Summary
Here, we present a protocol for setting three spectral flow cytometry panels for the characterization of human unconventional CD8+NKG2A/C+ T cells as well as other T and natural killer cell subsets. We describe steps for standardizing, preparing, and staining the cells, the experimental setup, and the final data analysis. This protocol should be advantageous in various settings including immunophenotyping of limited samples, immune function evaluation/monitoring, as well as research in oncology, autoimmune, and infectious diseases.
Subject areas: Flow Cytometry, Immunology, Antibody
Graphical abstract

Highlights
-
•
Protocol for deep phenotyping of human CD8+ T cells expressing NKG2A/NKG2C
-
•
Design and optimization of three spectral flow cytometry panels
-
•
Preparation, staining, and analysis of human PBMCs with spectral flow cytometry
-
•
Identification and analysis of NK, classical, and unconventional T cell populations
Publisher’s note: Undertaking any experimental protocol requires adherence to local institutional guidelines for laboratory safety and ethics.
Here, we present a protocol for setting three spectral flow cytometry panels for the characterization of human unconventional CD8+NKG2A/C+ T cells as well as other T and natural killer cell subsets. We describe steps for standardizing, preparing, and staining the cells, the experimental setup, and the final data analysis. This protocol should be advantageous in various settings including immunophenotyping of limited samples, immune function evaluation/monitoring, as well as research in oncology, autoimmune, and infectious diseases.
Before you begin
Rationalization and design of the spectral flow cytometry panels
CD8+ T cells expressing inhibitory NKG2A (CD159a) and activating NKG2C (CD159c) molecules (ligands of the non-classical molecule HLA-E) have been associated with several inflammatory conditions, such as inflammatory gut diseases, tumors, aging, autoimmune and infectious diseases.1,2,3,4,5,6,7,8,9,10,11 Previous data from our laboratory demonstrated that CD8+NKG2A/C+ T cells expressing cytotoxic and regulatory profiles expand in the gut of non-human primates (NHP) that naturally control SIV-induced disease as compared to those who develop chronic inflammation and disease.12
The main objective of this protocol is to phenotype human CD8+ T cells expressing inhibitory NKG2A (CD159a) and activating NKG2C (CD159c) markers amongst different classical T and natural killer (NK) cell subsets, and to distinguish them from other unconventional T cells such as yδ T, invariant natural killer T (iNKT), and mucosal-associated invariant T (MAIT) cells. These populations have been studied in the context of infectious, autoimmune diseases, allergy and cancer, are found in the blood at low frequencies as compared to the other classical CD8+ and CD4+ T cell major populations, have unique dynamics, and can express CD94, NKG2A, and NKG2C.13,14,15,16,17,18,19,20,21,22,23 We selected markers to separate all these populations and outlined a gating strategy for discriminating given cell subsets from each other.
Spectral flow cytometry allows the unambiguous simultaneous analysis of a high number of markers, and reduce the number of cells needed for a thorough phenotyping analysis. We built and standardized three panels: two focused on CD8+ T cells and one on CD4+ T cells. Based on literature reviews, and our own data from human donors and non-human primates (NHP), we selected markers of interest associated with the study of diverse features (homing, regulatory, cytotoxicity, exhaustion, etc.). We incorporated classical markers of differentiation (CD27, CCR7, CD45RA, and CD45RO), which is important for the study of T cell dynamics. Furthermore, we aimed to study molecules associated with the activity of NKG2A and NKG2C (such as the non-classical HLA-E), as well as other molecules expressed in additional T cell subsets of interest such as regulatory T cells (Treg) (such as CD4+CD25+FOXP3+ T cells), T helper 17 cells (Th17), and circulating T follicular helper cells (cTfh). Thus, the third panel (panel C), was simultaneously standardized.
We optimized the design of the panels using direct information from the reagents as well as online tools (described in the key resources table) to reduce the chances of non-compatibility between clones, host-species, and fluorochromes. For instance, we used the Cytek Full Spectrum Viewer (Cytek Biosciences 2019) (https://spectrum.cytekbio.com/) to verify that the combination of fluorochromes was maintained with the less similar dye signatures as possible. This tool provides a similarity index for every pair of fluorochromes and one complexity index for every full panel. For both type of indexes, lower values indicate low similarity between fluorochromes (Figure 1). For the final design, we took into account our gating strategies, the markers and fluorochromes available, as well as the configuration of the spectral cell analyzer. Afterwards, we standardized the panels with single stained controls (using beads), testing of regular stain buffer and brilliant stain buffer, blocking of human Fc receptors (FcR), antibody titration, different staining incubation temperature, different combinations of antibodies/fluorochromes, as well as the final fluorescence compensation (with the spectral unmixing tool), and data analysis. Table 1 shows a description of all markers included in the three panels.
Figure 1.
Overview of the panel’s design according to fluorochromes combination
(A) Spectrum viewer showing the expected emission for every dye across the channels for the panels (A, B, and C) and their corresponding complexity matrix representing the full spectrum signature similarities for every dye combination. Values inside the matrix (similarity index) closer to zero indicate very different signatures between the two dyes implicated while values closer to one indicate very similar signatures (which might be more difficult to discriminate).
(B) Spectrum viewer showing the expected dye emission for PE-Cy5 and PE-Fire 640, which are the ones with the more similar signatures according to the matrix in panel B (similarity index of 0.94). However, in the experimental standardization, both fluorochromes were properly identified and unmixed, allowing the identification of the markers (CD56 and granzyme B) in the final fully stained samples as shown in the FlowJo plot gated on CD3-CD56+ NK cells.
Table 1.
Target human lymphocyte cell markers
| Marker (common name) | Description (type/function) |
|---|---|
| CD195 (CCR5) | Chemokine receptor, differentiation |
| CD196 (CCR6) | Chemokine receptor, differentiation |
| CD197 (CCR7) | Central memory, differentiation |
| CD103 (integrin αE) | Cell adhesion, homing |
| CD16 (FcγRIII) | NK cell differentiation, antibody dependent cellular cytotoxicity (ADCC) |
| CD161 (NKR-P1A) | Th17 T-cell, differentiation |
| CD19 | B-cell marker |
| CD226 (DNAM-1) | Regulatory, co-stimulatory receptor |
| CD25 (IL2RA) | Regulatory T-cell, IL-2 receptor |
| CD26 (DPP-IV) | Co-stimulatory receptor, regulatory role |
| CD27 | Co-stimulatory receptor, T and B-cell differentiation |
| CD28 | Co-stimulatory receptor |
| CD3 (CD3ε) | T-cell co-receptor |
| CD38 (cyclic ADP ribose hydrolase) | T-cell activation, differentiation |
| CD39 (NTPDase1) | Differentiation, immunosuppressive marker |
| CD4 | CD4 T-cells |
| CD45RA (LCA isoform CD45RA) | Differentiation marker |
| CD45RO (LCA isoform CD45RO) | T-cell memory marker |
| CD49d (integrin α4) | Integrin α4, homing |
| CD56 (NCAM) | NK cell marker |
| CD73 (ecto-5′-nucleotidase) | Differentiation, immunosuppressive marker |
| CD8 | CD8 T-cells |
| CD94 (KLRD1) | NK cell receptor, NKG2/CD94 dimer |
| CD154 (CTLA-4) | Treg marker, immune checkpoint |
| CD183 (CXCR3) | Chemokine receptor, differentiation |
| CD185 (CXCR5) | Chemokine receptor, differentiation |
| CD186 (CXCR6) | Chemokine receptor |
| FOX-P3 | Treg marker |
| CD357 (GITR) | Glucocorticoid-induced TNFR-related protein, TNF receptor |
| Granzyme B | Cytotoxicity marker |
| HLA-DR | MHC class II, T-cell activation marker |
| HLA-E | Non-classical MHC class I |
| CD360 (IL-21R) | IL-21 receptor |
| IL-23R | IL-23 receptor |
| Ki-67 | Cell cycle marker |
| CD158 (KIR2DL1/S1/S3/S5) | Inhibitory/activating KIR receptors |
| CD158e1 (KIR3DL1) | Inhibitory KIR receptor |
| CD223 (LAG-3) | T cell activation, immune checkpoint |
| MICA | MHC class I chain-related protein A |
| CD159a (NKG2A) | NK cell inhibitory receptor |
| CD159c (NKG2C) | NK cell activating receptor |
| CD314 (NKG2D) | NK cell activating receptor |
| CD337 (NKP30) | NK cell activating receptor |
| CD335 (NKP46) | NK cell activating receptor |
| CLEC5C (NKP80) | NK cell activating receptor |
| CD279 (PD-1) | T-cell inhibitory receptor, immune checkpoint |
| TCRyδ | yδ T cells |
| CD336 (TIM-3) | Inhibitory receptor, regulatory marker |
| ULBP-2/5/6 | NKG2D ligand |
| TCRVα2.4-Jα18 | Invariant natural killer T cells (iNKT) |
| TCRVα7.2 | Mucosal-associated invariant T cells (MAIT) |
| Integrin β7 | Cell adhesion, homing marker |
Institutional permissions
Blood samples from healthy donors were obtained from the French blood bank (Etablissement Français du Sang) as part of an agreement with the Institut Pasteur (C CPSL UNT, number 15/EFS/023). The study was approved by the Ethics Review Committee (Comité de protection des personnes) of Île-de-France VII.
