Abstract
Porcine circoviruses (PCVs) are members of the genus Circovirus of the family Circoviridae, and four species of PCVs have been discovered and named PCV1–PCV4, respectively. With the first report of PCV3 in America in 2016, the pathogenic variant was found to be associated with various clinical features, called porcine circovirus associated disease (PCVAD), including multisystemic inflammation, porcine dermatitis and nephropathy syndrome (PDNS), reproductive disorders, respiratory or digestive disorders. Increasing experimental data have shown that PCV3 is widespread around the world, but the failure of virus isolation and propagation has put obstacles in the way of PCV3 research. Moreover, a large number of reports demonstrate that PCV3 usually co-infects with other pathogens in pigs. Thus, whether PCV3 alone causes clinical manifestations needs to be fully discussed. In addition, the host cell immune response was activated during PCV3 infection, and PCV3-encoded proteins may regulate immune responses to facilitate its replication. An in-depth understanding of PCV3 pathogenesis and immune regulation strategies is critical for PCVAD prevention. In this review, the advances in pathogenicity and innate immune modulation of PCV3 were summarized, which could deepen the understanding of this virus and PCV3-related diseases.
Keywords: porcine circovirus 3 (PCV3), porcine circovirus associated disease (PCVAD), advance, pathogenicity, immune modulation
1. Introduction
Porcine circoviruses (PCVs), the smallest known DNA viruses in mammals, are members of the genus Circovirus of the family Circoviridae. At least four species of PCVs have been discovered in pigs and named PCV1 to PCV4, respectively, based on the genome homology analysis. In the 1970s, PCV1 was first found in PK-15 cells and then proved non-pathogenic in subsequent studies (1, 2). Subsequently, a variant strain of PCV2 was found in North America and Europe and thought to be responsible for multiple clinical presentations, such as post-weaning multisystemic wasting syndrome (3–5). Therefore, extensive studies were carried out, and great progress was made on viral pathogenesis and immune regulation mechanisms in the following 20 years. In 2016, a new virus, causing similar symptoms to PCV2, was reported and named PCV3 (6). In addition, PCV4 was recently discovered in 2019 (7), and there is only limited information about the pathogenesis and clinical implications of PCV4.
PCV3 was initially detected in the United States in pigs suffering from porcine dermatitis and nephropathy syndrome (PDNS) using high-throughput sequencing technology (6). However, numerous retrospective studies suggest that the pathogen has been circulating among pigs for a long time in Asia, Europe, South America, and North America (8–14). Notably, most of the PDNS pigs are PCV3-positive but not PCV2, and the PDNS-like disease in piglets was successfully reproduced based on an infectious PCV3 DNA clone (15), indicating PCV3 is sufficient to cause PDNS solely, and much investigation should be done to unveil its biology and pathogenesis. Moreover, PCV3 was highly prevalent in wild boars (16–21). Thus, the wild populations may be an important reservoir and could represent a concrete risk of spreading to domestic populations.
In general, all PCVs share a similar morphology, consisting of a close circular single-stranded DNA genome (22). The genome consisted from about 1760 nucleotides (nt), 1767 nt, and 2000 nt in PCV1, PCV2, and PCV3 (23), respectively. In addition, the sequence of PCV3 was only 48% homologous with PCV2, although they share similar genome morphologies (24). Further phylogenetic analysis also demonstrated the low similarity between PCV3 and other PCVs, such as 45.5 and 43.2% for PCV1 and PCV4, respectively (25). Recent studies reported that the novel PCV3 might originate from bats due to its high homology to some bat circoviruses at both nucleotide and amino acid levels (26, 27).
PCVs consist of three major open reading frames (ORFs) arranged in the strands of the replicative form (28). ORF1 is located on the positive strand and identified as the most conserved region of the circovirus genome (29), encoding for Rep and Rep′ proteins involved in virus replication initiation (30); ORF2 is located on the negative DNA viral strand and encodes the only structural protein (Cap), which is the most immunogenic viral protein and possesses multiple functions (31); ORF3 is oriented in the negative strand, which encodes for a non-structural protein regulating host cell apoptosis in PCV1 and PCV2 (32, 33), while its function remains unknown in PCV3 (6, 34). Traditionally, PCV3 is divided into three main clades (PCV3a, PCV3b, and PCV3c) based on the amino acid variations (35, 36), and PCV3a could be further classified into subclades, including PCV3a-1, PCV3a-2, and one intermediate clade (PCV3a-IM) (26, 27, 35, 36).
Since the first report of PCV3, it has been widely detected in the lungs, heart, kidneys, and other organs or tissues (15) and has caused a variety of clinical manifestations (37). The current research proves that various confounding factors affect the clinical manifestations and pathological changes of diseased pigs (38). PCV3 can replicate in almost all tissues, especially immune cells, where it causes targeted cell damage such as cell apoptosis and immune suppression. In addition, PCV3 infection could significantly upregulate pro-inflammatory cytokines and cause multisystemic inflammation in piglets and sows (15). Therefore, the interplay between PCV3 infection and immune responses is critical to its pathogenicity. In this review, we will comb and organize the pathogenicity and immune modulation mechanisms of PCV3.
2. PCV3 pathogenicity and its associated diseases
A series of clinical symptoms caused by PCVs are collectively known as porcine circovirus-associated diseases (PCVAD). In the past 20 years, PCV2 has been regarded as the main cause of PCVAD. PCV2 mainly proliferates in lymph nodes and induces immune cell apoptosis to decrease the immunity of infected pigs. Exciting progress has also been made in PCV3 pathogenicity. Jiang et al. successfully reproduced PDNS-like disease based on PCV3 alone or PCV3 in combination with an immune stimulator in animals (15). It is clear from the evidence that PCV3 infection leads to a variety of clinical and pathological symptoms, including PDNS, reproductive disorders, systemic inflammatory diseases (39), respiratory disorders (34, 40, 41), diarrhea (40, 42), and central nervous system signs (43, 44). Here, we describe the recent advances in the most consistent clinical signs associated with PCV3 infection (Figure 1).
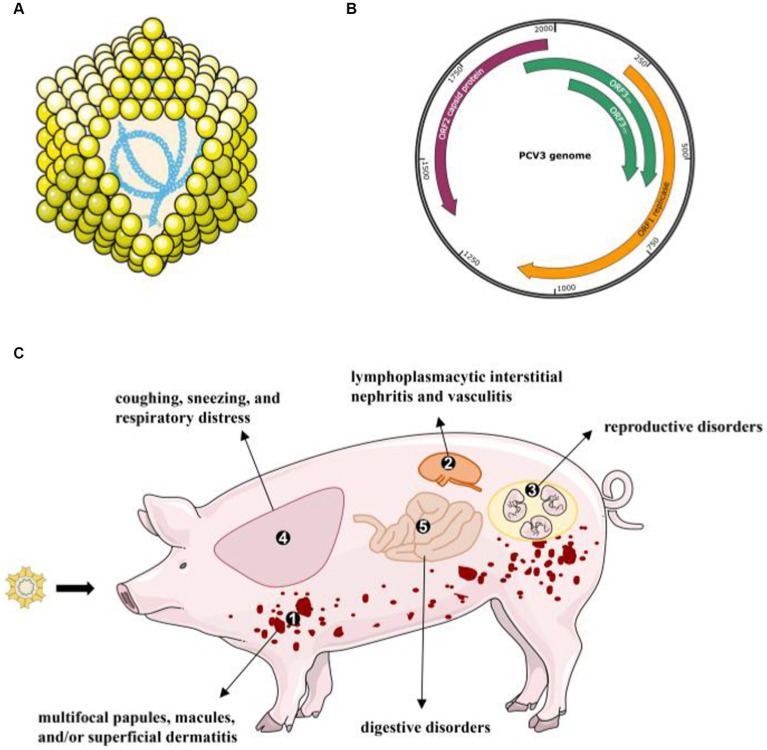
Figure 1.
