Summary
Mitochondrial morphology is an indicator of cellular health and function; however, its quantification and categorization into different subclasses is a complicated process. Here, we present a protocol for mitochondrial morphology quantification in the presence and absence of carbonyl cyanide m-chlorophenyl hydrazone stress. We describe steps for the preparation of cells for immunofluorescence microscopy, staining, and morphology quantification. The quantification protocol generates an aspect ratio that helps to categorize mitochondria into two clear subclasses.
For complete details on the use and execution of this protocol, please refer to Nag et al.1
Subject areas: Cell Biology, Cell culture, Metabolism, Microscopy, Molecular/Chemical Probes
Graphical abstract

Highlights
-
•
A quantification pipeline to study mitochondrial morphology
-
•
An unbiased pipeline has been created by using Fiji open-source image processing software
-
•
Applicable for characterizing mitochondrial morphology subclasses from confocal images
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Mitochondrial morphology is an indicator of cellular health and function; however, its quantification and categorization into different subclasses is a complicated process. Here, we present a protocol for mitochondrial morphology quantification in the presence and absence of carbonyl cyanide m-chlorophenyl hydrazone stress. We describe steps for the preparation of cells for immunofluorescence microscopy, staining, and morphology quantification. The quantification protocol generates an aspect ratio that helps to categorize mitochondria into two clear subclasses.
Before you begin
In healthy mammalian cells, mitochondria appear in the form of a network that in response to membrane depolarization stress undergoes severe fragmentation. In mitochondrial biology, CCCP is a highly used depolarization stress inducer. Under CCCP-induced stress, the mitochondrial fission pathway is activated resulting in severe fragmentation.
This protocol is divided into three major steps: 1. Preparation of cells for staining; 2. Staining; and imaging; 3. Quantification. By using the quantification pipeline, we divide mitochondrial morphology into two subclasses: network and puncta. The protocol below describes the steps for using U2OS cells. However, this protocol can be used for other cell types e.g., HeLa or HEK293T, and other stress-inducing conditions e.g., starvation.
Institutional permissions
Institutional permission is not needed.
Cell culture conditions and preparation of reagents
Timing: 30 min every week (for step 1)
Timing: 30 min (for step 2)
-
1.
Culture U2OS cells.
U2OS cells are cultured until passage 20 in McCoy’s 5A media with FBS in the 5% humidified CO2 incubator.-
a.During passaging, aspirate the media from the flask.
-
b.Wash the cells with 10 mL 1X PBS.
-
c.Add 1 mL trypsin and allow to sit in the incubator for 1–2 min.
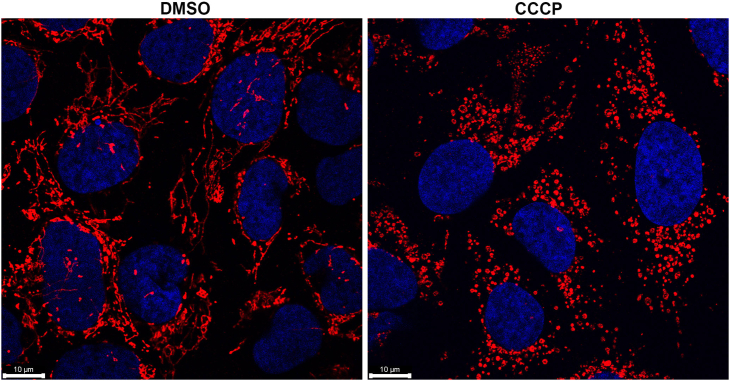
-
d.Check cells for trypsinization.
-
e.Add 9 mL media to the trypsinized cells.
-
f.Add 9 mL media to the fresh flask.
-
g.Add 1 mL of cells to each flask (1:10).
-
h.Keep the flask in the incubator.
 CRITICAL: Mitochondrial morphology is sensitive to various types of stress. Maintaining a clean and healthy cell culture is a pre-requirement of the experiment. Split the cells once a week in around 20% confluency to maintain a healthy culture. Monitor the cells periodically for mycoplasma contamination.
CRITICAL: Mitochondrial morphology is sensitive to various types of stress. Maintaining a clean and healthy cell culture is a pre-requirement of the experiment. Split the cells once a week in around 20% confluency to maintain a healthy culture. Monitor the cells periodically for mycoplasma contamination.
-
a.
-
2.Preparation of CCCP.
-
a.CCCP can be dissolved in either ethanol or DMSO. CCCP can be dissolved up to 100 mM in DMSO to prepare the stock solution.
-
b.Filter the stock solution with a 0.2 μM syringe filter.
