Abstract
Following a DNA double strand break (DSB), several nucleases and helicases coordinate to generate single-stranded DNA (ssDNA) with 3′ free ends, facilitating precise DNA repair by homologous recombination (HR). The same nucleases can act on stalled replication forks, promoting nascent DNA degradation and fork instability. Interestingly, some HR factors, such as CtIP and BRCA1, have opposite regulatory effects on the two processes, promoting end resection at DSB but inhibiting the degradation of nascent DNA on stalled forks. However, the reason why nuclease actions are regulated by different mechanisms in two DNA metabolism is poorly understood. We show that human HELQ acts as a DNA end resection regulator, with opposing activities on DNA end resection at DSBs and on stalled forks as seen for other regulators. Mechanistically, HELQ helicase activity is required for EXO1-mediated DSB end resection, while ssDNA-binding capacity of HELQ is required for its recruitment to stalled forks, facilitating fork protection and preventing chromosome aberrations caused by replication stress. Here, HELQ synergizes with CtIP but not BRCA1 or BRCA2 to protect stalled forks. These findings reveal an unanticipated role of HELQ in regulating DNA end resection at DSB and stalled forks, which is important for maintaining genome stability.
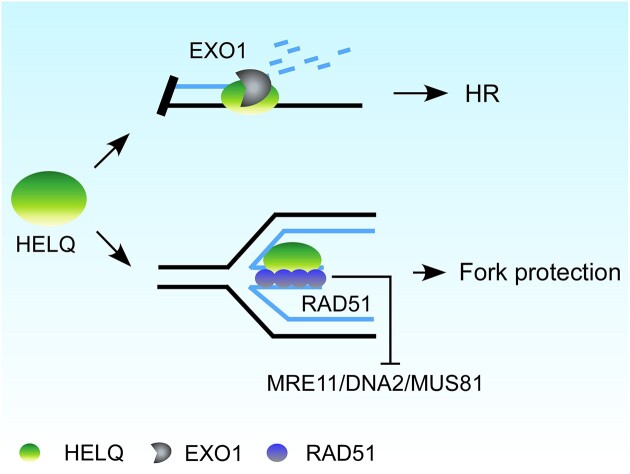
Graphical Abstract
Graphical Abstract.
Introduction
DNA double strand breaks (DSB) constitute a serious form of DNA damage and pose a threat to genome stability. Indeed, improper DSB repair can result in the generation of genetic mutations and/or gross chromosomal rearrangements that can precipitate various pathologies ranging from premature embryonic death, aging, immunodeficiency, neurological disorders and cancer (1–5). Nonhomologous end-joining (NHEJ) and homologous recombination (HR) are the two main pathways responsible for DSB repair: NHEJ repairs DSB by direct ligation of the DNA ends thus can readily generate mutations at the sites of joining; HR is highly accurate due to the use of the identical sister chromatid as a template (1,5,6).
DSB repair that is mediated by HR requires a series of interconnected molecular processes. During the early stages, DSB ends are degraded in the 5′-3′ direction to generate long 3′ single-stranded DNA (ssDNA) tails. In mammals, this so-called ‘end resection’ event is driven by evolutionarily conserved core resection machinery, comprising: MRE11-RAD50-NBS1 (MRN) complex, CtIP (CtBP-interacting protein), Bloom syndrome protein (BLM), DNA replication helicase/nuclease 2 (DNA2), and exonuclease 1 (EXO1) (7–9). Based on studies performed in yeast and in vitro biochemical data, it seems that end resection is carried out via two steps: the MRN (MRE11-RAD50-XRS2, in budding yeast) complex and CtIP (Sae2 in budding yeast) catalyze the initial processing of DNA ends, while the BLM helicase/DNA2 and EXO1 constitute two resection motors that drive extended resection and initiate HR (10–13).
The 3′-ssDNA generated by end resection provides a platform to recruit related protein factors to initiate HR (13–19). First, ssDNA is bound by replication protein A (RPA), which is then replaced by radiation sensitive 51 (RAD51) recombinase to form a nucleoprotein filament for subsequent strand invasion. The resulting RAD51-ssDNA filament pairs to complementary ssDNA regions within the homologous duplex. The 3′ invading end on the joint molecular intermediate then primes DNA synthesis using homologous sequences as templates to restore missing genetic information at the break site. The resolution of replication stress-induced Holliday Junction structures (formed as a result of fork reversal) (20,21) and end ligation completes HR (5,6,22–29).
Fork reversal occurs when stalled forks anneal to the two nascent strands behind the stalled fork (20,21). The annealed nascent strand of the reversed fork resembles a single-end DSB that can serve as an entry point for the resection enzymes to drive nucleolytic degradation. Fork reversal is mainly catalyzed by motor proteins including SMARCAL1, ZRANB3 and HLTF (30–35). Human RAD51, BRCA1, BRCA2, and additional factors harbor DSB repair-independent functions to protect stalled replication forks by preventing the degradation of nascent DNA (34,36–42). In the absence of these ‘fork protection’ factors, nascent DNA on the reversed fork can be degraded by resection nucleases such as MRE11, DNA2, EXO1 and MUS81, causing fork collapse and genome instability (10,43–47). RAD51 stabilization on the nascent DNA of the reversed fork is critical for fork protection-indeed, most protection factors prevent fork degradation by directly or indirectly promoting (and/or stabilizing) RAD51 recruitment to nascent DNA on the regressed arm of reversed forks or inhibiting the activities of resection nucleases on stalled forks. Meanwhile, RAD51 promotes fork reversal, triggering nuclease degradation at stalled forks, as described earlier (30,34,35,48–50).
Although the molecular mechanisms underlying DSB end resection and nascent DNA degradation on stalled forks differ, the two processes involve many of the same nucleases and regulators. For example, BRCA1 and CtIP promote DSB end resection but inhibit nascent DNA degradation on reversed forks (38,51–54). While DSB end resection always follows the 5′-3′ direction to generate 3′-ssDNA, both strands on the nascent DNA of the reversed fork are degraded in the absence of protection. How the same nucleases coordinate the degradation of the two strands of nascent DNA is unclear (10).
Human HELQ is a highly conserved superfamily 2 helicase (55,56) that when disrupted in murine or human cells, causes genomic instability and cancer susceptibility. Moreover, tumor cells that over-express HELQ exhibit low sensitivity to chemotherapy (57–60). HELQ acts independently from the Fanconi anemia pathway to overcome replication stress induced by DNA inter-strand crosslinks (ICLs) in human cells (57–59). Downstream from RAD51, HELQ and the MCM8/MCM9 helicase redundantly promote HR (61); but HELQ helicase activity can also remove RPA from and anneal ssDNA in vitro, allowing for annealing-mediated DSB repair, such as single-strand annealing (SSA) and microhomology-mediated end-joining (MMEJ) (62). It thus seems that HELQ is involved in various DSB repair pathways via its intrinsic enzyme activity, but the underlying mechanisms by which HELQ plays its role in DSB repair and other DNA metabolic processes is poorly understood. In this study, we reveal unexpected role of HELQ in regulating DNA end resection that occurs at DSBs and stalled replication forks. We show that HELQ promotes EXO1-mediated DSB end resection and blocks MRE11/DNA2/MUS81 nucleases-mediated nascent DNA degradation on stalled forks via its DNA binding capacity. HELQ functions in parallel with CtIP to protect stalled forks, the two genes show synergistic lethal effect.
Materials and methods
Cell culture, antibodies and reagents
Human U2OS cells and HCT116 cells were cultured at 37°C in Dulbecco's modified Eagle's medium with 10% fetal bovine serum in the presence of 1% penicillin/streptomycin (HyClone, SV30010) and 5% CO2. Both knockout (KO) and knockin (KI) cell lines were generated using CRISPR/Cas9 genome-editing technology (63,64). The following guide RNA (gRNA) sequences targeting the second exon of HELQ and the fourth exon of CtIP were designed using an optimized CRISPR design tool (https://zlab.squarespace.com/guide-design-resources) and used to edit the target genes: sgRNA-KO HELQ GTCGTCAACTTGAGCTATAA; sgRNA-KO CtIP TCACCAAAAATCAACAGCTG; and sgRNA-KI HELQ CTCACGTGACCTGCCGCGTA (used to knock in an N-terminal SFB tag at the HELQ gene locus). The resulting cells were designated HELQ KO, CtIP KO and HELQ KI-SFB, respectively.
Antibodies used in this study included the following: FLAG (F1804, Sigma-Aldrich), HELQ (PA5-65181, Sigma-Aldrich and 19436s, Cell Signaling Technology), Biotin (200-002-211, Jackson Immunoresearch and A150-109A, Bethyl), γH2AX (05–636, Millipore), CtIP (A300-488A, Bethyl), RPA2 S4/8p (A300-245A, Bethyl), RPA2 (A300-244A, Bethyl), BRCA1 (9010s, Cell Signaling Technology), BRCA2 (10741S, Cell Signaling Technology), MRE11 (ab208020, Abcam), RAD51 (ab133534, Abcam), BrdU (ab6326, Abcam and 347580, BD), MUS81(ab247136, Abcam), BLM (ab2179, Abcam), DNA2 (18727-1-AP, Proteintech), EXO1 (16253-1-AP, Proteintech), HLTF (Ab17984, Abcam), SMARCAL1 (sc-376377, Santa Cruze), ZRANB3 (A303-033A, Bethyl).
Etoposide (E1383), camptothecin (C9911), hydroxyurea (H8627), 4-OHT(H7904), IdU (l7125), Biotin-azide (Biotin-dPEG®7-azide, QBD10825), and CldU (C6891) were purchased from Sigma Aldrich. EdU (11590926) was purchased from Life Technologies.
Plasmids, mutagenesis and shRNA
EGFP-HELQ, EGFP-MRE11, EGFP-NBS1 and EGFP-EXO1 constructs were gifts from Dr Xiaohua Wu (The Scripps Research Institute). pBABE-ER-AsiSI, pCS2-mGP and pMD2G plasmids were provided by Dr Tanya T. Paull (The University of Texas at Austin) and Dr Gaëlle Legube (University of Toulouse). HELQ and EXO1 mutants were constructed by PCR-based site-directed mutagenesis according to standard procedures. HELQ wild-type and the indicated mutants were subcloned into pFastBAC-HTb (Invitrogen) and NBLV0051 (Novo Bio) vectors to generate recombinant baculoviruses and lentiviruses, respectively. EXO1 wild-type and mutant were cloned into a pTXB1vector (New England Biolabs) to allow for the expression of recombinant protein in bacteria.
Endogenous gene silencing was achieved via lentiviral infection using pDS-124 vectors to express the corresponding shRNAs (65,66). All the shRNA target sequences have been previously published and are listed as follows: MRE11, GAUGAGAACUCU UGGUUUAAC (67); NBS1, GAAGAAACGUGAACUCAAGUU (68);CtIP, GAGCAGACCUUUCUCAGUAUA (65); BRCA2, GAAGAAUGCAGGUUUAAUA (66); RAD51, GACUGCCAGGAUAAAGCUU (54); EXO1,GAAGUUUCGUUACAUGUGUAU (69); BLM, GAGCACAUCUGUAAAUUAAUU (70); DNA2, CAGUAUCUCCUCUAGCUAG (71); ZRANB3, GAGUUACCUUAUUGUGAAA; HLTF, GGAAUAUAAUGUUAACGAU; SMARCAL1, GCAGAAGAUCUACGACCUA (72); MUS81, CAGCCCUGGUGGAUCGAUA (73); RADX, CAUAGAGGCCAGCCGUAUA (74); BRCA1, CAACAUGCCCACAGAUCAACU (66); HELQ, CAAAGGAAGAUUUCCUCCAACUAAA (57).
Laser-induced microirradiation, live-cell imaging and immunofluorescence (IF)
U2OS cells were grown on a dish with a thin glass bottom and then irradiated locally with a 365 nm pulsed nitrogen UV laser (16 Hz pulse, 55% laser output) generated from a Micropoint System (Andor). Images were captured in real time every 20 s under a Nikon A1 confocal imaging system directly coupled to the Micropoint System. IF analyses were performed as previously described (65,66).
