Abstract
Cristae are high‐curvature structures in the inner mitochondrial membrane (IMM) that are crucial for ATP production. While cristae‐shaping proteins have been defined, analogous lipid‐based mechanisms have yet to be elucidated. Here, we combine experimental lipidome dissection with multi‐scale modeling to investigate how lipid interactions dictate IMM morphology and ATP generation. When modulating phospholipid (PL) saturation in engineered yeast strains, we observed a surprisingly abrupt breakpoint in IMM topology driven by a continuous loss of ATP synthase organization at cristae ridges. We found that cardiolipin (CL) specifically buffers the inner mitochondrial membrane against curvature loss, an effect that is independent of ATP synthase dimerization. To explain this interaction, we developed a continuum model for cristae tubule formation that integrates both lipid and protein‐mediated curvatures. This model highlighted a snapthrough instability, which drives IMM collapse upon small changes in membrane properties. We also showed that cardiolipin is essential in low‐oxygen conditions that promote PL saturation. These results demonstrate that the mechanical function of cardiolipin is dependent on the surrounding lipid and protein components of the IMM.
Keywords: cardiolipin, cristae, lipids, mechanics, mitochondria
Subject Categories: Membranes & Trafficking, Metabolism, Organelles
Together, the amount of lipid saturation and cardiolipin define mitochondrial ultrastructure.
Introduction
Mitochondria are ubiquitous eukaryotic organelles whose membrane architecture is required for their metabolic and non‐metabolic functions (Nunnari & Suomalainen, 2012). The inner mitochondrial membrane (IMM) is the site of the Electron Transport Chain (ETC) and consists of two regions defined by their curvature: the flat inner boundary membrane (IBM), adjacent to the outer mitochondrial membrane (OMM), and cristae membranes (CM), which invaginate into the matrix and are connected to the IBM by crista junctions (CJs) (Daems & Wisse, 1966; Perkins et al, 1997). CM structure is dependent on organism, tissue, and physiological state (Revel et al, 1963; Perkins et al, 1997) but is commonly composed of tubular and lamellar membranes (Revel et al, 1963; Mannella, 2006b; Zick et al, 2009; Pánek et al, 2020; Mendelsohn et al, 2022). Cristae effectively increase membrane surface area for ETC reactions and act as a “proton sink” where protons travel expeditiously to F1F0 adenosine–triphosphate (ATP) synthase (Davies et al, 2011; Rieger et al, 2014; Cogliati et al, 2016). Cristae could also serve as diffusion barriers for metabolites between the intracristal space (ICS) and the intermembrane space (IMS), controlling the flux of ADP/ATP through the adenine nucleotide translocase (ANT) (Mannella et al, 1994, 1997; Frey & Mannella, 2000; Mannella, 2006a). All these potential functions of cristae are dependent on their high intrinsic membrane curvature.
The IMM is shaped by proteins that drive assembly and maintenance of cristae. The molecular determinants of CM are best understood in Saccharomyces cerevisiae, where ATP synthases form ribbon‐like rows of dimers, which induce curvature along tubule and lamellar rims (Dudkina et al, 2005; Strauss et al, 2008; Davies et al, 2011; Blum et al, 2019). Loss of the dimerization subunit g, Atp20p, results in monomeric ATP synthases and onion‐like mitochondria with flat layers of IMM that run parallel with the OMM (Arnold et al, 1998; Paumard et al, 2002; Arselin et al, 2004; Rabl et al, 2009). Reconstituted ATP synthase dimers spontaneously assemble into rows driven by changes to elastic membrane bending energies (Anselmi et al, 2018) and are sufficient to form tubular liposomes (Blum et al, 2019). At the CJ, the dynamin‐related GTPase optic atrophy protein 1 (OPA1)/Mgm1p interacts with the mitochondrial contact site and cristae organizing system (MICOS) complex (Frezza et al, 2006; Harner et al, 2011; Hoppins et al, 2011; Patten et al, 2014; Glytsou et al, 2016; Hu et al, 2020). Cells lacking Mgm1p feature a completely flat IMM (Sesaki et al, 2003; Harner et al, 2016), while loss of the major MICOS subunit Mic60p results in elongated cristae sheets that do not contain CJs (Rabl et al, 2009; Harner et al, 2011).
In addition to their proteinaceous determinants, mitochondrial lipids are hypothesized to play key roles in shaping cristae. The predominant phospholipids (PLs) of the IMM are phosphatidylethanolamine (PE), phosphatidylcholine (PC), and cardiolipin (CL) (Zinser et al, 1991; Mejia & Hatch, 2016). IMM phospholipids, or their phosphatidylserine (PS) precursors in the case of PE, are imported from the ER at contact sites (Horvath & Daum, 2013), CL and its precursor phosphatidylglycerol (PG) are synthesized in and remain localized to the IMM. Among PLs, CL is unique in featuring four acyl chains whose larger cross‐sectional area contributes to an overall conical shape (LeCocq & Ballou, 1964; Beltrán‐Heredia et al, 2019). In liposomes, CL localizes to regions of high curvature and can drive pH‐dependent invaginations (Khalifat et al, 2008, 2011; Ikon & Ryan, 2017), suggesting a role in promoting curved membrane topologies. The curvature of CL itself varies depending on the local lipid and chemical environments (Chen et al, 2015; Beltrán‐Heredia et al, 2019); molecular simulations predict key roles for its ionization state (Dahlberg & Maliniak, 2010) and binding of counter ions (preprint: Konar et al, 2023). Despite these biophysical data, the fundamental mitochondrial functions of CL are not fully resolved. In the genetic disorder Barth syndrome, loss of the acyl chain remodeler Tafazzin causes reduced amounts and altered composition of CL (Adès et al, 1993; Bione et al, 1996) leading to abnormal cristae (Acehan et al, 2007), which have also been observed in cell lines lacking CL synthesis (Claypool & Koehler, 2012; Ren et al, 2014; Ikon & Ryan, 2017; Paradies et al, 2019). In yeast, however, loss of cardiolipin synthase (Crd1p) does not render a respiratory or morphological phenotype under regular growth temperatures (Jiang et al, 1997; Baile et al, 2014). It thus remains unknown if CL serves a mechanical role in the IMM, or has more organism‐specific functions relating to ETC enzymes (Xu et al, 2021) and their organization (Zhang et al, 2005).
The acyl chain composition of mitochondrial PLs also differs from other organelles (Harayama & Riezman, 2019) and broadly regulates membrane biophysical properties. The IMM is enriched in unsaturated and polyunsaturated PLs, which promote membrane fluidity, and lacks sterols and saturated sphingolipids, which promote membrane ordering (Filippov et al, 2003; Vance, 2015). During synthesis, lipid unsaturation is controlled by the activity of fatty acid desaturases, such as Ole1p in yeast (Bard, 1972; Stukey et al, 1989). OLE1 was discovered in genetic screens for both unsaturated fatty acid auxotrophy and mitochondrial distribution and morphology genes (MDM). Mutations in mdm2 resulted in abnormal mitochondrial morphology and inheritance (McConnell et al, 1990; Stewart & Yaffe, 1991) but were later identified as OLE1 alleles, indicating an unexplained link between desaturase activity and mitochondrial structure. In mammalian cells, addition of exogenous saturated fatty acids such as palmitic acid (PA) drives mitochondrial dysfunction (Sparagna et al, 2000; Penzo et al, 2002; Jheng et al, 2012) and can cause the progressive loss of CMs (Xue et al, 2019). Metabolic diseases, such as obesity and type 2 diabetes, have also been associated with both saturated fat accumulation and mitochondrial stress (Petersen et al, 2004; Lowell & Shulman, 2005).
Here, we combine experimental perturbations with multi‐scale modeling to elucidate new roles for conserved mitochondrial lipids in IMM morphology, using yeast as a model system. We first used genetic manipulation of PL saturation and observed a surprising lipidic breakpoint, in which the IMM becomes flat and mitochondria lose their ATP synthesis capacity. This transition is controlled by the IMM lipidome in two distinct ways: through modulation of ATP synthase oligomerization by PL saturation and through loss of intrinsic membrane curvature provided by CL. We develop a mathematical model to explain these effects by considering the energetics of lipid and protein‐mediated membrane curvature. We then show CL function is dependent on growth conditions that modulate PL saturation, most notably oxygenation, and that it has an essential role in natural yeast growth environments.
Results
Systematic modulation of the yeast PL double bond profile reveals a critical mitochondrial breakpoint in IMM curvature and ATP generation
Bulk PL membrane properties are in part controlled by the stoichiometry between saturated and unsaturated acyl chains. To modulate lipid saturation in budding yeast, we utilized a library of promoters controlling the expression of OLE1 (Fig 1A). We focused on four strains, saturated fatty acid (SFA) 1‐4, which showed a range in lipid saturation (Fig 1A). SFA1 features a wild‐type (WT) PL composition, while SFA2‐4 have consecutively increasing levels of PL acyl chain saturation due to lower levels of OLE1 expression (Appendix Fig S1A). Among PC and PE lipids, WT and SFA1 strains possess predominantly di‐unsaturated PLs, while SFA 2, 3, and 4 (weaker OLE1 expression) show an increasing ratio of mono‐ to di‐unsaturated species and incorporation of fully saturated PLs (Fig 1A). We observed potentially compensatory adaptations to increasing saturation in whole‐cell PLs (Appendix Fig S1, Dataset EV1), including a decrease in the PE/PC ratio (Appendix Fig S1B), used by several organisms to increase membrane fluidity (Janssen et al, 2000; Dawaliby et al, 2016), an increase in PI (Appendix Fig S1B), and shortening of acyl chains length (Appendix Fig S1C), which also occurs during yeast cold adaptation (Al‐Fageeh & Mark Smales, 2006).
Figure 1. Modulation of OLE1 expression results in a critical level of PL saturation driving loss of ATP synthase oligomerization and mitochondrial morphology.
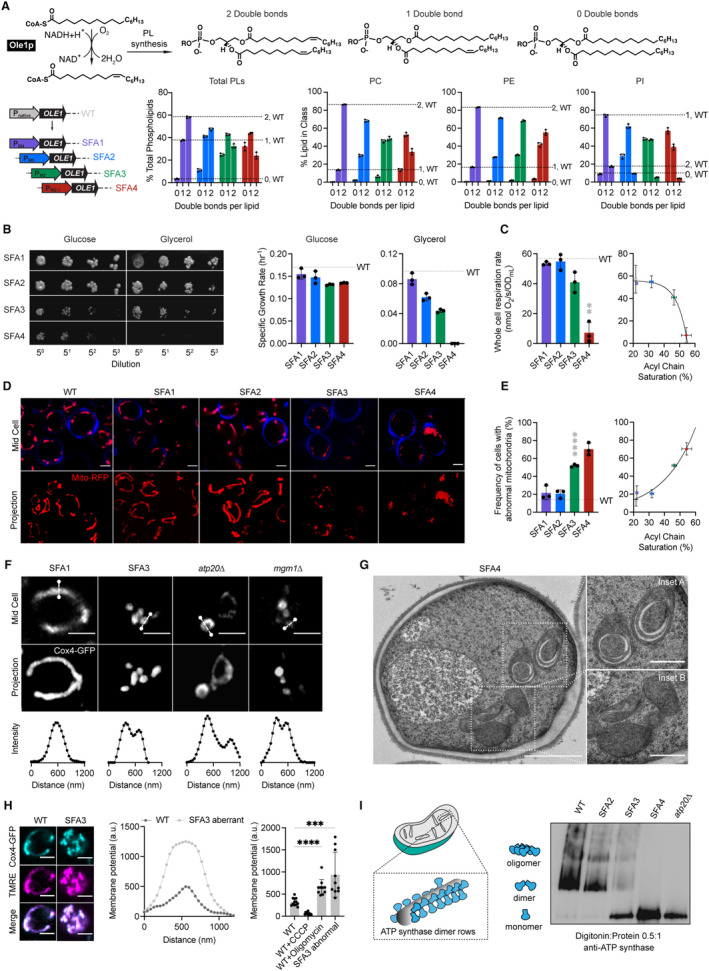
- The yeast desaturase, Ole1p, is an oxygen‐dependent enzyme that introduces cis double bonds at the C9 position (top left). SFA strains were generated via promoter substitution, resulting in progressively decreasing levels of OLE1 expression (bottom left). Lipidomics analysis showing double bond distributions of the total PL pool as well as of individual PLs within SFA strains; the wild‐type distribution is depicted with dotted lines. Error bars indicate SD from biological replicates n = 3.
- SFA4 cells lose viability under respiratory conditions. Shown are serial dilutions of yeast cells plated on media containing fermentable (glucose) and non‐fermentable (glycerol) carbon sources and specific growth rates for each in liquid cultures. Error bars indicate SD from n = 3 independent cultures.
- (Left) SFA3 and SFA4 cells show a drop in whole‐cell respiration, measured using a Clark electrode. Error bars indicate SD from n = 3 independent cultures. **P = 0.004, unpaired two‐tailed t‐test of SFA4 compared against wild‐type. (Right) Scatter plot depicting a fitted single exponential (R 2 = 0.99) for the decrease in respiration as a function of acyl chain saturation.
- SFA3 and SFA4 cells lose tubular mitochondrial morphology. Shown are representative Airyscan confocal micrographs of yeast expressing matrix‐localized RFP (mts‐RFP). Cells were stained with cell wall‐binding calcofluor white (blue) for clarity. Scale bars, 2 μm.
- Mitochondrial morphology changes between SFA2 and SFA3 strains, as assayed by confocal microscopy of cells harboring an mts‐RFP plasmid. N > 50 cells were counted in biological triplicate in each condition. Error bars indicate SD from n = 3 independent cultures. ****P < 0.0001, unpaired two‐tailed t‐test of SFA3 compared against wild‐type. (Right) Scatter plot depicting a fitted single exponential (R 2 = 0.97) for the increase in frequency of abnormal mitochondria as a function of acyl chain saturation.
- Airyscan confocal micrographs of yeast expressing IMM protein Cox4‐GFP showing hollow mitochondria in SFA3 cells, as is also observed in mutants of CM‐shaping proteins. Scale bars, 2 μm. Profiling analysis (below) depicts fluorescence intensity as a function of the distance across the indicated mitochondrion; two peaks indicate a lack of fenestrated IMM.
- Thin‐section TEM micrographs of high‐pressure frozen (HPF) SFA4 yeast showing the appearance of an onion‐like (Inset A) IMM and total loss of CM (Inset B). Scale bars, 1 μm (full image), 400 nm (insets A and B).
- Abnormal mitochondria show an increase in membrane potential, consistent with loss of ATP synthase, but not ETC activity. (Left) Representative micrographs are shown of individual cells expressing Cox4‐GFP and stained with TMRE. (Center) Example membrane potential line scan plots showing an example of an abnormal SFA3 cell with higher TMRE intensity compared to WT. (Right) Quantification of TMRE peak intensities from line profiling analysis of N > 10 cells per condition showing a higher membrane potential in aberrant mitochondria, as well as those where ATP synthase is inhibited by oligomycin (5 μM), and lower potential in cells treated with the uncoupler CCCP (20 μM). The line profile measured the intensity between two points crossing an individual mitochondrion. Scale bars, 2 μm. ***P < 0.0005, ****P < 0.0001 unpaired two‐tailed t‐test of SFA3 aberrant and WT + oligomycin compared against WT. Error bars indicate SD.
- ATP synthase oligomerization is lost with increasing PL saturation. SFA2 mitochondria lose higher order oligomers observed in WT cells, while SFA3 and SFA4 mitochondria possess predominantly monomeric ATP synthase, similar to atp20 cells. Shown are BN‐PAGE western blots of digitonin‐solubilized purified mitochondria, atp20 is the monomeric ATP synthase control.
Source data are available online for this figure.
