Abstract
Autophagy comprises a growing range of cellular pathways, which occupy central roles in response to energy deprivation, organelle turnover and proteostasis. Over the years, autophagy has been increasingly linked to governing several aspects of immunity, including host defence against various pathogens, unconventional secretion of cytokines and antigen presentation. While canonical autophagy-mediated antigen processing in thymic epithelial cells supports the generation of a self-tolerant CD4+ T cell repertoire, mounting evidence suggests that deregulated autophagy pathways contribute to or sustain autoimmune responses. In animal models of multiple sclerosis (MS), non-canonical autophagy pathways such as microtubule-associated protein 1 A/1 B-light chain 3 (LC3)-associated phagocytosis can contribute to major histocompatibility complex (MHC) class II presentation of autoantigen, thereby amplifying autoreactive CD4+ T cell responses. In systemic lupus erythematosus (SLE), increased type 1 interferon production is linked to excessive autophagy in plasmacytoid dendritic cells (DCs). In rheumatoid arthritis (RA), autophagy proteins contribute to pathological citrullination of autoantigen. Immunotherapies effective in autoimmune diseases modulate autophagy functions, and strategies harnessing autophagy pathways to restrain autoimmune responses have been developed. This review illustrates recent insights in how autophagy, distinct autophagy pathways and autophagy protein functions intersect with the evolution and progression of autoimmune diseases, focusing on MS, SLE and RA.
Keywords: Autophagy, Antigen presentation, Autoimmunity, Multiple sclerosis, Systemic lupus erythematosus, Rheumatoid arthritis
1. Autophagy – from yeast to human disease
In 1859 French physiologist M. Anselmier first coined the term “autophagy” [1]. Christian de Duve revived the expression during the Ciba Foundation Symposium on Lysosomes in 1963. Many years later, the Nobel Assembly at the Karolinska Institute in Stockholm awarded the 2016 Nobel Prize in Physiology or Medicine to Yoshinori Ohsumi for his outstanding role in identifying and functionally characterizing essential members of the autophagy machinery in yeast [2,3]. The various biological roles of autophagy are reflected by the tremendous expansion and diversification the field of autophagy research has undergone since the initial observation of the process and the subsequent identification of more than 40 autophagy-related genes/proteins (Atgs/ATGs). Initially, autophagy was mainly recognized for being a survival mechanism in response to cellular energy deficits and a housekeeping process for maintaining proteostasis. It has now become apparent that autophagy and related pathways are instrumental participants in physiological and pathological immune responses by tuning the secretion of proinflammatory molecules, limiting proliferation of pathogens and regulating adaptive immune responses through both loading and stabilization of antigens on and surface-expression of antigen-presenting molecules [4,5].
Although the exact molecular underpinnings remain unknown, it is widely accepted that certain genetic variants within the human leukocyte antigen (HLA) locus render individuals more susceptible to autoimmunity [6], which is commonly triggered by a specific adaptive immune response that is mounted against self-antigens. Pathways that facilitate antigen processing leading to MHC class II presentation of self-peptides include the classical MHC class II pathway, macroautophagy and LAP [7-10]. Because of their important function in antigen processing and presentation, autophagy pathways have been increasingly investigated in autoimmunity research. However, autophagy pathways may be linked to autoimmunity independently of antigen presentation. By taking the example of selected common autoimmune-related conditions, we will here provide a synoptic view of how the biological events during the development of autoimmunity may be influenced and regulated by autophagy.
2. Molecular characterization of autophagy and related processes
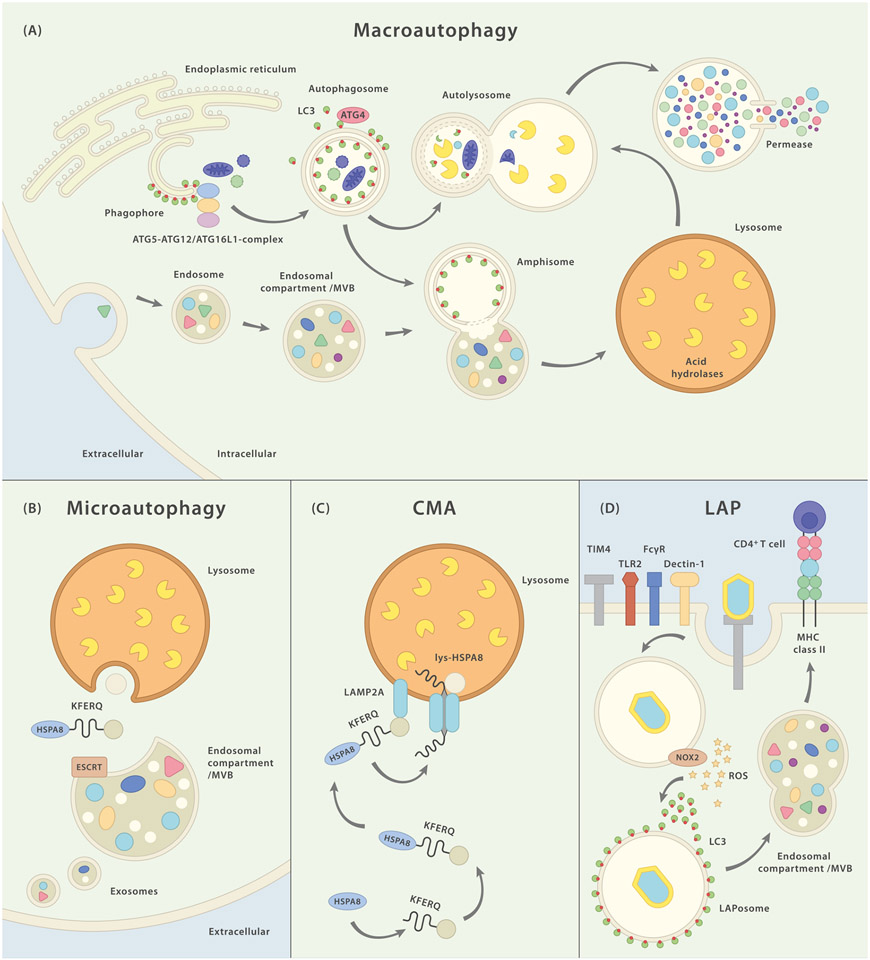
Bona fide autophagy comprises several cellular pathways that transport cytoplasmic material of both endogenous or exogenous origin to the endolysosomal compartment for enzymatic digestion [11]. According to this definition at least three distinct entities can be distinguished (Fig. 1). (i) During microautophagy (subtypes of which require known ATGs, but others do not) cytoplasmic material enters into lysosomes or late endosomes directly via membrane invaginations [12,13]. This process is endosomal sorting complex required for transport (ESCRT)-dependent if cargo enters late endosomes but occurs independently of ESCRT in case of lysosomal microautophagy [14,15]. (ii) Chaperone-mediated autophagy is the evolutionarily least conserved autophagy pathway, requires lysosome-associated membrane protein 2A (LAMP2A) and heat shock 70 kDa protein 8 (HSPA8) activity and highly selectively targets proteins containing a KFERQ-motif (Lys-Phe-Glu-Arg-Gln) in their amino acid sequence (which is also true for some forms of microautophagy) [16]. (iii) Finally, there is macroautophagy, the most broadly investigated autophagy pathway, during which neighbouring cytoplasmic constituents are sequestered by the formation of a double-membraned autophagosome – the morphological hallmark organelle that subsequently fuses with endolysosomal structures to form amphisomes or autolysosomes. This catabolically powerful degradation process, which is tightly controlled by several regulatory units (Fig. 2), can be both substrate specific or may occur unselectively in response to e.g. cellular energy deprivation.
Fig. 1.
Schematic representation of major autophagy pathways and their molecular components. (A) Macroautophagy (MA): PI3P-rich structures on the endoplasmic reticulum are believed to constitute the site of autophagogenesis and the primary origin of the phagophore. After recruitment of LC3 on the inner and outer membranes of the emanating autophagosome, the forming vesicle engulfs cytoplasmic constituents. Intraluminal LC3-II stays associated with the organelle while LC3-II on the outer membrane is removed by the hydrolase ATG4. The autophagosome closes and the completed organelle can now fuse with lysosomes to form autolysosomes in which the content is degraded and subsequently released back into the cytosol via permeases. Alternatively, autophagosomes may first merge with endosomal structures resulting in amphisomes which later on can fuse with lysosomes. (B) Microautophagy (MI): cytoplasmic material (some forms of MI appear to favor KFERQ-containing proteins) is targeted to endosomal vesicles in an HSPA8-, and ESCRT-dependent manner. Exosomes containing cytoplasmic material may be secreted into the extracellular space. “Classical” MI describes the direct invagination of cytoplasmic constituents into lysosomes. (C) Chaperone-mediated autophagy (CMA): With the help of chaperone HSPA8, this pathway targets KFERQ motif-bearing proteins to the lysosome. There, LAMP-2A unfolds the target protein and, (upon multimerization and) together with lysosomal HSPA8, transports it into the lumen. (D) LC3-associated phagocytosis (LAP): binding of a LAP-triggering cell surface receptor initiates receptor-mediated phagocytosis. Activation of membrane-bound NOX2 and the consecutive production of superoxide leads to recruitment and binding of LC3 to the outer membrane of the LAPosome. Fully assembled LAPosomes may either fuse with other endocytic vesicles or directly merge with lysosomes.
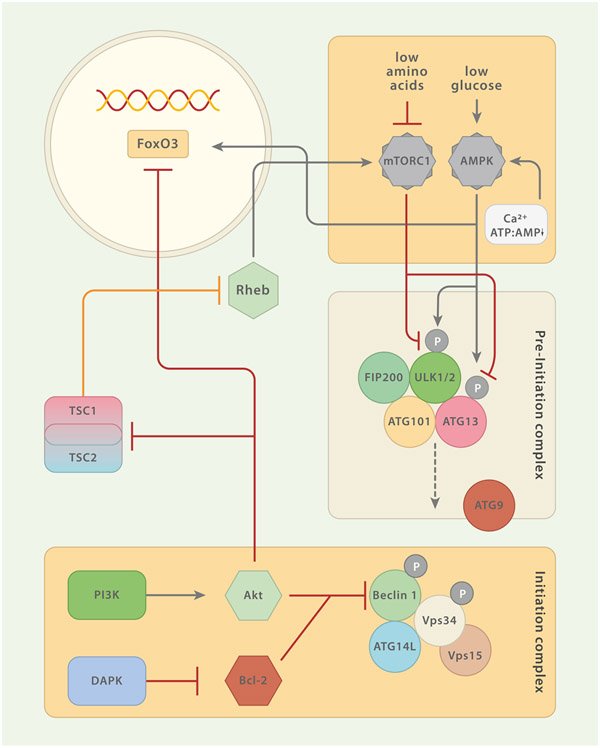
Fig. 2.
Regulatory Circuits of Macroautophagy (MA). The pre-initiation and the initiation complexes constitute the most proximal modules of MA. Several constituents of these complexes are under the control of regulatory elements. Both, AMPK and mTORC1 sense nutrients and are the two major regulatory units that control MA. mTORC1 inhibits MA by phosphorylation of pre-initiation complex member ULK1/2 (and/or ATG13). mTORC1 activity is promoted by amino acid sensing Rag GTPases which also mediate the translocation of mTORC1 to the lysosomal membrane. This translocation is important for the mTORC1-promoting activity of Ras homolog enriched in brain (Rheb). The tuberous sclerosis complex (TSC) 1/TSC2 can enhance MA by repressing Rheb. Akt reduces macroautophagic activity by inhibiting TSC1/TSC2 or via interaction with the transcription factor FoxO3. Moreover, growth factors may signal through phosphatidylinositide 3-kinase (PI3K) to Akt, which in turn inhibits the Beclin 1-containing class III PI3K complex. The calcium/calmodulin (Ca2+/CaM) serine/threonine kinase death-associated protein kinase (DAPK) activates MA via phosphorylation of Beclin 1 (initiation complex) which entails dissociation of Beclin 1 from Bcl-2. AMPK is a central positive regulator of MA. Phosphorylation of ULK1/2 (and/or ATG13) at residues different from those targeted by mTORC1 and direct interaction with FoxO3 augment MA activity. AMPK itself can be triggered by increasing levels of AMP relative to ATP and free Ca2+.
