同源重组(HR)是一种高度精确的DNA双链断裂(DSB)损伤修复方式。HR修复的关键步骤是重组酶RAD51通过包裹单链DNA形成核酸蛋白纤维丝,进行同源模板搜索,并进行DNA链入侵反应,从而启动DNA修复合成。本研究发现,纤维蛋白(FBL)是重要的HR调节因子。一旦发生DNA损伤,FBL就被招募到DSB位点,并直接与RAD51互作。细胞缺失FBL会导致RAD51在DNA损伤位点募集减少,HR修复效率降低。此外,细胞缺失FBL会导致染色体畸变增加,促使细胞对DNA损伤药物敏感。本研究提出了FBL介导的RAD51在DNA损伤位点的招募的新机制,并强调了FBL在癌症治疗中的潜在意义。
Keywords: 基因组不稳定性, DNA双链断裂, 同源重组, RAD51, 纤维蛋白
Eukaryotic organisms constantly face a wide range of internal and external factors that cause damage to their DNA. Failure to accurately and efficiently repair these DNA lesions can result in genomic instability and the development of tumors (Canela et al., 2017). Among the various forms of DNA damage, DNA double-strand breaks (DSBs) are particularly harmful. Two major pathways, non-homologous end joining (NHEJ) and homologous recombination (HR), are primarily responsible for repairing DSBs (Katsuki et al., 2020; Li and Yuan, 2021; Zhang and Gong, 2021; Xiang et al., 2023). NHEJ is an error-prone repair mechanism that simply joins the broken ends together (Blunt et al., 1995; Hartley et al., 1995). In contrast, HR is a precise repair process. It involves multiple proteins in eukaryotic cells, with the RAD51 recombinase being the key player, which is analogous to bacterial recombinase A (RecA) (Shinohara et al., 1992). The central event in HR is the formation of RAD51-single-stranded DNA (ssDNA) nucleoprotein filaments that facilitate homology search and DNA strand invasion, ultimately leading to the initiation of repair synthesis (Miné et al., 2007; Hilario et al., 2009; Ma et al., 2017).
Ribosomal RNA (rRNA) methyltransferase fibrillarin (FBL) serves as a key regulator in various early stages of ribosome biogenesis (Tollervey et al., 1993; Yoshikawa et al., 2011). It plays pivotal roles in processes such as ribosomal DNA (rDNA) synthesis and pre-rRNA cleavages (Schimmang et al., 1989; Snaar et al., 2000). FBL possesses methyltransferase activity, thus is responsible for the 2'-O-methylation (2'-O-Me) of rRNAs and the methylation of histone H2A at glutamine 104 (Omer et al., 2002; Tessarz et al., 2014). It is predominantly localized in the nucleus without significant tissue or immune cell expression specificity. FBL plays a vital role in early development, displaying heightened expression in pluripotent embryonic stem cells and contributing to stem cell pluripotency (Newton et al., 2003). The knockout of FBL results in reduced nucleolar size and lifespan extension (Tiku et al., 2017). Moreover, increased FBL expression is observed in mammalian oocytes with age-associated dysregulation of the protein metabolism. Interestingly, elevated FBL expression promotes cellular proliferation and confers resistance to chemotherapy in MCF-7 breast cancer cells (Marcel et al., 2013). In breast cancer patients, higher FBL expression correlates with lower survival rates (Marcel et al., 2013). However, whether FBL is involved in DNA repair remains to be elucidated.
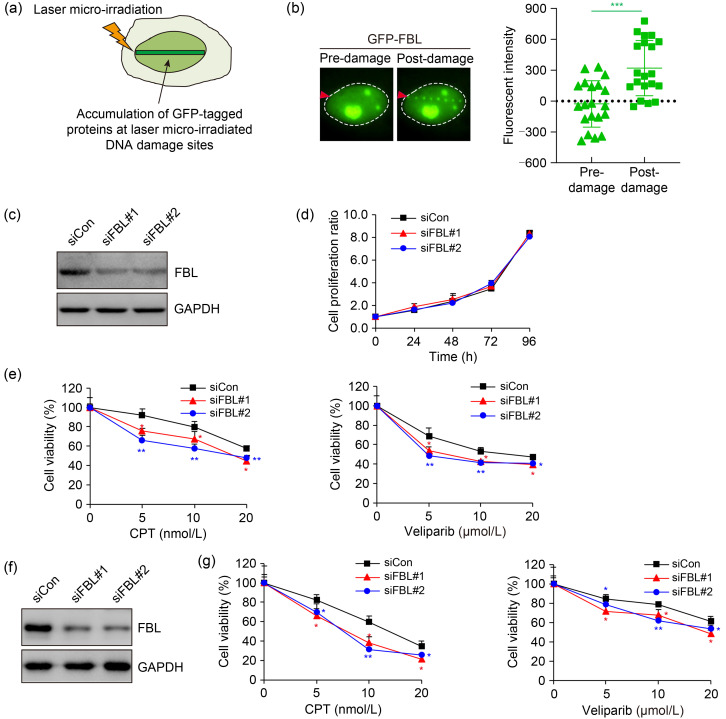
Relocalization to sites of DNA damage is characteristic of proteins involved in the DNA damage response (DDR). We utilized laser microirradiation to induce DNA breaks, which enables the monitoring of protein dynamics through fluorescence microscopy (Fig. 1a). Remarkably, we observed a remarkable accumulation of green fluorescent protein (GFP)-tagged FBL at laser-induced DNA damage stripes (Fig. 1b), suggesting its involvement in DDR.
