Abstract
Objective:
This study was to investigate whether annexin A7 (AnnexinA7, ANXA7) and its co-related protein tumor cell death domain silencer [suppressor of death domains (SODD)] regulates the migratory phenotype of liver cancer cells.
Materials and Methods:
In this experimental study, expression of ANXA7 in Hca-P cells, PANXA7 downregulated cells and PANXA7 unrelated sequence cells was detected by real-time quantitative polymerase chain reaction (PCR) at mRNA level and western blotting at protein level. Transwell migration and invasion assays were performed to determine the migratory phenotype.
Results:
After inhibition of ANXA7 expression, expression of SODD protein was also significantly decreased (P<0.05). Transwell cell transfer experiments showed that number of tumor cells that penetrated into the cell membrane was significantly reduced after ANXA7 silencing (P<0.05). Transwell cell invasion assay showed that number of tumor cells penetrating into Matrigel was significantly reduced after ANXA7 down-regulation (P<0.05). The CCK8 assay was measured at 0, 24 and 48 hours, and proliferation rate of PANXA7 lower weir cells was slower than that of Hca-P cells and PANXA7 non-related sequence cells (P<0.05).
Conclusion:
SODD expression was decreased with the down-regulation of ANXA7. Down-regulating ANXA7 in Hca-P cells decreased proliferation, migration and invasion of tumor cells.
Keywords: ANXA7, Lymph Node Metastasis, Transfection
Introduction
Formation and development of tumors are caused by various genes, and they are also affected by external factors, such as physical and chemical factors. Transformation of normal into precancerous cells is the initial step leading to tumor formation. In this process, abnormal expression of the related genes and their product proteins is the key point. Level of tumor malignancy mainly depends on its ability to proliferate, invade and metastasize. Abilities of proliferation, invasion and metastasis are also the fundamental reason for high mortality and poor prognosis of patients’ tumor. Primary liver cancer is a tumor derived from the liver epithelium, and one of its main risk factors is its early lymphatic metastasis (1). Therefore, studying occurrence and mechanism of lymphatic metastasis of liver cancer not only can help reduce mortality of liver cancer patients, but also lay a good foundation for preventing the early onset of the such diseases. In the recent years, several studies have been relatively performed on hematogenous metastasis of malignant tumors, but there are very few studies on lymphatic metastasis (2, 3). With the gradual deepening of the research on function and mechanism of genes related to lymphatic metastasis of liver cancer, new gene therapy methods can be developed, and clinical application prospect is very broad. Annexin A7 (AnncxinA7, ANXA7) belongs to the annexin family, as one of the earliest discovered members. ANXA7 is involved in mediating the Ca2+/GTP signaling pathway, and it is equivalent to the N-terminal tyrosine-, proline- and glycine-rich repeats of activated GTPase, providing a site for binding to the other proteins (4-7), due to the diversity of N-terminal amino acids of the annexin family, leading to the different properties of each annexin member (8-10). Proteins that bind or interact with ANXA7 have been discovered one after another. For example, suppressor of death domains (SODD) can assist ANXA7 to complete many biological functions. It binds to the TNF-R1 death domain and prevents TNF receptor 1 signaling activation (11). It was found that there was no relevant report on the mechanism of ANXA7 gene and its co-related protein SODD regulating lymphatic metastasis. Thus, we investigated whether ANXA7 and SODD are related to the migration phenotype of liver cancer cells in our study.
Materials and Methods
Materials
This is an experimental study, using hepatocarcinoma Hca-P cells as the in vitro model.
1. Cells: Hca-P cells with low lymphatic metastasis potential of mouse liver cancer, established and provided by the Department of Pathology, Dalian Medical University (Dalian, China).
