Significance
Type IV pili (T4P) are widely found in prokaryotes (bacteria and archaea). They are directly involved in a large range of functions, including surface sensing, biofilm formation, twitching motility, and uptake of DNA. T4P are essential virulence factors for many pathogenic bacteria. We show that the structural diversity in bacteria is greater than previously assumed, with the tight adhesion (tad) pili of Caulobacter crescentus more closely resembling archaeal T4P, supporting their acquisition from archaea via horizontal gene transfer. Although the pilus in Vibrio cholerae, the pathogen responsible for cholera, shows similar helical packing (axial rise and twist parameters) to other bacterial T4P, its structure reveals the direct relationship between sequence changes and structural rearrangements.
Keywords: cryo-EM, helical polymers, bacterial filaments
Abstract
Type IV pili (T4P) are ubiquitous in both bacteria and archaea. They are polymers of the major pilin protein, which has an extended and protruding N-terminal helix, α1, and a globular C-terminal domain. Cryo-EM structures have revealed key differences between the bacterial and archaeal T4P in their C-terminal domain structure and in the packing and continuity of α1. This segment forms a continuous α-helix in archaeal T4P but is partially melted in all published bacterial T4P structures due to a conserved helix breaking proline at position 22. The tad (tight adhesion) T4P are found in both bacteria and archaea and are thought to have been acquired by bacteria through horizontal transfer from archaea. Tad pilins are unique among the T4 pilins, being only 40 to 60 residues in length and entirely lacking a C-terminal domain. They also lack the Pro22 found in all high-resolution bacterial T4P structures. We show using cryo-EM that the bacterial tad pilus from Caulobacter crescentus is composed of continuous helical subunits that, like the archaeal pilins, lack the melted portion seen in other bacterial T4P and share the packing arrangement of the archaeal T4P. We further show that a bacterial T4P, the Vibrio cholerae toxin coregulated pilus, which lacks Pro22 but is not in the tad family, has a continuous N-terminal α-helix, yet its α1 s are arranged similar to those in other bacterial T4P. Our results highlight the role of Pro22 in helix melting and support an evolutionary relationship between tad and archaeal T4P.
Type IV pili are long thin retractile surface filaments that are extensively and broadly distributed in bacteria and archaea where they mediate diverse functions, including adhesion, motility, and DNA uptake and are critical for virulence in many Gram-negative bacterial pathogens (1). T4P belong to a broad class of filaments, the Type IV filament superfamily, that share the T4P architecture and core assembly components and include Type II secretion endopili and archaeal flagella (2). Common features of each system are listed in SI Appendix, Table S1, though exceptions continue to emerge as new Type IV filaments are discovered and characterized (3). Numerous studies have described the impressive diversity of bacterial (4) and archaeal (5, 6) T4P. Two main groupings have been proposed for bacterial T4P, T4aP and T4bP, based on distinct features of the primary building block, the major pilin, and on the composition and gene distribution of the pilus assembly components (7, 8). The major pilin has a canonical structure, with an extended ~50-residue N-terminal α-helix, α1, the C-terminal half of which is embedded in a globular C-terminal domain (1, 9). The N-terminal half of α1 (α1 N) is hydrophobic with the exceptions of a conserved serine/threonine at position 2 and a glutamate at position 5. The α1 N helix anchors the pilin in the cytoplasmic membrane prior to pilus assembly and is arranged in a helical array in the core of the assembled pilus. T4a pilins are typically smaller, 140 to 170 amino acids, with a smaller globular domain than those of T4b pilins, which are around 200 amino acids in length. The T4a major pilins have a conserved phenylalanine that becomes the first residue in the mature pilin following removal of a typically short leader peptide from a class III signal sequence. Though α1 is a continuous α-helix in X-ray crystal structures of full length pilins (10–13), cryo-electron microscopic reconstructions of T4a pili at near-atomic resolution reveal a melted segment of α1 N that is centered on the helix-disrupting proline at position 22 (14). Pro22 is essential for pilus assembly for the Neisseria meningitidis T4aP and is proposed to facilitate subunit packing and to confer elastic properties on these adhesive filaments by allowing further melting of the helix to withstand high shear forces (14, 15). In contrast to the T4a pilins, the T4b pilins have a long leader peptide, a variable residue at position 1, and lack a proline at position 22. T4b pilins do possess several glycines in α1 N that may, like Pro22, disrupt the α-helix. However, this region has not been visualized as there are no high-resolution structures available for T4bP filaments.
T4a and T4b pili also differ in the complexity of their biogenesis machinery. The T4b pili are comparatively simple, assembled by less than a dozen protein components spanning the inner and outer membrane and intervening periplasm. These components include a dedicated prepilin peptidase, a cytoplasmic assembly ATPase that powers pilus assembly, an inner membrane platform protein on which the pilus is built, an outer membrane secretin channel, and a single minor pilin. This minor pilin bears homology to the major pilin but is much lower in abundance and initiates pilus assembly as well as pilus retraction (16). The T4aP systems share the core assembly machinery found in the T4bP but possess additional assembly components that are typically encoded in clusters throughout the bacterial genome. These include a retraction ATPase, PilT, that coordinates with the assembly ATPase to produce highly dynamic filaments, plus multiple minor pilins that form a heterocomplex to prime pilus assembly (9).
Among the T4bP, a distinct subfamily has emerged (17), which have since been shown to be phylogenetically distinct from the T4bP (18). These are the tad pili, for tight adherence, which are present in most bacterial and archaeal species and mediate attachment, autoaggregation, surface sensing, and biofilm formation (19). While the major pilins of the tad family share the features of T4b pilins, i.e., a long leader peptide and lack of Phe1 and Pro22, they are remarkably small, 40 to 60 amino acids in length, comprising only α1. Accordingly, these pilins are referred to in some cases as flp, for fimbrial low-molecular-weight protein (20). Tad pilus assembly components are encoded on a single operon, which, like most T4b pili, lacks a gene encoding a retraction ATPase. Yet, tad pili differ from T4bP in their assembly components, including having two ATPases but no retraction ATPase, and two platform proteins, which are likely the result of gene fission (18). Some consider tad pili to be a separate class, T4c pili (21, 22). Rooted phylogenetic analysis and comparisons of the core assembly components show that the tad pili are more closely related to archaeal T4P than to bacterial T4b pili (18). In fact, the entire tad locus was proposed to have been transferred horizontally from archaea to bacteria. Archaeal major pilins are typically similar in size to bacterial T4P, but like the T4b and tad pilins they lack Phe1 and Pro22. Additionally, the globular C-terminal domain of archaeal T4 pilins lacks homology with those of bacterial T4P (23). With no available structures for tad pili, it has remained an enigma as to how these short pilin subunits form functional filaments that appear comparable in diameter to T4aP.
Another T4P, from the soil microbe Geobacter sulfurreducens, has a truncated major pilin of only 65 amino acids, comparable to flp pilins, yet it carries the hallmarks of T4a pili, i.e., a short leader peptide, Phe1 and Pro22, with pilus assembly components, including a retraction ATPase and multiple minor pilins encoded on several operons. Surprisingly the cryo-EM structure of the G. sulfurreducens T4P revealed a canonical T4aP with the α1 segments of the major pilin formed by the 65 amino acid PilA protein, now referred to as PilA-N, and the C-terminal globular domain formed by a second 104 residue polypeptide encoded by a gene immediately downstream of pilA-N, pilA-C (24, 25). Thus it was possible that the tad pilus, like the G. sulfurreducens T4P, might contain a second separately encoded C-terminal protein in addition to the flp pilin. In the tad system of Caulobacter crescentus (the Cpa pilus), the mature flp pilin, PilA, has only 45 residues, which are predicted to form a continuous α-helix. These pili act as surface contact sensors for C. crescentus as they adhere to surfaces and retract to bring the bacterium in direct contact with the surface, which promotes holdfast formation and long-term attachment (19, 21, 26). Atomic force microscopy measurements reveal limited spring-like mechanical properties for C. crescentus tad pili (27), which differ from the considerable elasticity observed for Neisseria gonorrhoeae T4aP (28). While it is possible that a second protein forming a globular domain may be present in these pili, SDS-PAGE from these C. crescentus filaments showed only a single low-molecular weight band that corresponds to the 45-amino acid PilA (29).
