Abstract
Hepatitis C virus (HCV) requires two cellular factors, microRNA-122 (miR-122) and poly(C) binding protein 2 (PCBP2), for optimal replication. These host factors compete for binding to the 5′ end of the single-stranded RNA genome to regulate the viral replication cycle. To understand how they interact with the RNA, we measured binding affinities of both factors for an RNA probe representing the 5′ 45 nucleotides of the HCV genome (HCV45). Isothermal titration calorimetry revealed two, unequal miR-122 binding sites in HCV45, high-affinity (S1) and low-affinity (S2), differing roughly 100-fold in binding affinity. PCBP2 binds a site overlapping S2 with affinity similar to miR-122 binding to S2. PCBP2 circularizes the genome by also binding to the 3′ UTR, bridging the 5′ and 3′ ends of the genome. By competing with PCBP2 for binding at S2, miR-122 disrupts PCBP2-mediated genome circularization. We show that the viral RNA-dependent RNA polymerase, NS5B, also binds to HCV45, and that the binding affinity of NS5B is increased in the presence of miR-122, suggesting miR-122 promotes recruitment of the polymerase. We propose that competition between miR-122 and PCBP2 for HCV45 functions as a translation-to-replication switch, determining whether the RNA genome templates protein synthesis or RNA replication.
Graphical Abstract
Graphical Abstract.
Introduction
Hepatitis C Virus (HCV) is a human hepacivirus that can persist for decades in the liver, greatly increasing the risk of liver cirrhosis and hepatocellular carcinoma (1). HCV is a member of the family Flaviviridae, which includes hepaciviruses, flaviviruses such as dengue virus, and pestiviruses such as bovine viral diarrhea virus. HCV is an enveloped virus with a single stranded, positive-sense RNA genome of ∼9600 nucleotides, consisting of a single open reading frame (ORF) flanked by two highly structured untranslated regions (UTRs) at the 5′ and 3′ ends. The ORF is translated into a single ∼3000-amino-acid-long polyprotein that is cleaved by cellular and viral proteases into three structural proteins (C, E1 and E2) and seven nonstructural proteins (p7, NS2, NS3, NS4A, NS4B, NS5A and NS5B) (Figure 1A). Functions of several nonstructural proteins are known. NS3 and NS4A form a protease that processes the HCV polyprotein (2). NS5B is the viral RNA-dependent-RNA-polymerase (RdRp), responsible for viral genome synthesis. As a positive-sense RNA virus, the HCV genome is used directly as a template for both protein synthesis and negative-strand RNA synthesis. The negative strand is then used as a template for synthesis of genomic positive-strand RNA (3).
Figure 1.
Location of miR-122 and PCBP2 binding sites at the 5′ end of the HCV genome. (A) Schematic of the HCV genome. The HCV genome consists of a single open reading frame (ORF) flanked by highly structured, untranslated regions at the 5′ and 3′ ends (5′ and 3′ UTRs). Stem-loop structures at the 5′ and 3′ UTRs are indicated. Structural proteins and non-structural proteins are colored by blue and orange, respectively. (B) The miR-122 and PCBP2 binding sites in the 5′ UTR of the HCV genome. Nucleotides 1–45 of the HCV genome contain two tandem miR-122 binding sites (blue) and a PCBP2 binding site (cyan) that overlaps with the second miR-122 binding site. (C) Two miR-122 molecules bind to the 45 nucleotides of HCV RNA. HCV45 (1 μM) was incubated with the increasing amounts of miR-122, 0–3 μM in steps of 0.5 μM in presence of 5 mM Mg2+, and analysed by non-denaturing gel electrophoresis mobility shift assay. The number of miR-122 in the HCV45 complex is indicated.
The 5′UTR is folded into four stem–loop structures and is involved in both viral translation and replication (Figure 1A). The 5′ terminal 120 nucleotides, containing stem–loops I and II, have been shown to be critical for RNA synthesis (4,5). Stem-loops II, III and IV form an internal ribosome entry site (IRES) critical for viral translation, as the HCV genome lacks a cap to otherwise interact with the host ribosome (4). Further, interactions of the 5′UTR with host factors microRNA-122 (miR-122) and poly(C) binding protein 2 (PCBP2) have been shown to be important for viral replication (3,6–8). miR-122 is an abundant liver-specific microRNA (miRNA) that is involved with the homeostasis of hepatocytes. miR-122 is associated with the Argonaute 2 (Ago2) protein as part of the RNA-induced silencing complex (RISC). Canonically, miR-122 binds to the 3′UTR of cognate mRNAs, guiding the RISC to repress translation of the target. However, two copies of miR-122 bind HCV at tandem binding sites (S1 and S2) near the 5′ terminus of the HCV genome (8–10). This noncanonical binding to HCV RNA has been shown to promote HCV replication through various mechanisms. The binding of miR-122 stabilizes the viral RNA by protecting it from 5′ exoribonuclease activity (11–15), promotes viral RNA synthesis (16,17) and promotes viral translation by preventing the formation of an alternative stem–loop II conformation, stabilizing the IRES (18–21). Each of these mechanisms is insufficient on its own to explain miR-122′s effect (22). Thus, the promotion of HCV replication by miR-122 appears to utilize several different mechanisms. It is also not clear how the interplay of these mechanisms changes over the course of the viral lifecycle. For instance, when miR-122 is supplemented into HCV infected cells depleted of the 5′ exoribonuclease XRN1, nascent viral RNA significantly accumulates within 1 hr, while nascent viral protein synthesis is delayed for several hours suggesting that miR-122′s relative contribution to translation and RNA synthesis may shift over time (16).
PCBP2 has also been described as a pro-viral HCV host factor and trans-activator of HCV IRES activity (7). PCBP2 consists of three hnRNP K homology (KH) domains, and binds to single stranded, C-rich RNA sequences (23). PCBP2 binds to the 5′UTR of HCV and regulates viral translation. HCV translation is inhibited in cells with suppressed PCBP2 levels (7). Additionally, miR-122 promotion of viral RNA synthesis requires the presence of PCBP2 in cells, as the pro-virus miR-122 effect was lost in PCBP2 knockdown cells (16). PCBP2 has been shown to mediate circularization of positive-strand HCV RNA by forming a protein bridge between the 5′ and 3′ UTRs (7,16). The PCBP2 binding site in the HCV 5′UTR overlaps with the second miR-122 binding site, S2 (Figure 1B). However, the mechanism by which the competition of PCBP2 and miR-122 promotes HCV replication is not known. We characterized the binding of miR-122 and PCBP2 to an RNA probe representing the 5′ terminal 45 nucleotides of the 5′UTR (HCV45) and how they affect viral replication. Our results indicate that miR-122 and PCBP2 bind to HCV45 with similar affinities. PCBP2 binding was inhibited in the presence of miR-122, indicating that miR-122 competes with PCBP2 for binding to HCV45. We show that miR-122 binding to 5′UTR reverses PCBP2-mediated genome circularization, and that the HCV RdRp NS5B binds the HCV45:miR-122 complex with higher affinity than HCV45 RNA alone. Our results suggest that the 5′UTR functions as a molecular switch that regulates whether the HCV genome is used as a template for translation or RNA synthesis by binding miR-122 or PCBP2, coordinating theses competing molecular processes. This provides a mechanism by which miR-122 promotes HCV replication.
Materials and methods
Reagents and buffers
HCV RNAs (GenBank accession no. NC_038882.1) containing the 5′ terminal 45 nucleotides of the HCV genome (HCV45) with and without a 5′ fluorescein tag, HCV45 with substitutions at positions 25 and 26 (HCVS1p34), HCV nts 21 through 65 (HCV21-65) and nucleotides 1–28 of HCV RNA followed by the reverse complement of nucleotides 29–45 (HCVS2,comp) were synthesized by Sigma-Adrich corporation (The Woodlands, TX) (Table 1). The 5′ terminal 47 nucleotides of the HCV genome (HCV47) and miR-122tandemS2m were synthesized by Integrated DNA Technologies (Coralville, IA) (Table 1). miR-122 (GenBank accession no. NR_029667), miR-124 (GenBank accession no. NR_029668), miR-122trunc, miR-122comp and miR-122tandem were also synthesized by Sigma Aldrich (Table 1). The concentrations of nucleic acids were determined using the nearest-neighbors model to calculate the extinction coefficients at 260 nm (24). An HCV 5′ end oligo (47 nt) was synthesized with biotin at its 3′ end (HCV47-biotin) by Integrated DNA Technologies (Coralville, IA).
Table 1.
RNA sequences used for ITC and anisotropy experiments
| Name | Sequence (5′ to 3′) |
|---|---|
| HCV45 | GCCAGCCCCCUGAUGGGGGCGACACUCCACCAUGAAUCACUCCCC |
| HCV47 | GCCAGCCCCCUGAUGGGGGCGACACUCCACCAUGAAUCACUCCCCUG |
| HCV21-65 | GACACUCCACCAUGAAUCACUCCCCUGUGAGGAACUACUGUCUUC |
| HCVS2-comp | GCCAGCCCCCUGAUGGGGGCGACACUCCAGGUACUUAGUGAGGGGAC |
| HCVS1p34 | GCCAGCCCCCUGAUGGGGGCGACACAGCACCAUGAAUCACUCCCC |
| miR-122 | UGGAGUGUGACAAUGGUGUUUG |
| miR-122comp | ACCUCACACUGUUACCACAAAC |
| miR-122trunc | UGGAGUGUGACAAUGGU |
| miR-122tandem | UGGAGUGUGACAAUGGUGGAGUGUGACAAUGGUGUUUG |
| miR-122tandemS2m | UGCUGUGUGACAAUGGUGGAGUGUGACAAUGGUGUUUG |
| miR-124 | CCGUGUUCACAGCGGACCUUG |
| U20 | UUUUUUUUUUUUUUUUUUUU |
| C20 | CCCCCCCCCCCCCCCCCCCC |
Expression and purification of PCBP2 and HCV NS5B
The pVP16 plasmid for expressing PCBP2, with a tobacco etch virus (TEV) protease cleavage site at the C-terminus, followed by maltose-binding protein (MBP) and His8-tag, was obtained from the DNASU depository (HsCD00084126) (25), originally deposited by the University of Wisconsin Madison, PSI, Center for Eukaryotic Structural Genomics. PCBP2 was expressed in Escherichia coli Rosetta (DE3) cells. The cells were grown at 37°C in Luria broth (LB) containing 50 μg/ml ampicillin to an OD600 of 0.6–0.8 and PCBP2 expression was induced by the addition of 1 mM isopropyl-β-d-1-thiogalactopyranoside. Cell Growth continued overnight at 18°C. Purification began with the pelleting of the cells and resuspension in lysis buffer (100 mM sodium phosphate, pH 8.0, 500 mM NaCl and 2 mM β-mercaptoethanol) and 1 tablet of an EDTA-free protease inhibitor (Roche Applied Science, Penzberg, Germany). The cells were lysed by sonication and the soluble fraction was collected after centrifugation and loaded to Talon metal affinity chromatography resin (Clontech, Mountain View, CA). The column was washed with equilibration buffer (100 mM sodium phosphate, pH 7.0, 500 mM NaCl and 2 mM β-mercaptoethanol), and PCBP2 was eluted on a gradient of 1–150 mM of imidazole in equilibration buffer. Fractions containing proteins were identified by SDS-PAGE. To remove the MBP-His8 tag, the pooled PCBP2 fractions were incubated with 10 μg TEV protease per 1 mg of the PCBP2-MBP fusion protein overnight at 4°C. The cleaved protein was loaded on a Talon metal affinity column and the flow-through containing the cleaved PCBP2 was collected and concentrated. The purity of the protein, estimated by SDS-PAGE, was >95%. The concentration of PCBP2 was determined by spectrophotometry with an extinction coefficient of ε280, of 16305 cm−1 M−1. The purified protein was used for fluorescence-based binding assays.