While all three of the aforementioned processes, micro-, macro- and chaperone-mediated autophagy, can be considered as bona fide autophagy (in that they direct cytoplasmic constituents of endogenous or exogenous origin to lysosomes), novel non-autodigestive pathways have emerged in recent years that require certain members of the autophagy machinery but ultimately defy the categorization as genuine autophagy. These autophagy-related processes may facilitate viral envelope acquisition during lytic infection [17], internalization of surface molecules [4, 5], secretion/exocytosis [18-20] and phagocytosis [21,22]. One pathway among the aforementioned that has been implicated in regulating antigen processing and presentation is LC3-associated phagocytosis (LAP). This specialized mode of phagocytosis is triggered by the engagement of extracellular receptors such as toll-like receptors (TLRs), Fc-receptors, C-type lectins, and phosphatidylserine (Ptd-L-Ser)-binding receptors [10,23].
2.1. Autophagogenesis
Different subtypes of macroautophagy vary in their dependence on certain autophagy core machinery proteins, autophagy adaptors and -receptors [24,25], but generally and true for most subtypes, five merging processes that are regulated by hierarchies of ATGs and other biochemical factors, pave the way to successful cargo break down in the lysosome. These canonical steps are nucleation/induction, elongation, maturation/closing, fusion and degradation.
Formation of the autophagosome is initiated by functionally distinct core modules such as the unc 51-like kinase (ULK) complex (I), the class III phosphatidylinositide 3-kinase (PI3K) complex (II), the ATG2/WD repeat domain phosphoinositide-interacting protein (WIPI) complex and the ATG9 cycling system (III), the ATG12-conjugation system (IV), and the LC3-conjugation system (V) [24].
The source of the emanating autophagosome is the cup-shaped phagophore, a double-membraned structure that emerges adjacent to endoplasmic reticulum (ER)-mitochondria contact sites and upon formation recruits the tetrameric ULK complex which consists of ULK1/2, FAK Family Kinase-Interacting Protein Of 200 kDa (FIP200), ATG13 and ATG101 and, as the most upstream core module, is under direct metabolic control via the kinases mammalian target of rapamycin complex 1 (mTORC1) and AMP-activated protein kinase (AMPK) [24,26]. Active ULK complex together with ATG9 subsequently recruits the PI3K complex (consisting of ATG14L, vacuolar protein sorting protein [Vps]34, Vps15, nuclear receptor binding factor 2 [NRBF2] and Beclin 1), which entails deposition of PI3P at designated membrane sites [27,28]. These PI3P-enriched areas function as platforms to which downstream PI3P-binding partners such as WIPI1/2 and ATG2 can now be recruited [29-31]. Their presence, in turn, is essential for autophagogenesis due to their role in recruitment and stabilization of lipidated LC3 (see below) as well as mediating the shuttling of the transmembrane protein ATG9 from peripheral membrane sources to autophagosomal initiation sites, where ATG9 aids in the provision of membrane material and removal of obsolete molecules. The ATG12-conjugation system is the first of two ubiquitin-like conjugation modules (Fig 3) that are essential for the successful generation of the autophagosome and is made up of ATG12, ATG7, ATG10, ATG5, and ATG16L1 [24]. Under the control of the E1-and E2-like enzymes ATG7 and ATG10 the ATG5-ATG12/ATG16L1 complex is assembled and then recruited via WIPI2 and FIP200 to the site of autophagosomal initiation where the complex executes its function as an E3-like enzyme for the LC3-conjugation system. This second and final conjugation system is made up of ATG4, ATG7, ATG3 and the ubiquitin-like LC3 which is lipidated by ATG3 and ATG7 with the assistance of the ATG5-ATG12/ATG16L1-complex. The lipidated LC3 or LC3-II localizes to the inner and outer membrane of the forming autophagosome where it facilitates membrane reorganization events and finally the closing of the organelle. Intraluminal LC3-II stays associated with the organelle while LC3-II on the outer membrane is removed by the hydrolase ATG4 [24]. Importantly, besides contributing during organelle elongation, LC3 mediates cargo selectivity by operating as an adaptor molecule. The interaction between LC3 and selected cargo is mediated via LC3-interacting regions (LIRs) containing the core consensus sequence [W/F/Y]xx [L/I/V] [32].
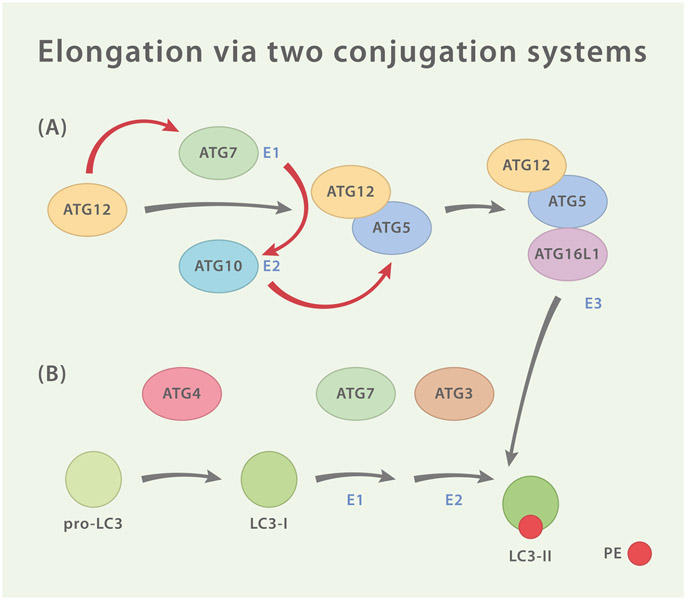
Fig. 3.
Two Ubiquitin-Like Conjugation Systems. (A) ATG12 is activated by homodimeric E1-like protein ATG7 and then transferred to E2-like protein ATG10 which catalyzes the covalent conjugation of ATG12 to ATG5. ATG16L1 associates with ATG5 and forms a tripartite conjugate which goes on to assemble tetrameric complexes with adjacent ATG5-ATG12/ATG16L1 conjugates (not shown). The complex is then recruited to the site of autophagogenesis by interaction of ATG16L1 with WIPI2 and/or FIP200. There, ATG5-ATG12/ATG16L1 functions as an E3-like enzyme for the second conjugation system (B) which consists of the ubiquitin-like LC3, the hydrolase ATG4, ATG7 and ATG3. ATG4 cleaves pro-LC3 resulting in LC3-I which exposes a C-terminal glycine. In order to generate LC3-II, this residue is then lipidated with phosphatidylethanolamine (PE) by ATG7 (E1-like), ATG3 (E2-like) and ATG5-ATG12/ATG16L1 (E3-like).
3. LAP – linking autophagy to phagocytosis
LAP is a recently discovered pathway that links phagocytosis of extracellular constituents and autophagy and has become increasingly recognized as a pivotal and versatile mechanism by which immune responses are regulated [10,23]. When LAP is triggered, a class III PI3PK complex consisting of Beclin 1, Vps34 and UV radiation resistance-associated gene protein (UVRAG) is recruited to the cytosolic membrane of the arising phagosome ensued by the recruitment of the NADPH oxidase (NOX)2 [21]. The PI3PK complex facilitates both the recruitment of the ATG12-and the LC3-conjugation systems as well as the stabilization of the NOX2 multiprotein unit on the phagosome. The resulting reactive oxygen species (ROS) and PI3P produced via the PI3PK complex are essential for the lipidation of LC3 and the decoration of the single-membrane phagosome with the resulting LC3-II, which assists in the fusion with the lysosome [21,33]. Although some components of the macroautophagy core machinery are essential for LAP (e.g., ATG5, ATG7, LC3, Beclin 1), others are dispensable (e.g., ULK, FIP200, ATG13, ATG101). Additionally, LAP requires certain factors that are expendable (e.g. NOX2) for macroautophagy or even function as an inhibitor thereof [21].
4. Autophagy in antigen presentation and central tolerance induction
Breakdown of immunological tolerance is widely acknowledged to be involved in the development of autoimmunity. During central tolerance, thymic epithelial cells (TECs) present self-antigen-derived epitopes on MHC class II molecules and ensure the generation of self-tolerant CD4+ T cell clones. TECs shape the T cell repertoire to recognize self-peptide/MHC complexes with low affinity by positive selection and weed out T cells that react to these self-structures with high affinity via negative selection [34]. Although TECs express MHC class II molecules, their endocytic capacity is low. Autophagosomes fuse frequently with MHC class II containing compartments (MIICs) in TECs [35]. Indeed, when positive selection of T cell receptor (TCR) transgenic T cells through macroautophagy-deficient thymi was investigated, only some T cell specificities were correctly selected, while others were deleted and loss of macroautophagy in TECs resulted in severe colitis and multiorgan inflammation [36]. Positive selection of CD8+ T cells was found to be macroautophagy-independent [36]. However, macroautophagy in medullary TECs (mTECs) participates in clonal deletion during thymic negative selection by facilitating the direct presentation of endogenous self-antigens [37]. These data suggest that macroautophagy processes certain self-ligands for MHC class II presentation that are involved in positive and negative T cell selection in the thymus and thereby contributes to the generation of a self-tolerant T cell repertoire. In addition to its function during the upkeep of central tolerance, macroautophagy is also found active in immature dendritic cells (DCs) [38], which have been implicated in peripheral tolerance induction [39]. Macroautophagy in DCs has recently been identified as one potential mechanism that underlies Foxp3+ regulatory T cells (Treg)-mediated immune suppression [40]. The authors show that Foxp3+ Tregs impair autolysosome formation and antigen presentation in DCs in a cytotoxic T lymphocyte associated protein 4 (CTLA4)-dependent manner. DCs in which macroautophagy had been damped via Tregs, exhibited reduced immunogenic potential and failed to prime autoantigen-specific CD4+ T cells to mediate CNS autoimmunity [40].
5. mTOR and autoimmunity
The highly conserved serine/threonine kinase mTOR belongs to the phosphatidylinositol 3-kinase-related kinase (PIKK) family and constitutes the catalytic unit of the two protein complexes mTORC1 and mTORC2 [41]. While mTORC2 has been implicated in regulation of the cytoskeleton and cell survival, mTORC1 has pivotal functions in governing cellular catabolic programs, such as autophagy [42].
Autoimmune processes regularly entail excessive tissue remodelling, cell proliferation and production of proinflammatory cytokines, all of which are metabolically demanding adaptations [43]. As a central regulator of cellular metabolism, mTOR integrates a wide range of environmental cues that include amino acid availability, mitochondrial transmembrane potential, proinflammatory mediators or pathogen-associated molecular patterns (PAMPs), thereby regulating adaptive and innate immune responses during inflammatory conditions [44,45]. Dysregulated mTOR activity in various cellular subsets has been reported in autoimmune conditions. In SLE mTOR is activated in T cells and mediates excessive IL-4 production [46,47]. While there is no evidence for significant dysregulation in T cells, fibroblast-like synoviocytes and osteoclasts depict increased mTOR activity in RA patients [48,49]. In RA-fibroblast-like synoviocytes mTOR has shown to facilitate TNF activation [50].
6. The role of autophagy in autoimmunity
That autophagy pathways orchestrate and maintain several key functions of both, the innate and the adaptive immune system is now widely acknowledged [51,52]. The growing amount of evidence that supports a pivotal role of autophagy during immune homeostasis is paralleled by mounting reports of linkage between autophagy defects and the development of autoimmunity. Notwithstanding that these defects are not limited to the process of antigen-presentation, it is this aspect of autophagy-assisted immune functions, that can be seen as exceptional for several reasons. For one, antigen presentation is a process at the very interface of innate and adaptive immunity and secondly, its role during antigen presentation is one of the first demonstrations that linked autophagy to immunity – a finding that ultimately lead to the elucidation of autophagy’s role in the establishing and upkeep of central tolerance [36-38,53,54]. In the following sections and by the example of three common autoimmune conditions, we will discuss how dysregulated autophagy is involved in the initiation or consolidation of autoimmunity.
6.1. Multiple sclerosis
Multiple sclerosis (MS) is a chronic inflammatory disease of the central nervous system (CNS) that is accompanied by progressive axonal demyelination and neurodegeneration [55,56] (Fig. 4). The strongest genetic risk factors are the HLA haplotypes HLA-DRB1*1501 and HLA-DRB5*0101 [57,58] which implies a central role for MHC-restricted antigen presentation in the disease etiology. Lesional and cerebrospinal fluid (CSF)-derived T lymphocytes from patients with MS show clonal expansion, indicating both involvement of yet unknown target antigens and a critical role for CD4+ T cells in the pathogenesis of the disease [59-62]. Possible implications for autophagy pathways in the regulation of CNS autoimmunity during MS are conceivable at both ends of the immunological synapse [63].
