Abstract
Exosomes are among the most puzzling vehicles of intercellular communication, but several crucial aspects of their biogenesis remain elusive, primarily due to the difficulty in purifying vesicles with similar sizes and densities. Here we report an effective methodology for labelling small extracellular vesicles (sEV) using Bodipy FL C16, a fluorescent palmitic acid analogue. In this study, we present compelling evidence that the fluorescent sEV population derived from Bodipy C16‐labelled cells represents a discrete subpopulation of small exosomes following an intracellular pathway. Rapid cellular uptake and metabolism of Bodipy C16 resulted in the incorporation of fluorescent phospholipids into intracellular organelles specifically excluding the plasma membrane and ultimately becoming part of the exosomal membrane. Importantly, our fluorescence labelling method facilitated accurate quantification and characterization of exosomes, overcoming the limitations of nonspecific dye incorporation into heterogeneous vesicle populations. The characterization of Bodipy‐labelled exosomes reveals their enrichment in tetraspanin markers, particularly CD63 and CD81, and in minor proportion CD9. Moreover, we employed nanoFACS sorting and electron microscopy to confirm the exosomal nature of Bodipy‐labelled vesicles. This innovative metabolic labelling approach, based on the fate of a fatty acid, offers new avenues for investigating exosome biogenesis and functional properties in various physiological and pathological contexts.
Keywords: exosome biogenesis, exosomes, extracellular vesicles, lipid metabolism, MVB, sEV
1. INTRODUCTION
Almost all cell types secrete a heterogeneous population of lipid bilayer vesicles, collectively called extracellular vesicles (EV) (Kalluri & LeBleu, 2020; Teng & Fussenegger, 2020; van Niel et al., 2018). Based on their biogenesis, EV can be classified into two main groups: microvesicles (MV), also known as ectosomes and exosomes. MVs are formed by direct outward budding of the plasma membrane (Clancy et al., 2021; Meldolesi, 2022), whereas exosomes are formed within late endocytic compartments or multivesicular bodies (MVB) by invagination of the limiting membrane into the lumen. Intraluminal vesicles (ILVs) accumulated in the MVB are then released into the extracellular space by the fusion of MVB with the plasma membrane. Exosomes play a crucial role in intercellular communication under normal and pathological conditions however, despite the extension of exosome studies to clinics, understanding important aspects of their biogenesis remains a longstanding goal, complicated by the heterogeneity among secreted vesicles with respect to their functionality (Kowal et al., 2016; Mathieu et al., 2021; Willms et al., 2018; Zhang et al., 2018).
Smaller vesicles that bud from the plasma membrane and share intrinsic characteristics with exosomes, such as molecular components or physical properties (size and floating density), have also been described, making their separation and characterization difficult (Jeppesen et al., 2019; van Niel et al., 2018; Zhang et al., 2018). To date, intracellular localization of exosome‐specific molecules has mainly been determined by microscopy‐based methods, but an actual understanding of late endosomes/MVB biogenesis requires the ability to more accurately establish the dynamics of organelle traffic leading to EV secretion. Different subpopulations of exosomes that coexist in the same MVB compartment can follow different intracellular pathways en route to release into the extracellular space, determining their molecular composition and ultimately the message delivered to target cells. Recently, new fluorescence labelling strategies ranging from novel lipid dyes to genetic labelling techniques have been developed to label EV or subtypes to track their biogenesis, biodistribution, and uptake (Lai et al., 2015; Liang et al., 2023; Liu et al., 2022; Mathieu et al., 2021; Verweij et al., 2021). However, the lack of suitable markers to specifically identify and separate EV subpopulations of different origins coexisting in secretory fluids complicates this process (Nieuwland et al., 2020; van Niel et al., 2022). Genetic EV labelling, which involves tagging EV with fluorescent reporter proteins, has proven to be a very promising technique as it targets known EV markers such as tetraspanins. This technique has several advantages over non‐specific labelling techniques, such as PKH26 or carboxyfluorescein diacetate succinimidyl ester (CFDA‐SE), and allows for the analysis and characterization of EV at the single‐vesicle level. However, tetraspanins, such as CD63, CD9, and CD81, have been identified in some EV subpopulations in different combinations (Jeppesen et al., 2019; Kowal et al., 2016; Zhang et al., 2018), while others lack these protein markers, suggesting that tetraspanins alone cannot be used to distinguish between different types of EV (Karimi et al., 2022; Mathieu et al., 2021). To address this issue, new reliable experimental strategies are needed to identify specific exosome populations originating from late endosomes/MVBs.
One of the less explored yet fundamental pathways of exosome biogenesis is the lipid pathway. With the aim of finding new strategies to obtain fluorescently labelled EV originating from an intracellular pathway, we previously exploited the lipid pathway using a fluorescent palmitic acid analogue, BODIPY FL C16 (Bodipy C16). Bodipy C16 is readily internalized by cells and transformed into phospholipid components of EV membranes, whose number and fluorescence intensity can then be precisely analyzed by flow cytometry (FC) (Coscia et al., 2016). In this study, we extended the characterization of fluorescent sEV (Bodipy sEV) to evaluate the general applicability of our method in obtaining a metabolically labelled subpopulation of sEV of intracellular origin, and excluding ectosomes contaminations. Our results show that Bodipy C16 labelling colocalizes with specific markers of intracellular compartments, such as mitochondria, lysosomes, Golgi, and late endosomes/MVB, but was not detected on the plasma membrane. The kinetics of Bodipy sEV release can be monitored over time, and a pure population of Bodipy sEV approximately 80 nm in size can be obtained by FACS sorting. Furthermore, we showed that individual Bodipy sEV colocalized with surface markers such as CD63, CD81, and CD9. Thus, our approach represents a new and effective strategy for labelling a discrete exosome population that is distinct from ectosomes and can be further exploited for biogenesis and functional studies.
2. METHODS
2.1. Cell cultures
The human melanoma cells Mel 501 were cultured in RPMI 1640; the human melanoma cells A375/M and Me665/1 (Me665), the human embryonic kidney 293T (HEK293T) and the human cervical adenocarcinoma HeLa cells, were cultured in Dulbecco's modified Eagle's medium. All media were supplemented with 10% heat‐inactivated fetal bovine serum (FBS), 100 units/ml penicillin, 100 µg/ml streptomycin and 2 mM L‐glutamine (complete media). Additional 1% MEM Non‐Essential Aminoacids were added to A375M cell media. All media and supplements were from Euroclone. When required for Nanoparticle Tracking Analysis (NTA) or CFSE staining of sEV, FBS was replaced with Exosome Depleted FBS (Euroclone) (EV‐depleted medium). Cell lines were grown at 37°C, under 5% CO2, in humidified incubators and routinely tested for mycoplasma contamination using the Mycoplasma PCR Detection Kit (ABM, #G238). All cell lines are commercially available (ATCC) with the exception of Me665/1 stabilized from surgical specimens at Istituto Nazionale Tumori (Milan, Italy) (Felicetti et al., 2008).
2.2. Cells and sEV labelling
BODIPY FL C16 (Bodipy C16) (4,4‐Difluoro‐5,7‐Dimethyl‐4‐Bora‐3a,4a‐Diaza‐s‐Indacene‐3‐Hexadecanoic Acid (ThermoFisher, #D3821) or Palmitic acid (Sigma, #P0500) were complexed with fatty acid‐free bovine serum albumin (BSA), (Sigma, #A8806) as described previously (Coscia et al., 2016). To isolate sEV, 60%–70% confluent monolayers of cells in exponential growth were incubated with 7 µM Bodipy C16 or palmitic acid at 37°C for 5 h, or for the times indicated in the figure legends, in medium containing antibiotics and glutamine and supplemented with 0.3% FBS (cell labelling medium). Cells were washed twice with Hank's balanced salt solution (HBSS) containing 0.1% (w/v) fatty acid‐free bovine serum albumin (H‐BSA) to remove excess probe and further incubated in a complete culture medium. The conditioned medium was either immediately processed for EV isolation or stored at 4°C for up to one week. Cells were detached from the plates with trypsin/EDTA, and their viability was assessed by Trypan Blue exclusion. The sEV release per cell was calculated for experiments with cells showing ≥ 75% viability. To determine the total cell‐associated fluorescence in pulse‐chase experiments, cells were detached at different time points, washed with PBS and fixed with 4% paraformaldehyde in PBS for 30 min in the dark at room temperature (RT). Next, cells were detached at different time points and the fluorescence (Mean Fluorence Intensity, MFI) was determined by FC.
To obtain sEV for generic fluorescent labelling, cells were treated as in pulse‐chase experiments with the difference that the complete medium was replaced by EV depleted medium. Freshly pelleted 100K sEV from conditioned medium were labelled with 10 µM 5‐(and‐6)‐Carboxyfluorescein Diacetate Succinimidyl Ester (CFDA‐SE, hereinafter referred to as CFSE) (ThermoFisher) for 30 min at RT in the dark. The reaction was stopped by adding 100 mM L‐glutamine, and the number of fluorescent sEV was quantified by FC.
2.3. sEV isolation by differential ultracentrifugation
Conditioned medium was collected 24 h after pulse media exchange or at the indicated chase time intervals and serially centrifuged at 2000 g for 20 min at 4°C to discard cells and large debris. The supernatant was centrifuged at 10,000 g for 20 min at 4°C for microvesicles and other debris. The supernatant was then ultracentrifuged 100,000 g for 90 min at 4°C and the pellet (100K pellet) was washed in 12 mL of PBS and centrifuged again at 100,000 g in an SW41 Ti swinging bucket rotor (Beckman Coulter). Pellets were resuspended in 100–150 µL of PBS.
2.4. NanoFACS analysis of Bodipy sEV
Quantification of fluorescent Bodipy sEV was evaluated as described (Coscia et al., 2016) with a conventional non‐customized CytoFLEX LX flow cytometer (Beckman Coulter) by exploiting their fluorescent emission. To establish the region containing the sEV, threshold and gain values were initially defined on the appropriate fluorescent channel with a bandpass of 525/40 by using fluorescent beads of known sizes ranging from 100 to 900 nm (Megamix Plus FSC/SSC, BioCytex, #7802/7803). The instrument was then set to include fluorescent‐size reference beads smaller than 100 nm. To obtain the number of sEV, the Cytoflex LX instrument utilized a sample peristaltic pump capable of calibrating the volume delivery of the sample, allowing for absolute EV counts without the need for bead‐based calibration.
