Abstract
The relationship between sterol uptake and heme competence in two yeast strains impaired in heme synthesis, namely, G204 and H12-6A, was analyzed. To evaluate heme availability, a heterologous 17α-hydroxylase cytochrome P-450 cDNA (P-450c17) was expressed in these strains, and its activity was measured in vivo. Heme deficiency in G204 led to accumulation of squalene and lethality. The heterologous cytochrome P-450 was inactive in this strain. The leaky H12-6A strain presented a slightly modified sterol content compared to that for the wild type, and the P-450c17 recovered partial activity. By analyzing sterol transfer on nongrowing cells, it was shown that the cells were permeable toward exogenous cholesterol when they were depleted of endogenous sterols, which was the case for G204 but not for H12-6A. It was concluded that the fully blocked heme mutant (G204) replenishes its diminishing endogenous sterol levels during growth by replacement with sterol from the outside medium. Endogenous sterol biosynthesis appears to be the primary factor capable of excluding exogenous sterol. Oleate but not palmitoleate was identified as a component that reduced but did not prevent sterol transfer. Sterol transfer was only slightly affected by a lack of esterification. It is described herein how avoidance of the potential cytotoxicity of the early intermediates of the mevalonate pathway could be achieved by a secondary heme mutation in erg auxotrophs.
Wild-type yeast cells are permeable toward exogenous sterols during anaerobiosis but not during aerobiosis. In the absence of oxygen, the metabolism of the yeast cell is affected at different levels. Indeed, oxygen is required for the synthesis of ergosterol, first at the level of 2,3-oxidosqualene formation and then for several demethylation and desaturation steps catalyzed by hemoproteins such as the ERG11, ERG25, ERG3, and ERG5 gene products (2, 4, 30, 42). Furthermore, heme is the prosthetic group of different hemoproteins or enzymes involved in important physiological functions of yeast cells, such as respiration (cytochromes of the mitochondrial respiratory chain), detoxification (peroxidase and catalases), sporulation (7), and sterol and unsaturated fatty acid (UFA) metabolism involving cytochrome b5 and P-450s (reviewed in references 23 and 24). Accordingly, apart from having an obligate fermentative metabolism, heme mutants have a requirement for sterol, methionine, and UFAs.
The isolation of mutants deficient in ergosterol biosynthesis (erg mutants) from yeast cells is possible only in the presence of a secondary heme mutation (21, 22). This has stimulated analysis of the relationship between the heme status and sterol uptake in erg mutants. When sterol uptake was analyzed in a hem1 erg7 double mutant (31), it could be shown that the sterol uptake is inversely correlated with the endogenous sterol content (sterol-dependent uptake).
Mutants blocked in the late steps of the ergosterol pathway (comprising erg6 [19], erg2 [1], erg3 [2], erg4 [27], and erg5 [42] and here designated late erg mutants) do not require any added sterol for their growth on glucose or at most low ergosterol levels for sparking of growth on oxidative substrates (43). The erg11, erg24, and erg25 mutants blocked, respectively, in cytochrome P-450 lanosterol 14"-demethylase, C14-sterol reductase, and C4-sterol demethylase have been desecribed as obligate anaerobes (29), which can also be grown when they are associated with suppressor mutations. In particular, an erg11 mutation can be relieved by an erg3 mutation (3, 46, 49), an erg24 mutation can be relieved by the suppressing effect of fen1 (25) and by complete synthetic medium (12), and erg25 can be relieved by the epistatic effect of an erg11 mutation associated with a supplementary leaky heme mutation required for ergosterol uptake (18). Mutations in the early genes specific for ergosterol biosynthesis (here designated early erg mutants), i.e., erg9 (16), erg1 (20, 28), and erg7 (11, 40), lead to strains which need the heme-deficient state to achieve the ergosterol uptake required for their growth (21, 22, 44). The term aerobic sterol exclusion has been coined to describe this process, which is independent of sterol-dependent uptake. When the hem1 strain (G204) was made heme competent by addition of δ-aminolevulinic acid (ALA), exogenous cholesterol uptake was reduced to a minimum (41). In the absence of heme precursors, this strain not only contained wild-type levels of membrane sterols, mostly ergosta-5,7-dienol, but was able to take up high levels of exogenous sterols (41). Aerobic sterol exclusion resulted in more than 70% reduction in the uptake of exogenous cholesterol when a hem1 erg1 double mutant was rendered heme competent, compared to the heme-deficient state (32).
The aim of the present study was to analyze the conditions for uptake of extracellular sterol in Saccharomyces cerevisiae. This study took advantage of mutants blocked in the heme biosynthetic pathway because they grow under aerobic conditions and are permeable to exogenous sterols when they are heme depleted. Heme and sterol biosynthesis can be restored by the addition of heme precursors to the growth medium. A number of physiological indicators, such as viability, sterol composition, and the extent of uptake or transfer, were monitored. The results led to a better understanding of factors governing sterol uptake by yeast cells.
MATERIALS AND METHODS
Yeast strains.
The heme status of the G204 mutant strain (MATα his4 hem1-5 [ρ+] gal− in an FL200 background) (48) is controversial. Although this strain was first described as a completely blocked hem1 mutant (48), it was later revealed to be a leaky mutant (41). In our hands, growth on glucose ceased at an optical density at 600 nm (OD600) of 2 (see Table 1) due to heme depletion and mortality (see Results); growth in the presence of excess hemin or ALA ceased at an OD600 of 20, whereas growth of the strain after addition of excess ergosterol, Tween 80 (as a source of UFA), and methionine ceased between 6 and 10 OD units. It was suggested that the difference in growth yields resulted from an oxidative or fermentative metabolism. A second strain, H12-6A (MATa ura2 leu1 hem13-3 [ρ+]), resulted from a cross between FL100 (MATα ura2 [ρ+]) and H12 (MATa leu1 arg4 hem13-3 [ρ−] in an SP4 background) (5). H12-6A was a leaky heme mutant, whose heme content was fivefold less than the parental SP4 heme content, namely, 12 to 14 nmol of heme/g (dry weight [DW]). The strain grows well without additions but lacks catalase and peroxidase activities.
TABLE 1.
Bioconversion as a probe of P-450c17 activity in heme mutants G204/pTG10710 and H12-6A/pTG10710
| Strain and supplement (concn [μg/ml]) | OD600
|
Bioconversion (%)a
|
||
|---|---|---|---|---|
| 24 h | 48 h | 24 h | 48 h | |
| G204/pTG10710 | ||||
| None | 2.1 | 2.8 | —c | — |
| Ergosterol (50), methionine (50), Tween 80b | 6.0 | 6.1 | 2 | 1 |
| Hemin (15) | 6.6 | 6.7 | 14 | 35 |
| Hemin (50) | 7.3 | 10.8 | 53 | 100 |
| ALA (50) | 6.0 | 21.1 | 98 | 90 |
| H12-6A/pTG10710 | ||||
| None | 8.2 | 8.4 | 10 | 24 |
| Ergosterol (50), methionine (50), Tween 80b | 7.8 | 9.4 | 8 | 22 |
| Hemin (15) | 7.7 | 7.7 | 23 | 45 |
| Hemin (50) | 8.0 | 8.5 | 40 | 100 |
Bioconversion was normalized to the maximum level (100%) obtained after 48 h under heme-competent conditions. The absolute values (expressed in micrograms of progesterone transformed into 17α-hydroxyprogesterone per milliliter of medium), normalized for 10 OD units, were 40 μg/ml for G204/pTG10710 and 25 μg/ml for H12-6A/pTG10710, after 48 h.
