Abstract
As a soil bacterium also found in estuarine and marine habitats, Bacillus subtilis has evolved various sensing and adaptation systems in order to face salt stress conditions. Among these regulatory mechanisms is the DegS-DegU signal transduction system, which was previously shown to be stimulated by high salt concentrations. A search for promoters regulated in response to salt stress led to the identification of wapA, encoding a wall-associated protein, which is strongly expressed at low salt concentrations and almost completely repressed in the presence of 0.7 M disodium succinate. Repression of wapA transcription by salt stress was shown to require the phosphorylated form of DegU. Moreover, DegU-mediated repression of wapA occurred only in high-salt medium. Alignment between the control region of wapA and other DegU-regulated promoters allowed the identification of a putative DegU target sequence, AGAAN11TTCAG. Mutation/deletion analyses of the wapA promoter region confirmed the role of the putative DegU control site in repression of wapA transcription at high salt concentrations and revealed a second site of repression located downstream from the transcription start site. Since residual negative control was observed at this second site in the absence of DegU, it seems likely that an additional repressor acts on the wapA control region to further downregulate wapA transcription under salt stress conditions.
In its natural environment, Bacillus subtilis spends most of its life in a starving or nongrowing state because of different growth-limiting and stress conditions. As a soil bacterium, B. subtilis is exposed to runoff into the sea, and it is largely found in coastal waters, estuarine sediments and other saline habitats. It dominates the marine flora to such an extent that it could be considered a primary inhabitant of the oceans (3, 27). To adapt to drastic variations of environmental conditions including increasing saline concentrations, B. subtilis has developed a highly sophisticated regulatory network that involves transcriptional modulation of large sets of genes controlling cellular differentiation (26) and the induction of a set of proteins called general stress proteins (GSPs) or stress-specific proteins (11).
At least three distinct mechanisms of salt stress induction have been identified in B. subtilis. Transcription of the so-called class II heat shock genes, encoding GSPs, is activated by the alternative sigma factor ςB. This regulation involves two separate pathways which respond either to environmental stresses, including salt stress, or energy limitation, through a dual multicomponent network (15, 32). Class II heat shock genes encode a ςB-independent group of GSPs including FtsH (5) and the ClpP, ClpC, and Lon proteases (16, 28, 33). Among these proteins involved in stress response, ClpC was clearly shown to be required for tolerance to salt stress (16). The DegS-DegU two-component system was suggested to be involved in sensing salt stress, as exemplified by the effect of high salt concentration on the production of degradative enzyme synthesis and the expression of comG, a late competence gene, known to be controlled by DegU (18). In contrast to the first two mechanisms, which provide nonspecific stress resistance, the DegS-DegU regulatory pair seems to be subjected to salt-specific induction. Interestingly, the DegS-DegU signal transduction system also plays a key role in the complex network that governs post-exponential-phase responses under growth-limiting conditions (21).
As a first step toward a better understanding of B. subtilis behavior in saline environments, we have sought promoters that are differentially expressed in low-salt and high-salt conditions. Here, we report the isolation of the B. subtilis wapA promoter, from which expression is relatively strong in low-salt medium and completely repressed by high salt concentrations. The wapA gene encodes a wall-associated protein which belongs to a large family of high-molecular-weight, surface-associated proteins involved in various cellular processes, including surface hydrophobicity, pathogenicity, wall metabolism, secretion, and cell adhesion (34). We further show that salt stress repression of wapA is mediated by the DegS-DegU two-component system and propose a tentative target site for DegU-mediated regulation based on mutation/deletion analysis of the wapA regulatory region together with comparison of promoters known to be controlled by DegU.
MATERIALS AND METHODS
Bacterial strains and genetic techniques.
All strains used in this study are listed in Table 1.
TABLE 1.
B. subtilis strains used in this study
| Strain | Genotype or descriptiona | Sourceb or reference |
|---|---|---|
| 168 | trpC2 | Laboratory stock |
| QB136 | trpC2 leuA8 degU32(Hy) | 17 |
| QB4414 | trpC2 degU146 | 4 |
| QB4487 | trpC2 degUΔBclI-EcoRI::erm | T. Msadek |
| QB4871 | trpC2 amyE::(wapA′-lacZ aphA3) | This work |
| QB4883 | trpC2 amyE::(wapA′-lacZ aphA3) ΔdegU::erm | QB4487→QB4871 |
| QB4950 | trpC2 amyE::(wapA′-lacZ aphA3) | pWP252→168 |
| QB4951 | trpC2 wapA::(wapA′-lacZ cat) | pWP253→168 |
| QB4955 | trpC2 amyE::(wapA′-lacZ aphA3) degU146 | QB4871→QB4414 |
| QB4959 | trpC2 amyE::(wapAΔD′-lacZ aphA3) | pWP263→168 |
| QB4961 | trpC2 amyE::(wapA[A−38G]′-lacZ aphA3) | pWP259.15→168 |
| QB4962 | trpC2 amyE::(wapA[A−38G]ΔD′-lacZ aphA3) | pWP265→168 |
| QB4963 | trpC2 amyE::(wapA[TT−23/−22AA]ΔD′-lacZ aphA3) | pWP267→168 |
| QB4964 | trpC2 amyE::(wapAΔD′-lacZ aphA3) ΔdegU::erm | QB4487→QB4959 |
| QB4965 | trpC2 amyE::(wapA[TT−23/−22AA]ΔD′-lacZ aphA3) ΔdegU::erm | QB4487→QB4963 |
| QB4966 | trpC2 amyE::(wapA[A−38G]ΔD′-lacZ aphA3) ΔdegU::erm | QB4487→QB4962 |
| QB4967 | trpC2 amyE::(wapAΔA′-lacZ aphA3) | pWP259→168 |
| QB4968 | trpC2 amyE::(wapA[TT−23/−22AA]ΔD′-lacZ aphA3) degU32(Hy) | pWP267→QB136 |
| QB4998 | trpC2 amyE::(wapAΔB′-lacZ aphA3) | pWP280→168 |
Escherichia coli transformations were performed by electroporation of the K-12 strain TG1 [F′ traD36 lacIq Δ(lacZ)M15 proA+B+/supE Δ(hsdM-mcrB)5 (rK− mK− McrB−) thi Δ(lac-proAB)] (9), using a Bio-Rad Gene Pulser as specified by the supplier. Selection was done on LB broth supplemented when appropriate with ampicillin (100 μg ml−1).
