Abstract
For medicine to fulfill its promise of personalized treatments based on a better understanding of disease biology, computational and statistical tools must exist to analyze the increasing amount of patient data that becomes available. A particular challenge is that several types of data are being measured to cope with the complexity of the underlying systems, enhance predictive modeling and enrich molecular understanding.
Here we review a number of recent approaches that specialize in the analysis of multimodal data in the context of predictive biomedicine. We focus on methods that combine different OMIC measurements with image or genome variation data. Our overview shows the diversity of methods that address analysis challenges and reveals new avenues for novel developments.
Keywords: Multimodal modeling, Predictive modeling, Multi-omics, Machine learning, Personalized medicine
Graphical abstract
1. Introduction
The development of personalized treatment for patients with any disease and condition is a current ambition in the medical research field. With our access to diverse molecular and phenotypic readouts of human bodies, their tissues, and cells, we hope to eventually understand their relationships and decipher all possible causal mechanisms behind diseases to act upon them. However, at the moment, this still seems like a distant goal. There is still much to be understood about the mechanisms of diseases and why certain drugs work the way they do.
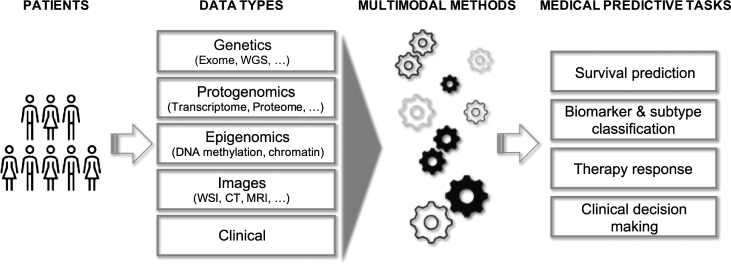
A big challenge along this path of discovery is the integration of patient data measured from multiple modalities (multimodal). In this review we summarize recent computational advances for multimodal analysis of data for the following tasks: (i) patient survival prediction, (ii) disease biomarker and subtype classification, (iii) therapy response prediction, and (iv) clinical decision making (Fig. 1).
Fig. 1.
Overview of multimodal data types and prediction tasks that are discussed in this review.
1.1. Multimodal data types
For these predictive tasks, we consider combinations of five high-level classes of data types. First, are genetic data that measure parts of the DNA sequence of patients, such as single nucleotide polymorphisms (SNPs) or copy-number variations (CNVs) using SNP arrays, enriching for regions of interest using panel-based, whole-exome, or whole-genome-sequencing (WGBS) approaches. These data allow us to assess DNA sequence variation and link them to disease [1].
Second, proteogenomic measurements of gene products from the human genome are a common source of measurement, as the activity of a subset of genes is often changed in diseased cells due to misregulation. Measuring the transcriptome of human cells is most commonly done using RNA-seq these days [2], [3]. It allows quantifying the expression activity of tens of thousands of transcripts made from cells. Many different types of RNAs can be quantified in this way for example messenger RNAs (mRNAs) or micro RNAs (miRNAs). After DNA transcription, mRNA transcripts are converted into proteins, which can be measured with Mass spectrometry (MassSpec) technologies some of which are directly applicable in a clinical setting [4]. MassSpec can be quantitative and can measure up to thousands of proteins in cells studying their protein synthesis dynamics [5].
Third, epigenomic data provides a useful source of information to investigate the function of genomic regions. Epigenome activity differs between human cell types albeit the DNA nucleotide sequence of these cells is the same. Epigenome activity can be measured by detecting histone modifications or accessible chromatin using different approaches [6], [7]. These measurements provide a genome-wide readout on accessible regions, where regulatory proteins, such as transcription factors, bind which play an important role in diseases. Another important epigenomic variation is DNA methylation (DNAm), a stable modification of the DNA, which can be measured from cells or liquid biopsies and with changes related to the occurrence of many diseases [8].
Fourth, imaging of human body parts and cells is routine in many clinical applications and different technologies exist. For example, thin tissue sections, stained with hematoxylin and eosin (H&E), are frequently used as a gold standard in pathology to confirm the presence of certain diseases and are thus available for most patients. To further solidify a diagnosis for some of the cases an IHC (immune histo chemistry) staining can be prepared in addition, where a certain protein is labeled by an antibody. These stained tissue sections can nowadays be digitalized with slide scanners and are called whole slide images (WSIs). Other technologies, such as Magnetic resonance Imaging (MRI), can be used to record living parts of patient bodies such as organs or individual blood vessels. Each method has different advantages concerning resolution, cost, and time involved and some clinical applications may involve the generation of multimodal images using different technologies [9].
Finally, clinical data that is compiled as part of medical examinations can contain a diverse set of measurements (e.g. blood pressure, blood glucose levels, or inflammatory markers) or patient characteristics (e.g. sex, age). It also may contain a record of a patient's drug prescription schedule or the history of previous therapies that may be utilized by models. Clinical data is used in daily routines to aid decisions and thus is vital to be considered in multimodal methods.
1.2. Resources for multimodal data
To be able to integrate data from different modalities one often needs to obtain large datasets from existing resources [10]. Many such resources are created from systematic datasets that are produced by large consortia. In the course of the papers discussed the following consortia are important. For example, The Functional ANnoTation Of the Mammalian genome (FANTOM) Consortium has generated many proteogenomic datasets for analysis [11]. The International Cancer Genome Consortium (ICGC) and The Cancer Genomics Atlas (TCGA) program [12] have gathered measurements of all five data types discussed here (Fig. 1). The international human epigenomics consortium (IHEC) [6] has gathered diverse epigenome and proteogenomic datasets. The Genotype-Tissue Expression (GTEx) [13] consortium has measured genetic and RNA expression data from diverse tissues. Consortia for pathological analyses that will collect and provide data are BigPicture (https://bigpicture.eu) and PathLAKE (https://www.pathlake.org/our-partners/). Finally, the UK Biobank [14] provides one of the largest datasets of genetic data with additional image and clinical data currently available.
There are resources specialized for the data types. For example, the NHGRI-EBI GWAS catalog harbors results from genome-wide association studies (GWAS) from human genetic studies [15]. Many processed epigenome datasets can be downloaded from the IHEC portal [16]. RadImageNet is a new resource specialized to enable transfer learning efforts using deep learning derived models for the analysis of radiology images [17]. The cancer imaging archive [18] holds mainly radiology images, but also whole WSIs and metadata and the NCI Imaging Data Commons [19] holds many different types of images from cancerous tissue.
Data from other resources that catalog interactions between gene products, i.e. STRING [20], or between regulatory regions and genes, i.e. EpiRegio [21], are sometimes used as prior information for integration.
1.3. Computational challenges in integrating multimodal data
A holistic characterization of patients and model organisms, which is key for progressing personalized medicine, requires the comprehensive integration of data from all available sources. However, the complex nature of these heterogeneous multi-modal data poses distinct challenges for successful integration in a predictive setting.
First, missing values are very common in omics data due to dropouts, the limited sensitivity of measuring instruments, and patients missing appointments.
Second, in predictive medicine clinical data is often available in addition to molecular data - the efficient integration of this typically low-dimensional data with high-dimensional omics data poses a challenge to many standard algorithms.
Third, often models need not only to have a high predictive power, but also be interpretable. In particular for deep-learning based methods there is a trade-off between discriminative power and interpretability, with those models with the highest predictive power often being black-box in nature. Related to this challenge is the existence of a wealth of prior knowledge in the form of gene set annotations or protein-protein-interaction networks. Integration of this knowledge into multi-omics models is challenging but can not only aid interpretability but also boost model performance.
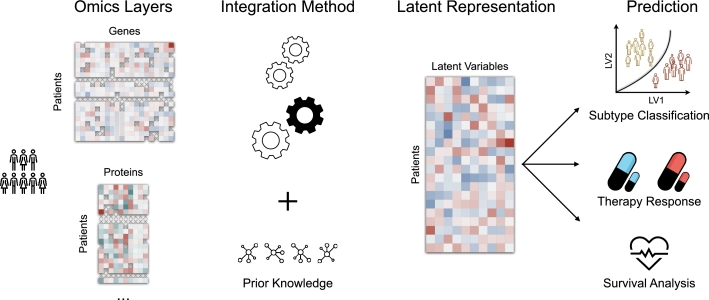
A common strategy to address these challenges is representation learning (Fig. 2). In this modeling paradigm, un-observable latent variables are inferred from observed high-dimensional data. For predictive tasks, these latent variables are learned such that they are associated with an outcome of interest (e.g. survival or therapy response). In multimodal representation learning, a joint representation across all modalities is inferred that paints a comprehensive picture of the underlying biological processes driving the outcome of interest. If available, prior knowledge e.g. in the form of protein-protein interaction networks can further guide the inference of interpretable latent representations. An explanation of all abbreviations used in the manuscript can be found in Table 3.
