Abstract
Ribosomes translating bacteriophage T4 gene 60 mRNA bypass 50 noncoding nucleotides from a takeoff site at codon 46 to a landing site just upstream of codon 47. A key signal for efficient bypassing is contained within the nascent peptide synthesized prior to takeoff. Here we show that this signal is insensitive to the addition of coding information at its N terminus. In addition, analysis of amino-terminal fusions, which allow detection of all major products synthesized from the gene 60 mRNA, show that 50% of ribosomes bypass the coding gap while the rest either terminate at a UAG stop codon immediately following codon 46 or fail to resume coding. Bypassing efficiency estimates significantly lower than 50% were obtained with enzymatic reporter systems that relied on comparing test constructs to constructs with a precise excision of the gap (gap deletion). Further analysis showed that these estimates are distorted by differences between test and gap deletion functional mRNA levels. An internal translation initiation site at Met12 of gene 60 (which eliminates part of the essential nascent peptide) also distorts these estimates. Together, these results support an efficiency estimate of ∼50%, less than previously reported. This estimate suggests that bypassing efficiency is determined by the competition between reading signals and release factors and gives new insight into the kinetics of bypassing signal action.
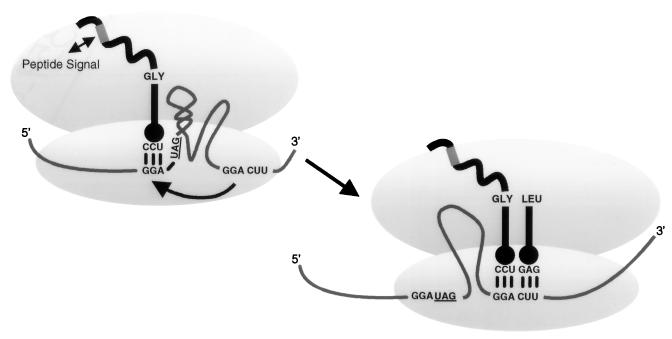
Bacteriophage T4 gene 60 mRNA, which encodes a subunit of the type II T4 DNA topoisomerase, contains a 50-nucleotide (nt) coding gap between codons 46 and 47 that is bypassed by ribosomes in order to synthesize the full-length protein (10). The cis-acting recoding signals required for this unusual event have been studied in detail (23). These signals are depicted in Fig. 1 and include (i) matching GGA codons bordering the gap (referred to as takeoff and landing sites), (ii) a stop codon immediately after the takeoff site, (iii) a short stem-loop structure in the 5′ end of the coding gap, (iv) an optimal 50-nt spacing, and (v) a region of the nascent peptide translated from codons preceding the gap.
FIG. 1.
Efficient T4 gene 60 translational bypassing requires five bypassing signals. These signals include matching GGA codons bordering the gap, a stop codon, a short stem-loop structure, an optimal 50-nt spacing, and a region of the nascent peptide.
Three steps describe the proposed mechanism of bypassing. The first involves dissociation of the codon-anticodon pairing between the peptidyl tRNA2Gly and the GGA at the takeoff site. The second is movement of the mRNA through the ribosome, which brings the landing site to the peptidyl tRNA. The third occurs when pairing is reestablished at the second GGA and coding resumes.
The critical part of the nascent peptide is amino acids 17 to 34, although positions on either side of this region are also important (23). It is unclear whether this signal exerts its effect within the peptide exit channel of the ribosome or in some other location. Although the spacing between the sequence encoding the critical region of the nascent peptide and the start of the coding gap is known to be important (23), the spacing between the amino terminus and the critical region of the nascent peptide has not been analyzed, nor has the importance of the identity of the amino terminus.
The efficiency of gene 60 bypassing has been determined in vivo with gene 60-lacZ reporter constructs driven by the tac promoter. In these experiments, efficiency was determined by comparing test constructs that contained 5′ fragments of gene 60 (including the coding gap) to constructs with a precise deletion of the coding gap (gap deletion). The first estimate of efficiency determined by Huang and coworkers was 70% (10). These constructs carried gene 60 sequence extending 24 codons downstream of the landing site. Weiss et al. demonstrated that similar constructs, carrying gene 60 sequence extending either 24 codons or 5 nt downstream of the landing site, gave efficiencies of 94 or 98%, respectively (23). This estimate is consistent with the inability to detect product due to termination at the stop codon following codon 46 (11). However, lability of the 46-amino-acid peptide may also account for this absence. Here, we reexamine the efficiency of bypassing and investigate the importance of the N terminus to the function of the nascent peptide signal.
MATERIALS AND METHODS
Bacteria.
Escherichia coli K-12 SU1675 {ara Δ(pro-lac) recA56 thi F′ [proAB+ lacIq]} is a recA derivative of CSH26 (22) and was used as a host strain in all experiments except those with the phoA fusions, where DH5α (phoA) (16) was used.
Plasmids.
All changes in the following constructs were verified by dideoxy sequencing as described previously (22). A variant with a precise excision of the coding gap was made in parallel with each test vector.
(i) Construction of gene 60-lacZ fusions.
The parent vector (4p101) used in the construction of the following lacZ fusions has been described previously (22). It is a pBR322-based vector that allows gene fusions to be made to the fifth codon of lacZ with unique HindIII and ApaI sites. A synthetic Shine-Dalgarno (SD) sequence, based on the lpp gene from E. coli, is cloned between the HindIII site and an upstream XbaI site. Translation begins at an AUG codon contained within this insert and results in the addition of 4 amino acids to the amino terminus of the nascent peptide of the downstream fusion. A tac promoter on a 269-bp fragment from pKK223-3, cloned into a BamHI site just upstream of the XbaI site, directs transcription of the gene fusion (4). Derivatives of this vector carrying fragments of gene 60 between the HindIII and ApaI sites have previously been used to estimate bypassing efficiency (10, 23). One of these, BA3 (23), was recreated by cloning an insert composed of four overlapping DNA oligonucleotides into the HindIII and ApaI sites of 4p101. This derivative was designated BX3*.
The tac promoter and SD sequence of 4p101 were replaced in all pGG vectors by the synthetic promoter and SD region indicated in Table 1 by cloning oligonucleotide inserts into the BamHI and HindIII sites. Changes in the coding sequence of gene 60 were constructed by using digested PCR products amplified from pT60.32 (10) and oligonucleotides with embedded HindIII and ApaI restriction sites.