Technical considerations
-
1.
Keep in mind that each panel is an individual experiment that needs to be set up and standardized independently. However, the three panels can be run in parallel.
-
2.Make sure to review the configurations (lasers, detectors, filters, mirrors, etc.) of the spectral flow cytometry equipment that will be used in order to adapt the fluorochromes of the panels if needed.
-
a.Control the compatibility of reagents and antibodies according to your objectives and settings.
-
b.Keep in mind the brightness, concentration and the use of fluorescence minus one (FMO) controls when constructing your panels.
-
c.Available online tools and other resources can be accessed to determine the similarity and complexity indexes amongst the fluorochromes in order to prevent or limit potential spectrum overlaps (https://spectrum.cytekbio.com/).
-
a.
-
3.
The use of compensation beads (used for this protocol) is recommended for the fluorescence compensation (unmixing) in the spectral cell analyzer (Sony ID7000) but preparation of single stained controls for testing the antibodies in the target cells is pertinent. Specific beads exist on the market; consider the bead-antibody compatibility factor.
-
4.
During the staining and further acquisition steps, protect the antibodies, master mixes, and stained samples from light and maintain them at 4°C of temperature.
-
5.
Acquire the samples immediately after staining in order to avoid loss of fluorescence and sample deterioration.
-
6.
For the preparation of your experiments, consider the amount of samples and cells to be prepared since this will decrease or increase the time and amount or reagents/consumables required.
-
7.
If you aim to include cytokines in the flow cytometry panels, consider and test if additional stimulation/activation is required for the cells in advance.
Sample preparation
Timing: 7–9 h
This section describes the preparation of the target PBMCs before the staining steps.
-
8.
Warm Roswell Park Memorial Institute (RPMI) 1640 medium to 37°C before thawing the frozen peripheral blood mononuclear cells (PBMCs).
-
9.Thaw the frozen PBMCs in a water bath at 37°C.
-
a.Just before full thawing of the cells, transfer the cryovials to the biosafety hood.
-
b.Slowly add warm RPMI medium for a final volume of twice the initial volume originally contained in the cryovial.
-
a.
-
10.
Transfer the cell suspension to a 50-mL polypropylene tube and complete the volume to 50-mL with warm RPMI medium.
-
11.
Centrifuge the cells at 400 g for 5 min.
-
12.
Decant and suspend the cells into fresh new warm RPMI medium.
-
13.
Count the cells using your local protocol (i.e., automated cell counter)
-
14.
Suspend the cells at 1 × 106 cells/mL.
-
15.
Incubate the cells between 6–8 h at 37°C and 5% CO2 in a flask container prior to the staining steps.
Note: We recommend to assess in advance, the amount of time for resting your target cells after thawing, given that the expression of some chemokine receptors may vary. We standardized this protocol by resting the PBMCs for 8 h after the thawing process since the detection of CCR7 significantly improved as compared to recently thawed cells. In any case, we recommend staining your samples at the same conditions each time. Incubation periods longer than 8 h might have a higher negative impact in the viability of PBMCs.
Note: Although these panels were standardized with frozen PBMCs, they could potentially be used for working with fresh blood cells or other tissue-isolated cells under their particular considerations. After the preparation of tissue and cell isolation, no major modifications are likely to be needed for antibody staining; nonetheless, we suggest reviewing specific provider recommendations on equipment and reagents for particular types of tissues.
Note: This protocol was standardized for staining up to 1 × 106 PBMCs in 100 μL of volume. If you plan to use a few or more cells, we suggest titrating and testing the panels in advance before conducting the experiments.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Zombie UV™ Fixable Viability Kit | BioLegend | Cat#423107 |
| APC/Fire™ 810 anti-human CD14 | BioLegend | Cat#367156 |
| APC/Fire™ 810 anti-human CD19 | BioLegend | Cat#302271 |
| Spark Blue™ 550 anti-human CD3 | BioLegend | Cat#344851 |
| BD OptiBuild™ BV480 Mouse Anti-Human γδ TCR | BD | Cat#746498 |
| Brilliant Violet 421™ anti-human TCR Vα24-Jα18 (iNKT cell) | BioLegend | Cat#342915 |
| APC/Cyanine7 anti-human TCR Vα7.2 | BioLegend | Cat#351713 |
| Brilliant Violet 510™ anti-human CD4 | BioLegend | Cat#317443 |
| Brilliant Violet 570™ anti-human CD8a | BioLegend | Cat#301037 |
| PE/Fire™ 640 anti-human CD56 (NCAM) Recombinant | BioLegend | Cat#392431 |
| Pacific Blue™ anti-human CD16 | BioLegend | Cat#302024 |
| FITC anti-human CD45RA | BioLegend | Cat#304105 |
| BD Horizon™ BUV395 Mouse Anti-Human CD45RO | BD | Cat#564291 |
| Brilliant Violet 650™ anti-human CD27 | BioLegend | Cat#302828 |
| PerCP/Cyanine5.5 anti-human CD28 | BioLegend | Cat#302921 |
| Brilliant Violet 711™ anti-human CD196 (CCR6) | BioLegend | Cat#353435 |
| PerCP anti-human CD197 (CCR7) | BioLegend | Cat#353242 |
| PE/Fire™ 810 anti-human CD183 (CXCR3) | BioLegend | Cat#353759 |
| Brilliant Violet 750™ anti-human CD185 (CXCR5) | BioLegend | Cat#356941 |
| BD OptiBuild™ BUV563 Mouse Anti-Human CXCR6 (CD186) | BD | Cat#748450 |
| PE/Cyanine7 anti-human/mouse integrin β7 | BioLegend | Cat#321241 |
| PE/Fire™ 700 anti-human CD103 (Integrin αE) | BioLegend | Cat#350239 |
| Alexa Fluor® 647 anti-human CD49d | BioLegend | Cat#304335 |
| BD Pharmingen™ PE Mouse Anti-Human CD94 | BD | Cat#555889 |
| Alexa Fluor® 700 anti-human CD159a (NKG2A) | BioLegend | Cat#375120 |
| BD OptiBuild™ BUV615 Mouse Anti-Human CD159C (NKG2C) | BD | Cat#751059 |
| BD OptiBuild™ BUV737 Mouse Anti-Human CD314 (NKG2D) | BD | Cat#748426 |
| PE/Dazzle™ 594 anti-human CD337 (NKp30) | BioLegend | Cat#325231 |
| Brilliant Violet 785™ anti-human CD335 (NKp46) | BioLegend | Cat#331945 |
| APC anti-human NKp80 | BioLegend | Cat#346708 |
| BD OptiBuild™ BV750 Mouse Anti-Human CD195 (CCR5) | BD | Cat#747224 |
| BD OptiBuild™ BUV395 Mouse Anti-Human CD26 | BD | Cat#744454 |
| PE/Fire™ 810 anti-human CD39 | BioLegend | Cat#328245 |
