Abstract
BACKGROUND:
One branch of the RHD phylogenetic tree is represented by the weak D type 4 cluster of alleles with F223V as the primordial amino acid substitution. F223V as well as a large number of further substitutions causing D variants are located at the extracellular RhD protein vestibule, which represents the entrance to the transmembraneous channel of the RhD protein.
STUDY DESIGN AND METHODS:
RHD and RHCE nucleotide sequences were determined from genomic DNA and cDNA. D epitope patterns were established with commercial monoclonal anti-D panels.
RESULTS:
The RHD alleles DOL-1 and DOL-2 had the two amino acid substitutions M170T (509T>C) and F223V (667T>G) in common. DOL-2 harbored the additional substitution L378V (1132C>G). Both alleles were observed in Africans and are probably evolutionary related. DMI carried M170I (510G>A), which differed from the DOL-typical substitution. DFW and DFL harbored the substitutions H166P (497A>C) and Y165C (494A>G). The antigen densities of DOL-1, DFL, and DFW were only moderately reduced.
CONCLUSION:
DOL-1 and DOL-2 belong to the weak D type 4 cluster of RHD alleles. Together with DMI, DFL, and DFW they represent D variants with amino acid substitutions located at extracellular loops 3 or 4 lining the RhD protein vestibule. These substitutions were of minor influence on antigen density while adjacent substitutions in the transmembraneous section caused weak D antigen expression. All these D variants were partial D and alloanti-D immunizations have been observed in DOL-1, DMI, and DFL carriers. The substitution at position 170 causes partial D although located deep in the vestibule.
A small fraction of D+ individuals have the potential to produce anti-D after exposure to D+ blood during transfusion or pregnancy. This apparent paradox was explained when the molecular bases underlying partial D were investigated.1 Findings at the genetic level confirmed the hypothesis of Tippett and coworkers2 that partial D phenotypes arise where part of the D antigen “mosaic” is lacking and that exposure to the complete D antigen could elicit an immune response to the missing parts of the mosaic. It was found that many partial D are variants of the prevalent RhD protein caused by single point mutations or by gene conversions between the RHCE and RHD genes. However, several partial D did not fit into this picture, because they harbor multiple point mutations that are dispersed throughout the RHD gene. The compilation of these alleles has been instrumental to construct an evolutionary tree of RHD alleles.3
The present phylogenetic model of RHD in humans discerns four allele clusters: the Eurasian D cluster with “normal” RHD as the primordial allele from which numerous alleles derive as well as three “African” groups of alleles designated DIVa, DAU, and weak D type 4 clusters.4–6 Each of these African clusters is characterized by a distinct primordial amino acid substitution relative to the normal RHD allele: T379M in the DAU cluster, N152T in the DIVa cluster, and F223V in the weak D type 4 cluster. The existence of the allele DFV, corresponding to the single-substitution F223V, has been postulated4 long before it was actually found in individuals of different ethnic origin. It represents the primordial allele of the weak D type 4 cluster, which comprises many clinically relevant alleles like weak D type 4.0, 4.1, and 4.2 (DAR) and RHDΨ.
We describe several partial D carrying amino acid substitutions at positions 170 and 223, which are located at the extracellular RhD protein vestibule. The African partial D DOL-17–10 and DOL-2 harbor the F223V substitutions in a cDe haplotype, which qualifies them as members of the weak D type 4 cluster. Both DOL types also carry the amino acid substitution M170T. These substitutions are located at the RhD vestibule, which represents the extracellular entrance to the transmembraneous protein channel recognized by homology modeling.11 The presently described DMI (M170I) and the known DFV (F223V), DCS-1 (F223V A226P), and DFR-1 (M169L, M170R, I172F) harbor substitutions at the amino acid positions 170 or 223. Furthermore, we describe the alleles DFL12,13 and DFW14 with single substitutions at positions 165 and 166, because of their proximity to position 170. We found that individuals carrying amino acid substitutions at the extracellular RhD protein vestibule were prone to making alloanti-D even if their substitution seemed to be located deep in the vestibule.
MATERIALS AND METHODS
Immunohematology
Serologic testing for agglutination was done by tube test with low-ionic-strength saline (LISS) and indirect antiglobulin test or in a gel matrix test (LISS-Coombs 37°C, DiaMed-ID Micro Typing System, DiaMed, Cressier sur Morat, Switzerland).15 Polyclonal antisera against RH10, RH20, RH23, and RH32 were from the International Blood Group Reference Laboratory (IBGRL, Bristol, UK) and the Australian Red Cross Blood Service (ARCBS, Sydney, Australia) collections. For phenotyping the C antigen, monoclonal anti-C clones MS24 (Ortho, Neckargmünd, Germany) and MS273 (Immucor, Rödermark, Germany) were used. Anti-LWa was obtained through the SCARF exchange program.
The mean D antigen density was determined by flow cytometry according to the protocol described previously16 with monoclonal immunoglobulin G (IgG) anti-D BS221, BS227, BS228, BS229, BS231, and H4111B7 (Biotest, Dreieich, Germany). The secondary antibody was goat anti-human IgG, Fab-fragment, fluorescein isothiocyanate conjugated (Jackson ImmunoResearch Laboratories, West Grove, PA).
Molecular analysis of genomic DNA
RHD nucleotide sequencing from genomic DNA for the RHD exons 1 through 10 including adjacent flanking intron regions was performed as described previously.5,12,15,17–19 The presence of the VS typical nucleotide substitution 733C>G was assessed by sequencing of RHCE exon 5; RHCE exon 1 was sequenced to investigate the concomitant presence of the nucleotide substitution 48G>C.20 The LWa molecular polymorphism was determined by sequencing of ICAM4 exon 1.21
Molecular analysis of cDNA
The nucleotide sequence of DOL-1, as part of the compound heterozygous DOL-1/(C)cdes and DOL-1/RHDΨ genotypes, was also determined from cDNA. Total RNA was isolated in one method from whole blood (RiboPure blood kit, Ambion, Austin, TX). RNA was reverse transcribed and cDNA was amplified in one-step reverse transcription buffer (SuperScript, Invitrogen, Carlsbad, CA), 2.5 mmol per L MgSO4, 500 nmol per L of each of the primers rh522 and rr3,22 and 1 μL of reverse transcriptase/polymerase (SuperScript II reverse transcriptase, platinum Taq DNA polymerase, Invitrogen) in a total volume of 50 μL. Incubation was carried out for 30 minutes at 55°C followed by 2 minutes at 94°C; thereafter 60 cycles were performed with 30 seconds at 92°C, 30 seconds at 62°C, and 3 minutes at 68°C followed by a final incubation for 10 minutes at 72°C. The mixture was then kept at 10°C. Amplified cDNA products were treated with exonuclease and alkaline phosphatase (ExoSAP, USB, Cleveland, OH). Nucleotide sequencing was performed with a cycle sequencing kit and a DNA sequencer (ABI 310, Applied Biosystems, Foster City, CA). Twenty-five cycles were performed, each cycle for 15 seconds at 94°C, 15 seconds at 58°C, and 4 minutes at 60°C; the mixture was finally kept at 10°C.