The virion structure of PCV3 and its clinical diseases or signs (A) PCV3 has a single-stranded, circular DNA genome, and the capsid consists of 12 pentagonal, trumpet-shaped pentamers. (B) The PCV3 genome is about 2000-nt in length and contains three major ORFs: ORF1, ORF2, and ORF3 (45). (C) A variety of clinical and pathological symptoms are associated with PCV3 infection: ① skin: multifocal papules, macules, and/or superficial dermatitis; ② kidney: lymphoplasmacytic interstitial nephritis and vasculitis; ③ reproductive disorders: abortion, stillbirth, and mummified fetuses; ④ lung: coughing, sneezing, and respiratory distress associated with lymphoplasmacytic interstitial pneumonia; ⑤ digestive disorders.
2.1. Multisystemic inflammation
PCV3 was initially discovered in sows and aborted fetuses that had PDNS-like clinical symptoms and reproductive dysfunction (6). PDNS is mainly harmful to nursery and finishing pigs, with the occasional onset of piglets. The disease has a low mortality rate but a high incidence and a long course and is clinically characterized by irregular erythema of multifocal papules and superficial dermatitis. Currently, studies have shown that PDNS-like disease is usually the most severe form of the disease that is present in pigs that are naturally and experimentally infected with PCV3 (15, 46, 47). Typical PDNS lesions such as necrotizing vasculitis and glomerulonephritis could be observed in sows (48, 49). PCV3-infected sows and aborted fetuses showed multiorgan inflammation (39) and multisystemic vasculitis with a prominent skin and kidney tropism caused by type III hypersensitivity reactions (50). A wide range of vasculitis, from local to systemic, has been reported in PCV3-infected pigs (6, 15, 39, 44, 46, 51). Furthermore, vasculitis has also been found in the heart, kidney, and intestinal tissues of PCV3-inoculated pigs (51, 52). In addition, hepatic pathology, such as granulomatous lymphadenitis, has been demonstrated in the PCV3 challenge experiment (15, 53). Moreover, a high level of chemokines and pro-inflammatory mediators was detected in the PCV3-infected pigs (15), and it may be responsible for multisystemic inflammation, and host immune responses, and the observed lesions described above.
2.2. Reproductive disorders
Although PCV3 did not always lead to symptomatic signs in sows, the virus was widely colonized in gilts, weak-born piglets, and in the fetuses and placenta (51, 54–58), PCV3 could also be transmitted vertically through the colostrum, semen, and placenta (59). Reproductive disorder is a disease that reduces the performance of sows, including infertility, abortion, stillbirth, and weakness or neonatal malformation (60), affecting the global pig industry’s health development. Existing studies have shown that high levels of PCV3 are detected in pig farms with reproductive failure by qPCR testing (40, 61), and high PCV3 titers could be found in the aborted fetuses (46, 57), indicating that PCV3 infection could directly affect the fetuses (44). Although PCV3 can be detected in sows with or without reproductive disorders, the positive rates of PCV3 are much higher in reproductive failure sows than those in healthy sows via qPCR assays (61). In addition, high PCV3 viral loads were detected using qPCR in the various tissues, including the fetal heart, thymus, lymph nodes, and placenta, suggesting additional tissue tropism of PCV3 (62). Interestingly, the virus could also be detected in multiple tissues of mummies or stillborn fetuses, such as the trophoblast cells, the placenta, and the umbilical cord (21, 44, 62, 63). Thus, infection of the fetuses was thought to contribute to the reproductive failure induced by PCV3. However, the mechanism underlying PCV3-induced reproductive failure remains unclear and requires further study.
2.3. Respiratory or digestive disorders
Pigs, especially weaned piglets, infected with PCV3 typically exhibit various clinical symptoms, including respiratory diseases and digestive disorders (6, 35, 40, 64). However, the association between PCV3 circulating status and clinical respiratory manifestation is not well understood. Zhai et al. systemically analyzed and found that the PCV3 positive rate was positively correlated with respiratory symptoms (40). Similarly, a much higher percentage of diarrheal-weaned pigs were PCV3 positive than non-diarrheal ones (40). Another report reached a similar conclusion, there was a close association between PCV3 infection and digestive or respiratory diseases (35). Although the results above proved that PCV3 was potentially associated with swine respiratory disease and diarrhea, direct evidence was imperative. To answer this question, specific-pathogen-free (SPF) piglets were intranasally inoculated with a PCV3 virus obtained from the infectious DNA clone by Jiang et al. and they found that the virus infection increased respiratory disease rates (15). A variety of clinical respiratory symptoms were observed in this study, including coughing, sneezing, and respiratory distress.
Previous studies proved that PCV3 was able to replicate in the lungs and that virus infection could activate the innate immune system and trigger the release of inflammatory cytokines (39, 65). Clinical respiratory signs are usually accompanied by interstitial pneumonia and pleuritis, as well as other types of pulmonary lesions (66). Pneumonia will lead to alveolar congestion and edema, and lots of inflammatory secretions in the alveolar cavity will affect oxygen exchange. However, there are some conflicting results. The pigs only demonstrated subclinical infection without any pathological lung changes and showed no evidence of PCV3 replication in the lung tissue (51, 52). Another study showed that rescued PCV3 virus infection in Kunming mice did not affect the tissues or organs of either control or infected groups (67). At the moment, there is not enough information regarding the factors contributing to the lung tropism and pathogenesis of PCV3.
A report shows that there is a tendency for diarrhea in PCV3-positive suckling and weaned piglets (42). Based on immunohistochemistry, it was determined that the PCV3 antigen was present in all organs, including the intestinal tract (65, 68). Consistently, the infected pigs developed diarrhea, with degeneration and necrosis of the small intestinal epithelium in the PDNS-like lesions study (15). The piglets inoculated with PCV3 showed a high level of PCV3-positive cells within their small intestine tissues (69). Experimental infection studies revealed that piglets inoculated with PCV3 showed a series of lesions in the small intestine, including villus and crypt atrophy, eosinophilic and lymphocyte infiltration, mucosal epithelial cells and lymphocyte necrosis, and small numbers of plasma cells (42, 69). Gut microbiota is made up of trillions of bacteria and plays critical roles in modulating host immunity; changes in the microbiota are closely linked to the progression of diseases. Therefore, it may be an effective way to prevent PCV3 infection through the regulation or changes of the gut microbiota.
