Abstract
Spiral roots are induced in germinated rice seeds through treatment with nanomolar brassinosteroids (BRs) but not with other plant hormones. Here, we determined the minimum effective concentration (MEC) of various BRs to induce spiral roots in germinated rice seeds. The reciprocal logarithm of MEC, pMEC, was used as the BL-like activity index, which was linearly correlated with the reciprocal logarithm of a 50% effective dose (pED50) as evaluated in the rice lamina inclination assay. Furthermore, a ligand-receptor docking simulation was performed against the BL receptor complex, Arabidopsis thaliana BRI1/SERK1, and the binding free energy (ΔGbind) was calculated for the tested BRs. The ΔGbind calculation was performed using the molecular mechanics/generalized Born surface area method on an ensemble of uncorrelated snapshots collected via molecular dynamics to predict biological activity.
Keywords: brassinosteroids, spiral root induction, docking simulation, MM/PBSA, MM/GBSA
Introduction
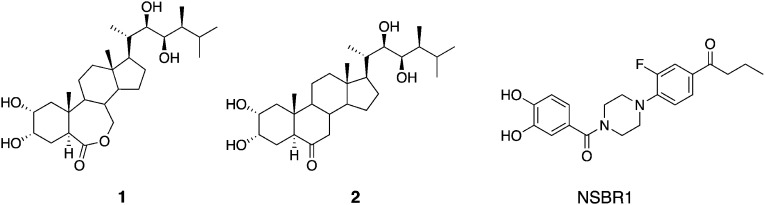
Brassinolide (BL, 1; Fig. 1) was discovered as a plant growth promoter in 1979 by a USDA group. BL has a steroidal skeleton constructed from four fused rings (A/B/C/D) and hydroxyalkyl chains at the C17 position.1) The B-ring of BL is an ε-caprolactone structure (seven-membered ring), but a few years later, castasterone (CS, 2; Fig. 1) with a six-membered cyclohexanone B-ring was isolated from chestnut insect gall.2) Various BL and CS analogs have been identified in nature and/or chemically synthesized, which are collectively called brassinosteroids (BRs).3) In previous structure–activity studies, the BL-specific bioassay systems described below were necessary to distinguish BRs from other plant hormones. To search for BL-like compounds, an in vitro rice lamina inclination assay (RLIA) was established,4) and later, an in vivo RLIA was developed.5) Previously, we synthesized various BL and CS analogs and quantitatively measured their BL-like activity using an in vivo RLIA.6–8) Other bioassay methods have been developed to measure the BL-like activity specifically, but many of them were not as simple.9)
Fig. 1. Structures of brassinolide (1), castasterone (2), and a non-steroidal BL agonist, NSBR1.
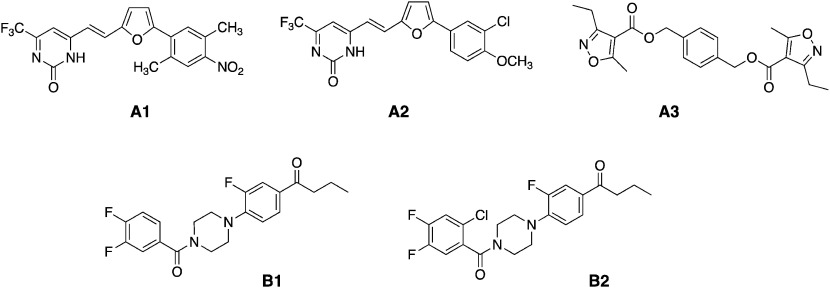
Whether BL is a genuine plant steroid hormone was disputed after the first characterization of BL from Brassica napus pollen, because many steroidal compounds are found in plants as secondary metabolites. Two decades later, however, BL was confirmed as a 6th plant hormone by the discovery of a BL-deficient mutant.10) Later, BR insensitive receptor kinase 1 (BRI1) was identified as the BL receptor,11) and the binding site of BL to BRI1 was confirmed by Kinoshita et al. in 2005.12) In 2011, the three-dimensional structures of BRI1 were worked out by two groups,13,14) and two years later, the crystal structure of BRI1-BL complexed with a kinase, somatic embryogenesis receptor-like kinase 1 (SERK1), was elucidated.15) The establishment of 3-D structures of the BL receptor opened the door for in silico screening of the BL-like compounds. We found three BL antagonists to the rice plant16) and two BL antagonists to Arabidopsis17) as shown in Fig. 2, using LigandScout18) in our in silico approach.
Fig. 2. Chemical structures of BL antagonists found through in silico screening against the rice plant (A1–A3) and Arabidopsis (B1 and B2).