TABLE 1.
Summary of gene 60-reporter fusionsa
| Plasmid | Promoter | RBSb | 5′ additionc | No. of codons after landing sited | U of activity withe:
|
Avg bypassing efficiency ± SD (%)f | |
|---|---|---|---|---|---|---|---|
| Test fusion | Δgap fusion | ||||||
| BX3* | tac | UAGAGGGUAUUAUG | AUGAAAAGCUUA(AUG) | 2 | 6,107 ± 490 | 24,270 ± 2,540 | 24.7 ± 1.4 |
| pSK31 | tac | Same as for BX3* | GST | 7 | 4,432 ± 85 | 31,648 ± 1,956 | 14.0 ± 0.8 |
| pPG33 | tac | Same as for BX3* | T7 gene 10 | 7 | 5,042 ± 120 | 42,156 ± 1,895 | 12.0 ± 0.5 |
| pPA31 | tac | Same as for BX3* | ProtA-CAT | 7 | 11,484 ± 590 | 49,852 ± 1,890 | 23.0 ± 1.4 |
| NH11 | λ pLpR | bla signal peptide | 24 | 81 ± 6 | 216 ± 12 | 37 ± 3 | |
| GLZ16 | Consensus | UCAGGAAGCUUAUG | 2 | 140 ± 3 | 2,273 ± 27 | 6.2 ± 0.1 | |
| GLZ22 | Consensus | Same as for GLZ16 | 24 | 99 ± 5 | 960 ± 28 | 10.3 ± 0.3 | |
| GLZ30 | tac | Same as for GLZ16 | 24 | 7,010 ± 1,400 | 18,580 ± 1,170 | 37 ± 4 | |
| GLZ32 | Consensus | UCCGGAAGCUUAUG | 24 | 69 ± 2 | 706 ± 20 | 9.8 ± 0.3 | |
| GLZ34 | Consensus | UGAGGAAGCUUAUG | 24 | 209 ± 16 | 1,320 ± 60 | 15.9 ± 0.6 | |
| GLZ40 | Consensus | AUGAGGUGUAUAAUG | 24 | 207 ± 24 | 1,370 ± 35 | 15.2 ± 1.4 | |
| GLZ49 | Consensus | UAGGAGGAAGCUUAUG | 24 | 131 ± 5 | 919 ± 45 | 14.3 ± 0.2 | |
| pGG34 | tac | CAGGAAACAGAAGCUUAUG | 2 | 19,220 ± 3,540 | 72,450 ± 6,232 | 26 ± 5 | |
| pGG36 | lacUV5 | Same as for pGG34 | 2 | 4,250 ± 820 | 23,620 ± 2,750 | 18 ± 4 | |
| pGG45 | tac | Same as for pGG34 | 7 | 10,940 ± 950 | 52,080 ± 3,820 | 21 ± 2 | |
| pGG47 | tac | Same as for pGG34 | 24 | 14,200 ± 1,620 | 50,640 ± 4,520 | 28 ± 4 | |
All reporter fusions were made to lacZ except that with NH11, which was made to phoA.
The underlined region is the SD region of the ribosome-binding sequence (RBS); the boldface region is the initiation codon in each construct.
In the sequence, the initiation codon is in boldface type, and the beginning of the gene 60 coding sequence is in parentheses. T7 gene 10, 5′ fragment of T7 gene 10; ProtA, immunoglobin-binding domain from protein A. BX3* carries two transversions at nt 87 and 89 that create an internal BglII site and that do not affect bypassing efficiency.
Number of gene 60 codons between the landing site and reporter fusion.
Activities are expressed in β-Gal units, except for that for NH11, which is expressed in phosphatase units. Values are averages of multiple independent measurements. Δgap, gap deletion.
Values are average efficiency estimates from several independent experiments.
The parent vector for all GLZ constructs described here (GLZ16) was made in two steps. First, the HindIII-EcoRV fragment of BX3* was cloned into a pBR322-based vector that carried the 3.1-kb SalI lacZ fragment of pMC1871 (18) in the SalI site oriented toward the origin of replication. Second, the −35 sequence of the tet promoter from the resulting plasmid was removed by digestion with EcoRI and HindIII. This sequence was replaced with an oligonucleotide insert that introduced (i) the ς70 E. coli consensus promoter (21) under the control of the lac operator (5) (flanked upstream by a KpnI site and downstream by an XbaI site) and (ii) a synthetic SD sequence (flanked by XbaI and HindIII sites). All subsequent promoter and SD sequence modifications were made by cloning oligonucleotide inserts into these three sites. Further changes to the coding sequence of gene 60 were made by the PCR cloning strategy mentioned above.
Vectors RW201 (a gift of R. Weiss, University of Utah), SKAGGS (a gift of S. Matsufuji, Jikei University, Tokyo, Japan), and pG10Z (a gift of O. Fayet, CNRS, Toulouse, France) are all derivatives of 4p101 and add the following respective coding sequences to the N terminus of the lacZ fusions: the immunoglobulin G-binding domain of protein A fused to a fragment of chloramphenicol acetyltransferase (CAT), Schistosoma japonicum glutathione S-transferase (GST), and T7 gene 10. Gene 60 sequences from the first codon to 7 codons after the landing site were introduced in frame with each N-terminal fusion by the PCR cloning strategy mentioned above.
(ii) Construction of gene 60-phoA fusions.
To make gene 60-phoA fusions, gene 60 sequence, from the initiation codon to 24 codons after the landing site, was amplified by PCR with oligonucleotides containing embedded NheI sites, digested with NheI, and cloned into pCGV1 (8). The forward primer includes sequence required to complete the bla signal peptide present in the vector. This inclusion ensures that the fusions are exported to the periplasm. Expression is driven by the pR and pL promoters of lambda phage and controlled by the thermosensitive repressor cI857, which is present on the vector.
(iii) Construction of gene 60 and GST-gene 60 plasmids.
To construct gene 60 expression plasmids, under the control of the tac promoter, the entire coding sequence of gene 60 plus an additional 45 nt upstream of the start codon was amplified by PCR from pT60.32 (10) with oligonucleotides containing embedded SmaI and HindIII sites, digested with SmaI and HindIII, and cloned into pKK223-3 (4). To construct GST-gene 60 expression plasmids, the coding sequence of gene 60 was amplified from pT60.32 with oligonucleotides containing embedded BamHI sites, digested with BamHI, and cloned into pGEX2-T in frame with GST (19). Exact gap deletion variants of these clones were constructed by amplifying DNA from a version of pT60.32 that already contained the coding gap deletion (1).
