Abstract
In Escherichia coli and Salmonella typhimurium, methylation and demethylation of receptors are responsible for chemotactic adaptation and are catalyzed by the methyltransferase CheR and the methylesterase CheB, respectively. Among the chemoreceptors of these species, Tsr, Tar, and Tcp have a well-conserved carboxy-terminal motif (NWET/SF) that is absent in Trg and Tap. When they are expressed as sole chemoreceptors, Tsr, Tar, and Tcp support good adaptation, but Trg and Tap are poorly methylated and supported only weak adaptation. It was recently discovered that CheR binds to the NWETF sequence of Tsr in vitro. To examine the physiological significance of this binding, we characterized mutant receptors in which this pentapeptide sequence was altered. C-terminally-mutated Tar and Tcp expressed in a receptorless E. coli strain mediated responses to aspartate and citrate, respectively, but their adaptation abilities were severely impaired. Their expression levels and attractant-sensing abilities were similar to those of the wild-type receptors, but the methylation levels of the mutant receptors increased only slightly upon addition of attractants. When CheR was overproduced, both the adaptation and methylation profiles of the mutant Tar receptor became comparable to those of wild-type Tar. Furthermore, overproduction of CheR also enhanced adaptive methylation of wild-type Trg, which lacks the NWETF sequence, in the absence of any other chemoreceptor. These results suggest that the pentapeptide sequence facilitates effective adaptation and methylation by recruiting CheR.
Escherichia coli and Salmonella typhimurium migrate toward or away from certain chemicals by controlling the direction of flagellar rotation (for reviews, see references 3 and 28). This chemotactic behavior consists of two essential aspects, excitation and adaptation. Without adaptation, cells cannot detect temporal changes, and hence spatial gradients of concentrations of chemoeffectors, even though they can respond to their absolute concentrations.
Most chemoeffectors are detected by a small family of closely related chemoreceptors localized in the cytoplasmic membrane (for reviews, see reference 48 and the reviews cited above), including Tsr (for serine), Tar (for aspartate), Trg (for ribose and galactose), Tap (for dipeptide), and Tcp (for citrate). Tsr, Tar, and Trg are present in both species, and Tap and Tcp are specific to E. coli and S. typhimurium, respectively. Tar (and presumably any other chemoreceptor as well) is a homodimeric protein (30) comprising subunits about 60 kDa in size, each consisting of an N-terminal periplasmic ligand-binding domain, a C-terminal cytoplasmic signaling domain, and two membrane-spanning segments.
Methylation and demethylation of these chemoreceptors are the key processes of adaptation (for reviews, see reference 44 and the reviews cited above). The cytoplasmic domain of a receptor monomer contains four to six methylatable glutamic acid residues located in two separate regions that are predicted to form antiparallel α-helical coiled coils (23, 26, 35, 50). The consensus sequence for the methylation sites is Glu-Glu-X-X-Ala-Ser/Thr (18, 35, 38, 50). This sequence is likely to be recognized by the active site of the methyltransferase CheR that catalyzes transfer of a methyl group from S-adenosylmethionine to the side chain of the glutamic acid residue indicated by underlining above (45). The methyl ester bond of a methylated glutamate residue is hydrolyzed by the methylesterase CheB (47). Some of the methylation sites are encoded as glutamine residues, which CheB converts posttranslationally to methylatable glutamic acid residues (19).
Although there is no evidence for any regulation of CheR activity, the activity of CheB is stimulated when the protein is phosphorylated by the histidine kinase CheA (25, 46). The CheA homodimer forms a ternary complex with the receptor homodimer and two molecules of the adapter protein CheW (14, 40). Binding of a repellent or an attractant to the receptor activates or inactivates autophosphorylation of CheA and thereby increases or decreases phosphotransfer from CheA to its substrate proteins, CheY and CheB. Phospho-CheY induces clockwise rotation of the flagellar motor to cause the cell to tumble. When it is not bound to phospho-CheY, the motor rotates counterclockwise to power smooth swimming. By regulating CheB activity, an attractant or a repellent stimulus increases or decreases methylation of the receptor to attenuate the initial counterclockwise or clockwise signal, respectively. Such adaptation presumably results from conformational changes in the signaling domain caused by the methylation state of the receptor (4, 9, 33–36, 41), although the methylation helices are not essential for production of the chemotactic signal itself (1). A further constraint on the role of methylation in adaptation is that covalent modification does not dramatically change the ligand-binding affinity of the receptor (4, 9, 17, 24).
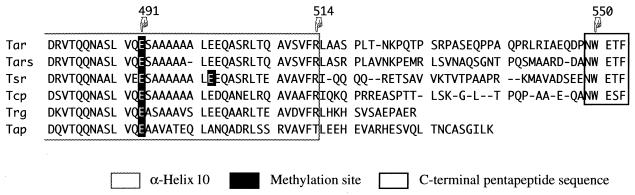
Among the chemoreceptors of E. coli and S. typhimurium, Trg and Tap are expressed at lower levels than the others (16, 42) and show weaker abilities to be methylated, to allow adaptation, and to support chemotaxis toward attractants when each of them is expressed as a sole chemoreceptor (12, 15, 29, 43, 51, 55). Trg and Tap are shorter and lack the C-terminal pentapeptide sequence (NWET/SF) that is highly conserved among Tsr, Tar, and Tcp (Fig. 1). A mutant form of S. typhimurium Tar, which has its C-terminal 35 residues removed and hence lacks the NWETF sequence, has been shown to be poorly methylated and not to support adaptation (39), and a mutant Tcp with an altered C-terminal sequence (NWESLA) also supports little adaptation (54). Recently, Wu et al. (53) demonstrated that CheR binds to E. coli Tsr in vitro at a 1:1 molar ratio through the NWETF sequence.
FIG. 1.
C-terminal amino acid sequences of chemoreceptors. The sequences of E. coli Tar, Tsr, Trg, and Tap and of S. typhimurium Tar (Tars) and Tcp as deduced from the nucleotide sequences are aligned. Gaps inserted to achieve the best match are indicated by dashes. The box with the solid black border indicates the C-terminal sequence conserved among Tar, Tsr, and Tcp, and the box with the gray border indicates the putative α-helical region (helix 10) proposed by Le Moual and Koshland (23). Methylation sites are indicated by white letters. Numbering of residues is for E. coli Tar.