| Brilliant Violet 711™ anti-human CD73 (Ecto-5'-nucleotidase) | BioLegend | Cat#344025 |
| BD OptiBuild™ BUV737 Mouse Anti-Human CD226 | BD | Cat#748428 |
| PE/Cyanine7 anti-human CD360 (IL-21R) | BioLegend | Cat#359513 |
| Human IL-23 R APC-conjugated Antibody | Bio-Techne | Cat#FAB14001A-100 |
| PE anti-human CD357 (GITR) | BioLegend | Cat#311603 |
| BD OptiBuild™ BUV805 Mouse Anti-Human KIR2DL1/S1/S3/S5 (CD158) | BD | Cat#752514 |
| PerCP/Cyanine5.5 anti-human CD158e1 (KIR3DL1, NKB1) | BioLegend | Cat#312717 |
| PE/Dazzle™ 594 anti-human CD152 (CTLA-4) | BioLegend | Cat#369615 |
| Brilliant Violet 785™ anti-human CD223 (LAG-3) | BioLegend | Cat#369321 |
| Brilliant Violet 650™ anti-human CD366 (Tim-3) | BioLegend | Cat#345027 |
| Alexa Fluor® 647 anti-human Ki-67 | BioLegend | Cat#350510 |
| PE/Fire™ 700 anti-human CD279 (PD-1) | BioLegend | Cat#621621 |
| PE/Cyanine5 anti-human/mouse Granzyme B Recombinant | BioLegend | Cat#372225 |
| Alexa Fluor® 700 anti-human CD27 | BioLegend | Cat#302814 |
| Alexa Fluor® 647 anti-mouse/rat/human FOXP3 | BioLegend | Cat#320013 |
| PE/Cyanine7 anti-human CD161 | BioLegend | Cat#339917 |
| PE anti-human CD25 | BioLegend | Cat#302605 |
| Brilliant Violet 650™ anti-human CD38 | BioLegend | Cat#356620 |
| PerCP/Cyanine5.5 anti-human HLA-DR | BioLegend | Cat#307629 |
| APC anti-human HLA-E | BioLegend | Cat#342605 |
| Brilliant Violet 421™ anti-human CD279 (PD-1) | BioLegend | Cat#329919 |
| BD OptiBuild™ BV605 Mouse Anti-Human ULBP-2/5/6 | BD | Cat#748131 |
| BUV805 Mouse Anti-Human MICA | BD | Cat#749768 |
| Experimental Models: Cell Lines | ||
| Frozen PBMCs from human donors isolated from fresh blood (information regarding sex, age, and other features of the donors is not available) | Établissement Français du Sang (France) | https://dondesang.efs.sante.fr/ |
| Other | ||
| Human TruStain FcX™ (Fc Receptor Blocking Solution) | BioLegend | Cat#422301 |
| Roswell Park Memorial Institute (RPMI) 1640 Medium | Life Technologies | Cat#21875034 |
| Dulbecco's Phosphate Buffered Saline (1X) | Life Technologies | Cat#14190144 |
| UltraComp eBeads™ Compensation Beads | Thermo Fisher Scientific | Cat#01-2222-42 |
| BD Pharmingen™ Stain Buffer (BSA) | BD Pharmingen | Cat#554656 |
| BD Horizon™ Brilliant Stain Buffer | BD Pharmingen | Cat#563794 |
| BD Cytofix/Cytoperm™ Fixation/Permeabilization Solution Kit | BD Pharmingen | Cat#554714 |
| Thermo Scientific™ Paraformaldehyde, 4% in PBS | Thermo Fisher Scientific | Cat#J61899 |
| Sony ID7000TM spectral cell analyzer | Sony | https://www.sonybiotechnology.com/us/instruments/id7000-spectral-cell-analyzer/ |
| Software and algorithms | ||
| Cytek Full Spectrum Viewer | Cytek® Biosciences 2019 | https://spectrum.cytekbio.com/ |
| FlowJo 10.7.1 | © Becton Dickinson & Company (BD) | https://www.flowjo.com/ |
| Spectral Flow Analysis (SFA) - Life Sciences Cloud Platform | Sony | https://www.sonybiotechnology.com/us/instruments/sfa-cloud-platform/ |
Note: This protocol was standardized in a Sony ID7000 spectral cell analyzer set with 6 lasers (deep ultra-violet 320 nm, ultra-violet 355 nm, violet 405 nm, blue 488 nm, yellow/green 561 nm, and red 637 nm). We advise that you follow the recommendation of the supplier for the appropriate use, configuration and handling of the cell analyzer instrument. For detailed information beyond the essentials specified in this protocol regarding the instrument, you can refer to the technical data and resources available online and directly contact the provider (sonybiotechnology.com).
Materials and equipment
Consumables and other resources
| Reagent or resource | Source | Identifier |
|---|---|---|
| Material and consumables | ||
| Falcon 5 mL Round Bottom Polystyrene Test Tube, with Snap Cap | Corning | Cat#352054 |
| Corning cell culture flasks | Corning | Cat#430639 |
| Corning 15 mL centrifuge tubes | Corning | Cat#430791 |
| Corning 50 mL centrifuge tubes | Corning | Cat#430829 |
| Half-Deep Well Plate, 0.5 mL, PP, sterile, round bottom | Cole-Parmer | Cat#FV-67104-57 |
| Other resources | ||
| Webinar: Spectral Cytometry Software Workflows and Tools that Enable Multiparametric Flow Cytometry | 2022 Sony Biotechnology Inc. | https://www.sonybiotechnology.com/us/blog/spectral-cytometry-software-workflows-and-tools-that-enable-multiparametric-flow-cytometry/ |
| Webinar: Panel Design Considerations for Spectral Flow Cytometry | 2022 Sony Biotechnology Inc. | https://www.sonybiotechnology.com/us/blog/webinar-panel-design-considerations-for-spectral-flow-cytometry/ |
Step-by-step method details
Preparation of single stained controls
Timing: 40 min
This section describes the preparation of control beads and cells for every marker. These controls are required for the creation of spectral references (signatures) for every fluorochrome. The controls and unmixing matrix will allow the correct identification and separation of markers in the fully stained samples.
-
1.Prepare single stained controls (except for the viability dye) with compensation beads (UltraComp eBeads, Invitrogen) following instructions from the manufacturer (https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFS-Assets%2FLSG%2Fmanuals%2FMAN0019374_UltraComp_eBeads_and_UltraComp_eBeads_Plus_Compensation_Beads_PI.pdf).
-
a.Mix the beads vigorously by pulse-vortexing and add 1 drop in a half-deep well plate (round bottom) (or single polystyrene tubes) for each antibody included in the panels.
- b.
-
c.Add 2 mL of flow cytometry stain buffer (BD) to each tube, and centrifuge at 400 g for 5 min.
-
d.Decant and fix the beads with 400 μL of 4% Paraformaldehyde (PFA) for 10 min at 4°C, and maintain protected from light.
-
e.Wash the beads with 1 mL of cold (4°C) 1× PBS by centrifuging at 400 g for 5 min.
-
f.Decant and suspend the beads in 500 μL of cold (4°C) 1× PBS and maintain them protected from light at 4°C.
-
a.
-
2.
For the viability dye (Zombie UV, BioLegend), prepare one reaction with PBMCs and stain following the sample staining steps (see below).
Note: It is recommended to run single stained controls each time that samples will be acquired, especially if there are changes of antibodies and/or reagent lot.
Optional: Add an extra sample of fully stained PBMCs (in each panel) to set the PMT configuration in the cytometer without losing events from your objective samples.
Table 2.