Tween 80 was added to a final concentration of 1% (vol/vol).
—, not determined.
The HEM1 gene was deleted by recombination with a hem1::LEU2 disruption cassette transformed into strains FY1679-28c (47) and CDS04 (FY1679-28c [are1::neo are2::HIS3]). Selection was for leucine prototrophy on synthetic minimal medium (39) supplemented with ALA at 50 μg/ml. Heme deficiency was confirmed by the absence of growth on yeast-peptone-dextrose (YPD), unless in the presence of ergosterol, oleate, and methionine. The hem1-deleted strains were named FYH04 and CDSH04, respectively.
Growth conditions.
For sterol uptake and in vivo progesterone bioconversion, the cells were grown in YPD medium (39), with additions as indicated in Tables 1 to 3. The culture was inoculated to an OD600 of 0.1 (106 cells/ml) with cells grown on YPD solid medium and, in the case of G204, on YPD medium containing 50 μg of ALA per ml.
TABLE 3.
Cholesterol uptake in hem mutants expressing the P450c17 cDNA
| Strain and supplement (concn [μg/ml]) | OD600 | Cholesterol uptake (μg/mg [DW])a |
|---|---|---|
| G204/pTG10710 | ||
| None | 2.4 | 8.2 |
| Ergosterol (50), methionine (50), Tween 80b | 8.7 | 0.3 |
| Hemin (50) | 10.0 | 0.2 |
| ALA (50) | 12.2 | 0.6 |
| H12-6A/pTG10710 | ||
| None | 10.2 | 0.3 |
| Ergosterol (50), methionine (50), Tween 80b | 10.6 | 0.02 |
| Hemin (50) | 10.5 | 0.03 |
Uptake was measured after 24 h of growth after cholesterol addition. (Similar uptake values were obtained after 46 h.)
Tween 80 was added to a final concentration of 1% (vol/vol).
For sterol transfer analysis, cultures were started with an inoculum (prepared as described above) at an OD600 of 0.05 and were shaken for 40 h at 28°C. Heme-depleting conditions were attained for the hem1 strains (G204, FYH04, and CDSH04) in YPD medium containing 50 μg of methionine per ml and 1% (vol/vol) Tergitol Nonidet P-40–95% ethanol (1:1 [vol/vol]) (Tergitol-ethanol), unless otherwise indicated. Heme competence was maintained for the wild-type FL200 by growth on YPD medium and for the hem1 strains by supplementation with 100 μg of ALA per ml. In all cases, the OD values were multiplied by 0.23 mg to obtain the corresponding numbers of milligrams (DW) per milliliter.
Cell survival.
Survival after 40 h of growth was measured by diluting the cells in distilled water and counting the numbers of CFU on YPD solid medium containing 50 μg of ALA per ml. Heme deficiency was then confirmed by the absence of growth on YPD.
Expression of the P-450c17 cDNA.
The bovine P-450c17 cDNA (15, 36, 54) was inserted under the control of the TEF1 promoter on a medium-copy-number plasmid associated with the neo gene of Tn5 as the selection marker. The framework of the plasmids and cloning by recombination in Escherichia coli (14) have been described elsewhere (13). The expression plasmid was called pTG10710.
Western blotting.
After 24 h of growth, the cells were harvested and lysed with glass beads. Protein amounts were determined according to the method described by Bradford (6) with a reagent purchased from Bio-Rad and bovine serum albumin as the reference. A 20-μg amount of protein per crude extract was resolved by sodium dodecyl sulfate-12% polyacrylamide gel electrophoresis performed according to the method described by Laemmli (26). Proteins were electroblotted onto a nitrocellulose (Millipore) membrane. Recombinant bovine P-450c17 was detected by probing with rabbit anti-P450c17 serum, followed by anti-rabbit immunoglobulin G (biotinylated) and streptavidin-horseradish peroxidase conjugate (Amersham). Immunoreactive proteins were visualized with the ECL detection system (Amersham) according to the manufacturer’s instructions.
In vivo P-450c17 activity.
P-450c17 activity was probed by analysis of in vivo conversion of progesterone to 17α-hydroxyprogesterone by growing cells in YPD medium supplemented with progesterone (solubilized in Tergitol-ethanol) at 100 μg/ml. The final Tergitol-ethanol concentration in the medium was 0.5%. After 24 or 48 h of growth, samples of the whole culture were taken and 250 μl was extracted twice with 5 ml of dichloromethane. Steroids were separated by reverse-phase high-performance liquid chromatography (HPLC; HP1090; Hewlett-Packard) under isocratic conditions with 60% acetonitrile in water with an Ultrasphere octyldecyl silane column (Beckman) at 45°C at a flow rate of 1 ml/min and were detected at 240 nm. Extraction efficiency was internally calibrated with 11-deoxycortisol as a standard.
Sterol uptake analysis.
Sterol uptake was measured for yeast strains grown in 30 ml of YPD medium with the addition of 10 μg of [4-14C]cholesterol (5,000 dpm/μg) per ml, prepared from a stock solution supplied by NEN (at 54 mCi/mmol and 40 μCi/ml of ethanol) or by Amersham (at 53 mCi/mmol and 50 μCi/ml of ethanol). Cholesterol was added from a 5-mg/ml stock solution in Tergitol-ethanol. Samples were collected by removing 10 ml of cultures after 22 and 46 h of growth. After centrifugation, the cells were washed three times with 0.5% (vol/vol) Tergitol NP-40 and once with water. The radioactivity accumulated in the cells was counted by liquid scintillation (41).
Sterol transfer analysis.
