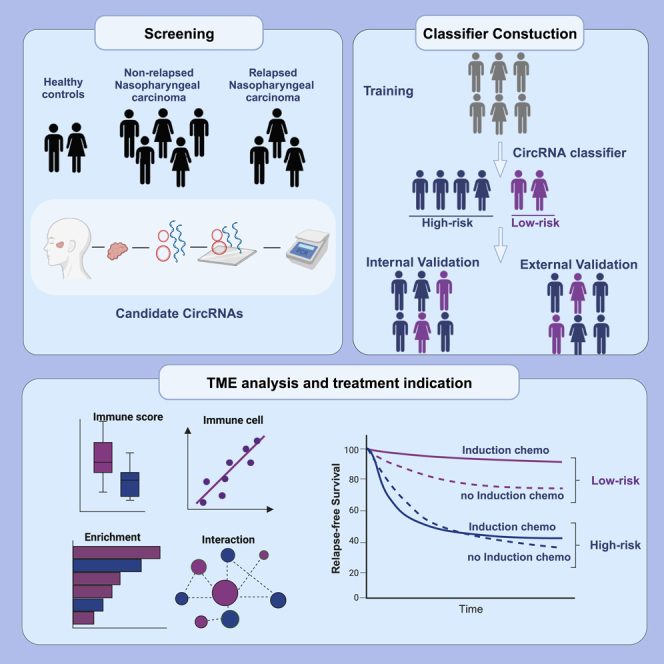
Summary
Accurate risk stratification for patients with locoregionally advanced nasopharyngeal carcinoma (LA-NPC) is crucial for prognosis and treatment decisions. Here, we develop a tumor microenvironment-associated circular RNA (circRNA) signature that can stratify LA-NPC patients with different risks of relapse and vulnerability to induction chemotherapy (IC). Relapsed-related circRNAs are identified by comparing expression profiles between patients with and without relapse, followed by quantitative validation in the training cohort (n = 170). A nine-circRNA signature is constructed to classify patients into high-risk and low-risk groups. Low-risk patients have significantly favorable clinical survivals, which is validated in the internal (n = 170) and external (n = 150) cohorts. They are characterized by an immune-active microenvironment and can derive benefits from IC. Meanwhile, high-risk patients characterized with pro-relapse and DNA repair-associated features, are vulnerable to chemoresistance. Overall, the circRNA-based classifier serves as a reliable prognostic tool and might guide chemotherapy decisions for patients with LA-NPC.
Subject areas: Health sciences, Medicine, Medical specialty, Internal medicine, Oncology, Natural sciences, Biological sciences, Systems biology, Cancer
Graphical abstract
Highlights
-
•
CircRNA classifier discriminates primary NPC with high and low risk of relapse
-
•
CircRNA classifier provides better separation of relapse than clinical indicators
-
•
The high- and low-risk group display discrepancy in the tumor microenvironment
-
•
Induction chemotherapy could improve survival in low-risk NPC
Health sciences; Medicine; Medical specialty; Internal medicine; Oncology; Natural sciences; Biological sciences; Systems biology; Cancer
Introduction
Nasopharyngeal carcinoma (NPC) is predominant in the east and Southeast Asia, with the highest incidence in southern China. Induction chemotherapy (IC) followed by concurrent chemoradiotherapy (CCRT) is the standard of care for locoregionally advanced NPC (LA-NPC).1,2 However, 20%–30% of patients still develop tumor relapse locoregionally or at distant organs.3,4 Currently, tumor-node-metastasis (TNM) staging and the plasma Epstein-Barr virus (EBV) DNA load enable prognostic prediction in patients with LA-NPC.5,6 However, the accuracy remains less satisfying, possibly due to a lack of association with tumor biological heterogeneity. Studies have reported that tumor-intrinsic properties such as epithelial-to-mesenchymal transition (EMT) would promote invasiveness and metastasis, serving as important prognostic factors.7 Apart from malignant cell characterization, intense immune cell infiltration is also a significant prognostic indicator in NPC.8,9 The interactions between cancer cells and immune cells in the tumor microenvironment (TME) profoundly influence tumor progression and the success of anti-cancer therapies.10,11 In this case, a comprehensive biomarker associated with TME heterogeneity would be valuable for more accurate prognostic assessment and individual treatment decisions in NPC.
Circular RNA (circRNA) is a unique RNA class generated by reverse splicing from both exons and introns, characterized by tissue-specific expression, evolutionary conservation, and high stability.12,13 In recent years, circRNAs have been found to play critical roles in tumor progression and immune evasion, including regulation of oncogene expression, promoting angiogenesis, and facilitating macrophage polarization and anti-PD-1 resistance.14,15,16,17 These heterogeneities and dynamic changes may lead to diverse clinical outcomes, and evidence is accumulating to support circRNAs as promising prognostic biomarkers in various cancers, such as esophageal cancer and colon cancer.18,19 However, the potential prognostic value of circRNAs for NPC has not yet been comprehensively investigated.
In this multicentre cohort study, we aimed to identify the prognostic and therapeutic stratification roles of circRNA signature in LA-NPC. A nine-circRNA classifier was built to distinguish patients with different tumor relapse risks through global circRNA profiles, followed by quantitative reverse transcription-polymerase chain reaction (RT-qPCR) validation and a penalized regression model construction in the validation cohort. Validations in the internal and external cohorts confirmed the prognostic accuracy of the signature, which is superior to clinical factors. Biological ecosystem analysis indicated an active TME in low-risk patients who could gain disease control benefits from IC, and a pro-relapse and DNA repair-related feature in high-risk patients resistant to chemotherapy. In summary, we develop and validate a circRNA classifier that could categorize patients with LA-NPC into distinct prognostic groups and determine stratified therapeutic strategies for each group.
Results
Clinicopathological features of patients
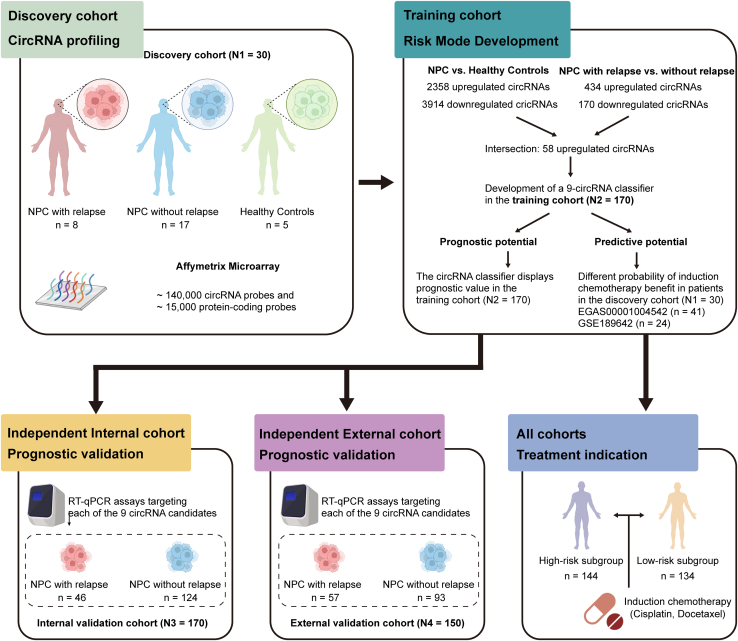
Fresh frozen samples from 515 patients with NPC and 5 healthy controls were obtained for this study (Figure 1). In the discovery cohort, circRNA profiling was conducted in 25 NPC samples and 5 healthy controls (N1 = 30). Prognostic classifier was developed and validated in Guangzhou training (N2 = 170) and internal validation (N3 = 170) cohorts, and Guilin external validation (N4 = 150) cohorts (Figure 1). The detailed clinicopathological characteristics of the patients are shown in Table 1. The median follow-up periods were 82.8 months (interquartile range (IQR) 60.8–96.5), 83.2 months (IQR 60.3–94.7), and 49.4 months (IQR 39.9–62.0) in the training, internal and external cohorts, respectively. Information on plasma EBV-DNA levels was available in the training and internal validation cohorts and was measured in the Department of Molecular Diagnosis at Sun Yat-sen University Cancer Center.