B. subtilis strains were transformed with chromosomal or plasmid DNA as follows. Cells were grown in LB liquid medium until they reached transition from exponential growth to the stationary phase. The culture was diluted 20-fold in GE medium, containing 1% glucose, 0.2% potassium l-glutamate, 100 mM potassium phosphate buffer (pH 7.0), 3 mM trisodium citrate, 3 mM MgSO4, 22 mg of ferric ammonium citrate per liter, and 50 mg of l-tryptophan per liter. After dilution, incubation was continued for 4 h at 37°C and DNA was added. Selection was carried out on erythromycin (1 μg ml−1; 10 μg ml−1 for pHT304 derivatives), chloramphenicol (5 μg ml−1), kanamycin (5 μg ml−1) and spectinomycin (100 μg ml−1).
Growth media.
LSM medium contains 1.7% Bacto Agar (Difco), 0.2% casein hydrolysate (Oxoid), 0.5% glucose, 100 mM potassium phosphate buffer (pH 7.0), 3 mM MgSO4, 22 mg of ferric ammonium citrate per liter, and 50 mg of l-tryptophan per liter, supplemented with 80 mg of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) per liter to detect β-galactosidase activity. HSM medium is LSM medium containing 0.7 M disodium succinate added from a 30% disodium succinate solution (pH 7.0). MB liquid medium contains 100 ml of a 10 × MB solution (tryptone, 100 g per liter; yeast extract, 50 g per liter) per liter, 3 mM MgSO4, and 50 mg of l-tryptophan per liter, supplemented with 100 mM disodium succinate (low-salt MB) or 0.7 M disodium succinate (high-salt MB). Bacterial growth was followed by measuring the optical density at 600 nm in liquid cultures.
Nucleic acid manipulations.
The B. subtilis genomic library used in this study was constructed as follows. Chromosomal DNA from wild-type strain 168 was partially digested with Sau3A, and fragments between 0.2 and 1.8 kb in size were ligated at the dephosphorylated BamHI site of the B. subtilis integrative vector pAZ7 to create transcriptional fusions with the lacZ reporter gene. E. coli TG1 was transformed with the ligation mixture, and plasmid DNA was extracted from a pool of 4,000 clones. After transformation of B. subtilis 168, integration into the chromosome at the amyE locus and screening on low-salt and high-salt media, the insert of the unique plasmid of interest was recovered by PCR using oligonucleotides complementary to regions located on each side of the insert and containing BamHI restriction sites. The resulting fragment was then cloned and sequenced.
Sequencing reactions were carried out on both strands of double-stranded DNA purified with QIAprep spin columns (Qiagen Inc., Chatsworth, Calif.), using a dideoxy-chain termination sequencing kit from Pharmacia and synthetic primers.
Plasmid constructions.
The following lacZ transcriptional fusions were constructed by using the pJM783 vector for integration at the original locus of the fused promoter and the pJM115 vector for double-crossover integration at the amyE locus (23). Plasmids pJM783 and pJM115 are chloramphenicol-resistant and kanamycin-resistant derivatives of pDH32, which harbors the lacZ reporter gene translated from the spoVG ribosome binding site (25). Plasmids pWP252 and pWP253 were obtained by subcloning the wapA promoter region and 5′ end of the coding sequence comprised between oligonucleotides OMD26 (5′-GAATGAATTCGCTGAGGGTACGGATATTG-3′) and OMD28 (5′-GTAGGGATCCCTAGTACATCGGCTGGCAC-3′) at the EcoRI and BamHI sites (underlined) of pJM115 and pJM783, respectively. Plasmid pWP259 is a variation of pWP252 that carries a shorter insert, ending near the wapA start codon, which was obtained by PCR with the oligonucleotides OMD26 and WP3BA (5′-GCAAGGATCCTTTTAAAGTTTCGCCTCTTTC-3′). Plasmid pWP259.15 is a pWP259 derivative harboring the A−38G mutation isolated from pWP266.15 after PCR random mutagenesis (see below). A wapA′-lacZ fusion devoid of sequences located downstream of the transcription start site (pWP263) was constructed in pJM115 by using oligonucleotides OMD26 and WP+1 (5′-AAAGGGATCCTTACTCAATAATCTTAACTAG-3′). Plasmid pWP265 is pWP263 carrying the A−38G mutation. Finally, plasmid pWP280 carries the wapA promoter region devoid of sequences located upstream from the putative DegU target site, obtained with oligonucleotides OMD261 (5′-TCGCGAATTCTTAAAATAAGATAAATTTTCTAGAAA-3′) and WP3BA.
Site-directed and PCR random mutagenesis of the wapA 5′ regulatory region.
Site-directed mutagenesis of the putative DegU binding site in the promoter region of wapA was carried out by PCR. A 150-bp upstream fragment (A) ending at the mutation site (TT−23/−22AA) was amplified by using oligonucleotides OMD26 and 5′-TTAAGAAGACTGTTATATCATTACAATATTTTTC-3′ and digested with EcoRI and BbsI to liberate 5′-TTAT-3′ asymmetric overhang carrying the mutations to be introduced. The downstream fragment (B) was amplified with the oligonucleotides 5′-TATTGAAGACATATAACAGTCTAGTTAACATTATTG-3′ and WP3BA. Digestion with BamHI and BbsI liberated 5′-ATAA-3′ cohesive ends. A three-way ligation was performed between the pJM115 integrational vector containing the lacZ reporter gene (23) cleaved with EcoRI and BamHI and fragments A and B described above. The recombinant clone was designated pWP267, and the insert was sequenced to verify that no additional mutation had been introduced in the course of PCR amplification and cloning processes.