Sugiura et al. modified the structure of the antagonists (B1 and B2; Fig. 2) successfully to get an agonist toward Arabidopsis, which was named NSBR1 (Fig. 1).17,19) However, the activity of NSBR1 was 30,000 times less potent than BL in terms of the 50% effective dose [ED50 (mol/plant)] in the RLIA. Watanabe et al. calculated the binding free energy (ΔGbind) of BL (1), CS (2), and their analogs to the BL receptor of Arabidopsis using molecular dynamics (MD) and the molecular mechanics/Poisson Boltzman surface area (MM/PBSA)20) and found the linear correlation between pED50 and ΔGbind.17) Sugiura et al. also executed a docking simulation for NSBR1 to calculate ΔGbind, which was predictable from the correlation between the ΔGbind and pED50 derived for BRs.17)
Even though the RLIA is a good bioassay system to determine the BL-like activity quantitatively, several processes are required to obtain reproducible data. Particularly, it is not easy to evaluate the activity of weak compounds, because they easily crystallize on the surface of the rice plant from the applied droplet of highly concentrated MeOH or EtOH solutions. Dimethyl sulfoxide (DMSO) is often used in bioassays to relieve the drawback of the property of alcohols, but DMSO cannot be used in the RLIA due to its toxic effect on the rice plant (personal communication). Therefore, the RLIA is difficult to use as a bioassay system to find leads and/or hits in a drug discovery study when the biological activity is generally very weak. To overcome the drawback of the RLIA, we developed a bioassay system named the spiral root induction assay (SRIA).21) The SRIA, regarding the spiral root induction as an index of BL-like activity is very simple and user friendly.
The aim of the present study is to measure the biological activity of various BRs quantitatively using the SRIA and discuss the structure–activity relationship of BRs. The minimum effective concentrations (MECs) required to induce the spiral root in the germinated rice seeds were determined for various BRs, and their reciprocal logarithm values (pMECs) were compared with corresponding pED50 values determined in the RLIA. Furthermore, the ΔGbind of ligands to the receptor is calculated using the molecular mechanics generalized Born surface area (MM/GBSA) method to examine the relationship with biological activity. While two popular methods, MM/PBSA and MM/GBSA, have been used to estimate the ΔGbind of small ligands for biological macromolecules,22–24) we previously employed MM/PBSA to calculate the ΔGbind of ecdysteroids25) and brassinosteroids8) and found good correlation between ΔGbind and biological activity. In this study, however, we used MM/GBSA, since MM/GBSA calculation is accurate and relatively simple, providing stable and reproducible results that are computationally effective for the in silico study.23)
Materials and methods
1. Observation of the root grown in rice seeds
The bioassay method is basically the same as that previously reported by our group.21) Rice seeds of Oryza sativa Koshihikari cultivated in Shiga Prefecture were soaked in running tap water for a few days to germinate. Bioassay solutions were prepared by adding an aliquot of DMSO or EtOH solution to 2 mL of water in a 50 mL beaker (Fig. S1). The 10 germinated seeds were put into beakers containing bioassay solutions. Germinated seeds were cultured for two days at 25°C, and the grown root was observed.
The EtOH and DMSO stock solutions for test compounds were prepared by roughly threefold dilutions (i.e., 10 mM, 3.33 mM, 1 mM, 0.33 mM, etc.), and 10 µL of each stock solution was added to the water (2 mL) to prepare the bioassay solutions. In some cases, the addition of an aliquot volume of stock solutions was changed to prepare the bioassay solutions, as shown in Fig. S2 and Fig. S3. In some cases, the volume of the additional aliquot of stock solutions was changed. Changing the stock solution aliquot volume is a convenient way to prepare the various bioassay solutions. To determine the MEC of BL precisely, the volume of the added aliquot of EtOH stock solutions (200 nM and 2 nM) was changed. As shown in Fig. S2, spiral roots were induced in all concentrations prepared from a 200 nM stock solution. However, when the diluted stock solution (2 nM) was used, the spiral roots were observed in the bioassay solution prepared by adding 30 µL of a 2 nM stock solution. The MEC of BL required to induce the spiral root in more than 50% of the treated seeds was determined to be 3×10−11 (M). We examined another concentration-response as shown in Fig. S3. The reciprocal logarithm of MEC, pMEC, was used as the activity index. We repeated this experiment three times and took the average of three pMEC values (10.7, 10.5, and 10.3) as 10.5±0.2 (n=3) for BL. The pMEC values of other BRs were obtained in the similar way, and they are listed in Table 1.