2.5. NanoFACS sorting of Bodipy sEV
To sort the Bodipy‐positive population of sEV, we utilized a MoFlo Astrios‐EQ instrument equipped with four lasers (405, 488, 561 and 640 nm) and set up as described in (Lanna et al., 2022). In detail, the threshold value was applied on the side scatter of the 488 nm blue laser (B‐SSC). Plotting B‐SCC Height signal versus B‐SCC Width signal allowed the exclusion of vesicle aggregates and other background noise from the target population. In order to determine the appropriate threshold value for sorting the Bodipy‐positive population of sEV, we utilized fluorescent size reference beads (110 nm to 1300 nm size beads, ApogeeMix, #1493). PBS and unlabelled sEV were used to assess the background noise and the not fluorescent signal, respectively. A gating strategy has also been applied to eliminate aggregated vesicles and background noise by plotting B‐SCC Height signal versus B‐SCC Width signal. The cells sorter is also equipped with an inline sheath filter with 40 nm pore size to decrease the background noise. Finally, the flow rate was settled to reduce the electronic and PBS background noises to less than 300 events/sec. Acquisitions, cell sorting, and analyses were performed by using the Summit 6.3.1 software (Beckman Coulter).
2.6. Nanoparticle tracking analysis
EVs secreted by the same initial number of cells, untreated or treated with Palmitic Acid or Bodipy C16, have been quantified by Nanoparticle tracking analysis (NTA) performed with a NanoSight NS300 system (Malvern Instruments, UK). Camera level was set at 16 for all recordings. Camera focus was adjusted to make the particles appear as sharp individual dots. Five 60‐second videos were recorded for each sample with a delay of 7 s between each recording at a fixed temperature of 25°C. Analysis was performed with NTA 3.4.4 software. Detection threshold was set at 5 and other settings were kept at default.
2.7. Iodixanol density gradient separation
Bodipy sEV (100K pellets) were bottom loaded in a discontinuous iodixanol gradient (35% (w/v), 25% (w/v), 15% (w/v) and 5% (w/v)) made by diluting OptiPrep (60% (w/v)) iodixanol (Sigma‐Aldrich, #D1556) with 60 mM Tris‐HCl (pH7.4), 0.25 M sucrose, 1 mM of EDTA from the bottom to the top of an open‐top polypropylene tube (Beckman Coulter, #328874). The gradient was centrifuged for 16 h at 33,000 RPM at 4°C (SW60 rotor, Beckman Coulter). A total of 13 fractions of 330 µL were manually collected from the top of the gradient and fraction density was assessed with a refractometer (Carl Zeiss). Bodipy sEV present in each fraction has been quantified by FC.
2.8. Confocal microscopy
To evaluate Bodipy C16 distribution, cells were seeded on 12 mm round coverslips in 24‐well plates at a density of 80,000 cells/well the day prior to experiments and then incubated with 1 µM Bodipy C16 in cell labelling medium for 2 h at 37°C. Subsequently, cells were washed twice with ice‐cold H‐BSA and then were fixed for 30 min with 4% paraformaldehyde in PBS at RT and stained with DAPI (Sigma) to visualize nuclei. For colocalization experiments with mitochondria and lysosomes, Mel501 cells on coverslips were incubated with 200 nM MitoTracker Deep Red or 50 nM LysoTracker Deep Red. MitoTracker incubation started 10 min before adding 1 µM Bodipy FL C16 whereas LysoTracker was added simultaneously. Incubation was carried out for 15 and 30 min after Bodipy C16 addition. Cells were then washed with PBS, fixed and mounted with DAPI as described above. For immunolocalization experiments, cells were pulsed with 1 µM Bodipy C16 for 2 h at 37°C and washed twice with ice‐cold H‐BSA. Cells were then fixed with 4% paraformaldehyde in PBS for 30 min at RT and permeabilized with 0.1% (w/v) saponin (Sigma) in PBS. Following blocking in PBS supplemented with 0.1% (w/v) saponin and 2.5% (w/v) BSA (blocking buffer), cells were incubated with primary antibodies diluted 1:100 in blocking buffer for 2 h at RT. Next, cells were washed twice with 0.1% (w/v) saponin in PBS for 10 min and successively incubated for 1 h at RT with secondary antibodies diluted 1:500 in blocking buffer. Cells were mounted with DAPI, as described above. For plasma membrane labelling, cells pulsed for 2 h with 1 µM Bodipy C16 and washed twice with ice‐cold H‐BSA were incubated with 50 µg/ml Concanavalin A (Con A) on ice for 30 min. Cells were then washed with PBS, fixed and mounted with DAPI as described above. Images were acquired on a Zeiss LSM 900 confocal microscope using a 63X objective. At least 20 fields were captured for each condition, and these were representative of at least 3 independent experiments. The percentage of colocalization was calculated by Zen Blu software as follows: first threshold values were established for each fluorescence channel by utilizing negative areas in control samples; the threshold values were then applied to count the positive pixels for each channel (converted in square microns). The software also provided the number of pixels that exhibited values above the threshold for both signals, representing the colocalized area. To normalize this measure, the number of Bodipy C16 pixels was used as the denominator in each image to calculate the percentage of colocalized area. Antibodies and reagents for immunofluorescence were: MitoTracker Deep Red FM (Invitrogen, #M22426), LysoTracker Deep Red, (Invitrogen, #L12492), Con A Alexa Fluor647 (Invitrogen, #C21421), mouse anti‐Phalloidin TRITC (ECM Biosciences, #PF7551), mouse anti‐BMP (Echelon, #Z‐SLBPA), mouse anti‐CD63 (BD Bioscience, #556019), mouse anti‐CD81 (BD Bioscience, # 555675), mouse anti‐CD9 (Santa Cruz Biotechnology, #sc‐13118), mouse anti‐GM130 (BD Bioscience, #610822), mouse anti‐Calnexin (BD Bioscience, #610524), goat anti‐Calregulin (Santa Cruz Biotechnology, #sc‐6467), goat anti‐mouse AlexaFluor555 (Invitrogen, #A21424) and donkey anti‐goat AlexaFluor 594 (Invitrogen, #11058).
2.9. High‐performance thin layer chromatography (HPTLC)
Lipids samples from cells and sEV were extracted by the Folch method (Folch et al., 1957). The extracted lipids were then resuspended in a small volume of 2:1 (v/v)CHCl3/MeOH and layered onto Silica gel 60 F₂₅₄ plates (Merck, Darmstadt, Germany). For separation of neutral lipids, a mixture of hexane/ethyl ether/acetic acid (70:30:1 v/v) was used, while for the separation of phospholipids, the solvent system used was chloroform/methanol/acetic acid/ water (50:37.5:3.5:2, v/v). Fluorescent lipids species were visualized with a Typhoon 9200 scanner (Amersham Biosciences). 10 µg each of the following standards were used: lyso(bis)phosphatidic acid (LBPA), phosphoethanolamine (PE), phosphatidilcholine (PC), phosphatidilserine (PS) (Echelon Biosciences); phosphatidylinositol (PI), diacylglycerol (DAG), triacylglycerol (TAG), cardiolipin (CL) (Avanti Polar Lipids); cholesterol (chol) (Sigma‐Aldrich). Lipid standards were detected with 3% copper acetate solution in 8% phosphoric acid and subsequent heating for 10 min at 120°C.
2.10. Western blot
EV proteins were separated by 10 or 12% sodium dodecyl sulphate (SDS) polyacrilamide gel electrophoresis (PAGE) under reducing conditions, with the exception of samples used to detect CD63, CD81 and CD9, and transferred on 0.22 µm nitrocellulose membranes (BioRad). Membranes were blocked for 5 min at RT in EveryBlot Blocking Buffer (BioRad) and then incubated for 1 h at RT with different primary antibodies: mouse anti‐CD63 (BD, #556019, 1: 300), mouse anti‐CD81 (BD, #555675, 1: 300), rabbit anti‐CD9 (CST, #13403, 1: 300), polyclonal anti‐Alix (Invitrogen, #PA5‐52873, 1:250), polyclonal anti‐Syntenin 1 (Invitrogen, #PA5‐92419, 1:200), mouse anti‐Annexin 1 (BD, #610066, 1:200). Secondary antibodies horseradish peroxidase (HRP)‐conjugated were used 1:3000 for 1 h at RT in EveryBlot. Western Blots were developed using Clarity Western ECL Substrate (BioRad). The presented immunoblots are representative of at least 3 independent experiments. Band analysis was performed using ImageLab software from BioRad.
2.11. Colocalization of Bodipy sEV with tetraspanins
Bodipy sEV isolated by ultracentrifugation were incubated with 5 µL of anti‐CD63 BV421 antibody (BD, #740080), 10 µL of anti‐CD81 APC antibody (BD, #551112), and 5 µL of anti‐CD9 PE antibody (BD, #555372) to determine colocalization between Bodipy sEV and the tetraspanins. As negative controls Bodipy sEV were incubated with isotype antibody IgG1,κ conjugated to BV421 (Biolegend, #400157), APC (BD, #555751), or PE (BD, #555749) for 45 min at RT in a HulaMixer Sample Mixer in PBS with 2% FBS. Samples were fixed with 4% PFA for 30 min at RT, and then washed with PBS at 500 × g for 20 min at 4°C. The supernatant was discarded, and vesicles were resuspended in PBS. The colocalization percentage of Bodipy C16 with anti‐CD63‐BV421, anti‐CD81‐APC, and anti‐CD9‐PE was determined by FC.
2.12. Electron microscopy
2.12.1. Transmission electron microscopy (TEM)
Negative staining was performed as described previously (Federici et al., 2020). Briefly, Bodipy sEV (>109 before and >107 after sorting as measured by NTA), were suspended in PBS (100 µL) and 10 µL were adsorbed on formvar‐carbon coated grids. Ammonium molybdate 4% pH 6.4 (5 µL) was added as contrasting solution for 30 s and adsorbed with filter paper. Samples were air dried and observed by PHILIPS EM208S TEM (FEI, ThermoFisher).
2.13. Immuno‐electron microscopy (IEM)
IEM was performed on the total sEV derived from Bodipy C16 pulsed cells, adsorbed on formvar‐carbon coated grids. Samples were air‐dried and transferred (membrane side down) to a drop of rabbit polyclonal anti‐Bodipy (Invitrogen, #A5770, 10 mg/ml;) or mouse monoclonal anti‐CD81 (Santa Cruz Biotechnology, #sc‐166029, 1:10) or mouse monoclonal anti‐CD63 (BD, #556019, 5 mg/ml) in PBS for 1 h at RT, rinsed in buffer, incubated on 10 nm gold‐conjugated goat anti‐rabbit IgG (Sigma, #G3779, 1:30,) or 10 nm gold‐conjugated goat anti‐mouse IgG (Sigma, #G777, 1:30) in buffer for an additional h. Double immunolabelling was performed with anti‐CD63 and anti‐Bodipy antibodies, applied one by one. Anti‐mouse IgG 10 nm and anti‐rabbit IgG 5 nm gold‐conjugated were chosen for showing anti‐CD63 and anti‐Bodipy antibodies, respectively. Samples were rinsed in buffer and successively incubated on a drop of 2% paraformaldehyde in PBS for 5 min. After a rapid final rinse with water, the grids were stained by ammonium molybdate as previously described and observed by a PHILIPS EM208S TEM (FEI, ThermoFisher).