Fig. 2.
Representation learning for multi-omics data integration. Low-dimensional latent variables (LVs, middle) are derived from multimodal, high-dimensional molecular data (omics layers, left). Based on different techniques including deep neural networks, autoencoders, or graph-based methods (integration method) and optionally leveraging existing prior knowledge, LVs are inferred such that they are associated with a clinical outcome of interest (right).
Table 3.
Abbreviations and their corresponding descriptions.
| Abbreviation | Description | |
|---|---|---|
| Data Types | SNP | Single Nucleotide Polymorphism |
| CNV | Copy Number Variation | |
| mRNA | messenger RNA | |
| miRNA | micro RNA | |
| DNAm | DNA methylation | |
| WSI | Whole Slide Image | |
| Resources | FANTOM | Functional ANnotation Of the Mammalian genome |
| ICGC | The International Cancer Genome Consortium | |
| TCGA | The Cancer Genomics Atlas | |
| CCLE | Cancer Cell Line Encyclopedia | |
| GDC | Genomic Data Commons | |
| IHEC | The International Human Epigenomics Consortium | |
| GTEx | Genotype-Tissue Expression | |
| GWAS | Genome-Wide Association Studies | |
| STRING | Search Tool for the Retrieval of Interacting Genes/Proteins | |
| ROSMAP | Religious Orders Study/Memory and Aging Project | |
| PORPOISE | Pathology-Omics Research Platform for Integrative Survival Estimation | |
| Modeling Approaches | LVM | Latent Variable Model |
| DL | Deep Learning | |
| VAE | Variational AutoEncoder | |
| DNN | Deep Neural Network | |
| CNN | Convolutional Neural Network | |
| GCN | Graph Convolutional Network | |
| Models | MOFA | Multi-Omics Factor Analysis |
| MOLI | Multi-Omics Late Integration | |
| MOSAE | Multi-omics Supervised Autoencoder | |
| MAE | Multi-view Factorization AutoEncoder | |
| MDNNMD | Multimodal Deep Neural Network by integrating Multi-dimensional Data | |
| MOGONET | Multi-Omics Graph cOnvolutional NETworks | |
| SALMON | Survival Analysis Learning with Multi-Omics Neural Networks | |
| HFBSurv | Hierarchical Factorized Bilinear fusion for cancer survival prediction | |
| DeepMOCCA | Deep Multi Omics CanCer Analysis | |
| MCAT | Multimodal Co-Attention Transformer |
2. Overview of multimodal analysis methods
2.1. Methods for multi-omics
With advances in high-throughput techniques for molecular profiling, omics data - molecular data that comprehensively assess a set of molecules - has been becoming more and more prevalent in the context of predictive biomedicine [22]. These data quantify different aspects of the proteogenomic and epigenomic makeup of patients and a plethora of methods has been developed to integrate these diverse data types. In the following, we will focus on algorithms that were specifically developed to solve one of the four predictive tasks outlined above.
2.1.1. Predictive multimodal data integration methods
Generally, we may group predictive multimodal data integration methods into two main categories – two-step or end-to-end approaches – depending on whether the discriminative task is optimized directly or in a post hoc manner. A two-step approach typically involves a preliminary decomposition of the observed data modalities into an integrated and lower dimensional latent space. Subsequently, this latent representation can be utilized to perform a supervised task in clinical settings such as subtype prediction or survival analysis. Multi-omics factor analysis (MOFA) [23] is a well-established statistical method for integrating single-cell multi-omics data. Inspired by group factor analysis [24], MOFA infers latent factors that capture sources of variability within and across different data modalities. MOFA was initially applied to patients of chronic lymphocytic leukemia profiled across multiple modalities such as RNA expression, DNA methylation, and drug response. A recent study on the proteogenomic characterization of acute myeloid leukemia (AML) applies a two-step approach based on MOFA to identify a subpopulation of patients exhibiting poor survival outcomes, that is characterized by a high expression of mitochondrial proteins [25]. More elaborate frameworks combine multiple unsupervised modules and statistical tests before generating the relevant features for the discriminative task such as survival analysis [26], [27], [28], [29]. Poirion et al. [30] propose an ensemble framework of deep learning and machine learning approaches for analyzing patient survival times. Normalized features of each modality are passed to the corresponding autoencoders, which transform the high-dimensional input into compact latent codes. A second set of modules applies a univariate Cox proportional hazards model [31] for each feature inferred from the bottleneck part of the autoencoders and selects only significant features based on a log-rank test. Next, a Gaussian mixture model detects patient subpopulations with clinical relevance regarding survival. Finally, a set of supervised classifiers predicts the disease subtype in new patients. We refer to [32], [33], [34] for a more comprehensive overview of unsupervised methods for multimodal integration.
Due to being very general in their approach, these methods perform relatively poorly in specific prediction tasks when compared to related end-to-end methods, which attempt to learn tailored representations for the task at hand. The main difference between end-to-end approaches when compared to two-step approaches is the association of the observed multimodal data with the ground truth targets during the optimization procedure.
In the next section, we focus exclusively on end-to-end methods that attempt to perform classification, e.g. mortality, short- and long-term survival, and therapy response, or perform time-to-event analysis such as survival prediction. There are, however, approaches that accommodate both classification and survival analysis tasks simultaneously. Zhang et al. [35], for instance, propose OmiEmbed, an end-to-end multi-task deep learning framework for performing supervised tasks in multimodal data. OmiEmbed is based on a variational autoencoder (VAE), which serves as an embedding module for mapping the observed modalities onto a lower dimensional and non-linear manifold. The inferred latent code from each encoder is then concatenated into a single latent code, which serves as the input for further downstream analysis tasks. The authors demonstrate the feasibility of their approach to disease subtype classification and prognosis prediction on a brain tumor multi-omics dataset and the Genomic Data Commons (GDC) pan-cancer dataset.
2.1.2. Classification
DNN-based approaches
Sun et al. [36] propose a deep learning framework for integrating multimodal data (MDNNMD) for the prognostic prediction of breast cancer. Their approach incorporates individual deep neural networks (DNNs) for extracting non-linear representations from each observed modality, which are then aggregated by a score-fusing module. The aggregation function implements a weighted linear combination of the output from each DNN to balance the contribution of each modality for the discriminative task. The authors validate their approach to classifying short- and long-term survivors on the METABRIC and TCGA dataset of breast cancer patients, which includes gene expression profiling, copy number variation (CNV), and clinical information. Similarly, Lin et al. [37] suggest concatenating the latent features inferred from each subnetwork before passing the resulting vector to a final classification network. A related model for drug response classification, MOLI [38], introduces an additional constraint in the objective function that encourages responders to the drug to be more similar to each other than to non-responders. Alternative approaches impose other statistical constraints on the latent space.
AE-based approaches
Lee and van der Schaar [39] introduce DeepIMV, a deep learning framework based on the principle of information bottleneck to learn a joint latent space that maximizes the mutual information between the latent code and the prediction target, while at the same time minimizing the mutual information between the observed modalities and the latent code. The reasoning behind this dual optimization objective is to ensure that the latent code depict a minimal sufficient statistic of the observed modalities for the observed label, while pruning all additional task-irrelevant information. The authors validate their approach on two real-world multi-omics datasets from TCGA and the Cancer Cell Line Encyclopedia, comprising multiple modalities such as mRNA expression, DNA methylation, DNA copy number, microRNA expression, and reverse phase protein array, where they attempt to predict the 1-year mortality and drug sensitivity of patients, respectively. Alternatively, MOSAE [40] apply a similar approach to DeepIMV but compute an average of the latent code inferred from each modality instead of employing a product of experts module. Alternative approaches rely on graph-based data structures to better capture feature- and sample-wise similarities. Moreover, sources of domain knowledge in computational biology typically support graph structures, e.g. protein-protein interaction networks [20], and can be effectively incorporated to further facilitate interpretability.
Graph-based approaches
Ma and Zhang [41] propose a multi-view factorization autoencoder (MAE) architecture that accommodates prior information in terms of molecular interaction networks across the observed features. This poses an additional constraint on the inferred feature representation by the decoder, by encouraging connected features to have similar numerical embeddings. The authors predict the progression-free interval (PFI) and the overall survival (OS) events on two patient cohorts from the TCGA database, namely the bladder urothelial carcinoma (BLCA) and brain lower-grade glioma (LGG), each comprising gene expression, miRNA expression, protein expression, and DNA methylation as well as clinical data. Wang et al. [42] introduce a multimodal graph convolutional network (GCN) framework for biomedical classification. After applying a preprocessing step for removing noisy and technical artifacts in each data modality, MOGONET utilizes GCNs for learning a sample-wise similarity graph, which serves as a basis for selecting discriminative features and better learning of relationships between nodes, i.e. samples. Finally, MOGONET learns the correlation structure across modalities by employing a view correlation discovery network (VCDN) to integrate the relevant information originating from each modality and provide the final features for the prediction task. The authors apply MOGONET on several classification tasks such as predicting patients with Alzheimer's Disease in the ROSMAP dataset, predicting the grade in low-grade glioma (LGG) patients, and classifying the cancer type in the kidney (KIPAN) and breast (BRCA) cancer patients. Each sample spans across three modalities: mRNA expression, DNA methylation, and miRNA expression.