To understand the molecular architecture and mechanical properties of the tad pilus in relation to other T4P, we determined the cryo-EM structure of the C. crescentus Cpa pilus, revealing a filament composed of PilA only. PilA is a continuous helix, consistent with the absence of Pro22, and forms a helical array that resembles the packing of α1 in archaeal T4P with respect to the rise and twist from one subunit to the next. We also determined the structure of the Vibrio cholerae toxin coregulated pilus (TCP), a T4bP whose major pilin, TcpA, like tad and other T4b pilins, lacks Pro22. As with C. crescentus PilA, α1 of TcpA is continuous. Yet, unlike PilA, the helical symmetry of TcpA falls in the range of existing bacterial T4aP structures. Our results are consistent with an archaeal origin for tad pili (18), and show that bacterial T4P structures can be even more diverse than originally imagined. These results have implications for understanding the diversity in mechanical properties of T4P with respect to strength, flexibility, and elasticity, properties that influence their functionality.
Results
Cryo-EM of C. crescentus Tad Pilus.
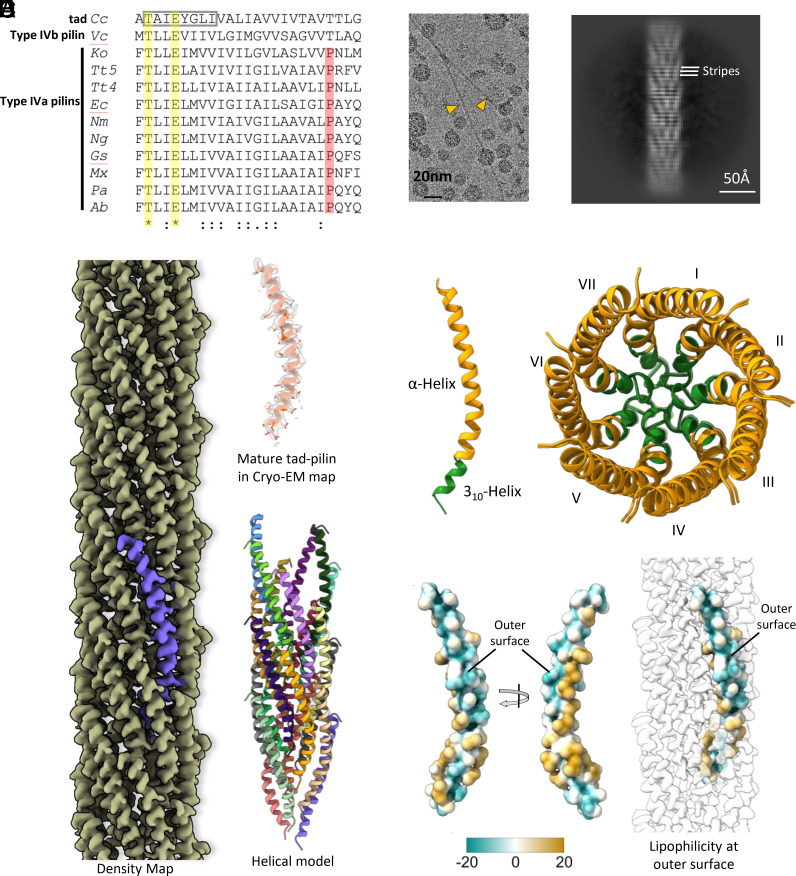
The mature pilin of the C. crescentus tad pilus, PilA, contains only 45 residues, in striking contrast to canonical T4 pilins of known structure, which contain 111 to 208 residues. Additionally, PilA lacks Pro22, conserved in all published bacterial T4aP structures (Fig. 1A). C. crescentus tad pili appear as thin (diameter ~45–50 Å) straight filaments in cryo-EM images (Fig. 1B). The absence of the C-terminal domain is evident from the 2D class average of tad filament segments, which clearly show an array of helices (Fig. 1C). The averaged power spectrum of tad pilus segments shows anomalously strong intensity at a spacing of 1/(4.9 Å) (SI Appendix, Fig. S1), reminiscent of amyloid filaments which show such strong intensity at ~1/(4.7 Å) representing β-strand separations (30). This extremely strong periodicity is also evident from the horizontal stripes at this frequency in the 2D class average (Fig. 1C). We reconstructed the filaments to a resolution of 3.4 Å (SI Appendix, Fig. S2) by imposing helical symmetry with an axial rise of 4.9 Å and a twist of 102.8°. The map was of sufficient quality, showing density for bulky side chains, to allow de novo model building for the entire mature tad pilin subunit [residues 1 to 45, (Fig. 1D)]. We did not observe any additional density, once the model was built, that might be due to glycosylation or other post-translational modification. The filament diameter is ~50 Å. Unlike all published bacterial T4aP structures (Fig. 1A), the helix in the tad pilus is fully intact and does not show any melting (Fig. 1D). However, it is curved, with its N-terminal end (residues 2 to 9) adopting a 310-helix secondary structure having three residues per helical turn, in contrast to an α-helix which has 3.6 residues per turn (Fig. 1E). The difference between the two secondary structure elements is due to a different pattern of hydrogen bonds. The 310-helix is viewed as having a lower stability than an α-helix, which may explain why 310 helices are relatively short (31). The 310-helical region forms the tightly packed core of the tad pilus (Fig. 1E). Interestingly the sequence of the 310-helix, which includes a helix-breaking glycine at position 7, is conserved in tad family members and was named the flp motif (17) (Fig. 1A and SI Appendix, Fig. S3). Of the three conserved polar residues in the N terminus (Thr2 and Glu5, conserved in all Type IV pilins and Tyr6, conserved in tad pilins), only Thr2 is involved in polar subunit:subunit interactions, forming a hydrogen bond with the N-terminal nitrogen of α1 of an adjacent subunit, as seen for T4aP (14). Additional polar interactions occur between conserved residues near the C-terminus of the tad subunit (SI Appendix, Fig. S4). Surprisingly, Glu5, which forms a salt bridge with the N-terminal nitrogen in published T4a pilus structures, is oriented toward the pilus surface and does interact with neighboring subunits. Glu5 is absolutely conserved in bacterial but not archaeal T4P. Instead, Tyr6 (of S0 subunit), which is conserved in all tad pili, is positioned below the Ala1 (N-terminal end) of S1 subunit maintaining the helical register (SI Appendix, Fig. S5). Furthermore, the flp motif hydrophobic residues (Val1, Ala3, Ile4, Gly7, Leu8, and Ile9) of S0 form extensive hydrophobic contacts with those of S−1, S1, and S2 and to residues 10 to 18 of S−3 stabilizing the core of the tad filament (SI Appendix, Fig. S5). These packing parameters plus the continuous helix for the C. crescentus tad pilus are reminiscent of the archaeal T4P, suggesting that tad pili are more closely related to the archaeal T4P rather than to the bacterial T4P.
Fig. 1.