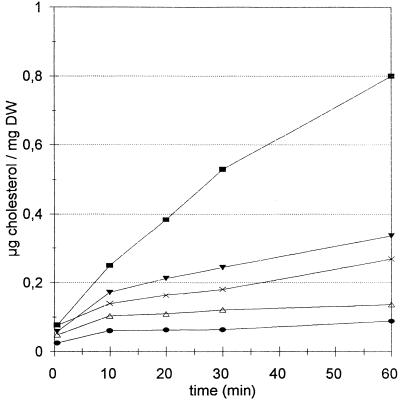
Sterol-depleted cells were collected by centrifugation and were washed once in 0.5% (vol/vol) Tergitol NP-40 and twice in Tris-EDTA (TE) buffer (pH 7.5) (37) and used for cholesterol transfer. Cells were suspended in TE buffer, and [4-14C]cholesterol, solubilized in Tergitol-ethanol, was added at a concentration of 100 μg per ml at a specific activity of 10,000 dpm/μg. The final concentration of Tergitol-ethanol was 4%. The transfer reaction was performed at 30°C with shaking and was started by the addition of a cell suspension at a final density of 20 OD U/ml. At 10-min intervals, samples of 100 μl were collected, centrifuged, and washed as described above for sterol uptake analysis. Controls showed that increasing the number of washes with Tergitol did not further decrease radioactivity, nor did a prolonged (1-h) treatment of the cells with 4% Tergitol. The amount of cholesterol transferred to the cells was expressed in micrograms per milligram (DW). The results are reproducible when the following two parameters are taken into account: (i) the time period of heme depletion (40 h was found necessary and sufficient and has been used throughout) and (ii) addition of ALA to the growth medium used for preparing the inoculum (see Results and Fig. 1). Nevertheless, it was technically difficult to avoid some sterol transfer at time zero (t0); i.e., the time required for washing the cells was significant compared to a fast initial cholesterol transfer reaction. In accordance with this, more cholesterol was transferred at t0 for more permeable cells (e.g., see Fig. 1). In impermeable cells, maximal transfer was completed within 20 to 60 min, whereas transfer lasted for more than 60 min in the permeable cells but declined.
FIG. 1.
Kinetics of cholesterol transfer. The G204 strain was grown for 40 h on YPD medium supplemented with 0, 0.5, 1, and 100 μg of ALA per ml (▪, ▾, ×, and ▵, respectively). •, wild-type FL200 included to show basal transfer level. FL200 was grown overnight. Survival and OD600 values, respectively, were 4% and 2.0, 55% and 5.7, 60% and 6.3, 100% and 20.2, and 100% and 10.5. Kinetics of cholesterol transfer were measured as described in Materials and Methods.
Sterol-ester analysis.
Lipids were extracted by dimethyl sulfoxide-dichloromethane treatment of the cells as described elsewhere (34). Samples were applied onto silica gel plates (Silica Gel 60; Merck), and the plates were developed with hexane-diethyl ether-acetic acid (70:30:2 [vol/vol/vol]). Sterols were detected by spraying with sulfuric acid (40%), followed by heating to 65°C for 20 min. The amounts of free and esterified [4-14C]cholesterol were quantified by scanning the thin-layer chromatography (TLC) plate with an Automatic TLC-linear analyzer (LB2832; Berthold).
Sterol analysis.
Sterol extraction after saponification of lyophilized cells and analysis were performed as described previously (33). Δ5-7 sterols (ergosterol and ergosta-5,7-dienol) were quantified by measuring the UV absorbance at 281.5 nm (38). Analysis of Δ5-7 sterols by HPLC was conducted as described elsewhere (50). For gas-chromatography (GC) analysis, a GC6000 (Carlo-Erba) supplied with a flame ionization detector and an on-column injector was employed. The capillary column was an RSL150 column (Alltech). Sterol composition was determined based on retention times relative to that of cholesterol, which was used as an internal standard (33).
RESULTS
Effect of heme status on progesterone bioconversion.
The activity of a heterologous cytochrome, P-450, was used to probe heme availability in yeast. Indeed, bovine P-450c17 was shown previously to be expressed from a high-copy-number vector to 0.4% of the total microsomal proteins (15). We hypothesized that the dispensable P-450c17 can compete with endogenous hemoproteins for their prosthetic group and, therefore, can reveal heme availability. The heme-requiring strains (G204 and H12-6A) were transformed with pTG10710 and analyzed for production of the P-450c17 antigen by Western blot analysis. The results show that a specific immunoreactive antigen with an estimated apparent molecular mass of 57 kDa, corresponding to that for P-450c17, was present. Moreover, whatever the growth conditions used (absence or presence of hemin or the heme precursor ALA), the P-450c17 antigen was present at roughly the same level. Thus, the endogenous heme level does not seem to have an effect on P-450c17 antigen stability.
As a probe of cytochrome P-450c17 activity, the bioconverting ability of the heme mutant G204, which was affected in the HEM1 gene encoding the ALA synthase (48), was tested. Transformants were submitted to a bioconversion assay with progesterone as the substrate (Table 1). When the G204/pTG10710 strain was supplemented with its growth requirements (ergosterol, Tween 80, and methionine), bioconversion was practically nil. Addition of hemin or ALA at 50 μg/ml allowed achievement of the level obtained with other strains (reference 15 and unpublished data). Maximal bioconversion was attained after 24 h of growth when the medium was supplemented with ALA, but only after 48 h of growth when it was supplemented with excess hemin. Thus, the results obtained with G204/pTG10710 show an absolute requirement for heme (hemin or its precursor ALA) for P-450c17 activity as measured by progesterone bioconversion. These results reveal that the G204 strain was fully blocked in its heme and hence ergosterol biosynthesis.
The leaky heme mutant strain H12-6A was affected in the HEM13 gene (5) and synthesized about 20% of the wild-type heme level (5). This is sufficient to provide the cell with functional cytochrome b5, P-450s, and hemoproteins to synthesize the methionine, UFA, and ergosterol necessary for growth (5). Bioconversion of progesterone by H12-6A/pTG10710 was obtained, but its level was low (25% of control) in medium lacking heme. When the strain was supplemented with heme-sparing products such as ergosterol, methionine, and fatty acids, the level of bioconversion of progesterone by P-450c17 was not increased. This indicates the absence of a feedback mechanism on the heme biosynthetic pathway involving these compounds. In the presence of a roughly constant apoprotein level, an increasing hemin supply correlated with an increase in the level of bioconversion (Table 1). Bioconversion activity is at least a semiquantitative measure of the heme status of cells and shows that P-450c17 is capable of acquiring heme, even under heme-limiting conditions.
Influence of competition for heme on sterol composition.
The extent of the competition for heme between P-450c17 and endogenous P-450s could be indirectly probed in the heme mutants by measuring their sterol compositions. Especially the extents of accumulation of squalene, lanosterol, or ergosta-5,7-dienol were able to reveal the activity of the hemoproteins involved in ergosterol biosynthesis.
The sterol composition of the heme-depleted G204 strain showed a strong squalene and lanosterol accumulation; this composition shifted toward that of the wild type upon addition of ALA (Table 2). These results are consistent with a complete block in heme biosynthesis (48).
TABLE 2.
