Significance
The tumor suppressor protein p53 is a transcription factor that orchestrates the expression of numerous genes involved in various pathways critical for cellular homeostasis. Accumulation of p53, induced by cellular stress, results in gene activation that depends upon the stress severity. Early in the response to stress, p53 causes cell-cycle arrest followed by attempts to repair the damage, and when this fails, p53 directs cells to apoptosis. The mechanistic basis for this p53-dependent selective gene expression is unknown. We demonstrate that flexibility of p53 response elements contributes to the first steps of selective p53-dependent gene expression by facilitating p53/REs interactions belonging to early-responsive genes. Thus, p53 REs’ flexibility plays a key role in cell-fate decisions in the p53 circuitry.
Keywords: p53, protein/DNA interactions, DNA twist flexibility, gene-expression regulation
Abstract
The tumor suppressor protein p53 accumulates in response to cellular stress and consequently orchestrates the expression of multiple genes in a p53-level and time-dependent manner to overcome stress consequences, for which a molecular mechanism is currently unknown. Previously, we reported that DNA torsional flexibility distinguishes among p53 response elements (REs) and that transactivation at basal p53 levels is correlated with p53 REs flexibility. Here, we calculated the flexibility of ~200 p53 REs. By connecting functional outcomes of p53-target genes’ activation to the calculated flexibility of their REs, we show that genes known to belong to pathways that are activated rapidly upon stress contain REs that are significantly more flexible relative to REs of genes known to be involved in pathways that are activated later in the response to stress. The global structural properties of several p53 REs belonging to different pathways were experimentally validated. Additionally, reporter-gene expression driven by flexible p53 REs occurred at lower p53 levels and with faster rates than expression from rigid REs. Furthermore, analysis of published endogenous mRNA levels of p53-target genes as a function of REs’ flexibility showed that early versus late genes differ significantly in their flexibility properties of their REs and that highly flexible p53 REs enable high-activation level exclusively to early-response genes. Overall, we demonstrate that DNA flexibility of p53 REs contributes significantly to functional selectivity in the p53 system by facilitating the initial steps of p53-dependent target-genes expression, thereby contributing to survival versus death decisions in the p53 system.
The tumor suppressor protein p53 is activated by various cellular-stress signals through its release from the complex with the HDM2 protein, which leads to its stabilization and thereby accumulation (1). Activated p53 functions mainly as a transcription factor, regulating the expression of numerous genes, involved not only in various cellular pathways critical for preventing cancer but also in pathways of “routine” cell activities (2, 3). Therefore, p53 is currently viewed as the “guardian of homeostasis” (4). p53 has a multi-domain modular structure (5, 6). The N terminus (NTD) contains a transactivation domain, the core domain contains a sequence-specific DNA binding domain (DBD), and the C-terminal domain (CTD) includes a tetramerization domain (TD) and a basic regulatory domain. The DBD of p53 binds to REs that consist of two decameric repeats of the form RRRCWWGYYY (R = A, G; W = A, T; Y = C, T), separated by 0 to 18 base pairs (bp) (7, 8). Base-pairs separating p53 half-sites (“spacer sequences”) are present in ∼50% of all validated p53 REs (7–9). p53 dimers are formed co-translationally (10) and p53 in unstressed cells is predominantly dimeric (11–13). The functional form of p53 is a tetramer, composed of a dimer of dimers (5, 14–16). Tetramerization occurs mostly on the DNA double helix, by stepwise binding of p53 dimers to DNA (17).
As the biological outcome of p53 activation depends on the function of p53 target genes, precise discrimination of p53 numerous target promoters is crucial for p53 to trigger correct cell-fate decisions. Broadly, two alternative models were proposed (18, 19). “The selective context model” suggests that p53 is not the crucial force behind target-gene selectivity, but that discrimination is achieved by other events at the cellular and genomic levels (18, 20–23). “The selective binding model” assumes that p53 REs are not equal, and discrimination is achieved at the level of DNA binding, through differences in binding affinity (24), in p53 PTMs (25–27), and in the binding of other p53 co-regulators (28). p53 response to stress has been termed “digital” (29) since the oscillation of the p53-HDM2 negative-feedback loop (30, 31) generates oscillations that have a constant amplitude but varying frequency (31–34). The digital nature of p53 response to stress ensures the release of controlled pulses of p53, sufficient for initial arresting of the cell cycle and ascertaining that the damage is being repaired, but not high enough to cause premature apoptosis. Indeed, the number of pulses per time unit has been found to correlate with the degree of cellular stress (31, 32), where few p53 pulses are released during normal growth (34) and for the transcription of genes required for rapid response upon acute DNA stress response (29). Proapoptotic genes are however activated only after a certain p53-dependent threshold is achieved, which takes multiple cycles of p53 pulses (29). The dependence of cell-faith decisions on p53 levels (35–37), the time elapsed since stress induction (early versus late response) (36), as well as the importance of differential binding affinities of p53 on its REs (24) are widely acknowledged. However, the mechanistic basis for selective p53-dependent gene expression is currently unknown. In our previous studies, we demonstrated experimentally that DNA torsional (twist) flexibility changes significantly between p53 REs, which leads to significant changes in DNA binding affinity and cooperativity (38, 39). Moreover, we previously showed that mutations that converted flexible REs to rigid REs significantly reduced transactivation under low p53 concentration (40). Furthermore, at low p53 levels, the torsional flexibility of p53 REs positively correlates with transactivation levels, whereas when p53 levels are highly induced, transactivation levels positively correlate with p53 binding affinity (40). These mechanistic observations support the selective binding model, as a major contributor to target-gene selectivity in the p53 system, but by themselves do not explain the unique manner by which p53 can coordinate its myriad target genes, in a selective fashion, to counteract an acute-stress signal or a routine cellular demand.
Here, we hypothesize that differential structural properties of p53 REs (“shape readout,” 41) contribute to target-gene promoter selection that leads to distinct cellular pathways. By connecting biological pathways of p53-target genes to the flexibility properties of their REs, and by using combined structural and functional analyses, we show that genes that are known to be expressed immediately upon stress have flexible REs, whereas genes that are known to be activated only later in the response to stress, or only at higher stress levels, harbor very rigid REs. We suggest that REs flexibility contributes to the initial stages of selective-gene expression in the p53 system by facilitating the upregulation of p53-target genes that need to be activated early in the response to stress, when p53 levels are low.
Results and Discussion
Organizing p53 Targets by Their Functional Outcome Yields Distinct Structural Features of Their REs.
To test our hypothesis, we have chosen only bona fide p53 targets (9) that are all upregulated by p53 and their REs reside in the promoter or the first exon/intron region, defined as the region from 5 kb upstream to the transcription start site to the end of the first intron (9, 39). Additionally, the REs of these genes lack spacer sequences to ensure that p53 binds specifically to both half-sites (“fully specific mode”), as opposed to REs with long spacer (>9 bp), where only one half-site is specifically bound to p53 (“hemi-specific mode”), and because of the observed reduced binding affinity of p53 REs with shorter spacer sequences (42). All in all, there are 236 genes that abide by these rules, as we previously published (39). However, a specific function is known for 210 out of these 236 genes, and therefore, the current analysis was focused on this set. The specific function of 210 genes, the biological process in which these genes participate, and the consequent cellular outcome of their activation are summarized in Dataset S1. For statistical analysis of possible relationships between functional outcome and RE flexibility, these genes were organized into supergroups based on the cellular outcome of their action (SI Appendix, Table S1).
We calculated the DNA flexibility of the REs belonging to the assembled set of the 210 p53-target genes using the energy parameter deformability (39, 43). This parameter [V(B) in units of °3Å3] is a quantitative measure for the overall flexibility of B-DNA base-pair steps (43, 44). We have previously determined that this rigorous measure of helical flexibility, which is dependent on all DNA helical degrees of freedom, has a strong and significant correlation with the torsional (twist) flexibility of p53 natural and consensus-like REs, measured using cyclization kinetics experiments (39), and can thus be used to estimate the flexibility properties of p53 REs. A minority of genes in our list (17 genes, Dataset S1) contained multiple REs (“REs clusters,” 7, 45), known to confer additional binding affinity and increase transactivation levels from the nearby gene. In cases of REs clusters, we chose for analyses the more flexible full-site RE, based on the assumption that the more flexible RE is the primary and dominant RE of the cluster (39). However, the analysis was unaltered when excluding all sites containing RE clusters.