Table 1. The BL-like activity of various BRs and the ΔGbind to the BL receptor protein.
| BL-like activity | ΔGbindc) | ||||
|---|---|---|---|---|---|
| No. | Compounds | pMECa) | pED50b) | A. thaliana | O. sativa |
| 1 | BL | 10.5±0.23 (4) | 13.6 | −78.7±5.0 | −79.2±5.8 |
| 2 | CS | 8.5±0 (3) | 12.3 | −75.1±5.3 | −90.1±5.3 |
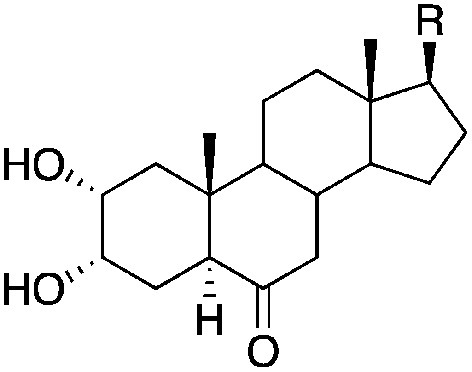
|
|||||
| R | |||||
| 3 |
|
8.0±0 (3) | 10.7 | −71.3±2.8 | −87.7±1.0 |
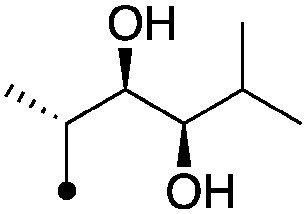
| 4 |
|
4.1±0.17 (3) | 8.5 | −67.3±3.7 | −80.9±3.7 |
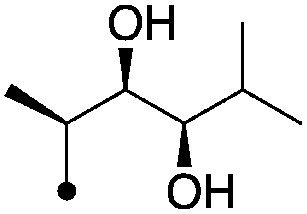
| 5 |
|
4.3±0 (3) | 8.7 | −64.9±5.0 | −74.9±8.5 |
| 6 |
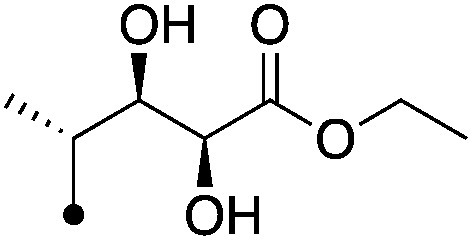
|
6.1±0 (3) | <10.0 (10%) | −72.4±3.1 | −75.2±10.8 |
| 7 |
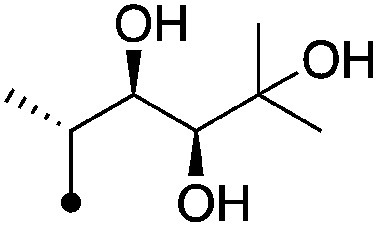
|
6.4±0.17 (3) | <9.0 (39%) | −69.2±3.8 | −77.6±1.3 |
| 8 |
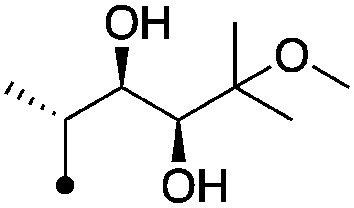
|
6.0±0.17 (3) | <9.2 (0.1%) | −71.8±2.1 | −84.1±2.0 |
| 9 |
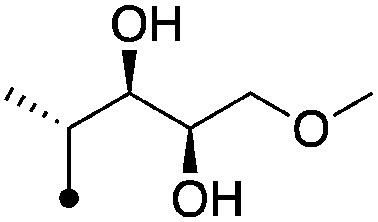
|
5.9±0.17 (3) | <9.3 (11%) | −67.5±3.3 | −85.3±4.0 |
| 10 |
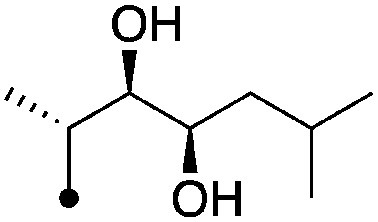
|
8.0±0.17 (3) | 10.2 | −72.6±3.9 | −87.4±6.4 |
| 11 |
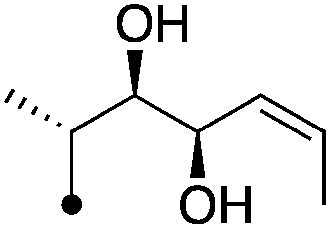
|
6.4±0.17 (3) | 9.5 | −70.0±6.7 | −82.2±9.0 |
| 12 |
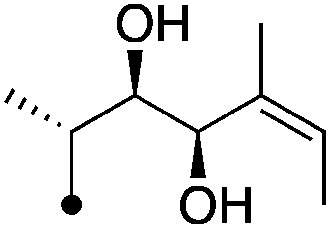
|
7.8±0 (3) | 10.7 | −75.0±6.1 | −80.8±1.4 |
| 13 |
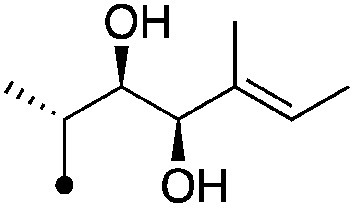
|