Random mutations in the control region of wapA leading to constitutive expression at high salt concentrations were isolated by PCR under conditions that reduce the fidelity of DNA synthesis by the Taq DNA polymerase (19). First, the wapA 5′ regulatory region was subcloned between the EcoRI and BamHI sites of pBluescriptSK, using oligonucleotides OMD26 and WP3BA, to give plasmid pWP258. PCRs reactions were then carried out in the presence of limiting dATP or dGTP (1:10 ratio), using the universal and reverse primers and pWP258 as the template, to obtain a collection of potentially mutated fragments. These were digested with HindIII and BamHI and ligated in front of lacZ in the replicative vector pHT304-18Z (1). E. coli TG1 was transformed by electroporation with the ligation mixture, and plasmid DNA was extracted from a pool of 12,000 clones. B. subtilis 168 was transformed with the resulting library, and transformants were screened on HSM medium. In parallel, the control plasmid pWP266 was obtained by subcloning the wild-type wapA promoter region in pHT304-18Z, using oligonucleotides OMD26 and WP3BA. The mutant derivatives were designated pWP266.1 to pWP266.48S.
β-Galactosidase assay.
B. subtilis strains harboring lacZ fusions were assayed for β-galactosidase activity as previously described (20).
Mapping of mRNA start site by primer extension.
The synthetic oligonucleotide OMD25 (5′-GCCAACACTAAAAATGCTGCAATGAACC-3′) was 5′-end labeled with [γ-32P]ATP (110 TBq mmol−1) by using T4 polynucleotide kinase. Forty micrograms of RNA and 1 pmol of labeled primer were annealed in a total volume of 18 μl of reverse transcriptase buffer (50 mM Tris-HCl, 8 mM MgCl2, 30 mM KCl, 1 mM dithiothreitol [pH 8.5]). The mixture was incubated for 3 min at 60°C and then slowly cooled at room temperature. One microliter (25 U) of avian myeloblastosis virus (Boehringer) reverse transcriptase and 1 μl of a deoxynucleoside triphosphate solution (20 mM each) were added. After 30 min at 37°C, reactions were stopped by the addition of 5 μl of 95% formamide, 10 mM EDTA, 0.3% xylene cyanol, and 3.3% bromophenol blue. Two microliters of each reaction was loaded on a 6% polyacrylamide sequencing gel.
RESULTS
Transcription of wapA is repressed under salt stress conditions.
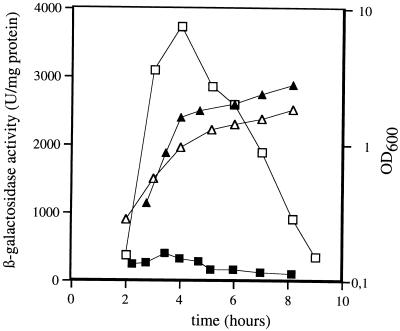
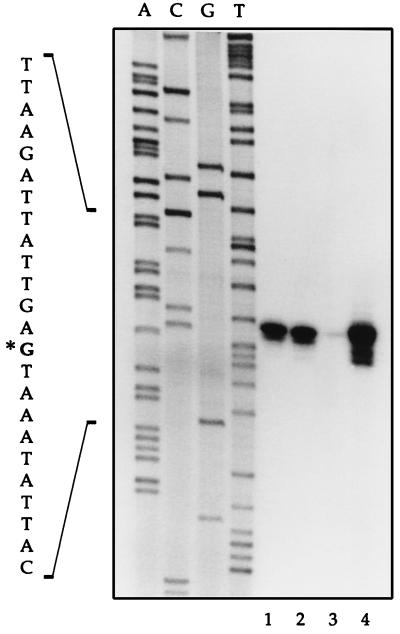
A B. subtilis genomic library was constructed by using the pAZ7 integrative vector that contains the lacZ reporter gene and genomic fragments between 0.5 and 1.8 kb in size to obtain a promoter library (see Materials and Methods for details). A preliminary screening of 150 clones was performed on high-salt and low-salt media to detect promoters affected by salt stress. One of the clones (strain QB4871) appeared to carry a promoter strongly expressed at low salt concentration as observed on LSM medium (minimal medium containing 100 mM disodium succinate) while significantly repressed in HSM (0.7 M disodium succinate). After PCR rescue and sequencing, the corresponding insert was found to harbor the promoter region and 5′ end of the coding sequence of wapA, encoding a wall-associated protein (8). Analysis of the wapA and adjacent chromosomal regions indicated that wapA is preceded by an open reading frame (ORF1) ending 165 bp upstream of the wapA start codon and that the ORF1-wapA intergenic region contained a putative ςA-dependent promoter. To determine whether the observed phenotype was conferred by a promoter located directly upstream of the wapA ORF, wapA′-lacZ fusions were made by using a PCR-amplified fragment encompassing the ORF1-wapA intergenic region. The resulting transcriptional fusions, carried on plasmids pWP252 and pWP253, were integrated in the chromosome of the wild-type strain 168 at the amyE and wapA loci to give strains QB4950 and QB4951, respectively. In both cases, wapA-driven expression of β-galactosidase was significantly decreased under salt stress conditions, as observed on HSM compared to LSM. This was further confirmed by monitoring the β-galactosidase activity of strain QB4950 grown in low-salt and high-salt MB liquid media. The results shown in Fig. 1 indicate that wapA is transcribed from a promoter located in the ORF1-wapA intergenic region and subjected to a drastic repression under high-salt conditions. The transcription start site, determined by primer extension analysis, was found to be located at nucleotide position −35 relative to the wapA start codon (Fig. 3, lane 1). Putative −35 and −10 boxes typical of ςA promoters were detected in the corresponding regions.
FIG. 1.
Effect of salt stress on β-galactosidase expression driven from a wapA′-lacZ transcriptional fusion integrated at the amyE locus. B. subtilis strain QB4950 was grown in MB medium (1% tryptone, 0.5% yeast extract, 50 μg of tryptophan per ml, 1 mM MgSO4) containing 100 mM disodium succinate (□) or 0.7 M disodium succinate (▪). The corresponding growth curves are shown with open (high salt) and closed (low salt) triangles. OD, optical density at 600 nm.
