Abstract
Epstein–Barr virus (EBV) establishes lifelong asymptomatic infection by replication of its chromatinized episomes with the host genome. EBV exhibits different latency-associated transcriptional repertoires, each with distinct three-dimensional structures. CTCF, Cohesin and PARP1 are involved in maintaining viral latency and establishing episome architecture. Epstein–Barr virus-associated gastric cancer (EBVaGC) represents 1.3–30.9% of all gastric cancers globally. EBV-positive gastric cancers exhibit an intermediate viral transcription profile known as ‘Latency II’, expressing specific viral genes and noncoding RNAs. In this study, we investigated the impact of PARP1 inhibition on CTCF/Cohesin binding in Type II latency. We observed destabilization of the binding of both factors, leading to a disrupted three-dimensional architecture of the episomes and an altered viral gene expression. Despite sharing the same CTCF binding profile, Type I, II and III latencies exhibit different 3D structures that correlate with variations in viral gene expression. Additionally, our analysis of H3K27ac-enriched interactions revealed differences between Type II latency episomes and a link to cellular transformation through docking of the EBV genome at specific sites of the Human genome, thus promoting oncogene expression. Overall, this work provides insights into the role of PARP1 in maintaining active latency and novel mechanisms of EBV-induced cellular transformation.
Graphical Abstract
Graphical Abstract.
Introduction
Epstein–Barr virus (or EBV) is a human gammaherpesvirus, that infects more than 90% of the adult population, establishing a lifelong asymptomatic condition (1) through the formation of circularized mini-chromosomes known as episomes, which replicate with the host genome. These episomes do not integrate into the host genome, thus making it essential for EBV to organize its chromatin in a way that allows access to essential genes for transcription and replication while maintaining genomic stability.
EBV exhibits different latency-associated transcriptional repertoires, each responsible for specific gene expressions during different stages of infection and characterized by different three-dimensional structures of the viral genome (2–4).
Our group and others have previously demonstrated that the cellular factor CTCF is involved in the maintenance of the viral latency and, together with Cohesin, in the establishment of the 3D architecture of the viral episomes (5–9).
Recent work from our group also demonstrated that Poly [ADP-ribose] Polymerase 1 (PARP1) is essential in maintaining an active latency program in Type III latency by stabilizing CTCF binding at CTCF/Cohesin co-localization sites (3,10,11). Moreover it has recently been demonstrated that PARP1 inhibition is a valuable approach to target EBV + Gastric Cancer (12).
Gastric cancers lead to approximately 780 000 deaths every year, making them the third most common cause of cancer-related deaths worldwide (13). Among the different types of gastric cancer, Epstein–Barr virus-associated gastric cancer (EBVaGC) is frequently observed (14). The prevalence of EBVaGC varies based on geographic regions, comprising approximately 1.3–30.9% of all gastric cancers (15–18), with an overall global average of 8.9% (15). This accounts for approximately 75 000 new cases diagnosed annually (14).
EBV-positive gastric cancer is associated with CpG island hypermethylation (19) and with distinct mutation patterns (20,21).
In EBV + gastric cancers, EBV establishes an intermediate transcriptional profile called ‘Latency II’ which is characterized by the expression of EBNA1, noncoding RNAs, and latent membrane proteins (LMP2A and 2B and occasionally LMP1).
Given all our previous findings, we aimed to investigate whether also in Type II latency CTCF/Cohesin binding is altered by PARP1 inhibition. Surprisingly, we observed a destabilization of both factors thus resulting in a completely disrupted three-dimensional architecture of the EBV episomes. As we expected, this genomic instability caused altered viral gene expression even though below statistical significance.
Moreover, using HiC assay we found varying structures in EBV + GC cells and B cells even though Type I, II and III latencies share the same CTCF binding profile across the viral genome. Notably, long-range chromatin interactions in the viral genome align with expression levels of latent genes, indicating a close connection between 3D viral chromatin structure and transcriptional activity of involved viral loci.
To further elucidate the link between EBV genomic architecture and its functional role we analyzed the H3K27ac-enriched EBV-EBV and EBV–human interactions. We found that Type II latency episomes show strong H3K27ac-rich sites on the Qp and BILF2 promoters differently from what is observed in Type III latency. Moreover, most EBV–human interactions occur between the BILF2 region and genes which are markers of gastric malignances.
Altogether, this work aims to shed light on the dependency of EBV latency on PARP1 to maintain an active latency program. Moreover, we wanted to further elucidate the molecular mechanism named ‘enhancer infestation’ by which EBV could induce cellular transformation by docking at specific sites of the Human genome both altering host epigenetic landscape and serving as an additional enhancer to promote oncogene expression (22).
All these findings may present opportunities for novel, targeted therapeutics tailored to specific latency types and associated malignancies.
Materials and methods
Cell culture and treatment
Cell lines were maintained in a humidified incubator containing 5% CO2 at 37 °C. YCCEL1 and SNU719 cell lines were cultured in RPMI 1640 supplemented with Fetal Bovine Serum at a concentration of 10% and supplemented with 1% penicillin–streptomycin.
Treatment with PARP inhibitor Olaparib (Selleck Chemicals, Catalog No. S1060) was given 72 h before collection at a concentration of 5 μM.
Dot blot
Dot Blot was carried out as per Abcam protocol. Briefly, three biological replicates of 2 × 106 YCCEL1/SNU719 cells per treatment group (with or without 5 μM Olaparib for 72 h) were extracted using radioimmunoprecipitation assay (RIPA) lysis buffer (50 mM Tris–HCl, pH 7.4, 150 mM NaCl, 0.25% deoxycholic acid, 1% NP-40, 1 mM EDTA; Millipore, Cat. No. 20–188) supplemented with 1X protease inhibitor cocktail (Sigma-Aldrich, Cat. No. P8340-5ML) and 1× PARG inhibitor (PDD00017273, Selleck Chemicals, Cat. No. S8862). Protein concentration was measured using a bicinchoninic acid (BCA) protein assay (Pierce, Cat. No. 23227) and 10 μg of protein were diluted in 50 μl of water and loaded onto the activated nitrocellulose membrane (Bio-Rad Cat. No. 1620113).
Red Ponceau staining (Ponceau S Staining Solution, Cell Signaling, Cat. No. 59803) was used as a loading control.
Membrane was blocked in 5% milk TBS-T and then incubated with anti-PAR antibody (R&D Systems, Cat. No. 4335-MC-100) as per manufacturer recommendation.
Membrane was washed 5 mins three times, incubated for 1 h with rabbit anti-mouse IgG-HRP (Jackson ImmunoResearch), at a dilution of 1:10000 and detected by enhanced chemiluminescence (SuperSignal West Dura Extended Duration Substrate, Cat. No. 34075).
Subcellular fractionation and western blot
Three biological replicates of 5 × 106 YCCEL1/SNU719 cells per treatment group (with or without 5 μM Olaparib for 72 h) were prepared using the Subcellular Protein Fractionation Kit for Cultured Cells (Thermo Scientific, Cat. No. 78840) as per manufacturer's protocol.
Protein concentration was measured using a bicinchoninic acid (BCA) protein assay (Pierce). Lysates were boiled with 2× Laemmli sample buffer (Bio-Rad, Cat. No. 1610737) containing 2.5% β-mercaptoethanol (Sigma-Aldrich, Cat. No. M6250-100ML).
Proteins were resolved by gel electrophoresis on a 4–20% polyacrylamide gradient Mini-Protean TGX pre-cast gel (Bio-Rad, Cat. No. 4561096) and transferred to an Immobilon-P membrane (Millipore, Cat. No. IPVH00010) using a Power Blotter XL System (Invitrogen).
Membranes were blocked in 5% milk PBS-T for 1 h at room temperature and incubated overnight at 4°C with primary antibodies against SMC3 (Bethyl Laboratories A300-060A), SMC1 (Bethyl Laboratories A300-055A), Rad21 (Bethyl Laboratories A300-080A), CTCF (Active Motif 61311), PARP1 N-terminal (Active Motif 39559) and Histone H3 (Abcam ab1791) as per manufacturer recommendation. Membranes were washed 5 mins three times, incubated for 1 h with the appropriate secondary antibody, either goat anti-rabbit IgG-HRP (Jackson ImmunoResearch) or rabbit anti-mouse IgG-HRP (Jackson ImmunoResearch), at a dilution of 1:10 000. Membranes were then washed and detected by enhanced chemiluminescence (SuperSignal West Dura Extended Duration Substrate).
Poly (ADP-ribose) ELISA assay
The ELISA assay to assess the levels of intracellular Poly (ADP-Ribose) (PAR) was carried out using 7 × 106 cells per sample using the poly(ADP-ribose) ELISA kit (Cell Biolabs, Cat. No. XDN-5114) as per manufacturer protocol.
RNA extraction and RNA-seq
Total RNA from SNU719 cell line was isolated from 2 × 106 cells using a Direct-zol RNA Kit (Zymo Research, Cat. No. R2050) according to the manufacturer's protocol. RNA samples were either used for downstream RT-qPCR or submitted to the Wistar Institute genomics core facility for RNA quality control and sequencing library preparation using the SENSE mRNA-Seq Library Prep Kit V2 (Lexogen) to generate Illumina-compatible sequencing libraries according to the manufacturer's instructions. Paired-end reads of 75 bp were obtained using a Illumina HiSeq 2500 sequencer. RNA-seq data was aligned using Bowtie2 (23) against hg19 version of the Human genome and all unaligned reads were then aligned against NC_007605.1 version of EBV genome and RSEM v1.2.12 software (24) was used to estimate raw read counts and RPKM for Human and EBV genes.
Datasets are available in Gene Expression Omnibus see Data Availability section for accession number.
RT-qPCR
For quantitative reverse transcription-PCR (RT-qPCR), SuperScript IV reverse transcriptase (Invitrogen, Cat. No. 18090200) was used to generate randomly primed cDNA from 1 μg of total RNA. 50 ng of cDNA sample was analyzed in triplicate by quantitative PCR using the ABI StepOnePlus system. Data were analyzed by the ΔCT method relative to 18S-ribosomal subunit control. Primer sequences are available in the Supplementary file 1.
Chromatin immunoprecipitation assays
Chromatin immunoprecipitation sequencing (ChIP-seq)
Chromatin immunoprecipitation with next-generation sequencing (ChIP-seq) was performed as previously described(3). Briefly, 25 × 106 cells per immunoprecipitation were collected and fixed with 1% formaldehyde for 15 min and then quenched with 0.25 M glycine for 5 min on ice. After 3 washes with 1× PBS, pellets were resuspended in 10 ml each of a series of two lysis buffers and resuspended in 1ml of the third lysis buffer before fragmentation in Covaris ME220 Ultrasonicator (peak power 75, duty factor 25, cycles/burst 1000, average power 18.8, time 720 s) to generate chromatin fragments roughly 200–500 bp in size. Chromatin was centrifuged to clear debris and a 1:20 of this cleared chromatin was kept as standard input for comparison against immunoprecipitations. Chromatin was incubated rotating at 4° 1 h with 25 μg H3K4me1 (Active Motif 39299) and 25 μg H3K27ac (Active Motif 39133), then chromatin–antibody complexes were precipitated using 50 μl of Dynabeads Protein A (ThermoFisher, product no. 10001D) incubated rotating at 4° overnight. DNA was purified using Promega Wizard SV Gel and PCR Clean-up Kit (product no. A9285). Libraries for sequencing were made using NEBNext Ultra II DNA Library Prep Kit (New England Biolabs, product no. E7103) and sequenced on the Illumina HiSeq 2500.