Chromatin immunoprecipitation (ChIP) and ER-AsiSI resection assay
HELQ-KI SFB U2OS cells expressing ER-AsiSI (75,76), in which the restriction enzyme AsiSI is fused to the estrogen receptor hormone-binding domain, were treated with 300 nM 4-OHT for 4 h to induce DSBs. Then, a ChIP assay was performed as previously described (67,70) using 200 μg chromatin immunoprecipitated with IgG and FLAG antibodies (2 μg). The immunoprecipitated DNA and input DNA were analyzed by qPCR using Taq pro Universal SYBR qPCR Master Mix (Vazyme). The following primers were used for qPCR, as previously described (76): DSB-F 5-GATTGGCTATGGGTGTGGAC and DSB-R 5-CATCCTTGCAAACCAGTCCT. The IP efficiency was calculated as the percentage of the input DNA immunoprecipitated. ER-AsiSI resection assay was performed as previously described (65,76).
Recombinant protein purification
HELQ and the indicated mutants were expressed in Sf9 insect cells via a Bac-to-Bac Baculovirus Expression System (Invitrogen). Briefly, the cDNA encoding human HELQ was constructed in a pFastBacHTb vector, which was then transformed into DH10Bac Escherichia coli to create the recombinant bacmid. After transfecting the resulting recombinant bacmid into Sf21 insect cells, the virus stock was acquired. A 400 ml spinner flask of Sf21 cells (1 × 106/ml) was infected with the HELQ baculovirus, and the infected cells were incubated at 27°C for 72 h with continuous agitation. The cells were then collected by centrifugation at 500 g for 10 min and washed with ice-cold PBS. The cells were lysed in 40 ml Buffer A [50 mM Tris, pH 8.0, 600 mM NaCl,1 mM phenyl methyl sulfonyl fluoride (PMSF), 0.5% NP-40, 10% glycerol, 20 mM imidazole, EDTA-free protease inhibitor mixture (Roche)] and incubated for 45 min with continuous agitation. Insoluble material was removed by 16 000 × g centrifugation for 20 min. Ni-NTA agarose (QIAGEN) was pre-equilibrated with buffer A and then added to 50 ml tubes containing the soluble extract before incubating at 4°C for 2 h with continuous rotation. After incubation, the Ni-NTA agarose was washed three times with 120 ml Buffer B (50 mM Tris, pH 8.0, 600 mM NaCl, 0.1% NP-40, 10% glycerol, 50 mM imidazole) by centrifugation at 2000 g for 2 min. The protein was eluted from the Ni-NTA agarose in Buffer C (50 mM Tris, pH 8.0, 150 mM NaCl, 0.1% NP-40, 10% glycerol, 400 mM imidazole), frozen in liquid nitrogen and stored at –80°C until further use. Recombinant human EXO1 and the indicated mutants were expressed in E.coli and purified as previously described (77).
DNA unwinding and DNA resection assays
The helicase unwinding assays were performed in 15 μl helicase buffer (25 mM Tris–HCl pH 7.5, 50 mM NaCl, 2 mM ATP, 2 mM MgCl2, 0.1 mg/ml bovine serum albumin (BSA). First, 25 nM FITC-labeled DNA substrate and the indicated concentrations of purified human HELQ were mixed and incubated at 37°C for 30 min before the reactions were stopped upon the addition of 5 μl stop buffer (30% glycerol, 150 mM EDTA, 0.1% bromophenol blue, 1 mg/ml proteinase K). To prevent re-annealing, the stop buffer was supplemented with a 20-fold excess of unlabeled oligos with the same sequence as the FITC-labeled oligo. The products were resolved on 10% native polyacrylamide gels by electrophoresis for 1 h at 100 V in 0.5 x TBE buffer. The gels were imaged using a GE Typhoon FLA 9500. The sequences of oligonucleotides used to generate substrates are indicated as follows: oligo 1 (5′FITC-AGCTACCATGCCTGCACGAATTAAGCAATTCGTAATCATGGTCATAGCT); oligo 2 (5′-AATTCGTGCAGGC ATGGTAGCT).
DNA resection assays were performed as previously described (78) with slight modifications. The nicked and 3′overhang substrates were respectively obtained by incubating the pUC19 plasmid derivative with Nt.BspQI (NEB) and KpnI (ThermoFisher). The substrates were then purified using the phenol-chloroform method. Exonuclease activity assays were performed in resection buffer (50 mM Tris–HCl pH 7.5, 2 mM ATP, 50 mM NaCl, 2 mM MgCl2 and 0.1 mg/ml BSA). First, DNA substrate and the indicated concentrations of purified recombinant proteins were mixed and incubated at 37°C. The reactions were terminated upon the addition of 5 μl stop buffer (30% glycerol, 150 mM EDTA, 0.1% bromophenol blue,1 mg/ml proteinase K). The DNA products were separated on an 0.8% agarose gel, which were then stained with Gel-Red (BIORIGIN) and imaged using a ChemiDoc XRS+ system (Bio-Rad). The sequences of oligonucleotides used to generate substrates are indicated as follows.
Electrophoretic mobility shift assay (EMSA)
EMSA was performed in a 15 μl reaction volume containing 25 nM Cy5 or FITC-labeled DNA substrate in binding buffer [25 mM Tris–HCl (pH 7.5), 50 mM NaCl, 2 mM ATP, 2 mM MgCl2 and 0.1 mg/ml BSA]. The recombinant proteins and substrate were gently mixed and incubated at 37°C for 10 min before being supplemented with 5 μl loading buffer (50% glycerol, 0.1% bromophenol blue). The proteins were resolved on a 6% native polyacrylamide gel by electrophoresis for 45 min at 80 V in 0.5× TBE buffer and imaged using a GE Typhoon FLA 9500 system.
Isothermal titration calorimetry (ITC)
ITC binding assays for HELQ and ssDNA were performed using an ITC200 titration calorimeter (MicroCal). Purified proteins were dialyzed overnight against ITC buffer (20 mM Tris–HCl, pH 7.5, 150 mM NaCl) before ITC testing. Then, 100 μM ssDNA was titrated into the calorimetric cell containing 14 μM wild-type HELQ or the indicated mutant. Titrations for all reactions were performed at 25°C, and all titrations required a 0.4 μl pre-injection of ssDNA and 19 consecutive 2 μl injections at 120 s intervals. The data obtained were processed with Origin software.
In situ proximity ligation assay (PLA)
In situ PLA was performed using Duolink PLA technology (Sigma-Aldrich), according to the manufacturer's instructions. Briefly, the drug-treated cells were washed three times with PBS at 37°C and fixed in 2% paraformaldehyde for 20 min. The cells were then permeabilized with 0.5% Triton X-100 for 10 min and blocked with 3% BSA for 30 min at room temperature. The cells were incubated with two primary antibodies at 4°C overnight and then subjected to the subsequent procedures following the manufacturer's instructions. Images were captured under an Olympus IX81 FL microscope. To test the binding of proteins to nascent DNA, the cells were labeled with 10 μM EdU for 20 min at 37°C, and then untreated or treated with 2 mM HU for 4 h prior to washing in PBS. Fixed and blocked cells were subjected to a Click-iT reaction to attach biotin to EdU. Antibodies against biotin and specific protein were used for the subsequent PLA assays.
DNA fiber analysis
DNA fiber analysis was performed as previously described (66,79). Briefly, cells were first labeled with 40 μM IdU for 30 min, followed by 100 μM CldU for a further 30 min before incubation with or without the indicated drugs. The cells were then harvested by trypsinization and then labeled and unlabeled cells were mixed in a ratio of 1:6 and loaded onto a slide for 3 min. A cell lysis solution (200 mM Tris–HCl, pH 7.5, 50 mM EDTA and 0.5% SDS) was added then added to lyse the cells for 3 min. The slide was angled to allow DNA to spread for 5 min. After air drying, the fibers were fixed in a 3:1 (vol/vol) methanol: acetic acid solution and denatured in 2.5 M HCl at 4°C overnight. Then, the slides were washed twice with 1 x PBS and blocked with 2% BSA before adding anti-BrdU and corresponding secondary antibodies. Images of the slides were captured under an Olympus IX81 FL microscope and analyzed using Image J software.
Chromosome analysis by metaphase spreading
Cells were treated with 2 mM HU for 4 h to induce replication stress, and then incubated with nocodazole (200 ng/ml) at 37°C for 16 h. Then, 50 mM KCl was used to swell the cells at 37°C for 15 min before they were fixed in a 3:1 (vol/vol) methanol: acetic acid solution for 20 min. Finally, the cells were dropped onto ice-cold wet glass slides, air dried and stained with 10% Giemsa for 10 min. Images of the slides were captured under an Olympus IX81 FL microscope. At least 50 cells were counted in each experiment; the mean values are shown.
Cell viability and colony formation assay
An MTS Cytotoxicity Assay was performed in accordance with the manufacturer's recommendations (Promega). Briefly, cells (3000 cells/well) were plated in 96 well plates, treated with HU or CPT for 72 h, and then incubated with MTS for 2 h. The cell viability was determined by measuring the emission at 490 nm on an EPOCH2 microplate reader (BioTek). Colony formation assays were performed as previously described. For these experiments, 5000 cells were plated in 10 cm plates and were grown in complete media for 15 days before fixing with cold methanol and staining with 1% crystal violet.
Statistical analysis
GraphPad Prism 9 and Microsoft Excel software were used to perform data analysis, as applicable. Three independent experiments were performed in all cases. Significant differences between two groups were determined by unpaired Student's t-test (RT-qPCR, ChIP, and cell viability assays) or a Mann–Whitney U test (IF, PLA, DNA fiber). In all cases: n.s., P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Results
HELQ is quickly recruited to DSBs
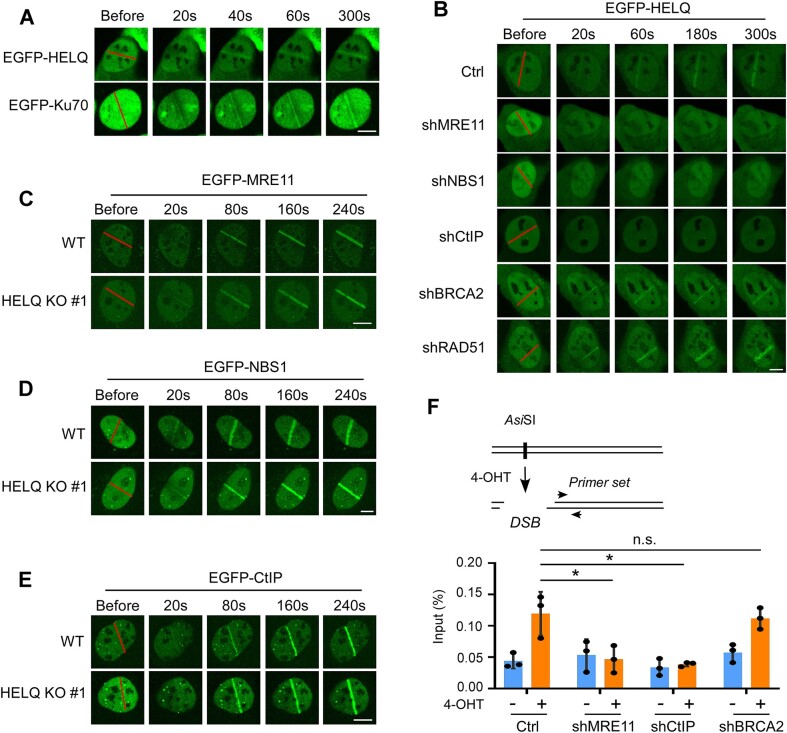
Live-cell imaging of fluorescently tagged proteins permits real-time monitoring of DNA repair factor recruitment to DNA damage sites. We leveraged this approach to address our first aim, to determine whether HELQ plays a role at the early stage of DSB repair. Specifically, we used CRISPR-Cas9 technology to introduce an EGFP fusion tag at the N terminal of endogenous Ku70, a widely studied early DSB response factor (Supplementary Figure S1A). As expected, EGFP-Ku70 was quickly recruited to laser-induced DSB damage sites, with a significant recruitment signal observed after 20 s (Figure 1A). Having demonstrated that EGFP-Ku70 is quickly recruited to DSBs, we compared these results with the recruitment pattern of HELQ, to assess whether HELQ can also be recruited to DSBs as quickly as Ku70. Interestingly, we observed that exogenously expressed EGFP-HELQ was also quickly recruited to DSB sites (Figure 1A), suggesting that HELQ serves a role at DSB sites during the early stages of DSB repair.