β-Gal activity assays.
Whole-cell assays were based on the procedure of Miller (14), and data obtained are expressed as β-galactosidase (β-Gal) units. Overnight cultures of constructs with the consensus promoter (Table 1) were diluted 1:100 in Luria broth (LB) plus 2 mM IPTG (isoproply-β-d-thiogalactopyranoside), grown at 37°C with rapid shaking, and chilled to 0°C when they had reached an optical density at 600 nm of 0.4 to 0.8. Overnight cultures of constructs with the tac promoter (Table 1) were diluted 1:50 in LB and were induced after 1 h of growth with 2 mM IPTG for an additional 1.5 h. Assays were done at 30°C in a total volume of 1 ml of Z buffer plus cell suspension (14). The substrate was added as 0.2 ml of 4-mg/ml O-nitrophenol-β-galactoside, and the reaction was stopped by the addition of 0.5 ml of 1 M Na2CO3. All assays were performed in duplicate with at least two independent clones on at least two separate occasions. The absolute β-Gal values and resulting bypassing efficiency estimates along with standard deviations are given in Table 1.
Phosphatase assays.
Gene 60-phoA fusions were partially derepressed at 39°C in DH5α (16). The expression of protein was estimated with the standard colorimetric phosphatase assay (3).
In vivo pulse-chase experiments.
All pulse-chase experiments were done in duplicate with at least two independent clones on at least two separate occasions.
(i) Bypass efficiency determination.
Overnight cultures were grown in VBMAM-AM medium (0.5% glucose; 0.1% Difco methionine assay medium; 60 μg [each] of isoleucine, leucine, and valine per ml; 8 mM NaOH; 1× E salts [20a]; 50 μg of ampicillin per ml [5a]) and diluted 1:20 in 1 ml of the same. After a 2-h incubation at 37°C, the cultures were induced with 2 mM IPTG for 8 min. The cells were pulsed with 7.5 μCi of [35S]methionine in 100 μl of VBMAM-AM for 30 s, chased with 100 μl of 4% cold methionine for 3 min, chilled on ice, and harvested by centrifugation. The cells were resuspended in 50 μl of cracking buffer (6 mol of urea per liter, and 1% sodium dodecyl sulfate [SDS] in 1× stacking buffer at Tris [pH 7.2]) (2), boiled 5 min, combined with 50 μl of 1× SDS loading buffer (17), and boiled an additional 5 min before 10-μl aliquots were loaded onto a Tris-glycine–SDS–13% polyacylamide gel. The resulting data were visualized with a Molecular Dynamics PhosphorImager. To control for loading differences, the amount of labeled gene 60 product was compared to that of the total labeled protein per lane. The resulting percentage was then used to estimate bypassing efficiency. Full-length GST-gene 60 fusions have 0.044 methionine per amino acid; the product due to termination following codon 46 has 0.0478 methionine per amino acid. This difference was used to correct the amount of the full-length test product before we calculated the final ratio.
(ii) Functional message stability determination.
The basic procedure outlined above was performed with the following modifications. Overnight VBMAM-AM cultures were diluted 1:20 in 7 ml of the same medium. Following 2 h of incubation, the samples were induced with 2 mM IPTG for 8 min. At 8 min, 0.5-ml aliquots of culture were transferred to separate tubes containing 50 μl of labeling mix, where they were chased as described above with 50 μl of 4% cold methionine. Rifampin (500 μg/ml) was added to the remaining cultures, and 0.5-ml aliquots were removed and assayed after 1, 2, 3.5, 5, and 10 min. The amount of labeled gene 60 product per lane was determined for three independent clones. To correct for loading differences among the clones, the result for each clone was multiplied by the ratio of the amount of total labeled protein in its lane to the average amount of total labeled protein in the lanes of all three clones. The resulting data points were then expressed as percentages of the amount of gene 60 synthesized prior to rifampin addition. Logarithmic plots of the resulting data points were made with Microsoft Excel. Exponential regression was used to derive the best-fit curve for the collection of data points over which exponential decay occurred. These curves were y = 146 × e−0.78x (r2 = 0.998) and y = 269 × e−0.76x (r2 = 0.985), respectively, for gene 60 (Fig. 2A, upper section) test and gap deletion constructs. For GLZ30 test and gap deletion constructs, these were y = 92 × e−0.67x (r2 = 0.992) and y = 244 × e−0.54x (r2 = 0.997), respectively.
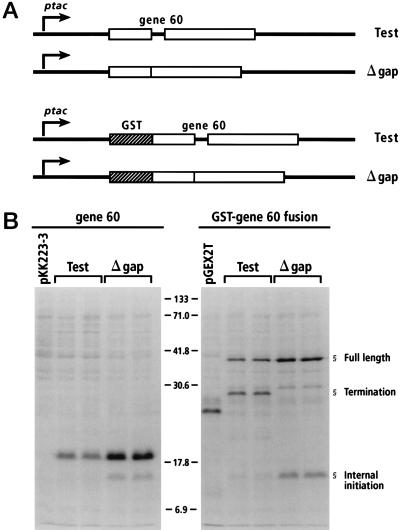
FIG. 2.
Bypassing efficiency with constructs containing entire gene 60 coding sequence. (A) Constructs. Abbreviations: Test, test construct; Δgap, gap deletion construct. (B) Pulse-chase analysis of two independent clones of each construct. Molecular mass indicators in the center are in kilodaltons. The product of bypassing in the left-hand section is 18 kDa. The product of bypassing in the right-hand section is 40 kDa. The left-hand lane of each section shows the product of strains carrying the parental plasmid.
(iii) Functional message accumulation determination.
The basic procedure outlined under “Bypass efficiency determination” was followed with the following modifications. Overnight VBMAM-AM cultures were diluted 1:20 into 7 ml of the same medium. Following a 2-h incubation at 37°C, the samples were induced with 2 mM IPTG. Samples (0.5 ml) were removed just prior to induction and then at 2, 5, 10, 30, and 60 min after induction and assayed as described above.
Protein purification and sequencing of internal initiation product.