To study the physiological significance of this pentapeptide sequence, we examined the effects of mutations in this sequence on various functions of E. coli Tar and S. typhimurium Tcp. We also examined the effects of CheR overproduction on the truncated Tar and on wild-type E. coli Trg. The results suggest that the C-terminal sequence, located far from the methylation sites in the primary sequence, facilitates effective receptor methylation by recruiting CheR to a receptor patch (27) that may enable trans methylation of a receptor dimer by CheR bound to another receptor dimer (53).
MATERIALS AND METHODS
Bacterial strains.
All strains used in this work are derivatives of E. coli K-12. Strain HCB339 [Δtsr-7021 Δ(tar-tap)5201 trg::Tn10 thr leu his met rpsL136] (52) lacks the four chemoreceptors, and strain CP553 [trg-100 Δtsr-7028 Δ(tar-cheB) leu his rpsL lac xyl ara tonA tsx thi zab::Tn5] (6) lacks CheB and CheR as well as the four chemoreceptors.
Plasmids.
A pBR322-based plasmid, pAK101 (21), carries the wild-type tar gene of E. coli (coding for wild-type Tar, i.e., Tar[NWETF]). Its derivative plasmid pAK101-W550Op (31), which carries a nonsense (opal) mutant tar gene (coding for Tar-W550Op, i.e., Tar[N]), was provided by K. Oosawa of Nagoya University. Another pAK101 derivative plasmid, pNI130 (32), carries the tar-Q295E,Q309E gene coding for Tar-EEEE, in which the two glutamine residues at the methylation sites are replaced by glutamate. A pACYC184-based plasmid, pRAR1 (32), carries the methyltransferase gene cheR and the chloramphenicol acetyltransferase gene cat (Cmr). Another pACYC184-based plasmid, pKB23 (4), in which the cheR gene is placed downstream of the tac promoter, was provided by M. I. Simon of California Institute of Technology. A pBR322-based plasmid, pCP31 (37), which carries the wild-type trg gene of E. coli, was provided by G. L. Hazelbauer of Washington State University.
Plasmids carrying the wild-type and mutant tcp genes of S. typhimurium were constructed as follows. Plasmid pAS101 carrying the wild-type tcp gene (coding for wild-type Tcp, i.e., Tcp[NWESF]) was constructed by subcloning the 6-kb EcoRI fragment of plasmid pKYP29 (54) into the unique EcoRI site of the vector pSU21 (2). Plasmid pAS103 carrying the tcp-F547LA gene (coding for Tcp-F547LA, i.e., Tcp[NWESLA]) was constructed by deleting the HindIII fragment from pAS101. Plasmid pAS103 was digested with HindIII, blunt-ended with T4 DNA polymerase, and ligated to yield plasmid pAS104 carrying the tcp-F547Am gene (coding for Tcp-F547Am, i.e., Tcp[NWES]) or ligated with the PacI 10mer linker (dGTTAATTAAC; New England BioLabs) to yield plasmid pAS105 carrying the tcp-F547C gene (coding for Tcp-F547C, i.e., Tcp[NWESC]). Plasmid pOKU106 carrying the tcp-F547A gene (coding for Tcp-F547A, i.e., Tcp[NWESA]) was obtained by site-directed mutagenesis by the method of Kunkel et al. (22). These mutations were verified by nucleotide sequencing.
Swarm assay.
Swarm assays were performed with tryptone semisolid agar (1% tryptone, 0.5% NaCl, 0.3% agar) or minimal semisolid agar [10 mM potassium phosphate buffer (pH 7.0), 1 mM (NH4)2SO4, 1 mM MgSO4, 1 mM glycerol, 1 mg of thiamine per ml, 0.1 mM threonine, 0.1 mM leucine, 0.1 mM histidine, 0.1 mM methionine] supplemented with 0.1 mM aspartate or 1 mM ribose. When necessary, 50 μg of ampicillin per ml, 25 μg of chloramphenicol per ml, and/or 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added. Cell suspensions (2 μl each, about 4 × 106 cells) were spotted onto a semisolid agar plate. The plate was then incubated at 30°C for 10 to 20 h.
Temporal stimulation assay.
Temporal stimulation assays for chemotaxis were carried out essentially as described previously (34). Cells were grown at 30°C in tryptone broth (1% tryptone, 0.5% NaCl) supplemented with 0.5% (wt/vol) glycerol and, when necessary, with 50 μg of ampicillin per ml, 25 μg of chloramphenicol per ml, and/or 1 mM IPTG. Cells were harvested in late exponential phase, washed with motility medium (10 mM potassium phosphate buffer [pH 7.0], 0.1 mM EDTA, 10 mM lactate, 0.1 mM methionine), resuspended in motility medium, and incubated at room temperature for 20 min. For dose-response assays, the swimming patterns of the cells were observed with a dark-field optical microscope immediately after the addition of a chemoeffector. For adaptation assays, a chemoeffector was added to the cell suspension and small aliquots were taken at intervals for microscopic observation. The smooth-swimming fraction (SSF) of the cells was measured with an Argus-10 image processor (Hamamatsu Photonics K.K., Hamamatsu, Shizuoka, Japan).
Immunoblotting.
Cells expressing a wild-type or mutant receptor were grown, harvested, and washed as described above (see “Temporal stimulation assay”). When necessary, chemoeffector was added to cells suspended in the motility medium and the suspension was incubated at room temperature for 20 min. The cells were collected by centrifugation and suspended in SDS loading buffer (67 mM Tris-HCl [pH 6.8], 8% glycerol, 1% sodium dodecyl sulfate [SDS], 0.003% bromophenol blue) supplemented with 7.7% 2-mercaptoethanol. Samples were boiled for 3 min and subjected to SDS-polyacrylamide gel electrophoresis. Proteins were transferred onto a polyvinylidene difluoride membrane (Millipore Japan, Tokyo) by using a semidry blotting apparatus (Biocraft, Tokyo, Japan). Anti-Tsr-T156C serum (17) was used as the first antibody, and alkaline phosphatase-conjugated goat anti-rabbit immunoglobulin G (Kirkegaard & Perry Laboratories, Gaithersburg, Md.) was used as the second antibody. The protein-antibody complexes were visualized in 5 ml of AP buffer (0.1 M Tris-HCl [pH 9.5], 1 M NaCl, 5 mM MgCl2) supplemented with 33 μl of nitroblue tetrazolium solution (50 mg of nitroblue tetrazolium per ml in 70% [vol/vol] dimethylformamide) and 16.5 μl of BCIP solution (50 mg of 5-bromo-4-chloro-3-indolylphosphate per ml in dimethylformamide).