Panel A (CD8+NKG2A/C+ T cells)
| # | Specificity | Clone | Fluorochrome | ng/mL |
|---|---|---|---|---|
| 1 | Live/Dead | - | Zombie UV | - |
| 2 | CD14 | 63D3 | APC-Fire 810 | 2.5 |
| 3 | CD19 | HIB19 | APC-Fire 810 | 0.625 |
| 4 | CD3 | SK7 | Spark Blue 550 | 2.5 |
| 5 | TCRyδ | 11F2 | BV480 | 2.5 |
| 6 | TCRVα2.4 | 6B11 | BV421 | 0.625 |
| 7 | TCRVα7.2 | 3C10 | APC-Cy7 | 1.25 |
| 8 | CD4 | OKT4 | BV510 | 1.25 |
| 9 | CD8 | RPA-T8 | BV570 | 1.25 |
| 10 | CD56 | QA17A16 | PE-Fire 640 | 1.25 |
| 11 | CD16 | 3G8 | Pacific Blue | 6.25 |
| 12 | CD45RA | HI100 | FITC | 2.5 |
| 13 | CD45RO | UCHL1 | BUV395 | 1.25 |
| 14 | CD27 | O323 | BV650 | 0.625 |
| 15 | CD28 | CD28.2 | PerCP/Cy5.5 | 2.5 |
| 16 | CCR6 | G034E3 | BV711 | 3 |
| 17 | CCR7 | G043H7 | PerCP | 8 |
| 18 | CXCR3 | G025H7 | PE-Fire 810 | 0.625 |
| 19 | CXCR5 | J252D4 | BV750 | 3 |
| 20 | CXCR6 | 13B 1E5 | BUV563 | 6 |
| 21 | Integrin β7 | FIB504 | PE-Cy7 | 2.5 |
| 22 | CD103 | Ber-ACT8 | PE-Fire 700 | 0.3125 |
| 23 | CD49d | 9F10 | AF647 | 1.25 |
| 24 | CD94 | HP-3D9 | PE | 0.06 |
| 25 | NKG2A | S19004C | AF700 | 5 |
| 26 | NKG2C | 134591 | BUV615 | 2.5 |
| 27 | NKG2D | 1D11 | BUV737 | 6 |
| 28 | NKP30 | P30-15 | PE-Dazzle 594 | 5 |
| 29 | NKP46 | 9E2 | BV785 | 5 |
| 30 | NKP80 | 5D12 | APC | 10 |
Table 3.
Panel B (CD8+NKG2A/C+ T cells)
| # | Specificity | Clone | Fluorochrome | ng/mL |
|---|---|---|---|---|
| 1 | Live/Dead | - | Zombie UV | - |
| 2 | CD14 | 63D3 | APC-Fire 810 | 2.5 |
| 3 | CD19 | HIB19 | APC-Fire 810 | 0.625 |
| 4 | CD3 | SK7 | Spark Blue 550 | 2.5 |
| 5 | TCRyδ | 11F2 | BV480 | 2.5 |
| 6 | TCRVα2.4 | 6B11 | BV421 | 0.625 |
| 7 | TCRVα7.2 | 3C10 | APC-Cy7 | 1.25 |
| 8 | CD4 | OKT4 | BV510 | 1.25 |
| 9 | CD8 | RPA-T8 | BV570 | 1.25 |
| 10 | CD56 | QA17A16 | PE-Fire 640 | 1.25 |
| 11 | CD16 | 3G8 | Pacific Blue | 6.25 |
| 12 | CD45RA | HI100 | FITC | 2.5 |
| 13 | CCR5 | 2D7/CCR5 | BV750 | 2.5 |
| 14 | CCR7 | G043H7 | PerCP | 8 |
| 15 | CD26 | M-A261 | BUV395 | 2.5 |
| 16 | CD39 | A1 | PE-Fire 810 | 1.25 |
| 17 | CD73 | AD2 | BV711 | 1.25 |
| 18 | CD226 | DX11 | BUV737 | 2.5 |
| 19 | IL-21R | 17A12 | PE-Cy7 | 2.5 |
| 20 | IL-23R | 218213 | APC | 3 μLb |
| 21 | GITR | 621 | PE | 12 |
| 22 | NKG2A | S19004C | AF700 | 5 |
| 23 | NKG2C | 134591 | BUV615 | 2.5 |
| 24 | KIR2DL1/S1/S3/S5 (CD158) | HP-MA4 | BUV805 | 2.5 |
| 25 | KIR3DL1 (CD158e1) | DX9 | PerCP/Cy5.5 | 2.5 |
| 26 | CTLA4a | BNI3 | PE-Dazzle 594 | 3 |
| 27 | LAG-3 | 11C3C65 | BV785 | 3 |
| 28 | TIM-3 | F38-2E2 | BV650 | 3 |
| 29 | PD-1 | A17188B | PE-Fire 700 | 0.15 |
| 30 | Granzyme Ba | QA16A02 | PE-Cy5 | 0.3125 |
| 31 | Ki-67a | Ki-67 | AF647 | 6 |
Intracellular marker.
Recommended concentration by the manufacturer: 10 μL/106 cells.
Table 4.
Panel C (CD4+ T cell subsets)
| # | Specificity | Clone | Fluorochrome | ng/mL |
|---|---|---|---|---|
| 1 | Live/Dead | - | Zombie UV | - |
| 2 | CD14 | 63D3 | APC-Fire 810 | 2.5 |
| 3 | CD19 | HIB19 | APC-Fire 810 | 0.625 |
| 4 | CD3 | SK7 | Spark Blue 550 | 2.5 |
| 5 | CD4 | OKT4 | BV510 | 1.25 |
| 6 | CD8 | RPA-T8 | BV570 | 1.25 |
| 7 | FOX-P3a | 150D | AF647 | 2.5 |
| 8 | CD161 | HP-3G10 | PE-Cy7 | 1.25 |
| 9 | CD45RA | HI100 | FITC | 2.5 |
| 10 | CCR5 | 2D7/CCR5 | BV480 | 2.5 |
| 11 | CCR6 | G034E3 | BV711 | 3 |
| 12 | CCR7 | G043H7 | PerCP | 8 |
| 13 | CXCR5 | J252D4 | BV750 | 3 |
| 14 | CXCR6 | 13B 1E5 | BUV563 | 6 |
| 15 | CD25 | M-A251 | PE | 0.625 |
| 16 | CD27 | O323 | AF700 | 6.25 |
| 17 | CD38 | HB-7 | BV650 | 1.25 |
| 18 | HLA-DR | L243 | PerCP/Cy5.5 | 6 |
| 19 | HLA-E | 3D12 | APC | 2.5 |
| 20 | PD-1 | EH12.2H7 | BV421 | 0.625 |
| 21 | ULBP-2/5/6 | 165903 | BV605 | 6 |
| 22 | CTLA4a | BNI3 | PE-Dazzle 594 | 3 |
| 23 | MICA | 159227 | BUV805 | 6 |
Intracellular marker.
Sample staining
Timing: 2 h
This section describes the preparation of antibody master mixes, as well as incubation and washing steps for the extracellular and intracellular staining of the target PBMCs. The concentration of antibodies for each panel is listed in Tables 2, 3, and 4.
-
3.Count, separate and wash the cells.
-
a.Centrifuge at 400 g for 5 min with 1× PBS.
-
b.Suspend 1 × 106 cells into 100 μL of cold stain buffer (1× PBS, 20% FBS) in a 96-half-deep well plate.
-
a.
-
4.
Add 5 μL of FcR-blocking reagent (Human TruStain FcX, BioLegend) directly in the stain buffer (1× PBS, 20% FBS) containing the cells, mix and incubate for 10 min at 18°C–20°C of temperature.
Note: This step is optional. Step 3 of the staining requires the suspension of the cells in 20% FBS-containing stain buffer, which helps to reduce non-specific FcRs binding and background fluorescence. In addition, we tested the use of an FcR-blocking solution and we did not find major differences in the markers except for the expression of CD3 in CD14+CD19+ cells (which are negatively selected in this protocol). Thus, we recommend performing an extra step with an Fc-blocking reagent to maximize the reduction of potential background signals especially when aiming to analyze cells containing high levels of Fc receptors such as monocytes, macrophages, and B-cells.
-
5.Wash the cells with cold (4°C) 1× PBS (complete to 500 μL of total volume).
-
a.Centrifuge at 400 g for 5 min,
-
b.Decant and suspend the cells in 100 μL of 1× PBS.
-
a.
-
6.
Add 1 μL of viability dye (Zombie UV, BioLegend) for each 1 × 106 cells in 100 μL cold 1× PBS (4°C) and incubate the cells for 20 min at 18°C–20°C of temperature in the dark.
-
7.In the meantime, prepare the different master mixes with the extracellular antibodies (panels A, B, and C) in cold (4°C) brilliant stain buffer (BD Biosciences) adjusting for a total volume of 100 μL/sample.Note: We recommend adding each one of the polymer-based antibodies directly into the sample and gently mixing instead of adding them all together in the master mix in order to reduce the chances of additional staining artifacts due to non-specific reactivity.
