Significance
Here, we present a large cohort study of peripheral blood TCR (T cell receptor) clonotype diversity in breast cancer patients diagnosed with either DCIS (ductal carcinoma in situ) or de novo stage IV disease at younger (<45) and older (≥45) ages. Peripheral blood TCR clonotype diversity is associated with age and intratumor immune status and might be linked to probability of disease progression.
Keywords: breast cancer, TCR clonotype, DCIS, diversity
Abstract
Immune escape is a prerequisite for tumor growth. We previously described a decline in intratumor activated cytotoxic T cells and T cell receptor (TCR) clonotype diversity in invasive breast carcinomas compared to ductal carcinoma in situ (DCIS), implying a central role of decreasing T cell responses in tumor progression. To determine potential associations between peripheral immunity and breast tumor progression, here, we assessed the peripheral blood TCR clonotype of 485 breast cancer patients diagnosed with either DCIS or de novo stage IV disease at younger (<45) or older (≥45) age. TCR clonotype diversity was significantly lower in older compared to younger breast cancer patients regardless of tumor stage at diagnosis. In the younger age group, TCR-α clonotype diversity was lower in patients diagnosed with de novo stage IV breast cancer compared to those diagnosed with DCIS. In the older age group, DCIS patients with higher TCR-α clonotype diversity were more likely to have a recurrence compared to those with lower diversity. Whole blood transcriptome profiles were distinct depending on the TCR-α Chao1 diversity score. There were more CD8+ T cells and a more active immune environment in DCIS tumors of young patients with higher peripheral blood TCR-α Chao1 diversity than in those with lower diversity. These results provide insights into the role that host immunity plays in breast cancer development across different age groups.
The immune landscape changes dynamically as breast cancer evolves from ductal hyperplasia to ductal carcinoma in situ (DCIS) and further to invasive and then metastatic disease (1). We previously described gradually increasing immunosuppression during breast tumor progression and identified the in situ to invasive carcinoma transition as a key step in immune escape, characterized by an increase of regulatory T cells and a decline in activated granzyme B positive CD8+ T cells and T cell receptor (TCR) clonotype diversity (2, 3). The tumor immune microenvironment is also impacted by tumor subtype [estrogen receptor–positive (ER–positive), HER2-positive, triple-negative] and histological grade (high or low), with HER2-positive and triple-negative breast cancer (TNBC) having a higher frequency of tumor-infiltrating lymphocytes (TILs) compared to estrogen receptor–positive (ER–positive) tumors. Similarly, high-grade tumors have more TILs than low-grade tumors, and this pattern is already evident at the DCIS stage (2). Differences in TILs correlate with responses to immune checkpoint inhibitor (ICI) therapy, and thus, ICI currently is only approved for a subset of patients with TNBC (4). However, there is increasing evidence that the efficacy of ICI is determined not only by the tumor immune microenvironment but also by characteristics of the host (i.e., patient), implying a link between peripheral immunity and anti-tumor immune responses (5).
Peripheral blood TCR clonotype diversity is a quantitative assessment of host immune status, which has been shown to be associated with the clinical outcome and the efficacy of immunotherapy in certain cancer types (6, 7). Besides pathogen exposure, TCR clonotype diversity is also strongly impacted by age, with decreasing richness (number of different TCR clonotypes) and increasing clonality (relative abundance of clonotypes) observed in older individuals (8, 9). Aging is also associated with an increase in chronic inflammatory conditions and immune dysfunction (10), which may diminish anti-tumor immune responses and favor tumor initiation, progression, and disease recurrence. These data suggest that peripheral blood TCR clonotype diversity might potentially be a clinically useful biomarker for predicting cancer risk and progression as well as treatment outcomes.
Here, we assessed peripheral blood TCR diversity in a large cohort of breast cancer patients of different ages diagnosed either with DCIS (pre-invasive) or with de novo stage IV (distant metastatic) disease to explore associations between TCR clonotype diversity and breast cancer progression. We also performed RNA-seq on whole blood and DCIS tumor tissues in a subset of cases to analyze how the transcriptome of peripheral blood and tumor tissue relates to TCR diversity.
Results
Peripheral TCR Clonotype Diversity in Breast Cancer Patients.
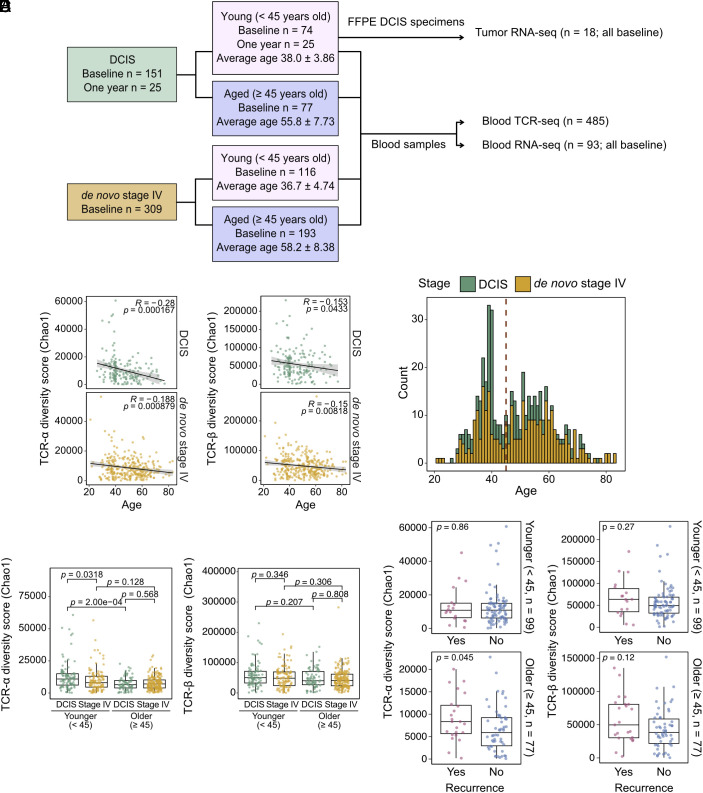
To assess peripheral blood TCR clonotype diversity, we extracted RNA from the peripheral blood of 485 breast cancer patients spanning a wide age distribution (mean age 48, range 21 to 83 y old), diagnosed with either DCIS (n = 176) or de novo stage IV disease (n = 309), and performed TCR-α and TCR-β chain sequencing (Fig. 1A and Dataset S1). We chose to profile patients with DCIS and de novo stage IV disease because they represent the earliest and the most advanced stages of breast cancer. In addition, comparing these two conditions facilitates the identification of tumor stage-related differences independent of therapeutic impact, since these patients did not have any treatment prior to diagnosis. Blood was collected at the time of diagnosis (baseline) from all patients with de novo stage IV disease (n = 309) and most (151 out of 176) DCIS cases. For 25 DCIS patients, blood was collected 1 y after diagnosis and following surgical resection of DCIS. For six DCIS cases, we profiled matched baseline and 1-y blood samples.
Fig. 1.