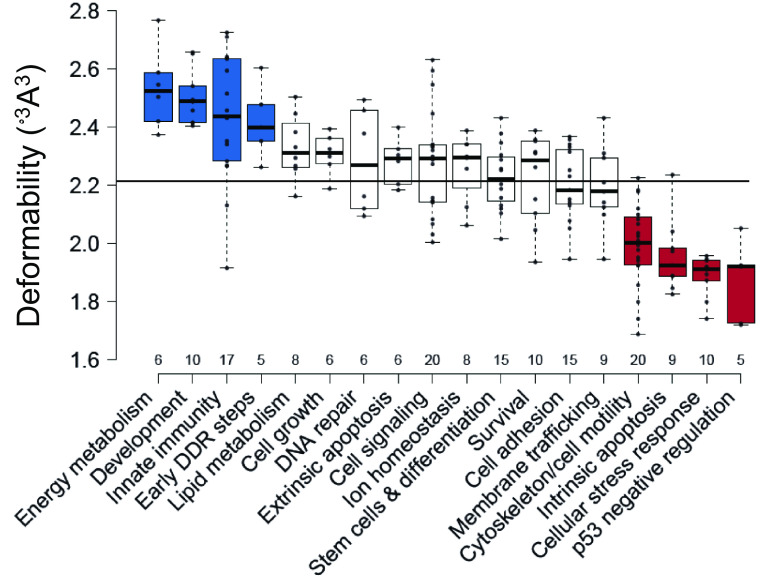
Next, we analyzed the relationship between calculated REs deformability and functional outcome supergroups that contain at least five genes each, to minimize noise from small groups (total 18 groups, 185 genes). One-way ANOVA of the variation in REs deformability between functional-outcome groups showed significant differences in their mean deformability values (F = 17.11, P = <0.0001, Fig. 1 and SI Appendix, Table S2). The results support our hypothesis that p53 target genes known to be induced early in response to cellular needs, or acute stress, are activated at low p53 levels and have REs with deformability values that are significantly above the mean (19, 35, 36, 46, 47). Genes in this category are known to participate in early DNA damage-response steps, innate immunity, development, and energy metabolism (colored blue in Fig. 1 and termed collectively as the “flexible-REs category”). On the other hand, p53 target genes known to be induced late in the stress response have REs with deformability values significantly below the mean. These genes belong to pathways that need to be more stringently regulated and are induced at relatively high p53 levels (19, 35, 36, 46, 47). These include genes that participate in intrinsic apoptosis, p53 negative regulation, cellular-stress response, and cytoskeleton/cell motility (colored red in Fig. 1 and termed collectively as the “rigid-REs category”). A post hoc test of the ANOVA established that each group within the flexible-REs category is significantly and solely different from each group within the rigid-REs category, but not from most of the remaining groups (SI Appendix, Table S2). Concerning the groups at the central part of Fig. 1, one can rationalize the intermediate range of the mean-deformability values of several groups but not all. For example, the extrinsic (death receptor) apoptosis pathway has above-average mean deformability, although it is a death-related functional-outcome group, and hence expected by our analysis to have deformability values that are below the mean. This may be related to the observations that extrinsic apoptosis is initiated by signals originating from outside the cell, such as natural-killer lymphocytes, or cytotoxic-T lymphocytes, and thus is intimately connected with the innate- and adaptive-immune systems (48), that by our analysis have above mean deformability values.
Fig. 1.
p53 REs belonging to genes in functional groups that are needed early in the response to stress are more deformable than REs belonging to genes needed later in the response to stress. Shown are functional groups with at least five genes each. All data points are shown as dots. The line across all groups marks the mean of means of the deformability of all analyzed groups. Center lines show the medians of each group. The figure was created by BoxPlotR (49). Box limits indicate the 25th and 75th percentiles, as determined by R software. Whiskers extend to maximum and minimum values. Boxes are arranged by decreasing group average values from left to right. Blue boxes denote the “flexible REs category,” and red boxes signify the “rigid REs category.” For significant differences between the groups, see SI Appendix, Table S2. DDR = DNA-damage response.
It is well established by reports from many groups (e.g., refs. 36, 50, and 51–55) that the kinetics of p53-dependent gene expression is differential and depends on p53 protein levels in the cell, the type and level of stress, and cell type. The underlying molecular mechanism, however, remains elusive. Here, we propose that flexibility characteristics of p53 REs play an important role in kinetics of p53-dependent gene expression. We arrived at the grouping shown in Fig. 1 based only on the flexibility characteristics of the REs of p53 target genes. Nonetheless, they show the known distinction between the kinetics of gene expression of early response genes (such as p21 and BTG2) in response to various stressors, versus the kinetics of late response genes, such as TP53AIP1, PMAIP1, and BBC3, to similar stressors (36, 50–55).
Functional Outcome Categories of p53-Activated Genes Have REs with a Unique Sequence Signature.
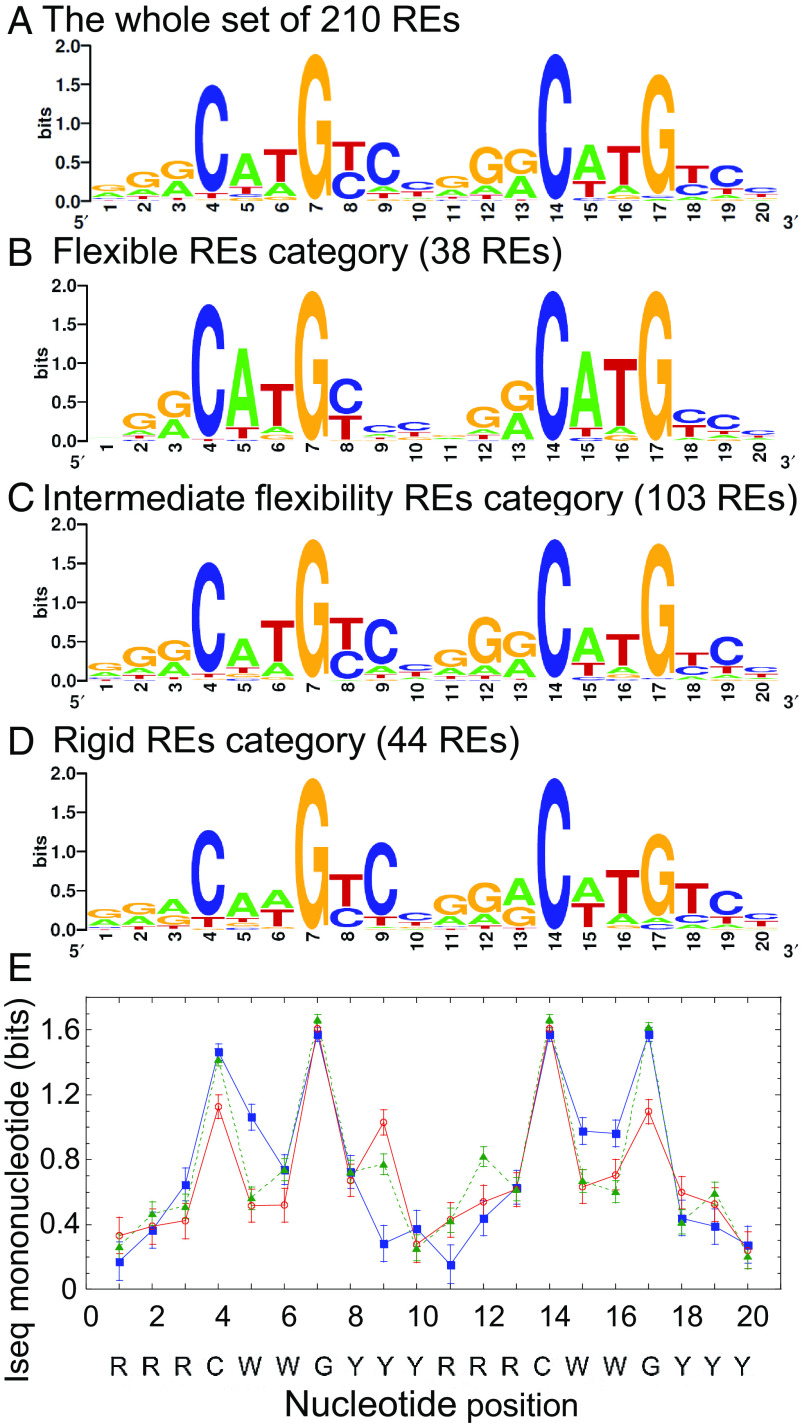
Sequence logos are a convenient way to depict base-pair preferences and information content of DNA binding sites (56–58). The sequence logo of the full set of 210 REs (Fig. 2A) shows a degenerate sequence pattern, similar to the pattern from previous studies (7, 8, 39, 59). Deconvoluting this overall sequence pattern to that of the two extreme categories (Fig. 2 B and D), one can observe an increase and decrease in nucleotide preferences in various positions, whereas the logo of REs with intermediate flexibility seems similar to that of the full set (Fig. 2C). To better evaluate quantitatively the differences in sequence preferences, we calculated the information content per nucleotide position (Iseql), of the set of aligned REs sequences per category (Fig. 2E). Information content is related to the thermodynamics of protein-DNA interactions because it is a measure of the discrimination, by a regulatory protein, between binding to a specific binding site versus binding to a random DNA sequence (60–63). Comparison of the information content of the two extreme-functional-outcome categories (Fig. 2E) shows that the 5′ CWW position and the 3′ WWG position (CWW on the opposite strand) of the flexible REs category have higher information content (i.e., the sequence space is more constrained and thus sequences vary less within this category in these positions) relative to the rigid REs category (~46% higher, respectively). On the other hand, the category with rigid REs is more constrained at the ninth nucleotide, in the middle of the inner YYY tract. The inner YYYRRR region of the rigid REs category has 39% higher information content than that of the flexible REs category. The intermediate category has relatively constrained C9 and G12 nucleotides. G12 nucleotide is in an equivalent position to that of the C9 position of the rigid category, both are inner quarter sites contacted by Lys120 residues from p53 to a G base. These changes in information content between nucleotide positions in the different structural categories are statistically significant (P < 1 × 10−10).