Figure 1.
HELQ is recruited to DSBs at the early stages of DSB repair. (A) Time-lapse imaging of EGFP-HELQ and EGFP-Ku70 in U2OS cells before and after microirradiation. The red line marks the damage region. (B) Time-lapse imaging of EGFP-HELQ in U2OS cells expressing vector control (Ctrl) or indicated shRNAs before and after microirradiation. C-E. EGFP-MRE11 (C), EGFP- NBS1 (D) and EGFP-CtIP (E) recruitment was monitored in U2OS (WT) and HELQ KO #1 cells. (F) Top, schematic of ChIP assay in ER-AsiSI U2OS cells. Bottom, SFB-HELQ enrichment at DSBs induced by AsiSI enzyme. ER-AsiSI U2OS cells expressing vector control (Ctrl) or the indicated shRNAs were treated with or without 4-OHT (300 nM, 4 h) to induce DSBs before a ChIP assay was performed using an anti-FLAG antibody. In panel A to E, scale bar = 5 μm. In panel F, the data are derived from three independent experiments and represent the means ± SD. *P < 0.05; n.s.: not significant. Student' s t-test was used.
Further analysis showed that individual shRNA-mediated depletion of end resection factors (MRE11, NBS1 or CtIP) but not recombination factors (RAD51 or BRCA2) significantly attenuated EGFP-HELQ recruitment to DSB sites (Figure 1B). By contrast, EGFP-MRE11, EGFP-NBS1 and EGFP-CtIP recruitment was unaffected in HELQ KO #1 cells prepared by CRISPR-Cas9 technology (Figure 1C–E, Supplementary Figure S1B), suggesting that HELQ is recruited to DSBs after resection factors but before recombination factors participate in DSB repair.
We next expressed ER-AsiSI (75,76) in U2OS cells, which would enable us to create DSBs at specific loci induced by the AsiSI restriction enzyme. These U2OS cells were engineered further such that endogenous HELQ contained a knocked-in S-protein-Flag-Streptavidin binding peptide (SFB) fusion tag (KI-SFB) at its N-terminal (Supplementary Figure S1C). Using this setup, we could investigate HELQ recruitment to the induced DSBs by chromatin immunoprecipitation (ChIP). We saw that SFB-HELQ was efficiently recruited to DSB-surrounding regions after 4-OHT induction, which induces the AsiSI enzyme to enter the nucleus and generate DSBs (Figure 1F). Consistent with our live-cell imaging data, MRE11 or CtIP but not BRCA2 depletion (by shRNA) impaired SFB-HELQ recruitment to DSBs (Figure 1F).
HELQ is required for the DSB end resection
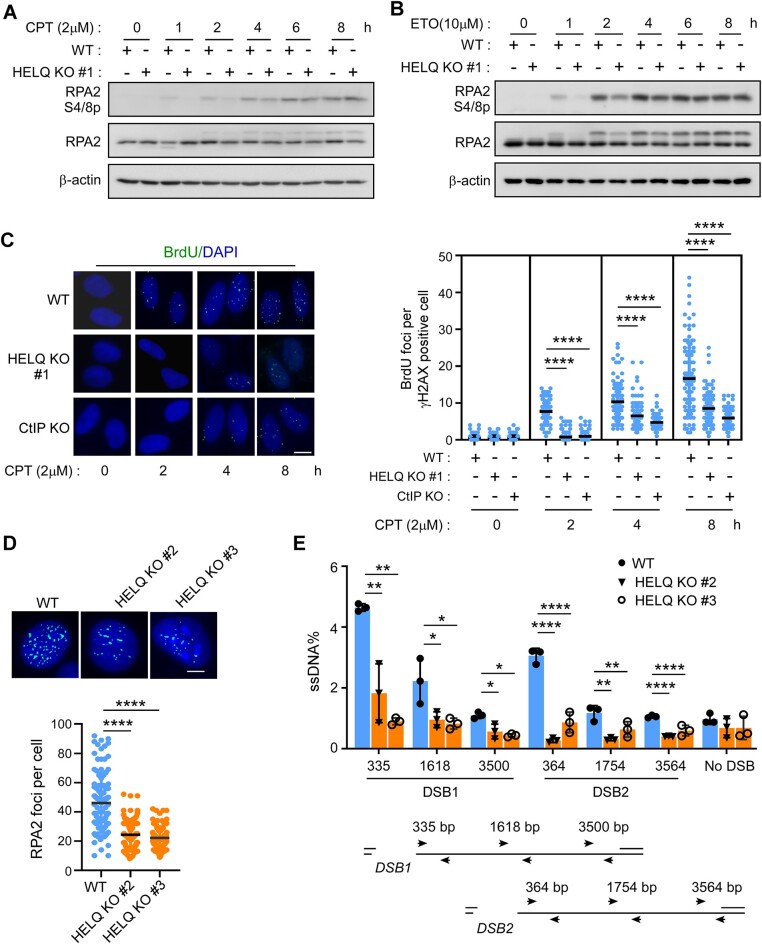
Having seen that HELQ is rapidly recruited to DSBs, we aimed to determine what role it plays during this early stage of DSB repair. As discussed, HR-mediated DSB repair begins with 5′-3′ end resection, with the resulting ssDNA attracting RPA binding and subsequent RPA2 S4/S8 phosphorylation (52). We therefore compared RPA2 phosphorylation levels after DSB induction between wild type and HELQ KO cells. Consistent with previous reports (62), long-term (> 4 h) camptothecin (CPT)- or etoposide (ETO)-induced RPA2 phosphorylation was not affected in HELQ KO #1 cells (Figure 2A and B). Surprisingly, however, short term (< 2 h) CPT- or ETO-induced RPA2 phosphorylation was affected, as evidenced by significantly lower phosphorylation levels in HELQ KO #1 cells than in wild-type cells (Figure 2A and B).
Figure 2.
HELQ is required for DSB end resection. (A, B) RPA2 phosphorylation was detected by western blotting after the indicated cells were treated with 2 μM camptothecin (CPT, A) or 10 μM etoposide (ETO, B) for the indicated times. (C) The cells were incubated with BrdU for 24 h, then treated with 2 μM CPT for the indicated times, fixed and then stained with BrdU and γH2AX antibodies under native conditions. Representative images (left) and the quantification of the average number of BrdU foci per γH2AX positive cell (right) are shown. scale bar = 10 μm. (D) Indicated cells were treated with CPT (2 μM, 1 h) followed by RPA2 foci formation analysis by immunostaining with the indicated antibodies. Representative images (top) and the quantification of the average number of RPA2 foci per cell (bottom) are shown. Scale bar = 5 μm. (E) Quantification and schematic of qPCR-based end resection assay in HELQ KO cells (#2 and #3). The data represent the means ± SD of three independent experiments. *P < 0.05, **P < 0.01, ****P < 0.0001. Mann–Whitney test was used in C and D; Student' s t-test was used in E.
CtIP is a classic DSB end resection factor, and the absence of CtIP can cause serious end resection defects (7,52). We prepared CtIP knockout cells (Supplementary Figure S2A) to compare the effects of deletion HELQ and deletion CtIP on DSB end resection. As shown in supplementary Figure S2B and C, knock-out of CtIP (CtIP KO) also only reduced RPA2 phosphorylation induced by short-term drug treatment. We reason that prolonged drug injury may cause global exhaustion of RPA (80), resulting in insufficient RPA recruitment to the DSB. RPA2 phosphorylation thus is no longer a reliable indicator for DSB end resection under prolonged drug treatment.
To verify this, we next labeled DNA with BrdU and measured DSB end resection by directly observing ssDNA. As predicted, ssDNA generation was significantly reduced in HELQ KO #1 cells to a comparable level as with CtIP KO, both after short-term and long-term drug treatment (Figure 2C and Supplementary Figure S2D). Next, we examined end resection after short-term CPT treatment in other two different HELQ KO cell lines (Supplementary Figure S1B). Here we saw that CPT-induced RPA2 phosphorylation was decreased in two other HELQ KO cell lines (Supplementary Figure S2E and F). DSB-induced RPA2 foci formation was also significantly reduced in these two HELQ KO cell lines (Figure 2D). Re-expression of HELQ (but not a vector control) in HELQ KO #1 cells could fully rescue CPT-induced-RPA2 phosphorylation (Supplementary Figure S2G), RPA2 foci formation (Supplementary Figure S2H) and we could thus confidently rule out the influence of off-target effects during gene editing.
To quantify the extent of DSB end resection, we generated ssDNA by DSB end resection at specific genomic AsiSI restriction-enzyme sites and then quantified them by q-PCR (76). To do so, we expressed ER-AsiSI in wild-type U2OS cells and our two HELQ KO cell lines (#2 and #3), generated DSB in these cells by 4-OHT induction and measured ssDNA production around two different DSB sites (DSB1 and DSB2). The resection levels at both DSB1 and DSB2 sites were reduced in the HELQ KO cells compared to the wild-type U2OS cells (Figure 2E). Together, these results established a role for HELQ in DSB end resection.
HELQ and EXO1 epistasis regulate DSB end resection
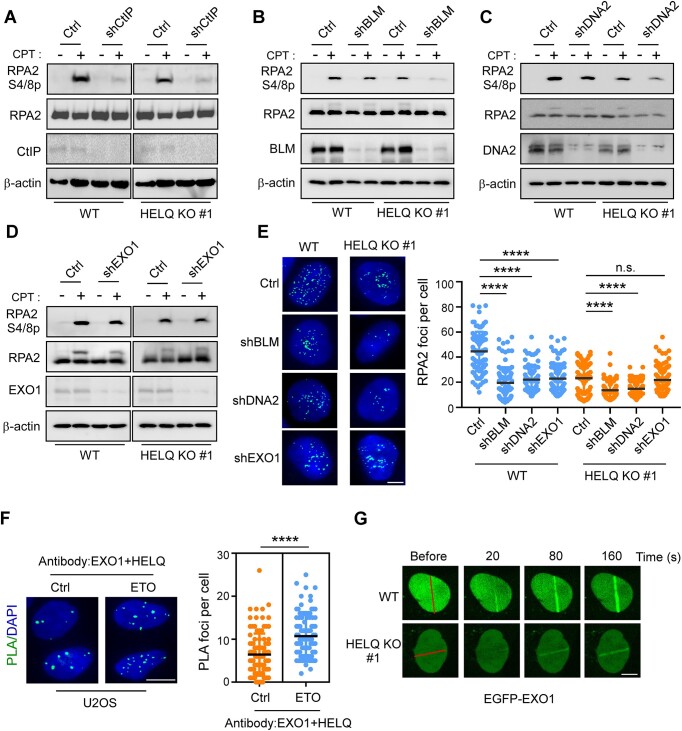
We next sought to ascertain whether HELQ promotes DSB end resection in collaboration with known resection molecular machinery comprising MRN/CtIP, BLM/DNA2 and EXO1. Consistent with previous reports (8,52), we observed DSB end resection defects in CtIP-, BLM-, DNA2-, or EXO1-depleted U2OS cells, as revealed by diminished RPA2 phosphorylation (Figure 3A–D) and RPA2 foci formation (Figure 3E). In HELQ KO #1 cells, CtIP, BLM or DNA2 depletion lead to a further decrease in RPA2 phosphorylation (Figure 3A–C) or RPA2 foci formation (Figure 3E). Yet notably, EXO1 depletion in HELQ KO #1 cells did not result in a further decrease in RPA2 phosphorylation (Figure 3D) or foci formation (Figure 3E) beyond the decrease elicited by HELQ alone. Further, we observed the formation of HELQ and EXO1 in situ PLA foci in U2OS cells, which significantly increased after DSB induction (Figure 3F), indicating that more HELQ and EXO1 would interact each other after DSB occurrence to cope with DSB damage. In addition, we observed that HELQ KO impaired EGFP-EXO1 recruitment to DSBs (Figure 3G). These data suggest that HELQ and EXO1 might operate in the same pathway to promote DSB end resection. Consistently, we found that HR was further reduced when BLM but not EXO1 was inactivated by shRNAs in the context of HELQ KO (Supplementary Figure S2I), indicating that HELQ and EXO1 also act on the same pathway to regulate HR.