Fig. 1. Fibrillarin (FBL) recruitment to DNA damage sites and its role in conferring resistance to DNA damage agents. (a) Schematic representation of the accumulation of green fluorescent protein (GFP)-tagged proteins at laser-induced DNA damage sites. (b) Fluorescence microscopy images demonstrating the translocation of GFP-tagged FBL to laser-induced DNA damage sites in U2OS cells. The dashed circles delineate the edges of nuclei, and the red arrowheads indicate laser microirradiation sites. A quantitative analysis of fluorescence intensity at the micro-irradiated site was conducted. (c) Verification of FBL-knockdown efficiency in HeLa cells by immunoblotting. (d) Assessment of cell proliferation in wild-type cells and FBL-knockdown HeLa cells via cell counting kit-8 (CCK8) assay at 0, 24, 46, 72, and 96 h. No significant statistical difference was observed between the control and FBL-knockdown cells using the Student's two-tailed t-test. (e) Colony formation assay illustrating the sensitivity of wild-type cells and FBL-knockdown HeLa cells to camptothecin (CPT) (left) and veliparib (also termed ABT-888) (right). Cells were exposed to varying doses of DNA damage agents and allowed to survive for 14 d before staining. (f) Verification of FBL-knockdown efficiency in Huh7 cells by immunoblotting. (g) Colony formation assay demonstrating the sensitivity of wild-type cells and FBL-deficient Huh7 cells to CPT (left) and veliparib (right). Cells were treated with the specified concentrations of DNA damage agents for 14 d, and then colonies were fixed and assessed using the Coomassie blue staining assay. Data are presented as mean±standard deviation (SD), n=3. * P<0.05, ** P<0.01, *** P<0.001 (Student's two-tailed t-test). siCon: control small interfering RNA; siFBL: FBL small interfering RNA; GAPDH: glyceraldehyde-3-phosphate dehydrogenase.
In order to gain deeper insights into the physiological significance of FBL in DDR, we investigated its requirement for cell survival upon DNA damage. Strikingly, while the partial knockdown of FBL did not have a significant impact on cell proliferation, it rendered cells more susceptible to camptothecin (CPT), a topoisomerase I inhibitor, and veliparib (also termed ABT-888), a poly-adenosine diphosphate (ADP)-ribose polymerase (PARP) inhibitor (Figs. 1c‒1e). We thus further extended our analysis to the hepatoma cell line Huh7 and observed similar results, with FBL depletion leading to increased cellular sensitivity to DNA damage-inducing agents (Figs. 1f and 1g). Collectively, these findings underscore the vital role of FBL in the DDR, highlighting its contribution to cellular resistance against DNA damage.