Fig. 2.
Sequence logos and information content of the studied REs. (A) All 210 p53 REs without spacers belonging to genes with the known functional outcome of p53 activation studied here. (B) REs belonging to the flexible REs category. (C) REs belonging to genes with intermediate flexible REs. (D) REs belonging to the rigid REs category. All included REs are those belonging to functional groups with at least five genes each, and the RE sequences were aligned by transcription direction before generating the sequence logos. Sequence logos were generated by Weblogo (57). (E) Information content per position (Iseql) on the mononucleotide level for the REs belonging to the flexible category (blue squares), rigid category (red circles), or REs with intermediate flexibility (green triangles).
The observation of high information content at the two CWWG motifs of the flexible REs category (Fig. 2B) is due to the almost exclusive preference for the flexible CATG sequences in both half-sites. p53 does not directly contact the WW doublet in the complex with its REs (64–70), and thus, DNA recognition in this region is by shape readout. Previously, we showed that CATG-containing p53 sites are more torsionally flexible than CAAG- and CTAG-containing sites (38). The high flexibility of CATG-containing REs is the reason for the high kinetic stability of p53 dimers on these sites, which facilitates a sequential assembly of functional p53 tetramers from DNA-bound p53 dimers even when p53 levels are low (40). The CAAG and the CTAG motifs are rigid and hence require higher p53 levels for forming functional tetramers, as the dimeric species are not stably bound to such sites (40). However, it is likely that this destabilization of the CAAG and CTAG motifs is partly compensated for by the stabilization of the p53-DNA interface via hydrogen bonds between Lys120 and the central G base in the opposite strand to the internal YYY tract in the rigid-category sequences (64–70). This may explain the stringent requirement for a C9 in the middle of the internal YYY tract in the rigid REs category and also the relatively high conservation of C9 and G12 in the YYY and RRR tracts of the sequences with intermediate deformability. Thus, our results suggest that various forces optimize p53–REs interactions selectively. In the flexible REs category, high information content (binding free-energy contributions of each base) in shape-readout positions comes together with high deformability (structural property of the DNA double helix). In the rigid REs category, and in sequences with intermediate deformability, a direct-readout contact (Lys120 to G base) compensates for low binding affinity and stability in the shape-readout positions.
It has been suggested that the distinction between pro-survival and pro-apoptotic genes resides at least in part in increased frequency of A/T versus G/C bps at positions 3, 8, 13, and 18 of p53 REs, respectively, regardless of the sequence of the CWWG core motif (71). This suggestion was based on the authors’ study of DNA binding by the Lys120Arg p53 mutant, and on an analysis of p53 REs of 16 pro-survival versus 16 pro-apoptotic genes (71). Indeed, it is known that when the sequence is G1G2A3, the binding affinity to p53 is higher than when the sequence is G1G2G3 or A1G2G3, where in the latter two cases, Lys120 binds to both G2 and G3 bases, and in the first case to G2 and the T base on the opposite strand to A3 (64). However, our results using 38 flexible REs of pro-survival genes and 44 rigid REs of pro-death genes show that other signals are as important for the distinction between survival and death. First, results from this study, as well as several previous studies (7, 24, 38), strongly suggest that the central CWWG varies between pro-survival genes (where it is CATG, shown to be more torsionally flexible, 38), to pro-death genes (where it is CAAG, shown to be more torsionally rigid, 38). Here, we add that a highly conserved C9 base at position 9 is important for optimal direct contact of Lys120 to the G base on the opposite strand, especially in the rigid REs category, where the contact to the central CAAG motif is sub-optimal.
Experimental Measurement of the Flexibility of Natural p53 REs by Cyclization Kinetics.
We have experimentally measured the global conformation of several natural p53 REs by cyclization kinetics of DNA minicircles to validate their calculated flexibility properties. Cyclization kinetics of DNA minicircles is a robust method to measure the global structural and dynamical properties of DNA molecules (72, 73), and it is carried out by following the rate of ligase-catalyzed closure of DNA molecules with cohesive ends into small rings. The REs studied here belong to genes that function at different cellular pathways (SI Appendix, Table S3) and were chosen to reflect a range of binding affinities (24) and deformability values (SI Appendix, Table S3, rightmost column). CCNG1 (cell growth), RRM2B (DNA repair), and p21-5′ (cell-cycle arrest) have REs with deformability values that are above the average of all 210 REs studied here (2.22 ± 0.01 Å3˚3), whereas PMAIP and TP53AIP1 (intrinsic apoptosis) have REs with deformability values below the average. Each sequence contained the 20-bp full site RE and 5-bp flanking sequence on both the 5′ and the 3′ sides of the test sequence, specific for each natural p53 site (SI Appendix, Table S3), because we have previously shown that sequences flanking p53 REs modulate p53 binding and transactivation, but that the influence of the flanks extends only to 4 to 5 bp from the RE (39). However, the global-structural characteristics of the sites (SI Appendix, Table S3 and Fig. S1) are those of the central 20-bp p53 sites without flanks, to concur with our previous studies (38, 39, 42). As in our previous studies on p53 binding sites (38, 39, 42), the only significant change in global-structural parameters of these sites is in the twist (torsional) flexibility of the DNA binding sites (SI Appendix, Table S3 and Fig. S1). Moreover, the results showed a similar trend to the values of the calculated deformability (SI Appendix, Table S3). Thus, experimentally derived torsional-flexibility values show that CCNG1, RRM2B, and p21-5’ are the most flexible sites in this set, whereas PMAIP1 and TP53AIP1 are the most rigid sites. These results support our hypothesis that genes that are activated earlier in the response to stress have more flexible REs than those that are activated later in the response to stress or at higher stress levels.
p53-Dependent Gene Expression As a Function of p53 Levels.