Figure 1.
Study design
LA-NPC, locoregionally advanced nasopharyngeal carcinoma.
Table 1.
Patient characteristics in the training, internal and external validation cohorts stratified according to the circRNA classifier
| Guangzhou Training cohort (n = 170) |
Guangzhou Internal Validation cohort (n = 170) |
Guilin External Validation cohort (n = 150) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| low-risk group | high-risk group | p | low-risk group | high-risk group | p | low-risk group | high-risk group | p | |
| Age | 0.877 | 0.709 | 0.861 | ||||||
| <45 | 48 (56.5) | 50 (58.8) | 52 (57.8) | 43 (53.8) | 23 (34.8) | 27 (32.1) | |||
| ≥45 | 37 (43.5) | 35 (41.2) | 38 (42.2) | 37 (46.2) | 43 (65.2) | 57 (67.9) | |||
| Sex | 0.863 | 0.250 | 0.999 | ||||||
| Male | 61 (71.8) | 63 (74.1) | 72 (80) | 57 (71.2) | 45 (68.2) | 57 (67.9) | |||
| Female | 24 (28.2) | 22 (25.9) | 18 (20) | 23 (28.8) | 21 (31.8) | 27 (32.1) | |||
| T stage | 0.999 | 0.604 | 0.442 | ||||||
| T1-2 | 11 (12.9) | 10 (11.8) | 9 (10.0) | 11 (13.8) | 10 (15.2) | 18 (21.4) | |||
| T3-4 | 74 (87.1) | 75 (88.2) | 81 (90.0) | 69 (86.2) | 56 (84.8) | 66 (78.6) | |||
| N stage | 0.759 | 0.108 | 0.149 | ||||||
| N0-1 | 46 (54.1) | 43 (50.6) | 47 (52.2) | 31 (38.8) | 33 (50.0) | 31 (36.9) | |||
| N2-3 | 39 (45.9) | 42 (49.4) | 43 (47.8) | 49 (61.3) | 33 (50.0) | 53 (63.1) | |||
| TNM stage | 0.434 | 0.495 | 0.715 | ||||||
| III | 54 (63.5) | 48 (56.5) | 53 (58.9) | 42 (52.5) | 39 (59.1) | 46 (54.8) | |||
| IV | 31 (36.5) | 37 (43.5) | 37 (41.1) | 38 (47.5) | 27 (40.9) | 38 (45.2) | |||
| EBV DNA (copies/ml) | 0.640 | 0.089 | |||||||
| <2000 | 33 (38.8) | 37 (43.5) | 37 (41.1) | 22 (27.5) | – | – | – | ||
| ≥2000 | 52 (61.2) | 48 (56.5) | 53 (58.9) | 58 (72.5) | – | – | – | ||
Abbreviations: TNM, tumor-node-metastasis; EBV, Epstein-Barr virus.
CircRNA profiling identified dysregulated relapse-related candidates
Using custom microarrays, we profiled circRNA expression in NPC tissues and normal nasopharyngeal tissues. The differential expression analysis between the tumor and normal nasopharyngeal tissues identified 6272 dysregulated circRNAs (2358 up-regulated and 3914 down-regulated) (Figures S1A and S1B). Furthermore, 604 dysregulated circRNAs (434 up-regulated and 170 down-regulated) were identified between the relapsed and non-relapsed NPC tissues (Figures S1C and S1D). The results indicated that the circRNAs experienced more remarkable changes between the normal and tumor samples than between the relapsed and non-relapsed samples. In total, 58 up-regulated circRNAs in the intersection of the two comparisons (Figure S1E). In addition, these circRNAs showed strong classification properties in differentiating each group (Figure 2A).
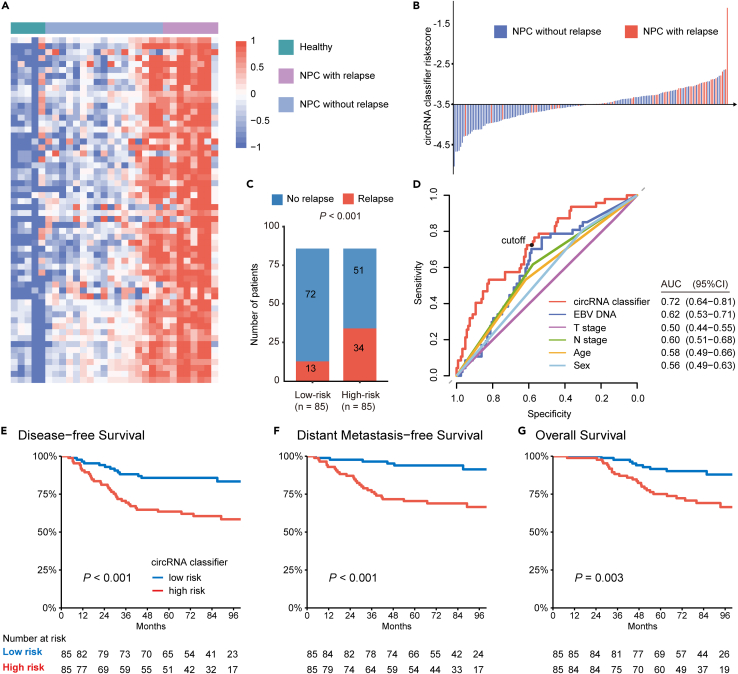
Figure 2.
Development of a circRNA classifier in Guangzhou training cohort
(A) Expression profiling of up-regulated circRNAs between NPC and healthy groups (n = 5), and between relapsed NPC (n = 8) and non-relapsed NPC groups (n = 17).
(B) Risk scores distribution in the training cohort (n = 170). Each line represents the risk score of a patient. The x axis is the cutoff value to divide patients into low- and high-risk groups.
(C) Bar plot showing the number of relapsed patients in the low- (n = 85) and high-risk (n = 85) group. p values were based on χ2 test.
(D) ROC analysis of circRNA classifier and other clinical characteristics for predicting relapse.
(E–G) Kaplan-Meier curves showing (E) disease-free survival, (F) distant metastasis-free survival and (G) overall survival according to the expression of the cricRNA classifier (n = 170). p values were based on the Log rank test.
A nine-circRNA classifier was constructed to predict tumor relapse in the training cohort
We then validated the expression of the candidate circRNAs by RT-qPCR assays. The least absolute shrinkage and selection operator (LASSO) Cox regression analysis was performed to construct a circRNA classifier for predicting posttreatment tumor relapse in the training cohort (Figure S1F). The circRNA-based classifier was constructed using a risk score formula that included nine circRNAs (Figures S1G and S1H) weighted by their regression coefficients in the penalized Cox model as follows: risk score =
Patients with a higher score than the median value (−3.506) of the risk score were classified into the high-risk group, and patients with lower scores were classified into the low-risk group (Figure 2B). The circRNA classifier showed a correct prediction of relapse in 34 out of 47 patients with posttreatment recurrence and metastasis (72.3%, Figure 2C). When employing receiver operating characteristic (ROC) curves as a measure of prognostic value, we found that the circRNA classifier gave the best performance, compared with other clinical variables (Figure 2D), including T stage, N stage, pretreatment EBV DNA, age, and sex, suggesting the circRNA classifier was a robust prognostic tool. Notably, patients in the high-risk group had a significantly higher risk of posttreatment relapse than those in the low-risk group (5-year disease-free survival (DFS) 63.5% vs. 85.7%, hazard ratio (HR) = 2.99, 95% confidence interval (CI) 1.58–5.66; p < 0.001; Figure 2E, Tables S1 and S2). Moreover, the 5-year distant metastasis-free survival (DMFS) and overall survival (OS) of patients in the low-risk group were consistently better than that of patients in the high-risk group (DMFS: HR = 5.09, 95% CI 2.10–12.33; p < 0.001; OS: HR = 2.85, 95% CI 1.37–5.91; p = 0.005; Figures 2F, 2G, S2, and S3).
