Summary
The complement receptors C3aR and C5aR1 are promising therapeutic targets. Here, we present a protocol to screen the effects of different agonists and antagonists on these receptors in vitro, using phosphorylated extracellular signal-regulated kinase (ERK) as a readout. We describe steps for isolating human monocyte-derived macrophages, culturing and preparing Chinese hamster ovary cells stably expressing human C5aR1 or C3aR, performing pharmacological assays, and detecting phospho-ERK1/2 in the cell lysate. This protocol can also be performed using other cell lines.
For complete details on the use and execution of this protocol, please refer to Li et al. (2020)1 and Li et al.2
Subject areas: Cell culture, Cell-based Assays, High-Throughput Screening, Immunology
Graphical abstract

Highlights
-
•
Steps for derivation of human monocyte-derived macrophages from peripheral blood
-
•
Detailed guide for efficient screening of ligands toward complement receptors in vitro
-
•
AlphaLISA-based detection of complement receptor-mediated ERK phosphorylation
-
•
Stepwise assay for agonist and antagonist testing toward C5a and C3a receptors
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
The complement receptors C3aR and C5aR1 are promising therapeutic targets. Here, we present a protocol to screen the effects of different agonists and antagonists on these receptors in vitro, using phosphorylated extracellular signal-regulated kinase (ERK) as a readout. We describe steps for isolating human monocyte-derived macrophages, culturing and preparing Chinese hamster ovary cells stably expressing human C5aR1 or C3aR, performing pharmacological assays, and detecting phospho-ERK1/2 in the cell lysate. This protocol can also be performed using other cell lines.
Before you begin
This protocol describes a cell-based screening assay to measure ERK1/2 phosphorylation as a readout for complement receptor activation. The protocol involves procedures performed by a human experimenter. These steps are, however, easily adaptable for automation.
Before you start, suitable cell models will need to be obtained for the experiment. This current protocol utilizes two transfected cell lines: Chinese hamster ovary (CHO) cells overexpressing human C5aR1 (CHO-C5aR1) or C3aR (CHO-C3aR) and the primary human monocyte-derived macrophages (HMDMs) isolated and cultured from human venous blood (procedures described below). However, choice of cell line and/or primary cell should be made based on individual requirements and availability.
We have successfully used the protocol in many cell types, with some examples being HEK293, CHO-K1, B16, THP-1, RAW264.7 and primary murine bone marrow derived macrophages. A suitable compound library can be prepared for screening (synthesized in house or obtained from external sources). We routinely use this protocol for small molecule peptidic and non-peptide compounds and larger sized molecules such as proteins and antibodies.
Institutional permissions (if applicable)
All studies involving human blood or primary human cell culture were approved by the University of Queensland Human Research Ethics Committee and the University of Queensland Biosafety Committee. Users should obtain appropriate approvals before working with human material.
Reagent preparation
Timing: 30 min (for step 1)
Timing: 30 min to days (for step 2)
-
1.
Preparation of buffers and culture reagents.
Prepare buffers for cell isolation, culturing and performing the assays:-
a.Ham’s F-12 Nutrient Mixture (F-12) (supplemented with 10% FBS, 1% Penicillin/Streptomycin).
-
b.Iscove’s Modified Dulbecco’s Medium (IMDM) (supplemented with 10% FBS, 1% Penicillin/Streptomycin, and 15 ng/ml recombinant human macrophage colony-stimulating factor (rhM-CSF)).
-
c.DPBS-based Separation Buffer (added to 2 mM EDTA and 0.5% BSA).
-
d.Serum free Ham’s F-12 (added to 0.1% BSA).
-
e.Serum-free IMDM (added to 0.1% BSA).
-
a.
-
2.
Preparation of compounds for screening.
For assay optimization/controls, obtain and utilize the following ligands:-
a.Suitable C5aR1 antagonist (e.g., PMX53, avacopan).
-
b.C3aR antagonist (e.g., SB290157).
-
c.Endogenous C5aR1 agonist human C5a.
-
d.Endogenous C3aR agonist human C3a.Note: For ligand selection, we refer readers to the IUPHAR/BPS Complement Peptide Receptor Guide to Pharmacology database: Complement peptide receptors in GtoPdb v. 2023.1.3For compound screening, relevant compounds libraries will need to be prepared (either synthesized in house or obtained from external sources). These can include smaller peptides or non-peptide compounds, or larger sized proteins or antibodies, which may demonstrate agonistic, antagonistic or other modulatory activities on C5aR1 and/or C3aR.Next, make up the stock solutions of the testing compounds. In general, a stock solution of 10 mM can be prepared in deionized water, PBS and/or DMSO (up to 100%). The stock concentration can be reduced if the solubility of the compound is low. However, the maximum testing concentration should be reduced accordingly to ensure the final DMSO concentration added to the cells does not exceed 0.5%.Note: Based our experience (with CHO and HMDM cells), adding a DMSO content of > 0.5% to the cells may reduce cell viability and/or ERK signaling, and should be avoided whenever possible. DMSO tolerance varies between cell types. We recommend to always include a vehicle-only control in the experiment (especially when the DMSO content in the final treatment solution exceeds 0.1%). Alternative solvents to DMSO can also be considered if required.
-
a.