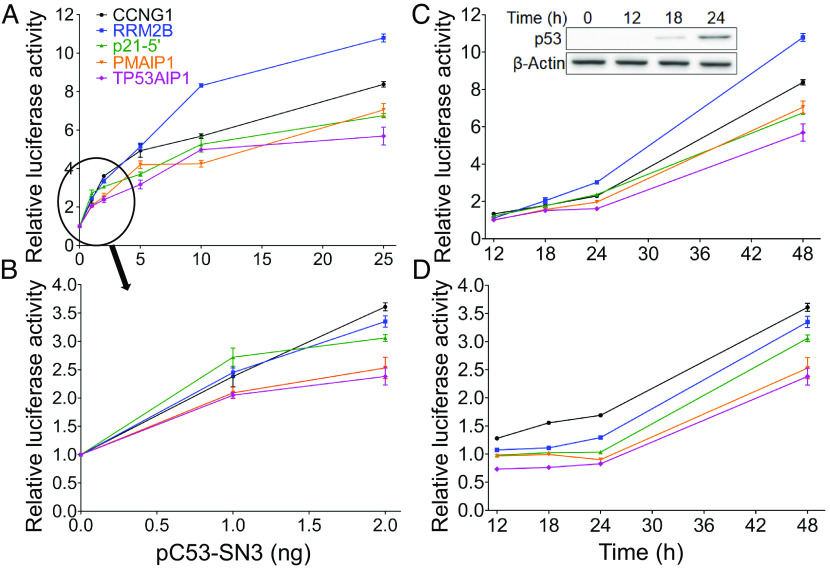
To study possible differences in activation levels of these five genes in cellular context, we carried out luciferase reporter-gene assays in the human non-small lung-carcinoma cell line (H1299), ectopically expressing p53 along with reporter plasmids, each carrying a distinct RE, surrounded by 5-bp natural-flanking sequence. First, we ascertained that p53 ectopic expression is linearly correlated with the amount of transfected p53-expression plasmid, pC53-SN3. This was done by transfecting cells with increasing amounts of p53-expression vector and immunoblotting the whole cell lysate (Fig. 3A). Densitometry analysis of the observed p53 band, normalized to the β-actin band, confirmed the linear relationship between p53 protein level and pC53-SN3 amount (SI Appendix, Fig. S2, r = 0.972, P < 0.0001). We then carried out reporter-gene assays for the five p53 binding sites described above (CCNG1, RRM2B, p21-5’, PMAIP1 and TP53AIP1) at various p53 levels (1, 2, 5, 10, 25, and 75 ng p53 plasmid), 48 h post transfection. Results show that transactivation levels rose as a function of p53 levels (Fig. 3 A and B and SI Appendix, Fig. S3). Transactivation levels of the five genes are significantly different from each other at 1 to 25 ng, but are similar at 75 ng of transfected p53-plasmid (see one-way ANOVA analysis in SI Appendix, Table S4A). At 1 ng p53 plasmid, such a distinction was found only for transactivation from p21-5’ compared to transactivation from the two rigid REs (Fig. 3B and SI Appendix, Table S4A). In cells transfected with 2 ng p53 plasmid (Fig. 3B), transactivation levels from each of the three flexible REs were significantly higher than that from the two rigid (SI Appendix, Table S4A). In cells transfected with higher p53 plasmid levels, individual pair-wise differences between REs are significantly different at all p53 levels only when comparing RRM2B to the two rigid REs—PMAIP1 and TP53AIP1 (Fig. 3A and SI Appendix, Table S4A). The transactivation from cells transfected with all tested p53 plasmid amounts, except 75 ng, was positively correlated with the experimental torsional (twist) flexibility of the five studied REs [Pearson r (P) = 0.35 (0.03), 0.74 (<0.0001), 0.64 (0.002), 0.84 (<0.0001) at 1, 5, 10, and 25 ng p53 plasmid, respectively], being most strongly at 2 ng plasmid (Pearson r = 0.91, P < 0.0001), further validating our previous results.
Fig. 3.
Luciferase activity from p53 REs as a function of the p53 expression level and of time. (A and B), Transactivation in cells transfected with the indicated p53-plasmid amounts measured 48 h post-transfection. (C and D) Fold increase in transactivation levels as a function of time, using 25 ng p53 plasmid (C) or 2 ng p53 plasmid (D). Kinetics of p53 accumulation (0 to 24 h) in cells transfected with 25 ng p53 plasmid are shown in panel C Inset. Luminescence values were normalized to the transfection efficiency of co-transfected, constitutively expressed Gaussia luciferase (Gluc). Results were further normalized first to the empty Cypridina secreted luciferase (CLuc) vector and then to results obtained without p53. Error bars represent the mean ± SD of three to eight independent experiments, each containing two technical replicas. The order of the REs in the legend within (A) is according to the measured torsional flexibility, decreasing from top to bottom.
Kinetics of p53-Dependent Gene Expression.
The higher transactivation levels observed 48 h post-transfection, from flexible REs relative to rigid REs, could be due to an earlier start of transactivation, or alternatively due to faster transactivation rates. To distinguish between these possibilities, we examined transactivation levels, at 2 and 25 ng p53 plasmid, at additional time points (12, 18, and 24 h post-transfection). Looking at the results of transactivation at 25 ng (Fig. 3C), one notes that p53 starts activating all five genes at the same time (12 h). One-way ANOVA at each time point post-transfection revealed a significant difference between the transactivation levels of the five REs (SI Appendix, Table S4B). A post hoc analysis indicated that at 12 h post-transfection, transactivation from CCNG1 and p21-5’ was significantly different from that of the rigid REs (SI Appendix, Table S4B). At 18 h post-transfection, post hoc analysis indicated that transactivation from RRM2B was significantly different from that from the two rigid REs (SI Appendix, Table S4B). At 24 h post-transfection, transactivation from all flexible REs was higher than transactivation by the two rigid REs (SI Appendix, Table S4B). A positive significant correlation is observed between p53-dependent transactivation level from the five studied REs, at all studied post-transfection time points, and their experimental twist flexibility (r = 0.50, 0.53, and 0.77, and P = 0.002, 0.001 and P < 0.0001, for 12, 18, and 24 h, respectively, for transactivation at 25 ng p53 plasmid.
Transactivation levels were lower in cells transfected with 2 ng p53 plasmid, as would be expected. Nevertheless, transactivation levels of the five studied genes were distinctly different from each other (Fig. 3D), as verified by one-way ANOVA analysis at each time point (SI Appendix, Table S4C). From post hoc pair-wise comparisons of group means (SI Appendix, Table S4C), it can be observed that all genes with flexible REs are distinctly different from those having rigid REs at all time points, except for p21-5’ versus PMAIP1 at 12 and 18 h. Transactivation values for the two most flexible REs (RRM2B and CCNG1) were consistently increased from 18 to 48 h post transfection. Transactivation levels from the remaining REs remained basal up to 24 h post transfection. However, at 48 h, transactivation levels from p21-5’ RE were comparable to those obtained with the most flexible REs (as observed also in Fig. 3B). In summary, the distinction between transactivation level arising from flexible versus rigid REs is maximal at 2-ng p53 plasmid and 48 h (Fig. 3 B and D), and the REs are ordered according to their measured flexibility properties in both 24- and 48-h time points; however, the kinetics of transactivation-level accumulation, prior to 48 h, are slower as compared to those obtained in cells transfected with 25 ng p53 plasmid.