ChIP-seq analysis
Reads were mapped against the human gammaherpesvirus 4 (HHV4) NC_007605.1 genome assembly using Burrows-Wheeler Aligner (BWA) (25). We used MACS2 (26,27) software packages to call peaks using input samples as control. deepTools (28) was used for data visualization.
For transcription factor binding motif analysis, Analysis of Motif Enrichment (AME) from the MEME-ChIP suite (29) was used using default options.
ChIP-seq data were deposited for public access at Gene Expression Omnibus see Data Availability section for accession number.
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation was performed as previously described (3) with minor changes. Briefly, after 72 h incubation with or without Olaparib, 1 × 106 cells per immunoprecipitation were collected and fixed with 1% formaldehyde for 15 min and then quenched with 0.125 M glycine for 5 min on ice. After centrifugation, the pellet was resuspended in 120 μl lysis buffer and sonicated using Covaris ME220 Ultrasonicator (peak power 75, duty factor 25, cycles/burst 1000, average power 18.8, time 600 s) to generate chromatin fragments roughly 100–200 bp in size. A sample of ‘input chromatin’ was collected at this point as a standard for comparison against immunoprecipitations (5% of total material). Chromatin was then incubated overnight rotating at 4° with 4 μg of antibody against CTCF, 5 μg of antibody against SMC1 and 5 μg of IgG antibody.
Real-time PCR was performed with a master mix containing 1X Maxima SYBR Green (Thermoscientific, REF No. K0223), 0.25 μM primers and 1:50 of ChIP or input DNA per well. Quantitative PCRs were carried out in triplicate using the ABI StepOnePlus PCR system. Data were analyzed by the ΔΔCT method (where CT is threshold cycle) relative to DNA input. Primer sequences are available in the Supplementary file 1.
Chromatin conformation capture assays
HiC assay
Hi-C assay was performed as previously described (3). Briefly, 5 × 106 cells per condition were collected for in-situ Hi-C. Libraries of total ligation products were produced using Ultralow Library Systems V2 (Tecan Genomics, part no. 0344NB-32) as per manufacturer's protocol.
Purified libraries were then enriched for only EBV genome ligation products using myBaits enrichment kit as per manufacturer's protocol. Libraries were sequenced using the Illumina HiSeq 2500 sequencing platform with paired-end 75bp read length. HiC data was preprocessed using HiC-Pro v2.10.0 pipeline (30) with default settings using NC_007605.1 version of the EBV genome at 1kb resolution. DESeq2 (31) was used to estimate significance of differential contact based on raw count matrix files. Significantly changed associations (FDR < 5%) were plotted as circos graph using the circlize package (version 0.4.12) of R (version 4.0.5) (32).
To identify cell-type specific loops we summed all the interactions that passed the FDR threshold based on cell type and further filtered them by CTCF binding. Then a differential analysis was performed using DESeq2 R package as described above.
The detailed protocol with all minor alterations will be happily supplied by corresponding author per request. Datasets are available in GEO see Data Availability section for the accession number.
HiChIP assay
HiChIP assay was performed as per protocol (33) with minor changes.
Briefly, 25 × 106 cells were fixed with 1% formaldehyde for 10 min and then quenched with 0.125 M glycine for 5 min at RT. Ligation products were immunoprecipitated with 25 μg H3K27ac (Active Motif 39133). 5 μl of Streptavidin C-1 beads were used for biotin pull-down. After the last wash with Tween Wash Buffer bead-bound DNA was used to prepare libraries with Ultralow Library Systems V2 (Tecan Genomics, part no. 0344NB-32) as per manufacturer's protocol.
Libraries were then deep sequenced using the Illumina HiSeq 2500 sequencing platform.
Data was preprocessed using HiC-Pro v2.10.0 pipeline (30) with default settings using NC_007605.1 version of the EBV genome at 5kb resolution and hichipper software (34) was used to perform restriction site bias modeling and interaction identification.
Only statistically significant interactions (FDR < 5%) were kept for downstream analysis.
Differential interactions between cell lines were identified using the diffloop Bioconductor R package (35). Significantly changed associations (FDR < 1%) were used for visualization in UCSC Genome Browser.
The detailed protocol with all minor alterations will be happily supplied by corresponding author per request.
To identify EBV–human interactions, the output of hichipper software was filtered for those interactions having more than 2 reads, than they were plotted using the circlize R package (32). H3K27ac peaks in proximity to EBV–human interactions where annotated using the HOMER software (36) and filtered for just those genes which Transcription Start Sites (TSS) were within 1 kb from the peak.
Transcription factor binding motif analysis was carried out using Analysis of Motif Enrichment (AME) from the MEME-ChIP suite (29) with default options.
Droplet digital PCR (ddPCR)
Multiplex DNA droplet PCR (ddPCR) was performed as previously described (37). DNA was isolated from 1 × 106 cells using the GeneJET Genomic DNA Purification Kit (Thermo Scientific) according to the manufacturer's protocol. 500 ng of DNA was digested with BamHI enzyme (10 U/μl, New England Biolabs) in a total volume of 10 μl for 1 h at 37°C. Digestion was diluted 1:20 in nuclease-free water. 10 μl of diluted DNA digest was mixed with 12.5 μl of 2× digital PCR supermix for probes (No dUTP) (Bio-Rad), 1.25 μl 20x FAM primers, 1.25 μl VIC primers for each reaction. The FAM primers sequence for EBV Lmp1 was Fw(5′-3′) AAGGTCAAAGAACAAGGCCAAG, Rv (5′-3′) GCATCGGAGTCGGTGG and FAM- AGCGTGTCCCCGTGGAGG.
Host control primer sequence for Ribonuclease P protein subunit 30 (Rpp30) was Fw (5′-3′) GATTTGGACCTGCGAGCG, Rv (5′-3′) GCGGCTGTCTCCACAAGT, and probe VIC-CTGACCTGAAGGCTCT. 20x primers contain 18 μM PCR primers and 5 μM probes for a final PCR reaction concentration of 900 nM PCR primers and 250 nM probe. Each sample was run in duplicate. The ddPCR plate was sealed with a foil heat seal using the PX1 PCR Plate Sealer (Bio-Rad) at 180°C for 5 s. The plate was vortexed and spun down at 1000 rpm for 1 min. Droplets were generated using the QX200 Droplet Digital PCR System (Bio-Rad) and transfer of emulsified samples to a PCR plate was performed according to manufacturer's instructions. PCR plate containing emulsified droplets was sealed with a foil heat seal. PCR reactions were performed on the C1000 Touch Thermal Cycler (Bio-Rad). The cycling protocol included an enzyme activation step at 95°C for 10 min and cycled 40 times between a denaturing step at 94°C for 30 s and an annealing and extension step at 60°C for 1 min, finally one enzyme deactivation step was performed at 98°C for 10. The ramp rate between these steps set at 2°C/s. Droplets were then counted using QX200 Droplet Reader (Bio-Rad). The absolute quantity of DNA per sample was determined using the QuantaSoft software.
Statistical analysis
All experiments presented were conducted at least in triplicate to ensure reproducibility of results. The Prism statistical software package (GraphPad) was used to identify statistically significant differences between experimental conditions and control samples, using Student's t test as indicated in the figure legends.
Results
The three-dimensional structure of the viral genome differs in all latency types and reflects the viral gene expression state
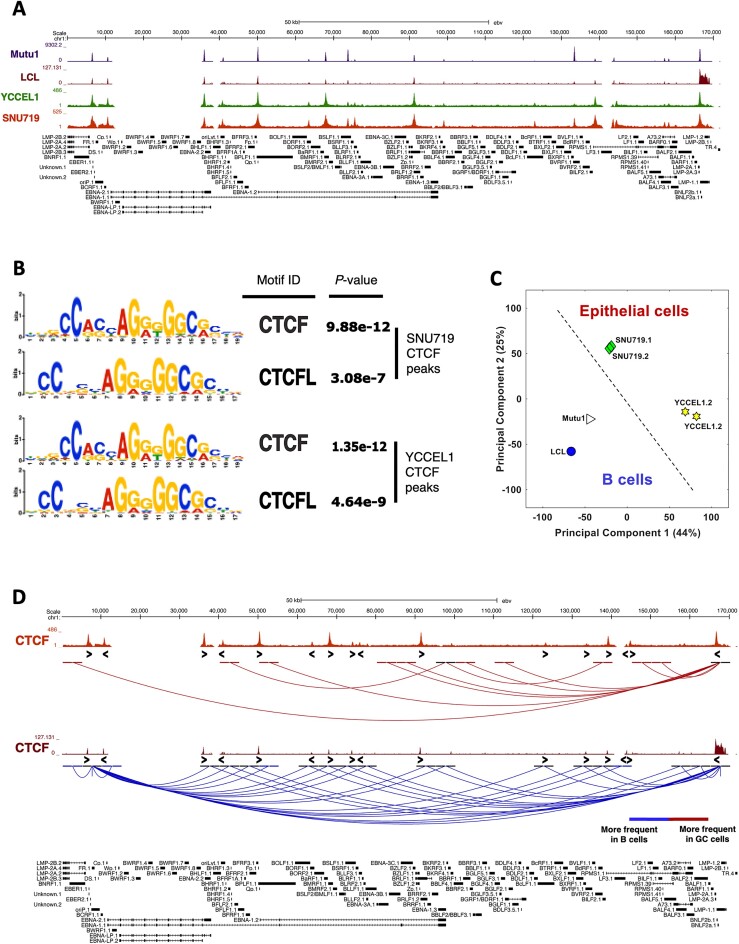
Recent works from our group and others have demonstrated that, following the establishment of latency in infected B cells, the transcription factor CTCF binds to the EBV genome, regulating its gene expression (5,7). Furthermore, in a recent study we identified that, despite the almost unchanged binding of CTCF between Type I and Type III latencies, the three-dimensional (3D) organization of the viral genome is markedly different between the two latency types and is tightly correlated with viral gene expression (3). To evaluate how the binding of CTCF differs between latencies I, III and II, which is typical of EBV-positive gastric tumors, first we conducted a Chromatin Immunoprecipitation followed by sequencing (ChIP-seq) experiment in the two EBV + gastric cancer (GC) cell lines YCCEL1 and SNU719, that both harbor a Type II EBV latency, and next we compared it with our previously published CTCF ChIP-seq data in EBV-positive B cells. For comparison purposes, the sequencing reads of this experiment and the previous ones were aligned to the NC_007605.1 version of the EBV genome. From the comparison of CTCF tracks (Figure 1A), we observed minimal differences with respect to CTCF occupancy between latency types, except for the peaks at ∼75 and ∼122 kb that are predominantly present in gastric cancer and the peak at ∼132 kb which is extremely variable between latencies. To further confirm that the peaks observed in the ChIP-seq experiment were specific of CTCF, we conducted a binding motif analysis in both cell lines, which revealed that the recognized motifs correspond to CTCF and CTCFL (Figure 1B).
Figure 1.