FIG. 3.
Determination of the transcription start site of wapA by primer extension analysis using an oligonucleotide complementary to the DNA sequence from positions +59 to +33 relative to the wapA start codon. The products of a sequencing reaction generated with the same oligonucleotide were run in parallel. mRNA was isolated from low-salt (lanes 1 and 2) and high-salt (lanes 3 and 4) cell cultures of the wild-type strain 168 and the degU strain QB4487, respectively. Equivalent amounts of total RNA were used in all primer extension experiments.
wapA belongs to the DegS-DegU regulon affected by salt stress.
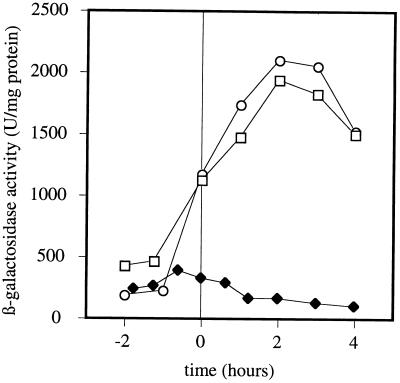
To determine whether wapA regulation could be categorized as one of the three salt stress induction mechanisms described above, expression of the wapA′-lacZ fusion in low-salt and high-salt media was tested in sigB, clpC, and degU deletional mutants. Only inactivation of the degU response regulator had an effect on the expression of wapA. More precisely, either deleting degU or replacing degU with the degU146 allele, which produces a nonphosphorylatable form of DegU, caused a strong derepression of wapA in high-salt medium (Fig. 2). These results confirmed that the DegS-DegU two-component system is involved in adaptation to salt stress and also indicated that transcription of wapA is repressed by the phosphorylated form of DegU under high-salt conditions. Interestingly, none of these two degU mutations (degU or degU146) had a significant effect on the expression of wapA in LB or low-salt medium (data not shown), indicating that DegU-mediated control of wapA occurs mostly under conditions of salt stress. These observations were confirmed by primer extension analysis using mRNA isolated from cultures of the wild-type and degU mutant strains in low-salt and high-salt media. As shown in Fig. 3, deletion of degU had no effect on either the position of the transcription start site or the apparent amount of wapA transcript under low-salt conditions (lanes 1 and 2). In agreement with the β-galactosidase results shown in Fig. 1 and 2, the apparent amount of wapA transcript in the wild-type strain was strongly decreased by high salt concentrations whereas wapA transcription in the degU null mutant was independent of the salt concentration.
FIG. 2.
Effects of degU mutations on the expression of wapA′-lacZ under salt stress conditions. B. subtilis QB4871, QB4883, and QB4955 were grown in high-salt MB medium. Symbols: ⧫, QB4871 (wild type); □, QB4883 (degU::erm); ○, QB4955 (degU146).
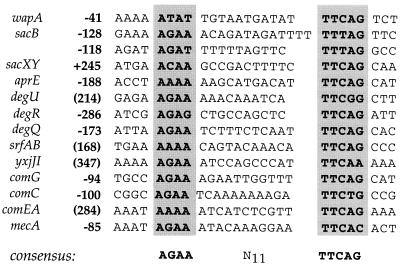
A consensus sequence upstream of DegU-regulated genes?
In an attempt to define a putative target sequence for DegU control, the promoter regions of genes known to be regulated by DegS and DegU were aligned, and the consensus sequence AGAAN11TTCAG was inferred (Fig. 4). To validate our observations, site-directed mutagenesis of the wapA promoter region was undertaken to transform the TTCAG conserved motif, which lies between the −35 and −10 boxes (Fig. 5), into AACAG, thereby modifying two invariant nucleotides in the putative DegU target site. The corresponding wapA[TT−22/−23AA]-lacZ fusion, carried on plasmid pWP267, was integrated at the amyE locus of strain 168 and tested for its ability to be repressed under salt stress conditions. The β-galactosidase phenotype observed on HSM solid medium as well as β-galactosidase assay in liquid high-salt MB medium revealed that expression of wapA[TT−22/−23AA]-lacZ was significantly derepressed in high-salt medium compared to the wild-type fusion (Table 2), indicating that the TT−23/−22AA mutations prevented full repression of wapA transcription under salt stress conditions. However, the degU32(Hy) mutation, which encodes a highly stable phosphorylated form of DegU (4), was able to restore full repression whereas inactivation of degU led to an additional derepression (Table 2), suggesting that the putative DegU target site had been only partially altered by the TT−23/−22AA modification and/or that another site within the wapA regulatory region is involved in DegU-mediated regulation.
FIG. 4.
Comparison of nucleotide sequences located upstream of genes reported to be regulated by DegU. Numbers preceding the sequences indicate positions of the leftmost nucleotide relative to the transcriptional start site; those in parentheses correspond to positions relative to the translational start site. The yxjJI operon was sequenced by Glaser et al. (10) in the framework of the genome sequencing project, and its transcription was shown to be repressed by DegU (4a).
FIG. 5.
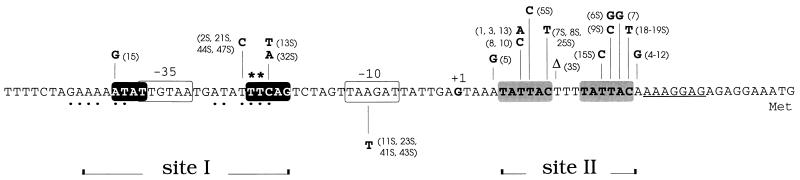
Localization of random mutations leading to high-salt-resistant expression of wapA. The mutations indicated above the nucleotide sequence conferred specific derepression under salt stress conditions, while mutations that exhibited a nonspecific increase of expression in both low- and high-salt media are indicated below the sequence. Asterisks mark nucleotides modified by site-directed mutagenesis. The black-boxed sequences correspond to the conserved motif found upstream of genes known to be regulated by DegU, also referred to as the upstream site or site I. The dots under nucleotides in the upstream region indicate a potential dyad symmetry. The gray boxes highlight the 6-bp direct repeats encompassing the second focus of mutations (site II).