Sterol composition of the hem1 (G204) and hem13 (H12-6A) mutantsa
| Strainb | Supplementc | OD600 after 24 h | Squalene (%)d | Lanosterol (%)d | Δ5-7 sterol (%)d | Ergosterol/ergosta-5,7-dienol ratioe |
|---|---|---|---|---|---|---|
| G204 | — | 3.3 | 69.0 | 13.9 | 0.4 | NA |
| ALA | 9.6 | ND | 10.4 | 74.2 | 0.8 | |
| H12-6A | — | 10.2 | ND | 8.6 | 66.4 | 1.4 |
| Hemin | 10.7 | ND | 3.9 | 62.3 | 3.1 | |
| H12-6A/pTG10710 | — | 10.0 | 1.0 | 19.8 | 58.7 | 0.5 |
| Hemin | 10.0 | ND | 6.1 | 62.3 | 2.4 | |
| FL200 | — | 12.2 | ND | 6.1 | 74.5 | 3.3 |
| SP4 | — | 14.6 | ND | 5.4 | 66.1 | 26.0 |
Data are mean values from three independent experiments, with a maximum mean deviation of 10%, except for H12-6A/pTG10710 with hemin, which had a maximum mean deviation of 19%. NA, not applicable, since there was no ergosterol; ND, not detectable.
Strains were grown as described in Materials and Methods for sterol uptake.
—, no additions. Hemin and ALA were added at a concentration of 50 μg/ml.
Percentage of total area after integration of GC peaks.
The ergosterol/ergosta-5,7-dienol ratios were determined by HPLC.
The sterol pattern of the heme-leaky H12-6A strain (Table 2) differs from those of its parentals (SP4 and FL200) and the hem1 strain. Although the levels of squalene and lanosterol were unchanged compared to those of the parent strains, a strong decrease in the ratio of ergosterol to ergosta-5,7-dienol was observed. This ratio was ≥3 for the wild-type strains but was 1.4 for H12-6A. The addition of hemin restored wild-type levels. This result demonstrates that both P-450s specific to the ergosterol pathway were affected by the leaky heme condition. When heterologous P-450 was overexpressed, the ratio of ergosterol to ergosta-5,7-dienol decreased from 1.4 to 0.5 with a slightly reduced Δ5-7 sterol amount. Addition of hemin to the culture medium modified the sterol pattern, reaching at least the level of the H12-6A strain (without heterologous P-450 overexpression) or even that of the wild-type strain.
In summary, our results confirm that the G204 strain is completely blocked in its heme biosynthesis. In contrast, overexpression of heterologous P-450 in H12-6A clearly competed with the endogenous P-450s for heme supply. Furthermore, the sterol composition of H12-6A paralleled, at least qualitatively, that of the G204 strain described by Shinabarger et al. (41); when the strain was grown with a limited supply or in the absence of heme precursors (ALA), ergosta-5,7-dienol accumulated. The ratio of ergosterol to ergosta-5,7-dienol decreased, yet the strain was capable of taking up high levels of exogenous sterol (41). The study of sterol uptake in strain H12-6A was of special interest, since not only did it contain sterols, but its heme content was significantly reduced compared to that of the wild type, especially upon coexpression of P-450c17.
Sterol uptake in heme mutants during growth.
Exogenous sterol uptake in both heme mutants expressing cytochrome P-450c17 cDNA was measured to exaggerate their heme deficiencies. The growth of the transformed mutants in medium containing cholesterol (Table 3) shows that addition of UFAs, methionine, and an excess of ergosterol, which leaves the heme status of the cells unchanged (see above), completely shut off the uptake of cholesterol in both strains. As expected, a maximal sterol uptake occurred for the G204 mutant grown on unsupplemented medium, which was reduced to 3 to 7% upon addition of ALA or hemin (Table 3). In contrast, the H12-6A mutant, with only 20% of the heme content of the wild type (48), showed a significant but low level of uptake, which was completely abolished upon addition of hemin. To measure sterol uptake while avoiding heme depletion during growth, we decided to study uptake kinetics in the absence of growth. This measurement offers an estimate of the uptake capacity of cells whose sterol composition is defined initially.
Cholesterol transfer in the absence of growth.
A kinetic method (see Materials and Methods) has been developed to measure sterol transfer at high cell density and during short time intervals for strain G204. The results show that the rate of sterol transfer in heme-depleted cells was maximal only in the absence of ALA (Fig. 1). Sterol transfer occurred only when ergosterol biosynthesis was arrested (Table 4) and when squalene, and to a lesser extent lanosterol, accumulated. The viability of the heme-depleted cells decreased to 4%. This is not per se incompatible with sterol uptake, since viability and cellular energy are not required (31) for this process.
TABLE 4.
Sterol composition of the heme-depleted G204 hem1 mutant
| Supplement (concn [μg/ml]) | Sterol composition (μg/mg [DW])a
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Erg. | 5,7-dienol | Zymo. | Feco. | Lano. | Squalene | Sterols 1 | Sterol 2 | Total sterol | |
| ALA (100) | 5.82 | 2.27 | 0.34 | 0.92 | 0.51 | ND | ND | ND | 9.86 |
| Methionine (50), Tergitolb | 0.21 | ND | ND | ND | 1.85 | 10.66 | 1.62 | 0.33 | 14.67 |
| Palmitoleate,c methionine (50), Tergitolb | ND | ND | ND | ND | 5.30 | 14.08 | 1.55 | 0.30 | 21.23 |
| Oleate,c methionine (50), Tergitolb | ND | ND | 0.16 | ND | 2.56 | 0.77 | 1.22 | 0.31 | 5.02 |
| Ergosterol (80), methionine (50), Tergitolb | 5.26 | ND | ND | ND | 3.75 | 8.50 | ND | 0.39 | 17.90 |
Sterol composition was determined by GC and calculated relative to the cholesterol standard, after 40 h of growth as described in Materials and Methods for sterol transfer. ND, not detectable; Erg., ergosterol; 5,7-dienol, ergosta-5,7-dienol; Zymo., zymosterol; Feco., fecosterol; Lano., lanosterol; Sterols 1, three unidentified sterols, each present in comparable amounts for the different heme-depleted growth conditions tested; Sterol 2, unidentified sterol whose levels vary independently of the levels of sterols 1.
Added to a final concentration of 1% (vol/vol) from a Tergitol-ethanol stock solution.
Added to a final concentration of 0.05% (vol/vol).
The time course of the sterol transfer is shown in Fig. 1. A value for the heme-depleted cells of 0.2 ± 0.02 μg/10 min/mg (DW) (mean of four independent measurements of four different cultures) was obtained. At this rate, the maximal cholesterol content of permissive cells, which was reported to attain 4 μg/mg (DW) (32), would be attained in 3 to 4 h. Comparison of sterol transfer in strains H12-6A and G204 showed that the heme-mediated sterol exclusion could be an indirect effect resulting from resumption of sterol biosynthesis when G204 was rendered heme competent.
Influence of ergosterol and UFAs on cholesterol transfer.