-
a.Add 50% overhead volume to account for potential pipetting errors.
-
b.Mix well by gently pipetting up and down.
-
a.
-
8.Wash the cells with cold 1× PBS (complete to 500 μL of total volume).
-
a.Centrifuge at 400 g for 5 min, and decant.
-
a.
-
9.Add the corresponding master mix of antibodies (100 μL) to the samples for extracellular staining (panels A, B, and C).
-
a.Mix well by gently pipetting up and down.
-
b.Incubate for 30 min at 18°C–20°C of temperature in the dark.
-
a.
-
10.Wash the cells with cold (4°C) 1× PBS (complete to 500 μL of total volume).
-
a.Centrifuge at 400 g for 5 min and decant.
-
a.
-
11.Fix the cells without intracellular staining markers (panel A).
-
a.Fix the cells with 500 μL of 4% Paraformaldehyde (PFA) for 10 min:
-
b.Wash the cells with cold (4°C) 1× PBS (complete to 500 μL of total volume).
-
c.Centrifuge at 400 g for 5 min.
-
d.Decant by inverting the plate on absorbent clean paper towel.
-
e.Suspend the cells in 500 μL of cold (4°C) 1× PBS.
-
a.
Pause point: These stained and fixed cells (panel A) are ready for acquisition (maintain protected from light at 4°C if these are not immediately acquired in the cytometer).
-
12.Continue with the intracellular staining of the corresponding samples (panels B and C):
-
a.Suspend the cells in 300 μL of cold (4°C) permeabilization buffer (Cytofix/Cytoperm solution, BD) and incubate for 20 min at 4°C in the dark.
-
b.Prepare the different master mixes with intracellular antibodies (Tables 2, 3, and 4).
-
i.Prepare the mixes in permeabilization/wash buffer (Perm/Wash Buffer, BD) previously diluted in distilled water (to make a 1× solution).
-
ii.Adjust for a total volume of 50 μL/sample and add 50% overhead volume to account for potential pipetting errors.
-
iii.Mix well by gently pipetting up and down.
-
i.
-
c.Wash the cells twice by adding 1× permeabilization/wash buffer (Perm/Wash Buffer, BD) (complete to 500 μL of total volume).
-
i.Centrifuge at 400 g for 5 min.
-
ii.Decant by inverting the plate on absorbent clean paper towel.
-
i.
-
d.Add the master mixes of antibodies (50 μL) to the samples.
-
i.Mix well by gently pipetting up and down.
-
ii.Incubate for 30 min at 4°C in the dark.
-
i.
-
a.
-
13.
Wash the cells with cold (4°C) 1× permeabilization/wash buffer (Perm/Wash Buffer, BD) (complete to 0.5 mL of total volume).
-
14.
Centrifuge at 400 g for 5 min at 4°C.
-
15.
Decant by inverting the plate on absorbent clean paper towel.
-
16.Fix the cells in 500 μL of cold (4°C) 4% PFA for 10 min.
-
a.Wash the cells by adding cold (4°C) 1× PBS (complete to 500 μL of total volume).
-
b.Centrifuge at 400 g for 5 min.
-
c.Decant by inverting the plate on absorbent clean paper towel.
-
d.Suspend the cells in 500 μL of cold (4°C) 1× PBS and maintain protected from light at 4°C.
-
a.
Note: Instead of a half-deep well plate, samples can be prepared in polystyrene tubes. The Sony ID7000 spectral cell analyzer supports 5-mL tube racks (24 tubes), 96-well standard height, 96-well half deep, 96-well deep plates, and 384-well standard flat bottom plates.
Note: The final volume for cell suspension should be adjusted depending on the number and type of cells, and whether tubes, deep-well or other plates will be used. Volumes in this protocol are standardized for 96-half-deep well plates.
Pause point: The stained and fixed cells are ready for acquisition (maintain protected from light at 4°C if they are not immediately acquired).
Instrument preparation and experiment set-up
Timing: 1 h
This section refers to the creation of a new experiment in the spectral cell analyzer system and preparing the instrument for sample acquisition. The steps include the calibration of the Sony ID7000 system, setting the characteristics of the experiment such as the template and groups of samples, indicating the fluorochromes in the panel, and setting the final PMT voltages to acquire the samples.
-
17.Set the spectral cell analyzer (Sony ID7000).
-
a.Run daily purge, flow rate and QC calibrations following the instructions of the manufacturer (https://vimeo.com/847254690/e408449a02).
-
a.
Note: The ID7000 spectral cell analyzer needs to be calibrated using the daily QC and performance 8-peak beads before setting-up the experiment and acquire the samples.
-
18.Create a new experiment (one individual experiment for each panel);
-
a.In the spectral cytometer system software, indicate the type of template (tubes and/or 96-half-deep-well plates).
-
b.Create a new sample group and add the samples to the group.
-
c.Name the tubes or wells with the samples and single-stained controls.
- d.
-
a.
-
19.Adjust the PMT voltages for the experiment.
-
a.Load the tube-rack or plate containing the single stained controls and/or the fully stained sample(s).
-
b.Acquire in Preview a full stained sample and at least one single stain for each laser for setting the PMT voltage for each laser.
-
i.Keep a homogeneous ratio between the lasers, and make sure dyes are higher in their corresponding excitation laser using the ribbon plots (Figure 2B).
-
i.
-
a.
Note: Do not change the voltage parameters after set-up; maintain the same values during acquisition of the samples and controls. PMT voltages and machine configuration for each panel must be independently set up.
-
20.Set the instrument settings for the experiments.
-
a.Adjust the number of events to record, stopping criteria, flow rate, cleaning mode, acquisition offset, agitation and other settings as needed (i.e., taking into account sample volumes).
-
a.
Note: More information from the manufacturer regarding the creation of a new experiment can be found online (https://vimeo.com/847255526/565a89c4bf).
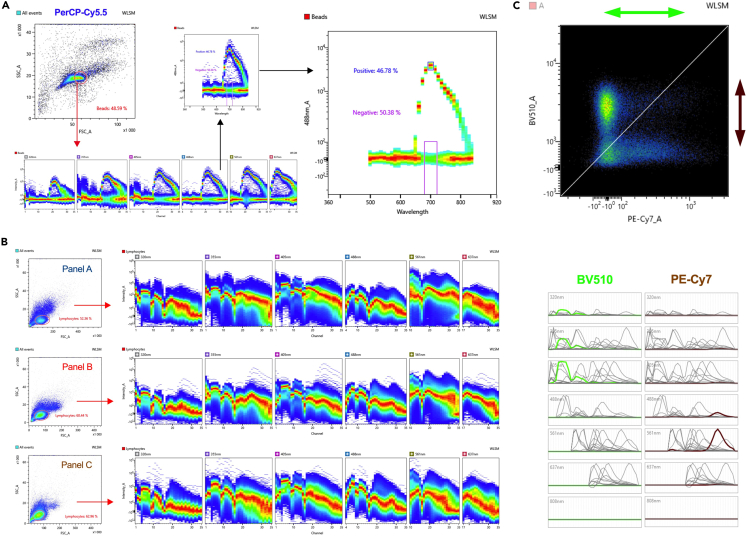
Figure 2.
Acquisition of single stained controls and fully stained samples
(A) Example of single stained (PerCP-Cy5.5) control beads; the emission is detected across the lasers and then the positive and negative populations can be assigned. This should be done for every single stained control (including the viability marker) in order to run the unmixing algorithm.
(B) Target population gated by morphology and ribbon plot examples of fully stained PBMCs showing data acquisition across the channels in every laser.
(C) Spectral reference viewer/adjuster (top figure) showing BV510 versus PE-Cy7 dyes. This tool allows (if required) to manually adjusting the positive populations (following the direction of the adjacent arrows) after data acquisition and unmixing. Correspondingly, emission signatures of both fluorochromes are simultaneously shown during adjustment (bottom figure).
Sample acquisition
Timing: 3–5 min/sample
This section describes the acquisition of single-stained controls and samples with auto-acquisition mode in the Sony ID7000 spectral cytometer system once the experiment settings have been fixed.