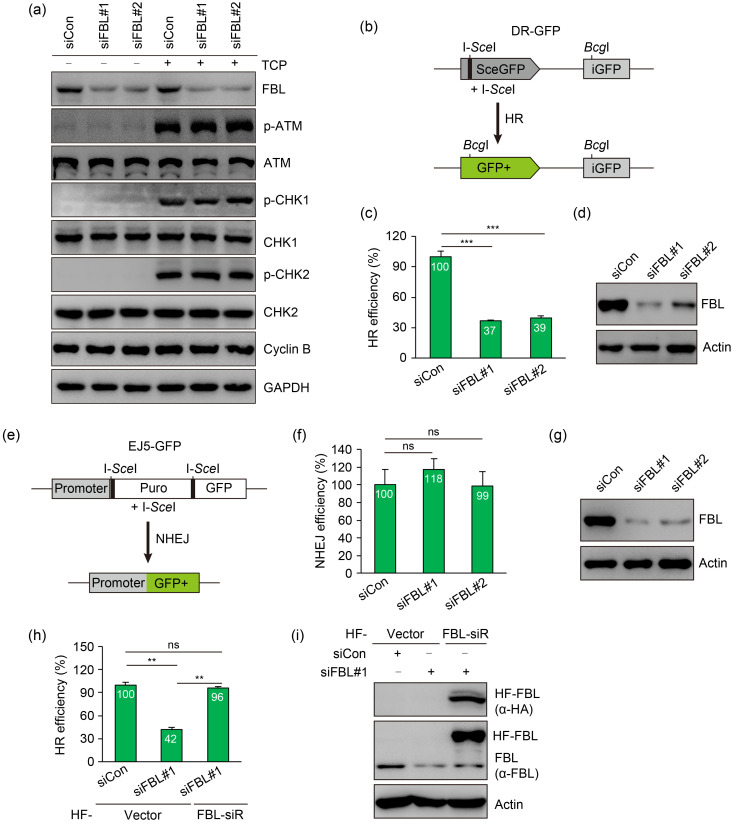
We next determined whether FBL is required for DNA damage-induced checkpoint activation. Interestingly, as shown in Fig. 2a, the activation of checkpoint kinase 1 (CHK1) and CHK2 was comparable between control and FBL-deficient cells following treatment with CPT for 1 h, indicating that FBL did not affect checkpoint regulation. Given that FBL depletion sensitizes cells to CPT and veliparib, we hypothesized that FBL is essential for DSB repair. To investigate the impact of FBL on DSB repair, we utilized U2OS cell lines containing direct repeat (DR)-GFP or end joining 5 (EJ5)-GFP reporter cassettes. The DR-GFP reporter allows for the accurate measurement of HR repair efficiency by monitoring the reconstitution of functional GFP following the repair of an I-SceI-induced DSB using intense GFP (iGFP) as a template (Fig. 2b). Remarkably, the knockdown of FBL resulted in a significant reduction in HR repair efficiency (Figs. 2c and 2d). In contrast, no significant differences were observed in the NHEJ repair frequency following FBL depletion, as assessed using the EJ5-GFP reporter system that measures the direct re-ligation of I-SceI-induced DSBs to restore GFP expression by joining the promoter to the GFP gene cassette (Figs. 2e‒2g). Importantly, the defect in HR repair caused by FBL knockdown was rescued when cells were reconstituted with small interfering RNA (siRNA)-resistant FBL (Figs. 2h and 2i), confirming the direct association between impaired HR repair and FBL silencing. Interestingly, the depletion of FBL had little impact on cell cycle distribution, indicating that the HR defect was not caused by changes in cell cycle progression (Fig. 3a).
Fig. 2. Fibrillarin (FBL) facilitates homologous recombination (HR) repair. (a) FBL silencing does not affect the DNA damage checkpoint activation. HeLa cells were transfected with control small interfering RNA (siRNA) or FBL-specific siRNAs. After 48 h, cells were left untreated or treated with camptothecin (CPT) (1 μmol/L) for 1 h and collected for immunoblotting using the indicated antibodies. (b) Schematic representation of the green fluorescent protein (GFP)-based HR reporter assay. (c, d) Reduced HR-directed DNA repair efficiency in FBL-depleted U2OS direct repeat (DR)-GFP cells. Cells were transfected with control siRNA or FBL-specific siRNAs, followed by electroporation with an I-SceI expression plasmid. After 48 h, cells were analyzed for GFP expression by flow cytometry. (e) Schematic representation of the GFP-based non-homologous end joining (NHEJ) reporter assay. (f, g) FBL depletion does not affect NHEJ-directed DNA repair efficiency in U2OS end joining 5 (EJ5)-GFP cells. Cells were transfected with control siRNA or FBL-specific siRNAs, followed by electroporation with an I-SceI expression plasmid. After 48 h, cells were analyzed for GFP expression by flow cytometry. (h, i) Restoration of HR efficiency in FBL-deficient U2OS DR-GFP cells by re-expression of siRNA-resistant FBL. FBL-deficient U2OS DR-GFP cells stably expressing an empty vector or hemagglutinin (HA) and flag (HF)-tagged wild-type FBL (HF-FBL) were electroporated with an I-SceI expression plasmid. After 48 h, cells were analyzed for HR efficiency. Data are presented as mean±standard deviation (SD), with n=3 (c, d, h, i) or n=4 (f, g). ** P<0.01; *** P<0.001; ns, not significant (Student's two-tailed t-test). ATM: ataxia telangiectasia mutated protein; p-ATM: phospho-ATM; CHK1: checkpoint kinase 1; GAPDH: glyceraldehyde-3-phosphate dehydrogenase; GFP+: wide-type GFP; iGFP: intense GFP; Puro: puromycin; siCon: control small interfering RNA; siFBL: FBL small interfering RNA; siR: small interfering RNA-resistant.