Prognostic accuracy of the nine-circRNA classifier was validated in multicenter independent cohorts
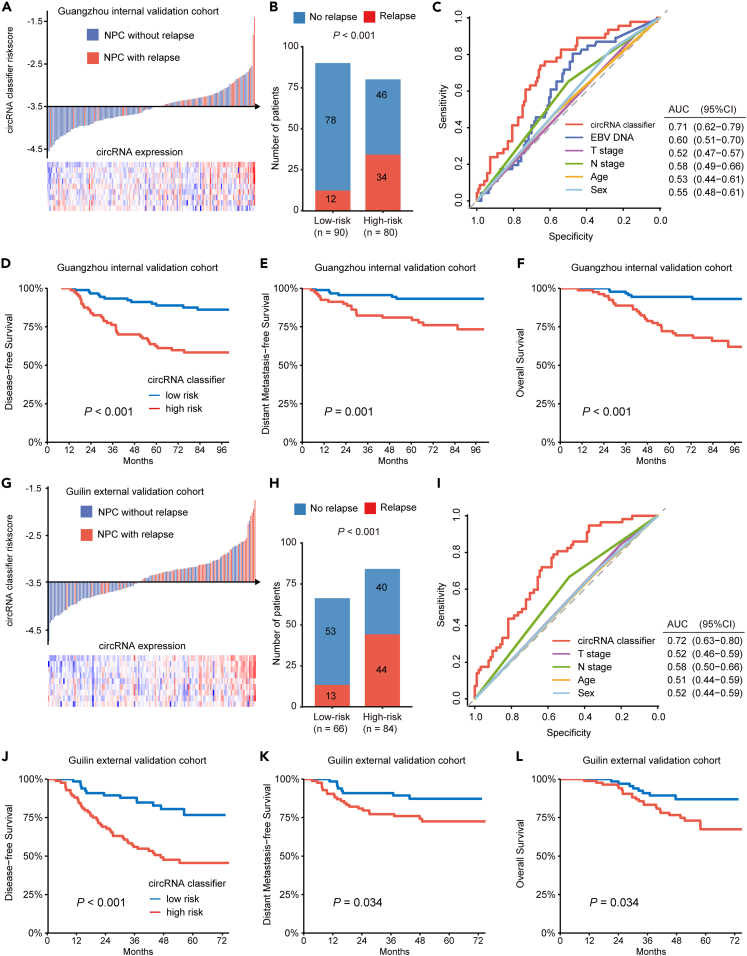
Using the formula and cutoff value from the training cohort, we applied the circRNA classifier in an independent internal cohort (Figure 3A). It showed a correct prediction of posttreatment relapse in 34 out of 46 patients with NPC (73.9%, Figure 3B) and outperformed other clinical variables (Figure 3C). The survival rate was significantly higher in low-risk group, with prolonged DFS, DMFS, and OS (DFS: HR 3.83, 95% CI 1.98–7.40; p < 0.001; DMFS: HR 4.03, 95% CI 1.61–10.09; p = 0.003; OS: HR 5.82, 95% CI 2.40–14.11; p < 0.001; Figures 3D–3F). Subsequently, the prognostic value of this circRNA classifier was further confirmed in an independent external cohort (Figure 3G), which displayed a correct prediction of relapse in 44 out of 57 primary NPC (73.2%) and the best relapse prediction performance (Figures 3H and 3I). The differences in survivals between low-risk and high-risk group were also significant (DFS: HR 3.32, 95% CI 1.78–6.16; p < 0.001; DMFS: HR 2.34, 95% CI 1.04–5.26, p = 0.039; OS: HR 2.33, 95% CI 1.04–5.21, p = 0.039; Figure 3J–3L).
Figure 3.
Validation of the circRNA classifier in two independent cohorts
(A) Risk scores distribution in the internal validation cohort (n = 170).
(B) Bar plot showing the number of relapsed patients in the low- (n = 90) and high-risk (n = 80) group in the internal validation cohort. p values were based on χ2 test.
(C) ROC analysis of circRNA classifier and other clinical characteristics for predicting relapse in the internal validation cohort.
(D–F) Kaplan-Meier curves showing (D) disease-free survival, (E) distant metastasis-free survival and (F) overall survival according to the expression of the cricRNA classifier in the internal validation cohort (n = 170). p values were based on the Log rank test.
(G) Risk scores distribution in the external validation cohort (n = 150).
(H) Bar plot showing the number of relapsed patients in the low- (n = 66) and high-risk (n = 84) group in the external validation cohort (n = 150). p values were based on χ2 test.
(I) ROC analysis of circRNA classifier and other clinical characteristics for predicting relapse in the external validation cohort.
(J–L) Kaplan-Meier curves showing (J) disease-free survival, (K) distant metastasis-free survival and (L) overall survival according to the expression of the cricRNA classifier in the internal validation cohort (n = 150). p values were based on the Log rank test.
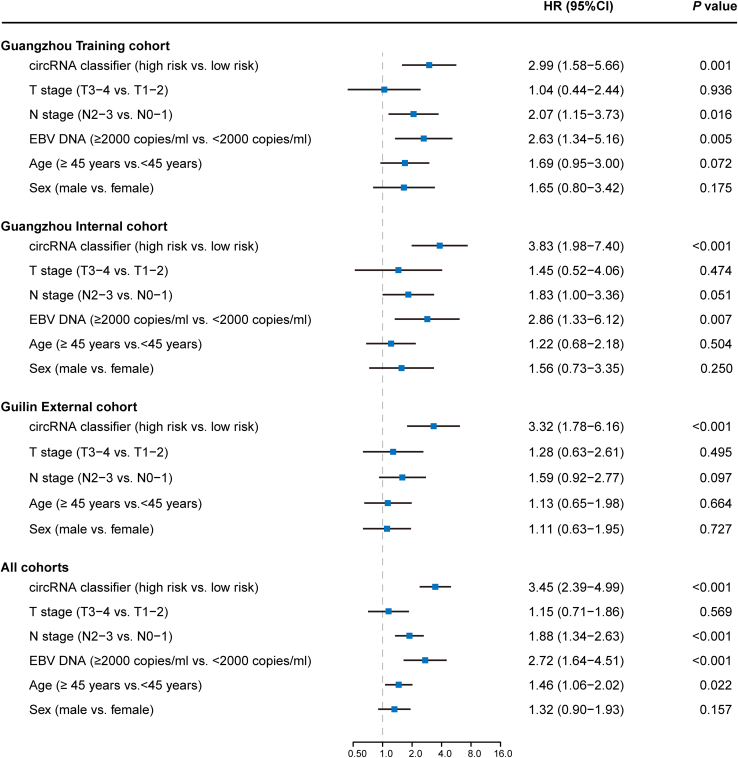
Univariate analysis indicated that the circRNA classifier was significantly associated with DFS across all three cohorts (Figure 4). Multivariable Cox regression further showed that the circRNA classifier was an independent and significant risk factor for DFS (Training cohort: HR 3.26, 95% CI 1.71–6.22, p < 0.001; internal cohort: HR 3.56, 95% CI 1.82–6.96, p < 0.001; external cohort: HR 3.29, 95% CI 1.76–6.14, p < 0.001; Figure S4). For DMFS and OS, the circRNA classifier remained a powerful independent factor after multivariable adjustment in all three cohorts (Figures S5 and S6).
Figure 4.
Univariate association of circRNA classifier and clinicopathological characteristics with disease-free survival
p values and hazard ratios were based on univariate Cox regression analyses.