-
c.Prepare a working stock solution (conc. 20 mM) by diluting the stock solution again with DMSO.
-
a.
CRITICAL: As CCCP is toxic to human health, weighing of the CCCP powder should be carried out inside the chemical hood.
CRITICAL: CCCP is light sensitive and prone to degradation with the freeze-thaw cycles. To prevent multiple freeze-thaw cycles, we recommend storing the working stock solution in small aliquots.
CRITICAL: Filtration of CCCP solution can be complicated as the filtration membrane may absorb a small fraction of CCCP from the solution, resulting in a slightly lower concentration in the filtrate. However, we discard the first 10% of the filtrate and then collect the remaining filtrate so that the effective CCCP concentration in the filtrate is the same as the target CCCP concentration. Hence, we do not anticipate any effect of the sterile filtration on the CCCP concentration.
Note: Stock and working solutions should be stored at ‒20°C. Stock solution can be stored until 3–4 freeze thaw cycles. Working solutions tend to degrade faster. More than one freeze-thaw cycle of the working solution is not recommended.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PGAM5 | Abcam | Cat#126534 |
| ATP5A | Abcam | Cat#14748 |
| Rabbit Alexa Fluor Plus 488 | Life Technologies | Cat#A32731; RRID: AB_2633280 |
| Mouse Alexa Fluor Plus 594 | Life Technologies | Cat#A32742; RRID: AB_2762825 |
| Chemicals, peptides, and recombinant proteins | ||
| McCoy’s 5A (modified) media | Gibco | Cat#16600082 |
| Trypsin | Gibco | Cat#25200056 |
| PBS | Gibco | Cat#14190144 |
| 16% Formaldehyde solution | Thermo Scientific | Cat#28908 |
| Goat serum | Gibco | Cat#16210072 |
| Fetal bovine serum | Gibco | Cat#26140079 |
| CCCP | Sigma | Cat#C2759 |
| DMSO | Sigma | Cat#41639 |
| Penicillin-streptomycin | Sigma | Cat#P4333 |
| Triton X-100 | Bioshop | Cat#TRX777 |
| BSA | Bioshop | Cat#ALB001 |
| Poly-D-lysine | Sigma | Cat#P6407 |
| Fluoromount-G | Invitrogen | Cat#00495802 |
| Experimental models: Cell lines | ||
| U2OS | Dr. Peter Kim’s lab, University of Toronto | N/A |
| Software and algorithms | ||
| Las X | Leica Microsystems | https://www.leica-microsystems.com/products/microscope-software/p/leica-las-x-ls/ |
| Fiji | GitHub | https://fiji.sc/ |
| ImageJ | NIH | https://imagej.nih.gov/ij///index.html |
| Other | ||
| Microscope slides | Fisherbrand | Cat#22-037-246 |
| 1.5 mm glass coverslips | Thermo Fisher Scientific | Cat#12 518 213 |
| Leica SP8 confocal microscope | Leica | N/A |
Materials and equipment
McCoy’s 5A media
| Reagent | Final concentration | Amount |
|---|---|---|
| McCoy’s 5A media | N/A | 445 mL |
| FBS | N/A | 50 mL |
| Penicillin-Streptomycin | 100 U/mL | 5 mL |
| Total | N/A | 500 mL |
CRITICAL: Prepare media using aseptic techniques. Store at 4°C for 1–2 months. Before use, warm media to 37°C using a water bath.
4% Formaldehyde solution
| Reagent | Final concentration | Amount |
|---|---|---|
| 16% Formaldehyde | 4% | 10 mL |
| H2O | N/A | 30 mL |
| Total | N/A | 40 mL |
CRITICAL: Formaldehyde is toxic to human health. Preparation of the formaldehyde solution should be done inside the chemical biosafety cabinet. 4% Formaldehyde solution can be stored at 4°C up to 1–2 months.
Alternatives: Alternatively, pre-made 4% Formaldehyde solution can also be used.
Permeabilization solution
| Reagent | Final concentration | Amount |
|---|---|---|
| Triton X-100 | 0.3% | 300 μL |
| 1X PBS | N/A | 100 mL |
| Total | N/A | 100 mL |
CRITICAL: Permeabilization buffer can be stored at 4°C up to one month.
Antibody dilution buffer
| Reagent | Final concentration | Amount |
|---|---|---|
| 1X PBS | N/A | 10 mL |
| BSA | 0.01 g/mL | 0.1 g |
| Total | N/A | 10 mL |
CRITICAL: Antibody dilution buffer can be stored at 4°C up to one month.