-
21.Acquire the samples and single-stained controls.
-
a.Load the plate or tube-rack with the samples and set Current Position in the spectral cytometer system software.
-
b.Select the wells deemed for acquisition.
-
c.Right-click and add to Auto Acquisition Target.
-
d.Run Auto Acquisition when ready.
-
a.
Note: Different groups of samples with different conditions can be recorded in the same experiment.
Unmixing analysis and file export
Timing: 1 h
This section describes how to create, set and apply the unmixing matrix for the experiments, and export the acquired sample files for further analysis. The unmixing matrix allows the correct identification of fluorescent signatures.
-
22.Assign the single stained controls to the corresponding beads/cells acquired.
-
a.Select the single stained beads/cells acquired; right-click and select the corresponding fluorochrome.
-
b.Assign the positive and negative populations using the gates (Figure 2A) (do this for every fluorochrome included in each panel).
-
c.Adjust the morphology gate in the FSC/SCC graph to select the target population (beads/cells) and verify that there is one single clear signal on all lasers using the ribbon plots (Figure 2B).
-
a.
-
23.Calculate the spectrum for each fluorochrome and apply the unmixing matrix.
-
a.After assigning the single stained controls, calculate the spectrum for all fluorochromes in the color palette by clicking on the option “calculate”.
-
b.For applying the new calculated unmixing matrix, go to the worksheet and select “apply” (unmixing matrix).
-
c.Control the results of the unmixing in the full stained samples using the flow cytometry and ribbon plots.
-
a.
Note: The unmixing matrix can be further adjusted directly in the plots under the Spectral Reference Viewer/Adjuster and applied to all samples (Figure 2C).
-
24.Export the resulting flow cytometry standard (FCS) files.
-
a.Select the samples to be exported and go to export files.
-
b.Select the folder, type (.fcs) and export the selected files (this will take time depending on the amount of data acquired).
-
a.
Data analysis
Timing: variable, objective dependent
This section describes the steps and features included in the data analysis, the gating strategy for the target populations, as well as the dimensionality reduction analysis in Figures 3, 4, and 5. These analyses were made in FlowJo (BD Biosciences), but other software and platforms compatibles with FCS files such as OMIQ (Dotmatics) and the SFA Life Sciences Cloud Platform (Sony) can be used.
-
25.Import and analyze the resulting files in the specialized software supporting FCS files.
-
a.Open the software (FlowJo, BD Biosciences) and import the files from folder in a new experiment.
-
a.
-
26.Analyze the data accordingly; the main general gating strategies defined for the panels are shown in Figures 3 and 4.
-
a.Run quality control plug-ins (recommended) for evaluating and eventually exclude bad flow cytometry quality data from your set (i.e., deviations of parameters during time of acquisition).
-
b.Transform and explore the data with other plug-ins for clustering and dimensionality reduction analysis (Figure 5).
- c.
-
a.
Note: If you detect a potential unmixing over/under compensation issue during analysis, you can go back to the spectral flow cytometry system software and adjust the unmixing matrix accordingly. After, apply the new unmixing matrix to the samples and export the files for further analysis.
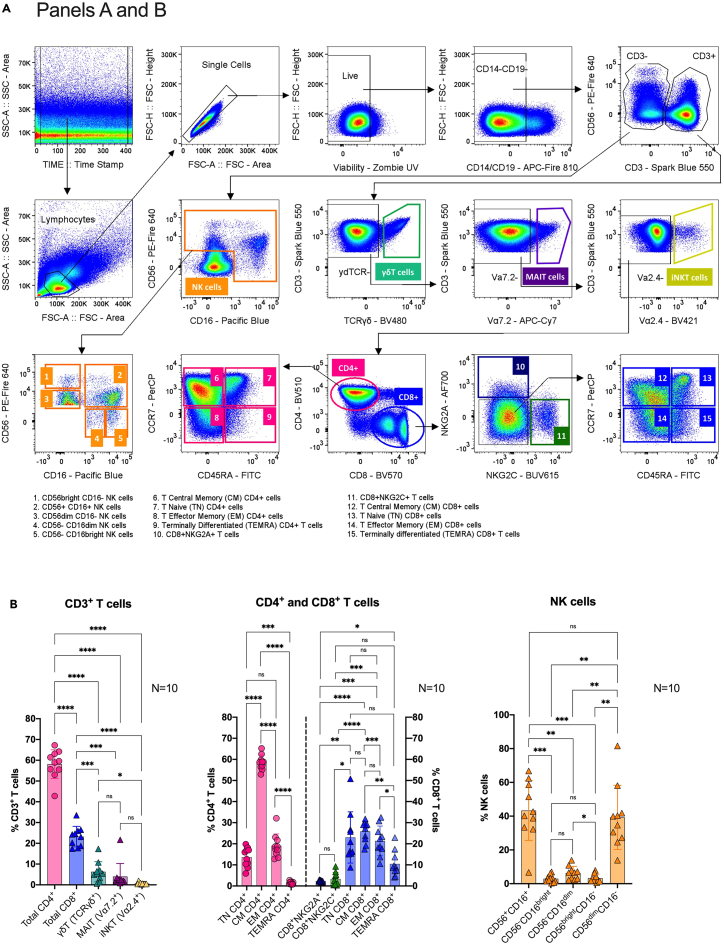
Figure 3.
Main general gating strategies for panels A and B
(A) After time lapse, lymphocyte morphology, single and live events selection, samples are gated to exclude CD14+ and CD19+ cells (monocytes and B cells). Subsequently, CD3-CD56+ cells are gated for further natural killer (NK) cells analysis using CD56 versus CD16 markers. On the other hand, CD3+ cells are gated to discriminate unconventional TCRγδ+ (γδT cells), TCRVα7.2+ (MAIT cells) and TCRVα2.4+ (iNKT cells) lymphocytes. From the remaining T cell population, cells are gated by CD4+ versus CD8+ cells. NKG2A+NKG2C- (CD8+NKG2A+) and NKG2A-NKG2C+ (CD8+NKG2C+) cells are then identified from the CD8+ T cell population. Then, the NKG2A-NKG2C- CD8+ subset as well as the total CD4+ cells are gated according to differentiation markers; CD45+CCR7+ T naïve (TN) cells, CD45-CCR7+ central memory (CM) cells, CD45RA-CCR7- effector memory (EM) cells, and CD45RA+CCR7- terminally differentiated (TEMRA) cells.
(B) Correspondingly, example graphs showing manually gated major populations (bottom figures) as proportions from their parent CD3+ T cell population (left), CD4+CD8- and CD4-CD8+ T cells (center), and NK cells (right).Bar graphs represent the means; error bars represent the standard deviation; and dots represent individual donors (N=10). One-way ANOVA with Tukey’s multiple comparison test. Significance is indicated as ∗∗∗∗P ≤ 0.0001, ∗∗∗P ≤ 0.001, ∗∗P ≤ 0.01, ∗P ≤ 0.05, or P > 0.05 (not significant (ns)).
Figure 4.
Main gating strategy for panel C
(A) Following the positive selection of time lapse, the lymphocyte morphology, and single and live events. Then, to exclude CD14+CD19+ cells. Then, CD3+ cells are gated by CD4+ versus CD8+ cells. Then, CD4+ lymphocytes are subsequently gated to identify CD4+CD25+FOXP3+ regulatory T cells (Treg), CD161+CCR6+ T helper 17 cells (Th17), and CD45RA-CXCR5+ circulating T follicular helper cells (cTfh). The remaining CD4+ cells are further gated on; CD45+CCR7+ T naive (TN) cells, CD45-CCR7+ central memory (CM) cells, CD45RA-CCR7- effector memory (EM) cells, and CD45RA+CCR7- terminally differentiated (TEMRA) cells.
(B) Correspondingly, example graph (right) showing manually gated populations as proportions from their CD4+CD8- T cell parent population. Bar graphs represent the means; error bars represent the standard deviation; and dots represent individual donors (N=10). One-way ANOVA with Tukey’s multiple comparison test. Significance is indicated as ∗∗∗∗P ≤ 0.0001, ∗∗∗P ≤ 0.001, ∗∗P ≤ 0.01, ∗P ≤ 0.05, or P > 0.05 (not significant (ns)).