NS5B, containing a C-terminal 21 amino acid deletion and a C-terminal His-tag was expressed from a pET22 plasmid, kindly provided by Dr Craig Cameron, as previously described (26). Briefly, the plasmid was transformed into Rosetta (DE3) competent cells and grown in an autoinduction media (LB media containing 50 μg/ml ampicillin, 1 mM MgSO4, 20 mM (NH4)2SO4, 40 mM KH2PO4, 40 mM Na2HPO4, 1% glycerol, 14 mM glucose and 30 mM lactose) at 37°C until reaching an OD600 of 1.0. Then the cells were grown at 25°C for additional 4 h and pelleted by centrifugation. The cell pellet was stored overnight at −20°C and resuspended in the lysis buffer (100 mM sodium phosphate, pH 8.0, 500 mM NaCl, 2 mM β-mercaptoethanol, 20% glycerol and 1 tablet of an EDTA-free protease inhibitor). Cells were lysed via sonication. Following centrifugation, polyethyleneimine was added to the soluble fraction to a final concentration of 0.25% and incubated at room temp for 30 min. The precipitate was removed by centrifugation and the soluble fraction was precipitated with 60% saturation of ammonium sulfate. The precipitated protein was resuspended in 20 ml of resuspension buffer (100 mM sodium phosphate, pH 8.0, 500 mM NaCl, 5 mM β-mercaptoethanol, 20% glycerol and 5 mM imidazole) and purified on a Talon metal affinity column using an imidazole gradient of 5–150 mM. The fractions containing NS5B were identified via SDS-PAGE and were pooled and concentrated. The purity of the protein, estimated by SDS-PAGE, was >95%.
Electrophoretic mobility shift assays with HCV RNA
For the electrophoretic mobility shift assay of HCV45 and miR-122, the HCV45 oligonucleotide (1 μM) was heated to 80°C for 2 min and cooled to 37°C, then mixed with miR-122 in molar ratios of 0, 0.5, 1.0, 1.5, 2.0 and 3.0. The mixture was incubated at 37°C for 30 min in a 10 μl volume containing 100 mM Tris (pH 7.5), 100 mM NaCl and 5 mM MgCl2. An equal volume of gel-loading dye (30% glycerol, 0.5× TBE and 5 mM MgCl2) was added to each mixture and the samples were loaded onto a 3% agarose gel and separated at 80 V for 2.5 h. RNA was visualized by staining with SYBR Gold (Invitrogen). For the electrophoretic mobility shift assay with PCBP2, 10 nM of IRDye-800 labelled HCV47 bait was incubated with 0–400 ng recombinant PCBP2 protein (0–500 nM) or 400 ng bovine serum albumin (BSA, 300 nM) for 30 min at room temperature. The resulting protein-RNA complex was resolved in a 5% native polyacrylamide gel and visualized by an Odyssey infra-red imager (Li-Cor). To determine competition between PCBP2 and miR-122 for interacting with HCV47, 10 nM of IRDye-800 labelled RNA bait was annealed with 50 nM of single-stranded miR-124 or miR-122 for 15 min in 20 μl binding buffer (20 mM Tris, pH 7.5, 100 mM KCl and 1 mM MgCl2) and then incubated with recombinant PCBP2 protein (0–500 nM) for 30 min at room temperature.
Isothermal titration calorimetry (ITC)
Interactions between HCV RNAs and miR-122 were measured using a MicroCal PEAQ-ITC (Malvern Panalytical). RNA samples were diluted to the desired concentration in a buffer containing 100 mM HEPES (pH 7.5), 100 mM NaCl and 5 mM MgCl2. HCV45 was folded by heating at 80°C for 2 min and cooled on ice for 10 min. 300 μl of 5 μM titrand RNA (HCV45, HCV47, HCVS1p34, HCV21-65, HCVS2-comp, or miR-122) was loaded into the cell, and 70 μl of 100 μM titrant RNA (miR-122, HCV45, miR-122comp, miR-122tandem or miR-122trunc) was loaded into the syringe. All ITC experiments were performed in triplicate at 37°C with a reference power of 3 μcal/s. Typically, experiments were performed with an initial injection of 0.4 μl followed by 39 0.8 μl injections with a duration of 1.6 s per injection and 150 s spacing. The first injection was discarded, and the data were fitted to either a single- or two-stie binding model using the MicroCal PEAQ-ITC Analysis Software (Malvern Panalytical). Fitted parameters are listed in Table 2 for each experiment.
Table 2.
HCV RNA interactions with miR-122 RNAs measured by ITC
| Site 1 | Site 2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Titrant | Titrand | K d (nM) | ΔH (kJ/mol) | –TΔSa (kJ/mol) | ΔG (kJ/mol) | K d (nM) | ΔH (kJ/mol) | –TΔS (kJ/mol) | ΔG (kJ/mol) |
| HCV45 | miR-122 | 11.1 ± 1.5 | −513 ± 2.0 | 466 ± 2.3 | −47.0 ± 0.3 | 979 ± 84 | −205 ± 2.9 | 169 ± 3.1 | −35.6 ± 0.2 |
| miR-122trunc | 9.45 ± 1.2 | −416 ± 11 | 369 ± 11 | −47.2 ± 0.1 | 187 ± 13 | −219 ± 7.0 | 179 ± 7.1 | −39.7 ± 0.1 | |
| miR-122tandem | 609 ± 42 | −455 ± 12 | 418 ± 12 | −36.9 ± 0.2 | – | – | – | – | |
| HCV21-65 | miR-122 | – | – | – | – | 712 ± 68 | −343 ± 13 | 306 ± 13 | −36.5 ± 0.5 |
| HCVS2-comp | miR-122 | 152 ± 31 | −127 ± 5.0 | 87 ± 5.8 | −40.0 ± 0.8 | – | – | – | – |
| miR-122comp | – | – | – | – | 567 ± 40 | −164 ± 6.9 | 127 ± 6.9 | −37.2 ± 1.0 | |
| HCVS1p34 | miR-122 | 54,300 ±12 000 | 20 ± 28 | −49 ± 28 | −29.0 ± 6.8 | 244 ± 9 | −685 ± 46 | 641 ± 46 | −44.1 ± 8.6 |
a All ITC experiments were carried out at 37°C.
HCV replication reporter assay
HCV replication was assessed using the HJ3-5/GLuc2A reporter virus (14,27). The parental ‘wild-type’ (WT) HJ3-5/GLuc2A and mutant reporter virus RNAs were transcribed using T7 MEGAscript transcription kit (Invitrogen, #AM1334). In vitro transcribed RNA (1.25 μg) was purified using the RNeasy Mini Kit (Qiagen, #74104) and transfected into 2.5 × 105 Huh-7.5 or miR-122 knockout Huh-7.5 cells (Huh-7.5-ΔmiR122) (kindly provided by Dr Charles Rice, Rockefeller University) using the TransIT mRNA kit (Mirus Bio, #MIR2250). Cell culture supernatant fluids were collected at intervals following RNA transfection, and cells re-fed with fresh media. Secreted GLuc activity was measured using the Biolux Gaussia Luciferase assay kit (New England Biolabs, #E3300S), as previously described (11). To deplete PCBP2 or Ago2 in infected cells, siRNA pools targeting PCBP2 (Dharmacon #L-012002-00-0010) or Ago2 (Dharmacon, #L-004639-00-0010) and control siRNA pools (Dharmacon, #D-001810-10) were transfected using siLentfect Lipid Reagent (Bio-Rad, # 1703360) at 20 nM concentration in 6-well plates 48 h before HCV RNA transfection. Where indicated, single- or double-stranded miR-122, miR-124 or miR-122tandem were transfected at 50 nM concentration into 24 h before HCV RNA transfection.
HCV translation reporter assay
HCV RNA translation was assessed using the HCV-ΔC/GLuc mini-genome reporter RNA in which an open reading frame containing the 5′ 36 nucleotides of core protein-coding sequence fused to GLuc sequence is flanked by 5′ and 3′ HCV UTRs (28). HCV-ΔC/GLuc and mutant reporter RNAs were transcribed as above, and 1.5 μg RNA transfected into 5 × 105 Huh-7.5 cells using the TransIT mRNA kit (Mirus Bio, #MIR2250). GLuc activity was measured in cell culture supernatant fluids 24–96 h later using the Biolux Gaussia Luciferase assay kit (New England Biolabs, #E3300S).
Cellular pull-down assay using biotinylated HCV RNA
The HCV 5′ end oligo, HCV47-biotin (10 pmol), was incubated alone or with indicated amount of single strand miR-122 oligo, heated at 75°C for 5 min then cooled down at room temperature. Annealed oligos were bound to magnetic streptavidin T1 beads (Invitrogen, #65606D) as recommended by the manufacturer, then incubated with Huh-7.5 cytoplasmic lysate for 1 h at 4°C. Materials bound to the beads were eluted in SDS-PAGE sample buffer, resolved by SDS-PAGE and subjected to Sypro-ruby staining or western blot. To identify pulled down proteins, immunoblotting was carried out using standard methods with the following antibodies: mouse mAb to β-actin (Sigma-Aldrich, clone AC-74), rat mAb to Ago2 (Sigma-Aldrich, SAB4200085) and mouse mAb to PCBP2 (Abnova, clone 5F12). Protein bands were visualized with an Odyssey Infrared Imaging System (Li-Cor Biosciences).
HCV 5′ and 3′ end RNA interaction
To probe 5′ and 3′ UTR interactions, HCV47-biotin (10 pmol) was incubated alone or with indicated amounts of single strand miR-122 oligo in binding buffer (20 mM Tris, pH 7.5, 100 mM KCl and 1 mM MgCl2). Recombinant PCBP2 protein (Abnova, H00005094-P01) and 32P-labelled HCV 3′UTR RNA (body-labeled, 40K cpm each reaction) were added to the reaction and incubated for 30 min at room temperature. Mixtures were then bound to magnetic streptavidin T1 beads (Invitrogen) according to manufacturer's recommended procedure. Materials bound to the beads were eluted in RNA gel loading buffer II (Ambion), resolved by 5% denaturing SDS-PAGE and visualized using a phosphorimager.