RESULTS
Mutations in the C-terminal sequences of Tar and Tcp cause impaired adaptation abilities.
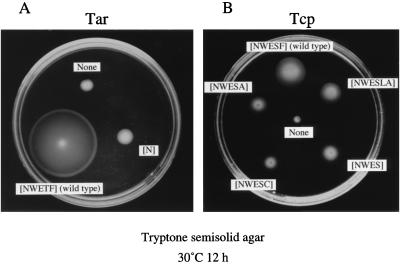
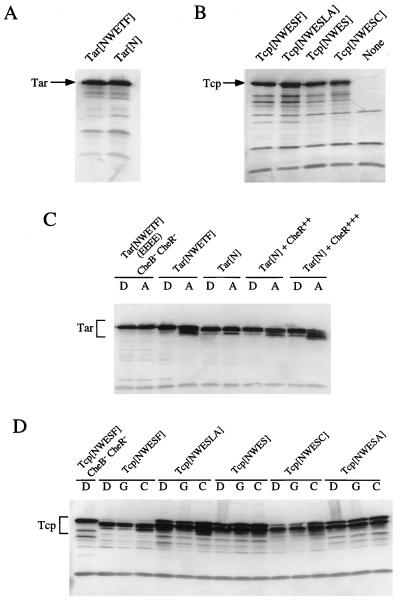
The nonsense mutant Tar protein Tar[N] does not allow the host strain RP4372 (tsr tar tap) to swarm in tryptone semisolid agar (31). The frameshift mutant Tcp protein Tcp[NWESLA] supports little adaptation in E. coli HCB339 (tsr tar tap trg) (54). We wanted to further characterize these mutant receptors as well as newly constructed mutant Tcp receptors (see Materials and Methods). Plasmids carrying the mutant tar or tcp genes were introduced into strain HCB399. The transformants were tested for their ability to swarm in tryptone semisolid agar (0.3%). Cells expressing the mutant Tar protein spread much slower than those expressing wild-type Tar (Fig. 2A). Similarly, cells expressing any mutant Tcp spread slower than cells expressing wild-type Tcp (Fig. 2B).
FIG. 2.
Swarming abilities of HCB339 cells expressing the mutant Tar (A) or Tcp (B) protein as a sole chemoreceptor. Two microliters of fresh overnight culture was spotted onto tryptone semisolid agar supplemented with 50 μg of ampicillin per ml (A) or 25 μg of chloramphenicol per ml (B), and the plates were then incubated at 30°C for 12 h. (A) Tar[NWETF] (wild type), cells carrying pAK101; Tar[N], cells carrying pAK101-W550Op; None, cells carrying pBR322. (B) Tcp[NWESF], (wild type), cells carrying pAS101; Tcp[NWESLA], cells carrying pAS103; Tcp[NWES], cells carrying pAS104; Tcp[NWESC], cells carrying pAS105; Tcp[NWESA], cells carrying pOKU106; None, cells carrying pSU21.
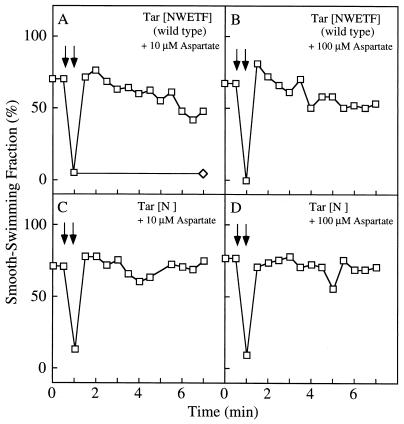
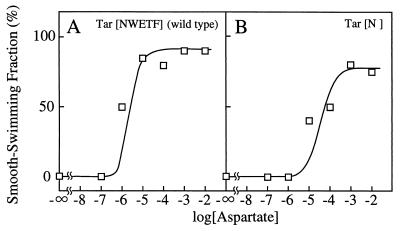
We then examined chemotactic responses directly by a temporal stimulation assay. The SSF of HCB339 cells expressing wild-type Tar was about 70% without chemotactic stimuli (Fig. 3A and B). When the repellent 10% glycerol was added, the SSF decreased to almost nothing, a response that persisted for at least 8 min. Immediately after 10 or 100 μM aspartate was added to cells tumbling in response to glycerol, the SSF jumped to 80% but then began to decrease gradually (Fig. 3A and B). This gradual decrease in SSF was not observed for cells expressing mutant Tar. They gave a persistent SSF in response to aspartate, demonstrating that the mutant Tar is defective in adaptation (Fig. 3C and D).
FIG. 3.
Adaptation of HCB339 cells expressing the mutant Tar protein. At the time indicated by the first downward arrow, glycerol was added to 10% (◊ and □), and at the time indicated by the second downward arrow, aspartate was added at 10 (A and C) or 100 (B and D) μM (□). (A and B) Tar[NWETF] (wild type), cells carrying pAK101; (C and D) Tar[N], cells carrying pAK101-W550Op.