2.13.1. Scanning electron microscopy (SEM)
SEM was performed as previously described (Federici et al., 2020). Bodipy sEV (before and after sorting, same amounts as above), were suspended in PBS (100 µL) and were left to adhere to polylisine‐treated round glass coverslips for 1 h. Samples were fixed with glutaraldehyde 2,5% in Na‐cacodylate buffer 0,1 M for 1 h and processed for SEM. Briefly, samples were post‐fixed with 1% OsO4 in 0.1 M sodium cacodylate buffer 0,1 M for 1 h at RT and were dehydrated through a graded series of ethanol solutions (from 30% to 100%). Then, absolute ethanol was gradually substituted by a 1:1 solution of hexamethyldisilazane (HMDS) and absolute ethanol for 30 min, and successively by pure HMDS for 1 h at RT. The final drying process was concluded removing completely the HMDS and leaving it to evaporate all the liquid phase on air, drying the samples at RT for 2 h and in a desiccator o.n. Finally, samples were mounted on stubs, lightly gold sputtered and analyzed in a field emission GeminiSEM450 (ZEISS). High magnification TEM images of total sEV or Bodipy‐sorted sEV were analyzed by Fiji, Image J open source (Schindelin et al., 2012) and vesicles more than 30 nm in diameter were taken into account for collecting manual measures (N > 100).
2.14. Statistical analysis
Statistical analyses in this study were conducted with Graphpad Prism 9.5 software. The data are presented as the mean ± SEM from at least three independent experiments. Individual group statistical comparisons were analyzed by the two‐tailed Student t‐test. A p‐value < 0.05 (two‐sided) was considered statistically significant.
3. RESULTS
3.1. Bodipy C16 internalization and sEV secretion in human cell lines
We previously showed (Coscia et al., 2016) that Bodipy C16, a fluorescent analog of palmitic acid, is rapidly internalized by cells and metabolized into phospholipids that enter the EV biogenesis pathway, ultimately producing fluorescent sEV (Figure 1a). To extend our observations we first evaluated the intracellular distribution of Bodipy C16 in a panel of five human cell lines, three melanoma (Mel501, Me665/1 and A375/M) and two cell lines frequently used in sEV studies (HEK293 and HeLa). Confocal images of all the tested cell lines showed intense fluorescence distributed in the perinuclear region and in discrete structures compatible with endolysosomal compartments after 2 h of Bodipy C16 uptake (Figure 1b), whereas the fluorescence signal appeared absent from the plasma membrane. No noticeable differences were observed among the different cell lines. We next compared the secretion of Bodipy sEV over a 24 h period to highlight any potential differences in secretion among the cell lines. Due to varying cell duplication rate, we evaluated the number of secreted Bodipy sEV per single cell, based on the effective number of cells recovered at the end of the 24 h period (Figure 1c). Cells exhibited different levels of sEV secretion with the highest amount generated by the Mel501 cell line that was chosen for further studies. Palmitic acid has been shown to be toxic to mammalian cells in a concentration‐dependent and time‐dependent manner, inducing cellular dysfunction, ER stress, and cell death (Akoumi et al., 2017; Malhi et al., 2006; Penke et al., 2017). In addition, it has been reported that palmitic acid treatment increases exosome production in hepatoma cell lines, although this effect was observed at an almost millimolar concentration range (Y. S. Lee et al., 2017). To rule out a potential effect of Bodipy C16 on sEV secretion, we treated Mel501 cells with 7 µM Bodipy C16 or equivalent concentrations of unlabelled palmitic acid for 5 h in cell labelling medium. Then, we chased the cells for 24 h in an EV‐depleted complete medium. The results indicated that there were no significant effects of palmitic acid or Bodipy C16 on the total amount of sEV secretion detected by NTA (Figure 1d) or on the levels of well‐known sEV markers such as CD63, CD81, CD9 and TSG101 (Figure 1e), compared to the untreated control cells. Therefore, we were able to rule out any effect of palmitic acid at the concentrations used on sEV secretion.
FIGURE 1.
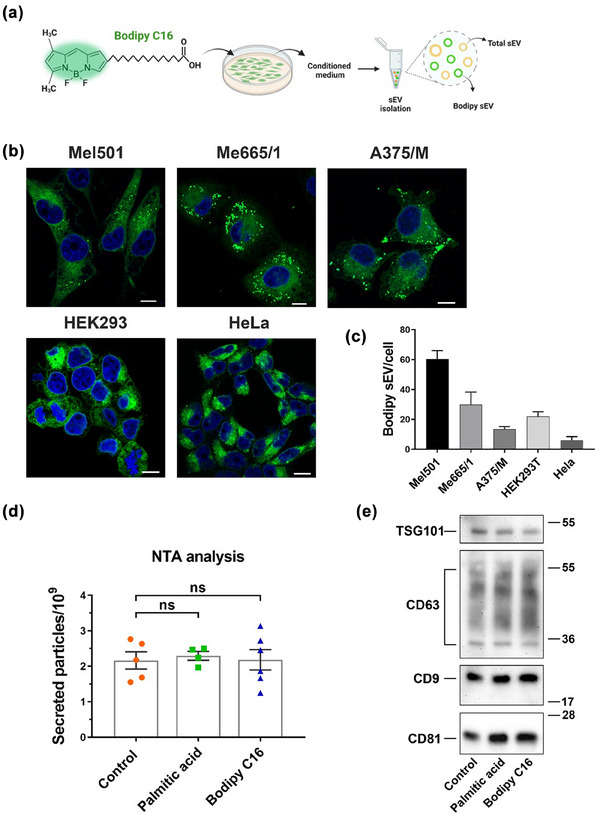
(a) Scheme of the structure of Bodipy C16 and experimental strategy. (b) Confocal microscopy images of cells labelled for 2 h with Bodipy C16 at steady state. (c) Quantification by Flow Cytometry (FC) of sEV secreted in the conditioned medium of different cell lines. (d) Measurement by Nanoparticle Tracking Analysis (NTA) of total sEV released by Mel501 cells untreated or treated with Bodipy C16 or with unlabelled palmitic acid for 5 h. SEV were purified from conditioned media after 24 h of chase. Bars represent the mean ± SEM (n > 3). (e) Western blot analysis of total sEV released by untreated or treated with Bodipy C16 or with unlabelled palmitic acid.
3.2. Bodipy C16 is transformed into phospholipids in melanoma cell
Palmitic acid plays a central role in the synthesis and maintenance of structural phospholipids and in cancer cells exogenous palmitate has been found to be incorporated into various lipids required for their proliferation and pro‐tumorigenic lipid signalling (Louie et al., 2013). In addition, various in vitro and in vivo studies have shown that palmitic acid overload causes increased triglyceride accumulation into lipid droplets (Olzmann & Carvalho, 2019) and it is often used in vitro to induce lipid droplet formation (Thomas et al., 2022).
To gain better insight into the metabolic transformation of Bodipy C16 within the cells, we first performed a time course experiment of Bodipy C16 uptake. We have previously reported that the optimal concentration of Bodipy C16 for probe incorporation is 7 µM (Coscia et al., 2016) which is much lower than that reported for the induction of lipid droplet formation or cytotoxic effects (Thomas et al., 2022). To further characterize the time course of Bodipy C16 uptake and metabolism we pulsed cells with the probe at different times and measured cell‐associated fluorescence by FC. Bodipy C16 uptake reached saturation at 5 h (Figure 2a). When the cells were incubated with the probe for 15 min and then washed and chased in fresh medium, a sharp decrease of total cell fluorescence was observed after 1 h, with 50% of the initial fluorescence reached at 6 h (Figure 2b). Subsequently, fluorescence levels decreased more slowly (Figure 2b). Next, we performed lipid extractions of cells that had been pulsed with Bodipy C16 for 15 min, washed and then chased in fresh media for up to 24 h and analyzed. As illustrated in Figure 2c, cells pulsed with Bodipy C16 for a short time, very rapidly metabolized the Bodipy C16 into phospholipids and neutral lipid intermediates, visible already at the end of the pulse time. These latter rapidly decreased over time and were significantly reduced by about 30 min of chase thus demonstrating that the observed intracellular fluorescence did not result from free probe diffusing in the cells either alone or bound to the BSA used as carrier. To identify all possible fluorescent lipid intermediates, we increased the pulse time to 5 h and collected cells and sEV after 2 h and 24 h of chase (Figure 3d). In these conditions, we observed in cell extracts the appearance of fluorescent phospholipids and neutral lipid intermediates but not of triacylglycerols (TAG), diacylglycerols (DAG) or cholesterol, which are the main constituents of the neutral lipid core of lipid droplets (Pol et al., 2014). This finding allowed us to conclude that at the low concentrations used, Bodipy C16‐derived lipids do not accumulate into lipid droplets. Notably, fluorescent lipids in sEV were mainly associated with phospholipids. Total lipids from both cells and sEV were then extracted and resolved for phospholipids (Figure 2e). In the cell extracts, phospholipids content greatly decreases with time, while the sEV extracts did not exhibit changes in phospholipids‐associated fluorescence intensity, even after 24 h. Our results show that in melanoma cells Bodipy C16 is a precursor of the de novo biosynthesis of phospholipids that enter membranes biosynthesis pathway.
FIGURE 2.