Electron microscopy
To facilitate visualization of linear and circular RNAs, we annealed HCV-ΔC/GLuc mini-genome RNA (28) to the RNA GLuc complement, resulting in a double-stranded GLuc duplex flanked by single-stranded HCV 5′ and 3′ UTRs. Complexes between the mini-genome duplex and PCBP2 were formed in the presence or absence of miR-122 (or a miR-124 control) in a buffer containing HEPES pH 7.5, followed by addition of 25% glutaraldehyde to 0.6% for 5 min at room temperature. The fixatives and unbound protein were removed by chromatography over 2 ml columns of 6% agarose beads equilibrated in 10 mM Tris pH 7.5 containing 0.1 mM EDTA. The peak fraction containing the nucleic acids was adsorbed to thin carbon foils in a buffer containing 2 mM spermidine, followed by dehydration in ethanol, air-drying and rotary shadow casting with tungsten (29). Samples were imaged using an FEI Tecnai 12 TEM at 40 kV equipped with a Gatan Orius CCD camera.
Fluorescence measurements
Fluorescence measurements were taken as previously described (30–33). Steady-state fluorescence titrations were performed using an ISS PC1 spectrofluorometer (ISS, Urbana, IL). Polarizers were placed in the excitation and emission channels set at 90° and 55° (magic angle), respectively, to avoid possible fluorescence anisotropy artifacts. All fluorescence binding experiments used the fluorescein signal (excitation wavelength [λex] = 495 nm; emission wavelength [λem] = 520 nm). Binding curves were fit by using SigmaPlot software (SYSTAT Software, CA). The relative fluorescence change was defined as  where
where  is the fluorescence of the solution at a given titration point,
is the fluorescence of the solution at a given titration point,  , and
, and  is the initial fluorescence of the sample. Both
is the initial fluorescence of the sample. Both  and
and  are corrected for background fluorescence at the applied excitation wavelength. Standard buffer B1, containing 50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 5 mM β-mercaptoethanol and 15% glycerol, was used for all binding experiments. All experiments were performed in triplicate.
are corrected for background fluorescence at the applied excitation wavelength. Standard buffer B1, containing 50 mM Tris–HCl, pH 7.5, 100 mM NaCl, 5 mM β-mercaptoethanol and 15% glycerol, was used for all binding experiments. All experiments were performed in triplicate.
To measure PCBP2 affinity for HCV45, PCBP2 (5 nM–8 μM), either with or without the MBP tag, was titrated into 30 nM of HCV45 or HCV47 with a 5′ fluorescein label. PCBP2 without the MBP tag was used for all further experiments. To measure the competition with miR-122 binding, PCBP2 (5 nM - 14 μM) was titrated into the solution containing 30 nM of fluorescein labeled HCV45 and 90 nM of unlabeled miR-122. As a control, PCBP2 (5 nM–14 μM) was also titrated into 30 nM of fluorescein labeled miR-122. To measure NS5B binding affinities, NS5B (5 nM–5.12 μM) was titrated into 30 nM of fluorescein labeled HCV45. This was repeated with 30 nM of fluorescein labeled U20 or fluorescein labeled C20 in place of the HCV45. To test whether the presence of miR-122 or PCBP2 modulated NS5B’s affinity to HCV45, NS5B (5 nM–3 μM) was titrated into 30 nM of fluorescein labeled HCV45 in the presence of either 90 nM of unlabeled miR-122, 90 nM of unlabeled miR-124, or 300 nM of PCBP2. NS5B (5 nM–3 μM) was also titrated with 30 nM of either HCVS1p23, HCV21-65 or HCVS2-comp either alone or in the presence of 90 nM of unlabeled miR-122. Additionally, NS5B (5 nM–3 μM) was titrated into either 30 nM of PCBP2 alone or 30 nM of fluorescein labeled miR-122 alone as controls. Experiments were conducted with the sample chamber heated to 37°C.
Fluorescence anisotropy signals were analyzed as follows. The binding constant  , characterizing the association of HCV45 with either PCBP2 or NS5B, is defined as:
, characterizing the association of HCV45 with either PCBP2 or NS5B, is defined as:
 |
(1) |
where  is the concentration of the formed complex. The observed fluorescence of the sample at any point of the titration is defined as:
is the concentration of the formed complex. The observed fluorescence of the sample at any point of the titration is defined as:
 |
(2) |
where  and
and  are the fluorescence anisotropy of the free HCV45 and the formed complex respectively. Thus,
are the fluorescence anisotropy of the free HCV45 and the formed complex respectively. Thus,
 |
(3) |
The mass conservation equation for the total HCV45 concentration  is
is
 |
(4) |
Using equations (3) and (4), the relative observed change of the HCV45 fluorescence,  is then
is then
 |
(5) |
where  is the maximum value for the observed relative fluorescence quenching.
is the maximum value for the observed relative fluorescence quenching.
Results
There are two unequal binding sites for miR-122 on the HCV 5′UTR
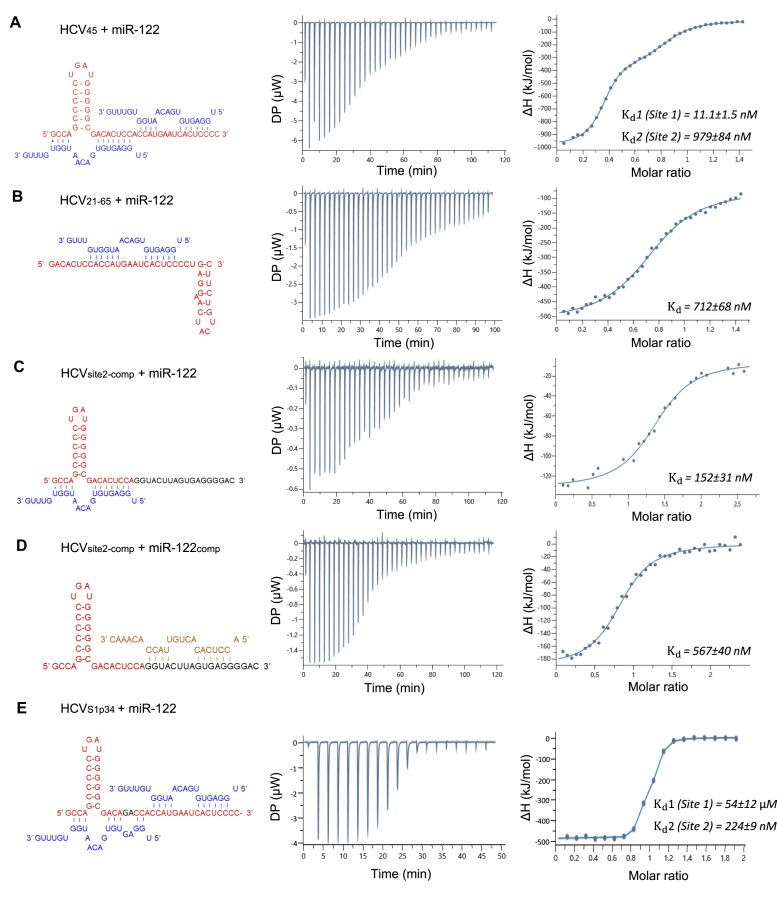
There are two miR-122 binding sites in the first 43 nucleotides at the 5′ end of the HCV genome (Figure 1B). The first binding site, S1, consists of nts 1 to 29, and the second site, S2, consists of nts 29 to 43, with A29 potentially base-pairing with miR-122 bound to either site (Figure 1B). To characterize these binding sites, an RNA molecule representing the first 45 nucleotides of the HCV 5′UTR (HCV45) was synthesized (Table 1), and its interactions with miR-122 were measured by electrophoretic mobility shift assay (EMSA) and isothermal titration calorimetry (ITC). EMSA showed that two copies of miR-122 bind to HCV45 sequentially (Figure 1C), suggesting unequal binding affinity for the two miR-122 sites. Next, the binding constants for the two miR-122 binding sites on HCV45 were determined by ITC. HCV45 at 100 μM was titrated into 5 μM of miR-122 in a HEPES buffer (100 mM HEPES, pH 7.5, 100 mM NaCl and 5 mM MgCl2) and the energy release from binding was analyzed to calculate the dissociation constant (Kd). The Kd values of the two binding sites were found to be 11.1 ± 1.5 and 979 ± 84 nM (Figure 2A and Table 2). The binding of miR-122 was highly enthalpic with the change in enthalpy of −513 ± 2.0 and −205 ± 2.9 kJ/mol for the first and second binding site respectively (Table 2). Previously published ITC data for an RNA consisting of the first 47 nucleotides of the HCV 5′UTR (HCV47) suggested high and low binding affinities of 90 ± 52 and 845 ± 354 nM, respectively (34). We thus additionally synthesized HCV47 and measured the binding affinities with miR-122. The Kd values for the high and low binding sites in HCV47 were 7.9 ± 2.3 and 1230 ± 260 nM (Supplementary Figure S1A). Thus, HCV45 and HCV47 have similar binding affinity to miR-122, indicating that the 3′ terminal two nucleotides of HCV47 do not participate in miR-122 binding.
Figure 2.
Isothermal calorimetry of 5′ HCV RNA and miR-122 variants. (A) Titration of HCV45 RNA into miR-122. Thermogram and resulting binding curve are shown. The sequences of HCV (red) and miR-122 (blue) are shown. The data were fit with two binding site model, and affinities of each binding site,  (Site 1) and
(Site 1) and  (Site 2), are indicated. (B) Titration of HCV21-65 RNA into miR-122. HCV21-65 RNA only contains Site 2. Single binding site model was used to fit the binding curve. (C) Titration of miR-122 into HCVS2-comp. Single binding site model was used to fit the binding curve. (D) Titration of miR-122comp into HCVS2-comp. Single binding site model was used to fit the binding curve. (E) Titration of miR-122 into HCVS1p34. Two binding site model was used to fit the binding curve. All data are representative of at least three independent replicas. The
(Site 2), are indicated. (B) Titration of HCV21-65 RNA into miR-122. HCV21-65 RNA only contains Site 2. Single binding site model was used to fit the binding curve. (C) Titration of miR-122 into HCVS2-comp. Single binding site model was used to fit the binding curve. (D) Titration of miR-122comp into HCVS2-comp. Single binding site model was used to fit the binding curve. (E) Titration of miR-122 into HCVS1p34. Two binding site model was used to fit the binding curve. All data are representative of at least three independent replicas. The  values are an average of three measurements ± errors propagated from individual fits.
values are an average of three measurements ± errors propagated from individual fits.