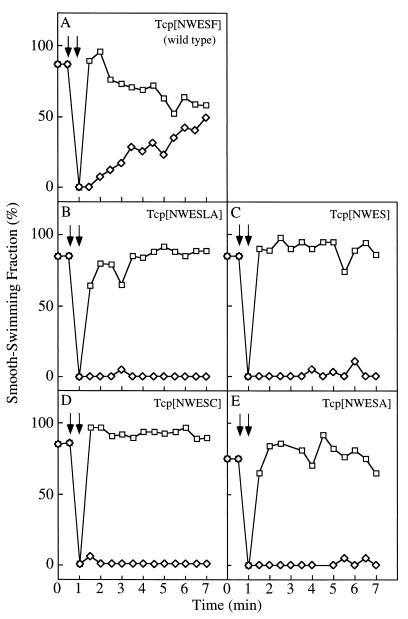
The SSF of cells expressing wild-type Tcp was about 80% without stimulation (Fig. 4). The addition of 10% glycerol reduced the SSF to almost nothing, but the SSF increased gradually to 40% after 7 min. Immediately after 10 mM citrate was added, the SSF increased to almost 100% and then decreased gradually to reach about 60% after 7 min. By contrast, cells expressing mutant Tcp proteins produced little or no change in the SSF after the initial response to glycerol or citrate (Fig. 4B to E). These results suggest that mutations in the C-terminal pentapeptide sequence of Tcp resulted in impaired adaptation after both attractant and repellent responses.
FIG. 4.
Adaptation of HCB339 cells expressing each mutant Tcp protein. Assays were carried out as described in the legend to Fig. 3, using 1 mM citrate instead of aspartate. (A) Tcp[NWESF] (wild type), cells carrying pAS101; (B) Tcp[NWESLA], cells carrying pAS103; (C) Tcp[NWES], cells carrying pAS104; (D) Tcp[NWESC], cells carrying pAS105; (E) Tcp[NWESA], cells carrying pOKU106.
Attractant-sensing abilities and expression levels of mutant Tar and Tcp proteins.
Impaired adaptation in the mutants might be an indirect result of an increased affinity to a ligand or increased signaling strength, either of which could mask adaptation. We therefore examined the dose-response characteristics of the mutant Tar (Fig. 5) and Tcp (data not shown) proteins. The swimming behavior of cells expressing each mutant receptor was observed just after the addition of various concentrations of an attractant. When 10% glycerol was added, the SSF of cells expressing either wild-type or mutant receptor was insignificant. Cells expressing wild-type Tar began to respond to 10−7 M aspartate and gave a maximum response at 10−5 M or higher concentrations of aspartate (Fig. 5A). The mutant Tar had a slightly weaker apparent affinity for aspartate than wild-type Tar (Fig. 5B). Cells expressing wild-type Tcp began to respond at 10−5 M citrate and showed a maximum response at 10−3 M or higher concentrations of citrate (data not shown). The concentrations of citrate required for attractant responses mediated by mutant Tcp proteins were similar to or slightly higher than those for wild-type Tcp (data not shown). These results indicate that the mutations in the C-terminal sequences did not increase ligand-binding affinity or signaling.
FIG. 5.
Dose-response relationship for the wild-type and mutant Tar proteins. HCB339 cells expressing the wild-type or mutant receptor were pretreated with 10% glycerol to reduce the SSF and were then exposed to various concentrations of aspartate. The SSF was measured within 1 min. (A) Tar[NWETF] (wild type), cells carrying pAK101; (B) Tar[N], cells carrying pAK101-W550Op.
We also examined the expression levels of the mutant receptors by immunoblotting with anti-Tsr serum (17), which cross-reacts with both Tar and Tcp. Strain CP553, which lacks the methyltransferase CheR, the methylesterase CheB, and the four chemoreceptors, was used as the plasmid host. Receptor proteins in this strain exist as single species because of the absence of posttranslational methylation or deamidation, a circumstance that makes comparison of their expression levels easier. Tar[N] was expressed at almost the same level as wild-type Tar (Fig. 6A). Tcp[NWESLA] and Tcp[NWESC] appeared to form slightly more intense and slightly less intense bands, respectively, than wild-type Tcp (Fig. 6B). The bands formed by all of the other mutant Tcp proteins were of almost the same intensity as that of wild-type Tcp (Fig. 6B; data for Tcp[NWESA] not shown). These results suggest that the mutations in the C-terminal sequences did not dramatically affect synthesis or degradation of the receptor proteins.
FIG. 6.
Expression levels (A and B) and methylation patterns (C and D) of the mutant Tar (A and C) and Tcp (B and D) proteins. (A and B) The wild-type or mutant receptors were expressed in the CheB− CheR− strain CP553 and detected by immunoblotting with anti-Tsr serum. Tar[NWETF] (wild type), cells carrying pAK101; Tar[N], cells carrying pAK101-W550Op; Tcp[NWESF] (wild type), cells carrying pAS101; Tcp[NWESLA], cells carrying pAS103; Tcp[NWES], cells carrying pAS104; Tcp[NWESC], cells carrying pAS105; None, cells carrying pSU21. (C and D) Cells expressing each receptor were preincubated with distilled deionized water (lanes D), 10 mM aspartate (lanes A), 10 mM citrate (lanes C), or 10% glycerol (lanes G). Chemoreceptors were detected as described for panels A and B. Tar[NWETF] (EEEE) CheB− CheR−, CP553 cells carrying pNI130; Tar[NWETF], HCB339 cells carrying pAK101; Tar[N], HCB339 cells carrying pAK101-W550Op; Tar[N] + CheR++, HCB339 cells carrying pAK101-W550Op and pRAR1 (a multicopy plasmid bearing cheR); Tar[N] + CheR+++, HCB339 cells carrying pAK101-W550Op and pKB23 (a CheR− overproducer plasmid), induced for CheR expression with 1 mM IPTG; Tcp[NWESF] CheB− CheR−, CP553 cells carrying pAS101; Tcp[NWESF], HCB339 cells carrying pAS101; Tcp[NWESLA], HCB339 cells carrying pAS103; Tcp[NWES], HCB339 cells carrying pAS104; Tcp[NWESC], HCB339 cells carrying pAS105; Tcp[NWESA], HCB339 cells carrying pOKU106.
Methylation patterns of mutant Tar and Tcp proteins.