Primers used
RHD cDNA nucleotide sequencing primers were D150r, 5′-AACTTGATAGGATGCCACGAGCCC-3′ (antisense, cDNA nucleotide position 150-127); DCE91f, 5′-TTTACCCACTATGACGCTTC-3′ (sense, 91-110); D4s, 5′-ACATGATGCACATCTACGTGTTCGC-3′ (sense, 503-527); and D1036f, 5′TTGCTGGTGCTTGATACC-3′ (sense, 1036-1053).
Nomenclature
The designation DOL was derived from the D antigen and the first two letters of the last name of the patient with DOL-1/RHDΨ.7 DOL-2 is molecularly similar to DOL-1. The designations DFW and DFL (RIR-16)23 were derived from DFR-like and Württemberg or Linköping, respectively, where the original observations were made. DMI represents RHD(M170I) and was named after its amino acid substitution M>I.
RESULTS
RHD alleles
In 1994 a D+ young male patient was recognized because he had developed an alloanti-D. His partial D did not fit into the classification of D categories. The underlying allele was DOL-1 (Table 1),13 which the patient carried in combination with (C)cdes. Since then, DOL-1 was discovered in two young female patients. One was hemizygous DOL-1, and the other was heterozygous DOL-1/RHDΨ. The name DOL was derived from the initials of the DOL-1/RHDΨ patient.7 DOL-2 was detected in a child due to weak D antigen expression. Her allele harbored an additional amino acid substitution compared to DOL-1. Another female patient carried the D variant DMI with a single substitution at amino acid position 170 (Table 1). DFW and DFL were two more variants with adjacent amino acid substitutions. DOL-2 was inherited from the father who carried DOL-2 in combination with a normal RHD (Fig. 1).
TABLE 1.
Partial D at the extracellular RhD protein vestibule described in this study
| Trivial name | Exon involved | Allele* | Nucleotide change | Effect on protein sequence | Membrane localization | Phenotype | Haplotype | Number of probands |
|---|---|---|---|---|---|---|---|---|
| DOL-1 | 4 | RHD(M170T, F223V) | 509T>C | Met to Thr at 170 | Rh vestibule | ccDee† | cDe | 3 |
| 5 | 667T>G | Phe to Val at 223 | Rh vestibule | |||||
| DOL-2 | 4 | RHD(M170T, F223V, L378V) | 509T>C | Met to Thr at 170 | Rh vestibule | ccDee | cDe | 1 |
| 5 | 667T>G | Phe to Val at 223 | Rh vestibule | |||||
| 8 | 1132C>G | Leu to Val at 378 | Transmembraneous | |||||
| DMI | 4 | RHD(M170I) | 510G>A | Met to Ile at 170 | Rh vestibule | CcDee | CDe | 1 |
| DFW | 4 | RHD(H166P) | 497A>C | His to Pro at 166 | Rh vestibule | CcDee | CDe | 2 |
| DFL‡ | 4 | RHD(Y165C) | 494A>G | Tyr to Cys at 165 | Rh vestibule | CcDee | CDe | 22 |
The nucleotide sequence data were deposited in EMBL under Accession Numbers AM087651 (cDNA) and FM201788 (genomic DNA) for DOL-1, AM072761 (genomic DNA) for DOL-2, and AM998551 for DMI.
Weak C antigen expression in one DOL-1 proband was caused by Ccdes in trans. The other two DOL-1 probands carried DOL-1/RHDΨ and hemizygous DOL-1, respectively.
Fig. 1.
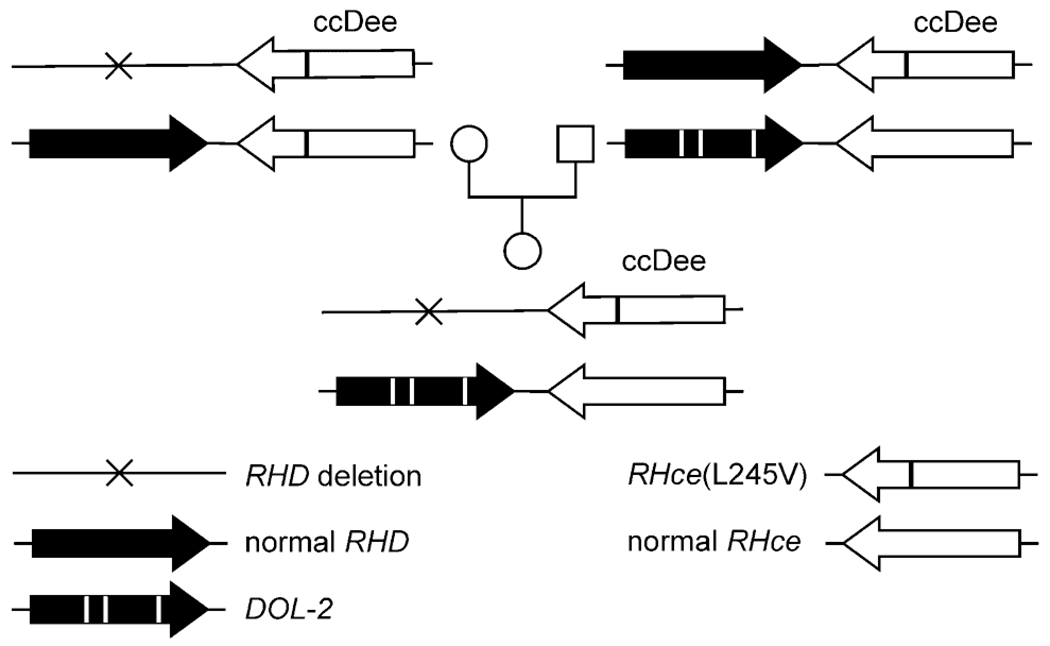
Inheritance of the DOL-2 allele. The Rhesus phenotype of the child with DOL-2 and her parents are shown along with their RHD and RHCE haplotypes. The normal RHce allele represents GenBank Accession Number DQ322275.