To identify which miR-122 binding site corresponded to the high and low affinity site, two alternative HCV RNAs were synthesized that individually target each binding site. First, a 45 nt RNA comprised of nts 21–65 of the 5′UTR (HCV21-65, Table 1) was synthesized and used in ITC. This RNA contains only S2. When HCV21-65 was titrated into miR-122, the binding constant was found to be 712 ± 68 nM, suggesting that the miR-122 S2 is the low affinity site (Figure 2B and Table 2). Second, a 45 nt RNA consisting of nts 1–28 of the 5′UTR and the reverse complementary sequence of nts 29–45 (HCVS2-comp, Table 1) was synthesized and used in additional ITC experiments. The HCVS2-comp was titrated with either wild-type (WT) miR-122, which can only bind to S1, or the reverse complement of miR-122 (miR-122comp), which would bind S2. WT miR-122 binds HCVS2-comp with a Kd of 152 ± 31 nM (Figure 2C and Table 2), while the miR-122comp and HCVS2-comp had a lower affinity with 567 ± 40 nM (Figure 2D and Table 2). Taken together, the first miR-122 binding site S1 on the HCV 5′UTR is the high affinity site, and the second site S2 is the low affinity site.
The truncation of miR-122 lowers the Kd of the S2 site but not the S1 site
It is not immediately clear why the Kd for the second S2 site is about two logs higher than the first site, as the number of complementary base pairs are conserved. Both binding sites are predicted to form ten or eleven base pairs between the miR-122 and HCV45, depending on whether a base pair is formed with A29 (Figure 1B); A29 on HCV45 can base pair with either U1 of the miR-122 at the first site or U17 of miR-122 at the second site. We thus investigated the effect of the 3′ overhang of miR-122 using a truncated miR-122, lacking the 3′ GUUUG overhang (miR-122trunc). When titrated with HCV45, a Kd of 9.45 ± 1.2 and 187 ± 13 nM was measured for the two binding sites (Figure 3A and Table 2). Thus, removal of the 5 nt overhang at the 3′ end of miR-122 did not change the affinity for S1, while it increased the affinity for S2 from 979 to 187 nM. This indicates that the unpaired bases at the 3′ end of miR-122 at the second site sterically hinder its binding to HCV45.
Figure 3.
Effects of miR-122 sequence on HCV interaction and replication. (A) Isothermal calorimetry of 5′ HCV RNA and truncated miR-122. The sequence of miR-122trunc is shown above. The data were fit with two binding site model, and affinities of each binding site,  (Site 1) and
(Site 1) and  (Site 2), are indicated. All data are representative of at least three independent replicas and
(Site 2), are indicated. All data are representative of at least three independent replicas and  values are an average of three measurements ± error propagated from individual fits. (B) Isothermal calorimetry of 5′ HCV RNA and tandem miR-122. The tandem miR-122 (miR-122tandem) was designed to occupy both miR-122 binding sites by removing the 3′ overhang between the sites. The data were fit into a single binding site model. (C) HCV reporter replication assay. Huh-7.5 miR-122 knockout cells were transfected with either ds-miR-122, ds-miR-124, ds-miR-122tandem, or ss-miR-122tandem for 16 h. Then an HCV reporter genome, HJ3-5/Gluc2A, was transfected to the cells and luciferase activity measured at 6, 24, 48 and 72 h post-transfection. Data were normalized to the GLuc activity expressed by cells transfected with ds-miR-122 at 6 h. P-value for miR-122 versus ds-miR-122tandem = 0.0024 by two-way ANOVA. Single stranded miR-122 and miR-122tandem were incapable of promoting viral replication. (D) HCV reporter replication assay in Ago2 knockdown cells. Huh-7.5 miR-122 knockout cells were transfected with Ago2 siRNA for 24 h then re-transfected with ds-miR-122 or ds-miR-122tandem for 16 h. HJ3-5/Gluc2A was transfected into the cells and luciferase activities measured at 6, 24, 48, 72 and 96 h post-infection, with GLuc activity normalized to that expressed by control cells transfected with ds-miR-122 at 6 h. Viral replication in the presence of either miR-122 or miR-122tandem requires Ago2. siCtrl: control, non-targeting siRNA. (E) HCV reporter replication assay in miR-122 knockout cells transfected with ds-miR-122tandem as in panel C, and ds-miR-122tandemS2m containing two base-substitutions (red font) that eliminate seed-sequence interactions at the S2 site (S2p3,4 mutant) (16). ds-miR124 and ds-miR122 included as positive and negative controls.
values are an average of three measurements ± error propagated from individual fits. (B) Isothermal calorimetry of 5′ HCV RNA and tandem miR-122. The tandem miR-122 (miR-122tandem) was designed to occupy both miR-122 binding sites by removing the 3′ overhang between the sites. The data were fit into a single binding site model. (C) HCV reporter replication assay. Huh-7.5 miR-122 knockout cells were transfected with either ds-miR-122, ds-miR-124, ds-miR-122tandem, or ss-miR-122tandem for 16 h. Then an HCV reporter genome, HJ3-5/Gluc2A, was transfected to the cells and luciferase activity measured at 6, 24, 48 and 72 h post-transfection. Data were normalized to the GLuc activity expressed by cells transfected with ds-miR-122 at 6 h. P-value for miR-122 versus ds-miR-122tandem = 0.0024 by two-way ANOVA. Single stranded miR-122 and miR-122tandem were incapable of promoting viral replication. (D) HCV reporter replication assay in Ago2 knockdown cells. Huh-7.5 miR-122 knockout cells were transfected with Ago2 siRNA for 24 h then re-transfected with ds-miR-122 or ds-miR-122tandem for 16 h. HJ3-5/Gluc2A was transfected into the cells and luciferase activities measured at 6, 24, 48, 72 and 96 h post-infection, with GLuc activity normalized to that expressed by control cells transfected with ds-miR-122 at 6 h. Viral replication in the presence of either miR-122 or miR-122tandem requires Ago2. siCtrl: control, non-targeting siRNA. (E) HCV reporter replication assay in miR-122 knockout cells transfected with ds-miR-122tandem as in panel C, and ds-miR-122tandemS2m containing two base-substitutions (red font) that eliminate seed-sequence interactions at the S2 site (S2p3,4 mutant) (16). ds-miR124 and ds-miR122 included as positive and negative controls.
Tandem miR-122 binds to HCV45 and promotes viral replication
To better understand the difference between the two miR-122 binding sites, a ‘tandem’ miR-122 sequence was used to bind HCV45. This tandem repeat miR-122 (miR-122tandem) consisted of two copies of the miR-122 sequence with the 3′ GUUUG removed from the 5′ copy (i.e. the copy binding to the S2 site) (Figure 3B). This RNA should form the same base pairs with HCV45 as the two separate miR-122 molecules (Figure 3B). The dissociation constant of miR-122tandem to HCV45 was 609 ± 42 nM, similar to the Kd obtained for the low affinity miR-122 binding site S2 (Figure 3B and Table 2). This was unexpected because the miR-122tandem should be capable of binding to either miR-122 site, so the binding affinity was expected to be similar to the high affinity site.
The miR-122tandem was also tested for its ability to promote HCV replication using a replication-competent reporter virus (HJ3-5/GLuc2A) that expresses Gaussia princeps luciferase (GLuc) from sequence inserted between the p7 and NS2A coding sequence (14,27). miR-122 depleted Huh-7.5 cells (Huh-7.5-ΔmiR122) were transfected with in vitro-transcribed reporter virus RNA together with either double-stranded (ds) or single-stranded (ss), miR-122, miR-122tandem or an unrelated brain-specific control miRNA, miR-124 (Table 1), and the luciferase activity was measured as a proxy for viral replication. As shown previously, double-stranded miR-122 promoted viral replication, whereas the double-stranded miR-124 did not (Figure 3C). miR-122tandem, provided in a double-stranded form, promoted viral replication with about 50% the activity of miR-122. Promotion by miR-122tandem also required Ago2, similar to miR-122, as it failed to promote viral replication in Ago2 knockdown cells (Figure 3D). Thus, miR-122tandem promotes replication in an Ago2-dependent manner similar to miR-122. To assess whether the promotion of viral replication by miR-122tandem required interactions with both seed-sequence binding sites, we modified the miRNA mimic to include two base substitutions known to disrupt binding at the low affinity S2 site (underlined bases in Figure. 3E) (16). This mutant, miR-122tandemS2m was as active as miR-122tandem itself in stimulating the replication of HCV, indicating that base pairing at the low affinity S2 site does not contribute to the promotion of virus replication by miR-122tandem (Figure. 3E). These data are consistent with the primacy of the high affinity S1 site in driving HCV replication.
PCBP2 binds to HCV45 directly and competes with miR-122 binding
Previous pull-down assays show that multiple cellular proteins, including PCBP2 and hnRNPL, bind to an RNA bait corresponding to the first 47 nts of the 5′ HCV genome (HCV47) (28). PCBP2 consists of three KH domains 1–3, each of which can bind ∼4 nts of single-stranded, C-rich sequence. PCBP2 likely binds the C-rich region at nts 40–45 (40UCCCC45), which overlaps with the miR-122 binding site S2 (Figure 1B) (16). We first confirmed that PCBP2 binds the RNA directly using EMSA. IRDye-800 labelled HCV47 was incubated with PCBP2, and the resulting protein–RNA complex resolved in a native polyacrylamide gel. The labeled HCV47 migrated as two distinct bands as previously observed (35), most likely reflecting monomer RNA and a predicted stable dimer formed by the 31CAUG34 sequence. The RNA band was shifted with increasing amounts of PCBP2 (as little as a 3:1 PCBP2/RNA molar ratio, Figure 4A). PCBP2 had greater affinity for the slower-migrating, dimeric RNA, but both RNA bands were shifted at the highest concentration tested (50:1 molar ratio) (Figure 4A). The PCBP2 interaction with HCV47 was specific, as HCV47 did not interact with a control protein, bovine serum albumin, even at 30-fold excess (Figure 4A). To test competition between PCBP2 and miR-122, we next pre-incubated HCV47 with miR-122 or miR-124 (as a control) and carried out an EMSA with PCBP2 (Figure 4B). The gel shift assay was repeated using a fixed amount of PCBP2 (250 nM) and increasing the amount of single-stranded miR-122 or miR-124 (0–200 nM). Preincubation with miR-122 greatly reduced PCBP2 binding to the RNA, whereas miR-124 had no such effect even at high concentrations (Figure 4C and Supplementary Figure S2).
Figure 4.