2. Main instruments and reagents: CO2 constant temperature incubator (ThermoFisher, USA), fluorescence inverted microscope from Olympus company (USA), microplate reader, UV-Vis spectrometer (both from ThemoFisher, USA), high-speed refrigerated centrifuge from Tomy kogyo company (Japan), ultra-low temperature refrigerator (Haier, China), gel imaging system (Shanfu Scientific Instrument, China), real-time polymerase chain reaction (PCR) instrument (Shanghai Fengling Biotechnology, China), fetal bovine serum (Wolcavi Biotech, Austria), RPMI-1640 medium (Gibco, USA), pGPU6/GFP/Neo (Shanghai Gema Pharmaceutical Technology, China), plasmid extraction kit (GE, USA), diethyl pyrocarbonate (DEPC, Invitrogen, USA), G418 (Gibco, USA), 24-wells cell plate and 96-wells cell plate (Coring,USA), the transfection reagent Sofast (Sunma Bioengineering, China), qRT-PCR kit (Qualit Yard, China), co-immunoprecipitation kit (ThermoFisher, USA), and ANXA7 antibody (Sigma, USA) and GAPDH antibodies (Beijing Quanshijin, China), SODD antibody (Abcam, USA), fluorescent secondary antibody (LI-COR,USA), BCA kit (Beijing Solarbio Life Sciences, China), and CCK8 kit was purchased from Tongjin Institute of Chemistry (Japan).
Methods
1. Synthesis of ANXA7 gene shRNA sequences and unrelated sequences
The ANXA7 gene sequence was searched in the gene bank (NM_009674.3), followed by analyzing the spatial accessibility and free energy properties of mRNA, while the off-target effects were excluded. Then, the RNA-interference efficiency prediction formulas were evaluated. shRNA and unrelated sequences were designed as negative controls.
TE buffer was used to dissolve DNA oligonucleotides in sense strand and antisense strand solutions at a concentration of 1000 μM. The annealing experiment of the shRNA template was carried out according to the annealing reaction system using a PCR machine. Linearization experiment of the vector pGPU6/GFP/ Neo was carried out according to the enzyme digestion reaction system, and constructing experiment of the expressing vector pGPU6/GFP/Neo-shRNA was carried out according to the ligation reaction system. ANXA7 and 0.6 μg of irrelevant sequence plasmids were used to dissolve them in 30 μl of serum-free RPMI-1640 solution and they were mixed well for transfection. Fluorescence microscope was used to observe transfection efficiency. If the expression of green fluorescent protein was seen under the fluorescence microscope, G418 could be used for screening and culture, while the screening concentration was 400 μg/ml. About 20 days, most of the cells died one after another, and after about 23 days, the cells began to gradually proliferate, expand, culture. They were then cryopreserved.
2. Quantitative reverse transcription PCR (qRT-PCR) was used to detect expression levels of PANXA7, ANXA7, and SODD in the cells.
After adding 0.2 ml chloroform to the cell samples and shaking slightly, they were centrifuged for 10 minutes at 4°C, 12000 rpm. The upper water phase was transferred to the eppendorf tube (EP) tube. Next, an equal volume of isopropanol was added to the samples and they were mixed. The samples were preserved at room temperature for 15 minutes, followed by centrifuging at 4°C and 12,000 rpm for 10 minutes.
Experiments were performed using a reverse transcription reaction system and a real-time quantitative PCR reaction system. Western blot was used to verify whether or not PAIlM7 down-regulated ANXA7, SODD and ALG in cells. Expression level of protein was collected by centrifugation to collect Hca-P cells, PANXA7 down-regulated cells, and PANXA7 unrelated sequence cells at density of 1×107, and three times the volume of cell lysis buffer was added to the cell pellet, and lysed on ice for 15 minutes. Cells were disrupted by sonication, three cycles of 5 seconds each, a total of five times, with an interval of 8 minutes. Using refrigerated centrifuge 4˚C, 12000 rpm, 10 minutes, the supernatant was aspirated and protein concentration was measured using a microplate reader.
3. Transwell chamber detected effect of down-regulation of ANXA7 gene expression in Hca-P cells on the migration ability of cells in vitro.
The cells were resuspended in serum-free RPMI-1640 culture media for starvation. The cells were collected and their density was adjusted to 1×104 cells/ml for 24 hours. The cell suspension was added to the chamber, and each group of cells was set to three duplicate wells, and 100 μl of cell suspension was added to each well. They were incubated in a 37°C, 5% CO2 incubator for 24 hours. The transwell chamber was taken out. The cells and a small amount of medium on the upper surface of the Transwell chamber were removed with a cotton swab and air-dried at room temperature for 30 minutes. Then, the cells were stained by adding 20 μl of crystal violet solution to each hole of the 24-well plate and incubation for 20 minutes. After that, PBS was added to each well for washing the membrane, for 10 minutes. The lower chamber surface was observed with a microscope, and 10 fields of view were randomly selected for each Transwell chamber to take pictures, and number of the starved cultured cells passing through the chamber was counted.