3. Immune modulation of PCV3
PCV3 has been widely reported around the world since its discovery and is characterized by typical PDNS lesions. Unfortunately, only one successful PCV3 isolation has been recently reported in cell culture (70), and there may be various unknown factors associated with the disease. Increasing results have shown that PCV3 triggers the host cells’ immune response when invading host cells (Figure 2). High levels of the PCV3 genome could be found in aborted and weak-born piglets, especially in the thymus and lymph nodes (62). Moreover, a few animal detections found that PCV3 exhibits persistent infection (18) or asymptomatic infection (63). Overall, the virus tends to replicate in almost all tissues in pigs, and strongly prefers to replicate in immune cells, and causes targeted cell damage.
Figure 2.
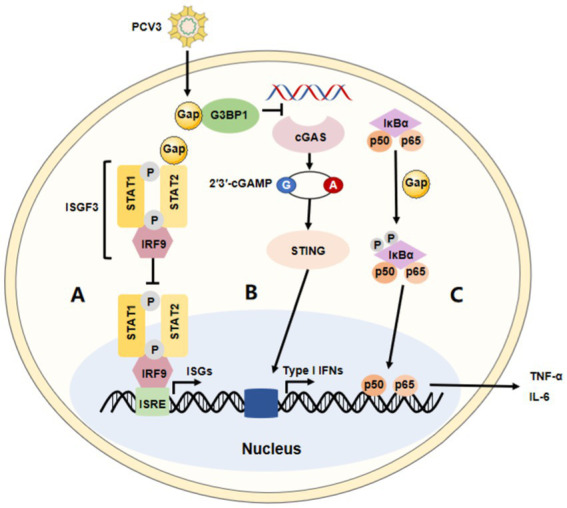
The immune modulation of PCV3. The summary of the major immune modulations of PCV3. (A) PCV3 Cap interacts with the transactivation domain of STAT2 and inhibits ISRE promoter activation induced by IRF9-S2C, helping PCV3 to escape type I interferon-meditated host innate immunity. (B) PCV3 Cap interacts with G3BP1 to prevent cGAS from recognizing DNA, affecting type I interferon production via the cGAS-STING pathway. (C) PCV3 Cap protein induces NF-κB activation and upregulates the expression of pro-inflammatory cytokines.
3.1. Innate immune response
3.1.1. Type I interferon response
A diverse range of defensive strategies are employed by different viruses to battle innate immunity. However, the immune evasion strategies of PCV3 are not fully understood. It has been reported that Cap protein could significantly inhibit the activity of the IFN-stimulated response element (ISRE) by interacting with the transcription (STAT2) transactivation domain (56, 71). Several studies have demonstrated PCV2 not only activates cGAS/STING and RIG-like receptor (RLR) signaling, but it also increases IFN-β expression, which promotes the proliferation of PCV2. Meanwhile, in this process, surprisingly, there seems no appearance to prevent its replication through the NF-κB signaling pathway (72–74). These findings are consistent with those in the studies on PCV3. It has been reported that the PCV3 Cap protein inhibits the type I interferon signaling pathway by inhibiting ISRE promoter activity. Mechanically, Cap interacts with the transactivation domain of STAT2 and binds to ISRE to prevent the binding of STAT2 and ISRE (71). Therefore, PCV3 could evade the host’s innate immunity mediated by IFN. In addition, PCV3 Cap can also interact with G3BP1 and inhibit the induction of type I interferon (75). According to proteomic analysis, PCV3 infection in SPF piglets infected with infectious clones of PCV3 increased several IFN-related factors, such as IFIT3, ISG15, and so on, in lung tissue (47). Another study performed on HEK-293 T cells found that, though it was not observed that PCV3 Cap contributed to the controlled expression of IFN-β, the mRNA level of RIG-I/MDA5 could significantly be increased by Cap protein in the RLRs pathway (76). Therefore, these studies indicate that the regulation of the type I interferon response may be an important strategy for host immune escape by PCV3. Moreover, a recent paper reported that the PCV3 Cap protein is involved in cell autophagy, another mechanism thought to be effectively used by viruses to enhance their replication and persistency (77). Mechanically, the protein induced complete autophagy by inhibiting the phosphorylation of the mammalian target of rapamycin in HEK-293 T cells, including the formation of autophagosomes and autophagic vesicles, as well as the transformation of LC3-I to LC3-II (78). Of course, the ubiquitin-proteasome pathway is also involved in this process. In all, information in this area is necessary to understand the role of pathogenicity and innate immunity during PCV3 infection.
3.1.2. Inflammatory responses
According to previous studies, a close association exists between PCV3 and multisystem inflammation, as well as respiratory disease, diarrhea, myocarditis, encephalitis, periarteritis, and so on (6, 34, 39, 44, 51, 67, 68, 79, 80). Studies have shown that uncontrolled inflammatory responses might contribute to significant damage in PCV3-infected pigs (51, 79). PCV3 infection was demonstrated to have regulatory effects on IL-8 expression by evaluating the innate immune response in vivo (52). Significantly, higher levels of pro-inflammatory mediators and chemokines were observed in PCV3-infected piglets (15). In a recent study, PCV3 Cap was shown to be a critical factor in activating NF-κB signaling by upregulating pro-inflammatory mediators such as RIG-I and MDA5 in HEK-293 T cells (76). This may provide a basis for the pathogenesis of PCV3 and the innate immunity of the host. Therefore, the induction of a chronic pro-inflammatory state of disease results in dysregulation of innate immunity in PCV3-infected pigs, which may provide a possible explanation for the subsequent clinical signs. PCV3 Cap protein appears to activate some signaling pathways, but the mechanism is still unclear and needs further investigation.
3.2. Cell-mediated immune responses
As a consequence of the innate immune subversion, reduced T-cell response, compromised antigen presentation ability, and the imbalance of immunosuppressive cytokine secretion, the adaptive immune response against PCV2 was severely influenced (81). For example, the proteins of PCV2 (such as Cap and Rep) stimulate antigen-specific IFN-γ secreting cells (82–84). Meanwhile, the infection of PCV2 could directly affect immature thymocytes, such as by inhibiting thymocyte selection, which results in disturbances of helper T cell immunity (85). Even when the immune response to PCV2 is low, pigs still display protective clinical symptoms, indicating that cell-mediated immune responses, specifically Th1-mediated responses, play crucial roles in protective immunity. A study using recombinant PCV3-infected conventional weaning piglets and PBMCs found that they were incapable of responding to mitogen stimulation, but it is unclear whether this effect is permanent or whether energy of lymphocytes can be reversed (15). As a result of proteomic analysis of PCV3-infected pigs, it was found that both SLA-I and II loci in lung tissue were significantly upregulated (47). At the same time, it has been reported that most of the influx of lymphocytes is associated with T-cell populations, regardless of peripheral response to PCV3 (52). However, a more detailed study of PCV3-mediated cell immunity is necessary to delve deeper into the pathogenesis of PCV3.