The low-risk group was characterized by immune-active TME
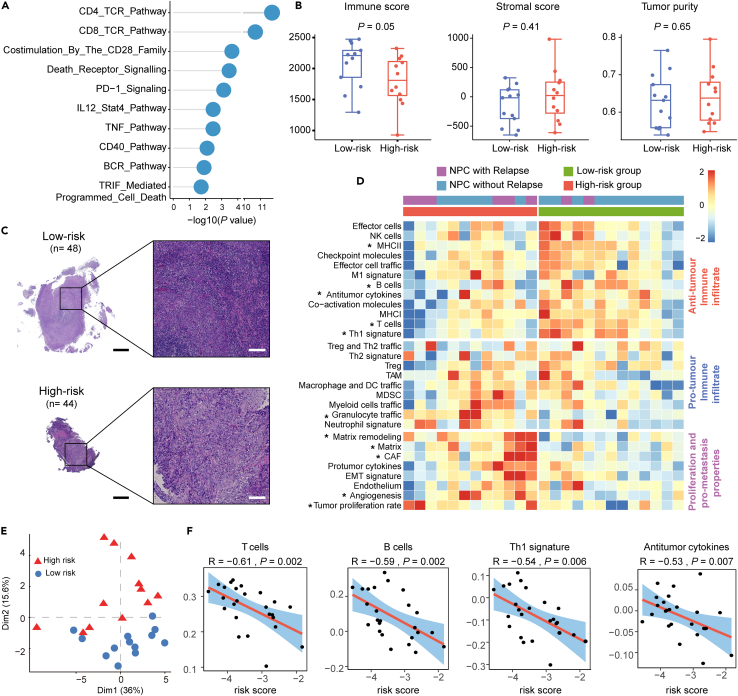
To gain insights into the biological ecosystems of different NPC groups classified by the nine-circRNA classifier, we conducted gene enrichment analysis with the data of circRNA microarray in canonical pathways. Genes up-regulated in the low-risk group were enriched in immune pathways (Figure 5A), including those related to T cell receptor (TCR) signaling, B cell receptor (BCR) signaling, and programmed death-1 (PD-1), which was validated with a public NPC data of Affymetrix Human Transcriptome Array (HTA, EGAS00001004542) analysis (Figure S7A). Inspired by these immune-related findings, we calculated the stromal-immune score with the ESTIMATE algorithm, and discovered that the immune score was higher in the low-risk group, while the stromal score and tumor purity were not significantly different (Figure 5B). Validation on H&E slides (low-risk, n = 48; high-risk, n = 44) confirmed more abundant lymphocyte infiltration in the low-risk group (Figures 5C and S7B).
Figure 5.
The low-risk group was characterized by active immune activity
(A) Pathway alterations enriched by differentially up-regulated genes in the low-risk group. p values were based on the hypergeometric distribution test.
(B) Immune, stromal scores, and tumor purity in groups classified by the circRNA classifier (n = 25), calculated by the ESTIMATE algorithm. In each boxplot, the centerline represents the median, the bounds represent the first and third quartiles and whiskers extend from the hinge to the largest value no further than 1.5 ∗ interquartile range (IQR) from the hinge. Each dot represents a data point for individual patients. p values were based on the Wilcoxon rank-sum test.
(C) Representative images of tumor infiltration lymphocytes in low- and high-group on H&E slides.
(D) Heatmap of TME ssGSEA scores in NPC samples classified into low- (n = 13) and high-group (n = 12) based on the circRNA-based classifier. ∗p < 0.05, p values were based on a simple linear model and moderate t-statistic.
(E) Principal component analysis based on TME ssGSEA scores (n = 25). (F) Spearman correlation between circRNA-based classifier and T cells, B cells, Th1 cells, and antitumor cytokines signatures. p values were based on Spearman correlation.
Next, we characterized various immune and malignant components in the TME, which discovered that the low-risk group was characterized by an anti-tumor immune microenvironment, with higher expression of signatures in T cells, Th1 cells, B cells, MHC II, and anti-tumor cytokines (Figure 5D). Similar results were also observed in the Affymetrix HTA and another RNA-seq data (GSE189642, Figure S7C). Relatively higher abundance levels of T cell and B cell infiltrates in the low-risk group were observed using the microenvironment cell populations (MCP)-counter cell deconvolution method in these three datasets (Figures S7D–S7F). Principal component analysis (PCA) based on the expression of the TME signatures revealed a clear distinction between the two groups defined by the circRNA classifier (Figure 5E). In addition, we observed a significant negative correlation between the circRNA-based risk score and immune features, such as T cells, B cells, Th1 cells, and anti-tumor cytokines (Figure 5F). The findings indicate that low-risk patients stratified by the nine-circRNA classifier are characterized by high immune infiltration and active immune TME.
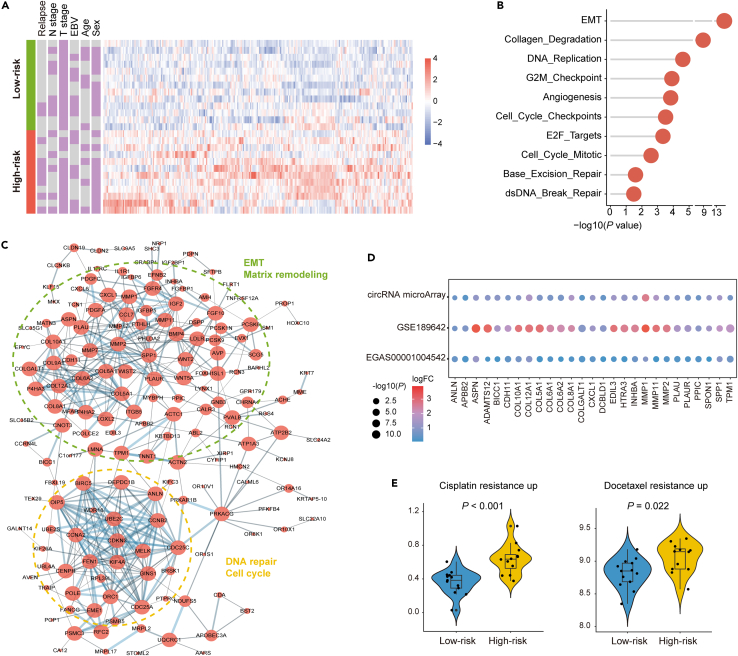
The high-risk group displayed characteristics indicative of tumor relapse and chemo-resistance
The up-regulated genes in the high-risk group showed no association with clinical variables (Figure 6A), but enrichment analysis showed that these genes were enriched in pro-relapse associated pathways, such as EMT, collagen degradation and angiogenesis (Figure 6B). The same findings could be noted with Affymetrix HTA data (Figure S8A), which collectively confirmed the results of TME characterization (Figure 5D). In addition, DNA repair pathways and cell cycle pathways were also enriched in patients with high tumor relapse risks (Figures 6B and S8B). The PPI network displayed two centered modules enriched in the high-risk group (Figure 6C). As expected, one module was predominant by EMT and matrix remodeling, and members in this module mainly encode collagen and matrix metalloproteinases. Another module was composed of molecules related to DNA repair and cell cycle. Key genes related to these pathways displayed consistent expressional alterations in the Affymetrix HTA and RNA-seq data (Figure 6D).
Figure 6.
The high-risk group was characterized by pro-relapse and chemo-resistant components
(A) Heatmap of up-regulated genes in the high-risk group.
(B) Pathway alterations enriched by differentially up-regulated genes in the high-risk group. p values were based on the hypergeometric distribution test.
(C) PPI network modules of the up-regulated genes in the high-risk group. Node sizes are proportional to node degrees. Dotted circles with different colors represent different network modules.
(D) Key genes relevant to enriched pathways showed consistently different expressions across three datasets.
(E) Violin plots showing expression of cisplatin resistance signature and docetaxel resistance signature between low-risk (n = 13) and high-risk (n = 12) groups according to the circRNA classifier. In each boxplot, the centerline represents the median, the bounds represent the first and third quartiles and whiskers extend from the hinge to the largest value no further than 1.5 ∗ interquartile range (IQR) from the hinge. Each dot represents a data points for individual patients. p values were based on the Wilcoxon rank-sum test.
Pathways that appeared up-regulated in the high-risk group included those related to the cell cycle and DNA repair (Figures 6B–6D), which were strongly correlated with chemotherapy resistance.20,21 Using single sample gene set enrichment analysis (ssGSEA), we discovered that the high-risk group displayed significantly higher scores in cisplatin and docetaxel resistance (Figure 6E). Conversely, the low-risk group exhibited a lower score in the chemoresistance pathway, indicating its potential chemosensitivity. Similar results could be validated in the data of Affymetrix HTA and RNA-seq (Figures S8C and S8D).