Methylation levels in the CheB+ CheR+ background were monitored by immunoblotting. Methylation of a receptor increases its mobility in SDS-polyacrylamide gel electrophoresis (5, 7, 8, 10). In the absence of aspartate, the methylation level of the mutant Tar protein was comparable to that of wild-type Tar (Fig. 6C). However, methylation of the mutant Tar was stimulated only slightly by aspartate. In the case of Tcp (Fig. 6D), all of the mutant proteins were significantly less methylated than wild-type Tcp in the absence of any chemoeffector (i.e., upper bands appeared). This seemed specific to Tcp, because the unstimulated methylation level of Tar[NWETA] was similar to that of Tar[N] (unpublished results). Their methylation levels increased little or only slightly when cells were incubated with citrate (NWESLA > NWES > NWESC ≈ NWESA). These results suggest that the mutations in the C-terminal sequence severely affect the attractant-stimulated methylation.
Suppression of the defect in adaptation and methylation of mutant Tar by overproduction of CheR.
While this work was in progress, in vitro results demonstrating that CheR binds to the NWETF sequence of Tsr were published (53). If the C-terminally-mutated receptors have weaker affinities to CheR, their lowered methylation levels might be increased by overproduction of CheR. Alternatively, the mutant receptors could be poor substrates for CheR, in which case overproduction of CheR would not compensate for loss of the pentapeptide sequence. We examined these possibilities with the mutant Tar protein.
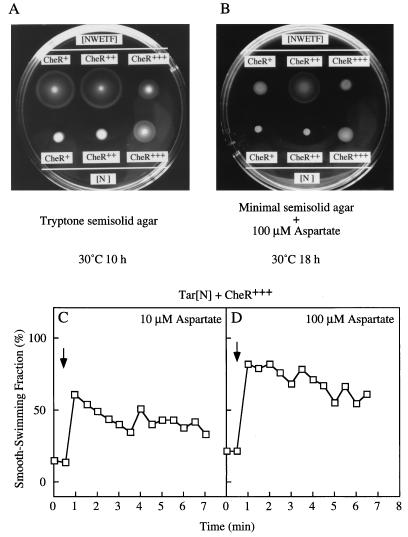
We examined whether CheR overproduction suppresses the defect in adaptation of the mutant Tar protein. Cells expressing mutant Tar were transformed with a CheR-expressing (pRAR1) or a CheR-overproducing (pKB23) plasmid. In both tryptone and aspartate-minimal semisolid agar, the swarming ability of HCB339 cells expressing the mutant Tar protein was enhanced by overproduction of CheR from the induced tac promoter (Fig. 7A and B; compare results for Tar[N] + CheR+ and Tar[N] + CheR+++). The resulting chemotactic rings were of comparable size to those of cells expressing wild-type Tar and containing the wild-type level of CheR (Fig. 7A and B; compare results for Tar[N] + CheR+++ and Tar [NWETF] + CheR+).
FIG. 7.
Effects of CheR overproduction on swarming (A and B) and adaptation (C and D) supported by mutant Tar. (A and B) Two microliters of fresh overnight culture of HCB339 cells carrying Tar-expressing and CheR-expressing plasmids was spotted onto tryptone semisolid agar or minimal semisolid agar containing 100 μM aspartate supplemented with ampicillin, chloramphenicol, and IPTG, and the plates were incubated at 30°C for 10 (A) or 18 (B) h. Receptor proteins and the plasmids bearing the genes encoding them: Tar[NWETF] (wild type), pAK101; Tar[N], pAK101-W550Op; CheR expression levels and the plasmid introduced: CheR+ (wild-type level), pACYC184 (vector); CheR++ (overproduced), pRAR1 (a multicopy plasmid bearing cheR); CheR+++ (highly overproduced), pKB23 (a CheR-overproducing plasmid). (C and D) Chemotactic responses to 10 (C) or 100 (D) μM aspartate were measured as described in the legend to Fig. 3 but without pretreatment with glycerol.
Adaptation times also were determined for cells overproducing CheR (Fig. 7C and D). Without any chemoeffector, the SSF of strain HCB339 expressing mutant Tar and overproducing CheR was only about 20%, although the resting methylation level was not much changed (see below). The reason for this reduced SSF is not clear, but one possibility may be that some stress results from carrying two different multicopy plasmids. The SSF reached 60 and 80% immediately after the addition of 10 and 100 μM aspartate, respectively, and then decreased gradually. These results suggest that the defect in adaptation in the mutant was suppressed by CheR overproduction.
Receptor methylation was then monitored by immunoblotting (Fig. 6C). With increasing levels of expression of CheR, the receptor became more methylated in the presence of aspartate, whereas the “resting” or unstimulated methylation level did not change very much. In cells carrying pKB23, in which CheR expression was induced by 1 mM IPTG, the aspartate-stimulated methylation level of the mutant Tar was comparable to that of wild-type Tar. These results suggest that impaired methylation was due to decreased affinity of the mutant Tar for CheR. Therefore, the pentapeptide sequence is likely to function as a CheR-binding site in vivo, and binding of CheR to this site is not a prerequisite for adaptive methylation.
Effects of CheR overproduction on the functions of wild-type Trg.
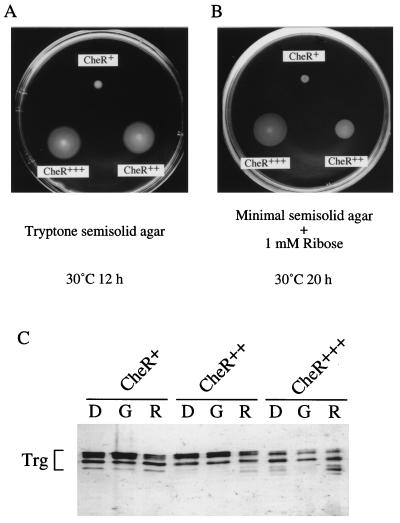
We also examined effects of CheR overproduction on methylation and adaptation of wild-type Trg, which naturally lacks the NWETF sequence. Both in tryptone and in ribose-minimal semisolid agar, cells expressing Trg as sole chemoreceptors swarmed better when CheR was overproduced from the tac promoter than when CheR was not overproduced (Fig. 8A and B). Overproduction of CheR enhanced stimulation of methylation by the Trg-specific attractant ribose and slightly increased methylation of unstimulated Trg (Fig. 8C). These results suggest that the weak adaptation and methylation phenotypes of wild-type Trg are mainly due to the lack of the primary CheR-binding site, a deficit which can be overcome by CheR overproduction.
