Abstract
Background
Aldehyde dehydrogenase 2 (ALDH2) is a crucial enzyme involved in endogenous aldehyde detoxification and has been implicated in tumor progression. However, its role in tumor immune evasion remains unclear.
Methods
Here, we analyzed the relationship between ALDH2 expression and antitumor immune features in multiple cancers. ALDH2 knockout tumor cells were then established using CRISPR/Cas9 system. In immunocompetent breast cancer EMT6 and melanoma B16-F10 mouse models, we investigated the impact of ALDH2 blockade on cytotoxic T lymphocyte function and tumor immune microenvironment by flow cytometry, mass cytometry, Luminex liquid suspension chip detection, and immunohistochemistry. Furthermore, RNA sequencing, flow cytometry, western blot, chromatin immunoprecipitation assay, and luciferase reporter assays were employed to explore the detailed mechanism of ALDH2 involved in tumor immune evasion. Lastly, the synergistic therapeutic efficacy of blocking ALDH2 by genetic depletion or its inhibitor disulfiram in combination with immune checkpoint blockade (ICB) was investigated in mouse models.
Results
In our study, we uncovered a positive correlation between the expression level of ALDH2 and T-cell dysfunction in multiple cancers. Furthermore, blocking ALDH2 significantly suppressed tumor growth by enhancing cytotoxic activity of CD8+ T cells and reshaping the immune landscape and cytokine milieu of tumors in vivo. Mechanistically, inhibiting ALDH2-mediated metabolism of aldehyde downregulated the expression of V-domain Ig suppressor of T-cell activation (VISTA) via inactivating the nucleotide oligomerization domain (NOD)/nuclear factor kappa-B (NF-κB) signaling pathway. As a result, the cytotoxic function of CD8+ T cells was revitalized. Importantly, ALDH2 blockade markedly reinforced the efficacy of ICB treatment.
Conclusions
Our data delineate that ALDH2-mediated aldehyde metabolism drives tumor immune evasion by activating the NOD/NF-κB/VISTA axis. Targeting ALDH2 provides an effective combinatorial therapeutic strategy for immunotherapy.
Keywords: Breast Neoplasms, Immunity, Immunotherapy, Melanoma
WHAT IS ALREADY KNOWN ON THIS TOPIC
Tumor metabolism contributes to tumor immune evasion, resulting in tumor progression. However, the underlying mechanism remains unclear.
WHAT THIS STUDY ADDS
We showed that ALDH2-mediated aldehyde metabolism induces tumor immune evasion by modulating the NOD/NF-κB/VISTA signaling pathway. Importantly, inhibiting ALDH2 enhances the therapeutic responses of ICB.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Our findings highlight the importance of ALDH2-mediated aldehyde metabolism in triggering tumor immune evasion, and present a potential therapeutic strategy to improve the effectiveness of ICB therapy.
Introduction
Immune evasion plays a key role in tumor progression, which implies that tumors have potential to establish conditions that resist immunosurveillance within the tumor microenvironment.1 2 Increasing studies reveal that tumor cells use various strategies to promote immune evasion, such as expressing immune checkpoints, altering human leukocyte antigen, developing tumor-specific mutations, and inducing chromosomal instability.3–5 Immune checkpoints are a classic suppressive mechanism used by cancer cells to evade T-cell recognition and elicit immune evasion. These checkpoints, which include but are not limited to programmed death-1 (PD-1), programmed cell death-Ligand 1 (PD-L1), and cytotoxic T lymphocyte-associated antigen 4 (CTLA-4), have been successfully applicated in the clinic.6 7 Nevertheless, the response rates of patients with cancer to immune checkpoint blockade (ICB) therapies are still low,8 underscoring that the deep mechanisms of ICB-related immune evasion needed to be further elucidated.
Metabolic reprogramming, which is a hallmark of tumor initiation and progression, plays a critical role in regulating tumor proliferation, survival, and metastasis.9 10 Moreover, emerging evidence exhibits that tumor metabolism might also have a significant impact on tumor immune evasion by affecting immune molecules expression, cytokine production and secretion, and immune cell composition.11–13 For instance, it has been shown that tumor cells could restrict glycolysis in tumor-infiltrating immune cells, resulting in T-cell anergy and depletion.14 Additionally, metabolic impairment of T cells can further promote the activation and differentiation of immunosuppressive cells in tumor microenvironment (TME), leading to immune evasion in multiple tumors.15 16 Nevertheless, while tumor metabolism has been identified as a key mediator of immune evasion, the detailed mechanism underlying this process is not yet clear.
Aldehyde dehydrogenase 2 (ALDH2) is a key enzyme involved in aldehyde metabolism and various biological processes, including fatty acid metabolism, oxidative stress, hypoxia, and DNA repair.17 18 Numerous studies suggest that ALDH2 serves as a biomarker for cancer stem cells and contributes to tumor progression in multiple cancers.19–21 A recent study indicates that ALDH2 could upregulate PD-L1 expression by inhibiting ubiquitin-dependent degradation in colorectal cancer, suggesting that ALDH2 might mediate alcohol-related immune evasion.22 However, the mechanisms underlying ALDH2-induced immune evasion have not been fully explored. In this study, we investigated the impact of ALDH2-mediated metabolism of aldehyde on tumor immune evasion and its potential therapeutic implications for ICB therapy.
Materials and methods
Supplemental materials and methods can be found in online supplemental materials.
jitc-2023-007487supp001.pdf (1MB, pdf)
Bioinformatics analyses
Functional gene expression signatures (Fges) are used to define different cell types in TME. The signature scores were calculated using an in-house python implementation of single-sample gene set enrichment analysis (ssGSEA).23 Then, the intensities were median-scaled (median-centered and median absolute deviation-scaled) for all the samples within the cohort groups. Melanoma samples were curated from The Cancer Genome Atlas (TCGA). Gene expression signatures of T-cell exhaustion were elucidated using a 5-gene signature (TIGIT, CTLA-4, HAVCR2, LAG3, PDCD1).24 T-cell dysfunction scores based on ALDH2 gene expression level in TCGA breast cancer (BRCA) and skin cutaneous melanoma (SKCM) were evaluated using the Tumor Immune Dysfunction and Exclusion (TIDE) system (http://tide.dfci.harvard.edu/faq/).25 The correlation between ALDH2 expression and cytotoxic T lymphocyte (CTL) infiltration was also analyzed through the TIDE system. Kaplan-Meier survival analysis was conducted to investigate the relationship between ALDH2 gene expression level and overall patient survival based on GEPIA 2.26 After quality filtering (p<0.05), only samples meeting the criteria were included in the analysis.
Cell lines and culture
The human BRCA cell lines MDA-MB-231 and MCF7, the BALB/c murine BRCA cell lines EMT6, and the C57BL/6 melanoma cell lines B16-F10 were obtained from the American Type Culture Collection. All cells were cultured in either Roswell Park Memorial Institute (RPMI)-1640 or Dulbecco's Modified Eagle Medium (DMEM) media supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin.
Generation of stable cells using lentiviral infection
The human ALDH2-targeting overexpression plasmid was purchased from GeneCopoeia Company and transfected into MCF7 cell lines using the Lipo2000 DNA in vitro Transfection Reagent (SignaGen Laboratories). Both human and mouse ALDH2-targeting sgRNAs were synthesized by BGI Genomics and cloned into the pLentiCRISP V.2 plasmid through denaturation, annealing, and fragment ligation post-digestion with BsmBI. All ligation products were transformed into DH5α competent cells and spread on solid Amp+ Luria broth medium. After single colonies were inoculated into liquid Amp+ Luria broth medium and sequenced, the validated plasmids were extracted and stored at −20°C. To obtain the system to knock out small guide ALDH2 (sgALDH2) in cell line, the lentiviral expression vector and lentivirus packing vectors were co-transfected into 293 T cells using the Lipo293T DNA in vitro Transfection Reagent. After 48–72 hours transfection, cancer cell lines were stably infected with viral particles. After transfection, the target cells were diluted to 1 cell/100 µL in medium, and seeded into a 96-well plate with a volume of 100 µL per well. Wells containing single cells were observed and preliminarily identified under a microscope 24 hours after puromycin filtration. After 5 days, monoclonal cells were transferred to 24-well plates. Once the cells reached approximately 70–80% confluency, a portion of cells were transferred to 6-well plates to confirm ALDH2 expression level through western blot. The CRISPR sgRNA sequences were listed in online supplemental table 1.
Mice and in vivo tumor studies
For the EMT6 and B16-F10 models, BALB/c or C57BL/6 mice were orthotopically injected with 4×105 or 1.5×105 tumor cells orthotopically, respectively. At day third post tumor induction, disulfiram (HY-B0240, MCE; 50 mg/kg) were intragastrically administered to the mice.27 Anti-CTLA-4 antibody (Bio X Cell, Clone:9D9, Catalog #: BP0164; 5 mg/kg) and anti-PD-1 antibody (Bio X Cell, Clone:29F.1A12, Catalog #: BP0273; 10 mg/kg) were administered intraperitoneally on days 7, 10, 13 and 16 after tumor inoculation.28 Tumor volume was measured using electronic calipers two or three times per week and was calculated according to the formula: volume=(length×width2/2). Female BALB/c and C57BL/6 mice aged between 6-8 weeks old were used in all experiments, and all mice were provided by the Laboratory Animal Center of Chongqing Medical University.
Preparation of single-cell suspension from tumors
After tumor inoculation, the tumor-bearing mice were sacrificed. EMT6 tumor samples were harvested, chopped with scissors, and incubated with 2 mg/mL collagenase A (Roche) for 40 min at 37°C. The dissociated cells were filtered through 70 µm filters (BD Biosciences) to obtain single-cell suspensions. B16-F10 tumor samples were directly harvested and filtered with a screen to obtain single-cell suspensions. Afterward, erythrocytes were lysed with red blood cell lysis buffer for 2 min on ice, and the resulting single-cell suspension was washed and re-suspended in either DMEM or phosphate-buffered saline (PBS) depending on further use.