Figure 3.
HELQ and EXO1 act on the same pathway to regulate DSB end resection. A-E. U2OS cells (WT) and HELQ KO #1 cells were infected with lentiviruses encoding the indicated shRNAs or a control vector (Ctrl). RPA2 phosphorylation analysis (A–D) and RPA2 foci formation analysis (E) were performed after CPT (2 μM, 1 h) treatment. Scale bar = 5 μm (E). (F) The HELQ-EXO1 interaction was analyzed by PLA in U2OS cells treated with or without ETO (10 μM, 1 h). Left, representative images. Right, quantification of the average number of PLA foci per nucleus. Scale bar = 10 μm. (G) Time-lapse imaging of EGFP-EXO1 in U2OS (WT) and HELQ KO #1 cells before and after microirradiation. The red line marks the damage region. Scale bar = 5 μm. In panel E and F, the data represent the means ± SD of three independent experiments. ****P < 0.0001, n.s.: not significant. Mann–Whitney test was used.
HELQ promotes EXO1 resection activity in vitro
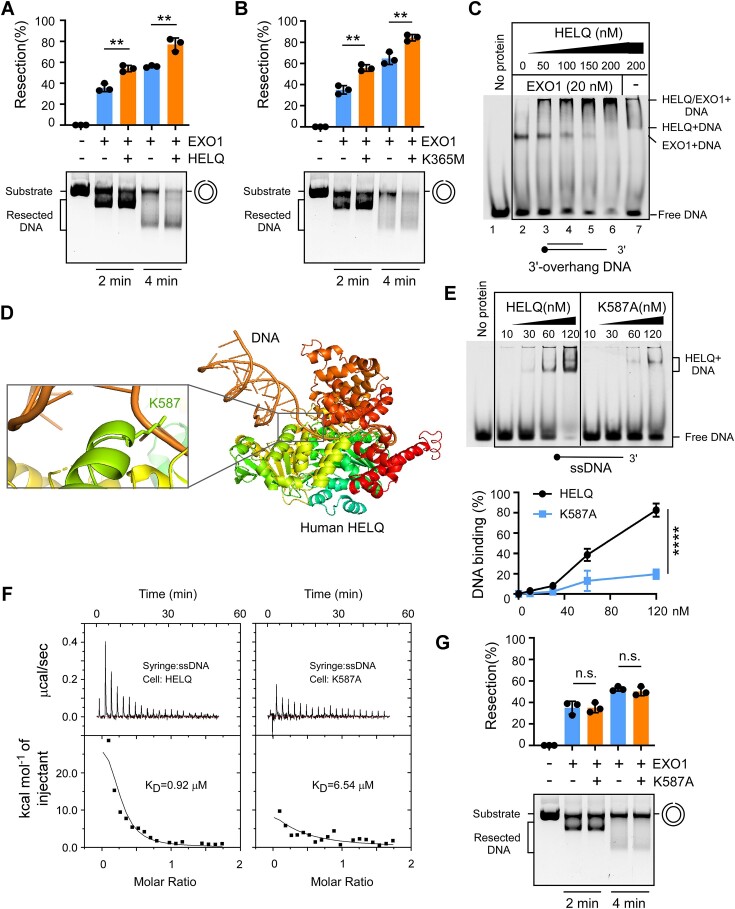
Thus far, our data suggest that HELQ promotes EXO1-mediated DSB end resection. We next probed further to understand whether HELQ directly affects 5′-3′ exonuclease activity, which drives end resection in cells. To do this, we purified human recombinant EXO1 and HELQ to homogeneity (Supplementary Figure S3A). We then monitored 5′-3′ EXO1 exonuclease activity using a nicked plasmid DNA substrate, as previously described (78). We saw that wild-type EXO1, but not a catalytically inactive mutant D173A showed exonuclease activity (Supplementary Figure S3B), indicating that there was no contaminant exonuclease activity in our preparation. We also detected EXO1 exonuclease activity on a linear double-stranded DNA substrate with a 3′ overhang (Supplementary Figure S3C). Consistent with the in vivo results, adding HELQ protein stimulated EXO1 exonuclease activity on both plasmid DNA and linear DNA substrate when compared to EXO1 alone (Figure 4A and Supplementary Figure S3C). Purified HELQ, but not a helicase-dead mutant of HELQ (K365M) (56), showed helicase activity (Supplementary Figure S3D and S3E). Interestingly, HELQ K365M still promoted EXO1 activity to the same extent as wild-type HELQ (Figure 4B), indicating that HELQ can promote EXO1 activity in a helicase activity-independent manner in vitro.
Figure 4.
HELQ promotes the nuclease activity of EXO1 in vitro. A-B. Nicked plasmid (7 nM) was incubated with purified wild type HELQ (A) or a helicase-dead HELQ mutant (K365M, (B)) in the presence or absence of EXO1 (15 nM) for the indicated times and the products were resolved on a 0.8% agarose gel. (C) 3′-overhang DNA substrate was incubated with the indicated concentration of HELQ and analyzed in 6% native polyacrylamide gel in the presence or absence of EXO1 (20 nM). Protein-DNA complexes are indicated. (D) A structural model of human HELQ-DNA, constructed by superimposing the structure of Archaeoglobus fulgidus HELQ-DNA with human HELQ using AlphaFold2. The enlarged area shows the interaction between human HELQ K587 and ssDNA. (E) Electrophoretic mobility shift assays such as (C) using HELQ variants and ssDNA substrate. The quantification of DNA binding capacity is shown in the bottom. (F) ITC titration and fitting curves of HELQ variants with ssDNA. (G) The effect of HELQ K587A on EXO1 nuclease as described in (A) and (B). In panels A, B, E and G, error bars represent means ± SD of three independent experiments. **P < 0.01, ****P < 0.0001, n.s.: not significant. Student's t-test.
We also performed gel mobility shift assays to investigate whether HELQ affects EXO1 DNA binding. We saw that both HELQ and EXO1 alone could bind to the 3′-overhang DNA (Figure 4C, line 2 and line 7). Upon adding HELQ protein, most of the labeled DNA migrated more slowly in the gel, forming a larger complex that was not observed with either protein preparation alone (Figure 4C, line 3 to line 6). HELQ thus seems to promote EXO1 binding to DNA and HELQ, DNA and EXO1 can form a complex.
We reasoned that HELQ binding to DNA may be important for EXO1 regulation, because we did not observe a strong interaction between purified HELQ and EXO1 protein in vitro. As no structure of human HELQ protein–DNA complex is available, we analyzed the binding of HELQ homologous proteins to DNA in other species. The crystal structure of HELQ from Archaeoglobus fulgidus (81) and Sulfolobus solfataricus (82) revealed five domains with a central cavity for ssDNA binding. Mutations in conserved arginine residues (such as R255 of HEL308 from Sulfolobus solfataricus) located in ssDNA through the central cavity substantially weakens HELQ binding to DNA (82). We modeled the three-dimensional human HELQ protein structure using AlphaFold 2.0. Superimposition of this structure and Archaeoglobus fulgidus HEL308 (PDB:2P6R) in PyMOL generated a root mean square deviation (rmsd) of 3.08 Å, indicating that the structure of human HELQ was similar to that of its homologue. We replaced HEL308 in the HEL308/ssDNA complex structure with human HELQ and obtained a simulated human HELQ/DNA complex structure from which we could observe HELQ binding to DNA. In this structure, K587 of human HELQ, corresponding to R252 in Archaeoglobus fulgidus HEL308 (Supplementary Figure S3F), was in close contact with ssDNA (Figure 4D), suggesting that K587 is essential for human HELQ binding to ssDNA.
We next purified the recombinant HELQ K587A mutant using the same procedure as the wild type (Supplementary Figure S3A). The HELQ K587A mutant was unable to function as a helicase (Supplementary Figure S3G) but showed weaker binding ability to ssDNA or 3′-overhang DNA than wild type HELQ, as demonstrated in gel shift assays (Figure 4E and Supplementary Figure S3H). We further assessed the ability of HELQ variants to bind to ssDNA using isothermal titration calorimetry (ITC). The KD values of wild type HELQ and K587A mutant binding with ssDNA were 0.92 and 6.54 μM, respectively (Figure 4F). This finding, consistent with the results of gel shift assays, indicates that mutating the K587 site weakens HELQ binding to DNA. Unlike the K365M mutant, the K587A mutant did not promote EXO1 activity (Figure 4G), suggesting that DNA binding ability, rather than helicase activity, is necessary for HELQ to promote EXO1 activity in vitro.
HELQ prevents resection of stressed replication fork
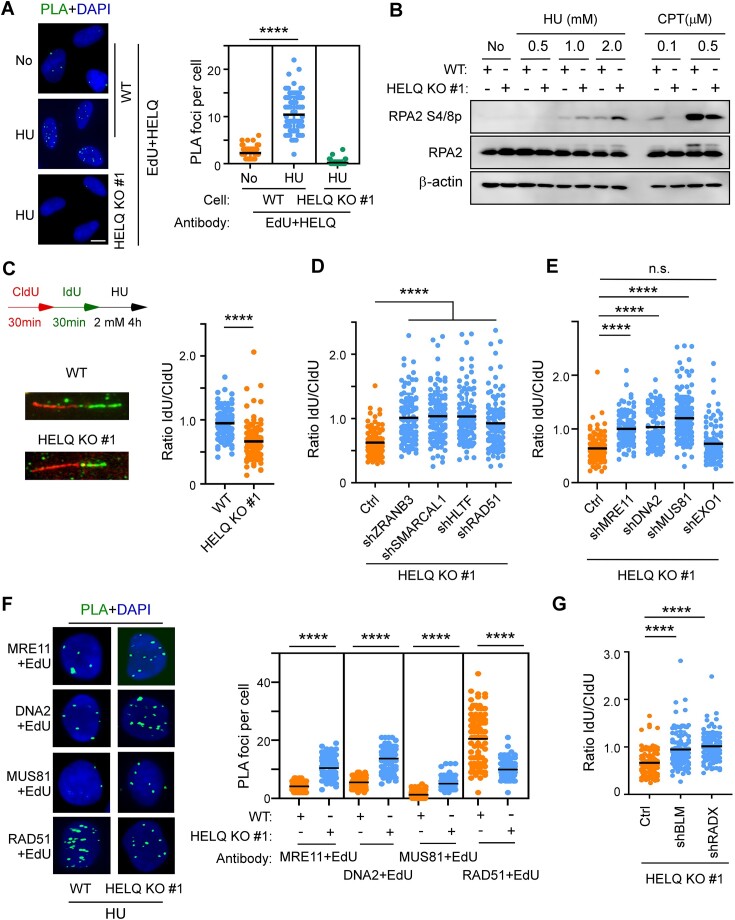
In response to replication stress, stalled forks undergo fork reversal, forming a ‘chicken foot’ structure containing a single end DSB that is susceptible to degradation by nucleases. DSB resection factors, such as MRE11, CtIP, BRCA1, participate in DNA metabolism at reversed forks either as protective factors or as degrading (non-protective) enzymes (10). We thus queried whether HELQ is also involved in degrading or protecting stalled forks as seen for other DSB resection factors. We first performed in situ PLA to test whether HELQ is recruited to stalled replication forks. Using antibodies against HELQ and RPA2, which binds and stabilizes ssDNA formed during DNA replication, we observed nuclear PLA foci in unstressed wild-type U2OS cells, which significantly increased in number upon treatment with hydroxyurea (HU), a drug that arrests replication forks (Supplementary Figure S4A). We next labeled cells with the thymidine analog EdU (5-ethynyl-29-deoxyuridine), then repeated the PLA this time using antibodies against HELQ and biotin, which labeled EdU by click reaction. HU treatment also increased the number of HELQ/EdU PLA foci in wild-type U2OS cells (Figure 5A), suggesting that HELQ is associated with replication proteins and nascent DNA during replication stress.