The three-dimensional structure of the viral genome differs in all latency types and reflects the viral gene expression state. (A) CTCF ChIP-seq profiles in latency Type I (Mutu, purple), III (LCL, brown) and II (YCCEL1, green and SNU719, dark red), normalized to input DNA. (B) Motif analysis for the CTCF ChIP-seq peaks identified in the latency Type II cell lines. (C) Principal Component Analysis (PCA) based on the 3D structure of the viral genome. (D) UCSC Genome Browser linearized visualization of the unique interactions found in HiC experiment in gastric cancer cell lines (dark red) and in B cells (blue) (FDR < 5%). CTCF motif directionality is represented by arrowheads under CTCF ChIP-seq peaks.
Considering the established role of CTCF in determining, along with the Cohesin complex, the formation of chromatin loops across the viral genome in B cells, we conducted a HiC experiment with the addition of an enrichment step for EBV-specific sequences, and identified the interactions present in the EBV genome in epithelial cells, and then compared them with those detected in EBV + B cells.
Based on the Principal Component Analysis (PCA) of the 3D architecture of the viral genome (Figure 1C), EBV-infected cells were separated according to their tissue of origin. Both B cells and epithelial cells clustered separately based on the two main components, 1 and 2. Moreover, our analysis reveals that the episomes in the YCCEL1 cell line significantly differ from those in B cells, while the episomes in SNU719 show a conformation of the EBV episome more closely resembling B-cell specific latencies. Next, the overall interactions were filtered based on CTCF binding and compared between B cells and GC cells. As shown in Figure 1D, there are some interactions that occur more frequently in GC cells (in dark red in Figure 1D) that originate from the CTCF binding site on the LMP promoter (∼166 kb) and terminate at the RPMS1 promoter region (∼140 kb) or the Qp promoter (∼50 kb), which is active in type II latency and transcribes for EBNA1 gene. We observed that the number of interactions specific to B cells is higher compared to that of GC cells (in blue in Figure 1D). These specific interactions originate from three strong CTCF binding sites, namely the LMP promoter, the origin of replication OriP (∼6–8 kb), and the Cp promoter (∼5–10 kb). Interestingly, the Cp promoter is active in B cells adopting the type III latency program and initiates the transcription for all the EBNAs.
These findings indicate that although CTCF binds similarly to the viral genome across the three latency types, each latency type exhibit distinct 3D architectures of the EBV episomes that are also specific to the infected cell type.
PARP1 inhibition alters CTCF/Cohesin binding and viral chromatin looping
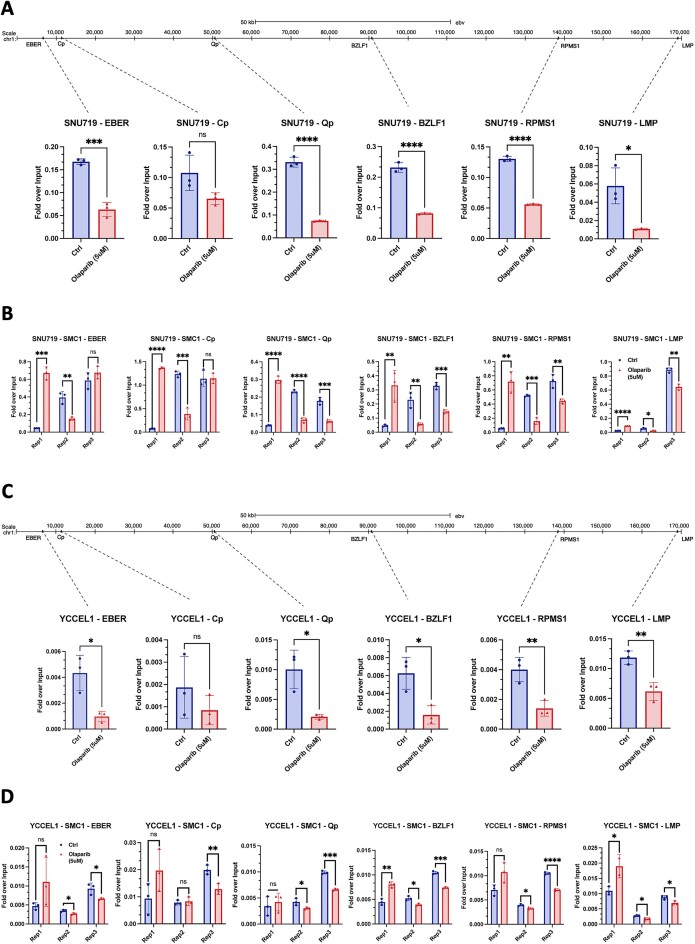
As previously demonstrated by our group (3,10,11), it has been shown that the enzymatic activity of PARP1 plays a crucial role in stabilizing the binding of CTCF to the viral genome. This mechanism is essential for maintaining an active latency program in the infected cells. Additionally, PARP1 inhibition affects the binding of the Cohesin complex, consequently altering the three-dimensional organization of the viral genome (3). To investigate whether the central role of PARP1 in latency maintenance is equally important in Type II latency, we evaluated the CTCF binding after inhibiting the enzymatic activity of PARP1 with a potent and specific FDA approved inhibitor, Olaparib (38). Following a 72-h inhibition period, we assessed the decrease in PARylation levels using Dot Blot (Supplemental Figure 1B) and conducted Chromatin Immunoprecipitation (ChIP) coupled with quantitative real-time PCR (qPCR) using antibodies against CTCF and the Cohesin subunit SMC1. In Type II EBV + GC cells, our observations revealed a general trend of reduced CTCF binding, indicating that the absence of PARylation by PARP1 leads to the destabilization of binding at the viral genome level (Figure 2A and C). This is consistent with our previous report that PARP1 activity is necessary to stabilize CTCF binding in B cells (11). However, only certain regions demonstrate a significant reduction in CTCF binding, such as the EBER promoter, the Qp promoter, the RPMS1 gene promoter, the lytic Zp promoter, and the LMP genes promoter. Interestingly, at the Cp promoter, which is silenced in latency type II cells, CTCF binding is unaffected by PARP1 inhibition (Figure 2A and C), in contrast to what we observed in type III latency (11). These results suggest that although PARP1 activity generally regulates CTCF binding across the entire EBV genome, the specific EBV regions that experience a loss of CTCF binding after PARP inhibition are dependent on the cell type and latency type. In other words, the impact of PARP inhibition on CTCF binding is context-specific and varies based on the characteristics of the infected cell and the type of latency exhibited by the EBV.
Figure 2.
PARP1 inhibition alters CTCF/Cohesin binding. (A) CTCF ChIP-qPCR in SNU719 cell line following PARP1 inhibition with 5uM Olaparib treatment for the main CTCF binding sites. (B) ChIP-qPCR in SNU719 cell line following PARP1 inhibition for SMC1 Cohesin subunit in all three replicates for the same CTCF binding sites. (C) CTCF ChIP-qPCR in YCCEL1 cell line following PARP1 inhibition with 5uM Olaparib treatment for the main CTCF binding sites. (D) ChIP-qPCR in YCCEL1 cell line following PARP1 inhibition for SMC1 Cohesin subunit in all three replicates for the same CTCF binding sites. Data are presented as %input. N = 3, Mean ± SD. The t test P values for the Olaparib/Ctrl comparison are indicated as asterisks (**** P ≤ 0.0001, *** P ≤ 0.001, ** P ≤ 0.01, * P ≤ 0.05).
Due to the close correlation observed between CTCF and Cohesin binding across the EBV genome in B cells, we assessed further the SMC1 binding at the same regions analyzed for CTCF. Surprisingly, in contrast to what observed in Type III B cells, Cohesin subunit binding shows random alterations at all the analyzed regions (Figure 2B and D) when PARP1 is inhibited. This suggests a potential destabilization of Cohesin binding induced by the inhibition of PARP1 activity.
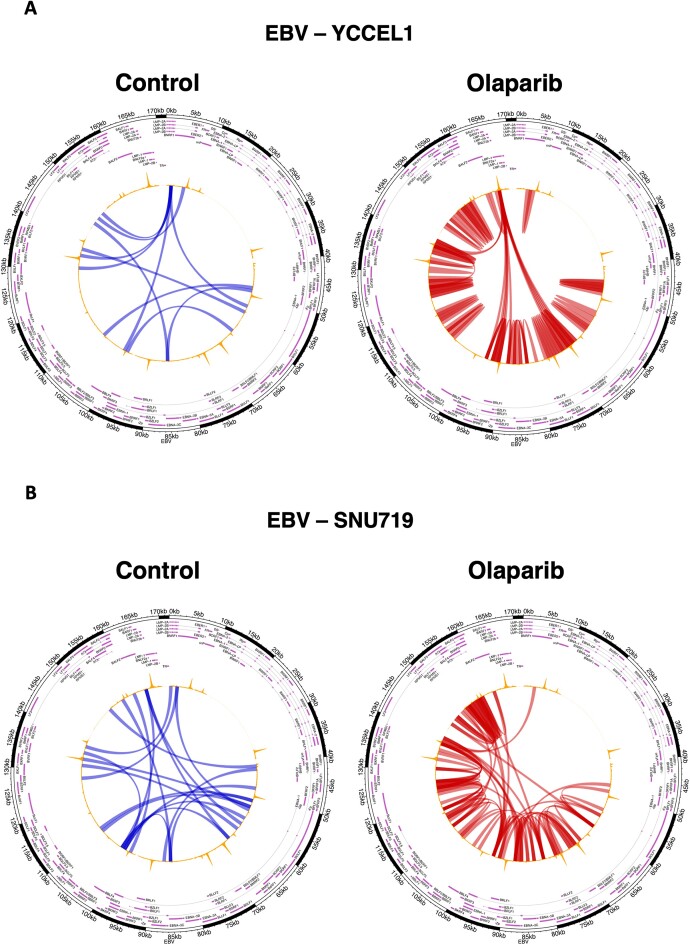
Considering the observed differences in CTCF and SMC1 binding across the EBV genome after PARP inhibition, and their role in promoting long-range chromatin loops (39,40), we next determined whether there were changes in the viral genome architecture. To do this, we performed a Hi-C experiment in both cell lines following a 72-h treatment with Olaparib. Similar to the observed alterations in B cells, EBV + GC cells also undergo changes in the 3D structure of the EBV episomes upon PARP1 inhibition. These changes include a reduction in the length of interactions, indicating that loops occur more frequently between adjacent regions. However, certain long-range interactions originating from the LMP1 gene promoter region remain unaffected and unchanged (Figure 3). Notably, the YCCEL1 cell line is more profoundly affected by PARP1 inhibition, displaying a significant decrease in the length of interactions and a shift in the origin of long-range loops, that now originate from the CTCF peak at the LMP gene promoter instead of the LMP2A/B gene region (Figure 3A).
Figure 3.
Viral chromatin looping is altered by PARP1 inhibition. (A) Circular visualization of the interactions derived from HiC matrices in YCCEL1 cell line. The arches represent the DNA-DNA interactions at 1 kb scale. The blue arches represent the interactions found more frequently in the control samples, while the red ones represent those found more frequently in the Olaparib treated samples (FDR < 5%). (B) Circular visualization (as described in A) of the interactions derived from HiC matrices in SNU719 cell line. In all plots CTCF ChIP-seq track is represented in yellow on top of the arches.