Cryo-EM structure and sequence analysis of the C. crescentus tad pilus. (A) Multiple sequence alignment of the N-terminal residues for all bacterial T4P and endopili whose structure has been determined. The abbreviations are: Cc, C. crescentus tad (PDB 8U1K); Vc, V. cholerae TCP (PDB 8U74); Ko, Klebsiella oxytoca PulG endopilin (PDB 5WDA); Tt5, Thermus thermophilus PilA5 (PDB 6XXE); Tt4, Thermus thermophilus PilA4 (PDB 6XXD); Ec, Enterohemorrhagic Escherichia coli (PDB 6GV9); Nm, Neisseria meningitidis (PDB 5KUA); Ng, N. gonorrhoeae (PDB 5VXX); Gs, Geobacter sulfurreducens (PDB 6VK9); Mx, Myxococcus xanthus (PDB 8TJ2); Pa, Pseudomonas aeruginosa PAK (PDB 5VXY); and Ab, Acinetobacter baumanii (PDB 8TOB). The symbols are (*) for completely conserved, (:) for strong conservation, and (.) for weak conservation. Sequence conservation is not evident beyond residue 25 and the alignment is truncated there. The characteristic 8-residue flp motif in the tad pilin is shown by the gray box. The only completely conserved residues in these structures are Thr2 and Glu5 (yellow highlighting). Aside from Thr2 and Glu5, all residues showing any degree of conservation are hydrophobic. Type IV pilins in all bacterial T4P structures published to date have a Pro22 (red highlighting), found in a partially melted region of the N-terminal helix. Both the C. crescentus tad pilus and V. cholerae TCP have a threonine at this position, and their helices are continuous. (B) Cryo-EM image showing two tad pili. (C) 2D class average, with horizontal stripes with a periodicity of 4.9 Å indicated. (D) Cryo-EM density map and structure of the C. crescentus tad pilus. (E) Secondary structure of Tad pilin (Left) and top view of the tad pilus in ribbon representation showing the 7-start helical organization. The flp motif forming a 310 helix is colored green, participating in the core interactions of the filaments. (F) Surface lipophilicity of tad pilin showing the polar patch on the C-terminus forming the filament outer surface. Polar and hydrophobic surfaces are represented by green and gold, respectively.
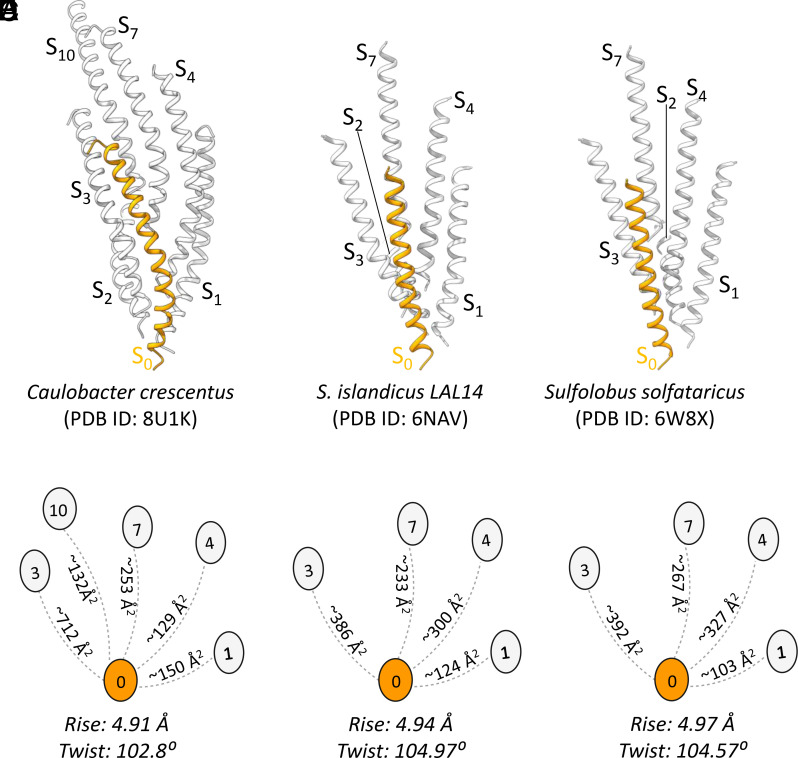
In all T4P structures published to date, the pilin globular C-terminal domain, or PilA-C in the case of the G. sulfurreducens T4P, forms the outer polar surface of the filament, burying hydrophobic residues of α1 that would otherwise be exposed (24, 32). The C. crescentus tad pilus structure departs radically from existing T4P structures as its major pilin, PilA, lacks this globular domain. Instead, PilA has a hydrophilic patch on the C-terminal end of the helix, which forms the outer surface of the filament (Fig. 1F and SI Appendix, Fig. S6). This atomic model of tad explains the very strong 4.9 Å periodicity seen in the power spectrum (SI Appendix, Fig. S1) and the 2D class average (Fig. 1C). PilA is predominantly α-helical, and the pitch of each turn in an α-helix is 5.4 Å. In the tad pilus, the PilA helices are tilted ~25° from the helical axis, thus the pitch of the α-helical turns projected onto the helical axis is 4.9 Å. This 4.9 Å is also the rise of each neighboring subunit, bringing all turns of the α-helices in the filament into register. Interestingly, the helical axial rise of subunits in the tad pilus (4.9 Å) falls in the range of the axial rise determined for existing archaeal T4P structures (4.9–5.5 Å, Fig. 2) (23). The interactions between subunits in the tad pilus are largely similar to that in archaeal T4aP as the tad subunit S0 interacts with subunits S1, S3, S4, and S7 as seen, for instance in the Sulfolobus islandicus and Sulfolobus solfataricus T4P structures (Fig. 2). Additionally, the packing within the tad pilus shows unique interactions between subunits S0 and S10, which are not observed in the archaeal T4P, nor in bacterial T4aP (SI Appendix, Fig. S7).
Fig. 2.
Comparison of the helical packing of (A and B) the C. crescentus tad pilus (PDB ID: 8U1K) with archaeal T4Ps (C and D) S. islandicus (PDB ID: 6NAV) and (E and F) S. solfataricus (PDB ID: 6W8X). The interacting subunits are connected by dashed lines with the interface area (in Å2) indicated between subunits. In all three cases, subunit S0 interacts with subunits S1, S3, S4, and S7 with similar interfacial areas revealing the similar packing of tad and archaeal T4P. Compared to the other archaeal T4P, subunit S0 has a larger interface with subunits S3 and S10 in tad, due to the curved helix of the tad pilin and the slight difference in the helical symmetry.
Cryo-EM of V. cholerae Toxin Coregulated Pilus (TCP).
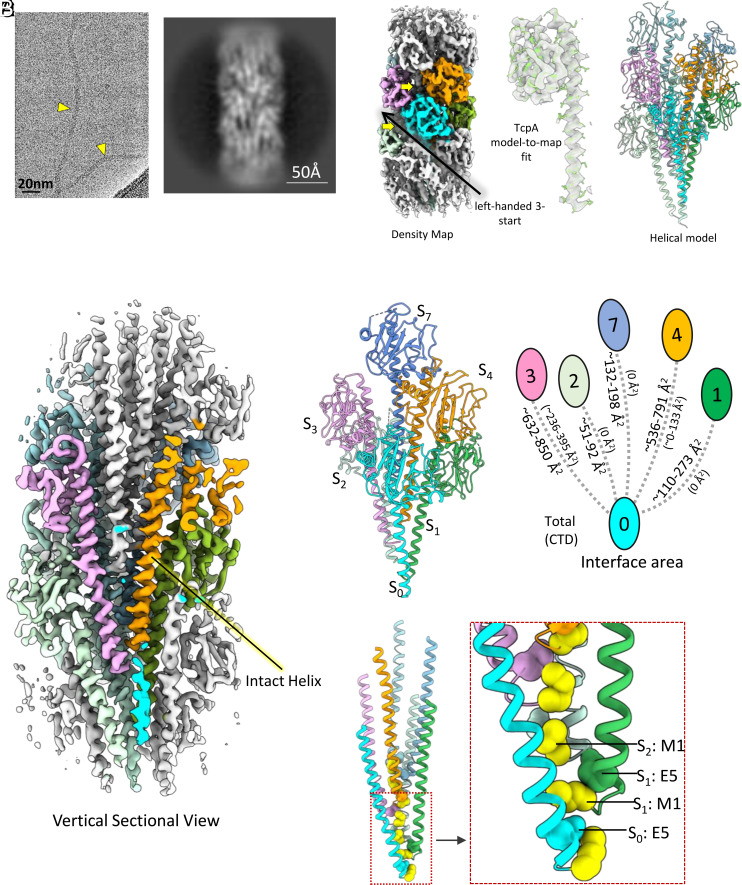
The T4b pili, like tad pili, lack the helix-breaking residue Pro22 (Fig. 1A). The only T4bP model available is from a low-resolution cryo-EM reconstruction of the V. cholerae TCP done in 2012 (33). The N-terminal α-helices are not resolved in this structure so it is not known whether these helices are melted as they are in the T4a pilins, perhaps due to multiple glycines in α1 N (residues 11, 14, and 19) or are continuous as seen for tad and archaeal T4P. With direct electron detectors providing much higher resolution than was possible in 2012, we obtained a new TCP cryoEM structure of considerably higher quality. Unlike typical T4aP and tad pili, which appear as rigid rods in vitrified ice, TCP appears to be extremely flexible under these conditions, and has a diameter of ~80 Å (Fig. 3A). Two-dimensional class averages of TCP segments show varying degrees of bending consistent with this flexibility (Fig. 3B and SI Appendix, Fig. S8). While other bacterial and archaeal T4P show clear features in their power spectra extending to high resolution [e.g., the very strong layer line from tad at 1/(4.9 Å)], the highest resolution feature in the averaged power spectrum from TCP segments is only a diffuse layer line at ~1/(21 Å) (SI Appendix, Fig. S1). Using helical symmetry with the axial rise/twist of 8.2 Å/97°, we obtained a TCP reconstruction at ~5.8 Å. This limited resolution, combined with the anfractuous morphology in the images and the lack of any high- or even medium-resolution information in the averaged power spectrum, indicates heterogeneity in the helical packing of the TCP filaments. To explore this further, we performed a reference-based classification of the TCP segments using models with different axial rise parameters and generated averaged power spectra for different classes. These spectra show differences in the position of the layer lines consistent with such structural heterogeneity (Movie S1). However, we were unable to find a homogeneous subset of segments from which we could generate a significantly higher-resolution helical reconstruction. We thus sought to use the asymmetric reconstruction approach as has been employed for other quasihelical filaments (34, 35).