Anti-D immunization
A woman with DFL was reported to the Rhesus Immunization Registry (RIR-16),23 because of an anti-D acquired by transfusion or pregnancy despite a D+ phenotype (Table 2). The patient with DOL-1/(C)cdes was double heterozygous for sickle trait (hemoglobin S) and β-thalassemia. Treatment of sickle thalassemia with two red blood cell (RBC) transfusions was the probable cause of anti-D immunization (RIR-27). An anti-D was found in a woman with DMI 5 months after D+ RBC transfusion (RIR-110). No anti-D was detected in the other 21 DFL and 3 DOL carriers.
TABLE 2.
Probands with alloanti-D
| Case* | RHD alleles | Phenotype | Anti-D titer | DAT | Possible immunization date | LW blood group | Ethnicity |
|---|---|---|---|---|---|---|---|
| RIR-16 | DFL | CcDee | 32 | Negative | Before 1997 | LW(a+) | Swedish |
| RIR-27 | DOL-1/(C)cdes | (C)cDee | 128 | Negative | 1992 | LW(a+) | Ewe (West Africa) |
| RIR-110 | DMI | CcDee | 4 | Positive | 2007 | LW*A/LW*A | German |
Rhesus Immunization Registry (RIR) entries 16, 27, and 110; data available online.23
Immunohematology
The D epitope (epD) patterns of D variants were determined using panels of monoclonal anti-D (Table 3).24 Results are shown for the hemizygous DOL-1 sample, for DOL-2, DMI, DFW, and DFL in comparison to the known patterns of DCS-1, DFR-1, and DFV.21 Weak D type 4.0 was used as a weak D control with cDe haplotype. A D+ phenotype was usually assigned to DOL-1, DMI, DFW, and DFL using the routine monoclonal anti-D (Table 3). The antigen densities determined for DOL-1, DFW, and DFL were typical for a D+ phenotype (Table 4).25 Based on its antigen density, DFR can be typed D+, but routine monoclonal anti-D often miss DFR-1 because it lacks several epitopes, especially parts of epD 6.
TABLE 3.
Serologic reactivity with commercial panels of anti-D
| Monoclonal anti-D |
Reactivity* |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Clone | Isotype | epD† | DOL-1 ccDee | DOL-2 ccDee | DMI CcDee | DFW CcDee | DFL CcDee | DCS-1 ccDEe | DFR-1 CcDee | DFV ccDEe | Weak D type 4.0 ccDee |
| BS226‡ | IgM | 6.4 | ++ | ++++ | +++ | ++ | +++ | ++ | − | ++++ | +++ |
| BS232‡ | IgM | 6.4 | ++ | ND | +++ | ++ | +++ | ++ | − | ++++ | +++ |
| RUM-1‡ | IgM | 6.1 | +++ | ND | +++ | +++ | +++ | ++ | − | ++++ | ++++ |
| D175-2‡ | IgM | 6.1 | +++ | ND | ++++ | ++++ | +++ | +++ | + | ++++ | +++ |
| HM10§ | IgM | 6.6 | + | (+) | ++++ | ++ | + | + | − | ++++ | +++ |
| HM16§ | IgG | 6.4 | +++ | ++++ | ND | +++ | +++ | +++ | − | ++++ | +++ |
| P3X61§ | IgM | 6.1 | ++ | ++++ | +++ | +++ | +++ | + | − | ++++ | ++++ |
| P3X35§ | IgG | 5.4 | +++ | + | ND | − | + | +++ | − | ++++ | +++ |
| P3X212 11 F1§ | IgM | 8.2 | − | − | − | − | − | − | − | ++++ | ++ |
| P3X212 23 B10§ | IgM | 9.1 | ++ | ++++ | +++ | +++ | +++ | ++ | +++ | +++ | + |
| P3X241§ | IgG | 5.4 | +++ | ++++ | ND | +++ | +++ | +++ | ++ | +++ | +++ |
| P3X249§ | IgG | 2.1 | ++++ | ++++ | ND | +++ | ++++ | ++++ | ++++ | ++++ | ++++ |
| P3X290§ | IgG | 3.1 | +++ | ++ | ND | ++++ | +++ | ++++ | +++ | ++++ | +++ |
| LHM76/58‖ | IgG1λ | ND | ++++ | ND | ND | +++ | ++++ | +++ | +++ | ++++ | ++++ |
| LHM76/59‖ | IgG1 | ND | ++++ | ND | ND | +++ | ++++ | ++++ | +++ | ++++ | +++ |
| LHM174/102‖ | IgG3κ | 1.2 | − | ND | ND | +++ | +++ | + | − | +++ | +++ |
| LHM50/2B‖ | IgG1λ | 6.3 | ++++ | ND | ND | ++++ | +++ | ++++ | +++ | ++++ | ++++ |
| LHM169/81‖ | IgG3κ | 1.1 | ++++ | ND | ND | ++++ | +++ | +++ | +++ | ++++ | ++++ |
| ESD1‖ | IgG1κ | ND | ++++ | ND | ND | +++ | ++++ | ++++ | +++ | ++++ | ++++ |
| LHM76/55‖¶ | IgG1κ | 3.1 | +++ | ++++ | ND | +++ | +++ | +++ | +++ | +++ | ++++ |
| LHM77/64‖¶ | IgG1κ | 9.1 | ++++ | ++++ | ND | ++++ | +++ | ++++ | ++++ | ++++ | ++++ |
| LHM70/45‖¶ | IgG1λ | 1.2 | − | − | ND | +++ | +++ | + | − | +++ | +++ |
| LHM59/19‖¶ | IgG3κ | 8.1 | − | − | ND | (+) | +++ | + | + | ++++ | +++ |
| LHM169/80‖¶ | IgG3λ | 6.3 | +++ | ++++ | ND | +++ | +++ | +++ | +++ | +++ | +++ |
| LHM57/17‖ | IgG1λ | 6.3 | ++ | ND | − | − | − | + | − | ++++ | +++ |
| LDM-1¶ | IgM | ND | +++ | ++++ | +++ | +++ | +++ | +++ | (+) | +++ | +++ |
| BS221** | IgG | 6.3 | +++ | ND | ND | +++ | +++ | ++++ | ++++ | +++ | +++ |
| BS227** | IgG | 2.2 | +++ | ND | ND | +++ | ++++ | ++ | − | +++ | +++ |
| BS228** | IgG | 6.3 | ++++ | ND | ND | +++ | ++++ | ++++ | ++++ | ++++ | +++ |
| BS229** | IgG | 5.4 | ++++ | ND | ND | ++++ | +++ | +++ | − | ++++ | ++++ |
| BS231** | IgG | 5.4 | ++++ | ND | ND | +++ | ++ | ++++ | − | +++ | ++++ |
| H41** | IgG | 3.1 | +++ | ND | ND | ++++ | ++++ | +++ | ++++ | ++++ | +++ |
Tests were performed in gel matrix test with antiglobulin if not stated otherwise. DOL-2 testing was done in tube, except for the six monoclonal anti-D from DiaMed.
epD patterns as described previously by Scott.24
Monoclonal anti-D approved for routine use in Germany (BS226 and BS232, Seraclone anti-D (RH1), Biotest; RUM-1, immuClone anti-D rapid and D175-2, immuClone anti-D fast, Immucor).
