Abstract
We have identified a gene in Escherichia coli that is required for both the normal decay of mRNA and RNA synthesis. Originally designated mrsC (mRNA stability), the mrsC505 mutation described here is, in fact, an allele of the hflB/ftsH locus (R.-F. Wang et al., J. Bacteriol. 180:1929–1938, 1998). Strains carrying the thermosensitive mrsC505 allele stopped growing soon after the temperature was shifted to 44°C but remained viable for several hours. Net RNA synthesis stopped within 20 min after the shift, while DNA and protein synthesis continued for over 60 min. At 44°C, the half-life of total pulse-labeled RNA rose from 2.9 min in a wild-type strain to 5.9 min in the mrsC505 single mutant. In an rne-1 mrsC505 double mutant, the average half-life was 19.8 min. Inactivating mrsC significantly increased the half-lives of the trxA, cat, secG, and kan mRNAs, particularly in an mrsC505 pnp-7 rnb-500 rne-1 multiple mutant. In addition, Northern analysis showed dramatic stabilizations of full-length mRNAs in a variety of mrsC505 multiple mutants at 44°C. These results suggest that MrsC, directly or indirectly, controls endonucleolytic processing of mRNAs that may be independent of the RNase E-PNPase-RhlB multiprotein complex.
Analysis of mRNA decay in Escherichia coli has focused on the structural features of mRNA molecules that affect stability (17, 33) and on a few enzymes that degrade RNA (13). Genetic and biochemical experiments have shown that four RNases in E. coli are involved in mRNA decay. These include the two 3′→5′ exonucleases polynucleotide phosphorylase (PNPase) and RNase II—encoded by the pnp and rnb genes, respectively—which have been hypothesized to degrade RNA oligonucleotides generated by endonucleolytic decay of larger RNA species (14). Although neither enzyme alone is essential for cell viability, in the absence of both, cells die (14).
Two riboendonucleases have also been characterized. RNase III (rnc), first identified as an rRNA-processing enzyme (16, 34), also cleaves many polycistronic mRNAs (41) and regulates both its own synthesis (8) and that of PNPase (38, 43, 47). Interestingly, mRNA decay is faster in RNase III-deficient strains (5).
RNase E (rne) is a second endonuclease that affects rRNA processing (20) and mRNA decay (37). Discrete trxA mRNA breakdown products can be visualized in an rne-1 pnp-7 rnb-500 multiple mutant (4). In addition, primer extension and S1 nuclease protection experiments showed that a series of endonucleolytic cleavages throughout the trxA mRNA produce these discrete breakdown products (3). RNase E has been implicated in the decay of a growing number of individual mRNAs, including those for ribosomal proteins S20 (27) and S15 (42), the dicB operon (18), several T4 genes (31, 32), and ompA (28). Recently, RNase E has been shown to be a constituent of a multiprotein complex including PNPase and the RhlB RNA helicase (11, 29, 39, 40).
When we observed that mRNA decay in a rncΔ38 rne-1 rnb-500 pnp-7 multiple mutant was only slightly slower than in a wild-type control (5), we set out to find additional genes that affect mRNA decay. Upon examining a series of conditionally lethal mutants that we had isolated while searching for temperature-sensitive alleles of pnp, we identified a series of new genes involved in mRNA turnover. We report here the in vivo characterization of mrsC (mRNA stability), a gene that maps near argG at 69 min on the E. coli chromosome (7). Inactivating mrsC quickly stopped cell growth and net RNA synthesis. DNA and protein synthesis continued normally for over 60 min after a shift to 44°C.
At the nonpermissive temperature, mrsC single mutants had longer half-lives for total pulse-labeled RNA as well as for the trxA, cat, and kan mRNAs; mRNA stability dramatically increased in mrsC505 rne-1 and mrsC505 rne-1 pnp-7 rnb-500 multiple mutants; and full-length trxA transcripts were stable for 60 min after shift to 44°C. While the mrsC505 allele only slightly affected cell morphology, the mrsC505 rne-1 double mutant looked very different at both 30 and 44°C.
In the accompanying report (49), we show that mrsC505 is an allele of the hflB/ftsH gene (23, 48). In addition, the mrsC505 allele confers a temperature-sensitive HflB phenotype, while the hflB29 mutation leads to significant alterations in the decay of specific mRNAs at both 30 and 44°C.
MATERIALS AND METHODS
Materials.
We obtained radioisotopes from the following suppliers: [2,6-3H]phenylalanine (72 Ci/mmol), Amersham; [methyl-3H]thymidine (35 Ci/mmol), ICN Biomedicals; [5,6-3H]uridine (40 Ci/mmol) and [α-32P]dATP, DuPont NEN Research Products. All materials used in RNA manipulations were molecular biology grade. All others were of reagent quality.
Media.
Luria (L) broth and K medium were prepared as described by Miller (30). For strains containing the thyA715 allele, the medium was supplemented with thymine (50 μg/ml). We added the antibiotics (all from Sigma) tetracycline (20 μg/ml), chloramphenicol (20 μg/ml), and kanamycin (50 μg/ml) as necessary. The incorporation medium contained 0.5 μg of thiamine per ml, 0.02 mM CaCl2, 1% glucose, 0.1 mM MgSO4, 10 mg of uridine per ml (as carrier), 10 mg of phenylalanine per ml (as carrier), and 80 mg of deoxyadenosine per ml (as carrier), plus thymine and drugs as required.
Bacterial strains and plasmids.
Table 1 lists the strains and plasmids used. Mutant alleles of RNase E (rne-1) (37), RNase II (rnb-500) (14), and MrsC (mrsC505) encode thermolabile proteins. The allele of PNPase (pnp-7) (44) encodes a nonsense mutation (25a). SK8232 (mrsC505) was constructed by P1-mediated transduction (51) with SK2262 (zgj-203::Tn10 Tcr) as the donor strain and SK6828 (mrsC505) as the recipient strain; the zgj-203::Tn10 insertion was 85 to 95% linked to mrsC. Subsequently, we constructed a series of isogenic, multiple-mutant strains containing mrsC505, using P1 transduction (51) and SK8232 (mrsC505 zgj-203::Tn10) as the donor strain and SK5665 (rne-1), SK5726 (rnb-500 pnp-7), or SK5704 (rne-1 rnb-500 pnp-7) as the recipient strain.
TABLE 1.