Overall, the results indicate that the inhibition of PARP1 has a significant impact on CTCF and Cohesin binding not only in B cells but also in EBV + gastric cancer cells. This disruption in the binding of these architectural proteins leads to alterations in the 3D structure of the viral genome. The changes observed in the interactions and loops within the viral episomes are likely to play a crucial role in regulating viral gene expression and may have implications for the development and progression of gastric cancer associated with EBV infection. These findings highlight the importance of PARP1 activity in maintaining the proper architecture and function of the EBV genome in different cell types and latency types.
PARPi affects viral gene expression
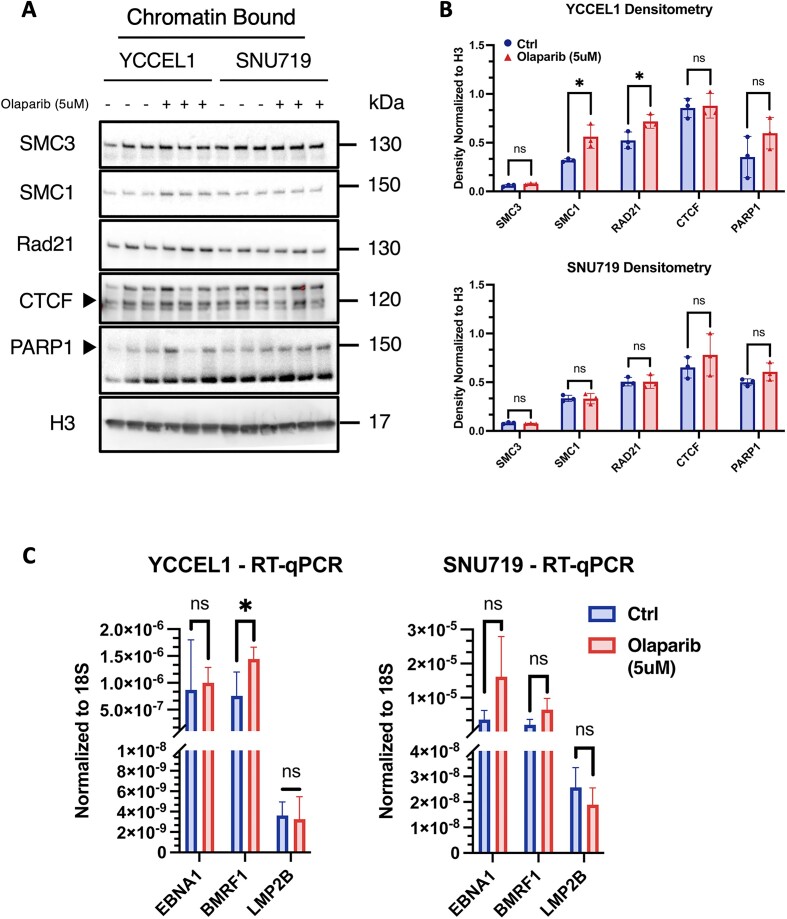
To determine whether the variations in CTCF and Cohesin binding observed after PARP1 inhibition were generalized throughout the host genome or specific to EBV episome, we performed subcellular fractionation and analyzed by western blot Cohesin subunits, CTCF and PARP1 for their presence in the chromatin-associated protein fraction (Figure 4A) in both cell lines. We observed that after PARP1 inhibition the YCCEL1 cells exhibit a noticeable increase in the chromatin-associated fraction of Cohesin subunits SMC3, SMC1A, and RAD21 (Figure 4A), with statistical significance observed for SMC1A and RAD21 (Figure 4B). However, the fractions of CTCF and PARP1 in YCCEL1 cells remain unchanged. Conversely, the SNU719 cell line displays consistent levels of chromatin-bound proteins without any notable variation (Figures 4A and B). To determine whether the differences observed in subcellular fractionation were linked to a widespread increase in the expression of genes encoding Cohesin subunits, we performed Reverse Transcription quantitative Real-Time PCR (RT-qPCR). However, in both cell lines, we did not find any significant increase in the expression of Cohesin subunits and CTCF, as illustrated in Supplemental Figure 1A.
Figure 4.
PARP1 inhibition alters viral gene expression. (A) Western Blot of chromatin-bound fraction of Cohesin subunits, CTCF and PARP1 proteins extracted from YCCEL1 and SNU719 cell lines following PARP1 inhibition (N = 3). Molecular weights are indicated on the side. (B) Densitometry analysis of the western blot described in (A). Data are normalized on the H3 histone density. The t test P values for the Olaparib/Ctrl comparison are indicated as asterisks (*P ≤ 0.05). (C) RT-qPCR of EBNA1, BMRF1 and LMP2B viral genes following Olaparib treatment. Bar graph represents the average expression of three biological replicates per treatment, each normalized to 18S expression, respectively (N = 3, mean ± SD). Paired Student's t test assuming equal variance (two-tailed) was used to compare the experiments (* P ≤ 0.05).
We recently have shown that alterations in CTCF and Cohesin binding, along with changes in the 3D architecture of the EBV genome, have significant effects on viral gene expression in B cells (3). These observations suggest a potential link between chromatin organization and viral gene regulation in infected cells. To gain further insights into the connection between chromatin organization and the regulation of viral genes within epithelial infected cells, we conducted RT-qPCR analysis focusing on specific latent genes expressed in Type II latency, including EBNA1 and LMP2, along with the viral DNA polymerase processivity factor BMRF1. By examining the expression levels of these genes, we aimed to gain a comprehensive understanding of the impact of chromatin organization on the transcriptional regulation of key EBV genes during Type II latency. Our analysis revealed intriguing findings regarding the YCCEL1 and SNU719 cell lines in response to Olaparib treatment. In the YCCEL1 cell line, we observed a significant increase in the expression of the BMRF1 gene, while the expression of the two analyzed latent genes, EBNA1 and LMP2, remained unchanged. In the SNU719 cell line, there was a non-significant change in the expression of the three genes analyzed. Moreover, the small differences observed in gene expression could be attributable to different activity of PARP1 between latency types (Supplementary Figure 3A). Remarkably, despite the increased expression of BMRF1 in YCCEL1 cell line, the overall copy number of EBV within the cells treated with Olaparib remained unaltered, as shown in Supplemental Figure 1D. This suggests that Olaparib treatment led to the destabilization of the viral genome, leading to unrestricted expression of viral latent and lytic genes without causing an increase in the viral copy number. These findings provide valuable insights into the mechanisms underlying Olaparib impact on EBV-infected cells, shedding light on the complex interplay between chromatin organization, viral gene expression, and viral genome stability.
CTCF occupies viral regions with an enhancer epigenetic signature
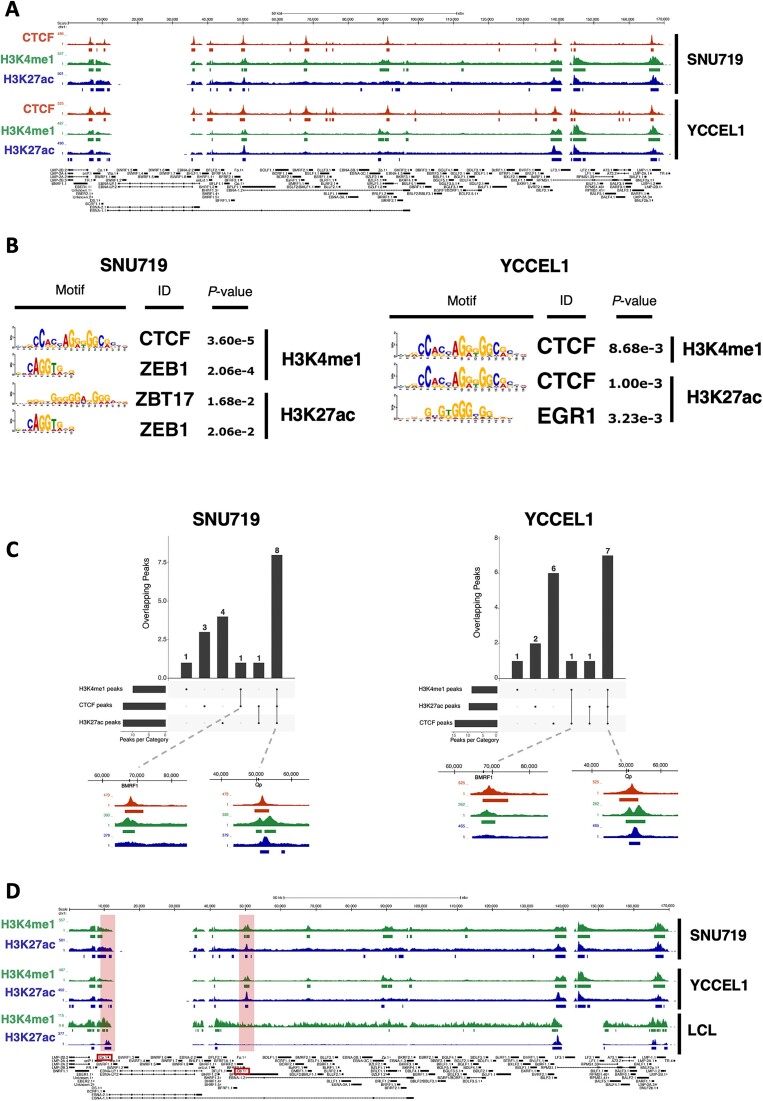
In our recent study, we have delved into the three-dimensional structure of the viral genome in two different latency types of EBV and presented experimental evidence that underscores the strong correlation between the architecture of episomes and viral gene expression (3). Moreover, recent research (41), has indicated the presence of viral genomic regions enriched with histone marks typical of enhancers, such as H3K4me1 and H3K27ac. To further understand the functional implications of the observed genomic architecture of EBV in these two cell lines, we conducted a ChIP-seq (Chromatin Immunoprecipitation followed by sequencing) experiment using antibodies targeting H3K4me1 and H3K27ac. These histone marks are known to respectively indicate poised and active enhancers. Our analysis revealed a notable enrichment of these two histone marks at specific regions of the viral genome, particularly at the origins of replication OriP and OriLyt, as well as the promoters Qp, Zp, BILF2 and LMP genes (Figure 5A).
Figure 5.
CTCF co-localizes with enhancer histone marks. (A) UCSC Genome Browser tracks for CTCF (dark red), H3K4me1 (green), H3K27ac (blue) on the EBV genome in both SNU719 (top) and YCCEL1 (bottom) cell lines. (B) Transcription factor motif analysis for H3K4me1 (top) and H3K27ac (bottom) peaks in SNU719 (left) and YCCEL1 (right) cell lines. (C) UpSet plot of the overlap between CTCF and H3K4me1 and H3K27ac histone marks. On the bottom, magnified ChIP-seq tracks for CTCF and both histone marks are shown. (D) H3K4me1 and H3K27ac ChIP-seq tracks on the EBV genome in SNU719 (top), YCCEL1 (middle) and LCL (bottom) cell lines.