Flow cytometry
Tumor cells from the mouse model were stained for Live/Dead with Fixable Viability Dye eFluor 450 (eBioscience) for 30 min at 4°C. After a single wash with flow cytometry buffer, cells were stained with various combinations of primary antibodies to cell surface markers, including CD45 (APC-CY7), CD11b (BV510), CD4 (PE-CY5.5), CD8 (PE-CY7/FITC), Gr-1 (PE), F4/80 (APC/BV605), V-domain Ig suppressor of T-cell activation (VISTA) (APC), LY6C (PE-CY7), LY6G (FITC), major histocompatibility complex-II (PerCP-CY5.5), T-cell immunoglobulin and mucin domain-3 (TIM-3, PE-CY7) and PD-1 (PerCP-CY5.5). After that, cells were washed with flow cytometry buffer. Intracellular staining was performed according to the manufacturer’s protocol, where cells were first fixed and permeabilized using the Foxp3 staining buffer kit (eBioscience), and then incubated with fluorochrome-conjugated antibodies to Ki67 (PE) from BioLegend. For intracellular cytokine staining, cells were stimulated with cell stimulation cocktail (eBioscience) for 4 hours at 37°C, followed by staining with CD206 (APC), perforin 1 (PRF1, PE), granzyme B (GZMB, PerCP-CY5.5), interleukin (IL)-2 (PE), interferon (IFN)-γ (APC) and tumor necrosis factor (TNF)-α (PE/Cyanine7). In vitro tumor cell lines were stained with dilutions of VISTA (PE or APC). The following agents were used in the study: Disulfiram (HY-B0240, MCE; 100 nM), 4-Hydroxynonenal (4-HNE, HY-113466, MCE; 100 nM), Alda1 (S5800, Selleck; 20 µM), Nodinitib-1 (ML130, HY-18639, MCE; 10 µM) and QNZ (S4902, Selleck; 10 µM). Cells were washed with flow cytometry buffer after staining and fixed with 4% paraformaldehyde. The dilution ratios for all antibodies were 1:100. Finally, cells were acquired using BD FACS Canto II and BD FACSDiva software (BD Biosciences) with subsequent analysis performed using FlowJo software.
Mass cytometry (CyTOF) and data analysis
EMT6 mouse tumor tissues were mechanically processed and digested as previously described.29 The resulting cell suspension was filtered through 70 µm (BD Biosciences), and cells were then incubated with 25 mM cisplatin for 1 min (viability staining), followed by staining with monoclonal antibody cocktails against intracellular proteins. Metal-tagged antibodies used in the mass cytometry analysis were purchased from Fluidigm. The cells suspension was diluted to approximately 106 cells per mL using ddH2O containing bead standards and then analyzed on a CyTOF 2 mass cytometer (Fluidigm). CyTOF data were normalized and manually gate using Cytobank software. After CD45+ immune cells were gated, the data were transformed using the cytofAsinh function before being applied to the downstream analysis. Immune subsets were generated during PhenoGraph clustering analysis using the R cytofkit package. FlowSOM, an unsupervised automated algorithm, was used to order cells based on their phenotypic similarities.
Heatmaps were generated, based on the mean value of each marker in clusters. To calculate the cell frequency of each cluster, the percent of each cell type was divided by the total CD45+ cells in the same sample. The antibodies used in the mass cytometry analysis were purchased from Fluidigm: 89Y-anti-CD45, 175Lu-anti-CD4, 141Pr-anti-PD1, 143Nd-anti-CD11b, 144Nd-anti-Siglec F, 145Nd-anti-CD69, 146Nd-anti-CD206, 148Nd-anti-Tbet, 114Sn-anti-CD103, 151Eu-anti-CD68, 152Gd-anti-CD3e, 156Gd-anti-CD14, 159Tb-anti-F4/80, 160Dy-anti-CD62L, 161Dy-anti-Ki67, 162Dy-anti-Ly-6C, 165Ho-anti-Foxp3, 149Sm-anti-CD19, 167Er-anti-GATA3, 142Ce-anti-NK1.1, 168Er-anti-CD8a, 172Yb-anti-CD86, 173Yb-anti-CD117, 174Yb-anti-ly-6G/Ly-6C (Gr-1), 209Bi-anti-I-A/I-E, 150Sm-anti-CD11c, 169Tm-CTLA-4.
Measurement of cytokine levels
Luminex liquid suspension chip detection was performed by Wayen Biotechnologies (Shanghai, China). The Bio-Plex Pro Human Chemo kine Panel 31-plex kit (Bio-Rad, Austin, Texas, USA) was used according to the manufacturer’s instructions. The results were assessed using Bio-Plex MAGPIX System (Bio-Rad, Austin, Texas, USA).
T cells proliferation and function assays in vitro
The splenic T cells were harvested from wild-type (WT) BALB/c mice and filtered through 70 µm filters (BD Biosciences) to generate a single-cell suspension. After red blood cell lysis, T cells were counted and plated in complete 1640 medium supplemented with 50 mM b-mercaptoethanol and 10 mM 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid onto 12-well plates coated with 2.5 mg/mL anti-CD3 (clone 145–2C11, BioLegend) and 3 mg/mL anti-CD28 (clone 37N, BioLegend) antibodies. A portion of T cells were labeled with 1 µM of CellTraceTM carboxyfluorescein succinimidyl ester. The splenic T cells were then activated for 48 hours before being co-cultured with myeloid cells sorted from system to knock out normal controls (sgNC) or sgALDH2 tumors or co-cultured with sgNC or sgALDH2 tumor cells treated with anti-VISTA antibody (Bio X Cell, Clone: 13F3, Catalog #: BE0310; 10 mg/mL). After co-cultured for another 24 or 48 hours, T cells were collected for proliferation and function assays by flow cytometry. The data was analyzed and performed using FlowJo software.
Metabolites assays
The levels of 4-HNE and malondialdehyde (MDA) in cell culture supernatant and tissues were detected by ELISA kits (D751041, BBI) and MDA assay kit (D799762, BBI) according to the manufacturer’s instructions.
Luciferase reporter assay
The pEZX-PL01‐VISTA was constructed by inserting the 2100 kb human VISTA promoter (−2000 to +100 nt) into the vector of pEZX-PL01‐basic (GeneCopoeia). The tumor cells were co-transfected with the above reporter plasmid with pM35-P65 overexpression plasmid using Lipo2000 DNA in vitro Transfection Reagent (SignaGen Laboratories). The luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega, USA).
Chromatin immunoprecipitation (ChIP)-quantitative polymerase chain reaction (qPCR) assay
We performed the ChIP assay using the SimpleChIP Enzymatic Chromatin IP Kit (CST), according to the manufacturer’s instructions. Antibodies against P65 (CST, 8242), anti-RNA polymerase II (positive control), or normal rabbit IgG (negative control) were used for incubation and precipitation. The immunoprecipitated DNA fragments were detected by qPCR analysis. The primer sequences used for the ChIP assay were listed in online supplemental table 1.
Immunohistochemistry (IHC) staining
The EMT6 tumor tissues were fixed in 4% formaldehyde solution (pH 7.0) and subsequently embedded in paraffin. Immunohistochemical studies were performed using the standard streptavidin-peroxidase method with the UltraSensitive TM SP Kit (Maixin-Bio, Fujian, China) following the manufacturer’s instructions. Tumor specimens were stained using antibodies against GZMB (Abcam, ab4059, 1:800) or CD8 (Abcam, ab199016, 1:200). Negative controls were performed by replacing the primary antibody with PBS. The immunostained slides were blindly evaluated by a trained pathologist.
RNA sequencing and data analysis
MDA-MB-231 tumor cells were collected, and total RNA was purified using TRIzol (Invitrogen). The RNA samples were then sent to the Novogene company for library construction and sequencing. Genes with adjusted p values<0.05, found by the DESeq2 R package, were designated as differential expression analysis (DEGs). Pathway analysis of Kyoto Encyclopedia of Genes and Genomes (KEGG) was conducted using R to identify which remarkably downregulated genes were notably enriched in pathways. The gene set enrichment analysis (GSEA) analysis was performed by GSEA software V.4.0.3.
Western blot
Western blot was performed as previously described.29 The following primary antibodies were used at a dilution ratio of 1:1000: ALDH2 (Abcam, ab108306), NOD1 (CST, 3545), P65 (CST, 8242), p-P65 (CST, 3033) and GAPDH (Proteintech, 60004–1-Ig). The following agents were used: Disulfiram (HY-B0240, MCE; 100 nM), 4-HNE (HY-113466, MCE; 100 nM) and Nodinitib-1 (ML130, HY-18639, MCE; 10 µM).
Statistical analysis
The flow cytometric data were analyzed using FlowJo software (V.10; Tree Star), and GraphPad Prism (V.8) was used to generate graphs and for statistical analysis. One-way or two-way analysis of variance (ANOVA) was used when comparing more than two groups, while unpaired two-tailed t-test was used when comparing only two groups. p<0.05 was considered statistically significant (*p<0.05, **p<0.01, ***p<0.001). Mouse survival was evaluated using the Kaplan-Meier method and analyzed by the Mantel-Cox log-rank test. All experiments were performed at least three times, and n refers to biological replicates.