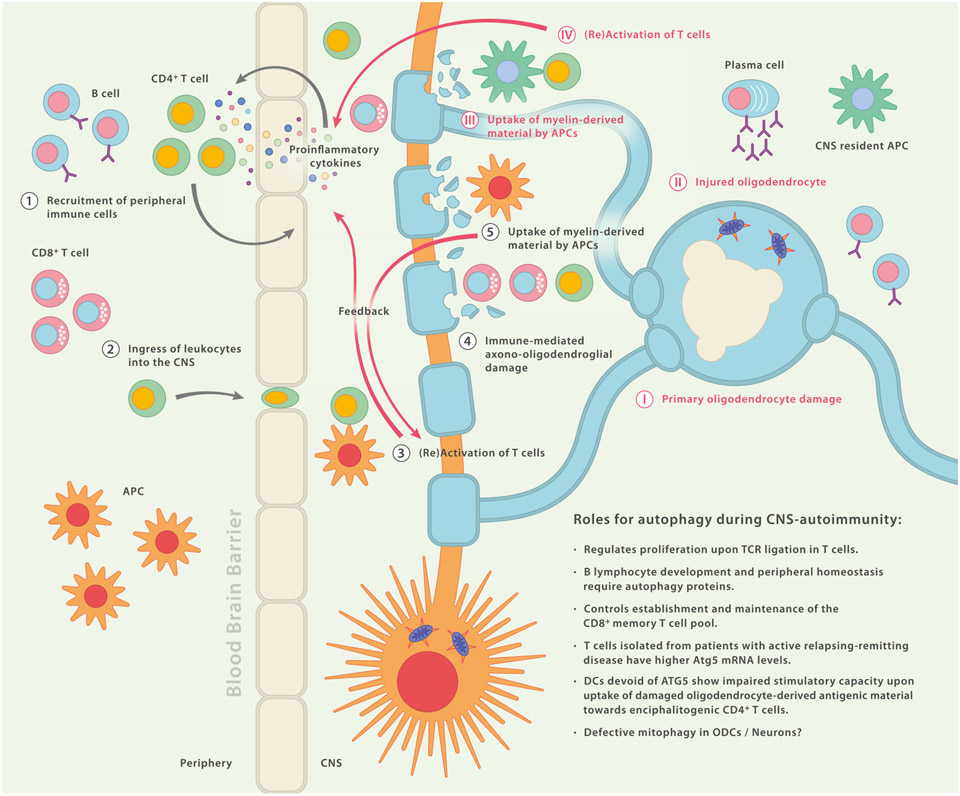
Fig. 4.
Possible Roles for Autophagy During CNS Autoimmunity. Simplified schematic diagram depicting a hypothetical sequence of events that may initiate disease (1–5): Peripheral leukocytes enter the central nervous system (CNS) where, together with resident leukocytes, they cause axono-oligodendroglial damage and trigger a vicious circle of self-perpetuating neuroinflammation (1–5). Alternatively, an initial disease triggering event occurs within the CNS (I–IV): primary oligodendrocyte damage of unknown cause may lead to injured and/or dying oligodendrocytes followed by phagocytosis of myelin-derived debris through CNS-resident antigen-presenting cells (APCs). These APCs can then present myelin-derived peptides to T cells which in turn initiate recruitment of peripheral immune cells by secretion of proinflammatory mediators. Reaching the CNS, infiltrating immune cells, including cytotoxic CD8+ T cells and myeloid cells, can cause secondary axonooligodendroglial damage. Impaired function of autophagy pathways may be relevant in several cell types and locations along these sequences.
Leaving thymic development largely unperturbed, cell-specific abrogation of autophagy has been shown to impede efficient proliferation upon TCR ligation in T lymphocyte subsets [64]. Mice lacking Beclin 1 in their T lymphocyte compartment depict complete protection from onset of experimental autoimmune encephalomyelitis (EAE), a commonly used CD4+ T cell-dependent animal model of MS [65]. Furthermore, autophagy proteins play a critical role in the establishing and maintenance of the CD8+ memory T cell pool [66,67]. Ample data support the notion that myelin-reactive T cells in MS depict a memory phenotype [68,69] and not only do patients with MS harbor elevated counts of circulating CD8+/CCR7+/CD45RA− central memory T cells (TCM) [70] but effector memory CD8+ T cells also constitute the predominant T cell subset in normal appearing white matter (NAWM) and white matter lesions (WML) of individuals with MS [71]. In line with this, circulating T cells isolated from patients with active relapsing-remitting disease exhibit significantly higher Atg5 mRNA levels in comparison to healthy controls. Moreover, elevated ATG5 transcripts and protein expression are detected in human post-mortem brain tissue from secondary progressive MS-derived specimens as compared to non-diseased control brain samples. While the aforementioned results do not unequivocally discern the cellular source of the detected mRNA and protein, immunofluorescent analyses suggest that in part these enhanced Atg5/ATG5 signals may be attributed to CD3+ T cells [72].
Supporting the notion that in general, autophagy is critical in the highly proliferative lymphocyte compartment, Miller and colleagues also found B lymphocyte development and peripheral homeostasis to require autophagy proteins [73]. Depletion of B cells via anti-CD20-directed monoclonal antibodies (mAbs) has proven to be clinically efficacious in people with relapsing-remitting MS [74,75], warranting the need for a deeper understanding of the pathoetiological role of this lymphocyte compartment during autoimmune neuroinflammation. The human γ-herpesvirus Epstein-Barr virus (EBV) infects epithelial cells and establishes latency in memory B lymphocytes and symptomatic primary EBV infection (infectious mononucleosis) in adolescents and young adults has shown to be associated with an elevated risk of developing MS [76-78]. CD4+ T cells specific for the EBV nuclear antigen 1 (EBNA1) are selectively expanded in MS patients, and a subpopulation of these virus-specific T cells cross-recognize MS-associated myelin antigens [79]. Data from studies in human B cells and from a nonhuman primate EAE model suggest that EBV- and EBV-related lymphocryptovirus-infection of B cells might render these cells’ antigen processing machinery more vulnerable to aberrant proteolysis via endolysosomal cathepsin G. This leads to the adverse protection of specific myelin protein-derived epitopes bearing a putative LC3-interacting region (LIR)-motif that allows for association with autophagosomes and potential delivery to MHC molecules [80-82].
Both the exact nature of inflammation-triggering epitopes and the identity of the cellular subsets that present these epitopes to encephalitogenic T cells within the diseased CNS during MS, remain elusive. However, a growing body of evidence supports a pivotal function of the myeloid compartment during capture, processing and presentation of disease-associated antigenic material [83-86]. In agreement with this, genome wide association studies (GWAS) revealed risk alleles for classical costimulatory molecules such as CD40, CD80 and CD86 that commonly equip professional APCs such as DCs, the prototypical immune cell for priming T cell responses [57]. During homeostasis CNS CD11c+ DCs are mainly located in proximity to entry sites that are permissive for immune cell trafficking (i.e. the choroid plexus and the juxtavascular area), thus placing them at a strategic checkpoint during the flare up of neuroinflammation [87-89]. Even though CD11c+ DCs are largely dispensable for the priming of T cells during EAE [90,91] and ablation of this subset even renders mice hypersusceptible to disease [91], presence of CNS CD11c+ DCs is an essential requirement for in situ reactivation of primed myelin-reactive T cells during EAE [83,92].
Apart from the classical endolysosomal MHC class II pathway, there are two autophagy pathways that may facilitate MHC class II presentation of autoantigens, i.e. macroautophagy and LAP. APC-intrinsic intracellular loading of endogenous antigenic cargo via canonical macroautophagy is unlikely to contribute to CNS reactivation of myelin-reactive T cells since myelin-derived candidate antigens are not known to be expressed in professional APCs. However, specialized phagocytosis pathways, such as LAP, that rely on certain autophagy modules, may well be involved. Not only is LAP known to help in the uptake of dead or injured cells, but this pathway also aids in the delivery of antigenic cargo to MHC class II loading compartments for subsequent recognition by cognate T cell clones [93,94]. It has been previously reported that conditional knockout (KO) of ATG7 in CD11c+ DCs ameliorates clinical disease severity in actively induced EAE and that targeted deletion of the essential autophagy protein ATG5 (which is known to be essential for functional LAP [21]) in DCs (including CNS DCs) leads to a significant reduction in encephalitogenic CD4+ T cell accumulation in the CNS and full protection of mice from onset of disease upon adoptive transfer EAE [84]. DCs devoid of ATG5 depict inferior stimulatory capacity upon uptake of injured oligodendrocyte-derived antigenic material towards encephalitogenic CD4+ T cells. These data therefore link ATG-dependent phagocytosis and MHC class II-mediated antigen presentation with oligodendrocyte damage and autoimmune T cell pathogenicity [84,95].
Aside from conceivable roles of autophagy pathways in resident or infiltrating hematopoietic system-arisen cell subsets during autoimmune neuroinflammation, implications for malregulated autophagy-related processes are also possible in a neuron- and neuroglia-autonomous manner. While there is preliminary evidence on the ultrastructural level for the former to promote synaptic loss during MS [96], the latter may influence several aspects in the disease etiology. Mitochondrial damage, particularly in oligodendrocytes and neurons is now believed to constitute a central and possibly early event in the disease development since aberrant versions of this organelle are frequently detected in active MS lesions [97,98]. Mitophagy-mediated clearance of dysfunctional and damaged mitochondria is a pivotal element in keeping mitostasis, thus, faulty execution of this removal may lead to a critical imbalance and to the further accumulation of ROS, which in turn may lead to further mitochondrial damage. As of now, robust evidence for dysfunctional mitophagy as a disease mechanism during MS is still lacking. A recent report found serum and CSF levels of Parkin, a ligase necessary for mitophagy, elevated in MS patients as compared to other inflammatory neurological disease controls [99]. It has yet to be clarified whether a potential defect in mitophagy is a causal and primary event or if the capacity of the process is reached downstream and due to an unknown trigger of potent mitochondrial damage.
6.2. Systemic lupus erythematosus
Systemic lupus erythematosus (SLE) is a complex multifactorial autoimmune condition with variable clinical manifestations. The disease is characterized by excessive production of autoantibodies against nucleic acids (ANA) and the spliceosome (Sm), immune complex deposition, sustained production of type I interferons (IFNs) and dysregulated T cell responses [100]. Single nucleotide polymorphisms (SNPs) that confer elevated risk to develop SLE are not limited to but include several autophagy-associated and -related genes such as Atg5 [101,102], Atg7 [101], LRRK2 [103], IRGM [101], DRAM1 [104], CDKN1B [104], MTMR3 [105], and APOL1 [106].
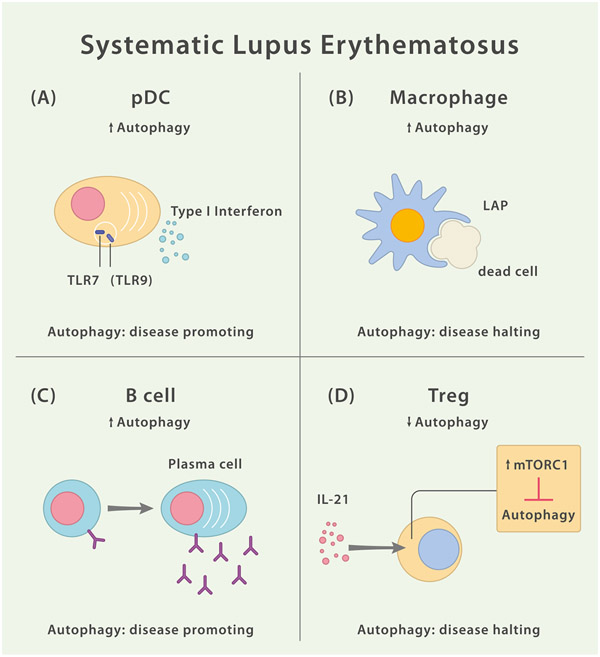
SLE shows a multifaceted clinical presentation and may affect a wide variety of organ systems throughout the disease course due to defects in both the innate and adaptive immune system. Accordingly, evidence for the involvement of autophagy pathways in the pathomechanism spans over several immune cell subsets (Fig. 5).
