Abstract
Variation of the pilus of Neisseria gonorrhoeae occurs by the recombination of silent pilin DNA sequences into the pilin expression locus. We have developed a quantitative, competitive reverse transcription-PCR assay which measures the frequency of pilin antigenic variation independently of changes in gonococcal colony morphology and have determined this frequency within a gonococcal population. We have also studied the frequency of antigenic variation during growth and have concluded that growth does not dramatically influence the frequency of pilin antigenic variation, although a reproducible, twofold increase is observed upon the transition into late log/stationary phase.
The gram-negative diplococcus Neisseria gonorrhoeae (the gonococcus) is the causative agent of the sexually transmitted disease gonorrhea. Pili, filamentous cell surface structures composed primarily of pilin monomers, mediate the initial attachment of the gonococcus to host mucosal epithelial cells (22) and are essential in the establishment of infection, as nonpiliated gonococci fail to cause disease (9, 10).
Pilin is encoded by pilE, the pilin expression gene (12). Gonococcal strain FA1090 contains one pilE gene (21), and 19 silent pilin copies distributed among five loci, with one silent copy associated with and located upstream of pilE (21). Variation of the gonococcal pilus results from the nonreciprocal transfer of partial pilin sequence information from one silent pilin copy into pilE (5, 6, 17). The resultant, altered pilE gene sequence may encode either an immunologically distinct pilin monomer which can be assembled into functional pili (antigenic variation) or a pilin monomer which is not produced or is inefficiently assembled, resulting in a switch from a piliated (P+) to a nonpiliated (P−) colony phenotype (colony morphology-based phase variation) (1, 6, 23).
Since pilin antigenic variation and changes in gonococcal colony morphology result from similar recombination-mediated processes, estimates of the frequency of antigenic variation have been based on that of colony morphology-based phase variation (5, 23, 24, 27), which has been reported to be about 10−4 to 10−2 colony morphology variants per total CFUs (1, 16, 27). However, changes in gonococcal colony morphology can also occur through processes distinct from those resulting in antigenic variation, such as the deletion of pilE or phase variation of PilC (7, 8, 16). Therefore, equating the frequency of pilin antigenic variation to that of colony morphology-based phase variation is not always appropriate. In order to quantitate pilS to pilE recombination independently of changes in colony morphology, we have combined a qualitative assay which specifically detects pilS to pilE recombination events (25), with a competitive reverse transcription-PCR (RT-PCR) strategy. By using this assay, we have determined the frequency of pilin antigenic variation during the growth of a gonococcal population.
Development of a quantitative RT-PCR assay to measure the frequency of pilin antigenic variation.
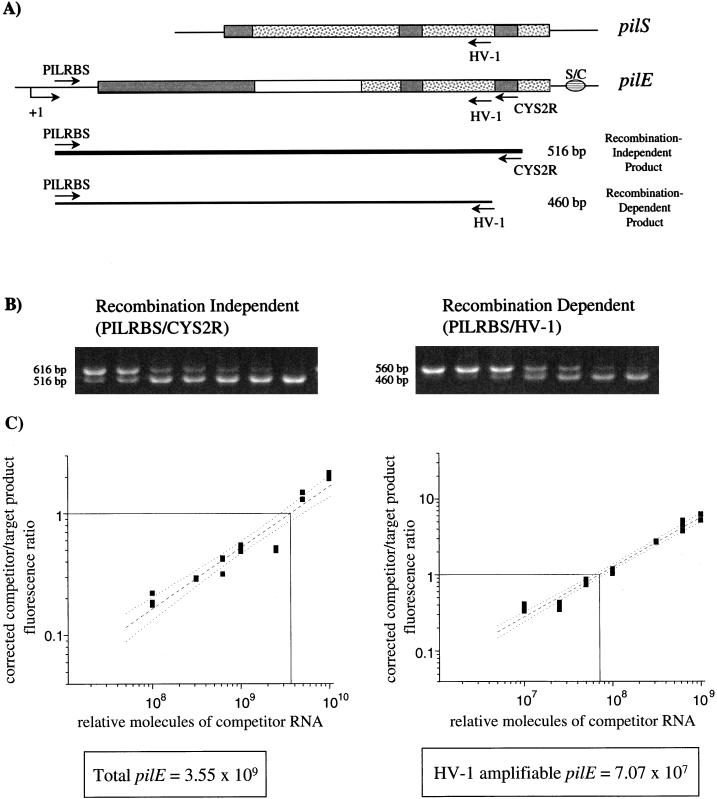
The quantitative RT-PCR assay involves two PCRs which amplify pilE (Fig. 1A): a recombination-independent reaction (primers PILRBS and CYS2R), which amplifies all pilE template present (25), and a recombination-dependent reaction (primers PILRBS and HV-1), which amplifies only those pilE genes into which HV-1 amplifiable silent sequences have recombined (11). To establish the assay, the frequency of pilin recombination in the FA1090 variant 1-81-S2 wild type (WT) (19), which does not initially contain HV-1 target sequences in pilE (20), was determined. A constant volume of total RNA isolated from a late-log-phase gonococcal population (target RNA) was mixed with a range of known concentrations of in vitro-derived competitor RNA. The cloned competitor contains PILRBS target sequences at the 5′ end, an extra 100 bp of nonpilin DNA, and both HV-1 and CYS2R target sequences at the 3′ end, downstream of the T7 polymerase promoter in pGEM-3 (pCDS2). This mixture of RNA was then reverse transcribed, and