D-Screen, Diagast, Loos, France.
Advanced partial RhD typing kit, Alba Bioscience, Edinburgh, UK. The results obtained with LHM76/58 and with the monoclonal antibody Number 74, documented as LHM76/58, in the Nantes workshop24 differed.
ID-Partial RhD-Typing Set, DiaMed.
Monoclonal anti-D panel, Biotest.
ND = not determined.
TABLE 4.
D variants with amino acid substitutions at loops 3 and 4
| D variant | D antigens/RBC* | Amino acid position and substitution |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Extracellular loop 3/transmembraneous helix 6 |
Transmembraneous helix 7/extracellular loop 4 |
|||||||||||||||
| 164 | 165 | 166 | 169 | 170 | 171 | 172 | 174 | 220 | 221 | 223 | 225 | 226 | 227 | 229 | ||
| DDN† | ND | N | Y | H | M | M | H | I | V | W | P | F | S | A | L | R |
| DFL | 7,499 | D | C | H | M | M | H | I | V | W | P | F | S | A | L | R |
| DFW | 10,019 | D | Y | P | M | M | H | I | V | W | P | F | S | A | L | R |
| DFR-1 | 4,895 | D | Y | H | L | R | H | F | V | W | P | F | S | A | L | R |
| DOL-1 | 7,158 | D | Y | H | M | T | H | I | V | W | P | V | S | A | L | R |
| DMI | ND | D | Y | H | M | I | H | I | V | W | P | F | S | A | L | R |
| DHQ | ND | D | Y | H | M | M | Q | I | V | W | P | F | S | A | L | R |
| Weak D type 33 | ND | D | Y | H | M | M | H | I | M | W | P | F | S | A | L | R |
| Weak D type 16 | 184 | D | Y | H | M | M | H | I | V | R | P | F | S | A | L | R |
| Weak D type 62 | ND | D | Y | H | M | M | H | I | V | W | T | F | S | A | L | R |
| Weak D type 27 | 409 | D | Y | H | M | M | H | I | V | W | S | F | S | A | L | R |
| DFV | 9,422 | D | Y | H | M | M | H | I | VV | W | P | V | S | A | L | R |
| DTO | ND | D | Y | H | M | M | H | I | V | W | P | V | F | A | L | R |
| DSF‡ | ND | D | Y | H | M | M | H | I | V | W | P | F | F | A | L | R |
| DCS-1 | 2,958 | D | Y | H | M | M | H | I | V | W | P | V | S | P | L | R |
| DCS-2 | 835 | D | Y | H | M | M | H | I | V | W | P | F | S | P | L | R |
| DBA | ND | D | Y | H | M | M | H | I | V | W | P | F | S | A | P | R |
| DHR | ND | D | Y | H | M | M | H | I | V | W | P | F | S | A | L | K |
| RhD consensus | NA | D | Y | H | M | M | H | I | V | W | P | F | S | A | L | R |
| RhCE consensus | NA | D | Y | H | L | R | H | F | V | W | P | V | S | P/A § | L | R |
Samples were tested by flow cytometry with six monoclonal anti-D (BS221, BS227, BS228, BS229, BS231, and H41; Biotest). Weak D type 4.0 RBC run as a control showed 1689 D antigens/RBC. DFL (RIR-16) shown, 6310 D antigens/RBC measured for DFL of German donor. Hemizygous DOL-1 shown, 6154 D antigens/RBC measured for DOL-1/Ccdes. The antigen densities of DCS-1 and DCS-2 were previously reported.21
The nucleotide sequence data were deposited in EMBL under Accession Number FM212439.
Originally described as D674.25
Amino acids P and A are typical for E and e antigens, respectively.
NA = not applicable; ND = not determined.
All DOL alleles observed in this study were associated with the cDe haplotype. The samples with hemizygous DOL-1, with DOL-1/RHDΨ and with DOL-2 had a ccDee phenotype. The weakened expression of the C antigen in the sample of the patient with DOL-1/(C)cdes was explained by the presence of the (C)cdes allele in trans: agglutination titers were similar for this sample and controls of known (C)cdes using 11 monoclonal anti-C (Table 5).26
TABLE 5.
Serologic reactivity with research-grade anti-C
| Agglutination titer in gel matrix test with antiglobulin* |
||||
|---|---|---|---|---|
| Monoclonal anti-C |
Controls |
|||
| Clone | Isotype | (C)cdes/cDe | (C)cdes/cdE | CDe/cde |
| C-93/44 | IgM | 0 | 0 | 32 |
| MS253† | IgM | 1 | 4 | 16 |
| HMR7 | IgG3 | 4 | 8 | 64 |
| MS242 | IgG3 | 16 | 32 | 64 |
| MS23 | IgG1 | 32 | 64 | 128 |
| 388F3 | IgM | 64 | 128 | 256 |
| MS24 | IgM | 64 | 256 | 256 |
| MS273 | IgG1 | 64 | 256 | 512 |
| MS257 | IgG | 128 | 256 | 256 |
| P3x255 13G8 | IgM | 256 | 128 | 512 |
| DGC02 | IgM | 1024 | 1024 | 4096 |
The phenotypes of the samples were (C)cDees (patient expressing a DOL-1 with anti-D, RIR-27), (C)cddEes and CcDee (controls); the derived genotypes are given in the table head.
Reported as anti-Cw.26
The DOL-1/RHDΨ sample was antigen G positive (RH12) and negative for the antigens V (RH10), VS (RH20), Dw (RH23), RH32 and FPTT (RH50); the DOL-1/(C)cdes sample was VS positive. According to molecular analysis the hemizygous DOL-1 carrier was VS negative while the two DOL-2 carriers were VS positive. However, DOL-2 was inherited independent of VS (Fig. 1). We concluded that the DOL-1 and DOL-2 alleles were not associated with VS.