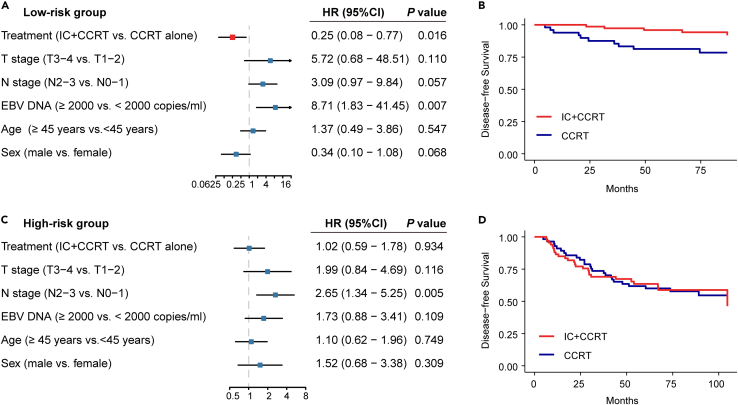
CircRNA classifier indicates distinct therapeutic strategies
The addition of IC containing cisplatin and docetaxel to CCRT has become standard care for patients with LA-NPC.1 Then, we investigated whether patients of different-risk groups would respond diversely to IC. A total of 278 patients were included (CCRT plus TP or TPF IC, n = 149; CCRT, n = 129). We observed that patients in the low-risk group could benefit from IC, after adjustment of other clinical variables using multivariate Cox analyses (HR 0.25, 95% CI 0.08–0.77, p = 0.016; Figures 7A and 7B). However, a similar result was not found in high-risk group (HR 1.02, 95% CI 0.59–1.78, p = 0.934; Figures 7C and 7D), suggesting that the addition of IC may not be enough to improve clinical outcomes in high-risk patients, probably due to their chemoresistance.
Figure 7.
Induction chemotherapy benefits in the high-risk and low-risk group stratified by the circRNA classifier
(A) Multivariate analysis of treatment strategy and clinicopathological characteristics with disease-free survival in the low-risk group (n = 134). p values were based on adjusted multivariate analysis.
(B) Kaplan-Meier curves showing disease-free survival in patients receiving IC plus CCRT versus CCRT alone in the low-risk group (n = 134).
(C) Multivariate analysis of treatment strategy and clinicopathological characteristics with disease-free survival in the high-risk group (n = 144). p values were based on adjusted multivariate analysis.
(D) Kaplan-Meier curves showing disease-free survival in patients receiving IC plus CCRT versus CCRT alone in the high-risk group (n = 144).
Discussion
CircRNA dysregulation can promote cancer development and progression through sophisticated interactions within the tumor ecosystem. In this multicenter, retrospective cohort study, we demonstrated the utility of circRNA as a prognostic classifier in LA-NPC patients. The proposed circRNA classifier improved the prognostic stratification of patients with LA-NPC by effectively distinguishing patients with different relapse risks. Patients in the low-risk group had prolonged DFS, whose TME was characterized by active immune compartments. By contrast, the high-risk patients, who had poor DFS, were predominant by upregulation of tumor-promoting signaling pathways and pro-relapse TME components.
Recent decades have seen great improvement in tumor control in patients with LA-NPC, with CCRT using a platinum-based agent constituting the cornerstone of treatment. However, 20%–30% of patients still develop relapse after radical treatment, suggesting an urgent need to revolutionize risk stratification and guide treatment decisions. Currently, circRNAs have captured the interest of researchers owing to their unique properties. Being evolutionarily conserved and more stable than their linear counterparts, circRNAs serve as reliable biomarkers in esophageal cancer and colon cancer.18,19 In the current study, we developed a circRNA-based classifier in large cohorts and subjected it to independent validation. Initially, we employed microarrays to detect global circRNA expression to discover circRNAs related to NPC tumorigenesis and progression. Next, we used RT-qPCR assays in the training cohort to quantify the candidate circRNAs and then developed a classifier by machine learning methods. We further confirmed its prognostic prediction performance in two independent cohorts. The proposed circRNA classifier effectively discriminated patients with an increased risk of relapse and remained an independent prognostic factor after adjustment for other clinical variables, serving as a promising tool for risk classification of patients.
Currently, tumor-node-metastasis (TNM) staging and the plasma EBV DNA load enable prognostic prediction in patients with LA-NPC.5,6 TNM stage reflects anatomy-based disease stage, and EBV DNA load is associated with tumor size as it consists of short DNA fragments released by EBV-infected NPC cells, but they reflect little about molecular heterogeneity. Here, the circRNA classifier distinguished patients with distinct biological characterization. The low-risk group presented with a brisk TME with abundant T cell and B cell infiltration, and activation of TCR and BCR signaling. In contrast, the high-risk group was characterized by an aggressive TME pattern, including tumor proliferation and metastasis-promoting components, such as elevated tumor proliferation rate, EMT, and angiogenesis. The association between poor prognosis and these TME risk factors in NPC has been recognized in previous studies.22,23,24 The different TME compartments and pathway regulations explain the underlying mechanism of varied prognoses of different groups.
One of the goals for prognostic biomarker development is to guide treatment decisions. Here, patients in the low-risk group, designated by the circRNA classifier, could acquire survival benefits from IC. In contrast, IC seemed insufficient to improve survival in high-risk patients who showed a low level of immune infiltration and a high tendency of chemoresistance. It is recently unveiled that chemotherapy and radiotherapy could repress the aggressiveness of cancer and reshape the TME,25 which may provide synergistic effects with immunotherapy in cancer treatment. Even in tumors with poor lymphocyte infiltration, immune checkpoint blockade (ICB) can take effect by enhancing cytotoxic T cell infiltration.26,27 We noticed that cells prohibiting T cell infiltration, including myeloid-derived suppressor cells (MDSCs) and tumor-associated macrophages (TAMs), displayed similar infiltration between the high- and low-risk group. It suggested that immunotherapy might offer benefits for high-risk patients.
In conclusion, the proposed circRNA-based classifier can effectively differentiate patients with LA-NPC into groups with distinct characteristics of the tumor ecosystem and different risks of posttreatment relapse. Specific to each group, we suggested tailored therapeutic strategies to facilitate future individualized treatment of NPC patients.
Limitations of the study
The limitation of the present study is that we were not able to determine the efficiency of the proposed circRNA classifier in early-stage patients, since we included late-stage patients in this study. In addition, our study was limited by its retrospective nature, which needs further validation in large-scale prospective studies. To ensure the generalizability of the circRNA classifier, we quantified the expression of circRNAs by RT-qPCR assays and performed validations in independent cohorts, which enables its extensive implementation in clinical practice.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Tissue samples of nasopharyngeal carcinoma | Sun Yat-sen University Cancer Center | Sun Yat-sen University Cancer Center |
| Tissue samples of nasopharyngeal carcinoma | Affiliated Hospital of Guilin Medical College | Affiliated Hospital of Guilin Medical College |
| Critical commercial assays | ||
| AllPrep DNA/RNA Mini Kit | QIAGEN | Cat# 80204 |
| SurePrint G3 Custom GE 4x180K Microarray | Agilent | 9G4862A |
| Deposited data | ||
| Raw circRNA microarray data of nasopharyngeal carcinoma | This paper | GEO: GSE190271 |
| Raw Affymetrix HTA microarray data of nasopharyngeal carcinoma | Chen et al.28 | EGA: EGAS00001004542 |
| Raw RNA-seq data of nasopharyngeal carcinoma | Qiao et al.29 | GEO: GSE189642 |
| Enriched gene lists | This paper | Table S3 |
| Key raw data | This paper | Research Data Deposit public platform (https://www.researchdata.org.cn/): RDDB2023454299 |
| Software and algorithms | ||
| LightCycler 480 PCR system | ROCHE | https://diagnostics.roche.com/global/en/products/instruments/lightcycler-480-ins-445.html |
| R software (4.0.3) | R Project | https://www.r-project.org/ |
| SVA | Leek et al.30 | https://bioconductor.org/packages/release/bioc/html/sva.html |
| limma | Ritchie et al.31 | https://bioconductor.org/packages/release/bioc/html/limma.html |
| glmnet | Friedman et al.32 | https://mirrors.tuna.tsinghua.edu.cn/CRAN/web/packages/glmnet/index.html |
| ESTIMATE | Yoshihara et al.33 | https://bioinformatics.mdanderson.org/public-software/estimate/ |
| MCP-counter | Becht et al.34 | https://github.com/ebecht/MCPcounter |
| Cytoscape | Shannon et al.35 | https://cytoscape.org/ |
| clusterProfiler | Yu et al.36 | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ying-qin Li (liyingq@sysucc.org.cn).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
The data of circRNA microarrays have been deposited at GEO and are publicly available as of the date of publication. This paper also analyses existing, publicly available data. The accession numbers for these datasets are listed in the key resources table. The key raw data have been deposited to the Research Data Deposit public platform (https://www.researchdata.org.cn/), with an approval number listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Sample collection
We collected fresh frozen nasopharyngeal samples from 515 patients with LA-NPC and 5 healthy controls. Samples obtained at Sun Yat-sen University Cancer Center (SYSUCC; Guangzhou, China) between July 2010 and December 2016 were assigned to the discovery cohort (N1 = 30, including 8 NPC with relapse, 17 NPC without relapse and 5 healthy control), Guangzhou training cohort (N2 = 170) and the Guangzhou internal validation cohort (N3 = 170). Additional samples were obtained at the Affiliated Hospital of Guilin Medical College (Guilin, China) between September 2014 and October 2017 as an external validation cohort (N4 = 150). The study was approved by the institutional ethical review boards of the Sun Yat-Sen University Cancer Center and the Affiliated Hospital of Guilin Medical University. The informed consent was obtained from all patients.