Fig. 5.
Dysfunctional Autophagy in Different Leukocytes Contributes to SLE Pathology. (A) Autophagy pathways may foster IFN-α release by pDCs in a TLR7 and TLR9 dependent manner. However, the role of TLR9 is complex. While both TLR7 and TLR9 signal via MyD88, TLR9 has B cell intrinsic protective functions independent of MyD88. (B) Macrophage-intrinsic LAP facilitates the efficient clearance of dead cells, thereby limiting inflammatory responses. Malfunction of this mechanism may contribute to SLE disease pathology. (C) Increased autophagy in SLE B cells appears to promote B cell differentiation into antibody-secreting plasma cells. (D) In SLE, Treg-intrinsic autophagy is inhibited via IL-21 mediated activation of mTORC1. IL-21 exposure leads to impaired Treg development and reduced suppressor function.
Dysfunctional T cell activation in SLE patients is characterized by excessive accumulation of mitochondria. Mitophagy is dependent on the translocation of dynamin-related protein 1 (Drp1) from the cytosol to mitochondria, resulting in lysosomal degradation of mitochondria [107]. The endosomal recycling regulator small GTPase HRES-1/Rab4 colocalizes with LC3 and mitochondria, and negatively regulates mitophagy [108]. In SLE patients disrupted mitophagy can be observed due to HRES-1/Rab4-mediated depletion of Drp1 [109]. Dysfunctional mitophagy in SLE is not confined to lymphocytes. Structured disposal of mitochondria physiologically occurs during erythropoiesis in a concerted process involving the ubiquitin-proteasome system (UPS) upstream of mitophagy. In SLE patients impairment of hypoxia-inducible factor (HIF)2α degradation hampers activations of the UPS leading to diminished mitophagy with accumulation of mitochondria in red blood cells [110].
SLE patients show elevated serum levels of type I IFNs (IFN-I) especially during active disease [111] and a persistent IFN-I-driven immune response is widely acknowledged to constitute a central part of the SLE disease mechanism [112]. One of the cellular sources of IFN-I is the plasmacytoid DC (pDC) which releases both IFN-α and IFN-β in response to viral and self-nucleic acids [113]. Autophagy has been implicated to facilitate the delivery of single-stranded RNA virus-derived replication intermediates to the endolysosomal compartment for efficient TLR7-mediated recognition as well as to be needed for the production of IFN-α via pDCs [114]. LAP is required to trigger release of IFN-α by pDCs in response to TLR9-mediated recognition of DNA-containing immune complexes [115]. However, the role of TLR9 is complex. While both TLR7 and TLR9 signal via MyD88, TLR9 has B cell intrinsic protective functions independent of MyD88 [116].
While autophagy appears to promote proinflammatory responses in pDCs upon integration of pattern recognition receptor-derived signals (leading to the release of IFN-α), autophagy pathways in other myeloid subsets seem to set bounds to an overshooting inflammatory reaction. The complexity of autophagy’s role during SLE pathogenesis, is corroborated by studies showing that ablation of DC- or B cell-intrinsic ATG5 in a mouse model of autoimmunity (mediated by overexpression of TLR7) leads to amelioration of the lupus-like disease course. Whereas the combined deletions of ATG5 in DCs and B cells provoke a fulminant autoantibody-driven lethal inflammation [117,118].
SLE patients-derived naïve B cells harbor higher amounts of LC3+ punctae and their amount does positively correlate with the SELENA-SLEDAI disease activity score [119]. Moreover, in vitro stimulated human B cells that are incubated in presence of an autophagy inhibitor fail to differentiate into plasmablasts. While the used agent to suppress autophagy (3-methyladenine [3-MA], a class III selective PI3K inhibitor) may have unspecific effects unrelated to the downmodulation of autophagy, mouse data using ATG7-deficient B cells show similar results in that B cells in absence of ATG7 largely fail to differentiate into CD138+ plasma cells [119]. These data imply an abnormal or increased autophagy activity in SLE B cells.
Under physiological conditions, impeccable B cell function is a requirement for the homeostasis of invariant natural killer T (iNKT) cells, an innate-like T cell subset that recognizes lipid antigens in the context of CD1d presentation. This crosstalk between B cells and iNKT cells is severely impaired in SLE patients who harbor significantly reduced peripheral iNKT cell numbers [120]. Bosma and colleagues demonstrated that the dysregulated interaction between B cells and iNKT cells is mainly founded in deranged CD1d trafficking within SLE B cells, which exhibit a marked reduction in CD1d surface levels [120]. Interestingly, in DCs, the autophagy protein ATG5 has been described to regulate CD1d surface levels and DCs devoid of ATG5 show significantly higher surface expression of CD1d and stimulate augmented iNKT cell responses in vivo [5].
Although B cells in SLE depict increased autophagy activity (thereby possibly contributing to dysregulated intracellular trafficking of critical surface molecules) and increased IFN-α levels in SLE patients can be linked to excessive autophagy in pDCs, recent evidence points towards diminished autophagy activity in SLE regulatory T cells, a phenotype that is accompanied by impaired differentiation and suppressive function of this immune cell subset [121].
6.3. Rheumatoid arthritis
Rheumatoid arthritis (RA) is a chronic autoimmune disease that affects multiple organ systems with a specific joint pattern as predilection sites and often accompanied by the presence of autoantibodies against citrullinated proteins and/or the Fc part of immunoglobulin G (= rheumatoid factor, RF). Similar to the autoimmune conditions discussed above, also in RA, the underlying pathology is complex and involves numerous cell types and tissues. Existing treatment strategies target the end stage of disease and follow a largely anti-inflammatory approach [122].
In RA, hyperproliferative synovial fibroblasts (RA synovial fibroblasts; RASF), a specialized mesenchyme-derived cell type located in the synovial intimal lining; also known as fibroblast-like synoviocytes, contribute to joint inflammation and the associated destruction of joint tissue [123].
Autophagy has a dual role in RASF by both promoting cell death induced by ER stress, and preventing apoptosis induced by proteasome inhibition [124]. This is particularly important in joint inflammation where tumor necrosis factor-α (TNF-α) exposure induces protein degradation and the ubiquitin/proteasome system is activated in order to cope with the ensuing ER stress and prolong survival [125]. Insufficient apoptosis of inflammatory cells present in the RA joint are a critical mechanism in the perpetuating inflammation and any regulation of inflammatory cell apoptosis by autophagic dysregulation is thus critical in the disease progression [126,127].
Increased autophagy following proteasomal inhibition is more pronounced in RASF compared to control-FS and inhibition of autophagy via 3-MA renders RASF more susceptible to apoptosis induced by proteasome inhibition [124] indicating that in RASF the two degradation systems work in concert to mitigate cellular stress signals for sustained viability. Recent data links IL-17-mediated disruption of mitochondrial respiration with the consecutive upregulation of autophagy and resistance towards apoptosis in RASF [128]. Taken together, there is an increasing body of evidence suggesting a possible role for dysregulated autophagy during synovial hyperplasia in RA in response to a proinflammatory microenvironment.
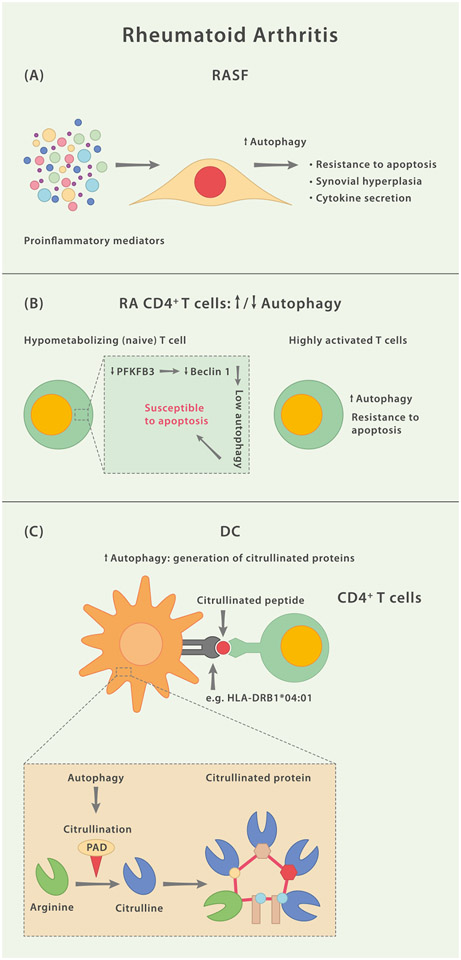
Autophagy also contributes to joint inflammation by altering the secretion of pro-inflammatory cytokines [51]. Specifically, autophagy inhibits IL-1β production by repressing inflammasome activation, and conversely, can facilitate the secretion of IL-1β [18]. Similarly, autophagy regulates IFNγ signaling and cellular inflammation by limiting ROS production [129], as well as IL-23 and transforming growth factor (TGF) β [130,131]. Proinflammatoiy cytokine signaling is critical in pathological bone loss via the activation of osteoclast precursors as previously described [132]. Autophagy not only can regulate cell survival but also activation of cells in the joint that contribute to joint inflammation and joint destruction. Additionally, autophagy can activate T cells by the regulation of antigen presentation in DCs [54,133]. In line with this, in vivo application of hydroxychloroquine (HCQ) ameliorates both arthritis incidence and disease severity in a collagen-induced animal model of arthritis [134]. Another study demonstrated that CD4+ T cells from RA patients exhibit a metabolically inept phenotype in that these cells were unable to induce the rate-limiting glycolytic enzyme 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase 3 (PFKFB3), leading to the hypometabolization of glucose. PFKFB3-deficient CD4+ cells were more prone to undergo apoptosis, suffered from reductive stress and displayed a clear inaptitude to utilize autophagy as an alternative source of energy due to the regulatory role for PFKFB3 upstream of Beclin 1 [135]. It was suggested that the hypoglycolytic phenotype in these T cells constitutes a pre-RA event upstream of inflammation. The perishing metabolically unfit and autophagy-defective T cells may open up a niche which only then, in response to lymphopenia, allows autoaggressive T cells to thrive, eventually leading to the clinical presentation of RA. While several questions remain unanswered, this model would support differential activity levels and functionalities of autophagy in distinct T helper cell subsets.
Tregs constitute a central lymphocyte subset whose major function is maintenance of immune tolerance. In doing so, Tregs interact with numerous other immune cells either via soluble mediators or through mechanisms that require direct cell-to-cell contact [136]. Patients with active RA show reduced numbers of Tregs in peripheral blood [137,138] and the typical proinflammatory environment of RA may debilitate regulatory properties of Tregs during this condition [139]. In RA synovium, Tregs colocalize with DCs in lymphoid aggregates, however as disease progresses, the DC population in aggregates appear to increase more severely than Tregs [140]. Intriguingly, one study outlines a novel CTLA4-dependent mechanism by which Foxp3+ Tregs suppress autoimmunity in vivo via inhibiting autophagy in DCs. Specifically, the binding of CTLA4 to CD80/CD86 on DCs leads to nuclear exclusion of the LC3-promoting transcription factor FoxO1 by activation of the PI3K/Akt/mTOR axis [40]. The selective costimulation modulator abatacept consists of an IgG1-derived Fc part fused to the extracellular domain of CTLA4 and is used for the treatment of RA. PBMC-derived DCs from patients with RA show a significant reduction in LC3 expression when RA patients had previously received abatacept as opposed to anti-TNF-α biologicals, suggesting that the clinical efficacy of this fusion protein may be in part due to modulation of DC-intrinsic autophagy [40].