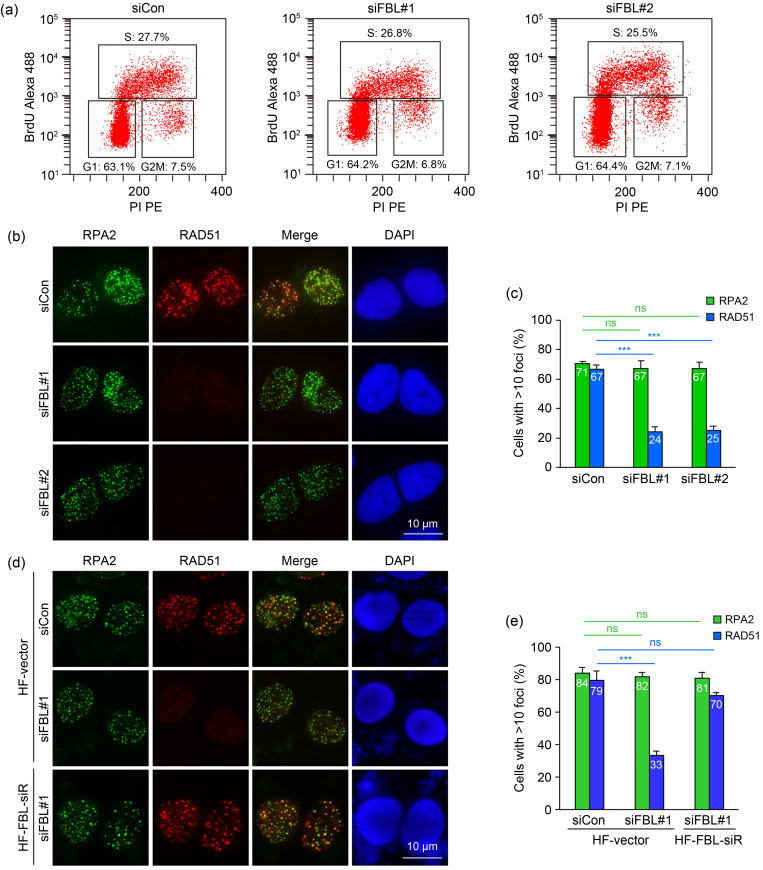
Fig. 3. Fibrillarin (FBL) promotes the recruitment of DNA repair protein RAD51 to DNA damage sites. (a) FBL deficiency has no effect on cell cycle distribution. BrdU incorporation assays were performed to assess cell cycle distribution as described in the Materials and Methods section. (b) FBL silencing in U2OS cells impairs RAD51 foci formation at DNA damage sites, while replication protein A2 (RPA2) recruitment remains unaffected. Cells were treated with 1 μmol/L camptothecin (CPT) for 3 h and subjected to RAD51 and RPA2 immunofluorescence. (c) Quantification of RAD51 and RPA2 foci formation. (d) Rescue of RAD51 recruitment at DNA damage sites by re-expressing wild-type FBL in FBL-depleted U2OS cells. Cells stably expressing an empty vector or hemagglutinin (HA) and flag (HF)-tagged wild-type FBL (HF-FBL) were transfected with the indicated siRNAs, then treated with 1 μmol/L CPT for 3 h and subjected to RAD51 and RPA2 immunofluorescence. (e) Quantification of RAD51 and RPA2 foci formation. Data are presented as mean±standard deviation (SD), n=3. *** P<0.001; ns, not significant (Student's two-tailed t-test). BrdU: bromodeoxyuridine; DAPI: 4',6-diamidino-2-phenylindole; PI: propidium iodide; PE: P-phycoerythrin; siCon: control small interfering RNA; siFBL: FBL small interfering RNA; siR: small interfering RNA-resistant.