When handling these strains, we observed that SFA3 and SFA4 cells were characterized by loss of viability in non‐fermentable carbon sources (Fig 1B), oxygen consumption (Fig 1C), and tubular mitochondrial networks (Fig 1D and E, Appendix Fig S2). In contrast, non‐mitochondrial physiology—assayed by growth under fermentation conditions (Fig 1B), morphology of other organelles (Appendix Fig S3A), and activation of the unfolded protein response (UPR) (Appendix Fig S3B)—remained unaffected within this range of OLE1 mutants. Thus, a modest increase in saturation conferred a sudden loss of mitochondrial function. We further observed that SFA3 and SFA4 mitochondria featured gaps in Cox4‐GFP, a subunit of cytochrome c oxidase in the IMM, similar to atp20 and mgm1 cells that lack CMs (Fig 1F, Appendix Table S4). Transmission electron microscopy (TEM) analysis of SFA4 cells prepared by high‐pressure freeze substitution (HPFS) (Fig 1G, Appendix Fig S4A and B) showed mitochondria with flat IMM membranes similar to those in atp20 (Paumard et al, 2002) or mgm1 cells (Harner et al, 2016). The moderate increase in saturation in SFA2 cells, which does not cause aberrancy independently, also showed epistasis with loss of cristae‐shaping proteins at both the cristae ridge (Atp20p) and rim (Mic60p of the MICOS complex) (Fig EV1A and B). We thus hypothesized that PL saturation has a specific effect on formation of CMs.
Figure EV1. Interactions between cristae‐shaping proteins, lipid saturation, and mitochondrial phenotypes.
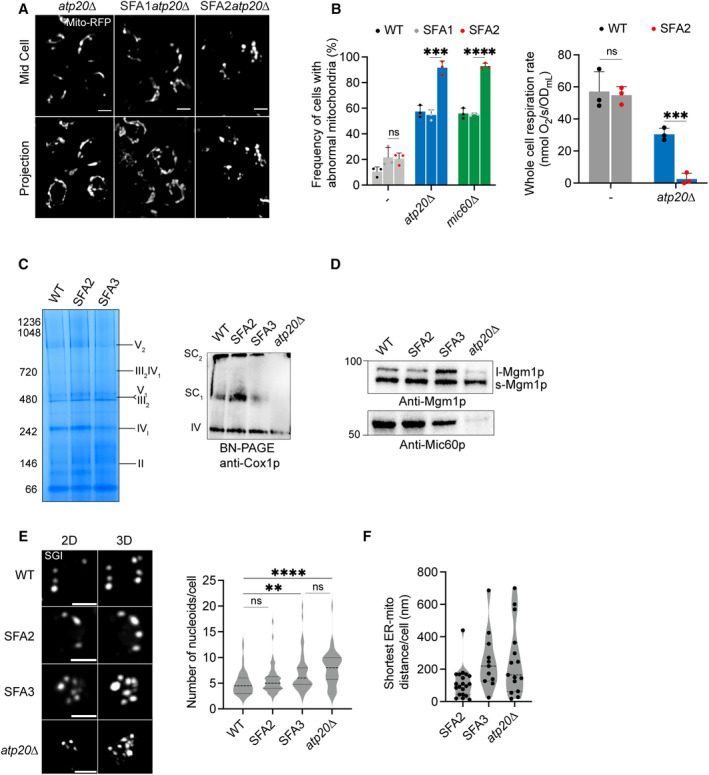
- Representative Airyscan confocal micrographs of yeast expressing matrix‐localized RFP (mts‐RFP). Scale bars, 2 μm.
- (Left) Frequency of mitochondrial abnormalities as assayed by analysis of mts‐RFP. N > 50 cells were counted in biological triplicate (n = 3) in each condition. Error bars indicate SD. ***P = 0.0006, ****P < 0.0001 unpaired two‐tailed t‐test compared SFA2atp20 and SFA2mic60 against SFA1atp20 and SFA1mic60 , respectively. (Right) Respiration rates in atp20 and SFA2atp20 cells measured in biological triplicate (n = 3) using a Clark electrode. Error bars represent SD. ***P < 0.0005, unpaired two‐tailed t‐test compared against atp20 .
- SFA2/3 mitochondria do not show defects in ETC complexes or SC formation. BN‐PAGE on digitonin‐solubilized isolated mitochondria from SFA strains and atp20 revealed no changes in ETC complex levels (left) or supercomplexes (top right). Mitochondria from atp20 cells show defects in formation of Complex IV‐containing supercomplexes.
- Increasing lipid saturation does not change the status of non‐ATP synthase cristae‐shaping proteins. To determine the presence of Mgm1p and Mic60p in SFA strains, 10 μg of isolated mitochondria was loaded and subjected to immunoblotting against anti‐Mgm1p (C‐term) and anti‐Mic60p antibodies, respectively.
- Both SFA3 and atp20 cells showed increased abundance of mtDNA nucleoids compared to WT. Nucleoids were visualized after staining with SYBR Green I (SGI), and the number of nucleoids per cell (N = 50 cells) were quantified. Error bars indicate SD. Scale bars, 1 μm. ****P < 0.0001, **P = 0.0020, unpaired two‐tailed t‐test against WT for atp20 and SFA3, respectively.
- SFA3 and atp20 cells showed increased ER‐mitochondria contact distances as analyzed by thin‐section TEM. In each condition, the ER‐mitochondria distances were measured using IMOD 3D software. Error bars represent SD. N = 15 micrographs were quantified for each condition.
We next asked how increasing lipid saturation resulted in the aberrant mitochondrial morphologies. SFA3 mitochondria retained normal levels of ETC complexes as well as respiratory chain supercomplexes (SCs) (Fig EV1C). Aberrant mitochondria in SFA3 cells retained membrane potential (Fig 1H) as measured by staining with tetramethylrhodamine ethyl ester (TMRE), which was also inconsistent with a loss of proton‐pumping reactions in the ETC. Furthermore, examination showed that TMRE fluorescence in aberrant mitochondria was elevated to a similar level as WT cells treated with the ATP synthase inhibitor oligomycin, suggesting that they were characterized by an impediment to ATP synthesis itself. We analyzed ATP synthase in isolated mitochondria with blue native PAGE (BN‐PAGE) western blotting, which can assay the organization of complexes. In mitochondria from SFA strains, increasing saturation progressively altered ATP synthase organization. WT mitochondria contained predominantly dimeric and oligomeric ATP synthase, while oligomers are lost in SFA2 cells. SFA3 and SFA4 mitochondria predominantly contained monomeric ATP synthases, similar to atp20 cells (Fig 1I). SFA3 cells notably retained other cristae‐shaping proteins, including Mic60p and the long and short forms of Mgm1p (Fig EV1D), indicating that this effect on ATP synthase was specific. Furthermore, all phenotypes observed in SFA3 mitochondria, including an increase in the number of mtDNA nucleoids (Fig EV1E) and the average contact distance with the ER (Fig EV1F), could also be phenocopied by loss of ATP synthase dimerization in atp20Δ. These data suggested that saturated lipids directly modulate ATP synthase organization and through this mechanism alter CM formation.
Modeling of transport processes in aberrant mitochondria suggest a mechanism for loss of ATP generation
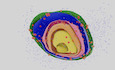
Loss of ATP synthase oligomerization provides a mechanism for loss of CM morphology, but did not fully explain the respiratory phenotypes of SFA3/4 and atp20Δ because monomeric ATP synthases retain ATPase activity (Arnold et al, 1998; Paumard et al, 2002). To explore the functional consequences of CM loss, we more extensively characterized aberrant IMM morphologies (Fig 2). Two types of structures were observed in wide‐field TEM micrographs of SFA4 cells: onion‐like mitochondria with multiple IMM layers, which were observed in at least 40% of cells, and flat IMMs with a single membrane running parallel to the OMM (Appendix Fig S5A). Tomography of onion‐like IMMs showed connection vertices and alternating layers of matrix and IMS (Appendix Figs S4C and S5B), as previously observed in atp20 cells (Paumard et al, 2002), suggesting a continuous IMM that could arise during membrane growth (Appendix Fig S5C) to resemble discrete IMM layers. We carried out 3D reconstructions of these morphologies from multi‐tilt TEM (Appendix Fig S5A, Movies [Link], [Link]) using the GAMer 2 platform (Lee et al, 2020) and computed two types of local membrane curvature across their surfaces, based on the maximum curvature (k1) and minimum curvature (k2) at each point. The mean between these two principal curvatures (k1 and k2) provides information about how the surface normal changes at a given point (mean curvature), while the difference between them captures the extent of anisotropy between the two directions (deviatoric curvature). This analysis showed that the IMM of SFA2 mitochondria was marked by regions of high (> 25 μm−1) mean and deviatoric curvatures (Fig 2A, C and D), but these were completely absent in both types of aberrant morphologies (Fig 2B–D).
Figure 2. PL saturation shifts the IMM to a regime of low membrane curvature, which reduces IMM surface area and modeled transport rates needed for ATP generation.
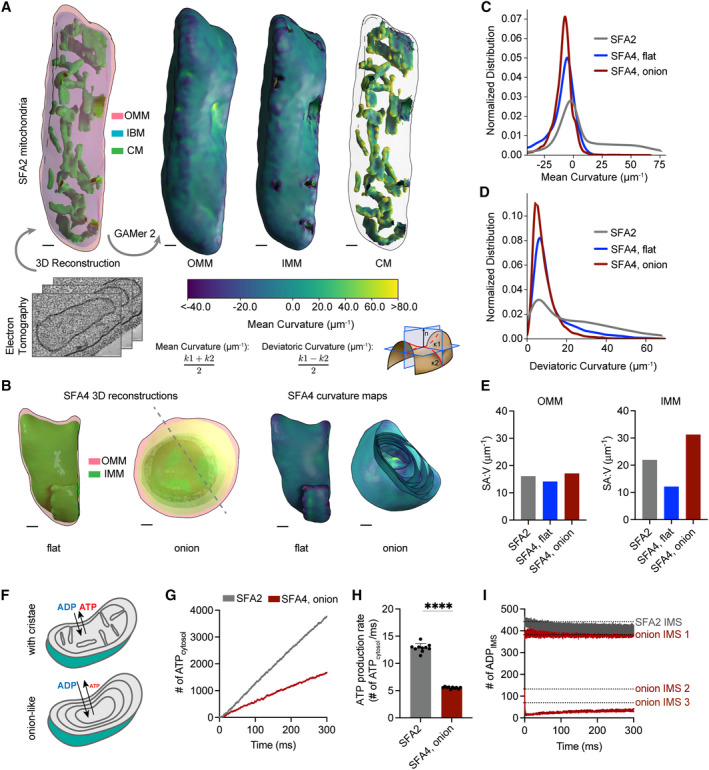
- 3D reconstructions of mitochondrial membrane topology and quantification of curvature using the GAMer 2 pipeline. Shown are electron tomograms of mitochondria from SFA2 cells, which show regular, tubular CM. CMs are highlighted alongside the OMM and the inner boundary membrane (IBM) in the 3D reconstruction. (Right) Maps of mean curvature of OMM, IMM, and CM computed using GAMer2. Scale bars, 50 nm.
- 3D reconstructions of SFA4 mitochondria showing flat (mgm1 ‐like) and onion (atp20 ‐like) abnormal morphologies. Also shown are the maps of mean curvature showing the IMM of these abnormal mitochondria, using the same color scale as in (A). The onion‐like IMM is sliced at an angle to illustrate the many layers; each layer is spherical in nature. Scale bars, 50 nm.
- Histograms of mean curvature distributions of inner membranes generated from SFA2 and SFA4 reconstructions, highlighting that high mean curvature areas in SFA2 IMM are lost in SFA4 cells.
- Histograms of deviatoric curvature distributions of inner membranes generated from SFA2 and SFA4 reconstructions, which are mechanical corollaries to curvature induced by ATP synthase dimerization, highlight that high deviatoric curvature areas in SFA2 IMM are lost in SFA4 cells.
- The IMMs of onion mitochondria show increased surface area:volume (SA:V) ratio compared to SFA2 while flat mitochondria show a decrease in the SA:V without major changes to the OMM.
- Schematic of ATP production in mitochondria containing normal CMs and those showing a multi‐layer, onion‐like IMM, highlighting how multiple membrane layers could impede in trafficking of ADP and ATP.
- Modeled cytosolic ATP generation from mitochondria with a CM‐containing morphology (taken from SFA2 tomogram) vs. an onion‐like (taken from SFA4 tomogram) IMM. ATP generated in the cytosol in each condition is an average from 10 simulations. Details of the model equations and simulations are provided in the Appendix.
- Comparison of cytosolic ATP generation rates derived from multiple Monte Carlo simulations shown in (G). Ten simulations were run for each morphology. Error bars indicate SD; ****P < 0.0001, unpaired two‐tailed t‐test.
- Modeled substrate depletion in onion‐like mitochondria. In the CM‐containing mitochondrion, the ADP level in the IMS remains constant at ~ 440 ADP throughout the 300 ms simulation. In the onion‐like SFA4 mitochondrion, ADP remains constant only in the first IMS layer and is rapidly depleted in layers 2 and 3, indicating that the multi‐layer structure could be limiting for ATP/ADP trafficking. Dotted lines represent initial values of ADP in each IMS layer.
Source data are available online for this figure.
Flat and onion‐like IMMs differed dramatically in their surface area, with the former showing a reduced surface area:volume ratio (SA:V) and the latter an increased one (Fig 2E). Previous modeling studies have shown that SA:V is an important determinant of flux of molecules between different compartments (Rangamani et al, 2013; Cugno et al, 2019; Calizo et al, 2020). We hypothesized that the onion‐like IMM structure would impede transport of ATP out of, and ADP into. To explore this hypothesis, we employed an MCell‐based reaction–diffusion simulation pipeline for modeling ATP generation using EM‐derived morphologies (Garcia et al, 2019). These simulations showed that mitochondria with multiple IMM layers cause lower ATP generation when cytosolic ADP concentration was kept constant (Fig 2F–H, Movies EV4 and EV5). We also observed that predicted ATP production was inhibited by depletion of ADP as a substrate in the inner layers of the onion‐like IMM, highlighting that ATP/ADP exchange by ANT could be limiting for high surface area IMM morphologies with low membrane curvature (Fig 2I). These simulations suggest that efficient substrate transport could be dependent on CMs and explain why low curvature IMM morphologies that retain high surface area could still impede ATP generation.
Lipid saturation modulates membrane mechanical properties which are buffered by mitochondrial‐specific headgroup changes
We next sought to define the lipid‐encoded membrane properties that drive CM loss. We first analyzed the lipidomes from mitochondria isolated from SFA strains by density ultracentrifugation (Appendix Fig S6). For our analyses of the IMM, we used whole mitochondrial lipidomes, as these were nearly identical to those of isolated IMMs from the same sample, with the notable exception of CL levels (Appendix Fig S1F, Dataset EV2). As in the whole‐cell lipidome, decreasing Ole1p activity in SFA strains increased mitochondrial PL saturation, but fewer fully saturated species were observed. Instead, the lipidic breakpoint in SFA3 cells corresponded to changes in the double bond distribution from predominantly di‐unsaturated to monounsaturated PLs, e.g. PE‐16:1/18:1 to PE‐16:0/16:1 (Fig 3A and B), a surprisingly modest shift given the magnitude of the morphological and functional change.
Figure 3. Modeling of lipid‐driven mechanical changes in the IMM and their homeostatic responses.
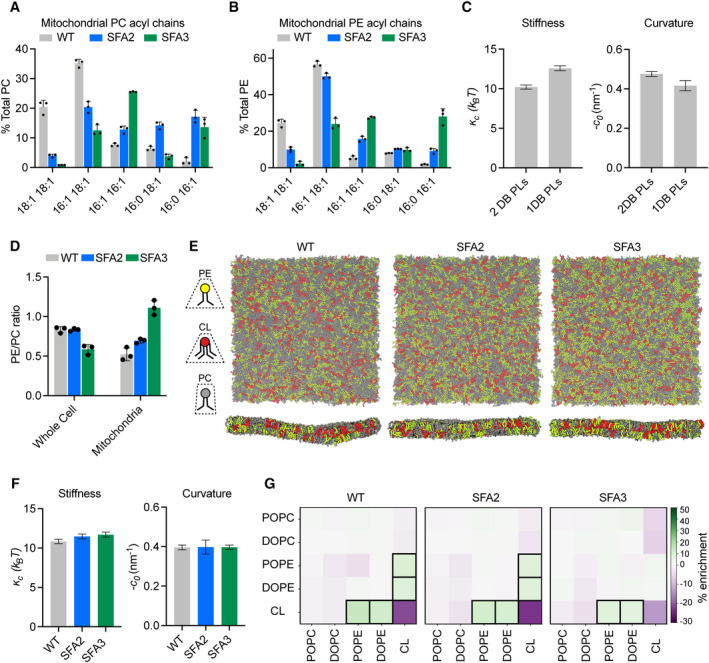
- Lipidomic profile of isolated mitochondria showing a transition from di‐unsaturated to monounsaturated PC, n = 3 biological replicates. Error bars represent SD.