Peripheral blood TCR diversity and patient age, tumor stage, and clinical outcome. (A) Schematic outline of the study. (B) Plot depicting correlation between peripheral blood TCR α- (Left) and β- (Right) chain Chao1 diversity with age in patients diagnosed with DCIS and de novo stage IV disease. Correlation is given by Pearson’s R. (C) Age distribution of DCIS and de novo Stage IV patients in the cohort. The red dashed line represents the age cutoff used to classify patients into younger (<45 y old) and older (≥45 y old) groups. (D) Graph showing differences in peripheral blood TCR α- (Left) and β- (Right) chain Chao1 diversity between patients diagnosed with DCIS and de novo stage IV disease in younger and older age groups using FDR-adjusted Wilcoxon rank sum test. (E) Dot plot illustrating differences in peripheral blood TCR α- (Left) and β- (Right) chain Chao1 diversity in DCIS patients with or without recurrence in younger and older age groups using the Wilcoxon rank sum test.
TCR diversity was calculated using multiple different metrics (Methods) assessing both the overall size of the TCR clonotype repertoire as well as the relative abundance of each clonotype. The richness measure is simply counting the number of unique TCR species in a sample, while the Chao1 index is a nonparametric estimate of the total unobserved richness. Shannon index, as well as Shannon and Simpson clonality, are hybrid indices jointly accounting for both the number of species and their relative abundance, via different mathematical formulations (11). All these metrics capture different biological aspects of the diversity of the TCR clonotype distribution.
The six matched blood samples from DCIS patients showed no statistically significant differences in any of the six diversity indices computed on both TCR-α and TCR-β chains between the baseline and 1-y time points (SI Appendix, Fig. S1A). This suggests that surgical resection of DCIS may not have a lasting impact on TCR clonotype diversity, although the sample size is very small. We excluded these six 1-y samples and only used the matched baseline samples for further analyses.
We first analyzed associations between TCR diversity and age. Increasing age was associated with decreasing Chao1 diversity (unobserved total richness) and increasing clonality (both Shannon and Simpson), for both DCIS and de novo stage IV patients and both TCR-α and TCR-β chains, with TCR-α diversity more highly correlated with age than TCR-β (Fig. 1B and SI Appendix, Fig. S1 B and C and Dataset S2). Due to the significant impact of age on TCR clonotypes and the clinical and molecular differences of breast tumors between younger and older patients in part due to menopause (12), we aimed to evaluate TCR diversity separately in these two groups. To this end, we used an age cutoff of 45 y old to separate patients into younger (<45) and older (≥45) age groups. The chosen cutoff is biologically relevant because it separates the patients into likely premenopausal (mean age in younger DCIS 38.0 ± 3.86 and stage IV 36.7 ± 4.74) and likely postmenopausal (mean age in older DCIS 55.8 ± 7.73 and stage IV 58.2 ± 8.38) groups. In addition, the age distribution of all the patients in our cohort is seemingly binomial (Fig. 1C).
Next, we compared TCR diversity among patients diagnosed with either DCIS or de novo stage IV disease to determine stage and age-specific differences. We focused primarily on the Chao1 index, which we considered to be a more robust assessment of TCR signal due to its overall association with age. Younger DCIS patients exhibited significantly higher TCR-α Chao1 diversity compared to either younger de novo stage IV patients or older patients with DCIS (Fig. 1D and Dataset S2). In contrast, TCR-α diversity was not significantly different between older DCIS patients and older de novo Stage IV patients (Fig. 1D and Dataset S2). No significant differences were observed when comparing categories using the TCR-β repertoire or other indices than Chao1 (Fig. 1D, SI Appendix, Fig. S1 D and E, and Dataset S2).
Next, we assessed potential associations between TCR diversity and clinical features. We found that TCR-α Chao1 diversity was significantly higher in older DCIS patients who experienced a subsequent cancer diagnosis (ipsi- or contralateral breast cancer or chest wall or distant recurrence, see Dataset S1) compared to those who did not (Fig. 1E and Dataset S2). In younger DCIS patients in contrast, recurrence was not associated with differences in TCR diversity, potentially due to the high frequency of bilateral mastectomies in these cases (Fig. 1E, SI Appendix, Fig. S2 A and B, and Datasets S1 and S2). We also evaluated whether TCR diversity is associated with overall survival in de novo stage IV patients by multivariate Cox regression and found that none of the diversity indices was significantly associated with survival in neither younger nor older patients (Dataset S2). Dataset S2 also contains the multiple testing corrected P-values across the different indices evaluated and tests performed.
Overall, we report that TCR-α Chao1 diversity is associated with age in patients with both DCIS and de novo stage IV disease and with tumor stage at diagnosis in younger women, as well as with the risk of subsequent cancer diagnosis in older patients with DCIS.
Unique Peripheral T Cell CDR3α Sequences among Patients.
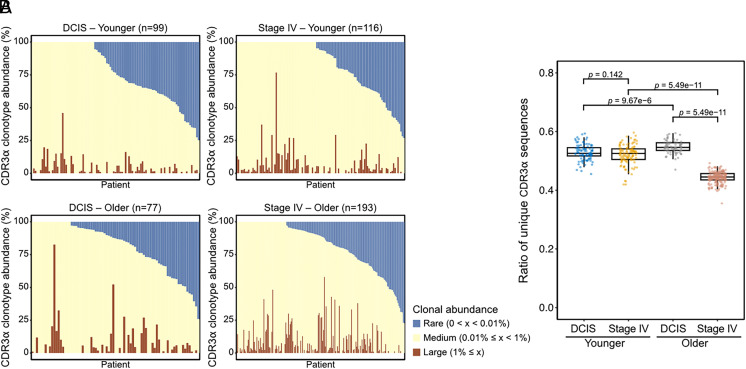
We next evaluated the frequencies of CDR3α sequences part of each patient’s clonotype to further explore the association between TCR clonotype diversity and patient groups or clinical outcomes. The frequency composition of the TCR clonotype, as assessed by the relative frequency of CDR3α sequences, was variable across patients (Fig. 2A and SI Appendix, Fig. S3A). The ratio of unique CDR3α sequences in individual patients (number of unique sequences divided by the total number of sequences in a single patient) was substantially lower in the older de novo stage IV group than in the other groups (Fig. 2B). As CDR3α sequences similar at the amino acid level may potentially recognize the same antigens, we clustered all CDR3α sequences from all patients using the Geometric Isometry-based TCR AligNment Algorithm (GIANA) tool (13) (Datasets S3 and S4). The younger age DCIS patients had significantly more CDR3α clusters per patient than stage IV cases (SI Appendix, Fig. S3B), implying higher richness of T cells potentially targeting different antigens in DCIS compared to stage IV patients. This is consistent with the observed differences in TCR-α Chao1 diversity (Fig. 1D). In contrast, in the older cohort, CDR3α sequences from both DCIS and stage IV patients clustered into similar numbers of clusters (SI Appendix, Fig. S3B).
Fig. 2.
CDR3 sequences of peripheral blood T cells and patient age or tumor stage. (A) The abundance of CDR3α sequences in the individual patients. (B) Graph depicting the number of CDR3α sequences which are unique or shared by two or more patients in each group, using one-way ANOVA with Tukey’s test.