the resulting cDNA was used as template for both the recombination-independent and recombination-dependent PCRs, which were performed in triplicate. By mixing target and competitor RNA prior to the reverse transcription step, the competior template acted as an internal control for the efficiencies of both the reverse transcription reactions and PCRs (15). Target and competitor PCR products were separated by gel electrophoresis (Fig. 1B), and the amount of each product present was measured by densitometry. The fluorescence intensities of the competitor products were then corrected for size, allowing for a direct comparison between competitor and target product molar amounts. The corrected competitor-to-target product fluorescence ratio was plotted as a function of the relative number of competitor RNA molecules present (Fig. 1C). The relative number of competitor RNA molecules at which equal molar amounts of target and competitor products are made reflects the concentration of target pilE RNA template present in the original sample (15). This calculation is independent of the level of transcription of total mRNA in the gonococcal cultures, since the amounts of both target RNAs are determined with regard to known concentrations of competitor RNA. The total number of pilE template molecules was calculated from the recombination-independent PCRs (primers PILRBS and CYS2R), while the number of pilE template molecules containing sequences amplified by HV-1 (due to recombination) was calculated from the recombination-dependent PCRs (primers PILRBS and HV-1) (Fig. 1C). Once the amounts of total and HV-1 amplifiable pilE template present were calculated with respect to the same competitor, they were used to determine the proportion of pilE genes containing HV-1 amplifiable sequences within the population. The frequency of HV-1 detection of pilE in variant 1-81-S2 WT at late log phase was 1.99 × 10−2 HV-1 amplifiable pilE per total pilE (HV-1 pilE/pilE). This is the first quantitative measurement of the frequency of pilin antigenic variation within a gonococcal population and independent of colony morphology-based phase variation.
FIG. 1.
Quantitation of pilin recombination in variant 1-81-S2 WT at late log phase using competitive RT-PCR. (A) Cartoon of PCR assay detecting pilS-to-pilE recombination. The major conserved regions of pilE and pilS are represented by the shaded boxes, and variable sequences are represented by either white or speckled boxes. S/C represents the conserved, 3′ Sma/Cla repeat. The transcription start site of pilE is indicated by +1. PILRBS targets the ribosomal binding site of the expression locus and is pilE specific (26). CYS2R targets the conserved cys2 region of all pilin copies (25), while HV-1 targets the HV sequences of pilS1 copy 4 and pilS6 copy 1 (19, 20). Recombination-independent primers PILRBS and CYS2R amplify all pilE templates present, generating a 516-bp product (thick line). Recombination-dependent primers PILRBS and HV-1 yield a 460-bp product only if HV-1 target sequences have recombined into pilE (thin line). (B) Representative agarose gels of the recombination-independent and -dependent PCR products. The products of one recombination-independent PCR and one recombination-dependent PCR are shown. In both gels, the top band is the competitor PCR product, and the bottom band is the target PCR product. (C) Graphs of the PCR data. The x axis shows the relative number of competitor RNA molecules added for each cDNA reaction, as determined by spectrophotometry at an optical density of 260 nm. The y axis shows the corrected competitor-to-target product fluorescence ratio. A line of least-squares fit (dashed) with 95% confidence level (dotted lines) is shown. The amount of competitor RNA present when equal molar amounts of competitor and target products are made (solid vertical and horizontal lines, respectively) equals the amount of target template present in the original reaction. Total pilE is calculated from the recombination-independent reactions, and HV-1 ampifiable pilE is calculated from the recombination-dependent reactions.