During HR repair, DNA end resection generates 3' ssDNA tails, which are initially bound by the replication protein A (RPA) complex. Subsequently, the recombinase RAD51 displaces RPA and forms RAD51-ssDNA filaments, facilitating strand invasion and homology search. To gain further insights into the role of FBL in HR repair, we examined the recruitment of RPA and RAD51 to DSBs. Strikingly, the depletion of FBL significantly impaired the formation of RAD51 foci in response to CPT-induced DNA damage (Figs. 3b and 3c). In contrast, FBL depletion had no observable effect on the formation of RPA2 foci (Figs. 3b and 3c). Furthermore, the defects in RAD51 foci formation in FBL-depleted cells were rescued when siRNA-resistant FBL was reintroduced (Figs. 3d and 3e). These observations collectively suggested that FBL is required for the recruitment of RAD51 to DSBs, highlighting its crucial role in facilitating RAD51-mediated HR repair.
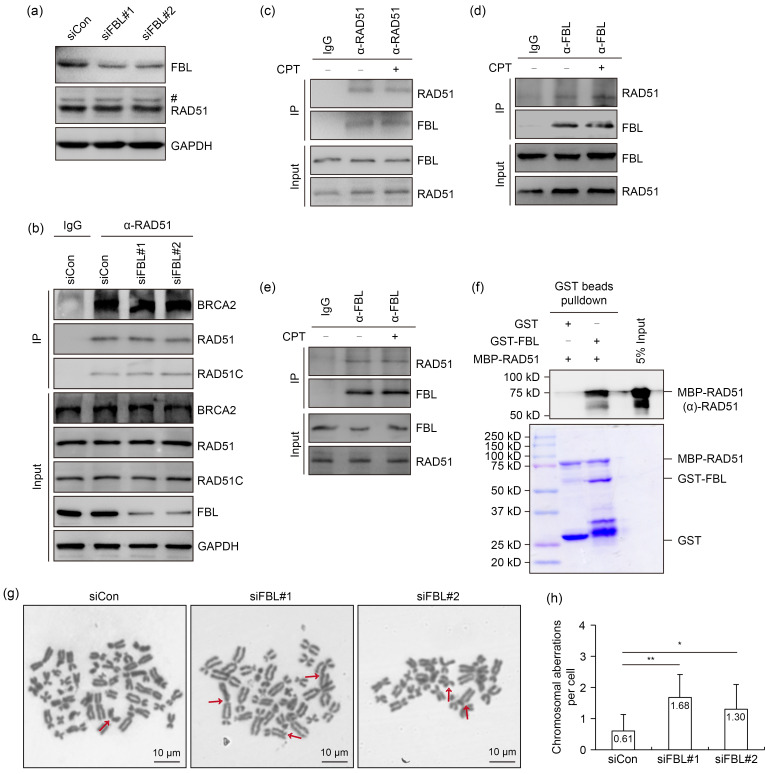
In order to elucidate the mechanism by which FBL promotes RAD51 foci formation, we initially assessed the protein level of RAD51 in FBL-depleted cells. As shown in Fig. 4a, a partial knockdown of FBL had no discernible impact on RAD51 expression. Moreover, the depletion of FBL did not affect the interaction between RAD51 and breast cancer 2 susceptibility protein (BRCA2) or RAD51 homolog C (RAD51C), both of which are essential for RAD51 recruitment to DSBs (Fig. 4b). We subsequently investigated the potential interaction between FBL and RAD51 by performing co-immunoprecipitation experiments using whole-cell extracts from HeLa cells. The immunoprecipitation of endogenous RAD51 using an anti-RAD51 antibody resulted in the detection of FBL in the immunoprecipitates, indicating the interaction between FBL and RAD51 (Fig. 4c). Reciprocal co-immunoprecipitation using an anti-FBL antibody further confirmed the association between FBL and RAD51 (Fig. 4d). Notably, the FBL–RAD51 interaction remained intact even in the presence of Benzonase® or following sonication treatment (Figs. 4c‒4e). Furthermore, the interaction between FBL and RAD51 was unaffected by CPT treatment, indicating that the association is not dependent on DNA damage (Figs. 4c‒4e).
