Dear Editor,
Ferroptosis is a form of non-apoptotic cell death characterized by the accumulation of iron-dependent lipid peroxides and oxidative damage to cell membrane.1 Discovered relative recently, ferroptosis is morphologically and mechanistically distinct from other forms of cell death, such as apoptosis, necroptosis, and pyroptosis.2 Ferroptosis has been implicated in various pathological conditions, including neurodegenerative diseases, cancer, and ischemia-reperfusion injury.3–5 Thus, studying ferroptosis could not only provide insights into the fundamental mechanisms of cell death and survival, but also identify new targets for therapeutic intervention in a variety of pathological conditions.
In recent years, several surveillance mechanisms have been discovered that suppress ferroptosis by preventing lipid peroxidation:2,4 (1) the cyst(e)ine-GSH-GPX4 system, in which glutathione-disulfide reductase (GSR) utilizes NADPH as a cofactor to generate GSH, which is then used by GPX4 to detoxify lipid hydroperoxides. (2) The GCH1-BH4 system, in which dihydrofolate reductase (DHFR) also utilizes NADPH to promote the formation of tetrahydrobiopterin (BH4), a radical-trapping antioxidant capable of suppressing ferroptosis. (3) The FSP1-CoQH2 system, in which flavoprotein ferroptosis suppressor protein 1 (FSP1) catalyzes the reduction of ubiquinone (CoQ10) or vitamin K, which in turn traps lipid peroxyl radicals and thus suppresses lipid peroxidation.6–8 Interestingly, FSP1, also known as apoptosis-inducing factor mitochondria-associated 2 (AIFM2), bears significant sequence similarity to AIFM1 (also called apoptosis-inducing factor, AIF), an established NADH-selective oxidoreductase.9 However, it remains unclear whether FSP1 utilizes NADH, NADPH, or both as electron donors.
To solve this puzzle, we purified the non-myristoylated FSP1 (residues 10–373) and determined its crystal structure at a resolution of 2.5 Å (Fig. 1a and Supplementary information, Table S1). The protein was crystalized in the P21 21 21 space group, with each asymmetric unit containing only copy of FSP1. Similar to AIFM1,10,11 FSP1 also contains three structural domains: NAD(P)H binding domain, FAD binding domain and C-terminal domain, in addition to their N-terminal sequences critical for subcellular localization (Fig. 1a and Supplementary information, Fig. S1). However, the structure of FSP1 differs from that of AIFM1 in three different aspects (Supplementary information, Figs. S1 and S2). First of all, their C-terminal domains are most dramatically different in terms of 3D structure and amino acid sequence (Supplementary information, Fig. S1c, d). The DALI search detects significant similarity between the NAD(P)H binding domains (2.4 Å RMSD for 149 Cα atoms) and between the FAD binding domains (2.3 Å RMSD for 114 Cα atoms), and no similarity between the C-terminal domains. Second, AIFM1 has been crystalized at the oxidized and reduced states, in which AIFM1 adopts monomeric and dimeric forms, respectively.10,11 Multiple elements critical for the dimerization of AIFM1, including the C-loop, E246 and R449, are not conserved in FSP1, indicating that FSP1 cannot dimerize in a similar manner (Supplementary information, Fig. S1e–g). Third, both FSP1 and AIFM1 harbor three pockets that can accommodate substrate or cofactor. The three pockets of FSP1 are interconnected, whereas the pocket III of AIFM1 is not connected with the other two pockets (Supplementary information, Fig. S1h, i). Pocket III in AIFM1 is known to accommodate the 2nd NADH molecule at the reduced state. The size of pocket III in FSP1 (3471 Å3), the putative substrate (CoQ10 or vitamin K) binding pocket, is significantly larger than that of AIFM1 (1148 Å3), consistent with the fact that FSP1 is capable of catalyzing the reduction of large substrates such as CoQ10. Altogether, our structural analysis supports the concepts that FSP1 is a genuine oxidoreductase, whereas AIFM1 functions as a NADH sensor to regulate mitochondrial activity and cell death.