E. coli strains and plasmids used
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| Strains | ||
| C600 | leu thr | B. Bachmann |
| JC158 | serA HfrH | A. J. Clark |
| JC1557 | argG6 hisG1 leuB7 metB1 rpsL309 | A. J. Clark |
| MG1693 | thyA715 | CGSC |
| SK2145 | argG6 hisG1 leuB7 metB1 rpsL309 [pVK88B (Tcrqa-2+)] | pVK88B tranformat of JC1557 |
| SK2262 | argG6 hisG1 leuB7 metB1 rpsL309 zgj-203::Tn10 | S. R. Kushner |
| SK2732 | his-61 leuB7 metB1 rpsL309 mrsC505 mrsB1 [pVK88B (Tcrqa-2+)] | This study |
| SK4900 | leu thr zgj-203::Tn10 | C600 × P1SK2262 Tcr transductant |
| SK5662 | argG6 thyA715 zgj-203::Tn10 | MG1693 × P1SK4900 Tcr transductant |
| SK5665 | thyA715 rne-1 | 4 |
| SK5704 | thyA715 rne-1 rnb-500 pnp-7 | 4 |
| SK5726 | thyA715 rnb-500 pnp-7 | 4 |
| SK6828 | thyA715 mrsC505 | SK5662 × P1SK2732 ArgG+ transductant |
| SK7961 | SK5704/pSYK103 | This laboratory |
| SK7967 | MG1693/pSYK103 | This laboratory |
| SK8232 | thyA715 mrsC505 zgj-203::Tn10 | SK6828 × P1SK2262 Tcr transductant |
| SK8236a | thyA715 rne-1 rnb-500 pnp-7 mrsC505, zgj-203::Tn10 | SK5704 × P1SK8232 Tcr transductant |
| SK8238a | thyA715 rne-1 rnb-500 pnp-7 mrsC505 zgj-203::Tn10 | SK5704 × P1SK8232 Tcr transductant |
| SK8239 | thyA715 rne-1 rnb-500 pnp-7 mrsC505 zgj-203::Tn10 [pWK912 (KmrmrsC+)] | This study |
| SK8244 | thyA715 rne-1 mrsC505 zgj-203::Tn10 | SK5665 × P1SK8232 Tcr transductant |
| SK8246 | SK8238/pSYK103 | This study |
| SK8247 | SK8244/pSYK103 | This study |
| SK8248 | SK8236/pSYK103 | This study |
| SK8249 | SK5665/pSYK103 | This study |
| Plasmids | ||
| pSYK103 | Kmr Cmr, pSC101 derivative | This study |
| pVK88B | qa-2+ Tcr, pBR322 derivative | 1 |
| pWK912 | mrsC+ Kmr, pSC101 derivative | 49 |
Independently derived transductant.
We constructed pSYK103 by inserting a 1.8-kb BamHI fragment containing Cmr from pSKS114 (45) into the BamHI site of pLG339 (46). The resulting low-copy-number plasmid (six to eight copies/cell) carried both Cmr and Kmr. pWK912 was derived as described by Wang et al. (49). Strains containing either pSYK103 or pWK912 were constructed by the transformation method of Kushner (25).
Mutant isolation.
To prepare bacteriophage P1, we used the plate lysate procedure (51) on strain JC158. Hydroxylamine mutagenesis on the P1 lysate was performed as described by Kushner et al. (26). Arg+ transductants of SK2145(pVK88B) were selected at 30°C on minimal medium plates. When the transductants were just visible to the naked eye, we replica plated them (12) to both minimal agar plates containing 0.1% Casamino Acids and L-agar plates. The minimal medium plates were then incubated at 42°C, while the L-agar plates were incubated at 44°C. After 24 h of growth at these temperatures, we compared the master and replica plates.
Those colonies visible at 30°C but not at 42 or 44°C were purified at 30°C on minimal medium plates containing 0.1% Casamino Acids. After two rounds of single-colony purifications, we retained as possible temperature-sensitive mutants those independent isolates unable to grow at 44°C. We used argG, gltB, and zgj-203::Tn10 as selected markers for three-factor crosses.
Curing cells containing pVK88B.
We added acridine orange to a final concentration of 50 μg/ml in each 5-ml culture (L broth) of the strains to be cured. We grew the cultures for 24 to 48 h at 30°C and then streaked each one on L-agar plates. Replica plating was used to test 100 colonies from each culture for the loss of Tcr and the retention of temperature sensitivity.
Growth curves and cell viability experiments.
Cells were grown aerobically at 30°C in either L broth or K medium supplemented with thymine and any necessary antibiotics. At a Klett reading of 40 (Klett 40; green filter, no. 42, mid-log phase), cells were switched to 44°C. Klett readings were taken every 20 min. As cells reached Klett 80, they were diluted 10-fold in fresh, prewarmed medium. Cell viabilities were determined by removing samples every 20 min, diluting them with fresh L broth, plating them on L-agar plates, and then incubating them at 30°C for 24 to 48 h.
Macromolecular synthesis.
Our procedure was a modification of Armstrong’s protocol (2). We grew bacterial strains in 10 ml of incorporation medium in a shaking water bath at 30°C. When cells reached Klett 8, either [3H]thymidine (60 μCi/10-ml cell culture), [3H]uridine (40μCi/10-ml cell culture), or [3H]phenylalanine (250 μCi/10-ml cell culture) was added. Klett readings were taken every 20 min, and duplicate 0.2-ml samples were removed and added to 4 ml of 10% ice-cold trichloroacetic acid. At Klett 20, the cells were transferred to a 44°C shaking water bath, and sampling continued every 20 min as described above. Precipitated cells were collected on Whatman GF/C filters (presoaked in 1 mM cold uridine–10% trichloroacetic acid). Filters were washed twice with 5 ml of 5% trichloroacetic acid. We determined radioactivity with liquid scintillation counting using Cytoscint (ICN Biomedicals).
Half-life determination for total pulse-labeled RNA.
We measured the chemical decay of total cellular RNA as described previously (14). Cells were grown at 30°C in K medium (plus thymine and drugs, as needed). At Klett 40, cells were pulse-labeled with [3H]uridine for 80 s. Labeling was stopped by adding rifampin (500 μg/ml), nalidixic acid (20 μg/ml), and cold uridine (200 μg/ml) to the cell culture. Aliquots (0.5 ml) of the cell cultures were removed at various times and added to 3.0 ml of ice-cold 20% trichloroacetic acid. The precipitates were then collected on Whatman GF/C filters, and radioactivity was determined as described above. We determined the half-lives by measuring the loss of acid-precipitable counts over time after transcription initiation was stopped.
RNA isolation.
Cells were grown in L broth at 30°C as described above. At Klett 40, cells were shifted to 44°C, and rifampin (500 μg/ml) and nalidixic acid (200 μg/ml) were added to stop transcription initiation. The cells were incubated for 80 s, 7-ml aliquots were removed at various times, and cells were harvested (24). Total cellular RNA was extracted (52) and treated with DNase I (RNase free; Boehringer Mannheim Biochemicals).
RNA-DNA hybridization.
Our dot blot technique was a modification of that of White and Bancroft (50). RNA samples were dissolved in 0.1× SSC (1× SSC is 0.15 M NaCl plus 15 mM sodium citrate) to a final volume of 100 μl, and then 300 μl of 10× SSC–50% formaldehyde was added. Samples were incubated at 65°C for 15 min to denature the RNA and then quick-chilled on ice. Using a Manifold dot blotting apparatus (Schleicher & Schuell, Inc.), we spotted 5-μg RNA samples onto Biotrans Plus nylon membranes (ICN Biomedicals) and fixed the RNA to the membranes by baking the membranes at 80°C for 1 h.
