We identified thresholds that separate normal baseline from abnormal heat stressed levels of heat shock protein 70 (HSP70) in Chinook salmon and coho salmon. The HSP70 biomarker can now be interpreted as lab work would be in a medical or veterinary setting as normal vs. abnormal.
Keywords: habitat, heat stress, mortality, Pacific salmon, sublethal, thermal stress, water temperature
Abstract
Rapid and accelerating warming of salmon habitat has the potential to lower productivity of Pacific salmon (Oncorhynchus species) populations. Heat stress biomarkers can indicate where warming is most likely affecting fish populations; however, we often lack clear classifications that separate individuals with and without heat stress needed to make these tools operational. We conducted a heat exposure experiment with trials lasting 12 or 36 h using juvenile Chinook salmon (Oncorhynchus tshawytscha) and coho salmon (Oncorhynchus kisutch) to validate heat stress biomarkers in white muscle. Following habituation to 13°C, individuals were exposed to water temperatures that increased to 15°C, 17°C, 19°C, 21°C or 23°C. Heat shock protein 70 abundance (HSP70 measured by ELISA) and transcription of 13 genes (mRNA measured by qPCR) including three heat shock protein genes (hsp70, hsp90, hsp27) were measured. A distinct heat stress response was apparent by 21°C in juvenile Chinook salmon and 23°C in juvenile coho salmon using HSP70. A threshold for heat stress classification in Chinook salmon of > 2 ng HSP70 mg.1 total protein identified heat stress in 100% of 21 and 23°C treated individuals compared to 4% in cooler treatments. For coho salmon, > 3 ng HSP70 mg.1 total protein identified heat stress in 100% of 23°C treated individuals compared to 4% in cooler treatments. Transcription from a panel of genes separated individuals between cooler and stressful temperature experiences (≥21°C for Chinook salmon and ≥23°C for coho salmon) with ~ 85% correct classification. Our findings indicate that juvenile Chinook salmon were more temperature-sensitive than juvenile coho salmon and support the use of a HSP70 threshold sampled from muscle for assessing heat stress in individual wild Pacific salmon with an option for non-lethal biopsies for spawning adults.
Introduction
Warming of Arctic and subarctic regions is nearly four times faster than the rest of Earth (Rantanen et al., 2022). Such rapid warming presents a threat to many species in the far north, even those who have temperature tolerances that allow them to range thousands of kilometers to the south such as Pacific salmon (Oncorhynchus species). Warmer and drier summers in the Arctic and subarctic can be stressful to some Pacific salmon populations and have been linked to mortality and declining productivity (Jones et al., 2020; von Biela et al., 2020, 2022; Carey et al., 2021; Howard and von Biela, 2023). Pacific salmon declines have been an area of focus for the U.S. Federal government because salmon are a key subsistence fishery on public lands and the Alaska National Interests Land Conservation Act of 1980 includes a commitment to manage land and resources to provide subsistence opportunities. Further development of existing tools would benefit research and monitoring of heat stress in Pacific salmon populations.
Existing tools for understanding salmon thermal sensitivity and tolerance span from cellular to population levels and include biomarkers that indicate temperature stress in cells (Iwama et al., 1998; Jeffries et al., 2021), physiological experiments using live wild fish to estimate critical thermal maximum temperature (CTmax) and optimal temperature (Topt) (Eliason et al., 2011; Turko et al., 2020) and spawning ground surveys to assess prespawn mortality outcomes (Spromberg and Scholz, 2011; Bowerman et al., 2021). Biomarkers can identify stressful (i.e. supraoptimal) water temperatures and individual stress when outcomes are either sublethal (e.g. higher energy requirements) and detect emerging drivers of population decline early or already lethal (e.g. prespawn mortality) to understand mortality mechanisms and do not require additional field efforts that are complicated by access to remote region. For example, existing monitoring projects collected hundreds of muscle biopsy punches that confirmed high prevalence of heat stress using the biomarker heat shock protein 70 (HSP70) or the mRNA of select genes among spawning adult Chinook salmon near their northern range extent in the Yukon River (von Biela et al., 2020). The existing experimental validation for identifying heat stress from muscle tissue used a small sample size (n = 22) of adult Chinook salmon (von Biela et al., 2020) so there is a need to reinforce this effort with additional experimental validation. Moreover, there is a need to consider different species of Pacific salmon. Here, we expand the validation of sampling skeletal muscle tissue as a heat stress research and monitoring tool by conducting a large (sample size >100 per species), comparative temperature manipulation experiment for Chinook salmon (Oncorhynchus tshawytscha) and coho salmon (Oncorhynchus kisutch), two species that co-occur in many salmon streams where they generally rear for 1 or 2 years, respectively, before migrating to sea (Quinn, 2018).
Chinook salmon are a species of conservation concern across their range. Multiple Chinook salmon evolutionarily significant units (ESU) and populations are listed as Threatened or Endangered under the U.S. Endangered Species Act (ESA) and the Canadian Species at Risk Act (SARA) across California, Oregon, Washington, Idaho and British Columbia. Near their northern range extent in Alaska and the Yukon Territory, Chinook salmon are associated with prolonged population declines, fishery disaster declarations and fishery closures (ADFG, 2013; Dorner et al., 2018; Welch et al., 2021). Several Chinook salmon stocks in western and southcentral Alaska are designed by the State of Alaska as ‘Stocks of Concern’ (e.g. Yukon designated in 2000 and multiple Cook Inlet and Southeast Alaska stocks designated between 2010 and 2021) indicating that abundance of the stock has often failed to meet harvestable yield or spawning escapement goals in recent years. Coho salmon are one of four additional Pacific salmon species that occur in Alaska and generally have lower management concerns in Alaska along with sockeye salmon (O. nerka), chum salmon (O. keta) and pink salmon (O. gorbuscha). For example, the State of Alaska stocks of concern include 15 Chinook salmon stocks compared to three sockeye salmon stocks, one chum salmon stock and zero coho salmon or pink salmon stocks as of April 2022 (https://www.adfg.alaska.gov/index.cfm?adfg=specialstatus.akfishstocks). The ability to contrast heat stress between Chinook salmon and coho salmon in wild settings would be particularly useful because of their differences in concern despite life history similarities that lead them to frequently co-occur: extended freshwater rearing in flowing waters (e.g. streams and rivers rather than lakes), ocean residency and similar marine diets (Quinn, 2018). Therefore, developing heat stress tools for both species will provide opportunity to assess whether differences in heat stress prevalence or thermal sensitivities could contribute to disproportionate Chinook salmon declines. Moreover, biomarker tools provide a non-lethal option, a necessity when examining species of conservation concern.
Thermal sensitivity of species and life stages can be inferred because activation of heat stress biomarkers at cooler temperatures indicates stronger temperature sensitivity and generally correlates with cooler optimal temperatures compared to activation at warmer temperatures (Dietz and Somero, 1993; Feder and Hofmann, 1999). Experiments validate biomarker response across water temperatures and can identify the separation between a ‘normal’ and ‘stressed’ biomarker signature. The absolute value or signature of stress in a biomarker is specific to a species and tissue given known differences in HSP concentrations (Fowler et al., 2009; Bowen et al., 2020). Within a species, the ‘stressed’ level of a biomarker is often similar enough across life stages (Fowler et al., 2009) that juvenile validation experiments are used to interpret stress biomarkers in adults (Carey et al., 2019). In other words, the absolute value of HSP70 that distinguished heat stress in a juvenile Chinook salmon would be presumed to be the same for adult Chinook salmon. Note that this assumption has no bearing on the water temperature that induces the stress response as differences in stressful water temperatures are well-known to be life stage specific (McCullough, 1999).
In this study, we focused on widely used heat shock proteins (HSPs) as biomarkers because they are a core component of the cellular heat shock response (Fowler et al., 2009; Akbarzadeh et al., 2018; Bowen et al., 2020; von Biela et al., 2020; Jeffries et al., 2021). A rapid production and elevation of heat shock proteins (HSPs) occurs in cells when warm temperatures begin to damage proteins that are required for normal cellular functions because HSPs are chaperones that help maintain proper folding, assist with repair of damage and prevent aggregation of other proteins (Feder and Hofmann, 1999). Induction of HSPs appears to extend the temperature tolerance range of an individual, although not indefinitely (Feder and Hofmann, 1999). If an individual cannot find suitable water temperatures in hours or days, then a range of sublethal (e.g. reduced growth or partial egg retention) to lethal outcomes follow the heat shock response (Bowen et al., 2020; Ern et al., 2023). HSPs can be measured at transcription as mRNA or after translation as abundance of proteins (Sagarin and Somero, 2006; Chadwick et al., 2015; Lewis et al., 2016). Transcription approaches often consider multiple HSP genes (e.g. hsp70, hsp90, hsp27), but protein approaches use primarily HSP70 as lab assays are not available for all proteins (note that lowercase italics denote mRNA from gene transcription and capitalized, plain font denotes protein in this paper). The increase in both mRNA and protein levels is associated with the high end of tolerable temperatures (Mottola et al., 2020). In some cases, transcription of other genes can also be useful at identifying heat stress (e.g. immune system genes) (Bowen et al., 2020; von Biela et al., 2020).
Our goal is to further develop heat stress biomarkers for Chinook salmon and coho salmon by using white muscle tissue samples to assess HSP70 concentrations and transcription of 13 genes including hsp27, hsp70 and hsp90. This is an experimental study that uses hatchery-reared juvenile salmon because they are more amenable to experimental manipulation compared to adults (Donnelly et al., 2020), they can be obtained in larger numbers and individuals have very similar past experiences that reduce concerns of latent variables compared to wild fish. Water temperature treatments at 15°C, 17°C, 19°C, 21°C and 23°C were designed to span the transition from preferred to stressful water temperatures for juvenile salmon (Brett, 1952; McCullough, 1999; Richter and Kolmes, 2005). This temperature range represents the warmer portion of water temperatures currently encountered by juvenile and spawning adult Pacific salmon during summer in Alaska’s freshwaters, but not the most extreme high temperatures (Mauger et al., 2017). We examined two exposure durations (12 h and 36 h) to ensure that biomarkers were robust to differences in the length of warm water exposure given that exposure length would not be known in future efforts to detect heat stress of wild-caught fish. Our analysis approach was divided into four steps: (1) compare HSP70 and gene transcription response among species, treatment temperatures and exposure durations to assess similarity in response across species and exposure durations; (2) compare hsp70 gene transcription levels to HSP70 protein concentrations within individuals to describe the relationship between the mRNA and its protein product for each species; (3) identify a threshold value of HSP70 protein that denotes elevation for identification of heat stress at an individual-level and identification of stressful water temperatures at the population-level; and (4) determine whether gene transcription signatures can separate individuals that were or were not exposed to stressful water temperatures. The ability to identify heat stress using thresholds and other classification approaches furthers the development of heat stress biomarkers from descriptive research to operational tools.