Amongst the established targets of the autoimmune response during RA are citrullinated peptides, the presence of which serves as a highly specific serological marker for diagnosis. Citrullination (deimination) constitutes a post-translational modification of arginine side chains via peptidylarginine deiminases (PADs) and may lead to changes in the 3-dimensional structure, function and antigenic potential of a given protein [141]. For example, citrullinated vimentin exhibits modified proteolytic cleavage, which leads to a much stronger propensity to bind to HLA-DRB1*04:01 (a known risk allele for RA) than vimentin, which harbors the corresponding arginine [142]. Ireland and Unanue have previously linked DC-intrinsic autophagy to the in vivo generation and presentation of citrullinated self-antigens via MHC class II in mice [143]. Not only are the isoforms PAD2 and PAD4 expressed in peritoneal macrophages and DCs, but PAD activity is also detected in purified autophagosome fractions. Moreover, the MHC class II-restricted presentation of citrullinated peptides, but not of unmodified versions thereof, is susceptible to inhibition via autophagy modulator 3-MA, whereas amino acid starvation enhances presentation of citrullinated peptides in DCs and macrophages [143]. These data were further corroborated by a study describing a role for autophagy during the PAD4-mediated generation of citrullinated peptides in RASF [144], supporting the notion that autophagy pathways facilitate the generation of pathological post-translational modifications of peptides rendering them immunogenic and thereby promote autoimmunity. Taken together, accumulating data indicate that cell type-specific dysfunction of autophagy pathways may be involved at several intersections during RA pathology (Fig. 6).
Fig. 6.
Mechanisms Associated With Faulty Autophagy Pathways During Rheumatoid Arthritis. (A) In response to the inflammatory milieu, fibroblast-like synoviocytes in rheumatoid arthritis (RASF) upregulate autophagy, which renders these cells less susceptible to undergo apoptosis. (B) Naïve CD4+ T cells in RA are less potent in generating ATP through metabolization of glucose due to a lower activity of the enzyme 6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (PFKFB3). PFKFB3 also functions as a positive regulator of autophagy via interaction with Beclin 1. Consequently, naïve RA CD4+ T cells isolated from RA patients and stimulated ex vivo depict lower activity of autophagy and a higher propensity to undergo apoptosis. On the other hand, highly activated CD4+ T cells isolated from RA patients exhibit increased autophagy activity and a relative resistance to apoptotic stimuli. (C) Autoantibodies against citrullinated proteins are a hallmark of RA. Peptidylarginine deiminases (PAD) 2 and PAD4, enzymes that mediate the citrullination of arginine residues, have been shown to be localized in autophagosomes of antigen presenting cells (APCs) and citrullination can be regulated via modulation of autophagy.
7. Concluding remarks and future perspectives
In recent decades research efforts have come a long way from focusing on the importance of autophagy in cellular housekeeping to acknowledging its vast implications in organismal integrity. Functions of autophagy pathways in immunity include metabolic sustenance of immune cells, antigen presentation or secretion of soluble inflammatory mediators. Distinct autophagy pathways and autophagy protein functions intersect with the evolution and progression of autoimmune diseases, such as MS, SLE and RA. Autophagy may be implicated in these conditions in many ways, ranging from regulating the presentation of cell-associated antigens in the CNS during neuroinflammation [84,95], or provision of TLR ligands into endolysosomal compartments and subsequent IFN-α secretion in the context of systemic autoimmunity [114], to mediating post-translational modifications of peptides that may contribute to the progressive destruction of synovial structures [143]. Dysregulated autophagy pathways which are involved in the pathogenesis of selected autoimmune conditions are heterogeneous (e.g. hyper- and hypoactive autophagy have been held accountable for contributing to disease) and located in diverse cell types (Table 1).
Table 1.
Key examples for tissue-specific roles of autophagy pathways during autoimmunity.
| Related Disease/ Disease Model |
Type of Autophagy Impairment/Regulation of Autophagy |
Tissue | Effect | Disease Promoting (DP) or Disease Halting (DH) |
Ref. |
|---|---|---|---|---|---|
| EAE | Beclin 1 KO | T cells | Protection from onset of disease. | DP | [65] |
| MS | ↑ Atg5 transcripts and ATG5 protein in circulating and CNS-infiltrating T cells | T cells | ? | DP? | [72] |
| MS | ↑ Autophagy activity through EBV infection | B cells | EBV infection of B cells modulates the processing of a disease-relevant myelin autoantigen through autophagy. | DP | [80] |
| EAE | ATG7 KO | DCs | Amelioration of active EAE course due to impaired antigen presentation. | DP | [145] |
| EAE | ATG5 KO | DCs | Protection from onset of adoptively transferred disease due to insufficient reactivation of T cells. | DP | [84] |
| MS | ↑ Autophagosomes | Neurons (cerebellar dentate and the pontine nuclei) | Autophagy as neuron-autonomous mechanism of synaptic loss? | DP? | [96] |
| SLE | SNPs of autophagy related and associated genes: Atg5, Atg7, LRRK2, IRGM, DRAM1, CDKN1B, MTMR3, and APOL1 | – | Increased susceptibility to develop SLE. | DP? | [101-106] |
| SLE-like disease (mouse model) | ATG5 KO | DCs or B cells | Amelioration of SLE-like disease. | DP | [117] |
| SLE-like disease (mouse model) | ATG5 KO | DCs and B cells | Exacerbation of SLE-like disease. | (DP) | [118] |
| SLE | ↑ Autophagy activity (↑ LC3+ punctae) | B cells | Amount of LC3+ punctae correlates with disease score. Pharmacological inhibition of autophagy impairs plasmablast differentiation. | DP | [119] |
| SLE | ↓ Autophagy activity (↓ LC3+ protein levels) | Tregs | Impaired Treg differentiation and suppressive capacity. | DH | [121] |
| RA | ↑ Autophagy activity (↑ LC3+ protein levels) | RASF | Possible role in hypoxia-mediated upregulation of proinflammatory cytokine IL-6 in RA-FLS. | DP | [146] |
| RA | ↑ Autophagy activity (↑ LC3+, Beclin 1 protein levels) | RASF | Inverse correlation of autophagy protein levels and fraction of apoptotic cells in synovial tissues | DP | [147] |
| RA | ↑ Autophagy activity | RASF | IL-17-mediated disruption of mitochondrial respiration leads to upregulation of autophagy and resistance towards apoptosis | DP | [128] |
| RA | ↑ Autophagy activity | CD4+ T cells (pre-activated) | T cell hyperactivation and apoptosis resistance | DP | [134] |
| RA | ↓ Autophagy activity (due to PFKFB3 insufficiency) | CD4+ T cells (naïve – ex vivo activated) | Autophagy is not able to compensate cellular energy deficit caused by insufficient PFKFB3-mediated glucose metabolization | DH | [135] |
| RA | ↑ Autophagy activity | DCs, RASF | PAD-mediated citrullination of disease-relevant peptides | DP | [143,144] |
To date, no robust method to reliably measure autophagy activity in vivo is available to monitor treatment, making the identification of autophagy biomarkers for use in a clinical setting a major challenge for the future [148]. It will be crucial to identify therapeutic strategies and appropriate monitoring tools to curb autoimmunity without interfering with immunity to infectious disease agents and tumorigenesis. Furthermore, systemic application of autophagy modulators may cause severe adverse events due to the abundant physiological functions of autophagy unrelated to the immune system. Future research will have to show if our increasing understanding of the autophagy network can eventually be translated into the development of efficacious and safe drugs that specifically modulate subtypes of autophagy for the treatment of autoimmune disorders.
Acknowledgments
The authors acknowledge support by the German Research Foundation (Collaborative Research Centre SFB-TR 128 ‘Initiating/Effector versus Regulatory Mechanisms in Multiple Sclerosis – Progress towards Tackling the Disease’ to J.D.L.).
Abbreviations
- AMPK
AMP-activated protein kinase
- ANA
antinuclear antibodies
- APC
antigen presenting cell
- Atgs/ATGs
autophagy-related genes/proteins
- CNS
central nervous system
- CSF
cerebrospinal fluid
- CTLA4
cytotoxic T lymphocyte-associated protein 4
- DC
dendritic cell
- Drp1
dynamin-related protein 1
- EAE
experimental autoimmune encephalomyelitis
- EBV
Epstein-Barr virus
- ER
endoplasmic reticulum
- ESCRT
endosomal sorting complex required for transport
- FIP200
FAK family kinase-interacting protein of 200 kDa
- FKBP
FK506-binding protein
- FRB
FKBP and rapamycin binding domain
- HCQ
hydroxychloroquine
- HIF
hypoxia-inducible factor
- HLA
human leukocyte antigen
- HRES
HTLV-1 related endogenous sequence
- HSPA8
heat shock 70 kDa protein 8
- IFN
interferon
- iNKT cell
invariant natural killer T cell
- LAMP2A
lysosome-associated membrane protein 2A
- LAP
LC3-associated phagocytosis
- LC3
microtubule-associated protein 1 A/1B-light chain 3 (LC3)
- LIR
LC3-interacting region
- mAbs
monoclonal antibodies
- MHC
major histocompatibility complex
- MIICs
MHC class II containing compartments
- MS
multiple sclerosis
- mTOR
mammalian target of rapamycin
- NAC
N-acetylcysteine
- NAWM
normal appearing white matter
- NOX2
NADPH oxidase
- NRBF2
nuclear receptor binding factor 2
- PAD
peptidylarginine deiminases
- PAMP
pathogen-associated molecular pattern
- pDC
plasmacytoid DC
- PFKFB3
6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3
- PI3K
class III phosphatidylinositide 3-kinase
- PIKK
phosphatidylinositol 3-kinase-related kinase
- Ptd-L-Ser
phosphatidylserine
- RA
rheumatoid arthritis
- RASF
RA synovial fibroblasts
- RF
rheumatoid factor
- ROS
reactive oxygen species
- SLE
systemic lupus erythematosus
- SNP
single nucleotide polymorphism
- TCM
central memory T cells
- TCR
T cell receptor
- TEC
thymic epithelial cell
- TGF
transforming growth factor
- TLR
toll-like receptor
- TNF
tumor necrosis factor
- ULK
unc 51-like kinase
- UPS
ubiquitin-proteasome system
- UVRAG
UV radiation resistance-associated gene protein
- Vps34
vacuolar protein sorting protein
- WIPI
ATG2/WD repeat domain phosphoinositide-interacting protein
- WML
white matter lesion
- 3-MA
3-methyladenine
Footnotes
Declaration of competing interest
CWK received travel support from UCB and Alexion.
JDL received speaker fees, research support, travel support, and/or served on advisory boards by Abbvie, Alexion, Argenx, Biogen, Merck, Novartis, Roche, Sanofi, Takeda.
IEA received speaker fees, research support, and/or served on advisory boards by Novartis, Pfizer, Merck, Abbvie, and Tanabe.
Data availability
Data will be made available on request.