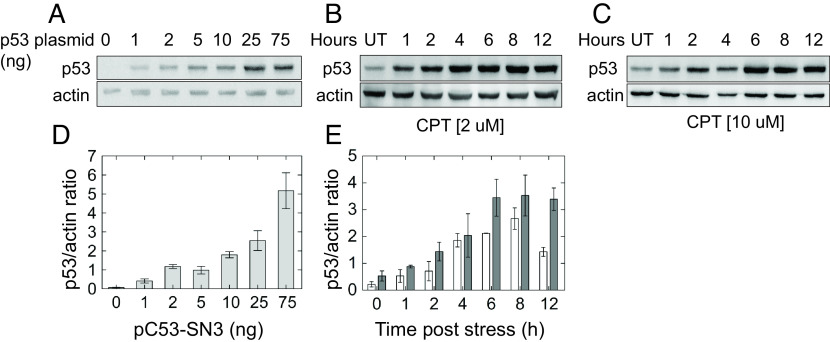
To ascertain the physiological relevance of ectopic p53 levels used in the reporter assays, we compared them to endogenous p53 protein levels obtained under stress conditions. For this, MCF-7 cells were treated with 2 µM or 10 µM camptothecin (CPT), and p53 levels were assessed at various time points post CPT application. As shown in Fig. 4, immunoblotting analysis showed that expressions of ectopic versus endogenous p53 are comparable. p53 levels in all cells transfected with 1 to 5 ng p53 plasmid are similar to endogenous p53 levels obtained at early time points post CPT application. Likewise, ectopic p53 levels in cells transfected with higher p53 plasmid concentrations (25, 75 ng) resemble the levels obtained at later time points post CPT application (≥6 h). Moreover, to show that the amounts of p53 plasmid used in our transfection experiments span the range from basal to stress-induced p53 levels in other cell types, we calibrated the transfection experiments to serially dilutions of purified recombinant p53 protein (SI Appendix, Fig. S4). In cells harboring 1 and 2 ng p53 plasmid, we estimated that p53 protein levels are approximately 4.2 × 10−4 and 9.2 × 10−4 pmoles/µg of whole-cell extract (WCE), respectively, which is similar to basal p53 levels previously found in HCT116 cells (~5 × 10−4 pmole/µg WCE) (74). At 75 ng p53 plasmid, there are 11.5 × 10−3 pmoles/µg WCE, which is similar to endogenous p53 levels found after stress induced by camptothecin treatment (23 ng/μg of WCE) in HCT116 cells (11 × 10−3 pmoles/µg WCE) (74). Thus, the results ascertain that the amount of p53 plasmid used in this transiently transfected plasmid study spans the same range of endogenous p53 protein levels induced by CPT also in HCT116 cells.
Fig. 4.
Comparison of ectopic p53 levels to its induced levels under stress. (A), p53 null H1299 cells were transfected with the indicated amounts of pC53-SN3 vector. p53 was immunodetected in whole cell lysates collected 48 h post transfection. (B and C) p53 levels in MCF-7 cells treated with 2 µM or 10 µM CPT for the indicated time points. Actin serves as a loading control. Representative gels are shown, UD = untreated. (D) Quantitative analysis for ectopic p53 protein levels shown in (A). (E) Quantitative analysis for p53 levels induced by CPT stress in MCF-7 cells shown in (B), white, and (C), gray. Intensity of bands was quantitated using Cliqs software. Error bars are the result of two to four independent experiments.
A Relationship between Deformability of p53 REs and mRNA Levels of p53 Target Genes.
To further support our observations that p53 REs belonging to genes known to be “early responders” to stress are more flexible than REs belonging to genes known to be activated late in the response to stress, we took advantage of RNA-seq studies that analyzed p53 target genes as a function of response time. The first study is that of Zhao et al. (36) on the kinetics of p53 induction by zinc, UV, or γ-irradiation. In this study (36), p53-responsive genes classified into broad functional categories were divided to early, intermediate, or late response, upon adding zinc chloride to EB-1 cells for various time spans. According to this study, p21 (CDKN1A), GADD45A1, TP53, and COL2A1 genes are induced early (2 h). Our calculation yielded high deformability values to their REs (2.263, 2.279, and 2.279 °3Å3, for p21, GADD45A, and TP53, respectively). COL2A1 RE has spacer sequences between its half-sites, and hence is not in our list, but COL7A1 in our list has the same functional classification and has a highly deformable RE [V(B) = 2.342 °3Å3]. We did not include GADD45A RE in our statistical analysis because it is an intragenic RE. Nonetheless, it does conform to our rules. Moreover, the FAS/APO1, Killer/DR5, and ACTA2 genes are specified as intermediate responders by Zhao et al. (36), and our calculations show them to have intermediate deformability values (2.184, 2.205, and 2.184 °3Å3, respectively).
Additional support to our observations is from the study by Freewoman et al. (47). In this study time course (6, 12, and 24 h) RNA-seq experiments were performed in HCT116 cells treated with fluorouracil (5-FU), and early versus late genes were identified. The authors created datasets specific for 6, 12, and 24 h treatment with 5-FU (47). Of note, 72 genes specific to 6 h and 31 genes specific to 24 h are common between our dataset and that of Freewoman et al. (47). By our hypothesis, we suggest that the deformability of the REs of early (6 h) genes should be higher than that of the late (24 h) genes, which is validated by a two-sample t test (t = 2.60; P = 0.005).
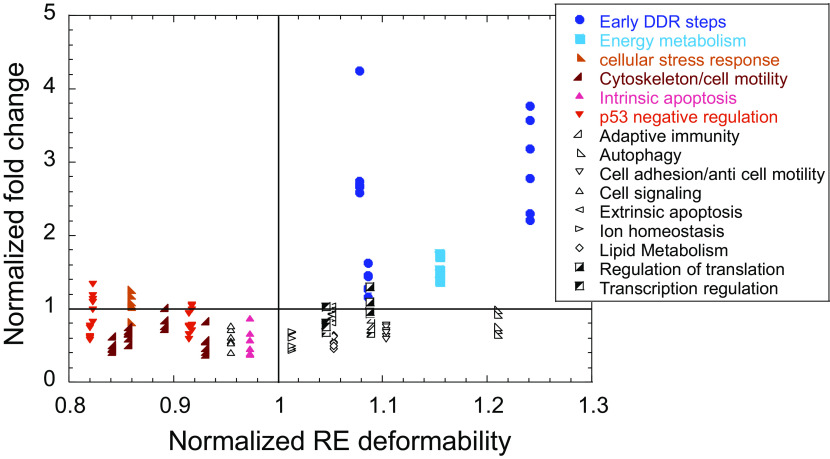
A third example to support our observations is from the time-dependent RNA-seq data study by Hafner et al. (75). These authors studied the relationship between p53 dynamics and p53-dependent gene expression following DNA damage caused by γ-irradiation in MCF-7 breast-cancer cell line, which triggers a series of p53 pulses. RNA-seq analysis was performed on samples taken every hour between 1 h and 12 h, that is during the first two pulses, each 6 h long, and another measurement at 24 h. Based on gene-expression dynamics, the studied genes were grouped into five distinct clusters (75). We analyzed mRNA levels of p53 target genes that belong to cluster 1 and compared them to the deformability of their respective REs. Cluster 1 contains the largest number of validated p53 targets and the largest number of genes that abide by the criteria of the current study (their REs lack spacer and are located either within the promoter or the first intron). Thus, the analysis was carried out on 23 out of 32 genes in cluster 1. Fig. 5 shows the normalized fold-change of mRNA levels as a function of normalized deformability. We normalized both values to the average value of all genes at each specific time point, so the genes’ expression levels could be plotted for all time points together. Fig. 5 shows that early DDR steps and energy metabolism genes (colored dark and light blue, respectively) have above-average flexible REs, and are expressed at above-average level at every time point. On the other hand, genes belonging to the rigid category (colored with various shades of red) are all below average rigidity and are all expressed at below average transactivation level. In spite of the small dataset, significant differences in expression levels were observed only between the flexible REs group and the rigid REs group, at all the studied time points (P < 0.003, where the maximum P value for 13 comparisons is 0.004). Thus, we conclude that at the stress level conditions (10 Gy γ-irradiation of MCF-7 cells) used in this study (75), the extra-flexibility of p53 REs belonging to early-response genes enable high expression level of these genes, even though only few pulsatile cycles are produced (4 pulses in 24 h, 74). Pro-apoptotic genes, and other genes that need to be carefully regulated, do not attain high level of expression at these stress levels and within the time frame (24 h) used in this study (75). Indeed, it has been shown that at these stress levels, a switch from oscillatory to elevated p53 levels is a late event that occurs more than 24 h after irradiation (48 to 72 h following irradiation, 76). All in all, these studies support our hypothesis that “early responder” to stress genes have REs with above-average deformability values, whereas REs of “late responder” genes have deformability values that are below average, also when the studies are conducted on endogenous genes in cellular context.