Methods
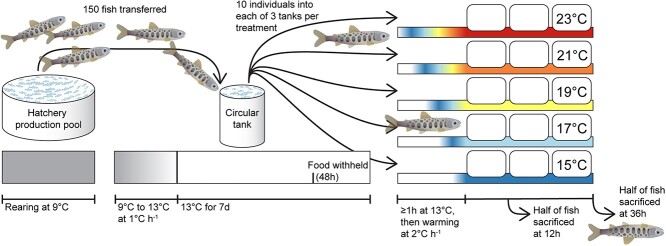
We conducted two identical experiments at the Alaska Department of Fish and Game’s William Jack Hernandez Sport Fish Hatchery in Anchorage, Alaska, USA (Figure 1). The Chinook salmon experiment was conducted in May 2020 using juveniles from naturally produced parents from Crooked Creek (southcentral Alaska, Kenai Peninsula), a stream with a long history of supplemental Chinook salmon stocking using both hatchery- and naturally produced parents (Gates, 2022). The coho salmon experiment was conducted in May 2021 using juveniles propagated from adults that returned to Ship Creek, Anchorage, Alaska, originally established from wild Little Susitna River (southcentral Alaska) parents. By conducting both experiments in May, we aimed to minimize seasonal differences in HSP expression (Feder and Hofmann, 1999). The fork length of juvenile Chinook salmon was ~100 mm and juvenile coho salmon were ~50 mm. For each experiment, approximately 150 individuals were transferred from a 9°C hatchery production pool (91 000 L) to a 9°C circular tank (1400 L) where water was warmed 1°C h−1to 13°C for one week. Food was withheld during the last 2 d of the week spent at 13°C and fasting continued over the next 1–2 d of the temperature manipulation experiment to reduce metabolic demands and nitrogenous waste during the experiment. Nitrogenous wastes were monitored using an ammonia test during each day of the experiment.
Figure 1.
Design of heat stress experiments for juvenile Chinook salmon and coho salmon. Approximately 150 individuals of each species were used in separate experiments and divided across 15 treatment tanks (three tanks for each of five temperature treatments).
Individuals were randomly divided among 15 experimental 75 L tanks filled with 13°C water oxygenated by aquarium bubblers. Each treatment tank was randomly assigned one of five treatment temperatures (15°C, 17°C, 19°C, 21°C and 23°C) such that each treatment was applied to three tanks that each contained 10 individual fish and one aquarium heater with a temperature controller (Inkbird Tech ITC-306 T or ITC-308) that activated the heater when water temperature dropped >0.2°C below the set point. Water temperatures were recorded by a Hobo Tidbit at 5-min intervals (Figure 2). Dissolved oxygen was monitored hourly using a handheld YSI meter when staff were present during the day. Each tank was covered by an opaque lid with two areas of black mesh to prevent fish from escaping and still allow light to enter tanks. After at least a 1-h habituation period, water temperatures were warmed at a rate of 2°C h−1. The rate of warming used across salmonid heat stress experiments has not been standardized, although attempts to standardize CTmax experiments using a rapid rate of 18°C h−1 were at least partially adopted with periodic calls for slower rates including a recommendation for the 2°C h−1 used here (Elliott and Elliott, 1995; Desforges et al., 2023). Ultimately, experimental warming rates balance being fast enough that individuals in warmer treatments have not acclimated to higher water temperatures than individuals from cooler treatments (some acclimation occurs within 24 to 72 h; Gilbert et al., 2022), but slow enough for ecological relevancy. Temperature increase in specific places can be as high as ~1°C h−1 (based on daily water temperature ranges up to ~10°C; (Gilbert et al., 2016; Mauger et al., 2017), but individual fish likely experience more rapid rates of water temperature change as they move across freshwater landscapes with heterogenous water temperatures horizontally and vertically. For example, juvenile coho salmon appear to voluntarily experience water temperature changes ≥2°C h−1 when they make quick (~1 h) movements between cooler areas with better feeding opportunities and areas ≥2°C warmer with faster digestion and energy assimilation (Armstrong et al., 2013). Half of the individuals in each tank were sacrificed after a 12 h exposure to the treatment temperature and the other half were sacrificed after 36 h. Maintaining a consistent rate of temperature increase and exposure to the treatment temperatures resulted in a longer time at elevated temperatures (> 13°C) for individuals in warmer treatments (e.g. an additional 4 h of warming time is needed for 23°C vs. 15°C treated individuals).
Figure 2.
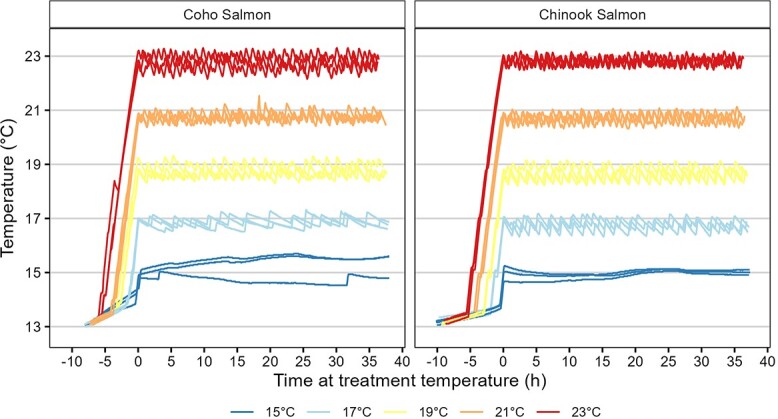
Water temperatures for each experiment tank recorded at 5-min intervals. Horizontal lines are labeled on the y-axis and denote habituation water temperature (13°C) and treatment water temperatures (15°C, 17°C, 19°C, 21°C and 23°C).
Individuals were dispatched by cranial concussions followed by cervical dislocation. Fork length was measured to the nearest millimeter. Two skeletal muscle tissue samples were removed from above the lateral line using a scalpel and immediately placed in 2 ml vials to flash freeze the sample in cryoshippers at temperatures below −80°C. One set of samples was sent to the USGS S.O. Conte Research Laboratory (Turners Falls, MA, USA) for measurement of HSP70 concentration. The other set of samples was sent to the USGS Western Ecological Research Center (Davis, CA, USA) to analyze the gene panel mRNA levels. This work was approved by the USGS Alaska Science Center Animal Care & Use Committee (ACUC 2020–07).
HSP70 protein assay
Frozen muscle samples were dissected away from subdermal fat and skin and weighed to the nearest mg. Twelve volumes of homogenization buffer (Phosphate Buffered Saline plus complete mini protease inhibitor, Sigma-Aldrich, USA) were added to each muscle sample at 4°C. Samples were homogenized with a Kontes Pestle Pellet handheld homogenizer (Fisher Scientific, USA) and centrifuged for 10 min at 10000 g. Protein concentration of the resulting supernatant was determined with bicinchoninic acid (BCA) protein assay (Pierce, Fisher Scientific, USA). Supernatants were used to compare our competitive enzyme-linked immunosorbent assay (ELISA), modified from (Faught et al., 2017), to our previously employed western blotting method of HSP quantitation (Bowen et al., 2020; von Biela et al., 2020). The primary antibody used in both western and ELISA methods specifically recognizes the inducible form of HSP70 (AS05061, Agrisera, Sweden) and results in a single specific band at ~70 kDa on western blots. Paired samples representing all temperature treatments showed a strong correlation between the two methods (R2 = 0.93). The linear relationship was greatest in the middle temperature range, as the western blot values tend to overestimate HSP near zero values with little resolution between 15°C and 17°C and plateau at the higher end of HSP induction at 23°C. Thus, our HSP ELISA expanded the linear range of detection, increased sensitivity of detection and was used for all further analysis as follows. High binding 96 well plates were coated with Chinook Salmon HSP (ADI-SPP-763-F, Enzo Life Sciences, USA) at 100 ng mL−1 in coating buffer (15 mM Sodium Carbonate, 35 mM Sodium Bicarbonate, pH 8.3) overnight at 4°C. In microcentrifuge tubes, muscle homogenates (final concentration 10 μL per well) were incubated with polyclonal primary antibody HSP70 in PBS plus 0.05% Tween 20 (PBS-Tw) overnight at 4°C. Standards ranging from 0–320 ng··mL−1 were diluted 1:1 in PBS-Tw and matrix (HSP70 negative muscle extract at 2.6 mgs·mL−1) and incubated overnight at 4°C. Plates were washed 5x with PBS-Tw and blocked with PBS-Tw + 1% BSA at room temperature for 1 h. Samples/antibody were added to wells in duplicate and standards/antibody were plated in triplicate. Plates were incubated overnight at 4°C, washed 5x with PBS-Tw and incubated with goat anti rabbit -HRP secondary antibody (5220-0283, Sera Care, MD, USA) at room temperature for 1 h. Plates were again washed and TMB (3,3′, 5, 5′—tetramethylbenzidine) substrate added (5120–0083, Sera Care, MD, USA). Plates were monitored at 650 nm and after 5 min the reaction was stopped and read at 450 nm (BioTek Synergy2, VT, USA). HSP70 values based on the standards ranging from 0.6 to 320 ng·mL−1 were calculated with a 4-parameter curve, and final data was corrected by total protein and expressed as ng HSP70 mg−1 total protein. The lower detection limit of the assay was 0.05 ng·mL−1. Inter-assay variation was 5.2% CV and intra-assay variation was 8.6% CV.
Gene transcription assay
Thirteen genes were selected to reflect potential influences of known stressors in the salmons’ environments. These include genes related to heat stress and other stressors associated with climate change (e.g. emerging infectious diseases, nutritional stress). The functionality and response of each gene have been validated in other studies and are well-documented in the literature (Table 1; for gene references and primer sequences see von Biela et al. (2020)Tables 1 and 2). Gene transcription was measured using quantitative real-time polymerase chain reaction (qPCR) assays of mRNA at the US Geological Survey Western Ecological Research Center in Davis, California, USA. Total RNA was extracted from ground muscle tissue using the RNeasy Lipid Tissue Mini Kit (Qiagen; www.qiagen.com). To remove contaminating genomic (g)DNA, extracted total RNA was treated with 10 U・μL–1 of RNase-free DNase I (DNase, Amersham Pharmacia Biotech Inc.) at room temperature (20–30°C) for 15 min. The extracted RNA was stored in a − 80°C freezer until analyzed. A standard cDNA synthesis was performed on 2 μg of RNA template from each Chinook salmon and coho salmon. Reaction conditions included four units reverse transcriptase (Omniscript, Qiagen, Valencia, California), 1 μmol・L−1 random hexamers, 0.5 mmol・L−1 each dNTP and 10 units RNase inhibitor, in reverse transcription (RT) buffer (Qiagen, Valencia, California). Reactions were incubated for 60 min at 37°C, followed by an enzyme inactivation step of 5 min at 93°C and then stored at −20°C until further analysis. Briefly, 1 μL of cDNA was added to a mix containing 12.5 μL of QuantiTect Fast SYBR Green Master Mix (5 mmol・L−1 Mg2+; Qiagen, Valencia, California), 0.5 μL each of forward and reverse sequence specific primers with a concentration of 10 μM (von Biela et al., 2020) and 10.5 μL of RNase-free water; total reaction mixture was 25 μL. The primers for hsp27, hsp70 and hsp90 are specific to the inducible forms of these genes. The reaction mixture cDNA samples for each gene of interest and reference genes were loaded into MicroAmp Fast Optical 96 well reaction plates in duplicate and sealed with optical sealing tape (Applied Biosystems, Foster City, California). Reaction mixtures containing water, but no cDNA, were used as negative controls. Amplifications were conducted on a QuantStudio 3 Real-time Thermal Cycler (Applied Biosystems, Foster City, California), using the QuantStudio 3 software. Reaction conditions were as follows: an initial hold stage of 95°C for 20 s, 40 cycles of 95°C for 1 s and 60°C for 20 s. The melt curve was 95°C for 1 s, 60°C for 20 s and 0.3°C・s−1 temperature increase, and then 95°C for 1 s.