Ethnic origin
The DOL-1/(C)cdes patient (Table 2) lived in Germany but was born in Togo27 and his family belonged to the Ewe for at least three generations. The Ewe people are with 22 percent the largest ethnic group of Togo and live in southeast Ghana, southern Togo, and southern Benin. The individual with DOL-1/RHDΨ lived in Australia but came from Botswana. The individual with hemizygous DOL-1 was Czech, but her father originated from Lebanon. The parents of the DOL-2 baby lived in Austria but were natives of Ghana. The ethnic origin was German for the DMI carrier and German and Sri Lankan for the DFW carriers. The original DFL carrier was Swedish; 1 German and 20 Austrian carriers have been observed since.
DFL population survey
DFL carriers were identified among blood donors of Upper Austria12 on the basis of weak reactions with monoclonal anti-D HM10. Between August 15, 2005, and July 31, 2008, we found 11 DFL carriers among 103,251 donors (frequency estimate 1 in 9386; 95% confidence interval, 1 in 5420 to 1 in 19,397, Poisson distribution). Considering this frequency in donors, at least 71 to 253 carriers of the DFL phenotype would be predicted to occur in the population of 1,370,000 inhabitants of Upper Austria. Twenty DFL carriers have already been identified at the Linz blood center among donor and patient reference blood samples, including one individual with the compound heterozygous alleles DFL/weak D type 3.
DISCUSSION
DOL-1 and DOL-2 are African alleles. The probands originated from Western and Southern Africa (three cases) or the Middle East (one case), although they were recognized in Germany, Australia, the Czech Republic, and Austria because of their variant D antigens. The three probands from Africa with DOL carried further RHD or RHCE alleles typical of African populations, such as (C)cdes,28 RHDΨ,18 and VS.28,29 People of African ancestry are frequently heterozygous for two different variants of RHD alleles. The exact molecular basis of such compound heterozygous RHD alleles is difficult to define without transcript analysis.
Phylogenetically DOL alleles belong to the weak D type 4 cluster, because they share the templated F223V substitution and occur in a cDe haplotype (Fig. 2).4 In a templated mutation, an isolated RHD-specific nucleotide is replaced by its RHCE-specific counterpart, the mechanism of which may be a short gene conversion or a single point mutation. Characteristic of DOL-1 is the additional M170T substitution and of DOL-2 the additional M170T and L378V substitutions, both of which are nontemplated. Hence, DOL-2 probably evolved from DOL-1. Compatible with this assumption were the origins of one DOL-1 carrier and the DOL-2 family, who came from the neighboring West African states of Togo and Ghana.
Fig. 2.
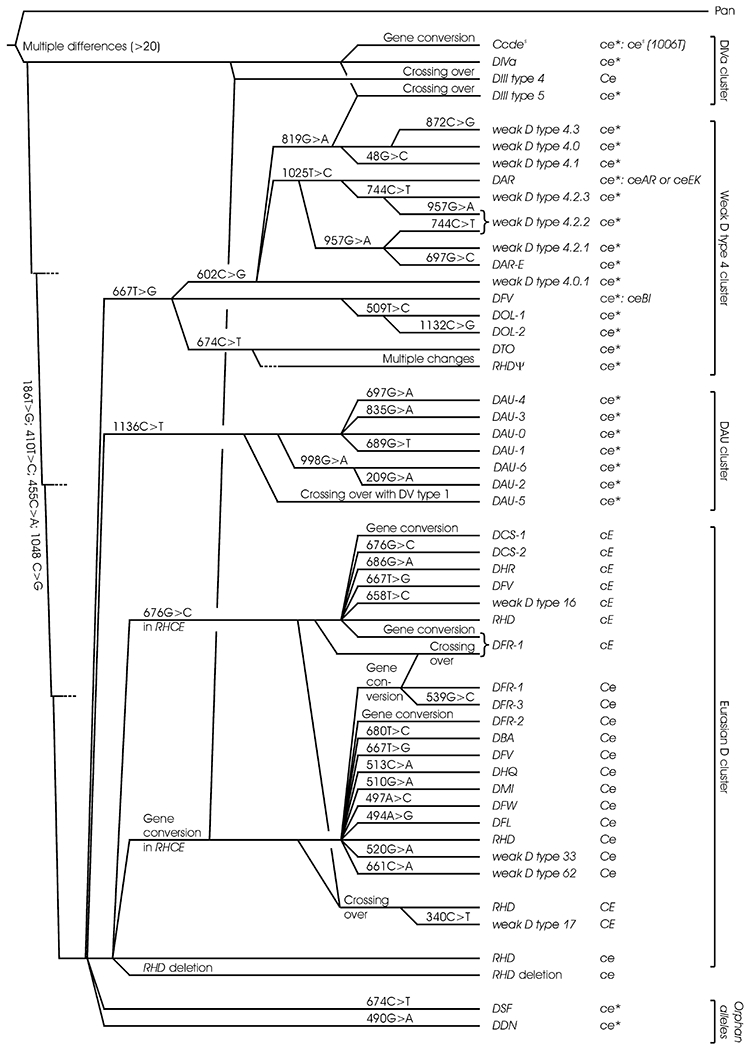
Phylogeny of Rhesus haplotypes. The phylogenetic tree is based on a previous tree for RHD20 taking into account the available data on Rhesus haplotypes. Four main clusters of RHD alleles have been described. The RHD alleles of the DIVa cluster share the three characteristic ancestral amino acids 62F, 137V, and 152T; those of the weak D type 4 cluster the F223V substitution; and those of the DAU cluster the T379M substitution. These three “African” clusters are segregated from the Eurasian D cluster with “normal” RHD as the primordial allele. The RHD alleles of the three African clusters generally occur in a cDe haplotype, which indicates that the cE and Ce alleles of RHCE evolved in the Eurasian branch after its divergence from the other branches. For each RHD allele, the accompanying RHCE allele is indicated. ce* is a general indicator of a ce-like allele which frequently may be a variant; if known, the typically associated RHCE alleles are given.