Fig. 5.
Endogenous mRNA levels from p53-dependent genes are grouped by their RE deformability. The data shown are based on the RNA-seq results from Hafner et al. (75). Fragments Per Kilobase Million (FPKM) values of each gene (i), at each time point (j) were normalized to their FPKM value at T = 0, arriving at fold-change values. Then, these values were further normalized to the average fold-change of all genes at each particular time point (j). That is, each point is calculated from the equation: (FPKMiTj/FPKMiT0)/ (FPKMiTj/FPKMiT0). Deformability of genes’ REs was normalized in the same manner. Plotted are the normalized fold-change values as a function of normalized deformability values at even numbers post irradiation. The lines dissecting each axis at 1 unit is the normalized average of each parameter, for visualization of values that are above/below the average value of these parameters.
General Discussion and Biological Implications
In this study, we showed that DNA flexibility of p53 REs enables selectivity in the initial steps of the digital response of p53 to stress. We suggest that highly flexible REs contribute to rapid response during normal growth and during the initial stages of acute stress, based on our previous findings (40) showing that when p53 levels are low, stable binding of p53 dimers to p53-dependent REs is facile only if the REs are torsionally flexible. Under these conditions, p53 tetramers can be formed sequentially (i.e., at low binding cooperativity) without the need for p53 accumulation over an extended time span. Thus, we would like to advance DNA-encoded flexibility as a molecular mechanism for selective “temporal-induction profiles” (29), and we hypothesize that evolutionary–selection pressure made sure that p53-dependent functionalities that need to be executed swiftly will be able to do so and that a key contributor to this need is coded in DNA structural properties. Hence, in addition to the statement that “p53 is smart” (19), the current data demonstrate that the DNA of p53 REs is “smart”—the REs base sequences are information-rich by virtue of their structural properties. Consequently, the initial step of gene-expression activation by p53 (and possibly other transcription factors as well) is another fundamental code within the layers of codes that co-exist within the DNA double helix.
Our observations that the initial step toward determining the functional outcome of p53 upregulation is encoded within the DNA double helix does not exclude other mechanisms shown to contribute to selectivity of p53-dependent gene regulation. The complex nature of p53 target-gene regulation needs multiple control steps for the process to trigger correct cell-fate decisions. We suggest that other mechanisms proposed to account for p53 selective gene regulation, such as mRNA stability of p53 target genes (75, 77), protein stability of p53 targets (78), and combinatorial control by co-activating TFs (28) occur at steps that follow this initial differential recognition by p53, which uses the structural properties of its REs.
Materials and Methods
Cyclization Kinetics and Simulation of Cyclization Data.
DNA constructs for cyclization kinetics were synthesized using the top library/bottom test sequence PCR scheme, described previously (38, 39, 42, 73), except that here we used as test sequences DNA sequences containing the full 20-bp long p53 sites plus 5-bp flanking sequences on both sides, specific for each natural p53 site (see SI Appendix, Table S3 for sequences). Cyclization kinetics measurements were carried out as described previously (38, 39, 42). We derived quantitative data on the conformational properties of the test sequences by simulating the cyclization data as previously described (38, 39, 42, 79), using the simulation program developed by Zhang and Crothers (79). The outcome of the simulations is the bend angle, twist angle, roll and tilt flexibility, and twist flexibility per DNA sequence.
Reporter Gene Assays (RGA).
H1299 cells were seeded in 24-well plates at a density of 5 × 104 cells/well and incubated for 24 h. Then, cells were co-transfected with 800 ng of either pCLuc Mini-TK 2 harboring p53 RE or a corresponding empty vector along with 50 ng of pCMV-GLuc and 0 to 75 ng (0, 1, 2, 5, 10, 25, and 75 ng) of p53 expression vector (pC53-SN3). The total DNA amount was adjusted using pcDNA 3.1 plasmid. Transfection was performed with jetPEI (Polyplus) reagent according to the manufacturer’s recommendation. Transfection efficiency was monitored by transfecting separate wells with 0.1 µg eGFP along with Gluc and pcDNA plasmids. An aliquot of growth medium (20 μL) was taken at various time points post-transfection, and luminescence was measured using the Pierce™ Cypridina Luciferase Flash Assay Kit and Pierce™ Gaussia Luciferase Flash Assay Kit (both from Thermo Fisher) according to the manufacturer’s protocol. Signal was detected by CLARIOstar 96 well plate reader (BMG LABTECH). Four to eight independent experiments were carried out, each containing two technical replicates.
Analysis of Transactivation from p53 REs.
To correct for transfection variability, all results from Cypridina Luciferase luminescence were normalized to Gaussia Luciferase luminescence. Next, the results were normalized in two steps. First, the results obtained with pCLuc Mini-TK 2 harboring p53RE (Cluc+RE) were divided by the results obtained with the corresponding empty vector (Cluc−RE), Second, the results obtained with p53 (1 to 75 ng of pC53-SN3) were divided by the results obtained without p53 (0 ng of pC53-SN3). Fold increase in transactivation is thus given by the equation: .
Functional Annotation of p53 Target Genes.
We used here a previously assembled set of 236 p53 target genes that are all upregulated by p53, are validated for binding in cellular context and for p53-dependent gene expression, and reside in the promoter or the first exon/intron region (39). We modified the previous dataset by deleting the DDIT4 REs because this gene was shown to be directly upregulated by RFX7 and not by p53 (80, 81) and added two genes RhoC and Scrib. We located each gene in the Genecards suite (https://www.genecards.org/) by their Entrez ID, gene symbol, and gene name. We then looked for functional annotations of these genes in the Gene Ontology (GO) project (under the “Biological process” and “Molecular function” categories) through UniProtKB/UniProt (https://www.uniprot.org). GO annotations were supplemented by publications retrieved through individual literature searches for each gene product, especially those centering on the role of these genes in the p53 system. This functional categorization was applicable to 207 out of the 210 genes for which a p53-dependent function is currently known. The remaining three genes (PRDM1, PTP4A1, and XPC) have two main functions, one of which is p53 negative regulation that is common to all three genes (82–84). In this p53-centric study, we chose to assign the above three genes to the p53-negative-regulation group in testing our hypothesis on the relationship between functional outcome and DNA flexibility. To avoid bias of the analyses of the relationship between DNA flexibility and functional outcome from p53-dependent gene activation, we took only one 20-bp RE per gene for structural and sequence analyses. This was done both when the cluster sites were separated by a long intervening region and when the sites were composed of more than two abutting decamers (Dataset S1).
Information Content Analysis.
Information content per DNA binding site (Iseq, refs. 60 and 61) was calculated separately for the whole set of REs, as well as for the REs of the flexible, the rigid functional outcome category, and the group with intermediate flexibility. All calculations were on the mononucleotide level, using the equations detailed in the SI Appendix, Materials and Methods.
Other Computational and Statistical Tests.
The values for the deformability [V(B) in units of °3Å3] of each DNA base-pair step were calculated as previously described (39), using the values of (43). Statistically significant differences in DNA deformability between p53-dependent functional outcome groups, as well as analysis of the differences in the transactivation level of the studied REs, were tested using one-way ANOVA. We used the Dunn method for joint ranking as a post hoc test to determine the groups that are significantly different from each other by their deformability, since this method is adjusted for multiple non-parametric comparisons. We used the Tukey HSD multiple comparison test as a post hoc test for the differences between RGA transactivation levels. Z (SI Appendix, Table S2) denotes the standardized test statistic, which has an asymptotic standard normal distribution under the null hypothesis of no difference in means. In testing the strength of the relationship between transactivation levels from RGA and deformability [V(B)], we used Pearson correlation and one-sided P values [because our hypothesis is directional—that there is a positive correlation between RGA results and V(B) values], using all data points from the biologically independent RGA experiments. All analyses were conducted using JMP®, Version 16 (SAS Institute Inc., 2021). P values for the differences between sequence motifs (Fig. 2) were calculated using TOMTOM (85), from within the MEME suite (https://meme-suite.org/meme/db/motifs).