4. Transwell chamber detected effect of down-regulation of ANXA7 gene expression in Hca-P cells on the invasion ability of cells cultured in vitro.
Extracellular matrix (ECM) gel was thawed at 4°C overnight, and the ECM gel and serum-free RPMI-l640 complete medium were mixed well at the ratio of 1. Next, 30 μ1 of the mixed ECM glue was added to each Transwell chamber, and the chamber was gently shaken; so that, the ECM glue was evenly and fully distributed on the surface of the upper chamber.
It was then incubated for 1 hour at 37°C. 10 μl of serum-free RPM-1640 medium was added to the lower chamber. Other steps were the same as the cell migration ability experiment.
5. Effect of down-regulating ANXA7 gene expression in Hca-P cells on the proliferation ability of in vitro cultured cells was detected by CCK8 assay.
Cells were adjusted to a density of 1×104 cells/ml, and plated in four 96-well plates. Six duplicate wells were set for each group of cells, and 100 ul of cell suspension was added to each well. They were incubated for 24, 48 and 72 hours in a 37°C, 5% CO2 incubator. Next, one of the 96-well plates was taken out, followed by adding 10 ul CCK8 solution to the cell suspension in each well, putting it back in the incubator and incubating it for 1 hour. Using a microplate reader, absorbance value of the cells was detected at 450 nm, and data of 5 replicate wells was recorded in each group. At different time-points of 0, 24, 48 and 72 hours, the numerical changes were observed. The experimental values analyzed by the microplate reader were statistically processed by repeated measures analysis of variance.
Ethics approval
This study was approved by Ethics Committee of Animal Ethical Care Committee of Qiqihar Medical University (Heilongjiang Province, China, No. QMUAECC- 2021-243).
Statistical analysis
The experimental data were processed with SPSS 23.0 statistical software (IBM, USA) Two-sample t test was used for analysis and comparison. LSD method was used for pairwise comparison of means. Chi-square test and Spearman rank correlation method were used for rate comparison and correlation analysis. P<0.05 indicated that the difference was statistically significant.
Results
Manipulating ANXA7 using shRNA mediated gene silencing
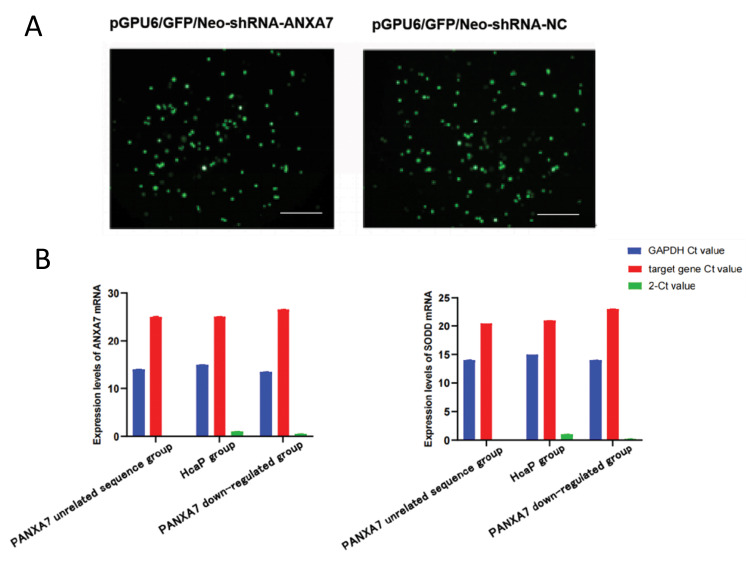
pGPU6/GFP/Neo-shRNA-ANXA7 and pGPU6/ GFP/Neo-shRNA-NC plasmids were transfected into Hca-P cells for 48 hours, and most of the cells showed green light under an inverted fluorescence microscope (Fig .1A). G418 was used to screen the cells, in terms of generating cells with stable PANXA7 down-regulation.
Fig.1.