Table 1.
F-statistics from an analysis of variance for mean differences in transcription of specific genes between species (coho salmon and Chinook salmon), among temperature treatments, an interaction between species and temperature treatment and collection time (12 or 36 h) nested within each species and treatment
| Biomarker | Gene name | Function | Species | Treatment | Species × treatment | Time |
|---|---|---|---|---|---|---|
| hsp27 | Heat shock protein 27 | Heat stress | 360*** | 8.30*** | 3.45** | 1.06 |
| hsp70 | Heat shock protein 70 | Heat stress | 177*** | 73.3*** | 4.08** | 0.628 |
| hsp90 | Heat shock protein 90 | Heat stress | 1610*** | 203*** | 11.0*** | 29.4*** |
| gata3 | GATA binding protein 3 | Immune | 85.4*** | 1.33 | 2.88* | 2.01* |
| ifna | Interferon alpha | Immune | 231*** | 3.36* | 0.665 | 3.36*** |
| ifng2 | Interferon gamma 2 | Immune | 363*** | 4.36** | 2.69* | 0.926 |
| mx1 | Myxovirus resistance 1 | Immune | 416*** | 0.913 | 0.384 | 1.49 |
| tbx21 | T-box transcription factor 21 | Immune | 443*** | 3.24* | 0.842 | 2.59** |
| ahr | Arylhydrocarbon receptor | Immune; detoxification | 183*** | 0.522 | 0.356 | 2.40** |
| cyp1A | Cytochrome P450 1A1 | Detoxification | 109*** | 0.201 | 1.32 | 1.9 |
| sod | Superoxide dismutase | Detoxification | 42.1*** | 1.12 | 0.844 | 1.06 |
| mt-a | Metallothionein a | Detoxification | 582*** | 3.20* | 0.632 | 3.53*** |
| leptin | leptin | Metabolism | 257*** | 3.25* | 1.73 | 2.16* |
*** P < 0.001.
** P < 0.01.
* P < 0.05.
Table 2.
Results from random forest models using transcription of all 13 genes examined (‘full model’) or a ‘reduced model’ of the most influential genes to identify heat stress
| Chinook salmon | Coho salmon | |||
|---|---|---|---|---|
| Full model | Reduced model | Full model | Reduced model | |
| Training data | ||||
| n | 61 | 61 | 31 | 31 |
| Correct classification | 90% | 92% | 87% | 88% |
| Sensitivity | 93% | 93% | 100% | 87% |
| Specificity | 87% | 90% | 75% | 88% |
| Validation data | ||||
| n | 90 | 90 | 110 | 110 |
| Correct classification | 86% | 87% | 89% | 87% |
| Sensitivity | 77% | 83% | 100% | 93% |
| Specificity | 90% | 88% | 88% | 86% |
Each model was fit with a subset of individuals (training data) and validated with all remaining individuals. Reported results include sample size (n), precent of individuals correctly classified to stressful temperature treatments (21 and 23° for Chinook salmon and 23°C for coho salmon), sensitivity and specificity.
We transformed the qPCR data according to the 2∧(-CT’) method (Livak and Schmittgen, 2001) as follows. First, we normalized values (threshold crossing (CT) of the housekeeping gene subtracted from the threshold crossing for the gene of interest). We then compared the normalized value of the target gene to the CT of the calibrator sample (the lowest level of transcription for each gene). This gave us normalized transcription values for each gene relative to the maximum observed CT across all samples. We then log-transformed the transcription values, and those log-transformed values served as the basis for all analyses going forward. Statistical analyses and plotting for HSP70 protein and gene transcription data were conducted in Rstudio (Poist, 2022). The reference genes selected, rpL8 and EF1a, were identified by Olsvik et al. (2005) and Veldhoen et al. (2010) as suitably stable reference genes. Briefly, stability of reference genes was evaluated and ranked using the web-based analysis tool RefFinder (https://www.heartcure.com.au/for-researchers/) (Xie et al., 2023). The gene rpL8 was found to be the more stable reference gene and was therefore used exclusively in normalization.
HSP70 comparison among species, temperature and treatment duration
We anticipated that HSP70 protein concentration values would violate parametric statistical analysis assumptions of normality and equal variance because temperature treatments that were not stressful would result in many individuals with values near zero and minimal variance while stressful treatments would result in higher values with more variation. Therefore, we used a non-parametric approach for HSP70 analyses and considered a single variable at a time. First, we determined that median HSP70 concentration were similar between the 12 h and 36 h time exposures within each species and treatment temperature using two-sample Wilcox tests and adjusted p-values with Benjamini-Hochberg corrections for multiple comparisons. We used the same approach to ensure that the HSP70 response was similar among the three tanks in each treatment (P > 0.05). Next, we described median differences in HSP70 concentrations between species and among treatments by combining species and treatment to a single category (e.g. Chinook salmon 15°C) and applying a Kruskal-Wallace one-way analysis of variance test with pairwise Benjamini-Hochberg corrections for multiple comparisons among the 10 groups (2 species x 5 temperatures).
Gene transcription comparison among species, temperature and treatment duration
We described mean differences for each gene between species, among treatment temperatures and between the two treatment durations using analysis of variance. Strong species and duration effects have implications for our classification analyses: a species effect would reinforce the need for species-specific heat stress classifications based on gene transcription data and a duration effect would signal potential complications if gene transcription data was used to classify heat stress in wild fish for which exposure durations will not be known. Analyses were conducted in R statistical software using the aov() function by specifying the following formula:
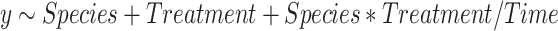 |
Where y is the gene of interest, Species is Chinook salmon or coho salmon, Treatment is a category for 15,17, 19, 21 or 23°C water temperature treatment, Species*Treatment is an interaction and Time is the 12 h or 36 h duration nested in the interaction because we are specifically interested in the possibility of a Time effect within a species and treatment. Similarity in gene transcription values among the three treatment tanks with the same temperature were also confirmed by replacing Time with Tank and testing the model for each gene (P > 0.05). Additional plots and post hoc tests were used to describe effects for genes most responsive to temperature treatments, as judged by the F-statistic for the treatment effect. Pairwise differences for treatment and interactions between species and treatment were identified with Tukey’s post hoc tests using the function TukeyHSD(). Pairwise differences between time exposure for each species and treatment were determined by fitting the same models with the lm() function because this analysis estimates a coefficient describing the difference between the two time exposures for each species and treatment combination while TukeyHSD() does not handle nested variables correctly. The t-value and P-value associated with each time coefficient in the lm() output identified where exposure time was related to differences in gene transcription for a specific combination of species and treatment.
Comparing gene transcription and protein concentration of HSP70
We examined the relationship between paired HSP70 gene transcription (mRNA) and the resulting HSP70 protein measured in the same individuals to clarify the extent to which transcription is coupled with protein abundance (Silver and Noble, 2012). We expect that higher levels of mRNA transcription are associated with higher levels of HSP70, either as a continuous or categorical variable. We considered if continuous values of HSP70 concentrations could be predicted from HSP70 mRNA values using a general additive model (GAM) that allows for linear and non-linear relationships, but transcription of hsp70 struggled to predict the specific HSP70 concentration once elevated above baseline HSP70 concentrations. Specifically, the GAM model overestimated the HSP70 concentration for 21°C treated fish and underestimated the protein concentrations for 23°C treated fish in Chinook salmon. Similarly, for coho salmon, the GAM model underestimated the HSP70 concentrations for 23°C treated fish. Because the model failed to meet the assumption of randomly distributed residuals among temperature treatments it is not presented in the paper. We tested whether levels of mRNA transcription predicted if HSP70 were elevated or not (categorical variable) using logistic regression (HSP70 concentrations greater than threshold value set to 1 and less than or equal to the threshold set to 0).
HSP70-based heat stress classification for individuals and water temperature treatments
To determine if a threshold value of HSP70 concentration can be used to classify individuals with and without heat stress, we examined a histogram of HSP70 concentration values for each species. We anticipated that many individuals would have low HSP70 values clustered around zero, reflecting maintenance level of HSP70, while elevated levels of HSP70 would form the remaining part of the histogram. We visually examined the histogram for a separation in the data between low and elevated levels that could be used as a classification threshold for heat stress. We calculated the percentage of individuals in each water temperature treatment that would be classified with elevated HSP70 indicative of heat stress under this visually assessed threshold and tested for differences among treatment temperatures using Chi-squared analysis for each species. We anticipated that cooler water temperature treatments would have virtually 0% elevated HSP70 and at least one of the warmest treatments would be associated with nearly 100% HSP70 elevation. A water temperature treatment was considered stressful if most (>50%) individuals had elevated HSP70 concentrations.
Gene transcription-based heat stress classification for individuals
To determine if transcription of the selected genes was related to stressful water temperatures, we used a Random Forest classification analysis that separated individuals between unstressful and stressful temperature treatments. Water temperature treatments were considered stressful if they elevated HSP70 in > 50% of individuals for a given species (i.e. 21 and 23° for Chinook salmon; 23° for coho salmon). Random Forest is a directed, machine learning approach where multiple versions of the training datasets are made through random-draws with replacement to identify the decision tree that is most common across the resampled training datasets. A subset of the samples was used to train the model and the remainder were used for validation. To ensure that the training data set was balanced between individuals that experienced stressful water temperatures or not, we used 50% of individuals from whichever group was smaller and then randomly selected a similar number of individuals from the other group. We considered a full model using all 13 genes examined and a stepwise (i.e. removed one gene at a time) reduced model with a subset of genes that had a ‘mean decrease in accuracy’ > 5 in the random forest model output. In each model we report the mean decreases in accuracy for each gene, percentage of individuals correctly classified, sensitivity (heat stress positives in stressful water temperature treatments/heat stress positives) as a measure of heat stress detection and specificity (heat stress negatives in unstressful water temperature treatments/heat stress negatives) as a measure of correctly identifying individuals that were not exposed to stressful temperatures. All data used in this study are available in Stanek et al. (2023).
Results
A total of 151 juvenile Chinook salmon and 141 juvenile coho salmon completed the experiment after a small number of individuals (<10 for each species) escaped experimental tanks. All fish that remained in the tanks survived the experiment. Sample size per treatment was 26 to 32 individuals per species. Of these, juvenile Chinook salmon had a fork length of 97 ± 7 mm (mean ± SD) and coho salmon had a fork length of 55 ± 6 mm. Water temperatures varied around the target water temperatures by ~0.25°C and maintained distinct separation among treatments (Figure 2). Dissolved oxygen concentrations were maintained at > 92% and > 8 mg L−1. Ammonia concentrations were maintained at ≤ 0.5 mg L−1.