Obtaining human blood samples
Timing: 30 min
In order to derive human monocyte-derived macrophages, human venous blood will need to be obtained. We utilize screened human buffy coat blood obtained through the Red Cross Blood Bank (Brisbane, Australia), which is donated by healthy human volunteers. Alternatively, anticoagulated human whole venous blood can be freshly collected and used, and the same protocol followed.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Buffy coat blood or venous whole blood from healthy human volunteers | Human donors | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant human C5a | Sino Biological | 10604-HNAE |
| Purified human C3a | Merck | US1204881 |
| C5aR1 antagonists (PMX53, PMX205, JPE-1375) | Synthesized in-house (Li et al., 2020) | https://doi.org/10.1016/j.bcp.2020.114156 |
| Bovine serum albumin | Sigma-Aldrich | A7906 |
| Trypsin-EDTA (0.5%), no phenol red | Thermo Fisher Scientific | 15400-054 |
| UltraPure 0.5 M EDTA, pH 8.0 | Thermo Fisher Scientific | 15575020 |
| Ham’s F12 | Thermo Fisher Scientific | 11765-062 |
| Iscove’s modified Dulbecco’s medium (IMDM) (with L-glutamine and HEPES) | Thermo Fisher Scientific | 12440053 |
| Fetal bovine serum (FBS), heat-inactivated | Thermo Fisher Scientific | 10082147 |
| Penicillin-Streptomycin (10,000 U/mL) | Thermo Fisher Scientific | 15140122 |
| G418 (Geneticin) | InvivoGen | ant-gn-5 |
| DPBS | Lonza | 181828 |
| 10× DPBS | Lonza | 181833 |
| Water for cell culture applications | Lonza | 181856 |
| Lymphoprep | STEMCELL Technologies | 07861 |
| CD14 microbeads | Miltenyi Biotec | 130-050-201 |
| Critical commercial assays | ||
| AlphaLISA SureFire Ultra p-ERK1/2 Assay Kit | PerkinElmer | ALSU-PERK-A10K |
| Experimental models: Cell lines | ||
| CHO-hC5aR1 | PerkinElmer | ES-731-C |
| CHO-hC3aR | PerkinElmer | ES-730-C |
| Software and algorithms | ||
| Prism 9 | GraphPad | https://www.graphpad.com/features |
| Other | ||
| Corning 96 well clear bottom TC-treated microplates | Merck | CLS3596 |
| ProxiPlate-384 Plus, white 384-shallow well microplate | PerkinElmer | 6008280 |
| SepMate PBMC isolation tubes | STEMCELL | 85450 |
| LS + positive selection column | Miltenyi Biotec | 130-042-401 |
| MidiMACS separator and multistand | Miltenyi Biotec | 130-042-302 |
| Sterilin 100 mm square Petri dishes | Thermo Fisher Scientific | 109-17TS |
| Corning 96 well storage microplate | Merck | CLS3363 |
| Falcon 40 μm cell strainer | Bio-Strategy | BDAA352340 |
| Falcon cell scraper with 25 cm handle and 1.8 cm blade | Bio-Strategy | BDAA353086 |
| Corning conical centrifuge tubes, sterile, 50 mL | Bio-Strategy | CORN430829 |
| Alpha assay-capable multiplate reader (e.g., Tecan Spark 20M) | Tecan | https://lifesciences.tecan.com/multimode-plate-reader |
| Automatic cell counter (e.g., LUNA-II automated cell counter by Logos Biosystems) | Logos Biosystems | https://logosbio.com/luna-ll/ |
| Suitable centrifuge (e.g., Beckman Coulter Allegra X-30 benchtop centrifuge) | Beckman Coulter | https://www.beckman.com/centrifuges/general-purpose/allegra-x-30# |
Materials and equipment
Separation buffer (pH 7.2)
| Reagent | Final concentration | Amount |
|---|---|---|
| EDTA (0.5 M) | 2 mM | 2 mL |
| BSA | 0. 5% | 2.5 g |
| 1 x DPBS | N/A | ∼ 490 mL |
| Total | N/A | 500 mL |
Note: Adjust the pH to 7.2 and sterile-filter the buffer. Store at 4°C for up to a month
Complete Ham’s F-12 medium for CHO cells
| Reagent | Final concentration | Amount |
|---|---|---|
| Heat-inactivated FBS | 10% | 50 mL |
| 100 X pen/strep | 1% | 5 mL |
| G418 (100 mg/mL) | 400 μg/mL | 2 mL |
| Ham’s F-12 nutrient mix | N/A | 443 mL |
| Total | N/A | 500 mL |
Note: Store at 4°C for up to one month and warm up to 37°C prior to use. Ham’s F-12 medium is prone to pH fluctuation. We recommend the medium to be made fresh and used promptly. Avoid prolonged cell handling at ambient conditions, and reduce exposure to air.
Complete IMDM medium for HMDMs:
| Reagent | Final concentration | Amount |
|---|---|---|
| Heat-inactivated FBS | 10% | 50 mL |
| 100 X pen/strep | 1% | 5 mL |
| Iscove’s Modified Dulbecco’s Medium (IMDM) (with L-glutamine and HEPES) | N/A | 445 mL |
| Total | N/A | 500 mL |
Note: Add an additional 15 ng/mL rhM-CSF directly to the cell solution when culturing. This commercially sourced IMDM contains 25 mM HEPES as an additive, which helps to stabilize the medium pH during cell handling at ambient conditions. Store at 4°C for up to one month, and warm up to 37°C prior to use.
Ligand diluents for assays:
-
•
IMDM / 0.1% BSA: add 50 mg BSA to a 50 mL conical tube and then top up to 50 mL with serum-free IMDM (with L-glutamine and HEPES), sterile-filter.
-
•
F-12 / 0.1% BSA: add 50 mg BSA to a 50 mL conical tube and then top up to 50 mL with serum-free Ham’s F-12 medium, sterile-filter.
Note: Ligand diluents should be stored at 4°C for up to 2 weeks, and warmed up to 37°C prior to use.
Step-by-step method details
Isolation and culturing of human monocyte-derived macrophages
Timing: 7 days
This section describes the procedure to isolate human CD14 monocytes from buffy coat blood and differentiate them in culture into monocyte-derived macrophages. Figure 1 provides an overview of the procedures. Please note: the separation buffer, cell culture water and 10× DPBS will need to be chilled to 4°C before commencing.
-
1.Isolating peripheral blood mononuclear cells using density centrifugation (Figure 1).
-
a.Empty 50 mL of buffy coat blood into a square 100 mm Sterilin dish.
-
b.Add 50 mL DPBS (1:1 dilution with blood), swirl to mix.
-
c.Add 15 mL Lymphoprep to each of four 50 mL SepMate PBMC Isolation Tubes.Note: SepMate tubes help to minimize the potential mixing between the Lymphoprep and blood layers during addition and centrifugation. These tubes are optional. A normal sterile 50 mL conical tube can also be used without compromising the experimental outcome. If smaller 15 mL conical/SepMate tubes are used instead, the volumes of blood and Lymphoprep should be reduced accordingly.
-
d.Gently layer 25 mL of blood/PBS mix onto Lymphoprep. Minimize any mixing between the blood and Lymphoprep layers.Note: This is best achieved using the gravity mode of an electric pipettor and leaning the serological pipet at an angle).
-
e.Transfer the SepMate tubes into a suitable centrifuge that allows for adjustable speeds (e.g., Beckman Coulter Allegra X-30 Benchtop Centrifuge).
-
f.Centrifuge (800 g) at 20°C–25°C for 30 min, with deceleration set to “Off” or “Slow”.Note: If a normal sterile conical tube is used instead of the SepMate tubes, the deceleration break must be set to “Off”.Note: After centrifuging, the blood samples will separate into distinct layers (Figure 2).
-
g.Remove the top plasma layer (∼12 mL) and discard, be careful not to disturb the white blood cell (peripheral blood mononuclear cell, PBMC) layer.
-
h.Transfer and combine the PBMC layers (from 4 SepMate tubes) into two sterile 50 mL conical tubes.Note: Avoid collecting the “red” layer as much as possible).