7.8±0 (3) | 11.3 | −69.4±6.0 | −79.4±4.5 |
| 14 |
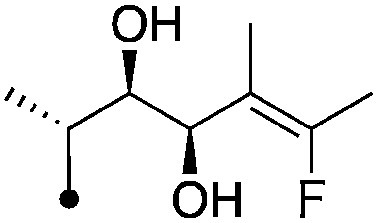
|
8.0±0.17 (3) | 10.8 | −73.0±5.0 | −86.7±6.6 |
| 15 |
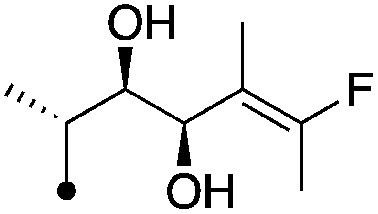
|
8.2±0.17 (3) | 11.2 | −68.4±2.2 | −83.5±0.4 |
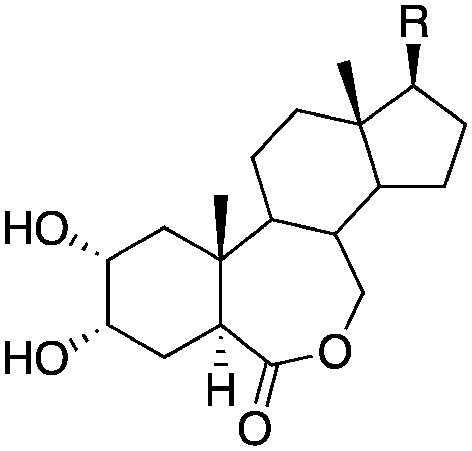
|
|||||
| R | |||||
| 16 |
|
4.15±0.21 (2) | <8.0 (13%) | −57.3±2.5 | −75.3±3.2 |
| 17 |
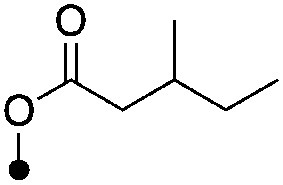
|
4.0±0.17 (3) | <8.0 (41%) | −66.1±1.6 | −78.6±1.6 |
| 18 |
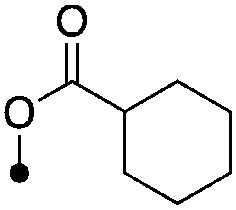
|
<4.3 (2) | <8.0 (48%) | −68.8±0.9 | −79.6±0.9 |
| 19 |
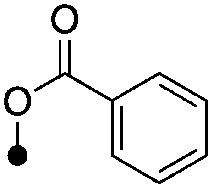
|
<4.3 (2) | <8.0 (44%) | −61.8±5.6 | −71.4±2.8 |
| 20 |
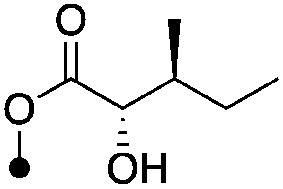
|
4.5±0.17 (3) | 9.3 | −60.6±4.9 | −72.8±4.9 |
| 21 |
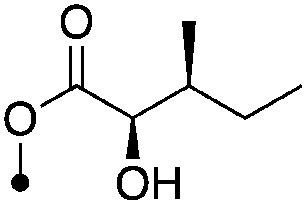
|
5.6±0 (3) | 10.5 | −69.6 ±2.7 | −82.5±6.6 |
| 22 |
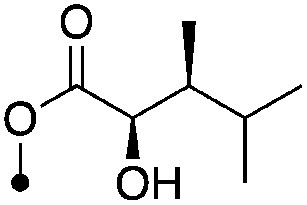
|
5.6±0 (3) | 10.1 | −64.5±3.1 | −83.7±6.5 |
| 23 |
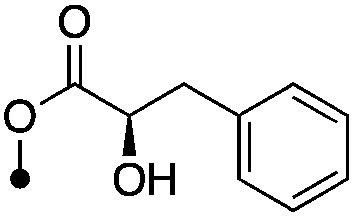
|
5.1±0 (3) | <8.7 (16%) | −67.8±4.8 | −72.8±4.4 |
| 24 |
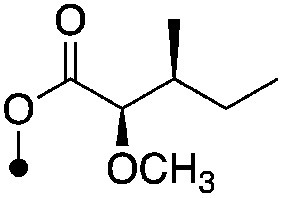
|
4.6±0 (3) | 9.1 | −62.1±5.5 | −81.4±2.5 |
a) Mean±standard deviation. The numbers in parentheses are the replication numbers. b) From Ref. 6–8. Measured by RLIA. The numbers in parentheses are the promotion of the lamina inclination at the corresponding dose. c) Mean±standard deviation for three calculations.