TABLE 2.
Effect of TT(−23/−22)AA and ΔdegU mutations on the repression of wapA transcription by salt stressa
| Strain | Relevant genotype | β-Galactosidase sp act (U/mg of protein)b |
|---|---|---|
| QB4950 | wapA′-lacZ | 100 |
| QB4883 | wapA′-lacZ degU | 1,900 |
| QB4963 | wapA[TT−23/−22AA]′-lacZ | 520 |
| QB4965 | wapA[TT−23/−22AA]′-lacZ degU | 1,950 |
| QB4968 | wapA[TT−23/−22AA]′-lacZ degU32(Hy) | <10 |
Cells were grown in MB liquid medium containing 0.7 M disodium succinate.
Determined 3 h after the end of the exponential growth phase.
Isolation of random mutations that prevent repression of wapA by salt stress.
To confirm the results obtained by site-directed mutagenesis and further define the target of wapA repression by salt stress, a PCR random mutagenesis was performed on the 5′ regulatory region of wapA as described in Materials and Methods. A library of potentially mutated PCR fragments was obtained and subcloned in the replicative vector pHT304-18Z (1) in front of the lacZ reporter gene. The resulting library was transferred into B. subtilis 168 and screened for clones that were no longer sensitive to high-salt repression and had therefore retained a Lac+ phenotype on HSM solid medium. Among 12,000 clones examined, over 50 colonies displayed a light to dark blue color; by comparison, the control strain harboring the wild-type wapA regulatory region (plasmid pWP266) remained white on HSM. Plasmid DNA was isolated from the positive clones, designated pWP266.1 to pWP266.48S; the corresponding inserts were sequenced, and only those clones that carried a single mutation were retained. Further screening allowed us to distinguish two classes of mutants. The majority (26 of 30) were specifically derepressed on HSM, while their expression remained unchanged on low-salt or sporulation medium. On the other hand, 4 clones of 30 exhibited a nonspecific increase of expression on all media tested. Accordingly, these mutants appeared to carry a promoter-up mutation (TAAGAT→TATGAT) that improved the −10 box recognized by ςA. A compilation of these mutations is presented in Fig. 5. Interestingly, the high-salt-specific mutations appeared to fall into two separate clusters: one corresponding to the conserved sequence thought to be involved in DegU binding, and a second focus of high mutation frequency located downstream from the transcription start site within the 5′ untranslated region (Fig. 5). Examination of the DNA sequence encompassing the second cluster of mutations allowed the detection of a direct repeat of two 6-bp motifs (TATTAC) separated by a three-nucleotide spacer. Shortening of this spacer by 1 bp (mutant 3S) also led to a derepressed phenotype. We selected a subset of representative mutants of each class in each region and determined the corresponding β-galactosidase activities in low-salt and high-salt media. The results presented in Table 3 and Fig. 6 confirm the phenotypes observed on plates. It must be noted that pWP266 plasmids are pHT304 derivatives which are present at about four copies per equivalent chromosome (2).
TABLE 3.
Effects of PCR random mutations on the repression of wapA transcription by salt stress
| Plasmida | β-Galactosidase sp act (U/mg of protein)b
|
|
|---|---|---|
| High salt | Low salt | |
| pWP266 (wild type) | 400 | 3,700 |
| pWP266.2S (T−24C) | 1,325 | 4,180 |
| pWP266.3S (T+12) | 2,460 | 4,780 |
| pWP266.7S (C+11T) | 3,570 | 4,780 |
| pWP266.19S (C+20T) | 4,500 | 2,800 |
| pWP266.32S (C−21A) | 2,125 | 3,900 |
See Fig. 5 for positions of the mutations.
Cells were grown in MB liquid medium containing 100 mM (low salt) or 0.7 M (high salt) disodium succinate. β-Galactosidase activities were determined 3 h after the end of the exponential growth phase.
FIG. 6.
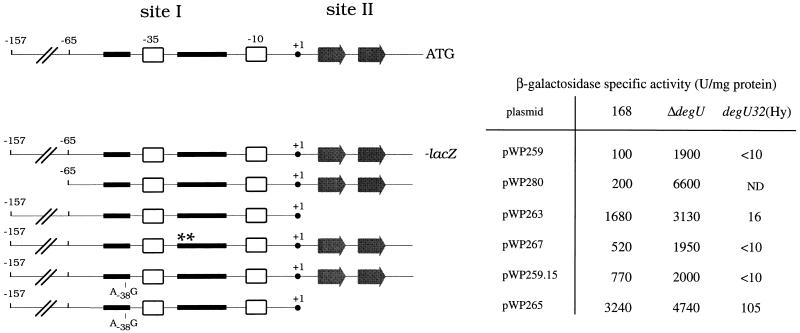
Schematic representation of the wapA regulatory region together with the different constructs designed to alter or delete each one or both of the putative sites involved in repression by salt stress. Plasmid pWP259 contains the entire wild-type 5′ regulatory region of wapA transcriptionally fused to lacZ. In pWP280, sequences located upstream of the two clusters of random mutations have been removed. Plasmid pWP263 is a pWP259 derivative that lacks the downstream site (site II). Plasmid pWP267 harbors the TT−22/−23AA site-directed mutation, pWP259.15 carries the A−38G mutation isolated in site I by random mutagenesis, and pWP265 combines deletion of site II and mutation A−38G in site I. Each plasmid was integrated at the chromosomal amyE locus of the wild-type, degU, and degU32(Hy) strains. The corresponding β-galactosidase activities were determined in high-salt liquid cultures as described for Table 3 and are shown at the right. The data relative to the wapA[TT−22/−23AA] mutant obtained by site-directed mutagenesis are shown for easier comparison.