Trustworthy approaches
Addressing crucial requirements on the trustworthiness of predictive models in safety-critical tasks, Han et al. [43] propose a deep learning framework for trustworthy multimodal classification in safety-critical applications. Their approach, termed Multimodal Dynamics, quantifies for each sample its corresponding feature-level and modality-level informativeness for the predictive task. They introduce a regularized gating module to achieve sparse feature representations via the -norm, and implement True Class Probability (TCP) as a criterion to assess the classification confidence of each modality. A low prediction confidence translates to higher uncertainty, meaning the corresponding modality provides little information and vice versa. The authors benchmark their approach against a comprehensive benchmark of competitive models including MOGONET [42], and demonstrate the utility of their method on several datasets from TCGA, and ROSMAP.
2.1.3. Survival
DNN-based approaches
Huang et al. [44] propose a deep learning framework for performing survival analysis with multi-omics neural networks (SALMON). In order to significantly reduce the number of features during the analysis, while preserving relevant information, the authors compute eigengene matrices of gene co-expression modules in an intermediate step, before passing the result to the neural network. Each neural network module learns a latent representation by performing consecutive non-linear transformations to the input features of its corresponding modality. The set of latent representations is then concatenated with additional clinical information into a feature vector, which serves as the input of a Cox regression module for predicting overall survival of the patients. The authors validate their method on breast cancer (BRCA) patients comprising several modalities like gene expression, miRNA, as well as demographic and clinical information such as estrogen or progesterone receptor status. A hierarchical factorized bilinear fusion strategy is proposed by Li et al. [45] with the HFBSurv for the integration of images, gene expression, CNV and clinical information in the context of breast invasive carcinoma survival prediction. The decomposition of the embedding problem into multiple levels reduces the trainable parameters and consequently the model complexity. A modality-specific attentional factorized bilinear module (MAFB) captures the modality-specific relations and a cross-modality attentional factorized bilinear module (CAFB) is used for describing the relations between modalities.
AE-based approaches
Tong et al. [46] extend the architecture in [44] by introducing two multimodal integration networks. Their first model, ConcatAE, is similar to SALMON in that it performs a concatenation of the inferred latent features during optimization. However, the authors employ an autoencoder pipeline which introduces a trade-off in the loss objective, by balancing the reconstruction error of the input features with the discriminative error generated by the survival prediction task. In addition, the authors omit the gene co-expression analysis, and instead perform a PCA or select highly variable features to reduce the input dimensionality. Their second proposed model is a cross-modality autoencoder, CrossAE, which encourages each data modality to reconstruct the input features of complementary modalities. The authors validate their approaches on synthetic and real data of breast cancer patients from the TCGA database, incorporating gene expression, DNA methylation, miRNA expression, and copy number variation.
Several other methods experiment with different approaches for integrating the intermediary features of each modality network into a single latent code representing each patient numerically. Cheerla and Gevaert [47] introduce a similarity loss that maximizes the cosine similarity between the latent feature vectors of the same sample, i.e. patient, while at the same time minimizing the cosine similarity of the latent feature vectors of different patients. Vale-Silva and Rohr [48] follow a simpler approach for encoding each patient by computing the maximum along each dimension in the latent space, effectively allowing only one modality to contribute in each latent dimension. While both models perform an end-to-end multimodal survival analysis, [47] employ a Cox-PH approach, whereas [48] rely on a discrete-time survival prediction method for cancer patient prognosis estimation. In recent work, Wissel et al. [49] attempt to compare different integration techniques in a unified benchmark, and suggest a hierarchical autoencoder architecture that outperforms the current state-of-the-art in survival prediction from multimodal data. Specifically, the authors compare several approaches in combining latent features such as simple concatenation, mean-pooling, and max-pooling. The authors find that incorporating more modalities during the analysis does not necessarily translate to better results. On the contrary, this may even diminish the overall performance. The experiments suggest the choice of the integration method is significantly less important than the choice of the right modalities to include in the analysis. However, when including all the available modalities, the hierarchical autoencoder architecture outperforms all other baselines. The authors claim that this approach casts the problem as a group-wise feature selection problem by introducing a soft modality selection mechanism, thereby focusing the optimization on the most informative modalities.
Graph-based approaches
In contrast to most of the aforementioned methods, which tackle the challenge of integrating multiple views by introducing individual feature-extracting modules followed by an aggregation step, Althubaiti et al. [50] rely on graph convolutional networks and graph-based domain knowledge to achieve data integration. The authors introduce DeepMOCCA, an end-to-end deep learning model that integrates multi-omics data by incorporating domain knowledge of cross-omic feature networks such as protein-protein interaction networks [20], followed by a Cox-PH module. DeepMOCCA relies on an attention mechanism that propagates predictions back to individual features, thereby identifying cancer drivers and prognostic markers of clinical relevance.
2.2. Methods for combining genetic and other data
Previously we introduced universal methods that combine multimodal data. Now we want to focus on an important type of data when it comes to medical analysis: the measurement of genetic mutations, such as those derived from SNP arrays, exome, or whole-genome sequencing. While the occurrence of SNPs can be modeled as a feature matrix and included in other methods mentioned thus far, there are other types of multimodal methods, that are specialized to handle properties of genetic data. For example, SNPs that are in the genomic vicinity often show genetic correlation due to linkage disequilibrium. While informative, the genome is, mostly, static in cells, and understanding the cellular context in which genomic mutations are relevant and which genes are important for a disease are challenges. Thus, other types of data need to be leveraged to address those questions. We will illustrate different methods that combine the interpretation of genome data in clever ways with image or epigenome data.
2.2.1. Genetic data and quantitative traits from images
Images of human body parts constitute a rich source of information to reveal parameters of disease, and despite many advances in automated analysis of images, there are novel ways in which images can be combined with genome data.
Commonly, two-step approaches are used to combine imaging-derived quantitative patient phenotypes with genetic mutation data if they are available in large quantities. Pirruccello et al. [51] use UK Biobank cardiac MRI images to train a deep learning classifier that can predict the diameter of human aortas. They predict the thickness of the aortas for 38K participants from the UK Biobank in order to conduct a genome-wide association analysis (GWAS) and identify correlated genetic markers. They are able to discover over 100 loci related to aorta size and use the combination of genetic markers and their predicted scores to derive new polygenic risk scores for aortic disease risk. A similar approach was done using brain imaging by deriving brain phenotypes, such as grey matter cortical thickness or structural connectivity from images. For each of these phenotypes associated SNPs are found using a GWAS, revealing novel genomic loci related to brain disorders [52].
Building on this general idea, Kirchler et al. introduce the concept of transferGWAS [53]. Instead of quantifying human-visible phenotypes from images, they argue that CNNs can learn features that may go beyond previously known patterns. They first learn a CNN feature extractor on an image training dataset. Then the image representation in the latent states of the CNN are interpreted as phenotypes and are used for a classical GWAS analysis using statistical methods, essentially transferring an image-derived quantity to reveal genetic associations. They show that this kind of approach reveals novel loci related to eye-related diseases and traits.
A different type of method that uses image data is the DeepGestalt and GestaltMatcher software [54], [55]. These are deep-learning based methods that predict the occurrence of rare diseases in patients from their facial images. Training machine learning models for diagnosing rare diseases is particularly hard, due to the lack of a large dataset. Computational prediction of a patient's disease from facial images can be combined with exome-seq data and assessment of the clinical phenotypes (from a physician's inspection) to improve the prediction of rare diseases by supervised classification [56] in a method called PEDIA.
As the above-mentioned approaches often need a large cohort of image and genetic data, which is not standard, the number of published studies is limited. However, with the availability of large paired datasets, e.g. the UK Biobank [14], more detailed methods for their analysis can be developed. Notably, the proposed methods for these problems are two-step approaches and do not employ tailored end-to-end approaches, thus providing ample opportunity for improvement.
2.2.2. Genetic and epigenome data
The cell-type specificity of epigenome data can be exploited to reveal the contribution of different cell types and mutations to diseases. For example, partitioned LD-score regression allows estimating the enrichment of genetic heritability of a phenotype using different genomic regions using statistical approaches [57]. These regions can be defined by epigenomic data or transcriptome data. Alternative formulations work particularly well for single-cell expression data in addition to the GWAS data [58].
