convolutional neural networks for generating male pelvic synthetic computed tomography from magnetic resonance imaging. Med. Phys. 2019;46:3788–3798. doi: 10.1002/mp.13672. [DOI] [PubMed] [Google Scholar]
- 12.Lee J.J., Yang H., Franc B.L., Iagaru A., Davidzon G.A. Deep learning detection of prostate cancer recurrence with (18)F-FACBC (fluciclovine, Axumin®) positron emission tomography. Eur. J. Nucl. Med. Mol. Imag. 2020;47:2992–2997. doi: 10.1007/s00259-020-04912-w. [DOI] [PubMed] [Google Scholar]
- 13.Papandrianos N., Papageorgiou E., Anagnostis A., Papageorgiou K. Efficient bone metastasis diagnosis in bone scintigraphy using a fast convolutional neural network architecture. Diagnostics. 2020;10 doi: 10.3390/diagnostics10080532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papandrianos N., Papageorgiou E., Anagnostis A., Feleki A. A deep-learning approach for diagnosis of metastatic breast cancer in bones from whole-body scans. Appl. Sci.-Basel. 2020;10:27. doi: 10.3390/app10030997. [DOI] [Google Scholar]
- 15.Pi Y., Zhao Z., Xiang Y., Li Y., Cai H., Yi Z. Automated diagnosis of bone metastasis based on multi-view bone scans using attention-augmented deep neural networks. Med. Image Anal. 2020;65 doi: 10.1016/j.media.2020.101784. [DOI] [PubMed] [Google Scholar]
- 16.Hsieh T.C., Liao C.W., Lai Y.C., Law K.M., Chan P.K., Kao C.H. Detection of bone metastases on bone scans through image classification with contrastive learning. J. Personalized Med. 2021;11:1248. doi: 10.3390/jpm11121248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo Y., Lin Q., Zhao S., et al. Automated detection of lung cancer-caused metastasis by classifying scintigraphic images using convolutional neural network with residual connection and hybrid attention mechanism. Insights Imaging. 2022;13:24. doi: 10.1186/s13244-022-01162-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han S., Oh J.S., Lee J.J. Diagnostic performance of deep learning models for detecting bone metastasis on whole-body bone scan in prostate cancer. Eur. J. Nucl. Med. Mol. Imag. 2022;49:585–595. doi: 10.1007/s00259-021-05481-2. [DOI] [PubMed] [Google Scholar]
- 19.Liu S., Feng M., Qiao T., et al. Deep learning for the automatic diagnosis and analysis of bone metastasis on bone scintigrams. Cancer Manag. Res. 2022;14:51–65. doi: 10.2147/CMAR.S340114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Terech K. Techradar; 2023. GPT-4 Is Bringing a Massive Upgrade to ChatGPT.https://www.techradar.com/news/gpt-4 [Google Scholar]
- 21.Panicek D.M., Hricak H. How sure are you, doctor? A standardized lexicon to describe the radiologist's level of certainty. AJR Am. J. Roentgenol. 2016;207:2–3. doi: 10.2214/ajr.15.15895. [DOI] [PubMed] [Google Scholar]
- 22.Kim H.E., Cosa-Linan A., Santhanam N., Jannesari M., Maros M.E., Ganslandt T. Transfer learning for medical image classification: a literature review. BMC Med. Imag. 2022;22:69. doi: 10.1186/s12880-022-00793-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Litjens G., Kooi T., Bejnordi B.E., et al. A survey on deep learning in medical image analysis. Med. Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 24.Bom H.H. Exploring the opportunities and challenges of ChatGPT in academic writing: a roundtable discussion. Nucl Med Mol Imaging. 2023;57:165–167. doi: 10.1007/s13139-023-00809-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghaffary S. Vox; 2023. The Makers of ChatGPT Just Released a New AI that Can Build Websites, Among Other Things.https://www.vox.com/2023/3/15/23640640/gpt-4-chatgpt-openai-generative-ai [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in our study has not been deposited into a publicly available repository. The datasets used and/or analyzed during the current study will be made available from the corresponding author upon reasonable request.