3.3. Immunopathogenesis
All the pathogenic PCV types are widespread and have been detected in both healthy and sick pigs. They have a variety of clinical manifestations and often cause chronic systemic infections. Current research results have suggested that the homeostasis disorder of the immune system may be the key factor leading to the pathogenesis of PCV infection. In PCV2 infection experiments, the virus could interact with components of the immune system, including immune cells, to impair innate and adaptive immunity. Dysregulation of the immune response can lead to a series of serious consequences, including rapid upregulation of pro-inflammatory factors, the formation of immune complexes, decreased antigen presentation ability, and necrosis of lymphocytes and immune cells. For example, several studies have demonstrated that the infections of PCV3 and PCV2 are connected to the immune complexes’ formation (15, 86). Although there are few reports on the immunosuppression of PCV3 to date, considering the similarity of clinical symptoms and pathogenicity between PCV3 and PCV2, we believe that PCV3 may also have strong immunosuppression, especially in the case of co-infection with other viruses, which is also worth exploring in the following study.
4. Others
4.1. Co-infection
In recent years, multi-pathogen mixed infection or co-infection has become more common in clinical practice, especially with a variety of pig viruses, including PCVs. Co-infection not only leads to more serious diseases than any single virus infection but also has a negative impact on the pig immune system and aggravates the complexity of pig farm diseases. There are reports that the co-infection rates of PCV3 with porcine reproductive and respiratory syndrome virus, classical swine fever virus, pseudorabies virus, porcine epidemic diarrhea virus (PEDV), and porcine parvovirus (PPV) were 36.36, 6.92, 14.53, 27.27, and 74.2% in some pig farms, respectively (87–91), indicating that the co-infection of PCV3 with other pathogens is common in pig farms. Moreover, co-infection of PCV2 and PCV3 was mostly reported in swine farms, although they share a similar genome and belong to the same genus. A survey of serum samples from clinically healthy pigs from major European countries showed a 3% positive rate of PCV2–PCV3 co-infection in fattening pigs (92). And a report found that the PCV2 and PCV3 co-infection rates gradually increased from 3.4% in 2016 to 16.1% in 2018 in the Midwest of the United States (93). In addition, studies have reported that the positive rate of PCV2 and PCV3 co-infection in different regions of China ranges from 6.78 to 19.7% (94, 95). These results indicate that the prevalence of PCV2 and PCV3 co-infection is widespread all over the world and has gradually increased in recent years.
4.2. Vaccination
Vaccination has been proven efficacious and successful for the prevention of PCV2 and other viral pathogens. Commercial PCV2 vaccines are mostly inactivated or Cap-based subunit vaccines, which provide favorable protective immunity against other types of PCV2 (68). ORF2 encodes the only structural protein capsid, and the identity between PCV2 and PCV3 was lower. Therefore, the vaccines based on PCV2 may only provide limited protection against PCV3. As expected, there was no correlation between PCV2 vaccination and PCV3 circulation, indicating the cross-protection between PCV2 and PCV3 was poor (96). However, there are no PCV3-based commercial vaccines to prevent PCV3 infection. Therefore, it is urgent to develop efficacious measures like universal antibodies and vaccines to prevent the disease in the future.
5. Conclusion and perspective
PCV3 was widespread in the world, including Asia, Europe, and America (8–13), according to retrospective epidemiological studies, although it was first discovered in pigs with PDNS-like clinical signs in 2016 (6). It is generally considered to be pathogenic and associated with various symptoms similar to those of PCV2, including PDNS, reproductive failure, and respiratory diseases. The symptoms described in this paper might not be the major clinical signs of PCV3, and there may be other contributing factors. Thus, the impact of PCV3 is still controversial to some extent, and further information is required to understand its potential pathogenicity. PK-15 and porcine testicular cells were susceptible to PCV2 and used for PCV2 isolation and propagation (97), but PCV3 mostly failed to be isolated and propagated in these passage cells (70). Furthermore, only one PCV3 infection model was reported. Therefore, the restrictions above hinder the research into PCV3 pathogenesis. Accumulating data indicated that PCV3 is often co-infected with PCV2, PPV, PEDV, and other pathogens, which not only lead to the severity of the disease, but also become a serious threat to the healthy development of the pig industry system (56, 58, 98). Moreover, PCV3 infection has a low mortality rate but a high morbidity and a long course of disease, which may be a great threat to the pork industry. Thus, it is imperative to further systematically evaluate and investigate the co-infection prevalence and pathogenicity of PCV3 with other pathogens and to develop an effective vaccine against PCV3.
Author contributions
DC: Data curation, Investigation, Methodology, Resources, Writing – original draft, Writing – review & editing. LZ: Conceptualization, Writing – review & editing. SX: Funding acquisition, Project administration, Resources, Supervision, Validation, Writing – review & editing.
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was supported by R&D Program of Beijing Municipal Education Commission (KM202310020004).
Conflict of interest
DC and LZ were employed by Zhongmu Institutes of China Animal Husbandry Industry Co., Ltd.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- 1.Tischer I, Rasch R, Tochtermann G. Characterization of papovavirus-and picornavirus-like particles in permanent pig kidney cell lines. Zentralbl Bakteriol Orig A. (1974) 226:153–67. PMID: [PubMed] [Google Scholar]
- 2.Tischer I, Mields W, Wolff D, Vagt M, Griem W. Studies on epidemiology and pathogenicity of porcine circovirus. Arch Virol. (1986) 91:271–6. doi: 10.1007/BF01314286 [DOI] [PubMed] [Google Scholar]
- 3.Allan GM, Mc Neilly F, Meehan BM, Kennedy S, Mackie DP, Ellis JA, et al. Isolation and characterisation of circoviruses from pigs with wasting syndromes in Spain, Denmark and Northern Ireland. Vet Microbiol. (1999) 66:115–23. doi: 10.1016/S0378-1135(99)00004-8, PMID: [DOI] [PubMed] [Google Scholar]