- Mitochondrial PE shows a similar transition from di‐unsaturated species to monounsaturated species, n = 3 biological replicates. Error bars represent SD.
- Changes to membrane mechanical properties predicted by Martini 2 CG‐MD simulations of mitochondrial‐like lipid mixtures that shift from di‐unsaturated (2 double bonds, DB) to monounsaturated (1 DB) PLs. Stiffness (bending modulus) increases while negative spontaneous curvature, derived from the first moment of the lateral pressure profile and bending modulus, decreases. Compositions of these “ideal” systems are shown in Fig EV2A. Error bars indicate SD. Error values were derived from one continuous simulation by quantification of statistical inefficiency, as described in the Materials and Methods.
- Increasing lipid saturation reduces the PE/PC ratio in whole cells, but increases it in isolated mitochondria, suggesting a curvature‐based adaptation to increasing saturation. Lipidomic analysis was conducted on biological triplicates (n = 3). Error bars indicate SD.
- Top‐down and side‐on snapshots of CG‐MD bilayers showing headgroup adaptation to increasing saturation in SFA strains.
- Simulations of the lipid systems derived from SFA mitochondria that incorporate homeostatic headgroup changes. In contrast to the systems in (C), the complex systems show only small changes to membrane mechanical properties, suggesting that headgroup adaptations may offset the changes to mechanical properties associated with increased saturation. Error bars indicate SD. Error values were derived from one continuous simulation by quantification of statistical inefficiency, as described in the Materials and Methods.
- Compositional enrichment within the local neighborhood of individual lipid types. Given a lipid type (y‐axis) the color indicates the percent enrichment in likelihood of observing an x‐axis labeled lipid within a 1.5‐nm radius neighborhood compared to random distribution. Boxes indicate conditions with > 5% enrichment from random. PO/DOPE are enriched around CL, indicating the association between conically shaped lipids.
Source data are available online for this figure.
To explore the effects of this change in acyl chains on membrane properties, we employed coarse‐grained molecular dynamics (CG‐MD) simulations of lipid bilayers. CG‐MD forgo atomistic detail in favor of improved sampling of complex mixtures (Marrink & Tieleman, 2013; Marrink et al, 2019), so we first tested whether they capture known mechanical properties of saturated PLs. For example, the stiffness (к c ) of saturated PL bilayers is higher than those with unsaturated (Appendix Table S1A and B) and polyunsaturated chains (Rawicz et al, 2000; Manni et al, 2018). A second key parameter is the monolayer spontaneous curvature, c 0 , a measure of lipid shape. Cylindrical lipids, like PC, feature c 0 values near zero, while conical lipids like PE or CL feature negative c 0 values (Dymond, 2021). PL acyl composition also modulates c 0 , with more voluminous unsaturated chains favoring negative spontaneous curvatures (Szule et al, 2002). We simulated large membranes (~ 40 nm × 40 nm, 5,000 lipids) containing a simplified mixture of IMM lipids (50:30:20 PC:PE:CL) with either monounsaturated or di‐unsaturated PC & PE. Thermally induced height undulations were analyzed to derive к c from each membrane composition. In parallel, small membranes (~ 15 nm × 15 nm, 700 lipids) of identical compositions were used to compute the lateral pressure profiles, whose first moment is equal to the product of к c and c 0 . Using к c values derived from the large systems, c 0 values can thus be extracted. The shift from diunsaturated to monounsaturated PLs led to a ~ 30% increase in stiffness, к c (Fig 3C), consistent with previous experimental measurements in monocomponent liposomes (Appendix Table S1A and B). Similarly, monounsaturated mixtures showed a ~ 25% increase in c 0, consistent with previous measurements by small angle x‐ray scattering (Appendix Table S1C). We thus concluded that CG‐MD using the Martini 2 force field can reproduce the expected changes to mechanical properties modulated by PL lipid saturation.
We then proceeded to analyze complex mixtures derived from mitochondrial lipidomes. In SFA strains, we observed a mitochondrial‐specific change in headgroup composition: PE levels increased with increasing saturation at the expense of PC, resulting in an increase in the PE/PC ratio (Fig 3D, Appendix Fig S1E). In contrast, the PE/PC ratio decreased in the corresponding whole‐cell lipidome (Fig 3D, Appendix Fig S1B). We also observed an increase in PS levels, which serve as an intermediate for PE synthesis by Psd1p (Voelker, 1997). Because PE has a higher melting temperature than PC (Dawaliby et al, 2016), its increase argues against a fluidity‐specific stress of saturation on the mitochondria. Instead, the high negative spontaneous curvature of PE suggested an adaptation to membrane spontaneous curvature itself. To understand the biophysical basis for this adaptation, we employed the CG‐MD workflow described on lipid mixtures that mimic the changes in headgroup composition in SFA strains (Fig 3E). In these systems, shifting from the WT mitochondrial lipidomes to those of the SFA3 mitochondria resulted in only a modest increase in stiffness (к c) and no change to the magnitude of c 0, despite the increase in PL saturation (Fig 3F). These findings suggested that the mitochondrial‐specific increase in conical lipids acts to buffer membrane mechanical properties relevant to curvature generation.
Beyond changes in the PE/PC ratio, our membrane simulations highlighted the key role of CL in dictating properties relevant to curvature generation (Fig EV2A and B). Replacement of CL with its precursor lipid PG resulted in an overall stiffening of simulated IMMs and loss of spontaneous curvature (Fig EV2D and E). Switching CL from the dianion form, predominant at neutral pH, to the monoanion form had the opposite effect, dramatically softening the bilayer and increasing spontaneous curvature (Fig EV2C–E). In simulations without CL, the increase in PE in SFA strains could only partially compensate for curvature lost when CL was removed (Fig EV2F). CL preferentially clustered with PE lipids in the simulated bilayers (Fig 3G), highlighting how conical lipids sort together in areas of high curvature (Callan‐Jones et al, 2011). This association was maintained in SFA lipidomes, though the extent of CL‐PE association was reduced in the saturated SFA3 bilayers (Fig 3G).
Figure EV2. Coarse‐grained molecular dynamics predicts an important role for CL in shaping membrane mechanical properties.
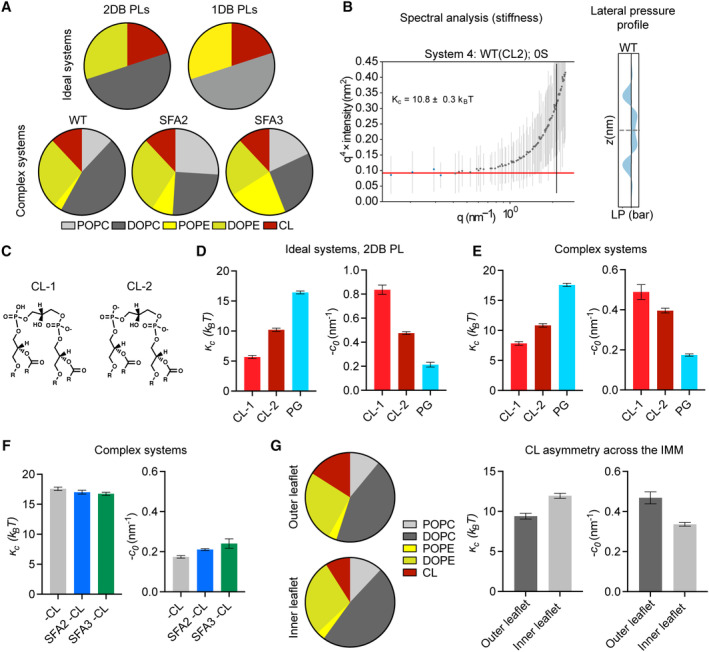
- Ideal systems contained lipid compositions of fixed abundance, while changing the unsaturation of the acyl chains from di‐unsaturated to monounsaturated. In contrast, the “complex” systems mimicking the mitochondrial lipidomes of SFA strains accounted for headgroup adaptations to increasing saturation, such as increasing PE. Full compositions are listed in Appendix Table S2.
- Example spectral analysis of thermal undulations, used to calculate bending moduli, and lateral pressure profiles, used to calculate spontaneous curvature.
- Chemical structures of CL in dianion (CL‐2) and monoanion (CL‐1) ionization states.
- In ideal systems, the changing of ionization state of CL from −1 to −2 causes a minor increase in membrane stiffness and a major reduction in spontaneous curvature. Absence of CL increases membrane stiffness and reduces spontaneous curvature as determined through Martini CG‐MD. Error bars indicate SD. Error values were derived from one continuous simulation by quantification of statistical inefficiency or analysis of non‐overlapping blocks, as described in the Appendix.
- Accounting for headgroup adaptations in complex systems, simulations still show the same trend with dianionic CL and loss of CL showing increases in membrane stiffness and a reduction in spontaneous curvature. Error bars indicate SD. Error values were derived from one continuous simulation by quantification of statistical inefficiency or analysis of non‐overlapping blocks, as described in the Appendix.
- While increasing lipid saturation has a minimal effect on membrane stiffness in the absence of CL, the increase in the magnitude of spontaneous curvature suggests that the presence of PE can partially, but not completely, compensate for loss of curvature provided by CL. Error bars indicate SD. Error values were derived from one continuous simulation by quantification of statistical inefficiency or analysis of non‐overlapping blocks, as described in the Appendix.
- Modeling of outer leaflet enrichment of CL in the yeast IMM. Simulated changes in CL concentrations previously reported (Gallet et al, 1997) results in membrane softening and increased spontaneous curvature. Two sets of simulations were set up with the estimated compositions of the outer and inner IMM leaflets shown in the pie charts. The simulated outer membrane systems were softer (lower stiffness) and had a larger negative spontaneous curvature. The difference in the outer and inner leaflet curvatures was 0.1 ± 0.04 nm−1, which is an estimation for asymmetry‐induced c 0. Error bars indicate SD. Error values were derived from one continuous simulation by quantification of statistical inefficiency or analysis of non‐overlapping blocks, as described in the Appendix.
An epistasis between PL saturation and CL abundance underlies IMM structure
The observation that mitochondria in SFA2 cells responded to increased PL saturation by increasing the abundance of high‐curvature PE and CL lipids compared to WT (Appendix Fig S1D) led us to consider the interplay of PL curvature and saturation. While PL saturation itself can modulate intrinsic lipid curvature (c 0 ) (Appendix Table S1C), there is no evidence that acyl chain composition differs across the two leaflets of the IMM, which would be needed to generate net membrane curvature across the bilayer (C 0 ). We instead hypothesized that the effects of CL on predicted membrane curvature and the established asymmetric localization of CL in the IMM (Gallet et al, 1997) could provide membrane curvature independent of cristae‐shaping proteins, such as ATP synthase dimers. To explore this possibility, we tested out compositions based on previously measured asymmetries of CL in the yeast IMM. The simulations predicted that enrichment of CL in the IMM outer leaflet increased c 0 and reduced к c (Fig EV2G). Based on values for the former, we estimated that CL asymmetry could contribute at least a −0.05 nm−1 net bilayer curvature to the IMM (C 0 ) (Fig EV2G).
We next asked if the IMM curvature imparted by CL could compensate for curvature lost by saturated PLs and loss of ATP synthase oligomerization. To test this hypothesis, we evaluated how loss of cardiolipin synthase, Crd1p, affects mitochondrial morphology in SFA1‐4 strains (Fig 4A). Lipidomics confirmed that CL was absent in crd1 strains and PL saturation was unaltered (Fig 4A). As previously observed (Baile et al, 2014), crd1 cells did not show defects in mitochondrial morphology or cellular respiration in WT (CRD1) or SFA1 backgrounds (Fig EV3A and B). However, loss of CL had a dramatic phenotype in the SFA2 background, which has increased PL saturation and reduced ATP synthase oligomerization but normal mitochondrial morphology (Fig 4B–E). SFA2crd1 exhibited low‐oxygen consumption rates (Fig 4B), aberrant mitochondria (Fig 4C), and loss of CMs (Fig 4D and E), similar to SFA3/SFA4. Loss of morphology and respiration in SFA2crd1 was rescued by supplementation with oleic acid (OA), demonstrating that crd1 has a specific interaction with PL saturation (Fig EV3C).
Figure 4. Epistasis between PL saturation and CL synthesis in shaping mitochondrial morphology.
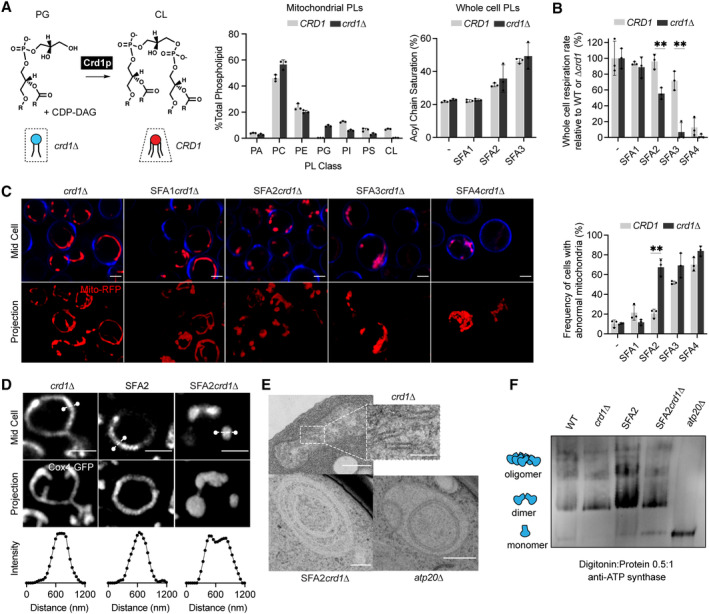
- Loss of Crd1p decreases mitochondrial CL content, but otherwise does not significantly affect the mitochondrial lipidome. (Left) Reaction schematic depicting cylindrical PG and CDP‐DAG converted into conical CL by Crd1p. (Right) Headgroup stoichiometry of lipidomes from isolated mitochondria from WT and crd1 cells and acyl chain saturation (% of acyl chains with a double bond) for CRD1 and crd1 cells across the SFA series. Error bars indicate SD from n = 3 biological replicates.
- Loss of CL causes a loss of respiration in SFA2 cells. Whole‐cell respiration rates are shown for SFA strains, comparing CRD1 with crd1 cells. Respirometry was conducted in biological triplicates (n = 3) by Clark electrode. Error bars indicate SD. **P < 0.005 unpaired two‐tailed t‐test between SFA2 and SFA3 strains and their corresponding crd1 mutants.
- The morphological breakpoint shifts to SFA2 upon loss of CL. (Left) Representative Airyscan confocal micrographs of yeast expressing matrix‐localized RFP (mts‐RFP). Cells were stained with cell wall‐binding calcofluor white (blue) for clarity. Scale bars, 2 μm. (Right) Frequency of mitochondrial abnormality, N > 50 cells were counted in n = 3 independent cultures for each condition. Error bars indicate SD. **P = 0.0011 unpaired two‐tailed t‐test between CRD1 and crd1 in the SFA2 background.
- SFA2crd1 cells show hollow mitochondria, imaged with Cox4‐GFP. Scale bars, 2 μm. Line profile analysis (below) depicts fluorescent intensity across the indicated mitochondria.
- Thin‐section TEM micrographs of crd1 show IMM with cristae invaginations (see inset, scale bars 100 nm), while SFA2crd1 cells show an onion‐like IMM similar to what has been observed in atp20 . Scale bars, 400 nm.
- SFA2crd1Δ retains ATP synthase dimerization. Shown are BN‐PAGE western blots run from digitonin‐solubilized isolated mitochondria of SFA strains and WT with and without CL, atp20 is the monomeric ATP synthase control.