Figure 5.
Clustering analysis and expression of markers between the CD3+NKG2A+ and CD3+NKG2C+ T lymphocytes
(A) We gated on CD3+NKG2A+NKG2C- (CD3+NKG2A+) and CD3+NKG2CA-NKG2C+ (CD3+NKG2C+) populations (shown in blue and green gates respectively), from single events of live CD14- and CD19- lymphocytes, with the aim to corroborate our previous exclusion gating strategy by running an unsupervised clustering analysis.
(B) Using such gating strategy, we concatenated ∼40,000 events for each population from fully stained PBMCs (N = 10). After, we run tSNE analyses with 3,000 iterations selecting all fluorescent parameters except for those previously gated (viability, CD3, CD14, CD19, NKG2A, and NKG2C). Relative expression of markers from panels A and B are shown across the maps for both analyzed subsets (CD3+NKG2A+ and CD3+NKG2C+ T cells). Clusters of CD8+ T cells independent from CD4+, TCRγδ+, TCRVα7.2+, and TCRVα2.4+ T cells can be identified.
Expected outcomes
The final analysis for identifying the target CD8+ T lymphocytes from other T cell populations in our data was completed in FlowJo (BD Biosciences). We concatenated the CD3+NKG2A+NKG2C- and CD3+ NKG2A-NKG2C+ T cell subsets from manually gated live single CD14-CD19- lymphocyte events and performed a dimensionality reduction analysis (tSNE) using all fluorescent parameters except viability, CD14, CD19, CD3, NKG2A and NKG2C (for panels A and B in parallel) (Figure 5A). Figure 5B display the analysis of defined groups of CD8+, CD4+, TCRVα2.4+, TCRVα7.2+, and TCRγδ+ cells as well as the corresponding expression of every other marker in panels A and B.
Figures 3 and 4 show the general gating strategy for the three panels, identifying target T and NK cell populations (top figures). Corresponding graphs (column charts) show a summary of the frequency of cell subsets from their corresponding parent population (N = 10). In order to analyze every cell population (based on their differentiation and functional profiles), we followed the same exclusion gating strategy to separate and concatenate each subset. Then, we evaluated the expression of each marker through the median fluorescence intensity (MFI) (Figure 6).
Figure 6.
Expression analysis of T and NK cell populations
Heat maps showing the relative expression of markers (individually normalized MFI) across every subpopulation (∼10,000-concatenated events each) of T and NK cells following the gating strategy from Figures 3 and 4. TN, T naïve cells; CM, central memory cells; EM, effector memory cells; TEMRA, terminally differentiated cells; iNKT, invariant natural killer cells; MAIT, mucosal-associated invariant T cells; γδT, TCRγδ+ T cells; Th17, helper 17 cells; cTfh, circulating T follicular helper cells.
All together, these deep-phenotyping analyses allow studying CD8+NKG2A+ and CD8+NKG2C+ T cells, examining their phenotype in depth, as well as screening other T and NK cell subsets in a platform providing high quality comprehensive data, with higher flexibility and sensitivity as compared to conventional flow cytometry.
Limitations
The number of markers that can be included in the panels is limited by the configuration of the spectral cell analyzer (the number of lasers and detectors), as well as the combination of available antibodies and fluorochromes. However, the ID7000, with a total of 184 detectors and 6 lasers, is the spectral analyzer with the highest number of detectors on the market and allows the detection of more than 40 parameters simultaneously.24
The number of available antibody clones also limits the analyses in other systems like non-human primate (NHP) cells. For instance, some clones do not differentiate between NKG2A and NKG2C in nonhuman primate cells. The clones included in this protocol for NKG2A (S19004C) and NKG2C (134591) have human reactivity and can differentiate between these markers on human cells.
Acquiring the samples under different conditions and times may affect the values of MFI, which can be problematic for longitudinal analyses. It is important to take into account the experimental design, the expected output data to be analyzed in advance and, if possible, plan to acquire all the samples at once.
It is well known that there might be variations in the expression of different markers depending on the conditions for the preparation of the samples (including, temperature, reagents, time of incubation, etc.). For instance, we found that the expression of markers like CCR7 was very limited at 4°C as compared to higher incubation temperatures. Thus, we recommend considering this for the target markers in the panels. We standardized this protocol using recently thawed cells, however, we advise taking into account these considerations in order to prepare appropriate controls for the standardization of frozen versus fresh samples. Also, the use of the brilliant stain buffer improve the background reduction during the standardization process, so we recommend to use it when using more than one polymer dye-conjugated antibody in order to prevent non-specific polymer interactions.
Troubleshooting
Problem 1
Markers too dim or no signal (data analysis, step 26).
Potential solution
This could be due to incorrect PMT voltage set-up, incorrect or no antibodies added to the mix and inadequate permeabilization in the case of intracellular markers. If the true signal is low, meaning low expression of such marker, the use of FMO controls is recommended. Keep in mind that the expression of some markers (in particular chemokine receptors) changes in response to handling of the cells such temperature variations, thus resting cells in warm culture medium is recommended especially with recently thawed cells. However, this should be considered according to the target markers to be investigated. For instance, if cytokines are included in the panels, the use of monensin and/or brefeldin A should be considered and tested during the incubation period.
Problem 2
Incorrect double positive events, spillover, under compensation and/or background artifacts (data analysis, step 26).
Potential solution
These can be due to different causes. Bad unmixing can arise due to single stain incorrect identification and saturated events. Biological exclusive markers should be kept in fluorochromes with close emissions to minimize false double-positive events. It is recommended the use special stain buffers (as the brilliant stain buffer described in this protocol) since some fluorescent dyes, in particular polymer-based, may cause staining artifacts due to non-specific reactivity when staining the cells. In addition, the Spectral Unmixing Adjuster mode in the Sony system software allows manually adjusting and correcting unmixing matrix if necessary.
Also, bad titration and cross-reactivity might be responsible for under compensation and/or artifacts. Thus, it is recommended to titrate the antibodies in advance and potentially pre-staining problematic markers.
Problem 3
Poor quality and low rate events (sample acquisition, step 21).
Potential solution
Poor quality events might be due to inadequate sample handling or cells with low viability. In addition, it is possible that compensation beads and cells fall to the bottom of the well during long periods of sample acquisition and thus lowering the rate events. In this case, activating the continuous mixing mode in the instrument might decrease this issue. However, if many samples are being acquired it is possible to make pause intervals between certain given amount of samples and manually re-mix the volume before continuing acquisition. When gating, it is recommended to include only events with stable acquisition over time, to exclude doublets, debris and dead cells.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be answered by the lead contact, Prof. Michaela Müller-Trutwin (michaela.muller-trutwin@pasteur.fr).
Materials availability
This protocol did not generate new unique reagents.
Data and code availability
This study did not generate datasets or code.
Acknowledgments
The authors would like to thank the Cytometry and Biomarkers (UTechS CB) and Flow Cytometry platforms at the Institut Pasteur for their advice and support for standardizing the protocol. Likewise, we would like to thank BioLegend, France, and Camille Baey for their support. This work was supported by funding from Sidaction, the ANRS-MIE, and the Institut Pasteur. A.O.-R. received a doctoral fellowship from the Université Paris Cité and the Ministère français de lʼEnseignement supérieur, de la Recherche et de lʼInnovation. C.P. received a grant from Sidaction (Financement aux jeunes chercheurs-13200). We would also like to thank the reviewers of the manuscript for their valuable comments and time.
Author contributions
M.M.-T. and A.O.-R. conceived the study. A.O.-R. designed and developed the methodology for the panels, standardized and optimized the protocol, performed the experiments, analyzed the data, and wrote the manuscript. S.S. and S.N. standardized the local protocol for the use of the spectral analyzer instrument and provided advice and support through the standardization process. C.P. and N.H. provided scientific advice for the setup of the panels. M.M.-T. acquired the funding. B.J. coordinated and supervised the respect of the institutional guidelines. All authors reviewed and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Aurelio Orta-Resendiz, Email: aurelio.orta-resendiz@pasteur.fr.
Sandrine Schmutz, Email: sandrine.schmutz@pasteur.fr.