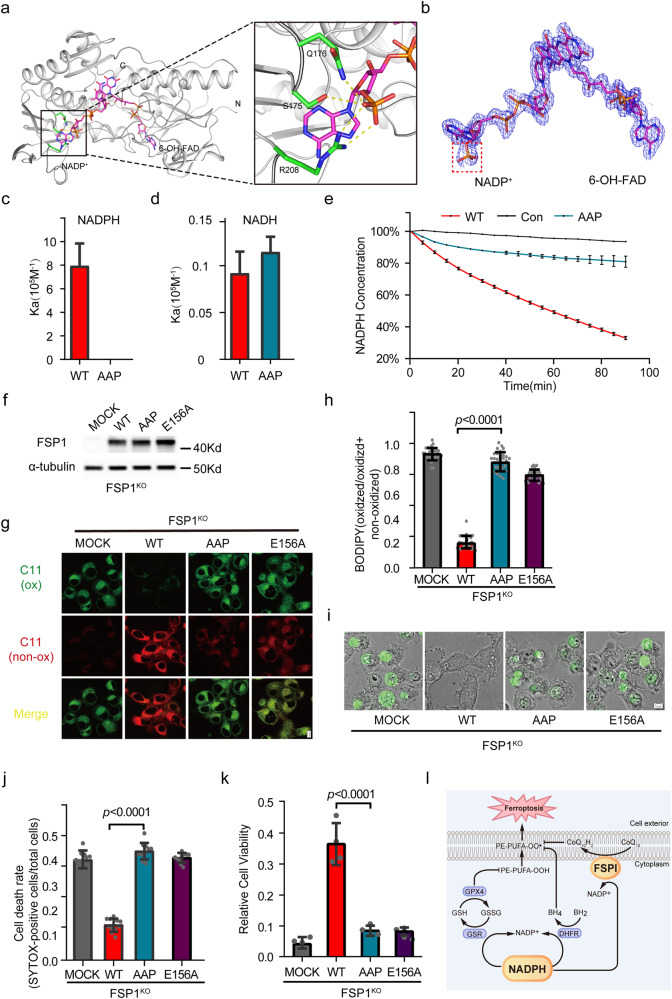
Fig. 1. FSP1 utilizes NADPH, but not NADH, to suppress ferroptosis.
a Overall structure of FSP1 bound to NADP+ and 6-OH-FAD. NADP+ and 6-OH-FAD are shown as sticks. Zoom view shows the interaction between 2′-phosphate of NADPH and S175, Q176 and R208 of FSP1. Yellow dotted lines, hydrogen bonds. b Overlay of NADP+ and 6-OH-FAD with a composite Simulated Annealing omit map at 2.5 Å resolution and contoured at 1σ. Red square highlights the 2′-phosphate group specific to NADPH. c Affinity between NADPH and FSP1 WT and mutant, as determined by isothermal titration calorimetry (ITC). d Affinity between NADH and FSP1 WT and mutant, as determined by ITC. e In vitro enzymatic activity of FSP1 WT and mutant. Purified FSP1 WT or mutant was incubated with NADPH and CoQ2, and absorbance at 340 nm was monitored over time. f Immunoblotting analysis of the expression levels of FSP1 WT and mutants. g FSP1KO HT1080 cells harboring FSP1 WT, AAP, E156A or empty vector were treated with 100 nM RSL3 for 2 h, and then stained with BODIPY 581/591 C11. Images were acquired using the Olympus scanR. ox oxidized, non-ox non-oxidized. Scale bar, 5 μm. h Quantitative analysis of lipid peroxidation levels in RSL3-treated FSP1KO HT1080 cells expressing the indicated FSP1 variants as in (f). i Live-cell imaging of FSP1KO HT1080 cells expressing the indicated FSP1 variants. Cells were treated with 300 nM RSL3 for 3 h, and then incubated with SYTOX Green Dead Cell Stain. Scale bar, 5 μm. j Quantitative analysis of cell death in (h). k FSP1KO HT1080 cells harboring FSP1 WT, AAP, E156A or empty vector were treated with 300 nM RSL3 for 4 h. Cell viability was assessed 24 h later using the CCK-8 kit. All experiments were performed in four replicates (g, i, k). Data are presented as mean ± SD (h, j, k). Statistical comparisons were performed using one-way ANOVA, and significant p values are indicated. l A model depicting how FSP1 suppresses ferroptosis in a NADPH-dependent manner. In addition to FSP1, the cyst(e)ine-GSH-GPX4 and GCH1-BH4 systems also utilize NADPH as a cofactor to prevent lipid peroxidation and to suppress ferroptosis.