Results
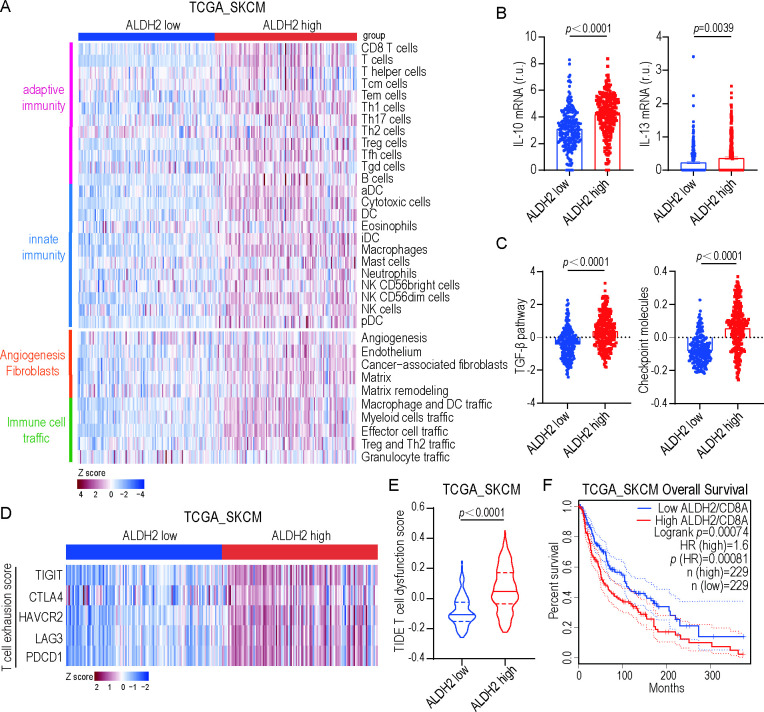
ALDH2 correlates with T-cell dysfunction in solid tumors
To classify the tumor immune microenvironment (TIME) in human samples, we used a list of Fges representing immune-enrich, immunosuppression and tumor stroma to reveal the molecular profiles of melanoma based on ssGSEA scores. Our analysis revealed that patients classified as ALDH2-low versus ALDH2-high exhibited different gene expression involved in adaptive and innate immunity, angiogenesis fibroblasts, and immune cell traffic (figure 1A). Interestingly, GSEA analysis also demonstrated that characteristic molecular components of immunosuppressive backgrounds were enriched in ALDH2-high versus ALDH2-low melanoma. Specifically, ALDH2-high group showed an increase in IL-10 and IL-13 expression (figure 1B), high transforming growth factor (TGF) -β and checkpoint molecules scores (figure 1C), indicating that ALDH2 is associated with an immunosuppressive TME.30 Furthermore, using a T-cell exhaustion signature, we found that ALDH2-high melanoma samples exhibited markedly higher T-cell exhaustion score compared with ALDH-low samples (figure 1D). Similarly, the ALDH2-high group had significantly higher T-cell dysfunction score than the ALDH2-low group in melanoma and BRCA based on the TIDE system (figure 1E and online supplemental figure 1A). Moreover, we analyzed CD8+ T cell-related survival in patients and found that the patients with high ALDH2 expression had significantly shorter overall survival (figure 1F). Consistently with this, high CTL infiltration (CTL-top) patients had prolonged survival compared with low CTL infiltration (CTL-bottom) patients in ALDH2-low group in multiple cancers based on the TIDE system (online supplemental figure 1B-C), suggesting that ALDH2 is correlated with T-cell dysfunction.
Figure 1.
ALDH2 correlates with T-cell dysfunction in solid tumors. (A) Heatmap of TCGA melanomas (TCGA-SKCM) classified into four distinct tumor microenvironment subtypes based on unsupervised dense clustering of the 29 functional gene expression signatures. P values were calculated with the χ2 test. (B) IL-10 (left) and IL-13 (right) mRNA levels in ALDH2 low versus ALDH2 high patients (low: n=229; high: n=243; t-test). r.u., relative units. (C) Enrichments for TGF-β score (left) and checkpoint molecules (right) in ALDH2 low versus ALDH2 high samples (low: n=229; high: n=243; t-test). (D) Heatmaps depicting ALDH2 expression and expression of genes associated with T-cell exhaustion across TCGA-SKCM samples. P values were calculated with the χ2 test. (E) T-cell dysfunction scores of ALDH2 low and ALDH2 high expression are assessed based on the TIDE system (low: n=198; high: n=215; t-test). (F) Kaplan-Meier curves of overall survival in ALDH2 low and ALDH2 high patient groups, as stratified by CD8 classification, selected in TCGA-SKCM database. P values correspond to two-sided log-rank analyses. Mean±SEM. DC, dendritic cell; HAVCR2, hepatitis A virus cellular receptor 2; LAG3, lymphocyte-activation gene 3; mRNA, messenger RNA; PDCD1, programmed cell death protein 1; TIGIT, T cell immunoglobulin and ITIM domain.
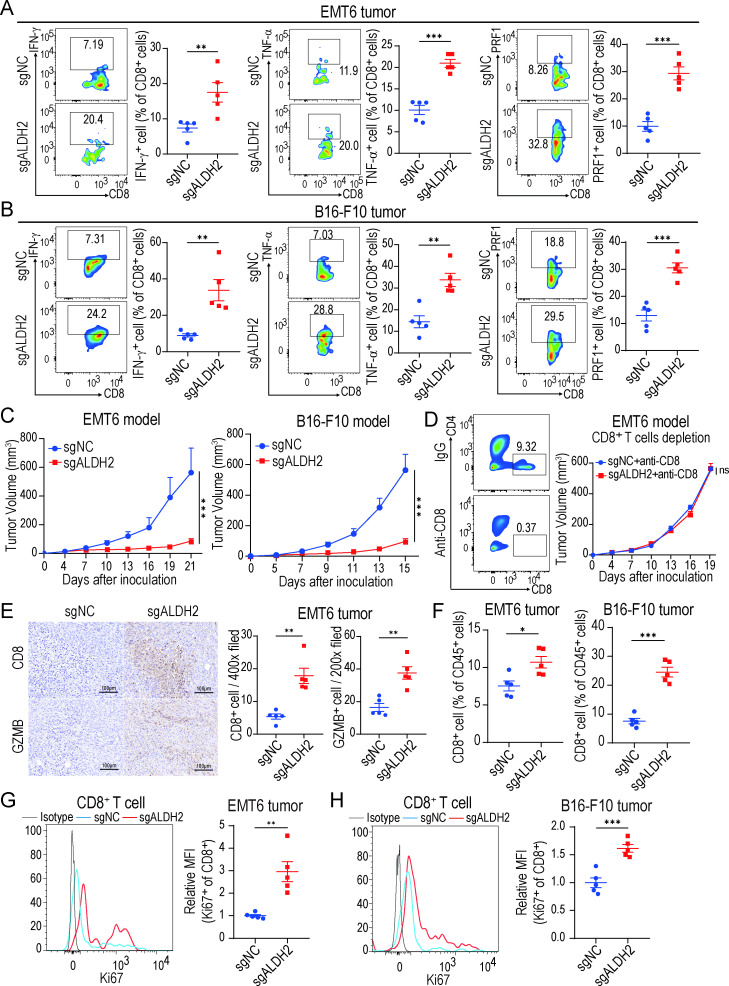
Blocking ALDH2 enhances antitumor activities of CD8+ T cells in vivo
To investigate the effect of ALDH2 on the cytotoxic activities of CD8+ T cells, we first used the CRISPER-CAS9 system to knock out ALDH2 (sgALDH2) in WT BRCA EMT6 tumor cells and melanoma B16-F10 tumor cells, with non-targeting constructs as normal control (sgNC). The reduction of ALDH2 at the protein level was confirmed by western blot (online supplemental figure 2A). Next, sgALDH2 or sgNC tumor cells were inoculated into immunocompetent mice to establish syngeneic mouse models. Our results showed that knocking out ALDH2 in tumors led to an increased release of effector cytokines including IFN-γ, TNF-α, PRF1, GZMB, and IL-2 from CD8+ T cells as well as retarded tumor growth (figure 2A–C and online supplemental figure 2B-C), suggesting that blocking ALDH2 could enhance the antitumor activities of CD8+ cytotoxic T cells. Consistently, IHC staining also demonstrated that ALDH2-knockout EMT6 tumors had higher expression of the effector molecule GZMB compared with control tumors (figure 2E), which provided further support for our hypothesis. To be noted, the inhibition of EMT6 tumor growth in the ALDH2-knockout group was blocked by depleting CD8+ T cells with anti-CD8 antibody (figure 2D), showing that ALDH2-related antitumor activity depended on CD8+ T cells. Furthermore, we found that the infiltration and proliferation (Ki67+) of CD8+ T cells was significantly increased in ALDH2-knockout tumors (figure 2F–H), indicating that more activated CD8+ T cell had been recruited after ALDH2 inhibition. Intriguingly, ALDH2-knockout also reduced the percentage of exhausted CD8+ T cells (PD-1+TIM-3+) in tumors, suggesting that the exhausted state of CD8+ T cells was reversed by ALDH2 blockade (online supplemental figure 2D). However, cell proliferation assay showed no statistically difference between sgALDH2 and sgNC tumor cells in vitro (online supplemental figure 2E), indicating that inhibiting ALDH2 did not directly affect tumor cell proliferation. Together, these findings demonstrate that ALDH2 inhibition might exert antitumor effect by reinvigorating cytotoxic activities of CD8+ T cells.
Figure 2.
Blocking ALDH2 enhances antitumor activities of CD8+ T cells in vivo. (A–B) Flow cytometry analysis of IFN-γ+, TNF-α+ and PRF1+ CD8+ T cells from EMT6 tumor (A) and B16-F10 tumor (B) (n=5, t-test). (C) EMT6 (left) and B16-F10 (right) tumor growth in ALDH2-knockout and control tumor-bearing mice (n=5 mice/group, two-way ANOVA). (D) EMT6 tumor growth with indicated treatment. CD8+ T cells were depleted by anti-CD8 antibodies (n=5 mice/group, two-way ANOVA). (E) Representative immunohistochemical images (left) and the quantity of positively stained cells (right) of CD8+ T cells and granzyme B in EMT6 tumors (n=5, t-test). Scale bar, 100 µm. (F) Representative quantification of CD8+ T cells ratios in EMT6 and B16-F10 tumors (n=5, t-test). (G–H) Flow cytometry analysis of Ki-67 expression on CD8+ cells in EMT6 (G) and B16-F10 (H) tumors (n=5, t-test). The MFI of each molecule was normalized to the MFI of the control group. Mean±SEM; *p<0.05; **p<0.01; ***p<0.001; ns, not significant. MFI, mean fluorescence intensity.