2. MD and the binding free energy calculation
We previously conducted a simulation of BR docking to the crystal structure of BRI1/SERK1 (PDB: 4LSX) using the FRED of OEDocking (OpenEye), then the calculated ΔGbind value using the MD-MM/PBSA of AMBER14 (http://ambermd/org/).8,17,25) In this study we used AMBER20. MD simulation was performed for 150 ps and 150 trajectories at 1.0 ps intervals were obtained. Energy calculation was performed using the MM/GBSA, because the MM/GBSA produces better performance in ranking the binding affinities for systems without metals and has a much lower computer-resource demand than the MM/PBSA method.22–24)
Results and discussion
1. BL-like activity of various BRs
We previously reported that BL induced the spiral root in germinated rice seeds at an extremely low concentration (5 nM) in the SRIA.21) We also reported that a BL antagonist unwound the spiral root induced by BL treatment, and this can be used as a bioassay method to detect both BL-like compounds and their antagonists. Since other plant hormones, such as gibberellin and auxin, did not induce the spiral root even at high concentrations (50 µM), the SRIA is a good bioassay method to find BL-like compounds. Some of the assay water solutions were prepared by changing the addition of stock solution. As shown in the supplementary figure (Fig. S1), the addition of 100 µL of EtOH to 2 mL of water did not affect the root growth, but 100 µL of DMSO significantly inhibited the root growth. The addition of 40 and 50 µL of EtOH and DMSO seemed to be nontoxic to rice seeds, but the maximum volume of the aliquot of stock solutions in 2 mL of water was kept below 30 µL (<1.5%) to avoid the toxic effect of the solvent.
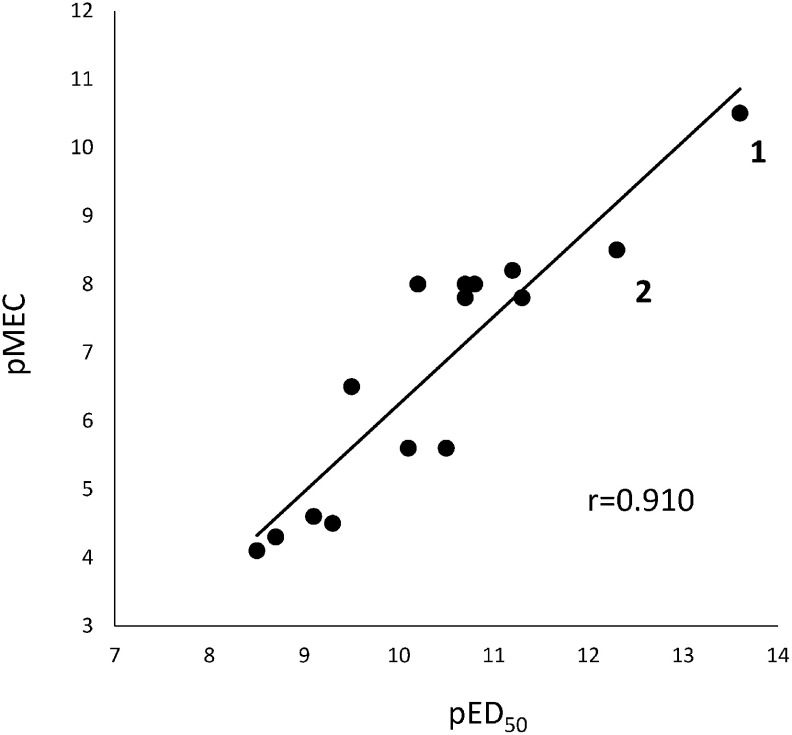
In this study, the MECs required to induce the spiral root were determined for 14 BL and 10 CS analogs, and their pMEC values are listed in Table 1. Among 24 tested BRs, only two compounds (18 and 19) were inactive in the SRIA. As shown in Table 1, the pMEC of BL (1) was determined to be 10.5, 100 times more potent than CS (2; pMEC=8.5). This is comparable to the result evaluated by in the RLIA (pED50: 13.6 vs. 12.3). Even though the pED50 values of CS analogs (6–9)8) and BL analogs (16–19, and 23)6) could not be obtained using an RLIA, their pMEC values were determined to be 4.0–6.4 in the SRIA. MEC values were not determined for compounds 18 and 19, but MEC values of 16 and 17 were determined at the lower concentrations than MECs of compounds 18 and 19. This is probably due to the difference of solubility among compounds. We compared the pMEC and pED50 values for 15 compounds to derive a good correlation (r=0.910), as shown in Fig. 3.