Fig. 4. Fibrillarin (FBL) directly interacts with RAD51. (a) FBL knockdown has no effect on the protein level of RAD51 under camptothecin (CPT) treatment. After FBL knockdown for 48 h, cells were treated with 1 μmol/L CPT for 3 h and whole-cell extracts were prepared to detect the protein level of RAD51. The pound (#) indicates a non-specific band. (b) The depletion of FBL does not disrupt the interaction between RAD51 and breast cancer 2 susceptibility protein (BRCA2) or RAD51 homolog C (RAD51C). After FBL knockdown for 48 h, HeLa cells were treated with 1 μmol/L CPT for 3 h. Cell lysates were incubated with protein A agarose beads conjugated with anti-RAD51 antibody. Western blot analysis was performed using the indicated antibodies. (c, d) Endogenous co-immunoprecipitation confirms the interaction between FBL and RAD51. (e) Lysates from nuclease-treated HeLa cell extracts were subjected to sonication and conducted for co-immunoprecipitation. (f) Recombinant glutathione-S-transferase (GST)-tagged FBL directly interacts with maltose-binding protein (MBP)-tagged RAD51 in vitro. An in vitro pull-down assay was performed using indicated recombinant proteins purified from Escherichia coli. Upper panel: RAD51 detected by immunoblotting; Lower panel: purified proteins visualized by Coomassie staining. (g, h) Metaphase spreads were prepared from wild-type cells and FBL-depleted HeLa cells treated with CPT (4 nmol/L) for 24 h. Representative images are shown in (g), with red arrows indicating chromosomal aberrations. Data are presented as mean±standard deviation (SD) (a minimum of 40 metaphase spreads were scored for each cell line). * P<0.05, ** P<0.01 (Student's two-tailed t-test). GAPDH: glyceraldehyde-3-phosphate dehydrogenase; IgG: immunoglobulin G; IP: immunoprecipitation; siCon: control small interfering RNA; siFBL: FBL small interfering RNA.
In order to determine whether the interaction between FBL and RAD51 is direct, we expressed and purified recombinant glutathione-S-transferase (GST)-tagged FBL and maltose-binding protein (MBP)-tagged RAD51 in Escherichia coli. In vitro pull-down assays demonstrated a direct binding between FBL and RAD51 (Fig. 4f). These findings provided compelling evidence for a direct physical interaction between FBL and RAD51, further supporting their functional association in DNA repair processes.
Considering the crucial role of FBL in HR repair, we hypothesized that the depletion of FBL may lead to genome instability. To investigate this matter, we examined the frequency of chromosomal aberrations in FBL-depleted cells under CPT treatment. Remarkably, these aberrations in FBL-depleted cells were significantly increased compared to the control (Figs. 4g and 4h). These findings suggested that FBL depletion compromises genome stability, highlighting the importance of FBL in maintaining genomic integrity through its involvement in HR repair.
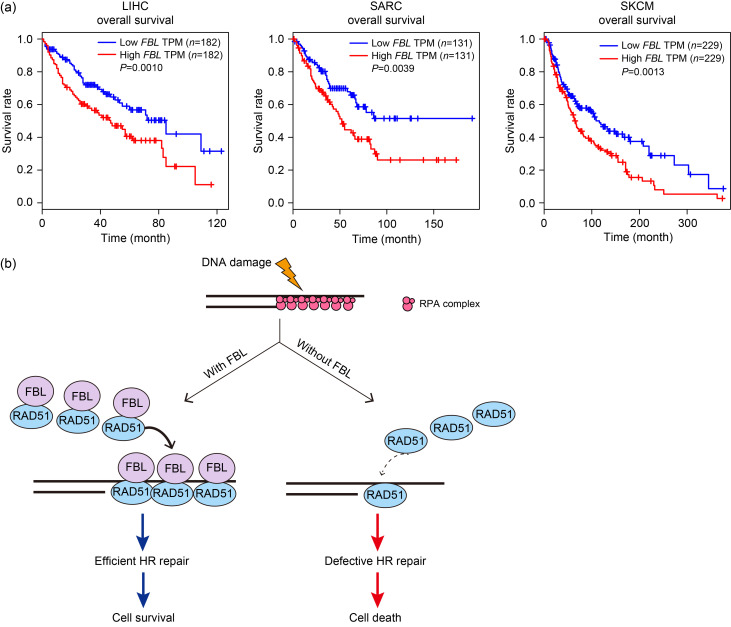
The loss of components involved in DDR pathway often leads to genomic instability and the development of cancer. These genetic defects in the DDR pathway make tumors more susceptible to specific DNA-targeted therapies (Hanahan, 2022). However, Yi et al. (2015) have revealed that an elevated DDR can contribute to therapeutic resistance. Intriguingly, the analysis of the Gene Expression Profiling Interactive Analysis (GEPIA) database showed a negative correlation between FBL RNA expression and the survival period of patients with liver hepatocellular carcinoma (LIHC), sarcoma (SARC), or skin cutaneous melanoma (SKCM) (Fig. 5a). These correlations suggested that FBL can serve as a valuable biomarker and a potential target for cancer therapy.