PCBP2 binds directly to 5′ terminal HCV RNA sequence in competition with miR-122. (A) Gel shift assay with HCV47 RNA and PCBP2. IRdye-800 labelled HCV47 RNA (10 nM) was incubated with recombinant PCBP2 protein or control BSA. RNA-protein complexes were resolved on a 5% native polyacrylamide gel and visualized with an Odyssey Imaging System. RNA monomers (*) and dimers (**) are evident as two discreet bands, along with more slowly migrating PCBP2-RNA complexes (arrow). (B) Gel shift assay with HCV47 and PCBP2 in the presence of miR-122. The IRdye-800 labelled HCV47 RNA was incubated with 50 nM of single-stranded miR-122 or miR-124 prior to addition of PCBP2. (C) MiR-122 prevents HCV and PCBP2 interaction. The gel shift assay shown in (B) was repeated using 250 nM PCBP2 and increasing quantities of single-stranded miR-122 or miR-124, and percent of RNA bound to PCBP2 was plotted. The EMSA image is shown in Supplementary Figure S2. (D) Coomassie stained SDS-PAGE gel with the purified recombinant PCBP2. (E) PCBP2 binding to HCV45 and the HCV45:miR-122 complex. PCBP2 was titrated into either fluorescein-labeled HCV45-F (30 nM), labeled miR-122-F (30 nM), or the HCV45-F (30 nM) and unlabeled miR-122 (90 nM) complex. Dissociation constants determined from the titration curves are listed on the right. (F) PCBP2 and Ago2 pull down assays with biotinylated RNA probes. Design of 3′-biotinylated HCV47 (HCV47-Bio) and related S2p6m-Bio, S2p3,4m-Bio, and Ins43AA-Bio RNA probes are shown (left). Potential PCBP2 binding site that overlaps with S2 is shown in green, and altered bases in the S2 seed sequence are shown in red. PCBP2 pull down assay was carried out with the biotinylated RNA probes and Huh-7.5 cell lysate. Co-precipitated PCBP2 or Ago2 protein was detected by immunoblot. Asterisk (*) indicates a non-specific Ago band. (G) Replication reporter assay for S2 mutant viruses. Luciferase activities were measured in supernatant fluids of cells transfected with the HJ3-5/GLuc2A or mutant reporter viral RNAs, with or without supplementation with complementary mutant miR-122s (p6 and p34). (H) Replication reporter virus assay for Ins43AA. Luciferase activities in supernatant fluids of cells transfected with the HJ3-5/GLuc2A and Ins43AA mutant reporter, with or without RNAi-mediated depletion of PCBP2. Immunoblots of PCBP2 and GAPDH (loading control) are shown on top. (I) Translation reporter assay for Ins43AA. The schematic of the HCV-ΔC/GLuc mini-genome translation reporter is shown on top. Luciferase activities were measured in PCBP2-depleted or control Huh-7.5 cells transfected with the HCV-ΔC/GLuc translation reporter or Ins43AA mutant RNA. Results shown represent the means from 3 independent experiments, each with 2–3 technical replicates, ±SD. P-value by two-sided t-test.
Next, we measured binding affinities of PCBP2 (Figure 4D) to HCV in the presence or absence of miR-122. The first 45 nts of HCV RNA was synthesized with a 5′ fluorescein tag (HCV45-F), and its interaction with PCBP2 was measured using fluorescence anisotropy titration (Figure 4E). Binding of PCBP2 protein to the HCV45-F led to an increase in the fluorescence anisotropy which was then fitted as a binding curve to determine the dissociation constant. PCBP2 binds to HCV45-F with a Kd of 667 ± 34 nM. We additionally tested PCBP2 binding to the fluorescein-labeled HCV47, and it had a similar affinity to HCV45-F (Supplementary Figure S1B). Next, the PCBP2 interaction with the HCV45-F and miR-122 complex was measured. Since HCV45-F has two miR-122 binding sites, HCV45-F was incubated with a 3-fold molar excess of unlabeled miR-122 prior to PCBP2 titrations (Figure 4E). PCBP2 bound the HCV45-F:miR-122 complex with an apparent Kd of 9.88 ± 1.6 μM, ∼18-fold lower affinity than to HCV45-F alone. PCBP2 was also titrated into 5′ fluorescein labeled miR-122 as a control (Figure 4E). PCBP2 bound miR-122 poorly with a Kd of 5.33 ± 0.47 μM. Thus, PCBP2 cannot effectively bind either the HCV45-F:miR-122 complex or free miR-122. Taken together, these results indicate that PCBP2 binds the 5′ terminal HCV sequence directly, and that miR-122 specifically blocks this interaction.
PCBP2 binding at the site overlapping with S2 is required for optimal HCV replication
To further define the PCBP2 binding site and its effects on viral replication, we created mutations in HCV47 (Figure 4F). We first substituted 41UC42, corresponding to positions 3 and 4 in the miR-122 seed sequence, to AG (S2p3,4m). This mutant was shown to disrupt both the miR-122 binding at S2 and PCBP2 binding to the poly(C) sequence (16). Next, to selectively disrupt miR-122 binding at S2 but not PCBP2 binding to the poly(C) sequence, A39, the position 6 in the seed sequence, was mutated to U (S2p6m). Finally, a two-base AA insertion between C43 and C44 (Ins43AA) was created to disrupt PCBP2 binding but not miR-122 binding to S2. Ins43AA would also preserve base pairing between 45CC46 and 117GG118, maintaining the SL-II and IRES structure (36). We first tested whether PCBP2 or Ago2 binding was disrupted in any of the mutants using pull-down assays with 3′-biotinylated HCV47 in Huh-7.5 cells. PCBP2 bound the WT RNA sequence (HCV47) and S2p6m, both of which were expected to have an intact PCBP2 binding site (Figure 4F). However, PCBP2 binding was ablated for S2p3,4m and Ins43AA, in which the poly(C) sequence at position 41–44 was disrupted. Additionally, S2p3,4m showed reduced Ago2 binding, while Ins43AA showed a similar level of Ago2 to the wild type in the pull-down assay (Figure 4F). There was no significant decrease in Ago2 binding to S2p6m (Figure 4F). This is likely because Ago2 is primarily pulled down by miR-122 interacting with S1, since S1 is a higher affinity site.
We next tested whether the mutations impacted viral replication using the HJ3-5/GLuc2A reporter virus. All three mutants demonstrated a significant reduction of HCV replication compared with the parental, ‘wild-type’ (WT) virus, likely due to the disruption of either miR-122 binding (S2p6m), PCBP2 binding (Ins43AA), or both (S2p3,4m) (Figure 4G and H). Accordingly, replication of the S2p6m mutant could be restored almost to WT levels by supplementing cells with a miR-122 mimic complementary to the A39U mutation (miR-122p6). By contrast, replication of the S2p3,4m mutant virus was more severely impaired and was only partially restored (∼10% of the WT GLuc activity) in the presence of complementary miR-122p3,4 (Figure 4G). Interestingly, replication of the Ins43AA mutant virus, which maintains the miR-122 binding site, was also severely impaired, reducing GLuc activity to 15% of the WT virus at 72–96 hrs (Figure 4H). Although replication of both the WT and the Ins43AA mutant virus was substantially restricted by PCBP2 depletion, the disparity in replication of the two viruses was reduced in cells that were partially depleted of PCBP2, in which the mutant virus expressed 31–35% as much GLuc as the parent virus (Figure 4H). We next tested if viral translation is affected by the Ins43AA mutation using an HCV mini-genome RNA (HCV-ΔC/GLuc) comprising truncated core protein-coding sequence fused to GLuc sequence, flanked by the 5′ and 3′ UTRs (Figure 4I). This RNA lacks the ability to replicate as it does not express nonstructural viral proteins. The Ins43AA mutation also reduced the translational activity of this mini-genome RNA. This suggests that PCBP2 binding at the S2 site is required for optimal HCV IRES activity as well as viral replication, even though the IRES sequence lies downstream of nt 45. Consistent with this, the magnitude of the reduction in translation caused by WT and Ins43AA mutation was no longer statistically significant in PCBP2-depleted cells (Figure 4I). Although depleting PCBP2 could have multiple effects on cellular metabolism due to the multi-functional nature of the protein, we noted no impact on cell viability over the course of the experiment. These data are consistent with previously published observations indicating that PCBP2 trans-activates the HCV IRES.
miR-122 blocks PCBP2-mediated circularization of the HCV genome
PCBP2 binds multiple sites within the HCV genome, both in the 5′UTR and 3′UTR, and self-interactions between PCBP2 protein molecules lead to circularization of HCV RNA (7,37). Genome circularization is known to be utilized by several positive-strand RNA viruses to coordinate and promote both RNA translation and genome replication (7,38,39). These observations led us to hypothesize that competition between miR-122 and PCBP2 may regulate the circular and linear forms of the genome and thus engagement of the positive-strand HCV RNA in translation versus RNA synthesis (3,16). Because miR-122 competes with PCBP2 for binding to the 5′end of the genome, miR-122 should counter PCBP2-mediated circularization. Using the approach described in Wang et al. (7), we first confirmed that PCBP2 binds both 5′ and 3′ UTRs of HCV. The biotin-labeled HCV47-biotin was incubated with PCBP2 and a 5′ [32P]-labeled 3′UTR, and then precipitated with streptavidin beads (Figure 5A). The radiolabeled 3′UTR RNA co-precipitated with HCV47-biotin in the presence of PCBP2, but not with a bovine serum albumin control (Figure 5B). We then repeated the experiment after pre-incubating HCV47-biotin with miR-122. The addition of miR-122, but not a miR-124 control, reduced the amount of co-precipitated 3′UTR RNA (Figure 5C). Thus, miR-122 blocks formation of a PCBP2 bridge between the two ends of the genome.
Figure 5.
miR-122 regulates PCBP2-mediated circularization of positive-strand HCV RNA. (A) Schematic showing experimental design of the 5′-3′ interaction assay with 3′-biotinylated HCV 5′ end sequence RNA (nts 1–47) and 5′ [32P]-labelled HCV 3′UTR RNA (nts 9378–9646) probes. (B) The 5′-3′ interaction of the HCV genome. Biotin-tagged 5′ end RNA (HCV47-Bio) was incubated with [32P]-labelled 3′ end RNA in the presence and absence of PCBP2, and pulled down with streptavidin beads. Pulled down [32P]-labelled 3′UTR was visualized by phosphoimager. BSA was used as unrelated control. The percent input 3′UTR RNA bound to the 5′RNA is shown below each lane. (C) Inhibition of 5′-3′ interaction by miR-122. Addition of ss-miR-122 mimic inhibits the PCBP2-mediated 5′-3′ interaction, whereas miR-124 does not. (D) Images of double-stranded mini-genome RNA in the presence of PCBP2. Design of HCV mini-genome duplex is shown on top. Electron micrographs showing linear and circular forms of the mini-genome duplex observed with TEM when incubated with or without PCBP2. (E) Inhibition of PCBP2-mediated circularization by miR-122. Mini-genome duplex RNAs were annealed with 100 ng of miR-122 or miR-124 before the addition of 100 or 50 ng of PCBP2 protein (approximate miRNA:protein molar ratios of 5:1 or 10:1), then visualized by TEM. 100 molecules in each preparation were manually classified as linear, circular or circular + linear. miR-122 versus miR-124 P-values by X2 test.