To quantitate the amount of hybridization, we scanned the autoradiograms with a model 300A computing densitometer (Molecular Dynamics). To determine the percentage of hybridization at each time point, we divided the optical density at that time by the optical density at time zero. These values were then plotted on a log scale, and half-lives were determined by linear regression. Only curves having a correlation coefficient of 0.9 or greater were used in half-life determinations. For the Northern analyses, RNA samples and molecular weight markers (0.16- to 1.77-kb RNA ladder; Bethesda Research Laboratories, Inc.) were dissolved in denaturing dye (deionized formamide with 0.3% xylene cyanol, 0.3% bromophenol blue, and 0.37% disodium EDTA) and incubated at 65°C for 15 min. Samples were then run on either 5 or 6% polyacrylamide gels containing 7 M urea. The RNA was then electroblotted onto Biotrans Plus membranes as described by the manufacturer.
RNA blots were prehybridized for at least 4 h at 42°C. All DNA fragments were labeled with [α-32P]dATP (DuPont NEN Research Products) (19). Following the hybridization step, the membrane was washed twice (15 min for each wash) under high-stringency conditions (0.1× SSC–0.4% sodium dodecyl sulfate at 50°C). Finally, the blots were autoradiographed at −70°C.
Photography of bacterial strains.
Strains were grown at 30°C to Klett 80 in a gyratory water-bath shaker in L broth and then switched to 44°C for further growth. Samples of 2 ml, removed before the temperature shift and at various times thereafter, were added to 40 μl of gluteraldehyde on ice. After centrifugation, cell pellets were resuspended in 0.5 ml of cold L broth and kept on ice, and 4 μl of each cell suspension was placed on a prewarmed (60°C) microscope slide; 4 μl of 4% low-melting-point agarose was added and mixed rapidly with a pipetman. A coverslip was placed on top of the slide and sealed with clear nail polish. The cells were photographed at a magnification of ×400 through a Zeiss Research Microscope, using Kodak Tmax black-and-white film.
RESULTS
Identification of mrsC.
The mrsC505 allele was identified fortuitously during a screen for temperature-sensitive alleles of pnp, the structural gene for PNPase (44). We were interested in PNPase because it helps regulate the expression of eukaryotic genes in E. coli (22). We observed that the presence of a plasmid containing a fragment of Neurospora crassa DNA [pVK88B (Tcr qa-2+)] caused cell inviability in pnp-7 mutants (24a). This property was therefore used to screen for temperature-sensitive mutations in PNPase by using P1-localized mutagenesis. Since the argG locus is closely linked to pnp, mutagenized bacteriophage P1 was transduced into SK2145 (argG6 [pVK88B]), and ArgG+ transductants were selected at 30°C.
Following replica plating to either minimal or L agar at 42 or 44°C, respectively, eight independent isolates out of more than 10,000 transductants exhibited a conditionally lethal growth phenotype. Surprisingly, all of the isolates contained normal PNPase activity and remained inviable at the elevated temperature even after they were cured of pVK88B (data not shown). Further characterization showed that one of the strains, SK2732, contained two independent mutations that both affected mRNA stability. These were designated mrsB1 and mrsC505. The mrsB1 mutation was shown to be unlinked to both argG6 and mrsC505 by P1 transduction. It appears to map near min 24 on the E. coli chromosome (data not shown).
We mapped the temperature-sensitive growth associated with mrsC505 by using three-factor cotransductional crosses with argG6, gltB, or zgj-203::Tn10, a Tcr insertion 80% linked to argG. A gene order of gltB–zgj-203::Tn10–mrsC505–argG-pnp was obtained where mrsC505 was 94% linked to argG and 82% linked to zgj-203::Tn10 (data not shown). For the subsequent experiments, the mrsC505 allele was transduced into MG1693 as described in Materials and Methods to generate SK8232 (thyA715 mrsC505).
Growth and viability of the mrsC505 mutant.
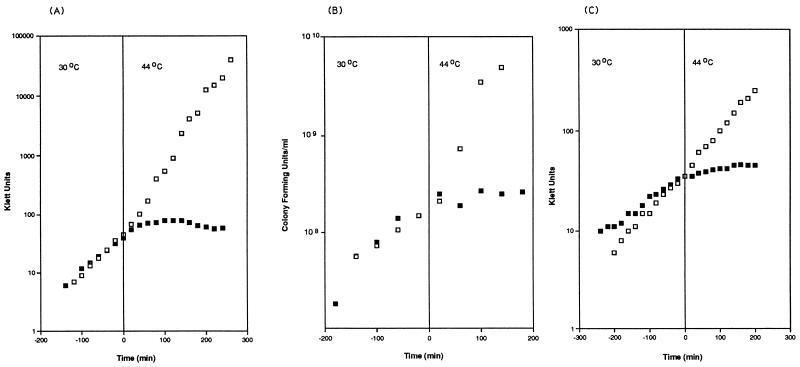
In L broth, the wild-type strain, MG1693, doubled in 35 min at 30°C; when the temperature was raised to 44°C, the generation time decreased to 24 min (Fig. 1A). At 30°C, SK8232 (mrsC505) grew more slowly (50 min) and stopped growing within 45 min after the temperature shift (Fig. 1A). In K medium, results were similar for MG1693 (40 min at 30°C and 35 min at 44°C [Fig. 1C]). The mrsC mutant strain grew much more slowly (83 min) at 30°C (Fig. 1C) and stopped growing immediately at 44°C.
FIG. 1.
Growth and viability of strains containing mrsC505. Cells were grown in either L broth (A and B) or K medium (C) (Materials and Methods). Viable counts (B) were determined by removing aliquots at the times indicated and plating them on L-agar plates at 30°C. □, MG1693; ▪, SK8232.
Cell viability counts were also determined for MG1693 and SK8232 grown in L broth (Fig. 1B). For MG1693 (wild type), the number of viable cells increased exponentially as the experiment progressed. For SK8232 (mrsC505), at 30°C the number of viable cells increased steadily at the same rate as seen for the wild type. When the temperature was shifted to 44°C, however, the number of viable cells leveled off almost immediately and remained constant throughout the remainder of the experiment (Fig. 1B). Of particular interest was that the mrsC505 strain stopped growing at 44°C, but the cells did not immediately die.
Macromolecular synthesis.
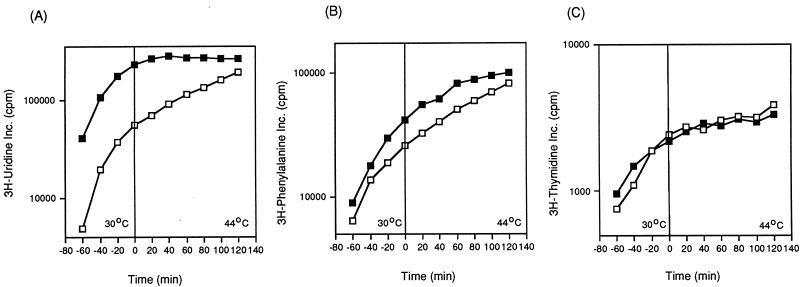
We monitored DNA, RNA, and protein synthesis in MG1693 (wild type) and SK8232 (mrsC505) by measuring the incorporation of either [3H]thymidine, [3H]uridine, or [3H]phenylalanine into cells growing in minimal medium. At 30°C, the rates of RNA, DNA, and protein synthesis in the two strains were comparable (Fig. 2). After the temperature shift to 44°C, DNA and protein synthesis in SK8232 continued normally for at least 60 min before leveling off. In contrast, net [3H]uridine incorporation remained constant by 20 min after the shift to 44°C compared to the wild-type control (MG1693).