Fig. 3. Relationship between pED50 (RLIA) and pMEC (SRIA).
Figure 3 indicates that the SRIA can be used as a suitable bioassay method to quantitatively measure BL-like activity against rice plants. The SRIA is also advantageous in that the person who is not familiar with the bioassay of BRs can perform it without training and quantitatively determine the BL-like activity of BRs within a few days. Moreover, germinated seeds for the SRIA can be easily prepared, whereas it takes a week or more to prepare the biological materials, such as a rice shoot with a second leaf, in an RLIA. Furthermore, DMSO can be used as a carrier solvent for the test compound in an SRIA, which is not the case in an RLIA.
2. In silico study
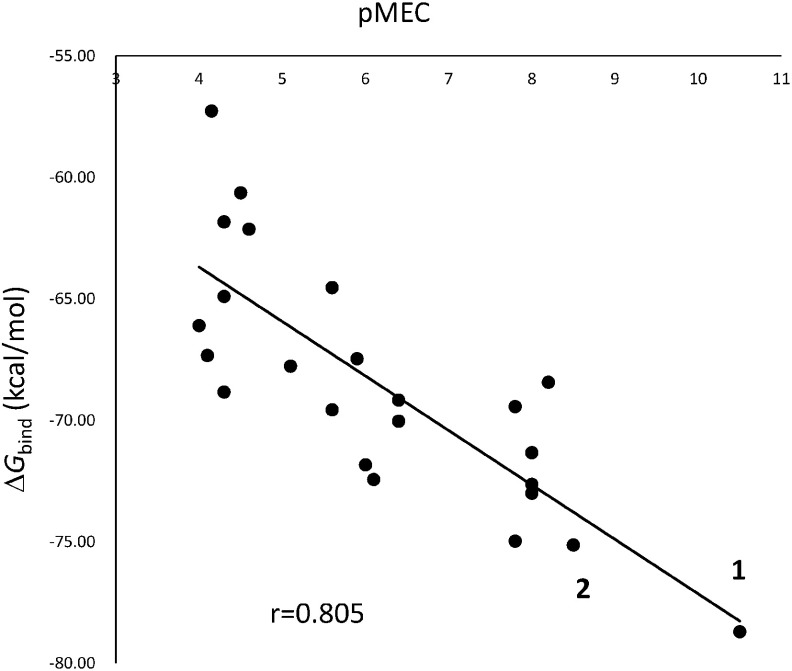
As shown in Table 1, we could quantitatively determine the biological activity of various BRs in an SRIA in terms of the pMEC. Here, we conducted MD for all 24 BRs and calculated their ΔGbind using the MM/GBSA. As stated above, the MM/GBSA was used in this study to calculate the ΔGbind, instead of the MM/PBSA, because the calculation cost of the MM/GBSA method is low for obtaining results comparable with those using an MM/PBSA. As shown in Fig. 4, the MM/GBSA worked well to calculate the ΔGbind to the Arabidopsis BL receptor to show a linear correlation with the pMEC. At present, MD simulation using the MM/PBSA or MM/GBSA is the most important tool for evaluating the affinity of ligand molecules to a receptor by calculating the free energy change in the binding process.
Fig. 4. Relationship between the pMEC and the ΔGbind to Arabidopsis receptor.
Yamamuro et al. isolated the rice BRI1 homologous gene, O. sativa BRI1 (OsBRI1), which was very similar to the Arabidopsis thaliana BRI1 (AtBRI1) gene.26) Since the bioassay was conducted using rice materials, we built a 3-D structure of OsBRI1 to calculate the ΔGbind to the rice receptor. The 3-D model of OsBRI1 is well superposed on the crystal structure of AtBRI1 (PDB: 4LSX). The ΔGbind values calculated using the MM/GBSA (Table 1) were linearly correlated with the pMEC, but the correlation (r=0.553) was not as good as that shown in Fig. 4. This is probably due to the poor prediction of the side chains of amino acid residues surrounding ligands in the 3-D model constructed in this study. It is very likely that a good correlation will be observed when the experimentally solved 3-D structure of OsBRI1 is available or the modelling system is further improved.