Fig. 3.
Cryo-EM structure of the V. cholerae toxin coregulated pilus (TCP). (A) Cryo-EM image, (B) 2D class average of V. cholerae TCP, (C) cryo-EM density map, TcpA model-to-map fit, helical model, and (D) vertical sectional view of density map of V. cholerae TCP. (E) Interaction between TCP subunits. Subunit:subunit interactions occur primarily between α1 helices. The S0 subunit interacts mainly with subunit S1, S3, and S4 and to a lesser extent with S2. Total interface area between subunits calculated by PISA server are shown to the left of the connecting lines, and interaction interface area between the C-terminal domains (CTD) are shown in parentheses to the right. (F) Ribbon model of the core of the TCP filament with Met1 (yellow) and Glu5 shown in sphere representation using van der Waals radii. Glu5 of S0 interacts with the amino-terminus of Met1 of the S1 neutralizing two charges in a hydrophobic environment.
With this approach, using a subset of the overlapping TCP segments (~115,000 out of the total ~553,000), we obtained a ~3.8 Å map of a curved TCP segment with well resolved α-helices in the filament core (Fig. 3 C and D and SI Appendix, Fig S2). The subset selected for the reconstruction is the largest subset of segments that were classified and shows the highest degree of curvature in the TCP filament among all subsets, ~30 radians/micron. To put this in perspective, the curvature seen in the rigid supercoil of bacterial flagellar filaments is ~1.0–1.5 radians/micron for the “normal” waveform and ~3.0 radians/micron for the “curly” waveform (36). The radius of curvature in this TCP reconstruction is ~330 Å. Based on the raw images this appears to be an extreme rather than an average curvature. It is perhaps because this is an extreme that the set is more homogeneous and thus yielded the highest resolution reconstruction of all subsets, as subsets with lesser curvature may have greater fluctuations due to inclusion of segments with both greater and lesser curvature.
The quality of the map allowed building an atomic model of a nine subunit TCP filament segment revealing a key difference between this T4b pilus and the T4a pilus structures solved to date: unlike the melting observed for the T4aP, the TCP α1 is continuous, as seen for the tad and archaeal T4P (Fig. 3 C and D). Subunit S0 mainly interacts with the subunits S1, S3, S4, and S7, and to a lesser extent with S2, determining the packing of the TCP filament (Fig. 3E). The main interactions in the filament core are between the helix of subunit S0 and helices of subunits S1, S3, S4, and S7 (Fig. 3E). Glu5 is positioned to form a salt bridge with the Met1 amine of the next subunit in the segment (e.g., S0 to S1, Fig. 3F and SI Appendix, Fig S9), consistent with this acidic residue being critical for TCP biogenesis (33).
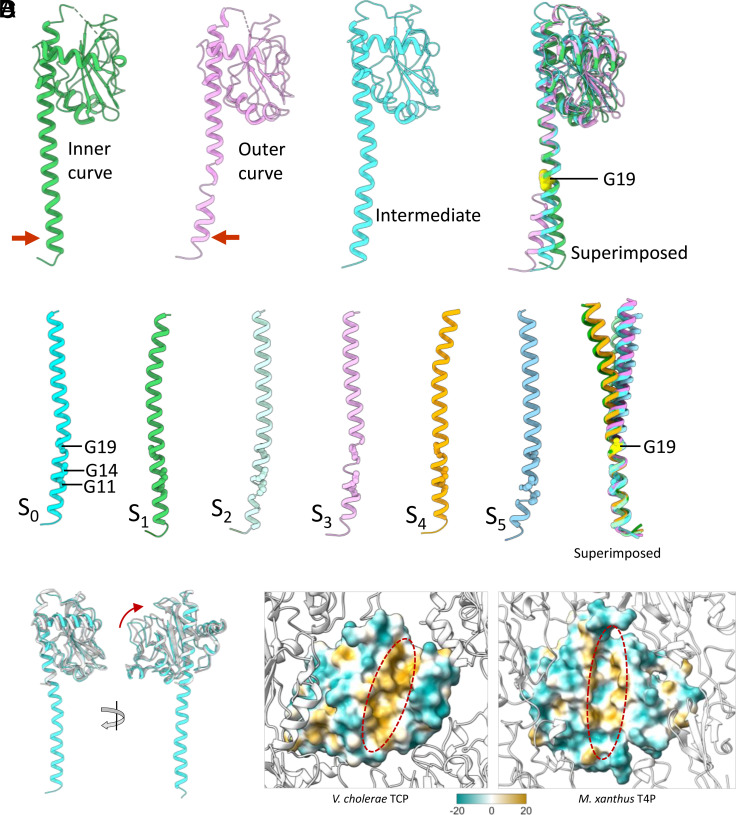
The TCP α1 appears flexible as its N-terminal end bends away from the core of filament in the subunits present at the inner curve and toward the core in subunits at the outer curve (Fig. 4A). Subunits located between the inner and outer curves show intermediate α1 bending. Flexibility of the helix apparently arises from the presence of glycines at positions 11, 14, and 19, which induce local deformations in the α-helix (Fig. 4 A and B). Superposition of S0 to S5 C-terminal domains (Fig. 4A) on α1 N-terminal ends (Fig. 4B) reveals the primary “hinge” point to be Gly19. While Gly14 is conserved among bacterial T4a and T4bP, Gly11 and Gly19 are specific to the Type IVb pilins of V. cholerae TCP (Fig. 1A), enterotoxigenic E. coli (CFA/III and longus pili), and Citrobacter rodentium (14), with enteropathogenic E. coli BfpA having only Gly19. This suggests that the TCP and possibly other T4bP achieve some flexibility via helix-deforming glycines rather than by proline-induced melting of α1. An interesting question is whether the deformations seen between the inner and outer curves occur mainly within a subunit (a change in secondary and/or tertiary structure) or between subunits (a change in quaternary structure). The axial rise between subunits on the inner curve is ~7.8 Å, while on the outer curve, it is ~9.2 Å. The contour length of the helix for the inner subunit is ~72 Å, while it is ~75 Å for the outer subunit. The change in quaternary structure (9.2/7.8 = 1.18) is much larger than the change in secondary/tertiary structure (75/72 = 1.04). Thus, changes in the subunit (compression/extension of the helix) play a relatively small role in flexibility. Most importantly, the resistance to bending in TCP will largely be determined by the outer domains and not the inner helices, and we can now show that these are the major source of the flexibility of the filaments.