Source data are available online for this figure.
Figure EV3. Interactions between cardiolipin synthase, lipid saturation, and ATP synthase dimerization.
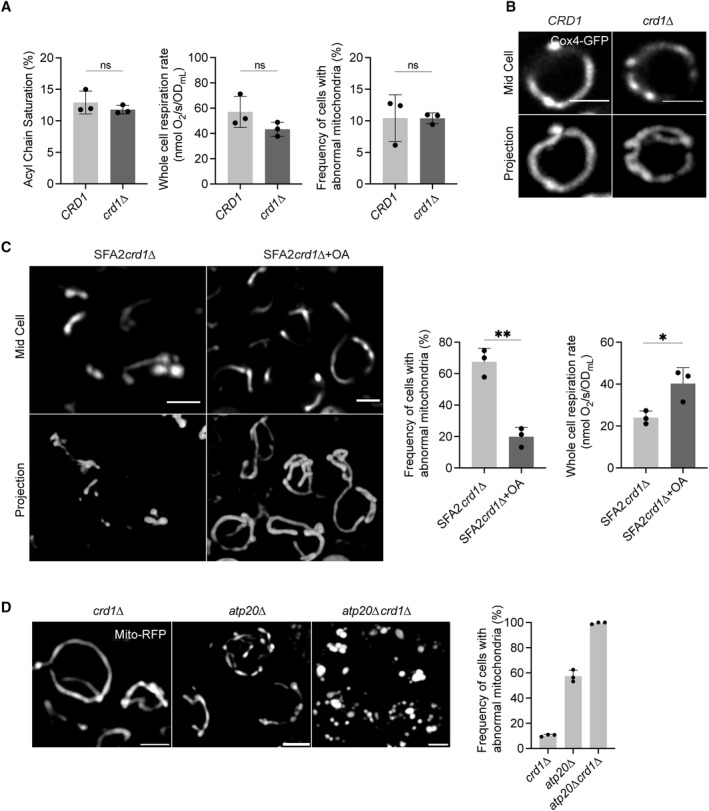
- Mitochondrial PL saturation, respiration rate, and mitochondrial abnormality measurements of CRD1 and crd1 cells. PL saturation was computed from lipidomics analysis on isolated mitochondria. Respiration measurements were performed using Clark electrode, and mitochondrial abnormalities were determined using confocal microscopy with yeast expressing a matrix‐localized mts‐RFP (N > 50 cells quantified per replicate). Measurements were taken from biological replicates (n = 3), error bars indicate SD. An unpaired t‐test was performed as the statistical test.
- Airyscan confocal micrographs depicting the mitochondrial morphology of aerobic wild‐type and crd1 ‐expressing IMM protein Cox4‐GFP. Scale bars, 2 μm.
- Representative Airyscan confocal micrographs of SFA2crd1 yeast, grown in the presence and absence of OA, expressing mts‐RFP. Scale bars, 2 μm. **P < 0.005, unpaired two‐tailed t‐test compared against SFA2crd1 . Respiration rates of SFA2crd1 cells in the presence and absence of OA were measured in biological replicates (n = 3) using a Clark electrode. Error bars indicate SD. *P < 0.05 unpaired two‐tailed t‐test.
- Loss of CL and ATP synthase dimerization results in complete ablation of mitochondrial morphology and structure as assayed by analysis with mts‐RFP. N > 50 cells were counted in biological triplicate (n = 3) in each condition. Error bars indicate SD. Individual deletion of Crd1p results in normal mitochondrial morphology, while half of the cells in atp20 still retain normal morphology. Scale bars, 2 μm.
While loss of CL modulated CM formation under increased PL saturation, it did so independently of ATP synthase dimerization: crd1 cells showed no defect in ATP synthase oligomerization compared to CRD1 cells, while SFA2crd1 cells maintained an identical level of oligomerization found in SFA2 cells (Fig 4F). However, loss of CL still showed a strong epistasis with the saturation induced loss of ATP synthase oligomerization in SFA2 cells, as well as with complete loss of dimerization in atp20 cells (Fig EV3D). Thus, CL acts orthogonally to ATP synthase oligomerization to modulate IMM morphology but is only required when PL saturation is increased.
Modeling predicts compensatory roles for lipid and protein‐encoded curvature in shaping cristae tubule formation
To understand the interaction between CL, which contributes to net membrane spontaneous curvature, and ATP synthase oligomers, whose induced curvature is localized specifically at cristae ridges, we employed a continuum modeling framework based on previous efforts to model membrane tubule formation (Mahapatra & Rangamani, 2023). We modeled the simplest CM structure—tubules—as axisymmetric tubes that bud from a flat membrane (Fig 5A). We added a pre‐defined coat area of ATP synthase oligomers that contribute an anisotropic curvature (D 0). An isotropic membrane curvature (C 0) was then applied across the entire membrane to model the effects of asymmetrically localized CL. In simulations, we varied the magnitude of D 0 and C 0 based on data from simulations of curvature induced by ATP synthase dimers (Anselmi et al, 2018) and CL effects on mitochondrial membrane compositions estimated by simulations of outer and inner IMM leaflets (Fig EV2G). In both cases, the approximate ranges were set from 0 to 0.035 nm−1. The stiffness of the membrane was also varied across biologically reasonable values (10–20 kBT), and the membrane tension was set at 0.01 pN/nm (Hassinger et al, 2017). Finally, to incorporate the stresses due to the proposed roles of the MICOS complex and Mgm1p at the CJs, we used a localized collar force density of 8 pN/nm around the base of the tubule neck. Additional details of the governing equations and parameters of the model can be found in the Appendix.
Figure 5. Continuum modeling of CM tubule formation reveals a snapthrough instability mediated by both lipid and ATP synthase generated curvatures.
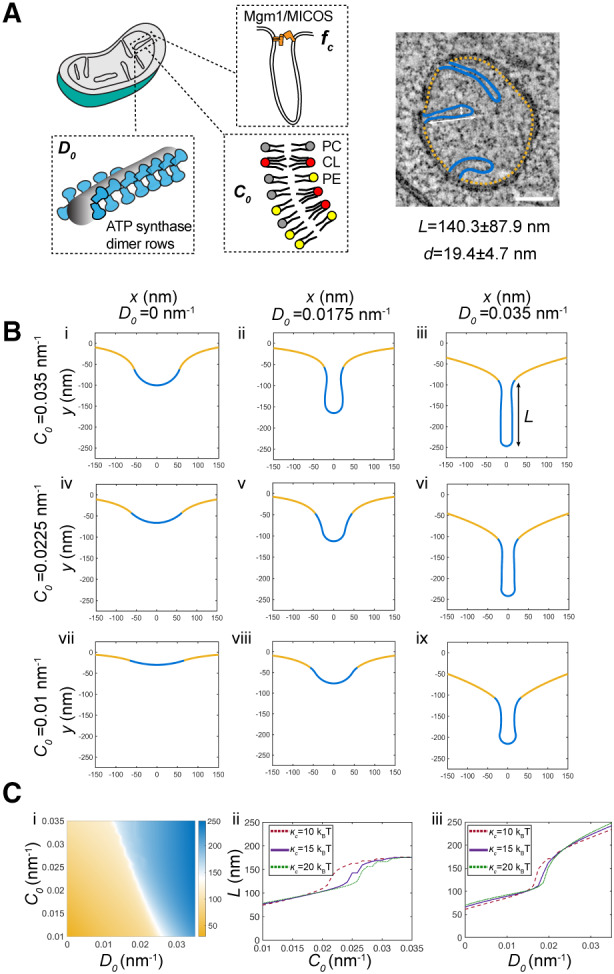
- Schematic depiction of yeast CM tubules containing a modeled neck force (f c) induced by Mgm1p and MICOS complexes, a deviatoric curvature imposed by ATP synthase at tubule ridges (D 0), and a spontaneous curvature along the entire membrane imposed by asymmetric distribution of CL across the IMM (C 0 ). (Right) TEM image showing typical yeast CM tubules in a single mitochondrion. Scale bars, 100 nm. The average tubule length (140.3 ± 87.9 nm) and diameter (19.4 ± 4.7 nm) of 15 cristae tubules were analyzed from tomograms of n = 3 SFA2 mitochondria.
- Changes in deviatoric and spontaneous curvatures modulates tubule morphology. Panels (i–ix) show shapes of the membranes from simulations of the continuum model. For these simulations, the bending modulus of the membrane was maintained at 15 kBT and the tension was set at 0.01 pN/nm. The total membrane area was set to 5.65 × 105 nm2, and the coated area was 1.413 × 104 nm2. The values of C 0 and D 0 were varied as shown, and f c was set to 8 pN/nm at the base of the coat.
- CM tubule formation shows a snapthrough behavior. (i) Length of the tube as a function of C 0 and D 0 for the same values of bending modulus, tension, and areas as shown in (B). The white region shows the transition from the short to the long tube. The color bar shows the length of the tube in nm. Line graphs in (ii) show the length of the tube as a function of C 0 for D 0 fixed at 0.0175 nm−1, and line graphs in (iii) show the length of the tube as a function of D 0 for C 0 fixed at 0.0225 nm−1 for three different values of кc . In both these graphs, the abrupt change in length is indicative of a snapthrough instability.
Source data are available online for this figure.
The model suggested that combination of D 0 and C 0 was sufficient to deform the membrane into different shapes reminiscent of flat membranes, buds that could be relevant for onion‐like IMM formation, and CM‐like tubules (Fig 5B). When tubules formed, their length (L) and diameter (d) were generally consistent with that of CMs in cells (Fig 5B). High values of D 0 (Fig 5B, third column) promoted the formation of tubules for all values of C 0 and bending moduli, and did so independently of the presence of the collar force (Appendix Fig S7). Low values of D 0, mimicking a loss of ATP synthase dimers in atp20 or SFA3/4 cells, did not allow for tubule formation and instead led to flat or bud‐like structures (Fig 5B, i–iii). The latter was observed even when C 0, potentially encoded by CL, remained high. For intermediate values of D 0, which could approximate the state in SFA2 cells that show partial loss of ATP synthase oligomerization, higher values of C 0 were required to form tubules (Fig 5B, ii and v), while lower values resulted in shallow invaginations. This dependence on isotropic spontaneous curvature when anisotropic spontaneous curvature is partially lost could explain the interaction experimentally observed between CL and PL saturation. We note that similar calculations in the absence of the collar force (Appendix Fig S7), show bud‐like and tubular structures with a wider neck, consistent with the model that local forces exerted by the MICOS and Mgm1p shape CJs.
Additional investigation into the parameter space for cristae‐like tubular structures revealed a transition from the short (low L) U‐shaped invaginations to long (high L) tubules in an abrupt manner (Fig 5Ci). This transition was observed both as C 0 (e.g. via CL‐encoded membrane spontaneous) increased for a fixed value of D 0 (Fig 5Cii, Movie EV6) or as D 0 (e.g. via ATP synthase oligomerization) was increased for a fixed value of C 0 (Fig 5Cii, Movie EV7) for the different values of bending moduli (see also Appendix Fig S7 for additional simulations). Such sudden changes to morphology, e.g. from a bud to a tube, in mechanical parameter space are termed a snapthrough instability or buckling event (Walani et al, 2015). In our model, the exact C 0 and D 0 values at which the snapthrough occurred was also modulated by the presence of a collar force at the tubule neck, suggesting that mechanics could underlie the functional interactions observed between Mic60p at the CJ and PL saturation (Fig EV1B, Appendix Fig S7).
CL is an essential mitochondrial lipid in low‐oxygen environments that promote saturated lipidomes
The lack of phenotype for crd1 strains under standard laboratory growth conditions has long been perplexing, given the demonstrated importance of CL for mitochondrial function in other organisms (Paradies et al, 2014). Natural yeast environments, such as rotting fruits or fermentation tanks, are intrinsically microaerobic and inhibit Ole1p, which like other desaturases requires oxygen as a electron acceptor. Oxygen binding to lipid desaturases is dependent on a low‐affinity di‐iron site (KM ~ 60 μM), and so, desaturase activity is sensitive to environmental oxygenation (Kwast et al, 1999; Vasconcelles et al, 2001). Yeast grown in microaerobic fermenters shows a lower level of di‐unsaturated PLs than those grown under highly aerated conditions (low volume shake flasks), with a lipidome that was intermediate between that of aerated SFA2 and SFA3 cells (Fig EV4A and B). We hypothesized that in these less aerated environments, CL metabolism would have evolved to support an essential mechanical function.
Figure EV4. Microaerobic cells exhibit an intermediate level of lipid saturation between SFA2/3 and retain mitochondrial volume and ATP synthase dimers.
- Lipidomic analysis of the double bond distribution of microaerobic yeast (n = 3 biological replicates) shows decreased di‐unsaturated PL chains and increased saturated PL chains. This is consistent with an intermediary increase in PL saturation between SFA2 and SFA3 levels. Error bars indicate SD.
- The major cellular PL classes, PC and PE, show increased lipid saturation under microaerobic conditions (n = 3 biological replicates). Also shown is the decrease in whole‐cell PE/PC ratio under microaerobic conditions, as observed in SFA strains (Fig 3D). Error bars indicate SD.
- CL content in microaerobic vs. aerobic conditions assayed by staining with 100 nM nonyl acridine orange (NAO). NAO intensities were quantified by line profile analysis from confocal micrographs of N > 15 cells in each condition. **P = 0.0021 unpaired two‐tailed t‐test against WT. Error bars indicate SD.
- Microaerobic growth conditions do not induce changes to mitochondrial volume or length. Mitograph software was used to measure the voxel volume and total length of mitochondrial tubules from confocal images taken from yeast cells grown in microaerobic vs. aerobic conditions. N = 10 cells were quantified from each condition. Error bars indicate SD. An unpaired t‐test was performed as the statistical test.
- Microaerobic cells still contain ATP synthase dimers. Digitonin‐solubilized crude mitochondria from microaerobic cultures were separated by BN‐PAGE and immunoblotted with anti‐ATP synthase antibodies. Crude mitochondria from microaerobic atp20 was used as a monomeric control.
To test the role of oxygenation on CL function, we grew CRD1 and crd1 yeast strains in microaerobic chambers, comparing their lipidomes and mitochondrial morphologies to those grown in highly aerated shake flasks (Fig 6A). Under limited oxygenation, CRD1 cells increased the abundance of CL twofold compared to highly aerated conditions (Fig 6B) and showed increased staining by the CL‐binding dye nonyl acridine orange (NAO) (Fig EV4C). This increase did not accompany any general changes to mitochondrial volume or length in aerobic vs. microaerobic cells (Fig EV4D). CRD1 and crd1 cells showed identical increases in the saturation of PLs under microaerobic conditions (Figs 6C and EV4B) and both retained ATP synthase dimers (Fig EV4E). After microaerobic growth, yeast containing CL still presented tubular mitochondrial morphologies, but crd1 cells predominantly displayed aberrant mitochondrial morphologies (Fig 6D and E). Ultrastructural analysis of IMM structure revealed that CRD1 cells grown under microaerobic conditions displayed tubular cristae morphologies, while crd1 cells displayed a mixture of flat and onion‐like IMM structures (Fig 6F). CL can thus be required for CMs in yeast, but not in highly oxygenated laboratory conditions that suppress PL saturation.
Figure 6. Cardiolipin synthesis is essential for mitochondria in yeast growth conditions characterized by low oxygenation.