Silencing ANXA7 gene by using shRNA. A. Hca-P cells were transfected with expressing vectors pGPU6/GFP/Neo-shRNA and pGPU6/GFP/NeoshRNA- ANXA7 (scale bar: 100 μm). B. Expression levels of ANXA7 and SODD mRNA in Hca-P cells, Hca-P cells transfected with pGPU6/GFP/Neo-shRNA or Hca-P cells transfected with pGPU6/GFP/Neo-shRNA-ANXA7.
qRT-PCR method was used to detect the downregulation of ANXA7 in Hca-P cells. Compared with Hca-P group and PANXA7 unrelated sequence group, mRNA expression levels of SODD and ANXA7 in the PANXA7 down-regulated group were decreased, by silencing ANXA7 gene expression; there was no statistical difference between the normal Hca-P cells and the PANXA7 unrelated sequence group (Fig .1B, Table 1). These results fully indicated that expression of SODD gene was also significantly reduced after inhibiting ANXA7 expression.
Table 1.
Expression levels of ANXA7 and SODD mRNA in each experimental group (X ± S)
|
| ||||
|---|---|---|---|---|
| Group | GAPDH OD value | Target gene OD value | IOD value (GAPDH OD value/target gene OD value) | Set irrelevant sequences to 1, normalize |
|
| ||||
| ANXA7 | ||||
| PANXA7 unrelated sequence group | 70.24 | 65.45 | 0.93 | |
| Hca-P group | 76.18 | 61.79 | 0.81 | 0.87 |
| PANXA7 down-regulated group | 69.31 | 14.85 | 0.21* | 0.23 |
| SODD | ||||
| PANXA7 unrelated sequence group | 70.24 | 76.32 | 1.09 | |
| Hca-P group | 76.18 | 80.84 | 1.06 | 0.98 |
| PANXA7 down-regulated group | 69.31 | 21.34 | 0.31* | 0.28 |
|
| ||||
SODD; Suppressor of death domains, OD; Optical density, IOD; Integrated optical density, and *; P<0.05, compared to the unrelated sequence group.
Protein levels of ANXA7 and SODD in PANXA7 downregulated cells
Protein levels of ANXA7 and SODD in Hca-P cells were determined by western blot. Findings showed that SODD level in the PANXA7 down-regulated group was lower than the Hca-P group and the PANXA7 unrelated sequence group. There was no significant difference between normal Hca-P cells and PANXA7 unrelated sequence group (Fig .2). These results fully indicated that expression of SODD protein was also significantly reduced after inhibiting ANXA7 expression.
Fig.2.
Western blot analysis of ANXA7 and SODD proteins in the indicated groups. Hca-P cells (a), Hca-P cells transfected with pGPU6/ GFP/NeoshRNA (b), and Hca-P cells transfected with pGPU6/GFP/NeoshRNAANXA7 (c). SODD; Suppressor of death domains.
Transwell assay to detect effect of down-regulation of ANXA7 on cell migration and invasion ability
Experiments showed that number of the cells penetrated into the membrane of the Transwell chamber was significantly reduced in the PANXA7 down-regulated group after 24 hours of starvation, compared to the Hca-P group and the PANXA7 unrelated sequence group (Fig .3A, B). This indicated that cell migration ability was decreased after down-regulation of ANXA7 gene expression. Transwell invasion assay showed that number of cells that penetrated into the membrane of the Transwell chamber was significantly reduced in the PANXA7 down-regulated group after 24 hours of starvation, compared to the cells of the Hca-P group and the PANXA7 unrelated sequence group. Additionally, invading cell number remained the same between the Hca-P group and the PANXA7 unrelated sequence group (Fig .4A, B). This indicated that the cell invasion ability was decreased after down-regulation of ANXA7 gene expression.
Fig.3.
The down-regulation effect of ANXA7 on cell migration was detected by Transwell assay. A. Representative images of transwell migration assay in Hca-P cells (a), Hca-P cells transfected with pGPU6/GFP/Neo-shRNA (b), and Hca-P cells transfected with pGPU6/GFP/Neo-shRNA-ANXA7 (c) (scale bar: 100 μm). B. Quantification of migrating cells in the above groups.
Fig.4.
The effect of silencing ANXA7 on cell invasion. A. Representative images of transwell invasion assay in Hca-P cells (a), Hca-P cells transfected with pGPU6/GFP/Neo-shRNA (b), and Hca-P cells transfected with pGPU6/GFP/Neo-shRNA-ANXA7 (c) (scale bar: 100 μm). B. Quantification of invading cells in the above groups.