An overnight culture containing the GST-gene 60 gap deletion construct was diluted 1:100 in LB with 2 mM IPTG and incubated for 2 h at 37°C with shaking. Cells (1.5 ml) were recovered by centrifugation and lysed under the conditions described above for the pulse-chase experiments. A 15-μl sample was then run on a Tris-glycine–SDS–15% polyacylamide gel. Following staining with Coomassie blue, the gel was electroblotted onto an Immobilon-P polyvinylidine difluoride 0.2-μm-pore-size transblot membrane. The band of interest was cut out, and the N terminus of the protein was sequenced through nine cycles with an Applied Biosystems model 477 protein sequencer.
Northern blot analysis.
Overnight cultures were diluted 1:100 in 10 ml of LB, grown for 2 h, and induced with 2 mM IPTG for 30 min. Total RNA was then purified as described previously (9). Thirty-microgram samples of RNA were loaded per lane, resolved by formamide agarose gel electrophoresis, and visualized by ethidium bromide staining (17). The RNA was transferred by capillary transfer to an Amersham Hybond N+ nylon membrane, cross-linked by UV irradiation as recommended by the manufacturer, and hybridized with probe prepared from the 3.1-kb ApaI-SalI lacZ and the 0.7-kb DraI β-lactamase fragments of GLZ22 with a Stratagene Prime It-II kit. The blots were then visualized and the amount of hybridization was quantified with a Molecular Dynamics PhosphorImager. The amount of hybridization to β-lactamase was used to normalize the amount of lacZ mRNA in each lane.
RESULTS
Amino-terminal fusions.
Previously, it has been shown (23) that a small addition of 4 amino acids to the amino terminus of the nascent peptide of gene 60 does not alter bypassing efficiency. We confirmed and extended this finding by adding 1 to 7 amino acids to the amino terminus without observing a significant effect (data not shown). To test if larger additions would alter the nascent peptide signal, GST, gene 10, and protein A-CAT coding regions were added separately to the 5′ end of gene 60-lacZ (to produce pSK31, pPG33, and pPA31, respectively) (Table 1). BX3*, which carries 4 codons at the amino terminus, serves as a control in these experiments. The efficiency estimates of 14, 12, 23, and 25% (Table 1) indicate that additions of GST and gene 10 reduce bypassing efficiency twofold. This may be the result of either a reduction in the effectiveness of the nascent peptide signal or simply differences in the levels of stability or functions of mRNAs or protein products of gap deletion constructs. (Note that β-Gal activities for pSK31, pPG33, and BX3* test constructs are comparable [4,395, 5,005, and 5,454 U, respectively] but that the values for the gap deletion controls differ [32,000, 41,440, and 23,709 U] [Table 1].)
The products of the amino-terminal fusions described thus far are all destined for the cytoplasm. To test if the action of the nascent peptide signal is altered by the process of export to the periplasm, gene 60-phoA fusions were made with the bla signal peptide fused to the N terminus (NH11) (Table 1) (8). These fusions gave a bypassing efficiency of 37% (Table 1). It is not clear, however, whether the increase in efficiency relative to that determined for BX3* should be interpreted as a minor stimulation of bypassing or a reflection of differences between the two reporter systems. The interpretation of this and the above-described results is further complicated by the observation that the bypassing efficiency reported here for BX3* (∼25%) is substantially lower than those of previous studies (70 and 98% [10, 23]) of similar lacZ fusions.
The following steps were taken to better define bypassing efficiency and the variables which affect it. First, a more accurate bypassing efficiency estimate, with and without a GST fusion, was made with gene 60 expression vectors containing the entire coding sequence without a downstream fusion. This step tests the hypothesis that the GST fusion in pSK31 (and by analogy the gene 10 fusion of pPG33) interferes with the nascent peptide signal. Second, the following variables that might influence estimates of bypassing efficiency were analyzed: (i) the distance between the landing site and the junction with the downstream fusion, (ii) the functional stability of test and control mRNAs, (iii) the level of transcription, and (iv) the level of ribosome loading. The first variable evaluates whether additional gene 60 information downstream of the landing site modulates bypassing. The second addresses whether functional mRNA stability differences between test and control constructs play a role in the variability seen in bypassing estimates. The third addresses whether high expression levels titrate a component of the translation machinery that either stimulates or represses bypassing. The fourth investigates whether increased numbers of translating ribosomes upstream of the takeoff site stimulate bypassing.
A new estimate of bypassing efficiency.
Test and gap deletion gene 60 constructs were made to measure bypassing efficiency in the absence of any fusion (Fig. 2A, upper section). In these constructs, 45 nt of the 5′ wild-type sequence were included so that translation begins from the natural initiation site of gene 60. GST-gene 60 fusions were made to reevaluate whether additions to the amino terminus alter the effectiveness of the nascent peptide signal (Fig. 2A, lower section). The constructs were analyzed by [35S]Met pulse-chase analysis (Fig. 2B), which assays protein synthesis within a 30-s window (see Materials and Methods). Three independent clones per construct were analyzed on multiple separate occasions. Shown here (Fig. 2B) are two representative clones for each construct alongside strains carrying the parental plasmids as controls.
Successful bypassing during translation of gene 60 in the absence of any fusions yields a product of 18 kDa. An efficiency of 54% (±4%) was estimated by comparing the amount of this product in the test lanes to that in the gap deletion control lanes (Fig. 2B, left gel). The product of bypassing translated from GST-gene 60 message is 40 kDa. Comparing the total amounts of this product in test and gap deletion lanes (Fig. 2B, right gel) gives a bypassing efficiency of 50% (±4%). Translation of test messages also produces a 26-kDa termination product resulting from the failure to bypass (Fig. 2B, right gel). The fraction of the product of bypassing (test, 40 kDa) to the total protein translated from test message (40 + 26 kDa) gives an efficiency estimate of 50% (±3%). These efficiency estimates are significantly less than those previously reported (10, 23) and indicate that GST does not substantially inhibit nascent peptide signaling. Furthermore, this revised estimate suggests that addition of the bla signal peptide does not drastically alter bypassing efficiency.
Internal initiation.