These findings provide compelling evidence that the genomic organization of EBV in these cell lines indeed serves a functional purpose, specifically in regulating viral gene expression through enhancer activity. Based on the location of the peaks for both histone modifications, we conducted a transcription factor binding motif analysis (Figure 5B). In both GC cell lines, the peaks of H3K4me1 were consistently associated with CTCF binding motifs. This suggests that CTCF may play a significant role in the regulatory activity of these enhancers in both cell lines. However, the peaks of H3K27ac exhibited greater variability between the two cell lines. In the SNU719 cell line, the most frequent binding motifs were identified as ZBT17 and ZEB1. This indicates that these transcription factors might be crucial for the enhancer activity and gene regulation specific to this cell line. In contrast, the YCCEL1 cell line showed predominant binding motifs for CTCF and EGR1 at the peaks of H3K27ac. This suggests a different transcription factor landscape in this cell line, possibly contributing to distinct enhancer activities and gene expression patterns. These findings indicate that the regulatory landscape of enhancers, marked by H3K4me1 and H3K27ac, varies between the two cell lines, and specific transcription factors may be pivotal in mediating the enhancer activity and subsequent gene expression profiles.
Next, we conducted an overlap analysis between CTCF peaks and the peaks associated with the enhancer chromatin signature. Analysis of CTCF, H3K4me1, and H3K27ac peaks (Figure 5C) revealed substantial overlap, with 8 peaks in SNU719 and 7 in YCCEL1, while the number of unique CTCF peaks was notably higher in YCCEL1, being twice that of SNU719. Notably, a shared region at the BMRF1 promoter in both cell lines exhibited only CTCF and H3K4me1, indicating a poised enhancer region. Comparing H3K4me1 and H3K27ac peaks between LCL and GC cell lines (Figure 5D), both histone marks showed fewer peaks in LCL compared to GC cells. These modifications were prominently enriched at the Cp promoter but completely absent at the Qp promoter, aligning with the distinct activities of these promoters in different latency types. These findings suggest functional relevance of the 3D structure of the EBV genome, bringing active enhancers into close proximity. CTCF, along with other transcription factors, likely plays a crucial role in mediating these processes across the EBV genome. This highlights the importance of chromatin organization and histone modifications in controlling EBV gene expression and may offer valuable insights into the mechanisms underlying EBV-associated diseases.
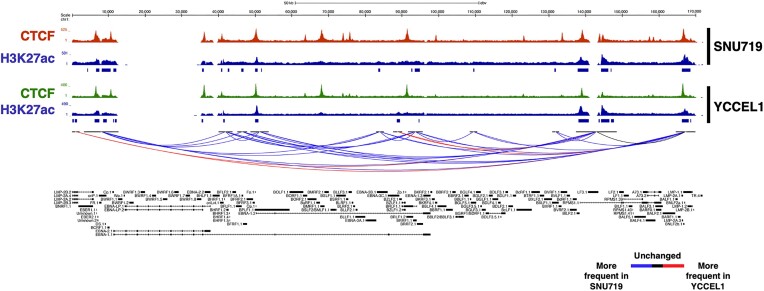
HiChIP analysis reveals distinct three-dimensional structures and enhancer interactions in SNU719 and YCCEL1 cell lines
To further strengthen the correlation between the three-dimensional structure and regions characterized by enhancer marks across the EBV genome, we conducted a HiChIP experiment for the H3K27ac histone modification. The Principal Component Analysis (Supplemental Figure 1C), indicates distinct clustering of the two cell lines, suggesting that the interactions between regions enriched for the H3K27ac histone mark vary between them. Building on this observation, we conducted a differential analysis of interactions occurring between different regions of the viral genome to explore how H3K27ac-enriched chromatin loop vary between the two cell lines. We observed that the SNU719 cell line exhibits a higher number of interactions that occur more frequently compared to the YCCEL1 cell line (Figure 6A). Conversely, the YCCEL1 cell line presents only three frequent interactions, specifically between the promoter regions of the LMP2A/B genes and the promoter of LMP1, as well as interactions involving the BILF2 region, and between the promoters of LMP1 and Zp (Figure 6A). However, both cell lines share a common interaction between the LMP promoter and a region enriched with H3K27ac, located approximately 20 kb upstream the promoter (Figure 6A). This interaction appears to be conserved between the two cell lines despite their distinct characteristics, further supporting the significance of this interaction in the regulation of viral gene expression.
Figure 6.
HiChIP analysis reveals distinct three-dimensional structures and enhancer interactions in SNU719 and YCCEL1 cell lines. UCSC Genome Browser tracks of CTCF (dark red and green) and H3K27ac (blue) on the EBV genome in SNU719 (top) and YCCEL1 (bottom) cell lines. On the bottom of the image are show the unique H3K27ac-rich interactions (FDR < 1%) in both cell lines (blue = more frequent in SNU719, red = more frequent in YCCEL1, black = no difference in frequency between cell lines).
EBV enhancer regions are tethered to specific loci across the host genome
In previous studies utilizing HiC or 4C assays, it was revealed that the EBV genome is connected to the host genome, suggesting a potential role in regulating host gene expression in infected cells (22,42). To further investigate this phenomenon, we delved into the possibility of specific physical connections between the identified viral enhancer regions and particular loci within the host genome. To achieve this, we analyzed H3K27ac HiChIP datasets for chromatin loops formed between viral and host genomic regions.
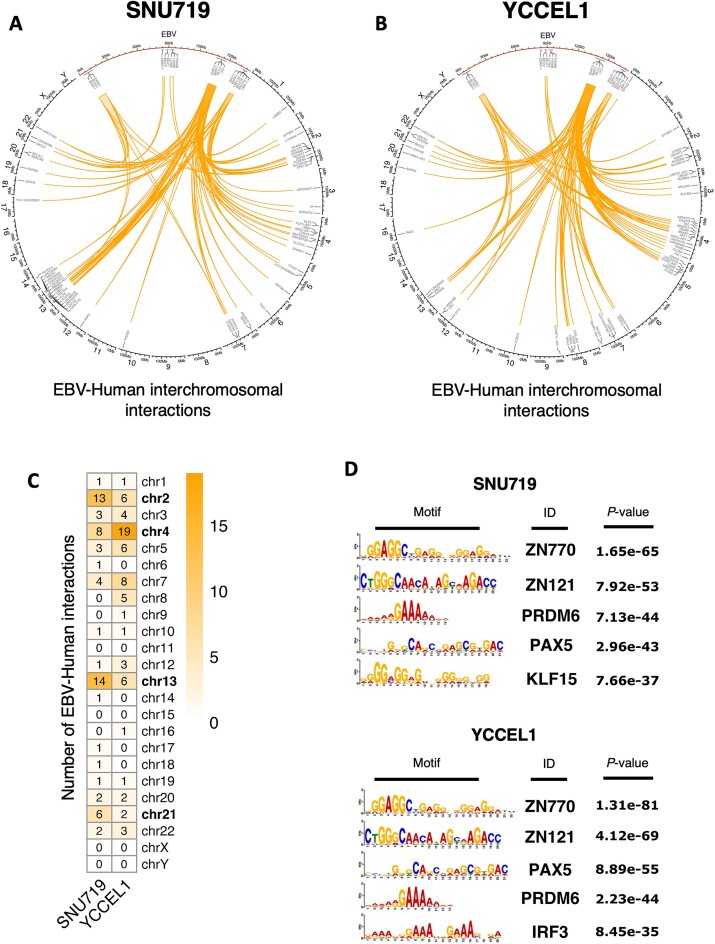
Through the HiChIP experiment, we were able to detect around 70 interactions taking place between the EBV genome and the human genome across both cell lines. To focus specifically on robust EBV-human interactions, we considered loops with more than 3 reads and visualized them in circos plots. In these plots, the EBV genome was represented in red and positioned at the top (see Figure 7A, B). This visualization allowed us to highlight the significant interactions between EBV and the human genome for further analysis and interpretation.
Figure 7.
EBV enhancer regions are tethered to specific loci across the host genome. (A) Circos plot of EBV–human interchromosomal interactions for SNU719 cell line. EBV is represented in red and enlarged on the top section of the plot. EBV–human interactions are represented as orange arches. Some of the genes near the interaction points are annotated. (B) Circos plot of EBV–human interchromosomal interactions for YCCEL1 cell line (as described in (A). (C) Heatmap showing the number of interactions occurring between EBV and the Human chromosomes in both cell lines. (D) Transcription factor motif analysis of EBV–human interaction regions on the Human genome for SNU719 (top) and YCCEL1 (bottom) cell lines.
An important observation we made was that, in contrast to recent studies conducted by Japanese researchers (22), most of the interactions we identified did not originate from the OriP region of the EBV genome (Supplemental Figure 2B). Instead, they predominantly arose from the BILF2 and LMP regions. This finding suggests that these two regions likely act as strong enhancers, a characteristic that had been previously highlighted by other research groups, particularly concerning Type II latency in B cells (41).
Furthermore, despite the involvement of the same regions of the EBV genome in the chromatin loops, the corresponding regions of the human genome differed between the two GC cell lines (SNU719 and YCCEL1). Notably, in the SNU719 cell line, chromosomes 2, 4, 13 and 21 exhibited the highest number of interactions with the EBV genome. In contrast, the YCCEL1 cell line displayed the highest number of interactions with chromosomes 2, 4, 5, 7 and 13 (Figure 7C). Moreover, there are differences in the number of interactions originating from the three EBV enhancer regions. By comparing the loops occurring between EBV and the Human chromosomes 2 and 13, which are common between the two cell lines, we observed that the SNU719 cell line exhibits more frequent interactions originating from BILF2 and LMP enhancer regions respect to the YCCEL1 cell line. On the other end YCCEL1 cell line shows more frequent interactions between BILF2, LMP and oriP and chromosome 4 (Supplemental Figure 2A).
Furthermore, we analyzed the transcription factor binding motifs present at this EBV docking sites (Figure 7D). Interestingly, the top 5 motifs identified were similar between the two cell lines, suggesting a possible key role for these transcription factor in the maintenance of these EBV–human interactions.
These distinctions in the human genome regions involved in interactions suggest cell line-specific differences in the regulatory mechanisms and potential implications for EBV-host interactions in the context of these two cell lines.
Indeed, the data obtained from the HiChIP experiment strongly indicate that the two cell lines (SNU719 and YCCEL1) not only exhibit differences in the 3D structure of the EBV genome but also in the specific interactions that take place between the EBV episomes and the Human genome. By forming such interactions, EBV enhancers can exert their regulatory effects on host genes, modulating their expression and influencing various cellular processes.
Functional role of EBV–human interactions: gene expression analysis reveals strong viral enhancers associated with gastric cancers
To determine if the interactions between EBV enhancer and the Human genome had functional significance, we evaluated the expression of cellular genes in proximity to the viral interaction sites. To achieve this, we focused on H3K27ac-mediated loops which were filtered based on the approximately 70 viral-human looping sites previously identified, and examined only the genes whose Transcription Start Site (TSS) fell within a 1 kb region from the H3K27ac peak considered. This approach allowed us to pinpoint genes that were in close proximity to the regions of active chromatin marked by H3K27ac and that might potentially be influenced by the EBV enhancers.