The templated 667T>G substitution causing F223V represents the primordial event in the evolution of the African weak D type 4 cluster (Fig. 2). As postulated when the phylogenetic tree was initially established,4 a DFV allele was found carrying F223V in isolated form and occurring in a cDe haplotype.25 Recently, two additional DFV alleles were observed associated with the haplotypes CDe6 and cDE.21 Based on the differing haplotype associations, these two DFV alleles may be grouped to the Eurasian D cluster; they probably evolved independently of the primordial allele of the weak D type 4 cluster (Fig. 2).21 Hence, a specific RHD allele may belong to more than one cluster. F223V is also found in the group of DV alleles, which are probably caused by RHD-CE-D gene conversions involving DNA stretches of the RHCE gene of different lengths. Likewise, the partial D DCS-1 carrying F223V and A226P may have arisen from a gene conversion.
Crossing over events may have caused the DAU-5 allele (recombination of DV type 1 with DAU-0)5 as well as the RHD-negative alleles of different haplotypes (Fig. 2). There are examples of RHD alleles, for which different evolutionary routes are probable. For instance, DFR-1 is often associated with Ce, rarely with cE. Hence, DFR-1 associated with cE may have derived from a chromosomal crossing over. Alternatively, DFR-1 associated with cE may have arisen from a gene conversion, where a stretch of amino acids including the three DFR-typical positions was transposed from the RHcE gene to the RHD gene. For some alleles relevant data are still missing and their definite position in the phylogeny is pending. RHCE associations may need reevaluation once more data will have accumulated, especially since there seems to be a relevant frequency of recombination events; for example, ceAR and ceEK may also be associated with RHD deletions.
The phylogenetic derivation of alleles may provide insight in older nucleotide substitutions that define whole groups of alleles, like D clusters, and are instrumental for devising efficient genotyping strategies. As exemplified by DFV, alleles can be predicted to occur before their actual observation. Systematic characterization will allow simplifying a hitherto confusing assortment of seemingly unrelated alleles.
The partial D DFL, DFW, and DMI with single amino acid substitutions at positions 165, 167, and 170, respectively, were allocated to the Eurasian D cluster, because the underlying nucleotide substitution occurred in the Eurasian RHD allele. In addition, their haplotype association was CDe and all probands encountered were from Europe or Asia.
As evident from three-dimensional homology modeling, the DOL typical amino acid substitutions F223V and M170T lie at the entrance to the transmembrane channel of the RhD protein, which is known as the extracellular RhD protein vestibule (Fig. 3).11 Amino acid residue 170, which seems to be located deep in the vestibule,11 is particularly variable with M170T in DOL-1 and DOL-2, M170I in DMI, and M170R in DFR. All of these alleles are partial D and several anti-D immunizations have been observed (Table 2).30 Moreover, the RHce(R170S) allele was described31 expressing the D antigen without D-specific amino acids.20,32 Hence, antigenic defects (Table 2)30 and neoantigens31 have been found in this region of the Rh protein. According to the Rh “antigenic vestibule” concept presented by Avent and colleagues,11,33 most RhC→RhD amino acid substitutions critical for Rh antigenicity lie in exofacial positions (i.e., localized at loops 3, 4, and 6) and line the boundary of the antigenic vestibule.33 Furthermore, several D variants harbor amino acid substitutions at these loops. So far, 18 D variants have been identified at loops 3 and 4 and the adjoining helices 6 and 7 (Table 4). D variants with substitutions in extracellular parts were partial D with normal or moderately depressed D antigen density. D variants with substitutions in transmembraneous sections were weak D with low D antigen expression. The unexpectedly low D antigen density of the partial D DCS-1 and DCS-2 was previously explained by the presence of proline at position 226.21
Fig. 3.
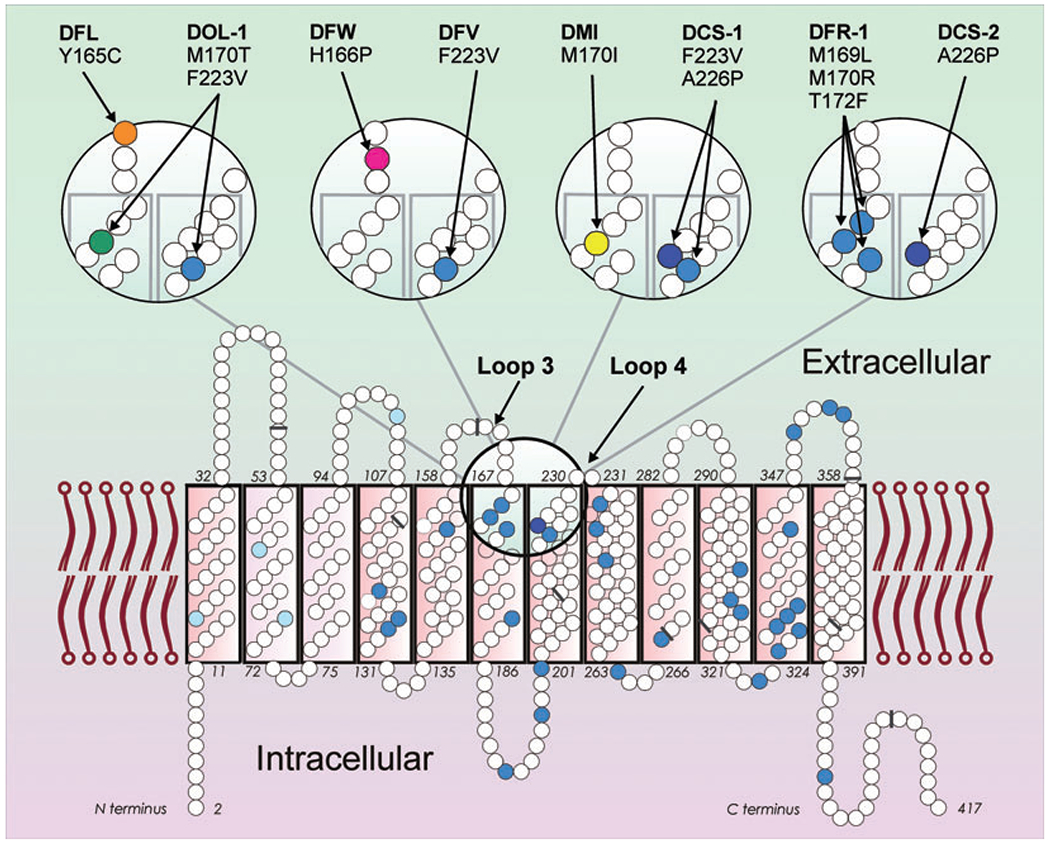
Partial D with amino acid substitutions at the extracellular RhD protein vestibule. The two-dimensional model of the RhD protein (bottom) with 417 amino acids (small circles) depicts amino acids that differ in RhCE (blue) with the four C-typical substitutions (light blue) and the one E-typical substitution A226P (dark blue). The vestibule is lined in part by amino acids of loops 3 and 4 (large circle). This region is shown in more detail (top). The amino acid substitutions characteristic of eight partial D are indicated (colored circles and arrows). Four partial D harbor RhCE-like substitutions (blue circles). The nine exon boundaries in the RHD cDNA, as reflected in the amino acid sequence, are indicated (gray bars).