Down-regulation of ANXA7 impaired cell proliferation of Hca-P cells
CCK-8 proliferation assay showed that after 0, 24, 48, 72 hour(s) of incubation, number of the Hca-P cells, PANXA7 unrelated sequence cells, and PANXA7 down-regulated cells increased significantly at 24 hours, while numbers of PANXA7 down-regulated cells’ proliferation ability was always lower than Hca-P and PANXA7 irrelevant sequence cells. There was no significant difference between Hca-P and PANXA7 irrelevant sequence cells (Fig .5). These data suggested that down-regulation of ANXA7 would impair cell proliferation of Hca-P cells.
Fig.5.
Inhibition of ANXA7 impaires the cell proliferation of Hca-P cells. CCK8 proliferation assay in Hca-P cells (blue group), Hca-P cells transfected with pGPU6/GFP/Neo-shRNA (yellow group), Hca-P cells transfected with pGPU6/GFP/Neo-shRNA-ANXA7 (grey group). h; Hour.
Discussion
Terminus of the ANXA7 containfour repeat sequences consisting of about 70 amino acids, each of which has phospholipid, Ca2+ and insulin sites. It can be phosphorylated by protein kinase C to become a substrate of protein kinase C (12). ANXA7 has also Ca2+-dependent membrane fusion activity and it is a Ca2+-dependent phospholipid-binding protein involved in mediating the Ca2+/GTP signaling pathway, equivalent to an activated GTPase. In the annexin family, ANXA7 has the longest N-terminus and it is rich of tyrosine, proline and glycine, providing binding sites for the other proteins. Experiments showed while ANXA7 and the N-terminus of ANXA7 were truncated, they did not bind to any protein (13). In terms of biological characteristics, ANXA7 gene is involved in many biological behaviors of cells. It can promote membrane binding, regulate function of membrane receptors, inhibit activity of phospholipase A in cells, promote cell secretion and exocytosis, and participate in interventional interventions. Activity of the cytoskeleton plays a significant role in the signal transduction process of cells. Numerous reports confirmed that expression of ANXA7 was various in different tumor tissues. For example, Srivastava et al. (14) found that ANXA7 had stably high expression in prostate cell lines, while expression of ANXA7 was significantly reduced in prostate cancer; when ANXA7 gene was transfected into prostate cancer cell lines, LNCaP and DU145 cells, colony formation and cell proliferation were observed.
In this study, proliferation was significantly reduced, leading to the conclusion that ANXA7 gene might belong to a tumor suppressor gene, which was consistent to the other study (15). However, the other study (16) found that expression of ANXA7 was significantly increased in 525 breast cancer tissues collected clinically. In this study, they demonstrated lower survival rate of patients in those who carried higher the expression of ANXA7. Ultimately, it was concluded that ANXA7 may be a tumorpromoting gene. There are also some related reports (17) that in gastric cancer, metastatic melanoma, cervical cancer and other tissue cells, while their conclusions on the role of ANXA7 as a tumor-promoting or a tumorsuppressing gene are controversial. In our previous transwell experiments of liver cancer Hca-P cells with ANXA7 down-regulation, migration ability of tumor cells was significantly decreased in comparison with the control cells. The PANXA7 unrelated sequence cells and PANXA7 down-regulated cells were injected into mouse footpads, respectively. After that, the metastasis rate of lymph nodes was counted. Therefore, we believe that ANAXA7 mechanism of action in tumors may be very complex, and it may be involved in regulation of different gene and protein pathways. So it may play different roles in different types or different stages of tumor.
Suppressor of death domains (SODD), also known as BAG4, is an apoptosis-regulating gene discovered by Jiang et al. (18), using yeast two-hybrid technology. Its molecular weight is 60 kD. SODD can specifically recognize and bind to cytoplasmic death domains (DD) such as TNF-R1 and Bcl-2 under physiological conditions, preventing activation of TNF apoptotic pathway. The DD protein sequence is an essential structure for generation of the intracellular toxicity signals and exerting TNF-R1 effects, providing a binding site for DD-binding proteins. Only when DD-containing proteins are involved, different death receptors (DRs) can be activated and play a role in inducing apoptosis. TNF-receptor (TNF-R) is divided into TNF-R1 and TNF-R2. TNF-R1 contains DD, activating NF-KB transcription factor after binding to the ligand and initiating apoptosis program. So TNF-R1 is also called DR. The emergence of SODD rationally explained mechanism of action of the TNF-R1 signal transduction pathway. The yeast two-hybrid assay showed that SODD could specifically recognize and combine with TNF-R1 and DR3 to form a complex, thereby preventing TNF-R1 from binding to the other proteins. SODD can combine with the DD structure of the TNF-R1 cytoplasmic segment to form a complex SODD. Once the TNF-R1 complex is formed, it can no longer interact with the other proteins of DD. Thus, the TNF-R1 signaling pathway was blocked and cell apoptosis was inhibited.