Contribution of HV-1 amplifiable silent copies to the frequency of antigenic variation.
The HV regions of pilS1 copy 4 and pilS6 copy 1 in FA1090 contain identical sequences that are specifically targeted by HV-1 (20). However, the HV regions of other silent copies contain sequences similar to HV-1 and therefore may have also been amplified during the recombination-dependent reaction. In order to determine the contribution of the HV-1-specific silent copies to the calculated frequency, recombination-dependent PCR products from the late log phase of preinduced 1-81-S2 recA6 and of 1-81-S2 recA6 induced at time zero (t0) (see below) were cloned and sequenced. None of the 54 clones sequenced contained the original pilS2 copy 1 sequence in the HV region, confirming that the assay detected the transfer of silent-copy HV sequences into pilE (Table 1). Fifty percent of pilE PCR products contained HV-1 target-specific pilS1 copy 4 or pilS6 copy 1 sequences in the HV region. The other clones sequenced contained HV sequences from a subset of other silent pilin copies (20), each of which contained two to five mismatches to the HV-1 primer. Therefore, although this RT-PCR assay does not detect recombination solely from pilS1 copy 4 or pilS6 copy 1 into pilE, it accurately and reproducibly quantitates the recombination frequency of HV-1 amplifiable silent pilin sequences into pilE.
TABLE 1.
Spectrum of sequences in recombination-dependent PCR product clones
| HV sequence | No. of clones (% total) | No. of mismatches to HV-1 |
|---|---|---|
| pilS1 copy 4 or pilS6 copy 1 | 27 (50) | 0 |
| pilS1 copy 5 or pilS2 copy 5 | 17 (31.5) | 2 |
| pilS1 copy 2 or pilS3 copy 1 | 5 (9.3) | 4 |
| pilS7 copy 1 | 4 (7.4) | 5 |
| pilS1 copy 3 | 1 (1.8) | 3 |
Effect of growth on the frequency of gonococcal pilin antigenic variation.
Since this competitive RT-PCR assay provides a reproducible measurement of the frequency of pilin antigenic variation within a gonococcal population, we used it to study the effect of growth. We used two FA1090 variants which do not initially contain HV-1 amplifiable sequences in pilE; 1-81-S2 WT (19) and 1-81-S2 recA6 (11). Variant 1-81-S2 recA6 contains the recA6 allele, in which lac regulatory sequences control the transcription of recA. In the absence of isopropyl-β-d-thiogalactopyranoside (IPTG), transcription of recA is undetectable, and recA6 variants are deficient in recombination. However, in the presence of IPTG, recA is transcribed, and recombination-mediated processes, such as pilin antigenic variation, can occur (18). The use of variant 1-81-S2 recA6 ensured that the initial pilE sequences within the gonococcal population did not contain HV-1 target sequences. Moreover, in the absence of IPTG, recombination-dependent product from the PCR with primers PILRBS and HV-1 was not detected at any point during growth (data not shown).