FIG. 2.
Macromolecular synthesis of wild-type and mrsC505 strains. Incorporation of [3H]uridine (A), [3H]phenylalanine (B), or [3H]thymidine (C) was carried out as described in Materials and Methods. □, MG1693; ▪, SK8232.
Effect of mrsC505 on the decay of total pulse-labeled RNA.
After shifting cells to 44°C, we pulse-labeled them with [3H]uridine and determined the RNA decay rate (Materials and Methods). The half-life of total pulse-labeled RNA in MG1693 (wild type) was 2.9 min (Table 2). The half-life calculated for SK8232 (mrsC505) was 5.9 min, almost double that for the wild-type control strain (Table 2). In comparison, the half-life for SK5665 (rne-1) was 8.5 min, similar to that reported by Arraiano et al. (4). For a mrsC505 rne-1 double mutant (SK8244), the half-life was 19.8 min, twice that for SK5665 (Table 2). The half-life of total pulse-labeled RNA in SK5704 (rne-1 pnp-7 rnb-500) was 11.4 min. When the mrsC505 mutation was added, the quadruple mutant, SK8238, had a bulk half-life of 25.7 min (Table 2). To show that this increase in half-life was caused by the mrsC505 mutation, we constructed SK8239, a quadruple mutant strain (rne-1 pnp-7 rnb-500 mrsC505) containing a low-copy-number plasmid (pWK912) that carries mrsC+ (49). The half-life of total pulse-labeled RNA in this strain was 12.3 min, comparable to that for SK5704 (11.4 min).
TABLE 2.
Half-lives of total pulse-labeled RNA
| Strain | Genotype | Half-life (min) ± SDa |
|---|---|---|
| MG1693 | Wild type | 2.9 ± 0.4 |
| SK8232 | mrsC505 | 5.9 ± 0.9 |
| SK5665 | rne-1 | 8.5 ± 0.5 |
| SK8244 | mrsC505 rne-1 | 19.8 ± 0.8 |
| SK5704 | pnp-7 rnb-500 rne-1 | 11.4 ± 0.4 |
| SK8238 | mrsC505 pnp-7 rnb-500 rne-1 | 25.7 ± 2.9 |
| SK8239 | mrsC505 pnp-7 rnb-500 rne-1 [pWK912 (mrsC+)] | 12.3 ± 0.3 |
Determined as described in Materials and Methods. Each result is the average of at least two experiments.
Chemical decay of specific mRNAs in mrsC505 strains.
To determine the chemical half-lives of specific mRNAs, we carried out a series of RNA-DNA dot blot assays using a variety of DNA probes. The half-lives of the trxA, cat, and kan mRNAs determined in a variety of genetic backgrounds are presented in Table 3. The trxA message had a half-life of 6.4 min in a mrsC single mutant (SK8248) and a half-life of 3.5 min in the wild-type genetic background (Table 3). Similar changes in half-life were seen in comparison of SK8249 (rne-1, 3.9 min) with SK8247 (rne-1 mrsC505, 8.2 min). Finally, in comparison of the triple mutant, SK7691 (pnp-7 rnb-500 rne-1) with SK8246 (pnp-7 rnb-500 rne-1 mrsC505), the half-life of the trxA transcript more than quadrupled, increasing from 10.4 min for SK7961 to 49.0 min for SK8246.
TABLE 3.
Chemical half-lives of specific mRNAs
| Strain | Genotype | Half-life (min) + SDa
|
||
|---|---|---|---|---|
| trxAb | catc | kanc | ||
| SK7967 | Wild type | 3.5 ± 0.1 | 2.7 ± 0.2 | 3.5 ± 0.6 |
| SK8248 | mrsC505 | 6.4 ± 0.2 | 4.6 ± 0.5 | 6.6 ± 1.0 |
| SK8249 | rne-1 | 3.9 ± 0.1 | 4.5 ± 1.5 | 6.5 ± 0.1 |
| SK8247 | mrsC505 rne-1 | 8.2 ± 0.1 | 6.5 ± 1.5 | 9.7 ± 1.6 |
| SK7961 | pnp-7 rnb-500 rne-1 | 10.4 ± 0.4 | 32.7 ± 5.0 | 22.0 ± 1.5 |
| SK8246 | mrsC505 pnp-7 rnb-500 rne-1 | 49.0 ± 7.0 | >100 | 26.2 ± 0.9 |
Determined as described in Materials and Methods, using RNA-DNA dot blots. Each result is the average of at least two independent experiments.
Determined for a single chromosomal copy of gene.
On a plasmid present at six to eight copies/cell.
In the case of cat (chloramphenicol acetyltransferase), inactivation of mrsC always increased the half-life of the cat mRNA (Table 3). There were similar changes in the half-life of kan (aminoglycoside 3′-phosphotransferase), except in the case of SK7961 versus SK8246 (Table 3).
Northern analysis of specific mRNAs.
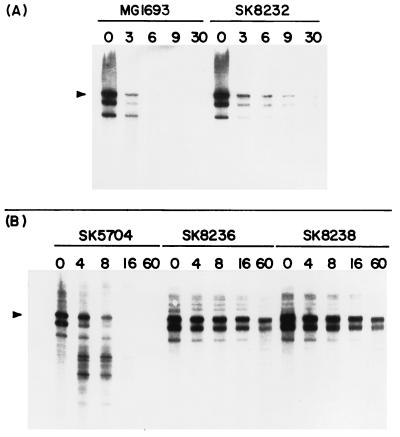
The decay patterns of four mRNAs, trxA, secG, cat, and kan, were investigated by Northern analysis. RNA was isolated at various times after shift to 44°C, separated on either 5 or 6% polyacrylamide gels containing 7 M urea, and probed (Materials and Methods). Shown in Fig. 3A are the trxA-specific Northern blot results for the wild-type strain MG1693 and for SK8232 (mrsC505). At time zero, three bands are present on the Northern blot in MG1693. The top band (493 nucleotides [nt]) is the full-length trxA transcript (3). A smaller fragment (453 nt) is a rapidly produced processing product shortened by 40 nt at the 5′ end (3). The third band is also a rapidly appearing processing product (3). No other, smaller decay products were evident in this strain. Decay occurred quickly, and by 6 min after the shift to the nonpermissive temperature, most of the trxA message was completely degraded. The decay process was slightly slower in SK8232 (mrsC505), with some full-length transcript still visible at 30 min after the shift.