Step-by-step method details
Preparing cells for staining
Timing: 2 days
This step explains the steps from seeding of the cells until the cell fixation.
Day: 1
Timing: 3 h
-
1.
Place coverslips in the wells of a 6-well dish.
-
2.
Incubate coverslips with Poly-D-lysine solution (0.1 mg/mL; 1.5 mL solution/well; dissolved in water) for 5 min.
-
3.
Remove Poly-D-lysine.
-
4.
Wash the wells with sterile distilled water.
-
5.
Keep the coverslips for drying for 2 hr. During this incubation time, the plates can be kept inside the hood by keeping the lids open.
-
6.
Seed U2OS cells at about 60%–70% (0.3–0.4∗106 cells/ well) confluency on the coverslips.
-
7.
Incubate the plate in 5% humidified CO2 incubator for 16–24 h.
Day: 2
Timing: 3 h
-
8.
Dilute CCCP working stock (conc. 20 mM) to a final concentration of 20 μM with McCoy’s 5A media.
-
9.
Use DMSO as a vehicle control.
-
10.
Aspirate the old media from the wells.
-
11.
Add 2 mL of DMSO/ CCCP containing media in each well.
-
12.
Incubate for 2 h in the incubator.
-
13.
After incubation, remove the media.
-
14.
Wash the wells with 2 mL 1X PBS.
-
15.
Add 4% Formaldehyde solution (2 mL/well) and incubate for 15 min.
-
16.
Wash three times with 1X PBS for 5 min each.
-
17.
Add 2 mL PBS to each well.
Note: After this step, the cells can be immediately used for staining or can be stored at 4°C until the next day.
Staining and imaging of the cells
Timing: 2 days
This step explains the steps from the staining of the cells to the imaging.
Day: 1
Timing: 4 h
-
18.
Remove the PBS.
-
19.
Add 2 mL of permeabilization buffer to each well and incubate for 15 min.
-
20.
Aspirate the permeabilization buffer.
-
21.
Wash three times in 1X PBS for 5 min each.
-
22.
Add 1 mL of Goat Serum to each well and incubate for 1–2 h.
-
23.
After incubation, wash the coverslips once with 1X PBS.
-
24.
Prepare the staining chamber by keeping parafilm on top of a wet blotting paper in closed environment.
-
25.
Dilute the primary antibodies in antibody dilution buffer and incubate for 2 hrs.
-
26.
Transfer the coverslips from the 6-well dish to the staining chamber by using a fine-tip tweezer and incubate it with 30‒40 μL primary antibody solution.
-
27.
After incubation with the primary antibody, transfer the coverslips back to the 6 well dish and wash them for 3 times with 1X PBS.
-
28.
Dilute the secondary antibodies in antibody dilution buffer and incubate for 1 hr.
-
29.
Wash coverslips for 3 times with 1X PBS.
-
30.
Clean the glass slides with 70% alcohol.
-
31.
Invert and mount the coverslips with Fluoromount G and leave it to dry overnight.
Note: We recommend the use of ATP5A (inner mitochondrial membrane marker) or Tom20 or Tom70 (outer mitochondrial membrane marker) antibodies (1:300 dilution) to check mitochondrial morphology.
Day: 2
Timing: 2 h
-
32.
Proceed for imaging. Images can be acquired either as individual images or Z-stacks.
-
33.
If the images are acquired as Z stacks, maximal projection images can be created and used for quantification. This quantification protocol can identify mitochondrial subclasses from both types of images.
-
34.
In DMSO-treated cells mitochondria appear as long, branched structures which create a network (Figure 1, left image). Comparatively CCCP treated cells mitochondria appear as punctate or dotted structures (Figure 1, right image).
-
35.
After imaging save/convert image files into the TIFF format for further analysis.
Figure 1.
U2OS cells treated with DMSO (used as vehicle control, left) or CCCP (20 μM for 2 h, right)
Mitochondria is stained with ATP5a antibody and nuclear staining is done with Hoechst. Scale bar 10 μM.
Figure 1: U2OS cells treated with DMSO (used as vehicle control, left) or CCCP (20 μM for 2 h, right). Mitochondria are stained with ATP5a antibody and nuclear staining is done with Hoechst. Scale bar 10 μM. Images were captured using 60x objective in Leica SP8 microscope.
Quantification analysis
Timing: 2 h
This step explains the image processing steps required to quantify the ratio of network and punctate mitochondria using Fiji software. Exact values may need to be modified based on quality images.
-
36.
Open the image stack in Fiji.
-
37.
Subtract background noise In the Fiji menu window, select Process > Subtract Background, and set the rolling ball radius to 50.0 pixels.