Another challenge is to predict disease genes for complex diseases from the genetic mutation data, meaning genes that are causally connected to the disease, and warrant further experimental investigation in relevant cell types and disease models to possibly aid clinical decision-making. A variety of approaches exists, such as methods that utilize expression-QTLs to reveal disease genes using different approaches [59] or exploit co-localization of eQTLS and SNPs [60]. Another example is the EPISPOT algorithm, which can be used to combine mutation data with epigenome data to predict molecular traits using a probabilistic graphical model [61] to investigate individual loci in more detail. This can be used to discern the complexity of hotspots of associated genetic mutations. Other methods specialize in using a large set of independent evidence in the form of multimodal information to prioritize genes most likely connected to the disease of interest. For example, the iRIGS risk gene prioritization method uses a Bayesian framework to combine several types of information. Their problem formulation returns the gene in a GWAS locus, a genomic window around a significant GWAS SNP, which has the strongest association among the multimodal data and is close within a gene-gene network [62].
2.3. Methods for combining histopathological images and other data
Histopathological whole slide images (WSIs) contain information on the tissue and cell morphology while they also provide information on the microenvironment together with information on cell neighborhoods. This information is completely distinct from genomic, radiology, and clinical data. So, intermediate and late fusion techniques are more suited for images in combination with other data types compared to early fusion techniques (no intermediate representation is learned, and pure features are used as an input for a single learner), as they have the advantage to handle feature imbalances, missing modalities and especially the huge heterogeneity between the different extracted features. Late fusion approaches combine decisions made by models trained by different data modalities and intermediate fusion techniques learn a representation, which is fused in a later branch of the network [63].
WSIs are very large in size (~1GB per image, despite jpeg compression), and contain tens of thousands of pixels, which makes it hard to process the whole image at once. In the case of deep learning the default strategy is to split the WSI into several tiles that are processed individually [64]. A patient-level diagnosis could e.g. be achieved by a simple consensus [65] or multiple instance learning(MIL) [66]. However, recent approaches process directly the complete WSI, but only with a minor performance benefit [67].
In general computational methods could help to overcome intra- and inter-observer variabilities and the integration of several datatypes has the potential to increase the performance of algorithms, which later can be used as decision support tools. In contrast to the well-established predictions, which are based solely on histopathological WSI data [65], [66], models based on multi-modal data are not yet well explored in the field of computational pathology at the moment [68]. But next to the fusion of different data modalities of course the fusion of different stainings is an upcoming question. For instance, Dwivedi et al. [69] proposed a method where a graph NN extracts embeddings from each stain, which are subsequently concatenated and used as an input for fully connected layers in order to predict the final score.
2.3.1. WSIs and molecular data
One of the big drawbacks of integrating WSIs with molecular data is the fact that molecular data is in many cases not available during routine diagnostics and is only generated for research or clinical trials. Thus multi-modal data integration of molecular data together with WSI data from pathology is at an early stage: For example, a recent review from 2022 of Schneider et al. [70] have only identified 11 relevant articles in the timespan of 2015 to 2020 in the field of combining CNN-processed WSI images, which is the state-of-the-art methodology in computational pathology, and molecular data (genomic and epigenomic DNA or transcriptome data). On the one hand, the combination of different data modalities improved the performance of all described algorithms in comparison to the individual data types. For data fusion, methods such as direct incorporation of features into fully connected layers or more advanced methods like tensor fusion or LSTM (long short term memory) are used. On the other hand, most of the 11 studies lack an external test dataset, indicating that little is known yet on the transferability and robustness of these algorithms.
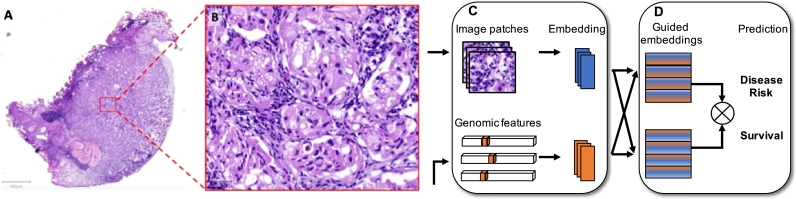
Chen et al. [71] propose an interpretable, multimodal learning framework called MCAT (Multimodal Co-Attention Transformer) that can learn a dense co-attention mapping between WSIs and genomics to predict survival outcomes and enable interpretability (Fig. 3). The proposed model embeds the different modalities via a genomic-guided co-attention (GCA) mechanism. Genomic features and image patches encoded as bags are forwarded to the attention layer and the set-based MIL Transformers. Tests on 7 different datasets reveal promising outcomes [71]. The same authors have also extended their method and now use a self-normalizing network (SNN) for molecular feature extraction and the Kronecker Product for fusion, which can recognize interactions between the different modalities. The results of 14 cancer types are stored in an open access database (PORPOISE) for further investigations and attention- and attribution-based interpretability are visualized [72]. Additional recent work by Vale-Silva and Rohr [48] attempts to integrate six different data modalities: tabular clinical data, mRNA expression, microRNA expression, DNA methylation, gene copy number variation, and WSI data with multiple neural networks. Every submodel (CNNs for the images and fully connected feed-forward networks for other data) is dedicated to one data type to extract feature representations, which are fused in an intermediate manner and passed into a common network that estimates survival. The MultiSurv model architecture achieves high prognostic accuracy for multiple cancer types, while being able to handle missing data.
Fig. 3.
Multimodal data fusion. Whole-slide images of a tissue (A) are segmented into smaller patches (B). (C) Image patch and genomic feature-specific embeddings are learned. (D) Multimodal-guided embeddings and concatenation allow prediction of survival or disease risk.
2.3.2. WSIs and radiology data
Combining WSIs with images from radiology is very promising, as macroscopic and microscopic features are combined. The importance of this data integration task is also reflected by the fact that recently a challenge was initiated on the subtype classification of brain cancers using 3D MRI images in combination with WSIs (CPM-RadPath 2019 and 2020): Within this challenge Yin et al. [73] was ranked first for the validation set. Here for every data modality a separate network is trained first to separate subtypes: the tumor is segmented before classification in the MRI images using a 3D-CNN and for the WSIs irrelevant regions (normal regions with a low number of cell nuclei) are excluded before classification. For the final classification, the features of both modalities are fused by a linear weighted module. A newer method, which was also tested against this data, needs no segmentation of the MRI images and performs better for the test set [74]. The authors trained separate CNNs for the subtype prediction and averaged the output probabilities to directly fuse their individual decisions. In addition, they performed a two-step approach for the classification, first, they separate Glioblastomas from the rest and then differentiated between the remaining Oligodendrogliomas and Astrocytomas, because the latter two are much more similar to each other in comparison to Glioblastomas.
Other recent works consider the integration of pathology and radiology data: already an early fusion approach with gradient-boosted decision trees using features directly extracted from MRI images and WSIs outperforms models that were exclusively trained on one data modality in predicting the tumor regression grade (TRG) in rectal cancer [75].
Boehm et al. [76] integrated not only image information from WSIs and CT images, they also included the HRD value, a clinicogenomic feature that is calculated based on gene panel sequencing, to stratify the patients according to their overall survival. The combination of these three data types improved the performance significantly compared to the individual modalities. However, using only two data modalities (radiology and pathology, but not the HRD value) also yielded similar results. Each datatype is processed individually to extract features and for data integration, a late fusion is chosen: For each data modality a Cox model is trained to infer the hazard for the individual patients and a multivariate Cox model integrates this information afterward. For the WSIs for example the tissue type of individual tiles is inferred by a CNN model and the cell nuclei were detected, based on that information the nuclei and tissue type features (serving as input for the Cox model) were calculated [76].
Another example by Schulz et al. [77] uses an intermediate fusion to integrate radiology (CT/MRI scans) and WSI data: individual images are all processed by a CNN, the individual network outputs are concatenated by an attention layer and a fully connected layer is used to e.g. perform binary classification of 5-year disease-specific survival in renal cancer. Special for this approach is, that two CNNs for histopathological data are used to capture information at different resolutions. Similar to the previous paper the addition of simple genomic data (presence and absence of the 10 most frequent mutations) did not increase model performance further [77].
2.3.3. WSIs and clinical data
Similar to molecular and radiology data, the addition of clinical data to WSIs can increase model performance. For these kinds of data integration tasks, it is impressive that already few clinical features may improve the predictions. For example to identify the origin of a cancer with an unknown primary (CUP) a multiclass, multitask, multiple-instance learning approach using a CNN encoder and an attention module was suggested. Before the final classification layer clinical data is concatenated: in this case, only the sex as a binary variable is added, adding additional features (biopsy site) showed again reduced performance [78].
Nevertheless, other reports use many clinical features in addition to the WSIs successfully. Yan et al. [79] used 29 clinical features, including information on sex, personal and family disease history, and the potential tumor itself. Features of the clinical data are extracted using a denoising autoencoder and image features are extracted by a richer fusion network, a CNN where after several convolution blocks an average pooling is performed and features are concatenated. The final decision is made via an intermediate fusion by three fully connected layers using all generated features as input [79].