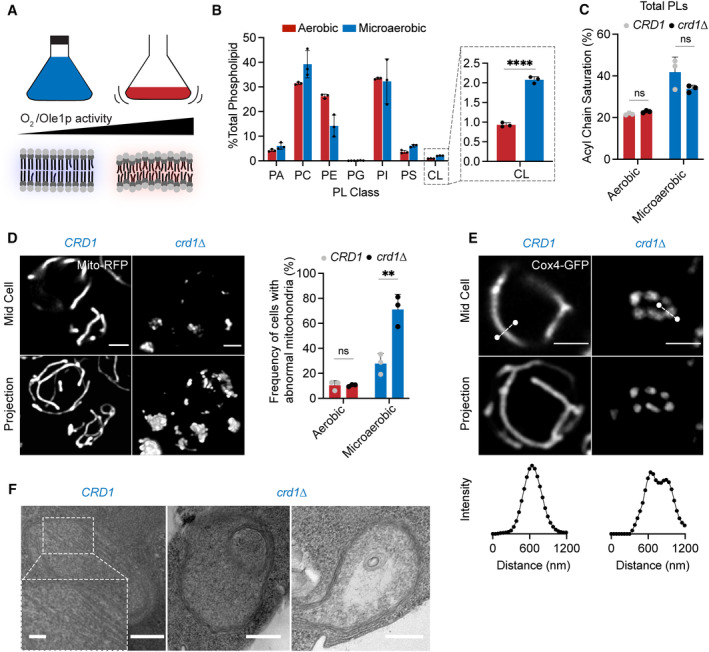
- Schematic representation of different oxygen concentrations in different yeast growth environments. Microaerobic conditions cause increased saturation of membranes due to lower desaturate activity of Ole1p, an oxygen‐dependent enzyme.
- WT cells increase the abundance of CL under microaerobic conditions. Shown are abundances of each PL class, n = 3 biological replicates. Error bars indicate SD. ****P < 0.0001, unpaired two‐tailed t‐test compared against wild‐type.
- Microaerobic growth conditions cause an increase in acyl chain saturation in the total PL pool, which is not affected by loss of CL in crd1 strains as determined by an unpaired t‐test. Error bars indicate SD from n = 3 biological replicates.
- Under microaerobic conditions, loss of CL (crd1 ) causes loss of tubular mitochondrial structure. WT (CRD1) and crd1 cells were grown in microaerobic chambers for 48 h prior to imaging, n = 3 biological replicates. Scale bars, 2 μm. **P < 0.005, unpaired two‐tailed t‐test compared against WT.
- Microaerobic crd1 show hollow mitochondria, imaged with Cox4‐GFP. Scale bars, 2 μm. Line profile analysis (below) depicts fluorescent intensity across the indicated mitochondria.
- Under microaerobic conditions, CRD1 cells contain long, sheet‐like cristae structures while crd1 cells lack cristae, and display both onion‐like and flat abnormal IMM structures as visualized by thin‐section TEM. Scale bars, 250 nm. Inset showing abundant cristae sheets in CRD1 cells, scale bar, 125 nm.
Source data are available online for this figure.
Discussion
In this study, we explored an initial observation that small changes to the composition of yeast PLs cause the loss of CMs in mitochondria. We found that saturated PLs inhibit ATP synthase oligomerization, which is progressively lost when either the expression or activity of the lipid desaturase Ole1p decreases. Based on molecular modeling of IMM lipidomes, we hypothesized that conical lipids buffer against loss of ATP synthase as a cristae‐shaping protein. We tested the interaction between the curvature at cristae ridges induced by ATP synthase oligomerization and modulated by PL saturation, and lipid‐encoded curvature across the IMM, which is promoted by conical lipids like CL. Both factors contribute to whether the IMM exists in a high or low curvature state, which in turns controls ATP generation and mitochondrial fitness. The sharp transition we initially observed between these states can be understood through a continuum model that highlights a snapthrough instability, a phenomenon in which a system can shift between two morphological states via modest perturbations in its material properties or forces applied. Also termed mechanical buckling, such events are intrinsic to membrane bending mechanics (Ou‐Yang et al, 1999; Vasan et al, 2020) and have been proposed to act in other curved cellular structures, such as endocytic buds (Walani et al, 2015; Hassinger et al, 2017).
Based on our observations, we propose that the intrinsic membrane curvature needed to form CMs can be generated both by cristae‐shaping proteins and by asymmetric distributions of lipids across the bilayer, which contributes net spontaneous curvature between the two leaflets. CL is a negatively curved PL in the IMM for which there are multiple potential sources of asymmetry in the IMM. Staining with NAO suggests that the yeast IMM features up to twofold more CL in the outer leaflet (Petit et al, 1994; Gallet et al, 1997), which would optimize its negative curvature at cristae ridges. The differences in local pH between the matrix and IMS could also favor the monoanion CL species in the IMS‐facing leaflet, which would also promote negative curvature at cristae ridges. Finally, CL and other conical lipids segregate into deformed membrane regions, further promoting curvature. Such a phenomenon has been observed experimentally when CL localized to thin, high‐curvature tubules that have been pulled from large, low curvature vesicles (Beltrán‐Heredia et al, 2019). In simulations, this effect is apparent in localized CL concentrations in thermal fluctuations (Fig 3G) and those induced by compression (Dahlberg & Maliniak, 2010; Boyd et al, 2017). In the IMM, it is likely that local deformations induced by ATP synthase (Blum et al, 2019) cause the local concentration of conical lipids (e.g. CL and di‐unsaturated PE), which further act to deform the IMM. The action of membrane‐shaping lipids and proteins is therefore likely to be intrinsically linked.
The interplay of CL with PL saturation provides a biophysical rationale for long‐standing questions regarding its role in the IMM. The consequences of CL loss, widely explored to understand the pathophysiology of Barth syndrome, differ strikingly across experimental systems. CL is essential in both mice (Kasahara et al, 2020; Xu et al, 2021) and mammalian cell lines (Choi et al, 2007), but not in yeast or Drosophila, where its loss has more subtle effects on respiration (Xu et al, 2021). The mechanical functions of CL function could be dependent on the surrounding lipid environment in the IMM, which can change depending on growth conditions. Oxygenation is one natural modulator of PL saturation due to the intrinsic enzymology of lipid desaturases. In yeast, growth under microaerobic conditions leads to lipidomes with predominantly monounsaturated PLs that require CL to generate CMs in the IMM. In contrast, growth under highly aerated laboratory conditions show an unusually high level of di‐unsaturated PLs (Appendix Fig S1G) and does not necessitate CL.
The role of oxygenation in controlling PL saturation is not exclusive to yeast. In cancer cells, hypoxia reduces activity of SCD1 (stearoyl CoA desaturase 1), which also drives lipidome remodeling (Kamphorst et al, 2013; Ackerman et al, 2018). Specific tissue environments, such as in the gut, are also microaerobic (Zheng et al, 2015), and a recent genome‐wide screen implicated CL metabolism in low‐oxygen fitness of intestinal T‐cells (Reina‐Campos et al, 2023). In human embryonic kidney (HEK) 293 cells, the morphological and respiratory effects of knocking down cardiolipin synthase (CRLS1) are potentiated by increased saturation caused by either mild PA treatment or low‐oxygen growth (Fig EV5). In PA‐treated CRLS1 knockdown cells, the density and length of cristae sheets are strongly reduced (Fig EV5D and E). Thus, the interaction between CL and PL saturation is likely to extend beyond yeast mitochondria.
Figure EV5. Epistasis between lipid saturation and cardiolipin metabolism in HEK293 cells.
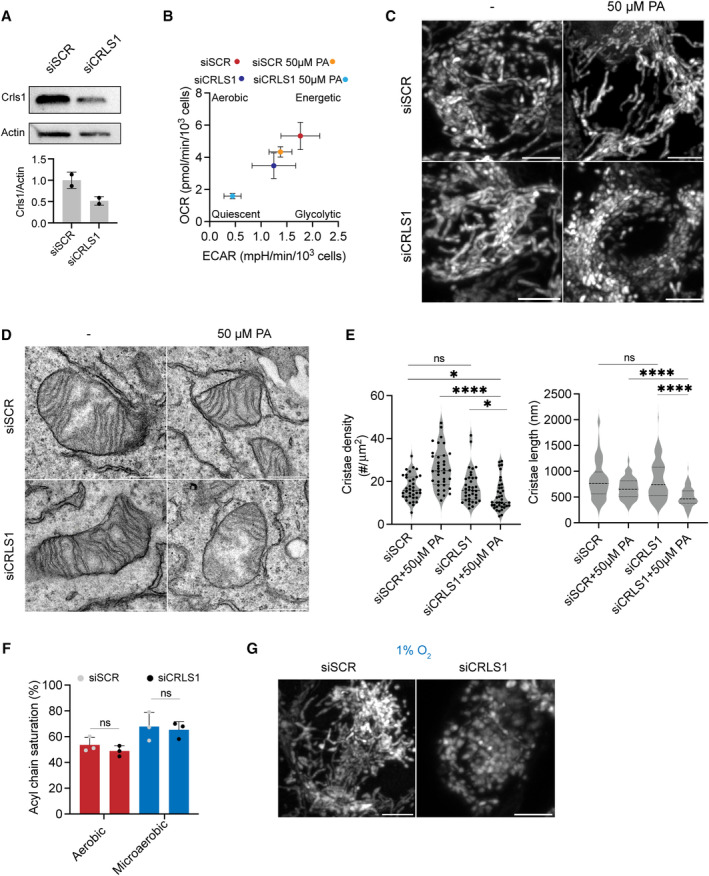
- Knockdown of CRLS1 by siRNA (siCRLS1) results in a ~ 50% decrease in CRLS1 intensity as normalized to β‐Actin intensity compared to the scrambled control (siSCR). Error bars indicate SD from n = 2 independent transfections.
- Coordinated increase in saturated fatty acid and depletion of CL result in decreased oxygen consumption rate (OCR) and extracellular acidification rate (ECAR). OCR and ECAR were measured from 10,000 cells in triplicates (n = 3) in each condition assayed using Seahorse respirometry. Cells treated with both mild concentrations (50 μM) of PA and siCRLS1 showed a quiescent metabolic phenotype.
- Knockdown of CRLS1 results in loss of tubular mitochondrial morphology upon treatment with 50 μM PA. Representative images of single cells transfected with scrambled (siSCR) or CRLS1 (siCRLS1) siRNAs and subsequently treated with BSA or BSA complexed with 50 μM PA. Cells were stained with Mitotracker DeepRed. Images show individual cells. Scale bars, 5 μm.
- CRLS1 knockdown cells treated with 50 μM PA are bereft of abundant cristae sheets observed in control cells or those treated with only siCRLS1 or 50 μM PA. Representative TEMs of individual mitochondria are shown from each condition. Scale bars, 500 nm.
- CRLS1 knockdown cells treated with 50 μM PA show decreased cristae density and cristae length. Cristae density was quantified as the number of cristae per mitochondrial area and assessed from N = 35 mitochondria. ****P < 0.0001 unpaired t‐test of siCRLS1 with 50 μM PA against siSCR with 50 μM PA, *P < 0.025 unpaired t‐test of siCRLS1 with 50 μM PA against siSCR and siCRLS1. Cristae length was measured in N = 100 cristae in each condition. ****P < 0.0001 unpaired t‐test of siCRLS1 with 50 μM PA against siSCR with 50 μM PA and siCRLS1. Error bars indicate SD.
- Knockdown of CRLS1 does not change the acyl chain saturation in aerobic or microaerobically grown cells, but the latter feature higher lipid saturation due to reduced oxygenation. Error bars indicate SD. Samples were analyzed in biological triplicates (n = 3). An unpaired t‐test was performed as the statistical test.
- Under 1% oxygen growth conditions, knockdown of CRLS1 results in loss of tubular mitochondrial morphology. Cells were incubated in microaerobic chambers for 72 h or normoxic chambers for 48 h prior to staining with Mitotracker DeepRed. Images show individual cells. Scale bars, 5 μm.
An outstanding question of this work is how specific lipid perturbations modulate the dimerization and higher order organization of ATP synthases. PL saturation, which influences both membrane stiffness and curvature, is a unique regulator of ATP synthase organization in cells: yeast strains that have drastically altered mitochondrial PC, PE, and CL levels all retain dimers (Claypool et al, 2008; Baile et al, 2014; Baker et al, 2016). Among membrane protein dimers, the yeast ER sensors Mga2p/Spt23p show a similar dimer to monomer transition when their host membrane shifts di‐unsaturated to monounsaturated PLs (Covino et al, 2016; Ballweg et al, 2020). While Mga2p/Spt23p are small, single‐pass transmembrane proteins, the CLC Cl−/H+ antiporter dimer has a similar buried surface area as ATP synthase (~ 2,400 Å2) and a modest dimerization free energy (19 kBT) (Chadda et al, 2016) that is also highly sensitive to its lipidic environment (Chadda et al, 2021). In CLC, short‐chain PLs have been shown to stabilize the monomer conformation and through this mechanism weaken its dimerization. Unlike CLC, dimerization of ATP synthase is associated with an extreme local membrane deformation—a ~ 90o bend in S. cerevisiae (Guo et al, 2017). One hypothesis is that the changes in mechanical properties encoded by PL saturation, especially stiffness, mediate local membrane deformation, reducing the dimer:monomer equilibrium by increasing its elastic cost. Distinguishing between these models will require future experiments and modeling, which would be aided by structures of monomeric ATP synthases with resolved dimerization subunits.
During the evolution of eukaryotic cells, the machinery for ATP generation in the ETC adopted a secondary function in the shaping of the IMM. The emergence of ATP synthase dimers and other cristae‐shaping protein complexes occurred alongside a specialization in the inner membrane lipidome, including the proliferation of CL from the proteobacterial inner membrane (Sohlenkamp & Geiger, 2016; Rowlett et al, 2017). Among extant eukaryotes, ATP synthase organization varies widely; for example, in mammalian mitochondria, ATP synthases range from primarily monomeric (Galber et al, 2019) to mixtures of monomers and dimers (Bisetto et al, 2007). It is notable that CL is essential for proper IMM structure in these systems, unlike in aerobic yeast in which dimers are predominant. The composition of other cristae‐shaping proteins, such as the MICOS complex (Huynen et al, 2016), also differs across eukaryotes, despite the ubiquity of CMs in mitochondria. Such variability in IMM‐shaping proteins could have necessitated the utilization of alternative forms of curvature generation encoded by the mitochondrial lipidome.
Materials and Methods
Strains and growth media
The yeast strains used in this study can be found in Appendix Table S3. Yeast cells were grown in YPEG (1% Bacto yeast extract, 2% Bacto peptone, 2% Ethanol, 2.5% Glycerol), YPD medium (1% Bacto yeast extract, 2% Bacto peptone, 2% glucose), or complete supplement mixture (CSM, 0.5% Ammonium Sulfate, 0.17% yeast nitrogen base without amino acids and 2% glucose) lacking appropriate amino acids for selection. Yeast mutants were generated by PCR‐based homologous recombination, ORFs of the gene of interest were replaced by either KanMX or His3MX6 cassettes. For OLE1 promoter substitution, a set of previously generated mutant TEF1 promoters were utilized (Alper et al, 2005). An additional, weaker promoter (Pm1) was generated by error‐prone PCR and added to this set. Promoter substitution was completed on the haploid base strain W303a as previously described (Degreif et al, 2017).
Yeast physiology
Growth on non‐fermentable carbon sources was assayed using growth curves on 24‐well plates (Avantor) sealed with a gas permeable film (Diversified Biotech). Cells were first grown in biological triplicate overnight in complete synthetic medium (CSM) containing 2% glucose. Cells were back‐diluted 1:100 in fresh CSM containing 2% glucose or 3% glycerol and shaken in a plate reader (Tecan) for 48 h. Specific growth rates were extracted from the exponential phase of growth. For viability assays, cells were first grown in the same fashion as for yeast growth curves but were serially diluted onto CSM plates (1:5 successive dilutions) containing either 2% glucose or 3% glycerol and 2% ethanol. Plates were grown for 3 days on glucose plates and for 4 days for ethanol/glycerol plates.
For microaerobic growth, cells were pre‐incubated overnight in CSM without uracil (2% glucose) in a controlled temperature shaker at 30°C. Cells were then back‐diluted into fresh synthetic medium and grown to stationary phase in home‐built microaerobic chambers with limited oxygen supply. Chambers consisted of glass culture tubes with tight fitting rubber caps; tubing allowed for gas efflux into an attached bubbler. Live cell imaging was conducted on aliquots, and cell pellets were resuspended in sterile water, lysed, and flash frozen for lipidomics analysis.