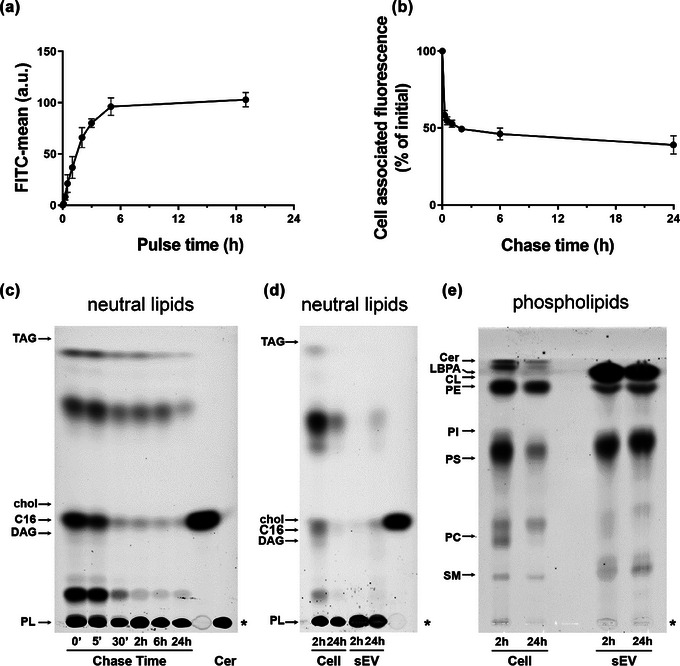
Dynamics of Bodipy C16 uptake and transformation into phospholipids. (a) Mel501 cells were treated with 7 µM Bodipy C16 for the indicated times or (b) treated for 15 min, washed with H‐BSA and chased in complete medium. Cell associated fluorescence was measured by FC. Data are expressed as mean ± SEM (n = 3). (c) Total lipids from cells pulsed with Bodipy C16 for 15 min and chased for different times were extracted and analyzed by HPTLC. The amount of lipid spotted per lane was equivalent to 5 × 104 cells. (d) Lipids from 2 × 104 cells pulsed with Bodipy C16 for 5 h and chased for 2 h and 24 h and Bodipy sEV isolated from the conditioned medium were extracted and analyzed by HPTLC for neutral lipids and (e) phospholipids. The amount of lipid spotted per lane was equivalent to 4 × 104 cells and 8,6 × 108 sEV. Asterisk indicates the sample origin line. Lipids were identified based on comparison to the respective lipid standards. Neutral lipids: TAG, triacylglycerols; DAG, diacylglycerol; chol, cholesterol; C16, Bodipy C16; PL, Polar Lipids. Phospholipids: Cer, ceramide; LBPA, Lysobisphosphatidic acid; CL, cardiolipin; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PS, phosphatidylserine; PC phosphatidylcholine; SM, sphingomyelin.
3.3. Bodipy C16 labels membranes of the ER/endolysosomal compartment
To investigate the intracellular journey of Bodipy C16, which leads to fluorescent sEV release, we initially examined whether Bodipy C16 is transported to mitochondria and lysosomes, organelles involved in fatty acid metabolism. We pulsed cells with Bodipy C16 for short periods (15 min and 30 min) and tracked its colocalization with the mitochondrial marker MitoTracker and LysoTracker, a cell permeable weak base dye that accumulates in acidified organelles. As shown in Figure 3a and b, the Bodipy green fluorescent signal colocalized with MitoTracker (red) by about 40% and with LysoTracker (red) by about 30% after 15 min of Bodipy C16 uptake. At 30 min, we observed persistent colocalization of the fluorescent signal with mitochondria, while colocalization with lysosomes showed a significant decrease to about 41% of the initial value. These experiments suggest that upon uptake, Bodipy C16 is quickly targeted to organelles where lipid transformation may commence. To assess the degree of colocalization between Bodipy FL C16 and endogenous marker proteins targeting different locations on the endocytic pathway, we conducted confocal immunofluorescence experiments. As a first step, we aimed to establish whether lipids derived from Bodipy C16 colocalized with the plasma membrane, in order to exclude the possibility of fluorescent sEV originating from it. For this purpose, we pulsed cells with Bodipy C16 for 2 h (Figure 3c) and examined the colocalization with the lectin Concanavalin A (Con A), which labels the plasma membrane. The analysis of colocalization, as shown in Figure 3d, indicated that there was almost no colocalization, as only 0.8 ± 0.1% SEM of Bodipy fluorescence was found at the plasma membrane. We then established the colocalization with Con A as a baseline for comparison with other markers. Next, we utilized Phalloidin, which stains polymerized actin, to outline the cell periphery. The colocalization with this marker was poor (3.7 ± 0.4%), further suggesting that Bodipy C16‐derived phospholipids are not trafficked to the plasma membrane but follow different intracellular routes. Since the trans‐Golgi network and endoplasmic reticulum (ER), in addition to being main sites of membrane lipid synthesis (Fagone & Jackowski, 2009), are organelles known to contribute to the formation of the early‐sorting endosome that eventually generates MVB (Kalluri & LeBleu, 2020), we analyzed colocalization with ER and Golgi markers. As expected, we found significant colocalization with calregulin (14.4 ± 2.3%) and calnexin (19.7 ± 2.7%), and GM130 (24.1 ± 3.3%) (Figure 3d), indicating that 2 h after the addition of Bodipy C16, fluorescent lipids are present in these compartments presumably en route to compartments from which intraluminal vesicles are generated. Finally, we determined colocalization with the protein markers CD63, CD81, and CD9, which are commonly used to identify sEV, and with LBPA, an atypical lipid highly enriched in late endosomes/MVB (Gruenberg, 2020; Kobayashi et al., 1998). As shown in Figure 3c and d we found high colocalization with CD63 (17.8 ± 1.8%), and CD9 (14.4 ± 1.5%) and lower but significant colocalization with LBPA (9.1 ± 0.7%). It has to be noted that CD9 staining exhibited a punctate distribution pattern within intracellular compartments, which differs from the predominantly plasma membrane localization observed in HeLa cells (Mathieu et al., 2021). Conversely, colocalization with CD81 was low (2.4 ± 0.3%) consistent with previous observations that CD81 is mostly localized to the plasma membrane (Fordjour et al., 2022; Larios et al., 2020) and further supporting the hypothesis that Bodipy lipids are not targeted to the plasma membrane. Taken together these results provide evidence that upon internalization Bodipy C16 rapidly localizes in lipid metabolism sites where transformation in phospholipids likely occurs and that the lipids derived from Bodipy C16 become a part of the late endosome membranes with the exclusion of the plasma membrane.
FIGURE 3.
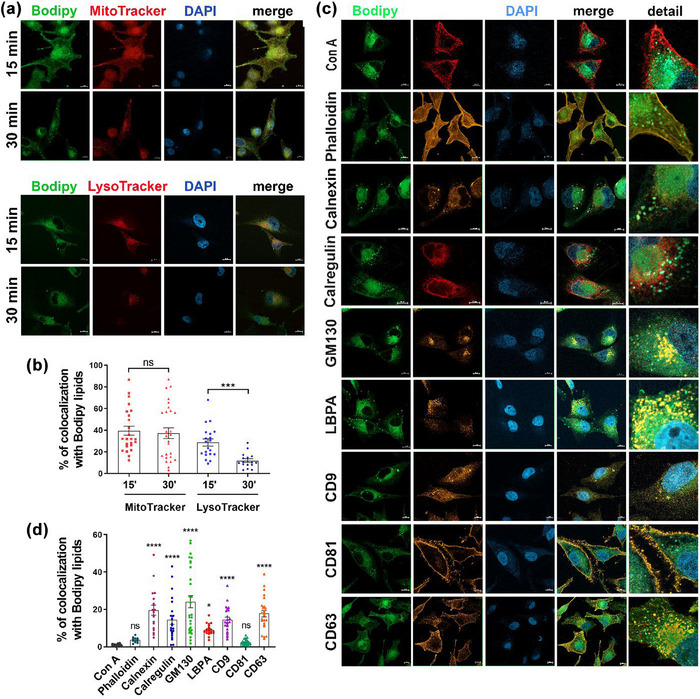
Co‐localization analysis of Bodipy lipids with intracellular markers. (a) Colocalization with mitochondria and lysosomes at different pulse time. Cells were labelled with 1 µM Bodipy C16 and Mitotracker Deep Red or Lysotracker Deep Red for 15 or 30 min, fixed and mounted with DAPI. (b) Quantification of colocalization with Bodipy lipids. Quantification of 3 independent experiments showing the mean ± SEM (≥20 fields). (c) Colocalization of Bodipy lipids with intracellular organelles markers and tetraspanins after a 2 h pulse with Bodipy C16 at steady‐state. Fixed cells were permeabilized with 0,1% saponin prior to incubation with antibodies or non permeabilized when incubated with Con A. Percentage of colocalization is calculated by summing the pixels in the colocalized region and then dividing by the sum of Bodipy C16 pixels. All data are expressed as means ± SEM (n ≥ 3) (n ≥ 20 fields). Individual unpaired t‐test was performed to compare two groups. Significance of colocalization in (d) was calculated by comparing percent of individual markers colocalization with Con A colocalization. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, ns, no statistical significance. Scale bar = 10 µm.
3.4. Bodipy sEV are a homogeneous subpopulation of EV
Previous reports have shown that cells release subpopulations of exosomes differing in molecular and biophysical properties such as density range of sedimentation, and that these can be separated by density gradient centrifugation (Kowal et al., 2016; Willms et al., 2016; Zhang et al., 2018). To address the question of whether Bodipy sEV represent a specific subpopulation of sEV we subjected the conditioned media of Mel501 cells pulsed with Bodipy C16 and chased for 24 h, to differential centrifugations to obtain an ultracentrifugation pellet (100,000 g = 100K), classically considered as containing exosomes. The resulting pellet was loaded at the bottom of an iodixanol gradient, followed by ultracentrifugation for 16 h to allow vesicles to float into the gradient. The distribution of fluorescent sEV within the gradient fractions was analyzed by FC. Bodipy sEV were separated as a discrete peak in fractions 4–5 (density range 1.067‐1.081 g/ml, as measured by refractometry) (Figure 4a and c). These fractions contained the exosome marker proteins CD63, CD81, CD9, Alix, and Syntenin 1, as well as Annexin 1, a marker of microvesicles shed from the plasma membrane (Jeppesen et al., 2019), which showed a similar distribution (Figure 4d). To be noted, the density range of the Bodipy sEV was lower than that previously reported for exosomes (Colombo et al., 2014; Willms et al., 2016). To rule out the possibility that Bodipy sEV were an artifact, we added to the gradient fractions 60 mM octyl‐glucoside, a nonionic detergent widely used for membrane protein solubilization, in order to discriminate between protein aggregates and membrane vesicles (Inglis et al., 2015; Jeppesen et al., 2019; Ricklefs et al., 2019). As shown in Figure 4a, OG completely abolished fluorescent particles detection in the low‐density fractions, thus confirming that Bodipy sEV are indeed membrane‐enclosed vesicles. Since floatation of sEV bottom loaded into Iodixanol gradients has been widely used to separate distinct subpopulations of sEV, we incubated 100K pellets containing the total population of sEV derived from conditioned media of untreated Mel501 cells, with CFSE, a cell‐permeable protein‐binding dye commonly used to obtain fluorescently labelled sEV preparations (Kormelink et al., 2015; Morales‐Kastresana et al., 2017; Pospichalova et al., 2015; Woud et al., 2022). The total CFSE sEV pellets were subjected to iodixanol density gradient centrifugation and the fractions analyzed by FC. As shown in Figure 4b, CFSE vesicles separated into two distinct peaks, a lower density peak in the same density range as Bodipy sEV and a higher density peak that was only partially soluble with detergent, consisting mostly of protein aggregates. This suggests that the higher‐density peak contains a small proportion of membrane vesicles along with non‐vesicular contaminants that co‐isolated with the sEV during the purification process.