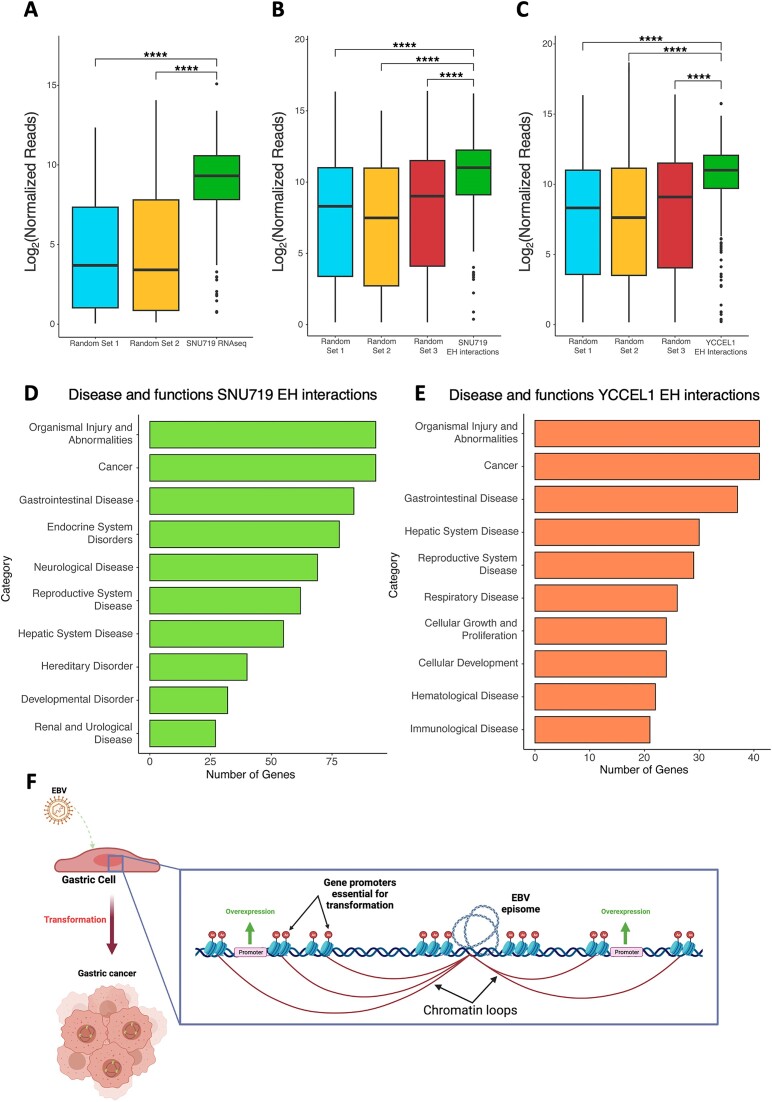
In the YCCEL1 cell line, approximately 300 genes were identified near the EBV-human interaction sites, while in SNU719, around 200 genes were identified (as shown in Supplemental Figure 2C). Interestingly, only 53 of these genes were found to be common between the two cell lines, indicating a considerable degree of cell line-specificity in the genes influenced by the EBV enhancers. To further investigate the functional impact of these interactions, we analyzed the expression levels of these identified genes using RNA sequencing (RNAseq) data obtained from the SNU719 cell line (as depicted in Figure 8A). Comparing the expression levels of these genes to two sets of randomly selected genes, we observed that the genes located near the EBV interaction sites displayed significantly higher expression levels. This difference in expression between the genes near EBV interaction sites and the randomly selected genes was found to be statistically significant, underscoring the potential regulatory role of the EBV enhancers in influencing the expression of nearby host genes.
Figure 8.
Functional role of EBV–human interactions: gene expression analysis reveals strong viral enhancers associated with gastric cancers. (A) Boxplot comparing the normalized reads for genes near EBV–human interactions to two different random gene sets in SNU719 cell line (****P ≤ 0.0001). (B) Boxplot comparing the normalized reads for genes near EBV–human interactions found in SNU719 to three different random gene sets in TCGA datasets from biopsies of EBV + gastric malignances (****P ≤ 0.0001). (C) Boxplot (as described in B) for EBV–human interactions found in YCCEL1 cell line (****P ≤ 0.0001). (D) Ingenuity Pathway Analysis (IPA) of genes found near EBV–human interaction sites in SNU719 cell line. (E) IPA (as described in D) of genes found near EBV–human interaction sites in YCCEL1 cell line. (F) Model of the supposed mechanism of EBV-driven cell transformation. The figure was created with BioRender.com.
To investigate whether similar gene expression patterns were present in patients with EBV-positive gastric tumors, we examined the expression of the two sets of genes identified in the cell lines, along with three sets of random genes, using The Cancer Genome Atlas (TCGA) database. Once again, we observed that all the genes located near the EBV–human interactions exhibited significantly higher expression levels (as depicted in Figure 8B, C).
To further assess if the overexpression of these genes was correlated to the presence of EBV episomes in proximity to those regions we conducted a bioinformatic analysis on publicly available RNA-seq datasets from Okabe et al. (22). In particular, we reanalyzed the RNA-seq experiments conducted on normal gastric epithelial cells (GES) before and after EBV infection. Similarly to what we observed in SNU719, the genes near EBV tethering regions appeared significantly upregulated upon EBV infection respect to the uninfected condition (Supplementary Figure 3B).
Subsequently, to explore the potential role of these genes in the neoplastic transformation of EBV-infected cells, we performed an Ingenuity Pathway Analysis (IPA) focusing on the genes located near the viral genome in both cell line (Figure 8D, E). The analysis of disease and functions associated with these genes revealed prominent associations with ‘Cancer’ and ‘Gastrointestinal Disease’, implying that these genes may serve as crucial markers of gastric cancers.
Collectively, these findings strongly suggest that the presence of strong viral enhancers in proximity to specific genes leads to their overexpression (as depicted in Figure 8F). This, in turn, may play a critical role in the neoplastic transformation of the cells. The data provide valuable insights into the molecular mechanisms by which EBV enhancers can influence the host gene expression and potentially contribute to the development of gastric cancers in EBV-infected individuals.
Discussion
In recent years, a growing body of research, including a recent study conducted by our group, has highlighted the significance of the three-dimensional structure of the EBV genome in governing viral gene expression (2,3,43). However, most of these earlier studies have primarily focused on characterizing the viral genome architecture within B cells, where EBV expresses either the highly restricted latency I program or the more permissive latency III program. Consequently, remains a considerable knowledge gap concerning the essential role, if any, played by the 3D EBV structure in epithelial cells, such as EBV + gastric cancer cells, wherein the virus adopts a type II latency program. To address this gap, our present study aimed to explore the extent of this dependence in epithelial infected cells that exhibit a type II latency program. Through our studies, we aimed to shed light on the significance of the three-dimensional EBV genome structure in this particular cellular context.
Our findings reveal the crucial role of the 3D structure of the viral episome in EBV latency, with each EBV latency program characterized by a distinct three-dimensional conformation of the viral episome. Moreover, the 3D structure changes according to the type of infected cell and to the type of gastric cancer considered as showed by Principal Component Analysis of the HiC assay. The YCCEL1 cell line, in fact, presents a structure markedly different from B cells, but not entirely identical to that of SNU719, which, on the contrary, presents a structure more similar to that found in EBV + B cells. An intriguing observation we report is the correlation between the network of long-distance chromatin interactions occurring throughout the viral genome and the expression levels of viral latent genes. This suggests an intimate link between the 3D viral chromatin structure and the transcriptional state of underlying viral loci engaged in chromatin loops. These observations align with recent studies by Zhang et al. and Barshad et al., which demonstrate how RNA Pol II activity and transcription levels can influence the formation of chromatin loops (44,45). According to these studies, RNA Pol II pausing can facilitate Cohesin loading and blocking, thereby promoting the formation of enhancer-promoter loops. Notably, research from Dr West's group (46) has shown that RNA Pol II stalling occurs at the Cp promoter, and this stalling is essential for Cp activation and B cell immortalization. Considering these findings, it is tempting to speculate that differences in transcriptional activation and RNA Pol II activity across the EBV genome, coupled with Cohesin complex and CTCF binding at viral loci, may enable viral enhancer regions to actively scan neighboring chromatin regions for functional elements and generate different sets of chromatin loops. Such interplay between the 3D viral chromatin structure and transcriptional regulation could play a pivotal role in EBV’s ability to maintain latency and influence its pathogenicity.
Chromatin loops connect functional genomic regions. Thus, our study further delved into the presence of functional elements across the EBV genome in epithelial cells, leading to the identification of novel potential viral enhancers. In both EBV + gastric cancer cell lines, we observed an enrichment of histone modifications typical of enhancers, namely H3K4me1 and H3K27ac, at specific loci of the viral genome. An intriguing finding emerged from our analysis, as we not only identified enhancer signatures at well-known EBV functional regions like OriP, Cp and Qp but also uncovered a chromatin enhancer signature at viral loci that are usually inactive during latency, such as Zp, OriLyt and the LMP promoter adjacent to the TR regions. The use of HiChIP assay, which allowed us to assess chromatin loops mediated by H3K27ac, indicative of enhancer-promoter interactions, revealed that these putative novel viral enhancers are indeed engaged in chromatin loops that connect with viral promoters and are associated with CTCF occupancy. Specifically, we detected strong enhancer regions at the BILF2 gene promoter and the Qp promoter, both active in Type I and II latency programs. However, one surprising result was the observed differences in enhancer-promoter loops between the YCCEL1 and SNU719 cell lines, as evidenced by PCA analysis. The SNU719 cell line displayed multiple interactions, whereas the YCCEL1 cell line only exhibited three frequent interactions. These discrepancies might be attributed to differences in the origin of the cells (i.e. primary tumor versus metastatic lesion). Nevertheless, these findings indicate that 3D chromatin studies unveil novel and unexplored functional regions across the viral genome in epithelial cells. Our results align with the recent work of Dr Zhao group, who conducted a similar HiChIP analysis in EBV + B cells and identified novel potential enhancer-promoter loops involving similar regions (41). Collectively, these findings enhance our understanding of the dynamic interplay between chromatin architecture and viral gene regulation in different cellular contexts, providing valuable insights into EBV pathogenesis in epithelial cells. However, how different enhancer-promoter chromatin loops are established and maintained between different EBV latency type and EBV-infected cells of different cellular origin is still unknow and we plan to investigate it in the future.
Regarding the mechanisms controlling EBV chromatin, this study reaffirms the crucial role of PARP1 activity in the regulation of EBV chromatin architecture. Notably, our findings demonstrate that inhibiting PARP1 in both epithelial EBV + cell lines resulted in the destabilization of CTCF binding across the viral genome, leading to significant alterations in the 3D chromatin structure of the EBV episome. These observations are consistent with our previous work on EBV + B cells, where PARP1 inhibition was shown to epigenetically destabilize type III latency (3,10,11). However, some divergences were observed between EBV + B cells and EBV + epithelial cells. In EBV + gastric cancer cells, we noticed that Cohesin binding seemed to be randomly deregulated, suggesting a direct dependence between PARP1 activity and Cohesin binding. Unlike Type I and III latencies observed in B cells, in EBV + GC cells, we observed an overall increase in short-range interactions in both cell lines, with a particularly prominent effect in the YCCEL1 cell line compared to SNU719 which could potentially be explained by their distinct origins (47,48). Similar to our observations in EBV + B cells, the changes in 3D viral chromatin structure resulting from PARP1 inhibition in EBV + epithelial cells also led to the repression of latent viral gene expression. Notably, these changes did not induce lytic reactivation, indicating that alterations in the 3D chromatin viral architecture are necessary but not sufficient to trigger EBV lytic replication (49,50). Further research is necessary to unravel the intricate relationship between epigenetic modifications and chromatin loop formation, and their role in regulating the lytic reactivation of EBV.