The inclusion criteria were as follows: histologic confirmation of NPC; diagnosed stage III-IVA according to the 8th edition of the American Joint Committee on Cancer (AJCC) staging system37; no previous treatment for cancer; treated with intensity-modulated radiation therapy (IMRT)38 and platinum-based chemotherapy; pretreatment EBV DNA levels available (for training and internal cohorts); follow-up information available. Key exclusion criteria were the following: a history of cancer; lactation or pregnancy; or severe coexisting illness.
Method details
Study design
In this study, we screened circRNAs related to relapse of NPC using custom circRNA microarray assays and validated the candidate circRNAs with RT–qPCR assays. SurePrint G3 Custom GE 4x180K Microarray (Agilent, 9G4862A, 4×180K) has high coverage of probes that can measure coding and noncoding transcripts, containing > 170,000 probes covering > 140,000 circRNA transcripts and > 15,000 coding transcripts. First, 5 normal nasopharyngeal tissue samples and 25 primary LA-NPC tissue samples, among which 8 samples were derived from patients with posttreatment relapse, were subjected to circRNA microarray detection. We then detected the relapse-related circRNAs with RT–qPCR assays and developed a circRNA-based classifier for prognosis prediction in the training cohort, which was validated in the Guangzhou internal validation cohort and the Guilin external validation cohort. Based on the presented classifier, patients were classified into two groups with different risks of disease relapse. Among all patients, we had previously profiled gene expression using Affymetrix HTA 2.0 in 41 participants, and profiled gene expression using RNA-seq in 24 participants, respectively.28,29 In addition, 92 participants in Guangzhou cohorts had H&E staining slides available. To characterize the biological characteristics of the two groups, we investigated the variations in the TME and pathway alterations, which indicated distinct therapeutic vulnerabilities for patients with LA-NPC (Figure 1).
RNA extraction, microarray analysis and RT–qPCR assay
Total RNA was extracted from fresh frozen tissues using the AllPrep DNA/RNA Mini Kit (QIAGEN GmbH, Hilden, Germany) according to the manufacturer’s instructions. RNA profiles were detected using a SurePrint G3 Custom GE 4x180K Microarray (Agilent, 9G4862A, 4×180K), which was designed for the global profiling of human circRNAs, as well as protein-coding and noncoding RNA transcripts.
After normalization, probes with raw expression > 4 in more than 5 samples (Healthy controls group, n = 5) were included in further analysis. Then, we used the “ComBat” algorithm in R package ‘SVA’ to remove batch effects.30 Then, for probes representing coding transcripts, we calculated the median of the probe expression values of each transcript. For probes representing circRNA, we only included those with raw average expression of samples in either group (healthy controls, relapsed NPC and non-relapsed NPC) >300 for detection convenience. Differentially expressed circRNAs were discovered by the R package ‘limma’,31,39 and statistical significance between the two groups was identified with a cutoff P value < 0.01 and fold change > 1.5.
For RT–qPCR assays, total RNA was reverse-transcribed using the GoScript™ Reverse Transcription System (Promega Corporation, Madison, Wisconsin, USA), according to the manufacturer’s protocol. The complementary DNA was analyzed by RT–qPCR with SYBR reagent using a LightCycler 480 PCR system (ROCHE Ltd., Rotkreuz, Switzerland). Data were analyzed using the ΔCt method with β-actin as the internal control gene.
Quantification and statistical analysis
Construction and validation of the circRNA classifier
To construct a circRNA classifier in the training cohort, we first selected circRNAs based on the following criteria: (i) up-regulated circRNAs in NPC compared with normal tissues (fold change > 1.5, P-value < 0.01); (ii) up-regulated circRNAs in NPC with relapse compared with NPC without relapse (fold change > 1.5, P-value < 0.01); (iii) circRNAs with an average raw expression >300 in the relapsed NPC group. In total, 58 circRNAs were qualified and 11 of them could be detected with designable RT-qPCR primers. Then, the expression levels of candidate circRNAs were quantified by RT–qPCR in the training cohort (presented by Δct) and subjected to the least absolute shrinkage and selection operator (LASSO) method with 10-fold cross-validation,40,41 generating a risk classifier based on a penalized Cox model. In short, the 170 LA-NPC patients in the training cohort were randomly partitioned into 10 equal-sized subsamples. A series of different λ values for LASSO was generated by the “glmnet” package in R software.32 For each λ, 9 subsamples were used as training data to generate a model, and the remaining 1 subsample was retained to validate the model. The partial likelihood deviance was calculated to evaluate the efficacy variation between the training and validation subsamples. The cross-validation process is then repeated 10 times, with each of the 10 subsamples used exactly once as the validation data. In this way, for each λ, the mean and estimated standard error of the partial likelihood deviances in ten times were calculated. We choose λ via minimum criteria,42 i.e. the optimal λ is value for which the partial likelihood deviance is the smallest. Based on this λ value, we could obtain the variables whose beta coefficients were not zero and constructed the circRNA-based classifier with these circRNAs.
Using the circRNA-based classifier, we classified patients into the low-risk and high-risk group based on the median value. We next tested the classifier on the samples from the internal and external validation cohorts by RT-qPCR, using the formula and cutoff value developed in the training cohort. The performance of the classifier was assessed by receiver operating characteristic (ROC) analyses.43
Tumor microenvironment (TME) characterization and pathway enrichment
To characterize the biological ecosystem of the NPC groups based on the circRNA classifier, we used single sample gene set enrichment analysis (ssGSEA) with 29 functional gene expression signatures,44,45 representing the major cell populations and functional elements to decompose TME features. The mRNA expression data from NPC samples were subjected to gene set variation analysis to calculate the ssGSEA scores for each gene set. We also employed Spearman correlation analysis to explore the relationship between TME features and the circRNA-based classifier. The significance of the enrichment scores of the NPC groups was estimated using a simple linear model and moderated with the t-statistic with the R package ‘limma’. In addition, the principal component analysis (PCA) was performed to visualize the patient classification based on the TME ssGSEA scores and the circRNA-based classifier. Immune infiltration was measured using the ESTIMATE algorithm and the MCP-counter algorithm,33,34 as well as evaluated by a pathologist on H&E slides. Protein-protein interaction (PPI) network was obtained from STRING website (https://string-db.org/) and constructed in the Cytoscape.35
Canonical pathways enriched in different groups were estimated by enrichment analysis of the differentially expressed genes.36 The gene sets were obtained from the MsigDB database (http://www.gsea-msigdb.org/gsea/index.jsp), including those from hallmark gene sets, PID and Reactome. Gene sets used in this study were listed in the key resources table.