Figure 5.
HELQ prevents excessive resection of nascent DNA on the stalled forks. (A) Cells were labelled with EdU for 10 min prior to the addition of 2 mM HU for 4 h. The HELQ-nascent DNA (EdU) interaction was analyzed by PLA. Quantification of the average number of PLA foci per nucleus is shown on the right. Scale bar = 10 μm. (B) U2OS (WT) and HELQ KO #1 cells were treated with the indicated drugs (HU and CPT for 4 h) followed by RPA2 phosphorylation analysis by western blotting. (C) Fork degradation assays were performed in U2OS (WT) and HELQ KO #1 cells. Left, schematic of the assays and representative fiber images for the indicated samples. Right, the scatterplot of IdU-/CldU- tract length ratios for individual replication fork was shown. (D and E) The same fork degradation assays as in C were performed in HELQ KO #1 cells expressing vector control (Ctrl) or indicated shRNAs. The scatterplot of IdU-/CldU- tract length ratios for individual replication forks is shown. (F) Interactions between the indicated proteins and nascent DNA (EdU) were detected as described in A. Representative PLA images and quantification of the average number of PLA foci per nucleus detected by the indicated antibodies are shown. Scale bar = 5 μm. (G) Fork degradation assays as in C were performed in HELQ KO #1 cells expressing vector control (Ctrl) or the indicated shRNAs. A scatterplot of IdU-/CldU- tract length ratios for individual replication forks is shown. In panels A, C, D, E, F and G, the data are representative of at least three independent experiments. **** P < 0.0001, n.s.: not significant. Mann–Whitney test.
We probed the RPA2 S4/S8 phosphorylation levels to measure the resection on stalled replication forks induced by HU treatment or replication-associated DSBs induced by CPT (52,54,83). In contrast to CPT-induced stress, we saw an unexpected increase in RPA2 phosphorylation in HELQ KO (#1 and #2) cells exposed to HU-induced stress (Figure 5B and Supplementary Figure S4B). These findings suggest that HELQ limits DNA resection at stressed replication forks, which opposes its function in promotion DSB end resection as revealed earlier (Figure 2).
We next sought to investigate whether newly synthesized DNA is prone to being degraded by nucleases during replication stress. To this aim, we utilized a DNA fiber-labeling approach (36,49,79) to monitor the stability of nascent DNA. After HU treatment, we observed nascent DNA degradation in HELQ KO #1 (Figure 5C), suggesting that HELQ is involved in protecting stalled forks.
The SNF2 family DNA translocases, including ZRANB3, HLTF, SMARCAL1 and RAD51 (45,84–86), catalyze the reversal of stalled forks. We showed that depletion of each individual fork remodeler by shRNA reversed HU-induced fork degradation in HELQ KO #1 cells (Figure 5D and Supplementary Figure S4C), indicating that fork reversal is a pre-requisite for triggering nascent DNA degradation when HELQ is absent. We also silenced individual nucleases by shRNA in HELQ KO #1 cells to determine which nuclease is responsible for fork degradation. Here, down-regulation of MRE11, DNA2 or MUS81 but not EXO1, restored fork protection (Figure 5E and Supplementary Figure S4D), indicating that MRE11, DNA2 and MUS81 nucleases degrade nascent DNA in the absence of HELQ. Consistently, using PLA technique, we found that the binding of MRE11, DNA2 and MUS81 nucleases to nascent DNA (EdU) was also increased in HELQ KO #1 cells compared to wild-type U2OS cells under replication stress (Figure 5F).
RAD51 recruitment and stabilization at stalled replication forks is a central mechanism for replication fork protection (35). We thus asked whether HELQ has a role in recruiting or stabilizing RAD51 at stalled forks, like other fork protectors. In contrast to the nucleases, RAD51 recruitment to nascent DNA upon replication stress was reduced in HELQ KO #1 cells compared with wild-type cells (Figure 5F). Further, depleting anti-recombinase BLM or RADX, which antagonizes RAD51 accumulation at unprotected forks (87,88), restored RAD51 recruitment to nascent DNA (Supplementary Figure S4E and F) and fork protection in HELQ KO #1 cells (Figure 5G). These data suggest that HELQ modulates fork stability by suppressing the eviction of RAD51 from nascent DNA, which is mediated by an anti-recombinase or RADX.
HELQ ssDNA-binding ability is required for DSB end resection and stalled fork protection
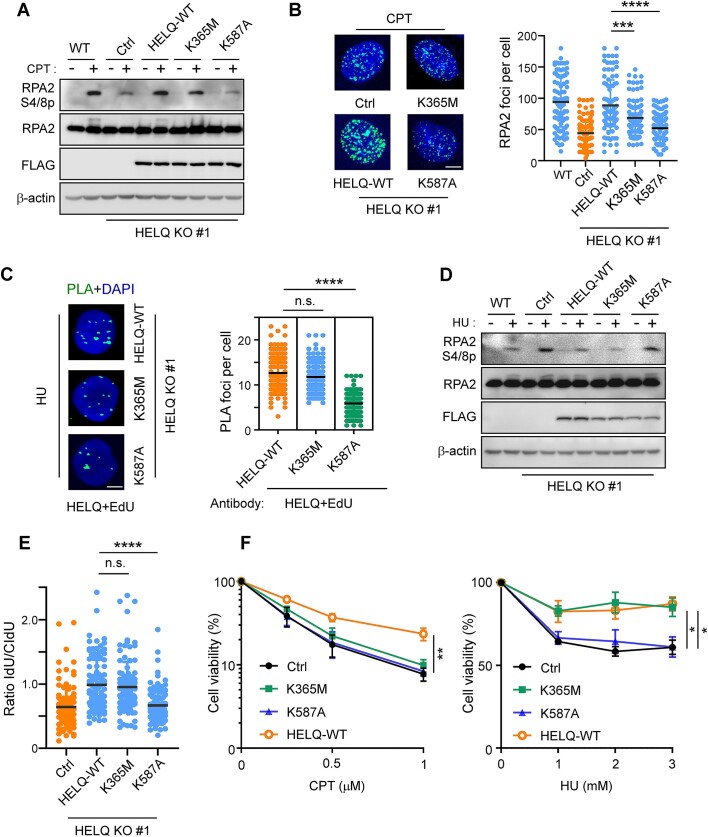
Thus far, we have shown that HELQ is involved in DSB end resection and replication fork protection, but the biochemical activities through which HELQ plays a role in these processes remained unclear. To address this gap, we first performed live-cell imaging to examine the recruitment of EGFP-tagged HELQ variants to DSBs upon laser microirradiation. HELQ K365M (helicase-dead) mutant, recruitment to DSBs was comparable to HELQ WT, but HELQ K587A (ssDNA binding deficient) mutant recruitment was significantly reduced (Supplementary Figure S5A). We then expressed FLAG-tagged HELQ wild-type, HELQ K365M or HELQ K587A mutants in HELQ KO #1 cells and based on CPT-induced RPA2 phosphorylation analysis (Figure 6A), RPA foci formation (Figure 6B), saw that DSB end resection was defective in the HELQ K365M and HELQ K587A mutant cells. Consistently, we introduced FLAG-tagged HELQ variants into EGFP-HR-HELQ KO cells and found that re-expression of HELQ-WT but not HELQ K365M or HELQ K587A mutant in HELQ KO cells could rescue HR defects (Supplementary Figure S5B). These data indicate that ssDNA-binding ability, but not helicase activity, is necessary for HELQ to recruit to DSB and promote DSB end resection and subsequent HR. Meanwhile, HELQ helicase activity promotes DSB end resection and HR. Thus while HELQ helicase is not necessary to promote EXO1 activity in vitro (Figure 4B), it is required to promote DSB end resection in cells. While the direct effects of HELQ on EXO1 can be revealed in vitro, some regulatory mechanisms may still affect EXO1 activity and end resection in vivo. We reason that HELQ helicase activity might promote end resection in cells through other indirect mechanisms.
Figure 6.
The ssDNA binding activity of HELQ is important for DSB end resection and fork protection. A-B. HELQ KO #1 cells expressing vector control or the indicated HELQ variants were subjected to CPT (2 μM, 2 h) induced RPA2 phosphorylation (A) and RPA2 foci formation (B) assays. Scale bar = 5 μm (B). (C–E) HELQ-EdU PLA assay, RPA2 phosphorylation (D) and fork degradation assay (E) (C-E: 2 mM HU for 4 h,) were performed in HELQ KO #1 cells expressing the indicated HELQ variants. Scale bar = 5 μm (C). F. HELQ KO #1 cells expressing vector control or the indicated HELQ variants were treated with the indicated concentrations of CPT (left) or HU (right) for 48 h, then a cell viability assay was performed. In panels B, C, E and F, data represent the means ± SD from three independent experiments. *P < 0.05, **P < 0.01, ****P < 0.0001. n.s.: not significant. Mann–Whitney test was used in B, C and D; Student's t-test was used in F.
We next determined the roles of HELQ variants in fork protection. We found that HELQ K587A recruitment to stalled forks also significantly reduced while the recruitment of HELQ K365M to stalled forks was comparable to that of HELQ WT (Figure 6C). HU induced RPA2 S4/S8 phosphorylation and nascent DNA degradation in HELQ KO #1 cells could be suppressed upon the re-expression of either HELQ WT or HELQ K365M but not HELQ K587A (Figure 6D and E). These data indicate that ssDNA-binding ability, but not helicase activity, is necessary for HELQ to recruit to stalled forks and suppress nascent DNA degradation.
Consistently, HELQ KO #1 cells expressing HELQ K587A mutant exhibited strong sensitivity to both HU and CPT, while HELQ KO #1 cells expressing HELQ K365M mutant exhibited strong sensitivity to CPT but similar HU sensitivity with HELQ KO #1 cells expressing HELQ-WT (Figure 6F). Further, knocking down HELQ (by CRISPR/Cas9) in HCT116 cell elevated chromosome aberrations induced by HU, while re-expression of HELQ-WT and HELQ K365M mutant but not HELQ K587A mutant suppressed chromosome aberrations (Supplementary Figure S5C).
Collectively, our data suggest that the ssDNA binding ability of HELQ is important for HELQ recruitment to stalled forks, protecting stalled forks from degradation and avoiding chromosome aberrations during replication stress. Meanwhile, HELQ helicase activity does not contribute to fork protection, differing from its roles in DSB end resection.