FIG. 3.
Northern analysis of trxA mRNA decay in various strains containing mrsC505. RNA was isolated from the various strains (Materials and Methods) and separated in 6% polyacrylamide gels containing 7 M urea. The number above each lane indicates the time (minutes) after the temperature shift when RNA was extracted; 7 μg of RNA was loaded in each lane. The full-length trxA mRNA (493 nt) is marked with an arrow. (A) MG1693 (wild type) and SK8232 (mrsC505); (B) SK5704 (pnp-7 rnb-500 rne-1), SK8236 (mrsC505 pnp-7 rnb-500 rne-1), and SK8238 (mrsC505 pnp-7 rnb-500 rne-1). SK8236 and SK8238 are independent isolates of the quadruple mutant.
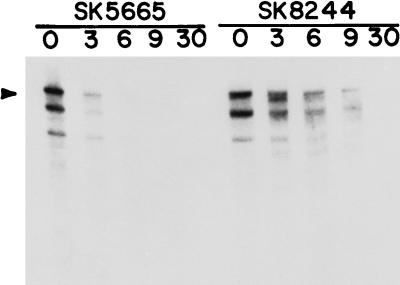
The decay pattern of the trxA transcript was also evaluated in a rne-1 single-mutant strain (SK5665) as well as an rne-1 mrsC505 double-mutant strain (SK8244). The rne-1 allele had little effect on the decay pattern (Fig. 4). In SK5665, as in MG1693, by 6 min after the temperature shift most of the full-length transcript was gone, correlating with the half-lives presented in Table 3. In contrast, in SK8244 (rne-1 mrsC505), full-length transcript could still clearly be seen 9 min after the shift. In addition, several smaller bands representing discrete decay products appeared at later times.
FIG. 4.
trxA mRNA decay in rne-1 (SK5665) and mrsC505 rne-1 (SK8244) mutants. RNA was isolated and separated as described in the legend to Fig. 3; 7 μg of RNA was loaded in each lane. The number above each lane indicates the time (minutes) after the temperature shift when RNA was extracted.
Finally, we examined the trxA decay patterns in SK5704 (pnp-7 rnb-500 rne-1), SK8236 (pnp-7 rnb-500 rne-1 mrsC505), and SK8238 (pnp-7 rnb-500 rne-1 mrsC505). mRNA degradation is much slower in SK5704, and the observed decay products generated in the absence of RNase E, RNase II, and PNPase have been extensively analyzed (3, 4). As expected, we obtained the decay pattern typically seen for the trxA message in SK5704 (Fig. 3B), in which a series of smaller discrete bands appeared, while the full-length transcript disappeared over time after the shift to 44°C. For the quadruple mutants SK8236 and SK8238, the expected full-length bands were visible at time zero. However, no endonucleolytic cleavage products appeared over the course of the experiment. Full-length transcripts could still be seen 60 min after the shift. The intensity of the full-length bands decreased only slightly over time, confirming the half-life data shown in Table 3.
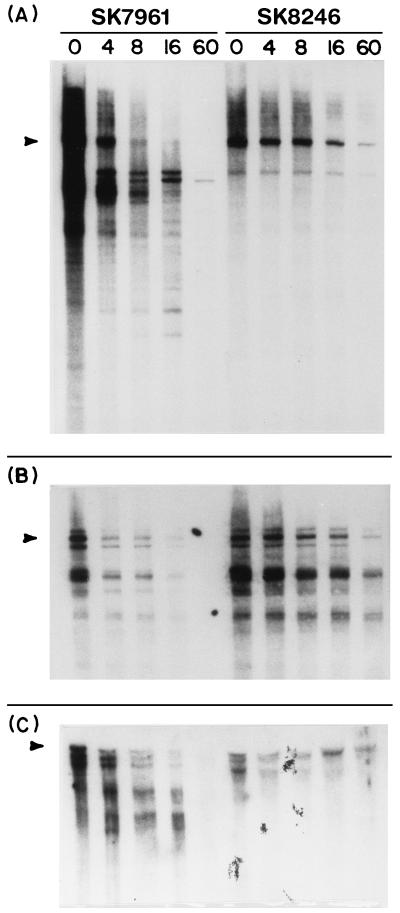
To test the generality of mRNA stabilization in mrsC mutants, the cat, secG, and kan transcripts were tested in both SK7961 (pnp-7 rnb-500 rne-1) and SK8246 (pnp-7 rnb-500 rne-1 mrsC505). The cat probe revealed a strong band at approximately 1,020 nt in SK7961, which appears to be the full-length transcript (Fig. 5A). Also visible at time zero were a series of discrete decay products. As time progressed, the full-length transcript disappeared and the smaller processing products became more intense. Eight minutes after the shift, the full-length transcript was almost completely gone. In contrast, the cat mRNA in SK8246 was dramatically stabilized. At time zero, full-length mRNA was present, as were two smaller processing products. No other discrete bands were seen. As time passed, there was a gradual loss both of full-length transcript and of the smaller bands. Full-length transcript was still visible 60 min after the temperature shift.
FIG. 5.
Effects of mrsC505 on decay of the cat, kan, and secG mRNAs. RNA was isolated as described in the legend to Fig. 3. For analysis of the cat (A) and kan (C) transcripts, RNA was separated on 5% polyacrylamide gels. For secG (B), analysis on a 6% gel was used. The number above each lane indicates the time (minutes) after the temperature shift. (A) cat analysis, 7 μg of RNA/well; (B) secG analysis, 7 μg of RNA/well; (C) kan analysis, 7 μg of RNA/well for SK7961 (pnp-7 rnb-500 rne-1 [pSYK103 cat kan]) and 11 μg/well for SK8246 (mrsC505 pnp-7 rnb-500 rne-1 [pSYK103 cat kan]). The respective full-length transcripts for each mRNA are marked. Full-length transcripts are indicated by arrowheads on the left.
Next, we examined the decay of the secG transcript. secG encodes a protein involved in protein translocation (15, 35, 36) and maps near the argG locus (69 min) on the E. coli chromosome (35). A series of three bands (approximately 450 to 560 nt) were present at time zero in SK7961 (pnp-7 rnb-500 rne-1) (Fig. 5B). The intense band at 530 nt appeared to correspond to the full-length transcript. The secG mRNA decayed quickly in SK7961. Most of the mRNA was fully degraded in 16 min. In contrast, the full-length secG transcript was visible at 16 min in SK8246 (pnp-7 rnb-500 rne-1 mrsC505). Smaller decay products also accumulated in the quadruple mutant and were clearly visible at 60 min.