Fig. 5. Fibrillarin (FBL) overexpression is associated with poor survival in patients. (a) The Kaplan-Meier curves demonstrate an inverse relationship between the quantity of FBL transcripts and the overall survival rate of patients with liver hepatocellular carcinoma (LIHC), sarcoma (SARC), or skin cutaneous melanoma (SKCM). (b) Mechanism model by which FBL facilitates homologous recombination (HR) repair. Upon DNA damage, FBL is recruited to the sites of DNA breaks. FBL directly interacts with recombinase RAD51, promoting its recruitment to DNA damage sites. This interaction enables the formation of RAD51-single-stranded DNA (ssDNA) filaments, which are crucial for strand invasion and homology search. TPM: transcripts per million; RPA: replication protein A.
In conclusion, our study identified FBL as a regulator of HR. Upon DNA damage, FBL is recruited to DSBs and directly interacts with the recombinase RAD51. The depletion of FBL leads to impaired formation of RAD51 foci and reduced HR efficiency. In addition, FBL depletion results in an increase in chromosome aberrations and enhances cellular sensitivity to DNA-damaging agents. These findings shed light on the mechanism of RAD51 recruitment mediated by FBL and highlight the significant role of FBL in cancer therapy (Fig. 5b). Further investigations are warranted to elucidate whether FBL influences the DNA binding properties, adenosine triphosphatase (ATPase) activity, or interactions of RAD51 with other HR-involved proteins.
Materials and methods
Detailed methods are provided in the electronic supplementary materials of this paper.
Supplementary information
Acknowledgments
This work was supported by the National Key Research and Development Program of China (Nos. 2022YFA1302800 and 2021YFA1101000), the National Natural Science Foundation of China (Nos. 31961160725, 31730021, 31971220, 32270769, and 32170730), the Fok Ying Tung Education Foundation, the Fundamental Research Funds for the Provincial Universities of Zhejiang (No. 2021XZZX039), the Natural Science Foundation of Shaanxi Province (No. 2023-JC-QN-0942), the Research Fund of Xi’an Jiaotong University (No. YXJLRH2022098), and the Research Fund of the Second Affiliated Hospital of Xi’an Jiaotong University (Nos. 2020YJ(ZYTS)546-10 and YJ(QN)202014), China. We thank all the members of HUANG groups (Zhejiang Hospital and Zhejiang University, Hangzhou, China) and WU groups (Zhejiang University, Hangzhou, China) for insightful discussions.
Author contributions
Yanhua MU, Jinhua HAN, and Mingjie WU performed most of the experiments with help from Zongfang LI, Ke DU, and Yameng WEI. Jun HUANG and Mengjie WU designed and supervised the project. Yanhua MU, Jinhua HAN, and Jun HUANG wrote and revised the manuscript. All authors have read and approved the final manuscript, and therefore, have full access to all the data in the study and take responsibility for the integrity and security of the data.
Compliance with ethics guidelines
Jun HUANG is an associate editor-in-chief for Journal of Zhejiang University-SCIENCE B (Biomedicine & Biotechnology) and was not involved in the editorial review or the decision to publish this article. Mengjie WU is a young scientist committee member for Journal of Zhejiang University-SCIENCE B (Biomedicine & Biotechnology) and was not involved in the editorial review or the decision to publish this article. Yanhua MU, Jinhua HAN, Mingjie WU, Zongfang LI, Ke DU, Yameng WEI, Mengjie WU, and Jun HUANG declare that they have no conflict of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
Data availability statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
- Blunt T, Finnie NJ, Taccioli GE, et al. , 1995. Defective DNA-dependent protein kinase activity is linked to V(D)J recombination and DNA repair defects associated with the murine scid mutation. Cell, 80(5): 813-823. 10.1016/0092-8674(95)90360-7 [DOI] [PubMed] [Google Scholar]
- Canela A, Maman Y, Jung S, et al. , 2017. Genome organization drives chromosome fragility. Cell, 170(3): 507-521.e18. 10.1016/j.cell.2017.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, 2022. Hallmarks of cancer: new dimensions. Cancer Discov, 12(1): 31-46. 10.1158/2159-8290.CD-21-1059 [DOI] [PubMed] [Google Scholar]
- Hartley KO, Gell D, Smith GCM, et al. , 1995. DNA-dependent protein kinase catalytic subunit: a relative of phosphatidylinositol 3-kinase and the ataxia telangiectasia gene product. Cell, 82(5): 849-856. 10.1016/0092-8674(95)90482-4 [DOI] [PubMed] [Google Scholar]