Lipidomics
Lipid compositions of whole cells and isolated mitochondria from yeast strains were conducted at Lipotype GmbH (Dresden, Germany). Mass spectrometry‐based lipid analysis was performed as previously described (Ejsing et al, 2009; Klose et al, 2012). Lipids were extracted using a two‐step chloroform/methanol procedure (Ejsing et al, 2009). Samples were spiked with an internal lipid standard mixture containing: CL 14:0/14:0/14:0/14:0, ceramide 18:1;2/17:0 (Cer), diacylglycerol 17:0/17:0 (DAG), lyso‐phosphatidate 17:0 (LPA), lyso‐phosphatidyl‐choline 12:0 (LPC), lysophosphatidylethanolamine 17:1 (LPE), lyso‐phosphatidylinositol 17:1 (LPI), lysophosphatidylserine 17:1 (LPS), phosphatidate 17:0/14:1 (PA), phosphatidylcholine 17:0/14:1 (PC), PE 17:0/14:1, PG 17:0/14:1, PI 17:0/14:1, phosphatidylserine 17:0/14:1 (PS), ergosterol ester 13:0 (EE) and triacylglycerol 17:0/17:0/17:0 (TAG). After extraction, the organic phase was transferred to an infusion plate and dried in a speed vacuum concentrator. 1st step dry extract was resuspended in 7.5 mM ammonium acetate in chloroform/methanol/propanol (1:2:4, v:v:v) and 2nd step dry extract in 33% ethanol solution of methylamine in chloroform/methanol (0.003:5:1; v:v:v). All liquid handling steps were performed using Hamilton Robotics STARlet robotic platform with the Anti Droplet Control feature for organic solvents pipetting. Samples were analyzed by direct infusion on a QExactive mass spectrometer (Thermo Scientific) equipped with a TriVersa NanoMate ion source (Advion Biosciences). Samples were analyzed in both positive and negative ion modes with a resolution of R m/z=200 = 280,000 for MS and R m/z=200 = 17,500 for MSMS experiments, in a single acquisition. MSMS was triggered by an inclusion list encompassing corresponding MS mass ranges scanned in 1 Da increments (Surma et al, 2015). Both MS and MSMS data were combined to monitor EE, DAG, and TAG ions as ammonium adducts; PC as an acetate adduct; and CL, PA, PE, PG, PI, and PS as deprotonated anions. MS only was used to monitor LPA, LPE, LPI, and LPS as deprotonated anions; Cer and LPC as acetate adducts. Data were analyzed with in‐house developed lipid identification software based on LipidXplorer (Herzog et al, 2011, 2012). Data post‐processing and normalization were performed using an in‐house developed data management system. Only lipid identifications with a signal‐to‐noise ratio > 5 and a signal intensity fivefold higher than in corresponding blank samples were considered for further data analysis.
The acyl chain composition of HEK293 cells was analyzed by Bligh‐Dyer extraction followed by Gas Chromatography coupled to Mass Spectrometry (GC–MS) of transesterified fatty acid methyl esters (FAME) on an Agilent 8890‐5977B GC–MS, as previously described (Winnikoff et al, 2021). Quantification was performed in Agilent MassHunter using external FAME standards (Cayman Chemical #20503).
Mitochondrial purification
Yeast mitochondria were isolated from 1 L of yeast cells grown in YPEG (1% Bacto yeast extract, 2% Bacto peptone, 2% Ethanol, 3% Glycerol), YPD medium (1% Bacto yeast extract, 2% Bacto peptone, 2% glucose), or CSM with 2% glucose (microaerobic conditions) at 30°C as previously described (Meisinger et al, 2006; Gregg et al, 2009). Cells were grown to stationary phase and harvested in a buffer consisting of 100 mM Tris/H2SO4 (pH 9.4) and 10 mM dithiothreitol. Spheroplasts were formed from digestion of the cell wall using zymolyase 20‐T (MP Biomedicals) in a buffer containing 20 mM potassium phosphate (pH 7.4) and 1.2 M Sorbitol. Spheroplasts were lysed by homogenization using a glass homogenizer and subsequently centrifuged to remove unbroken cells, large debris, and nuclei. Enriched mitochondria were pelleted and resuspended in SEM buffer (10 mM MOPS/KOH (pH 7.2), 250 mM sucrose, and 1 mM EDTA), snap frozen, and stored for up to 1 month at −80°C. To obtain purified mitochondria, bereft of contamination from other organelles, such as microsomes and vacuoles, the crude mitochondrial fraction was subjected to sucrose density cushion ultracentrifugation. Cushions are poured containing 60 and 32% (w/v) sucrose concentrations in EM buffer (10 mM MOPS/KOH [pH 7.2], 1 mM EDTA). Density cushions containing crude mitochondrial samples in SEM buffer are subjected to ultracentrifugation in a SW32 Ti swinging‐bucket rotor for 1 h at 100,000 g at 4°C. The yellow/brown band at the 60/32% (w/v) sucrose interface is removed and centrifuged to pellet purified mitochondria for subsequent analysis. Mitochondrial protein quantity was determined via BCA assay.
Mitoplasts containing an intact inner mitochondrial membrane stripped of the outer membrane were isolated as previously described (Zhang et al, 2005). After initial incubation of isolated mitochondria in a hypotonic buffer followed by centrifugation for 10 min at 14,000 g at 4°C. Mitoplasts were resuspended in SEM buffer prior to lipidomic analysis.
Respirometry
Oxygen consumption rates (OCR) of whole‐cell yeast strains were measured with a Clark electrode (YSI 5300A Biological Oxygen Monitor System). Cells were grown in biological replicates overnight in CSM containing 0.4% glucose (starvation conditions). Cells were then back‐diluted into fresh CSM and grown to OD 0.4–0.6. OCRs were quantified after initial stirring of culture for 3 min in a thermostatically controlled chamber at 30°C. Respiration rates were determined after normalization to OD of the sample. Respirometry on HEK293 cells was performed on a SeaHorse XF Pro (Agilent) using the real‐time ATP rate assay kit (103591‐100) on 10,000 cells per replicate per condition.
Live cell microscopy
All live cell microscopy was conducted using Plan‐Apochromat 63×/1.4 Oil DIC M27 on the Zeiss LSM 880 with Airyscan (default processing settings); image acquisition and processing were performed using ZEN software. In yeast experiments, cells were plated on 8‐well chambered coverglass (Nunc Lab‐Tek) pre‐incubated with concanavalin A (MP Biomedicals).
To assess mitochondrial morphology in yeast, cells were initially grown in biological replicates overnight in CSM containing 0.4% glucose under selection conditions. Cells were then back‐diluted and imaged in exponential phase. For analysis, yeast cells were split into three groups based on mitochondrial morphology: “aberrant” groups display a punctate cluster at the center of the cell, while “fragmented” groups display a lack of tubular morphology and consist of individual mitochondrial puncta separated within the cell (Appendix Fig S2). Normal mitochondrial morphology is associated with tubes spanning the length of the cell that display regular lateral motion. Percentage “mitochondrial abnormality” is derived from combining the amount of cells with an aberrant or fragmented morphology divided by the total number of cells. At least 50 cells were quantified in each sample, and replicates were of individually grown cultures. To visualize the yeast cell wall, cells were stained with calcofluor white (Sigma‐Aldrich), which binds chitin within the cell wall. Mitochondrial volume and length was quantified using Mitograph software as previously described (Viana et al, 2015). Analysis of HEK293 mitochondria was done by staining cells grown on glass bottom dishes (MatTek) with MitoTracker Deep Red (ThermoFisher Scientific M22426) for 30 min prior to imaging.
Membrane potential was assayed by growing yeast to exponential phase in CSM containing 0.4% glucose followed by staining with 200 nM TMRE (ThermoFisher Scientific T669) for 20 min. Cells were then washed twice with water prior to imaging. For uncoupled conditions, cells were incubated with 20 μM carbonyl cyanide m‐chlorophenyl hydrazone (CCCP, Sigma‐Aldrich) for 10 min prior to incubation with TMRE. For analysis of membrane potential in ATP synthase inhibited conditions, cells were incubated with 5 μM oligomycin for 45 min prior to incubation with TMRE. To analyze yeast mtDNA nucleoids, cells were grown to exponential phase in CSM containing 0.4% glucose and washed once with PBS prior to staining with SYBR Green I (SGI, ThermoFisher Scientific S7563) for 10 min. Cells were then washed three times with PBS prior to imaging. Relative yeast CL content in aerobic and microaerobic growth conditions was determined by staining cells with NAO (ThermoFisher Scientific A1372). Cells were stained with 100 nM NAO for 20 min and then washed three times with water prior to imaging. The maximum intensity per cell was determined using profile analysis in ImageJ.
Blue native PAGE analysis
Isolated mitochondria solubilized in digitonin (0.5:1 g/g protein) were assayed by BN‐PAGE as previously described (Timón‐Gómez et al, 2020), with minor modifications. 200–400 μg of mitochondria was incubated with digitonin for 10 min prior to centrifugation at 20,000 g for 30 min at 4°C. The subsequent supernatant was mixed with native PAGE buffer and glycerol and loaded onto precast native PAGE gels (Invitrogen). ATP synthase dimerization state was probed using an anti‐ATP synthase primary antibody (Rak & Tzagoloff, 2009) (1:1,000) and anti‐rabbit IgG secondary antibody (Thermo Fisher Scientific). Supercomplex formation was assessed using an anti‐Cox1p (CIV) antibody and anti‐mouse secondary antibody (Thermo Fisher Scientific).
Immunoblot analysis
Whole‐cell yeast lysates were grown in YPEG and 2.5 OD units were subjected to protein extraction and SDS–PAGE as previously described (Kushnirov, 2000). After transfer to PVDF membranes, western blot analysis was completed using the following primary antibodies at stipulated dilutions in blocking buffer (5% BSA in TBST): 1:1,000 for Cox4p and Dpm1p and 1:250 for Pho8p. For analysis of isolated mitochondria, 10 μg of total protein was loaded on SDS–PAGE gels and transferred to PVDF membranes prior to western blotting with the aforementioned antibodies as well as with the Mgm1p and Mic60p antibodies (Rabl et al, 2009). In HEK293 cells, siCRLS1 knockdowns were verified by immunoblotting with an anti‐CRLS1 polyclonal antibody and a polyclonal Actin loading control. All antibodies are listed in Appendix Table S5.
Electron microscopy
Blocks of late exponential phase yeast cells were prepared either by high‐pressure freezing/freeze substitution (HPF‐FS) (McDonald & Müller‐Reichert, 2002) (SFA4, SFA2crd1 , atp20 ) or chemical fixation followed by partial cell wall digestion (Bauer et al, 2001) (SFA2, SFA4, crd1 , and microaerobic cells) as previously described. Cell wall digestion was required for high contrast staining of WT‐like tubular CM for 3D segmentation. For microaerobic cells, chemical digestion was performed with 0.25 mg/ml zymolyase 20T for 1 h at room temperature. Aerobically grown mutant cells lacking CMs showed sufficient membrane contrast in HPFS samples. Thin sections about 60 nm thick were cut from the blocks of yeast with a Leica ultramicrotome and placed on 200‐mesh uncoated thin‐bar copper grids. A Tecnai Spirit (FEI, Hillsboro, Oregon) electron microscope operated at 120 kV was used to record images with a Gatan Ultrascan 4 K × 4 K CCD camera at 6.0, 2.9, and 1.9 nm/pixel. For TEM on HEK 293 mitochondria, cells were grown to confluency in MatTek dishes coated with fibronectin, prepared, and recorded as previously described (Darshi et al, 2011).
Tomography
Semi‐thick sections of thickness about 300 nm were cut from the blocks of yeast with a Leica ultramicrotome and placed on 200‐mesh uncoated thin‐bar copper grids. 20‐nm colloidal gold particles were deposited on each side of the grid to serve as fiducial cues. The specimens were irradiated for about 20 min to limit anisotropic specimen thinning during image collection at the magnification used to collect the tilt series before initiating a dual‐axis tilt series. During data collection, the illumination was held to near parallel beam conditions and the beam intensity was kept constant. Tilt series were captured using SerialEM (University of Colorado, Boulder) software on a Tecnai HiBase Titan (FEI) electron microscope operated at 300 kV and 0.81 nm/pixel. Images were recorded with a Gatan 4 K × 4 K CCD camera. Each dual‐axis tilt series consisted of first collecting 121 images taken at 1 degree increment over a range of −60 to +60 degrees followed by rotating the grid 90 degrees and collecting another 121 images with the same tilt increment. To improve the signal‐to‐noise ratio, 2× binning was performed on each image by averaging a 2 × 2 x‐y pixel box into 1 pixel using the newstack command in IMOD (University of Colorado, Boulder). The IMOD package was used for tilt‐series alignment, reconstruction, and volume segmentation. R‐weighted back projection was used to generate the reconstructions.
Mesh generation and analysis
3D in silico reconstructions of mitochondria were generated from electron‐tomographic images. The software IMOD was used to trace the mitochondrial membranes in 2D: the outer leaflet of the OM, and the inner leaflet of the IBM and CM was manually traced as separate objects, following procedures previously described (Mendelsohn et al, 2022). Subsequently, 2D traces were imported into Blender using the NeuropilTools module in CellBlender. The program Contour Tiler (Edwards et al, 2014)—integrated with NeuropilTools—was used to generate 3D triangulated meshes in Blender. The triangulation was performed individually for each membrane object in each mitochondrion. Afterward, the Boolean Difference Modifier was used to subtract the CM object from the IBM object, generating in this manner the CJs in the IBM. The meshes were refined with the Smooth and Normal Smooth improvement tools from GAMer2 (Lee et al, 2020). Curvature calculations were carried out with GAMer2, using the MDSB algorithm. For all curvature analysis, the smooth curvature after one iteration was considered. This smoothing represents the average curvature of a vertex and its neighbors. Surface areas and volumes were calculated using the CellBlender add‐on in Blender.
Mammalian cell culture
HEK293 cells (Sigma‐Aldrich) were cultured in DMEM (Gibco) supplemented with 10% FBS (Gibco) at 37°C in humidified air containing 5% CO2. For silencing experiments, Lipofectamine RNAiMAX was used per manufactures' instructions for an siRNA final concentration of 10 nM, and cells were imaged or treated 24 h after transfection. The siRNA constructs (ThermoFisher Scientific) included a non‐targeting control (Catalog #4390843) and a previously validated CRLS1‐targeting sequence (siRNA ID: s29306) (Ohlig et al, 2018; Yang et al, 2023). For PA treatment, cells were transfected into complete DMEM containing specified 50 μM PA complexed to BSA and analyzed 24 h after transfection. For aerobic/microaerobic incubations, media was replaced with DMEM containing delipidated FBS before incubation in either normoxic or microaerobic conditions 24 h after transfection; the latter was maintained by continual flushing with nitrogen as previously described (Doedens et al, 2013). Microaerobic grown cells (1% oxygen) were incubated for 72 h and aerobic grown cells (21% oxygen) were incubated for 48 h before analysis to allow for two doublings each.
Author contributions
Kailash Venkatraman: Conceptualization; formal analysis; investigation; visualization; writing – original draft; writing – review and editing. Christopher T Lee: Data curation; software; formal analysis; investigation; visualization; methodology; writing – original draft; writing – review and editing. Guadalupe C Garcia: Conceptualization; data curation; software; formal analysis; investigation; visualization; methodology; writing – original draft; writing – review and editing. Arijit Mahapatra: Data curation; formal analysis; investigation; methodology; writing – review and editing. Daniel Milshteyn: Resources; formal analysis; investigation; writing – review and editing. Guy Perkins: Resources; investigation; methodology; writing – review and editing. Keun‐Young Kim: Resources; investigation; methodology; writing – review and editing. H Amalia Pasolli: Resources; software; investigation; methodology. Sebastien Phan: Resources; software; investigation; methodology. Jennifer Lippincott‐Schwartz: Resources; funding acquisition; methodology; project administration. Mark H Ellisman: Conceptualization; resources; supervision; funding acquisition; methodology; writing – original draft; project administration; writing – review and editing. Padmini Rangamani: Conceptualization; resources; data curation; formal analysis; supervision; funding acquisition; validation; investigation; visualization; writing – original draft; project administration; writing – review and editing. Itay Budin: Conceptualization; resources; data curation; formal analysis; supervision; funding acquisition; validation; investigation; visualization; writing – original draft; project administration; writing – review and editing.