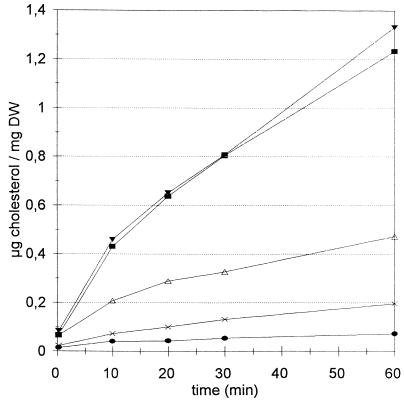
As shown in Fig. 1, heme-depleted cells are largely inviable but are nevertheless able to transfer cholesterol efficiently. Sterol transfer in the absence of growth allows assessment of the influence of all requirements resulting from heme deficiency (methionine, UFA, and ergosterol) on transfer. Sterol transfer on G204 cells which were loaded with oleate or palmitoleate with or without ergosterol during heme depletion was measured. Whenever the cells were loaded with ergosterol (without or with oleate or palmitoleate), transfer was very low compared to that for the wild-type strain or the G204 strain made fully heme competent (Fig. 2). The viability of these cells was somewhat less than the level obtained with heme-competent cells (Fig. 2). It is to be concluded that the sterol content defines the rate and extent of sterol transfer or uptake, independently of heme status.
FIG. 2.
Cholesterol transfer on the heme-depleted G204 strain loaded with oleate. Cholesterol transfer was analyzed for G204 strains previously grown for 40 h in YPD medium supplemented with 50-μg/ml methionine and 1% (vol/vol) Tergitol-ethanol (▪); 0.05% (vol/vol) palmitoleate, 50-μg/ml methionine, and 1% (vol/vol) Tergitol-ethanol (▾); 0.05% (vol/vol) oleate, 50-μg/ml methionine, and 1% (vol/vol) Tergitol-ethanol (▵); and 0.05% (vol/vol) palmitoleate, 80-μg/ml ergosterol, 50-μg/ml methionine, and 1% (vol/vol) Tergitol-ethanol (×). Wild-type FL200 (•) was included to show basal transfer level. Survival and OD600 values, respectively, were 7% and 1.9, 8% and 1.0, 73% and 8.1, 64% and 5.8, and 100% and 21.6. Kinetics of cholesterol transfer were measured as described in Materials and Methods. Repeat experiments show essentially the same results. Cholesterol transfer rates measured for the G204 strain grown on medium supplemented with ALA (100 μg/ml); with ergosterol, methionine, and Tergitol-ethanol; and with oleate, ergosterol, methionine, and Tergitol-ethanol have not been included because their rates were superposable to that of FL200. Survival and OD600 values, respectively, were 100% and 22.1, 47% and 3.4, and 74% and 7.1. Similarly, the kinetics obtained for 1% (vol/vol) Tween 80 and methionine were superposable to those of oleate, methionine, and Tergitol-ethanol.
On the other hand, heme-depleted cells or palmitoleate-loaded, heme-depleted cells showed maximal sterol transfer rates (Fig. 2). Viability decreased to very low levels. Sterol transfer measured on oleate (or Tween 80 [data not shown])-loaded, heme-depleted cells decreased, whereas cell yield and viability improved over those for the completely heme-depleted cells. The reduction (measured over a 60-min period) amounted to three- to fourfold compared to fully heme-depleted cells. The difference is similar to that observed for uptake between a heme-competent and a heme-deficient hem1 erg1 strain (32). It may be concluded that one aspect of the aerobic sterol exclusion is the cells’ capacity to synthesize oleate.
Sterol analysis in heme-depleted cells.
Sterol composition in heme-depleted cells prepared under different conditions was analyzed. When cells contained ergosterol, low sterol transfer rates were observed (Table 4). However, a comparable amount of lanosterol (Table 4) did not prevent sterol transfer. The presence of high amounts of squalene (Table 4) correlated with an increased rate of sterol transfer in the absence of ergosterol. The sterol composition of the oleate-loaded, heme-depleted cells did not explain their low sterol transfer capacity.
A number of observations allowed the correlation of sterol content and lethality observed under the different culture conditions. A low rate of survival was correlated not with the presence of unidentified peaks or with the accumulation of lanosterol (Fig. 2; Table 4) but with the presence of squalene, in the absence of ergosterol. If the presence of ergosterol or heme competency protected the cells from dying, interestingly, a reduction of squalene content, due to the presence of oleate but not palmitoleate, had the same positive effect on viability.
Cholesterol transfer is independent of sterol esterification.
Esterification of absorbed sterol might influence the kinetics of the transfer reaction (44). This possibility would be in agreement with the fact that in yeast, cholesterol is deposited mainly in the steryl ester fraction (35). Recently, two genes, ARE1 and ARE2, whose products are implicated in the esterification of sterols in yeast, were identified (51, 52). Strain CDS04 contains null alleles of both ARE1 and ARE2 genes and consequently no longer accumulates either steryl esters or cholesteryl esters (reference 51 and data not shown). A hem1 deletion was introduced into strain CDS04 and its parent, FY1679-28c, by gene replacement.
The results (Table 5) show that the absolute amount of sterol transferred by the heme-depleted CDSH04 (hem1 are1 are2) strain was reduced by about 20% compared to the ARE parent strain. During the first hour of transfer, no evidence for the appearance of esterified cholesterol was found. It follows that the effect of oleate on sterol transfer did not operate through its esterification.
TABLE 5.
Comparison of the in vitro kinetic of sterol transfer between a hem1 (FYH04) strain and a hem1 are1 are2 (CDSH04) strain
| Strain and supplement (concn [μg/ml]) | Amt of cholesterol transferred (μg/mg [DW]/time unit)
|
||
|---|---|---|---|
| 10 min | 1 h | 24 h | |
| FYH04 | |||
| Methionine (50) and Tergitola | 0.18 | 0.46 | 3.86 |
| ALA (100) | 0.16 | 0.19 | 0.27 |
| CDSH04 | |||
| Methionine (50) and Tergitola | 0.14 | 0.35 | 3.07 |
| ALA (100) | 0.07 | 0.09 | 0.18 |
Added to a final concentration of 1% (vol/vol) from a Tergitol-ethanol stock solution.
DISCUSSION
In this study, we analyzed the role of heme competence with respect to three aspects of cell metabolism: (i) generation of active P-450 enzymes, (ii) alteration in sterol composition, and (iii) permeability toward cholesterol.
Coexpression of a heterologous P-450 as an indicator of heme status.
Physiological differences between a heme auxotroph, G204, and a leaky heme mutant, H12-6A, were uncovered. The G204 strain under aerobiosis conditions behaved as a wild-type yeast cultured under anaerobiosis. It took up considerable amounts of sterol (Table 3) as described by Shinabarger et al. (41). The heterologous P-450c17 antigen levels remained unchanged independently of the exogenous heme supply. However, P-450c17 was inactive, in agreement with the data of Urban-Grimal and Labbe-Bois (48), indicating a lack of heme pigments.
On the other hand, the leaky heme H12-6A mutant showed various modifications of its sterol pattern resulting from lowered heme-dependent endogenous enzyme activities. Moreover, overexpression of heterologous P-450c17 (H12-6A/pTG10710) led to a reduced ratio of ergosterol to ergosta-5,7-dienol (Table 2). The heterologous P-450c17 presents partial activity, as probed by a fast and sensitive bioassay. Both of these observations indicate that a competition between hemoproteins for the incorporation of heme exists. Clearly, yeast cell enzymes have a hierarchy in the fulfillment of their heme requirements (28).