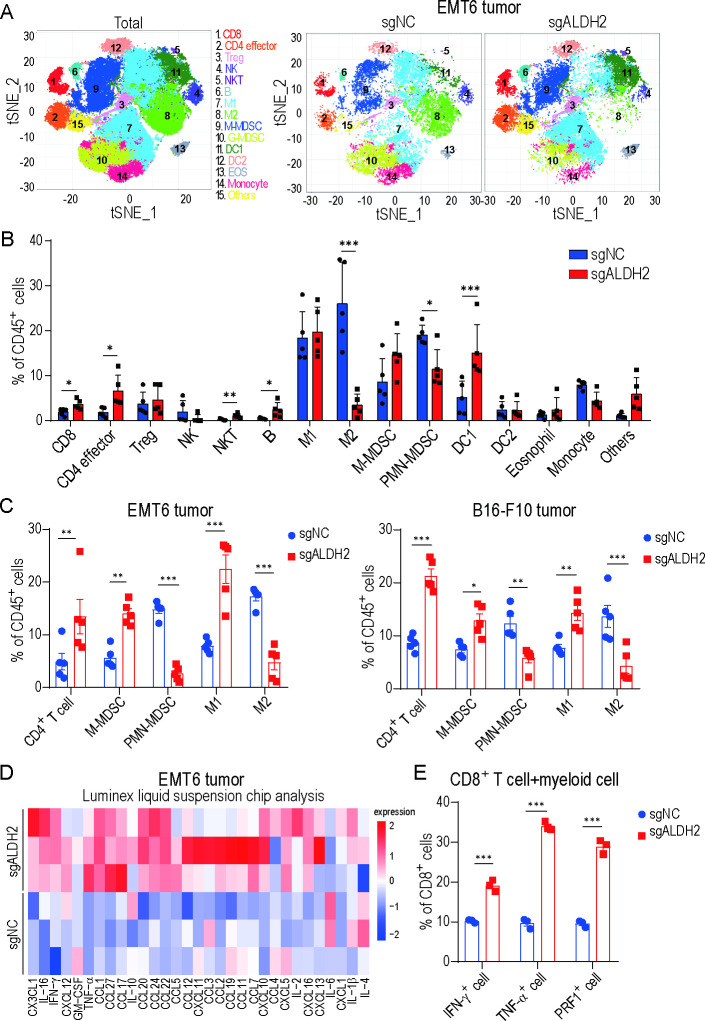
Inhibiting ALDH2 reshapes tumor immune landscape
To gain deeper insights into the global impact of ALDH2 blockade on TIME, we conducted a comprehensive investigation to assess the overall alteration of tumor immune landscape after ALDH2 inhibition. Using CyTOF, we analyzed CD45+ immune cells in EMT6 tumors, which revealed the difference in 16 distinct cell clusters (figure 3A–B and online supplemental figure 3A-B). Among these indicated clusters, ALDH2-knockout tumors demonstrated significant expansion of CD8+ T cell (cluster 1), CD4+ effector T cell (cluster 2), natural killer T cell (NKT) (cluster 5), B cell (cluster 6) and dendritic cell (DC) 1 (cluster 11), while showing fewer M2-like macrophage (cluster 8) and polymorphonuclear (PMN)-myeloid-derived suppressor cell (MDSC) (cluster 10). Consistently, similar changes as observed in CyTOF results were detected in EMT6 and B16-F10 tumors by flow cytometry. The analysis showed that the infiltration of CD4+ T cell and M1-like macrophage was prominently increased, accompanied by a notable decrease in the proportion of immunosuppressive cells including PMN-MDSC and M2-like macrophage after ALDH2 inhibition (figure 3C and online supplemental figure 3C). These findings indicated that inhibiting ALDH2 has the potential to induce a shift from an immunosuppressive to an immune-activating microenvironment.
Figure 3.
Inhibiting ALDH2 reshapes tumor immune landscape. (A) T-SNE plot of tumor-infiltrating leukocytes overlaid with color-coded clusters from ALDH2-knockout and control EMT6 tumors. (B) Frequency of clusters in indicated immune cell subsets from ALDH2 knockout and control tumors (n=5, t-test). (C) Representative quantification of CD4+ T cell, M-MDSC, PMN-MDSC, M1-like macrophage and M2-like macrophage of CD45+ live cells in EMT6 (left) and B16-F10 (right) tumors (n=5, two-way ANOVA). (D) Heatmap showing the average relative secretion of cytokines/chemokines enriched in ALDH2 knockout tumors compared with controls (n=3/group). (E) Representative quantification of IFN-γ+ cell, TNF-α+ cell and PRF1+ cell of CD8+ T cell co-cultured with myeloid cell isolated from ALDH2 knockout or control tumors (n=3, two-way ANOVA). Mean±SEM; *p<0.05; **p<0.01; ***p<0.001. GM-CSF, granulocyte-macrophage colony-stimulating factor; Tregs, regulatory T cells; t-SNE, t-distributed Stochastic Neighbor Embedding.
Furthermore, we used Luminex liquid suspension chip detection to compare the cytokine profile differences between ALDH2-knockout tumors and control tumors. The majority of pro-inflammatory cytokines/chemokines, such as IL-2, IL-16, IFN-γ and TNF-α, showed significantly higher levels in the ALDH2-knockout tumors compared with the control group (figure 3D). In contrast, the levels of several known anti-inflammatory/immunosuppressive cytokines were decreased. Several of these tested cytokines, such as IL-4 and IL-6, had been implicated in recruiting MDSCs and influencing on the differentiation of tumor-associated macrophages (TAMs),31–33 deeply emphasizing the significance role of cytokines in reshaping TIME. Moreover, to investigate the potential inhibitory effects of immunosuppressive cells such as MDSCs and TAMs on T-cell activities, we co-cultured CD8+ T cells with myeloid cells isolated from sgALDH2 tumors or sgNC tumors. The proliferation and cytotoxic activity of CD8+ T cells were enhanced in sgALDH2 tumors compared with control tumors (figure 3E and online supplemental figure 3D). In conclusion, these findings indicate that ALDH2 blockade remodels the TIME, potentially leading to enhanced antitumor immunity.
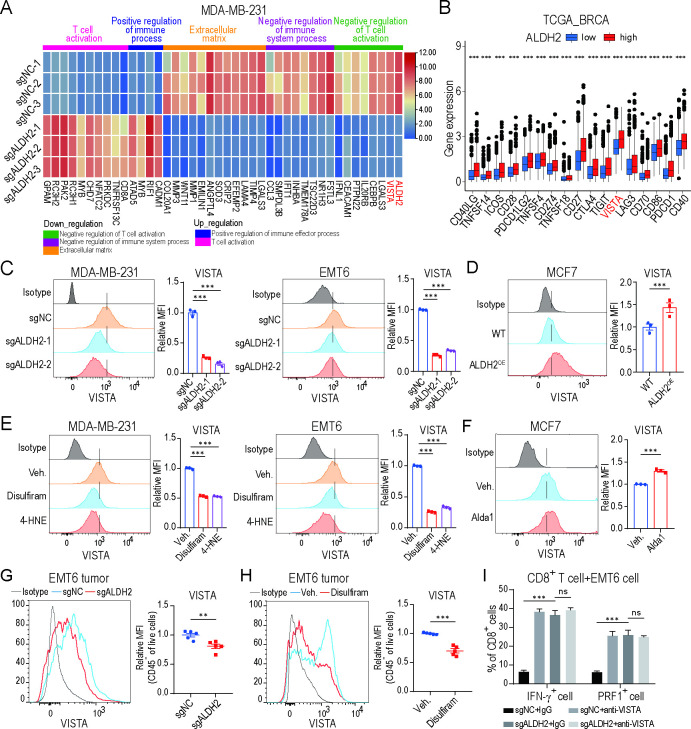
Blocking ALDH2 downregulates VISTA expression on tumor cells
To explore the potential mechanism involved in ALDH2-mediated tumor evasion, we knocked out ALDH2 gene expression in the human MDA-MB-231 BRCA cell line, which has a high ALDH2 expression level (online supplemental figure 4A). Furthermore, we assessed the levels of aldehydes, including 4-HNE and MDA, which are the major metabolites of ALDH2-mediated aldehyde metabolism. It showed a significant increase in ALDH2-knockout MDA-MB-231, EMT6 and B16-F10 tumor cells compared with their respective control medium (online supplemental figure 4B). Similarly, the elevated metabolite levels were also detected in ALDH2-knockout EMT6 and B16-F10 tumors compared with respective control tumors (online supplemental figure 4C), underscoring the impact of ALDH2 blockade on aldehyde metabolism.
Then, we compared global transcriptomic differences between sgALDH2 and control MDA-MB-231 cells using RNA sequencing. Knocking out ALDH2 downregulated the expression of genes related to negative regulation of the immune system, extracellular matrix synthesis and T-cell activation. Conversely, genes associated with positive regulation of T-cell activation and the immune system were upregulated in ALDH2-knockout tumor cells (figure 4A). Strikingly, VISTA was one of the major downregulated immune checkpoint molecules after ALDH2 inhibition (figure 4A). In addition, we calculated the expression levels of immune checkpoints in BRCA and melanoma using bioinformatics analysis based on the TCGA data, which showed that immune checkpoints, especially VISTA, were highly expressed in ALDH-high samples compared with ALDH2-low samples (figure 4B and online supplemental figure 4D), suggesting VISTA might be a key factor implicated in ALDH2-mediated tumor evasion.
Figure 4.
Blocking ALDH2 downregulates VISTA expression on tumor cells. (A) Gene ontology analysis by RNA sequencing of ALDH2 knockout and control MDA-MB-231 tumor cells (n=3/group). Heatmap shows the DEGs and associated signatures. (B) Expression of immune checkpoints among ALDH2 low and ALDH2 high groups in patients with breast cancer based on the TCGA database. (C) Flow cytometry analysis of VISTA expression on ALDH2 knockout and control breast cancer cells (one-way ANOVA). (D) Flow cytometry analysis of VISTA expression on ALDH2 overexpression and control breast cancer cells (t-test). (E) Flow cytometry analysis of VISTA expression on breast cancer cells treated with ALDH2 inhibitors (disulfiram and 4-HNE) or vehicle (one-way ANOVA). (F) Flow cytometry analysis of VISTA expression on breast cancer cells treated with alda1 or vehicle (t-test). (G) Flow cytometry analysis of VISTA expression on CD45− cells in ALDH2 knockout and control EMT6 tumors (n=5, t-test). (H) Flow cytometry analysis of VISTA expression on CD45− cells in disulfiram or vehicle-treated EMT6 tumors (n=5, t-test). (I) Percentages of IFN-γ+ cell and PRF1+ cell of CD8+ T cells co-cultured with EMT6 tumor cells pretreated with IgG or anti-VISTA (10 mg/mL) antibodies (n=3, two-way ANOVA). All in vitro experiments were performed at least three times. Mean±SEM; **p<0.01; ***p<0.001, ns, not significant. ICOS, inducible Co-Stimulator.