-
i.Wash the cell by adding ice-cold cell culture water to the 45 mL mark, invert to mix continuously for 1 min.Note: This will enable hypotonic lysis of contaminating red blood cells.
-
j.Quickly add 5 mL cold 10× DPBS and mix to restore tonicity.
-
k.Centrifuge at 800 g (10 min, 20°C–25°C).
-
l.Carefully pour off supernatant and discard.
-
m.Repeat Step i to l 2–3 times until no more red blood cells are visible in the pellet.
-
a.
-
2.Isolate CD14+ monocytes using magnetic separation.
-
a.Resuspend the cell pellet (in each of the 2 conical tubes) in 2.5 mL cold Separation Buffer
-
b.Break the cell pellet as much as possible by gently pipetting up and down.
-
c.Combine the resuspended cell pellets (5 mL in total).
-
d.Pass the combined cells through a cell strainer (40 μm) placed over a 50 mL conical tube.
-
e.Add a further of 10 mL Separation buffer to wash the filter for maximum cell recovery.
-
f.Count cells using a hemacytometer or automatic cell counter (e.g., LUNA-II Automated Cell Counter by Logos Biosystems).
-
g.Total cell number = cell concentration (per mL) 5 mL.
-
h.Determine the following volumes required:
-
i.Separation buffer volume: 80 μL/107 cells.
-
ii.CD14 microbeads volume: 20 μL/107 cells.
-
i.
-
i.Centrifuge cells (800 g) at 20°C–25°C for 6 min.
-
j.Resuspend the cell pellet in calculated volume of the separation buffer (from 2.h(i)).
-
k.To the cell solution, add the required volume of CD14 microbeads (from 2.h(ii)) (under dimmed lighting).
-
l.Cover the cell tubes with aluminum foil and incubate at 4°C for 15 min.
-
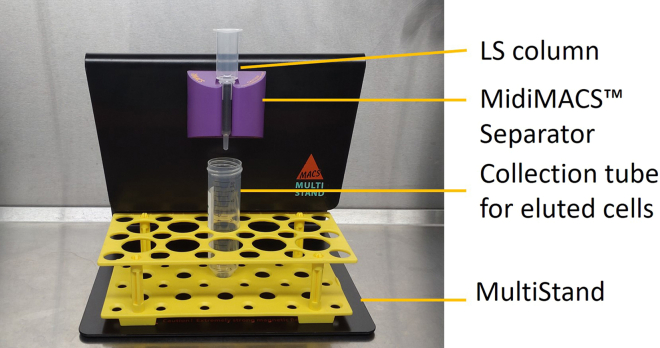
m.While waiting, set up a MACS separator.Note: Different MACS separators are available from the supplier with variable throughput (refer to manufacturer’s instructions for separator specific protocols).
-
n.Using MidiMACS as an example, insert a LS column into a MACS separator (Figure 3).Note: Ensuring the column is vertical and pushed in all the way.
-
o.Following 15 min incubation, add 30 mL of Separation Buffer to the conical tube containing labeled cells.
-
p.Gently pipette up and down to wash the cells.
-
q.Centrifuge cells (800 g) at 20°C–25°C for 5 min.
-
r.While waiting, prime the LS column using 3 mL of separation buffer.Note: A beaker/empty conical tube can be placed beneath the column to catch the flow-through.
-
s.After centrifugation, resuspend the cell pellet in Separation Buffer (500 μL per 108 cells, based on the counting result from 2.g).Note: Any cell clump can be removed by passing the cell solution through another 40 μm cell strainer. This is a necessary for samples with high cell numbers. Cell clumps can lead to blockade of LS columns.
-
t.Load the cell solution to the LS column. Any CD14+ cells will bind to the column, while the remaining cells will flow through.Note: According to the manufacturer’s specification (https://static.miltenyibiotec.com/asset/150655405641/document_5be6o7abnt1kd79gscorsrvd2t?content-disposition=inline), a maximum number of 108 labeled cells in up to 2 × 109 total cells can be handled by each column.
-
u.After all the cell solutions have flowed through, add 3 mL Separation Buffer to the column to wash the column.
-
v.Repeat the wash two more times to remove any residual non-CD14+ cells.
-
w.For elution, detach column from magnet and place over a clean sterile 50 mL conical tube.
-
x.Add 5 mL of separation buffer; attach the column plunger (included) to the column and firmly apply to immediately flush out labeled cells.
-
y.Count cells using a hemacytometer or automatic cell counter to determine the total cell number.Note: Trypan blue dye exclusion can be used to assess cell viability. However, some reversible membrane damage may result from the centrifugation and separation process, resulting in cells stained with trypan blue, despite remaining viable.4 As such, the number of viable cells may be under-estimated.
-
a.
-
3.Differentiate human CD14+ monocytes in culture into monocyte-derived macrophages.
-
a.Pellet the cells by centrifuging (800 × g, 5 min, 20°C–25°C).
-
b.Resuspend cells (∼1.2 × 106/ mL) in complete IMDM medium (supplemented with 10% FBS, 1% penicillin/streptomycin, and 15 ng/mL rhM-CSF).
-
c.Add cell solution to sterile 100 mm square Petri dishes (12 mL per dish). An approximate number of 1.2–1.5 × 107 can be added to each dish.Note: The seeding density can be adjusted based on the differentiation outcome.
-
d.Transfer the dishes to a cell culture incubator (37°C, 5% CO2) and culture for 7 days.Note: No medium change or supplementation is required during this period. However, if the culture medium is turning visibly acidic (in the later stages of differentiation, day 5–7), add additional fresh complete medium containing 15 ng/mL rhM-CSF (3 mL per dish).
-
a.
-
4.Harvesting of human monocyte-derived macrophages (HMDMs).
-
a.Harvest the cells when most of the cells attach to the plate (ranging from Day 5 to Day 7 post seeding).Note: Upon differentiation, HMDMs increase in size and display an irregular (“fried-egg”) morphology and adhere to the dish. This can be observed from Day 3 and continue up to Day 7. The rate of differentiation and the cell morphology varies between individuals, and significant variations in morphology are often expected between donors (Figure 4). The efficiency of differentiation varies between donors. In general, we obtain an efficiency between 30%-50% of plated cell number.
-
b.To harvest HMDMs, aspirate the medium containing non-adherent cells, and then gently add DPBS (5 mL/dish) to wash the adherent macrophages.
-
c.Upon removing DPBS, add 5 mL/dish complete IMDM medium (without rhM-CSF).
-
d.Gently lift the cells using a cell scraper.