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Acknowledgments
We thank Varda Rotter (Weizmann Institute of Science, Rehovot, Israel) for the p53 expression plasmid, pC53-SN3. We thank Alberto Inga (CIBIO, Trento, Italy) for stimulating discussions of the manuscript. We thank Chaya Zilberstein for help in statistical analysis of information content. We thank the biomedical core facility (BCF, Technion) for Sanger sequencing. This work was supported by the Israel Science Foundation (Grants #1517/14 and 875/22 to T.E.H.).
Author contributions
D.R. and T.E.H. designed research; J.S., A.C., H.S., and P.V. performed research; J.S., A.C., Y.D.-P., D.R., and T.E.H. analyzed data; T.E.H. obtained funding for the project; and J.S., Y.D.-P., D.R., and T.E.H. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
Preprint Server (a prior version of this manuscript, not updated to the current one): BioRxiv (CC BY-NC-ND).
This article is a PNAS Direct Submission.
Contributor Information
Dina Ron, Email: dinar@technion.ac.il.
Tali E. Haran, Email: bitali@technion.ac.il.
Data, Materials, and Software Availability
All study data are included in the article and/or supporting information.
Supporting Information
References
- 1.Michael D., Oren M., The p53-Mdm2 module and the ubiquitin system. Semin. Cancer Biol. 13, 49–58 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Vousden K. H., Prives C., Blinded by the light: The growing complexity of p53. Cell 137, 413–431 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Kastenhuber E. R., Lowe S. W., Putting p53 in context. Cell 170, 1062–1078 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aylon Y., Oren M., The paradox of p53: What, how, and why? Cold Spring Harb. Perspect. Med. 6, a026328 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joerger A. C., Fersht A. R., Structural biology of the tumor suppressor p53. Annu. Rev. Biochem. 77, 557–582 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Joerger A. C., Fersht A. R., The p53 pathway: Origins, inactivation in cancer, and emerging therapeutic approaches. Annu. Rev. Biochem. 85, 375–404 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Riley T., Sontag E., Chen P., Levine A., Transcriptional control of human p53-regulated genes. Nat. Rev. Mol. Cell Biol. 9, 402–412 (2008). [DOI] [PubMed] [Google Scholar]
- 8.Menendez D., Inga A., Resnick M. A., The expanding universe of p53 targets. Nat. Rev. Cancer. 9, 724–737 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Nguyen T. T., et al. , Revealing a human p53 universe. Nucleic Acids Res. 46, 8153–8167 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nicholls C. D., McLure K. G., Shields M. A., Lee P. W., Biogenesis of p53 involves cotranslational dimerization of monomers and posttranslational dimerization of dimers. Implications on the dominant negative effect. J. Biol. Chem. 277, 12937–12945 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Weinberg R. L., Veprintsev D. B., Fersht A. R., Cooperative binding of tetrameric p53 to DNA. J. Mol. Biol. 341, 1145–1159 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Rajagopalan S., Huang F., Fersht A. R., Single-Molecule characterization of oligomerization kinetics and equilibria of the tumor suppressor p53. Nucleic Acids Res. 39, 2294–2303 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaglia G., Guan Y., Shah J. V., Lahav G., Activation and control of p53 tetramerization in individual living cells. Proc. Natl. Acad. Sci. U.S.A. 110, 15497–15501 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clore G. M., et al. , Refined solution structure of the oligomerization domain of the tumour suppressor p53. Nat. Struct. Biol. 2, 321–333 (1995). [DOI] [PubMed] [Google Scholar]
- 15.Jeffrey P. D., Gorina S., Pavletich N. P., Crystal structure of the tetramerization domain of the p53 tumor suppressor at 1.7 angstroms. Science 267, 1498–1502 (1995). [DOI] [PubMed] [Google Scholar]
- 16.Lee W., et al. , Solution structure of the tetrameric minimum transforming domain of p53. Nat. Struct. Biol. 1, 877–890 (1994). [DOI] [PubMed] [Google Scholar]
- 17.Natan E., Hirschberg D., Morgner N., Robinson C. V., Fersht A. R., Ultraslow oligomerization equilibria of p53 and its implications. Proc. Natl. Acad. Sci. U.S.A. 106, 14327–14332 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Espinosa J. M., Mechanisms of regulatory diversity within the p53 transcriptional network. Oncogene 27, 4013–4023 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vousden K. H., p53: Death star. Cell 103, 691–694 (2000). [DOI] [PubMed] [Google Scholar]
- 20.Espinosa J. M., Emerson B. M., Transcriptional regulation by p53 through intrinsic DNA/chromatin binding and site-directed cofactor recruitment. Mol. Cell. 8, 57–69 (2001). [DOI] [PubMed] [Google Scholar]
- 21.Gomes N. P., et al. , Gene-specific requirement for P-TEFb activity and RNA polymerase II phosphorylation within the p53 transcriptional program. Genes Dev. 20, 601–612 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morachis J. M., Murawsky C. M., Emerson B. M., Regulation of the p53 transcriptional response by structurally diverse core promoters. Genes Dev. 24, 135–147 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zilfou J. T., Lowe S. W., Tumor suppressive functions of p53. Cold Spring Harb. Perspect. Biol. 1, a001883 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weinberg R. L., Veprintsev D. B., Bycroft M., Fersht A. R., Comparative binding of p53 to its promoter and DNA recognition elements. J. Mol. Biol. 348, 589–596 (2005). [DOI] [PubMed] [Google Scholar]
- 25.Appella E., Anderson C. W., Post-translational modifications and activation of p53 by genotoxic stresses. Eur. J. Biochem. 268, 2764–2772 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Gu B., Zhu W. G., Surf the post-translational modification network of p53 regulation. Int. J. Biol. Sci. 8, 672–684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Y., Regulation of p53 responses by post-translational modifications. Cell Death Differ. 10, 400–403 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Catizone A. N., et al. , Locally acting transcription factors regulate p53-dependent cis-regulatory element activity. Nucleic Acids Res. 48, 4195–4213 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ma L., et al. , A plausible model for the digital response of p53 to DNA damage. Proc. Natl. Acad. Sci. U.S.A. 102, 14266–14271 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lev Bar-Or R., et al. , Generation of oscillations by the p53-Mdm2 feedback loop: A theoretical and experimental study. Proc. Natl. Acad. Sci. U.S.A. 97, 11250–11255 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lahav G., et al. , Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat. Genet. 36, 147–150 (2004). [DOI] [PubMed] [Google Scholar]
- 32.Geva-Zatorsky N., et al. , Oscillations and variability in the p53 system. Mol. Syst. Biol. 2, 2006 0033 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Batchelor E., Mock C. S., Bhan I., Loewer A., Lahav G., Recurrent initiation: A mechanism for triggering p53 pulses in response to DNA damage. Mol. Cell 30, 277–289 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loewer A., Batchelor E., Gaglia G., Lahav G., Basal dynamics of p53 reveal transcriptionally attenuated pulses in cycling cells. Cell 142, 89–100 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen X., Ko L. J., Jayaraman L., Prives C., p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 10, 2438–2451 (1996). [DOI] [PubMed] [Google Scholar]
- 36.Zhao R., et al. , Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 14, 981–993 (2000). [PMC free article] [PubMed] [Google Scholar]