In our previous study, the ΔGbind to the Arabidopsis receptor was calculated for BRs using the MD-MM/PBSA method and linearly correlated with the pED50 values measured in the RLIA.8) Sugiura et al. also calculated the ΔGbind of NSBR1, which is predicted well by the relationship between ΔGbind and pED50 for BRs. However, some of the ΔGbind values of weak compounds, including NSBR1, were positive, which is not adequate to express the specific ligand-receptor binding. In the MM/PBSA, the exterior dielectric constant is set as 78.5, but the interior dielectric constant (εin) is changeable from 1 to 4, and 1 was used in the previous calculation.8,17) According to Wang,24) the higher solute dielectric constant (εin=2 or 4) is preferable to improve the rescoring accuracy of the MM/PBSA. A value of 2 was assigned for the dielectric constant of non-polar residues, a value of 3 for polar residues, and a value of 4 charged residues. Matsuo recalculated ΔGbind using 2 in the MM/PBSA and obtained negative ΔGbind for all active compounds (unpublished). The correlation between the pED50 and ΔGbind calculated using εin=2 was slightly better than the previous one (Fig. S4). The ΔGbind values of all BL-like compounds calculated in the MM/GBSA were negative. MM/PBSA calculations are very time consuming, but the MM/GBSA requires far fewer computer resources than the MM/PBSA method. It is said that the accuracy of the calculated energy using the MM/GBSA is compromised at the expense of computational speed.24) The correlation shown in Fig. 4 means that the MM/GBSA calculation is sufficient for practical use, such as in silico drug design.
Conclusion
The SRIA is a simple method to detect BL-like activity. The MEC of BL was determined to be 3×10−11 M (pMEC=10.5). The BL-like activity in terms of the pMEC was linearly correlated with the pED50 measured in the RLIA. DMSO can constitute up to 1.5% of the assay solution in the SRIA. Therefore, the concentration of test compounds can be enhanced in the SRIA. This constitutes an advantage over the RLIA, in which DMSO cannot be used. The docking and MD simulations were performed for BRs against the crystal structure of the BRI1/SERK1 complex of Arabidopsis and the modelled receptor structure of the rice plant. The ΔGbind calculated in the MD-MM/GBSA was linearly correlated with the pMEC determined in the SRIA. This study contributes to the search for and rational design of BL-like compounds.
Acknowledgements
This work was supported in part by Japan Society of the Promotion of Science JSPS KAKENHI Grant Number JP16K07625.
Electronic supplementary materials
The online version of this article contains supplementary materials (Fig. S1–S4) which are available at https://www.jstage.jst.go.jp/browse/jpestics/.
Supplementary Data
References
- 1) M. D. Grove, G. F. Spencer, W. K. Rohwedder, N. Mandava, J. F. Worley, J. D. Warthen Jr., G. L. Steffens, J. L. Flippen-Anderson and J. C. Cook Jr.: Brassinolide, a plant growth-promoting steroid isolated from Brassica napus pollen. Nature 281, 216–217 (1979). [Google Scholar]
- 2) T. Yokota, M. Arima and N. Takahashi: Castasterone, a new phytosterol with plant-hormone potency, from chestnut insect gall. Tetrahedron Lett. 23, 1275–1278 (1982). [Google Scholar]
- 3) S. Fujioka: Natural occurrence of brassinosteroids in the plant kingdom. In “Brassinosteroids,” ed by. A. Sakurai, T. Yokota, and S. D. Clouse, Springer, Tokyo, pp. 21–45, 1999.
- 4) K. Wada, S. Marumo, N. Ikekawa, M. Morisaki and K. Mori: Brassinolide and homobrassinolide promotion of lamina inclination of rice seedlings. Plant Cell Physiol. 22, 323–325 (1981). [Google Scholar]
- 5) S. Fujioka, T. Noguchi, S. Takatsuto and S. Yoshida: Activity of brassinosteroids in the dwarf rice lamina inclination bioassay. Phytochemistry 49, 1841–1848 (1998). [Google Scholar]
- 6) S. Uesusuki, B. Watanabe, S. Yamamoto, J. Otsuki, Y. Nakagawa and H. Miyagawa: Synthesis of brassinosteroids of varying acyl side chains and evaluation of their brassinolide-like activity. Biosci. Biotechnol. Biochem. 68, 1097–1105 (2004). [DOI] [PubMed] [Google Scholar]
- 7) S. Yamamoto, B. Watanabe, J. Otsuki, Y. Nakagawa, M. Akamatsu and H. Miyagawa: Synthesis of 26,27-bisnorcastasterone analogs and analysis of conformation–activity relationship for brassinolide-like activity. Bioorg. Med. Chem. 14, 1761–1770 (2006). [DOI] [PubMed] [Google Scholar]
- 8) B. Watanabe, S. Yamamoto, T. Yokoi, A. Sugiura, S. Horoiwa, T. Aoki, H. Miyagawa and Y. Nakagawa: Brassinolide-like activity of castasterone analogs with varied side chains against rice lamina inclination. Bioorg. Med. Chem. 25, 4566–4578 (2017). [DOI] [PubMed] [Google Scholar]
- 9) K. Wada: Brassinosteroid. In “Experimental Method for Phytochemical Regulation,” ed. by N. Takahashi., The Society for Plant Growth Regulation of Plants, Sasatoku Printing Co., Aichi Japan, pp. 80–88, 1989. (in Japanese).