Upon stimulating by TNF receptors, SODD sheds the TNFR-I-TRADD-FADD-caspase-8 death-inducing signaling complex (DISC) from the SODD-TNF-R1 complex, resulting in apoptosis (19). Studies showed that SODD was stably and highly expressed in cervical cancer, liver cancer and the other cell lines (20).
It was also seen in our experiments that by reduction of ANXA7 expression level, SODD expression, proliferation, migration and invasion ability of tumor cells cultured in vitro was also decreased. We speculated that the possible mechanisms are as follows: First, ANXA7 gene participates in or regulates transcription and translation of SODD. At the transcriptional level, when ANXA7 was down-regulated, expression level of SODD gene was decreased, resulting in the down-regulation of SODD protein expression, which in turn affected cell function; at the translational level, ANXA7 may be a cis-translation agent of SODD. Part of the acting element, either ANXA7 positively regulates the cis-acting element of SODD or negatively regulates its trans-acting element, participates in regulating the translation level of SODD. The second possible mechanism is that ANXA7 protein can act as a regulatory factor and participate in its own signal transduction pathway by binding to SODD. In the third possible mechanism, after ANXA7 forms a complex with SODD, the latter can no longer bind to the receptors in the original signal transduction pathway. So, it can no longer participate in its signal transduction pathway, resulting in destruction of the apoptosis mechanism of cells.
This study confirmed that SODD was stably expressed in Hca-P cells. In addition, we determined that SODD and ANXA7 were co-associated proteins in tumor cytoplasm. When ANXA7 was decreased, protein expression and mRNA level of SODD were decreased, indicating that ANXA7 gene was involved in or regulated transcription and translation of SODD. At the transcriptional level, when ANXA7 was down-regulated, expression level of SODD gene was decreased, and reduction of mRNA template resulted in down-regulation of cellular SODD protein expression, thus affecting the function of cells. The results of CCK8 showed that proliferative ability of tumor cells was decreased by down-regulation of ANXA7. High expression of SODD can also inhibit TNFR1- mediated apoptosis, resulting in decreased sensitivity of tumor cells to DR-mediated apoptosis; if the expression of intracellular SODD protein is inhibited, the sensitivity to apoptosis can be restored (21-28). Our experiments also demonstrated that down-regulating ANXA7 expression inhibited migration and metastasis of the Hca-P cell. Therefore, it can be considered that ANXA7 and SODD proteins are not only related to cell apoptosis, but also to cell migration. The limitation of this study was lack of the prospective studies. In addition, results of the animal experiments may have errors and contingencies. So it needs to be confirmed by repeated experiments.
Conclusion
Both mRNA and protein levels of SODD were decreased after ANXA7 knockdown. ANXA7 silencing suppressed proliferation, migration and the invasion of hepatocarcinoma Hca-P cells. These data suggest that ANXA7 and SODD may be implicated in the invasiveness of hepatocarcinoma cells.
Acknowledgments
This study was supported by Item of Scientific Research Fund for Doctor of Qiqihar Medical University (No. QMSI2017B-12). The authors declare that they have no competing interest.
Author’s Contributions
X.W.; Was dedicated to the integrity of the entire study, study concepts, study design, literature research, and manuscript review. Y.S.; Review and editing the manuscript and statistical analysis. S.W.; Was dedicated to the experimental studies, manuscript preparation and manuscript editing. X.Y.; Was involved in the definition of intellectual content and data analysis. F.G.; Original draft preparation and clinical studies. Q.B.; Methodology. All authors read and approved the final manuscript.