Considering that TCR richness was slightly higher in younger DCIS patients than younger de novo stage IV patients and was also associated with recurrence in older DCIS patients, we anticipated that the abundance of unique CDR3α sequences in DCIS cases may be relevant to DCIS recurrence. To this end, we compared the numbers of total CDR3α clusters in each of the four patient groups (younger/older and DCIS/de novo stage IV) and the overlap among them (SI Appendix, Fig. S3C). We investigated whether the numbers of CDR3α clusters unique in each case is different in DCIS patients with or without recurrence. The younger age group DCIS patients without recurrence had relatively more unique CDR3α clusters than those with recurrence [10.89 (871/80) per patients without recurrence compared to 7.41 (126/17) per patients with recurrence] while both the older age group DCIS patients with or without recurrence had a similar number of unique CDR3α clusters [5.32 (266/50) per patients without recurrence compared to 5.43 (123/23) patients with recurrence] (SI Appendix, Fig. S3 C and D). We further explored whether the categories of the predicted antigens recognized by these unique CDR3α clusters might be different depending on DCIS recurrence status. The antigen of CDR3α sequences predicted by a manually curated catalogue of pathology-associated T cell receptor sequences (McPAS-TCR) (14) exhibited differences in the relative frequencies of the clusters between younger DCIS patients with and without recurrence (SI Appendix, Fig. S3E and Dataset S5). While these associations with specific CDR3α clusters might potentially be related to clinical outcomes especially in younger DCIS patients, further analysis and experimental validation would be required to ensure the integrity and accuracy of these predictions. Overall, the CDR3 sequences show substantial variability across individuals, patient groups, and patients with different clinical outcomes, with a high number of sequences in the younger DCIS patients.
Peripheral Blood TCR Diversity and Peripheral Blood Transcriptomes.
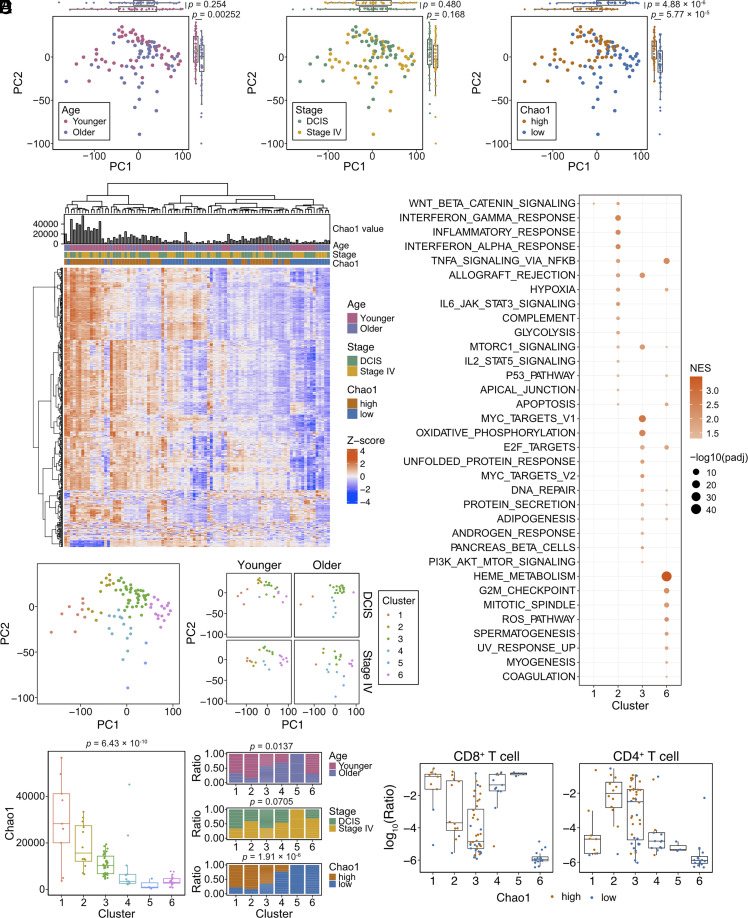
We next investigated whether differences in peripheral blood TCR diversity reflect differences in peripheral blood transcriptomes. We performed bulk RNA-seq of peripheral blood in 100 patients, 25 from each of the four age/tumor stage groups (93 samples passed QC; Dataset S6). Due to the observed associations between the TCR-α Chao1 index and age and stage at diagnosis, we assessed associations between TCR-α Chao1 index and peripheral blood transcriptomes. All patients were divided into Chao1-high and -low groups based on the median score of the TCR-α Chao1 index of 93 samples (Dataset S6). Principal component and hierarchical clustering analysis showed superior separation of RNA-seq samples based on the TCR-α Chao1 index of diversity than based on tumor stage or patient age demonstrating that the peripheral blood TCR diversity has a strong association with gene expression patterns. (Fig. 3 A and B).
Fig. 3.
Peripheral blood TCR diversity and total blood transcriptomes. (A) Principal component analysis plots of peripheral blood RNA-seq samples (n = 93) colored based on age (Left; younger and older), tumor stage (Middle; DCIS and stage IV), and peripheral blood TCR α-chain Chao1 index (Right; Chao1-high and -low), using Welch’s t-test. (B) Heatmap depicting unsupervised clustering of samples based on the top 500 differentially expressed genes. (C) PCA plots of all samples together (Left) or subdivided by age and stage (Right) colored based on hierarchical clustering analysis. (D) Peripheral blood TCR α-chain Chao1 index of the different RNA-seq clusters (Left), using one-way ANOVA. Ratio of samples according to age (Top), stage (Middle), and peripheral blood TCR α-chain Chao1 index (Bottom) in each RNA-seq cluster, using the Chi-squared test. (E) GSEA Hallmark analysis of each RNA-seq cluster. All enriched pathways (adjusted P-value ≤ 0.05) are indicated. (F) Plots depicting results of digital immune cytometry analysis using BayesPrism. The ratio of the indicated immune cell types to all cells was estimated based on the Azimuth PBMC celltype.l1 (15). Samples are colored based on their peripheral blood TCR Chao1 index.
To explore the potential functional relevance of these transcriptomic differences, we performed GSEA (geneset enrichment analysis) C2 core pathway analysis on the different patient groups (younger and older, DCIS and de novo stage IV, and Chao1-high and Chao1-low). Signatures related to innate immunity (REACTOME_NEUTROPHIL_DEGRANULATION and REACTOME_INNATE_IMMUNE_SYSTEM) were enriched in de novo stage IV patient samples compared to DCIS (SI Appendix, Fig. S4A). Terms related to general cellular functions, including translation, transcription, mitochondrial function, and cell cycle were enriched in younger, DCIS, and Chao1 high groups.