Next, we investigated the effect of ALDH2 on VISTA expression in multiple tumor cells. We overexpressed ALDH2 (ALDH2OE) in MCF7 BRCA cell, which has low ALDH2 expression (online supplemental figure 4A). Blocking ALDH2 significantly reduced VISTA transcriptional and membrane expression in MDA-MB-231, EMT6 and B16-F10 tumor cells, whereas VISTA expression was increased in ALDH2OE MCF7 cells, compared with respective controls (figure 4C–D and online supplemental figure 4E-H). Consistently, treatment with ALDH2 inhibitors, disulfiram and 4-HNE, markedly inhibited VISTA expression in tumor cells (figure 4E and online supplemental figure 4I). In contrast, Alda-1, a selective ALDH2 agonist, enhanced VISTA expression (figure 4F). Similarly, we demonstrated the relationship between ALDH2 and VISTA expression in EMT6 and B16-F10 mouse models, where reduced VISTA expression was detected on ALDH2-knockout or disulfiram-treated tumors compared with respective controls (figure 4G–H and online supplemental figure 4J). Functionally, when sgALDH2 tumor cells were co-cultured with T cells, the antitumor activity of cytotoxic CD8+ T cell was significantly increased compared with those co-cultured with sgNC tumor cells. However, it had no additional effect after pretreating tumor cells with VISTA blocking antibody (figure 4I and online supplemental figure 4K). These results suggested that VISTA acts as a major downstream factor of ALDH2 to affect the function of T cells. Collectively, all these data indicate that ALDH2 might upregulate VISTA expression in tumor cells.
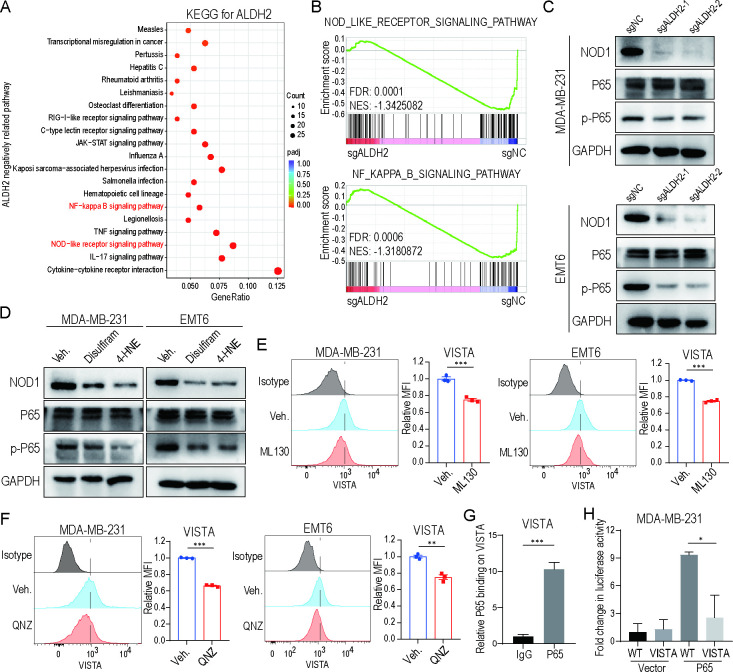
ALDH2 maintains VISTA expression by activating the NOD/NF-κB signaling pathway
In order to identify the regulatory mechanism involved in ALDH2-mediated VISTA expression, we reanalyzed RNA sequencing results by KEGG and GSEA. We found that NOD-like receptor signaling pathway and NF-κB signaling pathway were significantly correlated with ALDH2 expression (figure 5A–B). The inactivation of the NOD1 pathway was confirmed in ALDH2-knockout or ALDH2 inhibitors (disulfiram and 4-HNE)-treated tumor cells compared with respective controls (figure 5C–D and online supplemental figure 5A). Consistently, the expression of phosphorylated-P65 (p-P65) was significantly decreased after the ALDH2 blockade while total P65 expression showed no change (figure 5C–D and online supplemental figure 5A), suggesting that ALDH2 could activate the NF-κB signaling pathway in tumor cells. Furthermore, ML130, an inhibitor of NOD1, downregulated the expression of VISTA and p-P65 in tumor cells (figure 5E and online supplemental figure 5B-D). The reduction of VISTA expression was also validated by adding QNZ, an NF-κB activation inhibitor (figure 5F and online supplemental figure 5B,E). According to the JASPAR database, the VISTA gene promoter was predicted to cover the P65 binding site (online supplemental figure 5F). To identify this prediction, we performed ChIP assay and observed direct bounding of NF-κB to VISTA promoter (figure 5G). Additionally, through the luciferase reporter assay, we demonstrated that the expression of luciferase activity driven by the promoter was upregulated on overexpression of P65 (figure 5H). However, when mutating the P65 binding site in the VISTA promoter, the activity remarkably decreased (figure 5H), suggesting that the p65 binding site is critical for the basal activity of the VISTA promoter. Altogether, these findings demonstrate that ALDH2 upregulates VISTA expression by stimulating NOD/NF-κB signaling pathway activation.
Figure 5.
ALDH2 maintains VISTA expression by activating NOD/NF-κB signaling pathway. (A) KEGG enrichment analysis of downregulated genes in ALDH2 knockout tumor cells compared with control tumor cells. (B) Enrichment plot of NOD-like receptor signaling pathway and NF-κB signaling pathway, identified by gene set enrichment analysis from ALDH2 knockout and control tumor cells. (C–D) Western blot analysis of NOD1, total P65 and phosphorylated-P65 expression on ALDH2 knockout (C) and ALDH2 inhibitors (disulfiram and 4-HNE)-treated (D) tumor cells. GAPDH was used as loading control. (E) Flow cytometry analysis of VISTA expression on breast cancer cells treated with ML130 or vehicle (t-test). (F) Flow cytometry analysis of VISTA expression on breast cancer cells treated with QNZ or vehicle (n=3, t-test). All in vitro experiments were performed at least three times. (G) Chromatin immunoprecipitation analysis of P65 binding to VISTA promoters in tumor cells overexpressing P65 (n=3, t-test). (H) Dual luciferase analysis of the effect of P65 expression on the VISTA-mutant promoter in tumor cells (n=3, one-way analysis of variance). Mean±SEM; *p<0.01; **p<0.01; ***p<0.001.FDR, false discovery rate; NES, Nintendo entertainment system.
Targeting ALDH2 synergizes with ICB therapy
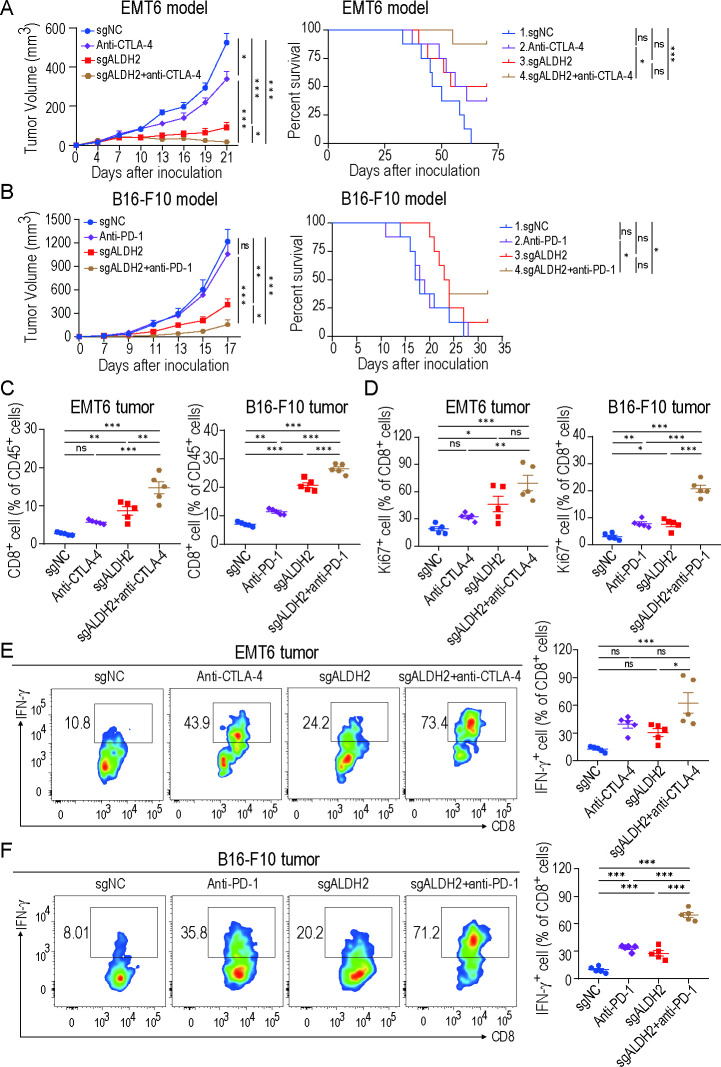
Based on our in vitro evidence that ALDH2 induced immune evasion, we sought to investigate whether ALDH2 blockade could enhance response to immunotherapy. In the EMT6 BRCA mouse model, ICB-treated ALDH2-knockout tumors exhibited effective inhibition of tumor growth compared with other groups (figure 6A). Remarkably, mice survival was prolonged compared with ICB alone (figure 6A). The B16-F10 melanoma model showed similar results, where combination treatment inhibited tumor growth and extended mice survival (figure 6B). Furthermore, ALDH2 blockade plus ICB therapy drastically increased the infiltration and proliferation of CD8+ T cells (figure 6C–D). Notably, the secretion of IFN-γ in CD8+ T cells was significantly increased in combinational therapy (figure 6E–F). Additionally, in both EMT6 and B16-F10 tumors, CD4+ T cells were also markedly increased and MDSCs were decreased with combinational therapy compared with control group or ICB-treated group (online supplemental figure 6A,B). These results indicate that ALDH2 inhibition could synergize with ICB to enhance antitumor activity.