Fig. 4.
Structural variability of α1 helix of TCP subunits. (A) Ribbon models of the TCP subunits present at the inner curve, outer curve, the intermediate position, and their superimposed view showing the variation in the N-terminal half of the exposed part of α1, with the hinge point located at Gly19. (B) The α1 helices of S0, S1, S2, S3, S4, and S5 and their superimposed view showing different curvature in them stemming from Gly19. (C) Superimposed view of TcpA cryo-EM (cyan) and crystal structure (white) (PDB id: 1OQV) showing the minor swinging movement of C-terminal domain relative to α1 helix. (D) The lipophilicity of the α1 helix binding groove on the C-terminal domain in V. cholerae TCP and M. xanthus T4a pilus. The binding groove of the V. cholerae TCP is highly hydrophobic in comparison with M. xanthus T4a. Polar and hydrophobic surfaces are represented by green and gold, respectively.
The C-terminal domain (CTD) of the TCP major pilin, TcpA, is considerably larger than that of almost all published T4aP filament structures, and thus might be expected to interact tightly with neighboring outer domains, as seen for the unusually large (208 residue) pilin subunit of the Myxococcus xanthus T4a pilus (37). But instead, the interactions among the TcpA globular domains are minimal, occurring mainly among the subunits along the left-handed 3-start helices (i.e., between S0 and S3 subunits) and to a lesser extent among the subunits related by right-handed 4-start helices (i.e., between subunits S0 and S4, Fig. 3 C and E). PISA analysis indicates that 3-start and 4-start CTD interfaces vary between the inner and outer curved region of the filament, in the range of 236 to 395 Å2 and 0 to 133 Å2, respectively (Fig. 3E). In addition, 60 to 70% of the 3-start and 4-start interfacial area is hydrophobic in nature. The globular domains do not contact one another along the right-handed 1-start helix (i.e., S0 and S1), leaving substantial grooves between 3-start helices (Fig. 3 C and E), and exposing a segment of α1. This exposure is supported by deuterium exchange analysis (38). These grooves can be readily compressed (on the inside of a curve) or expanded (on the outside of a curve). This surface malleability, together with the glycine-mediated suppleness of α1, provides the basis for the flexibility observed for these filaments in vitreous ice (Fig. 3A).
We do not see ordered density for the linker connecting the α1 and CTD in six out of nine subunits, suggesting that it is flexible. Furthermore, the orientation of the CTD relative to α1 exhibits significant variability among subunits modeled in our map and in a previously reported crystal structure (PDB is: 1OQV) (Fig. 4 A and C). Almost all of the variability among subunits in the map can be traced to their different environments, i.e., on the inside versus the outside of the curvature. Significantly, the α1 helix-binding groove on the CTD is largely hydrophobic (Fig. 4D) implying that the interactions between α1 and CTD are nonspecific and, consequently, permit minor sliding or swinging motions of the CTD in relation to α1. In contrast, this groove in the M. xanthus T4a pilus, having a CTD of comparable size, possesses substantial polar patches (37) providing specific interactions between α1 and the CTD (Fig. 4D). Subunit packing observed in this 3.8 Å TCP reconstruction is similar to the model from the ~20 Å resolution cryo-EM structure of 2012 (33). As with the tad pilus structure, we did not see any additional density that might be due to post-translational modifications.
Comparisons with Existing Bacterial and Archaeal T4P Structures.
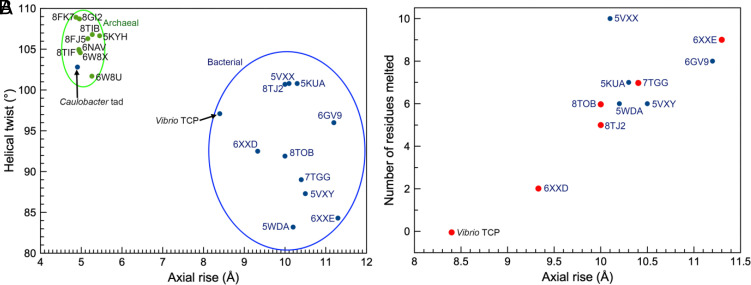
Although both the tad pilus and TCP lack a Pro22 residue and have a continuous N-terminal helix in the polymerized filament, the helical parameters (rise and twist) place tad in the cluster of helical symmetries reported for archaeal T4P, while TCP groups with bacterial T4aP and endopili (Fig. 5A). The short rise of the tad pilus, 4.9 Å, means greater overlap among the subunits, giving a diameter of ~50 Å, comparable to that of the P. aeruginosa T4aP (52 Å), which possesses a globular domain but has a rise of 10.3 Å. The packing of helices is also very similar between the tad and archaeal pili and distinct from TCP and other bacterial T4P (Fig. 2). However, the 310-helix appears to be a unique feature of the tad pili.
Fig. 5.
Comparison of symmetry parameters from archaeal and bacterial T4P structures. (A) The axial rise and the helical twist are plotted for all published atomic structures of archaeal (green dots) and bacterial (blue dots) T4P. The C. crescentus tad pilus has a twist (~103°) and rise (~5 Å) that clusters with the archaeal T4P. In contrast, all other bacterial T4P whose structures have been determined have a significantly larger rise (~10 Å). V. cholerae TCP falls into this loose cluster, with the smallest rise (~8.4 Å). (B) The degree of melting of the N-terminal helix in bacterial T4Ps is plotted versus the axial rise per subunit (there is no melting for the archaeal T4Ps or the C. crescentus tad pilus). The points in red are from structures determined at better than 4.0 Å resolution where the models are more reliable and thus the number of melted residues is more accurate. These points imply a linear relationship between rise and number of melted residues.
We proposed previously that partial melting of α1 was necessary for the packing of these helices in the filament core (14, 32). Yet, we show here that V. cholerae TcpA form filaments with similar symmetry and packing to the T4aP but with a continuous α1. To understand how these very different α1 conformations can fit into a helical filament, we plotted the axial rise versus the number of residues melted in the N-terminal helices of existing bacterial T4P structures (Fig. 5B). The axial rise can be determined to a very high degree of precision even at modest resolution. Our analysis suggests that the axial rise in bacterial T4P is linearly dependent on the number of residues melted. This relationship can be understood if one considers that the helical rise is determined both by the interactions among the subunits, which are driven largely by α1, but also by the packing of the globular domains on the filament surface. Melting may be a mechanism for α1 to stretch to accommodate differences in globular domain packing arrangements while maintaining the interactions among the conserved residues in the most N-terminal portion of α1, including the Glu5-to-amino terminus salt bridge and the Thr/Ser-to-amino terminus hydrogen bond. Thus, filaments with a larger axial rise would require more stretching to allow subunit packing. It is important to note that the TCP α1 s have the capacity to deform in the region of the three glycines, which corresponds to the melted region of T4aP. Thus, while α1 is melted in all subunits in the T4aP, relatively small deformations appear to be dynamic in TCP, occurring in response to bending. The TCP α1 s actually provide relatively little resistance to bending, as can be seen by the fact that a “canonical” α-helix (with no helix-breaking residues) has a persistence length estimated to only be ~150 Å (39) while an anomalously rigid myosin α-helix has a persistence length estimated to be ~224 Å (40). In contrast, the radius of curvature in the very curved TCP reconstruction is ~330 Å. By definition, the persistence length is defined so that a polymer of length L when bent through a radius of curvature L will have a stored bending energy of kT. Thus, if one ignores the outer domains an energy significantly less than kT would be needed to induce a bend with a radius of curvature of 330 Å in an α-helix, even in the absence of the glycines. As discussed above, the glycine-centered deformations have minimal impact on the length of α1, with the gaps between the globular domains providing most of the basis for filament flexibility. In contrast, the proline-centered melting in the T4aP significantly increases the length of α1. For example, in the T. thermophilus PilA5 pilus (PDB 6XXE), the length of 11 residues spanning the melted region is 27 Å, while this length would be 15 Å in an α-helical coil. This extension facilitates subunit packing and provides filament elasticity.