References
- [1].Ktistakis NT, In praise of M. Anselmier who first used the term “autophagie” in 1859, Autophagy 13 (2017) 2015, 10.1080/15548627.2017.1367473. –2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Klionsky DJ, Autophagy revisited: a conversation with Christian de Duve, Autophagy 4 (2008) 740–743. [DOI] [PubMed] [Google Scholar]
- [3].Tsukada M, Ohsumi Y, Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae, FEBS Lett. 333 (1993) 169–174. [DOI] [PubMed] [Google Scholar]
- [4].Loi M, Müller A, Steinbach K, Niven J, Barreira da Silva R, Paul P, et al. , Macroautophagy proteins control MHC class I levels on dendritic cells and shape anti-viral CD8(+) T cell responses, Cell Rep. 15 (2016) 1076–1087, 10.1016/j.celrep.2016.04.002. [DOI] [PubMed] [Google Scholar]
- [5].Keller CW, Loi M, Ewert S, Quast I, Theiler R, Gannagé M, et al. , The autophagy machinery restrains iNKT cell activation through CD1D1 internalization, Autophagy 13 (2017) 1025–1036, 10.1080/15548627.2017.1297907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Matzaraki V, Kumar V, Wijmenga C, Zhernakova A, The MHC locus and genetic susceptibility to autoimmune and infectious diseases 18 (2017) 76, 10.1186/S13059-017-1207-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Münz C, Autophagy proteins in antigen processing for presentation on MHC molecules, Immunol. Rev 272 (2016) 17–27, 10.1111/imr.12422. [DOI] [PubMed] [Google Scholar]
- [8].Münz C, Autophagy beyond intracellular MHC class II antigen presentation, Trends Immunol. 37 (2016) 755–763, 10.1016/j.it.2016.08.017. [DOI] [PubMed] [Google Scholar]
- [9].Münz C, Canonical and non-canonical functions of the autophagy machinery in MHC restricted antigen presentation, Front. Immunol 13 (2022), 868888, 10.3389/fimmu.2022.868888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Peña-Martinez C, Rickman AD, Heckmann BL, Beyond autophagy: LC3-associated phagocytosis and endocytosis, Sci. Adv 8 (2022), eabn1702, 10.1126/sciadv.abn1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhang Z, Yue P, Lu T, Wang Y, Wei Y, Wei X, Role of lysosomes in physiological activities, diseases, and therapy, J. Hematol. Oncol 14 (2021) 79, 10.1186/s13045-021-01087-1, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Yamamoto H, Zhang S, Mizushima N, Autophagy genes in biology and disease, Nat. Rev. Genet (2023) 1–19, 10.1038/s41576-022-00562-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wang L, Klionsky DJ, Shen H-M, The emerging mechanisms and functions of microautophagy, Nat. Rev. Mol. Cell Biol (2022) 1–18, 10.1038/S41580-022-00529-z. [DOI] [PubMed] [Google Scholar]
- [14].Li W-W, Li J, Bao J-K, Microautophagy: lesser-known self-eating, Cell. Mol. Life Sci 69 (2012) 1125–1136, 10.1007/s00018-011-0865-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Galluzzi L, Baehrecke EH, Ballabio A, Boya P, Bravo-San Pedro JM, Cecconi F, et al. , Molecular definitions of autophagy and related processes, EMBO J. 36 (2017) 1811–1836, 10.15252/embj.201796697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Tasset I, Cuervo AM, Role of chaperone-mediated autophagy in metabolism, FEBS J. 283 (2016) 2403–2413, 10.1111/febs.13677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Nowag H, Guhl B, Thriene K, Romao S, Ziegler U, Dengjel J, et al. , Macroautophagy proteins assist Epstein barr virus production and get incorporated into the virus particles, EBioMedicine 1 (2014) 116–125, 10.1016/j.ebiom.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Dupont N, Jiang S, Pilli M, Ornatowski W, Bhattacharya D, Deretic V, Autophagy-based unconventional secretory pathway for extracellular delivery of IL-1β, EMBO J. 30 (2011) 4701–4711, 10.1038/emboj.2011.398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Saitoh T, Fujita N, Jang MH, Uematsu S, Yang B-G, Satoh T, et al. , Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production, Nature 456 (2008) 264–268, 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- [20].Zhang M, Kenny SJ, Ge L, Xu K, Schekman R, Translocation of interleukin-1β into a vesicle intermediate in autophagy-mediated secretion, Elife 4 (2015) 1463, 10.7554/eLife.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Martinez J, Malireddi RKS, Lu Q, Cunha LD, Pelletier S, Gingras S, et al. , Molecular characterization of LC3-associated phagocytosis reveals distinct roles for Rubicon, NOX2 and autophagy proteins, Nat. Cell Biol 17 (2015) 893–906, 10.1038/ncb3192. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [22].Sanjuan MA, Dillon CP, Tait SWG, Moshiach S, Dorsey F, Connell S, et al. , Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis, Nature 450 (2007) 1253–1257, 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- [23].Heckmann BL, Boada-Romero E, Cunha LD, Magne J, Green DR, LC3-Associated phagocytosis and inflammation, J. Mol. Biol 429 (2017) 3561–3576, 10.1016/j.jmb.2017.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Mizushima N, Yoshimori T, Ohsumi Y, The role of Atg proteins in autophagosome formation, Annu. Rev. Cell Dev. Biol 27 (2011) 107–132, 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- [25].Kimura T, Mandell M, Deretic V, Precision autophagy directed by receptor regulators - emerging examples within the TRIM family, J. Cell Sci 129 (2016) 881–891, 10.1242/jcs.163758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Feng Y, He D, Yao Z, Klionsky DJ, The machinery of macroautophagy, Cell Res. 24 (2014) 24–41, 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Karanasios E, Walker SA, Okkenhaug H, Manifava M, Hummel E, Zimmermann H, et al. , Autophagy initiation by ULK complex assembly on ER tubulovesicular regions marked by ATG9 vesicles, Nat. Commun 7 (2016), 12420, 10.1038/ncomms12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Suzuki H, Osawa T, Fujioka Y, Noda NN, Structural biology of the core autophagy machinery, Curr. Opin. Struct. Biol 43 (2017) 10–17, 10.1016/j.sbi.2016.09.010. [DOI] [PubMed] [Google Scholar]
- [29].Itakura E, Mizushima N, Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins, Autophagy 6 (2010) 764–776, 10.4161/auto.6.6.12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Obara K, Sekito T, Niimi K, Ohsumi Y, The Atg18-Atg2 complex is recruited to autophagic membranes via phosphatidylinositol 3-phosphate and exerts an essential function, J. Biol. Chem 283 (2008) 23972–23980, 10.1074/jbc.M803180200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Velikkakath AKG, Nishimura T, Oita E, Ishihara N, Mizushima N, Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets, Mol. Biol. Cell 23 (2012) 896–909, 10.1091/mbc.E11-09-0785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Birgisdottir ÅB, Lamark T, Johansen T, The LIR motif - crucial for selective autophagy, J. Cell Sci 126 (2013) 3237–3247, 10.1242/jcs.126128. [DOI] [PubMed] [Google Scholar]
- [33].Wild P, McEwan DG, Dikic I, The LC3 interactome at a glance, J. Cell Sci 127 (2014) 3–9, 10.1242/jcs.140426. [DOI] [PubMed] [Google Scholar]
- [34].Kyewski B, Klein L, A central role for central tolerance, Annu. Rev. Immunol 24 (2006) 571–606, 10.1146/annurev.immunol.23.021704.115601. [DOI] [PubMed] [Google Scholar]
- [35].Kasai M, Tanida I, Ueno T, Kominami E, Seki S, Ikeda T, et al. , Autophagic compartments gain access to the MHC class II compartments in thymic epithelium, J. Immunol 183 (2009) 7278–7285, 10.4049/jimmunol.0804087. [DOI] [PubMed] [Google Scholar]
- [36].Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L, Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance, Nature 455 (2008) 396–400, 10.1038/nature07208. [DOI] [PubMed] [Google Scholar]
- [37].Aichinger M, Wu C, Nedjic J, Klein L, Macroautophagy substrates are loaded onto MHC class II of medullary thymic epithelial cells for central tolerance, J. Exp. Med 210 (2013) 287–300, 10.1084/jem.20122149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Schmid D, Pypaert M, Münz C, Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes, Immunity 26 (2007) 79–92, 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Waisman A, Lukas D, Clausen BE, Yogev N, Dendritic cells as gatekeepers of tolerance, Semin. Immunopathol 39 (2017) 153–163, 10.1007/S00281-016-0583-z. [DOI] [PubMed] [Google Scholar]
- [40].Alissafi T, Banos A, Boon L, Sparwasser T, Ghigo A, Wing K, et al. , Tregs restrain dendritic cell autophagy to ameliorate autoimmunity, J. Clin. Invest 127 (2017) 2789–2804, 10.1172/JCI92079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kim J, Guan K-L, mTOR as a central hub of nutrient signalling and cell growth, Nat. Cell Biol 21 (2019) 63–71, 10.1038/s41556-018-0205-1. [DOI] [PubMed] [Google Scholar]
- [42].Kim J, Guan K-L, Amino acid signaling in TOR activation, Annu. Rev. Biochem 80 (2011) 1001–1032, 10.1146/annurev-biochem-062209-094414. [DOI] [PubMed] [Google Scholar]
- [43].Gaber T, Strehl C, Buttgereit F, Metabolic regulation of inflammation, Nat. Rev. Rheumatol 13 (2017) 267–279, 10.1038/nrrheum.2017.37. [DOI] [PubMed] [Google Scholar]
- [44].Weichhart T, Hengstschläger M, Linke M, Regulation of innate immune cell function by mTOR, Nat. Rev. Immunol 15 (2015) 599–614, 10.1038/nri3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Suto T, Karonitsch T, The immunobiology of mTOR in autoimmunity, J. Autoimmun 110 (2020), 102373, 10.1016/j.jaut.2019.102373. [DOI] [PubMed] [Google Scholar]
- [46].Fernandez DR, Telarico T, Bonilla E, Li Q, Banerjee S, Middleton FA, et al. , Activation of mammalian target of rapamycin controls the loss of TCRzeta in lupus T cells through HRES-1/Rab4-regulated lysosomal degradation, J. Immunol 182 (2009) 2063–2073, 10.4049/jimmunol.0803600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lai Z-W, Borsuk R, Shadakshari A, Yu J, Dawood M, Garcia R, et al. , Mechanistic target of rapamycin activation triggers IL-4 production and necrotic death of double-negative T cells in patients with systemic lupus erythematosus, J. Immunol 191 (2013) 2236–2246, 10.4049/jimmunol.1301005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Perl A, Activation of mTOR (mechanistic target of rapamycin) in rheumatic diseases, Nat. Rev. Rheumatol 12 (2016) 169–182, 10.1038/nrrheum.2015.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Wyman B, Perl A, Metabolic pathways mediate pathogenesis and offer targets for treatment in rheumatic diseases, Curr. Opin. Rheumatol 32 (2020) 184, 10.1097/BOR.0000000000000687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Karonitsch T, Kandasamy RK, Kartnig F, Herdy B, Dalwigk K, Niederreiter B, et al. , mTOR senses environmental cues to shape the fibroblast-like synoviocyte response to inflammation, Cell Rep. 23 (2018) 2157–2167, 10.1016/j.celrep.2018.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Cadwell K, Crosstalk between autophagy and inflammatory signalling pathways: balancing defence and homeostasis, Nat. Rev. Immunol 16 (2016) 661–675, 10.1038/nri.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Münz C, Autophagy in immunity, Prog Mol Biol Transl Sci 172 (2020) 67–85, 10.1016/bs.pmbts.2020.03.005. [DOI] [PubMed] [Google Scholar]
- [53].Nimmerjahn F, Milosevic S, Behrends U, Jaffee EM, Pardoll DM, Bornkamm GW, et al. , Major histocompatibility complex class II-restricted presentation of a cytosolic antigen by autophagy, Eur. J. Immunol 33 (2003) 1250–1259, 10.1002/eji.200323730. [DOI] [PubMed] [Google Scholar]
- [54].Paludan C, Schmid D, Landthaler M, Vockerodt M, Kube D, Tuschl T, et al. , Endogenous MHC class II processing of a viral nuclear antigen after autophagy, Science 307 (2005) 593–596, 10.1126/science.1104904. [DOI] [PubMed] [Google Scholar]
- [55].Mahad DH, Trapp BD, Lassmann H, Pathological mechanisms in progressive multiple sclerosis, Lancet Neurol. 14 (2015) 183–193, 10.1016/S1474-4422(14)70256-X. [DOI] [PubMed] [Google Scholar]