References
- 1.Zanetto A, Campello E, Pelizzaro F, Farinati F, Burra P, Simioni P, et al. Haemostatic alterations in patients with cirrhosis and hepatocellular carcinoma: laboratory evidence and clinical implications. Liver Int. 2022;42(6):1229–1240. doi: 10.1111/liv.15183. [DOI] [PubMed] [Google Scholar]
- 2.Haschemi R, Kobelt D, Steinwarz E, Schlesinger M, Stein U, Bendas G. Insulin-like growth factor binding protein-2 (IGFBP2) is a key molecule in the MACC1-mediated platelet communication and metastasis of colorectal cancer cells. Int J Mol Sci. 2021;22(22):12195–12195. doi: 10.3390/ijms222212195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang WJ, Zhang GL, Cao KX, Yang GW. A hypercoagulable hematological metastasis breast cancer model. Biomed Res Int. 2021;2021:5473959–5473959. doi: 10.1155/2021/5473959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leighton X, Eidelman O, Jozwik C, Pollard HB, Srivastava M. ANXA7-GTPase as tumor suppressor: mechanisms and therapeutic opportunities. Methods Mol Biol. 2017;1513:23–35. doi: 10.1007/978-1-4939-6539-7_3. [DOI] [PubMed] [Google Scholar]
- 5.Manke MC, Geue S, Coman C, Peng B, Kollotzek F, Münzer P, et al. ANXA7 regulates platelet lipid metabolism and Ca2+ release in arterial thrombosis. Circ Res. 2021;129(4):494–507. doi: 10.1161/CIRCRESAHA.121.319207. [DOI] [PubMed] [Google Scholar]
- 6.Voelkl J, Alesutan I, Pakladok T, Viereck R, Feger M, Mia S, et al. Annexin A7 deficiency potentiates cardiac NFAT activity promoting hypertrophic signaling. Biochem Biophys Res Commun. 2014;445(1):244–249. doi: 10.1016/j.bbrc.2014.01.186. [DOI] [PubMed] [Google Scholar]
- 7.Monastyrskaya K, Babiychuk EB, Draeger A. The annexins: spatial and temporal coordination of signaling events during cellular stress. Cell Mol Life Sci. 2009;66(16):2623–2642. doi: 10.1007/s00018-009-0027-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simpkins B, Donohue MP, Li Y. Molecular dynamic studies on the impact of mutations on the structure, stability, and N-terminal orientation of annexin A1: implications for membrane aggregation. Proteins. 2014;82(12):3327–3334. doi: 10.1002/prot.24684. [DOI] [PubMed] [Google Scholar]
- 9.Stuqui B, de Paula-Silva M, Carlos CP, Ullah A, Arni RK, Gil CD, et al. Ac2-26 mimetic peptide of annexin A1 inhibits local and systemic inflammatory processes induced by bothrops moojeni venom and the Lys-49 phospholipase A2 in a rat model. PLoS One. 2015;10(7):e0130803–e0130803. doi: 10.1371/journal.pone.0130803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vago JP, Tavares LP, Sugimoto MA, Lima GL, Galvão I, de Caux TR, et al. Proresolving actions of synthetic and natural protease inhibitors are mediated by annexin A1. J Immunol. 2016;196(4):1922–1932. doi: 10.4049/jimmunol.1500886. [DOI] [PubMed] [Google Scholar]
- 11.Singh PR, Priya ES, Balakrishnan S, Arunkumar R, Sharmila G, Rajalakshmi M, et al. Nimbolide inhibits androgen independent prostate cancer cells survival and proliferation by modulating multiple pro-survival signaling pathways. Biomed Pharmacother. 2016;84:1623–1634. doi: 10.1016/j.biopha.2016.10.076. [DOI] [PubMed] [Google Scholar]
- 12.Caohuy H, Pollard HB. Activation of annexin 7 by protein kinase C in vitro and in vivo. J Biol Chem. 2001;276(16):12813–12821. doi: 10.1074/jbc.M008482200. [DOI] [PubMed] [Google Scholar]
- 13.Liu S, Li X, Lin Z, Su L, Yan S, Zhao B, et al. SEC-induced activation of ANXA7 GTPase suppresses prostate cancer metastasis. Cancer Lett. 2018;416:11–23. doi: 10.1016/j.canlet.2017.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Srivastava M, Torosyan Y, Raffeld M, Eidelman O, Pollard HB, Bubendorf L. ANXA7 expression represents hormone-relevant tumor suppression in different cancers. Int J Cancer. 2007;121(12):2628–2636. doi: 10.1002/ijc.23008. [DOI] [PubMed] [Google Scholar]