In an attempt to better elucidate the role of each of the two apparent regulatory regions in DegU-mediated regulation, we designed several lacZ fusions to (i) remove sequences located upstream of the two clusters of random mutations, (ii) alter the upstream site (site I), and/or (iii) delete the downstream site (site II) as schematically represented in Fig. 6 and further described in Materials and Methods. They were integrated as a single copy at the amyE locus of strains 168, QB4883 (degU), and QB136 [degU32(Hy)], and the resulting strains were assayed for β-galactosidase activity in high-salt medium. Our data first indicate that the region starting from −65, relative to the transcription start site, and extending downstream the initiation codon (pWP280) is repressed under high-salt conditions except in the absence of phosphorylated DegU, indicating that pWP280 carries the determinant(s) for DegU-mediated repression under salt stress. Second, altering or deleting site I or site II leads to a partial derepression of wapA transcription in high-salt medium, and deleting degU causes an additional derepression of the corresponding constructs (plasmids pWP263, -267, and -259.15, [Fig. 6]). Third, combining a mutation in site I with the deletion of site II led to a strong derepression of wapA, indicating that negative control at the two sites is cumulative. In the degU32(Hy) mutant, a drastic repression of wapA transcription was observed for each construct tested. The efficiency of this negative control was reduced when both sites I and II were altered.
DISCUSSION
As a widespread soil bacterium also found in coastal and estuarine sediments, as well as in marine and freshwater habitats (27), B. subtilis must be able to adapt to transient or permanent high salt concentrations in its environment. Extensive studies on general and salt stress-specific proteins (see reference 11 for a review) together with the recent advances in functional analysis of the newly sequenced B. subtilis genome indeed indicate that B. subtilis modulates the expression of an impressive number of genes in response to salt stress (2a). However, the molecular mechanisms that govern salt stress-specific regulation remain to be fully elucidated. Recently, expression of genes that are controlled by the DegS-DegU two-component system was shown to be affected under salt stress conditions, such as 1 M NaCl or 1 M KCl. The stimulatory or inhibitory effect proved to be DegS-DegU dependent, as well as salt specific since it could not be obtained during growth at high osmolarity such as 0.5 M lactose (18). A similar effect was also obtained when NaCl was replaced with disodium succinate, indicating that chloride anions were not involved in this salt-mediated regulation (16a).
In the course of a screening for novel promoters modulated by salt stress, the wapA gene has been isolated and shown to undergo negative regulation by the DegS-DegU regulatory pair in high-salt medium. Since inactivation of degU had no effect on wapA transcription at low salt concentrations (data not shown), we propose that the negative control exerted on wapA under salt stress conditions is mediated at least partially by DegU. We also observed that the DegU(Hy)32 protein, of which the phosphorylated form is highly stable, remains able to repress wapA transcription at low salt concentrations (data not shown). This finding supports the hypothesis that salt stress increases the amount of phosphorylated DegU in the cell, by either increasing its synthesis or stimulating DegS-mediated phosphorylation of DegU. As already pointed out, DegU is a pleiotropic regulator involved in various post-exponential-phase responses. It positively controls the synthesis of certain exoenzymes such as the sacB-encoded levansucrase, which produces branched polymers called levans (17). Accordingly, it has been observed that salt stress increases sacB transcription (18). In addition, DegU is a positive regulator of competence and DNA uptake. Thus, the DegS-DegU signal transduction system could be viewed as a means to modulate the synthesis, degradation, or entry into the cells of various macromolecules, thereby participating to osmoregulation and adaptation upon osmotic upshock. In the case of B. subtilis WapA, no precise function could be assigned to the protein based on the absence of any particular phenotype of a wapA null mutant (8). Nevertheless, wall-associated proteins are generally involved in pathogenicity (e.g., the major antigen A from Streptococcus mutans) (7), wall metabolism, secretion, adhesion, or other cell surface-associated properties (34). It could be hypothesized that WapA increases cell wall permeability by functioning as a sieve, or an ion pore or channel, and that turning off its synthesis may prevent excessive exchanges between the outside medium and the cytoplasmic compartment.
Based on a conserved region previously detected by Dubnau upstream from the comC, comG, and recA genes (6), alignment of several promoters known to be controlled by the DegS-DegU system allowed the identification of the conserved sequence AGAAN11TTCAG. It must be pointed out that in the case of sacB, the two detected motifs (Fig. 4) are centered around a 20-bp region shown to be strictly required for DegU-mediated activation, by successive 5′ deletions in the control region of sacB (4a, 12). A computer search of the B. subtilis genome has revealed sequences that perfectly match the consensus presented above in promoter regions not known to be regulated by DegU to date, such as rapA, encoding an aspartate phosphatase that functions to prevent sporulation (24), and paiB, a negative regulator of sporulation and degradative enzyme synthesis (13). Since both rapA and paiB null mutants are able to sporulate in glucose-enriched medium (13, 22) as is the degU32(Hy) mutant, one could hypothesize that phosphorylated DegU lies upstream of rapA and paiB in the regulatory cascade leading to catabolite repression of sporulation.
Site-directed mutagenesis of the putative DegU target, by changing the two invariant T residues in the TTCAG motif to obtain AACAG, led to a partial derepression of wapA transcription in high-salt medium compared to the transcriptional levels of the wild-type wapA′-lacZ in a degU null mutant (about 1900 Miller units). Deleting degU in the wild-type or wapA′ [TT−23/−22AA] wapA′-lacZ strain had a similar effect, supporting the hypothesis that the elevated levels observed in strain QB4963 (wapA[TT−23/−22AA]) were due to a decreased efficiency of DegU repression. At least two reasons can be envisioned to explain the partial effect of the TT−23/−22AA mutation: (i) it may only slightly decrease the binding capability of DegU to the modified target or (ii) DegU could bind to some additional site(s) located in the wapA control region to further downregulate its transcription. The observation that DegU(Hy)32, a mutant version of DegU that exhibits a hyperstable phosphorylated state, remains able to fully repress transcription of wapA [TT−23/−22AA] is compatible with either of these explanations. Complete elimination or drastic modification of the putative target of DegU proved impossible since it overlaps the −35 box that specifies ςA binding.