-
38.
Apply a low median filter to smooth intensity variation within structures: Process >Filters > Median and set the radius to 1 pixel.
-
39.
Binarize: Process > Binary > Make Binary > Default, select “Calculate threshold for each image”
-
40.
Despeckle to remove excess noise: Process > Noise > Despeckle.
-
41.
Pause and manually check each image to ensure effective binarization. If features are lost, try a different filter and pixel size, or thresholding method for binarization.
-
42.
Open classifier trainer: Plugins > Segmentation > Trainable Weka Segmentation.
-
43.
Make 3 classes by selecting “Create new class”, and label them network, puncta, and background.
-
44.
Using the wand tool from the Fiji menu, select 5 networks and puncta, one at a time, and add to their respective label. Avoid using shapes with holes. Select 5 areas of background by drawing shapes in the background and add them to label.
-
45.
Select “Train classifier”, with default settings.
-
46.
Refine the classifier if needed by adding more traces to labels. Repeat until satisfied with the classifier.
-
47.
Save the classifier, load the binarized image stack, and select “Apply classifier”. Reuse the same classifier for all subsequent image stacks.
-
48.
Convert the classified image stack to RGB: Image > Type > RGB Color.
-
49.
Duplicate the image stack.
-
50.
Color threshold for each stack to keep only networks in one and puncta in the other: Image > Adjust > Color threshold > move the bars under “Hue” to the threshold and select “Stack” to apply to all images.
-
51.
Binarize each stack: Process > Binary > Make Binary > Default.
-
52.
Quantify the total area of networks and puncta: Analyze > Analyze particles, select “Display results”.
-
53.
Save, open in Excel, and calculate the sum of all particles from each label. Add labels together to get the total area of mitochondria. Divide each labeled area by the total area to get the percentage of the total.
Note: If planning on repeating protocol multiple times, record the pipeline as a macro on Fiji for automation.
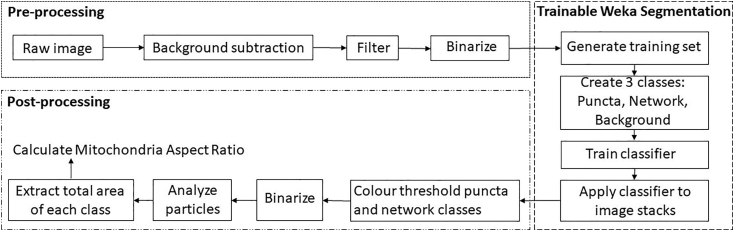
Figure 2: Quantification pipeline.
Figure 2.
Quantification pipeline
Expected outcomes
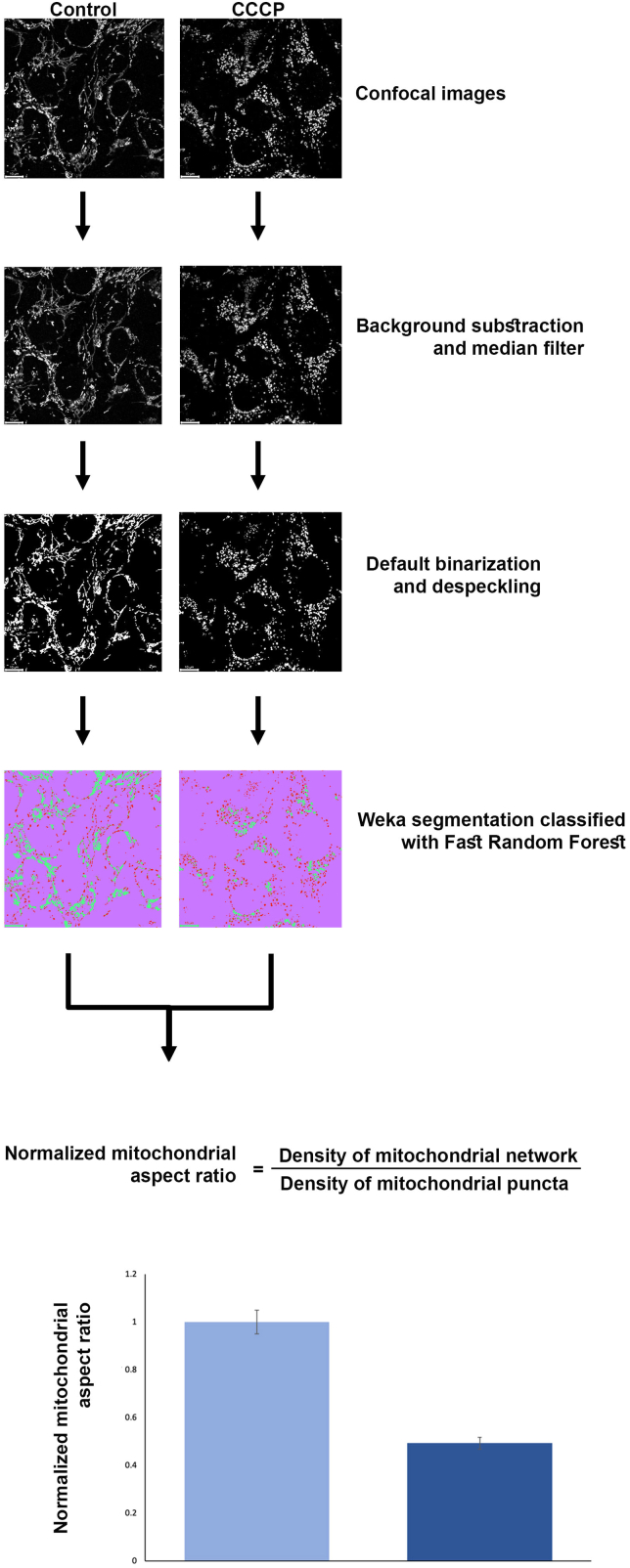
According to this protocol, we calculated the mitochondrial aspect ratio of the U2OS cells treated with DMSO and CCCP. A higher aspect ratio represents the mitochondrial network whereas a lower aspect ratio represents punctate mitochondria.
Limitations
This image quantification method works best with high-resolution images. We recommend mitochondrial imaging with a confocal microscope for better resolution and clarity of images.
Figure 3: Steps of the quantification pipeline.
Figure 3.
Steps of the quantification pipeline
Troubleshooting
Problem 1
Improper fragmentation initiation after CCCP treatment.
Potential solution