However, reports on data integration of WSIs and clinical data are not always successful. For example, it is reported that for the skin cancer classification a WSI-based CNN classifier performed better than those where the image information was fused to clinical data (e.g. sex, age, site of the lesion) using the concatenation or Squeeze-and-Excitation approach. In this case, a naive approach with a late fusion performed best, where the result of a CNN classifier, using single tiles of the WSI, was simply replaced by a random forest classifier using the clinical data in case the output score of the CNN is below a certain threshold [80].
3. Summary and outlook
In this review, we are not able to comprehensively reconstruct the historic use of multimodal methods for any of the data combinations studied. Instead, we want to highlight recent developments and interesting methods for combining multimodal data, as we believe this is a universal challenge for all diseases or other phenotype studies of interest. To summarize the key models that have emerged from those recent developments, we give an overview in Table 1, Table 2, where we group algorithms by application and characterize them with a brief method description, as well as the specific data types they integrate.
Table 1.
Overview of different algorithms for multimodal data integration for biomarker and subtype prediction and clinical decision making. Methods are grouped by application tasks, characterized with a brief method description, as well as the specific data types they integrate.
| Application | Method Description | Data Types |
Reference | ||||
|---|---|---|---|---|---|---|---|
| Genetic | Proteogenomic | Epigenomic | Images | Clinical | |||
| Biomarker and Subtype Prediction | |||||||
| Prediction of aortic sizes and aortic disease risk | CNN | ✓ | ✓ | Pirruccello et al. [51] | |||
| Fine mapping of genetic loci | Probabilistic graphical model | ✓ | ✓ | ✓ | Ruffieux et al. [61] | ||
| Prediction of disease risk genes in Schizophrenia | Bayesian model | ✓ | ✓ | ✓ | Wang et al. [62] | ||
| Prediction of retinal related genes | CNN | ✓ | ✓ | Kirchler et al. [53] | |||
| Classification of breast cancer subtypes | DL, latent feature concatenation | ✓ | ✓ | ✓ | Lin et al. [37] | ||
| Classification of multiple disease subtypes | AE, uncertainty quantification | ✓ | ✓ | Han et al. [43] | |||
| Clinical Decision Making | |||||||
| Prediction of rare diseases | DL, SVM | ✓ | ✓ | ✓ | Hsieh et al. [56] | ||
| Classification of brain cancers | CNN, linear weighted module | ✓ | Yin et al. [73] | ||||
| Prediction of the cancer origin of unknown primary | CNN, multiple instance learning | ✓ | ✓ | Lu et al. [78] | |||
| Classification of tumor type and survival prediction | AE, multi-task learning | ✓ | ✓ | ✓ | Zhang et al. [35] | ||
| Classification of multiple clinical outcomes | AE, latent feature averaging | ✓ | ✓ | ✓ | Tan et al. [40] | ||
| Classification of multiple clinical outcomes | AE, feature interaction network | ✓ | ✓ | ✓ | ✓ | Ma and Zhang [41] | |
| Classification of patients with Alzheimer's disease | GCN, correlation discover network | ✓ | ✓ | Wang et al. [42] | |||
Table 2.
Overview of different algorithms for multimodal data integration for survival prediction and therapy response prediction. Methods are grouped by application tasks, characterized with a brief method description, as well as the specific data types they integrate.
| Application | Method Description | Data Types |
Reference | ||||
|---|---|---|---|---|---|---|---|
| Genetic | Proteogenomic | Epigenomic | Images | Clinical | |||
| Survival Prediction | |||||||
| Prediction of survival for cancer patients | DL, attention mechanism | ✓ | ✓ | Chen et al. [71] | |||
| Prediction of breast invasive carcinoma survival | DL, factorized bilinear model | ✓ | ✓ | ✓ | ✓ | Li et al. [94] | |
| Prediction of overall survival | CNN, Cox model | ✓ | ✓ | Boehm et al. [76] | |||
| Prediction of 5-year survival in renal cancer | CNN, attention mechanism | ✓ | ✓ | Schulz et al. [77] | |||
| Classification of breast cancer patient survival | DL, linear weighted module | ✓ | ✓ | Sun et al. [36] | |||
| Prediction of survival in breast invasive carcinoma | DL, feature selection | ✓ | ✓ | ✓ | Huang et al. [44] | ||
| Prediction of survival in breast invasive carcinoma | AE, feature selection | ✓ | ✓ | ✓ | Tong et al. [46] | ||
| Prediction of survival in pan-cancer data | DL, CNN, Cox-PH | ✓ | ✓ | ✓ | ✓ | Cheerla and Gevaert [47] | |
| Prediction of survival in pan-cancer data | DL, CNN, discrete-time surv | ✓ | ✓ | ✓ | ✓ | Vale-Silva and Rohr [48] | |
| Prediction of survival in bladder cancer and sarcoma | AE, hierarchical | ✓ | ✓ | ✓ | ✓ | Wissel et al. [49] | |
| Prediction of survival in pan-cancer data | GCN, feature interaction network | ✓ | ✓ | ✓ | Althubaiti et al. [50] | ||
| Therapy Response Prediction | |||||||
| Classification of drug response in cancer patients | DL, triplet loss objective | ✓ | Sharifi-Noghabi et al. [38] | ||||
| Classification of drug response and mortality | AE, information bottleneck, PoE | ✓ | ✓ | ✓ | Lee and van der Schaar [39] | ||
Tremendous progress has been made in recent years in developing new algorithms for multimodal data integration in predictive modeling tasks, mainly by leveraging advances in modern deep learning. However, the gap between the in silico modeling bench and bedside remains wide [81], [82]. To narrow this gap and bring multimodal predictive models into the clinic, several challenges have to be overcome. First, to facilitate a translation into clinical application, models need to be trustworthy [83]. That is, practitioners need to be able to rely on a model's predictions throughout its life cycle. This does not only imply a good in-distribution generalization performance on data from the same patient cohort but also transferability to other cohorts that may exhibit some degree of distribution shift [84] as well as robustness to erroneous inputs. Importantly, for tools to be applied in a clinical setting, they must be able to reliably estimate their uncertainty and communicate to a practitioner when they “don't know” [85].
A second, related challenge is explainability. For many biomedical applications, a good predictive power of a black-box model is not sufficient: practitioners also need to know why a model has made a specific prediction, a prerequisite for ensuring human oversight and facilitating accountability. While this is currently mainly being addressed via co-attention mechanisms [71], [72], a plethora of algorithms for explainable AI has been developed in the context of single-view methods; the general mechanisms of these models are also applicable in many multi-modal modeling approaches and we refer to a recent survey on explainable AI approaches in medicine for a detailed overview [86]. A third challenge for translational multimodal modeling is for analyses to be privacy-preserving. While the use of multimodal data can often lead to improved predictive performance, caution must be taken when storing or allowing access to those. Individual data types could be exploited to re-identify the patient [87], [88], [89], which may then be used to investigate molecular details of a patient in other data layers. Thus a topic that will gain more attention in the future will be the data-privacy secure analysis of multimodal data.
While method development has focused on common data modalities, such as proteogenomics and imaging, new technologies have led to a rise in novel data types including spatial transcriptomics [90], proteome sequencing [91] and single-cell proteomics [92]. To date, only the first attempts have been made to develop tools integrating such data in an unsupervised manner via a clustering approach [93]. Developing novel predictive algorithms for data integration that generalize to these novel data types will lead to ever more powerful tools in many areas of predictive biomedicine.
However more data does not always lead to better models. It is important to choose the modalities wisely and to ensure that they (at least have the potential) to contain complementary information for the question at hand. Therefore, future research must also address in which data analysis scenarios extension to more modalities may be helpful, as additional data leads to increased costs and analysis time. Even so, given that many processes and genes remain unknown for the majority of diseases, we believe that multimodal data integration methods will play an important role in future discoveries.
Declaration of Competing Interest
Florian Buettner is employed by Siemens AG. He reports funding from Merck KGaA and renumeration from Albireo.
Arber Qoku, Nicoletta Katsaouni, Dr Nadine Flinner, and Prof. Dr Marcel H. Schulz do not report any conflicts of interest.
Acknowledgements
This work has been supported by the DZHK (German Centre for Cardiovascular Research, 81Z0200101) and the Cardio-Pulmonary Institute (CPI) [EXC 2026] ID: 390649896 (NK,MHS), DFG SFB (TRR 267) Noncoding RNAs in the cardiovascular system, Project-ID 403584255 (MHS), DFG SFB 1531 Schadenskontrolle durch das Stroma-vaskuläre Kompartiment, Project-ID:456687919 (MHS) and DFG Proteo-genomische Charakterisierung des diffus großzelligen B-Zell-Lymphoms, Project-ID: 496906589 (FB). We acknowledge funding from the Alfons und Gertrud Kassel-Stiftung as part of the center for data science and AI (NK). NF has been supported by the Mildred Scheel Career Center (MSNZ) Frankfurt.