FIGURE 4.
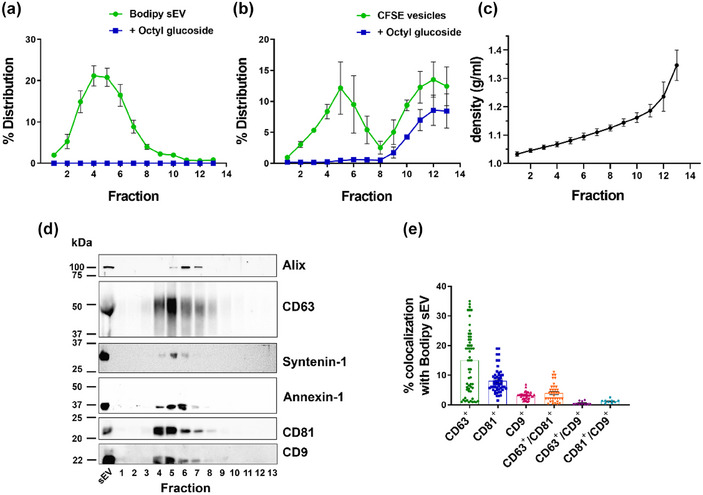
Bodipy sEV separate as a single peak enriched in tetraspanins. (a) Bodipy sEV 100K pellets were loaded at the bottom of an iodixanol density gradient and ultracentrifuged for 16 h. Fractions were collected from the top of the gradient and were analyzed for Bodipy sEV number by FC before and after the addition of Octyl‐beta‐glucoside (OG). Data are expressed as mean ± SEM (n = 14). (b) 100K pellets from conditioned media of untreated Mel501 cells were incubated with CFSE to non specifically label the total population of sEV and loaded at the bottom of an iodixanol density gradient. Data are expressed as mean ± SEM (n = 4). (c) Density of the fractions was measured by refractometry. Data are expressed as mean ± SEM (n = 17). (d) The total volume of each fraction was analyzed by Western blot. Data shown are representative of three independent experiments. (d) Analysis of colocalization of tetraspanins fluorescent antibodies with Bodipy sEV.
In recent years, several reports have shown that sEV of endosomal origin is enriched on their surface in CD63, CD81 and CD9 tetraspanins although in different combinations and amounts and depending on the cell type (Hurwitz et al., 2016; Kowal et al., 2016; Mathieu et al., 2021; Willms et al., 2016). To characterize the Bodipy sEV population comprised in the 100K pellet among microvesicles and other particles we performed colocalization experiments of Bodipy sEV with fluorescent‐labelled antibodies against CD63, CD81 and CD9 (Ricklefs et al., 2019). Results show that 14.2% of Bodipy sEV were CD63+, whereas CD81+ and CD9+ amounted respectively to 8.1 % and 3.1%. Double positive CD63+/CD81+ were 3.9%, CD81+/CD9+ were 1.2% while CD63+/CD9+ were only 0.6 % of the total positive vesicles. Taken together these results support the hypothesis that Bodipy sEV represent a distinct low density subpopulation of sEV of endosomal/MVB origin.
3.5. Kinetic of secretion of sEV
The regulation and kinetics of exosome secretion have not yet been well characterized due to the lack of suitable methods to distinguish between subpopulations of sEV. However, our methodology allows for the analysis of a distinct population of vesicles of intracellular origin. Therefore, to trail the dynamics of Bodipy sEV release we set up experiments to determine the kinetics of Bodipy sEV secretion in Mel501 cells and determine if there is a correlation between cell fluorescence decrease (Figure 2b) and fluorescent sEV secretion. We pulsed cells with Bodipy C16 for 2 h, washed the cells, and chased them in fresh medium. Conditioned medium was harvested at different time points to determine the number of Bodipy sEV secreted. The results show that at 6 h of chase, the number of Bodipy sEV released in the conditioned media reaches a plateau, and there is no further increase at 24 h (Figure 5a). As a control, we determined the kinetics of Bodipy sEV release in conditioned media in comparison with the total particles present in the same 100K pellet. NTA quantification of the total particles in the same sample showed a similar kinetics up to 6 h, steeply increasing at later time points (Figure 5b). This is supported by the ratio of Bodipy sEV secretion versus total particles, which is constant up to 6 h with approximately 4.5 times more particles than Bodipy sEV (Figure 5c). However, at 24 h, this ratio is 11, indicating that the intracellular Bodipy C16 reservoir is depleted in the first few hours after the Bodipy C16 pulse, as Bodipy phospholipids are trafficked to sites where ILV are formed and readily secreted. Once Bodipy labelled sEV are released, the secretion of unlabelled sEV takes over. Western blot analysis of 100k pellets collected at different time points show the increase over time of CD63, CD81, and CD9 tetraspanins (Figure 5d) reflecting the kinetics of Bodipy sEV secretion. CD63, which is highly enriched on ILVs in late endosomal MVB, is detected first at 30 min followed by CD81, whereas CD9 a marker that in recent studies is more often associated with ectosomes (Mathieu et al., 2021) becomes detectable at later time points. In conclusion, the data presented collectively indicate that the metabolic labelling methodology described in this study provides the means to distinguish between various subpopulations of sEV and accurately monitor the release dynamics of Bodipy‐labelled sEV.
FIGURE 5.
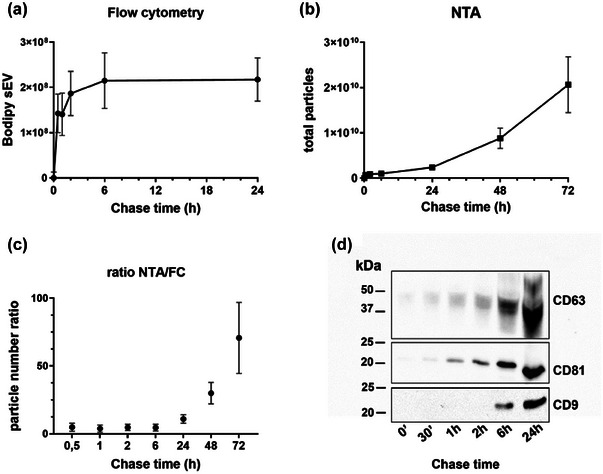
Secretion kinetics of Bodipy sEV. Cells were pulsed with Bodipy C16 for 2 h and chased for different times. The number of purified sEV purified by differential ultracentrifugation was measured by (a) FC or by (b) Nanoparticle Tracking Analysis (NTA). Results are expressed as means ± SEM of at least 6 independent experiments. (c) Ratio between the total number of sEV counted by NTA and Bodipy sEV counted by FC. A constant ratio is showed up to 6 h, while there is an increase of total vesicles over the time. Results are expressed as means ± SEM of at least six independent experiments. (d) Representative Western blot analysis of tetraspanins expression in Bodipy sEV secreted at different chase time. The total sEV released at each time point has been analyzed.
3.6. Bodipy sEV are a subpopulation of small exosomes
Recently, it has been shown that high‐sensitivity flow cytometers can discriminate sEV in the range of 100 nm that can be efficiently sorted (Morales‐Kastresana et al., 2019). Analysis and sorting of both sEV and larger EVs with multiplex fluorescent detection requires fluorescent labelling of the vesicles that can be obtained in multiple ways such as generic labelling of the EV surface (Morales‐Kastresana et al., 2019), incorporation of fluorescence protein tags (i.e., GFP) or fluorescent immunostaining using antibodies to specific surface biomarkers on EV. However, all these methodologies still need to be implemented since no generic marker can stain all sEV or more specifically a specific subpopulation of sEV like exosomes. We took advantage of our metabolically labelled Bodipy sEV to investigate the possibility of obtaining a pure population of Bodipy‐labelled sEV by sorting the Bodipy‐positive population using a MoFlo Astrios‐EQ instrument.
To assess the background noise, we used PBS, while unlabelled sEV were used to evaluate the non‐fluorescent signal (Figure 6a, left dot plot). The representative sorting in Figure 6a shows the total EVs, with the middle dot plot indicating that the Bodipy+ sEV account for 50% of the total EVs. After sorting, the percentage of purified Bodipy+ sEVs increases to 66% (Figure 6a, right dot plot). It is important to note that the background noise cannot be completely eliminated as it is intrinsic to the system, as previously described (Morales‐Kastresana et al., 2017). Nonetheless, the sorting procedure allowed for the isolation of the Bodipy‐positive sEV population, which can be used for further analysis.
FIGURE 6.

Bodipy sEV can be sorted in a homogenous subpopulation of small exosomes. (a) Fluorescent sEV pellets have been sorted with a MoFlow AstriosCell Sorter. Dot plot on the left shows unlabelled EVs. The dot plot in the middle shows total Bodipy+ sEV and the arrow indicates the region used for sort decision. The post sorting analysis shows purified Bodipy+ sEV (dot plot on the right). Dot plots are representative of three independent experiments. (b) Unlabelled sEV and total Bodipy+ sEV were analyzed by TEM showing the presence of a discrete population of membrane vesicles in both samples. (c) FACS sorted Bodipy+ sEV analyzed by SEM revealed a homogeneous population of small vesicles in a very low background (right), particularly appreciable when compared directly to the total Bodipy‐labelled sEV population. (left). (d) Total sEV size was determined by NTA and electron microscopy (EM) The semi‐quantitative EM analysis (N > 100) showed that the sorted Bodipy+ sEV had a median size of 80 nm, significantly smaller than total Bodipy‐labelled sEV population (*** p < 0.001). (e) Total sEV were immunolabeled with anti CD81, CD63 and Bodipy antibodies, revealed by 10 nm colloidal gold conjugated secondary antibodies (upper left, upper right and lower left TEM micrographs). Colocalization of CD63 (10 nm) and Bodipy (5 nm, white arrow) antigens on the same vesicle is shown in the lower right image.