- 15.Leighton X, Bera A, Eidelman O, Eklund M, Puthillathu N, Pollard HB, Srivastava M. High ANXA7 potentiates eucalyptol toxicity in hormone-refractory prostate cancer. Anticancer Res. 2018;38(7):3831–3842. doi: 10.21873/anticanres.12667. [DOI] [PubMed] [Google Scholar]
- 16.Huang Y, Wang H, Yang Y. Annexin A7 is correlated with better clinical outcomes of patients with breast cancer. J Cell Biochem. 2018;119(9):7577–7584. doi: 10.1002/jcb.27087. [DOI] [PubMed] [Google Scholar]
- 17.Guo C, Liu S, Greenaway F, Sun MZ. Potential role of annexin A7 in cancers. Clin Chim Acta. 2013;423:83–89. doi: 10.1016/j.cca.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 18.Jiang Y, Woronicz JD, Liu W, Goeddel DV. Prevention of constitutive TNF receptor 1 signaling by silencer of death domains. Science. 1999;283(5401):543–546. doi: 10.1126/science.283.5401.543. [DOI] [PubMed] [Google Scholar]
- 19.Cisterne A, Baraz R, Khan NI, Welschinger R, Basnett J, Fung C, et al. Silencer of death domains controls cell death through tumour necrosis factor-receptor 1 and caspase-10 in acute lymphoblastic leukemia. PLoS One. 2014;9(7):e103383–e103383. doi: 10.1371/journal.pone.0103383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lv W, Wu C, Lin S, Wang X, Wang Y. Integrated utilization strategy for soybean oil deodorizer distillate: synergically synthesizing biodiesel and recovering bioactive compounds by a combined enzymatic process and molecular distillation. ACS Omega. 2021;6(13):9141–9152. doi: 10.1021/acsomega.1c00333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duell ER, D’Agostino PM, Shapiro N, Woyke T, Fuchs TM, Gulder TAM. Direct pathway cloning of the sodorifen biosynthetic gene cluster and recombinant generation of its product in E.coli. Microb Cell Fact. 2019;18(1):32–32. doi: 10.1186/s12934-019-1080-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nielsen KO, Jacobsen KS, Mirza AH, Winther TN, Størling J, Glebe D, et al. Hepatitis B virus upregulates host microRNAs that target apoptosis-regulatory genes in an in vitro cell model. Exp Cell Res. 2018;371(1):92–103. doi: 10.1016/j.yexcr.2018.07.044. [DOI] [PubMed] [Google Scholar]
- 23.Zhang M, Yao F, Qin T, Hou L, Zou X. Identification, expression pattern and functional characterization of As-kip2 in diapause embryo restarting process of Artemia sinica. Gene. 2017;608:28–40. doi: 10.1016/j.gene.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 24.Li D, Hong J, Cao W. Silencer-of-death domain mediates acidinduced decrease in cell apoptosis in barrett’s associated esophageal adenocarcinoma cells. J Pharmacol Exp Ther. 2017;360(1):14–22. doi: 10.1124/jpet.116.236620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davidson B, Valborg Reinertsen K, Trinh D, Reed W, Bøhler PJ. BAG-1/SODD, HSP70, and HSP90 are potential prognostic markers of poor survival in node-negative breast carcinoma. Hum Pathol. 2016;54:64–73. doi: 10.1016/j.humpath.2016.02.023. [DOI] [PubMed] [Google Scholar]
- 26.Tao H, Hu Q, Fang J, Liu A, Liu S, Zhang L, et al. Expression of SODD and P65 in ALL of children and its relationship with chemotherapeutic drugs. J Huazhong Univ Sci Technolog Med Sci. 2007;27(3):326–329. doi: 10.1007/s11596-007-0328-2. [DOI] [PubMed] [Google Scholar]
- 27.Tao HF, Hu Q, Liu SY, Liu AG, Hu Y, Jiang Y, et al. The significance and correlation of SODD and Bcl-2 protein expression in acute leukemia of children. Chinese Journal of Clinical Oncology. 2006;3(5):332–336. [Google Scholar]
- 28.Reuland SN, Smith SM, Bemis LT, Goldstein NB, Almeida AR, Partyka KA, et al. MicroRNA-26a is strongly downregulated in melanoma and induces cell death through repression of silencer of death domains (SODD) J Invest Dermatol. 2013;133(5):1286–1293. doi: 10.1038/jid.2012.400. [DOI] [PMC free article] [PubMed] [Google Scholar]