In the crystal structure, we also observed that each FSP1 protein bound one NADP+ molecule, the oxidized form of NADPH, and one 6-OH-FAD (FAD derivative) molecule, with strong π–π interaction between the nicotinamide of NADP+ and the Flavin nucleotide portion of 6-OH-FAD (Fig. 1b). The models of NADPH and 6-OH-FAD are well supported by their respective electron density (Fig. 1b). As we added neither molecule during protein purification and crystallization steps, the two molecules were likely carried from the medium and tightly associated with FSP1. Our structure indicates that FSP1 functions to catalyze electron transfer from NADPH to 6-OH-FAD and then to CoQ10 or vitamin K. FSP1 is a member of Type II NADH:quinone oxidoreductases (NDH-2s), which also encompasses AIFM1 and many enzymes from all three kingdoms of life. Among them, Ndi1 from Saccharomyces cerevisiae (Sc.Ndi1) and NDH-2 from Caldalkalibacillus thermarum (Ct.NDH-2) display the closest structural similarity to FSP1,12,13 with the Dali’s Z-score being 36 and 31, respectively (Supplementary information, Fig. S3). Remarkably, all of them, except for FSP1, bind NAD(H) (Supplementary information, Fig. S3).
NADH and NADPH are highly similar to each other, with the only difference being the phosphate group at the 2′ position of the ribose ring (Supplementary information, Fig. S3a, b). To understand how FSP1 specifically recognizes NADPH, we compared structures and sequences around the NAD(P)H-binding pocket. The NAD(P)H-binding pocket of FSP1 is more positively charged than those of other proteins, consistent with its role in contacting the negatively-charged phosphate group (Supplementary information, Fig. S3c). Specifically, the side chain of R208 of FSP1 form two short hydrogen bonds (2.2–2.3 Å) with the oxygen atoms from the phosphate group specific to NADPH (Fig. 1a). S175 and Q176 of FSP1 further contribute to the recognition of the 2′-phosphate by forming additional hydrogen bonds (Fig. 1a). Importantly, although amino acids recognizing nicotinamide and adenine of NAD(P)H are largely conserved in all four proteins, these three residues are substituted by other residues in AIFM1, Sc.Ndi1 and Ct.NDH-2 (Supplementary information, Fig. S3d).
To investigate whether FSP1 selectively binds NADPH via these amino acids, we replaced S175, Q176, and R208 with corresponding residues in AIFM1 or Ct.NDH-2, and generated a triple (S175A/Q176A/R208P, referred to as AAP) mutant. FSP1 displayed a tighter binding toward NADPH, with a dissociation constant (KD) of 1.26 ± 0.29 μM, which was ~80-fold of that toward NADH (Fig. 1c, d and Supplementary information, Fig. S4). No binding was detected between FSP1 AAP and NADPH (Fig. 1c). Critically, these mutations did not dramatically alter the binding toward NADH (Fig. 1d). To assess the importance of these residues for FSP1 enzyme activity, we performed in vitro enzymatic activity assay using NADPH and CoQ2 as substrates. The enzymatic activity was determined by the reduction of NADPH. FSP1 AAP almost completely lost the enzymatic activity toward NADPH (Fig. 1e). Notably, FSP1 AAP showed a slightly decreased enzymatic activity toward NADH, consistent with the binding experiment results (Supplementary information, Fig. S5).
To investigate how the binding toward NADPH affects FSP1’s ability in suppressing ferroptosis, we generated the FSP1KO cell using the CRISPR/Cas9 technology (Supplementary information, Fig. S6a). Consistent with previous studies,6,7 control and FSP1KO HT1080 cells exhibited similar levels of lipid peroxidation, as determined by BODIPY 581/591 C11 staining, under basal conditions (Supplementary information, Fig. S6b–d). However, the FSP1KO cells displayed greatly increased lipid peroxidation when induced by RSL3, a ferroptosis inducer targeting GPX4 (Supplementary information, Fig. S6b–d). The lipid peroxidation level could be restored when FSP1 WT was re-introduced (Fig. 1f–h). Cells with FSP1 E156A, a mutant known to abolish the interaction with its cofactor FAD,7 displayed a significantly higher level of lipid peroxidation relative to those with FSP1 WT (Fig. 1f–h). Importantly, cells with FSP1 AAP showed a similar lipid peroxidation level to those with FSP1 E156A, indicating that the binding of NADPH, but not NADH, was critical for FSP1 to suppress lipid peroxidation (Fig. 1f–h). Similar to HT1080 cells, the major cell model in ferroptosis research, we observed a strong inverse correlation between lipid peroxidation level and the affinity toward NADPH in HeLa and A549 cells (Supplementary information, Fig. S7).