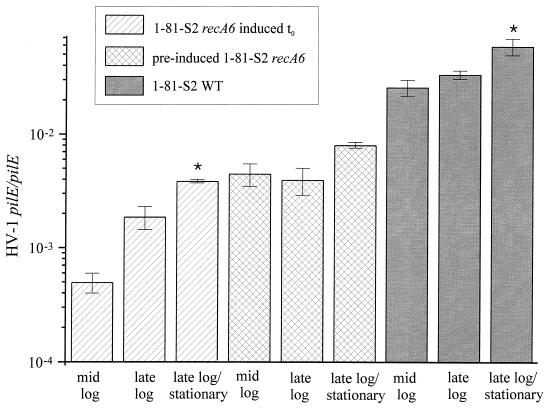
The frequency of pilin antigenic variation was determined for mid-log, late log, and late log/stationary phases of growth of these two FA1090 variants. Following the induction of recA with IPTG at t0, there was a considerable increase in the frequency by which HV-1 sequences recombined into pilE over time in 1-81-S2 recA6 (Fig. 2). A 3.8-fold increase, from 4.97 × 10−4 to 1.87 × 10−3 HV-1 pilE/pilE, was observed between mid- and late log phases, followed by a 2-fold increase, from 1.87 × 10−3 to 3.86 × 10−3 HV-1 pilE/pilE, between late log and late log/stationary phases. Rates of variation, which reflect the number of antigenic variants per generation, were calculated during exponential growth of the gonococcal population and were 2.76 × 10−4 variants per generation (var/gen) from t0 to mid-log, 4.90 × 10−4 var/gen from mid-log to late log, and 3.36 × 10−4 var/gen from t0 to late log. The increase in the frequency over time suggested that the number of gonococci within the population containing recombinant, HV-1 amplifiable pilE accumulated from the time recombination could occur with the induction of recA through to late log/stationary phase of growth.
FIG. 2.
Average frequency of pilin antigenic variation with respect to growth phase. The frequency of antigenic variation is expressed as HV-1 pilE/pilE for mid-log, late log, and late log/stationary phases of 1-81-S2 recA6 induced at t0, preinduced 1-81-S2 recA6 and 1-81-S2 WT. Six to 15 individual sets of PCRs were performed for each time point, and the standard errors of the means are shown by the error bars. The asterisk indicates a significant difference relative to the value at late log phase by the Student t test with P < 0.05.
In order to determine whether the level of HV-1 amplifiable pilE in variant 1-81-S2 recA6 reaches an equilibrium within a population undergoing continual pilin recombination, this variant was preinduced with IPTG prior to the growth curve assayed. The frequency of pilin recombination in variant 1-81-S2 recA6 preinduced with IPTG remained relatively constant throughout early growth and with repeats of growth, with 4.48 × 10−3 HV-1 pilE/pilE at mid-log and 3.97 × 10−3 HV-1 pilE/pilE at late log. There was a twofold increase in this frequency upon late log/stationary phase relative to late log from 3.97 × 10−3 to 8.05 × 10−3 HV-1 pilE/pilE (Fig. 2). The consistency in the frequency over time suggested the presence of an equilibrium subpopulation of HV-1 amplifiable pilE-expressing gonococci within the population which was not drastically influenced by growth.
With variant 1-81-S2 WT, the frequency of pilin antigenic variation also remained relatively constant throughout the exponential phase of growth, with 2.59 × 10−2 HV-1 pilE/pilE at mid-log and 3.35 × 10−2 HV-1 pilE/pilE at late log (Fig. 2). The rate of variation over this period of growth was 7.92 × 10−3 var/gen. Overall, the frequency by which HV-1 target sequences recombined into pilE was approximately 10-fold higher in the 1-81-S2 WT variant than in the 1-81-S2 recA6 variant, presumably due to the lower levels of recA mRNA present in fully induced recA6 variants compared to those in the WT counterparts (18). The frequency of pilin antigenic variation in the WT variant was consistent both throughout early growth and throughout repeated growth curves assayed. This further suggested the presence of an equilibrium subpopulation of variant gonococci within the population which was not affected by growth. In contrast, there was a significant twofold increase in the frequency of pilin recombination upon late log/stationary phase relative to late log from 3.35 × 10−2 to 5.92 × 10−2 HV-1 pilE/pilE. This suggested that some event during the transition into the late log/stationary phase of growth may have influenced pilin recombination. Stationary-phase effects have been reported in N. gonorrhoeae, such as an enhancement in gonococcal autolysis (2, 3, 13). This increased autolysis, coupled with the natural transformation competence of gonococci, has previously been proposed to affect pilin sequence changes (4, 14). It is possible that an increase in autolysis and the subsequent uptake of released DNA by intact, unlysed gonococci may account for the increase in the frequency of pilin antigenic variation observed upon late log/stationary phase.