We next separated all samples into six clusters by hierarchical clustering (Methods) for further characterization of transcriptomes and cellular composition (Fig. 3C and SI Appendix, Fig. S4B). The distribution of samples with high and low Chao1 diversity was significantly different among clusters (Fig. 3D). GSEA Hallmark analysis revealed that signatures related to inflammatory response (e.g., INTERFERON_GAMMA_RESPONSE and INFLAMMATORY_ RESPONSE) were significantly enriched in cluster 2, which has a high proportion of patients with a high Chao1 index. On the other hand, several signatures related to proliferation (e.g., MYC_TARGETS and G2M_CHECKPOINT) and metabolic features (e.g., OXIDATIVE_PHOSPHORYLATION and UNFOLDED_PROTEIN_RESPONSE) were enriched in clusters 3 and 6 implying higher proliferation (Fig. 3E). To determine to what degree the observed differences in gene expression profiles might be due to differences in cells composing each cluster, we performed cell type and gene expression deconvolution using BayesPrism (16) (Dataset S6). We observed high heterogeneity in the predicted ratios of various immune cell populations among clusters (Fig. 3F and SI Appendix, Fig. S4C). However, potential contamination with red blood cells and platelets is likely, as the fraction of inferred T and B cells, dendritic cells, monocytes, and NK cells are overall small. For example, samples in cluster 6 are almost entirely made up of erythrocytes (SI Appendix, Fig. S4D). The ratio of monocytes was the highest in cluster 2 compared to the other clusters (SI Appendix, Fig. S4C), which may explain the strong inflammatory signature characterizing this cluster (Fig. 3E). Overall, these results suggest that the peripheral blood TCR diversity reflects global transcriptional differences defined by a combination of cellular composition and functional activity.
Peripheral TCR Diversity and DCIS Immune Microenvironment.
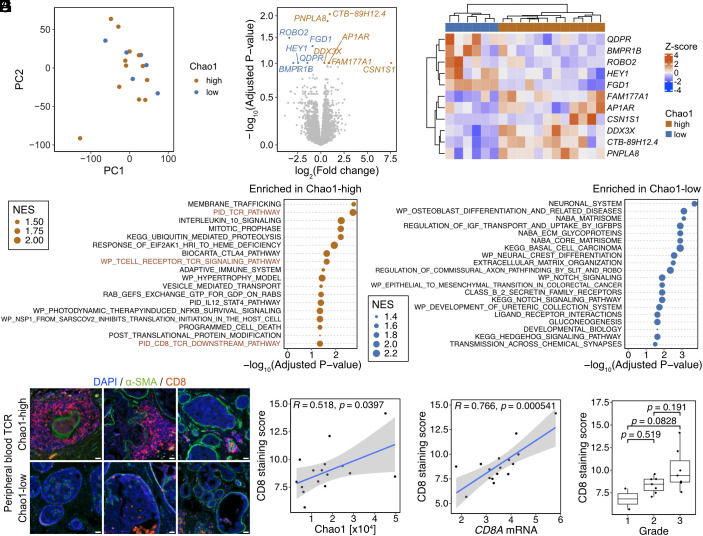
Last, we investigated potential associations between peripheral blood TCR diversity and DCIS tumor transcriptomes by performing RNA-seq on formalin-fixed paraffin-embedded (FFPE) tumor specimens of 18 younger DCIS cases (Dataset S7). We focused on DCIS to avoid organ site of metastasis-related differences in de novo stage 4 patients. In these samples, the peripheral blood TCR-α Chao1 index did not show significant association with tumor grade (SI Appendix, Fig. S5A) nor seem to impact global DCIS tumor transcriptomes (Fig. 4A and SI Appendix, Fig. S5B), although the sample size is too small for conclusive observations. Comparison of transcriptomes of DCIS from patients with a high and low peripheral blood TCR-α Chao1 index revealed 11 differentially expressed genes (DEGs) (Fig. 4 B and C). FGD1, encoding a protein required for invadopodia biogenesis and extracellular matrix degradation (17), had consistently high expression in tumors from patients with low peripheral blood TCR-α Chao1 index, while PNPLA8 (phospholipase A2) and DDX3X (DEAD-box RNA helicase) were highly expressed in cases with a high peripheral blood TCR-α index. Both PNPLA8 and DDX3X play a role in inflammation (18, 19) potentially implying an inflammatory microenvironment in DCIS of patients with high peripheral blood TCR-α Chao1 diversity. In line with this, GSEA pathway (C2 core pathway) analysis demonstrated that DCIS from patients with high peripheral blood TCR-α Chao1 index were enriched in pathways related to the adaptive immune system and TCR signaling (Fig. 4D). In contrast, DCIS from cases with a low peripheral blood TCR-α Chao1 index displayed enriched pathways related to neuronal system and ECM-related pathways, as represented by high expression of FGD1, HEY1, and BMPR1B (mediators of these pathways) in these tumors (Fig. 4 B–D). Consistently, GSEA hallmark analysis demonstrated enrichment for TNFA and WNT-BETA CATENIN signaling as the most significantly enriched pathways in DCIS from patients with high and low peripheral blood TCR-α Chao1 diversity, respectively (SI Appendix, Fig. S5C).
Fig. 4.
Peripheral blood TCR diversity and DCIS immune microenvironment in younger patients. (A) Principal component analysis of DCIS tumor transcriptomes (n = 18). Samples are colored based on peripheral blood TCR α-chain Chao1 index (Chao1-high or -low). (B and C) DEG analysis of tumor transcriptomes between peripheral blood TCR α-chain Chao1 index-high and -low patients. (B) Volcano plot depicting DEGs (adjusted P-value ≤ 0.05; orange, DEGs in Chao1-high cases; blue, DEGs in Chao1-low cases). (C) Heatmap depicting the expression of significant DEGs in individual cases. (D) GSEA C2 pathway analysis of tumors between peripheral blood TCR α-chain Chao1 index-high and low patients. NES, Normalized enrichment score. (E) Representative images of immunostaining for CD8 and the α-SMA myoepithelial cell marker. (F) Plot depicting correlation between CD8+ cell accumulation and peripheral blood TCR α-chain Chao1 index, using Pearson’s R. (G) Plot depicting correlation between CD8 immunofluorescence score and CD8A mRNA in RNA-seq data, using Pearson’s R. (H) CD8 immunofluorescence score in DCIS tumors with different grade, using one-way ANOVA with Tukey’s test.
To characterize the DCIS immune microenvironment in more detail, we performed BayesPrism cellular decomposition analysis using the breast tumor immune microenvironment as reference (16). Immune cell composition was overall very similar between tumors from DCIS patients with a high and low peripheral TCR-α Chao1 index with the exception of some T cell subsets (e.g., CD8 IFNG), which showed a higher ratio in tumors of patients with high compared to those with low peripheral blood TCR-α Chao1 diversity (SI Appendix, Fig. S5 D and E). To experimentally validate these predictions, we performed immunofluorescence for CD8 and found high frequency of CD8+ T cells around ducts in DCIS from patients with high peripheral blood TCR-α Chao1 diversity (Fig. 4E). The CD8 staining score positively correlated with peripheral blood TCR-α Chao1 diversity (Fig. 4F). The CD8 staining score also significantly correlated with CD8 mRNA levels in our tumor RNA-seq data, suggesting that the enrichment of CD8+ T cell-related pathways in tumor RNA-seq of peripheral blood TCR-α Chao1-high patients is likely due to the higher abundance of CD8+ T cells in these DCIS (Fig. 4G). The CD8 staining score also showed an increasing trend with increasing DCIS grade, although there were very few low-grade cases (Fig. 4H).