-
e.Transfer the cell suspension into a sterile 50 mL conical tube, then add a further 5 mL to the dish to wash the residual cells off the dish.
-
f.Mix the cell solution by pipetting up and down, and then perform a cell count using a hemacytometer or automatic cell counter.Note: Once differentiated, we recommend harvesting the cells and using them for experiments within 3 days. Prolonged culture will reduce cell viability and cause changes in cell morphology and potentially function.
-
a.
Figure 1.
Schematic diagram showing the general processes in derivation of human monocyte-derived macrophages
Figure 2.
Density centrifugation separates the buffy coat blood into distinct layers
Figure 3.
Set up of MACS separation using a MidiMACS Separator from Miltenyi Biotec
Figure 4.
Examples of differentiated human macrophages from two separate donors
A clear difference in cell morphology can be seen between donors (inverted microscopy, 10×). Scale bars represent 20 μm.
Alternative: Culturing of transfected CHO cells stably expressing human C3aR or C5aR1
Timing: 1 h
This section describes the general culture and maintenance of commercially sourced CHO cells stably expressing human C5aR1 (CHO-C5aR1) or C3aR (CHO-C3aR).
-
5.
Maintain CHO cells (both CHO-C3aR and CHO-C5aR2) in complete Ham’s F-12 medium (supplemented with 10% FBS, 1% P/S, 400 μg/mL G418) in T75 or T175 cell culture flasks.
-
6.
Passage the cells routinely when they reach 80%–90% confluency.
Note: In general, for both CHO-C5aR1 and CHO-C3aR, perform a 1:5 passage every two days or 1:10 passage every 3 days.
-
7.The general procedure of cell passage in a T75 flask is as follows:
-
a.Remove cell medium by aspirating.
-
b.Add 4 mL DPBS to gently wash the cells, discard.
-
c.Add 4 mL of 0.05% Trypsin-EDTA. Incubate the flask at 20°C–25°C for 5 min to detach the cells.
-
d.After incubation, add 6 mL of complete F-12 medium to neutralize the trypsinNote: the FBS in the medium will stop Trypsin from further action)
-
e.Transfer 2 mL of the cell mixture to a new T75 flask containing 10 mL fresh F-12 medium (i.e., 1:5 split). The remaining (8 mL) cell solution can be discarded or seeded into microplates for assays.Note: G418 is used as a selection antibiotic to maintain C3aR and C5aR1 expression in CHO cells. The identity of the required antibiotic varies between expression plasmids and systems (check with the manufacturer for specifics). If the cells are to be seeded for assays with no passaging required, after Step 3.d., the cell solution can be counted using a hemacytometer or automatic cell counter and used.
-
a.
Seeding of CHO cells or HMDMs for assays
Timing: 30 min to 1 h
This step describes seeding cells into 96-well microplates for assay.
-
8.
Using cell solutions obtained in the above sections (refer to HMDMs—Step 4.f., CHO cells—Step 3.d), perform a cell count using a hemacytometer or automated cell counter (with trypan blue dye exclusion to account for viable cells only). Then, pellet the cells by centrifugation (400 × g, 20°C–25°C, 5 min).
-
9.
Resuspend the cell pellet in complete F-12 medium (for CHO cells) or complete IMDM medium (for HMDMs, supplemented 15 ng/mL rhM-CSF).
-
10.
We recommend using the following cell concentrations for plating: 500,000 cells/ mL for HMDMs, 400,000 cells/ mL for CHO-C5aR1 and 500,000 cells/ mL for CHO-C3aR.
-
11.
Use a multichannel pipette to transfer 100 μL cells/ well into a tissue-treated clear-bottom 96-well plate. This will give a final concentration of 50,000 cells/ well for CHO-C3aR and HMDMs, and 40,000 cells/well for CHO-C5aR1.
Note: The optimal seeding density varies between cell types and will need to be optimized. In general, for an immortal dividing cell line, seeding cells at full confluency encourages contact inhibition, helps to dampen background ERK signaling and thus reduces well-to-well variations.
-
12.
Incubate the plates at 37°C, 5% CO2 for 24 h for the cells to settle.
Serum starvation of seeded HMDMs or CHO cells
Timing: 30 min
This step describes serum starvation of seeded cells (CHOs or HMDMs).
-
13.
After the cells have adhered to the plate, use a multichannel pipette or vacuum aspirator to gently remove the complete culture medium, avoiding disturbing the cell monolayer.
-
14.
Add 100 μL per well of serum-free IMDM (for HMDM) or F-12 (for CHO-C5aR1/C3aR). No additional supplements are required for these media.
-
15.
Incubate at 37°C/5% CO2 for 8–16 h.
Note: Serum starvation helps to reduce background ERK signaling and reduce potential interference by other proteins in the serum. The optimal duration of serum starvation will need to be empirically determined for each cell type. In general, a serum-starvation duration of 6–18 h is effective for most cell types. A minimum duration of 4-h starvation can be used for HMDMs and CHO cells. As a longer seeding + starvation time may also increase well-to-well variation due to possible edge effects and differential cell proliferation (for CHO cells),5 we do not recommend keeping cells in serum-free medium for longer than 24 hrs.
Performing the pharmacological assay
Timing: 30 min to 1.5 h
This section describes the general steps involved in performing the pharmacological assay. This includes preparation of ligand dilutions, pre-treatment with inhibitors, stimulation with agonists and cell lysate preparation. The overall assay scheme is shown in Figure 5.
Figure 5.
Schematic diagram showing the general procedure of AlphaLISA SureFire Ultra p-ERK 1/2 (Thr202/Tyr204) assay conducted in adherent cells
Importantly, this protocol describes the general procedure only of ligand testing using the phospho-ERK1/2 assay; the specific ligands used will depend on the specific aim of the experiment: whether it is conducted in CHO-C3aR/C5aR1 or HMDMs, to test for agonists or antagonists.
-
16.
Determine the suitable agonist/antagonist to be used for the assay.
Note: We have successfully utilized this assay to a wide range of ligands to measure agonist/antagonistic activities on CHO-C5aR1, CHO-C3aR and HMDMs, among others. The details can be found in our prior publications.1,2,6,7,8,9,10,11,12,13 Based on our work, we recommend the following ligand concentrations and treatment schemes in CHO cell lines (Table 1). Any hits can then be progressed for validation in HMDMs. However, HMDMs express a multitude of receptors (both C3aR and C5aR1, among others). As such, it is important to verify in HMDMs, whether the induced ERK signaling is driven through the receptor of interest, like C5aR1 or C3aR (Table 2).
-
17.Prepare ligand dilutions and set up a source plate.