Co-funded by the European Union (ERC, TAIPO, 101088594 to FB). Views and opinions expressed are however those of the authors only and do not necessarily reflect those of the European Union or the European Research Council. Neither the European Union nor the granting authority can be held responsible for them.
Contributor Information
Nadine Flinner, Email: nadine.flinner@kgu.de.
Florian Buettner, Email: florian.buettner@dkfz-heidelberg.de.
Marcel H. Schulz, Email: marcel.schulz@em.uni-frankfurt.de.
References
- 1.Findlay Gregory M. Linking genome variants to disease: scalable approaches to test the functional impact of human mutations. Hum Mol Genet. 2021;30(R2):187–197. doi: 10.1093/hmg/ddab219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sultan Marc, Schulz Marcel H., Richard Hugues, Magen Alon, Klingenhoff Andreas, Scherf Matthias, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. aug 2008;321(5891):956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 3.Conesa Ana, Madrigal Pedro, Tarazona Sonia, Gomez-Cabrero David, Cervera Alejandra, McPherson Andrew, et al. A survey of best practices for RNA-seq data analysis. Genome Biol. dec 2016;17(1):13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou Xiaoyu, Zhang Wenpeng, Ouyang Zheng. Recent advances in on-site mass spectrometry analysis for clinical applications. TrAC, Trends Anal Chem. apr 2022;149 doi: 10.1016/j.trac.2022.116548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klann Kevin, Tascher Georg, Münch Christian. Functional translatome proteomics reveal converging and dose-dependent regulation by mTORC1 and eIF2α. Mol Cell. 2020;77(4):913–925. doi: 10.1016/j.molcel.2019.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stunnenberg Hendrik G., Hirst Martin, International Human Epigenome Consortium The international human epigenome consortium: a blueprint for scientific collaboration and discovery. Cell. nov 2016;167(5):1145–1149. doi: 10.1016/j.cell.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 7.Nordström Karl J.V., Schmidt Florian, Gasparoni Nina, Salhab Abdulrahman, Gasparoni Gilles, Kattler Kathrin, et al. Pfeifer DEEP consortium Unique and assay specific features of NOMe-, ATAC- and DNase I-seq data. Nucleic Acids Res. 2019;47(20):10580–10596. doi: 10.1093/nar/gkz799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenberg Maxim V.C., Bourc'his Deborah. The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol. oct 2019;20(10):590–607. doi: 10.1038/s41580-019-0159-6. [DOI] [PubMed] [Google Scholar]
- 9.Yankeelov Thomas E., Abramson Richard G., Quarles C. Chad. Quantitative multimodality imaging in cancer research and therapy. Nat Rev Clin Oncol. nov 2014;11(11):670–680. doi: 10.1038/nrclinonc.2014.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kannan Lavanya, Ramos Marcel, Re Angela, El-Hachem Nehme, Safikhani Zhaleh, Gendoo Deena M.A., et al. Public data and open source tools for multi-assay genomic investigation of disease. Brief Bioinform. 2016;17(4):603–615. doi: 10.1093/bib/bbv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abugessaisa Imad, Ramilowski Jordan A., Lizio Marina, Severin Jesicca, Hasegawa Akira, Harshbarger Jayson, et al. FANTOM enters 20th year: expansion of transcriptomic atlases and functional annotation of non-coding RNAs. Nucleic Acids Res. jan 2021;49(D1):892–898. doi: 10.1093/nar/gkaa1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.ICGC/TCGA pan-cancer analysis of whole genomes consortium. Pan-cancer analysis of whole genomes. Nature. 2020;578(7793):82–93. doi: 10.1038/s41586-020-1969-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao Xu, Chai Xiaoran, Shi Xingjie, Chen Lin S., Liu Jin. The statistical practice of the GTEx project: from single to multiple tissues. Quant Biol. aug 2020:1–17. [Google Scholar]
- 14.Bycroft Clare, Freeman Colin, Petkova Desislava, Band Gavin, Elliott Lloyd T., Sharp Kevin, et al. The UK biobank resource with deep phenotyping and genomic data. Nature. oct 2018;562(7726):203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buniello Annalisa, MacArthur Jacqueline A.L., Cerezo Maria, Harris Laura W., Hayhurst James, Malangone Cinzia, et al. The NHGRI-EBI GWAS catalog of published genome-wide association studies, targeted arrays and summary statistics 2019. Nucleic Acids Res. jan 2019;47(D1):1005–1012. doi: 10.1093/nar/gky1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bujold David, Grégoire Romain, Brownlee David, Zaytseva Ksenia, Bourque Guillaume. Practical guide to life science databases. Springer Nature; Singapore, Singapore: 2021. IHEC data portal; pp. 77–94. [Google Scholar]
- 17.Mei Xueyan, Liu Zelong, Robson Philip M., Marinelli Brett, Huang Mingqian, Doshi Amish, et al. An open radiologic deep learning research dataset for effective transfer learning. Radiol Artif Intell. sep 2022;4(5) doi: 10.1148/ryai.210315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prior Fred W., Clark Ken, Commean Paul, Freymann John, Jaffe Carl, Kirby Justin, et al. Annual international conference of the IEEE engineering in medicine and biology society. 2013. An information resource to enable open science; pp. 1282–1285. ISSN: 2694-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fedorov Andrey, Longabaugh William J.R., Pot David, Clunie David A., Pieper Steve, Aerts Hugo J.W.L., et al. NCI Imaging Data Commons Cancer Res. 2021;81(16):4188–4193. doi: 10.1158/0008-5472.CAN-21-0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szklarczyk Damian, Gable Annika L., Nastou Katerina C., Lyon David, Kirsch Rebecca, Pyysalo Sampo, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):605–612. doi: 10.1093/nar/gkaa1074. https://doi.org/10.1093/nar/gkaa1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baumgarten Nina, Hecker Dennis, Karunanithi Sivarajan, Schmidt Florian, List Markus, Schulz EpiRegio Marcel H. Analysis and retrieval of regulatory elements linked to genes. Nucleic Acids Res. 2021;48(W1):193–199. doi: 10.1093/nar/gkaa382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hasin Yehudit, Seldin Marcus, Lusis Aldons. Multi-omics approaches to disease. Genome Biol. 2017;18(1):1–15. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Argelaguet Ricard, Velten Britta, Arnol Damien, Dietrich Sascha, Zenz Thorsten, Marioni John C., et al. Multi-omics factor analysis—a framework for unsupervised integration of multi-omics data sets. Mol Syst Biol. 2018;14(6) doi: 10.15252/msb.20178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klami Arto, Virtanen Seppo, Leppäaho Eemeli, Kaski Samuel. Group factor analysis. IEEE Trans Neural Netw Learn Syst. 2014;26(9):2136–2147. doi: 10.1109/TNNLS.2014.2376974. [DOI] [PubMed] [Google Scholar]
- 25.Kumar Jayavelu Ashok, Wolf Sebastian, Buettner Florian, Alexe Gabriela, Häupl Björn, Comoglio Federico, et al. The proteogenomic subtypes of acute myeloid leukemia. Cancer Cell. 2022;40(3):301–317. doi: 10.1016/j.ccell.2022.02.006. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Li, Lv Chenkai, Jin Yaqiong, Cheng Ganqi, Fu Yibao, Yuan Dongsheng, et al. Deep learning-based multi-omics data integration reveals two prognostic subtypes in high-risk neuroblastoma. Front Genet. 2018;9:477. doi: 10.3389/fgene.2018.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaudhary Kumardeep, Poirion Olivier B., Lu Liangqun, Garmire Lana X. Deep learning–based multi-omics integration robustly predicts survival in liver cancerusing deep learning to predict liver cancer prognosis. Clin Cancer Res. 2018;24(6):1248–1259. doi: 10.1158/1078-0432.CCR-17-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poirion Olivier B., Chaudhary Kumardeep, Garmire Lana X. Deep learning data integration for better risk stratification models of bladder cancer. AMIA Summits Transl Sci Proc. 2018;2018:197. [PMC free article] [PubMed] [Google Scholar]
- 29.Baek Bin, Lee Hyunju. Prediction of survival and recurrence in patients with pancreatic cancer by integrating multi-omics data. Sci Rep. 2020;10(1):1–11. doi: 10.1038/s41598-020-76025-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poirion Olivier B., Jing Zheng, Chaudhary Kumardeep, Huang Sijia, Garmire Lana X. DeepProg: an ensemble of deep-learning and machine-learning models for prognosis prediction using multi-omics data. Gen Med. 2021;13(1):1–15. doi: 10.1186/s13073-021-00930-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cox David R. Regression models and life-tables. J R Stat Soc, Ser B, Methodol. 1972;34(2):187–202. [Google Scholar]
- 32.Baltrušaitis Tadas, Ahuja Chaitanya, Morency Louis-Philippe. Multimodal machine learning: a survey and taxonomy. IEEE Trans Pattern Anal Mach Intell. 2018;41(2):423–443. doi: 10.1109/TPAMI.2018.2798607. [DOI] [PubMed] [Google Scholar]
- 33.Ma Anjun, McDermaid Adam, Xu Jennifer, Chang Yuzhou, Ma Qin. Integrative methods and practical challenges for single-cell multi-omics. Trends Biotechnol. 2020;38(9):1007–1022. doi: 10.1016/j.tibtech.2020.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Argelaguet Ricard, SE Cuomo Anna, Stegle Oliver, Marioni John C. Computational principles and challenges in single-cell data integration. Nat Biotechnol. 2021;39(10):1202–1215. doi: 10.1038/s41587-021-00895-7. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Xiaoyu, Xing Yuting, Sun Kai, Guo Yike. Omiembed: a unified multi-task deep learning framework for multi-omics data. Cancers. 2021;13(12):3047. doi: 10.3390/cancers13123047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun Dongdong, Wang Minghui, Li Ao. A multimodal deep neural network for human breast cancer prognosis prediction by integrating multi-dimensional data. IEEE/ACM Trans Comput Biol Bioinform. 2018;16(3):841–850. doi: 10.1109/TCBB.2018.2806438. [DOI] [PubMed] [Google Scholar]