- [56].Attfield KE, Jensen LT, Kaufmann M, Friese MA, Fugger L, The immunology of multiple sclerosis, Nat. Rev. Immunol 22 (2022) 734–750, 10.1038/S41577-022-00718-z. [DOI] [PubMed] [Google Scholar]
- [57].International Multiple Sclerosis Genetics Consortium, Wellcome Trust Case Control Consortium 2, Sawcer S, Hellenthal G, Pirinen M, Spencer CCA, et al. , Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis, Nature 476 (2011) 214–219, 10.1038/nature10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].International Multiple Sclerosis Genetics Consortium (IMSGC), Beecham AH, Patsopoulos NA, Xifara DK, Davis MF, Kemppinen A, et al. , Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis, Nat. Genet 45 (2013) 1353–1360, 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Babbe H, Roers A, Waisman A, Lassmann H, Goebels N, Hohlfeld R, et al. , Clonal expansions of CD8(+) T cells dominate the T cell infiltrate in active multiple sclerosis lesions as shown by micromanipulation and single cell polymerase chain reaction, J. Exp. Med 192 (2000) 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Jacobsen M, Cepok S, Quak E, Happel M, Gaber R, Ziegler A, et al. , Oligoclonal expansion of memory CD8+ T cells in cerebrospinal fluid from multiple sclerosis patients, Brain 125 (2002) 538–550. [DOI] [PubMed] [Google Scholar]
- [61].Sospedra M, Muraro PA, Stefanová I, Zhao Y, Chung K, Li Y, et al. , Redundancy in antigen-presenting function of the HLA-DR and -DQ molecules in the multiple sclerosis-associated HLA-DR2 haplotype, J. Immunol 176 (2006) 1951–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Planas R, Metz I, Ortiz Y, Vilarrasa N, Jelcic I, Salinas-Riester G, et al. , Central role of Th2/Tc2 lymphocytes in pattern II multiple sclerosis lesions, Ann Clin Transl Neurol 2 (2015) 875–893, 10.1002/acn3.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Keller CW, Münz C, Lünemann JD, Autophagy pathways in CNS myeloid cell immune functions, Trends Neurosci. 43 (2020) 1024–1033, 10.1016/j.tins.2020.09.003. [DOI] [PubMed] [Google Scholar]
- [64].Pua HH, Dzhagalov I, Chuck M, Mizushima N, He Y-W, A critical role for the autophagy gene Atg5 in T cell survival and proliferation, J. Exp. Med 204 (2007) 25–31, 10.1084/jem.20061303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Kovacs JR, Li C, Yang Q, Li G, Garcia IG, Ju S, et al. , Autophagy promotes T-cell survival through degradation of proteins of the cell death machinery, Cell Death Differ. 19 (2012) 144–152, 10.1038/cdd.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Puleston DJ, Zhang H, Powell TJ, Lipina E, Sims S, Panse I, et al. , Autophagy is a critical regulator of memory CD8(+) T cell formation, Elife 3 (2014) 2516, 10.7554/eLife.03706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Xu X, Araki K, Li S, Han J-H, Ye L, Tan WG, et al. , Autophagy is essential for effector CD8(+) T cell survival and memory formation, Nat. Immunol 15 (2014) 1152–1161, 10.1038/ni.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Allegretta M, Nicklas JA, Sriram S, Albertini RJ, T cells responsive to myelin basic protein in patients with multiple sclerosis, Science 247 (1990) 718–721. [DOI] [PubMed] [Google Scholar]
- [69].Markovic-Plese S, Cortese I, Wandinger KP, McFarland HF, Martin R, CD4+ CD28− costimulation-independent T cells in multiple sclerosis, J. Clin. Invest 108 (2001) 1185–1194, 10.1172/JCI12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Liu G-Z, Fang L-B, Hjelmström P, Gao X-G, Increased CD8+ central memory T cells in patients with multiple sclerosis, Mult. Scler 13 (2007) 149–155, 10.1177/1352458506069246. [DOI] [PubMed] [Google Scholar]
- [71].van Nierop GP, van Luijn MM, Michels SS, Melief M-J, Janssen M, Langerak AW, et al. , Phenotypic and functional characterization of T cells in white matter lesions of multiple sclerosis patients, Acta Neuropathol. 134 (2017) 383–401, 10.1007/s00401-017-1744-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Alirezaei M, Fox HS, Flynn CT, Moore CS, Hebb ALO, Frausto RF, et al. , Elevated ATG5 expression in autoimmune demyelination and multiple sclerosis, Autophagy 5 (2009) 152–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Miller BC, Zhao Z, Stephenson LM, Cadwell K, Pua HH, Lee HK, et al. , The autophagy gene ATG5 plays an essential role in B lymphocyte development, Autophagy 4 (2008) 309–314. [DOI] [PubMed] [Google Scholar]
- [74].Hauser SL, Waubant E, Arnold DL, Vollmer T, Antel J, Fox RJ, et al. , B-cell depletion with rituximab in relapsing-remitting multiple sclerosis, N. Engl. J. Med 358 (2008) 676–688, 10.1056/NEJMoa0706383. [DOI] [PubMed] [Google Scholar]
- [75].Barun B, Bar-Or A, Treatment of multiple sclerosis with anti-CD20 antibodies, Clin. Immunol 142 (2012) 31–37, 10.1016/j.clim.2011.04.005. [DOI] [PubMed] [Google Scholar]
- [76].Thacker EL, Mirzaei F, Ascherio A, Infectious mononucleosis and risk for multiple sclerosis: a meta-analysis, Ann. Neurol 59 (2006) 499–503, 10.1002/ana.20820. [DOI] [PubMed] [Google Scholar]
- [77].Lanz TV, Brewer RC, Ho PP, Moon J-S, Jude KM, Fernandez D, et al. , Clonally expanded B cells in multiple sclerosis bind EBV EBNA1 and GlialCAM, Nature 603 (2022) 321–327, 10.1038/s41586-022-04432-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Bjornevik K, Cortese M, Healy BC, Kuhle J, Mina MJ, Leng Y, et al. , Longitudinal analysis reveals high prevalence of Epstein-Barr virus associated with multiple sclerosis, Science 375 (2022) 296–301, 10.1126/science.abj8222. [DOI] [PubMed] [Google Scholar]
- [79].Lünemann JD, Jelcic I, Roberts S, Lutterotti A, Tackenberg B, Martin R, et al. , EBNA1-specific T cells from patients with multiple sclerosis cross react with myelin antigens and co-produce IFN-gamma and IL-2, J. Exp. Med 205 (2008) 1763–1773, 10.1084/jem.20072397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Morandi E, Jagessar SA, ’t Hart BA, Gran B, EBV infection empowers human B cells for autoimmunity: role of autophagy and relevance to multiple sclerosis, J. Immunol 199 (2017) 435–448, 10.4049/jimmunol.1700178. [DOI] [PubMed] [Google Scholar]
- [81].Jagessar SA, Holtman IR, Hofman S, Morandi E, Heijmans N, Laman JD, et al. , Lymphocryptovirus infection of nonhuman primate B cells converts destructive into productive processing of the pathogenic CD8 T cell epitope in myelin oligodendrocyte glycoprotein, J. Immunol 197 (2016) 1074–1088, 10.4049/jimmunol.1600124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].’tHart BA, Kap YS, Morandi E, Laman JD, Gran B, EBV infection and multiple sclerosis: lessons from a marmoset model, Trends Mol. Med 22 (2016) 1012–1024, 10.1016/j.molmed.2016.10.007. [DOI] [PubMed] [Google Scholar]
- [83].Greter M, Heppner FL, Lemos MP, Odermatt BM, Goebels N, Laufer T, et al. , Dendritic cells permit immune invasion of the CNS in an animal model of multiple sclerosis, Nat. Med 11 (2005) 328–334, 10.1038/nm1197. [DOI] [PubMed] [Google Scholar]
- [84].Keller CW, Sina C, Kotur MB, Ramelli G, Mundt S, Quast I, et al. , ATG-dependent phagocytosis in dendritic cells drives myelin-specific CD4+ T cell pathogenicity during CNS inflammation, Proc. Natl. Acad. Sci. U.S.a 114 (2017) E11228–E11237, 10.1073/pnas.1713664114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Keller CW, Kotur MB, Mundt S, Dokalis N, Ligeon L-A, Shah AM, et al. , CYBB/NOX2 in conventional DCs controls T cell encephalitogenicity during neuroinflammation, Autophagy 43 (2020) 1–15, 10.1080/15548627.2020.1756678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Mundt S, Mrdjen D, Utz SG, Greter M, Schreiner B, Becher B, Conventional DCs sample and present myelin antigens in the healthy CNS and allow parenchymal T cell entry to initiate neuroinflammation, Sci Immunol 4 (2019), eaau8380, 10.1126/sciimmunol.aau8380. [DOI] [PubMed] [Google Scholar]
- [87].Prodinger C, Bunse J, Krüger M, Schiefenhövel F, Brandt C, Laman JD, et al. , CD11c-expressing cells reside in the juxtavascular parenchyma and extend processes into the glia limitans of the mouse nervous system, Acta Neuropathol. 121 (2011) 445–458, 10.1007/s00401-010-0774-y. [DOI] [PubMed] [Google Scholar]
- [88].Mohammad MG, Tsai VWW, Ruitenberg MJ, Hassanpour M, Li H, Hart PH, et al. , Immune cell trafficking from the brain maintains CNS immune tolerance, J. Clin. Invest 124 (2014) 1228–1241, 10.1172/JCI71544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Schläger C, Körner H, Krueger M, Vidoli S, Haberl M, Mielke D, et al. , Effector T-cell trafficking between the leptomeninges and the cerebrospinal fluid, Nature 530 (2016) 349–353, 10.1038/nature16939. [DOI] [PubMed] [Google Scholar]
- [90].Isaksson M, Lundgren BA, Ahlgren KM, Kämpe O, Lobell A, Conditional DC depletion does not affect priming of encephalitogenic Th cells in EAE, Eur. J. Immunol 42 (2012) 2555–2563, 10.1002/eji.201142239. [DOI] [PubMed] [Google Scholar]
- [91].Yogev N, Frommer F, Lukas D, Kautz-Neu K, Karram K, Ielo D, et al. , Dendritic cells ameliorate autoimmunity in the CNS by controlling the homeostasis of PD-1 receptor(+) regulatory T cells, Immunity 37 (2012) 264–275, 10.1016/j.immuni.2012.05.025. [DOI] [PubMed] [Google Scholar]
- [92].Paterka M, Siffrin V, Voss JO, Werr J, Hoppmann N, Gollan R, et al. , Gatekeeper role of brain antigen-presenting CD11c+ cells in neuroinflammation, EMBO J. 35 (2016) 89–101, 10.15252/embj.201591488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Martinez J, Almendinger J, Oberst A, Ness R, Dillon CP, Fitzgerald P, et al. , Microtubule-associated protein 1 light chain 3 alpha (LC3)-associated phagocytosis is required for the efficient clearance of dead cells, Proc. Natl. Acad. Sci. U.S.a 108 (2011) 17396–17401, 10.1073/pnas.1113421108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [94].Romao S, Gasser N, Becker AC, Guhl B, Bajagic M, Vanoaica D, et al. , Autophagy proteins stabilize pathogen-containing phagosomes for prolonged MHC II antigen processing, J. Cell Biol 203 (2013) 757–766, 10.1083/jcb.201308173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Keller CW, Lünemann JD, Noncanonical autophagy in dendritic cells triggers CNS autoimmunity, Autophagy 14 (2018) 560–561, 10.1080/15548627.2018.1427397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Albert M, Barrantes-Freer A, Lohrberg M, Antel JP, Prineas JW, Palkovits M, et al. , Synaptic pathology in the cerebellar dentate nucleus in chronic multiple sclerosis, Brain Pathol. 27 (2017) 737–747, 10.1111/bpa.12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Mahad D, Ziabreva I, Lassmann H, Turnbull D, Mitochondrial defects in acute multiple sclerosis lesions, Brain 131 (2008) 1722–1735, 10.1093/brain/awn105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Mahad DJ, Ziabreva I, Campbell G, Lax N, White K, Hanson PS, et al. , Mitochondrial changes within axons in multiple sclerosis, Brain 132 (2009) 1161–1174, 10.1093/brain/awp046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Patergnani S, Castellazzi M, Bonora M, Marchi S, Casetta I, Pugliatti M, et al. , Autophagy and mitophagy elements are increased in body fluids of multiple sclerosis-affected individuals, J. Neurol. Neurosurg. Psychiatr 89 (2018) 439–441, 10.1136/jnnp-2017-316234. [DOI] [PubMed] [Google Scholar]
- [100].Lo MS, Tsokos GC, Recent developments in systemic lupus erythematosus pathogenesis and applications for therapy, Curr. Opin. Rheumatol 30 (2018) 222–228, 10.1097/BOR.0000000000000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Zhou X-J, Lu X-L, Lv J-C, Yang H-Z, Qin L-X, Zhao M-H, et al. , Genetic association of PRDM1-ATG5 intergenic region and autophagy with systemic lupus erythematosus in a Chinese population, Ann. Rheum. Dis 70 (2011) 1330–1337, 10.1136/ard.2010.140111. [DOI] [PubMed] [Google Scholar]