- 4.Wellenberg GJ, Pesch S, Berndsen FW, Steverink PJ, Hunneman W, Van der Vorst TJ, et al. Isolation and characterization of porcine circovirus type 2 from pigs showing signs of post-weaning multisystemic wasting syndrome in the Netherlands. Vet Q. (2000) 22:167–72. doi: 10.1080/01652176.2000.9695049, PMID: [DOI] [PubMed] [Google Scholar]
- 5.Allan GM, McNeilly F, Kennedy S, Daft B, Clarke EG, Ellis JA, et al. Isolation of porcine circovirus-like viruses from pigs with a wasting disease in the USA and Europe. J Vet Diagn Investig. (1998) 10:3–10. doi: 10.1177/104063879801000102 [DOI] [PubMed] [Google Scholar]
- 6.Palinski R, Pineyro P, Shang P, Yuan F, Guo R, Fang Y, et al. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. J Virol. (2017) 91:e01879. doi: 10.1128/JVI.01879-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang HH, Hu WQ, Li JY, Liu TN, Zhou JY, Opriessnig T, et al. Novel circovirus species identified in farmed pigs designated as porcine circovirus 4, Hunan province. China Transbound Emerg Dis. (2020) 67:1057–61. doi: 10.1111/tbed.13446, PMID: [DOI] [PubMed] [Google Scholar]
- 8.Faccini S, Barbieri I, Gilioli A, Sala G, Gibelli LR, Moreno A, et al. Detection and genetic characterization of porcine circovirus type 3 in Italy. Transbound Emerg Dis. (2017) 64:1661–4. doi: 10.1111/tbed.12714, PMID: [DOI] [PubMed] [Google Scholar]
- 9.Franzo G, Legnardi M, Hjulsager CK, Klaumann F, Larsen LE, Segales J, et al. Full-genome sequencing of porcine circovirus 3 field strains from Denmark, Italy and Spain demonstrates a high within-Europe genetic heterogeneity. Transbound Emerg Dis. (2018) 65:602–6. doi: 10.1111/tbed.12836, PMID: [DOI] [PubMed] [Google Scholar]
- 10.Ku X, Chen F, Li P, Wang Y, Yu X, Fan S, et al. Identification and genetic characterization of porcine circovirus type 3 in China. Transbound Emerg Dis. (2017) 64:703–8. doi: 10.1111/tbed.12638, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon T, Yoo SJ, Park CK, Lyoo YS. Prevalence of novel porcine circovirus 3 in Korean pig populations. Vet Microbiol. (2017) 207:178–80. doi: 10.1016/j.vetmic.2017.06.013, PMID: [DOI] [PubMed] [Google Scholar]
- 12.Saraiva GL, Vidigal PMP, Assao VS, Fajardo MLM, Loreto ANS, Fietto JLR, et al. Retrospective detection and genetic characterization of porcine circovirus 3 (PCV3) strains identified between 2006 and 2007 in Brazil. Viruses. (2019) 11:201. doi: 10.3390/v11030201, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stadejek T, Wozniak A, Milek D, Biernacka K. First detection of porcine circovirus type 3 on commercial pig farms in Poland. Transbound Emerg Dis. (2017) 64:1350–3. doi: 10.1111/tbed.12672, PMID: [DOI] [PubMed] [Google Scholar]
- 14.Chen D, Huang Y, Guo Y, Wang L, Zhang Y, Zhou L, et al. Prevalence and evolution analysis of porcine circovirus 3 in China from 2018 to 2022. Animals. (2022) 12:1588. doi: 10.3390/ani12121588, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang H, Wang D, Wang J, Zhu S, She R, Ren X, et al. Induction of porcine dermatitis and nephropathy syndrome in piglets by infection with porcine circovirus type 3. J Virol. (2019) 93:e02018. doi: 10.1128/JVI.02045-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amoroso MG, Serra F, Esposito C, D’Alessio N, Ferrara G, Cioffi B, et al. Prevalence of infection with porcine circovirus types 2 and 3 in the wild boar population in the Campania region (southern Italy). Animals. (2021) 11:3215. doi: 10.3390/ani11113215, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franzo G, Tucciarone CM, Drigo M, Cecchinato M, Martini M, Mondin A, et al. First report of wild boar susceptibility to porcine circovirus type 3: high prevalence in the Colli Euganei Regional Park (Italy) in the absence of clinical signs. Transbound Emerg Dis. (2018) 65:957–62. doi: 10.1111/tbed.12905, PMID: [DOI] [PubMed] [Google Scholar]
- 18.Klaumann F, Dias-Alves A, Cabezon O, Mentaberre G, Castillo-Contreras R, Lopez-Bejar M, et al. Porcine circovirus 3 is highly prevalent in serum and tissues and may persistently infect wild boar (Sus scrofa scrofa). Transbound Emerg Dis. (2019) 66:91–101. doi: 10.1111/tbed.12988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prinz C, Stillfried M, Neubert LK, Denner J. Detection of PCV3 in German wild boars. Virol J. (2019) 16:25. doi: 10.1186/s12985-019-1133-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Souza T, Gava D, Schaefer R, Leme RA, Porto GDS, Alfieri AA. Porcine circovirus 3a field strains in free-living wild boars in Parana state, Brazil. Animals. (2021) 11:1634. doi: 10.3390/ani11061634, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dei Giudici S, Franzoni G, Bonelli P, Angioi PP, Zinellu S, Deriu V, et al. Genetic characterization of porcine circovirus 3 strains circulating in Sardinian pigs and wild boars. Pathogens. (2020) 9:344. doi: 10.3390/pathogens9050344, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tischer I, Gelderblom H, Vettermann W, Koch MA. A very small porcine virus with circular single-stranded DNA. Nature. (1982) 295:64–6. doi: 10.1038/295064a0 [DOI] [PubMed] [Google Scholar]
- 23.Zhang D, He K, Wen L, Fan H. Genetic and phylogenetic analysis of a new porcine circovirus type 2 (PCV2) strain in China. Arch Virol. (2015) 160:3149–51. doi: 10.1007/s00705-015-2615-8 [DOI] [PubMed] [Google Scholar]
- 24.Yuzhakov AG, Raev SA, Alekseev KP, Grebennikova TV, Verkhovsky OA, Zaberezhny AD, et al. First detection and full genome sequence of porcine circovirus type 3 in Russia. Virus Genes. (2018) 54:608–11. doi: 10.1007/s11262-018-1582-z [DOI] [PubMed] [Google Scholar]
- 25.Guo Z, Li X, Deng R, Zhang G. Detection and genetic characteristics of porcine circovirus 3 based on oral fluids from asymptomatic pigs in Central China. BMC Vet Res. (2019) 15:200. doi: 10.1186/s12917-019-1952-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li G, He W, Zhu H, Bi Y, Wang R, Xing G, et al. Origin, genetic diversity, and evolutionary dynamics of novel porcine circovirus 3. Adv Sci. (2018) 5:1800275. doi: 10.1002/advs.201800275, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li G, Wang H, Wang S, Xing G, Zhang C, Zhang W, et al. Insights into the genetic and host adaptability of emerging porcine circovirus 3. Virulence. (2018) 9:1301–13. doi: 10.1080/21505594.2018.1492863, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosario K, Breitbart M, Harrach B, Segales J, Delwart E, Biagini P, et al. Revisiting the taxonomy of the family Circoviridae: establishment of the genus Cyclovirus and removal of the genus Gyrovirus. Arch Virol. (2017) 162:1447–63. doi: 10.1007/s00705-017-3247-y, PMID: [DOI] [PubMed] [Google Scholar]