Multiple freeze-thaw cycles reduce CCCP activity. Store CCCP in smaller aliquots. Also, CCCP is light-sensitive. Therefore, CCCP aliquots should be preserved in the dark.
Problem 2
Non-specific background staining of cells.
Potential solution
Non-specific staining is a common problem in the IFM (immune fluorescence microscopy). It can also interfere with the quantification result. There can be multiple problems behind that.
-
•
Antibody binding is non-specific: For every antibody, dilution ratio should be optimized.
-
•
Increasing the time for blocking also can be helpful to troubleshoot this problem.
-
•
Contaminations in cells can appear as non-specific background staining. Therefore, if this type of problem is noticed, cells in the culture should be checked for contamination.
-
•
PBS, Permeabilization buffer, and antibody staining buffer should be checked for contamination before use.
Problem 3
Loss of features during the binarization step of image processing.
Potential solution
-
•
Test different background subtraction radius, filters, and thresholding methods to optimize image processing for the images.
-
•
Increase fluorescence signal to detector. If using a confocal microscope, try using a high magnification and numerical aperture objective, increasing the image resolution, the pixel dwell time, averaging, or laser power (but beware of bleaching).
Problem 4
The trained classifier is making errors when applied to image stacks.
Potential solution
-
•
Continue refining classifier.
-
•
If many iterations of refining have been done and the classifier keeps getting worse, it might be over-refined, try restarting.
-
•
Depending on the errors it is making, select either a wider variety or more specific shapes for each label.
Resource availability
Lead contact
Further information about all reagents mentioned in this article should be directed to and will be fulfilled by the lead contact, Sudeshna Nag (sudeshna.nag@mail.utoronto.ca).
Materials availability
This study did not generate any new reagents.
Data and code availability
This study did not generate any new dataset/code.
Acknowledgments
This work was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC). S.N. is supported by NSERC.
Author contributions
S.N. performed cell culture, staining, and imaging. K.S. and C.M.Y. designed the quantification pipeline. S.N. and K.S. wrote the manuscript, which was edited by C.M.Y. and G.A.M.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Sudeshna Nag, Email: sudeshna.nag@mail.utoronto.ca.
Kaitlin Szederkenyi, Email: k.szederkenyi@mail.utoronto.ca.
G. Angus McQuibban, Email: angus.mcquibban@utoronto.ca.
References
- 1.Nag S., Szederkenyi K., Gorbenko O., Tyrrell H., Yip C.M., McQuibban G.A. PGAM5 is an MFN2 phosphatase that plays an essential role in the regulation of mitochondrial dynamics. Cell Rep. 2023;42:112895. doi: 10.1016/j.celrep.2023.112895. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate any new dataset/code.