HELQ synergizes with CtIP to protect stalled forks
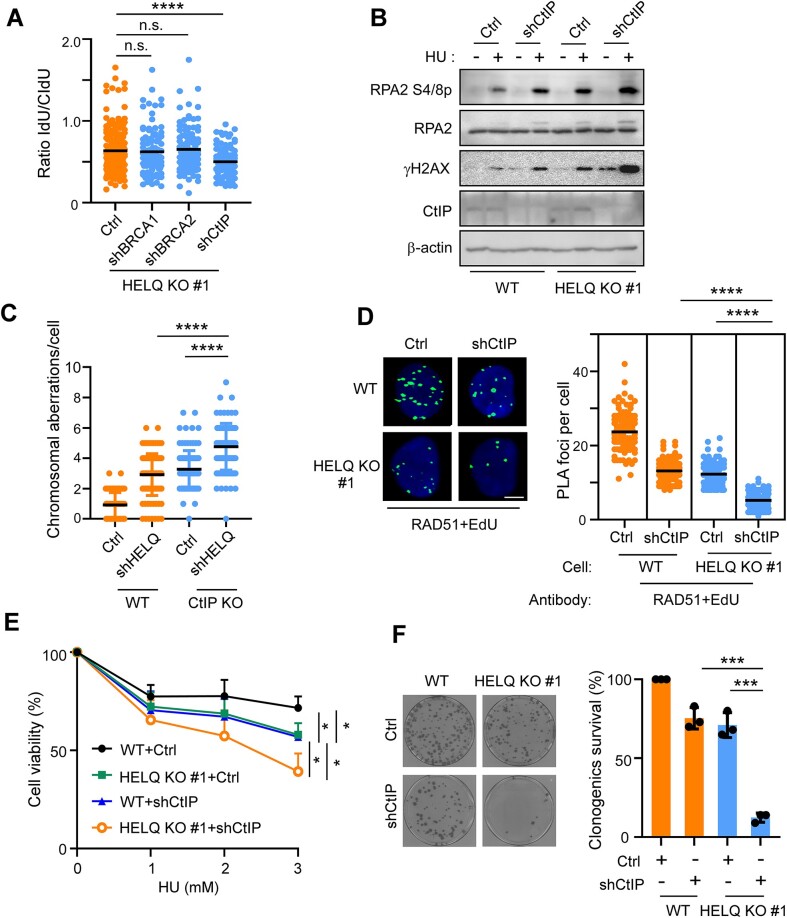
We finally sought to determine whether HELQ cooperates with other reported fork protectors in safeguarding stalled forks. Knocking down (by shRNA) BRCA1 or BRCA2 in HELQ KO #1 cells did not further aggravate the degradation of stalled forks, while knocking down of CtIP in HELQ KO #1 cells resulted in a further reduction of the CIdU-/IdU-tract length ratio (Figure 7A, Supplementary Figure S6A and B). Consistently, we observed that CtIP synergistically increased stalled forks resection in HELQ KO #1 cells (Figure 7B) and HELQ synergistically increased the extent of chromosome aberrations in CtIP KO cells upon replication stress (Figure 7C and Supplementary Figure S6C). These findings imply an additive genetic interaction between HELQ and CtIP, but not between HELQ and BRCA1 or BRCA2, in protecting stalled forks.
Figure 7.
HELQ functions in parallel to CtIP to preserve fork integrity. (A) Fork degradation assays were performed in HELQ KO #1 cells expressing the indicated shRNAs. (B) HU (2 mM, 4 h) induced RPA2 phosphorylation assays were performed in wild type (WT) and HELQ KO #1 cells expressing shCtIP or vector (Ctrl). (C) HCT116 cells (WT) and HCT116 CtIP KO cells (CtIP KO) expressing shHELQ or vector (Ctrl) were treated with 2 mM HU for 4 h and subjected to metaphase spread assay. The chromosomal aberrations per chromosome spread are plotted. (D) The HU (2 mM, 4 h) induced RAD51-nascent DNA (EdU) interaction was analyzed by PLA in U2OS cells (WT) and HELQ KO #1 cells expressing shCtIP or vector (Ctrl). Representative images and quantification of the average number of PLA foci per nucleus are shown. Scale bar = 5 μm. (E) WT and HELQ KO #1 cells expressing shCtIP or vector (Ctrl) were treated with the indicated concentration of HU for 48 h, then a cell viability assay was performed. (F) The viability of WT and HELQ KO #1 cells expressing shCtIP or vector (Ctrl) was analyzed by colony formation assay. Representative images (left) and quantifications of the relative survival (right) relative to vector (Ctrl) infected wild type U2OS cells (WT), which was arbitrarily set to 100%, are shown. In panels A, C, D, E and F, the data represent the means ± SD from at least three independent experiments. *P < 0.05, ***P < 0.001, ****P < 0.0001, n.s.: not significant. Mann–Whitney test was used in A, C and D; Student's t-test was used in E and F.
We also observed that knock down of CtIP (by shRNA) in wild-type U2OS cells attenuated RAD51 binding to nascent DNA under replication stress, as revealed by RAD51/EdU PLA foci formation. This finding is consistent with a recent report showing that CtIP protects stalled forks against enhanced fork degradation by promoting RAD51 nucleofilament stability (89). Knock down of CtIP in HELQ KO #1 cells further exacerbated the reduction of RAD51/EdU PLA foci caused by HELQ depletion (Figure 7D). These data support that HELQ functions in parallel with CtIP to stabilize RAD51 nucleofilaments on stalled forks, thereby protecting forks from nuclease degradation upon replication stress. Co-depletion of HELQ and CtIP lead to a higher sensitivity to HU (Figure 7E) and a significant reduction in clonogenic survival (Figure 7F) compared with cells depleted of either factor alone during unperturbed growth. These data indicate that HELQ and CtIP act on the different pathways to counteract endogenous replication stress.
Discussion
Dysregulated end resection at DSBs and stalled replication forks cause genome instability. In this study, we aimed to delineate the roles of HELQ in DSB end resection and replication forks protection. To do so, we conducted a series of in vitro and in vivo experiments. To this end, we demonstrated that HELQ has two opposing regulatory roles in DNA end resection occurring at DSB damage sites and stalled replication forks. We show that while HELQ promotes DSB end resection via modulating EXO1 nuclease activity, it also prevents fork degradation by stabilizing RAD51 binding to nascent DNA on reversed forks (Figure 8). Our findings are therefore consistent with previous demonstrations that DNA metabolism on DSB and stalled forks differ greatly in their regulation and underlying mechanisms.
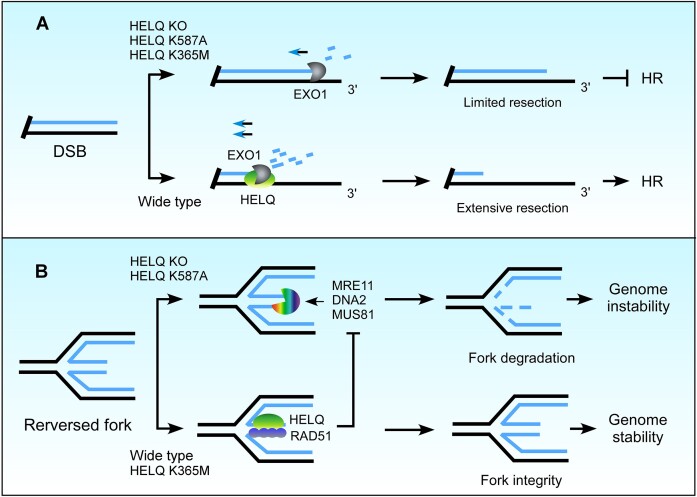
Figure 8.
A model illustrating the roles of HELQ in DSB end resection and fork protection. (A) HELQ promotes EXO1-mediated DSB end resection. In the presence of HELQ, EXO1 drives extensive end resection, which is necessary for HR. Without HELQ, EXO1 activity is not fully stimulated, and the ssDNA generated by end resection is insufficient to initiate HR. Both HELQ helicase activity and ssDNA binding ability are needed to promote EXO1 activity in cells. (B) HELQ prevents degradation of nascent DNA on reversed replication forks. HELQ stabilizes RAD51 at reversed forks and limits fork resection by nucleases. Loss of HELQ or its ssDNA binding capacity leads to fork degradation and genomic instability.
By performing live-cell imaging analyses, we showed that HELQ is recruited to DSB damage sites during the early stage of DSB repair where it promotes EXO1-mediated end resection and HR. EXO1 possesses 5′-3′ exonuclease activity that degrades 5′-terminated DNA strands within dsDNA to expose ssDNA (78). Human EXO1 is a processive nuclease (90), and its DSB end resection activity can be promoted by its interacting proteins such as MRN complex (9), BLM (9) proliferating cell nuclear antigen (PCNA) (91) or by the single-stranded DNA-binding protein sensor of single-stranded DNA complex 1 (SOSS1) (92). Although earlier reports suggested that RPA could also promote EXO1 resection activity in vitro (9), more recent studies have indicated that RPA is more likely to act as an EXO1 suppressor, reducing EXO1 recruitment to DSBs and inhibiting its DSB end resection activity (90,93,94). The DSB end resection activity of EXO1 thus seems to be modulated either by direct contact with the enzyme and substrate or by the removal of RPA. Consistent with this notion, plant homeodomain finger 11 (PHF11) can promote EXO1 activity only in the presence of RPA (93). As the HELQ K587A mutant, with no ssDNA binding ability, cannot stimulate EXO1 activity in vitro or in vivo (Figures 4G and 6A to B), our data support that HELQ represents another EXO1 cofactor that similar to SOSS1, binds to ssDNA and helps EXO1 perform efficient DSB end resection.
Interestingly, however, we saw that the HELQ K365M mutant, which loses helicases activity (56), behaves differently between in vitro and in vivo contexts. This mutant promoted EXO1 activity as efficiently as wild-type HELQ in vitro (Figure 4B), but in cells, it only partially restored DSB end resection activity that was lost due to HELQ KO (Figure 6A and B). This finding suggests that HELQ helicase activity is still required for intracellular DSB end resection, but it might act through some indirect mechanism. Others have reported that wild-type HELQ but not the K365M mutant can strip RPA from ssDNA in the presence of ATP in vitro (62). It is therefore reasonable to speculate that HELQ helicase activity might also be able to strip some RPA from DSB sites in cells to reduce the restriction of RPA on EXO1 activity. HELQ thus seems to promote EXO1-mediated end resection through two different pathways. Further work is now warranted to clarify how the different EXO1 regulators coordinate DSB end resection and promote genome stability.
Unlike for DSB end resection, the mechanisms underlying nascent DNA degradation at stalled forks are poorly understood. Our results shed light on this mechanism, showing that HELQ is recruited to DSBs and stalled replication forks, where like CtIP and BRCA1, it has opposing regulatory effects. This finding further demonstrates that DNA resection at the two types of damage sites are mediated by different mechanisms. Fork reversal is a pre-request for the degradation of forks that have lost protection. DSBs assembled by the annealing of nascent DNA on reversed forks attract nucleases and trigger DNA degradation. We saw that there is still substantial fork degradation in HELQ KO cells, and that silencing any one of several fork reversal enzymes (including SMARCAL1, ZRANB3, HLTF and even RAD51) restores fork protection to HELQ KO cells (Figure 5D). Thus, fork remodelling persists in HELQ KO cells and HELQ is not required for fork reversal. The formation of stable RAD51 nucleoprotein filaments on the regressed arm of reversed forks is essential for fork protection. HELQ KO resulted in increased ssDNA on stalled forks (Figure 5B) but significantly reduced RAD51 binding to nascent DNA (Figure 5F), indicating that HELQ could protect forks by stabilizing RAD51 nucleoprotein filaments. This effect is similar to that of other known fork protectors such as CtIP (89), BOD1L (95) and FANCD2 (36). Moreover, inactivating RADX or BLM, which destabilizes RAD51 nucleoprotein filaments (87,88), restored fork protection in HELQ KO cells further confirming that HELQ protects forks via RAD51 in vivo. Interestingly, however, our data also show that HELQ KO increases the binding of MRE11, DNA2 and MUS81 to nascent DNA on stalled forks (Figure 5F), suggesting that HELQ could also protect forks directly by preventing nuclease attack. Notably, the helicase activity of HELQ is not required for fork protection (Figure 6D and E) indicating that HELQ protects stalled forks through ssDNA binding activity or other biochemical activities related to ssDNA binding rather than through helicase activity.