- 29.Mankertz A, Caliskan R, Hattermann K, Hillenbrand B, Kurzendoerfer P, Mueller B, et al. Molecular biology of porcine circovirus: analyses of gene expression and viral replication. Vet Microbiol. (2004) 98:81–8. doi: 10.1016/j.vetmic.2003.10.014, PMID: [DOI] [PubMed] [Google Scholar]
- 30.Rosario K, Duffy S, Breitbart M. A field guide to eukaryotic circular single-stranded DNA viruses: insights gained from metagenomics. Arch Virol. (2012) 157:1851–71. doi: 10.1007/s00705-012-1391-y, PMID: [DOI] [PubMed] [Google Scholar]
- 31.Nawagitgul P, Harms PA, Morozov I, Thacker BJ, Sorden SD, Lekcharoensuk C, et al. Modified indirect porcine circovirus (PCV) type 2-based and recombinant capsid protein (ORF2)-based enzyme-linked immunosorbent assays for detection of antibodies to PCV. Clin Diagn Lab Immunol. (2002) 9:33–40. doi: 10.1128/CDLI.9.1.33-40.2002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Karuppannan AK, Kwang J. ORF3 of porcine circovirus 2 enhances the in vitro and in vivo spread of the of the virus. Virology. (2011) 410:248–56. doi: 10.1016/j.virol.2010.11.009, PMID: [DOI] [PubMed] [Google Scholar]
- 33.Lin WL, Chien MS, Wu PC, Lai CL, Huang C. The porcine circovirus type 2 nonstructural protein ORF3 induces apoptosis in porcine peripheral blood mononuclear cells. Open Virol J. (2011) 5:148–53. doi: 10.2174/1874357901105010148, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim SH, Park JY, Jung JY, Kim HY, Park YR, Lee KK, et al. Detection and genetic characterization of porcine circovirus 3 from aborted fetuses and pigs with respiratory disease in Korea. J Vet Sci. (2018) 19:721–4. doi: 10.4142/jvs.2018.19.5.721, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi S, Su M, Guo D, Li C, Wei S, Feng L, et al. Molecular detection and phylogenetic analysis of porcine circovirus type 3 in 21 provinces of China during 2015-2017. Transbound Emerg Dis. (2019) 66:1004–15. doi: 10.1111/tbed.13125, PMID: [DOI] [PubMed] [Google Scholar]
- 36.Fu X, Fang B, Ma J, Liu Y, Bu D, Zhou P, et al. Insights into the epidemic characteristics and evolutionary history of the novel porcine circovirus type 3 in southern China. Transbound Emerg Dis. (2018) 65:e296–303. doi: 10.1111/tbed.12752 [DOI] [PubMed] [Google Scholar]
- 37.Kroeger M, Temeeyasen G, Pineyro PE. Five years of porcine circovirus 3: what have we learned about the clinical disease, immune pathogenesis, and diagnosis. Virus Res. (2022) 314:198764. doi: 10.1016/j.virusres.2022.198764, PMID: [DOI] [PubMed] [Google Scholar]
- 38.Sirisereewan C, Thanawongnuwech R, Kedkovid R. Current understanding of the pathogenesis of porcine circovirus 3. Pathogens. (2022) 11:64. doi: 10.3390/pathogens11010064, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Phan TG, Giannitti F, Rossow S, Marthaler D, Knutson TP, Li L, et al. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol J. (2016) 13:184. doi: 10.1186/s12985-016-0642-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhai SL, Zhou X, Zhang H, Hause BM, Lin T, Liu R, et al. Comparative epidemiology of porcine circovirus type 3 in pigs with different clinical presentations. Virol J. (2017) 14:222. doi: 10.1186/s12985-017-0892-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kedkovid R, Woonwong Y, Arunorat J, Sirisereewan C, Sangpratum N, Lumyai M, et al. Porcine circovirus type 3 (PCV3) infection in grower pigs from a Thai farm suffering from porcine respiratory disease complex (PRDC). Vet Microbiol. (2018) 215:71–6. doi: 10.1016/j.vetmic.2018.01.004 [DOI] [PubMed] [Google Scholar]
- 42.Zhang F, Yuan W, Li Z, Zhang Y, Zeng X, Zhao M, et al. Porcine circovirus type 3 in pig farms experiencing diarrhea in Jiangxi, China: prevalence, genome sequence and pathogenicity. Animals. (2020) 10:2324. doi: 10.3390/ani10122324, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen GH, Mai KJ, Zhou L, Wu RT, Tang XY, Wu JL, et al. Detection and genome sequencing of porcine circovirus 3 in neonatal pigs with congenital tremors in South China. Transbound Emerg Dis. (2017) 64:1650–4. doi: 10.1111/tbed.12702, PMID: [DOI] [PubMed] [Google Scholar]
- 44.Arruda B, Pineyro P, Derscheid R, Hause B, Byers E, Dion K, et al. PCV3-associated disease in the United States swine herd. Emerg Microbes Infect. (2019) 8:684–98. doi: 10.1080/22221751.2019.1613176, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fux R, Sockler C, Link EK, Renken C, Krejci R, Sutter G, et al. Full genome characterization of porcine circovirus type 3 isolates reveals the existence of two distinct groups of virus strains. Virol J. (2018) 15:25. doi: 10.1186/s12985-018-0929-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bera BC, Choudhary M, Anand T, Virmani N, Sundaram K, Choudhary B, et al. Detection and genetic characterization of porcine circovirus 3 (PCV3) in pigs in India. Transbound Emerg Dis. (2020) 67:1062–7. doi: 10.1111/tbed.13463 [DOI] [PubMed] [Google Scholar]
- 47.Jiang H, Wei L, Wang D, Wang J, Zhu S, She R, et al. ITRAQ-based quantitative proteomics reveals the first proteome profiles of piglets infected with porcine circovirus type 3. J Proteome. (2020) 212:103598. doi: 10.1016/j.jprot.2019.103598 [DOI] [PubMed] [Google Scholar]
- 48.Wellenberg GJ, Stockhofe-Zurwieden N, de Jong MF, Boersma WJ, Elbers AR. Excessive porcine circovirus type 2 antibody titres may trigger the development of porcine dermatitis and nephropathy syndrome: a case-control study. Vet Microbiol. (2004) 99:203–14. doi: 10.1016/j.vetmic.2004.01.001, PMID: [DOI] [PubMed] [Google Scholar]
- 49.Opriessnig T, Meng XJ, Halbur PG. Porcine circovirus type 2 associated disease: update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J Vet Diagn Investig. (2007) 19:591–615. doi: 10.1177/104063870701900601, PMID: [DOI] [PubMed] [Google Scholar]
- 50.Thibault S, Drolet R, Germain MC, D'Allaire S, Larochelle R, Magar R. Cutaneous and systemic necrotizing vasculitis in swine. Vet Pathol. (1998) 35:108–16. doi: 10.1177/030098589803500204, PMID: [DOI] [PubMed] [Google Scholar]
- 51.Mora-Diaz J, Pineyro P, Shen H, Schwartz K, Vannucci F, Li G, et al. Isolation of PCV3 from perinatal and reproductive cases of PCV3-associated disease and in vivo characterization of PCV3 replication in CD/CD growing pigs. Viruses. (2020) 12:219. doi: 10.3390/v12020219, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Temeeyasen G, Lierman S, Arruda BL, Main R, Vannucci F, Gimenez-Lirola LG, et al. Pathogenicity and immune response against porcine circovirus type 3 infection in caesarean-derived, colostrum-deprived pigs. J Gen Virol. (2021) 102:1502. doi: 10.1099/jgv.0.001502 [DOI] [PubMed] [Google Scholar]