Sterol uptake during growth occurs in the absence of ergosterol.
We have shown that G204 is fully permissive for uptake during growth (41) or transfer in nongrowing cells. In contrast, H12-6A is much less competent for uptake of exogenous sterols. Importantly, H12-6A contains the same levels of ergosterol and ergosta-5,7-dienol previously reported for the G204 (hem1) strain (41). Is the G204 strain able to take up sterols despite the presence of cyclic sterols, whereas H12-6A is much less able to do so? In order to solve this discrepancy, a method to measure sterol transfer (see below) on nongrowing cells was developed. The cells were grown either under complete heme depletion conditions or with small amounts of ALA. When ALA was present in the growth medium, the cells contained ergosterol and ergosta-5,7-dienol, and cholesterol transfer was reduced (Fig. 1). On the other hand, completely heme-depleted G204 cells contained only squalene and a small amount of lanosterol instead of ergosterol and ergosta-5,7-dienol. These cells were inviable. The mortality observed with the hem1 strain explains the discrepancy between the sterol profile found in this study and that reported by Shinabarger et al. (41). Indeed, the latter authors studied sterol uptake during growth, which requires viable cells which were not fully depleted of sterol. The remaining level of sterols is comparable to that of the leaky hem13 strain analyzed in the present study. It follows that the partially heme-depleted (but viable) G204 strain is capable of counterbalancing the dilution of its endogenous sterols during growth by replacement with exogenous sterols.
Esterification is not a driving force in sterol transfer.
It has been shown that uptake and esterification are used to control the types of sterols in the free sterol fraction, leading to a relative enrichment of ergosterol-like sterols (35, 45). In particular, no esterification occurs when ergosterol is supplied in the outside medium, whereas other sterols (such as cholesterol) are esterified (45). Ergosterol is localized to the plasma membrane (53), whereas esterification occurs in the microsomal fraction (53) and could be a driving force for (chole)sterol transfer. Therefore, it was of interest to analyze whether the mutants unable to esterify sterols (51, 52) were still capable of sterol transfer. The contribution of the ester synthases to the transfer reaction was small. It follows that the leveling off of the initial transfer reaction did not reflect a saturation of the steryl ester pools. The membranes incorporated ergosterol more readily than any other sterol tested in competition experiments (45). Furthermore, as shown here, cholesterol transfer was virtually abolished when the cells were loaded with ergosterol prior to (cholesterol) transfer. Finally, it was shown that cholesterol transfer is not affected by a lanosterol content equivalent to the level obtained at ergosterol saturation.
Oleate but not palmitoleate can reduce sterol transfer rates.
In contrast to palmitoleate, oleate-fed, heme-depleted cells incorporated cholesterol at lower rates. This could indicate the modified physical properties of the (plasma) membrane, in which oleate could influence sterol insertion, as suggested by others (8), while palmitoleate had no effect. The reduction in the rate of sterol transfer observed in oleate-fed, heme-depleted cells correlated with that found during growth of a hem1 erg1 strain rendered heme competent with ALA. This suggests that the oleate formed during heme competency is responsible for the reduction in sterol uptake in the hem1 erg1 strain. The estimated amount (0.4 μg/mg [DW]) taken up during the first hour was close to the minimal vital amount, which was estimated to be 0.3 to 0.6 μg/mg (DW) (31, 32). However, the extent of the reduction in sterol uptake by oleate suggests that this is not the mechanism which explains the anaerobic nature of early erg mutants. Why, then, are early erg mutants obligate anaerobes?
Viability of early erg mutants could depend on adjusted hydroxymethylglutaryl-coenzyme A reductase (HMGR) levels.
Hampton et al. (19) suggested that ergosterol biosynthesis represents a sink which could reduce or prevent buildup of the early intermediates of the mevalonate pathway, involved in the conversion of acetyl coenzyme A into farnesyl pyrophosphate, and of squalene. These intermediates are potentially toxic due to the physical properties of these molecules (8, 10, 19).
The need for a secondary heme mutation to rescue early erg mutants is understood by the physiological consequence of heme deficiency. This reduces the accumulation of toxic mevalonate pathway intermediates. Indeed, Casey et al. (9) have shown that heme deficiency dramatically reduces mevalonate-derived nonsaponifiable lipid production in yeast cells by a decreased flux through HMGR1. During heme competence, yeast cells have a stable and high flux through HMGR1, which in early erg mutants—grown under aerobioc conditions—is no longer feedback regulated by ergosterol (9). The predictable accumulation of cytotoxic intermediates causes cell death. Under conditions of heme depletion, synthesis of HMGR2 increases (9, 19), while HMGR1 remains at a minimum. Heme-depleted cells lose their viability and accumulate high levels of squalene, indicating that the mevalonate pathway is not coordinately regulated. Furthermore, oleate-fed, heme-depleted cells, in comparison to heme-depleted cells or palmitoleate-fed, heme-depleted cells, contain less sterol, in particular squalene, and remain viable (references 8 and 9 and the present work). The reduction in squalene content is in accord with the feedback inhibition of HMGR2 by oleate (or derivatives) (9), which in the absence of heme activation of HMGR1 allows the cell to remain viable. This hypothesis agrees with the effect of a leaky erg12-2 mutation, which affects mevalonate kinase while reducing the overall flux through the mevalonate pathway and rescues erg20 mutants defective in farnesyl diphosphate synthase from aerobic death (10).
ACKNOWLEDGMENTS
We thank R. Labbe-Bois for her kind gift of the G204 and H12-6A strains, B. Guiard for the plasmid construct containing the hem1::LEU2 deletion block, G. Cauet and C. Ledoux for help with steroid analysis, and D. Pompon for helpful advice in obtaining the ARE knockout strains and critical reading of the paper.
This work was supported by Hoechst Marion Roussel.