Michaela Müller-Trutwin, Email: michaela.muller-trutwin@pasteur.fr.
References
- 1.Braud V.M., Allan D.S., O’Callaghan C.A., Söderström K., D’Andrea A., Ogg G.S., Lazetic S., Young N.T., Bell J.I., Phillips J.H., et al. HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature. 1998;391:795–799. doi: 10.1038/35869. [DOI] [PubMed] [Google Scholar]
- 2.Walsh C.E., Ryan E.J., O’Farrelly C., Golden-Mason L., FitzGerald O., Veale D.J., Bresnihan B., Fearon U. Differential expression of NK receptors CD94 and NKG2A by T cells in rheumatoid arthritis patients in remission compared to active disease. PLoS One. 2011;6 doi: 10.1371/journal.pone.0027182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Katsurada T., Kobayashi W., Tomaru U., Baba T., Furukawa S., Ishizu A., Takeda K., Sakamoto N., Asaka M., Takeda H., Kasahara M. Decrease of peripheral and intestinal NKG2A-positive T cells in patients with ulcerative colitis. PLoS One. 2012;7 doi: 10.1371/journal.pone.0044113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li W.-X., Pan H.-F., Hu J.-L., Wang C.-Z., Zhang N., Li J., Li X.-P., Xu J.-H., Ye D.-Q. Assay of T- and NK-cell subsets and the expression of NKG2A and NKG2D in patients with new-onset systemic lupus erythematosus. Clin. Rheumatol. 2010;29:315–323. doi: 10.1007/s10067-009-1322-9. [DOI] [PubMed] [Google Scholar]
- 5.Kasakovski D., Zeng X., Lai J., Yu Z., Yao D., Chen S., Zha X., Li Y., Xu L. Characterization of KIR + NKG2A + Eomes- NK-like CD8+ T cells and their decline with age in healthy individuals. Cytometry B Clin. Cytom. 2021;100:467–475. doi: 10.1002/cyto.b.21945. [DOI] [PubMed] [Google Scholar]
- 6.Bian Y., Shang S., Siddiqui S., Zhao J., Joosten S.A., Ottenhoff T.H.M., Cantor H., Wang C.-R. MHC Ib molecule Qa-1 presents Mycobacterium tuberculosis peptide antigens to CD8+ T cells and contributes to protection against infection. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sottile R., Panjwani M.K., Lau C.M., Daniyan A.F., Tanaka K., Barker J.N., Brentjens R.J., Sun J.C., Le Luduec J.-B., Hsu K.C. Human cytomegalovirus expands a CD8+ T cell population with loss of BCL11B expression and gain of NK cell identity. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abe6968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zeddou M., Rahmouni S., Vandamme A., Jacobs N., Frippiat F., Leonard P., Schaaf-Lafontaine N., Vaira D., Boniver J., Moutschen M. Downregulation of CD94/NKG2A inhibitory receptors on CD8+ T cells in HIV infection is more pronounced in subjects with detected viral load than in their aviraemic counterparts. Retrovirology. 2007;4:72. doi: 10.1186/1742-4690-4-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng M., Gao Y., Wang G., Song G., Liu S., Sun D., Xu Y., Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell. Mol. Immunol. 2020;17:533–535. doi: 10.1038/s41423-020-0402-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y., Xin Z., Huang L., Zhao L., Wang S., Cheng J., Wu P., Chai Y. Cd8+ t cells form the predominant subset of nkg2a+ cells in human lung cancer. Front. Immunol. 2019;10:3002. doi: 10.3389/fimmu.2019.03002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ducoin K., Oger R., Bilonda Mutala L., Deleine C., Jouand N., Desfrançois J., Podevin J., Duchalais E., Cruard J., Benlalam H., et al. Targeting NKG2A to boost anti-tumor CD8 T-cell responses in human colorectal cancer. OncoImmunology. 2022;11 doi: 10.1080/2162402X.2022.2046931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huot N., Rascle P., Tchitchek N., Wimmer B., Passaes C., Contreras V., Desjardins D., Stahl-Hennig C., Le Grand R., Saez-Cirion A., et al. Role of NKG2a/c+CD8+ T cells in pathogenic versus non-pathogenic SIV infections. iScience. 2021;24 doi: 10.1016/j.isci.2021.102314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wagner J.A., Rosario M., Romee R., Berrien-Elliott M.M., Schneider S.E., Leong J.W., Sullivan R.P., Jewell B.A., Becker-Hapak M., Schappe T., et al. CD56bright NK cells exhibit potent antitumor responses following IL-15 priming. J. Clin. Invest. 2017;127:4042–4058. doi: 10.1172/JCI90387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Jonge K., Ebering A., Nassiri S., Maby-El Hajjami H., Ouertatani-Sakouhi H., Baumgaertner P., Speiser D.E. Circulating CD56bright NK cells inversely correlate with survival of melanoma patients. Sci. Rep. 2019;9:4487. doi: 10.1038/s41598-019-40933-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X., Feng J., Chen S., Yang H., Dong Z. Synergized regulation of NK cell education by NKG2A and specific Ly49 family members. Nat. Commun. 2019;10:5010. doi: 10.1038/s41467-019-13032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ram D.R., Manickam C., Hueber B., Itell H.L., Permar S.R., Varner V., Reeves R.K. Tracking KLRC2 (Nkg2c)+ memory-like NK cells in SIV+ and rhCMV+ rhesus macaques. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huot N., Rascle P., Petitdemange C., Contreras V., Stürzel C.M., Baquero E., Harper J.L., Passaes C., Legendre R., Varet H., et al. SIV-induced terminally differentiated adaptive NK cells in lymph nodes associated with enhanced MHC-E restricted activity. Nat. Commun. 2021;12:1282. doi: 10.1038/s41467-021-21402-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foley B., Cooley S., Verneris M.R., Pitt M., Curtsinger J., Luo X., Lopez-Vergès S., Lanier L.L., Weisdorf D., Miller J.S. Cytomegalovirus reactivation after allogeneic transplantation promotes a lasting increase in educated NKG2C+ natural killer cells with potent function. Blood. 2012;119:2665–2674. doi: 10.1182/blood-2011-10-386995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gondois-Rey F., Chéret A., Granjeaud S., Mallet F., Bidaut G., Lécuroux C., Ploquin M., Müller-Trutwin M., Rouzioux C., Avettand-Fenoël V., et al. NKG2C+ memory-like NK cells contribute to the control of HIV viremia during primary infection: Optiprim-ANRS 147. Clin. Transl. Immunology. 2017;6:e150. doi: 10.1038/cti.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chien Y.h., Meyer C., Bonneville M. γδ T cells: First line of defense and beyond. Annu. Rev. Immunol. 2014;32:121–155. doi: 10.1146/annurev-immunol-032713-120216. [DOI] [PubMed] [Google Scholar]
- 21.Carding S.R., Egan P.J. Gammadelta T cells: Functional plasticity and heterogeneity. Nat. Rev. Immunol. 2002;2:336–345. doi: 10.1038/nri797. [DOI] [PubMed] [Google Scholar]
- 22.Godfrey D.I., Koay H.-F., McCluskey J., Gherardin N.A. The biology and functional importance of MAIT cells. Nat. Immunol. 2019;20:1110–1128. doi: 10.1038/s41590-019-0444-8. [DOI] [PubMed] [Google Scholar]
- 23.Brennan P.J., Brigl M., Brenner M.B. Invariant natural killer T cells: An innate activation scheme linked to diverse effector functions. Nat. Rev. Immunol. 2013;13:101–117. doi: 10.1038/nri3369. [DOI] [PubMed] [Google Scholar]
- 24.Application Note: Forty-Two Color Flow Cytometry Panel Data Collected. Forty-two Color Flow Cytometry Panel Data Collected with the ID7000™ Spectral Cell Analyzer for Identifying Cellular Subsets in Human Peripheral Blood. Sony Biotechnology Inc. https://s3.amazonaws.com/creative.sonybiotechnology.com/ID7000/Sony+ID7000+Application+Note+42-Color+Flow+Cytometry+Panel+Data+Collected+with+the+ID7000+Spectral+Cell+Analyzer.pdf
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study did not generate datasets or code.