-
a.Warm up the dilution buffer (for CHO cells: serum-free F-12 + 0.1% BSA; for HMDMs: IMDM + 0.1% BSA), which is used to prepare serial dilutions for all ligands.Note: 0.1% BSA is added to the diluent as a stabilizer, which helps to minimize ligand loss due to nonspecific binding to the microplate (this is especially important for C3a and C5a). The requirement of additional stabilizer varies depending on testing ligands. 0.5% gelatin can also be used in place of BSA.14
-
b.Prepare all ligands at the final concentration (1×) to be added to the cells.Note: For a 96-well plate, triplicates of 50 μL/well is typically used for the assay. Taking into account the dead volume, we recommend having at least 180 μL per concentration/condition.
-
c.If required, prepare serial dilutions for the test ligands.Note: For example, prepare a working solution of 10 μM as the highest concentration, and then serially dilute 1/10 by sequentially transferring 20 μL of ligand solution to 180 μL of diluent for the next concentration. Pipette up and down to mix well following each transfer.Note: The highest concentration of ligand tested will need to be adjusted based on the ligand solubility and maximum DMSO content allowed (Refer to “Preparation of compounds for screening” above).
-
d.Transfer all dilutions into a 96 well polypropylene V-bottom source plate and pipette out in layout used for assay.Note: Alternatively, all dilutions can be directly performed in the V-bottom source plate using a multichannel pipette for automation and/or increasing throughput.
-
e.Cover the plate with lid or sealing film to minimize evaporation.Note: The above serial dilution procedure can be used for both agonist and antagonist testing. If pre-treatment with inhibitors is performed, agonists (e.g. C5a or C3a) will need to be prepared at 10 X concentration and added on top of the inhibitors. For instance, to screen for C3a inhibitors, a 10 X concentration (3 nM) of recombinant/purified human C3a (1×: 0.3 nM, 10×: 3 nM) can be prepared and used to stimulate the cells following inhibitor pretreatment. We recommend using EC80-90 concentrations of agonist to give sufficient assay window whilst minimizing the deviation of IC50 values from Ki of the compound (Table 3).15,16
-
a.
-
18.Pre-treat cells with antagonists (if applicable).
-
a.Take out the cell plate from the incubator, use a vacuum aspirator or multichannel pipette to gently remove the serum-free medium, avoid touching the cell monolayer.
-
b.Use a manual or electronic multichannel pipettor to add 45 μL/well of known inhibitor or testing compound (from the source plate) into the cell plate.
-
c.Return the plate to the tissue culture incubator and incubate at 37°C for 30 min.
-
a.
Note: A 30-min pre-treatment is generally sufficient for small-molecule and protein/antibody testing ligands. This duration can be altered if the binding kinetics of the testing compound is known to be slower/faster.
-
19.Stimulate cells with agonists.
-
a.For an inhibition assay, where pretreatment with inhibitors has been conducted (Step 2 above): directly add 5 μL/well of 10 X agonist (C5a or C3a) to the cells, without removing the pretreatment solution.
-
b.For agonist assay (no pretreatment performed): use a vacuum aspirator or multichannel pipette to gently remove the serum-free medium. Then, transfer 50 μL/ well of the testing agonist solution to the cells.
-
c.Return the cell plate to the incubator (37°C, 5% CO2) and incubate for 10 min (stimulation).Note: This step can also be performed at 20°C–25°C.
-
d.While waiting, prepare 1 X Ultra lysis buffer by mixing 1 part of 5 X Ultra lysis buffer (supplied in the kit) to 4 volumes of deionized water, invert to mix.
-
e.Upon 10 min stimulation, take out the cell plate from the incubator.
-
f.Quickly remove the ligand treatment solution using a multichannel pipette or vacuum aspirator and then add 50 μL /well of 1× lysis buffer.
-
g.Place the plate on an orbital shaker and shake for 15 min (450 rpm, 20°C–25°C).
-
h.The plate can now be used for detection in the next step.Note: Alternatively, for detection at a later time. The plate can be sealed using a plate sealer or parafilming the sides and storing −20°C or below.
 CRITICAL: Anaphylatoxin mediated pERK signaling is highly time sensitive,17 we recommend obtaining all the required equipment, reagents, ligands and buffers before commencing the assay. It is important to keep the stimulation duration consistent across experimental repeats.
CRITICAL: Anaphylatoxin mediated pERK signaling is highly time sensitive,17 we recommend obtaining all the required equipment, reagents, ligands and buffers before commencing the assay. It is important to keep the stimulation duration consistent across experimental repeats.
-
a.
Table 1.
Screening of ligand (X) activity in CHO-C5aR1/C3aR cells
| Cells/ receptor | To screen for | Treatment scheme |
|---|---|---|
| CHO-C5aR1 | Agonist | Directly stimulate cells with ligand X Positive control: hC5a (e.g., 100 nM) |
| Antagonist | Pretreat cells with ligand X, then stimulate with 0.3 nM hC5a (∼EC80) | |
| CHO-C3aR | Agonist | Directly stimulate cells with ligand X Positive control: hC3a (e.g., 100 nM) |
| Antagonist | Pretreat cells with ligand X, then stimulate with 0.3 nM hC3a (∼EC80) | |
| Counter-screening: Can similar effect be observed on other receptors? | ||
Note: Human C5aR1 and C3aR demonstrate high degree of structural similarity and ligand promiscuity,13 as such, we recommend all potential hits for human C5aR1 to be counter-screened against human C3aR and vice versa. This test should be performed first prior to other more comprehensive selectivity screens.
Table 2.
Validation of ligand activity in HMDMs
| Receptor of interest | Validation method | |
|---|---|---|
| Agonist | C5aR1 | Pre-treat cells with PMX53 (10 μM), does it block ligand X induced ERK signaling? |
| C3aR | Pre-treat cells with SB290157 (10 μM), does it block ligand X induced ERK signaling? | |
| Antagonist | C5aR1 | Pre-treat cells with ligand X, does it block hC5a (1 nM) induced ERK signaling? |
| C3aR | Pre-treat cells with ligand X, does it block hC3a (5 nM) induced ERK signaling? |
Note: Alternative specific C5aR1 inhibitors (e.g., Avacopan, PMX205, JPE-1375, etc.) can also be used. A list of potent and commonly used C5aR1 inhibitors can be found in our previous publication1 and the IUPHAR/BPS Complement Peptide Receptor Guide to Pharmacology database.3 SB290157 is the only commercially-available C3aR inhibitor and has acknowledged off-target activity,2,18 however, in HMDMs at this concentration, it acts as a full-antagonist for C3aR.2,19
Table 3.