- 10) S. D. Clouse: Molecular genetic studies confirm the role of brassinosteroids in plant growth and development. Plant J. 10, 1–8 (1996). [DOI] [PubMed] [Google Scholar]
- 11) Z. Y. Wang, H. Seto, S. Fujioka, S. Yoshida and J. Chory: BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature 410, 380–383 (2001). [DOI] [PubMed] [Google Scholar]
- 12) T. Kinoshita, A. Cano-Delgado, H. Seto, S. Hiranuma, S. Fujioka, S. Yoshida and J. Chory: Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433, 167–171 (2005). [DOI] [PubMed] [Google Scholar]
- 13) M. Hothorn, Y. Belkhadir, M. Dreux, T. Dabi, J. P. Noel, I. A. Wilson and J. Chory: Structural basis of steroid hormone perception by the receptor kinase BRI1. Nature 474, 467–471 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14) J. She, Z. Han, T.-W. Kim, J. Wang, W. Cheng, J. Chang, S. Shi, J. Wang, M. Yang, Z.-Y. Wang and J. Chai: Structural insight into brassinosteroid perception by BRI1. Nature 474, 472–476 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15) J. Santiago, C. Henzler and M. Hothorn: Molecular mechanism for plant steroid receptor activation by somatic embryogenesis co-receptor kinases. Science 341, 889–892 (2013). [DOI] [PubMed] [Google Scholar]
- 16) S. Takimoto, A. Sugiura, S. Minami, T. Tasaka, Y. Nakagawa and H. Miyagawa: In silico exploration for agonists/antagonists of brassinolide. Bioorg. Med. Chem. Lett. 26, 1709–1714 (2016). [DOI] [PubMed] [Google Scholar]
- 17) A. Sugiura, S. Horoiwa, T. Aoki, S. Takimoto, A. Yamagami, T. Nakano, Y. Nakagawa and H. Miyagawa: Discovery of a nonsteroidal brassinolide-like compound, NSBR1. J. Pestic. Sci. 42, 105–111 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18) G. Wolber and T. Langer: LigandScout: 3-D pharmacophores derived from protein-bound ligands and their use as virtual screening filters. J. Chem. Inf. Model. 45, 160–169 (2005). [DOI] [PubMed] [Google Scholar]
- 19) Y. Nakagawa, A. Sugiura, S. Takimoto, S. Horoiwa, H. Miyagawa and M. Matsuo (Kyoto University): Production of non-steroid compound having plant steroid hormone (brassinolide)-like activity. WO 2018/159827 A1 (2018).
- 20) P. A. Kollman, I. Massova, C. Reyes, B. Kuhn, S. Huo, L. Chong, M. Lee, T. Lee, Y. Duan, W. Wang, O. Donini, P. Cieplak, J. Srinivasan, D. A. Case and T. E. Cheatham, III: Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc. Chem. Res. 33, 889–897 (2000). [DOI] [PubMed] [Google Scholar]
- 21) Y. Nakagawa, B. Nishikawa and H. Miyagawa: Effects of brassinolide on the growing of rice plants. J. Pestic. Sci. 46, 274–277 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22) G. Rastelli, A. D. Rio, G. Degliesposti and M. Sgobba: Fast and accurate predictions of binding free energies using MM-PBSA and MM-GBSA. J. Comput. Chem. 31, 797–810 (2010). [DOI] [PubMed] [Google Scholar]
- 23) S. Genheden and U. Ryde: The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 10, 449–461 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24) E. Wang, H. Sun, J. Wang, Z. Wang, H. Liu, J. Z. H. Zhang and T. Hou: End-point binding free energy calculation with MM/PBSA and MM/GBSA: strategies and applications in drug design. Chem. Rev. 119, 9478–9508 (2019). [DOI] [PubMed] [Google Scholar]
- 25) S. Horoiwa, T. Yokoi, S. Masumoto, S. Minami, C. Ishizuka, H. Kishikawa, S. Ozaki, S. Kitsuda, Y. Nakagawa and H. Miyagawa: Structure-based virtual screening for insect ecdysone receptor ligands using MM/PBSA. Bioorg. Med. Chem. 27, 1065–1075 (2019). [DOI] [PubMed] [Google Scholar]
- 26) C. Yamamuro, Y. Ihara, X. Wu, T. Noguchi, S. Fujioka, S. Takatsuto, M. Ashikari, H. Kitano and M. Matsuoka: Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12, 1591–1605 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.