A secondary product of ca. 16 kDa is apparent in both gene 60 and GST-gene 60 gap deletion lanes (Fig. 2B). The same product is also clearly visible in all test lanes after a longer exposure. To learn its identity, the gap deletion GST-gene 60 construct was overexpressed, the 16-kDa product was purified, and the N terminus was sequenced by Edman degradation. The results (Fig. 3) show that this product results either from internal initiation at Met12 of gene 60 or from proteolysis of the N-terminal region. Previous studies have shown that similarly sized N-terminal deletions of gene 60 reduce bypassing efficiency 10-fold (23). The test 16-kDa product was 5% of the 16-kDa gap deletion product. This finding is consistent with a 10-fold reduction in bypassing efficiency and suggests that this product results from internal initiation. The region around Met12 lacks a strong SD sequence or downstream box, yet the 16-kDa gap deletion band is 30% of the 18-kDa main initiation band, indicating that ribosome loading is fairly efficient from this site. This result implies that internal initiation may distort efficiency estimates in systems where the test-to-gap deletion control ratio is determined by comparing the enzymatic activities of downstream reporter genes.
FIG. 3.
Amino-terminal sequencing of the gel-purified internal initiation product. The amount of product in each cycle is in picomoles. The derived sequence is boxed.
Distance between landing site and junction with lacZ.
While being less than those from previous reports, the above-mentioned estimates are still greater than those obtained with the lacZ fusions described in the first section. One possible variable that may account for these differences is the amount of gene 60 coding information downstream of the landing site. Almost all of the fusions tested above, except for BX3* and the gene 60-phoA fusions, carry 7 codons downstream of the landing site instead of the 2 or 24 codons previously reported (Table 1) (23). To address the issue of whether bypassing efficiency is independent of the distance between the landing site and the junction with lacZ, we compared three isogenic constructs that had 2, 7, and 24 codons between the landing site and lacZ (pGG34, pGG45, and pGG47, respectively) (Table 1). These had bypassing efficiencies of 26, 21, and 28%, respectively, indicating that there are no strict spacing requirements.
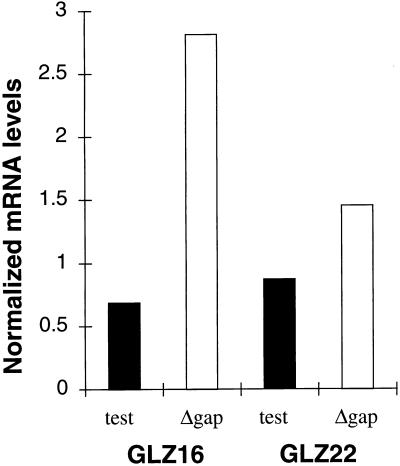
A second set of constructs (GLZ16 and GLZ22) (Table 1), driven by the E. coli consensus promoter, tested the spacing of 2 and 24 codons. Test constructs show similar β-Gal activity levels (142 and 101 U), while levels for the gap deletion control constructs are different (2,255 and 950 U). To try to account for this difference, steady-state levels of the four messages were assayed by Northern blot analysis with a probe for the lacZ portion of the message. Shown (Fig. 4) are the results from a single experiment. The GLZ16 gap deletion control has nearly twice the amount of lacZ mRNA as the GLZ22 gap deletion control. Even more striking, both gap deletion controls have substantially more lacZ mRNA than their test counterparts. All constructs under the control of the consensus promoter (Table 1) were tested and displayed similar differences between test and gap deletion constructs (data not shown).
FIG. 4.
Steady-state mRNA levels of GLZ16 and GLZ22 determined by Northern blot analysis. GLZ16 and GLZ22 are isogenic constructs that differ only in their amounts of gene 60 sequence between the landing site and the junction with lacZ (Table 1). Test, test construct; Δgap, gap deletion construct. Amounts of lacZ mRNA per lane were normalized to the amount of bla mRNA per lane. The data shown represent results of a single experiment (see the text).
These data may be explained by differences in either mRNA physical decay rates or steady-state functional mRNA levels. lacZ mRNA physical stability is determined by the inherent susceptibility of the message to degradation and, as shown by McCormick and coworkers, the number of translating ribosomes per message (increased translation is correlated with increased physical stability) (13). This same study observed a uniform functional mRNA decay rate regardless of translation efficiency. This and other results led the investigators to propose a pathway of degradation for lacZ mRNA which does not substantially interfere with translating ribosomes (13). Thus, if the proportion of ribosomes which bypass successfully is low enough, there may be a corresponding reduction in the stability of test messages that does not alter bypassing efficiency estimates.
Functional mRNA levels are determined by three components: transcription initiation frequency, transcription efficiency (the percentage of nascent transcripts transcribed to completion), and functional message decay rate (the rate protein synthesis capacity disappears). Differences between test and gap deletion constructs in any of the three will distort bypassing efficiency estimates. Below we assess whether differences in functional stability exist between test and gap deletion constructs and if these differences result in different functional mRNA levels.
Gene 60 functional mRNA stability.
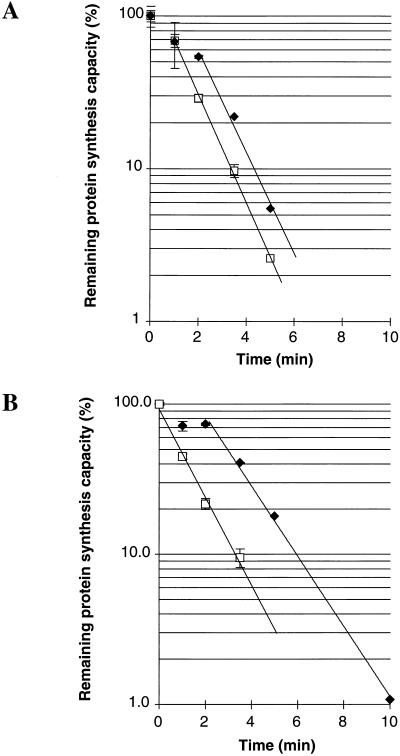
Functional message half-lives of test and gap deletion gene 60 mRNAs (Fig. 2A, top section) were determined by pulse-chase analysis of total protein with [35S]Met following rifampin addition (Fig. 5A). Previous work has shown that a lag in the loss of protein synthesis is often observed following rifampin addition (13). This lag is attributed to an increased translation efficiency of longer-lived transcripts as ribosomes released from faster-decaying transcripts increase the available ribosome pool. Following this lag, protein synthesis rates decay exponentially and represent the actual functional half-life of the mRNA (13). Exponential regression was used to model the exponential decay seen from 1 to 3.5 min for test mRNA (Fig. 5A) and from 2 to 5 min for gap deletion mRNA (Fig. 5A). From these tests, functional message half-lives of 0.89 min for test mRNA and 0.91 min for gap deletion mRNA were calculated. Thus, bypassing estimates from these constructs are not distorted by differences in functional mRNA decay rates.