Both DSB end resection and fork protection play a critical role in genome maintenance and tumor suppression. As such, our findings are useful in explaining how HELQ acts as a ‘caretaker’ (96) to reduce the incidence of mutations and tumors. It will now be interesting to identify which germline mutations in the HELQ gene are associated with cancer and determine whether these mutations affect DSB end resection and/or fork protection. This information will help us to clarify the underlying mechanisms by which HELQ suppresses tumorigenesis. So far, our data reveal functionally synergistic effects between HELQ and CtIP in reducing genomic instability and increasing cell viability upon replication stress (Figure 7C–F); this information will be helpful for developing new clinical therapeutic strategies for breast cancer containing CtIP mutations that do not affect DSB end resection efficiency, but lead to reduced fork protection (89). We posit that inhibitors that target HELQ-induced replication fork protection but not HELQ helicase activity would render such tumors more sensitive to chemotherapeutic agents that serve to induce replication stress. By the same principle, the development of a specific CtIP inhibitor should also facilitate the clinical treatment of patients with HELQ mutations.
Supplementary Material
Acknowledgements
We thank Dr Donyi Xu (School of Life Sciences, Peking University), Dr Tanya T. Paull (The University of Texas at Austin) and Dr Gaëlle Legube (University of Toulouse) for kindly providing reagents and Dr Xiaowei Pan (Capital Normal University) for her help in the simulation of protein structure. We also thank all members of the H.W. laboratory for their help and useful discussions.
Contributor Information
Yuqin Zhao, Beijing Key Laboratory of DNA Damage Response and College of Life Sciences, Capital Normal University, Beijing 100048, China.
Kaiping Hou, Beijing Key Laboratory of DNA Damage Response and College of Life Sciences, Capital Normal University, Beijing 100048, China.
Youhang Li, Beijing Key Laboratory of DNA Damage Response and College of Life Sciences, Capital Normal University, Beijing 100048, China.
Shuailin Hao, Beijing Key Laboratory of DNA Damage Response and College of Life Sciences, Capital Normal University, Beijing 100048, China.
Yu Liu, Beijing Key Laboratory of DNA Damage Response and College of Life Sciences, Capital Normal University, Beijing 100048, China.
Yinan Na, Beijing Key Laboratory of DNA Damage Response and College of Life Sciences, Capital Normal University, Beijing 100048, China.
Chao Li, Beijing Key Laboratory of DNA Damage Response and College of Life Sciences, Capital Normal University, Beijing 100048, China.
Jian Cui, Beijing Key Laboratory of DNA Damage Response and College of Life Sciences, Capital Normal University, Beijing 100048, China.
Xingzhi Xu, Guangdong Key Laboratory for Genome Stability & Disease Prevention and Carson International Cancer Center, Marshall Laboratory of Biomedical Engineering, China Shenzhen University School of Medicine, Shenzhen, Guangdong 518060, China.
Xiaohua Wu, Department of Molecular Medicine, The Scripps Research Institute, La Jolla, CA 92037, USA.
Hailong Wang, Beijing Key Laboratory of DNA Damage Response and College of Life Sciences, Capital Normal University, Beijing 100048, China.
Data availability
The data underlying this article are available in the article and in its online supplementary material.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Natural Science Foundation of China [32371354 and 31971221 to H.W.]; Beijing Natural Science Foundation [5182003 to H.W.]. Funding for open access charge: National Natural Science Foundation of China [grant number: 31971221 to H.W.].
Conflict of interest statement. None declared.
References
- 1. Aguilera A., Gomez-Gonzalez B.. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008; 9:204–217. [DOI] [PubMed] [Google Scholar]
- 2. White R.R., Vijg J.. Do DNA double-strand breaks drive aging?. Mol. Cell. 2016; 63:729–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gennery A.R., Cant A.J., Jeggo P.A.. Immunodeficiency associated with DNA repair defects. Clin. Exp. Immunol. 2000; 121:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McKinnon P.J. DNA repair deficiency and neurological disease. Nat. Rev. Neurosci. 2009; 10:100–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moynahan M.E., Jasin M.. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat. Rev. Mol. Cell Bio. 2010; 11:196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lieber M.R. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu. Rev. Biochem. 2010; 79:181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen L., Nievera C.J., Lee A.Y., Wu X.. Cell cycle-dependent complex formation of BRCA1.CtIP.MRN is important for DNA double-strand break repair. J. Biol. Chem. 2008; 283:7713–7720. [DOI] [PubMed] [Google Scholar]
- 8. Gravel S., Chapman J.R., Magill C., Jackson S.P.. DNA helicases Sgs1 and BLM promote DNA double-strand break resection. Genes Dev. 2008; 22:2767–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nimonkar A.V., Genschel J., Kinoshita E., Polaczek P., Campbell J.L., Wyman C., Modrich P., Kowalczykowski S.C.. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011; 25:350–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cejka P., Symington L.S.. DNA end resection: mechanism and control. Annu. Rev. Genet. 2021; 55:285–307. [DOI] [PubMed] [Google Scholar]
- 11. Gnugge R., Symington L.S.. DNA end resection during homologous recombination. Curr. Opin. Genet. Dev. 2021; 71:99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Symington L.S. End resection at double-strand breaks: mechanism and regulation. Cold Spring Harb. Perspect. Biol. 2014; 6:a016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao F., Kim W., Kloeber J.A., Lou Z.. DNA end resection and its role in DNA replication and DSB repair choice in mammalian cells. Exp. Mol. Med. 2020; 52:1705–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Symington L.S., Gautier J.. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011; 45:247–271. [DOI] [PubMed] [Google Scholar]
- 15. Wang H.L., Xu X.Z.. Microhomology-mediated end joining: new players join the team. Cell Biosci. 2017; 7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shibata A. Regulation of repair pathway choice at two-ended DNA double-strand breaks. Mutat. Res-Fund Mol. M. 2017; 803:51–55. [DOI] [PubMed] [Google Scholar]
- 17. Aparicio T., Baer R., Gautier J.. DNA double-strand break repair pathway choice and cancer. DNA Repair (Amst.). 2014; 19:169–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu T., Huang J.. DNA end resection: facts and mechanisms. Genom Proteom Bioinf. 2016; 14:126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huertas P. DNA resection in eukaryotes: deciding how to fix the break. Nat. Struct. Mol. Biol. 2010; 17:11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Higgins N.P., Kato K., Strauss B.. A model for replication repair in mammalian cells. J. Mol. Biol. 1976; 101:417–425. [DOI] [PubMed] [Google Scholar]
- 21. Neelsen K.J., Lopes M.. Replication fork reversal in eukaryotes: from dead end to dynamic response. Nat. Rev. Mol. Cell Biol. 2015; 16:207–220. [DOI] [PubMed] [Google Scholar]
- 22. Haber J.E., Ira G., Malkova A., Sugawara N.. Repairing a double-strand chromosome break by homologous recombination: revisiting Robin Holliday's model. Philos. Trans. Roy. Soc. B. 2004; 359:79–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Orrweaver T.L., Szostak J.W.. Yeast recombination - the association between double-strand gap repair and crossing-over. Proc. Natl. Acad. Sci. Biol. 1983; 80:4417–4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Filippo J.S., Sung P., Klein H.. Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 2008; 77:229–257. [DOI] [PubMed] [Google Scholar]
- 25. Szostak J.W., Orrweaver T.L., Rothstein R.J., Stahl F.W.. The double-strand-break repair model for recombination. Cell. 1983; 33:25–35. [DOI] [PubMed] [Google Scholar]
- 26. Thompson L.H., Schild D.. Homologous recombinational repair of DNA ensures mammalian chromosome stability. Mutat. Res-Fund. Mol. M. 2001; 477:131–153. [DOI] [PubMed] [Google Scholar]
- 27. Jasin M., Rothstein R.. Repair of Strand breaks by homologous recombination. Csh. Perspect. Biol. 2013; 5:a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wright W.D., Shah S.S., Heyer W.D.. Homologous recombination and the repair of DNA double-strand breaks. J. Biol. Chem. 2018; 293:10524–10535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ranjha L., Howard S.M., Cejka P.. Main steps in DNA double-strand break repair: an introduction to homologous recombination and related processes. Chromosoma. 2018; 127:187–214. [DOI] [PubMed] [Google Scholar]
- 30. Berti M., Cortez D., Lopes M.. The plasticity of DNA replication forks in response to clinically relevant genotoxic stress. Nat. Rev. Mol. Cell Biol. 2020; 21:633–651. [DOI] [PubMed] [Google Scholar]
- 31. Quinet A., Lemacon D., Vindigni A.. Replication fork reversal: players and guardians. Mol. Cell. 2017; 68:830–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liao H., Ji F., Helleday T., Ying S.. Mechanisms for stalled replication fork stabilization: new targets for synthetic lethality strategies in cancer treatments. EMBO Rep. 2018; 19:e46263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kondratick C.M., Washington M.T., Spies M.. Making choices: DNA replication fork recovery mechanisms. Semin. Cell Dev. Biol. 2021; 113:27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bhat K.P., Cortez D.. RPA and RAD51: fork reversal, fork protection, and genome stability. Nat. Struct. Mol. Biol. 2018; 25:446–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Thakar T., Moldovan G.L.. The emerging determinants of replication fork stability. Nucleic Acids Res. 2021; 49:7224–7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schlacher K., Wu H., Jasin M.. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012; 22:106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zadorozhny K., Sannino V., Belan O., Mlcouskova J., Spirek M., Costanzo V., Krejci L.. Fanconi-anemia-associated mutations destabilize RAD51 Filaments and impair replication fork protection. Cell Rep. 2017; 21:333–340. [DOI] [PubMed] [Google Scholar]
- 38. Ray Chaudhuri A., Callen E., Ding X., Gogola E., Duarte A.A., Lee J.E., Wong N., Lafarga V., Calvo J.A., Panzarino N.J.et al.. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 2016; 535:382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Feng W., Jasin M.. Homologous recombination and replication fork protection: BRCA2 and more!. Cold Spring Harbor Symp. Quant. Biol. 2017; 82:329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Billing D., Horiguchi M., Wu-Baer F., Taglialatela A., Leuzzi G., Nanez S.A., Jiang W., Zha S., Szabolcs M., Lin C.S.et al.. The BRCT domains of the BRCA1 and BARD1 tumor suppressors differentially regulate homology-directed repair and stalled fork protection. Mol. Cell. 2018; 72:127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Daza-Martin M., Starowicz K., Jamshad M., Tye S., Ronson G.E., MacKay H.L., Chauhan A.S., Walker A.K., Stone H.R., Beesley J.F.J.et al.. Isomerization of BRCA1-BARD1 promotes replication fork protection. Nature. 2019; 571:521–527. [DOI] [PubMed] [Google Scholar]