Recommend agonist concentrations to be used for inhibitor screening
| Cell type/Target receptor | Endogenous agonist | Concentration (1 X) |
|---|---|---|
| CHO-C5aR1 | hC5a | 0.3 nM |
| CHO-C3aR | hC3a | 0.3 nM |
| HMDM (C5aR1) | hC5a | 1 nM |
| HMDM (C3aR) | hC3a | 5 nM |
Detection of phosho-ERK1/2 in cell lysate
Timing: 1 h (addition of reagents) to 17 h (incubation)
This section describes the detection of phosphorylated ERK1/2 in the cell lysate using the AlphaLISA SureFire Ultra p-ERK1/2 (Thr202/Tyr204) Assay Kit.
-
20.
If the samples had been frozen, thaw the cell lysate by gently shaking the plate on an orbital shaker.
Note: Allow 30 min to ensure all samples have been completely thawed and warmed to 20°C–25°C before commencing detection.
-
21.
Calculate the volume of the reaction mixes (2.5 μL per sample) needed for the assay. The required volumes can be calculated as:
Note: We have halved the per well reaction volume of this assay from that specified in the manufacturer’s protocol (https://resources.perkinelmer.com/lab-solutions/resources/docs/MAN_AlphaLISA_SureFire_Ultra.pdf). Based on our experience, this does not compromise the assay performance in a 384-well ProxiPlate. However, if other microplate types (e.g., standard 384 Opti-plate) or a one-plate protocol are to be used, we recommend using the assay volume as per the manufacture’s recommendations.
-
22.
Prepare AlphaLISA acceptor and donor mixes (under dimmed lighting), as per Table 4.
Note: Store the reaction mixes at 20°C–25°C while performing the sample transfer.
-
23.
Use a multichannel pipette to transfer 5 μL of cell lysate from each well of the 96-well cell plate to a 384-well ProxiPlate.
Note: This protocol adopts a 2-plate assay scheme (cell treatment and detection are conducted in two separate plates). If a 1-plate protocol is preferred instead (i.e. both cell treatment and detection are performed in the same plate), this step of lysate transfer can be omitted.
-
24.
Prepare the detection assay control samples as follows and add to the ProxiPlate in triplicates: negative control - 1× lysis buffer only, positive control - phospho-ERK positive control lysate (supplied by the kit).
-
25.
Add 2.5 μL/well of Acceptor mix followed by 2.5 μL/well of the Donor mix.
Note: The reaction mixes should be added separately in the sequence of Acceptor mix then Donor mix. The reaction mixes should not be mixed before adding to the cells.
-
26.
Gently tap the plate to ensure proper mixing.
-
27.
Seal plate with TopSeal-A plate seal and cover with aluminum foil to protect from light.
-
28.
Incubate the plate for at least 2 h (up to 16 h) at 20°C–25°C.
Note: Based on our experience, 2-h incubation is generally sufficient for CHO cells, for HMDMs, we recommend incubating the plate for at least 3 h (up to 16 h) before reading.
-
29.
Without removing the TopSeal-A film, place the plate in an Alpha-capable plate reader (e.g., a Tecan Spark M20).
Note: Allow the plate to equilibrate to the plate reader temperature for at least 5 min prior to reading.
-
30.
Read the plate using standard AlphaLISA settings.
Note: If the instrument does not have dedicated settings for AlphaLISA, a AlphaScreen script can be modified and used by changing the detection wavelength bandwidth to be centered around 615 nm.
Table 4.
Composition of the AlphaLISA reaction mixes
| Acceptor mix components | For 100 μL | Donor mix components | For 100 μL |
|---|---|---|---|
| Reaction buffer 1 | 47 μL | Dilution buffer | 98 μL |
| Reaction buffer 2 | 47 μL | Donor bead | 2 μL |
| Activation buffer | 4 μL | ||
| Acceptor bead | 2 μL |
Expected outcomes
The phospho-ERK1/2 signal (measured by counts per second) varies widely between cell types, receptors, incubation time, instrument used, detection temperature, among other variables. Figure 6A shows a sample measurement of a kit-supplied lysis buffer (negative control) and positive control lysate, where a signal-to-noise ratio of ∼1600-fold can be seen.
Figure 6.
Sample data and analysis for p-ERK1/2 assay performed in CHO-C5aR1 and HMDM cells
(A) Sample data obtained with AlphaLISA SureFire Ultra p-ERK 1/2 (Thr202/Tyr204) assay kit (lysis buffer as a negative control; positive control lysate supplied by the kit). The means of triplicate measurements are shown above the bars (mean + S.E.M.).
(B) Representative raw data for C5a and the linear peptidic agonist EP54 inducing p-ERK1/2 activity in CHO-C5aR1 cells.
(C) The data in B was normalized to the top (100%) and bottom (0%) values of C5a-induced activity and re-plotted.
(D) Representative data of an inhibitor assay performed in HMDMs using the C5aR1 inhibitors PMX205 and JPE-1375 (30-min pre-incubation followed by 10-min stimulation with 1 nM hC5a). All data are triplicate measurements performed in a single assay (mean ± S.E.M.).
All cells possess some degree of background ERK signaling, which is typically higher in immortal proliferative cell lines (e.g., CHO cells) relative to terminal primary cells (e.g., HMDMs). As such, the expected signal-to-noise ratio varies between cell lines. Representation data for CHO-C5aR1 (Figures 6B and 6C) and HMDMs (Figure 6D) are shown, which will be further discussed in the next section.
Quantification and statistical analysis
The detailed method used for data analysis is experiment-specific. As a general guide for pharmacological screening assays, we recommend performing at least 3 repeats (on separate occasions/days) of triplicates to confidently determine the ligand activities. In the following sections, we briefly describe some examples of data processing/analysis using the software GraphPad Prism 9. Figure 6 shows sample results obtained in CHO-C5aR1 and HMDMs, for activation or inhibition assays.
Determine the percentage of activation/inhibition of compounds following single point screening
-
1.
Plot all data using a column graph table format.
-
2.Perform the following normalization:
-
a.For agonists:
-
i.Normalize all ligand-induced signals to the maximum endogenous agonist (e.g., C5a or C3a)-induced response (100%) and vehicle only-induced response (0%).
-
ii.Determine the percentage of activation (100% indicates full activation).Note: A full agonist or agonist of interest can also be used in place of the endogenous agonist during normalization.
-
i.
-
b.For inhibitors:
-
i.Normalize the data to the agonist only-induced response (100%, e.g., 0.3 nM for CHO-C5aR1), and the serum-free medium only response (0%).
-
ii.Compute the percentage of inhibition (0% indicates full inhibition).
-
i.
-
a.
-
3.