FIG. 5.
Functional message stability of gene 60 and gene 60-lacZ mRNA. Plots represent the amounts of protein synthesis determined by pulse-chase analysis of induced clones at different times following rifampin addition. Each data point represents the percentage of product synthesized at the given time point relative to the amount synthesized prior to rifampin addition. Error bars represent the standard deviation at each point from results of three independent experiments. Exponential regression was used to generate the curves for each set of data points (given in Materials and Methods). (A) Test (open squares) and gap deletion (filled diamonds) gene 60 (Fig. 2A, upper section) functional mRNA decay. Half lives are 0.89 and 0.91 min, respectively. (B) Test (open squares) and gap deletion (filled diamonds) GLZ30 (Table 1) functional mRNA decay. Half lives are 1.0 and 1.3 min, respectively.
Functional message half-lives were also determined for gene 60-lacZ (GLZ30) (Table 1) test and gap deletion mRNAs (Fig. 5B). Exponential regression was used to determine the best-fit curves for the decay seen between 0 and 3.5 min for test mRNA (Fig. 5B) and 2 and 10 min (Fig. 5B) for gap deletion mRNA. Functional half-lives of 1 and 1.3 min were calculated for test and gap deletion messages, respectively, from the resulting equations. This difference in levels of functional mRNA stability leads to differences in functional mRNA levels that likely lower any estimates of bypassing efficiency made with gene 60-lacZ fusions.
Levels of transcription and accumulation of gap deletion lacZ mRNA.
All studies previous to this one have relied on gene 60-lacZ fusions driven by the tac promoter to derive bypassing efficiency estimates. It is possible that high levels of expression titrate a component of the translational machinery that either stimulates or represses bypassing. The constructs discussed above (GLZ16 and pGG34) suggest that efficiency increases with expression levels. To test this suggestion further, we made two sets of constructs that were isogenic except for the promoters. These are GLZ22, GLZ30, pGG36, and pGG34 (Table 1). Bypassing efficiencies for the first two (driven by either the consensus or the tac promoter) are 10 and 32%, respectively. Bypassing efficiencies for the last two (driven by either the lacUV5 or tac promoter) are 18 and 26%, respectively.
From the percentages alone, one might conclude that a factor that represses bypassing is being titrated at higher levels of expression. Caution should be used here, as expression of gene 60-lacZ gap deletion fusions, under the control of the tac promoter, severely compromises cell growth. This compromised cell growth may result from toxicity due to high intracellular concentrations of the β-Gal fusion or from titration of essential components required for growth. For example, the functional stability difference noted above for GLZ30 may lead to an accumulation of gap deletion mRNA that exceeds the translational capacity of the cell.
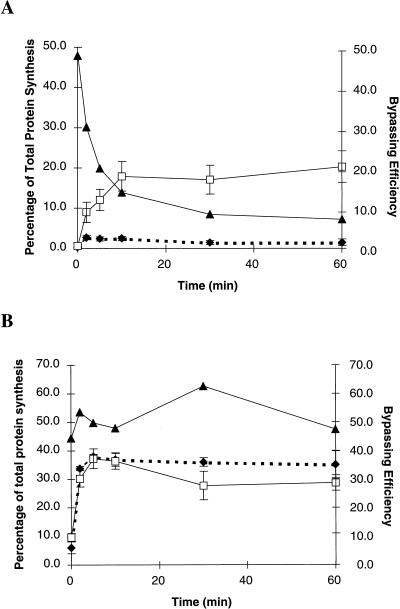
To examine the dynamics of translation during high levels of expression, translation of GLZ30 test and gap deletion mRNAs was monitored by [35S]Met pulse-chase analysis at different time points after induction (Fig. 6A). The amount of product is presented as the percentage of total protein synthesis. Translation from gap deletion mRNA (Fig. 6A) increases steadily over the first 10 min, whereas the amount of test product synthesis (Fig. 6A) reaches steady state by 2 minutes. As a result, bypassing efficiency drops sharply during the first 10 min (Fig. 6A). This dramatic difference between test and gap deletion translation levels suggests that gap deletion functional mRNA accumulates rather than reaches steady state. In the gap deletion lanes, increasing amounts of aggregated protein in the wells is observed with respect to time (data not shown). If this protein is considered to be largely β-Gal, the level of functional gap deletion mRNA continues to increase until it accounts for nearly 50% of all protein synthesis by 60 min.
FIG. 6.
Accumulation of functional mRNA following IPTG induction. Amounts of protein from test mRNA (filled diamonds) and gap deletion mRNA (open squares) were determined by pulse-chase analysis at different time points after IPTG induction and analyzed as percentages of the total cellular protein synthesized. The standard deviation from results of three independent experiments was used to estimate the error for each data point. These data are plotted relative to the scale on the left. Bypassing efficiency (filled triangles) is plotted relative to the scale on the right. (A) GLZ30 (Table 1). Note that this plot does not include the aggregation in the wells mentioned in the text. (B) GST-gene 60. Amounts of product synthesized from test mRNA include both full-length and termination products (Fig. 2A).
It appears from this result that GLZ30 gap deletion mRNA does not reach steady state. In contrast, Northern blot analysis of GLZ22 gap deletion mRNA at 10 and 30 min after induction indicates that it does reach steady state (data not shown). Thus, the ratio of test-to-gap deletion functional mRNA is constant for GLZ22 but varies for GLZ30, making meaningful comparisons of bypassing efficiency difficult. The GLZ30 bypassing estimate is further complicated by the possibility that aggregation in gap deletion-expressing cells reduces enzyme activity. Given this possibility, it seems likely that the increase in bypassing efficiency seen with high levels of expression is the result of a combination of artifacts.
To address if differences in functional mRNA concentration exist in the GST-gene 60 constructs (Fig. 2A, lower section), protein synthesis following induction was monitored as described above. Total levels of translation from GST-gene 60 test mRNA (Fig. 6B) and gap deletion mRNA (Fig. 6B) appear to be similar, indicating that the functional message levels are the same and remain relatively constant. In addition, there is a lack of aggregation in the wells, suggesting that the estimate of the amount of product synthesized is accurate. Similar results were obtained when the test and gap deletion gene 60 constructs described above (Fig. 2A, upper section) were tested (data not shown).