Next, we visualized PCBP2-mediated circularization of HCV RNA using transmission electron microscopy (TEM) (7). Recombinant PCBP2 protein was added to mini-genome RNA annealed to a complement of the GLuc sequence inserted between its single-stranded 5′UTR and 3′ UTR segments (Figure 5D). The presence of the duplex RNA in the center of the annealed mini-genome RNA facilitated TEM visualization of its structure. In the absence of PCBP2, >98% of the HCV mini-genome duplexes were linear, consistent with a previous report (7) (Figure 5E). The addition of PCBP2 (6-fold molar excess) resulted in 84% of the mini-genome RNAs adopting a circular structure, occasionally in association with a second linear molecule (Figure 5E). Thus, PCBP2 alone can circularize the mini-genome by binding to both the 5′ and 3′UTRs. Pre-annealing the mini-genome with miR-122 before the addition of PCBP2 reduced the proportion of circular genomes in a concentration-dependent manner (Figure 5E). At a 5-fold molar excess of miR-122 over PCBP2, the circular form was reduced to ∼50% and at 10-fold excess of miR-122 over PCBP2, the circular form was reduced to 20% (P< 0.0001). The reduction of circular RNA molecules was specific for miR-122 since pre-annealing the mini-genome with miR-124 had relatively little effect (Figure 5E). Thus, miR-122 disrupts interactions between the 5′ and 3′ ends mediated by PCBP2, preventing circularization of an HCV mini-genome.
NS5B binds the 5′UTR and miR-122 complex with high affinity
Several positive-strand RNA viruses, including flaviviruses and enteroviruses, have been shown to have an RNA promoter structure at the 5′ end of the genome that the cognate viral RdRp specifically interacts with to initiate negative-strand RNA synthesis from the 3′ end of a circularized viral genome (40–43). For example, flaviviruses such as dengue and West Nile viruses have a branched stem–loop, called stem–loop A at the 5′ end of the genome, with which the flavivirus NS5 polymerase specifically interacts (41,42). Similarly, enteroviruses such as poliovirus and coxsackievirus have a cloverleaf structure at the 5′ end of the genome, which interacts with the viral 3CD protein, the precursor protein to the viral 3Dpol polymerase (40). Previous studies using an HCV replicon showed that the stem–loop I structure at the 5′ end is required for viral replication (7), suggesting that the 5′ stem–loop may function similarly as an RNA promoter. We thus tested whether the HCV polymerase, NS5B, could specifically recognize the 5′ HCV RNA sequence and whether the binding affinity of NS5B to the RNA is modulated by the addition of the host factors PCBP2 or miR-122. We determined the binding affinity of recombinant HCV NS5B (Figure 6A) to HCV45 and other RNAs (C20 and U20) using fluorescence anisotropy. We found that NS5B binds the fluorescein-labeled HCV45-F with a Kd of 71.2 ± 7.8 nM (Figure 6B). In comparison, NS5B bound to fluorescein-labeled U20 and C20 (i.e. non-specific RNAs) with Kd’s of 273 ± 11 and 284 ± 24 nM, respectively (Figure 6D). We next tested if NS5B recognizes the S1, S2 or both sites using RNAs containing a single miR-122 binding site. HCVS2-comp contains only the S1 site (nts 1–29, Table 1). NS5B bound the fluorescein-labeled HCVS2-comp with a reduced Kd of 253 ± 16 nM, similar to the binding affinities of U20 and C20 (Figure 6C). HCV21-65 was comprised of nts 21–65, and thus contains only the S2 site (nts 21–43, Table 1). NS5B bound fluorescein-labeled HCV21-65 with a Kd of 89.6 ± 12.0 nM, similar to the NS5B affinity for HCV45 which contains both sites.
Figure 6.
Fluorescence anisotropy measurement of NS5B binding to the HCV45 RNA. (A) Coomassie stained SDS-PAGE gel with the purified recombinant NS5B. (B) NS5B binding to HCV45 in the presence of miR-122, miR-124 or PCBP2. NS5B was titrated into either HCV45-F (30 nM), the HCV45-F and PCBP2 mixture (30 and 300 nM, respectively), or the HCV45-F and unlabeled miR-122 complex (30 and 90 nM, respectively). The dissociation constants were determined from the binding curve fits and are listed in (E). (C) NS5B interactions with S1 or S2 HCV mutants. NS5B was titrated with either HCVS1p34-F, HCVS2,comp-F or HCV21-65-F (30 nM each) with or without unlabeled miR-122 (90 nM) and the binding affinities were measured. The dissociation constants were determined from the binding curve fits and are listed in (E). (D) NS5B interaction with non-specific RNAs. Binding affinity of NS5B to labeled C20-F, U20-F (30 nM each) or U20-F in the presence of 90 nM unlabeled miR-122 were measured. (E) Dissociation constants for NS5B interactions with HCV RNAs. Dissociation constants are the mean of three replicates with error propagated from the individual fits.
Since miR-122 and PCBP2 bind the 5′UTR, we next tested whether these host factors modulate the binding of NS5B to HCV45. When NS5B was titrated with HCV45-F in the presence of a 10-fold molar excess of PCBP2, the Kd was found to be 105 ± 18 nM (Figure 6C). This value is not significantly different from that of HCV45 alone, and thus PCBP2 does not affect the affinity of NS5B to the HCV45. Surprisingly, NS5B bound to the HCV45:miR-122 complex with a much higher affinity than HCV45 alone. In the presence of unlabeled miR-122 (3-fold excess), NS5B bound to HCV45-F with a Kd of 14.5 ± 0.7 nM, a ∼5-fold increase over HCV45 alone (Figure 6B). To ascertain whether miR-122 specifically increases the affinity of NS5B’s for RNA, the NS5B and HCV45 interaction was measured in the presence of a 3-fold molar excess of unlabeled miR-124. The Kd of NS5B binding to HCV45 in the presence of miR-124 was 75.3 ± 11.9 nM, nearly identical to HCV45 alone (Figure 6B). Additionally, NS5B was titrated into U20-F with a 3-fold molar excess of unlabeled miR-122. The Kd of NS5B binding to U20-F in the presence of miR-122 was 221 ± 8.6 nM (Figure 6D), similar to the Kd for U20-F alone. Hence, NS5B specifically recognizes the HCV45:miR-122 complex.
Next, we determined whether the increase in NS5B binding affinity is dependent upon the interaction of miR-122 at S1 or S2, or both, using RNAs containing a single miR-122 binding site. As discussed previously, HCVS2-comp contains only S1, and HCV21-65 contains only S2 (Table 1, Figure 2B and C). The presence of miR-122 increased the affinity of NS5B for HCVS2-comp, lowering the Kd from 253 ± 16 to 12.6 ± 0.5 nM, a 20-fold increase in binding affinity (Figure 6C). This is similar to its affinity for the HCV45:miR-122 complex (Figure 6B). In contrast, NS5B bound to HCV21-65 with a Kd of 90.8 ± 9.2 nM in the presence of miR-122, similar to its affinity in the absence of miR-122 (89.6 ± 12.0 nM) (Figure 6C). Thus, the increase in NS5B binding affinity derives primarily from interaction of miR-122 with the HCV RNA at S1. The miR-122 binding site S1 differs from S2 in that the stem–loop I structure is a part of the HCV and miR-122 complex (Figure 1B). To determine whether NS5B recognizes stem–loop I, or if additional duplex regions formed by the binding of HCV and miR-122 are required for the NS5B interaction, we generated an HCV45 S1 mutant containing substitutions at nt 25 and 26 that base pair with positions 3 and 4 of the miR-122 seed sequence (HCVS1p34). This mutant is known to abolish the S1 interaction with miR-122, and significantly reduces viral replication (16). We first confirmed the impact of the mutation on miR-122 binding at S1 using ITC (Figure 2E), showing that miR-122 bound HCVS1p34 with Kd of 224 ± 9 nM and 54 ± 12 μM. Thus, the micromolar affinity site is likely to be S1. We next measured the NS5B interaction with labeled HCVS1p34, alone and in the presence of unlabeled miR-122 (Figure 6C). NS5B bound HCVS1p34 with a Kd of 62.9 ± 9.3 nM in the absence of miR-122, and 74.5 ± 9.0 nM in the presence of miR-122. These values are almost identical to the NS5B interaction with HCV45 alone. Thus, the stem–loop I structure alone is not sufficient to enhance NS5B affinity for the 5′ UTR of the HCV genome. The data suggest NS5B recognizes both stem–loop I and the dsRNA formed by the viral genome and miR-122 within the HCV:miR-122 complex.
Discussion
Liver-specific miR-122 binding to the HCV genome promotes viral replication by protecting the genome from 5′ dependent exonuclease-mediated decay (11–15) and stabilizing the IRES structure by preventing the formation of an alternative SLII structure (18,19). miR-122 has also been shown to promote HCV RNA synthesis in a PCBP2-dependent manner (16). Both miR-122 and PCBP2 have known binding sites within the first 45 nts of the HCV genome, and binding of one component prevents the binding of the other (28). To understand the interplay between miR-122 and PCBP2, we determined the binding affinities of miR-122 and PCBP2 to the 5′ terminal 45 nts of the HCV genome (HCV45). We observed that this segment of the genome has two miR-122 binding sites, S1 and S2, which have ∼100-fold difference in affinity (11 and 979 nM, Figures 1 and 2). We determined that S1 has a higher affinity than S2, using HCV constructs with which miR-122 can specifically bind either S1 or S2 (HCVS2-comp and HCV21-65). These results are in agreement with Chahal et al. (19), but distinct from those of Mortimer and Doudna (34) who identified S2 as having the higher affinity. This discrepancy is likely due to differences in the approach taken by mutations by Mortimer and Doudna which involved introducing mutations into the miR-122 sequence rather than the 5′ HCV sequence. These miR-122 mutations would affect binding to both S1 and S2, making the results difficult to interpret. Additionally, due to the overlapping binding sites for two miR-122 molecules, the miR-122 RNA representing S2-only would form additional base pairs with HCV RNA, leading to a higher binding affinity for S2 (34).
We then used a truncated miR-122 mutant (deletion of 5 nts at the 3′ end, miR-122trunc) to investigate the overlap in miR-122 binding to the S1 and S2 sites. Surprisingly, the miR-122trunc bound HCV45 with a 5-fold higher affinity for S2 (187 nM) than WT miR-122, without significantly changing the affinity for S1 (9.5 nM). Thus, miR-122 occupying the high-affinity site seems to hinder a second miR-122 molecule binding to S2, contributing to the difference in affinities between S1 and S2. Finally, we determined that the tandem miR-122 (miR-122tandem) bound HCV45 with a 609 nM affinity. This was lower affinity than expected, since miR-122tandem contains the intact miR-122 sequence, complementary to the high affinity S1 binding site on HCV45. Nevertheless, when HCV replication was tested after supplementing miR-122 depleted cells with miR-122tandem, miR-122tandem promoted viral replication to roughly half of the miR-122 activity (Figure 3C-D). Previously, disruption of S1 (the high-affinity site) has been shown to be more detrimental to viral replication than S2 (22,44,45), suggesting that high-affinity binding of miR-122 to S1 may be essential for effective viral replication. However, even with a miR-122tandem having a 55-fold lower affinity for HCV45 than miR-122 to S1, there was not a significant decrease in viral replication. Thus, high-affinity binding of miR-122 is not required for viral replication, and as long as it remains above some threshold, replication may not be greatly impacted. This is similar to the observations where small RNAs that anneal to the HCV sequence overlapping S1 and S2 promote HCV replication (17,20,46).