Disclosure and competing interests statement
The authors declare that they have no conflict of interest.
Supporting information
Appendix S1
Expanded View Figures PDF
Movie EV1
Movie EV2
Movie EV3
Movie EV4
Movie EV5
Movie EV6
Movie EV7
Dataset EV1
Dataset EV2
PDF+
Source Data for Figure 1
Source Data for Figure 2
Source Data for Figure 3
Source Data for Figure 4
Source Data for Figure 5
Source Data for Figure 6
Acknowledgements
José Faraldo‐Gómez, Edward Lyman, Nicolas‐Frédéric Lipp, and Yi‐Ting Tsai provided helpful discussions. The Herzik lab provided biochemistry assistance. Daniel Degreif and Sterling Ramsey assisted in strain development. Miguel Reina‐Campos assisted with microaerobic cell culture experiments. Alexander Tzagoloff, Leticia Franco, Mário Barros, and Andreas Reichert supplied antibodies. The National Institutes of Health (NIH) (R35‐GM142960 to IB, R01AG065549, R24GM137200, and U24NS120055 to MHE), the Office of Naval Research (ONR N00014‐20‐1‐2469 to PR), the National Science Foundation (DBI‐2014862 to MHE), the Department of Energy (DE‐SC0022954 to IB), and the Moore–Simons Project on the Origin of the Eukaryotic Cell (GBMF‐9734 to IB, PR, and MHE) provided financial support. KV was supported by the NIH Molecular Biophysics Training Grant (T32‐GM008326C). CTL was supported by a Kavli Institute for Brain and Mind Postdoctoral Fellowship. Howard Hughes Medical Institute supported initial project conception through the Janelia Visiting Scientist Program. The UCSD‐CMM‐EM Core (RRID: SCR_022039), partly supported by NIH S10OD023527, provided technical assistance and equipment access. Molecular dynamics simulations were run on hardware hosted by the Triton Shared Computing Cluster.
The EMBO Journal (2023) 42: e114054
Contributor Information
Padmini Rangamani, Email: prangamani@ucsd.edu.
Itay Budin, Email: ibudin@ucsd.edu.
Data availability
All strains and plasmids are available upon request to the corresponding author. This study includes no data deposited in external repositories.
References
- Acehan D, Xu Y, Stokes DL, Schlame M (2007) Comparison of lymphoblast mitochondria from normal subjects and patients with Barth syndrome using electron microscopic tomography. Lab Invest 87: 40–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ackerman D, Tumanov S, Qiu B, Michalopoulou E, Spata M, Azzam A, Xie H, Simon MC, Kamphorst JJ (2018) Triglycerides promote lipid homeostasis during hypoxic stress by balancing fatty acid saturation. Cell Rep 24: 2596–2605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adès LC, Gedeon AK, Wilson MJ, Latham M, Partington MW, Mulley JC, Nelson J, Lui K, Sillence DO (1993) Barth syndrome: clinical features and confirmation of gene localisation to distal Xq28. Am J Med Genet 45: 327–334 [DOI] [PubMed] [Google Scholar]
- Al‐Fageeh MB, Mark Smales C (2006) Control and regulation of the cellular responses to cold shock: the responses in yeast and mammalian systems. Biochem J 397: 247–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alper H, Fischer C, Nevoigt E, Stephanopoulos G (2005) Tuning genetic control through promoter engineering. Proc Natl Acad Sci USA 102: 12678–12683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anselmi C, Davies KM, Faraldo‐Gómez JD (2018) Mitochondrial ATP synthase dimers spontaneously associate due to a long‐range membrane‐induced force. J Gen Physiol 150: 763–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold I, Pfeiffer K, Neupert W, Stuart RA, Schägger H (1998) Yeast mitochondrial F1F0‐ATP synthase exists as a dimer: identification of three dimer‐specific subunits. EMBO J 17: 7170–7178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arselin G, Vaillier J, Salin B, Schaeffer J, Giraud M‐F, Dautant A, Brèthes D, Velours J (2004) The modulation in subunits e and g amounts of yeast ATP synthase modifies mitochondrial cristae morphology. J Biol Chem 279: 40392–40399 [DOI] [PubMed] [Google Scholar]
- Baile MG, Sathappa M, Lu Y‐W, Pryce E, Whited K, Michael McCaffery J, Han X, Alder NN, Claypool SM (2014) Unremodeled and remodeled cardiolipin are functionally indistinguishable in yeast. J Biol Chem 289: 1768–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CD, Basu Ball W, Pryce EN, Gohil VM (2016) Specific requirements of nonbilayer phospholipids in mitochondrial respiratory chain function and formation. Mol Biol Cell 27: 2161–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballweg S, Sezgin E, Doktorova M, Covino R, Reinhard J, Wunnicke D, Hänelt I, Levental I, Hummer G, Ernst R (2020) Regulation of lipid saturation without sensing membrane fluidity. Nat Commun 11: 756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bard M (1972) Biochemical and genetic aspects of nystatin resistance in Saccharomyces cerevisiae . J Bacteriol 111: 649–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer C, Herzog V, Bauer MF (2001) Improved technique for electron microscope visualization of yeast membrane structure. Microsc Microanal 7: 530–534 [DOI] [PubMed] [Google Scholar]
- Beltrán‐Heredia E, Tsai F‐C, Salinas‐Almaguer S, Cao FJ, Bassereau P, Monroy F (2019) Membrane curvature induces cardiolipin sorting. Commun Biol 2: 225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bione S, D'Adamo P, Maestrini E, Gedeon AK, Bolhuis PA, Toniolo D (1996) A novel X‐linked gene, G4.5. is responsible for Barth syndrome. Nat Genet 12: 385–389 [DOI] [PubMed] [Google Scholar]
- Bisetto E, Di Pancrazio F, Simula MP, Mavelli I, Lippe G (2007) Mammalian ATPsynthase monomer versus dimer profiled by blue native PAGE and activity stain. Electrophoresis 28: 3178–3185 [DOI] [PubMed] [Google Scholar]
- Blum TB, Hahn A, Meier T, Davies KM, Kühlbrandt W (2019) Dimers of mitochondrial ATP synthase induce membrane curvature and self‐assemble into rows. Proc Natl Acad Sci USA 116: 4250–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd KJ, Alder NN, May ER (2017) Buckling under pressure: curvature‐based lipid segregation and stability modulation in cardiolipin‐containing bilayers. Langmuir 33: 6937–6946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calizo RC, Bell MK, Ron A, Hu M, Bhattacharya S, Wong NJ, Janssen WGM, Perumal G, Pederson P, Scarlata S et al (2020) Cell shape regulates subcellular organelle location to control early Ca2+ signal dynamics in vascular smooth muscle cells. Sci Rep 10: 17866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callan‐Jones A, Sorre B, Bassereau P (2011) Curvature‐driven lipid sorting in biomembranes. Cold Spring Harb Perspect Biol 3: a004648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadda R, Krishnamani V, Mersch K, Wong J, Brimberry M, Chadda A, Kolmakova‐Partensky L, Friedman LJ, Gelles J, Robertson JL (2016) The dimerization equilibrium of a ClC Cl(−)/H(+) antiporter in lipid bilayers. Elife 5: e17438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadda R, Bernhardt N, Kelley EG, Teixeira SC, Griffith K, Gil‐Ley A, Öztürk TN, Hughes LE, Forsythe A, Krishnamani V et al (2021) Membrane transporter dimerization driven by differential lipid solvation energetics of dissociated and associated states. Elife 10: e63288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y‐F, Tsang K‐Y, Chang W‐F, Fan Z‐A (2015) Differential dependencies on [Ca2 ] and temperature of the monolayer spontaneous curvatures of DOPE, DOPA and cardiolipin: effects of modulating the strength of the inter‐headgroup repulsion. Soft Matter 11: 4041–4053 [DOI] [PubMed] [Google Scholar]
- Choi S‐Y, Gonzalvez F, Jenkins GM, Slomianny C, Chretien D, Arnoult D, Petit PX, Frohman MA (2007) Cardiolipin deficiency releases cytochrome c from the inner mitochondrial membrane and accelerates stimuli‐elicited apoptosis. Cell Death Differ 14: 597–606 [DOI] [PubMed] [Google Scholar]
- Claypool SM, Koehler CM (2012) The complexity of cardiolipin in health and disease. Trends Biochem Sci 37: 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool SM, Oktay Y, Boontheung P, Loo JA, Koehler CM (2008) Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J Cell Biol 182: 937–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogliati S, Enriquez JA, Scorrano L (2016) Mitochondrial cristae: where beauty meets functionality. Trends Biochem Sci 41: 261–273 [DOI] [PubMed] [Google Scholar]
- Covino R, Ballweg S, Stordeur C, Michaelis JB, Puth K, Wernig F, Bahrami A, Ernst AM, Hummer G, Ernst R (2016) A eukaryotic sensor for membrane lipid saturation. Mol Cell 63: 49–59 [DOI] [PubMed] [Google Scholar]
- Cugno A, Bartol TM, Sejnowski TJ, Iyengar R, Rangamani P (2019) Geometric principles of second messenger dynamics in dendritic spines. Sci Rep 9: 11676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daems WT, Wisse E (1966) Shape and attachment of the cristae mitochondriales in mouse hepatic cell mitochondria. J Ultrastruct Res 16: 123–140 [DOI] [PubMed] [Google Scholar]
- Dahlberg M, Maliniak A (2010) Mechanical properties of coarse‐grained bilayers formed by cardiolipin and zwitterionic lipids. J Chem Theory Comput 6: 1638–1649 [DOI] [PubMed] [Google Scholar]
- Darshi M, Mendiola VL, Mackey MR, Murphy AN, Koller A, Perkins GA, Ellisman MH, Taylor SS (2011) ChChd3, an inner mitochondrial membrane protein, is essential for maintaining crista integrity and mitochondrial function. J Biol Chem 286: 2918–2932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies KM, Strauss M, Daum B, Kief JH, Osiewacz HD, Rycovska A, Zickermann V, Kühlbrandt W (2011) Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc Natl Acad Sci USA 108: 14121–14126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawaliby R, Trubbia C, Delporte C, Noyon C, Ruysschaert J‐M, Van Antwerpen P, Govaerts C (2016) Phosphatidylethanolamine is a key regulator of membrane fluidity in eukaryotic cells. J Biol Chem 291: 3658–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degreif D, de Rond T, Bertl A, Keasling JD, Budin I (2017) Lipid engineering reveals regulatory roles for membrane fluidity in yeast flocculation and oxygen‐limited growth. Metab Eng 41: 46–56 [DOI] [PubMed] [Google Scholar]
- Doedens AL, Phan AT, Stradner MH, Fujimoto JK, Nguyen JV, Yang E, Johnson RS, Goldrath AW (2013) Hypoxia‐inducible factors enhance the effector responses of CD8+ T cells to persistent antigen. Nat Immunol 14: 1173–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudkina NV, Heinemeyer J, Keegstra W, Boekema EJ, Braun H‐P (2005) Structure of dimeric ATP synthase from mitochondria: an angular association of monomers induces the strong curvature of the inner membrane. FEBS Lett 579: 5769–5772 [DOI] [PubMed] [Google Scholar]
- Dymond MK (2021) Lipid monolayer spontaneous curvatures: a collection of published values. Chem Phys Lipids 239: 105117 [DOI] [PubMed] [Google Scholar]
- Edwards J, Daniel E, Kinney J, Bartol T, Sejnowski T, Johnston D, Harris K, Bajaj C (2014) VolRoverN: enhancing surface and volumetric reconstruction for realistic dynamical simulation of cellular and subcellular function. Neuroinformatics 12: 277–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ejsing CS, Sampaio JL, Surendranath V, Duchoslav E, Ekroos K, Klemm RW, Simons K, Shevchenko A (2009) Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc Natl Acad Sci USA 106: 2136–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippov A, Orädd G, Lindblom G (2003) The effect of cholesterol on the lateral diffusion of phospholipids in oriented bilayers. Biophys J 84: 3079–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey TG, Mannella CA (2000) The internal structure of mitochondria. Trends Biochem Sci 25: 319–324 [DOI] [PubMed] [Google Scholar]
- Frezza C, Cipolat S, Martins de Brito O, Micaroni M, Beznoussenko GV, Rudka T, Bartoli D, Polishuck RS, Danial NN, De Strooper B et al (2006) OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 126: 177–189 [DOI] [PubMed] [Google Scholar]
- Galber C, Valente G, von Stockum S, Giorgio V (2019) Purification of functional F‐ATP synthase from blue native PAGE. Methods Mol Biol 1925: 233–243 [DOI] [PubMed] [Google Scholar]
- Gallet PF, Petit JM, Maftah A, Zachowski A, Julien R (1997) Asymmetrical distribution of cardiolipin in yeast inner mitochondrial membrane triggered by carbon catabolite repression. Biochem J 324: 627–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia GC, Bartol TM, Phan S, Bushong EA, Perkins G, Sejnowski TJ, Ellisman MH, Skupin A (2019) Mitochondrial morphology provides a mechanism for energy buffering at synapses. Sci Rep 9: 18306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glytsou C, Calvo E, Cogliati S, Mehrotra A, Anastasia I, Rigoni G, Raimondi A, Shintani N, Loureiro M, Vazquez J et al (2016) Optic atrophy 1 is epistatic to the core MICOS component MIC60 in mitochondrial cristae shape control. Cell Rep 17: 3024–3034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregg C, Kyryakov P, Titorenko VI (2009) Purification of mitochondria from yeast cells. J Vis Exp 30: e1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Bueler SA, Rubinstein JL (2017) Atomic model for the dimeric FO region of mitochondrial ATP synthase. Science 358: 936–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama T, Riezman H (2019) Author correction: understanding the diversity of membrane lipid composition. Nat Rev Mol Cell Biol 20: 715 [DOI] [PubMed] [Google Scholar]
- Harner M, Körner C, Walther D, Mokranjac D, Kaesmacher J, Welsch U, Griffith J, Mann M, Reggiori F, Neupert W (2011) The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J 30: 4356–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harner ME, Unger A‐K, Geerts WJ, Mari M, Izawa T, Stenger M, Geimer S, Reggiori F, Westermann B, Neupert W (2016) An evidence based hypothesis on the existence of two pathways of mitochondrial crista formation. Elife 5: e18853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassinger JE, Oster G, Drubin DG, Rangamani P (2017) Design principles for robust vesiculation in clathrin‐mediated endocytosis. Proc Natl Acad Sci USA 114: E1118–E1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog R, Schwudke D, Schuhmann K, Sampaio JL, Bornstein SR, Schroeder M, Shevchenko A (2011) A novel informatics concept for high‐throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol 12: R8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog R, Schuhmann K, Schwudke D, Sampaio JL, Bornstein SR, Schroeder M, Shevchenko A (2012) LipidXplorer: a software for consensual cross‐platform lipidomics. PLoS One 7: e29851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppins S, Collins SR, Cassidy‐Stone A, Hummel E, Devay RM, Lackner LL, Westermann B, Schuldiner M, Weissman JS, Nunnari J (2011) A mitochondrial‐focused genetic interaction map reveals a scaffold‐like complex required for inner membrane organization in mitochondria. J Cell Biol 195: 323–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath SE, Daum G (2013) Lipids of mitochondria. Prog Lipid Res 52: 590–614 [DOI] [PubMed] [Google Scholar]
- Hu C, Shu L, Huang X, Yu J, Li L, Gong L, Yang M, Wu Z, Gao Z, Zhao Y et al (2020) OPA1 and MICOS regulate mitochondrial crista dynamics and formation. Cell Death Dis 11: 940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynen MA, Mühlmeister M, Gotthardt K, Guerrero‐Castillo S, Brandt U (2016) Evolution and structural organization of the mitochondrial contact site (MICOS) complex and the mitochondrial intermembrane space bridging (MIB) complex. Biochim Biophys Acta 1863: 91–101 [DOI] [PubMed] [Google Scholar]