Level of ribosome loading.
To test if bypassing efficiency is dependent upon translation initiation frequency, we constructed isogenic fusions that differed only in their degrees of Watson and Crick base-pairing potential to the anti-SD region of 16S rRNA (CGGAAGCUUCCUCCAC, GLZ32; AGGAAGCUUCCUCCAC, GLZ22; UUGAGGAAUCCUCCAC, GLZ34; and AGGAGGUGUCCUCCAC, GLZ49; where bases with pairing potential appear in boldface type and anti-SD region sequences appear above SD region sequences) (Table 1). In addition, we made a set of constructs that replaced the synthetic SD region with the same 45 nt of the upstream wild-type sequence used for the gene 60 construct shown in Fig. 2A (AUGAGGUGUCCUCCAC, GLZ140).
The bypassing efficiencies (Table 1) separate the five SD variants into two groups that correlate with anti-SD sequence base-pairing potential: those with efficiencies of 10% (GLZ32 and GLZ22) and those with efficiencies of 14 to 16% (GLZ34, GLZ40, and GLZ49). β-Gal activities (Table 1) provide an indication of whether increased base-pairing potential is correlated with increased ribosome loading. An increase in pairing potential by a single base pair (viz., the difference between GLZ32 and GLZ22) increases ribosome loading yet does not change bypassing efficiency. In contrast, a single base change between the SD sequences of GLZ22 and GLZ34 (that increases the strength and changes the positioning of the predicted pairing) increases apparent bypassing efficiency by 60% while enhancing ribosome loading. A similar increase in bypassing efficiency and ribosome loading is seen between GLZ22 and GLZ40, where the SD sequence change mirrors that of GLZ34. The apparent link between, ribosome loading and bypassing efficiency is supported less well by GLZ49, where ribosome loading appears equivalent to that with GLZ22 yet bypassing efficiency is higher. These data indicate that bypassing efficiency moderately increases once a threshold of SD sequence strength is surpassed.
DISCUSSION
We have shown here that the nascent peptide signal does not require a particular spacing from the amino terminus of the protein to function. Although bypassing efficiency determined from GST-gene 60 fusions (50%) is slightly lower than the efficiency determined from gene 60 that lacks an N-terminal fusion (54%), there are a number of possible explanations for this. Because amino-terminal fusions allow quantification of both failed and successful products of bypassing synthesized from the same mRNA, they will be valuable tools for assaying the effects of various mutations on bypassing efficiency in future studies.
The mechanism by which the nascent peptide signal stimulates bypassing is unknown. It may be that it interacts with the ribosome in the peptide exit channel (7). Interestingly, cryoelectron microscopy experiments have hinted that the 50S subunit may contain more than one exit channel (6, 20) raising the possibility that there are alternate exit paths depending on the character of the growing peptide chain. For instance, proteins destined for export may exit by a different channel than that used for a cytoplasmic protein. In light of this possibility, altering the N-terminal sequence of gene 60 might relocate the emerging nascent peptide within the large subunit. The gene 60-phoA fusions targeting the protein for export suggest that if this is true, it has little bearing on the activity of the nascent peptide signal.
The data presented here support a revised bypassing efficiency estimate of around 50% based on the data from gene 60 and GST-gene 60 constructs (Fig. 2B). These constructs are not subject to the variable functional mRNA levels (Fig. 5 and 6) or internal initiation at Met12 (Fig. 3) that appears to distort efficiency estimates made with gene 60-lacZ fusions. The lack of accumulation of functional gene 60-lacZ test mRNA (expressed from the tac promoter) (Fig. 6A) indicates that the gap may disrupt transcription-translational coupling. This disruption may, in turn, lead to either premature transcription termination or to riboendonuclease cleavage downstream of ribosomes translating lacZ. In either case, ribosomes translating these truncated messages (after bypassing the coding gap successfully) would produce a truncated product not detected by β-Gal assays or included in the quantification of protein by pulse-chase analysis. In fact, IPTG-inducible products smaller than the full-length β-Gal fusion are present in higher concentrations in clones expressing gene 60-lacZ test constructs than in control constructs (data not shown).
There may be evidence for yet another difference between test and gap deletion constructs that we have not addressed directly. Both gene 60 gap deletion and gene 60-lacZ gap deletion clones have longer lag phases before exponential decay than their test counterparts (Fig. 5). The length of the lag phase has been correlated with translation efficiency (13). This correlation may be an indication of differential ribosome loading rates between test and gap deletion constructs. Alternatively, this might reflect the degree of ribosome protection of downstream message.
The observation that bypassing is significantly less than 100% has two interpretations. The first is that release factors have access to the UAG stop codon just 3′ of the takeoff site and that efficiency is determined by the competition between bypassing and termination. Since 1990 (23), any model dealing with bypassing has had to postulate that the UAG codon was inaccessible due to a kinetic or physical barrier erected by the recoding signals. Mechanistically, bypass signals could compete either by interfering with stop codon recognition or by enhancing the dissociation between the peptidyl tRNA and the message. The second interpretation is that all ribosomes initiate bypassing but that only a portion of transiting ribosomes resume translation in the correct reading frame. The termination product (Fig. 2B, right gel) may reflect either a high rate of ribosome dissociation from the message or reinitiation in a different reading frame followed by termination. Experiments to distinguish between these two possibilities are under way.
The results addressing the strength of the ribosome-binding site are intriguing. The increase in bypassing efficiency of 60% upon alteration of a single base pair in the SD sequence suggests that the number of ribosomes upstream of the takeoff site influences the efficiency of bypassing. One possible interpretation is that an upstream ribosome enhances the dissociation of a downstream ribosome complex poised at the takeoff site. Direct ribosome-ribosome contact or application of a force on the mRNA in the 5′ direction might be sufficient to stimulate bypassing. There are several alternative explanations. It may be that increased ribosome loading stabilizes test functional mRNA more than gap deletion mRNA by reducing the effects of uncoupling translation from transcription, as postulated above. A second alternative is that strong ribosome initiation at the main initiation site reduces internal initiation at Met12 (Fig. 3), increasing the percentage of bypassing-competent ribosomes. A final alternative requires two assumptions: first, the mRNA can slip in either direction through the ribosome after initiation of bypassing and, second, the slipping back through the ribosome results in unproductive bypassing events. Given these suppositions, upstream ribosomes may limit message movement through the bypassing ribosome to a forward direction and thereby increase the frequency of successful events.