Bodipy‐labelled sEV were validated using transmission (TEM) and scanning (SEM) electron microscopy. Fresh preparations of total unlabelled sEV or total Bodipy‐labelled sEV were examined by TEM to assess their morphology and integrity (Figure 6b). Moreover, SEM micrographs of total Bodipy‐labelled sEV show a heterogeneous mixture of vesicles with a median size of the major diameter of 98 nm (Figure 6d). NTA analysis of the same samples gave an even higher mode size of 120 nm, consistent with the fact that NTA cannot distinguish between effective vesicles and other types of aggregates that are present in the vesicle preparations. SEM analysis of total Bodipy‐labelled sEV analyzed before (Figure 6c, left) and after sorting (Figure 6c, right) exclusively reveal that sorted Bodipy positive sEV are a more homogeneous population of vesicles emerging in a very low background (Figure 6c, right). The semi‐quantitative EM analysis of the size distribution of these population of vesicles (n > 100) shows a median value of 80 nm, significantly smaller than that of the total EV (p = 0.0008) (Figure 6d).
The immunogold labelling of total EVs with anti‐Bodipy FL antibodies shows low levels of the gold signal in the Bodipy+ sEV (Figure 6e, middle right) suggesting that the Bodipy epitope is not as readily exposed as the tetraspanin antigens, indicating potential differences in the accessibility of the antigens on the sEV membrane. In contrast, the anti‐CD81 antibodies as well as anti‐CD63 easily recognize the respective antigens around the exosome surface (Figure 6e, left and middle left). The image presented in Figure 6e (right) provides evidence of colocalization between CD63 (10 nm) and Bodipy (5 nm) antigens on the same vesicle, confirming that the Bodipy sEV analyzed by immune‐electron microscopy indeed exhibit Bodipy positivity in accordance with the relative percentage observed in flow cytometry data. Taken together these results indicate that the Bodipy labelled sEV are a discrete subpopulations of small exosomes valuable for further characterization and exosome biogenesis studies.
4. DISCUSSION
Exosomes play a crucial role in intercellular communication by delivering bioactive payloads that modify the phenotype of recipient cells. It is widely accepted that exosomes are generated through an endosome‐dependent pathway of biogenesis, although multiple intracellular pathways likely contribute to EV biogenesis. However, under specific microenvironmental conditions such as those encountered during tumor progression (Garcia‐Silva et al., 2021; Maas et al., 2017), exosomes originating from the late endosomal/MVB compartment exhibit dynamic variations in number, size, molecular composition, and ultimately the message conveyed to target cells. Additionally, the intracellular pathway followed en route to release into the extracellular space further influences exosome heterogeneity, and the discovery in recent years of subsets of exosome populations referred to as Exo‐S and Exo‐L which differ in size (small exosome vesicles, Exo‐S, 60−80 nm, large exosome vesicles, Exo‐L, 90−120 nm) (Zhang et al., 2018), and present unique biophysical properties has further complicated the labelling of specific sEV subpopulations with complete specificity. Nevertheless, comprehending these complexities is crucial for understanding the intricate mechanisms underlying intercellular communication.
With the aim to develop an effective strategy for labelling exosomes that could be distinguished from microvesicles or ectosomes we previously reported a novel methodology by using Bodipy C16, a fluorescent palmitic acid analogue. We have already shown that Bodipy C16 is mainly metabolized into constitutive lipids that become part of the sEV membrane bilayer producing fluorescent sEV that can be readily counted by FC (Coscia et al., 2016). In this study, we provide evidence that the population of fluorescent sEV released from Bodipy C16 labelled cells is a discrete subpopulation of small exosomes originating from an intracellular pathway. Cellular uptake of Bodipy C16 is very rapid and we here show that the probe is promptly metabolized to polar lipids and very little free Bodipy C16 can still be detected as early as 30 min of chase. Fluorescent exosomes, regardless of the secretion time, contain Bodipy lipids almost completely in the form of phospholipids with very little residual unmetabolized Bodipy C16. Newly formed Bodipy lipids can be tracked in phospholipid biosynthesis sites such as the ER and mitochondria, and in definite intracellular organelles where they colocalize with established markers of exosomes such as LBPA, CD63 and CD9 but not with CD81 that in melanoma cells, as also reported by others for other cell types (Fordjour et al., 2022) we found was mainly localized on the plasma membrane. Absence of colocalization of Bodipy with the lectin Con A used to label the plasma membrane was a further proof that Bodipy phospholipids are excluded from the plasma membrane.
In order to establish a more direct relationship between fluorescent exosome secretion and the intracellular route of ILV formation, we conducted a detailed analysis of the physicochemical and molecular characteristics of Bodipy‐labelled exosomes. The fluorescence labelling procedure we developed represents a significant improvement over current methodologies used for generic labelling of sEV. Despite recent advancements in achieving high‐resolution quantitative and qualitative analysis of cell‐derived vesicles using FC (Higginbotham et al., 2016; Kormelink et al., 2015; Morales‐Kastresana et al., 2019; Pospichalova et al., 2015), it is important to note that all the dyes utilized in these approaches are nonspecifically incorporated into ultracentrifugation pellets, which consist of heterogeneous populations of vesicles. Consequently, this poses a challenge in accurately quantifying and characterizing the vesicles. Our methodology enabled us to track fluorescent vesicles originating from the metabolic site of phospholipid synthesis that could be accurately monitored after isolation using current protocols involving differential centrifugation combined with iodixanol density gradient floatation. Bodipy exosomes separated as a discrete peak (1.07‐1.08 g/ml), positive for the exosome markers CD63, CD81, CD9, Alix, and Syntenin 1. In contrast, CFSE‐labelled sEV displayed an additional fluorescent peak at a higher density (>1.16 g/ml), which only in part was detergent soluble suggesting the presence of non‐detergent soluble aggregates. Annexin 1, identified as a specific marker for ectosomes shed from the plasma membrane (Jeppesen et al., 2019), was present in the same fractions as Bodipy‐labelled exosomes. This finding aligns with previous reports indicating that Annexin 1 can be found in both Exo‐S and Exo‐L preparations (Zhang et al., 2018). Given that the ratio of Bodipy‐labelled exosomes to total sEV in our samples prepared from 24 h chase conditioned media is 1:10, it is plausible that vesicles of different origins, such as exosomes and ectosomes, may coexist within the density gradient fractions corresponding to the fluorescent peak.
Tetraspanins proteins, particularly CD63, CD81, and CD9, play a critical role in vesicle biogenesis and cargo sorting and have been widely used as markers for exosomes although it is now evident that these tetraspanins are also expressed on ectosomes and other subtypes of sEV (Fordjour et al., 2022; Mathieu et al., 2021; Zhang et al., 2018). Recent advances in imaging flow cytometry that enable the multiparameter characterization of single EV combined with novel EV labelling strategies, now allow us to investigate the phenotypic heterogeneity within exosomes and other EV populations with high precision (K. Lee et al., 2018). Nevertheless, the exploration of heterogeneity within tetraspanin‐positive EV subpopulations is currently limited to a few studies that primarily rely on bulk vesicle preparations (Ricklefs et al., 2019; Woud et al., 2022). While bulk vesicle preparations are useful for studying total circulating EVs in an in vivo setting, their applicability in vitro is restricted, especially when more advanced techniques such as immunoisolation, asymmetric flow field‐flow fractionation or microfluidics methods are necessary for enriching specific EV subtypes (Jeong et al., 2016; Zhang et al., 2018). We exploited the unique features of our methodology to accurately determine the tetraspanins surface profiles on Bodipy‐labelled exosomes. Our data indicate that CD63+ Bodipy labelled exosomes are the most abundant followed by those positive for CD81. In contrast, CD9‐positive exosomes represent a minor fraction, despite observing a higher degree of intracellular colocalization between Bodipy lipids and CD9 compared to CD81. This suggests that although there is a significant intracellular association between CD9 and Bodipy lipids, CD9 may be selectively sorted or retained within the cell, resulting a lower abundance of CD9+ Bodipy labelled exosomes. Double positive CD63+/CD81+ are also present while CD63+/CD9+ exosomes were detected in smaller proportions. The differential sorting and release mechanisms of tetraspanins may contribute to the observed variations in their presence on exosomal membranes. It has been proposed that exosomes are primarily enriched in CD63, which serves as a late endosome and MVB marker, while EVs bearing only CD9 or CD81, without CD63, may not form within endosomes (Mathieu et al., 2021; Zhang et al., 2018). We cannot rule out the possibility that single‐positive CD81+ Bodipy‐labelled exosomes may include small vesicles directly budded from the plasma membrane, where CD81 is predominantly localized in Mel501 cells. However, in our study, we did not observe colocalization of CD81 with Bodipy lipids on the plasma membrane, despite observing it at steady state. A potential explanation for the observed discrepancy has already been raised in previous work in a human B‐cell line where it has been observed that CD81 is the most highly enriched protein in exosomes, even though it is primarily localized to the plasma membrane (Escola et al., 1998). This discrepancy was attributed to the inefficient detection of intracellular CD81 molecules by the relevant antibody, potentially due to epitope masking. Our data also show that despite the significant intracellular colocalization of CD9 with Bodipy lipids, CD9+ Bodipy‐labelled exosomes were present in lower percentage compared to other tetraspanin‐positive exosomes suggesting an active mechanism of intracellular Bodipy lipids sorting. However, the absence of a particular tetraspanin marker does not necessarily indicate the mode of sEV biogenesis, as some sEV may lack certain markers despite forming in endosomes.
By applying a nanoFACS sorting strategy we obtained a pure subpopulation of exosomes that could be further examined by electron microscopy (TEM). The sorted subpopulation of Bodipy exosomes had a mean size of 80 nm significantly smaller that the bulk of sEV present in the presorted samples analyzed by both SEM and NTA. The size of Bodipy labelled exosomes coincides with the Exo‐S subpopulation of exosomes reported recently as the most likely bona fide/canonical exosomes (Zhang et al., 2018). Furthermore, immunogold staining revealed the colocalization of the Bodipy moiety and CD63 within the same vesicle, providing conclusive evidence for the exosome nature of the Bodipy‐labelled vesicles.
By studying the release dynamics of Bodipy exosome release, we determined that they accounted for approximately 20 percent of the total sEV secretion. The kinetic profile of Bodipy exosomes overlapped with that of total sEV for the first 6 h of the chase period. After this time, it is presumed that the intracellular reservoir of Bodipy lipids was depleted, although residual fluorescence associated with the cells could still be detected. This observation suggests that Bodipy‐derived phospholipids might be incorporated into other intracellular membranes that do not contribute to the formation of ILVs.
In conclusion, our study introduces an innovative methodology for metabolically labelling fluorescent exosomes using a fluorescent palmitic acid that is readily internalized by cells and is transformed into phospholipids which will form part of the lipid bilayer of the secreted exosomes. The strong evidence presented in this study confirms that the Bodipy‐labelled sEV are indeed exosomes.