Figure 6.
Targeting ALDH2 synergizes with ICB therapy. (A–B) EMT6 (A) and B16-F10 (B) tumor growth (left) and survival analysis (right) of control, ALDH2 knockout, ICB-treated (anti-PD-1 antibody or anti-CTLA-4 antibody) or ICB-treated ALDH2-knockout mice. n=5 mice/group for tumor volume analysis; n=8 mice/group for survival analysis; two-way ANOVA for tumor volume comparison; log-rank test for survival comparison. (C) Quantification of CD8+ T-cell ratios in EMT6 (left) and B16-F10 (right) tumors (n=5, one-way ANOVA). (D) Percentages of Ki-67+ CD8+ cells in EMT6 (left) and B16-F10 (right) tumors (n=5, one-way ANOVA). (E–F) Flow cytometry analysis of IFN-γ+CD8+ T cells from EMT6 tumor (E) and B16-F10 tumor (F) (n=5, one-way ANOVA). Mean±SEM; *p<0.05; **p<0.01; ***p<0.001; ns, not significant.
Disulfiram enhances the efficacy of ICB therapy
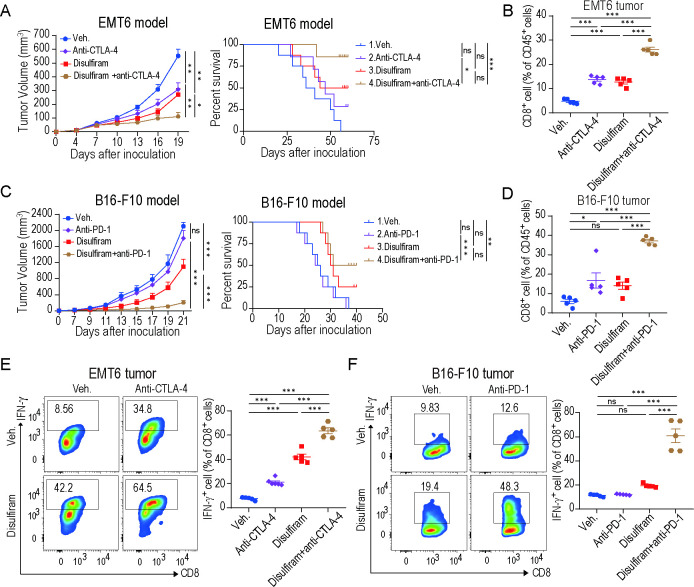
Disulfiram, an inhibitor of ALDH2, has been approved by the US Food and Drug Administration for clinical application.34 Accumulating studies support that disulfiram can be repurposed as an adjunctive drug for cancer treatment.35–37 Given the synergistic efficacy of ALDH2 blockade with ICB therapy, we questioned whether disulfiram would have a similar influence on immunotherapy response. In the EMT6 and B16-F10 tumor-bearing mice models, we treated the animals with vehicle, ICB, disulfiram, and combinatorial treatment of disulfiram plus ICB, respectively. Disulfiram or anti-CTLA-4 treatment both delayed tumor growth in EMT6 tumor model, and combinatorial treatment of disulfiram plus anti-CTLA-4 achieved better efficacy (figure 7A). Remarkably, disulfiram prolonged mice survival in the EMT6 tumor model, and its combination with ICB further extended it (figure 7A). Moreover, in the B16-F10 tumor model, disulfiram combined with anti-PD-1 also achieved higher therapeutic response, which dramatically suppressed tumor growth and prolonged mice survival compared with disulfiram or ICB alone (figure 7C). Then, we investigated the TIME of these tumor models and found that CD8+ T-cell infiltration was significantly increased in the disulfiram-treated group compared with the vehicle group, and combination therapy further improved this effect (figure 7B and D). Notably, the combination therapy remarkably enhanced the secretion of IFN-γ in CD8+ T cells compared with other groups (figure 7E–F). In addition, total CD4+ T cells were significantly increased, while MDSCs and TAMs were decreased in the combination therapy compared with the other groups (online supplemental figure 7A,B). In summary, these results suggest that disulfiram could enhance the antitumor immune response of ICB therapy.
Figure 7.
Disulfiram enhances the efficacy of immune checkpoint blockade therapy. (A) EMT6 tumor growth (left) and survival analysis (right) of mice treated with vehicle, disulfiram, anti-CTLA-4, or disulfiram+anti-CTLA-4. n=6 mice/group for tumor volume analysis; n=8 mice/group for survival analysis; two-way ANOVA for tumor volume comparison; log-rank test for survival comparison. (B) Quantification of CD8+ T cells ratios in EMT6 tumors treated with indicated treatments (n=5, one-way ANOVA). (C) B16-F10 tumor growth (left) and survival analysis (right) of mice treated with vehicle, disulfiram, anti-PD-1, or disulfiram+anti-PD-1. n=6 mice/group for tumor volume analysis; n=8 mice/group for survival analysis; two-way ANOVA for tumor volume comparison; log-rank test for survival comparison. (D) Quantification of CD8+ T cells ratios in B16-F10 tumors treated with indicated treatments (n=5, one-way ANOVA). (E–F) Representative flow cytometry images and percentages of IFN-γ+ CD8+ T cells from EMT6 (E) and B16-F10 (F) tumors (n=5, one-way ANOVA). Mean±SEM; *p<0.05; **p<0.01; ***p<0.001.
Discussion
ALDH2 is a metabolic enzyme of aldehyde that plays a vital role in regulating various biological processes.17 18 Accumulating studies indicate that ALDH2 has a bidirectional effect on cancer progression. On the one hand, aldehydes can activate antigen presentation of immune cells and help eliminate tumor cells, which is beneficial for tumor regression.38 On the other hand, several other lines of evidence support that ALDH2 deficiency may promote the activation of oncogenic signaling pathway and cause cancer progression.39 40 In our study, we demonstrated that ALDH2 is critical for tumor growth in BRCA and melanoma mouse models. Since T cell-dependent antitumor immunity plays a key role in defensing against cancer,41 we found that cancer cell-intrinsic ALDH2 has a powerful immune suppressive effect on T-cell function. Moreover, inhibiting ALDH2 could remodel the TIME, leading to a shift in TME from immunosuppression to immune activation, potentially resulting in the inhibition of T-cell proliferation and function. Altogether, ALDH2 inhibition improved prognosis via reversing T-cell suppression and promoting tumor immunity in multiple tumors, suggesting that ALDH2 could serve as a promising therapeutic target for cancer.
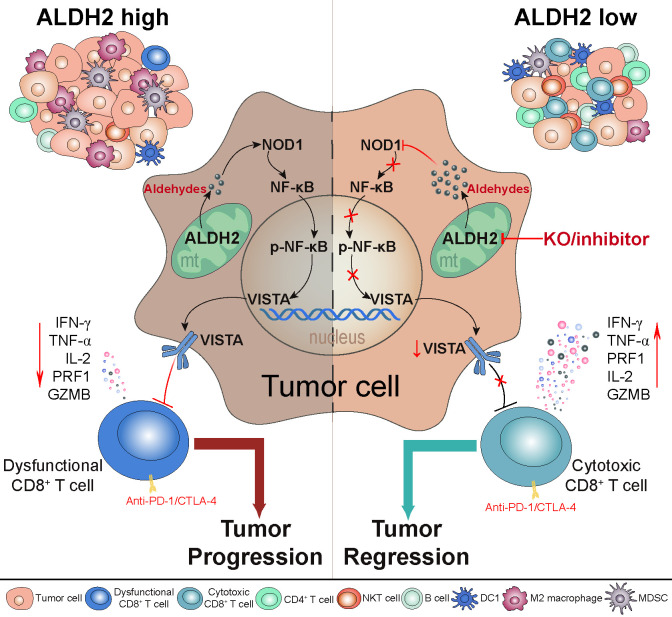
The immune checkpoint is a well-known mechanism used by cancer cells to evade immune surveillance.42 VISTA, an important immune checkpoint molecule resembling PD-L1, has been identified as a selective ligand for immune co-inhibitory receptor on T cells, which blocks the activation of T cells.43 44 Moreover, VISTA expression correlates with poor overall survival in cancer, and might be a novel diagnostic biomarker and immunotherapy target for cancer.45 46 However, the detailed mechanisms involved in VISTA regulation remain unclear. Studies have suggested that VISTA can be induced by JAK/STAT pathway-mediated IFN-γ signaling, or regulated by NF-kB signaling pathway.47 48 In addition, hypoxia can upregulate the expression of VISTA, leading to immune suppression and tumor progression.49 In our previous study, we proved that histamine, an allergic mediator, promotes VISTA membrane localization by binding to histamine receptor H1 on macrophages.28 Here, we found that ALDH2 knockout and disulfiram treatment resulted in a reduction of both transcriptional and membrane levels of VISTA expression, suggesting that ALDH2 could be a crucial factor in sustaining VISTA expression on tumors. To be noted, we confirmed a significant increase in the essential endogenous aldehydic products of ALDH2, such as 4-HNE and MDA, following ALDH2 inhibition. Additionally, 4-HNE also demonstrated an inhibitory effect on VISTA expression, emphasizing that ALDH2-mediated aldehyde metabolism facilitates VISTA expression (figure 8).
Figure 8.
Schematic illustration of the mechanism by which ALDH2 drives immune evasion. ALDH2-mediated aldehyde metabolism drives tumor immune evasion by modulating the expression of VISTA immune checkpoint in tumors. Blocking ALDH2 downregulates VISTA expression by suppressing the activation of NOD/NF-κB signaling pathway, resulting in improving cytotoxic activity of CD8+ T cell and reshaping the global immune landscape, thereby curbing tumor growth. Notably, inhibiting ALDH2 enhances the antitumor immune response of ICB therapy. KO, knock out.