FIG. 8.
Effects of CheR overproduction on swarming (A and B) and methylation (C) supported by wild-type Trg. HCB339 cells carrying pCP31 (trg+) were transformed with plasmid pACYC184 (vector; CheR+), pRAR1 (a multicopy plasmid bearing cheR; CheR++), or pKB23 (a CheR-overproducing plasmid; CheR+++). (A and B) Swarming was assayed as described in the legend to Fig. 7, using 10 mM ribose instead of aspartate. (C) Receptor methylation was assayed as described in the legend to Fig. 6, using 10 mM ribose (R) as an attractant. Lanes labeled D and G are defined in the legend to Fig. 6.
DISCUSSION
We here investigated the effects of mutations altering the C-terminal NWET/SF sequence which is conserved among the major chemoreceptors of E. coli and S. typhimurium (Tsr, Tar, and Tcp) but not among the minor chemoreceptors (Trg and Tap). The recent in vitro study showed that the NWETF sequence of Tsr is a primary site for CheR binding (53). Consistent with this, mutations in Tar and Tcp impaired their adaptation and methylation abilities without much affecting their cellular levels and apparent ligand-binding affinities. Overproduction of CheR suppressed the adaptation- and methylation-defective phenotypes of the mutant Tar protein as well as those of wild-type Trg. These in vivo results demonstrate that the NWET/SF sequence is not a prerequisite for receptor methylation itself but suggest that the sequence facilitates effective adaptive methylation by recruiting CheR.
Although aspartate-stimulated methylation of Tar[N] was restored by overproduction of CheR, its unstimulated methylation level was almost the same as that of cells expressing the normal level of chromosomally encoded CheR (Fig. 6C). Thus, the NWETF sequence might be responsible for allowing the high initial rate of methylation upon stimulation with an attractant but dispensable for the resting steady-state level of methylation. The situation is similar for wild-type Trg, but its unstimulated methylation levels increased slightly with increasing levels of CheR (Fig. 8A). This difference might be due to either the absence of more of the carboxy-terminal sequence of the major receptors from wild-type Trg (Fig. 1) than from Tar[N] or to other intrinsic differences between Trg and Tar.
Wu et al. (53) proposed that CheR bound to a receptor dimer may catalyze methylation of a neighboring receptor dimer. This interdimer methylation model can explain why Trg is more efficiently methylated when Tsr and/or Tar is present than when Trg is expressed alone (20, 55). The possibility of interdimer interaction in signal production has also been argued, since Tar dimers with only one intact cytoplasmic domain have attractant signaling abilities (13, 49). Although these possibilities remain to be tested experimentally, it is quite possible that receptor dimers encounter each other in the cytoplasmic membrane, because the receptors have been shown to cluster at the poles of E. coli cells (27).
Unlike the situation with CheB, there is no evidence that the activity of CheR is regulated. Identification of a CheR-binding site on the major receptors distinct from their methylation sites suggests the following possible mechanisms for regulation of CheR activity, however: (i) CheR might associate with the receptor under some conditions and dissociate from it under others; (ii) ligand binding might change the configuration of bound CheR relative to the methylation sites (possibly in another receptor dimer as discussed above). Alternatively, the NWETF sequence might exist only to provide a docking site for CheR. Recruitment of CheR to the receptor will increase the local concentration of the enzyme and should thereby increase the probability of its collision with the methylation sites.
In any case, a multiprotein complex containing the receptor, CheA, CheW, and CheR clearly plays a central role in chemotactic signal transduction. Schuster et al. (40) showed that CheY binds stably to the receptor-CheW-CheA ternary complex and that CheY is released from the complex when phosphorylated. It is reasonable to speculate that CheB behaves in a similar fashion. Thus, the whole chemotactic signal transduction system is apt to be spatially well organized rather than to consist of freely diffusible components encountering each other stochastically.
Such higher-order structures have been implicated in eukaryotic signal transduction. For example, the yeast STE5 protein serves as a scaffold for members of the mitogen-activated protein kinase cascade, and the mammalian A-kinase anchoring protein AKAP79 tethers the type II cyclic AMP-dependent protein kinase, protein kinase C, and calcineurin to the postsynaptic densities (11). These complexes may guarantee rapid and discrete flows of information by placing their components in the correct sequence to promote signal transduction and to prevent cross talk between homologous but distinct systems.
ACKNOWLEDGMENTS
We thank G. L. Hazelbauer, K. Oosawa, and M. I. Simon for providing us with plasmids. We especially thank M. D. Manson of Texas A & M University for critically reading the manuscript.
This work was supported in part by a grant-in-aid for scientific research to I.K. from the Ministry of Education, Science, and Culture of Japan.
ADDENDUM IN PROOF
Recently, Li et al. (J. Li, G. Li, and R. M. Weis, Biochemistry 36:11851–11857, 1997) and Le Moual and Koshland (H. Le Moual and D. E. Koshland, Jr., Biochemistry 36:13441–13448, 1997) demonstrated interdimer methylation by in vitro assays.