HSP70 protein comparison among species, temperature and treatment duration
HSP70 concentration differed as a function of species and treatment (Kruskal-Wallace chi-squared = 207, df = 9, P < 0.001; Figure 3). Median HSP70 concentrations were lowest for both species in the 15°C, 17°C and 19°C treatments and elevated in the 21°C and 23°C treatments (pairwise Dunn tests, adjusted P < 0.05; Figure 3). In the 21°C treatment, HSP70 concentrations were higher in Chinook salmon than coho salmon (adjusted P = 0.03). The highest HSP70 concentrations occurred in 23°C treated Chinook salmon. HSP70 concentrations for 23°C treated coho salmon were statistically similar to 21°C and 23°C Chinook salmon, although the mean values for at 23°C were 10-fold higher in Chinook than in coho salmon.
Figure 3.
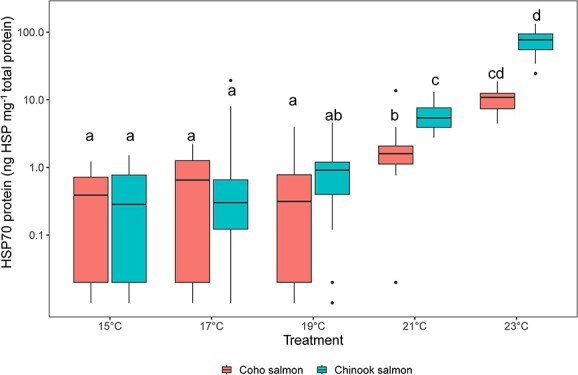
Boxplots of heat shock protein 70 (HSP70) protein concentrations (ng HSP70 mg−1 total protein) among coho salmon (left) and Chinook salmon (right) across five different treatment temperatures. Individuals from the 12- and 36-h exposure are combined for this plot because HSP70 concentrations did not differ between the time exposures within a treatment and species. Different letters indicate significant differences (P < 0.05).
Gene transcription comparison among species, temperature and treatment duration
Transcription of each gene was considered relative to the influence of species, temperature and exposure time simultaneously in an analysis of variance model that also included interactions of species and temperature to allow for different thermal sensitivities between species (Table 1). Across all 13 genes examined, transcription was higher in Chinook salmon compared to coho salmon. Among the genes examined, the three HSP genes (hsp70, hsp90 and hsp27) were most responsive to the water temperature treatment (F = 8.3–203; < 4.4 for all other genes; Table 1, Figure 4). For hsp70, an increase was evident at 21 and 23°C for coho salmon and at 19°C for Chinook salmon with continued increases in the 21°C and 23°C treatments. For hsp27, gene transcription did not increase until the 23°C treatment in Chinook salmon and there was no difference for any treatment for coho salmon. For hsp90, Chinook salmon gene transcription elevation began at 19°C and increased further at both 21°C and 23°C. Elevation of coho salmon hsp90 began at 21°C and increased further at 23°C. Of the three HSP genes, the time exposure effect nested within the interaction of species and treatment was not related to the transcription of hsp27 or hsp70 (Figure 4). Exposure time did influence transcription of hsp90, with lower mRNA levels at 36 h than at 12 h in the 19°C, 21°C and 23°C treatments for both species as well as the 15°C treatment for coho salmon (Figure 5). Among the genes with weak temperature treatment effects (P < 0.05 and F < 4.4), pairwise differences in post-hoc tests were not significant after adjustments for multiple comparisons (ifna, tbx21, mt-a, or leptin), except for ifng2 where transcription was lower in 21°C treated coho salmon compared to coho salmon in other treatments.
Figure 4.
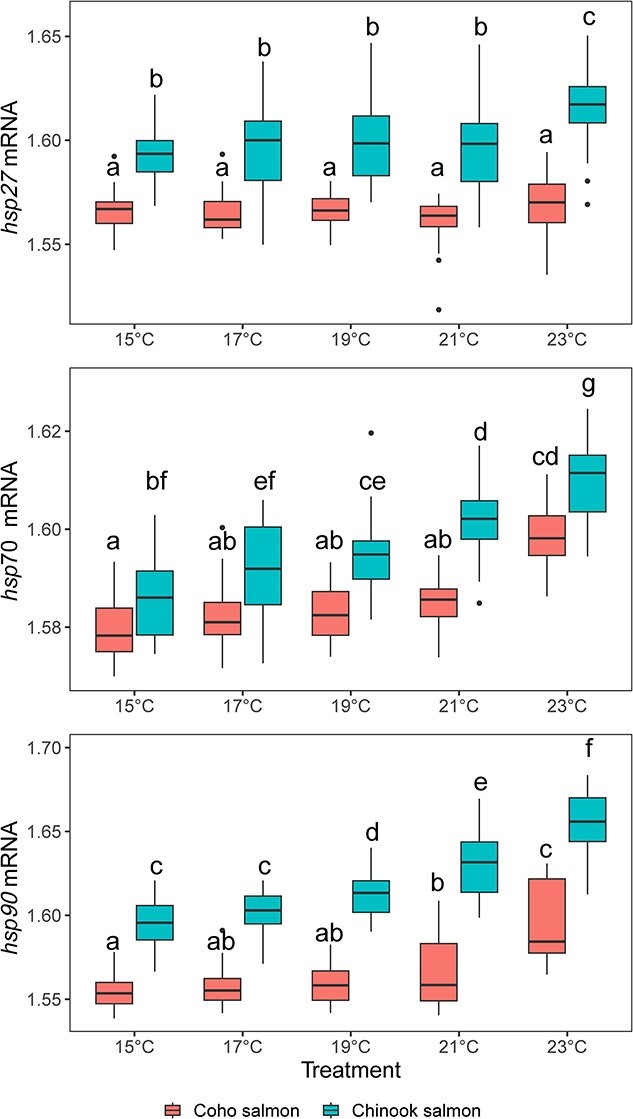
Relative abundance of mRNA from three heat shock protein genes (HSP27, HS70 and HSP90) across five different temperature treatments measured in coho salmon (left) and Chinook salmon (right) exposed to each treatment for 12 or 36 h. Letters indicate significant differences from the analysis of variance test presented in Table 1.
Figure 5.
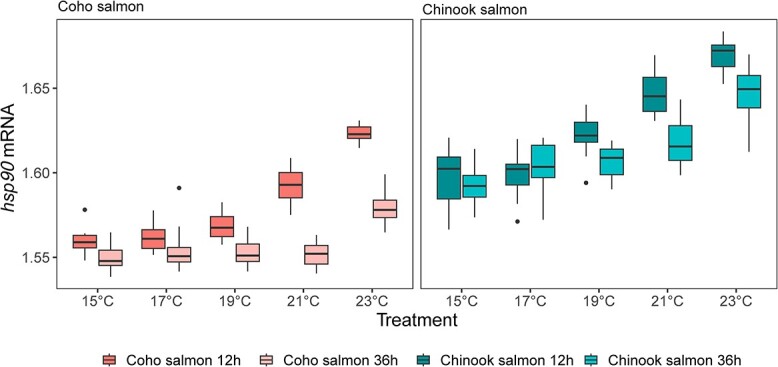
The response of HSP90 gene transcription (mRNA) between species, water temperature treatments and time exposure (12 or 36 h). Stars indicate significant differences between the time exposures for specific species and treatment combinations as follows: ***P < 0.001, **P < 0.01 or *P < 0.05.
Comparing gene transcription and protein concentration of HSP70
Increased transcription of hsp70 was related to the categorical presence/absence of elevated HSP70 concentrations for Chinook salmon (logistic regression, deviance = −63.8, df = 1, P < 0.001) and coho salmon (deviance = −81.4, df = 1, P < 0.001; Figure 6). The hsp70 gene transcription value associated with a 50% probability of HSP70 elevation as determined by the logistic regression was 1.600 (transformed values are unitless see methods) for Chinook salmon and 1.593 for coho salmon. Plots of paired mRNA compared to protein were divided into four quadrants that represent the possible categorical groups of mRNA and protein elevation (Figure 6). In both species, most (78% of Chinook salmon and 88% of coho salmon) individuals had either low mRNA and low protein (46% of Chinook salmon and 72% of coho salmon) or elevated mRNA and elevated protein (32% of Chinook salmon and 16% of coho salmon).
Figure 6.
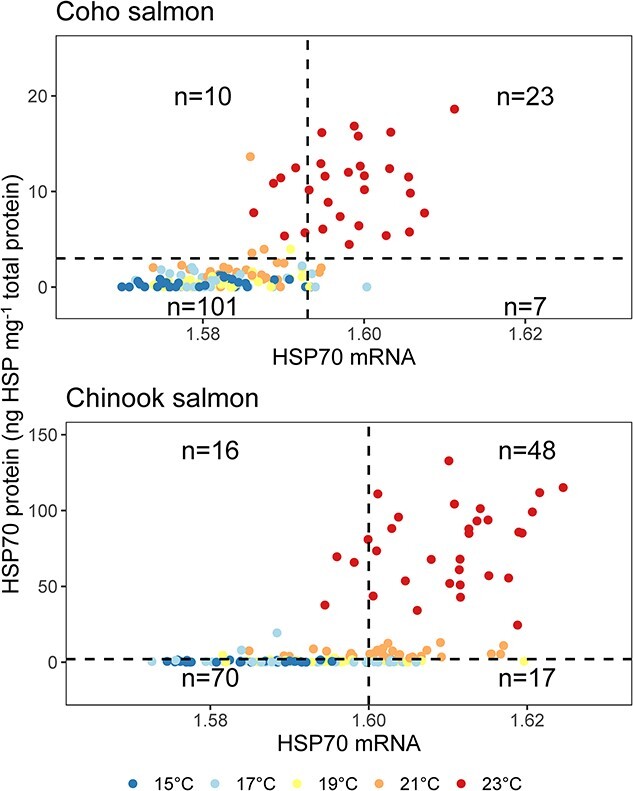
Relationship between the levels of HSP70 transcription (mRNA) and the resulting HSP70 concentration (ng HSP70 mg−1 total protein). Colors indicate water temperature treatment. Horizontal dashed lines are the HSP70 threshold used to identify heat stress in Chinook salmon (2 ng HSP70 mg−1 total protein) and coho salmon (3 ng HSP70 mg−1 total protein). Verticle dashed lines are HSP70 transcription value associated with the 0.5 probability of elevated protein concentrations in Chinook salmon (1.600) and coho salmon (1.593). Sample size notations indicate the number individual fish that occur in each quadrant of the plot out of an overall sample size of 151 Chinook salmon and 141 coho salmon.