- [102].Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, et al. , A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus, Nat. Genet 41 (2009) 1228–1233, 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Zhang Y-M, Zhou X-J, Cheng F-J, Qi Y-Y, Hou P, Zhao M-H, et al. , Autophagy-related gene LRRK2 is likely a susceptibility gene for systemic lupus erythematosus in northern Han Chinese, Oncotarget 8 (2017) 13754–13761, 10.18632/oncotarget.14631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Yang W, Tang H, Zhang Y, Tang X, Zhang J, Sun L, et al. , Meta-analysis followed by replication identifies loci in or near CDKN1B, TET3, CD80, DRAM1, and ARID5B as associated with systemic lupus erythematosus in Asians, Am. J. Hum. Genet 92 (2013) 41–51, 10.1016/j.ajhg.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Zhou X-J, Nath SK, Qi Y-Y, Cheng F-J, Yang H-Z, Zhang Y, et al. , Brief Report: identification of MTMR3 as a novel susceptibility gene for lupus nephritis in northern Han Chinese by shared-gene analysis with IgA nephropathy, Arthritis Rheumatol. 66 (2014) 2842–2848, 10.1002/art.38749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Freedman BI, Langefeld CD, Andringa KK, Croker JA, Williams AH, Garner NE, et al. , End-stage renal disease in African Americans with lupus nephritis is associated with APOL1, Arthritis Rheumatol. 66 (2014) 390–396, 10.1002/art.38220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Gomes LC, Di Benedetto G, Scorrano L, During autophagy mitochondria elongate, are spared from degradation and sustain cell viability, Nat. Cell Biol 13 (2011) 589–598, 10.1038/ncb2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Talaber G, Miklossy G, Oaks Z, Liu Y, Tooze SA, Chudakov DM, et al. , HRES-1/Rab4 promotes the formation of LC3(+) autophagosomes and the accumulation of mitochondria during autophagy, PLoS One 9 (2014), e84392, 10.1371/journal.pone.0084392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Caza TN, Fernandez DR, Talaber G, Oaks Z, Haas M, Madaio MP, et al. , HRES-1/Rab4-mediated depletion of Drp1 impairs mitochondrial homeostasis and represents a target for treatment in SLE, Ann. Rheum. Dis 73 (2014) 1888–1897, 10.1136/annrheumdis-2013-203794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Caielli S, Cardenas J, de Jesus AA, Baisch J, Walters L, Blanck JP, et al. , Erythroid mitochondrial retention triggers myeloid-dependent type I interferon in human SLE, Cell 184 (2021) 4464–4479.e19, 10.1016/j.cell.2021.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Hooks JJ, Moutsopoulos HM, Geis SA, Stahl NI, Decker JL, Notkins AL, Immune interferon in the circulation of patients with autoimmune disease, N. Engl. J. Med 301 (1979) 5–8, 10.1056/NEJM197907053010102. [DOI] [PubMed] [Google Scholar]
- [112].Crow MK, Type I interferon in the pathogenesis of lupus, J. Immunol. 192 (2014) 5459–5468, 10.4049/jimmunol.1002795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Elkon KB, Stone VV, Type I interferon and systemic lupus erythematosus, J. Interferon Cytokine Res 31 (2011) 803–812, 10.1089/jir.2011.0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A, Autophagy-dependent viral recognition by plasmacytoid dendritic cells, Science 315 (2007) 1398–1401, 10.1126/science.1136880. [DOI] [PubMed] [Google Scholar]
- [115].Henault J, Martinez J, Riggs JM, Tian J, Mehta P, Clarke L, et al. , Noncanonical autophagy is required for type I interferon secretion in response to DNA-immune complexes, Immunity 37 (2012) 986–997, 10.1016/j.immuni.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Leibler C, John S, Elsner RA, Thomas KB, Smita S, Joachim S, et al. , Genetic dissection of TLR9 reveals complex regulatory and cryptic proinflammatory roles in mouse lupus, Nat. Immunol 23 (2022) 1457–1469, 10.1038/S41590-022-01310-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Weindel CG, Richey LJ, Bolland S, Mehta AJ, Kearney JF, Huber BT, B cell autophagy mediates TLR7-dependent autoimmunity and inflammation, Autophagy 11 (2015) 1010–1024, 10.1080/15548627.2015.1052206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Weindel CG, Richey LJ, Mehta AJ, Shah M, Huber BT, Autophagy in dendritic cells and B cells is critical for the inflammatory state of TLR7-mediated autoimmunity, J. Immunol 198 (2017) 1081–1092, 10.4049/jimmunol.1601307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Clarke AJ, Ellinghaus U, Cortini A, Stranks A, Simon AK, Botto M, et al. , Autophagy is activated in systemic lupus erythematosus and required for plasmablast development, Ann. Rheum. Dis 74 (2015) 912–920, 10.1136/annrheumdis-2013-204343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Bosma A, Abdel-Gadir A, Isenberg DA, Jury EC, Mauri C, Lipid-antigen presentation by CD1d(+) B cells is essential for the maintenance of invariant natural killer T cells, Immunity 36 (2012) 477–490, 10.1016/j.immuni.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Kato H, Perl A, Blockade of Treg cell differentiation and function by the interleukin-21-mechanistic target of rapamycin Axis via suppression of autophagy in patients with systemic lupus erythematosus, Arthritis Rheumatol. 70 (2018) 427–438, 10.1002/art.40380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Weyand CM, Goronzy JJ, The immunology of rheumatoid arthritis, Nat. Immunol 22 (2021) 10–18, 10.1038/s41590-020-00816-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Huber LC, Distler O, Tarner I, Gay RE, Gay S, Pap T, Synovial fibroblasts: key players in rheumatoid arthritis, Rheumatology 45 (2006) 669–675, 10.1093/rheumatology/kel065. [DOI] [PubMed] [Google Scholar]
- [124].Kato M, Ospelt C, Gay RE, Gay S, Klein K, Dual role of autophagy in stress-induced cell death in rheumatoid arthritis synovial fibroblasts, Arthritis Rheumatol. 66 (2014) 40–48, 10.1002/art.38190. [DOI] [PubMed] [Google Scholar]
- [125].Connor AM, Mahomed N, Gandhi R, Keystone EC, Berger SA, TNFα modulates protein degradation pathways in rheumatoid arthritis synovial fibroblasts, Arthritis Res. Ther 14 (2012) R62, 10.1186/ar3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Young CL, Adamson TC, Vaughan JH, Fox RI, Immunohistologic characterization of synovial membrane lymphocytes in rheumatoid arthritis, Arthritis Rheum. 27 (1984) 32–39. [DOI] [PubMed] [Google Scholar]
- [127].Pope RM, Apoptosis as a therapeutic tool in rheumatoid arthritis, Nat. Rev. Immunol 2 (2002) 527–535, 10.1038/nri846. [DOI] [PubMed] [Google Scholar]
- [128].Kim EK, Kwon J-E, Lee S-Y, Lee E-J, Kim DS, Moon S-J, et al. , IL-17-mediated mitochondrial dysfunction impairs apoptosis in rheumatoid arthritis synovial fibroblasts through activation of autophagy, Cell Death Dis. 8 (2017) e2565, 10.1038/cddis.2016.490. –e2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Chang Y-P, Tsai C-C, Huang W-C, Wang C-Y, Chen C-L, Lin Y-S, et al. , Autophagy facilitates IFN-gamma-induced Jak2-STAT1 activation and cellular inflammation, J. Biol. Chem 285 (2010) 28715–28722, 10.1074/jbc.M110.133355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Peral de Castro C, Jones SA, Ní Cheallaigh C, Hearnden CA, Williams L, Winter J, et al. , Autophagy regulates IL-23 secretion and innate T cell responses through effects on IL-1 secretion, J. Immunol 189 (2012) 4144–4153, 10.4049/jimmunol.1201946. [DOI] [PubMed] [Google Scholar]
- [131].Ding Y, Kim SL, Lee S-Y, Koo JK, Wang Z, Choi ME, Autophagy regulates TGF-β expression and suppresses kidney fibrosis induced by unilateral ureteral obstruction, J. Am. Soc. Nephrol 25 (2014) 2835–2846, 10.1681/ASN.2013101068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Adamopoulos IE, Mellins ED, Alternative pathways of osteoclastogenesis in inflammatory arthritis, Nat. Rev. Rheumatol 11 (2015) 189–194, 10.1038/nrrheum.2014.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Lee HK, Mattei LM, Steinberg BE, Alberts P, Lee YH, Chervonsky A, et al. , In vivo requirement for Atg5 in antigen presentation by dendritic cells, Immunity 32 (2010) 227–239, 10.1016/j.immuni.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].van Loosdregt J, Rossetti M, Spreafico R, Moshref M, Olmer M, Williams GW, et al. , Increased autophagy in CD4+T cells of rheumatoid arthritis patients results in T-cell hyperactivation and apoptosis resistance, Eur. J. Immunol 46 (2016) 2862–2870, 10.1002/eji.201646375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Yang Z, Fujii H, Mohan SV, Goronzy JJ, Weyand CM, Phosphofructokinase deficiency impairs ATP generation, autophagy, and redox balance in rheumatoid arthritis T cells, J. Exp. Med 210 (2013) 2119–2134, 10.1084/jem.20130252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Cooles FAH, Isaacs JD, Anderson AE, Treg cells in rheumatoid arthritis: an update, Curr. Rheumatol. Rep 15 (2013) 352, 10.1007/s11926-013-0352-0. [DOI] [PubMed] [Google Scholar]
- [137].Wang W, Shao S, Jiao Z, Guo M, Xu H, Wang S, The Th17/Treg imbalance and cytokine environment in peripheral blood of patients with rheumatoid arthritis, Rheumatol. Int 32 (2012) 887–893, 10.1007/s00296-010-1710-0. [DOI] [PubMed] [Google Scholar]
- [138].Niu Q, Cai B, Huang Z-C, Shi Y-Y, Wang L-L, Disturbed Th17/Treg balance in patients with rheumatoid arthritis, Rheumatol. Int 32 (2012) 2731–2736, 10.1007/s00296-011-1984-x. [DOI] [PubMed] [Google Scholar]
- [139].van Amelsfort JMR, van Roon JAG, Noordegraaf M, Jacobs KMG, Bijlsma JWJ, Lafeber FPJG, et al. , Proinflammatory mediator-induced reversal of CD4+,CD25+ regulatory T cell-mediated suppression in rheumatoid arthritis, Arthritis Rheum. 56 (2007) 732–742, 10.1002/art.22414. [DOI] [PubMed] [Google Scholar]
- [140].E XQ, Meng HX, Cao Y, Zhang SQ, Bi ZG, Yamakawa M, Distribution of regulatory T cells and interaction with dendritic cells in the synovium of rheumatoid arthritis, Scand. J. Rheumatol 41 (2012) 413–420, 10.3109/03009742.2012.696135. [DOI] [PubMed] [Google Scholar]
- [141].Wegner N, Lundberg K, Kinloch A, Fisher B, Malmström V, Feldmann M, et al. , Autoimmunity to specific citrullinated proteins gives the first clues to the etiology of rheumatoid arthritis, Immunol. Rev 233 (2010) 34–54, 10.1111/j.0105-2896.2009.00850.x. [DOI] [PubMed] [Google Scholar]
- [142].Scally SW, Petersen J, Law SC, Dudek NL, Nel HJ, Loh KL, et al. , A molecular basis for the association of the HLA-DRB1 locus, citrullination, and rheumatoid arthritis, J. Exp. Med 210 (2013) 2569–2582, 10.1084/jem.20131241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Ireland JM, Unanue ER, Autophagy in antigen-presenting cells results in presentation of citrullinated peptides to CD4 T cells, J. Exp. Med 208 (2011) 2625–2632, 10.1084/jem.20110640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Sorice M, Iannuccelli C, Manganelli V, Capozzi A, Alessandri C, Lococo E, et al. , Autophagy generates citrullinated peptides in human synoviocytes: a possible trigger for anti-citrullinated peptide antibodies, Rheumatology 55 (2016) 1374–1385, 10.1093/rheumatology/kew178. [DOI] [PubMed] [Google Scholar]
- [145].Bhattacharya A, Parillon X, Zeng S, Han S, Eissa NT, Deficiency of autophagy in dendritic cells protects against experimental autoimmune encephalomyelitis, J. Biol. Chem 289 (2014) 26525–26532, 10.1074/jbc.M114.575860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Yang R, Zhang Y, Wang L, Hu J, Wen J, Xue L, et al. , Increased autophagy in fibroblast-like synoviocytes leads to immune enhancement potential in rheumatoid arthritis, Oncotarget 8 (2017) 15420–15430, 10.18632/oncotarget.14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Xu K, Xu P, Yao J-F, Zhang Y-G, Hou W-K, Lu S-M, Reduced apoptosis correlates with enhanced autophagy in synovial tissues of rheumatoid arthritis, Inflamm. Res 62 (2013) 229–237, 10.1007/s00011-012-0572-1. [DOI] [PubMed] [Google Scholar]
- [148].Mizushima N, Murphy LO, Autophagy assays for biological discovery and therapeutic development, Trends Biochem. Sci 45 (2020) 1080–1093, 10.1016/j.tibs.2020.07.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.