- 37.Kracikova M., Akiri G., George A., Sachidanandam R., Aaronson S. A., A threshold mechanism mediates p53 cell fate decision between growth arrest and apoptosis. Cell Death Differ. 20, 576–588 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beno I., Rosenthal K., Levitine M., Shaulov L., Haran T. E., Sequence-dependent cooperative binding of p53 to DNA targets and its relationship to the structural properties of the DNA targets. Nucleic Acids Res. 39, 1919–1932 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Senitzki A., et al. , The complex architecture of p53 binding sites. Nucleic Acids Res. 49, 1364–1382 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jordan J. J., et al. , Low-level p53 expression changes transactivation rules and reveals superactivating sequences. Proc. Natl. Acad. Sci. U.S.A. 109, 14387–14392 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rohs R., et al. , Origins of specificity in protein-DNA recognition. Annu. Rev. Biochem. 79, 233–269 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vyas P., et al. , Diverse p53/DNA binding modes expand the repertoire of p53 response elements. Proc. Natl. Acad. Sci. U.S.A. 114, 10624–10629 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Balasubramanian S., Xu F., Olson W. K., DNA sequence-directed organization of chromatin: Structure-based computational analysis of nucleosome-binding sequences. Biophys. J. 96, 2245–2260 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Olson W. K., Gorin A. A., Lu X. J., Hock L. M., Zhurkin V. B., DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl. Acad. Sci. U.S.A. 95, 11163–11168 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bourdon J. C., et al. , Further characterisation of the p53 responsive element–identification of new candidate genes for trans-activation by p53. Oncogene 14, 85–94 (1997). [DOI] [PubMed] [Google Scholar]
- 46.Vousden K. H., Lu X., Live or let die: The cell’s response to p53. Nat. Rev. Cancer 2, 594–604 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Freewoman J. M., Snape R., Cui F., Temporal gene regulation by p53 is associated with the rotational setting of its binding sites in nucleosomes. Cell Cycle 20, 792–807 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ricci M. S., El-Deiry W. S., “The extrinsic pathway of apoptosis” in Apoptosis, Senescence, and Cancer. Cancer Drug Discovery and Development series, Gewirtz D. A., Holt S. E., Grant S., Eds. (Humana Press, 2007). [Google Scholar]
- 49.Spitzer M., Wildenhain J., Rappsilber J., Tyers M., BoxPlotR: A web tool for generation of box plots. Nat. Methods 11, 121–122 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abedin M. J., Wang D., McDonnell M. A., Lehmann U., Kelekar A., Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 14, 500–510 (2007). [DOI] [PubMed] [Google Scholar]
- 51.Duriez C., Falette N., Cortes U., Moyret-Lalle C., Puisieux A., Absence of p53-dependent induction of the metastatic suppressor KAI1 gene after DNA damage. Oncogene 19, 2461–2464 (2000). [DOI] [PubMed] [Google Scholar]
- 52.Espinosa J. M., Verdun R. E., Emerson B. M., p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol. Cell 12, 1015–1027 (2003). [DOI] [PubMed] [Google Scholar]
- 53.Oda K., et al. , p53AIP1, a potential mediator of p53-dependent apoptosis, and its regulation by Ser-46-phosphorylated p53. Cell 102, 849–862 (2000). [DOI] [PubMed] [Google Scholar]
- 54.Amundson S. A., et al. , Differential responses of stress genes to low dose-rate gamma irradiation. Mol. Cancer Res. 1, 445–452 (2003). [PubMed] [Google Scholar]
- 55.Kannan K., et al. , DNA microarrays identification of primary and secondary target genes regulated by p53. Oncogene 20, 2225–2234 (2001). [DOI] [PubMed] [Google Scholar]
- 56.Schneider T. D., Stormo G. D., Gold L., Ehrenfeucht A., Information content of binding sites on nucleotide sequences. J. Mol. Biol. 188, 415–431 (1986). [DOI] [PubMed] [Google Scholar]
- 57.Schneider T. D., Stephens R. M., Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 18, 6097–6100 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Crooks G. E., Hon G., Chandonia J. M., Brenner S. E., WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hafner A., Bulyk M. L., Jambhekar A., Lahav G., The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell. Biol. 20, 199–210 (2019). [DOI] [PubMed] [Google Scholar]
- 60.Berg O. G., von Hippel P. H., Selection of DNA binding sites by regulatory proteins. Statistical- mechanical theory and application to operators and promoters. J. Mol. Biol. 193, 723–750 (1987). [DOI] [PubMed] [Google Scholar]
- 61.Berg O. G., von Hippel P. H., Selection of DNA binding sites by regulatory proteins. II. The binding specificity of cyclic AMP receptor protein to recognition sites. J. Mol. Biol. 200, 709–723 (1988). [DOI] [PubMed] [Google Scholar]
- 62.Stormo G. D., Probing information content of DNA-binding sites. Methods Enzymol. 208, 458–468 (1991). [DOI] [PubMed] [Google Scholar]
- 63.Hertz G. Z., Stormo G. D., Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics 15, 563–577 (1999). [DOI] [PubMed] [Google Scholar]
- 64.Kitayner M., et al. , Structural basis of DNA recognition by p53 tetramers. Mol. Cell 22, 741–753 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Kitayner M., et al. , Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs. Nat. Struct. Mol. Biol. 17, 423–429 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ho W. C., Fitzgerald M. X., Marmorstein R., Structure of the p53 core domain dimer bound to DNA. J. Biol. Chem. 281, 20494–20502 (2006). [DOI] [PubMed] [Google Scholar]
- 67.Malecka K. A., Ho W. C., Marmorstein R., Crystal structure of a p53 core tetramer bound to DNA. Oncogene 28, 325–333 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen Y., Dey R., Chen L., Crystal structure of the p53 core domain bound to a full consensus site as a self-assembled tetramer. Structure 18, 246–256 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen Y., et al. , Structure of p53 binding to the BAX response element reveals DNA unwinding and compression to accommodate base-pair insertion. Nucleic Acids Res. 41, 8368–8376 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Golovenko D., et al. , New insights into the role of DNA shape on its recognition by p53 proteins. Structure 26, 1–14 (2018). [DOI] [PubMed] [Google Scholar]
- 71.Farkas M., et al. , Distinct mechanisms control genome recognition by p53 at its target genes linked to different cell fates. Nat. Commun. 12, 484 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Crothers D. M., Drak J., Kahn J. D., Levene S. D., DNA bending, flexibility, and helical repeat by cyclization kinetics. Methods Enzymol. 212, 3–29 (1992). [DOI] [PubMed] [Google Scholar]
- 73.Zhang Y., Crothers D. M., High-throughput approach for detection of DNA bending and flexibility based on cyclization. Proc. Natl. Acad. Sci. U.S.A. 100, 3161–3166 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lokshin M., Tanaka T., Prives C., Transcriptional regulation by p53 and p73. Cold Spring Harb. Symp. Quant. Biol. 70, 121–128 (2005). [DOI] [PubMed] [Google Scholar]
- 75.Hafner A., et al. , p53 pulses lead to distinct patterns of gene expression albeit similar DNA-binding dynamics. Nat. Struct. Mo.l Biol. 24, 840–847 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tsabar M., et al. , A switch in p53 Dynamics marks cells that escape from DSB-induced cell cycle arrest. Cell Rep. 33, 108392 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Porter J. R., Fisher B. E., Batchelor E., p53 pulses diversify Target gene expression dynamics in an mRNA half-life-dependent manner and delineate co-regulated target gene subnetworks. Cell Syst. 2, 272–282 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hanson R. L., Porter J. R., Batchelor E., Protein stability of p53 targets determines their temporal expression dynamics in response to p53 pulsing. J. Cell Biol. 218, 1282–1297 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y., Crothers D. M., Statistical mechanics of sequence-dependent circular DNA and its application for DNA cyclization. Biophys. J. 84, 136–153 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Coronel L., et al. , p53-mediated AKT and mTOR inhibition requires RFX7 and DDIT4 and depends on nutrient abundance. Oncogene 41, 1063–1069 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Coronel L., et al. , Transcription factor RFX7 governs a tumor suppressor network in response to p53 and stress. Nucleic Acids Res. 49, 7437–7456 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou X., Cao B., Lu H., Negative auto-regulators trap p53 in their web. J. Mol. Cell Biol. 9, 62–68 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Min S. H., et al. , New p53 target, phosphatase of regenerating liver 1 (PRL-1) downregulates p53. Oncogene 28, 545–554 (2009). [DOI] [PubMed] [Google Scholar]
- 84.Krzeszinski J. Y., et al. , XPC promotes MDM2-mediated degradation of the p53 tumor suppressor. Mol. Biol. Cell 25, 213–221 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gupta S., Stamatoyannopolous J. A., Bailey T., Noble W. S., Quantifying similarity between motifs. Genome Biol. 8, R24 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Data Availability Statement
All study data are included in the article and/or supporting information.