- Hilario J, Amitani I, Baskin RJ, et al. , 2009. Direct imaging of human Rad51 nucleoprotein dynamics on individual DNA molecules. Proc Natl Acad Sci USA, 106(2): 361-368. 10.1073/pnas.0811965106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuki Y, Jeggo PA, Uchihara Y, et al. , 2020. DNA double-strand break end resection: a critical relay point for determining the pathway of repair and signaling. Genome Instab Dis, 1(4): 155-171. 10.1007/s42764-020-00017-8 [DOI] [Google Scholar]
- Li YH, Yuan J, 2021. Role of deubiquitinating enzymes in DNA double-strand break repair. J Zhejiang Univ-Sci B (Biomed & Biotechnol), 22(1): 63-72. 10.1631/jzus.B2000309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma CJ, Gibb B, Kwon Y, et al. , 2017. Protein dynamics of human RPA and RAD51 on ssDNA during assembly and disassembly of the RAD51 filament. Nucleic Acids Res, 45(2): 749-761. 10.1093/nar/gkw1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcel V, Ghayad SE, Belin S, et al. , 2013. p53 acts as a safeguard of translational control by regulating fibrillarin and rRNA methylation in cancer. Cancer Cell, 24(3): 318-330. 10.1016/j.ccr.2013.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miné J, Disseau L, Takahashi M, et al. , 2007. Real-time measurements of the nucleation, growth and dissociation of single Rad51-DNA nucleoprotein filaments. Nucleic Acids Res, 35(21): 7171-7187. 10.1093/nar/gkm752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton K, Petfalski E, Tollervey D, et al. , 2003. Fibrillarin is essential for early development and required for accumulation of an intron-encoded small nucleolar RNA in the mouse. Mol Cell Biol, 23(23): 8519-8527. 10.1128/MCB.23.23.8519-8527.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer AD, Ziesche S, Ebhardt H, et al. , 2002. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc Natl Acad Sci USA, 99(8): 5289-5294. 10.1073/pnas.082101999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmang T, Tollervey D, Kern H, et al. , 1989. A yeast nucleolar protein related to mammalian fibrillarin is associated with small nucleolar RNA and is essential for viability. EMBO J, 8(13): 4015-4024. 10.1002/j.1460-2075.1989.tb08584.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara A, Ogawa H, Ogawa T, 1992. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell, 69(3): 457-470. 10.1016/0092-8674(92)90447-K [DOI] [PubMed] [Google Scholar]
- Snaar S, Wiesmeijer K, Jochemsen AG, et al. , 2000. Mutational analysis of fibrillarin and its mobility in living human cells. J Cell Biol, 151(3): 653-662. 10.1083/jcb.151.3.653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessarz P, Santos-Rosa H, Robson SC, et al. , 2014. Glutamine methylation in histone H2A is an RNA-polymerase-I-dedicated modification. Nature, 505(7484): 564-568. 10.1038/nature12819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiku V, Jain C, Raz Y, et al. , 2017. Small nucleoli are a cellular hallmark of longevity. Nat Commun, 8: 16083. 10.1038/ncomms16083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tollervey D, Lehtonen H, Jansen R, et al. , 1993. Temperature-sensitive mutations demonstrate roles for yeast fibrillarin in pre-rRNA processing, pre-rRNA methylation, and ribosome assembly. Cell, 72(3): 443-457. 10.1016/0092-8674(93)90120-F [DOI] [PubMed] [Google Scholar]
- Xiang ZY, Liu H, Hu Y, 2023. DNA damage repair and cancer immunotherapy. Genome Instab Dis, 4(4): 210-226. 10.1007/s42764-023-00098-1 [DOI] [Google Scholar]
- Yi YH, Ma TH, Lee LW, et al. , 2015. A genetic cascade of let-7-ncl-1-fib-1 modulates nucleolar size and rRNA pool in Caenorhabditis elegans . PLoS Genet, 11(10): e1005580. 10.1371/journal.pgen.1005580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa H, Komatsu W, Hayano T, et al. , 2011. Splicing factor 2-associated protein p32 participates in ribosome biogenesis by regulating the binding of Nop52 and fibrillarin to preribosome particles. Mol Cell Proteomics, 10(8): M110. 006148. 10.1074/mcp.M110.006148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gong ZH, 2021. Regulation of DNA double-strand break repair pathway choice: a new focus on 53BP1. J Zhejiang Univ-Sci B (Biomed & Biotechnol), 22(1): 38-46. 10.1631/jzus.B2000306 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.