These results highlight the association between peripheral blood TCR-α Chao1 diversity and the DCIS immune microenvironment and imply that high CD8+ T cell content in pre-invasive tumors might be linked to more diverse host immunity.
Discussion
In this study, we describe associations between peripheral blood TCR clonotype diversity and clinico-pathologic features of breast cancer in defined age groups of patients diagnosed with DCIS or de novo stage IV disease. We confirm previous findings of TCR diversity decreasing with age and show that this is independent of stage of breast cancer at diagnosis. In the younger age group, estimated unobserved total richness of the alpha chain, as assessed by Chao1, is significantly higher in patients diagnosed with DCIS vs. de novo stage IV disease and correlates with intratumoral CD8+ T cell accumulation in DCIS. In older DCIS patients, higher peripheral blood TCR diversity is associated with a higher risk of a subsequent cancer diagnosis (breast cancer in ipsi- or contralateral breast and distant site combined).
Aging is strongly associated with cancer initiation and progression (20), with 62 being the median age of breast cancer diagnosis in the United States (SEER 2012 to 2016 data). While aging is widely thought to contribute to increased cancer risk due to time-dependent mutation accumulation (21–23), key facets of anti-tumor immune function also decline with age (24). The relative frequencies of naïve T and B cell populations decrease with age with a concomitant increase in memory and exhausted T and B cells and impaired cytokine production by CD4+ and CD8+ T cells, all of which have the potential to affect anti-tumor immune responses (24–26). Peripheral blood TCR clonotype diversity is a quantitative measure of host immune status that may better reflect biological than chronological age. Peripheral blood TCR richness expands at an early post-natal stage and decreases throughout an individual’s lifetime, while T cell clonal expansion in older individuals leads to increased peripheral TCR clonality (5). Our data showing decreasing diversity and increasing clonality with age are in line with these prior findings and show that this also occurs both in patients diagnosed with DCIS and de novo stage IV disease.
Peripheral host immune status, defined largely by life-long pathogen exposure, has been proposed to influence anti-tumor immunity through a variety of mechanisms. Activation of toll-like receptor 3, an innate immune signaling pathway that can be activated by microbial infection, has been shown to mediate cross-priming of CD8+ T cells, which has been proposed to enhance anti-tumor immunity (27). Importantly, reports of T cell cross-reactivity between common bacterial and viral antigens and tumor-associated antigens also support a role for infectious history in long-term host immune status and anti-tumor immunity (28). Epitope mimicry may also occur between microbial antigens and tumor-associated antigens, leading to T and B cell cross-reactivity and a potentially enhanced anti-tumor immune response (29). For example, mumps and influenza infections have been found to enhance CD8+ T cell recognition of tumor-associated antigens and enhance overall anti-tumor immunity (30, 31). Similarly, extended antibiotic use is associated with higher breast cancer risk (32). This extensive interplay between host immune status and anti-tumor immunity raises a possibility that the peripheral TCR diversity shaped by infections and vaccinations may impact the intratumor immune microenvironment and breast tumor evolution.
Overtreatment of DCIS is a major clinical issue; thus, it is critical to identify clinically useful biomarkers that predict the risk of invasive progression (33). Higher-grade DCIS exhibits greater immune infiltration, but it is also associated with higher risk of recurrence (34). Younger age is also one of the risk factors of DCIS progression and recurrence (35, 36) highlighting the impact of age on tumor evolution. Here, we show that peripheral blood TCR diversity is closely associated with peripheral blood global transcriptomes and the intratumor DCIS microenvironment including CD8+ T cell infiltration, and it might predict the risk of tumor progression in an age-dependent manner. The finding that in older DCIS patients, higher peripheral blood TCR diversity is associated with higher risk of recurrence (both ipsi- and contralateral breast cancer) may seem paradoxical. This may be explained by the balance between anti-tumor immunity and tumor-supportive immunity, i.e., abundance of immunosuppressive cells like regulatory T cells (Tregs). Antitumor immunity can also be a selection pressure that shapes tumor evolution—while stronger selection may delay tumor initiation, once tumors arose, they may progress faster as they have already passed an evolutionary bottleneck (37). Further studies using much larger cohorts are needed to determine whether and how host immunity and intra-tumoral immune cell composition can determine DCIS tumor evolution and whether TCR diversity per se or specific TCR sequences may be most relevant for determining outcomes. Assessing diversity of purified T cell subpopulations (e.g., CD4+, CD8+, and Treg) would also be important due to the distinct biological functions of different T cells that can positively (e.g., Treg) or negatively (e.g., CD8+) impact tumor progression. The interpretation of TCR sequencing data is hampered by under-sampling issues. We used the Chao1 index throughout this manuscript to measure the unobserved size of the TCR repertoire considering the strong correlation with patient age. The Chao1 index specifically corrects for under-sampling, making it particularly suitable in our context. Deriving biological insights on a population level from higher-order diversity measures such as Shannon or Simpson clonality based on TCR sequencing data will likely require much larger patient datasets.
In summary, our results highlight the importance of peripheral immunity and age in tumor evolution and suggest further exploration of blood TCR diversity as a biomarker of disease progression in larger patient cohorts.
Materials and Methods
Patient Samples.
All patient samples were collected following informed consent using protocols approved by the Dana-Farber Cancer Institute Institutional Review Board, protocol # 06-169 (Young Women’s Breast Cancer Study, YWS) and Project SHARE (Specimens Help Research Efforts) (DF-HCC #93-085) (all other cases). All samples used in this study are listed in the Dataset S1. YWS is the largest cohort of women with young-onset breast cancer designed to conduct regular medical record review and collect biospecimens and patient-reported outcomes. The original objectives were to 1) enroll a cohort of women age ≤40 newly diagnosed with breast cancer; 2) characterize the cohort at diagnosis and in follow-up regarding disease and psychosocial outcomes; and 3) archive tumor and blood specimens for future studies. From 2006 to 2016, participants enrolled from 13 North American academic and community-based sites with high accrual of those approached (60%) and high participant engagement (average survey completion rate: 86%). This engagement has facilitated extended follow-up to investigate longer-term survivorship issues and outcomes. We have completed medical record review on 100% of participants through 18 mo post-diagnosis, collected ≥1 blood sample on 92%, and have centrally reviewed primary tumor pathology on 97%, with blocks archived on 84% and tissue microarrays created on 89%. At a median follow-up of 10.0 y in April 2023, the YWS is uniquely positioned for investigations of age-related tumor and host biology as well as for studies evaluating the impact of hereditary predisposition, post-diagnosis pregnancy, premature menopause, psychosocial, lifestyle, and care delivery on cancer endpoints and comorbidities. The EMBRACE (Ending Metastatic Breast Cancer for Everyone) Research Cohort Study (DF-HCC #09-204) prospectively collects baseline whole blood and serial plasma samples along with clinical, pathologic, treatment, and outcomes data of patients with metastatic breast cancer treated at Dana-Farber Cancer Institute since 2009. In addition to data from YWS and EMBRACE, we also captured additional clinically annotated blood samples from Stage IV de novo breast cancer patients and patients with DCIS disease treated at DFCI under Project SHARE (Specimens Help Research Efforts) (DF-HCC #93-085). Since its inception in 1993, Project SHARE has allowed us to collect a large biorepository of blood samples and a clinical outcomes database for future research studies. Clinical data elements used for the analyses included: time from initial breast cancer diagnosis date to date of blood draw, stage at initial diagnosis, histology, age at time of diagnosis, recurrence, and survival status.