Furthermore, our aim was to gain a deeper understanding of the correlation between the presence of EBV and the transformation process leading to the tumorigenic fate of infected cells (22,42,51). Employing the HiChIP assay, we not only identified EBV-host genomic interactions mediated by H3K27ac but also investigated the potential impact of strong active enhancers of EBV on neighboring oncogenes, possibly resulting in their overexpression and subsequent cell transformation. In our analysis, we discovered a prominent enhancer region near the BILF2 gene, along with the well-known OriP and LMP regions, as major sites of EBV-host interactions. Notably, genes near these interaction points were found to be markers of gastric tumors based on Ingenuity Pathway Analysis (IPA). Comparative assessment with a random set of control genes revealed significantly higher expression levels of these genes, in the SNU719 RNA-seq data, GES RNA-seq datasets and in biopsy samples from patients deposited in The Cancer Genome Atlas (TCGA) (52). These findings indicate a clear association between these tumors and EBV infection, even though similar overexpression of these genes is observed in EBV-negative gastric tumors, serving as a characteristic marker of this tumor type. It is plausible to hypothesize that the etiologic agent responsible for inducing their marked overexpression differs in these two cases.
This study reinforces the importance of the CTCF/Cohesin complex in governing the three-dimensional structure of the viral genome during EBV infection. This observation may have broader implications beyond EBV biology and could extend to other herpes viruses. Dr Bloom and Dr Neumann's research on HSV-1, along with Dr. Izumiya's work on KSHV, and findings from other groups, suggest that CTCF/Cohesin plays a role in regulating the 3D viral structure in various viruses, including HCMV, HPV, and HTLV-1 (53–68). Understanding if CTCF acts similarly across these viruses or if there are differences presents an intriguing area for further research particularly in the DNA viruses field.
However, it is important to acknowledge some limitations in our work. A major constraint of this study is the relatively small scale at which the CTCF/Cohesin/PARP1 crosstalk is observed, spanning only approximately 170 kb. This limited scope may not fully capture the complexities present in the Human genome at a megabase scale (39,40,69). Additionally, the Hi-C assay, while providing valuable insights, is performed on a population of EBV + cells, where each cell contains multiple episomes. This makes it challenging to discern the individual three-dimensional structures of each individual episome. Furthermore, our study is complicated by recent findings from our group (70), which reveal the presence of two populations of episomes in these EBV + gastric cancer cell lines, each exhibiting distinct methylation patterns. This divergence could potentially account for the marked dissimilarities we observed in Cohesin binding and the extensive genome reorganization following treatment with a PARP1 inhibitor. Given these limitations, future investigations on larger genomic scales and single-cell level analyses may offer a more comprehensive understanding of the complex interactions involving CTCF, Cohesin, and PARP1 in EBV-infected cells. Moreover, addressing the impact of distinct episome populations and their methylation patterns could lead to a deeper appreciation of the observed effects in the context of EBV-associated gastric cancer.
In summary, our study underscores the significance of PARP1 activity in shaping EBV chromatin architecture, providing valuable insights into the mechanisms underlying viral latency regulation. Moreover, the differences observed between EBV + B cells and EBV + epithelial cells emphasize the complexity of chromatin dynamics and its context-specific nature in different cell types, shedding light on the intricate interplay between PARP1, CTCF, and Cohesin in the control of EBV gene expression. In addition, our research reveals the intricate interplay between EBV infection, chromatin interactions, and oncogene expression in the context of gastric tumors. The identification of potential mechanisms underlying the transformation process enhances our understanding of EBV-associated tumorigenesis and opens new avenues for further investigation into the role of viral enhancers in promoting oncogene expression and cellular transformation.
Supplementary Material
Acknowledgements
The results shown here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. We thank Dr. Samantha S. Soldan for providing technical support for ddPCR. We are grateful to The Wistar Institute's Genomics, and Bioinformatics core facilities for providing technical support. Funding support for The Wistar Institute core facilities was provided by Cancer Center Support Grant P30 CA010815. IT is supported by R01 AI130209, GM124449, P01 CA269043, and P30 CA010815; SPA is supported by T32 CA09171.
Author contributions. D.M. and I.T. designed study; D.M., G.N., S.P.A., L.B.C. performed research; D.M., A.K. analyzed data; D.M. and I.T. wrote the paper; D.M. and I.T. final edited the paper.
Contributor Information
Davide Maestri, The Wistar Institute, Philadelphia, PA 19104, USA; Department of Pharmacy and Biotechnology, University of Bologna, 40126 Bologna, Italy.
Giorgia Napoletani, The Wistar Institute, Philadelphia, PA 19104, USA.
Andrew Kossenkov, The Wistar Institute, Philadelphia, PA 19104, USA.
Sarah Preston-Alp, The Wistar Institute, Philadelphia, PA 19104, USA.
Lisa B Caruso, The Wistar Institute, Philadelphia, PA 19104, USA.
Italo Tempera, The Wistar Institute, Philadelphia, PA 19104, USA.
Data availability
The data that support this study are available from the corresponding author upon reasonable request. The data for the HiC assay, ChIP-seq assay for H3K27ac, H3K4me1, RNA-seq for SNU719 and the H3K27ac HiChIP have been deposited in the Gene Expression Omnibus database under the following accession code GSE239995. ChIP-seq experiments for CTCF in Mutu1, LCL and Gastric Cancer cells were obtained by publicly available sequencing datasets GSE115829, GSE160973, GSE234603, generated in our previous works. RNA-seq datasets on GES cells before and after EBV infection obtained by publicly available work from Okabe et al. are available on Gene Expression Omnibus under the accession code GSE147152.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Wistar Institute core facilities was provided by Cancer Center Support Grant [P30 CA010815]; I.T. is supported by R01 AI130209, GM124449, P01 CA269043, and P30 CA010815; S.P.A. is supported by T32 CA09171. Funding for open access charge: NIH, Institutional Funding.
Conflict of interest statement. None declared.
References
- 1. Smatti M.K., Al-Sadeq D.W., Ali N.H., Pintus G., Abou-Saleh H., Nasrallah G.K.. Epstein–Barr virus epidemiology, serology, and genetic variability of LMP-1 oncogene among healthy population: an update. Front. Oncol. 2018; 8:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Caruso L.B., Maestri D., Tempera I.. Three-dimensional chromatin structure of the EBV genome: a crucial factor in viral infection. Viruses. 2023; 15:1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Morgan S.M., Tanizawa H., Caruso L.B., Hulse M., Kossenkov A., Madzo J., Keith K., Tan Y., Boyle S., Lieberman P.M.et al.. The three-dimensional structure of Epstein–Barr virus genome varies by latency type and is regulated by PARP1 enzymatic activity. Nat. Commun. 2022; 13:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Price A.M., Luftig M.A.. To be or not IIb: a multi-step process for Epstein–Barr virus latency establishment and consequences for B cell tumorigenesis. PLoS Pathog. 2015; 11:e1004656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tempera I., Wiedmer A., Dheekollu J., Lieberman P.M.. CTCF prevents the epigenetic drift of EBV latency promoter Qp. PLoS Pathog. 2010; 6:e1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Holdorf M.M., Cooper S.B., Yamamoto K.R., Miranda J.L.. Occupancy of chromatin organizers in the Epstein–Barr virus genome. Virology. 2011; 415:1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arvey A., Tempera I., Tsai K., Chen H.-S., Tikhmyanova N., Klichinsky M., Leslie C., Lieberman P.M.. An atlas of the Epstein–Barr virus transcriptome and epigenome reveals host-virus regulatory interactions. Cell Host Microbe. 2012; 12:233–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chau C.M., Zhang X.-Y., McMahon S.B., Lieberman P.M.. Regulation of Epstein–Barr virus latency type by the chromatin boundary factor CTCF. J. Virol. 2006; 80:5723–5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Day L., Chau C.M., Nebozhyn M., Rennekamp A.J., Showe M., Lieberman P.M.. Chromatin profiling of Epstein–Barr virus latency control region. J. Virol. 2007; 81:6389–6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lupey-Green L.N., Moquin S.A., Martin K.A., McDevitt S.M., Hulse M., Caruso L.B., Pomerantz R.T., Miranda J.L., Tempera I.. PARP1 restricts Epstein Barr Virus lytic reactivation by binding the BZLF1 promoter. Virology. 2017; 507:220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lupey-Green L.N., Caruso L.B., Madzo J., Martin K.A., Tan Y., Hulse M., Tempera I.. PARP1 stabilizes CTCF binding and chromatin structure to maintain Epstein–Barr virus latency type. J. Virol. 2018; 92:e00755–e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moon S.H., Park N.-S., Noh M.H., Kim Y.S., Cheong S.H., Hur D.Y.. Olaparib-induced apoptosis through EBNA1-ATR-p38 MAPK signaling pathway in Epstein–Barr virus-positive gastric cancer cells. Anticancer Res. 2022; 42:555–563. [DOI] [PubMed] [Google Scholar]
- 13. Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A.. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018; 68:394–424. [DOI] [PubMed] [Google Scholar]
- 14. Young L.S., Rickinson A.B.. Epstein–Barr virus: 40 years on. Nat. Rev. Cancer. 2004; 4:757–768. [DOI] [PubMed] [Google Scholar]
- 15. Murphy G., Pfeiffer R., Camargo M.C., Rabkin C.S.. Meta-analysis shows that prevalence of Epstein–Barr virus-positive gastric cancer differs based on sex and anatomic location. Gastroenterology. 2009; 137:824–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Camargo M.C., Murphy G., Koriyama C., Pfeiffer R.M., Kim W.H., Herrera-Goepfert R., Corvalan A.H., Carrascal E., Abdirad A., Anwar M.et al.. Determinants of Epstein–Barr virus-positive gastric cancer: an international pooled analysis. Br. J. Cancer. 2011; 105:38–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Naseem M., Barzi A., Brezden-Masley C., Puccini A., Berger M.D., Tokunaga R., Battaglin F., Soni S., McSkane M., Zhang W.et al.. Outlooks on Epstein–Barr virus associated gastric cancer. Cancer Treat. Rev. 2018; 66:15–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cristescu R., Lee J., Nebozhyn M., Kim K.-M., Ting J.C., Wong S.S., Liu J., Yue Y.G., Wang J., Yu K.et al.. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat. Med. 2015; 21:449–456. [DOI] [PubMed] [Google Scholar]
- 19. Stanland L.J., Luftig M.A.. The role of EBV-induced hypermethylation in gastric cancer tumorigenesis. Viruses. 2020; 12:1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang K., Yuen S.T., Xu J., Lee S.P., Yan H.H.N., Shi S.T., Siu H.C., Deng S., Chu K.M., Law S.et al.. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat. Genet. 2014; 46:573–582. [DOI] [PubMed] [Google Scholar]
- 21. Wang K., Kan J., Yuen S.T., Shi S.T., Chu K.M., Law S., Chan T.L., Kan Z., Chan A.S.Y., Tsui W.Y.et al.. Exome sequencing identifies frequent mutation of ARID1A in molecular subtypes of gastric cancer. Nat. Genet. 2011; 43:1219–1223. [DOI] [PubMed] [Google Scholar]