- 37.Lin Yuqi, Zhang Wen, Cao Huanshen, Li Gaoyang, Du Wei. Classifying breast cancer subtypes using deep neural networks based on multi-omics data. Genes. 2020;11(8):888. doi: 10.3390/genes11080888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharifi-Noghabi Hossein, Zolotareva Olga, Collins Colin C., Ester Martin. MOLI: multi-omics late integration with deep neural networks for drug response prediction. Bioinformatics. 2019;35(14) doi: 10.1093/bioinformatics/btz318. i501–i509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee Changhee, van der Schaar Mihaela. International conference on artificial intelligence and statistics. PMLR; 2021. A variational information bottleneck approach to multi-omics data integration; pp. 1513–1521. [Google Scholar]
- 40.Tan Kaiwen, Huang Weixian, Hu Jinlong, Dong Shoubin. A multi-omics supervised autoencoder for pan-cancer clinical outcome endpoints prediction. BMC Med Inform Decis Mak. 2020;20(3):1–9. doi: 10.1186/s12911-020-1114-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ma Tianle, Zhang Aidong. Integrate multi-omics data with biological interaction networks using multi-view factorization autoencoder (mae) BMC Genomics. 2019;20(11):1–11. doi: 10.1186/s12864-019-6285-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Tongxin, Shao Wei, Huang Zhi, Tang Haixu, Zhang Jie, Ding Zhengming, et al. Mogonet integrates multi-omics data using graph convolutional networks allowing patient classification and biomarker identification. Nat Commun. 2021;12(1):1–13. doi: 10.1038/s41467-021-23774-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Han Zongbo, Yang Fan, Huang Junzhou, Zhang Changqing, Yao Jianhua. Proceedings of the IEEE/CVF conference on computer vision and pattern recognition. 2022. Multimodal dynamics: dynamical fusion for trustworthy multimodal classification; pp. 20707–20717. [Google Scholar]
- 44.Huang Zhi, Zhan Xiaohui, Xiang Shunian, Johnson Travis S., Helm Bryan, Yu Christina Y., et al. SALMON: survival analysis learning with multi-omics neural networks on breast cancer. Front Genet. 2019;10:166. doi: 10.3389/fgene.2019.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li Ruiqing, Wu Xingqi, Li Ao, Wang Minghui. Hfbsurv: hierarchical multimodal fusion with factorized bilinear models for cancer survival prediction. Bioinformatics. 2022;38(9):2587–2594. doi: 10.1093/bioinformatics/btac113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tong Li, Mitchel Jonathan, Chatlin Kevin, Wang May D. Deep learning based feature-level integration of multi-omics data for breast cancer patients survival analysis. BMC Med Inform Decis Mak. 2020;20(1):1–12. doi: 10.1186/s12911-020-01225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheerla Anika, Gevaert Olivier. Deep learning with multimodal representation for pancancer prognosis prediction. Bioinformatics. 2019;35(14) doi: 10.1093/bioinformatics/btz342. i446–i454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vale-Silva Luís A., Rohr Karl. Long-term cancer survival prediction using multimodal deep learning. Sci Rep. 2021;11(1):1–12. doi: 10.1038/s41598-021-92799-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wissel David, Rowson Daniel, Boeva Valentina. Hierarchical autoencoder-based integration improves performance in multi-omics cancer survival models through soft modality selection. 2022. bioRxiv.
- 50.Althubaiti Sara, Kulmanov Maxat, Liu Yang, Gkoutos Georgios V, Schofield Paul, DeepMOCCA Robert Hoehndorf. A pan-cancer prognostic model identifies personalized prognostic markers through graph attention and multi-omics data integration. 2021. bioRxiv.
- 51.Pirruccello James P., Chaffin Mark D., Chou Elizabeth L., Fleming Stephen J., Lin Honghuang, Nekoui Mahan, et al. Deep learning enables genetic analysis of the human thoracic aorta. Nat Genet. jan 2022;54(1):40–51. doi: 10.1038/s41588-021-00962-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith Stephen M., Douaud Gwenaëlle, Chen Winfield, Hanayik Taylor, Alfaro-Almagro Fidel, Sharp Kevin, et al. An expanded set of genome-wide association studies of brain imaging phenotypes in UK biobank. Nat Neurosci. may 2021;24(5):737–745. doi: 10.1038/s41593-021-00826-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kirchler Matthias, Konigorski Stefan, Norden Matthias, Meltendorf Christian, Kloft Marius, Schurmann Claudia, et al. GWAS of images using deep transfer learning. Bioinformatics. jul 2022;38(14):3621–3628. doi: 10.1093/bioinformatics/btac369. [DOI] [PubMed] [Google Scholar]
- 54.Gurovich Yaron, Hanani Yair, Bar Omri, Nadav Guy, Fleischer Dekel Gelbman Nicole, Basel-Salmon Lina, et al. Identifying facial phenotypes of genetic disorders using deep learning. Nat Med. jan 2019;25(1):60–64. doi: 10.1038/s41591-018-0279-0. [DOI] [PubMed] [Google Scholar]
- 55.Hsieh Tzung-Chien, Bar-Haim Aviram, Moosa Shahida, Ehmke Nadja, Gripp Karen W., Tori Pantel Jean, et al. GestaltMatcher facilitates rare disease matching using facial phenotype descriptors. Nat Genet. mar 2022;54(3):349–357. doi: 10.1038/s41588-021-01010-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hsieh Tzung-Chien, Mensah Martin A., Pantel Jean T., Aguilar Dione, Bar Omri, Bayat Allan, et al. Prioritization of exome data by image analysis. Genet Med. dec 2019;21(12):2807–2814. doi: 10.1038/s41436-019-0566-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Finucane Hilary K., Bulik-Sullivan Brendan, Gusev Alexander, Trynka Gosia, Reshef Yakir, Loh Po-Ru, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. nov 2015;47(11):1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang Rujin, Lin Dan-Yu, Jiang Yuchao. EPIC: inferring relevant cell types for complex traits by integrating genome-wide association studies and single-cell RNA sequencing. PLoS Genet. jun 2022;18(6) doi: 10.1371/journal.pgen.1010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu Zhihong, Zhang Futao, Hu Han, Bakshi Andrew, Robinson Matthew R., Powell Joseph E., et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. may 2016;48(5):481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 60.Giambartolomei Claudia, Zhenli Liu Jimmy, Zhang Wen, Hauberg Mads, Shi Huwenbo, Boocock James, et al. Pasaniuc CommonMind consortium, bogdan pasaniuc, and panos roussos. A Bayesian framework for multiple trait colocalization from summary association statistics. Bioinformatics. 2018;34(15):2538–2545. doi: 10.1093/bioinformatics/bty147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ruffieux Hélène, Fairfax Benjamin P., Nassiri Isar, Vigorito Elena, Wallace Chris, Richardson Sylvia, et al. An epigenome-driven approach for detecting and interpreting hotspots in molecular QTL studies. Am J Hum Genet. 2021;108(6):983–1000. doi: 10.1016/j.ajhg.2021.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Quan, Chen Rui, Cheng Feixiong, Wei Qiang, Ji Ying, Yang Hai, et al. A Bayesian framework that integrates multi-omics data and gene networks predicts risk genes from schizophrenia GWAS data. Nat Neurosci. may 2019;22(5):691–699. doi: 10.1038/s41593-019-0382-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stahlschmidt Sören Richard, Ulfenborg Benjamin, Synnergren Jane. Multimodal deep learning for biomedical data fusion: a review. Brief Bioinform. 2022;23(2) doi: 10.1093/bib/bbab569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bera Kaustav, Schalper Kurt A., Rimm David L., Velcheti Vamsidhar, Madabhushi Anant. Artificial intelligence in digital pathology - new tools for diagnosis and precision oncology. Nat Rev Clin Oncol. 2019;16(11):703–715. doi: 10.1038/s41571-019-0252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kather Jakob Nikolas, Pearson Alexander T., Halama Niels, Jäger Dirk, Krause Jeremias, Loosen Sven H., et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med. 2019;25(7):1054–1056. doi: 10.1038/s41591-019-0462-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Campanella Gabriele, Hanna Matthew G., Geneslaw Luke, Miraflor Allen, Werneck Krauss Silva Vitor, Busam Klaus J., et al. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. 2019;25(8):1301–1309. doi: 10.1038/s41591-019-0508-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen Chi-Long, Chen Chi-Chung, Yu Wei-Hsiang, Chen Szu-Hua, Chang Yu-Chan, Hsu Tai-I., et al. An annotation-free whole-slide training approach to pathological classification of lung cancer types using deep learning. Nat Commun. 2021;12(1):1193. doi: 10.1038/s41467-021-21467-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boehm Kevin M., Khosravi Pegah, Vanguri Rami, Gao Jianjiong, Shah Sohrab P. Harnessing multimodal data integration to advance precision oncology. Nat Rev Cancer. 2022;22(2):114–126. doi: 10.1038/s41568-021-00408-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dwivedi Chaitanya, Nofallah Shima, Pouryahya Maryam, Iyer Janani, Leidal Kenneth, Chung Chuhan, et al. Multi stain graph fusion for multimodal integration in pathology. 2022.