The second miR-122 binding site S2 overlaps with the PCBP2 binding site. In gel shift and pull-down assays using Huh-7.5 cells, miR-122 inhibits PCBP2 binding to HCV RNA, consistent with previous reports that miR-122 and PCBP2 compete for binding to HCV RNA (16) (Figure 4B, C and E). Gebert et al. recently demonstrated that Ago2-loaded miR-122 can also compete with PCBP2 for binding to 5′ HCV RNA, and that the viral RNA can form a ternary complex with miR-122 and PCBP2 where miR-122 binds at S1 and PCBP2 binds at S2 (35). We did not observe the mixed complex in our EMSA with miR-122 and PCBP2 (Figure 4A), likely due to the amount of PCBP2 used. Gebert et al used PCBP2 at a 20 to 500 molar excess over miR-122:Ago2, while we used ratios of 2.5 to 10 fold molar excess of PCBP2 to miR-122. Thus, it seems likely that higher concentrations of PCBP2 relative to miR-122 are needed to create a significant population of the mixed complex to visualize on an EMSA.
We determined that PCBP2 binds to HCV45 with a Kd of 667 nM, similar to that of miR-122 to S2 or miR-122tandem to HCV45 (979 and 609 nM). The presence of excess miR-122 decreased the apparent affinity of PCBP2 to HCV45 by ∼15-fold, confirming that there is direct competition between the two factors for binding to HCV45. The 41UCCCC45 sequence within S2 is critical for PCBP2 interaction, as the S2p3,4m and Ins43AA mutants had reduced co-precipitation with PCBP2 in pull-down assays and severely reduced viral replication levels compared to the wild-type reporter virus (Figure 4E-G). Ins43AA, which maintains both miR-122 binding sites, demonstrated reduced GLuc expression levels in experiments with a replication-incompetent mini-genome reporter (HCV-ΔC/GLuc), indicating that PCBP2 binding at nts 41–45 promotes viral translation, although this binding site resides upstream of the minimal IRES sequence (Figure 4H).
In positive-strand RNA viruses, the viral genome functions as a template for both viral protein synthesis and negative-strand RNA synthesis, requiring a mechanism to regulate these mutually incompatible processes. Flaviviruses and enteroviruses use complex RNA structure at the 5′ ends of their genomes to regulate the engagement of the RNA in translation versus genome replication (40–43). For example, the flavivirus polymerase NS5 specifically interacts with stem–loop A (SLA) at the 5′ end of the positive-sense RNA, reducing viral translation (47). Further, the SLA structure is required as an RNA promoter for the viral polymerase to initiate negative-strand RNA synthesis at the 3′ end of the genome (41,48). The transition from translation to replication is accompanied by changes in the viral genome from a linear to circularized form bridged at the 5′ and 3′ ends (40,41,48). A similar 5′ RNA structure that functions as a translation to replication switch has not been described in hepaciviruses such as HCV. However, mutating the 5′ stem–loop I (SL-I) in the HCV 5′ UTR has been shown to abolish viral replication, suggesting that the SL-I structure may function similarly in viral replication (7). Since miR-122 and PCBP2 compete for binding to the 5′ end of HCV genome, their interactions with the 5′UTR could regulate whether the genome engages in protein synthesis or genome replication. We thus investigated how miR-122 affects circularization of the genome mediated by PCBP2 binding to both the 5′ and 3′ UTRs. When PCBP2 was added to linear HCV mini-genome RNA, circular forms were observed in vitro by electron microscopy, consistent with the previous report that PCBP2 mediates HCV genome circularization (7). Importantly, prior annealing of the RNA with miR-122 greatly reduced the circular forms, indicating that miR-122 binding to the 5′UTR displaces PCBP2 and disrupts HCV genome circularization (Figure 5).
To function as an RNA promoter, RNA structure should be specifically recognized by the viral polymerase. We thus asked whether the HCV polymerase, NS5B, specifically recognizes 5′UTR sequence. We observed that NS5B has a binding preference for HCV45, binding HCV45 with higher affinity (71 nM) than other ssRNA such as U20 and A20 (273–285 nM). Surprisingly, NS5B bound HCV45 with significantly higher affinity (15 nM) in the presence of miR-122, while the binding affinity in the presence of miR-124 did not increase (Figure 6B). This suggests that NS5B specifically recognizes the miR-122:HCV45 complex. We then determined that the affinity of NS5B for the miR-122:HCV45 complex is mediated by miR-122 binding to S1. NS5B bound to an HCV RNA:miR-122 complex containing only S1 (HCVS2,comp) with high affinity, similar to that of the HCV45:miR-122 complex, whereas its affinity for HCV RNA containing only S2 (HCV21-65 and HCVS1p34) was not enhanced by miR-122. The high affinity of NS5B for the 5′ end of HCV RNA in the presence of miR-122 suggests that the 5′UTR:miR-122 complex may function as an RNA promoter to promote negative-strand RNA synthesis (see Figure 7).
Figure 7.
Proposed mechanism of HCV genome replication. Competition between miR-122 and PCBP2 for binding to the 5′UTR determines whether the genome is engaged in translation or RNA synthesis. At the initial stage of HCV infection, PCBP2 binding to the 5′UTR promotes a closed-loop form of the viral genome by bridging the 5′ and 3′ UTRs that favors translation (left). By contrast, miR-122 binding to the 5′UTR both disrupts the closed-loop, circular form of the genome, and enhances affinity of the 5′ end of the RNA for the RNA-dependent NS5B RNA polymerase, thereby promoting viral RNA synthesis (right). The interaction of miR-122 with the HCV 5′UTR additionally protects the genome from 5′ dependent cellular exonucleases and stabilizes RNA structure optimal for IRES activity. Continued competition between miR-122 and PCBP2 with similar binding affinities for S2 allows for the dynamic regulation of viral protein versus viral RNA synthesis required for persistent infection with a nonlytic virus.
Taken together, our data provide further support for the concept that the 5′UTR of HCV functions as a molecular switch and that competition between miR-122 and PCBP2 for the 5′UTR regulates whether the genome is engaged in protein synthesis or genome replication (3,16). Because miR-122 and PCBP2 have similar affinities for overlapping binding sites on the 5′UTR, there is a low barrier for the genome to switch between binding to miR-122 or PCBP2 (Figure 7). As such, the fraction of genome population bound to either miR-122 or PCBP2 should closely reflect the relative concentration of the two factors, which could be in flux over the course of infection (49,50). Thus, switching between the two factors would be sensitive to the cellular environment and would allow HCV to tune the balance between replication and translation over its lifecycle. When PCBP2 binds to the 5′UTR, the genome would be circularized, and promote viral translation, possibly by facilitating ribosome re-entry (Figure 7). By contrast, binding of miR-122 to 5′UTR would convert viral genome into a linear form and recruit viral polymerase NS5B for RNA synthesis. This would provide a mechanism by which miR-122 and PCBP2 together regulate both negative-strand RNA synthesis and viral translation. The interaction of miR-122 with the HCV 5′UTR thus serves three distinct functions: (a) protecting the genome from 5′ dependent cellular exonucleases (11–15), (b) stabilizing RNA structure optimal for IRES activity (18–21) and (c) regulating viral RNA synthesis by both modulating closed-loop circularization of the genome and enhancing the affinity of NS5B for 5′ RNA structure. Interestingly, PCBP2 is also an important factor in regulating translation versus replication in the poliovirus life cycle. Cleavage of PCBP2 protein by the viral 3Cpro protease halts viral translation and transitions the RNA into the replication stage (40). Such an ‘on-and- off’ switch is not feasible for HCV due to the need to maintain continuous translation and replication during a lengthy period of persistent infection. Therefore, HCV has adopted an alternative strategy to use miR-122, in competition with PCBP2, to fine-tune the balance between translation and replication.
Supplementary Material
Acknowledgements
We thank the Sealy Center for Structural Biology and Molecular Biophysics for the assistance provided. We thank Luis Holthauzen for his technical help with the ITC experiments, Pawel Bujalowski for his advice on the fluorescence anisotropy experiments and Junji Iwahara for generously allowing the use of his fluorescence spectrophotometer.
Notes
Present address: Oya Bermek, Genome Integrity and Structural Biology Laboratory, National Institute of Environmental Health Sciences, Research Triangle Park, NC 27709, USA.
Contributor Information
Seth Scott, Department of Biochemistry and Molecular Biology, Sealy Center for Structural Biology and Molecular Biophysics, University of Texas Medical Branch, Galveston, TX 77555, USA.
You Li, Department of Medicine, The University of North Carolina at Chapel Hill, Chapel Hill, NC 27517, USA; Lineberger Comprehensive Cancer Center, The University of North Carolina at Chapel Hill, Chapel Hill, NC 27517, USA.
Oya Bermek, Lineberger Comprehensive Cancer Center, The University of North Carolina at Chapel Hill, Chapel Hill, NC 27517, USA; Department of Microbiology and Immunology, The University of North Carolina at Chapel Hill, Chapel Hill, NC 27517, USA.
Jack D Griffith, Lineberger Comprehensive Cancer Center, The University of North Carolina at Chapel Hill, Chapel Hill, NC 27517, USA; Department of Microbiology and Immunology, The University of North Carolina at Chapel Hill, Chapel Hill, NC 27517, USA.
Stanley M Lemon, Department of Medicine, The University of North Carolina at Chapel Hill, Chapel Hill, NC 27517, USA; Lineberger Comprehensive Cancer Center, The University of North Carolina at Chapel Hill, Chapel Hill, NC 27517, USA; Department of Microbiology and Immunology, The University of North Carolina at Chapel Hill, Chapel Hill, NC 27517, USA.
Kyung H Choi, Department of Biochemistry and Molecular Biology, Sealy Center for Structural Biology and Molecular Biophysics, University of Texas Medical Branch, Galveston, TX 77555, USA; Department of Molecular and Cellular Biochemistry, Indiana University, Bloomington, IN 47405, USA.
Data availability
The data underlying this article are available in the article and in its online supplementary material.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Institutes of Health [R01 AI087856 and R21 AI157336 to K.H.C., R01 AI 095690 to S.M.L., ESO13773 and ESO31635 to J.D.G., and T32 GM 008280 to SS]. Funding for open access charge: Indiana University funds.
Conflict of interest statement. None declared.