- 53.Zhang J, Liu Z, Zou Y, Zhang N, Wang D, Tu D, et al. First molecular detection of porcine circovirus type 3 in dogs in China. Virus Genes. (2018) 54:140–4. doi: 10.1007/s11262-017-1509-0 [DOI] [PubMed] [Google Scholar]
- 54.Saporiti V, Martorell S, Cruz TF, Klaumann F, Correa-Fiz F, Balasch M, et al. Frequency of detection and phylogenetic analysis of porcine circovirus 3 (PCV-3) in healthy primiparous and multiparous sows and their mummified fetuses and stillborn. Pathogens. (2020) 9:533. doi: 10.3390/pathogens9070533, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vargas-Bermudez DS, Vargas-Pinto MA, Mogollon JD, Jaime J. Field infection of a gilt and its litter demonstrates vertical transmission and effect on reproductive failure caused by porcine circovirus type 3 (PCV3). BMC Vet Res. (2021) 17:150. doi: 10.1186/s12917-021-02862-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dal Santo AC, Cezario KC, Bennemann PE, Machado SA, Martins M. Full-genome sequences of porcine circovirus 3 (PCV3) and high prevalence in mummified fetuses from commercial farms in Brazil. Microb Pathog. (2020) 141:104027. doi: 10.1016/j.micpath.2020.104027 [DOI] [PubMed] [Google Scholar]
- 57.Saporiti V, Valls L, Maldonado J, Perez M, Correa-Fiz F, Segales J, et al. Porcine circovirus 3 detection in aborted fetuses and stillborn piglets from swine reproductive failure cases. Viruses. (2021) 13:264. doi: 10.3390/v13020264, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Serena MS, Cappuccio JA, Barrales H, Metz GE, Aspitia CG, Lozada I, et al. First detection and genetic characterization of porcine circovirus type 3 (PCV3) in Argentina and its association with reproductive failure. Transbound Emerg Dis. (2021) 68:1761–6. doi: 10.1111/tbed.13893, PMID: [DOI] [PubMed] [Google Scholar]
- 59.Chen S, Zhang L, Li X, Niu G, Ren L. Recent Progress on epidemiology and pathobiology of porcine circovirus 3. Viruses. (2021) 13:1944. doi: 10.3390/v13101944, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanchez RE, Nauwynck HJ, McNeilly F, Allan GM, Pensaert MB. Porcine circovirus 2 infection in swine foetuses inoculated at different stages of gestation. Vet Microbiol. (2001) 83:169–76. doi: 10.1016/S0378-1135(01)00425-4 [DOI] [PubMed] [Google Scholar]
- 61.Zou Y, Zhang N, Zhang J, Zhang S, Jiang Y, Wang D, et al. Molecular detection and sequence analysis of porcine circovirus type 3 in sow sera from farms with prolonged histories of reproductive problems in Hunan. China Arch Virol. (2018) 163:2841–7. doi: 10.1007/s00705-018-3914-7, PMID: [DOI] [PubMed] [Google Scholar]
- 62.Deim Z, Dencso L, Erdelyi I, Valappil SK, Varga C, Posa A, et al. Porcine circovirus type 3 detection in a Hungarian pig farm experiencing reproductive failures. Vet Rec. (2019) 185:84. doi: 10.1136/vr.104784 [DOI] [PubMed] [Google Scholar]
- 63.Zheng S, Wu X, Zhang L, Xin C, Liu Y, Shi J, et al. The occurrence of porcine circovirus 3 without clinical infection signs in Shandong Province. Transbound Emerg Dis. (2017) 64:1337–41. doi: 10.1111/tbed.12667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang K, Zhang M, Liu Q, Cao Y, Zhang W, Liang Y, et al. Epidemiology and evolution of emerging porcine circovirus-like viruses in pigs with hemorrhagic dysentery and diarrhea symptoms in Central China from 2018 to 2021. Viruses. (2021) 13:2282. doi: 10.3390/v13112282, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.De Conti ER, Resende TP, Marshall-Lund L, Rovira A, Vannucci FA. Histological lesions and replication sites of PCV3 in naturally infected pigs. Animals. (2021) 11:1520. doi: 10.3390/ani11061520, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Saporiti V, Cruz TF, Correa-Fiz F, Nunez JI, Sibila M, Segales J. Similar frequency of porcine circovirus 3 (PCV-3) detection in serum samples of pigs affected by digestive or respiratory disorders and age-matched clinically healthy pigs. Transbound Emerg Dis. (2020) 67:199–205. doi: 10.1111/tbed.13341, PMID: [DOI] [PubMed] [Google Scholar]
- 67.Jiang Z, Wu J, Jiang M, Xie Y, Bu W, Liu C, et al. A novel technique for constructing infectious cloning of type 3 porcine circovirus. Front Microbiol. (2020) 11:1067. doi: 10.3389/fmicb.2020.01067, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Opriessnig T, Karuppannan AK, Castro A, Xiao CT. Porcine circoviruses: current status, knowledge gaps and challenges. Virus Res. (2020) 286:198044. doi: 10.1016/j.virusres.2020.198044 [DOI] [PubMed] [Google Scholar]
- 69.Hou L, Wang J, Zhang W, Quan R, Wang D, Zhu S, et al. Dynamic alterations of gut microbiota in porcine circovirus type 3-infected piglets. Front Microbiol. (2020) 11:1360. doi: 10.3389/fmicb.2020.01360, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Oh T, Chae C. First isolation and genetic characterization of porcine circovirus type 3 using primary porcine kidney cells. Vet Microbiol. (2020) 241:108576. doi: 10.1016/j.vetmic.2020.108576, PMID: [DOI] [PubMed] [Google Scholar]
- 71.Shen H, Liu X, Zhang P, Wang S, Liu Y, Zhang L, et al. Porcine circovirus 3 Cap inhibits type I interferon signaling through interaction with STAT2. Virus Res. (2020) 275:197804. doi: 10.1016/j.virusres.2019.197804 [DOI] [PubMed] [Google Scholar]
- 72.Medzhitov R. Recognition of microorganisms and activation of the immune response. Nature. (2007) 449:819–26. doi: 10.1038/nature06246 [DOI] [PubMed] [Google Scholar]
- 73.Dixit V, Mak TW. NF-kappaB signaling. Many roads lead to Madrid. Cells. (2002) 111:615–9. doi: 10.1016/S0092-8674(02)01166-2 [DOI] [PubMed] [Google Scholar]
- 74.Shen H, Liu X, Zhang P, Wang L, Liu Y, Zhang L, et al. Genome characterization of a porcine circovirus type 3 in South China. Transbound Emerg Dis. (2018) 65:264–6. doi: 10.1111/tbed.12639 [DOI] [PubMed] [Google Scholar]
- 75.Zhang P, Shen H, Liu X, Wang S, Liu Y, Xu Z, et al. Porcine circovirus type 3 Cap inhibits type I interferon induction through interaction with G3BP1. Front Vet Sci. (2020) 7:594438. doi: 10.3389/fvets.2020.594438, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu X, Shen H, Zhang X, Liang T, Ban Y, Yu L, et al. Porcine circovirus type 3 capsid protein induces NF-kappaB activation and upregulates pro-inflammatory cytokine expression in HEK-293T cells. Arch Virol. (2021) 166:2141–9. doi: 10.1007/s00705-021-05104-z, PMID: [DOI] [PubMed] [Google Scholar]