REFERENCES
- 1.Arthington B A, Hoskins J, Skatrud P L, Bard M. Nucleotide sequence of the gene encoding yeast C-8 sterol isomerase. Gene. 1991;107:173–174. doi: 10.1016/0378-1119(91)90314-2. [DOI] [PubMed] [Google Scholar]
- 2.Arthington B A, Bennett L G, Skatrud P L, Guynn C J, Barbuch R J, Ulbright C E, Bard M. Cloning, disruption and sequence of the gene encoding yeast C-5 sterol desaturase. Gene. 1991;102:39–44. doi: 10.1016/0378-1119(91)90535-j. [DOI] [PubMed] [Google Scholar]
- 3.Bard M, Lees N D, Turi T, Craft D, Cofrin L, Barbuch R, Koegel C, Loper J C. Sterol synthesis and viability of erg11 (cytochrome P450 lanosterol demethylase) mutations in Saccharomyces cerevisiae and Candida albicans. Lipids. 1993;28:963–967. doi: 10.1007/BF02537115. [DOI] [PubMed] [Google Scholar]
- 4.Bard M, Bruner D A, Pierson C A, Lees N D, Biermann B, Frye L, Koegel C, Barbuch R. Cloning and characterization of ERG25, the Saccharomyces cerevisiae gene encoding C-4 sterol methyl oxidase. Proc Natl Acad Sci USA. 1996;93:186–190. doi: 10.1073/pnas.93.1.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bilinski T, Litwinska J, Lukaskiewicz J, Rytka J, Simon M, Labbe-Bois R. Characterization of two mutant strains of Saccharomyces cerevisiae deficient in coproporphyrinogen oxidase activity. J Gen Microbiol. 1981;122:79–87. doi: 10.1099/00221287-122-1-79. [DOI] [PubMed] [Google Scholar]
- 6.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 7.Briza P, Eckerstoffer M, Breitenbach M. The sporulation-specific enzymes encoded by the DIT1 and DIT2 genes catalyze a two-step reaction leading to a soluble LL-dityrosine-containing precursor of the yeast spore wall. Proc Natl Acad Sci USA. 1994;91:4524–4528. doi: 10.1073/pnas.91.10.4524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buttke T M, Pyle A L. Effects of unsaturated fatty acid deprivation on neutral lipid synthesis in Saccharomyces cerevisiae. J Bacteriol. 1982;152:747–756. doi: 10.1128/jb.152.2.747-756.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casey W M, Keesler G A, Parks L W. Regulation of partitioned sterol biosynthesis in Saccharomyces cerevisiae. J Bacteriol. 1992;174:7283–7288. doi: 10.1128/jb.174.22.7283-7288.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chambon C, Ladeveze V, Servouse M, Blanchard L, Javelot C, Vladescu B, Karst F. Sterol pathway in yeast. Identification and properties of mutant strains defective in mevalonate diphosphate decarboxylase and farnesyl diphosphate synthase. Lipids. 1991;26:633–636. doi: 10.1007/BF02536428. [DOI] [PubMed] [Google Scholar]
- 11.Corey E J, Matsuda S P, Bartel B. Molecular cloning, characterization, and overexpression of ERG7, the Saccharomyces cerevisiae gene encoding lanosterol synthase. Proc Natl Acad Sci USA. 1994;91:2211–2215. doi: 10.1073/pnas.91.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crowley J H, Smith S J, Leak F W, Parks L W. Aerobic isolation of an ERG24 null mutant of Saccharomyces cerevisiae. J Bacteriol. 1996;178:2991–2993. doi: 10.1128/jb.178.10.2991-2993.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Degryse E, Dumas B, Dietrich M, Laruelle L, Achstetter T. In vivo cloning by homologous recombination in yeast using a two-plasmid-based system. Yeast. 1995;11:629–640. doi: 10.1002/yea.320110704. [DOI] [PubMed] [Google Scholar]
- 14.Degryse E. In vivo intermolecular recombination in Escherichia coli: application to plasmid constructions. Gene. 1996;170:45–50. doi: 10.1016/0378-1119(95)00858-6. [DOI] [PubMed] [Google Scholar]
- 15.Dumas B, Cauet G, Degryse E, Spagnoli R, Achtetter T. Expression of a bovine P450c17 cDNA in the yeast Saccharomyces cerevisiae. In: Lechner M C, editor. 8th International Conference on Cytochrome P450. Paris, France: John Libbey Eurotext; 1994. pp. 527–530. [Google Scholar]
- 16.Fegueur M, Richard L, Charles N D, Karst F. Isolation and primary structure of the ERG9 gene of Saccharomyces cerevisiae encoding squalene synthetase. Curr Genet. 1991;20:365–372. doi: 10.1007/BF00317063. [DOI] [PubMed] [Google Scholar]
- 17.Gaber R F, Copple D M, Kennedy B K, Vidal M, Bard M. The yeast gene ERG6 is required for normal membrane function but is not essential for biosynthesis of the cell-cycle-sparking sterol. Mol Cell Biol. 1989;9:3447–3456. doi: 10.1128/mcb.9.8.3447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gachotte D, Pierson C A, Lees N D, Barbuch R, Koegel C, Bard M. A yeast sterol auxotroph (erg25) is rescued by addition of azole antifungals and reduced levels of heme. Proc Natl Acad Sci USA. 1997;94:11173–11178. doi: 10.1073/pnas.94.21.11173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hampton R, Dimster-Denk D, Rine J. The biology of HMG-CoA reductase: the pros of contra-regulation. Trends Biochem Sci. 1996;21:140–145. [PubMed] [Google Scholar]
- 20.Jandrowitz A, Turnowsky F, Hogenauer G. The gene encoding squalene epoxidase from Saccharomyces cerevisiae: cloning and characterization. Gene. 1991;107:155–160. doi: 10.1016/0378-1119(91)90310-8. [DOI] [PubMed] [Google Scholar]
- 21.Karst F, Lacroute F. Isolation of pleiotropic yeast mutants requiring ergosterol for growth. Biochem Biophys Res Commun. 1973;52:741–747. doi: 10.1016/0006-291x(73)90999-6. [DOI] [PubMed] [Google Scholar]
- 22.Karst F, Lacroute F. Ergosterol biosynthesis in Saccharomyces cerevisiae. Mutants deficient in the early steps of the pathway. Mol Gen Genet. 1977;154:269–277. doi: 10.1007/BF00571282. [DOI] [PubMed] [Google Scholar]
- 23.Labbe-Bois R, Labbe P. Tetrapyrrole and heme biosynthesis in the yeast Saccharomyces cerevisiae. In: Dailey H A, editor. Biosynthesis of heme and chlorophylls. New York, N.Y: MacGraw Hill; 1990. pp. 235–286. [Google Scholar]
- 24.Labbe-Bois R, Camadro J M. Ferrochelatase in Saccharomyces cerevisiae. In: Winkelmann G, Winge W R, editors. Metal ions in fungi. New York, N.Y: Marcel Dekker, Inc.; 1994. pp. 413–453. [Google Scholar]
- 25.Ladevèze V, Marcireau C, Delourme D, Karst F. General resistance to sterol biosynthesis inhibitors in Saccharomyces cerevisiae. Lipids. 1993;28:907–912. doi: 10.1007/BF02537499. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Lai M H, Bard M, Pierson C A, Alexander J F, Goebl M, Carter G T, Kirsch D R. The identification of a gene family in Saccharomyces cerevisiae ergosterol biosynthesis pathway. Gene. 1994;140:41–49. doi: 10.1016/0378-1119(94)90728-5. [DOI] [PubMed] [Google Scholar]