TCR-Sequencing.
Total RNA of peripheral blood was extracted from 400 μL of each blood sample using the Quick-DNA/RNA miniprep plus kit (Zymo Research) based on the manufacturer’s protocol for mammalian whole blood samples. The total amount of RNA was measured using the Qubit Fluorometer. Bulk TCR-sequencing was performed with 200 ng of total RNA. The sequencing and data preprocessing were performed by the Dana-Farber Cancer Institute Translational Immunogenomics Laboratory following the protocol of RNase H-dependent PCR enabled T-cell receptor sequencing (38). The fastq files generated from the Illumina MiSeq count the total number of reads. BLAST48 separates them into alpha and beta by aligning to chain-specific primers that were used in amplification. MIXCR (39) is used to assemble and annotate the reads for alignment of each TCR, known V and J regions and identification of CDR3s and merges TRBV genes. The output of this pipeline was all unique clonotypes with V identity, CDR3 amino acid sequence, read count, and frequency.
RNA-Sequencing.
Total RNA of peripheral blood collected for the TCR-sequencing was used for RNA-sequencing. Library preparation and sequencing on an Illumina Nova-seq 6000 with paired-end reads was performed by the Dana-Farber Cancer Institute Molecular Biology Core Facilities. Library preparation was done using the KAPA mRNA HyperPrep Kit. The concentrations and fragment sizes of the libraries were measured using the Agilent TapeStation 2200 according to the manufacturer’s protocols. FFPE specimens (n = 18) were obtained from Brigham and Women’s Hospital, Massachusetts General Hospital, Faulkner Hospital, as well as a variety of other outside institutions part of the Young Women’s Study. The presence of DCIS was confirmed by hematoxylin-eosin staining of sections adjacent to the scrolls used for RNA preparation. Total RNA was extracted from 40-μm scroll of FFPE specimens (n = 18) using the MagMAX™ FFPE DNA/RNA Ultra Kit (Thermo Fisher Scientific) based on the manufacturer’s protocol. FFPE RNA-sequencing was performed by Novogene.
Immunostaining.
FFPE specimens were obtained from Brigham and Women’s Hospital, Massachusetts General Hospital, Faulkner Hospital, as well as a variety of other outside institutions part of the Young Women’s Study. All specimens stained (n = 16) were matched with samples used for FFPE tumor transcriptome analysis. Specimens were deparaffinized in xylene and ethanol and washed with tap water. Heat-induced antigen retrieval was performed in ethylenediaminetetraacetic acid (EDTA)-based solution (pH 9.0). Specimens were blocked with 10% normal goat serum in phosphate buffered saline (PBS) at room temperature for 15 min. Primary antibodies were diluted in 5% normal goat serum in PBS and incubated at 4 °C for overnight. Anti-CD8 antibody (clone C8/144B) and anti-α-SMA antibody (clone 1A4) were obtained from Thermo Fisher Scientific. Secondary antibodies (Alexa Flour 555-conjugated anti-mouse IgG1 antibody and Alexa Flour 488-conjugated anti-mouse IgG2a antibody; Thermo Fisher Scientific) were diluted in 5% normal goat serum in PBS and incubated at room temperature for 1 h. After washing with PBS, specimens were mounted with VECTASHIELD® Vibrance™ Antifade Mounting Medium with DAPI (Vector Laboratories). Images were captured by the Nikon Ti/E inverted microscope and analyzed with QuPath. For the analysis, we manually selected DCIS tumors demarcated by α-SMA staining and their surrounding regions in which CD8+ cells existed. The number of CD8+ cells was automatically counted and normalized by the automatically calculated size of the DCIS tumor. The CD8 staining index was calculated based on the following formula: log2(number of surrounding CD8+ cells/mm2 DCIS tumor).
Data Analysis.
Calculation of TCR diversity.
TCR diversity can be quantified and assessed in multiple different ways. As discussed in ref. 11, commonly used measures are related to the family of Hill numbers, also known as effective numbers of species. If N is the number of unique clonotypes in a sampled repertoire and pi are the respective frequencies of the N unique clonotypes, then:
Taking the logarithm of Dα leads to Hα, the generalized measure of entropy, or the Renyi entropy:
Different values of α correspond to metrics which weight differently the relative abundance of the TCR species, further capturing different aspects of the immune system. Here, we report TCR diversity measures of order 0 (α = 0, richness), 1 (α → 0, Shannon entropy), 2 (α = 2, Simpson index), as well as two additional measures: Pielou’s index and Chao1.
Richness (α = 0) is defined as simply the number of unique clonotypes, independent of their abundance.
Next, α → 1 corresponds to the Shannon entropy. Shannon entropy reaches its maximum when a repertoire consists of equally distributed TCR sequences (11):
Further, α = 2 in the Renyi entropy leads to , which represents the logarithm of the probability of two randomly sampled TCRs with replacement to belong to the same clonotype (also known as the Simpson Index). Based on H2, we computed here the Simpson Clonality Index as follows:
In addition to the above entropy measures, the clonality of the TCR repertoire can also be assessed using Pielou’s Index, a scaled version of the Shannon entropy, where the scaling factor equals the logarithm of the total number of species:
Equal clonotype frequencies correspond to a Shannon clonality of 0, and Shannon clonality scores closer to 1 suggest stronger clonal dominance.
All the 4 diversity measures introduced above assume that the observed clonotype distribution is also the real one. The human TCR repertoire is however much more diverse than molecular sequencing can currently capture, leading to undersampling. In order to address the problem of unseen clonotypes which are real, but not present in the sequencing sample, we report and discuss the Chao1 Index, an abundance-based nonparametric estimator for the total unobserved richness in a sample.
where n1 and n2 are the number of singletons and doubletons (i.e., number of unique TCR sequences whose counts are one and two in the samples, respectively) (40).
All graphs were generated using R 4.3.0. The Cox regression analysis was performed with the survival package (v3.5-5) and the survminer package (v0.4.9).
Multiple correction testing.
We performed multiple correction testing to calculate adjusted P-values in Fig. 1 and SI Appendix, Figs. S1 and S2. The P-values obtained in the analysis of each diversity index were combined and adjusted using the Benjamini–Hochberg method for the multiple correction testing of Fig. 1B and SI Appendix, Fig. S1 B and C. For Fig. 1D and SI Appendix, Fig. S1 D and E as well as Fig. 1E and SI Appendix, Fig. S2 A and B, the P-values obtained by pairwise t tests of each group of comparison were combined and jointly false discovery rate (FDR)-corrected. All results of multiple correction testing were listed in Dataset S2.
CDR3 sequence analysis.