The antigenic relationship of DOL-1 and DOL-2 with DFV DCS-1, DMI, and DFR was investigated, because these D variants share amino acid substitutions either at positions 223 or 170. F223V seems to have little influence on the protein structure, because DFV was almost undistinguishable from normal D (Table 3).21 In contrast, DOL and DCS-1 phenotypes showed distinct serologic profiles (Table 3). Therefore, the additional M170T and A226P substitutions, respectively, seem to induce structural changes reflected in serologically recognizable differences in antigenicity. The serologic profile of DOL-1 was similar to that of DOL-2 and DCS-1. Compared to DOL-1, DOL-2 carried an additional conservative substitution located in the transmembraneous helix 12; this substitution was of minor influence on antigenicity. The serologic profiles of DOL, DMI, DFW, and DFL were compatible with partial D. Despite some similarity between these D variants and DFR, the overall epD pattern showed considerable differences (Table 3). DFR expresses the low prevalence antigen FPTT (RH50),30 which was absent in DOL-1.
DOL, DFL, and DMI qualified as partial D also by the presence of alloanti-D after D+ transfusion (Table 2). DFL carriers are quite frequent in Upper Austria with 20 cases detected so far. Nevertheless, only one anti-D immunization in a Swedish individual (RIR-16) was reported to the Rhesus Immunization Registry.23 Therefore, anti-D immunizations in DFL carriers are probably infrequent. DOL carriers are typed D+ by standard monoclonal anti-D reagents (e.g., BS226 and RUM-1, Table 3) and will be transfused with D+ RBC units and not receive prophylactic anti-D in pregnancy. It will be important to determine the frequency of the DOL allele in the African population to assess the risk for anti-D formation after transfusion or pregnancy. With DOL-1 and DOL-2, DFL, DFW, and DMI, further alleles are added to the growing list of RH variants that are difficult to discern by current serologic methods, but are clearly amenable to specific detection by blood group genotyping.
ACKNOWLEDGMENTS
We thank Barbara Diefenbach (Hamburg, Germany) and Birgit Ladewig and Christa Das Gupta (Dreieich, Germany) for procuring blood samples and clinical data of the DOL-1/(C)cdes patient with anti-D; Julie M. Watt and Hayley C. Bruce for referring a blood sample with DOL-1/RHDΨ; and Belinda K. Singleton and Geoff L. Daniels (Bristol, UK) for identifying that the original DOL patient was heterozygous for DOL-1 and RHDΨ. One DFW sample was contributed by the late Nicole I. Eicher (Bern, Switzerland). Anti-LWa antisera were provided by Tom Kristensen (Odense, Denmark) and Janis Hamilton (Detroit, MI). We acknowledge the technical assistance of Marianne Lotsch, Anita Link, and Hedwig Erne in Ulm, Germany.
This work was supported by intramural grants from the DRK-Blutspendedienst Baden-Württemberg–Hessen, Mannheim, and by a grant from the Ministry of Health of the Czech Republic (Scientific Project No. 23736).
Footnotes
CONFLICT OF INTEREST
W.A.F., I.v.Z., and F.F.W are current or former employees of the DRK Blutspendedienst Baden-Württemberg–Hessen. D.R.K. and W.A.F. are holding patents or have patents pending on nucleotide sequences and their use in molecular genetics of the Rhesus blood group.
N.D.A. is a member of the scientific advisory board of Progenika Biopharma, which developed BloodChip that tests for the known DOL alleles.
All other authors do not declare any conflict of interest.
REFERENCES
- 1.Avent ND, Reid ME. The Rh blood group system: a review. Blood 2000;95:375–87. [PubMed] [Google Scholar]
- 2.Tippett P, Lomas-Francis C, Wallace M. The Rh antigen D: partial D antigens and associated low incidence antigens. Vox Sang 1996;70:123–31. [DOI] [PubMed] [Google Scholar]
- 3.Flegel WA, Wagner FF. Molecular genetics of RH. Vox Sang 2000;78:109–15. [PubMed] [Google Scholar]
- 4.Wagner FF, Ladewig B, Angert KS, Heymann GA, Eicher NI, Flegel WA. The DAU allele cluster of the RHD gene. Blood 2002;100:306–11. [DOI] [PubMed] [Google Scholar]
- 5.Chen Q, Flegel WA. Random survey for RHD alleles among D+ European persons. Transfusion 2005;45:1183–91. [DOI] [PubMed] [Google Scholar]
- 6.Grootkerk-Tax MG, van Wintershoven JD, Ligthart PC, van Rhenen DJ, van der Schoot CE, Maaskant-van Wijk PA. RHD (T201R, F223V) cluster analysis in five different ethnic groups and serological characterization of a new Ethiopian variant DARE, the DIII type 6 and the RHD(F223V). Transfusion 2006;46:606–15. [DOI] [PubMed] [Google Scholar]
- 7.Avent ND, Poole J, Singleton BK, Chabert T, Romeiras MC, Rodrigues MJ, Watt J, Bruce H. Studies of two partial Ds: DMH and DOL [abstract]. Transfus Med 1999;9Suppl:33. [Google Scholar]
- 8.Písacka M, Vytisková J, Zavadil J, Karasová R, Králová M, Prosická M, Flidrová H, Flegel WA, Wagner FF. Slabé a variantní RhD antigeny—charakteristiky a frekvence typu, vyţetrovaných v letech 1997–2002 v Referencní laboratori pro imunohematologii [Weak and variant RhD antigens—characteristics and frequency of types investigated in 1997-2002 in the Reference Laboratory for Immunohaematology]. Transfuze a Hematologie Dnes 2003;9:37–47. [Google Scholar]
- 9.Písacka M, Vytisková J, Králová M, Flegel WA. Variant D antigens and weak D types in the Czech Republic (Central European Population)—serologic and molecular analysis of 285 cases. Transfusion 2005;45 Suppl:120A. [Google Scholar]