- 70.Schneider Lucas, Laiouar-Pedari Sara, Kuntz Sara, Krieghoff-Henning Eva, Hekler Achim, Kather Jakob N., et al. Integration of deep learning-based image analysis and genomic data in cancer pathology: a systematic review. Eur J Cancer. 2022;160:80–91. doi: 10.1016/j.ejca.2021.10.007. [DOI] [PubMed] [Google Scholar]
- 71.Chen Richard J., Lu Ming Y., Weng Wei-Hung, Chen Tiffany Y., Williamson Drew F.K., Manz Trevor, et al. 2021 IEEE/CVF international conference on computer vision (ICCV) IEEE; October 2021. Multimodal co-attention transformer for survival prediction in gigapixel whole slide images. [Google Scholar]
- 72.Chen R.J., Lu M.Y., Williamson D.F.K., Chen T.Y., Lipkova J., Noor Z., et al. Pan-cancer integrative histology-genomic analysis via multimodal deep learning. Cancer Cell. 2022;40(8):865–878. doi: 10.1016/j.ccell.2022.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yin Baocai, Cheng Hu, Wang Fengyan, Wang Zengfu. In: Brainlesion: glioma, multiple sclerosis, stroke and traumatic brain injuries. Crimi Alessandro, Bakas Spyridon., editors. Springer International Publishing; Cham: 2021. Brain tumor classification based on mri images and noise reduced pathology images; pp. 465–474. [Google Scholar]
- 74.Wang Xiyue, Wang Ruijie, Yang Sen, Zhang Jun, Wang Minghui, Zhong Dexing, et al. Combining radiology and pathology for automatic glioma classification. Front Bioeng Biotechnol. 2022;10 doi: 10.3389/fbioe.2022.841958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shao Lizhi, Liu Zhenyu, Feng Lili, Lou Xiaoying, Li Zhenhui, Zhang Xiao-Yan, et al. Multiparametric MRI and whole slide image-based pretreatment prediction of pathological response to neoadjuvant chemoradiotherapy in rectal cancer: a multicenter radiopathomic study. Ann Surg Oncol. 2020;27(11):4296–4306. doi: 10.1245/s10434-020-08659-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boehm Kevin M., Aherne Emily A., Ellenson Lora, Nikolovski Ines, Alghamdi Mohammed, Vázquez-García Ignacio, et al. MSK MIND Consortium Multimodal data integration using machine learning improves risk stratification of high-grade serous ovarian cancer. Nat Cancer. 2022;3(6):723–733. doi: 10.1038/s43018-022-00388-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schulz Stefan, Woerl Ann-Christin, Jungmann Florian, Glasner Christina, Stenzel Philipp, Strobl Stephanie, et al. Multimodal deep learning for prognosis prediction in renal cancer. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.788740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lu Ming Y., Chen Tiffany Y., Williamson Drew F.K., Zhao Melissa, Shady Maha, Lipkova Jana, et al. AI-based pathology predicts origins for cancers of unknown primary. Nature. 2021;594(7861):106–110. doi: 10.1038/s41586-021-03512-4. [DOI] [PubMed] [Google Scholar]
- 79.Yan Rui, Zhang Fa, Rao Xiaosong, Lv Zhilong, Li Jintao, Zhang Lingling, et al. Richer fusion network for breast cancer classification based on multimodal data. BMC Med Inform Decis Mak. 2021;21(Suppl 1):134. doi: 10.1186/s12911-020-01340-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Höhn Julia, Krieghoff-Henning Eva, Jutzi Tanja B., von Kalle Christof, Utikal Jochen S., Meier Friedegund, et al. Combining CNN-based histologic whole slide image analysis and patient data to improve skin cancer classification. Eur J Cancer. 2021;149:94–101. doi: 10.1016/j.ejca.2021.02.032. [DOI] [PubMed] [Google Scholar]
- 81.Kelly Christopher J., Karthikesalingam Alan, Suleyman Mustafa, Corrado Greg, King Dominic. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 2019;17(1):1–9. doi: 10.1186/s12916-019-1426-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goecks Jeremy, Jalili Vahid, Heiser Laura M., Gray Joe W. How machine learning will transform biomedicine. Cell. 2020;181(1):92–101. doi: 10.1016/j.cell.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Eshete Birhanu. Making machine learning trustworthy. Science. 2021;373(6556):743–744. doi: 10.1126/science.abi5052. [DOI] [PubMed] [Google Scholar]
- 84.Hosseini Mahdi S., Chan Lyndon, Huang Weimin, Wang Yichen, Hasan Danial, Rowsell Corwyn, et al. European conference on computer vision. Springer; 2020. On transferability of histological tissue labels in computational pathology; pp. 453–469. [Google Scholar]
- 85.Ghahramani Zoubin. Probabilistic machine learning and artificial intelligence. Nature. 2015;521(7553):452–459. doi: 10.1038/nature14541. [DOI] [PubMed] [Google Scholar]
- 86.Tjoa Erico, Guan Cuntai. A survey on explainable artificial intelligence (xai): toward medical xai. IEEE Trans Neural Netw Learn Syst. 2020;32(11):4793–4813. doi: 10.1109/TNNLS.2020.3027314. [DOI] [PubMed] [Google Scholar]
- 87.Erlich Yaniv, Shor Tal, Pe'er Itsik, Carmi Shai. Identity inference of genomic data using long-range familial searches. Science. nov 2018;362(6415):690–694. doi: 10.1126/science.aau4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.von Thenen Nora, Ayday Erman, Cicek A. Ercument. Re-identification of individuals in genomic data-sharing beacons via allele inference. Bioinformatics. feb 2019;35(3):365–371. doi: 10.1093/bioinformatics/bty643. [DOI] [PubMed] [Google Scholar]
- 89.Venkatesaramani Rajagopal, Malin Bradley A., Vorobeychik Yevgeniy. Re-identification of individuals in genomic datasets using public face images. Sci Adv. nov 2021;7(47) doi: 10.1126/sciadv.abg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rao Anjali, Barkley Dalia, França Gustavo S., Yanai Itai. Exploring tissue architecture using spatial transcriptomics. Nature. 2021;596(7871):211–220. doi: 10.1038/s41586-021-03634-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Richards Alicia L., Merrill Anna E., Coon Joshua J. Proteome sequencing goes deep. Curr Opin Chem Biol. 2015;24:11–17. doi: 10.1016/j.cbpa.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kelly Ryan T. Single-cell proteomics: progress and prospects. Mol Cell Proteomics. 2020;19(11):1739–1748. doi: 10.1074/mcp.R120.002234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hu J., Li X., Coleman K., Schroeder A., Ma N., Irwin D.J., et al. Integrating gene expression, spatial location and histology to identify spatial domains and spatially variable genes by graph convolutional network. Nat Methods. 2021;18(11):1342–1351. doi: 10.1038/s41592-021-01255-8. [DOI] [PubMed] [Google Scholar]
- 94.Li Ruiqing, Wu Xingqi, Li Ao, Wang Minghui. HFBSurv: hierarchical multimodal fusion with factorized bilinear models for cancer survival prediction. Bioinformatics. apr 2022;38(9):2587–2594. doi: 10.1093/bioinformatics/btac113. [DOI] [PMC free article] [PubMed] [Google Scholar]