- 42. Rickman K.A., Noonan R.J., Lach F.P., Sridhar S., Wang A.T., Abhyankar A., Huang A., Kelly M., Auerbach A.D., Smogorzewska A.. Distinct roles of BRCA2 in replication fork protection in response to hydroxyurea and DNA interstrand cross-links. Genes Dev. 2020; 34:832–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rickman K., Smogorzewska A.. Advances in understanding DNA processing and protection at stalled replication forks. J. Cell Biol. 2019; 218:1096–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Thangavel S., Berti M., Levikova M., Pinto C., Gomathinayagam S., Vujanovic M., Zellweger R., Moore H., Lee E.H., Hendrickson E.A.et al.. DNA2 drives processing and restart of reversed replication forks in human cells. J. Cell Biol. 2015; 208:545–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mijic S., Zellweger R., Chappidi N., Berti M., Jacobs K., Mutreja K., Ursich S., Ray Chaudhuri A., Nussenzweig A., Janscak P.et al.. Replication fork reversal triggers fork degradation in BRCA2-defective cells. Nat. Commun. 2017; 8:859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kolinjivadi A.M., Sannino V., De Antoni A., Zadorozhny K., Kilkenny M., Techer H., Baldi G., Shen R., Ciccia A., Pellegrini L.et al.. Smarcal1-Mediated fork reversal triggers Mre11-dependent degradation of nascent DNA in the absence of Brca2 and stable Rad51 nucleofilaments. Mol. Cell. 2017; 67:867–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rondinelli B., Gogola E., Yucel H., Duarte A.A., van de Ven M., van der Sluijs R., Konstantinopoulos P.A., Jonkers J., Ceccaldi R., Rottenberg S.et al.. EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat. Cell Biol. 2017; 19:1371–1378. [DOI] [PubMed] [Google Scholar]
- 48. Mason J.M., Chan Y.L., Weichselbaum R.W., Bishop D.K.. Non-enzymatic roles of human RAD51 at stalled replication forks. Nat. Commun. 2019; 10:4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M.. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011; 145:529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Halder S., Sanchez A., Ranjha L., Reginato G., Ceppi I., Acharya A., Anand R., Cejka P.. Double-stranded DNA binding function of RAD51 in DNA protection and its regulation by BRCA2. Mol. Cell. 2022; 82:3553–3565. [DOI] [PubMed] [Google Scholar]
- 51. Cruz-Garcia A., Lopez-Saavedra A., Huertas P.. BRCA1 accelerates CtIP-mediated DNA-end resection. Cell Rep. 2014; 9:451–459. [DOI] [PubMed] [Google Scholar]
- 52. Sartori A.A., Lukas C., Coates J., Mistrik M., Fu S., Bartek J., Baer R., Lukas J., Jackson S.P.. Human CtIP promotes DNA end resection. Nature. 2007; 450:509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schlegel B.P., Jodelka F.M., Nunez R.. BRCA1 promotes induction of ssDNA by ionizing radiation. Cancer Res. 2006; 66:5181–5189. [DOI] [PubMed] [Google Scholar]
- 54. Przetocka S., Porro A., Bolck H.A., Walker C., Lezaja A., Trenner A., von Aesch C., Himmels S.F., D’Andrea A.D., Ceccaldi R.et al.. CtIP-mediated fork protection synergizes with BRCA1 to suppress genomic instability upon DNA replication stress. Mol. Cell. 2018; 72:568–582. [DOI] [PubMed] [Google Scholar]
- 55. Fairman-Williams M.E., Guenther U.P., Jankowsky E.. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 2010; 20:313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Marini F., Wood R.D.. A human DNA helicase homologous to the DNA cross-link sensitivity protein Mus308. J. Biol. Chem. 2002; 277:8716–8723. [DOI] [PubMed] [Google Scholar]
- 57. Adelman C.A., Lolo R.L., Birkbak N.J., Murina O., Matsuzaki K., Horejsi Z., Parmar K., Borel V., Skehel J.M., Stamp G.et al.. HELQ promotes RAD51 paralogue-dependent repair to avert germ cell loss and tumorigenesis. Nature. 2013; 502:381–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Luebben S.W., Kawabata T., Akre M.K., Lee W.L., Johnson C.S., O'Sullivan M.G., Shima N.. Helq acts in parallel to Fancc to suppress replication-associated genome instability. Nucleic Acids Res. 2013; 41:10283–10297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Takata K., Reh S., Tomida J., Person M.D., Wood R.D.. Human DNA helicase HELQ participates in DNA interstrand crosslink tolerance with ATR and RAD51 paralogs. Nat. Commun. 2013; 4:2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhu F., Yang S., Lei M., He Q., Wu L., Zhang Y.. DNA repair protein HELQ and XAB2 as chemoresponse and prognosis biomarkers in ascites tumor cells of high-grade serous ovarian cancer. J. Oncol. 2022; 2022:7521934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hustedt N., Saito Y., Zimmermann M., Alvarez-Quilon A., Setiaputra D., Adam S., McEwan A., Yuan J.Y., Olivieri M., Zhao Y.et al.. Control of homologous recombination by the HROB-MCM8-MCM9 pathway. Genes Dev. 2019; 33:1397–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Anand R., Buechelmaier E., Belan O., Newton M., Vancevska A., Kaczmarczyk A., Takaki T., Rueda D.S., Powell S.N., Boulton S.J.. HELQ is a dual-function DSB repair enzyme modulated by RPA and RAD51. Nature. 2022; 601:268–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Cong L., Ran F.A., Cox D., Lin S., Barretto R., Habib N., Hsu P.D., Wu X., Jiang W., Marraffini L.A.et al.. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339:819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M.. RNA-guided human genome engineering via Cas9. Science. 2013; 339:823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang H., Qiu Z., Liu B., Wu Y., Ren J., Liu Y., Zhao Y., Wang Y., Hao S., Li Z.et al.. PLK1 targets CtIP to promote microhomology-mediated end joining. Nucleic Acids Res. 2018; 46:10724–10739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hao S., Wang Y., Zhao Y., Gao W., Cui W., Li Y., Cui J., Liu Y., Lin L., Xu X.et al.. Dynamic switching of crotonylation to ubiquitination of H2A at lysine 119 attenuates transcription-replication conflicts caused by replication stress. Nucleic Acids Res. 2022; 50:9873–9892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wang H., Li S., Oaks J., Ren J., Li L., Wu X.. The concerted roles of FANCM and Rad52 in the protection of common fragile sites. Nat. Commun. 2018; 9:2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang H., Shi L.Z., Wong C.C., Han X., Hwang P.Y., Truong L.N., Zhu Q., Shao Z., Chen D.J., Berns M.W.et al.. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS Genet. 2013; 9:e1003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Truong L.N., Li Y., Shi L.Z., Hwang P.Y., He J., Wang H., Razavian N., Berns M.W., Wu X.. Microhomology-mediated end joining and homologous recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:7720–7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang H., Li S., Zhang H., Wang Y., Hao S., Wu X.. BLM prevents instability of structure-forming DNA sequences at common fragile sites. PLoS Genet. 2018; 14:e1007816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Wu X., Wang B.. Abraxas suppresses DNA end resection and limits break-induced replication by controlling SLX4/MUS81 chromatin loading in response to TOP1 inhibitor-induced DNA damage. Nat. Commun. 2021; 12:4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tian T., Bu M., Chen X., Ding L., Yang Y., Han J., Feng X.H., Xu P., Liu T., Ying S.et al.. The ZATT-TOP2A-PICH axis drives extensive replication fork reversal to promote genome stability. Mol. Cell. 2021; 81:198–211. [DOI] [PubMed] [Google Scholar]
- 73. Di Marco S., Hasanova Z., Kanagaraj R., Chappidi N., Altmannova V., Menon S., Sedlackova H., Langhoff J., Surendranath K., Huhn D.et al.. RECQ5 Helicase cooperates with MUS81 endonuclease in processing stalled replication forks at common fragile sites during mitosis. Mol. Cell. 2017; 66:658–671. [DOI] [PubMed] [Google Scholar]
- 74. Adolph M.B., Mohamed T.M., Balakrishnan S., Xue C., Morati F., Modesti M., Greene E.C., Chazin W.J., Cortez D.. RADX controls RAD51 filament dynamics to regulate replication fork stability. Mol. Cell. 2021; 81:1074–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Iacovoni J.S., Caron P., Lassadi I., Nicolas E., Massip L., Trouche D., Legube G.. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010; 29:1446–1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zhou Y., Caron P., Legube G., Paull T.T.. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic Acids Res. 2014; 42:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. El-Shemerly M., Janscak P., Hess D., Jiricny J., Ferrari S.. Degradation of human exonuclease 1b upon DNA synthesis inhibition. Cancer Res. 2005; 65:3604–3609. [DOI] [PubMed] [Google Scholar]
- 78. Eid W., Steger M., El-Shemerly M., Ferretti L.P., Pena-Diaz J., Konig C., Valtorta E., Sartori A.A., Ferrari S.. DNA end resection by CtIP and exonuclease 1 prevents genomic instability. EMBO Rep. 2010; 11:962–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Nieminuszczy J., Schwab R.A., Niedzwiedz W.. The DNA fibre technique - tracking helicases at work. Methods. 2016; 108:92–98. [DOI] [PubMed] [Google Scholar]
- 80. Toledo L.I., Altmeyer M., Rask M.B., Lukas C., Larsen D.H., Povlsen L.K., Bekker-Jensen S., Mailand N., Bartek J., Lukas J.. ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell. 2013; 155:1088–1103. [DOI] [PubMed] [Google Scholar]
- 81. Buttner K., Nehring S., Hopfner K.P.. Structural basis for DNA duplex separation by a superfamily-2 helicase. Nat. Struct. Mol. Biol. 2007; 14:647–652. [DOI] [PubMed] [Google Scholar]
- 82. Richards J.D., Johnson K.A., Liu H., McRobbie A.M., McMahon S., Oke M., Carter L., Naismith J.H., White M.F.. Structure of the DNA repair helicase hel308 reveals DNA binding and autoinhibitory domains. J. Biol. Chem. 2008; 283:5118–5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Petermann E., Orta M.L., Issaeva N., Schultz N., Helleday T.. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 2010; 37:492–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Betous R., Mason A.C., Rambo R.P., Bansbach C.E., Badu-Nkansah A., Sirbu B.M., Eichman B.F., Cortez D.. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 2012; 26:151–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ciccia A., Nimonkar A.V., Hu Y., Hajdu I., Achar Y.J., Izhar L., Petit S.A., Adamson B., Yoon J.C., Kowalczykowski S.C.et al.. Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol. Cell. 2012; 47:396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kile A.C., Chavez D.A., Bacal J., Eldirany S., Korzhnev D.M., Bezsonova I., Eichman B.F., Cimprich K.A.. HLTF’s ancient HIRAN domain binds 3' DNA ends to drive replication fork reversal. Mol. Cell. 2015; 58:1090–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Dungrawala H., Bhat K.P., Le Meur R., Chazin W.J., Ding X., Sharan S.K., Wessel S.R., Sathe A.A., Zhao R., Cortez D.. RADX promotes genome stability and modulates chemosensitivity by regulating RAD51 at replication forks. Mol. Cell. 2017; 67:374–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Patel D.S., Misenko S.M., Her J., Bunting S.F.. BLM helicase regulates DNA repair by counteracting RAD51 loading at DNA double-strand break sites. J. Cell Biol. 2017; 216:3521–3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zarrizi R., Higgs M.R., Vossgrone K., Rossing M., Bertelsen B., Bose M., Kousholt A.N., Rosner H., Network T.C., Ejlertsen B.et al.. Germline RBBP8 variants associated with early-onset breast cancer compromise replication fork stability. J. Clin. Invest. 2020; 130:4069–4080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Myler L.R., Gallardo I.F., Zhou Y., Gong F., Yang S.H., Wold M.S., Miller K.M., Paull T.T., Finkelstein I.J.. Single-molecule imaging reveals the mechanism of Exo1 regulation by single-stranded DNA binding proteins. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:E1170–E1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Chen X., Paudyal S.C., Chin R.I., You Z.. PCNA promotes processive DNA end resection by Exo1. Nucleic Acids Res. 2013; 41:9325–9338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Yang S.H., Zhou R., Campbell J., Chen J., Ha T., Paull T.T.. The SOSS1 single-stranded DNA binding complex promotes DNA end resection in concert with Exo1. EMBO J. 2013; 32:126–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Gong Y., Handa N., Kowalczykowski S.C., de Lange T.. PHF11 promotes DSB resection, ATR signaling, and HR. Genes Dev. 2017; 31:46–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Genschel J., Modrich P.. Mechanism of 5'-directed excision in human mismatch repair. Mol. Cell. 2003; 12:1077–1086. [DOI] [PubMed] [Google Scholar]
- 95. Higgs M.R., Reynolds J.J., Winczura A., Blackford A.N., Borel V., Miller E.S., Zlatanou A., Nieminuszczy J., Ryan E.L., Davies N.J.et al.. BOD1L Is required to suppress deleterious resection of stressed replication forks. Mol. Cell. 2015; 59:462–477. [DOI] [PubMed] [Google Scholar]
- 96. Kinzler K.W., Vogelstein B.. Cancer-susceptibility genes. Gatekeepers and caretakers. Nature. 1997; 386:761. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.