Increasing evidence has expanded the concept that inflammation is a hallmark of cancer, contributing to the proliferation, survival, and migration of cancer cells.50 The NOD-like receptor (NLR) plays a vital role in inflammation and immunity.51 It participates in the NOD-like receptor signaling pathway, which leads to the activation of NF-κB and promotes the production of pro-inflammatory cytokines.52 53 Moreover, the NOD/NF-κB pathway has been implicated in the regulation of immune checkpoints such as PD-L1.54 55 Here, our study demonstrated that inhibiting ALDH2-mediated aldehyde metabolism inactivated the NOD/NF-κB signaling pathway and resulted in the downregulation of VISTA expression. Subsequently, we explored the impact of NF-κB on VISTA expression. It has been demonstrated that the absence of NF-κB reduced the expression of checkpoints such as PD-L1.56 Using ChIP and luciferase assays, we identified a functional NF-κB binding site in the core promoter region of the human VISTA gene, and disrupting this binding site significantly reduced the VISTA promoter activity. These results highlighted the crucial involvement of inflammatory-related signaling pathways, especially the NOD/NF-κB pathway, in regulating immune checkpoint expression and inducing tumor immune evasion.
Now, ICB has been widely used in cancer therapy, but unfortunately, most patients with cancer get limited benefit from this treatment.8 Accumulating studies are focusing on combination treatments to surmount immunotherapy resistance and broaden the clinical utility of ICB.57 58 In our study, we found that targeting ALDH2 might be an effective strategy for improving ICB response via decreasing VISTA expression. Disulfiram is a well-established drug that targets ALDH2-mediated aldehyde metabolism and has been used for decades to treat alcohol dependence due to its safety and tolerability.34 It has been reported that the combination of disulfiram and copper could inhibit cancer progression by modulating cancer stem cell-like properties, inducing oxidative stress, or enhancing DNA damage.59–61 Furthermore, it has been demonstrated to enhance biological activity of chemotherapeutic or molecular targeted drugs and improve the treatment efficacy.62–64 Our data showed that disulfiram sensitizes BRCA and melanoma to ICB therapy. It not only significantly improved the infiltration of antitumor immune cells but also boosted the cytotoxic activities of CD8+ T cells. These findings underscore that the potential of ALDH2 as a therapeutic target for overcoming immunotherapy resistance.
Conclusions
In conclusion, ALDH2-mediated aldehyde metabolism in tumors promotes VISTA expression by activating NOD/NF-κB signaling pathway, leading to tumor immune evasion. Targeting ALDH2 could enhance CD8+ T-cell function, retard tumor growth, and improve the response to ICB therapy.
Acknowledgments
Publicly available data from TCGA, TIDE, and GEPIA was used in this study, and we would like to thank the authors for making their data available. We thank all the laboratory members for discussion and suggestions.
Footnotes
YC and JS contributed equally.
Contributors: YC and JS contributed equally to this paper. Conceptualization: YC, HL, XW. Flow and mass cytometry analysis: YC, JS, HL. Performed Bioinformatics studies: YC, JS, JL. Performed in vitro assays: YC, JS. Performed in vivo studies: YC, JS, JL, XW, HF. IHC and histopathology: YC. Writing manuscript: YC, HL. Resources and conceptual: YW, HD, JH, QL, HL, XW, GR. HL is responsible for the overall content as the guarantor. Manuscript review and approval: All authors.
Funding: This work was supported by the National Natural Science Foundation of China (No. 82173166, 81472475, 82273282, 82372886 and 31420103915), Natural Science Foundation of Chongqing (cstc2021jcyj-msxmX0015 and CSTB2022NSCQ-MSX0783), and Chongqing medical scientific research project/Joint project of Chongqing Health Commission and Science and Technology Bureau (NO.2021MSXM033 and 2023QNXM039), Chongqing Graduate Tutor Team Construction Project, Chongqing Education Commission Foundation (cqmudstd202216), and CQMU Program for Youth Innovation in Future Medicine (NO. W0094).
Competing interests: No, there are no competing interests.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Publicly available data sets were analyzed in study. The JASPAR database (http://jaspar.genereg.net/) was used to predict the p65 binding sites in the VISTA gene promoter.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
Not applicable.
References
- 1. Li J, Stanger BZ. How tumor cell Dedifferentiation drives immune evasion and resistance to Immunotherapy. Cancer Res 2020;80:4037–41. 10.1158/0008-5472.CAN-20-1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Clara JA, Monge C, Yang Y, et al. Targeting signalling pathways and the immune Microenvironment of cancer stem cells - a clinical update. Nat Rev Clin Oncol 2020;17:204–32. 10.1038/s41571-019-0293-2 [DOI] [PubMed] [Google Scholar]
- 3. Dersh D, Hollý J, Yewdell JW. A few good peptides: MHC class I-based cancer Immunosurveillance and Immunoevasion. Nat Rev Immunol 2021;21:116–28. 10.1038/s41577-020-0390-6 [DOI] [PubMed] [Google Scholar]
- 4. Jhunjhunwala S, Hammer C, Delamarre L. Antigen presentation in cancer: insights into tumour Immunogenicity and immune evasion. Nat Rev Cancer 2021;21:298–312. 10.1038/s41568-021-00339-z [DOI] [PubMed] [Google Scholar]
- 5. Tripathi R, Modur V, Senovilla L, et al. Suppression of tumor antigen presentation during Aneuploid tumor evolution contributes to immune evasion. Oncoimmunology 2019;8:1657374. 10.1080/2162402X.2019.1657374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bassez A, Vos H, Van Dyck L, et al. A single-cell map of Intratumoral changes during anti-Pd1 treatment of patients with breast cancer. Nat Med 2021;27:820–32. 10.1038/s41591-021-01323-8 [DOI] [PubMed] [Google Scholar]
- 7. Adams S, Gatti-Mays ME, Kalinsky K, et al. Current landscape of Immunotherapy in breast cancer: A review. JAMA Oncol 2019;5:1205–14. 10.1001/jamaoncol.2018.7147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. He X, Xu C. Immune Checkpoint signaling and cancer Immunotherapy. Cell Res 2020;30:660–9. 10.1038/s41422-020-0343-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Faubert B, Solmonson A, DeBerardinis RJ. Metabolic Reprogramming and cancer progression. Science 2020;368:eaaw5473. 10.1126/science.aaw5473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lian X, Yang K, Li R, et al. Immunometabolic Rewiring in tumorigenesis and anti-tumor Immunotherapy. Mol Cancer 2022;21:27. 10.1186/s12943-021-01486-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xia L, Oyang L, Lin J, et al. The cancer metabolic Reprogramming and immune response. Mol Cancer 2021;20:28. 10.1186/s12943-021-01316-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Leone RD, Powell JD. Metabolism of immune cells in cancer. Nat Rev Cancer 2020;20:516–31. 10.1038/s41568-020-0273-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Biswas SK. Metabolic Reprogramming of immune cells in cancer progression. Immunity 2015;43:435–49. 10.1016/j.immuni.2015.09.001 [DOI] [PubMed] [Google Scholar]
- 14. DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci Adv 2016;2:e1600200. 10.1126/sciadv.1600200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dey P, Kimmelman AC, DePinho RA. Metabolic Codependencies in the tumor Microenvironment. Cancer Discov 2021;11:1067–81. 10.1158/2159-8290.CD-20-1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fan C, Zhang S, Gong Z, et al. Emerging role of metabolic Reprogramming in tumor immune evasion and Immunotherapy. Sci China Life Sci 2021;64:534–47. 10.1007/s11427-019-1735-4 [DOI] [PubMed] [Google Scholar]
- 17. Gao Y, Zhou Z, Ren T, et al. Alcohol inhibits T-cell glucose metabolism and hepatitis in Aldh2-deficient mice and humans: roles of acetaldehyde and glucocorticoids. Gut 2019;68:1311–22. 10.1136/gutjnl-2018-316221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kimura M, Yokoyama A, Higuchi S. Aldehyde Dehydrogenase-2 as a therapeutic target. Expert Opin Ther Targets 2019;23:955–66. 10.1080/14728222.2019.1690454 [DOI] [PubMed] [Google Scholar]
- 19. Zhang H, Fu L. The role of Aldh2 in tumorigenesis and tumor progression: targeting Aldh2 as a potential cancer treatment. Acta Pharmaceutica Sinica B 2021;11:1400–11. 10.1016/j.apsb.2021.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen C-H, Ferreira JCB, Gross ER, et al. Targeting Aldehyde dehydrogenase 2: new therapeutic opportunities. Physiol Rev 2014;94:1–34. 10.1152/physrev.00017.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li R, Zhao Z, Sun M, et al. Aldh2 gene polymorphism in different types of cancers and its clinical significance. Life Sci 2016;147:59–66. 10.1016/j.lfs.2016.01.028 [DOI] [PubMed] [Google Scholar]
- 22. Zhang H, Xia Y, Wang F, et al. Aldehyde dehydrogenase 2 mediates alcohol-induced colorectal cancer immune escape through stabilizing PD-L1 expression. Advanced Science 2021;8:10. 10.1002/advs.202003404 Available: https://onlinelibrary.wiley.com/toc/21983844/8/10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bagaev A, Kotlov N, Nomie K, et al. Conserved Pan-cancer Microenvironment subtypes predict response to Immunotherapy. Cancer Cell 2021;39:845–65. 10.1016/j.ccell.2021.04.014 [DOI] [PubMed] [Google Scholar]