Discussion
The packing of the helices in T4P filaments, whether bacterial or archaeal, is driven primarily by hydrophobic interactions. The largely hydrophobic N-terminal half of α1 s, which are integral membrane domains before filament polymerization, are commonly surrounded by globular soluble C-terminal domains in both bacteria and archaea, burying hydrophobic residues that would be otherwise exposed. Our C. crescentus tad pilus reconstruction shows that a pilus can form without these globular domains, with polar residues in the C-terminal half of the tad helix forming the surface of the filament (Fig. 1F and SI Appendix, Fig. S6). The tad pilus from another Caulobacter species, C. crescentus bNY30a, has 92% sequence identity with our tad pilus, and is also devoid of the globular domains (PDB 8U2B). The similarity of the subunit packing in the bacterial tad pilus to all existing archaeal T4P structures (Fig. 5A) strongly supports the proposal that the tad genes came from archaea to bacteria via horizontal transfer (18). Additionally, its unique structure supports its classification as a distinct pilus type, Type IVc, rather than as a subset of the T4bP.
Since the hydrophobic effect is, by definition, nonspecific, we expect that some of the specificity in the interactions that hold T4P together comes from the globular outer domains for non-tad pili as well as the interactions of Thr2 and Glu5 with neighboring chains. For the bacterial T4aP, the helices have many polar residues that make specific interactions in the core of the filaments (SI Appendix, Fig. S7). In the case of tad, there are interactions between the polar residues near both the N- and C-terminus of each subunit that generate some of this specificity (SI Appendix, Fig. S4). Beside the specific interaction of Thr2 in S0 with Ala1 at the N terminus of S1, the interaction of Asn27 in S3 with the backbone of Gly36 in S0, and the interaction of Lys34 in S3 with the backbone of Ala43 near the C-terminus of S0 might provide specific interactions to keep the tad pilins in helical register (SI Appendix, Fig. S4). The lack of specificity for the largely hydrophobic interactions can be seen in the structures of two different forms of a pilus formed by the same archaeal pilin (41), where different packing arrangements of the outer domains cause the inner helical domains to change their twist and axial rise from 106.8°, 5.3 Å to 104.8°, 5.0 Å. While this shift may sound small, after 20 subunits it would be a difference of 40° and 6 Å.
Though both tad/flp pilins and G. sulfurreducens PilA-N are short, and lack the C-terminal globular domains, the C. crescentus tad pilus structure establishes these filaments as being a class of their own, forming a pilus composed only of helices, in contrast to G. sulfurreducens T4P, which uses a second polypeptide, PilA-C, as the C-terminal domain. While the presence of two genes in G. sulfurreducens, one for the N-terminal helical domain and one for the C-terminal globular domain, may be interpreted as the result of a gene fission, it is also possible that this represents a more ancestral form of this protein, which eventually evolved by a gene fusion separately for archaeal and T4a pilins. The existence of a Gram-positive T4P containing an all helical C-terminal domain (42) and the lack of any homology between archaeal T4P C-terminal domains and those in bacteria support the speculation that ancestral T4P proteins may have existed as two separate genes. From this point of view, the tad proteins may reflect a more ancestral relative of all T4P before such a gene fusion occurred, or alternatively, reflect a subsequent loss of the second gene in evolution.
The surprising diversity in T4P filaments revealed here shows that a relatively conserved building block, type IV pilin, can undergo small changes in sequence (such as a Pro22 to a Thr22) that cause large changes in the quaternary structure. This has been shown in vitro using a peptide model, where the semi-conservative substitution of one residue (arginine to lysine) in a 29 residue peptide led to a filament with twice the diameter and two peptides in the asymmetric unit, rather than one (43). These evolutionary changes may reflect mechanical adaptations to specific biological niches. C. crescentus is an aqueous bacterium that uses tad pili to adhere to surfaces, but the holdfast, which is stimulated by tad pilus binding, provides secure attachment (26, 44, 45). The N. gonorrhoeae T4a pili must withstand high flow forces in the urogenital tract, facilitated by considerable elasticity in their T4aP (28) due to the extensibility of the melted portion of α1 N. However, T4bP and tad pili may not require such elasticity. V. cholerae colonize the base of villi in the small intestine (46) where they form microcolonies and are thus somewhat protected from flow forces. Though TCP lacks Pro22 and the melted portion of α1, which would be expected to decrease the axial compliance (the extent to which the filament can stretch under an applied force), they demonstrate a large degree of bending flexibility mainly due to the grooves between globular domains on their surface. Additionally, our data indicate considerable heterogeneity in the axial rise per subunit for TCP (Movie S1), possibly arising from the ability of subunits to slide past each other. In summary, a relatively simple building block, type IV pilin, can create a large number of T4P with very different mechanical properties arising from adaptations to very different environments.
Methods
Caulobacter Tad Pilus Preparation.
Eight liters of C. crescentus strain bNY304 cells were grown to an OD600 of 0.6. Cells were pelleted at 10,000×g for 15 min. Cells were resuspended in PBS buffer (Corning) supplemented with protease inhibitor cocktail (Millipore Sigma). Specifically, the cell pellet from 500 mL of culture was resuspended in 2 mL supplemented PBS buffer until clumps were no longer visible. This suspension was transferred to an additional cell pellet from 500 mL of culture, the cell pellet was resuspended as described previously, and the process was repeated for a third cell pellet from 500 mL of culture before transferring the resuspended pellet (from 1.5 L) to a cold 50 mL Falcon tube. Cells were passed through a 25-gauge needle 5 times to shear pili. These steps were repeated for all 8 L of cells and the sheared cell resuspensions were combined. Sheared cells were pelleted at 17,000×g for 10 min and the supernatant (containing pili) was collected. To precipitate pili from the supernatant solution, 100% saturated ammonium sulfate was added dropwise to the supernatant solution to a final concentration of 30% saturated ammonium sulfate. The solution was allowed to mix at room temperature for 15 min under gentle stirring followed by centrifugation at 16,000×g for 10 min to pellet the pili precipitate. The pellet was resuspended in 10 mL of PBS buffer, added to a 20,000 MWCO dialysis cassette (Thermo Fisher Scientific), and dialyzed overnight against 2 L of PBS under gentle stirring at 4 °C. The dialyzed product was recovered from the cassette and centrifuged at 125,000×g for 4.5 h. The pili pellet was resuspended in 1 mL of PBS by shaking overnight at 4 °C. To further remove contaminants, the resuspended pellet was centrifuged at 16,000×g for 10 min, the supernatant was collected and centrifuged at 150,000×g for 1 h. The final pili pellet was resuspended in 50 μL of PBS under gentle shaking overnight at 4 °C.
V. cholerae Toxin Coregulated Pilus Preparation.
V. cholerae strain RT4225 (47) was grown in 200 mL lysogeny broth (1% tryptone, 0.5% yeast extract and 86 mM NaCl), pH 6.5 with 0.04 mM IPTG for 16 h at 30 °C shaking at 225 rpm. Strain RT4225 has a mutation in the tcpA gene encoding a His181Ala substitution that disrupts pilus:pilus interactions, producing exceptionally long pili that are released from the cells and can be isolated from the culture supernatant (33). These pili appear as individual filaments when imaged in a frozen-hydrated state, making them amenable to reconstruction. Overexpression of the tcp transcriptional activator ToxT on plasmid pMT561 increases the pilus yield. Overnight cultures were centrifuged to remove cells, and TCP were precipitated from the supernatant using 30% ammonium sulfate, resuspended in PBS with 10 mM EDTA, and dialyzed exhaustively to remove the ammonium sulfate. Purification steps were performed on ice or at 4 °C.
Negative-staining Transmission Electron Microscopy (NS-TEM).
A 3-μL pilus sample was applied to glow-discharged carbon film grids, excessive sample was blotted away, and stained with 1 to 2% uranyl acetate. The stained grids were imaged on a Tecnai T12 electron microscope. Negatively stained electron micrographs were used to analyze the quality of pilus preparations.
Grid Preparation for Cryo-Electron Microscopy.