Treatment response in circRNA-based groups
For analysis of the benefit of induction chemotherapy, patients who underwent concurrent chemoradiotherapy (CCRT, at least 200 mg/m2 cumulative cisplatin),46 with or without induction chemotherapy (IC) containing docetaxel and cisplatin were selected for subgroup analysis. IC consisted of cisplatin with docetaxel (TP regime), or cisplatin with docetaxel and fluorouracil (TPF regime).
Statistical analysis
The primary endpoint of our study was disease-free survival (DFS), and the secondary endpoints were distant metastasis-free survival (DMFS) and overall survival (OS). We defined DFS as the period from the start of treatment to the first relapse at any site or death from any cause, whichever occurred first; DMFS as the period from the start of treatment to the first distant relapse; and OS as the period from the start of treatment to death from any cause.
We applied univariate Cox regression analyses to distinguish clinical features associated with clinical prognosis, and multivariate Cox regression analyses were performed to select independent prognostic factors. The Kaplan-Meier method and the log-rank test were used to estimate the survival probability of patients among different groups, and Cox regression analyses were applied to calculate the hazard ratios (HRs). Receiver operating characteristic (ROC) curves were generated to test the efficiency of our classifier and other clinical factors. The χ2 test or Fisher’s exact test was used to compare categorical variables, while the t test or the Wilcoxon rank sum test was used to compare continuous variables. All statistical tests were performed in R software (version 4.0.3), and a P value < 0.05 (∗) was considered significant.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (82173286 and 81930072), Guangdong Basic and Applied Basic Research Foundation (2023B1515020013), Fundamental Research Funds for the Central Universities, Sun Yat-sen University (23ykbj009), Young Talents Program of Sun Yat-sen University Cancer Center (YTP-SYSUCC-0032), Chih Kuang Scholarship for Outstanding Young Physician-Scientists of Sun Yat-sen University Cancer Center (CKS-SYSUCC-2023004), Natural Science Foundation of Guangdong Province (2017A030312003), Overseas Expertise Introduction Project for Discipline Innovation (111 Project, B14035).
Author contributions
Conception and design: Y.-Q.L., J.M., and N.L.; development of methodology: Y.-L.L., Y.-H.Z., C.D., and S.-W.H.; experiment implement: Y.-L.L., Y.-H.Z., C.D., S.-W.H., and Q.-M.H.; acquisition of sample and data: W.J., Q.L., C.-M.Z., L.-L.T., Y.-P.M., L.C., W.-F.L., G.-Q.Z., and C.D.; analysis and interpretation of data: Y.-L.L., Y.-H.Z., C.D., and S.-W.H.; writing the manuscript: Y.-L.L., Y.-H.Z., C.D., S.-W.H., and W.J.; revision of the manuscript: Y.-Q.L., J.M., N.L., C.-M.Z., Q.-M.H., L.-L.T., Y.-P.M., L.C., W.-F.L., G.-Q.Z., C.D., and Q.L.; administrative, technical, or material support: Y.-Q.L. and J.M.; study supervision: Y.-Q.L. and J.M. All authors reviewed and approved the final manuscript.
Declaration of interests
All authors declare no conflict of interest.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: November 14, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.108467.
Contributor Information
Wei Jiang, Email: weijiang@glmc.edu.cn.
Jun Ma, Email: majun2@mail.sysu.edu.cn.
Ying-Qin Li, Email: liyingq@sysucc.org.cn.
Supplemental information
References
- 1.Sun Y., Li W.F., Chen N.Y., Zhang N., Hu G.Q., Xie F.Y., Sun Y., Chen X.Z., Li J.G., Zhu X.D., et al. Induction chemotherapy plus concurrent chemoradiotherapy versus concurrent chemoradiotherapy alone in locoregionally advanced nasopharyngeal carcinoma: a phase 3, multicentre, randomised controlled trial. Lancet Oncol. 2016;17:1509–1520. doi: 10.1016/S1470-2045(16)30410-7. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Y., Chen L., Hu G.Q., Zhang N., Zhu X.D., Yang K.Y., Jin F., Shi M., Chen Y.P., Hu W.H., et al. Gemcitabine and Cisplatin Induction Chemotherapy in Nasopharyngeal Carcinoma. N. Engl. J. Med. 2019;381:1124–1135. doi: 10.1056/NEJMoa1905287. [DOI] [PubMed] [Google Scholar]
- 3.Hui E.P., Leung S.F., Au J.S.K., Zee B., Tung S., Chua D., Sze W.M., Law C.K., Leung T.W., Chan A.T.C. Lung metastasis alone in nasopharyngeal carcinoma: a relatively favorable prognostic group. A study by the Hong Kong Nasopharyngeal Carcinoma Study Group. Cancer. 2004;101:300–306. doi: 10.1002/cncr.20358. [DOI] [PubMed] [Google Scholar]
- 4.Chen L., Zhang Y., Lai S.Z., Li W.F., Hu W.H., Sun R., Liu L.Z., Zhang F., Peng H., Du X.J., et al. 10-Year Results of Therapeutic Ratio by Intensity-Modulated Radiotherapy Versus Two-Dimensional Radiotherapy in Patients with Nasopharyngeal Carcinoma. Oncol. 2019;24:e38–e45. doi: 10.1634/theoncologist.2017-0577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang L.Q., Li C.F., Li J., Chen W.H., Chen Q.Y., Yuan L.X., Lai X.P., He Y., Xu Y.X.X., Hu D.P., et al. Establishment and Validation of Prognostic Nomograms for Endemic Nasopharyngeal Carcinoma. J. Natl. Cancer Inst. 2016;108 doi: 10.1093/jnci/djv291. [DOI] [PubMed] [Google Scholar]
- 6.Guo R., Tang L.L., Mao Y.P., Du X.J., Chen L., Zhang Z.C., Liu L.Z., Tian L., Luo X.T., Xie Y.B., et al. Proposed modifications and incorporation of plasma Epstein-Barr virus DNA improve the TNM staging system for Epstein-Barr virus-related nasopharyngeal carcinoma. Cancer. 2019;125:79–89. doi: 10.1002/cncr.31741. [DOI] [PubMed] [Google Scholar]
- 7.Wan L., Pantel K., Kang Y. Tumor metastasis: moving new biological insights into the clinic. Nat. Med. 2013;19:1450–1464. doi: 10.1038/nm.3391. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y.Q., Chen Y.P., Zhang Y., Jiang W., Liu N., Yun J.P., Sun Y., He Q.M., Tang X.R., Wen X., et al. Prognostic significance of tumor-infiltrating lymphocytes in nondisseminated nasopharyngeal carcinoma: A large-scale cohort study. Int. J. Cancer. 2018;142:2558–2566. doi: 10.1002/ijc.31279. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y.P., Yin J.H., Li W.F., Li H.J., Chen D.P., Zhang C.J., Lv J.W., Wang Y.Q., Li X.M., Li J.Y., et al. Single-cell transcriptomics reveals regulators underlying immune cell diversity and immune subtypes associated with prognosis in nasopharyngeal carcinoma. Cell Res. 2020;30:1024–1042. doi: 10.1038/s41422-020-0374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen D.S., Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 11.Fridman W.H., Pagès F., Sautès-Fridman C., Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat. Rev. Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 12.Chen L.L., Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12:381–388. doi: 10.1080/15476286.2015.1020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qu S., Yang X., Li X., Wang J., Gao Y., Shang R., Sun W., Dou K., Li H. Circular RNA: A new star of noncoding RNAs. Cancer Lett. 2015;365:141–148. doi: 10.1016/j.canlet.2015.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Hong X., Liu N., Liang Y., He Q., Yang X., Lei Y., Zhang P., Zhao Y., He S., Wang Y., et al. Circular RNA CRIM1 functions as a ceRNA to promote nasopharyngeal carcinoma metastasis and docetaxel chemoresistance through upregulating FOXQ1. Mol. Cancer. 2020;19:33. doi: 10.1186/s12943-020-01149-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang X.Y., Huang Z.L., Huang J., Xu B., Huang X.Y., Xu Y.H., Zhou J., Tang Z.Y. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J. Exp. Clin. Cancer Res. 2020;39:20. doi: 10.1186/s13046-020-1529-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan S., Wang S., Huang T., Wang J., Yuan X. circRNAs: Insight Into Their Role in Tumor-Associated Macrophages. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.780744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen D.L., Sheng H., Zhang D.S., Jin Y., Zhao B.T., Chen N., Song K., Xu R.H. The circular RNA circDLG1 promotes gastric cancer progression and anti-PD-1 resistance through the regulation of CXCL12 by sponging miR-141-3p. Mol. Cancer. 2021;20:166. doi: 10.1186/s12943-021-01475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fan L., Cao Q., Liu J., Zhang J., Li B. Circular RNA profiling and its potential for esophageal squamous cell cancer diagnosis and prognosis. Mol. Cancer. 2019;18:16. doi: 10.1186/s12943-018-0936-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ju H.Q., Zhao Q., Wang F., Lan P., Wang Z., Zuo Z.X., Wu Q.N., Fan X.J., Mo H.Y., Chen L., et al. A circRNA signature predicts postoperative recurrence in stage II/III colon cancer. EMBO Mol. Med. 2019;11 doi: 10.15252/emmm.201810168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alimbetov D., Askarova S., Umbayev B., Davis T., Kipling D. Pharmacological Targeting of Cell Cycle, Apoptotic and Cell Adhesion Signaling Pathways Implicated in Chemoresistance of Cancer Cells. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19061690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhu Y., Hu J., Hu Y., Liu W. Targeting DNA repair pathways: a novel approach to reduce cancer therapeutic resistance. Cancer Treat Rev. 2009;35:590–596. doi: 10.1016/j.ctrv.2009.06.005. [DOI] [PubMed] [Google Scholar]
- 22.Yip T.T., Lau W.H., Chan J.K., Ngan R.K., Poon Y.F., Lung C.W., Lo T.Y., Ho J.H. Prognostic significance of DNA flow cytometric analysis in patients with nasopharyngeal carcinoma. Cancer. 1998;83:2284–2292. [PubMed] [Google Scholar]
- 23.Cheng H., Zhou L., Long Y., Xiang J., Chen L. MACC1 Is Associated With Epithelial-Mesenchymal Transition and Can Predict Poor Prognosis in Nasopharyngeal Carcinoma. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.644120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J.X., Cai M.B., Wang X.P., Duan L.P., Shao Q., Tong Z.T., Liao D.Z., Li Y.Y., Huang M.Y., Zeng Y.X., Shao J.Y. Elevated DLL4 expression is correlated with VEGF and predicts poor prognosis of nasopharyngeal carcinoma. Med. Oncol. 2013;30:390. doi: 10.1007/s12032-012-0390-x. [DOI] [PubMed] [Google Scholar]
- 25.Lu Y., Zhao Q., Liao J.Y., Song E., Xia Q., Pan J., Li Y., Li J., Zhou B., Ye Y., et al. Complement Signals Determine Opposite Effects of B Cells in Chemotherapy-Induced Immunity. Cell. 2020;180:1081–1097.e24. doi: 10.1016/j.cell.2020.02.015. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Diaz A., Shin D.S., Moreno B.H., Saco J., Escuin-Ordinas H., Rodriguez G.A., Zaretsky J.M., Sun L., Hugo W., Wang X., et al. Interferon Receptor Signaling Pathways Regulating PD-L1 and PD-L2 Expression. Cell Rep. 2017;19:1189–1201. doi: 10.1016/j.celrep.2017.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riaz N., Havel J.J., Makarov V., Desrichard A., Urba W.J., Sims J.S., Hodi F.S., Martín-Algarra S., Mandal R., Sharfman W.H., et al. Tumor and Microenvironment Evolution during Immunotherapy with Nivolumab. Cell. 2017;171:934–949.e16. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Y.P., Lv J.W., Mao Y.P., Li X.M., Li J.Y., Wang Y.Q., Xu C., Li Y.Q., He Q.M., Yang X.J., et al. Unraveling tumour microenvironment heterogeneity in nasopharyngeal carcinoma identifies biologically distinct immune subtypes predicting prognosis and immunotherapy responses. Mol. Cancer. 2021;20:14. doi: 10.1186/s12943-020-01292-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qiao H., Tan X.R., Li H., Li J.Y., Chen X.Z., Li Y.Q., Li W.F., Tang L.L., Zhou G.Q., Zhang Y., et al. Association of Intratumoral Microbiota With Prognosis in Patients With Nasopharyngeal Carcinoma From 2 Hospitals in China. JAMA Oncol. 2022;8:1301–1309. doi: 10.1001/jamaoncol.2022.2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Friedman J., Hastie T., Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Software. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W., Treviño V., Shen H., Laird P.W., Levine D.A., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becht E., Giraldo N.A., Lacroix L., Buttard B., Elarouci N., Petitprez F., Selves J., Laurent-Puig P., Sautès-Fridman C., Fridman W.H., de Reyniès A. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:218. doi: 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS A J. Integr. Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee A.W.M., Lydiatt W.M., Colevas A.D., Glastonbury C.M., Le Q.T.X., O’Sullivan B., Weber R.S., Shah J.P. Nasopharynx. AJCC Cancer Staging Manual. 8th edition. Springer; 2016. [Google Scholar]
- 38.Chen Y.P., Chan A.T.C., Le Q.T., Blanchard P., Sun Y., Ma J. Nasopharyngeal carcinoma. Lancet. 2019;394:64–80. doi: 10.1016/S0140-6736(19)30956-0. [DOI] [PubMed] [Google Scholar]
- 39.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 40.Tibshirani R. The lasso method for variable selection in the Cox model. Stat. Med. 1997;16:385–395. doi: 10.1002/(sici)1097-0258(19970228)16:4<385::aid-sim380>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 41.Simon N., Friedman J., Hastie T., Tibshirani R. Regularization Paths for Cox's Proportional Hazards Model via Coordinate Descent. J. Stat. Software. 2011;39:1–13. doi: 10.18637/jss.v039.i05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liang W., Liang H., Ou L., Chen B., Chen A., Li C., Li Y., Guan W., Sang L., Lu J., et al. Development and Validation of a Clinical Risk Score to Predict the Occurrence of Critical Illness in Hospitalized Patients With COVID-19. JAMA Intern. Med. 2020;180:1081–1089. doi: 10.1001/jamainternmed.2020.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robin X., Turck N., Hainard A., Tiberti N., Lisacek F., Sanchez J.C., Müller M. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinf. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barbie D.A., Tamayo P., Boehm J.S., Kim S.Y., Moody S.E., Dunn I.F., Schinzel A.C., Sandy P., Meylan E., Scholl C., et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bagaev A., Kotlov N., Nomie K., Svekolkin V., Gafurov A., Isaeva O., Osokin N., Kozlov I., Frenkel F., Gancharova O., et al. Conserved pan-cancer microenvironment subtypes predict response to immunotherapy. Cancer Cell. 2021;39:845–865.e7. doi: 10.1016/j.ccell.2021.04.014. [DOI] [PubMed] [Google Scholar]
- 46.Peng H., Chen L., Zhang Y., Li W.F., Mao Y.P., Zhang F., Guo R., Liu L.Z., Lin A.H., Sun Y., Ma J. Prognostic Value of the Cumulative Cisplatin Dose During Concurrent Chemoradiotherapy in Locoregionally Advanced Nasopharyngeal Carcinoma: A Secondary Analysis of a Prospective Phase III Clinical Trial. Oncol. 2016;21:1369–1376. doi: 10.1634/theoncologist.2016-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The data of circRNA microarrays have been deposited at GEO and are publicly available as of the date of publication. This paper also analyses existing, publicly available data. The accession numbers for these datasets are listed in the key resources table. The key raw data have been deposited to the Research Data Deposit public platform (https://www.researchdata.org.cn/), with an approval number listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.