Conclusions.
This quantitative RT-PCR assay measures pilin antigenic variation independently of changes in gonococcal colony morphology, thus taking into account the fact that pilin antigenic and colony morphology-based phase variation are separable events. Furthermore, this assay can be used to quantitate and to compare the frequency of gonococcal pilin antigenic variation between different variants, growth conditions, or mutants and is adaptable for the detection and quantitation of recombination from other silent pilin copies into pilE. Using this quantitative assay to examine pilin antigenic variation frequencies during growth, we can conclude that growth does not dramatically affect the frequency of pilin antigenic variation in a gonococcal population but that a reproducible, twofold increase in the frequency does occur upon transition into the late log/stationary phase of growth.
Acknowledgments
This work was supported by grant RO1AI33493 from the National Institutes of Health.
We thank Joe Dillard, Becky Howell-Adams, Cindy Long, and Ian Mehr for critical reading of this manuscript.
REFERENCES
- 1.Bergström S, Robbins K, Koomey J M, Swanson J. Piliation control mechanisms in Neisseria gonorrhoeae. Proc Natl Acad Sci USA. 1986;83:3890–3894. doi: 10.1073/pnas.83.11.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dillard J P, Seifert H S. A peptidoglycan hydrolase similar to bacteriophage endolysins acts as an autolysin in Neisseria gonorrhoeae. Mol Microbiol. 1997;25:893–902. doi: 10.1111/j.1365-2958.1997.mmi522.x. [DOI] [PubMed] [Google Scholar]
- 3.Elmros T, Burman L G, Bloom G D. Autolysis of Neisseria gonorrhoeae. J Bacteriol. 1976;126:969–976. doi: 10.1128/jb.126.2.969-976.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gibbs C P, Reimann B, Schultz E, Kaufmann A, Haas R, Meyer T F. Reassortment of pilin genes in Neisseria gonorrhoeae occurs by two distinct mechanisms. Nature. 1989;338:651–652. doi: 10.1038/338651a0. [DOI] [PubMed] [Google Scholar]
- 5.Haas R, Meyer T F. The repertoire of silent pilus genes in Neisseria gonorrhoeae: evidence for gene conversion. Cell. 1986;44:107–115. doi: 10.1016/0092-8674(86)90489-7. [DOI] [PubMed] [Google Scholar]
- 6.Hagblom P, Segal E, Billyard E, So M. Intragenic recombination leads to pilus antigenic variation in Neisseria gonorrhoeae. Nature. 1985;315:156–158. doi: 10.1038/315156a0. [DOI] [PubMed] [Google Scholar]
- 7.Jonsson A, Ilver D, Falk P, Pepose J, Normark S. Sequence changes in the pilus subunit lead to tropism variation of Neisseria gonorrhoeae to human tissue. Mol Microbiol. 1994;13:403–416. doi: 10.1111/j.1365-2958.1994.tb00435.x. [DOI] [PubMed] [Google Scholar]
- 8.Jonsson A, Pfeifer J, Normark S. Neisseria gonorrhoeae PilC expression provides a selective mechanism for structural diversity of pili. Proc Natl Acad Sci USA. 1992;89:3204–3208. doi: 10.1073/pnas.89.8.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kellogg D S, Jr, Cohen I R, Norins L C, Schroeter A L, Reising G. Neisseria gonorrhoeae. II. Colonial variation and pathogenicity during 35 months in vitro. J Bacteriol. 1968;96:596–605. doi: 10.1128/jb.96.3.596-605.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kellogg D S, Jr, Peacock W L, Jr, Deacon W E, Brown L, Pirkle C I. Neisseria gonorrhoeae. I. Virulence genetically linked to clonal variation. J Bacteriol. 1963;85:1274–1279. doi: 10.1128/jb.85.6.1274-1279.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehr I J, Seifert H S. Random shuttle mutagenesis: gonococcal mutants deficient in pilin antigenic variation. Mol Microbiol. 1997;23:1121–1131. doi: 10.1046/j.1365-2958.1997.2971660.x. [DOI] [PubMed] [Google Scholar]