We next examined how the affinity between FSP1 and NADPH affected cell death. The FSP1KO HT1080 cells were complemented with FSP1 WT or mutants, and then treated with 300 nM of RSL3 for 3 h. RSL3 induced dramatically more cell death in cells transfected with AAP and E156A, relative to FSP1 WT (Fig. 1i, j). To further investigate the importance of NADPH binding in ferroptosis, we examined cell viability under RSL3 treatment. Cells with FSP1 WT showed significantly higher cell viability than those with AAP or E156A (Fig. 1k). Thus, binding NADPH, but not NADH, is critical for FSP1 to suppress ferroptosis.
In summary, we combined biochemical, structural, and cellular studies and showed that the ferroptosis suppressor FSP1 is a NADPH-selective enzyme, in contrast with AIFM1 that specifically interacts with NADH. More importantly, the anti-ferroptosis activity of FSP1 strictly depends on its affinity toward NADPH. Thus, all three ferroptosis surveillance systems utilizes NADPH, but not NADH, as electron donors (Fig. 1l). Our results are in line with previous studies that suggested NADPH as a marker for ferroptosis sensitivity.14,15 However, to the best of our knowledge, our study is the first one that quantitatively demonstrate this relationship. Given the important role of FSP1 in cancer therapy, the elucidation of its structure and reaction mechanism may greatly aid the discovery and development of novel therapeutic approaches.
Supplementary information
Acknowledgements
We thank members of our laboratory for helpful discussions. This work was supported by the National Key R&D Program of China (2022YFA1105200), the National Natural Science Foundation of China (92254302), and the National Science Fund for Distinguished Young Scholars (32125012).
Author contributions
S.Z. performed biochemical and structural studies together with Q.Z. S.G. performed cellular studies, and X.Y. provided technical assistance. B.G. provided intellectual assistance and edited the manuscript. D.J. supervised the project and prepared the manuscript with input from every author.
Data availability
Structure factor and atomic coordinates were deposited to the Protein Data Bank with accession code 8JSC.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Sitao Zhang, Shengsong Gou, Qian Zhang.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-023-00879-z.
References
- 1.Dixon SJ, et al. Cell. 2012;149:1060–1072. doi: 10.1016/j.cell.2012.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lei G, Zhuang L, Gan B. Nat. Rev. Cancer. 2022;22:381–396. doi: 10.1038/s41568-022-00459-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stockwell BR, Jiang X, Gu W. Trends Cell Biol. 2020;30:478–490. doi: 10.1016/j.tcb.2020.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang X, Stockwell BR, Conrad M. Nat. Rev. Mol. Cell Biol. 2021;22:266–282. doi: 10.1038/s41580-020-00324-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, et al. Cell Death Dis. 2020;11:88. doi: 10.1038/s41419-020-2298-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doll S, et al. Nature. 2019;575:693–698. doi: 10.1038/s41586-019-1707-0. [DOI] [PubMed] [Google Scholar]
- 7.Bersuker K, et al. Nature. 2019;575:688–692. doi: 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mishima E, et al. Nature. 2022;608:778–783. doi: 10.1038/s41586-022-05022-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herrmann JM, Riemer J. Biol. Chem. 2021;402:289–297. doi: 10.1515/hsz-2020-0254. [DOI] [PubMed] [Google Scholar]
- 10.Brosey CA, et al. Structure. 2016;24:2067–2079. doi: 10.1016/j.str.2016.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferreira P, et al. Biochemistry. 2014;53:4204–4215. doi: 10.1021/bi500343r. [DOI] [PubMed] [Google Scholar]
- 12.Feng Y, et al. Nature. 2012;491:478–482. doi: 10.1038/nature11541. [DOI] [PubMed] [Google Scholar]
- 13.Heikal A, et al. Mol. Microbiol. 2014;91:950–964. doi: 10.1111/mmi.12507. [DOI] [PubMed] [Google Scholar]
- 14.Shimada K, Hayano M, Pagano NC, Stockwell BR. Cell Chem. Biol. 2016;23:225–235. doi: 10.1016/j.chembiol.2015.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding CC, et al. Nat. Metab. 2020;2:270–277. doi: 10.1038/s42255-020-0181-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Structure factor and atomic coordinates were deposited to the Protein Data Bank with accession code 8JSC.