- 28.Landl K M, Klosch B, Turnowsky F. ERG1, encoding squalene epoxidase, is located on the right arm of chromosome VII of Saccharomyces cerevisiae. Yeast. 1996;12:609–613. doi: 10.1002/(SICI)1097-0061(199605)12:6%3C609::AID-YEA949%3E3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 29.Lees N D, Skaggs B, Kirsch D R, Bard M. Cloning of the late genes in the ergosterol biosynthetic pathway of Saccharomyces cerevisiae—review. Lipids. 1995;30:221–226. doi: 10.1007/BF02537824. [DOI] [PubMed] [Google Scholar]
- 30.Loper J. Cytochrome P450 lanosterol 14-demethylase (CYP51): insights from molecular genetic analysis of the ERG11 gene in Saccharomyces cerevisiae. J Steroid Biochem Mol Biol. 1992;43:1107–1116. doi: 10.1016/0960-0760(92)90339-K. [DOI] [PubMed] [Google Scholar]
- 31.Lorenz R T, Rodriguez R J, Lewis T A, Parks L W. Characteristics of sterol uptake in Saccharomyces cerevisiae. J Bacteriol. 1986;167:981–985. doi: 10.1128/jb.167.3.981-985.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lorenz R T, Parks L W. Involvement of heme components in sterol metabolism of Saccharomyces cerevisiae. Lipids. 1991;26:598–603. doi: 10.1007/BF02536423. [DOI] [PubMed] [Google Scholar]
- 33.Marcireau C, Guilloton M, Karst F. In vivo effects of fenpropimorph on the yeast Saccharomyces cerevisiae and determination of the molecular basis of the antifungal property. Antimicrob Agents Chemother. 1990;34:989–992. doi: 10.1128/aac.34.6.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parks L W, Bottema C D K, Rodriguez R J, Lewis T A. Yeast sterols: yeast mutants as tools for the study of sterol metabolism. Methods Enzymol. 1985;111:333–346. doi: 10.1016/s0076-6879(85)11020-7. [DOI] [PubMed] [Google Scholar]
- 35.Parks L W, Casey W M. Physiological implication of sterol biosynthesis in yeast. Annu Rev Microbiol. 1995;49:95–116. doi: 10.1146/annurev.mi.49.100195.000523. [DOI] [PubMed] [Google Scholar]
- 36.Sakaki T, Shibata M, Yabusaki Y, Murakami H, Ohkawa H. Expression of bovine P450c17 cDNA in Saccharomyces cerevisiae. DNA. 1989;8:409–418. doi: 10.1089/dna.1.1989.8.409. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Servouse M, Karst F. Regulation of early enzymes of ergosterol biosynthesis in S. cerevisiae. Biochem J. 1986;240:541–547. doi: 10.1042/bj2400541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 40.Shi Z, Buntel C J, Griffin J H. Isolation and characterization of the gene encoding 2,3-oxidosqualene-lanosterol cyclase from Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1994;91:7370–7374. doi: 10.1073/pnas.91.15.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shinabarger D L, Keesler G A, Parks L W. Regulation by heme of sterol uptake in Saccharomyces cerevisiae. Steroids. 1989;53:607–623. doi: 10.1016/0039-128x(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 42.Skaggs B A, Alexander J F, Pierson C A, Schweitzer K S, Chun K T, Koegel C, Barbuch R, Bard M. Cloning and characterization of the Saccharomyces cerevisiae C-22 sterol desaturase gene, encoding a second cytochrome P-450 involved in ergosterol biosynthesis. Gene. 1996;169:105–109. doi: 10.1016/0378-1119(95)00770-9. [DOI] [PubMed] [Google Scholar]
- 43.Smith S J, Parks L W. The ERG3 gene in Saccharomyces cerevisiae is required for the utilization of respiratory substrates and in heme-deficient cells. Yeast. 1993;9:1177–1187. doi: 10.1002/yea.320091104. [DOI] [PubMed] [Google Scholar]
- 44.Taylor F R, Parks L W. Adaptation of Saccharomyces cerevisiae to growth on cholesterol: selection of mutants defective in the formation of lanosterol. Biochem Biophys Res Commun. 1980;95:1437–1445. doi: 10.1016/s0006-291x(80)80058-1. [DOI] [PubMed] [Google Scholar]
- 45.Taylor F R, Parks L W. An assessment of the specificity of sterol uptake and esterification in Saccharomyces cerevisiae. J Biol Chem. 1981;256:13048–13054. [PubMed] [Google Scholar]
- 46.Taylor F R, Rodriguez R J, Parks L W. Requirement for a second sterol biosynthetic mutation for viability of a sterol C-14 demethylation defect in Saccharomyces cerevisiae. J Bacteriol. 1983;155:64–68. doi: 10.1128/jb.155.1.64-68.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thierry A, Gaillon L, Galibert F, Dujon B. Construction of a complete genomic library of Saccharomyces cerevisiae and physical mapping of chromosome XI at 3.7 kb resolution. Yeast. 1995;11:121–135. doi: 10.1002/yea.320110204. [DOI] [PubMed] [Google Scholar]
- 48.Urban-Grimal D, Labbe-Bois R. Genetic and biochemical characterization of mutants of Saccharomyces cerevisiae blocked in six different steps of heme biosynthesis. Mol Gen Genet. 1981;183:85–92. doi: 10.1007/BF00270144. [DOI] [PubMed] [Google Scholar]
- 49.Watson P F, Rose M E, Ellis S W, England H, Kelly S L. Defective sterol C5-6 desaturation and azole resistance: a new hypothesis for the mode of action of azole antifungals. Biochem Biophys Res Commun. 1989;164:1170–1175. doi: 10.1016/0006-291x(89)91792-0. [DOI] [PubMed] [Google Scholar]
- 50.Xu S, Norton R A, Crumley F G, Nes W D. Comparison of the chromatographic properties of sterols, select additional steroids and terpenoids: gravity-flow column liquid chromatography, gas-liquid chromatography, and high-performance liquid chromatography. J Chromatogr. 1988;452:377–398. doi: 10.1016/s0021-9673(01)81462-x. [DOI] [PubMed] [Google Scholar]
- 51.Yang H, Bard M, Bruner D A, Gleeson A, Deckelbaum R J, Aljinovic G, Pohl T M, Rothstein R, Sturley S L. Sterol esterification in yeast: a two-gene process. Science. 1996;272:1353–1356. doi: 10.1126/science.272.5266.1353. [DOI] [PubMed] [Google Scholar]
- 52.Yu C, Kennedy N J, Chang C C Y, Rothblatt J A. Molecular characterization of two isoforms of Saccharomyces cerevisiae acyl-coA: sterol acyltransferase. J Biol Chem. 1996;271:24157–24163. doi: 10.1074/jbc.271.39.24157. [DOI] [PubMed] [Google Scholar]
- 53.Zinser E, Paltauf F, Daum G. Sterol composition of yeast organelle membranes and subcellular distribution of enzymes involved in sterol metabolism. J Bacteriol. 1993;175:2853–2858. doi: 10.1128/jb.175.10.2853-2858.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zuber M X, John M E, Okamura T, Simpson E R, Waterman M R. Bovine adrenocortical cytochrome P45017α: regulation of gene expression by ACTH and elucidation of primary sequence. J Biol Chem. 1986;261:2475–2482. [PubMed] [Google Scholar]