- 12.Meyer T F. Pilus expression in Neisseria gonorrhoeae involves chromosomal rearrangement. Cell. 1982;30:45–52. doi: 10.1016/0092-8674(82)90010-1. [DOI] [PubMed] [Google Scholar]
- 13.Morse S A, Bartenstein L. Factors affecting autolysis of Neisseria gonorrhoeae. Proc Soc Exp Biol Med. 1974;145:1418–1421. doi: 10.3181/00379727-145-38025. [DOI] [PubMed] [Google Scholar]
- 14.Norlander L, Davies J, Norqvist A, Normark S. Genetic basis for colonial variation in Neisseria gonorrhoeae. J Bacteriol. 1979;138:762–769. doi: 10.1128/jb.138.3.762-769.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Piatak M J, Luk K, Williams B, Lifson J D. Quantitative competitive polymerase chain reaction for accurate quantitation of HIV DNA and RNA species. BioTechniques. 1993;14:70–80. [PubMed] [Google Scholar]
- 16.Segal E, Billyard E, So M, Storzbach S, Meyer T F. Role of chromosomal rearrangement in N. gonorrhoeae pilus phase variation. Cell. 1985;40:293–300. doi: 10.1016/0092-8674(85)90143-6. [DOI] [PubMed] [Google Scholar]
- 17.Segal E, Hagblom P, Seifert H S, So M. Antigenic variation of gonococcal pilus involves assembly of separated silent gene segments. Proc Natl Acad Sci USA. 1986;83:2177–2181. doi: 10.1073/pnas.83.7.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Seifert H S. Insertionally inactivated and inducible recA alleles for use in Neisseria. Gene. 1997;199:215–220. doi: 10.1016/s0378-1119(96)00810-4. [DOI] [PubMed] [Google Scholar]
- 19.Seifert H S, Wright C J, Jerse A E, Cohen M S, Cannon J G. Multiple gonococcal pilin antigenic variants are produced during experimental human infections. J Clin Invest. 1994;93:2744–2749. doi: 10.1172/JCI117290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snodgrass, T. L., and J. G. Cannon. 1997. Personal communication.
- 21.Snodgrass T L, Dempsey J A F, Cannon J G. Neisseria 94—Proceedings from the Ninth International Pathogenic Neisseria Conference. 1994. The repertoire of silent pilin gene copies and their chromosomal location is similar but not identical in gonococcal strains FA1090 and MS11; pp. 113–115. [Google Scholar]
- 22.Swanson J. Studies on gonococcus infection. IV. Pili: their role in attachment of gonococci to tissue culture cells. J Exp Med. 1973;137:571–589. doi: 10.1084/jem.137.3.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Swanson J, Bergström S, Robbins K, Barrera O, Corwin D, Koomey J M. Gene conversion involving the pilin structural gene correlates with pilus+↔pilus− changes in Neisseria gonorrhoeae. Cell. 1986;47:267–276. doi: 10.1016/0092-8674(86)90449-6. [DOI] [PubMed] [Google Scholar]
- 24.Swanson J, Morrison S, Barrera O, Hill S. Piliation changes in transformation-defective gonococci. J Exp Med. 1990;171:2131–2139. doi: 10.1084/jem.171.6.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wainwright L A, Pritchard K H, Seifert H S. A conserved DNA sequence is required for efficient gonococcal pilin antigenic variation. Mol Microbiol. 1994;13:75–87. doi: 10.1111/j.1365-2958.1994.tb00403.x. [DOI] [PubMed] [Google Scholar]
- 26.Wright C J, Jerse A E, Cohen M S, Cannon J G, Seifert H S. Nonrepresentative PCR amplifications of variable gene sequences in clinical specimens containing dilute, complex mixtures of microorganisms. J Clin Microbiol. 1994;32:464–468. doi: 10.1128/jcm.32.2.464-468.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Q Y, DeRyckere D, Lauer P, Koomey J M. Gene conversion in Neisseria gonorrhoeae: evidence for its role in pilus antigenic variation. Proc Natl Acad Sci USA. 1992;89:5366–5370. doi: 10.1073/pnas.89.12.5366. [DOI] [PMC free article] [PubMed] [Google Scholar]