A 3-microliter pilus sample was applied to glow discharged, R2/1 200 mesh copper Quantifoil and lacey carbon grids for C. crescentus tad and V. cholerae TCP, respectively. Grids were plunge-frozen in liquid ethane using a Vitrobot Mark IV (Thermo Fisher Scientific, Hillsboro, Oregon).
Cryo-Electron Microscopy and Analysis.
Data collection and image preprocessing.
Cryo-EM data collection was performed on a Titan Krios (Thermo Fisher Scientific) operated at 300 keV, and equipped with a Gatan K3 direct electron detector (Gatan, Inc., Eindhoven, Netherlands). Total exposure per movie was ~50 e Å–2 with a pixel size of ~0.83 Å. Cryo-EM raw movies were motion corrected and CTF-estimated by “patch motion correction” and “patch CTF estimation” jobs in cryoSPARC (48).
Reconstructions of Caulobacter tad pilus volume map.
The 3D reconstruction was achieved by helical processing in cryoSPARC (48). Tad pili segments were manually boxed from several micrographs to generate 2D class averages for automatic template-based particle picking by Template picker. Bad picks were removed by iterative 2D-classification and selection of 2D-classes. The averaged power spectrum of selected tad segments was generated for the indexing of the helical symmetry parameters. Different symmetry parameters were tested, and the final reconstruction was achieved by the “helix refine” job. The map was further improved by iterative cycle of refinements with different options like “local CTF refinement,” and “nonuniform refinement.”
Reconstructions of Vibrio TCP volume map.
The stack of good segments, averaged power spectrum, and initial helical reconstruction for V. cholerae TCP were obtained through the same procedures used for C. crescentus tad pili. Due to the anfractuous morphology of TCP, the helical reconstruction failed to give a higher resolution map than 5.8 Å, showing separated density rods in the core of the filament corresponding to the α1 helix, but no helical pitch. We thus used the asymmetric reconstruction approach. The stack of ~553,000 segments was subjected to the “ab-initio reconstruction” to generate the initial model. The initial model was refined by “heterogenous refinement” with five classes. The class of ~115,000 segments and corresponding map having homogenous curvature was further subjected to “local refinement” giving the refined map. The postprocessing of the locally refined map by EMReady (49) yielded a final map of ~3.8 Å resolution showing the hand of the core α1 helices and bulky side chains. While the density of the CTD for several subunits is well resolved, it is poorly resolved for others. The data processing workflow is summarized in SI Appendix, Fig. S10.
Cryo-EM data collection and processing statistics are provided in SI Appendix, Table S2. The averaged power spectra and Fourier shell correlation curves for both pili are provided in SI Appendix, Figs. S1 and S2, respectively.
Model Building.
The C. crescentus tad pilin structure (Uniport Id: WP_010920785) predicted by AlphaFold (50), and crystal structure of V. cholerae TCP (PDB id: 1OQV) were used as initial models for rigid-body-fitting in the respective maps. The models underwent iterative cycles of model building in COOT (51) and automatic refinement by Phenix Real Space Refinement (52). Models were built for 10 subunits in the tad filament and nine subunits in the TCP filament. The TCP filament model contains three subunits where the entire chain could be built (residues 1 to 198) except for the C-terminal Ser199 where no density is present. In six subunits, the linker (residues 53 to 60) is absent due to poor density. The model building statistics are provided in SI Appendix, Table S2.
Structural and Sequence Analysis.
Sequence similarity analysis was done with NCBI-BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Structural interface analysis was performed by PISA (53). All molecular figures were prepared with ChimeraX (54).
Supplementary Material
Appendix 01 (PDF)
Animation of the averaged power spectra of segments from two different classes. The change in the spacing of the strongest layer line (n=-3) corresponds to a change in the pitch of the 3-start helices from ~116 Å to ~127 Å. A reference-based classification of the TCP segments using models with different axial rise values was employed to obtain these classes.
Acknowledgments
We thank Mart Krupovic for valuable discussions. This work was supported by NIH GM122510 (to E.H.E.), GM104540, and U24 GM139168 (to E.R.W.), by NSERC RGPIN-2023-03556 (to L.C.), and by the Department of Biochemistry at the University of Wisconsin, Madison. This work was also supported in part by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research DE-SC0018409 (to E.R.W.). J.C.S. was supported in part by the Biotechnology Training Program at the University of Wisconsin, Madison, T32 GM135066. We are grateful for the use of facilities and instrumentation at the Cryo-EM Research Center in the Department of Biochemistry at the University of Wisconsin, Madison, and at the University of Virginia Molecular Electron Microscopy Core facility.
Author contributions
E.R.W., L.C., and E.H.E. designed research; R.R.S., J.C.S., J.K.B., S.K., E.R.W., and L.C. performed research; R.R.S. and E.H.E. analyzed data; and R.R.S., L.C., and E.H.E. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Reviewers: V.A.G., University of Exeter; and V.P., Laboratoire de Chimie Bacterienne.
Data, Materials, and Software Availability
Atomic models have been deposited in the Protein Data Bank: 8U1K (55) and 8U74 (56). Cryo-EM maps have been deposited in the Electron Microscopy Data Bank: EMD-41815 (57), EMD-42279 (58) and EMD-41968 (59).
Supporting Information
References
- 1.Craig L., Forest K. T., Maier B., Type IV pili: Dynamics, biophysics and functional consequences. Nat. Rev. Microbiol. 17, 429–440 (2019). [DOI] [PubMed] [Google Scholar]
- 2.Berry J. L., Pelicic V., Exceptionally widespread nanomachines composed of type IV pilins: The prokaryotic Swiss Army knives. FEMS Microbiol. Rev. 39, 134–154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pelicic V., Mechanism of assembly of type 4 filaments: Everything you always wanted to know (but were afraid to ask). Microbiology (Reading) 169, 001311 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pelicic V., Type IV pili: E pluribus unum? Mol. Microbiol. 68, 827–837 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Makarova K. S., Koonin E. V., Albers S. V., Diversity and evolution of type IV pili systems in Archaea. Front. Microbiol. 7, 667 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pohlschroder M., Ghosh A., Tripepi M., Albers S. V., Archaeal type IV pilus-like structures–evolutionarily conserved prokaryotic surface organelles. Curr. Opin. Microbiol. 14, 357–363 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Giron J. A., Gomez-Duarte O. G., Jarvis K. G., Kaper J. B., Longus pilus of enterotoxigenic Escherichia coli and its relatedness to other type-4 pili—A minireview. Gene 192, 39–43 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Craig L., Li J., Type IV pili: Paradoxes in form and function. Curr. Opin. Struct. Biol. 18, 267–277 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCallum M., Burrows L. L., Howell P. L., The dynamic structures of the type IV pilus. Microbiol. Spectr. 7, PSIB-0006-2018 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Parge H. E., et al. , Structure of the fibre-forming protein pilin at 2.6 A resolution. Nature 378, 32–38 (1995). [DOI] [PubMed] [Google Scholar]
- 11.Craig L., et al. , Type IV pilin structure and assembly: X-ray and EM analyses of Vibrio cholerae toxin-coregulated pilus and Pseudomonas aeruginosa PAK pilin. Mol. Cell 11, 1139–1150 (2003). [DOI] [PubMed] [Google Scholar]
- 12.Craig L., et al. , Type IV pilus structure by cryo-electron microscopy and crystallography: Implications for pilus assembly and functions. Mol. Cell 23, 651–662 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Hartung S., et al. , Ultrahigh resolution and full-length pilin structures with insights for filament assembly, pathogenic functions, and vaccine potential. J. Biol. Chem. 286, 44254–44265 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolappan S., et al. , Structure of the Neisseria meningitidis Type IV pilus. Nat. Commun. 7, 13015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang F., et al. , Cryoelectron microscopy reconstructions of the Pseudomonas aeruginosa and Neisseria gonorrhoeae type IV pili at sub-nanometer resolution. Structure 25, 1423–1435.e1424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng D., et al. , The Vibrio cholerae minor pilin TcpB initiates assembly and retraction of the toxin-coregulated pilus. PLoS Pathog. 12, e1006109 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kachlany S. C., et al. , flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 40, 542–554 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Denise R., Abby S. S., Rocha E. P. C., Diversification of the type IV filament superfamily into machines for adhesion, protein secretion, DNA uptake, and motility. PLoS Biol. 17, e3000390 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pu M., Rowe-Magnus D. A., A Tad pilus promotes the establishment and resistance of Vibrio vulnificus biofilms to mechanical clearance. npj Biofilms Microbiomes 4, 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomich M., Planet P. J., Figurski D. H., The tad locus: Postcards from the widespread colonization island. Nat. Rev. Microbiol. 5, 363–375 (2007). [DOI] [PubMed] [Google Scholar]