- 22. Okabe A., Huang K.K., Matsusaka K., Fukuyo M., Xing M., Ong X., Hoshii T., Usui G., Seki M., Mano Y.et al.. Cross-species chromatin interactions drive transcriptional rewiring in Epstein–Barr virus–positive gastric adenocarcinoma. Nat. Genet. 2020; 52:919–930. [DOI] [PubMed] [Google Scholar]
- 23. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Li B., Dewey C.N.. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinf. 2011; 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Li H., Durbin R.. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010; 26:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Feng J., Liu T., Qin B., Zhang Y., Liu X.S.. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012; 7:1728–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W.et al.. Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 2008; 9:R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ramírez F., Ryan D.P., Grüning B., Bhardwaj V., Kilpert F., Richter A.S., Heyne S., Dündar F., Manke T.. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016; 44:W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bailey T.L., Johnson J., Grant C.E., Noble W.S.. The MEME suite. Nucleic Acids Res. 2015; 43:W39–W49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Servant N., Varoquaux N., Lajoie B.R., Viara E., Chen C.-J., Vert J.-P., Heard E., Dekker J., Barillot E.. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015; 16:259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gu Z., Gu L., Eils R., Schlesner M., Brors B.. circlize implements and enhances circular visualization in R. Bioinformatics. 2014; 30:2811–2812. [DOI] [PubMed] [Google Scholar]
- 33. Mumbach M.R., Rubin A.J., Flynn R.A., Dai C., Khavari P.A., Greenleaf W.J., Chang H.Y.. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods. 2016; 13:919–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lareau C., Aryee M.. hichipper: a preprocessing pipeline for assessing library quality and DNA loops from HiChIP data Bioinformatics. 2017; [DOI] [PMC free article] [PubMed]
- 35. Lareau C.A., Aryee M.J.. diffloop: a computational framework for identifying and analyzing differential DNA loops from sequencing data. Bioinformatics. 2018; 34:672–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., Glass C.K.. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 2010; 38:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lin C.-T.M., Leibovitch E.C., Almira-Suarez M.I., Jacobson S.. Human herpesvirus multiplex ddPCR detection in brain tissue from low- and high-grade astrocytoma cases and controls. Infect Agents Cancer. 2016; 11:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bochum S., Berger S., Martens U.M.. Martens U.M. Olaparib. Small Molecules in Oncology, Recent Results in Cancer Research. 2018; 211:Cham: Springer International Publishing; 217–233. [DOI] [PubMed] [Google Scholar]
- 39. Rao S.S.P., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S.et al.. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014; 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang K., Li N., Ainsworth R.I., Wang W.. Systematic identification of protein combinations mediating chromatin looping. Nat. Commun. 2016; 7:12249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ding W., Wang C., Narita Y., Wang H., Leong M.M.L., Huang A., Liao Y., Liu X., Okuno Y., Kimura H.et al.. The Epstein–Barr virus enhancer interaction landscapes in virus-associated cancer cell lines. J. Virol. 2022; 96:e00739-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim K.-D., Tanizawa H., De Leo A., Vladimirova O., Kossenkov A., Lu F., Showe L.C., Noma K., Lieberman P.M.. Epigenetic specifications of host chromosome docking sites for latent Epstein–Barr virus. Nat. Commun. 2020; 11:877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tempera I., Klichinsky M., Lieberman P.M.. EBV latency types adopt alternative chromatin conformations. PLoS Pathog. 2011; 7:e1002180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhang S., Übelmesser N., Barbieri M., Papantonis A.. Enhancer–promoter contact formation requires RNAPII and antagonizes loop extrusion. Nat. Genet. 2023; 55:832–840. [DOI] [PubMed] [Google Scholar]
- 45. Barshad G., Lewis J.J., Chivu A.G., Abuhashem A., Krietenstein N., Rice E.J., Ma Y., Wang Z., Rando O.J., Hadjantonakis A.-K.et al.. RNA polymerase II dynamics shape enhancer–promoter interactions. Nat. Genet. 2023; 55:1370–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Palermo R.D., Webb H.M., West M.J.. RNA polymerase II stalling promotes nucleosome occlusion and pTEFb recruitment to drive immortalization by Epstein–Barr virus. PLoS Pathog. 2011; 7:e1002334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Oh S.T., Seo J.S., Moon U.Y., Kang K.H., Shin D.-J., Yoon S.K., Kim W.H., Park J.-G., Lee S.K.. A naturally derived gastric cancer cell line shows latency I Epstein–Barr virus infection closely resembling EBV-associated gastric cancer. Virology. 2004; 320:330–336. [DOI] [PubMed] [Google Scholar]
- 48. Kim D.N., Seo M.K., Choi H., Kim S.Y., Shin H.J., Yoon A.-R., Tao Q., Rha S.Y., Lee S.K.. Characterization of naturally Epstein–Barr virus-infected gastric carcinoma cell line YCCEL1. J. Gen. Virol. 2013; 94:497–506. [DOI] [PubMed] [Google Scholar]
- 49. Tempera I., Deng Z., Atanasiu C., Chen C.-J., D’Erme M., Lieberman P.M. Regulation of Epstein–Barr virus OriP replication by poly(ADP-ribose) polymerase 1. J. Virol. 2010; 84:4988–4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mattiussi S., Tempera I., Matusali G., Mearini G., Lenti L., Fratarcangeli S., Mosca L., D’Erme M., Mattia E.. Inhibition of Poly(ADP-ribose)polymerase impairs Epstein Barr Virus lytic cycle progression. Infect. Agents Cancer. 2007; 2:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kumar A., Lyu Y., Yanagihashi Y., Chantarasrivong C., Majerciak V., Salemi M., Wang K.-H., Inagaki T., Chuang F., Davis R.R.et al.. KSHV episome tethering sites on host chromosomes and regulation of latency-lytic switch by CHD4. Cell Rep. 2022; 39:110788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. The Cancer Genome Atlas Research Network Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014; 513:202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Campbell M., Chantarasrivong C., Yanagihashi Y., Inagaki T., Davis R.R., Nakano K., Kumar A., Tepper C.G., Izumiya Y.. KSHV topologically associating domains in latent and reactivated viral chromatin. J. Virol. 2022; 96:e00565–e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Elder E.G., Krishna B.A., Poole E., Perera M., Sinclair J.. Regulation of host and viral promoters during human cytomegalovirus latency via US28 and CTCF. J. Gen. Virol. 2021; 102:001609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ertel M.K., Cammarata A.L., Hron R.J., Neumann D.M.. CTCF occupation of the Herpes Simplex Virus 1 genome is disrupted at early times postreactivation in a transcription-dependent manner. J. Virol. 2012; 86:12741–12759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kang H., Wiedmer A., Yuan Y., Robertson E., Lieberman P.M.. Coordination of KSHV latent and lytic gene control by CTCF-cohesin mediated chromosome conformation. PLoS Pathog. 2011; 7:e1002140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kang H., Lieberman P.M.. Cell Cycle Control of Kaposi's sarcoma-associated herpesvirus latency transcription by CTCF-Cohesin interactions. J. Virol. 2009; 83:6199–6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Li D.-J., Verma D., Mosbruger T., Swaminathan S.. CTCF and Rad21 act as host cell restriction factors for Kaposi's sarcoma-associated herpesvirus (KSHV) lytic replication by modulating viral gene transcription. PLoS Pathog. 2014; 10:e1003880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Li D., Mosbruger T., Verma D., Swaminathan S.. Complex interactions between cohesin and CTCF in regulation of Kaposi's sarcoma-associated herpesvirus lytic transcription. J. Virol. 2020; 94:e01279-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Martinez M.P., Cheng X., Joseph A., Al-Saleem J., Panfil A.R., Palettas M., Dirksen W.P., Ratner L., Green P.L.. HTLV-1 CTCF-binding site is dispensable for in vitro immortalization and persistent infection in vivo. Retrovirology. 2019; 16:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Martínez F.P., Cruz R., Lu F., Plasschaert R., Deng Z., Rivera-Molina Y.A., Bartolomei M.S., Lieberman P.M., Tang Q.. CTCF binding to the first intron of the major immediate early (MIE) gene of human cytomegalovirus (HCMV) negatively regulates MIE gene expression and HCMV replication. J. Virol. 2014; 88:7389–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Mehta K., Gunasekharan V., Satsuka A., Laimins L.A.. Human papillomaviruses activate and recruit SMC1 cohesin proteins for the differentiation-dependent life cycle through association with CTCF insulators. PLoS Pathog. 2015; 11:e1004763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Paris C., Pentland I., Groves I., Roberts D.C., Powis S.J., Coleman N., Roberts S., Parish J.L.. CCCTC-binding factor recruitment to the early region of the human papillomavirus 18 genome regulates viral oncogene expression. J. Virol. 2015; 89:4770–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pentland I., Campos-León K., Cotic M., Davies K.-J., Wood C.D., Groves I.J., Burley M., Coleman N., Stockton J.D., Noyvert B.et al.. Disruption of CTCF-YY1–dependent looping of the human papillomavirus genome activates differentiation-induced viral oncogene transcription. PLoS Biol. 2018; 16:e2005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Stedman W., Kang H., Lin S., Kissil J.L., Bartolomei M.S., Lieberman P.M.. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 2008; 27:654–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Washington S.D., Edenfield S.I., Lieux C., Watson Z.L., Taasan S.M., Dhummakupt A., Bloom D.C., Neumann D.M.. Depletion of the insulator protein CTCF results in herpes simplex virus 1 reactivation in vivo. J. Virol. 2018; 92:e00173-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Washington S.D., Musarrat F., Ertel M.K., Backes G.L., Neumann D.M.. CTCF binding sites in the herpes simplex virus 1 genome display site-specific CTCF occupation, protein recruitment, and insulator function. J. Virol. 2018; 92:e00156-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Watson Z.L., Washington S.D., Phelan D.M., Lewin A.S., Tuli S.S., Schultz G.S., Neumann D.M., Bloom D.C.. In vivo knockdown of the herpes simplex virus 1 latency-associated transcript reduces reactivation from latency. J. Virol. 2018; 92:e00812-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rowley M.J., Corces V.G.. Organizational principles of 3D genome architecture. Nat. Rev. Genet. 2018; 19:789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Preston-Alp S., Caruso L.B., Su C., Keith K., Soldan S.S., Maestri D., Madzo J., Kossenkov A., Napoletani G., Gewurz B.et al.. Decitabine disrupts EBV genomic epiallele DNA methylation patterns around CTCF binding sites to increase chromatin accessibility and lytic transcription in gastric cancer. mBio. 2023; 10.1128/mbio.00396-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support this study are available from the corresponding author upon reasonable request. The data for the HiC assay, ChIP-seq assay for H3K27ac, H3K4me1, RNA-seq for SNU719 and the H3K27ac HiChIP have been deposited in the Gene Expression Omnibus database under the following accession code GSE239995. ChIP-seq experiments for CTCF in Mutu1, LCL and Gastric Cancer cells were obtained by publicly available sequencing datasets GSE115829, GSE160973, GSE234603, generated in our previous works. RNA-seq datasets on GES cells before and after EBV infection obtained by publicly available work from Okabe et al. are available on Gene Expression Omnibus under the accession code GSE147152.