All CDR3 sequences were used for analysis. The Venn diagrams were plotted using the ggvenn package (v0.1.9) and VennDiagram package (v1.7.3). Fig. 2B data represent the count of clusters of CDR3 sequences generated using GIANA clustering (v1.2.0) without the TRBV variable gene option (13). GIANA was run in a linux-64 environment with python 3.10.4. CDR3α clustering data by GIANA are listed in Dataset S3. The antigen prediction analysis was performed using McPAS-TCR database via a web browser interface (14). Max Levenshtein distance was set to two (14). The output of database is listed in Dataset S4. In SI Appendix, Fig. S3E, we categorized the cluster in which either of predicted antigens were annotated to cancer antigen as “Cancer” while other clusters as “Allergy/Autoimmune/Pathogens/Others”.
Processing bulk RNA FASTQ files for both blood and tumor.
Both the blood and tumor bulk RNA FASTQ files were processed using the RIMA (RNA-seq Immune Analysis) pipeline (41). RIMA performs integrative computational analysis of the tumor microenvironment from bulk tumor RNA-seq data. In RIMA, the raw reads are aligned to a reference genome (hg38) from GDC using the STAR (Spliced Transcripts Alignment to a Reference) tool. This tool converts the FASTQ to BAM files. The gene transcripts are quantified using Salmon which outputs TPM (transcripts per million). TPMs were converted to a matrix using the tximport package and log normalized for further statistical analysis (15).
Bulk RNA-sequencing of peripheral blood.
All graphs were generated using R 4.3.0. Hierarchical clustering for the PCA plot was performed based on PC1 and PC2 using hclust. The number of clusters was then decided based on second differences D index values calculated by NbClust package (v3.0.1), which suggested two clusters as a first choice, followed by six clusters. We chose six clusters considering the evenness of the samples in each cluster and separation based on TCR diversity. Pre-ranked GSEA was performed using the FGSEA package (v1.24.0), while the DESeq2 package (v1.38.3) was used for the pre-ranking of gene expression. The MSigDB C2 pathway collection (v7.5.1) was used for two-group comparisons. The MSigDB hallmark collection (v7.5.1) was used for cluster analysis. Enriched pathways in each cluster were analyzed for each cluster based on the one-by-one comparison between samples in one cluster and all other samples in the other clusters. Digital immune cytometry was performed by the BayesPrism algorithm with the reference of the Azimuth PBMC celltype.l1 or celltype.l2 (16). GSEA and BayesPrism analysis results are listed in Dataset S5.
Bulk FFPE RNA-sequencing of younger DCIS tumors.
All graphs were made using R 4.3.0. DEG analysis was performed using the DESeq2 package (v1.38.3) with a cut-off of adjusted P-values ≤ 0.05 to define the DEGs. Hierarchical clustering in the heatmap of DEGs was performed by the Ward.D2 method in the ComplexHeatmap package (v2.14.0). Pre-ranked GSEA was performed by the FGSEA package (v1.24.0), and the DESeq2 package (v1.38.3) was used for the pre-ranking of gene expression. Each collection of the MSigDB C2 pathway subsets used were the Reactome subset (v2022.1), the WikiPathways subset (v2022.1), the PID subset (v2022.1), and the KEGG subset (v2022.1). The MSigDB hallmark collection (v7.5.1) was also used. Digital immune cytometry was performed by the BayesPrism algorithm with the reference of previously published breast tumor immune microenvironment data (42). GSEA and BayesPrism analysis results are listed in Dataset S6.
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Acknowledgments
We thank members of our laboratories for their critical reading of this manuscript and useful discussions. We thank the Dana-Farber Cancer Institute Molecular Biology Core Facilities and Translational Immunogenomics Laboratory for their outstanding service. This research was supported by the National Cancer Institute R35 CA197623 (K.P.), DF/HCC SPORE P50CA168504 (M.H., N.U.L., and K.P.), the Cancer Immunological Data Commons (NCI U24CA224613, to F.M.), Breast Cancer Research Foundation (A.H.P., N.U.L., and K.P.), Susan G. Komen (A.H.P. and K.P.), American Cancer Society (K.P.), Louise Sandberg Fund - Mad About Louise (N.U.L.), and the Ludwig Center at Harvard (F.M. and K.P.).
Author contributions
K.P. designed research; J.N., J.P., and A.P. performed research; S.C., A.P., M.H., C.S., V.B., K.J.R., L.C.C., A.-M.F., K.S., V.B., D.D., N.U.L., and A.H.P. contributed new reagents/analytic tools; J.N., S.C., S.B., G.B., and F.M. analyzed data; and J.N., S.C., S.B., F.M., and K.P. wrote the paper.
Competing interests
K.P. serves on the Scientific Advisory Boards of Novartis, Ideaya Biosciences, and Scorpion Therapeutics, holds equity options in Scorpion Therapeutics and Ideaya Biosciences, and receives sponsored research funding through Dana-Farber from Novartis. F.M. is a cofounder of and has equity in Harbinger Health, has equity in Zephyr AI, and serves as a consultant for Harbinger Health, and Zephyr AI. She is also on the board of directors of Exscientia Plc. F.M. declares that none of these relationships are directly or indirectly related to the content of this manuscript. D.D. receives research funding from Canon, Inc. J.P. is currently an employee of Merck Sharp & Dohme LLC, a subsidiary of Merck & Co., Inc., Rahway, NJ, USA. J.P. contributed to this work prior to their employment at Merck Sharp & Dohme LLC. The opinions or perspectives expressed herein do not represent the opinions or perspectives of Merck Sharp & Dohme LLC.
Footnotes
Reviewers: T.A.C., Cleveland Clinic; and O.J.F., University of Pittsburgh School of Medicine.
Contributor Information
Simona Cristea, Email: scristea@jimmy.harvard.edu.
Franziska Michor, Email: michor@jimmy.harvard.edu.
Kornelia Polyak, Email: kornelia_polyak@dfci.harvard.edu.
Data, Materials, and Software Availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or supporting information. The RNA-seq and TCR-seq data generated in this study have been deposited to the NCBI GEO database under accession number GSE239935 (43). The code used for data analyses can be found at https://github.com/Michorlab/TCRSeqBC (44).