HSP70-based heat stress classification for individuals and water temperature treatments
HSP70 concentration histograms revealed a separation between low and high values in both species (Figure 7). For Chinook salmon, the separation in HSP70 concentrations occurred at 2 ng HSP70 mg−1 total protein and values higher than this were considered HSP70 elevation and indicative of heat stress. Among Chinook salmon, HSP70 concentrations were always elevated in individual fish from 21 and 23°C treatments and rarely elevated in individual fish when exposed to 15 to 19°C and (0% of individuals in 15°C treatment, 7% of individuals in the 17°C treatment and 6% of individuals in the 19°C treatment compared to 100% of individuals in the 21°C and 23°C treatments). The differences in the percentage of individual Chinook salmon with elevated HSP70 among treatment temperatures were statistically significant (χ2 = 136, df = 4, P < 0.001). The HSP70 concentrations for individual Chinook salmon exposed to 21°C and 23°C separated near 20 ng HSP70 mg−1 total protein. Temperature treatments of 21°C and 23°C were considered stressful for these hatchery-reared juvenile Chinook salmon. The baseline HSP70 concentration was 0.56 ± 0.50 (mean ± SD) ng HSP70 mg−1 total protein among the individuals without elevated HSP70 (n = 87).
Figure 7.
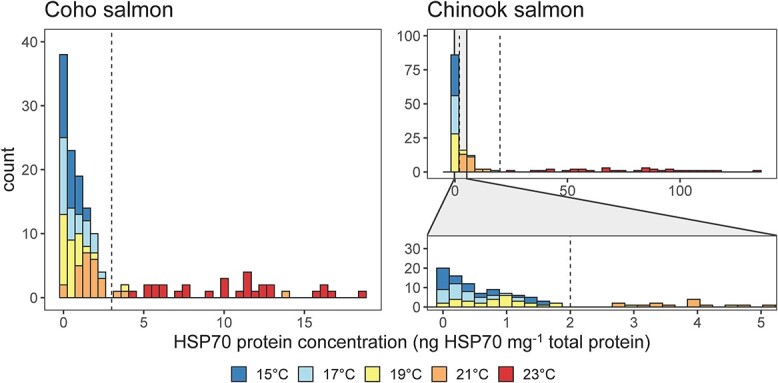
Histograms of HSP70 concentrations (ng HSP70 mg−1 total protein) measured in the skeletal muscle tissue of juvenile coho salmon (left) and Chinook salmon (right). This figure combines individuals in 12 and 36 h exposures because HSP70 concentrations were similar within species and treatments. Note the wider range of HSP70 concentrations across juvenile Chinook salmon examined with a lower panel zoomed in to see separation in the distribution of HSP70 concentrations. Dashed lines are the thresholds used to designate heat stress. For coho salmon, the separation between off/baseline and elevated with heat stress was apparent at 3 ng HPS70 mg−1 total protein. For Chinook salmon, there are two lines denoting separation between off/baseline and elevated values and elevated levels of HSP70 also separate between an elevated-lower (21°C treatment) and an elevated higher (23°C treatment) at 2 and 20 ng HSP70 mg−1 total protein, respectively.
For coho salmon, separation in the histogram at 3 ng HSP70 mg−1 total protein denoted the threshold for HSP70 elevation and heat stress (Figure 7). Among coho salmon, HSP70 concentrations were always elevated among individuals exposed to 23°C and rarely elevated in individuals from 15 to 21°C water temperature treatments (0% of individuals in the 15°C and 17°C treatments, 4% of individuals in the 19°C treatment, 12% of individuals in the 21°C treatment and 100% of individuals in the 23°C treatment). The differences in the percentage of individual coho salmon with elevated HSP70 was related to treatment temperature (χ2 = 121, df = 4, P < 0.001). Only the 23°C temperature treatment was considered stressful for these hatchery-reared juvenile coho salmon. The baseline HSP70 concentration was 0.72 ± 0.70 (mean ± SD) ng HSP70 mg−1 total protein among the individuals without elevated HSP70 (n = 108).
Gene transcription-based heat stress classification
Random forest classification models were able to separate individuals exposed to stressful and unstressful water temperatures in Chinook salmon and coho salmon based on transcription of all 13 genes or the reduced subset of genes when judged by ‘training data’ used to fit models and ‘validation data’ that was independent of model fitting (Tables 2 and 3; Figure 8). Among Chinook salmon, gene transcription classified exposure to stressful water temperatures (≥21°C) correctly in 90% (n = 61) of individuals in the training data set and 86% (n = 90) of individuals in the validation dataset of the full model with all 13 genes included. When the model was reduced to the most influential genes, only six genes were retained (hsp70, hsp90, gata3, tbx21, cyp11a and mt-a) (Table 1) and exposure to stressful temperatures was correctly classified in 92% of individuals used in the training data set and 87% of individuals in the validation data set.
Table 3.
Mean decrease in accuracy for each gene used in a random forest model to identify heat stress using transcription of all 13 genes examined (full model) or a reduced model of the most influential genes to identifying heat stress
| Chinook salmon | Coho salmon | |||
|---|---|---|---|---|
| Gene | Full model | Reduced model | Full model | Reduced model |
| hsp27 | 3.02 | 4.69 | ||
| hsp70 | 24.7 | 29.1 | 14.58 | 16.3 |
| hsp90 | 17.4 | 21.8 | 13.43 | 17.0 |
| gata3 | 4.44 | 8.35 | −1.49 | |
| ifna | 4.33 | 1.36 | ||
| ifng2 | 0.85 | −1.08 | ||
| mx1 | 1.24 | −0.31 | ||
| tbx21 | 1.47 | 5.17 | −0.16 | |
| ahr | 0.56 | −0.71 | ||
| cyp1a | 4.27 | 8.05 | 3.53 | 7.63 |
| sod | 4.18 | −1.36 | ||
| mt-a | 5.19 | 5.58 | 0.60 | |
| leptin | 4.74 | 5.22 | 5.06 | |
Higher values indicate genes that improve model accuracy more, values near zero indicate genes that have little influence on model accuracy and negative values indicate genes that reduce model accuracy. Models are all fit using the training data from 61 Chinook salmon and 31 coho salmon sampled after a controlled heat stress experiment. Stressful temperature treatments were defined as 21 and 23° for Chinook salmon and 23°C for coho salmon.
Figure 8.
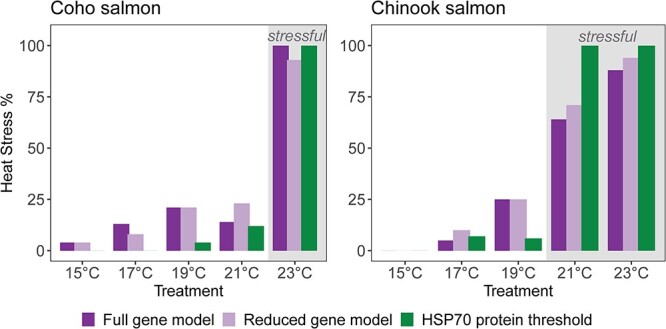
The percentage of individuals with the heat stress in each temperature treatment by classification approach and species. Classification approaches are shown as separate bars: full gene model (left bar), reduced gene model (middle bar) and HSP70 concentration threshold (right bar). This figure combines individuals in 12 and 36 h exposures because exposure time would not be known in future applications. Results from the full and reduced gene models are based on the validation data set (n = 90 Chinook salmon; n = 110 coho salmon). Results from the HSP70 threshold use all individuals (n = 151 Chinook salmon; n = 141 coho salmon). Shaded treatments are water temperatures deemed stressful based on > 50% elevation of HSP70 protein.
Among coho salmon, gene transcription correctly classified exposure to stressful water temperatures (≥23°C) in 87% of individuals in the training data set (n = 31) and 89% of individuals in the validation dataset (n = 110) of the full model with all 13 genes included (Table 2; Figure 8). When the model was reduced to the most influential genes, only four genes were retained (hsp70, hsp90, cyp1a and leptin). The reduced model identified heat stress correctly in 88% of coho salmon in the training data set and 87% in the validation data set.
Discussion
Major findings
There was a clear effect of water temperature on both the HSP70 concentration and gene transcription (mRNA) of hsp70 and hsp90 in Chinook salmon and coho salmon. Our findings are consistent with other studies of heat stress in Pacific salmon, Atlantic salmon (Salmo salar), brook trout (Salvelinus fontinalis) and Arctic charr (Salvelinus alpinus) (Evans et al., 2011; Chadwick et al., 2015; Lewis et al., 2016; Akbarzadeh et al., 2018; Houde et al., 2019; Bowen et al., 2020; von Biela et al., 2020) that find increases in hsp transcription and HSP70 protein with warmer water temperatures. Stressful water temperature treatments in this study were 21°C and 23°C for juvenile Chinook salmon, but only 23°C for juvenile coho salmon based on distinctly elevated HSP70 (Figure 7). Our results generally agree with past studies of juvenile Pacific salmon that indicate temperatures > 20°C are stressful for juveniles (reviewed by Mayer, 2022). Threshold temperatures for induction of the inducible forms of heat shock protein, such as the ones measured here, have been found for most organisms examined, and the threshold temperature generally increases with increasing thermal tolerance of the species (Feder and Hofmann, 1999). Therefore, the differences in water temperatures that resulted in elevated HSP70 suggests that juvenile Chinook salmon are more temperature sensitive than juvenile coho salmon. Our results provided several indications that transcription and translation-based heat stress assessments are related, but not necessarily directly correlated as noted elsewhere (Lund et al., 2002; Buckley et al., 2006; Lewis et al., 2016; Mottola et al., 2020). First, we saw an increase in hsp70 and hsp90 transcription at water temperatures 2°C cooler than the increase in energy costly translation of HSP70 protein for Chinook salmon (19°C vs 21°C; Figures 3 and 4), similar to differences noted in other studies (Lund et al., 2002; Lewis et al., 2016; Mottola et al., 2020). Relatedly, the heat stress classification results differed where transcription-based models estimated higher heat stress percentages for Chinook salmon at 19°C and coho salmon at 17–19°C compared to HSP70-based percentages (Figure 8). Last, there were instances of individuals with elevated HSP70 protein, but not hsp70 transcription, indicating that transcription can be intermittent during heat stress (Figure 6). Poor correlation between mRNA produced by transcription and protein from translation at a particular snapshot can be related to the lag between transcription and translation (Buckley et al., 2006; Lewis et al., 2016). However, that lag is thought to be shorter (1–3 h depending on tissue type in Lewis et al., 2016) than the > 12 h exposure used in this experiment. Moreover, there was no indication that elevated hsp70 transcription from the 12 h exposure was followed by elevated HSP70 translation at 36 h. Instead, HSP70 protein was similar between 12 h and 36 h in all temperature treatments and suggested that our time course was sufficient to capture transcription and translation. Thus, the data are more consistent with the notion of post-transcriptional regulation that allows HSP70 transcription and translation to be disconnected in certain temperature ranges and/or lengths of exposure (Lund et al., 2002; Lewis et al., 2016; Mottola et al., 2020). Lewis et al. (2016) predicted that an increase in both HSP70 mRNA and protein occurs only at the high end of temperature tolerance when there is a threat of mortality to balance preparedness with the high energetic cost of translation. When transcription of mRNA and translation of protein are considered separate steps in the larger-tiered stress response (see Figure 1Bowen et al., 2020) alongside the more dramatic responses of cell death and salmon mortality, anticipating step-specific activation temperature becomes intuitive.