REFERENCES
- 1.Ames P, Yu Y A, Parkinson J S. Methylation segments are not required for chemotactic signaling by cytoplasmic fragments of Tsr, the methyl-accepting serine chemoreceptor of Escherichia coli. Mol Microbiol. 1996;19:737–746. doi: 10.1046/j.1365-2958.1996.408930.x. [DOI] [PubMed] [Google Scholar]
- 2.Bartolomé B, Jubete Y, Martinez E, Cruz F D. Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives. Gene. 1991;102:75–78. doi: 10.1016/0378-1119(91)90541-i. [DOI] [PubMed] [Google Scholar]
- 3.Blair D F. How bacteria sense and swim. Annu Rev Microbiol. 1995;49:489–522. doi: 10.1146/annurev.mi.49.100195.002421. [DOI] [PubMed] [Google Scholar]
- 4.Borkovich K A, Alex L A, Simon M I. Attenuation of sensory receptor signaling by covalent modification. Proc Natl Acad Sci USA. 1992;89:6756–6760. doi: 10.1073/pnas.89.15.6756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd A, Simon M I. Multiple electrophoretic forms of methyl-accepting chemotaxis proteins generated by stimulus-elicited methylation in Escherichia coli. J Bacteriol. 1980;143:809–815. doi: 10.1128/jb.143.2.809-815.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burrows G G, Newcomer M E, Hazelbauer G L. Purification of receptor protein Trg by exploiting a property. J Biol Chem. 1989;264:17309–17315. [PubMed] [Google Scholar]
- 7.Chelsky D, Dahlquist F W. Structural studies of methyl-accepting chemotaxis proteins of Escherichia coli: evidence for multiple methylation sites. Proc Natl Acad Sci USA. 1980;77:2434–2438. doi: 10.1073/pnas.77.5.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeFranco A L, Koshland D E., Jr Multiple methylation in processing of sensory signals during bacterial chemotaxis. Proc Natl Acad Sci USA. 1980;77:2429–2433. doi: 10.1073/pnas.77.5.2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunten P, Koshland D E., Jr Tuning the responsiveness of a sensory receptor via covalent modification. J Biol Chem. 1991;266:1491–1496. [PubMed] [Google Scholar]
- 10.Engström P, Hazelbauer G L. Multiple methylation of methyl-accepting chemotaxis proteins during adaptation of E. coli to chemical stimuli. Cell. 1980;20:165–171. doi: 10.1016/0092-8674(80)90244-5. [DOI] [PubMed] [Google Scholar]
- 11.Faux M C, Scott J D. Molecular glue: kinase anchoring and scaffold proteins. Cell. 1996;85:9–12. doi: 10.1016/s0092-8674(00)81075-2. [DOI] [PubMed] [Google Scholar]
- 12.Feng X, Baumgartner J W, Hazelbauer G L. High- and low-abundance chemoreceptors in Escherichia coli: differential activities associated with closely related cytoplasmic domains. J Bacteriol. 1997;179:6714–6720. doi: 10.1128/jb.179.21.6714-6720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gardina P J, Manson M D. Attractant signaling by an aspartate chemoreceptor dimer with a single cytoplasmic domain. Science. 1996;274:425–426. doi: 10.1126/science.274.5286.425. [DOI] [PubMed] [Google Scholar]
- 14.Gegner J A, Graham D R, Roth A F, Dahlquist F W. Assembly of an MCP, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell. 1992;70:975–982. doi: 10.1016/0092-8674(92)90247-a. [DOI] [PubMed] [Google Scholar]
- 15.Hazelbauer G L, Engström P. Parallel pathways for transduction of chemotactic signals in Escherichia coli. Nature. 1980;283:98–100. doi: 10.1038/283098a0. [DOI] [PubMed] [Google Scholar]
- 16.Hazelbauer G L, Engström P, Harayama S. Methyl-accepting chemotaxis protein III and transducer gene trg. J Bacteriol. 1981;145:43–49. doi: 10.1128/jb.145.1.43-49.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iwama T, Homma M, Kawagishi I. Uncoupling of ligand-binding affinity of the bacterial serine chemoreceptor from methylation- and temperature-modulated signaling states. J Biol Chem. 1997;272:13810–13815. doi: 10.1074/jbc.272.21.13810. [DOI] [PubMed] [Google Scholar]
- 18.Kehry M R, Engström P, Dahlquist F W, Hazelbauer G L. Multiple covalent modifications of Trg, a sensory transducer of Escherichia coli. J Biol Chem. 1983;258:5050–5055. [PubMed] [Google Scholar]
- 19.Kehry M R, Bond M W, Hunkapiller M W, Dahlquist F W. Enzymatic deamidation of methyl-accepting chemotaxis proteins in Escherichia coli catalyzed by the cheB gene product. Proc Natl Acad Sci USA. 1983;80:3599–3603. doi: 10.1073/pnas.80.12.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondoh H, Ball C B B, Adler J. Identification of a methyl-accepting chemotaxis protein for the ribose and galactose chemoreceptors of Escherichia coli. Proc Natl Acad Sci USA. 1979;76:260–264. doi: 10.1073/pnas.76.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krikos A, Conley M P, Boyd A, Berg H C, Simon M I. Chimeric chemosensory transducers of Escherichia coli. Proc Natl Acad Sci USA. 1985;82:1326–1330. doi: 10.1073/pnas.82.5.1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunkel T A, Roberts J D, Zokour R A. Rapid and efficient site-specific mutagenesis without phenotype selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 23.Le Moual H, Koshland D E., Jr Molecular evolution of the C-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J Mol Biol. 1996;261:568–585. doi: 10.1006/jmbi.1996.0483. [DOI] [PubMed] [Google Scholar]
- 24.Lin L-N, Li J, Brandts J F, Weis R M. The serine receptor of bacterial chemotaxis exhibits half-of-site saturation for serine binding. Biochemistry. 1994;33:6564–6570. doi: 10.1021/bi00187a025. [DOI] [PubMed] [Google Scholar]
- 25.Lupas A, Stock J. Phosphorylation of an N-terminal regulatory domain activates the CheB methyltransferase in bacterial chemotaxis. J Biol Chem. 1989;264:17337–17342. [PubMed] [Google Scholar]
- 26.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 27.Maddock J R, Shapiro L. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science. 1993;259:1717–1723. doi: 10.1126/science.8456299. [DOI] [PubMed] [Google Scholar]
- 28.Manson M D. Bacterial motility and chemotaxis. Adv Microb Physiol. 1992;33:277–346. doi: 10.1016/s0065-2911(08)60219-2. [DOI] [PubMed] [Google Scholar]
- 29.Manson M D, Blank V, Brade G, Higgins C F. Peptide chemotaxis in E. coli involves the Tap signal transducer and the peptide permease. Nature. 1986;321:253–256. doi: 10.1038/321253a0. [DOI] [PubMed] [Google Scholar]