Supporting Information
References
- 1.Gil Del Alcazar C. R., Aleckovic M., Polyak K., Immune escape during breast tumor progression. Cancer Immunol. Res. 8, 422–427 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gil Del Alcazar C. R., et al. , Immune escape in breast cancer during in situ to invasive carcinoma transition. Cancer Discov. 7, 1098–1115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stover D. G., et al. , Phase II study of ruxolitinib, a selective JAK1/2 inhibitor, in patients with metastatic triple-negative breast cancer. NPJ Breast Cancer 4, 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Debien V., et al. , Immunotherapy in breast cancer: An overview of current strategies and perspectives. NPJ Breast Cancer 9, 7 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hiam-Galvez K. J., Allen B. M., Spitzer M. H., Systemic immunity in cancer. Nat. Rev. Cancer 21, 345–359 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin Y. B., et al. , TCR repertoire profiling of tumors, adjacent normal tissues, and peripheral blood predicts survival in nasopharyngeal carcinoma. Cancer Immunol. Immunother. 67, 1719–1730 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X., et al. , T cell repertoire in peripheral blood as a potential biomarker for predicting response to concurrent cetuximab and nivolumab in head and neck squamous cell carcinoma. J. Immunother. Cancer 10, e004512 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lanfermeijer J., Borghans J. A. M., van Baarle D., How age and infection history shape the antigen-specific CD8(+) T-cell repertoire: Implications for vaccination strategies in older adults. Aging Cell 19, e13262 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krishna C., Chowell D., Gonen M., Elhanati Y., Chan T. A., Genetic and environmental determinants of human TCR repertoire diversity. Immun. Ageing 17, 26 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrucci L., Fabbri E., Inflammageing: Chronic inflammation in ageing, cardiovascular disease, and frailty. Nat. Rev. Cardiol. 15, 505–522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiffelle J., et al. , T-cell repertoire analysis and metrics of diversity and clonality. Curr. Opin. Biotechnol. 65, 284–295 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Li C. H., Haider S., Boutros P. C., Age influences on the molecular presentation of tumours. Nat. Commun. 13, 208 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang H., Zhan X., Li B., GIANA allows computationally-efficient TCR clustering and multi-disease repertoire classification by isometric transformation. Nat. Commun. 12, 4699 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tickotsky N., Sagiv T., Prilusky J., Shifrut E., Friedman N., McPAS-TCR: A manually curated catalogue of pathology-associated T cell receptor sequences. Bioinformatics 33, 2924–2929 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Stuart T., et al. , Comprehensive integration of single-cell data. Cell 177, 1888–1902.e21 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chu T., Wang Z., Pe’er D., Danko C. G., Cell type and gene expression deconvolution with BayesPrism enables Bayesian integrative analysis across bulk and single-cell RNA sequencing in oncology. Nat. Cancer 3, 505–517 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ayala I., et al. , Faciogenital dysplasia protein Fgd1 regulates invadopodia biogenesis and extracellular matrix degradation and is up-regulated in prostate and breast cancer. Cancer Res. 69, 747–752 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Samir P., Kanneganti T. D., DEAD/H-Box helicases in immunity, inflammation, cell differentiation, and cell death and disease. Cells 11, 1608 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ramanadham S., et al. , Calcium-independent phospholipases A2 and their roles in biological processes and diseases. J. Lipid. Res. 56, 1643–1668 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laconi E., Marongiu F., DeGregori J., Cancer as a disease of old age: Changing mutational and microenvironmental landscapes. Br. J. Cancer 122, 943–952 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martincorena I., Campbell P. J., Somatic mutation in cancer and normal cells. Science 349, 1483–1489 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Milholland B., Suh Y., Vijg J., Mutation and catastrophe in the aging genome. Exp. Gerontol. 94, 34–40 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Risques R. A., Kennedy S. R., Aging and the rise of somatic cancer-associated mutations in normal tissues. PLoS Genet. 14, e1007108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X., Meng X., Chen Y., Leng S. X., Zhang H., The biology of aging and cancer: Frailty, inflammation, and immunity. Cancer J. 23, 201–205 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Derhovanessian E., Solana R., Larbi A., Pawelec G., Immunity, ageing and cancer. Immun. Ageing 5, 11 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon A. K., Hollander G. A., McMichael A., Evolution of the immune system in humans from infancy to old age. Proc. Biol. Sci. 282, 20143085 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumoto M., Takeda Y., Tatematsu M., Seya T., Toll-like receptor 3 signal in dendritic cells benefits cancer immunotherapy. Front. Immunol. 8, 1897 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sioud M., T-cell cross-reactivity may explain the large variation in how cancer patients respond to checkpoint inhibitors. Scand. J. Immunol. 87, e12643 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Sioud M., How does autoimmunity cause tumor regression? A potential mechanism involving cross-reaction through epitope mimicry Mol. Med. 8, 115–119 (2002). [PMC free article] [PubMed] [Google Scholar]
- 30.Cramer D. W., et al. , Mumps and ovarian cancer: Modern interpretation of an historic association. Cancer Causes Control 21, 1193–1201 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iheagwara U. K., et al. , Influenza virus infection elicits protective antibodies and T cells specific for host cell antigens also expressed as tumor-associated antigens: A new view of cancer immunosurveillance. Cancer Immunol. Res. 2, 263–273 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Velicer C. M., et al. , Antibiotic use in relation to the risk of breast cancer. JAMA 291, 827–835 (2004). [DOI] [PubMed] [Google Scholar]
- 33.van Seijen M., et al. , Ductal carcinoma in situ: To treat or not to treat, that is the question. Br. J. Cancer 121, 285–292 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Visser L. L., et al. , Predictors of an invasive breast cancer recurrence after DCIS: A systematic review and meta-analyses. Cancer Epidemiol. Biomarkers Prev. 28, 835–845 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Elshof L. E., et al. , Cause-specific mortality in a population-based cohort of 9799 women treated for ductal carcinoma in situ. Ann. Surg. 267, 952–958 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cronin P. A., Olcese C., Patil S., Morrow M., Van Zee K. J., Impact of age on risk of recurrence of ductal carcinoma in situ: Outcomes of 2996 women treated with breast-conserving surgery over 30 years. Ann. Surg. Oncol. 23, 2816–2824 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dunn G. P., Bruce A. T., Ikeda H., Old L. J., Schreiber R. D., Cancer immunoediting: From immunosurveillance to tumor escape. Nat. Immunol. 3, 991–998 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Li S., et al. , RNase H-dependent PCR-enabled T-cell receptor sequencing for highly specific and efficient targeted sequencing of T-cell receptor mRNA for single-cell and repertoire analysis. Nat. Protoc. 14, 2571–2594 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bolotin D. A., et al. , MiXCR: Software for comprehensive adaptive immunity profiling. Nat. Methods 12, 380–381 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Hughes J. B., Hellmann J. J., Ricketts T. H., Bohannan B. J., Counting the uncountable: Statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67, 4399–4406 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang L., et al. , Tutorial: Integrative computational analysis of bulk RNA-sequencing data to characterize tumor immunity using RIMA. Nat. Protoc. 18, 2404–2414 (2023), 10.1038/s41596-023-00841-8. [DOI] [PubMed] [Google Scholar]
- 42.Wu S. Z., et al. , A single-cell and spatially resolved atlas of human breast cancers. Nat. Genet. 53, 1334–1347 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Polyak K., Peripheral blood TCR clonotype diversity as an age-associated marker of breast cancer progression. NCBI GEO. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE239935. Deposited 2 August 2023. [DOI] [PMC free article] [PubMed]
- 44.Bodapati S., Michorlab/TCRSeqBC. Github. https://github.com/Michorlab/TCRSeqBC. Deposited 1 November 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Dataset S02 (XLSX)
Dataset S03 (XLSX)
Dataset S04 (XLSX)
Dataset S05 (XLSX)
Dataset S06 (XLSX)
Dataset S07 (XLSX)
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper and/or supporting information. The RNA-seq and TCR-seq data generated in this study have been deposited to the NCBI GEO database under accession number GSE239935 (43). The code used for data analyses can be found at https://github.com/Michorlab/TCRSeqBC (44).