Our approach to classifying a temperature treatment as “stressful” required a distinct HSP70 protein elevation, rather than an increase in the mean or median of any HSP biomarker considered (gene transcription or protein translation). Distinct HSP70 elevation in muscle tissue was apparent in the 21 and 23°C treatments as > 2 ng HSP70.mg−1 total protein in Chinook salmon and the 23°C treatment as > 3 ng HSP70.mg−1 total protein in Coho salmon. The water temperature needed to induce HSP70 elevation can vary by species, population, exposure duration and life stage (Iwama et al., 1998; Fowler et al., 2009; Lewis et al., 2016). For example, HSP70 was elevated at a lower water temperature treatment (18°C) in the muscle tissue of migrating adult Chinook salmon (von Biela et al., 2020) in line with a lower thermal tolerance for migrating (spawning) adults compared to juveniles as previously established (McCullough, 1999; Evans et al., 2011; Miller et al., 2011; Jeffries et al., 2012; Teffer et al., 2019; Dahlke et al., 2020; Hinch et al., 2021; Mayer, 2022).
Species comparison
The higher levels of HSP70 and hsp transcription at 21 and 23°C in Chinook salmon compared to coho salmon indicates that juvenile Chinook salmon in our study were more temperature sensitive than juvenile coho salmon. Our results agree with conclusions from Mayer (2022) that juvenile coho salmon appear to have a higher upper thermal tolerance range than juvenile Chinook salmon based on literature review. However, direct comparison of thermal tolerance of these (and other) species of Pacific salmon using the same methods would be a valuable addition to our ability to predict differential responses to climate change. Our comparative experimental effort lays the groundwork for future research projects to assess whether wild Chinook salmon have evidence of heat stress at lower water temperatures or at a higher prevalence compared to co-occurring wild coho salmon. If wild Chinook salmon are consistently shown to have stronger temperature sensitivity than co-occurring coho salmon, then it may help explain why Chinook salmon populations have more widespread declines and conservation concerns. Furthermore, differences in heat stress prevalence among spawning adults could arise because northern populations of Chinook salmon are mostly summer-run and overlap with seasonal water temperature peaks more so than fall-run coho salmon.
Development of heat stress biomarkers in muscle tissue
We took advantage of recent developments that allow HSP70 concentration to be assessed using an ELISA that provided finer resolution data at lower concentrations than western blots and enables faster and more reliable HSP70 protein assessment. Detection of low abundance proteins or detection of proteins from small quantity samples can be a major challenge when performing western blotting, resulting in faint or undetectable band(s) during the imaging step and the possibility of inconclusive data analysis. White skeletal muscle has not been the primary focus for stress biomarker development in fishes (Fowler et al., 2009; Jeffries et al., 2021) because the concentration of HSP70 tends to be lower than other options such as liver or gill (Iwama et al., 1998; Fowler et al., 2009; Bowen et al., 2020). Disadvantages of other tissues for heat stress detection include the necessity of lethal sampling for liver tissue, questionable reliability for non-lethal fin clips (Madeira et al., 2017) and anesthesia considerations to collect non-lethal gill tissue (McCormick, 1993; Cooke et al., 2005). Anesthesia complicates non-lethal sampling of salmon because of strict requirements for fishes that might be consumed by people (e.g. 21 day hold time following use of approved anesthetics) (Priborsky and Velisek, 2018). Cooke et al. (2005) provides a protocol that includes gill biopsy of adult salmonids without anesthesia due to human consumption after release, but it was carried out by those with a high level of fish handling experience to ensure fish and researcher safety. Muscle tissue now provides another reliable measure of HSP70 abundance with a non-lethal option among adults (Bøe et al., 2020) that can be accomplished by technicians stationed at monitoring projects like weirs (von Biela et al., 2020).
HSP70 concentrations demonstrated a threshold pattern with distinct separation between cooler and warmer water temperature treatments that was fundamentally different from the gradual and overlapping values for transcription of hsp genes. As a result, HSP70 concentrations could identify heat stress alone, but transcription from an individual gene could not. Baseline HSP70 concentrations in muscle tissue of juvenile salmon held in cooler temperature treatments were near zero, similar to muscle tissue of wild adult Chinook salmon captured along their spawning migration and held as ‘controls’ (i.e. ~15°C) during a previous heat stress experiment (Donnelly et al., 2020; von Biela et al., 2020). A caveat of comparing data presented in this paper and data presented in von Biela et al. (2020) is the difference in the laboratory approaches used to measure HSP70 concentrations (ELISA vs western blot). Improved resolution of HSP70 concentrations with the ELISA method does impede a direct comparison between HSP70 heat stress thresholds between the two studies, but not the generalization that baseline HSP70 levels were near zero for juveniles and adults. Similarity in baseline HSP70 concentrations is consistent with the ability to use the same HSP70 thresholds to identify heat stress across life history stages. Nevertheless, studies should apply HSP70 thresholds with caution because any life stage can have events that temporarily and minimally elevate HSP70 for reasons unrelated to water temperature (e.g. river entry was suspected to elevate HSP70 in von Biela et al., 2020).
Identifying heat stress with a gene transcription approach required multiple genes with the combination of hsp70 and hsp90 being particularly helpful out of the genes examined. With the suite of genes considered here, transcription was able to correctly identify exposure to stressful temperatures > 85% of individuals with a good balance between detecting heat stress (i.e. sensitivity) and the absence of heat stress (i.e. specificity). Other studies that rely on transcription to identify physiological stressors also use a combination of genes to identify stress exposure in fish, bird and mammal species (Jeffries et al., 2021; Trego et al., 2021). The gene transcription signature associated with heat stress in this study was generally similar to other examples in the literature in that one or more hsp genes were highly influential, but genes related to other cellular functions could also improve predictions (Buckley et al., 2006; Evans et al., 2011; Anttila et al., 2014; Tomalty et al., 2015; Akbarzadeh et al., 2018; Houde et al., 2019; Dettleff et al., 2020). In this example, genes related to immune system (gata3 and tbx21 for Chinook salmon) and detoxification (cyp1a) were helpful at classifying heat stress when considered in tandem with hsp70 and hsp90, even when they were not strongly influenced by water temperature when considered alone. For Chinook salmon, the three most influential genes (hsp70, hsp90, gata3) for identifying heat stress in this study were consistent with those identified during an experiment of wild Yukon River Chinook salmon spawning adults (von Biela et al., 2020). This consistency between juvenile and adult spawning Chinook salmon heat stress gene transcription signatures reinforces the idea that biomarkers developed from juveniles can be applied to adults. The 13 genes examined here represent one possible gene panel out of thousands of known Chinook and coho salmon genes. Full transcriptome studies of muscle tissue from individuals exposed to different water temperatures can help direct the selection of an improved selection of genes for heat stress identification.
Some individuals had a mismatch between their biomarkers and water temperature experiences that may reflect individual variation in temperature tolerance (i.e. biomarkers consistent with heat stress even when they were not in a stressful temperature treatment for HSP70 and gene transcription and vice versa for gene transcription). Temperature tolerance is malleable with age, body condition, pathogen exposure, temperature acclimation and stress history (Bruneaux et al., 2017; Gallant et al., 2017; Turko et al., 2020; Rodgers and Gomez Isaza, 2022). Individual differences in thermal tolerance have been found in brook trout (Salvelinus fontinalis) and other species and are repeatable over most of the animal’s adult stage (O’Donnell et al., 2020), indicating a possible genetic basis as well. Beyond individual variation, HSP70 concentrations and gene transcription can be elevated by latent (i.e. unidentified) abiotic factors including pollution (Iwama et al., 1998; Köhler et al., 2001) and social stress (Currie et al., 2010). In this study, individual variation in temperature tolerance appears the most likely explanation of biomarker and temperature mismatch because the patterns observed in the mismatches: there were almost no mismatches in the coolest temperature treatment and mismatches were most common when approaching stressful temperatures suggesting that these individuals simply experienced heat stress at slightly lower water temperatures than their conspecifics. Classification analyses make the simplifying assumptions that temperature tolerances are the same across individuals within a species and consider these mismatched individuals to be misclassifications. While we cannot definitively separate true misclassifications from individual differences in temperature tolerances, these individuals comprise only 3% of cases for HSP70 elevation and such a low rate is encouraging for future application to wild Pacific salmon when individual variation and other stressors will have more influence on HSP70.
Recommendations for future studies
The results of this experiment support the use of HSP70 protein elevation measured in white muscle tissue as a Chinook salmon and coho salmon heat stress indicator for individuals during research and monitoring with an option for non-lethal muscle biopsy for adults (Bøe et al., 2020). The simple threshold that distinguishes normal from elevated HSP70 protein levels is transparent, does not require statistical classification analyses and is understandable to other researchers and the public alike. If other researchers choose to apply our HSP70 thresholds to identify heat stress, we strongly encourage some level of indirect validation to ensure threshold applicability (i.e. plotting HSP70 concentrations compared to recent maximum water temperatures to verify that HSP70 elevation is temperature-related where the range of water temperatures spans optimal to supraoptimal). The two distinct levels of HSP70 elevation from Chinook salmon treated with 21°C and 23°C water raises the possibility that HSP70 concentrations could confer some information about mortality risk that could be examined in future studies. Gene transcription-based tools for group-level research and monitoring were also supported by our results. Individual-level heat stress detections based on gene transcription remain promising and require less tissue, allowing for non-lethal sampling of juvenile fishes (Jeffries et al., 2021), but are inherently more complex and require further study in Pacific salmon muscle tissue.
Acknowledgements
We thank the Alaska Department of Fish and Game staff at the William Jack Hernandez Sport Fish Hatchery in Anchorage for providing fish and space for this experiment and fish holding expertise, especially Gary George, Charles Pratt and Diane Loopstra. We thank Erin Faught and Matt Vijayan for sharing their rainbow trout ELISA protocol, Sarah Laske for helping process fish following the experiment and Daniel Hall for his help processing samples at the USGS S.O. Conte Research Laboratory. Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Contributor Information
Vanessa R von Biela, U.S. Geological Survey, Alaska Science Center, 4210 University Drive, Anchorage, AK 99508, USA.
Amy M Regish, U.S. Geological Survey, Eastern Ecological Science Center at the S.O. Conte Research Laboratory, One Migratory Way, Turners Falls, MA 01376, USA.
Lizabeth Bowen, U.S. Geological Survey, Western Ecological Science Center, One Shields Avenue, Davis, CA 95616, USA.
Ashley E Stanek, U.S. Geological Survey, Alaska Science Center, 4210 University Drive, Anchorage, AK 99508, USA.
Shannon Waters, U.S. Geological Survey, Western Ecological Science Center, One Shields Avenue, Davis, CA 95616, USA.
Michael P Carey, U.S. Geological Survey, Alaska Science Center, 4210 University Drive, Anchorage, AK 99508, USA.
Christian E Zimmerman, U.S. Geological Survey, Alaska Science Center, 4210 University Drive, Anchorage, AK 99508, USA.
Jonathon Gerken, Anchorage Field Office, U.S. Fish and Wildlife Service, 4700 BLM Road, Anchorage, AK 99507, USA.
Daniel Rinella, Anchorage Field Office, U.S. Fish and Wildlife Service, 4700 BLM Road, Anchorage, AK 99507, USA.