Random PCR mutagenesis of the control region of wapA confirmed the role of the putative DegU binding site (site I) in repression of wapA transcription at high salt concentrations and allowed the detection of a second site of repression located downstream from the transcription start site (site II [Fig. 5]). The relatively low occurrence of mutations that were obtained in site I could be due to possible interference of such mutations with the binding of ςA to the −35 promoter box. Using constructs designed to alter site I and/or delete site II, we have shown that the negative control at the two sites is cumulative in the wild-type background as well as in the degU null mutant. This suggests the existence of a second mechanism of repression by salt stress occurring at site II and DegU independent, since deletion of site II causes an additional derepression in a degU background (compare β-galactosidase activities conferred by constructs pWP259 and pWP263 or by pWP259.15 and pWP265 in the degU strain). Furthermore, derepression observed in the absence of the downstream operator site proved to be salt specific and could not be observed at low salt concentrations (data not shown). It therefore seems unlikely that the removal of site II, located downstream from the transcription start site, generally affects transcription efficiency by altering the 5′ untranslated sequence of the wapA transcript in pWP263 and derived plasmids. The hypothesis of a second repressor is also reinforced by the observation that transcriptional levels of wapA′-lacZ in low-salt medium (Fig. 1) are twofold higher than those observed at high salt concentrations in the absence of active DegU (Fig. 2), suggesting the existence of residual negative control in the absence of DegU.
However, the degU32(Hy)-encoded protein remained able to fully repress transcription of wapA′-lacZ fusions that carried either an intact or a modified site I (Fig. 6). In addition to the wapA mutant promoters presented in Fig. 6, regulation by the degU32(Hy)-encoded protein was examined on each PCR mutant in the putative operator site I (pWP259.21S, -13S, and -32S [Fig. 5]). None of these mutations was able to eliminate or otherwise affect the ability of DegU(hy) to repress wapA transcription at high salt concentrations. Since DegU(Hy) only partially represses wapA at low salt concentrations (not shown), the effect of each mutation located in site I was examined in low-salt medium in the degU32(Hy) background. A slight resistance to DegU(Hy) repression could be observed with pWP259.21S and 32S compared to the wild-type promoter (twofold decrease), suggesting that at least those two mutations affect indeed the ability of DegU(Hy) to repress wapA. On the other hand, deletion of site II also appeared to slightly affect repression by the DegU(Hy) protein (Fig. 6). Although the significance of these last data might be questionable, the possibility that DegU itself is involved directly or indirectly in regulation at the downstream site cannot be ruled out. A computer search of the B. subtilis genome revealed sequences that match the TATTACN3TATTAC motif upstream from aprE, srfA, and sacX, suggesting that the site I-site II combination may not be unique to the wapA regulatory region. Transposon mutagenesis will be undertaken in an attempt to identify an additional repressor and characterize the second mechanism of salt stress repression exerted on wapA in combination with DegU.
ACKNOWLEDGMENTS
We are grateful to Tarek Msadek for helpful comments and Ivan Moszer for computer analysis.
This study was supported by research funds from the Institut Pasteur and the Centre National de la Recherche Scientifique. Véronique Dartois holds a Biotech research grant from the European Commission (contract BIO4 CT965028).
REFERENCES
- 1.Agaisse H, Lereclus D. Structural and functional analysis of the promoter region involved in full expression of the cryIIIA toxin gene of Bacillus thuringiensis. Mol Microbiol. 1994;13:97–107. doi: 10.1111/j.1365-2958.1994.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 2.Arantes O, Lereclus D. Construction of cloning vectors for Bacillus thuringiensis. Gene. 1991;108:115–119. doi: 10.1016/0378-1119(91)90495-w. [DOI] [PubMed] [Google Scholar]
- 2a.Arnaud, M., and R. Gardan. Personal communication.
- 3.Bonde G J. Bacillus from marine habitats, allocation to phena established by numerical techniques. In: Berkeley R C W, Goodfellow M, editors. The aerobic endospore-forming bacteria, classification and identification. London, England: Academic Press; 1981. pp. 181–215. [Google Scholar]
- 4.Dahl M K, Msadek T, Kunst F, Rapoport G. Mutational analysis of the Bacillus subtilis DegU regulator and its phosphorylation by the DegS protein kinase. J Bacteriol. 1991;173:2539–2547. doi: 10.1128/jb.173.8.2539-2547.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4a.Dartois, V. Unpublished data.
- 5.Deuerling E, Paeslack B, Schumann W. The ftsH gene of Bacillus subtilis is transiently induced after osmotic and temperature upshift. J Bacteriol. 1995;177:4105–4112. doi: 10.1128/jb.177.14.4105-4112.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubnau D. Genetic competence in Bacillus subtilis. Microbiol Rev. 1991;55:395–424. doi: 10.1128/mr.55.3.395-424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feretti J J, Russell R R B, Dao M L. Sequence analysis of the wall-associated protein precursor of Streptococcus mutans antigen A. Mol Microbiol. 1989;3:469–478. doi: 10.1111/j.1365-2958.1989.tb00193.x. [DOI] [PubMed] [Google Scholar]
- 8.Foster S J. Molecular analysis of three major wall-associated proteins of Bacillus subtilis 168: evidence for processing of the product of a gene encoding a 258 kDa precursor two-domain ligand-binding protein. Mol Microbiol. 1993;8:299–310. doi: 10.1111/j.1365-2958.1993.tb01574.x. [DOI] [PubMed] [Google Scholar]
- 9.Gibson T J. Ph.D. thesis. Cambridge, England: University of Cambridge; 1984. [Google Scholar]
- 10.Glaser P, Kunst F, Arnaud M, Coudart M-P, Gonzales W, Hullo M-F, Ionescu M, Lubochinsky B, Marcelino L, Moszer I, Presecan E, Santana M, Schneider E, Schweizer J, Vertès A, Rapoport G, Danchin A. Bacillus subtilis genome project: cloning and sequencing of the 97 kilobases region from 325° to 333°. Mol Microbiol. 1993;10:371–384. [PubMed] [Google Scholar]
- 11.Hecker M, Schumann W, Völker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 12.Henner D J, Yang M, Band L, Shimotsu H, Ruppen M, Ferrari E. Genes of Bacillus subtilis that regulate the expression of degradative enzymes. In: Alacevic M, Hranueli D, Toman Z, editors. Genetics of industrial microorganisms. Proceedings of the Fifth International Symposium on the Genetics of Industrial Microorganisms. Karlovac, Yugoslavia: Ognjen Prica Printing Works; 1987. pp. 81–90. [Google Scholar]
- 13.Honjo M, Nakajama A, Fukazawa K, Kawamura K, Ando K, Hori M, Furutani Y. A novel Bacillus subtilis gene involved in negative control of sporulation and degradative enzyme production. J Bacteriol. 1990;172:1783–1790. doi: 10.1128/jb.172.4.1783-1790.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horinouchi S, Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982;150:815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kang C M, Brody M S, Akbar S, Yang X, Price C W. Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor ςB in response to environmental stress. J Bacteriol. 1996;178:3846–3853. doi: 10.1128/jb.178.13.3846-3853.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krüger E, Völker U, Hecker M. Stress induction of clpC in Bacillus subtilis and involvement in stress tolerance. J Bacteriol. 1994;176:3360–3367. doi: 10.1128/jb.176.11.3360-3367.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16a.Kunst, F. Unpublished data.