References
- 1. Chen S.L., Morgan T.R.. The natural history of hepatitis C virus (HCV) infection. Int. J. Med. Sci. 2006; 3:47–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li H.C., Yang C.H., Lo S.Y.. Hepatitis C viral replication Complex. Viruses. 2021; 13:520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li Y., Yamane D., Masaki T., Lemon S.M.. The yin and yang of hepatitis C: synthesis and decay of hepatitis C virus RNA. Nat. Rev. Microbiol. 2015; 13:544–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Luo G., Xin S., Cai Z.. Role of the 5′-proximal stem–loop structure of the 5′ untranslated region in replication and translation of hepatitis C virus RNA. J. Virol. 2003; 77:3312–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Friebe P., Lohmann V., Krieger N., Bartenschlager R.. Sequences in the 5′ nontranslated region of hepatitis C virus required for RNA replication. J. Virol. 2001; 75:12047–12057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Henke J.I., Goergen D., Zheng J., Song Y., Schuttler C.G., Fehr C., Junemann C., Niepmann M.. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008; 27:3300–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang L., Jeng K.S., Lai M.M.. Poly(C)-binding protein 2 interacts with sequences required for viral replication in the hepatitis C virus (HCV) 5′ untranslated region and directs HCV RNA replication through circularizing the viral genome. J. Virol. 2011; 85:7954–7964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jopling C.L., Yi M., Lancaster A.M., Lemon S.M., Sarnow P.. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005; 309:1577–1581. [DOI] [PubMed] [Google Scholar]
- 9. Chang J., Guo J.T., Jiang D., Guo H., Taylor J.M., Block T.M.. Liver-specific microRNA miR-122 enhances the replication of hepatitis C virus in nonhepatic cells. J. Virol. 2008; 82:8215–8223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Machlin E.S., Sarnow P., Sagan S.M.. Masking the 5′ terminal nucleotides of the hepatitis C virus genome by an unconventional microRNA-target RNA complex. Proc. Natl. Acad. Sci. U.S.A. 2011; 108:3193–3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shimakami T., Yamane D., Jangra R.K., Kempf B.J., Spaniel C., Barton D.J., Lemon S.M.. Stabilization of hepatitis C virus RNA by an Ago2-miR-122 complex. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:941–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li Y., Masaki T., Yamane D., McGivern D.R., Lemon S.M.. Competing and noncompeting activities of miR-122 and the 5′ exonuclease Xrn1 in regulation of hepatitis C virus replication. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:1881–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sedano C.D., Sarnow P.. Hepatitis C virus subverts liver-specific miR-122 to protect the viral genome from exoribonuclease Xrn2. Cell Host Microbe. 2014; 16:257–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li Y., Yamane D., Lemon S.M.. Dissecting the roles of the 5′ exoribonucleases Xrn1 and Xrn2 in restricting hepatitis C virus replication. J. Virol. 2015; 89:4857–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Amador-Canizares Y., Bernier A., Wilson J.A., Sagan S.M.. miR-122 does not impact recognition of the HCV genome by innate sensors of RNA but rather protects the 5′ end from the cellular pyrophosphatases, DOM3Z and DUSP11. Nucleic Acids Res. 2018; 46:5139–5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Masaki T., Arend K.C., Li Y., Yamane D., McGivern D.R., Kato T., Wakita T., Moorman N.J., Lemon S.M.. miR-122 stimulates hepatitis C virus RNA synthesis by altering the balance of viral RNAs engaged in replication versus translation. Cell Host Microbe. 2015; 17:217–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Amador-Canizares Y., Panigrahi M., Huys A., Kunden R.D., Adams H.M., Schinold M.J., Wilson J.A.. miR-122, small RNA annealing and sequence mutations alter the predicted structure of the Hepatitis C virus 5′ UTR RNA to stabilize and promote viral RNA accumulation. Nucleic Acids Res. 2018; 46:9776–9792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schult P., Roth H., Adams R.L., Mas C., Imbert L., Orlik C., Ruggieri A., Pyle A.M., Lohmann V.. microRNA-122 amplifies hepatitis C virus translation by shaping the structure of the internal ribosomal entry site. Nat. Commun. 2018; 9:2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chahal J., Gebert L.F.R., Gan H.H., Camacho E., Gunsalus K.C., MacRae I.J., Sagan S.M.. miR-122 and ago interactions with the HCV genome alter the structure of the viral 5′ terminus. Nucleic Acids Res. 2019; 47:5307–5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kunden R.D., Ghezelbash S., Khan J.Q., Wilson J.A.. Location specific annealing of miR-122 and other small RNAs defines an Hepatitis C Virus 5′ UTR regulatory element with distinct impacts on virus translation and genome stability. Nucleic Acids Res. 2020; 48:9235–9249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Diaz-Toledano R., Ariza-Mateos A., Birk A., Martinez-Garcia B., Gomez J.. In vitro characterization of a miR-122-sensitive double-helical switch element in the 5′ region of hepatitis C virus RNA. Nucleic Acids Res. 2009; 37:5498–5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jangra R.K., Yi M., Lemon S.M.. Regulation of hepatitis C virus translation and infectious virus production by the microRNA miR-122. J. Virol. 2010; 84:6615–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Makeyev A.V., Liebhaber S.A.. The poly(C)-binding proteins: a multiplicity of functions and a search for mechanisms. RNA. 2002; 8:265–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cavaluzzi M.J., Borer P.N.. Revised UV extinction coefficients for nucleoside-5′-monophosphates and unpaired DNA and RNA. Nucleic Acids Res. 2004; 32:e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seiler C.Y., Park J.G., Sharma A., Hunter P., Surapaneni P., Sedillo C., Field J., Algar R., Price A., Steel J.et al.. DNASU plasmid and PSI:biology-materials repositories: resources to accelerate biological research. Nucleic Acids Res. 2014; 42:D1253–D1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ferrari E., Wright-Minogue J., Fang J.W., Baroudy B.M., Lau J.Y., Hong Z.. Characterization of soluble hepatitis C virus RNA-dependent RNA polymerase expressed in Escherichia coli. J. Virol. 1999; 73:1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yamane D., McGivern D.R., Wauthier E., Yi M., Madden V.J., Welsch C., Antes I., Wen Y., Chugh P.E., McGee C.E.et al.. Regulation of the hepatitis C virus RNA replicase by endogenous lipid peroxidation. Nat. Med. 2014; 20:927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li Y., Masaki T., Shimakami T., Lemon S.M.. hnRNP L and NF90 interact with hepatitis C virus 5′-terminal untranslated RNA and promote efficient replication. J. Virol. 2014; 88:7199–7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Al-Turki T.M., Griffith J.D.. Mammalian telomeric RNA (TERRA) can be translated to produce valine-arginine and glycine-leucine dipeptide repeat proteins. Proc. Natl. Acad. Sci. U.S.A. 2023; 120:e2221529120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Szymanski M.R., Jezewska M.J., Bujalowski P.J., Bussetta C., Ye M., Choi K.H., Bujalowski W.. Full-length Dengue virus RNA-dependent RNA polymerase-RNA/DNA complexes: stoichiometries, intrinsic affinities, cooperativities, base, and conformational specificities. J. Biol. Chem. 2011; 286:33095–33108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bujalowski P.J., Bujalowski W., Choi K.H.. Interactions between the dengue virus polymerase NS5 and stem–loop A. J. Virol. 2017; 91:e00047-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bujalowski P.J., Bujalowski W., Choi K.H.. Identification of the viral RNA promoter stem loop A (SLA)-binding site on Zika virus polymerase NS5. Sci. Rep. 2020; 10:13306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee E., Bujalowski P.J., Teramoto T., Gottipati K., Scott S.D., Padmanabhan R., Choi K.H.. Structures of flavivirus RNA promoters suggest two binding modes with NS5 polymerase. Nat. Commun. 2021; 12:2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mortimer S.A., Doudna J.A.. Unconventional miR-122 binding stabilizes the HCV genome by forming a trimolecular RNA structure. Nucleic Acids Res. 2013; 41:4230–4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gebert L.F.R., Law M., MacRae I.J.. A structured RNA motif locks Argonaute2:miR-122 onto the 5′ end of the HCV genome. Nat. Commun. 2021; 12:6836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Honda M., Beard M.R., Ping L.H., Lemon S.M.. A phylogenetically conserved stem–loop structure at the 5′ border of the internal ribosome entry site of hepatitis C virus is required for cap-independent viral translation. J. Virol. 1999; 73:1165–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Flynn R.A., Martin L., Spitale R.C., Do B.T., Sagan S.M., Zarnegar B., Qu K., Khavari P.A., Quake S.R., Sarnow P.et al.. Dissecting noncoding and pathogen RNA-protein interactomes. RNA. 2015; 21:135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Khromykh A.A., Meka H., Guyatt K.J., Westaway E.G.. Essential role of cyclization sequences in flavivirus RNA replication. J. Virol. 2001; 75:6719–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lopez-Manriquez E., Vashist S., Urena L., Goodfellow I., Chavez P., Mora-Heredia J.E., Cancio-Lonches C., Garrido E., Gutierrez-Escolano A.L.. Norovirus genome circularization and efficient replication are facilitated by binding of PCBP2 and hnRNP A1. J. Virol. 2013; 87:11371–11387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Perera R., Daijogo S., Walter B.L., Nguyen J.H., Semler B.L.. Cellular protein modification by poliovirus: the two faces of poly(rC)-binding protein. J. Virol. 2007; 81:8919–8932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Choi K.H. The role of the stem–loop A RNA promoter in flavivirus replication. Viruses. 2021; 13:1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. You S., Padmanabhan R.. A novel in vitro replication system for Dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J. Biol. Chem. 1999; 274:33714–33722. [DOI] [PubMed] [Google Scholar]
- 43. Gamarnik A.V., Andino R.. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998; 12:2293–2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shimakami T., Yamane D., Welsch C., Hensley L., Jangra R.K., Lemon S.M.. Base pairing between hepatitis C Virus RNA and microRNA 122 3′ of its seed sequence is essential for genome stabilization and production of infectious virus. J. Virol. 2012; 86:7372–7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thibault P.A., Huys A., Amador-Canizares Y., Gailius J.E., Pinel D.E., Wilson J.A.. Regulation of hepatitis C virus genome replication by Xrn1 and microRNA-122 binding to individual sites in the 5′ untranslated region. J. Virol. 2015; 89:6294–6311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ono C., Fukuhara T., Li S., Wang J., Sato A., Izumi T., Fauzyah Y., Yamamoto T., Morioka Y., Dokholyan N.V.et al.. Various miRNAs compensate the role of miR-122 on HCV replication. PLoS Pathog. 2020; 16:e1008308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Fajardo T., Sanford T.J., Mears H.V., Jasper A., Storrie S., Mansur D.S., Sweeney T.R.. The flavivirus polymerase NS5 regulates translation of viral genomic RNA. Nucleic Acids Res. 2020; 48:5081–5093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Filomatori C.V., Lodeiro M.F., Alvarez D.E., Samsa M.M., Pietrasanta L., Gamarnik A.V.. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev. 2006; 20:2238–2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Choi Y., Dienes H.P., Krawczynski K.. Kinetics of miR-122 expression in the liver during acute HCV infection. PLoS One. 2013; 8:e76501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Luna J.M., Scheel T.K., Danino T., Shaw K.S., Mele A., Fak J.J., Nishiuchi E., Takacs C.N., Catanese M.T., de Jong Y.P.et al.. Hepatitis C virus RNA functionally sequesters miR-122. Cell. 2015; 160:1099–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.