Stephen D McCormick, U.S. Geological Survey, Eastern Ecological Science Center at the S.O. Conte Research Laboratory, One Migratory Way, Turners Falls, MA 01376, USA.
Author Contributions
All authors participated in conceptualization, funding acquisition, and writing – review & editing. Data Curation by A.E.S. and S.W.; formal analysis by V.R.V.; investigation by V.R.V., A.E.S., A.M.R., L.B. and S.W.; Methodology by V.R.V., A.M.R., L.B., M.P.C., and S.D.M.; project administration and supervision by V.R.V, L.B., J.G., and S.D.M.; resources by V.R.V., L.B., A.M.R, S.D.M., and William Jack Hernandez Sport Fish Hatchery Staff; software by the R Core Team; validation by L.B., S.W., and A.M.R; visualization by V.R.V. and A.E.S.; writing – origional draft by V.R.V.
Conflict of Interest
The authors declare no competing interest.
Funding
This work was supported by the USGS Science Support Partnership Program, USGS Ecosystem Mission Area and U.S. Fish and Wildlife Service.
Data Availability
The data presented in this study are available in Stanek et al. (2023).
References
- ADFG (2013) Chinook Salmon Stock Assessment and Research Plan, 2013. In Alaska Department of Fish and Game, Special Publication No. 130-01, Anchorage. https://www.adfg.alaska.gov/static/home/news/ho ttopics/pdfs/chinook_research_plan.pdf. [Google Scholar]
- Akbarzadeh A, Günther OP, Houde AL, Li S, Ming TJ, Jeffries KM, Hinch SG, Miller KM (2018) Developing specific molecular biomarkers for thermal stress in salmonids. BMC Genomics 19: 749. 10.1186/s12864-018-5108-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anttila K, Eliason EJ, Kaukinen KH, Miller KM, Farrell AP (2014) Facing warm temperatures during migration: cardiac mRNA responses of two adult Oncorhynchus nerka populations to warming and swimming challenges. J Fish Biol 84: 1439–1456. 10.1111/jfb.12367. [DOI] [PubMed] [Google Scholar]
- Armstrong JB, Schindler DE, Ruff CP, Brooks GT, Bentley KE, Torgersen CE (2013) Diel horizontal migration in streams: juvenile fish exploit spatial heterogeneity in thermal and trophic resources. Ecology 94: 2066–2075. 10.1890/12-1200.1. [DOI] [PubMed] [Google Scholar]
- Bøe K, Robertson MJ, Fleming IA, Power M (2020) Evaluating the effect of dorsal muscle biopsies on adult Atlantic salmon growth and marine return rates. Conserv Physiol 8: 1–10. 10.1093/conphys/coz099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen L, Biela VR, McCormick SD, Regish AM, Waters SC, Durbin-Johnson B, Britton M, Settles ML, Donnelly DS, Laske SMet al. (2020) Transcriptomic response to elevated water temperatures in adult migrating Yukon River Chinook salmon (Oncorhynchus tshawytscha). Conserv Physiol 8: 1–22. 10.1093/conphys/coaa084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowerman TE, Keefer ML, Caudill CC (2021) Elevated stream temperature, origin, and individual size influence Chinook salmon prespawn mortality across the Columbia River basin. Fish Res 237: 105874. 10.1016/j.fishres.2021.105874. [DOI] [Google Scholar]
- Brett JR (1952) Temperature tolerance in Young Pacific Salmon, genus Oncorhynchus. J Fish Res Bd Can 9: 265–323. 10.1139/f52-016. [DOI] [Google Scholar]
- Bruneaux M, Visse M, Gross R, Pukk L, Saks L, Vasemägi A (2017) Parasite infection and decreased thermal tolerance: impact of proliferative kidney disease on a wild salmonid fish in the context of climate change. Funct Ecol 31: 216–226. 10.1111/1365-2435.12701. [DOI] [Google Scholar]
- Buckley BA, Gracey AY, Somero GN (2006) The cellular response to heat stress in the goby Gillichthys mirabilis: a cDNA microarray and protein-level analysis. J Exp Biol 209: 2660–2677. 10.1242/jeb.02292. [DOI] [PubMed] [Google Scholar]
- Carey MP, Keith KD, Schelske M, Lean C, McCormick SD, Regish A, Zimmerman CE (2019) Energy depletion and stress levels in sockeye salmon migrating at the northern edge of their distribution. Trans Am Fish Soc 148: 785–797. 10.1002/tafs.10172. [DOI] [Google Scholar]
- Carey MP, Biela VR, Dunker A, Keith KD, Schelske M, Lean C, Zimmerman CE (2021) Egg retention of high-latitude sockeye salmon (Oncorhynchus nerka) in the Pilgrim River, Alaska, during the Pacific marine heatwave of 2014–2016. Polar Biol 44: 1643–1654. 10.1007/s00300-021-02902-8. [DOI] [Google Scholar]
- Chadwick JG, Nislow KH, McCormick SD (2015) Thermal onset of cellular and endocrine stress responses correspond to ecological limits in brook trout, an iconic cold-water fish. Conserv Physiol 3: cov017. 10.1093/conphys/cov017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke SJ, Crossin GT, Patterson DA, English KK, Hinch SG, Young JL, Alexander RF, Healey MC, Van Der Kraak G, Farrell AP (2005) Coupling non-invasive physiological assessments with telemetry to understand inter-individual variation in behaviour and survivorship of sockeye salmon: development and validation of a technique. J Fish Biol 67: 1342–1358. 10.1111/j.1095-8649.2005.00830.x. [DOI] [Google Scholar]
- Currie S, LeBlanc S, Watters MA, Gilmour KM (2010) Agonistic encounters and cellular angst: social interactions induce heat shock proteins in juvenile salmonid fish. Proc R Soc B Biol Sci 277: 905–913. 10.1098/rspb.2009.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlke FT, Wohlrab S, Butzin M, Pörtner H-O (2020) Thermal bottlenecks in the life cycle define climate vulnerability of fish. Science 369: 65–70. 10.1126/science.aaz3658. [DOI] [PubMed] [Google Scholar]
- Desforges JE, Birnie-Gauvin K, Jutfelt F, Gilmour KM, Eliason EJ, Dressler TL, McKenzie DJ, Bates AE, Lawrence MJ, Fangue Net al. (2023) The ecological relevance of critical thermal maxima methodology for fishes. J Fish Biol 102: 1000–1016. 10.1111/jfb.15368. [DOI] [PubMed] [Google Scholar]
- Dettleff P, Zuloaga R, Fuentes M, Gonzalez P, Aedo J, Estrada JM, Molina A, Valdés JA (2020) Physiological and molecular responses to thermal stress in red cusk-eel (Genypterus chilensis) juveniles reveals atrophy and oxidative damage in skeletal muscle. J Therm Biol 94: 102750. 10.1016/j.jtherbio.2020.102750. [DOI] [PubMed] [Google Scholar]
- Dietz TJ, Somero GN (1993) Species- and tissue-specific synthesis patterns for heat-shock proteins HSP70 and HSP90 in several marine teleost fishes. Physiol Zool 66: 863–880. 10.1086/physzool.66.6.30163744. [DOI] [Google Scholar]
- Donnelly DS, Von Biela VR, McCormick SD, Laske SM, Carey MP, Waters S, Bowen L, Brown RJ, Larson S, Zimmerman CE (2020) A manipulative thermal challenge protocol for adult salmonids in remote field settings. Conserv Physiol 8: coaa074. 10.1093/conphys/coaa074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner B, Catalano MJ, Peterman RM (2018) Spatial and temporal patterns of covariation in productivity of chinook salmon populations of the northeastern pacific ocean. Can J Fish Aquat Sci 75: 1082–1095. 10.1139/cjfas-2017-0197. [DOI] [Google Scholar]
- Eliason EJ, Clark TD, Hague MJ, Hanson LM, Gallagher ZS, Jeffries KM, Gale MK, Patterson DA, Hinch SG, Farrell AP (2011) Differences in thermal tolerance among sockeye salmon populations. Science 332: 109–112. 10.1126/science.1199158. [DOI] [PubMed] [Google Scholar]
- Elliott JM, Elliott JA (1995) The effect of the rate of temperature increase on the critical thermal maximum for parr of Atlantic salmon and brown trout. J Fish Biol 47: 917–919. 10.1111/j.1095-8649.1995.tb06014.x. [DOI] [Google Scholar]
- Ern R, Andreassen AH, Jutfelt F (2023) Physiological mechanisms of acute upper thermal tolerance in fish. Phys Ther 38: 141–158. 10.1152/physiol.00027.2022. [DOI] [PubMed] [Google Scholar]
- Evans TG, Hammill E, Kaukinen K, Schulze AD, Patterson DA, English KK, Curtis JMR, Miller KM (2011) Transcriptomics of environmental acclimatization and survival in wild adult Pacific sockeye salmon (Oncorhynchus nerka) during spawning migration. Mol Ecol 20: 4472–4489. 10.1111/j.1365-294X.2011.05276.x. [DOI] [PubMed] [Google Scholar]
- Faught E, Henrickson L, Vijayan MM (2017) Plasma exosomes are enriched in Hsp70 and modulated by stress and cortisol in rainbow trout. J Endocrinol 232: 237–246. [DOI] [PubMed] [Google Scholar]
- Feder ME, Hofmann GE (1999) Heat-shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu Rev Physiol 61: 243–282. 10.1146/annurev.physiol.61.1.243. [DOI] [PubMed] [Google Scholar]
- Fowler SL, Hamilton D, Currie S (2009) A comparison of the heat shock response in juvenile and adult rainbow trout (Oncorhynchus mykiss): implications for increased thermal sensitivity with age. Can J Fish Aquat Sci 66: 91–100. 10.1139/F08-192. [DOI] [Google Scholar]
- Gallant MJ, LeBlanc S, MacCormack TJ, Currie S (2017) Physiological responses to a short-term, environmentally realistic, acute heat stress in Atlantic salmon, Salmo salar. Facets 2: 330–341. 10.1139/facets-2016-0053. [DOI] [Google Scholar]
- Gates JL (2022) Operational Plan: Crooked Creek Chinook Salmon Enhancement Project, 2022–2024. In ALaska Department of Fish and Game, Division of Sport Fish, Regional Operational Plan No. ROP.SF.2A.2022.25, Anchorage, AK. https://www.adfg.alaska.gov/FedAidPDFs/ROP.SF.2A.2022.25.pdf. [Google Scholar]
- Gilbert MJH, Adams OA, Farrell AP (2022) A sudden change of heart: warm acclimation can produce a rapid adjustment of maximum heart rate and cardiac thermal sensitivity in rainbow trout. Curr Res Physiol 5: 179–183. 10.1016/j.crphys.2022.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert MJH, Donadt CR, Swanson HK, Tierney KB (2016) Low annual fidelity and early upstream migration of anadromous Arctic char in a variable environment. Trans Am Fish Soc 145: 931–942. 10.1080/00028487.2016.1173095. [DOI] [Google Scholar]
- Hinch S, Bett NN, Eliason EJ, Farrell AP, Cooke SJ, Patterson DA (2021) Exceptionally high mortality of adult female salmon: a large-scale pattern and a conservation concern. Can J Fish Aquat Sci 78: 639–654. 10.1139/cjfas-2020-0385. [DOI] [Google Scholar]