Finally, we examined the kan mRNA in SK7961 (pnp-7 rnb-500 rne-1) and SK8248 (pnp-7 rnb-500 rne-1 mrsC505) (Fig. 5C). To study the kan mRNA, we had to load 11 μg of RNA for the quadruple mutant strain to see any bands on an autoradiograph. To rule out a problem with the copy number of plasmid pSYK103 in SK8246, plasmid DNA was isolated from SK8246 and SK7961 (10). An equal volume of each preparation was run on a 0.8% agarose gel, and the DNA was visualized by using ethidium bromide. The plasmid bands were of equal intensity for both strains (data not shown), indicating that all strains had retained the plasmid at comparable copy numbers (six to eight copies per cell). For SK7961, a series of three bands was present at time zero. These bands ranged from 1,000 to 1,300 nt long, corresponding to the size expected for the full-length kan transcript. Over time, these bands disappeared with the appearance of a series of smaller degradation products. By 16 min, little full-length message was present; by 60 min everything was fully degraded. In contrast, for SK8246, the full-length message was present at time zero, but over time, the intensity of the full-length bands decreased slightly, with a full-length transcript still clearly visible at 60 min. Furthermore, there was no evidence of smaller decay products.
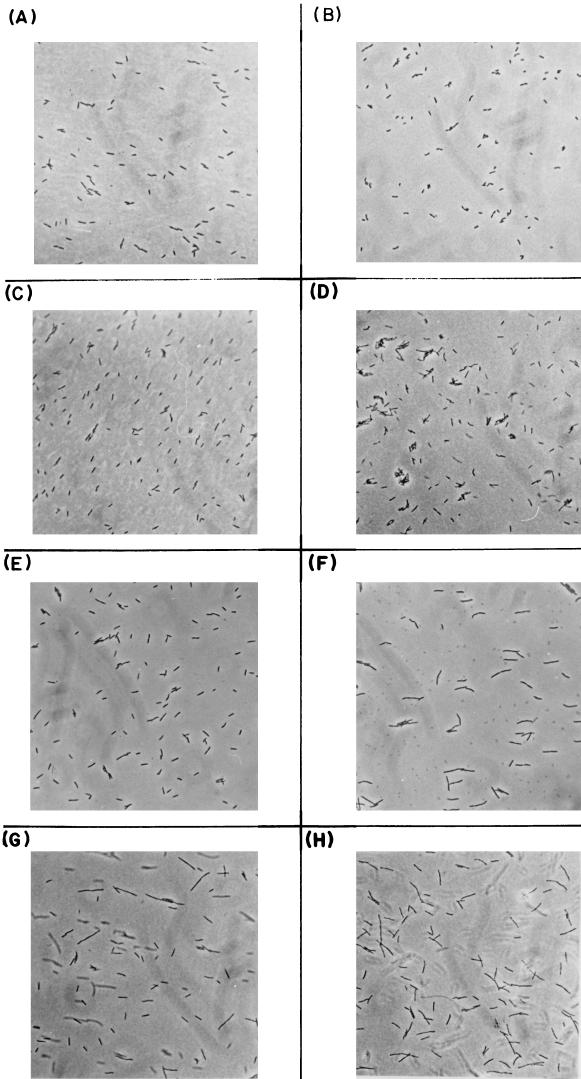
Effect of mrsC505 on cell division.
MG1693 (wild type), SK8232 (mrsC505), SK5665 (rne-1), and SK8244 (mrsC505 rne-1) were grown in L broth at 30°C to mid-log phase and then shifted to 44°C. Cells were examined before shift and at times up to 3 h after the shift. The morphology of the wild-type cells (Fig. 6A and B) was the same at both temperatures. The mrsC505 allele caused the cells to elongate slightly at 44°C, but this change became visible only 3 h after the shift (Fig. 6C and D). In contrast, the rne-1 allele resulted in much longer cells within 2 h after the temperature shift (Fig. 6E and F). Dramatic changes in cell morphology were apparent at both temperatures in the mrsC505 rne-1 double mutant (Fig. 6G and H).
FIG. 6.
Photographs of various E. coli strains. The strains were grown in L broth at 30°C to mid-log phase and then shifted to 44°C. Samples were prepared as described in Materials and Methods and visualized with a Zeiss Research Microscope, using Kodak Tmax black-and-white film. Magnification, ×400. (A) MG1693 (wild type), 30°C; (B) MG1693 (wild type), 180 min at 44°C; (C) SK8232 (mrsC505), 30°C; (D) SK8232 (mrsC505), 180 min at 44°C; (E) SK5665 (rne-1), 30°C; (F) SK5665 (rne-1), 120 min at 44°C; (G) SK8244 (mrsC505 rne-1), 30°C; (H) SK8244 (mrsC505 rne-1), 180 min at 44°C.
DISCUSSION
We report here the identification and characterization of an E. coli gene (mrsC) that is essential for cell growth, normal mRNA decay, and RNA synthesis. Located near the argG locus at 69 min on the chromosome, the mrsC505 allele was identified among a group of temperature-sensitive alleles originally isolated as potential mutations in pnp. The mrsC505 mutant revealed several interesting properties. Its growth in rich and minimal medium stopped quickly after the shift to the nonpermissive temperature (Fig. 1A and C), but the cells did not die (Fig. 1B). In addition, net RNA synthesis stopped shortly after growth ceased (Fig. 2A). The effect on RNA synthesis appeared specific since protein and DNA synthesis continued normally for at least 60 min after the temperature shift (Fig. 2B and C).
The second critical feature of the mrsC505 allele is its ability to alter the half-lives of both total pulse-labeled RNA (Table 2) and various individual mRNAs (Table 3). It is significant that half-lives were affected in both a mrsC505 single mutant and mrsC505 multiple mutants. It is also worth comparing SK8232 (mrsC505) with SK5665. This isogenic strain carries a thermosensitive mutation in RNase E (rne-1), an enzyme that plays an important role in mRNA decay (3, 4, 6, 37). mrsC’s effect on the half-lives of the trxA, cat, and kan mRNAs was greater than rne’s effect (Table 3). In addition, when the two mutations were combined (SK8244), the half-lives were further increased (Tables 2 and 3), suggesting that the two genes function in distinct pathways of mRNA decay. In addition, we observed long half-lives for both total pulse-labeled RNA (25.7 min [Table 2]) and individual mRNAs (26 to 100 min [Table 3]) in a mrsC505 pnp-7 rnb-500 rne-1 quadruple mutant.
Northern analysis of the trxA, secG, kan, and cat mRNAs (Fig. 3 to 5) confirmed our chemical half-life data (Table 3). While the decay pattern of the trxA mRNA did not change significantly in SK8232 (mrsC505), the decay rate slowed (Fig. 3A). In contrast, full-length transcripts in the multiple mutants (SK8236 and SK8238) were the major species present even 60 min after the shift to the nonpermissive temperature (Fig. 3B). This is one of the most significant examples that we have seen of mRNA stabilization in RNA turnover mutants of E. coli. Comparable stabilizations were also seen with cat, secG, and kan mRNAs (Fig. 5). Taken together, our data support the hypothesis that the mrsC-encoded protein may be part of a different mRNA decay pathway that does not involve rne and pnp, two major components of the so-called RNA degradasome (39, 40). Alternatively, the MrsC protein could affect the level of RNase E at the nonpermissive temperature. Furthermore, the absence of the MrsC protein, directly or indirectly, prevents endonucleolytic cleavages of numerous mRNAs.