- 21.Ellison C. K., et al. , Obstruction of pilus retraction stimulates bacterial surface sensing. Science 358, 535–538 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Del Medico L., Cerletti D., Schachle P., Christen M., Christen B., The type IV pilin PilA couples surface attachment and cell-cycle initiation in Caulobacter crescentus. Proc. Natl. Acad. Sci. U.S.A. 117, 9546–9553 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kreutzberger M. A. B., et al. , The evolution of archaeal flagellar filaments. Proc. Natl. Acad. Sci. U.S.A. 120, e2304256120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu Y., et al. , Structure of Geobacter pili reveals secretory rather than nanowire behaviour. Nature 597, 430–434 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang F., et al. , Cryo-EM structure of an extracellular Geobacter OmcE cytochrome filament reveals tetrahaem packing. Nat. Microbiol. 7, 1291–1300 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Curtis P. D., Brun Y. V., Getting in the loop: Regulation of development in Caulobacter crescentus. Microbiol. Mol. Biol. Rev. MMBR 74, 13–41 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mignolet J., et al. , AFM unravels the unique adhesion properties of the Caulobacter Type IVc pilus nanomachine. Nano Lett. 21, 3075–3082 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Biais N., Higashi D. L., Brujic J., So M., Sheetz M. P., Force-dependent polymorphism in type IV pili reveals hidden epitopes. Proc. Natl. Acad. Sci. U.S.A. 107, 11358–11363 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skerker J. M., Shapiro L., Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J 19, 3223–3234 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sachse C., Fändrich M., Grigorieff N., Paired β-sheet structure of an Aβ (1–40) amyloid fibril revealed by electron microscopy. Proc. Natl. Acad. Sci. U.S.A. 105, 7462–7466 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calladine C. R., Construction of bacterial flagella. Nature 255, 121–124 (1975). [DOI] [PubMed] [Google Scholar]
- 32.Wang F., Craig L., Liu X., Rensing C., Egelman E. H., Microbial nanowires: Type IV pili or cytochrome filaments? Trends Microbiol. 31, 384–392 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li J., Egelman E. H., Craig L., Structure of the Vibrio cholerae Type IVb Pilus and stability comparison with the Neisseria gonorrhoeae type IVa pilus. J. Mol. Biol. 418, 47–64 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anger R., et al. , Structure of a heteropolymeric type 4 pilus from a monoderm bacterium. Nat. Commun. 14, 7143 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kreutzberger M. A. B., et al. , Convergent evolution in the supercoiling of prokaryotic flagellar filaments. Cell 185, 3487–3500.e3414 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calladine C. R., Change of waveform in bacterial flagella: The role of mechanics at the molecular level. J. Mol. Biol. 118, 457–479 (1978). [Google Scholar]
- 37.Treuner-Lange A., et al., Large pilin subunits provide distinct structural and mechanical properties for the Myxococcus xanthus type IV pilus. bioRxiv 2023.07.22.550172 [Preprint] (2023). 10.1101/2023.07.22.550172 (Accessed 22 July 2023). [DOI] [PMC free article] [PubMed]
- 38.Li J., et al. , Vibrio cholerae toxin-coregulated pilus structure analyzed by hydrogen/deuterium exchange mass spectrometry. Structure 16, 137–148 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sivaramakrishnan S., et al. , Combining single-molecule optical trapping and small-angle X-ray scattering measurements to compute the persistence length of a protein ER/K α-helix. Biophys. J. 97, 2993–2999 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barnes C. A., et al. , Remarkable rigidity of the single α-helical domain of myosin-VI as revealed by NMR spectroscopy. J. Am. Chem. Soc. 141, 9004–9017 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liu J., et al. , Two dramatically distinct archaeal type IV pili structures formed by the same pilin. bioRxiv 2023.08.07.552285 [Preprint] (2023). 10.1101/2023.08.07.552285 (Accessed 7 August 2023). [DOI]
- 42.Sheppard D., et al. , The major subunit of widespread competence pili exhibits a novel and conserved type IV pilin fold. J. Biol. Chem. 295, 6594–6604 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Egelman E. H., et al. , Structural plasticity of helical nanotubes based on coiled-coil assemblies. Structure 23, 280–289 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Govers S. K., Jacobs-Wagner C., Caulobacter crescentus: Model system extraordinaire. Curr. Biol. 30, R1151–R1158 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Bodenmiller D., Toh E., Brun Y. V., Development of surface adhesion in Caulobacter crescentus. J. Bacteriol. 186, 1438–1447 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Millet Y. A., et al. , Insights into Vibrio cholerae intestinal colonization from monitoring fluorescently labeled bacteria. PLoS Pathog. 10, e1004405 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kirn T. J., Lafferty M. J., Sandoe C. M., Taylor R. K., Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol. Microbiol. 35, 896–910 (2000). [DOI] [PubMed] [Google Scholar]
- 48.Punjani A., Rubinstein J. L., Fleet D. J., Brubaker M. A., cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 49.He J., Li T., Huang S.-Y., Improvement of cryo-EM maps by simultaneous local and non-local deep learning. Nat. Commun. 14, 3217 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mirdita M., et al., ColabFold: Making protein folding accessible to all. Nat. Methods 19, 679–682 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Cryst. D 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Afonine P. V., et al. , Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Cryst. Sect. D, Struct. Biol. 74, 531–544 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krissinel E., Protein interfaces, surfaces and assemblies service PISA at European Bioinformatics Institute. J. Mol. Biol. 372, 774 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Goddard T. D., et al. , UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 27, 14–25 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sonani R. R., Sanchez J. C., Baumgardt J. K., Wright E. R., Egelman E. H., 8U1K, Cryo-EM of Caulobacter crescentus tad pilus. Protein Data Bank. https://www.rcsb.org/structure/8U1K. Deposited 1 September 2023.
- 56.Sonani R. R., Kundra S., Craig L., Egelman E. H., 8U74, Cryo-EM of Vibrio cholerae toxin co-regulated pilus (TCP). Protein Data Bank. https://www.rcsb.org/structure/8U74. Deposited 9 October 2023.
- 57.Sonani R. R., Sanchez J. C., Baumgardt J. K., Wright E. R., Egelman E. H., 41815, Cryo-EM of Caulobacter crescentus tad pilus. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-41815. Deposited 1 September 2023.
- 58.Sonani R. R., Kundra S., Craig L., Egelman E. H., 42279, Cryo-EM of Vibrio cholerae toxin co-regulated pilus - asymmetric reconstruction. Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-42279. Deposited 9 October 2023.
- 59.Sonani R. R., Craig L., Egelman E. H., EMD-41968, Cryo-EM of Vibrio cholerae toxin co-regulated pilus (TCP). Electron Microscopy Data Bank. https://www.ebi.ac.uk/emdb/EMD-41968. Deposited 14 September 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Animation of the averaged power spectra of segments from two different classes. The change in the spacing of the strongest layer line (n=-3) corresponds to a change in the pitch of the 3-start helices from ~116 Å to ~127 Å. A reference-based classification of the TCP segments using models with different axial rise values was employed to obtain these classes.
Data Availability Statement
Atomic models have been deposited in the Protein Data Bank: 8U1K (55) and 8U74 (56). Cryo-EM maps have been deposited in the Electron Microscopy Data Bank: EMD-41815 (57), EMD-42279 (58) and EMD-41968 (59).