- 30.Milligan D L, Koshland D E., Jr Site-directed cross linking: establishing the dimeric structure of the aspartate receptor of bacterial chemotaxis. J Biol Chem. 1988;263:6268–6275. [PubMed] [Google Scholar]
- 31.Mutoh N, Oosawa K, Simon M I. Characterization of Escherichia coli chemotaxis receptor mutants with null phenotypes. J Bacteriol. 1986;167:992–998. doi: 10.1128/jb.167.3.992-998.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nara T, Kawagishi I, Nishiyama S, Homma M, Imae Y. Modulation of the thermosensing profile of the Escherichia coli aspartate receptor Tar by covalent modification of the methyl-accepting site. J Biol Chem. 1996;271:17932–17936. doi: 10.1074/jbc.271.30.17932. [DOI] [PubMed] [Google Scholar]
- 33.Ninfa E G, Stock A, Mowbray S, Stock J. Reconstitution of the bacterial chemotaxis signal transduction system from purified components. J Biol Chem. 1991;266:9764–9770. [PubMed] [Google Scholar]
- 34.Nishiyama S, Nara T, Imae Y, Homma M, Kawagishi I. Thermosensing properties of mutant aspartate receptors having methyl-accepting sites substituted multiply or singly with alanine. J Bacteriol. 1997;179:6573–6580. doi: 10.1128/jb.179.21.6573-6580.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nowlin D M, Bollinger J, Hazelbauer G L. Site-directed mutations altering methyl-accepting residues of a sensory transducer protein. Proteins. 1988;3:102–112. doi: 10.1002/prot.340030205. [DOI] [PubMed] [Google Scholar]
- 36.Park C, Dutton D P, Hazelbauer G L. Effects of glutamines and glutamates at sites of covalent modification of a methyl-accepting transducer. J Bacteriol. 1990;172:7179–7187. doi: 10.1128/jb.172.12.7179-7187.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park C, Hazelbauer G L. Transfer of chromosomal mutations to plasmids via Hfr-mediated conduction. J Bacteriol. 1986;165:312–314. doi: 10.1128/jb.165.1.312-314.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rice M S, Dahlquist F W. Sites of deamidation and methylation in Tsr, a bacterial chemotaxis sensory transducer. J Biol Chem. 1991;266:9746–9753. [PubMed] [Google Scholar]
- 39.Russo A F, Koshland D E., Jr Separation of signal transduction and adaptation functions of the aspartate receptor in bacterial sensing. Science. 1983;220:1016–1020. doi: 10.1126/science.6302843. [DOI] [PubMed] [Google Scholar]
- 40.Schuster S C, Swanson R V, Alex L A, Bourret R B, Simon M I. Assembly and function of a quaternary signal transduction complex monitored by surface plasmon resonance. Nature. 1993;365:343–347. doi: 10.1038/365343a0. [DOI] [PubMed] [Google Scholar]
- 41.Shapiro M J, Chakrabarti I, Koshland D E., Jr Contributions made by individual methylation sites of the Escherichia coli aspartate receptor to chemotactic behavior. Proc Natl Acad Sci USA. 1995;92:1053–1056. doi: 10.1073/pnas.92.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Slocum M K, Parkinson J S. Genetics of methyl-accepting chemotaxis proteins in Escherichia coli: organization of the tar region. J Bacteriol. 1983;155:565–577. doi: 10.1128/jb.155.2.565-577.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Springer M S, Goy M F, Adler J. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc Natl Acad Sci USA. 1977;74:3312–3316. doi: 10.1073/pnas.74.8.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Springer M S, Goy M F, Adler J. Protein methylation in behavioural control mechanisms and in signal transduction. Nature. 1979;280:279–284. doi: 10.1038/280279a0. [DOI] [PubMed] [Google Scholar]
- 45.Springer W R, Koshland D E., Jr Identification of a protein methyltransferase as the cheR gene product in the bacterial sensing system. Proc Natl Acad Sci USA. 1977;74:533–537. doi: 10.1073/pnas.74.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stock A M, Wylie D C, Mottonen J M, Lupas A M, Ninfa E G, Ninfa A J, Schutt C E, Stock J B. Phospho-proteins involved in bacterial signal transduction. Cold Spring Harbor Symp Quant Biol. 1988;53:49–57. doi: 10.1101/sqb.1988.053.01.009. [DOI] [PubMed] [Google Scholar]
- 47.Stock J B, Koshland D E., Jr A protein methylesterase involved in bacterial sensing. Proc Natl Acad Sci USA. 1978;75:3659–3663. doi: 10.1073/pnas.75.8.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stock J B, Surette M G. Chemotaxis. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 1103–1129. [Google Scholar]
- 49.Tatsuno I, Oosawa K, Homma M, Kawagishi I. Signaling by the Escherichia coli aspartate chemoreceptor Tar with a single cytoplasmic domain per dimer. Science. 1996;274:423–425. doi: 10.1126/science.274.5286.423. [DOI] [PubMed] [Google Scholar]
- 50.Terwilliger T C, Wang J Y, Koshland D E., Jr Surface structure recognized for covalent modification of the aspartate receptor in chemotaxis. Proc Natl Acad Sci USA. 1986;83:6707–6710. doi: 10.1073/pnas.83.18.6707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weerasuriya S, Schneider B M, Manson M D. Chimeric chemoreceptors in Escherichia coli: signaling properties of Tar-Tap and Tap-Tar hybrids. J Bacteriol. 1998;180:914–920. doi: 10.1128/jb.180.4.914-920.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wolfe A J, Conley M P, Kramer T J, Berg H C. Reconstitution of signaling in bacterial chemotaxis. J Bacteriol. 1987;169:1878–1885. doi: 10.1128/jb.169.5.1878-1885.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wu J, Li J, Li G, Long D G, Weis R M. The receptor binding site for the methyltransferase of bacterial chemotaxis is distinct from the sites of methylation. Biochemistry. 1996;35:4984–4993. doi: 10.1021/bi9530189. [DOI] [PubMed] [Google Scholar]
- 54.Yamamoto K, Imae Y. Cloning and characterization of the Salmonella typhimurium-specific chemoreceptor Tcp for taxis to citrate and from phenol. Proc Natl Acad Sci USA. 1993;90:217–221. doi: 10.1073/pnas.90.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamamoto K, Macnab R M, Imae Y. Repellent response functions of the Trg and Tap chemoreceptors of Escherichia coli. J Bacteriol. 1990;172:383–388. doi: 10.1128/jb.172.1.383-388.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]