During the course of our experiments, Tomoyasu et al. reported the nucleotide sequence for ftsH (48), and it was identical to the sequence for mrsC (49). Accordingly, we examined the morphology of the mrsC505 and the rne-1 strains at both 30 and 44°C, since we knew that rne mutants also alter normal cell division (21). Indeed, the rne-1 cells became much longer within 2 h after the shift to 44°C (Fig. 6F). In contrast, the cell shape in the mrsC505 strain was only slightly changed 3 h after the shift. Our results suggest that the mrsC/ftsH-encoded protein has no significant effect on cell division and support the observation that the original ftsH1 allele isolated in Y16 altered cell morphology because the strain also carried an ftsl mutation (9).
In the accompanying report (49), we demonstrate that mrsC505 is an allele of the hflB/ftsH locus (23, 48). In addition, we show that mrsC translation starts at an UUG codon, a rare start codon in E. coli. Furthermore, we demonstrate that mrsC505 confers a temperature-sensitive HflB phenotype, while hflB29 confers a MrsC phenotype at both 30 and 44°C.
ACKNOWLEDGMENTS
We thank C. Ingle and D. Crater for advice on the manuscript.
This work was supported in part by NIHGMS grant GM28760 to S.R.K.
REFERENCES
- 1.Alton N K, Hautala J A, Giles N H, Kushner S R, Vapnek D. Transcription and translation in E. coli of hybrid plasmids containing the catabolic dehydroquinase gene from Neurospora crassa. Gene. 1978;4:241–259. doi: 10.1016/0378-1119(78)90021-5. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong K. M.S. thesis. Athens, Ga: University of Georgia; 1982. [Google Scholar]
- 3.Arraiano C M, Yancey S D, Kushner S R. Identification of endonucleolytic cleavage sites involved in decay of Escherichia coli trxA mRNA. J Bacteriol. 1993;175:1043–1052. doi: 10.1128/jb.175.4.1043-1052.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arraiano C M, Yancey S D, Kushner S R. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J Bacteriol. 1988;170:4625–4633. doi: 10.1128/jb.170.10.4625-4633.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babitzke P, Granger L, Kushner S R. Analysis of mRNA decay and rRNA processing in Escherichia coli multiple mutants carrying a deletion in RNase III. J Bacteriol. 1993;175:229–239. doi: 10.1128/jb.175.1.229-239.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babitzke P, Kushner S R. The Ams (altered mRNA stability) protein and ribonuclease E are encoded by the same structural gene of Escherichia coli. Proc Natl Acad Sci USA. 1991;88:1–5. doi: 10.1073/pnas.88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bachmann B J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990;54:130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bardwell J C A, Regnier P, Chen S-M, Nakamura Y, Grunberg-Manago M, Court D L. Autoregulation of RNase III operon by mRNA processing. EMBO J. 1989;8:3401–3407. doi: 10.1002/j.1460-2075.1989.tb08504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Begg K J, Tomoyasu T, Donachie W D, Khattar M, Niki H, Yamanaka K, Hiraga S, Ogura T. Escherichia coli mutant Y16 is a double mutant carrying thermosensitive ftsH and ftsl mutations. J Bacteriol. 1992;173:2416–2417. doi: 10.1128/jb.174.7.2416-2417.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1520. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carpousis A J, Van Houwe G, Ehretsmann C, Krisch H M. Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell. 1994;76:889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- 12.Clark A J, Margulies A D. Isolation and characterization of recombination-deficient mutants of Escherichia coli K12. Proc Natl Acad Sci USA. 1965;53:451–459. doi: 10.1073/pnas.53.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deutscher M P. The metabolic role of RNases. Trends Biochem Sci. 1988;13:136–139. doi: 10.1016/0968-0004(88)90070-9. [DOI] [PubMed] [Google Scholar]
- 14.Donovan W P, Kushner S R. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci USA. 1986;83:120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Douville K, Leonard M, Brundage L, Nishiyama K, Tokuda H, Mizushima S, Wickner W. Band q subunit of Escherichia coli preprotein translocase and intergral membrane export factor P12 are the same protein. J Biol Chem. 1994;269:18705–18707. [PubMed] [Google Scholar]
- 16.Dunn J J, Studier F W. T7 early RNAs and Escherichia coli ribosomal RNAs are cut from large precursor RNAs in vivo by ribonuclease III. Proc Natl Acad Sci USA. 1973;70:3296–3300. doi: 10.1073/pnas.70.12.3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emory S A, Bouvet P, Belasco J G. A 5′-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 1992;6:135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- 18.Faubladier M, Cam K, Bouche J P. Escherichia coli cell division inhibitor dicF-RNA of the dicB operon. J Mol Biol. 1990;32:461–471. doi: 10.1016/0022-2836(90)90325-G. [DOI] [PubMed] [Google Scholar]
- 19.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 20.Ghora B K, Apirion D. Structural analysis and in vitro processing to p5 (5S) rRNA of a 9S RNA molecule isolated from an rne mutant of E. coli. Cell. 1978;15:1055–1066. doi: 10.1016/0092-8674(78)90289-1. [DOI] [PubMed] [Google Scholar]
- 21.Goldblum K, Apirion D. Inactivation of the ribonucleic acid-processing enzyme ribonuclease E blocks cell division. J Bacteriol. 1981;146:128–132. doi: 10.1128/jb.146.1.128-132.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hautala J A, Bassett C L, Giles N H, Kushner S R. Increased expression of a eukaryotic gene in Escherichia coli through stabilization of its messenger RNA. Proc Natl Acad Sci USA. 1979;76:5774–5778. doi: 10.1073/pnas.76.11.5774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herman C, Ogura T, Tomoyasu T, Hiraga S, Akiyama Y, Ito K, Thomas R, D’Ari R, Bouloc P. Cell growth and λ phage development controlled by the same essential Escherichia coli gene, ftsH/hflB. Proc Natl Acad Sci USA. 1993;90:10861–10865. doi: 10.1073/pnas.90.22.10861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krzyzek R, Rogers P. Arginine control of transcription of argECBH messenger ribonucleic acid in Escherichia coli. J Bacteriol. 1972;110:945–954. doi: 10.1128/jb.110.3.945-954.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Kushner, S. R. Unpublished results.
- 25.Kushner S R. An improved method for transformation of Escherichia coli with ColE1-derived plasmids. In: Boyer H W, Nicosia S, editors. Genetic engineering. Amsterdam, The Netherlands: Elsevier/North-Holland Biomedical Press; 1978. pp. 17–23. [Google Scholar]
- 25a.Kushner, S. R., and R. Ivarie. Unpublished results.