Defining the range of gene 60 bypassing confirms that it is one of the most efficient of all programmed recoding events (which also include programmed frameshifting and stop codon redefinition [7]). Why it occurs at all is still a mystery. T2 and T6 phages have a gene fusion between gene 39 (an upstream topoisomerase subunit gene) and gene 60. While a portion of the coding sequence upstream of the gap remains, the amino terminus (including the critical region) as well as the coding gap are missing (12). This suggests that bypassing is dispensable for phage development. However, it seems that T4 topisomerase II activity is important for high-efficiency replication, as T4 amber mutants defective for gene 60 display cold-sensitive and DNA delay phenotypes (15). There remains the intriguing possibility that bypassing is regulated during T4 infection as a means of enhancing replication; so far, all measurements of bypass efficiency have been done only in uninfected E. coli cells.
ACKNOWLEDGMENTS
Both authors contributed equally to this work.
We thank Ray Gesteland and John Atkins for encouraging this effort with invaluable advice during both the experiments and the writing of the manuscript. We also thank Mike Howard for helpful discussions and Bob Shackman for assistance with protein sequencing.
This work was supported by the Howard Hughes Medical Institute (via Ray Gesteland) and a grant (to John Atkins) from the NIH (RO1-GM48152-05).
REFERENCES
- 1.Adamski, F. Unpublished data.
- 2.Adamski F. Ph.D. dissertation. Dundedin, New Zealand: University of Otago; 1992. [Google Scholar]
- 3.Brickman E, Bechwith J. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and phi80 transducing phages. J Mol Biol. 1975;96:307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- 4.Brosius J, Holy A. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc Natl Acad Sci USA. 1984;81:6929–6933. doi: 10.1073/pnas.81.22.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dickson R C, Abelson J, Barnes W M, Reznikoff W S. Genetic regulation: the Lac control region. Science. 1975;187:27–35. doi: 10.1126/science.1088926. [DOI] [PubMed] [Google Scholar]
- 5a.Fayet, O. Personal communication.
- 6.Frank J, Zhu J, Penczek P, Li Y, Srivastava S, Verschoor A, Radermacher M, Grassuccl R, Lata R K, Agrwal R K. A model of protein synthesis based on cryo-electron microscopy of the E. coli ribosome. Nature. 1995;376:441–444. doi: 10.1038/376441a0. [DOI] [PubMed] [Google Scholar]
- 7.Gesteland R F, Atkins J F. Recoding: dynamic reprogramming of translation. Annu Rev Biochem. 1996;65:741–768. doi: 10.1146/annurev.bi.65.070196.003521. [DOI] [PubMed] [Google Scholar]
- 8.Guzman C A, Piatti G, Walker M J, Guardati M C, Pruzzo C. A novel Escherichia coli expression-export vector containing alkaline phosphatase as an insertional inactivation screening system. Gene. 1994;148:171–172. doi: 10.1016/0378-1119(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 9.Hajnsdorf E, Steier O, Coscoy L, Teysset L, Regnier P. Roles of RNase E, RNase II and PNPase in the degradation of the rpsO transcripts of Escherichia: stabilizing function of RNase II and evidence for efficient degradation in an ams pnp rnb mutant. EMBO J. 1994;13:3368–3377. doi: 10.1002/j.1460-2075.1994.tb06639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang W M, Ao S Z, Casjens S, Orlandi R, Zeikus R, Weiss R, Winge D, Fang M. A persistent untranslated sequence within bacteriophage T4 DNA topoisomerase gene 60. Science. 1988;239:1005–1012. doi: 10.1126/science.2830666. [DOI] [PubMed] [Google Scholar]
- 11.Larsen, B. Unpublished data.
- 12.Maldonado, R. Unpublished sequence alignments.
- 13.McCormick J R, Zengel J M, Lindahl L. Correlation of translation efficiency with the decay of lacZ mRNA in Escherichia coli. J Mol Biol. 1994;239:608–622. doi: 10.1006/jmbi.1994.1403. [DOI] [PubMed] [Google Scholar]
- 14.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 15.Mufti S, Bernstein H. The DNA-delay mutants of bacteriophage T4. J Virol. 1974;14:860–871. doi: 10.1128/jvi.14.4.860-871.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rodriguez-Quinones F, Hernandez-Alles S, Alberti S, Escriba P V, Benedi V J. A novel plasmid series for in vitro production of phoA translational fusions and its use in the construction of Escherichia coli PhoE::PhoA hybrid fusions. Gene. 1994;151:125–130. doi: 10.1016/0378-1119(94)90642-4. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 18.Shapira S K, Chou J, Richaud F V, Casadaban M J. New versatile plasmid vectors for expression of hybrid proteins coded by a cloned gene fused to lacZ gene sequences encoding an enzymatically active carboxy-terminal portion of beta-galactosidase. Gene. 1983;25:71–82. doi: 10.1016/0378-1119(83)90169-5. [DOI] [PubMed] [Google Scholar]
- 19.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 20.Stark H, Mueller F, Orlova E V, Schatz M, Dube P, Erdemir T, Zemlin F, Brimacombe R, van Heel M. The 70S ribosome at 23 Å resolution; fitting the ribosomal RNA. Structure. 1995;8:815–821. doi: 10.1016/s0969-2126(01)00216-7. [DOI] [PubMed] [Google Scholar]
- 20a.Vogel H J, Bonner D M. Acetylornithase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 21.Watson J D, Tooze J, Kurtz D T, editors. Recombinant DNA: a short course. W. H. New York, N.Y: Freeman; 1983. [Google Scholar]
- 22.Weiss R B, Dunn D M, Atkins J F, Gesteland R F. Slippery runs, shifty stops, backward steps, and forward hops: −2, −1, +1, +2, +5, and +6 ribosomal frameshifting. Cold Spring Harbor Symp Quant Biol. 1987;52:687–693. doi: 10.1101/sqb.1987.052.01.078. [DOI] [PubMed] [Google Scholar]
- 23.Weiss R B, Huang W M, Dunn D M. A nascent peptide is required for ribosomal bypass of the coding gap in bacteriophage T4 gene 60. Cell. 1990;62:117–126. doi: 10.1016/0092-8674(90)90245-A. [DOI] [PMC free article] [PubMed] [Google Scholar]