If needed, progress ligands that demonstrate receptor activation or inhibition above a predetermined threshold (e.g., 50%) onto the next stage of compound testing (derivation of a dose-response curve and determination of EC50 or IC50 values).
Determine the dose-response curve of active compounds
-
4.
Plot a dose-response curve of the compounds of interest using a data handling software (e.g., GraphPad Prism or Excel).
Note: We suggest labeling the y-axis to be Alpha signal and the x-axis to be Log ligand concentration.
-
5.
Fit a non-linear regression curve (three parameter curve recommended).
-
6.
Use the best-fit non-linear regression curve to determine the EC50 (for agonists) or IC50 values (for antagonists).
Note: For illustration, Figure 6B shows the non-linear regression analysis of an agonist assay performed in CHO-C5aR1 cells, and Figure 6D shows the result of an inhibition assay performed in HMDMs.
Combining data across multiple repeats
-
7.
For combining data between different experimental repeats, normalize the results from each repeat to the respective agonist only-induced response (100%) and vehicle only induced response (0%).
Note: For illustration, an example of such normalized data is shown in Figure 6C.
-
8.
Combine the data from different repeats as needed.
Limitations
This protocol is written based on CHO cells and HMDMs using a “two-plate” assay protocol. Further optimization is needed to adapt this protocol into a one-plate protocol or for suspension cells. In addition, this protocol does not include a control for total ERK expression in the cells. Total ERK expression can be used to help control for any changes in cell number or major changes in cell functionality. We recommend measuring total ERK expression when setting up new experiments or working with ligands that may induce significant cell proliferation/death. This can be done using AlphaLISA SureFire Ultra Total ERK 1/2 Assay Kit to measure the level of total ERK in the same cell lysate.
Troubleshooting
Problem 1
During passing PBMCs through the LS column (Step 2.t-v, Isolate CD14+ monocytes using magnetic separation) the flow rate of the cell solution through the LS column is very low. The typical flow rate of cells through the column is ∼1 mL/min. In some cases, the flow rate may become too slow or the column is blocked.
Potential solution 1
-
•
According to the manufacturer’s specification (https://static.miltenyibiotec.com/asset/150655405641/document_5be6o7abnt1kd79gscorsrvd2t?content-disposition=inline), a maximum number of 10⁸ labeled cells in up to 2 × 109 total cells can be handled by each column. Depending on the number of PBMCs, two or more columns may need to be used per buffy coat sample to avoid overloading the column
-
•
If the column is blocked, remove the LS Column from the separator and place it on a new collection tube. Pipette 5 mL of Separation buffer onto the LS Column, and then immediately flush out the fraction with the magnetically labeled cells by firmly applying the plunger supplied with the column. Filter the cell solution by passing through a 40 μm cell strainer to eliminate any cell clumps.
-
•
Insert a new column into the magnet, and then reload the cell solution onto the new column.
Problem 2
Significant cell death observed within 24 h after plating isolated CD14 monocytes into Sterilin dishes (see Differentiate human CD14+ monocytes in culture into monocyte-derived macrophages).
Potential solution 2
This is likely caused by significant cell injury and/or culture contamination incurred during Isolate CD14+ monocytes using magnetic separation. This can be resolved by gentler cell handling (e.g., reduce the duration and times of pipetting during cell resuspension and prevent the formation of bubbles), as well as adopting strict sterile techniques. In addition, human primary monocytes/ macrophages are sensitive cells, quality IMDM medium, FBS and rhM-CSF should be used to culture these cells.
Problem 3
During Serum starvation of seeded HMDMs or CHO cells, HMDMs from some donors dislodge following medium change.
Potential solution 3
The issue with cell dislodgement can be improved by adopting gentler pipetting techniques during medium change. In addition, based on our experience, HMDMs from some donors may need longer time to securely adhere to the plate. As such, during Seeding of CHO cells or HMDMs for assays, a resting time of up to 48 h can be used for HMDMs.
Problem 4
For the detection of phosho-ERK1/2 in cell lysate, the AlphaLISA signals obtained with some cell types are very low. For example, the highest counts per second measurement is below 5,000, versus the 100,000 we may usually expect. This is less common with HMDMs and CHO cells, but can be seen in hard-to-lyse cells (e.g., primary human neutrophils) or cells with low intrinsic ERK signaling.
Potential solution 4
The following protocol modifications can be trialed.
-
•
Seed cells at a higher density. This will enable a more concentrated lysate to be generated.
-
•
Allow a longer incubation time (up to 16 h) for the detection step.
-
•Follow a “2-addition” detection protocol (instead of the 1-addition as depicted in Step 25), which can help to improve the detection sensitivity.
-
○Add Acceptor mix to the cell lysate.
-
○Cover the plate using TopSeal-A and incubate at 20°C–25°C for 1 h.
-
○Add donor mix to the plate, incubate the plate at 20°C–25°C for 2 h up to 16 h.
-
○
-
•Perform additional controls to check for the presence of a “Hook effect”.
-
○The Hook effect occurs when the beads are saturated with analyte. Excess analyte disrupts associations between Donor and Acceptor beads beyond the hook point, and thus causing a reduction in Alpha signal.20
-
○This can be controlled for by diluting the cell lysate 1/10th and observing if the signal increases.
-
○
Problem 5
For the detection of phosho-ERK1/2 in cell lysate, the AlphaLISA signals obtained are highly variable across replicates.
Potential solution 5
The following steps/precautions can be taken.
-
•
Significant variations are observed between replicate measurements across the whole plate: this can be caused by inconsistent cell lysis. The lysis outcome can be improved by increasing the lysing time (Step 16.g) to 30 min. For hard-to-lyse cells (e.g., primary human neutrophils), freeze-thawing the cell plate may help to liberate the phosphorylated ERK into the solution and create a more homogenous cell solution, and thereby reducing the background noise.
-
•
Significant variations are observed mainly along the edges of the 96-well plate: this is likely caused by non-uniform cell growth, which is particularly prominent for fast proliferative cells such as CHO cells. To improve this, the edges of the 96-well plate can be avoided when seeding the cells during Seeding of CHO cells or HMDMs for assays.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Prof Trent Woodruff (t.woodruff@uq.edu.au).
Materials availability
This study did not generate any unique materials or reagents.
Data and code availability
This study did not generate any unique datasets or code.
Acknowledgments
This work was supported by the National Health and Medical Research Council (grant 2009957 to T.M.W.).
We would like to acknowledge Australian Red Cross Lifeblood and human donors for providing the cells used in these studies.
Selected figures were created using BioRender.com.
Author contributions
Conceptualization and visualization, T.M.W. and X.X.L.; methodology, data acquisition, and writing – initial draft, X.X.L.; funding acquisition and writing – review and editing, T.M.W.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Xaria X. Li, Email: xaria.li@uq.edu.au.