- 17.Kunst F, Pascal M, Lepesant-Kejzlarová J, Lepesant J-A, Billault A, Dedonder R. Pleiotropic mutations affecting sporulation conditions and the synthesis of extracellular enzymes in Bacillus subtilis 168. Biochimie. 1974;56:1481–1489. doi: 10.1016/s0300-9084(75)80270-7. [DOI] [PubMed] [Google Scholar]
- 18.Kunst F, Rapoport G. Salt stress is an environmental signal affecting degradative enzyme synthesis in Bacillus subtilis. J Bacteriol. 1995;177:2403–2407. doi: 10.1128/jb.177.9.2403-2407.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leung D W, Chen L, Goeddel D V. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Methods Cell Mol Biol. 1989;1:11–15. [Google Scholar]
- 20.Msadek T, Kunst F, Henner D, Klier A, Rapoport G, Dedonder R. Signal transduction pathway controlling synthesis of a class of degradative enzymes in Bacillus subtilis: expression of the regulatory genes and analysis of mutations in degS and degU. J Bacteriol. 1990;172:824–834. doi: 10.1128/jb.172.2.824-834.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Msadek T, Kunst F, Rapoport G. A signal transduction network in Bacillus subtilis includes the DegS/DegU and ComP/ComA two-component systems. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. pp. 447–471. [Google Scholar]
- 22.Mueller J P, Sonenshein A L. Role of the Bacillus subtilis gsiA gene in regulation of early sporulation gene expression. J Bacteriol. 1992;174:4374–4383. doi: 10.1128/jb.174.13.4374-4383.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Perego M. Integrational vectors for genetic manipulation in Bacillus subtilis. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 615–624. [Google Scholar]
- 24.Perego M, Hanstein C, Welsh K M, Djavakhishvili T, Glaser P, Hoch J A. Multiple protein-aspartate phosphatases provide a mechanism for the integration of diverse signals in the control of development in B. subtilis. Cell. 1994;79:1047–1055. doi: 10.1016/0092-8674(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 25.Perkins J B, Youngman P J. Construction and properties of Tn917-lac, a transposon derivative that mediates transcriptional gene fusions in Bacillus subtilis. Proc Natl Acad Sci USA. 1986;83:140–144. doi: 10.1073/pnas.83.1.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Piggot P J, Moran C P Jr, Youngman P, editors. Regulation of bacterial differentiation. Washington, D.C: American Society for Microbiology; 1993. [Google Scholar]
- 27.Priest F G. Systematics and ecology of Bacillus. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 3–16. [Google Scholar]
- 28.Riethdorf S, Völker U, Gerth U, Winkler A, Engelmann S, Hecker M. Cloning, nucleotide sequence and expression of the Bacillus subtilis lon gene. J Bacteriol. 1994;176:6518–6527. doi: 10.1128/jb.176.21.6518-6527.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sheehan B, Klarsfeld A, Msadek T, Cossart P. Differential expression of virulence gene expression by PrfA, the Listeria monocytegenes virulence regulator. J Bacteriol. 1995;177:3771–3780. doi: 10.1128/jb.177.22.6469-6476.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trieu-Cuot P, Courvalin P. Nucleotide sequence of the Streptococcus faecalis plasmid gene encoding the 3′5‘-aminoglycoside phosphotransferase type III. Gene. 1983;23:331–341. doi: 10.1016/0378-1119(83)90022-7. [DOI] [PubMed] [Google Scholar]
- 31.Trieu-Cuot P, Poyart-Salmeron C, Carlier C, Courvalin P. Nucleotide sequence of the erythromycin resistance gene of the conjugative transposon Tn1545. Nucleic Acids Res. 1990;18:3660. doi: 10.1093/nar/18.12.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voelker U, Voelker A, Maul B, Hecker M, Dufour A, Haldenwang W G. Separate mechanisms activate sigmaB of Bacillus subtilis in response to environmental and metabolic stresses. J Bacteriol. 1995;177:3771–3780. doi: 10.1128/jb.177.13.3771-3780.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Völker U, Mach H, Schmid R, Hecker M. Stress proteins and cross-protection by heat shock and salt stress in Bacillus subtilis. J Gen Microbiol. 1992;138:2125–2135. doi: 10.1099/00221287-138-10-2125. [DOI] [PubMed] [Google Scholar]
- 34.Ward J B, Williamson R. Bacterial autolysins: specificity and function. In: Nombela C, editor. Microbial wall synthesis and autolysis. Amsterdam, The Netherlands: Elsevier Biomedical Press; 1984. pp. 159–166. [Google Scholar]