- 10.Doescher A, Vogt C, das Gupta C, Wagner FF, Petershofen E. Anti-D antibody induction in a patient of African orign with 3 single mutations in the RHD gene [abstract]. Transfus Med Hemother 2007;32:58. [Google Scholar]
- 11.Conroy MJ, Bullough PA, Merrick M, Avent ND. Modeling the human Rhesus proteins: implications for structure and function. Br J Haematol 2005;131:543–51. [DOI] [PubMed] [Google Scholar]
- 12.Polin H, Danzer M, Hofer K, Gassner W, Gabriel C. Effective molecular RHD typing strategy for blood donations. Transfusion 2007;47:1350–5. [DOI] [PubMed] [Google Scholar]
- 13.Le Marechal C, Guerry C, Benech C, Burlot L, Cavelier B, Porra V, Delamaire M, Ferec C, Chen JM. Identification of 12 novel RHD alleles in western France by denaturing high-performance liquid chromatography analysis. Transfusion 2007;47:858–63. [DOI] [PubMed] [Google Scholar]
- 14.Wagner FF, Gassner C, Eicher NI, Lonicer C, Flegel WA. Characterization of D category IV type IV, DFW, and DNB [abstract]. Transfusion 1998;38:63S. [Google Scholar]
- 15.Flegel WA, Eicher NI, Doescher A, Hustinx H, Gowland P, Monsouri Taleghani B, Petershofen EK, Bauerfeind U, Ernst M, von Zabern I, Schrezenmeier H, Wagner FF. In-frame triplet deletions in RHD alter the D antigen phenotype. Transfusion 2006;46:2156–61. [DOI] [PubMed] [Google Scholar]
- 16.Flegel WA, Curin-Serbec V, Delamaire M, Donvito B, Ikeda H, Jorgensen J, Kumpel BM, Le Pennec PY, Pisacka M, Tani Y, Uchikawa M, Wendel S, Wagner FF. Section 1B: Rh flow cytometry. Coordinator’s report. Rhesus index and antigen density: an analysis of the reproducibility of flow cytometric determination. Transfus Clin Biol 2002;9:33–44. [DOI] [PubMed] [Google Scholar]
- 17.Wagner FF, Gassner C, Müller TH, Schönitzer D, Schunter F, Flegel WA. Molecular basis of weak D phenotypes. Blood 1999;93:385–93. [PubMed] [Google Scholar]
- 18.Singleton BK, Green CA, Avent ND, Martin PG, Smart E, Daka A, Narter-Olaga EG, Hawthorne LM, Daniels G. The presence of an RHD pseudogene containing a 37 base pair duplication and a nonsense mutation in Africans with the Rh D-negative blood group phenotype. Blood 2000;95:12–8. [PubMed] [Google Scholar]
- 19.Doescher A, Flegel WA, Petershofen EK, Bauerfeind U, Wagner FF. Weak D type 1.1 exemplifies another complexity in weak D genotyping. Transfusion 2005;45:1568–73. [DOI] [PubMed] [Google Scholar]
- 20.Wagner FF, Ladewig B, Flegel WA. The RHCE allele ceRT: D epitope 6 expression does not require D-specific amino acids. Transfusion 2003;43:1248–54. [DOI] [PubMed] [Google Scholar]
- 21.Flegel WA, von Zabern I, Doescher A, Wagner FF, Vytiskova J, Pisacka M. DCS-1, DCS-2 and DFV share amino acid substitutions at the extracellular RhD protein vestibule. Transfusion 2008;48:25–33. [DOI] [PubMed] [Google Scholar]
- 22.Wagner FF, Gassner C, Müller TH, Schönitzer D, Schunter F, Flegel WA. Three molecular structures cause Rhesus D category VI phenotypes with distinct immunohematologic features. Blood 1998;91:2157–68. [PubMed] [Google Scholar]
- 23.Flegel WA. The Rhesus Site. Ulm: DRK-Blutspendedienst Baden-Württemberg-Hessen; 1998-2009. Available from: http://www.uni-ulm.de/~wflegel/RH/ [Google Scholar]
- 24.Scott M. Section 1A: Rh serology. Coordinator’s report. Transfus Clin Biol 2002;9:23–9. [DOI] [PubMed] [Google Scholar]
- 25.Noizat-Pirenne F, Lee K, Pennec PY, Simon P, Kazup P, Bachir D, Rouzaud AM, Roussel M, Juszczak G, Menanteau C, Rouger P, Kotb R, Cartron JP, Ansart-Pirenne H. Rare RHCE phenotypes in black individuals of Afro-Caribbean origin: identification and transfusion safety. Blood 2002;100:4223–31. [DOI] [PubMed] [Google Scholar]
- 26.Rouger P, Muller JY. Third international workshop and symposium on monoclonal antibodies against human red cells and related antigens: section RH. Transfus Clin Biol 1996;3:329–541. [PubMed] [Google Scholar]
- 27.Segbéna AY, Fétéké L, Haudréchy D, Bigey F, Waller C, North ML. Situation of the blood transfusion sector in Togo in 2007. Transfus Today 2007;73:26–7. [Google Scholar]
- 28.Daniels GL, Faas BH, Green CA, Smart E, Maaskant-van Wijk PA, Avent ND, Zondervan HA, von dem Borne AE, van der Schoot CE. The VS and V blood group polymorphisms in Africans: a serologic and molecular analysis. Transfusion 1998;38:951–8. [DOI] [PubMed] [Google Scholar]
- 29.Flegel WA, Wagner FF, Chen Q, Schlanser G, Frame T, Westhoff CM, Moulds MK. The RHCE allele ceCF: the molecular basis of Crawford (RH43). Transfusion 2006;46:1334–42. [DOI] [PubMed] [Google Scholar]
- 30.Lomas C, Grassmann W, Ford D, Watt J, Gooch A, Jones J, Beolet M, Stern D, Wallace M, Tippett P. FPTT is a low-incidence Rh antigen associated with a “new” partial Rh D phenotype, DFR. Transfusion 1994;34:612–6. [DOI] [PubMed] [Google Scholar]
- 31.Just B, Legler TJ, Doescher A. A previously unknown polymorphism in RHCE exon 4 is associated with a D-positive phenotype [abstract]. Transfus Med Hemother 2007;34:51. [Google Scholar]
- 32.Chen Q, Hustinx H, Flegel WA. The RHCE allele ceSL: the second example for D antigen expression without D-specific amino acids. Transfusion 2006;46:766–72. [DOI] [PubMed] [Google Scholar]
- 33.Avent ND. New insight in the Rh system: structure and function. ISBT Sci Ser 2007;2:35–43. [Google Scholar]