This novel strategy of metabolically labelling exosomes with fluorescence offers significant advantages in terms of experimental versatility and can be applied to studies focusing on exosome biogenesis. The uniqueness of this approach lies in the utilization of the metabolic fate of a fatty acid, rather than relying on protein markers, to track exosome formation. This innovative technique opens up new avenues for investigating the biogenesis and functional properties of exosomes in different physiological and pathological contexts.
AUTHOR CONTRIBUTIONS
Valeria Barreca, Zaira Boussadia, Deborah Polignano and Lorenzo Galli performed the experiments. Valeria Barreca designed the experiments and analyzed the data. Massimo Sanchez and Valentina Tirelli performed and analyzed the FC experiments. Mario Falchi performed the confocal microscopy imaging and analyzed the data. Lucia Bertuccini and Francesca Iosi performed the electron microscopy experiments and analyzed the data. Massimo Tatti performed the lipid analysis experiments. Maria Luisa Fiani and Massimo Sargiacomo conceived, designed the experiments, interpreted the data and wrote the article with the imput of all authors. Maria Luisa Fiani supervised all aspects of the project.
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
ACKNOWLEDGEMENTS
This work was supported by a grant from Italian Ministry of Health, Ricerca Finalizzata RF‐2019‐12369719 to M.Sargiacomo.
Open access funding provided by BIBLIOSAN.
Barreca, V. , Boussadia, Z. , Polignano, D. , Galli, L. , Tirelli, V. , Sanchez, M. , Falchi, M. , Bertuccini, L. , Iosi, F. , Tatti, M. , Sargiacomo, M. , & Fiani, M. L. (2023). Metabolic labelling of a subpopulation of small extracellular vesicles using a fluorescent palmitic acid analogue. Journal of Extracellular Vesicles, 12, e12392. 10.1002/jev2.12392
Contributor Information
Massimo Sargiacomo, Email: massimo.sargiacomo@iss.it.
Maria Luisa Fiani, Email: maria.fiani@iss.it.
REFERENCES
- Akoumi, A. , Haffar, T. , Mousterji, M. , Kiss, R. S. , & Bousette, N. (2017). Palmitate mediated diacylglycerol accumulation causes endoplasmic reticulum stress, Plin2 degradation, and cell death in H9C2 cardiomyoblasts. Experimental Cell Research, 354(2), 85–94. 10.1016/j.yexcr.2017.03.032 [DOI] [PubMed] [Google Scholar]
- Clancy, J. W. , Schmidtmann, M. , & D'Souza‐Schorey, C. (2021). The ins and outs of microvesicles. FASEB BioAdvances, 3(6), 399–406. 10.1096/fba.2020-00127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo, M. , Raposo, G. , & Thery, C. (2014). Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annual Review of Cell and Developmental Biology, 30, 255–289. 10.1146/annurev-cellbio-101512-122326 [DOI] [PubMed] [Google Scholar]
- Coscia, C. , Parolini, I. , Sanchez, M. , Biffoni, M. , Boussadia, Z. , Zanetti, C. , Fiani, M. L. , & Sargiacomo, M. (2016). Generation, quantification, and tracing of metabolically labeled fluorescent exosomes. In Federico M., (ed.) Lentiviral vectors and exosomes as gene and protein delivery tools. pp. 217–235. Springer New York. [DOI] [PubMed] [Google Scholar]
- Escola, J. M. , Kleijmeer, M. J. , Stoorvogel, W. , Griffith, J. M. , Yoshie, O. , & Geuze, H. J. (1998). Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B‐lymphocytes. Journal of Biological Chemistry, 273(32), 20121–20127. 10.1074/jbc.273.32.20121 [DOI] [PubMed] [Google Scholar]
- Fagone, P. , & Jackowski, S. (2009). Membrane phospholipid synthesis and endoplasmic reticulum function. Journal of Lipid Research, 50, (Suppl), S311–S316. 10.1194/jlr.R800049-JLR200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federici, C. , Shahaj, E. , Cecchetti, S. , Camerini, S. , Casella, M. , Iessi, E. , Camisaschi, C. , Paolino, G. , Calvieri, S. , Ferro, S. , Cova, A. , Squarcina, P. , Bertuccini, L. , Iosi, F. , Huber, V. , & Lugini, L. (2020). Natural‐killer‐derived extracellular vesicles: Immune sensors and interactors. Frontiers in immunology, 11, 262. 10.3389/fimmu.2020.00262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felicetti, F. , Errico, M. C. , Bottero, L. , Segnalini, P. , Stoppacciaro, A. , Biffoni, M. , Felli, N. , Mattia, G. , Petrini, M. , Colombo, M. P. , Peschle, C. , & Carè, A. (2008). The promyelocytic leukemia zinc finger–MicroRNA‐221/‐222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer Research, 68(8), 2745–2754. 10.1158/0008-5472.Can-07-2538 [DOI] [PubMed] [Google Scholar]
- Folch, J. , Lees, M. , & Sloane Stanley, G. H. (1957). A simple method for the isolation and purification of total lipides from animal tissues. Journal of Biological Chemistry, 226(1), 497–509. [PubMed] [Google Scholar]
- Fordjour, F. K. , Guo, C. , Ai, Y. , Daaboul, G. G. , & Gould, S. J. (2022). A shared, stochastic pathway mediates exosome protein budding along plasma and endosome membranes. Journal of Biological Chemistry, 298(10), 102394. 10.1016/j.jbc.2022.102394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Silva, S. , Benito‐Martin, A. , Nogues, L. , Hernandez‐Barranco, A. , Mazariegos, M. S. , Santos, V. , Hergueta‐Redondo, M. , Ximénez‐Embún, P. , Kataru, R. P. , Lopez, A. A. , Merino, C. , Sánchez‐Redondo, S. , Graña‐Castro, O. , Matei, I. , Nicolás‐Avila, J. Á. , Torres‐Ruiz, R. , Rodríguez‐Perales, S. , Martínez, L. , Pérez‐Martínez, M. , … Peinado, H. (2021). Melanoma‐derived small extracellular vesicles induce lymphangiogenesis and metastasis through an NGFR‐dependent mechanism. Nature Cancer, 2(12), 1387–1405. 10.1038/s43018-021-00272-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenberg, J. (2020). Life in the lumen: The multivesicular endosome. Traffic (Copenhagen, Denmark), 21(1), 76–93. 10.1111/tra.12715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginbotham, J. N. , Zhang, Q. , Jeppesen, D. K. , Scott, A. M. , Manning, H. C. , Ochieng, J. , Franklin, J. L. , & Coffey, R. J. (2016). Identification and characterization of EGF receptor in individual exosomes by fluorescence‐activated vesicle sorting. Journal of Extracellular Vesicles, 5, 29254. 10.3402/jev.v5.29254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz, S. N. , Conlon, M. M. , Rider, M. A. , Brownstein, N. C. , & Meckes, D. G., Jr. (2016). Nanoparticle analysis sheds budding insights into genetic drivers of extracellular vesicle biogenesis. Journal of Extracellular Vesicles, 5, 31295. 10.3402/jev.v5.31295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglis, H. C. , Danesh, A. , Shah, A. , Lacroix, J. , Spinella, P. C. , & Norris, P. J. (2015). Techniques to improve detection and analysis of extracellular vesicles using flow cytometry. Cytometry Part A: the journal of the International Society for Analytical Cytology, 87(11), 1052–1063. 10.1002/cyto.a.22649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong, S. , Park, J. , Pathania, D. , Castro, C. M. , Weissleder, R. , & Lee, H. (2016). Integrated magneto‐electrochemical sensor for exosome analysis. ACS Nano, 10(2), 1802–1809. 10.1021/acsnano.5b07584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen, D. K. , Fenix, A. M. , Franklin, J. L. , Higginbotham, J. N. , Zhang, Q. , Zimmerman, L. J. , Liebler, D. C. , Ping, J. , Liu, Q. , Evans, R. , Fissell, W. H. , Patton, J. G. , Rome, L. H. , Burnette, D. T. , & Coffey, R. J. (2019). Reassessment of exosome composition. Cell, 177(2), 428–445 e418. 10.1016/j.cell.2019.02.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri, R. , & LeBleu, V. S. (2020). The biology, function, and biomedical applications of exosomes. Science, 367(6478), eaau6977. 10.1126/science.aau6977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, N. , Dalirfardouei, R. , Dias, T. , Lotvall, J. , & Lasser, C. (2022). Tetraspanins distinguish separate extracellular vesicle subpopulations in human serum and plasma—Contributions of platelet extracellular vesicles in plasma samples. Journal of Extracellular Vesicles, 11(5), e12213. 10.1002/jev2.12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, T. , Stang, E. , Fang, K. S. , de Moerloose, P. , Parton, R. G. , & Gruenberg, J. (1998). A lipid associated with the antiphospholipid syndrome regulates endosome structure and function. Nature, 392(6672), 193–197. 10.1038/32440 [DOI] [PubMed] [Google Scholar]
- Kormelink, T. G. , Arkesteijn, G. J. , Nauwelaers, F. A. , van den Engh, G. , Nolte‐’t Hoen, E. N. , & Wauben, M. H. (2015). Prerequisites for the analysis and sorting of extracellular vesicle subpopulations by high‐resolution flow cytometry. Cytometry. Part A: the journal of the International Society for Analytical Cytology, 89(2), 135–147. 10.1002/cyto.a.22644 [DOI] [PubMed] [Google Scholar]