- 24. Tirosh I, Izar B, Prakadan SM, et al. Dissecting the Multicellular Ecosystem of metastatic Melanoma by single-cell RNA-Seq. Science 2016;352:189–96. 10.1126/science.aad0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fu J, Li K, Zhang W, et al. Large-scale public data Reuse to model Immunotherapy response and resistance. Genome Med 2020;12:21. 10.1186/s13073-020-0721-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li C, Tang Z, Zhang W, et al. Gepia2021: integrating multiple Deconvolution-based analysis into GEPIA. Nucleic Acids Res 2021;49:W242–6. 10.1093/nar/gkab418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Skrott Z, Mistrik M, Andersen KK, et al. Alcohol-abuse drug disulfiram targets cancer via P97 Segregase Adaptor Npl4. Nature 2017;552:194–9. 10.1038/nature25016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li H, Xiao Y, Li Q, et al. The allergy mediator histamine confers resistance to Immunotherapy in cancer patients via activation of the macrophage histamine receptor H1. Cancer Cell 2022;40:36–52. 10.1016/j.ccell.2021.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu Y, Yi Z, Li J, et al. FGFR blockade BOOSTS T cell infiltration into triple-negative breast cancer by regulating cancer-associated fibroblasts. Theranostics 2022;12:4564–80. 10.7150/thno.68972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cerezo-Wallis D, Contreras-Alcalde M, Troulé K, et al. Midkine Rewires the Melanoma Microenvironment toward a Tolerogenic and immune-resistant state. Nat Med 2020;26:1865–77. 10.1038/s41591-020-1073-3 [DOI] [PubMed] [Google Scholar]
- 31. Mantovani A, Marchesi F, Malesci A, et al. Tumour-associated Macrophages as treatment targets in oncology. Nat Rev Clin Oncol 2017;14:399–416. 10.1038/nrclinonc.2016.217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Arina A, Corrales L, Bronte V. Enhancing T cell therapy by overcoming the immunosuppressive tumor Microenvironment. Semin Immunol 2016;28:54–63. 10.1016/j.smim.2016.01.002 [DOI] [PubMed] [Google Scholar]
- 33. Marigo I, Bosio E, Solito S, et al. Tumor-induced tolerance and immune suppression depend on the C/Ebpbeta transcription factor. Immunity 2010;32:790–802. 10.1016/j.immuni.2010.05.010 [DOI] [PubMed] [Google Scholar]
- 34. Suh JJ, Pettinati HM, Kampman KM, et al. The status of disulfiram: a half of a century later. J Clin Psychopharmacol 2006;26:290–302. 10.1097/01.jcp.0000222512.25649.08 [DOI] [PubMed] [Google Scholar]
- 35. Lu C, Li X, Ren Y, et al. Disulfiram: a novel Repurposed drug for cancer therapy. Cancer Chemother Pharmacol 2021;87:159–72. 10.1007/s00280-020-04216-8 [DOI] [PubMed] [Google Scholar]
- 36. Xu L, Sun Y, Li Y, et al. Disulfiram: A food and Drug Administration-approved Multifunctional role in synergistically drug delivery systems for tumor treatment. Int J Pharm 2022;626:122130. 10.1016/j.ijpharm.2022.122130 [DOI] [PubMed] [Google Scholar]
- 37. Li H, Wang J, Wu C, et al. The combination of disulfiram and copper for cancer treatment. Drug Discovery Today 2020;25:1099–108. 10.1016/j.drudis.2020.04.003 [DOI] [PubMed] [Google Scholar]
- 38. Allison ME, Fearon DT. Enhanced Immunogenicity of Aldehyde-bearing antigens: a possible link between innate and adaptive immunity. Eur J Immunol 2000;30:2881–7. [DOI] [PubMed] [Google Scholar]
- 39. Yuan B, El Dana F, Ly S, et al. Bone marrow Stromal cells induce an ALDH+ stem cell-like phenotype and enhance therapy resistance in AML through a TGF-Β-P38-Aldh2 pathway. PLoS One 2020;15:e0242809. 10.1371/journal.pone.0242809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Feng D, Zhu W, You J, et al. Mitochondrial Aldehyde dehydrogenase 2 represents a potential biomarker of biochemical recurrence in prostate cancer patients. Molecules 2022;27:6000. 10.3390/molecules27186000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Paijens ST, Vledder A, de Bruyn M, et al. Tumor-infiltrating lymphocytes in the Immunotherapy era. Cell Mol Immunol 2021;18:842–59. 10.1038/s41423-020-00565-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang Y, Zheng J. Functions of immune Checkpoint molecules beyond immune evasion. Adv Exp Med Biol 2020;1248:201–26. 10.1007/978-981-15-3266-5_9 [DOI] [PubMed] [Google Scholar]
- 43. Nowak EC, Lines JL, Varn FS, et al. Immunoregulatory functions of VISTA. Immunol Rev 2017;276:66–79. 10.1111/imr.12525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mulati K, Hamanishi J, Matsumura N, et al. VISTA expressed in tumour cells regulates T cell function. Br J Cancer 2019;120:115–27. 10.1038/s41416-018-0313-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Burugu S, Dancsok AR, Nielsen TO. Emerging targets in cancer Immunotherapy. Semin Cancer Biol 2018;52(Pt 2):39–52. 10.1016/j.semcancer.2017.10.001 [DOI] [PubMed] [Google Scholar]
- 46. Yum J-EI, Hong Y-K. Terminating cancer by blocking VISTA as a novel Immunotherapy: Hasta La vista, baby Front Oncol 2021;11:658488. 10.3389/fonc.2021.658488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Katabathula R, Joseph P, Singh S, et al. Multi-scale Pan-cancer integrative analyses identify the Stat3-VSIR axis as a key immunosuppressive mechanism in head and neck cancer. Clin Cancer Res 2022;28:984–92. 10.1158/1078-0432.CCR-21-1978 [DOI] [PubMed] [Google Scholar]
- 48. Xu W, Dong J, Zheng Y, et al. Immune-Checkpoint protein VISTA regulates antitumor immunity by controlling myeloid cell-mediated inflammation and immunosuppression. Cancer Immunol Res 2019;7:1497–510. 10.1158/2326-6066.CIR-18-0489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Deng J, Li J, Sarde A, et al. Hypoxia-induced VISTA promotes the suppressive function of myeloid-derived Suppressor cells in the tumor Microenvironment. Cancer Immunol Res 2019;7:1079–90. 10.1158/2326-6066.CIR-18-0507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Coussens LM, Werb Z. Inflammation and cancer. Nature 2002;420:860–7. 10.1038/nature01322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wen H, Miao EA, Ting J-Y. Mechanisms of NOD-like receptor-associated Inflammasome activation. Immunity 2013;39:432–41. 10.1016/j.immuni.2013.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Scott MJ, Chen C, Sun Q, et al. Hepatocytes express functional Nod1 and Nod2 receptors: a role for Nod1 in hepatocyte CC and CXC Chemokine production. J Hepatol 2010;53:693–701. 10.1016/j.jhep.2010.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ashton JJ, Boukas K, Stafford IS, et al. Deleterious genetic variation across the NOD signaling pathway is associated with reduced NFKB signaling transcription and upregulation of alternative inflammatory transcripts in pediatric inflammatory bowel disease. Inflamm Bowel Dis 2022;28:912–22. 10.1093/ibd/izab318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kravtsova-Ivantsiv Y, Goldhirsh G, Ivantsiv A, et al. Excess of the NF-ĸB P50 subunit generated by the Ubiquitin Ligase Kpc1 suppresses tumors via PD-L1- and Chemokines-mediated mechanisms. Proc Natl Acad Sci U S A 2020;117:29823–31. 10.1073/pnas.2019604117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lim S-O, Li C-W, Xia W, et al. Deubiquitination and stabilization of PD-L1 by Csn5. Cancer Cell 2016;30:925–39. 10.1016/j.ccell.2016.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rong D, Sun G, Zheng Z, et al. MGP promotes Cd8+ T cell exhaustion by activating the NF-ΚB pathway leading to liver metastasis of colorectal cancer. Int J Biol Sci 2022;18:2345–61. 10.7150/ijbs.70137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jiao S, Xia W, Yamaguchi H, et al. PARP inhibitor Upregulates PD-L1 expression and enhances cancer-associated immunosuppression. Clin Cancer Res 2017;23:3711–20. 10.1158/1078-0432.CCR-16-3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ye Y, Kuang X, Xie Z, et al. Small-molecule Mmp2/Mmp9 inhibitor SB-3Ct modulates tumor immune surveillance by regulating PD-L1. Genome Med 2020;12:83. 10.1186/s13073-020-00780-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yip NC, Fombon IS, Liu P, et al. Disulfiram modulated ROS-MAPK and NFκB pathways and targeted breast cancer cells with cancer stem cell-like properties. Br J Cancer 2011;104:1564–74. 10.1038/bjc.2011.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Choi SA, Choi JW, Wang K-C, et al. Disulfiram modulates Stemness and metabolism of brain tumor initiating cells in atypical Teratoid/Rhabdoid tumors. Neuro Oncol 2015;17:810–21. 10.1093/neuonc/nou305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chu M, An X, Zhang D, et al. Combination of the 6-Thioguanine and disulfiram/cu synergistically inhibits proliferation of triple-negative breast cancer cells by enhancing DNA damage and disrupting DNA damage Checkpoint. Biochim Biophys Acta Mol Cell Res 2022;1869:119169. 10.1016/j.bbamcr.2021.119169 [DOI] [PubMed] [Google Scholar]
- 62. Li H, Wang J, Wu C, et al. The combination of disulfiram and copper for cancer treatment. Drug Discov Today 2020;25:1099–108. 10.1016/j.drudis.2020.04.003 [DOI] [PubMed] [Google Scholar]
- 63. Komarova TV, Sheshukova EV, Kosobokova EN, et al. The biological activity of Bispecific Trastuzumab/Pertuzumab plant Biosimilars may be drastically boosted by disulfiram increasing Formaldehyde accumulation in cancer cells. Sci Rep 2019;9:16168. 10.1038/s41598-019-52507-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zheng X, Liu Z, Mi M, et al. Disulfiram improves the anti-PD-1 therapy efficacy by regulating PD-L1 expression via Epigenetically reactivation of Irf7 in triple negative breast cancer. Front Oncol 2021;11:734853. 10.3389/fonc.2021.734853 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2023-007487supp001.pdf (1MB, pdf)
Data Availability Statement
Publicly available data sets were analyzed in study. The JASPAR database (http://jaspar.genereg.net/) was used to predict the p65 binding sites in the VISTA gene promoter.