- 26.Kushner S R, Maples V F, Champney W S. Conditionally lethal ribosomal protein mutants: characterization of a locus required for modification of 50S subunit proteins. Proc Natl Acad Sci USA. 1977;74:467–471. doi: 10.1073/pnas.74.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mackie G A. Specific endonucleolytic cleavage of the mRNA for ribosomal protein S20 of Escherichia coli requires the products of the ams gene in vivo and in vitro. J Bacteriol. 1991;173:2488–2497. doi: 10.1128/jb.173.8.2488-2497.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melefors Ö, von Gabain A A. Genetic studies of cleavage-initiated mRNA decay and processing of ribosomal 9S RNA show that the Escherichia coli ams and rne loci are the same. Mol Microbiol. 1991;5:857–864. doi: 10.1111/j.1365-2958.1991.tb00759.x. [DOI] [PubMed] [Google Scholar]
- 29.Miczak A, Kaberdin V R, Wei C-L, Lin-Chao S. Proteins associated with RNase E in a multicomponent ribonucleolytic complex. Proc Natl Acad Sci USA. 1996;93:3865–3869. doi: 10.1073/pnas.93.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 31.Mudd E A, Carpousis A J, Krisch H M. Escherichia coli RNAse E has a role in the decay of bacteriophage T4 mRNA. Genes Dev. 1990;4:873–881. doi: 10.1101/gad.4.5.873. [DOI] [PubMed] [Google Scholar]
- 32.Mudd E A, Prentki P, Belin D, Krisch H M. Processing of unstable bacteriophage T4 32 mRNAs into a stable species requires Escherichia coli ribonuclease E. EMBO J. 1988;7:3601–3607. doi: 10.1002/j.1460-2075.1988.tb03238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Newbury S F, Smith N H, Robinson E C, Hiles I D, Higgins C F. Stabilization of translationally active mRNA by prokaryotic REP sequences. Cell. 1987;48:297–310. doi: 10.1016/0092-8674(87)90433-8. [DOI] [PubMed] [Google Scholar]
- 34.Nikolaev N, Silengo L, Schlessinger D. Synthesis of a larger precursor to ribosomal RNA in a mutant of Escherichia coli. Proc Natl Acad Sci USA. 1973;70:3361–3365. doi: 10.1073/pnas.70.12.3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nishiyama K, Hanada M, Tokuda H. Disruption of the gene encoding p12 (secG) reveals the direct involvement and important function of SecG in the protein translocation of Escherichia coli at low temperature. EMBO J. 1994;13:3272–3277. doi: 10.1002/j.1460-2075.1994.tb06628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishiyama K, Mizushima S, Tokuda H. A novel membrane protein involved in protein translocation across the cytoplasmic membrane of Escherichia coli. EMBO J. 1993;12:3409–3415. doi: 10.1002/j.1460-2075.1993.tb06015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ono M, Kuwano M. A conditional lethal mutation in an Escherichia coli strain with a longer chemical lifetime of mRNA. J Mol Biol. 1979;129:343–357. doi: 10.1016/0022-2836(79)90500-x. [DOI] [PubMed] [Google Scholar]
- 38.Portier C, Dondon L, Grunberg-Manago M, Regnier P. The first step in the functional inactivation of the Escherichia coli polynucleotide phosphorylase messenger is ribonuclease III processing at the 5′ end. EMBO J. 1987;6:2165–2170. doi: 10.1002/j.1460-2075.1987.tb02484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Py B, Causton H, Mudd E A, Higgins C F. A protein complex mediating mRNA degradation in Escherichia coli. Mol Microbiol. 1994;14:717–729. doi: 10.1111/j.1365-2958.1994.tb01309.x. [DOI] [PubMed] [Google Scholar]
- 40.Py B, Higgins C F, Krisch H M, Carpousis A J. A DEAD-box RNA helicase in the Escherichia coli RNA degradosome. Nature. 1996;381:169–172. doi: 10.1038/381169a0. [DOI] [PubMed] [Google Scholar]
- 41.Regnier P, Grunberg-Manago M. RNase III cleavages in non-coding leaders of Escherichia coli transcripts control mRNA stability and genetic expression. Biochimie. 1990;72:825–834. doi: 10.1016/0300-9084(90)90192-j. [DOI] [PubMed] [Google Scholar]
- 42.Regnier P, Hajnsdorf E. Decay of mRNA encoding ribosomal protein S15 of Escherichia coli is initiated by an RNase E-dependent endonucleolytic cleavage that removes the 3′ stabilizing stem and loop structure. J Mol Biol. 1991;187:23–32. doi: 10.1016/0022-2836(91)90542-e. [DOI] [PubMed] [Google Scholar]
- 43.Regnier P, Portier C. Initiation attenuation and RNase III processing of transcripts from the Escherichia coli operon encoding ribosomal protein S15 and polynucleotide phosphorylase. J Mol Biol. 1986;187:23–32. doi: 10.1016/0022-2836(86)90403-1. [DOI] [PubMed] [Google Scholar]
- 44.Reiner A M. Characterization of polynucleotide phosphorylase mutants of Escherichia coli. J Bacteriol. 1969;97:1437–1443. doi: 10.1128/jb.97.3.1437-1443.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shapira S K, Chou J, Richaud F V, Casadaban M. New versatile vectors for expression of hybrid proteins coded by a cloned gene fused to lacZ gene sequences encoding an enzymatically active carboxy-terminal portion of β-galactosidase. Gene. 1983;25:71–82. doi: 10.1016/0378-1119(83)90169-5. [DOI] [PubMed] [Google Scholar]
- 46.Stoker N G, Fairweather N F, Spratt B G. Versatile low-copy-number plasmid vectors for cloning in Escherichia coli. Gene. 1982;18:335–341. doi: 10.1016/0378-1119(82)90172-x. [DOI] [PubMed] [Google Scholar]
- 47.Takata R, Mukai T, Hori K. RNA processing by RNase III is involved in the synthesis of Escherichia coli poynucleotide phosphorylase. Mol Gen Genet. 1987;209:28–32. doi: 10.1007/BF00329832. [DOI] [PubMed] [Google Scholar]
- 48.Tomoyasu T, Yuki T, Morimura S, Mori H, Yamanaka K, Niki J, Hiraga S. The Escherichia coli FtsH protein is a prokaryotic member of a protein family of putative ATPases involved in membrane functions, cell cycle control, and gene expression. J Bacteriol. 1993;175:1344–1351. doi: 10.1128/jb.175.5.1344-1351.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang R-F, O’Hara E B, Aldea M, Bargmann C I, Gromley H, Kushner S R. Escherichia coli mrsC is an allele of hflB, encoding a membrane associated ATPase and protease that is required for mRNA decay. J Bacteriol. 1998;180:1929–1938. doi: 10.1128/jb.180.7.1929-1938.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.White B A, Bancroft F C. Cytoplasmic dot hybridization. J Biol Chem. 1982;257:8569–8572. [PubMed] [Google Scholar]
- 51.Willetts N S, Mount D W. Genetic analysis of recombination deficient mutants of Escherichia coli K-12 carrying rec mutations cotransducing with thyA. J Bacteriol. 1969;100:923–934. doi: 10.1128/jb.100.2.923-934.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Williams M G, Rogers P. Expression of arg genes in Escherichia coli during arginine limitation dependent upon stringent control of translation. J Bacteriol. 1987;169:1644–1650. doi: 10.1128/jb.169.4.1644-1650.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]