- Kowal, J. , Arras, G. , Colombo, M. , Jouve, M. , Morath, J. P. , Primdal‐Bengtson, B. , Dingli, F. , Loew, D. , Tkach, M. , & Théry, C. (2016). Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proceedings of the National Academy of Sciences, 113(8), E968–E977. 10.1073/pnas.1521230113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, C. P. , Kim, E. Y. , Badr, C. E. , Weissleder, R. , Mempel, T. R. , Tannous, B. A. , & Breakefield, X. O. (2015). Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nature Communications, 6, 7029. 10.1038/ncomms8029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanna, A. , Vaz, B. , D'Ambra, C. , Valvo, S. , Vuotto, C. , Chiurchiu, V. , Devine, O. , Sanchez, M. , Borsellino, G. , Akbar, A. N. , De Bardi, M. , Gilroy, D. W. , Dustin, M. L. , Blumer, B. , & Karin, M. (2022). An intercellular transfer of telomeres rescues T cells from senescence and promotes long‐term immunological memory. Nature Cell Biology, 24(10), 1461–1474. 10.1038/s41556-022-00991-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larios, J. , Mercier, V. , Roux, A. , & Gruenberg, J. (2020). ALIX‐ and ESCRT‐III‐dependent sorting of tetraspanins to exosomes. Journal of Cell Biology, 219(3), e201904113. 10.1083/jcb.201904113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K. , Fraser, K. , Ghaddar, B. , Yang, K. , Kim, E. , Balaj, L. , Chiocca, E. A. , Breakefield, X. O. , Lee, H. , & Weissleder, R. (2018). Multiplexed profiling of single extracellular vesicles. ACS Nano, 12(1), 494–503. 10.1021/acsnano.7b07060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Y. S. , Kim, S. Y. , Ko, E. , Lee, J. H. , Yi, H. S. , Yoo, Y. J. , Je, J. , Suh, S. J. , Jung, Y. K. , Kim, J. H. , Seo, Y. S. , Yim, H. J. , Jeong, W. I. , Yeon, J. E. , Um, S. H. , & Byun, K. S. (2017). Exosomes derived from palmitic acid‐treated hepatocytes induce fibrotic activation of hepatic stellate cells. Scientific Reports, 7(1), 3710. 10.1038/s41598-017-03389-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, Y. , Iqbal, Z. , Lu, J. , Wang, J. , Zhang, H. , Chen, X. , Duan, L. , & Xia, J. (2023). Cell‐derived nanovesicle‐mediated drug delivery to the brain: Principles and strategies for vesicle engineering. Molecular Therapy, 31(5), 1207–1224. 10.1016/j.ymthe.2022.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Q. , Huang, J. , Xia, J. , Liang, Y. , & Li, G. (2022). Tracking tools of extracellular vesicles for biomedical research. Frontiers in Bioengineering and Biotechnology, 10, 943712. 10.3389/fbioe.2022.943712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louie, S. M. , Roberts, L. S. , Mulvihill, M. M. , Luo, K. , & Nomura, D. K. (2013). Cancer cells incorporate and remodel exogenous palmitate into structural and oncogenic signaling lipids. Biochimica Et Biophysica Acta, 1831(10), 1566–1572. 10.1016/j.bbalip.2013.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maas, S. L. N. , Breakefield, X. O. , & Weaver, A. M. (2017). Extracellular vesicles: Unique intercellular delivery vehicles. Trends in Cell Biology, 27(3), 172–188. 10.1016/j.tcb.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhi, H. , Bronk, S. F. , Werneburg, N. W. , & Gores, G. J. (2006). Free fatty acids induce JNK‐dependent hepatocyte lipoapoptosis. Journal of Biological Chemistry, 281(17), 12093–12101. 10.1074/jbc.M510660200 [DOI] [PubMed] [Google Scholar]
- Mathieu, M. , Nevo, N. , Jouve, M. , Valenzuela, J. I. , Maurin, M. , Verweij, F. J. , Palmulli, R. , Lankar, D. , Dingli, F. , Loew, D. , Rubinstein, E. , Boncompain, G. , Perez, F. , & Théry, C. (2021). Specificities of exosome versus small ectosome secretion revealed by live intracellular tracking of CD63 and CD9. Nature Communications, 12(1), 4389. 10.1038/s41467-021-24384-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meldolesi, J. (2022). Unconventional protein secretion dependent on two extracellular vesicles: Exosomes and ectosomes. Frontiers in Cell and Developmental Biology, 10, 877344. 10.3389/fcell.2022.877344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales‐Kastresana, A. , Musich, T. A. , Welsh, J. A. , Telford, W. , Demberg, T. , Wood, J. C. S. , Bigos, M. , Ross, C. D. , Kachynski, A. , Dean, A. , Felton, E. J. , Van Dyke, J. , Tigges, J. , Toxavidis, V. , Parks, D. R. , Overton, W. R. , Kesarwala, A. H. , Freeman, G. J. , Rosner, A. , … Jones, J. C. (2019). High‐fidelity detection and sorting of nanoscale vesicles in viral disease and cancer. Journal of Extracellular Vesicles, 8(1), 1597603. 10.1080/20013078.2019.1597603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales‐Kastresana, A. , Telford, B. , Musich, T. A. , McKinnon, K. , Clayborne, C. , Braig, Z. , Rosner, A. , Demberg, T. , Watson, D. C. , Karpova, T. S. , Freeman, G. J. , DeKruyff, R. H. , Pavlakis, G. N. , Terabe, M. , Robert‐Guroff, M. , Berzofsky, J. A. , & Jones, J. C. (2017). Labeling extracellular vesicles for nanoscale flow cytometry. Scientific Reports, 7(1), 1878. 10.1038/s41598-017-01731-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwland, R. , Falcon‐Perez, J. M. , Thery, C. , & Witwer, K. W. (2020). Rigor and standardization of extracellular vesicle research: Paving the road towards robustness. Journal of Extracellular Vesicles, 10(2), e12037. 10.1002/jev2.12037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann, J. A. , & Carvalho, P. (2019). Dynamics and functions of lipid droplets. Nature Reviews Molecular Cell Biology, 20(3), 137–155. 10.1038/s41580-018-0085-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penke, M. , Schuster, S. , Gorski, T. , Gebhardt, R. , Kiess, W. , & Garten, A. (2017). Oleate ameliorates palmitate‐induced reduction of NAMPT activity and NAD levels in primary human hepatocytes and hepatocarcinoma cells. Lipids in Health and Disease, 16(1), 191. 10.1186/s12944-017-0583-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pol, A. , Gross, S. P. , & Parton, R. G. (2014). Review: Biogenesis of the multifunctional lipid droplet: Lipids, proteins, and sites. Journal of Cell Biology, 204(5), 635–646. 10.1083/jcb.201311051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pospichalova, V. , Svoboda, J. , Dave, Z. , Kotrbova, A. , Kaiser, K. , Klemova, D. , Ilkovics, L. , Hampl, A. , Crha, I. , Jandakova, E. , Minar, L. , Weinberger, V. , & Bryja, V. (2015). Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer. Journal of Extracellular Vesicles, 4, 25530. 10.3402/jev.v4.25530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklefs, F. L. , Maire, C. L. , Reimer, R. , Duhrsen, L. , Kolbe, K. , Holz, M. , Schneider, E. , Rissiek, A. , Babayan, A. , Hille, C. , Pantel, K. , Krasemann, S. , Glatzel, M. , Heiland, D. H. , Flitsch, J. , Martens, T. , Schmidt, N. O. , Peine, S. , Breakefield, X. O. , … Lamszus, K. (2019). Imaging flow cytometry facilitates multiparametric characterization of extracellular vesicles in malignant brain tumours. Journal of Extracellular Vesicles, 8(1), 1588555. 10.1080/20013078.2019.1588555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin, J. , Arganda‐Carreras, I. , Frise, E. , Kaynig, V. , Longair, M. , Pietzsch, T. , Preibisch, S. , Rueden, C. , Saalfeld, S. , Schmid, B. , Tinevez, J. Y. , White, D. J. , Hartenstein, V. , Eliceiri, K. , Tomancak, P. , & Cardona, A. (2012). Fiji: An open‐source platform for biological‐image analysis. Nature Methods, 9(7), 676–682. 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng, F. , & Fussenegger, M. (2020). Shedding light on extracellular vesicle biogenesis and bioengineering. Advanced Science (Weinh), 8(1), 2003505. 10.1002/advs.202003505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, P. , Arden, C. , Corcoran, J. , Hacker, C. , Welters, H. J. , & Morgan, N. G. (2022). Differential routing and disposition of the long‐chain saturated fatty acid palmitate in rodent vs human beta‐cells. Nutrition & Diabetes, 12(1), 22. 10.1038/s41387-022-00199-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Niel, G. , Carter, D. R. F. , Clayton, A. , Lambert, D. W. , Raposo, G. , & Vader, P. (2022). Challenges and directions in studying cell‐cell communication by extracellular vesicles. Nature Reviews Molecular Cell Biology, 23(5), 369–382. 10.1038/s41580-022-00460-3 [DOI] [PubMed] [Google Scholar]
- van Niel, G. , D'Angelo, G. , & Raposo, G. (2018). Shedding light on the cell biology of extracellular vesicles. Nature Reviews Molecular Cell Biology, 19(4), 213–228. 10.1038/nrm.2017.125 [DOI] [PubMed] [Google Scholar]
- Verweij, F. J. , Balaj, L. , Boulanger, C. M. , Carter, D. R. F. , Compeer, E. B. , D'Angelo, G. , El Andaloussi, S. , Goetz, J. G. , Gross, J. C. , Hyenne, V. , Krämer‐Albers, E. M. , Lai, C. P. , Loyer, X. , Marki, A. , Momma, S. , Nolte‐'t Hoen, E. N. M. , Pegtel, D. M. , Peinado, H. , Raposo, G. , …, van Niel, G. (2021). The power of imaging to understand extracellular vesicle biology in vivo. Nature Methods, 18(9), 1013–1026. 10.1038/s41592-021-01206-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willms, E. , Cabanas, C. , Mager, I. , Wood, M. J. A. , & Vader, P. (2018). Extracellular vesicle heterogeneity: Subpopulations, isolation techniques, and diverse functions in cancer progression. Frontiers in immunology, 9, 738. 10.3389/fimmu.2018.00738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willms, E. , Johansson, H. J. , Mager, I. , Lee, Y. , Blomberg, K. E. , Sadik, M. , Alaarg, A. , Smith, C. I. , Lehtiö, J. , El Andaloussi, S. , Wood, M. J. , & Vader, P. (2016). Cells release subpopulations of exosomes with distinct molecular and biological properties. Scientific Reports, 6, 22519. 10.1038/srep22519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woud, W. W. , van der Pol, E. , Mul, E. , Hoogduijn, M. J. , Baan, C. C. , Boer, K. , & Merino, A. (2022). An imaging flow cytometry‐based methodology for the analysis of single extracellular vesicles in unprocessed human plasma. Communications Biology, 5(1), 633. 10.1038/s42003-022-03569-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Freitas, D. , Kim, H. S. , Fabijanic, K. , Li, Z. , Chen, H. , Mark, M. T. , Molina, H. , Martin, A. B. , Bojmar, L. , Fang, J. , Rampersaud, S. , Hoshino, A. , Matei, I. , Kenific, C. M. , Nakajima, M. , Mutvei, A. P. , Sansone, P. , Buehring, W. , … Lyden, D. (2018). Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field‐flow fractionation. Nature Cell Biology, 20(3), 332–343. 10.1038/s41556-018-0040-4 [DOI] [PMC free article] [PubMed] [Google Scholar]