- Houde ALS, Akbarzadeh A, Günther OP, Li S, Patterson DA, Farrell AP, Hinch SG, Miller KM (2019) Salmonid gene expression biomarkers indicative of physiological responses to changes in salinity and temperature, but not dissolved oxygen. J Exp Biol 222: jeb198036. 10.1242/jeb.198036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard KG, Biela VR (2023) Adult spawners: a critical period for subarctic Chinook salmon in a changing climate. Glob Chang Biol 29: 1759–1773. 10.1111/gcb.16610. [DOI] [PubMed] [Google Scholar]
- Iwama GK, Thomas PT, Forsyth RB, Vijayan MM (1998) Heat shock protein expression in fish. Rev Fish Biol Fish 8: 35–56. 10.1023/A:1008812500650. [DOI] [Google Scholar]
- Jeffries KM, Hinch SG, Sierocinski T, Clark TD, Eliason EJ, Donaldson MR, Li S, Pavlidis P, Miller KM (2012) Consequences of high temperatures and premature mortality on the transcriptome and blood physiology of wild adult sockeye salmon (Oncorhynchus nerka). Ecol Evol 2: 1747–1764. 10.1002/ece3.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffries KM, Teffer A, Michaleski S, Bernier NJ, Heath DD, Miller KM (2021) The use of non-lethal sampling for transcriptomics to assess the physiological status of wild fishes. Comp Biochem Physiol Part B Biochem Mol Biol 256: 110629. 10.1016/j.cbpb.2021.110629. [DOI] [PubMed] [Google Scholar]
- Jones LA, Schoen ER, Shaftel R, Cunningham CJ, Mauger S, Rinella DJ, St. Saviour A (2020) Watershed-scale climate influences productivity of Chinook salmon populations across south central Alaska. Glob Chang Biol 26: 4919–4936. 10.1111/gcb.15155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler HR, Bartussek C, Eckwert H, Farian K, Gränzer S, Knigge T, Kunz N (2001) The hepatic stress protein (hsp70) response to interacting abiotic parameters in fish exposed to various levels of pollution. J Aquat Ecosyst Stress Recover 8: 261–279. 10.1023/A:1012935931161. [DOI] [Google Scholar]
- Lewis M, Götting M, Anttila K, Kanerva M, Prokkola JM, Seppänen E, Kolari I, Nikinmaa M (2016) Different relationship between hsp70 mRNA and hsp70 levels in the heat shock response of two salmonids with dissimilar temperature preference. Front Physiol 7: 1–12. 10.3389/fphys.2016.00511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408. 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lund SG, Caissie D, Cunjak RA, Vijayan MM, Tufts BL (2002) The effects of environmental heat stress on heat-shock mRNA and protein expression in Miramichi Atlantic salmon (Salmo salar) parr. Can J Fish Aquat Sci 59: 1553–1562. 10.1139/f02-117. [DOI] [Google Scholar]
- Madeira C, Madeira D, Diniz MS, Cabral HN, Vinagre C (2017) Comparing biomarker responses during thermal acclimation: a lethal vs non-lethal approach in a tropical reef clownfish. Comp Biochem Physiol Part A Mol Integr Physiol 204: 104–112. 10.1016/j.cbpa.2016.11.018. [DOI] [PubMed] [Google Scholar]
- Mauger S, Shaftel R, Leppi JC, Rinella DJ (2017) Summer temperature regimes in southcentral Alaska streams: watershed drivers of variation and potential implications for Pacific salmon. Can J Fish Aquat Sci 74: 702–715. 10.1139/cjfas-2016-0076. [DOI] [Google Scholar]
- Mayer NEB (2022) Thermal Tolerance of Pacific Salmon (Oncorhynchus Spp.): Methodological and Inherent Variability in Upper Thermal Tolerance Limits and Their Use in Assessing Vulnerability to Climate Change. Master’s Thesis. University of British Columbia, Vancouver, B.C., Canada [Google Scholar]
- McCormick SD (1993) Methods for non biopsy and measurement of Na+, K+-ATPase activity. Can J Aquat Sci 50: 656–658. 10.1139/f93-075. [DOI] [Google Scholar]
- McCullough DA (1999) A review and synthesis of effects of alterations to the water temperature regime on freshwater life stages of salmonids, with special reference to Chinook Salmon. Reg 10 Water Resour Assess Rep No 910-R-99-010, p 279. [Google Scholar]
- Miller KM, Li S, Kaukinen KH, Ginther N, Hammill E, Curtis JMR, Patterson DA, Sierocinski T, Donnison L, Pavlidis Pet al. (2011) Genomic signatures predict migration and spawning failure in wild Canadian salmon. Science 331: 214–217. 10.1126/science.1196901. [DOI] [PubMed] [Google Scholar]
- Mottola G, Nikinmaa M, Anttila K (2020) Hsp70s transcription-translation relationship depends on the heat shock temperature in zebrafish. Comp Biochem Physiol Part A Mol Integr Physiol 240: 110629. 10.1016/j.cbpa.2019.110629. [DOI] [PubMed] [Google Scholar]
- O’Donnell MJ, Regish AM, McCormick SD, Letcher BH (2020) How repeatable is CTmax within individual brook trout over short- and long-time intervals? J Therm Biol 89: 102559. 10.1016/j.jtherbio.2020.102559. [DOI] [PubMed] [Google Scholar]
- Olsvik PA, Lie KK, Jordal AE, Nilsen TO, Hordvik I (2005) Evaluation of potential reference genes in real-time RT-PCR studies of Atlantic salmon. BMC Mol Biol 6: 21. 10.1186/1471-2199-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poist T (2022) RStudio: integrated development environment for R.
- Priborsky J, Velisek J (2018) A review of three commonly used fish anesthetics. Rev Fish Sci Aquac 26: 417–442. 10.1080/23308249.2018.1442812. [DOI] [Google Scholar]
- Quinn TP (2018) The Behavior and Ecology of Pacific Salmon and Trout, Ed2nd. University of Washington Press, Seattle, Washington [Google Scholar]
- Rantanen M, Karpechko AY, Lipponen A, Nordling K, Hyvärinen O, Ruosteenoja K, Vihma T, Laaksonen A (2022) The Arctic has warmed nearly four times faster than the globe since 1979. Commun Earth Environ 3: 168. 10.1038/s43247-022-00498-3. [DOI] [Google Scholar]
- Richter A, Kolmes SA (2005) Maximum temperature limits for Chinook, coho, and chum salmon, and steelhead trout in the Pacific Northwest. Rev Fish Sci 13: 23–49. 10.1080/10641260590885861. [DOI] [Google Scholar]
- Rodgers EM, Gomez Isaza DF (2022) Stress history affects heat tolerance in an aquatic ectotherm (Chinook salmon, Oncorhynchus tshawytscha). J Therm Biol 106: 103252. 10.1016/j.jtherbio.2022.103252. [DOI] [PubMed] [Google Scholar]
- Sagarin RD, Somero GN (2006) Complex patterns of expression of heat-shock protein 70 across the southern biogeographical ranges of the intertidal mussel Mytilus californianus and snail Nucella ostrina. J Biogeogr 33: 622–630. 10.1111/j.1365-2699.2005.01403.x. [DOI] [Google Scholar]
- Silver JT, Noble EG (2012) Regulation of survival gene hsp70. Cell Stress Chaperones 17: 1–9. 10.1007/s12192-011-0290-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spromberg JA, Scholz NL (2011) Estimating the future decline of wild coho salmon populations resulting from early spawner die-offs in urbanizing watersheds of the Pacific northwest, USA. Integr Environ Assess Manag 7: 648–656. 10.1002/ieam.219. [DOI] [PubMed] [Google Scholar]
- Stanek AE, Bowen L, Regish AM, Waters S, Biela VR (2023) Gene transcription and heat shock protein 70 abundance in juvenile hatchery reared coho salmon and Chinook salmon during a manipulative thermal experiment, Anchorage, Alaska 2020-2021. US Geol Surv data release . 10.5066/P9GPHHTC. [DOI] [Google Scholar]
- Teffer AK, Hinch S, Miller K, Jeffries K, Patterson D, Cooke S, Farrell A, Kaukinen KH, Li S, Juanes F (2019) Cumulative effects of thermal and fisheries stressors reveal sex-specific effects on infection development and early mortality of adult coho salmon (Oncorhynchus kisutch). Physiol Biochem Zool 92: 505–529. 10.1086/705125. [DOI] [PubMed] [Google Scholar]
- Tomalty KMH, Meek MH, Stephens MR, Rincón G, Fangue NA, May BP, Baerwald MR (2015) Transcriptional response to acute thermal exposure in juvenile Chinook salmon determined by RNAseq. G three 5: 1335–1349. 10.1534/g3.115.017699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trego ML, Brown CA, Dubansky B, Hess CD, Galvez F, Whitehead A (2021) Transcriptome profiling in conservation physiology and ecotoxicology: mechanistic insights into organism—environment interactions to both test and generate hypotheses. In Madliger CL, Franklin CE, Love OP, Cooke SJ, eds, Conservation Physiology: Applications for Wildlife Conservation and Management. Oxford University Press, Oxford, UK, pp. 109–124 [Google Scholar]
- Turko AJ, Nolan CB, Balshine S, Scott GR, Pitcher TE (2020) Thermal tolerance depends on season, age and body condition in imperilled redside dace Clinostomus elongatus. Conserv Physiol 8: 1–15. 10.1093/conphys/coaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen N, Ikonomou MG, Dubetz C, MacPherson N, Sampson T, Kelly BC, Helbing CC (2010) Gene expression profiling and environmental contaminant assessment of migrating Pacific salmon in the Fraser River watershed of British Columbia. Aquat Toxicol 97: 212–225. 10.1016/j.aquatox.2009.09.009. [DOI] [PubMed] [Google Scholar]
- Biela VR, Bowen L, McCormick SD, Carey MP, Donnelly DS, Waters S, Regish AM, Laske SM, Brown RJ, Larson Set al. (2020) Evidence of prevalent heat stress in Yukon River Chinook salmon. Can J Fish Aquat Sci 77: 1878–1892. 10.1139/cjfas-2020-0209. [DOI] [Google Scholar]
- Biela VR, Sergeant CJ, Carey MP, Liller Z, Russell C, Quinn-Davidson S, Rand PS, Westley PAH, Zimmerman CE (2022) Premature mortality among Alaska’s Pacific Salmon during record heat and drought in 2019. Fisheries 47: 157–168. 10.1002/fsh.10705. [DOI] [Google Scholar]
- Welch DW, Porter AD, Rechisky EL (2021) A synthesis of the coast-wide decline in survival of west coast Chinook Salmon (Oncorhynchus tshawytscha, Salmonidae). Fish Fish 22: 194–211. 10.1111/faf.12514. [DOI] [Google Scholar]
- Xie F, Wang J, Zhang B (2023) RefFinder: a web-based tool for comprehensively analyzing and identifying reference genes. Funct Integr Genomics 23: 125. 10.1007/s10142-023-01055-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available in Stanek et al. (2023).