- Ikon N, Ryan RO (2017) Cardiolipin and mitochondrial cristae organization. Biochim Biophys Acta Biomembr 1859: 1156–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen MJFW, Janssen MJF, Koorengevel MC, de Kruijff B, de Kroon AIPM (2000) The phosphatidylcholine to phosphatidylethanolamine ratio of Saccharomyces cerevisiae varies with the growth phase. Yeast 16: 641–650 [DOI] [PubMed] [Google Scholar]
- Jheng H‐F, Tsai P‐J, Guo S‐M, Kuo L‐H, Chang C‐S, Su I‐J, Chang C‐R, Tsai Y‐S (2012) Mitochondrial fission contributes to mitochondrial dysfunction and insulin resistance in skeletal muscle. Mol Cell Biol 32: 309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Rizavi HS, Greenberg ML (1997) Cardiolipin is not essential for the growth of Saccharomyces cerevisiae on fermentable or non‐fermentable carbon sources. Mol Microbiol 26: 481–491 [DOI] [PubMed] [Google Scholar]
- Kamphorst JJ, Cross JR, Fan J, de Stanchina E, Mathew R, White EP, Thompson CB, Rabinowitz JD (2013) Hypoxic and Ras‐transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc Natl Acad Sci USA 110: 8882–8887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara T, Kubota‐Sakashita M, Nagatsuka Y, Hirabayashi Y, Hanasaka T, Tohyama K, Kato T (2020) Cardiolipin is essential for early embryonic viability and mitochondrial integrity of neurons in mammals. FASEB J 34: 1465–1480 [DOI] [PubMed] [Google Scholar]
- Khalifat N, Puff N, Bonneau S, Fournier J‐B, Angelova MI (2008) Membrane deformation under local pH gradient: mimicking mitochondrial cristae dynamics. Biophys J 95: 4924–4933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalifat N, Fournier J‐B, Angelova MI, Puff N (2011) Lipid packing variations induced by pH in cardiolipin‐containing bilayers: the driving force for the cristae‐like shape instability. Biochim Biophys Acta Biomembr 1808: 2724–2733 [DOI] [PubMed] [Google Scholar]
- Klose C, Surma MA, Gerl MJ, Meyenhofer F, Shevchenko A, Simons K (2012) Flexibility of a eukaryotic lipidome – insights from yeast lipidomics. PLoS One 7: e35063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konar S, Arif H, Allolio C (2023) Mitochondrial membranes: model lipid compositions, material properties and the changing curvature of cardiolipin. bioRxiv 10.1101/2023.02.06.527315 [PREPRINT] [DOI] [PMC free article] [PubMed]
- Kushnirov VV (2000) Rapid and reliable protein extraction from yeast. Yeast 16: 857–860 [DOI] [PubMed] [Google Scholar]
- Kwast KE, Burke PV, Staahl BT, Poyton RO (1999) Oxygen sensing in yeast: evidence for the involvement of the respiratory chain in regulating the transcription of a subset of hypoxic genes. Proc Natl Acad Sci USA 96: 5446–5451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeCocq J, Ballou CE (1964) On the structure of cardiolipin*. Biochemistry 3: 976–980 [DOI] [PubMed] [Google Scholar]
- Lee CT, Laughlin JG, Angliviel de La Beaumelle N, Amaro RE, McCammon JA, Ramamoorthi R, Holst M, Rangamani P (2020) 3D mesh processing using GAMer 2 to enable reaction‐diffusion simulations in realistic cellular geometries. PLoS Comput Biol 16: e1007756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell BB, Shulman GI (2005) Mitochondrial dysfunction and type 2 diabetes. Science 307: 384–387 [DOI] [PubMed] [Google Scholar]
- Mahapatra A, Rangamani P (2023) Formation of protein-mediated bilayer tubes is governed by a snapthrough transition. Soft Matter 19: 4345–4359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannella CA (2006a) The relevance of mitochondrial membrane topology to mitochondrial function. Biochim Biophys Acta Mol Cell Res 1762: 140–147 [DOI] [PubMed] [Google Scholar]
- Mannella CA (2006b) Structure and dynamics of the mitochondrial inner membrane cristae. Biochim Biophys Acta 1763: 542–548 [DOI] [PubMed] [Google Scholar]
- Mannella CA, Marko M, Penczek P, Barnard D, Frank J (1994) The internal compartmentation of rat‐liver mitochondria: tomographic study using the high‐voltage transmission electron microscope. Microsc Res Tech 27: 278–283 [DOI] [PubMed] [Google Scholar]
- Mannella CA, Marko M, Buttle K (1997) Reconsidering mitochondrial structure: new views of an old organelle. Trends Biochem Sci 22: 37–38 [DOI] [PubMed] [Google Scholar]
- Manni MM, Tiberti ML, Pagnotta S, Barelli H, Gautier R, Antonny B (2018) Acyl chain asymmetry and polyunsaturation of brain phospholipids facilitate membrane vesiculation without leakage. Elife 7: e34394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrink SJ, Tieleman DP (2013) Perspective on the Martini model. Chem Soc Rev 42: 6801–6822 [DOI] [PubMed] [Google Scholar]
- Marrink SJ, Corradi V, Souza PCT, Ingólfsson HI, Tieleman DP, Sansom MSP (2019) Computational modeling of realistic cell membranes. Chem Rev 119: 6184–6226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell SJ, Stewart LC, Talin A, Yaffe MP (1990) Temperature‐sensitive yeast mutants defective in mitochondrial inheritance. J Cell Biol 111: 967–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald K, Müller‐Reichert T (2002) Cryomethods for thin section electron microscopy. Methods Enzymol 351: 96–123 [DOI] [PubMed] [Google Scholar]
- Meisinger C, Pfanner N, Truscott KN (2006) Isolation of yeast mitochondria. In Yeast Protocols, Xiao W (ed), pp 33–40. Totowa, NJ: Humana Press; [DOI] [PubMed] [Google Scholar]
- Mejia EM, Hatch GM (2016) Mitochondrial phospholipids: role in mitochondrial function. J Bioenerg Biomembr 48: 99–112 [DOI] [PubMed] [Google Scholar]
- Mendelsohn R, Garcia GC, Bartol TM, Lee CT, Khandelwal P, Liu E, Spencer DJ, Husar A, Bushong EA, Phan S et al (2022) Morphological principles of neuronal mitochondria. J Comp Neurol 530: 886–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Suomalainen A (2012) Mitochondria: in sickness and in health. Cell 148: 1145–1159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlig T, Le DV, Gardemann A, Wolke C, Gürtler S, Peter D, Schild L, Lendeckel U (2018) Effects of siRNA‐dependent knock‐down of cardiolipin synthase and tafazzin on mitochondria and proliferation of glioma cells. Biochim Biophys Acta Mol Cell Biol Lipids 1863: 379–387 [DOI] [PubMed] [Google Scholar]
- Ou‐Yang Z‐C, Liu J‐X, Xie Y‐Z, Yu‐Zhang X (1999) Geometric methods in the elastic theory of membranes in liquid crystal phases. Singapore: World Scientific; [Google Scholar]
- Pánek T, Eliáš M, Vancová M, Lukeš J, Hashimi H (2020) Returning to the fold for lessons in mitochondrial crista diversity and evolution. Curr Biol 30: R575–R588 [DOI] [PubMed] [Google Scholar]
- Paradies G, Paradies V, De Benedictis V, Ruggiero FM, Petrosillo G (2014) Functional role of cardiolipin in mitochondrial bioenergetics. Biochim Biophys Acta Bioenerg 1837: 408–417 [DOI] [PubMed] [Google Scholar]
- Paradies G, Paradies V, Ruggiero FM, Petrosillo G (2019) Role of cardiolipin in mitochondrial function and dynamics in health and disease: molecular and pharmacological aspects. Cell 8: 728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten DA, Wong J, Khacho M, Soubannier V, Mailloux RJ, Pilon‐Larose K, MacLaurin JG, Park DS, McBride HM, Trinkle‐Mulcahy L et al (2014) OPA1‐dependent cristae modulation is essential for cellular adaptation to metabolic demand. EMBO J 33: 2676–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paumard P, Vaillier J, Coulary B, Schaeffer J, Soubannier V, Mueller DM, Brèthes D, di Rago J‐P, Velours J (2002) The ATP synthase is involved in generating mitochondrial cristae morphology. EMBO J 21: 221–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penzo D, Tagliapietra C, Colonna R, Petronilli V, Bernardi P (2002) Effects of fatty acids on mitochondria: implications for cell death. Biochim Biophys Acta Bioenerg 1555: 160–165 [DOI] [PubMed] [Google Scholar]
- Perkins G, Renken C, Martone ME, Young SJ, Ellisman M, Frey T (1997) Electron tomography of neuronal mitochondria: three‐dimensional structure and organization of cristae and membrane contacts. J Struct Biol 119: 260–272 [DOI] [PubMed] [Google Scholar]
- Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI (2004) Impaired mitochondrial activity in the insulin‐resistant offspring of patients with type 2 diabetes. N Engl J Med 350: 664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit JM, Huet O, Gallet PF, Maftah A, Ratinaud MH, Julien R (1994) Direct analysis and significance of cardiolipin transverse distribution in mitochondrial inner membranes. Eur J Biochem 220: 871–879 [DOI] [PubMed] [Google Scholar]
- Rabl R, Soubannier V, Scholz R, Vogel F, Mendl N, Vasiljev‐Neumeyer A, Körner C, Jagasia R, Keil T, Baumeister W et al (2009) Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J Cell Biol 185: 1047–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rak M, Tzagoloff A (2009) F1‐dependent translation of mitochondrially encoded Atp6p and Atp8p subunits of yeast ATP synthase. Proc Natl Acad Sci USA 106: 18509–18514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangamani P, Lipshtat A, Azeloglu EU, Calizo RC, Hu M, Ghassemi S, Hone J, Scarlata S, Neves SR, Iyengar R (2013) Decoding information in cell shape. Cell 154: 1356–1369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawicz W, Olbrich KC, McIntosh T, Needham D, Evans E (2000) Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys J 79: 328–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina‐Campos M, Heeg M, Kennewick K, Mathews IT, Galletti G, Luna V, Nguyen Q, Huang H, Milner JJ, Hu KH et al (2023) Metabolic programs of T cell tissue residency empower tumour immunity. Nature 621: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren M, Phoon CKL, Schlame M (2014) Metabolism and function of mitochondrial cardiolipin. Prog Lipid Res 55: 1–16 [DOI] [PubMed] [Google Scholar]
- Revel JP, Fawcett DW, Philpott CW (1963) Observations on mitochondrial structure. J Cell Biol 16: 187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieger B, Junge W, Busch KB (2014) Lateral pH gradient between OXPHOS complex IV and F0F1 ATP‐synthase in folded mitochondrial membranes. Nat Commun 5: 3103 [DOI] [PubMed] [Google Scholar]
- Rowlett VW, Mallampalli VKPS, Karlstaedt A, Dowhan W, Taegtmeyer H, Margolin W, Vitrac H (2017) Impact of membrane phospholipid alterations in Escherichia coli on cellular function and bacterial stress adaptation. J Bacteriol 199: e00849‐16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sesaki H, Southard SM, Yaffe MP, Jensen RE (2003) Mgm1p, a dynamin‐related GTPase, is essential for fusion of the mitochondrial outer membrane. Mol Biol Cell 14: 2342–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohlenkamp C, Geiger O (2016) Bacterial membrane lipids: diversity in structures and pathways. FEMS Microbiol Rev 40: 133–159 [DOI] [PubMed] [Google Scholar]
- Sparagna GC, Hickson‐Bick DL, Maximilian Buja L, McMillin JB (2000) A metabolic role for mitochondria in palmitate‐induced cardiac myocyte apoptosis. Am J Physiol Heart Circ Physiol 279: H2124–H2132 [DOI] [PubMed] [Google Scholar]
- Stewart LC, Yaffe MP (1991) A role for unsaturated fatty acids in mitochondrial movement and inheritance. J Cell Biol 115: 1249–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss M, Hofhaus G, Schröder RR, Kühlbrandt W (2008) Dimer ribbons of ATP synthase shape the inner mitochondrial membrane. EMBO J 27: 1154–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stukey JE, McDonough VM, Martin CE (1989) Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae . J Biol Chem 264: 16537–16544 [PubMed] [Google Scholar]
- Surma MA, Herzog R, Vasilj A, Klose C, Christinat N, Morin‐Rivron D, Simons K, Masoodi M, Sampaio JL (2015) An automated shotgun lipidomics platform for high throughput, comprehensive, and quantitative analysis of blood plasma intact lipids. Eur J Lipid Sci Technol 117: 1540–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szule JA, Fuller NL, Rand RP (2002) The effects of acyl chain length and saturation of diacylglycerols and phosphatidylcholines on membrane monolayer curvature. Biophys J 83: 977–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timón‐Gómez A, Pérez‐Pérez R, Nyvltova E, Ugalde C, Fontanesi F, Barrientos A (2020) Protocol for the analysis of yeast and human mitochondrial respiratory chain complexes and supercomplexes by blue native electrophoresis. STAR Protoc 1: 100089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance JE (2015) Phospholipid synthesis and transport in mammalian cells. Traffic 16: 1–18 [DOI] [PubMed] [Google Scholar]
- Vasan R, Rudraraju S, Akamatsu M, Garikipati K, Rangamani P (2020) A mechanical model reveals that non‐axisymmetric buckling lowers the energy barrier associated with membrane neck constriction. Soft Matter 16: 784–797 [DOI] [PubMed] [Google Scholar]
- Vasconcelles MJ, Jiang Y, McDaid K, Gilooly L, Wretzel S, Porter DL, Martin CE, Goldberg MA (2001) Identification and characterization of a low oxygen response element involved in the hypoxic induction of a family of Saccharomyces cerevisiae genes. J Biol Chem 276: 14374–14384 [DOI] [PubMed] [Google Scholar]
- Viana MP, Lim S, Rafelski SM (2015) Quantifying mitochondrial content in living cells. Methods Cell Biol 125: 77–93 [DOI] [PubMed] [Google Scholar]
- Voelker DR (1997) Phosphatidylserine decarboxylase. Biochim Biophys Acta 1348: 236–244 [DOI] [PubMed] [Google Scholar]
- Walani N, Torres J, Agrawal A (2015) Endocytic proteins drive vesicle growth via instability in high membrane tension environment. Proc Natl Acad Sci USA 112: 201418491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnikoff JR, Haddock SHD, Budin I (2021) Depth‐ and temperature‐specific fatty acid adaptations in ctenophores from extreme habitats. J Exp Biol 224: jeb242800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Erdjument‐Bromage H, Phoon CKL, Neubert TA, Ren M, Schlame M (2021) Cardiolipin remodeling enables protein crowding in the inner mitochondrial membrane. EMBO J 40: e108428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue R‐Q, Zhao M, Wu Q, Yang S, Cui Y‐L, Yu X‐J, Liu J, Zang W‐J (2019) Regulation of mitochondrial cristae remodelling by acetylcholine alleviates palmitate‐induced cardiomyocyte hypertrophy. Free Radic Biol Med 145: 103–117 [DOI] [PubMed] [Google Scholar]
- Yang D, Li J, Li Z, Zhao M, Wang D, Sun Z, Wen P, Gou F, Dai Y, Ji Y et al (2023) Cardiolipin externalization mediates prion protein (PrP) peptide 106‐126‐associated mitophagy and mitochondrial dysfunction. Front Mol Neurosci 16: 1163981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Mileykovskaya E, Dowhan W (2005) Cardiolipin is essential for organization of complexes III and IV into a supercomplex in intact yeast mitochondria. J Biol Chem 280: 29403–29408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Kelly CJ, Colgan SP (2015) Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A review in the theme: cellular responses to hypoxia. Am J Physiol Cell Physiol 309: C350–C360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zick M, Rabl R, Reichert AS (2009) Cristae formation—linking ultrastructure and function of mitochondria. Biochim Biophys Acta Mol Cell Res 1793: 5–19 [DOI] [PubMed] [Google Scholar]
- Zinser E, Sperka‐Gottlieb CD, Fasch EV, Kohlwein SD, Paltauf F, Daum G (1991) Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J Bacteriol 173: 2026–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]