- 77.Parzych KR, Klionsky DJ. An overview of autophagy: morphology, mechanism, and regulation. Antioxid Redox Signal. (2014) 20:460–73. doi: 10.1089/ars.2013.5371, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Geng SC, Li XL, Fang WH. Porcine circovirus 3 capsid protein induces autophagy in HEK293T cells by inhibiting phosphorylation of the mammalian target of rapamycin. J Zhejiang Univ Sci B. (2020) 21:560–70. doi: 10.1631/jzus.B1900657, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sun J, Wei L, Lu Z, Mi S, Bao F, Guo H, et al. Retrospective study of porcine circovirus 3 infection in China. Transbound Emerg Dis. (2018) 65:607–13. doi: 10.1111/tbed.12853, PMID: [DOI] [PubMed] [Google Scholar]
- 80.Franzo G, Delwart E, Fux R, Hause B, Su S, Zhou J, et al. Genotyping porcine circovirus 3 (PCV-3) nowadays: does it make sense? Viruses. (2020) 12:265. doi: 10.3390/v12030265, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yang N, Li J, Yang Q, Qiao J, Cui D, Liu F, et al. Reduced antigen presentation capability and modified inflammatory/immunosuppressive cytokine expression of induced monocyte-derived dendritic cells from peripheral blood of piglets infected with porcine circovirus type 2. Arch Virol. (2018) 163:1231–9. doi: 10.1007/s00705-018-3735-8, PMID: [DOI] [PubMed] [Google Scholar]
- 82.Fort M, Sibila M, Nofrarias M, Perez-Martin E, Olvera A, Mateu E, et al. Porcine circovirus type 2 (PCV2) Cap and rep proteins are involved in the development of cell-mediated immunity upon PCV2 infection. Vet Immunol Immunopathol. (2010) 137:226–34. doi: 10.1016/j.vetimm.2010.05.013, PMID: [DOI] [PubMed] [Google Scholar]
- 83.Jung BK, Kim HR, Lee YH, Jang H, Chang KS. Comparison of immune responses to the PCV2 Replicase-capsid and capsid virus-like particle vaccines in mice. J Microbiol Biotechnol. (2019) 29:482–8. doi: 10.4014/jmb.1809.09032 [DOI] [PubMed] [Google Scholar]
- 84.Koinig HC, Talker SC, Stadler M, Ladinig A, Graage R, Ritzmann M, et al. PCV2 vaccination induces IFN-gamma/TNF-alpha co-producing T cells with a potential role in protection. Vet Res. (2015) 46:20. doi: 10.1186/s13567-015-0157-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Klausmann S, Sydler T, Summerfield A, Lewis FI, Weilenmann R, Sidler X, et al. T-cell reprogramming through targeted CD4-coreceptor and T-cell receptor expression on maturing thymocytes by latent Circoviridae family member porcine circovirus type 2 cell infections in the thymus. Emerg Microbes Infect. (2015) 4:e15. doi: 10.1038/emi.2015.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Langohr IM, Stevenson GW, Nelson EA, Lenz SD, HogenEsch H, Wei H, et al. Vascular lesions in pigs experimentally infected with porcine circovirus type 2 serogroup B. Vet Pathol. (2010) 47:140–7. doi: 10.1177/0300985809352793 [DOI] [PubMed] [Google Scholar]
- 87.Zheng LL, Chai LY, Tian RB, Zhao Y, Chen HY, Wang ZY. Simultaneous detection of porcine reproductive and respiratory syndrome virus and porcine circovirus 3 by SYBR green capital I, Ukrainian-based duplex real-time PCR. Mol Cell Probes. (2020) 49:101474. doi: 10.1016/j.mcp.2019.101474, PMID: [DOI] [PubMed] [Google Scholar]
- 88.Zheng HH, Zhang SJ, Cui JT, Zhang J, Wang L, Liu F, et al. Simultaneous detection of classical swine fever virus and porcine circovirus 3 by SYBR green I-based duplex real-time fluorescence quantitative PCR. Mol Cell Probes. (2020) 50:101524. doi: 10.1016/j.mcp.2020.101524 [DOI] [PubMed] [Google Scholar]
- 89.Tian RB, Jin Y, Xu T, Zhao Y, Wang ZY, Chen HY. Development of a SYBR green I-based duplex real-time PCR assay for detection of pseudorabies virus and porcine circovirus 3. Mol Cell Probes. (2020) 53:101593. doi: 10.1016/j.mcp.2020.101593, PMID: [DOI] [PubMed] [Google Scholar]
- 90.Han HY, Zheng HH, Zhao Y, Tian RB, Xu PL, Hou HL, et al. Development of a SYBR green I-based duplex real-time fluorescence quantitative PCR assay for the simultaneous detection of porcine epidemic diarrhea virus and porcine circovirus 3. Mol Cell Probes. (2019) 44:44–50. doi: 10.1016/j.mcp.2019.02.002, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ha Z, Xie CZ, Li JF, Wen SB, Zhang KL, Nan FL, et al. Molecular detection and genomic characterization of porcine circovirus 3 in pigs from Northeast China. BMC Vet Res. (2018) 14:321. doi: 10.1186/s12917-018-1634-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Saporiti V, Huerta E, Correa-Fiz F, Grosse Liesner B, Duran O, Segales J, et al. Detection and genotyping of porcine circovirus 2 (PCV-2) and detection of porcine circovirus 3 (PCV-3) in sera from fattening pigs of different European countries. Transbound Emerg Dis. (2020) 67:2521–31. doi: 10.1111/tbed.13596, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang Y, Noll L, Lu N, Porter E, Stoy C, Zheng W, et al. Genetic diversity and prevalence of porcine circovirus type 3 (PCV3) and type 2 (PCV2) in the Midwest of the USA during 2016–2018. Transbound Emerg Dis. (2020) 67:1284–94. doi: 10.1111/tbed.13467, PMID: [DOI] [PubMed] [Google Scholar]
- 94.Xia D, Huang L, Xie Y, Zhang X, Wei Y, Liu D, et al. The prevalence and genetic diversity of porcine circovirus types 2 and 3 in Northeast China from 2015 to 2018. Arch Virol. (2019) 164:2435–49. doi: 10.1007/s00705-019-04336-4, PMID: [DOI] [PubMed] [Google Scholar]
- 95.Xu T, Zhang YH, Tian RB, Hou CY, Li XS, Zheng LL, et al. Prevalence and genetic analysis of porcine circovirus type 2 (PCV2) and type 3 (PCV3) between 2018 and 2020 in Central China. Infect Genet Evol. (2021) 94:105016. doi: 10.1016/j.meegid.2021.105016, PMID: [DOI] [PubMed] [Google Scholar]
- 96.Woźniak A, Miłek D, Bąska P, Stadejek T. Does porcine circovirus type 3 (PCV3) interfere with porcine circovirus type 2 (PCV2) vaccine efficacy? Transbound Emerg Dis. (2019) 66:1454–61. doi: 10.1111/tbed.13221, PMID: [DOI] [PubMed] [Google Scholar]
- 97.Roca M, Balasch M, Segales J, Calsamiglia M, Viaplana E, Urniza A, et al. In vitro and in vivo characterization of an infectious clone of a European strain of porcine circovirus type 2. J Gen Virol. (2004) 85:1259–66. doi: 10.1099/vir.0.79836-0, PMID: [DOI] [PubMed] [Google Scholar]
- 98.Mai J, Wang D, Zou Y, Zhang S, Meng C, Wang A, et al. High co-infection status of novel porcine parvovirus 7 with porcine circovirus 3 in sows that experienced reproductive failure. Front Vet Sci. (2021) 8:695553. doi: 10.3389/fvets.2021.695553, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]