Trent M. Woodruff, Email: t.woodruff@uq.edu.au.
References
- 1.Li X.X., Lee J.D., Massey N.L., Guan C., Robertson A.A.B., Clark R.J., Woodruff T.M. Pharmacological characterisation of small molecule C5aR1 inhibitors in human cells reveals biased activities for signalling and function. Biochem. Pharmacol. 2020;180 doi: 10.1016/j.bcp.2020.114156. [DOI] [PubMed] [Google Scholar]
- 2.Li X.X., Kumar V., Clark R.J., Lee J.D., Woodruff T.M. The "C3aR Antagonist" SB290157 is a Partial C5aR2 Agonist. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.591398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cianciulli A., Coulthard L., Hawksworth O., Lee J.D., Li X.X., Mitolo V., Monk P., Panaro M.A., Woodruff T.M. 2023. Complement Peptide Receptors in GtoPdb V. 2023.1. IUPHAR/BPS Guide to Pharmacology CITE 2023. [Google Scholar]
- 4.Tran S.L., Puhar A., Ngo-Camus M., Ramarao N. Trypan blue dye enters viable cells incubated with the pore-forming toxin HlyII of Bacillus cereus. PLoS One. 2011;6 doi: 10.1371/journal.pone.0022876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mansoury M., Hamed M., Karmustaji R., Al Hannan F., Safrany S.T. The edge effect: A global problem. The trouble with culturing cells in 96-well plates. Biochem. Biophys. Rep. 2021;26 doi: 10.1016/j.bbrep.2021.100987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X.X., Lee J.D., Lee H.S., Clark R.J., Woodruff T.M. TLQP-21 is a low potency partial C3aR activator on human primary macrophages. Front. Immunol. 2023;14:1086673. doi: 10.3389/fimmu.2023.1086673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gorman D.M., Li X.X., Lee J.D., Fung J.N., Cui C.S., Lee H.S., Rolfe B.E., Woodruff T.M., Clark R.J. Development of Potent and Selective Agonists for Complement C5a Receptor 1 with In Vivo Activity. J. Med. Chem. 2021;64:16598–16608. doi: 10.1021/acs.jmedchem.1c01174. [DOI] [PubMed] [Google Scholar]
- 8.Xu W., Kumar V., Cui C.S., Li X.X., Whittaker A.K., Xu Z.P., Smith M.T., Woodruff T.M., Han F.Y. Success in Navigating Hurdles to Oral Delivery of a Bioactive Peptide Complement Antagonist through Use of Nanoparticles to Increase Bioavailability and in Vivo Efficacy. Advanced Therapeutics. 2022;5 [Google Scholar]
- 9.Pandey S., Li X.X., Srivastava A., Baidya M., Kumari P., Dwivedi H., Chaturvedi M., Ghosh E., Woodruff T.M., Shukla A.K. Partial ligand-receptor engagement yields functional bias at the human complement receptor. J. Biol. Chem. 2019;294:9416–9429. doi: 10.1074/jbc.RA119.007485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brennan F.H., Jogia T., Gillespie E.R., Blomster L.V., Li X.X., Nowlan B., Williams G.M., Jacobson E., Osborne G.W., Meunier F.A., et al. Complement receptor C3aR1 controls neutrophil mobilization following spinal cord injury through physiological antagonism of CXCR2. JCI Insight. 2019;4 doi: 10.1172/jci.insight.98254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorman D.M., Li X.X., Payne C.D., Cui C.S., Lee J.D., Rosengren K.J., Woodruff T.M., Clark R.J. Development of Synthetic Human and Mouse C5a: Application to Binding and Functional Assays In Vitro and In Vivo. ACS Pharmacol. Transl. Sci. 2021;4:1808–1817. doi: 10.1021/acsptsci.1c00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X.X., Clark R.J., Woodruff T.M. C5aR2 Activation Broadly Modulates the Signaling and Function of Primary Human Macrophages. J. Immunol. 2020;205:1102–1112. doi: 10.4049/jimmunol.2000407. [DOI] [PubMed] [Google Scholar]
- 13.Li X.X., Clark R.J., Woodruff T.M. Anaphylatoxin receptor promiscuity for commonly used complement C5a peptide agonists. Int. Immunopharmacol. 2021;100 doi: 10.1016/j.intimp.2021.108074. [DOI] [PubMed] [Google Scholar]
- 14.Chenoweth D.E., Goodman M.G., Weigle W.O. Demonstration of a specific receptor for human C5a anaphylatoxin on murine macrophages. J. Exp. Med. 1982;156:68–78. doi: 10.1084/jem.156.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyllie D.J.A., Chen P.E. Taking The Time To Study Competitive Antagonism. Br. J. Pharmacol. 2007;150:541–551. doi: 10.1038/sj.bjp.0706997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neale P.A., Leusch F.D.L. Considerations when assessing antagonism in vitro: Why standardizing the agonist concentration matters. Chemosphere. 2015;135:20–23. doi: 10.1016/j.chemosphere.2015.03.054. [DOI] [PubMed] [Google Scholar]
- 17.Seow V., Lim J., Iyer A., Suen J.Y., Ariffin J.K., Hohenhaus D.M., Sweet M.J., Fairlie D.P. Inflammatory responses induced by lipopolysaccharide are amplified in primary human monocytes but suppressed in macrophages by complement protein C5a. J. Immunol. 2013;191:4308–4316. doi: 10.4049/jimmunol.1301355. [DOI] [PubMed] [Google Scholar]
- 18.Lee J.D., Taylor S.M., Woodruff T.M. Is the C3a receptor antagonist SB290157 a useful pharmacological tool? Br. J. Pharmacol. 2020;177:5677–5678. doi: 10.1111/bph.15264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rowley J.A., Reid R.C., Poon E.K.Y., Wu K.-C., Lim J., Lohman R.-J., Hamidon J.K., Yau M.-K., Halili M.A., Durek T., et al. Potent Thiophene Antagonists of Human Complement C3a Receptor with Anti-Inflammatory Activity. J. Med. Chem. 2020;63:529–541. doi: 10.1021/acs.jmedchem.9b00927. [DOI] [PubMed] [Google Scholar]
- 20.Yasgar A., Jadhav A., Simeonov A., Coussens N.P. In: High Throughput Screening: Methods and Protocols. Janzen W.P., editor. Springer New York; 2016. AlphaScreen-Based Assays: Ultra-High-Throughput Screening for Small-Molecule Inhibitors of Challenging Enzymes and Protein-Protein Interactions; pp. 77–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate any unique datasets or code.