Abstract
Alterations of the brain-gut-microbiome system (BGM) have been implicated in the pathophysiology of irritable bowel syndrome (IBS), yet bowel habit-specific alterations have not been elucidated. In this cross-sectional study, we apply a systems biology approach to characterize BGM patterns related to predominant bowel habit. Fecal samples and resting state fMRI were obtained from 102 premenopausal women (36 constipation-predominant IBS (IBS–C), 27 diarrhea-predominant IBS (IBS-D), 39 healthy controls (HCs)). Data integration analysis using latent components (DIABLO) was used to integrate data from the phenome, microbiome, metabolome, and resting-state connectome to predict HCs vs IBS-C vs IBS-D. Bloating and visceral sensitivity, distinguishing IBS from HC, were negatively associated with beneficial microbes and connectivity involving the orbitofrontal cortex. This suggests that gut interactions may generate aberrant central autonomic and descending pain pathways in IBS. The connection between IBS symptom duration, key microbes, and caudate connectivity may provide mechanistic insight to the chronicity of pain in IBS. Compared to IBS-C and HCs, IBS-D had higher levels of many key metabolites including tryptophan and phenylalanine, and increased connectivity between the sensorimotor and default mode networks; thus, suggestingan influence on diarrhea, self-related thoughts, and pain perception in IBS-D (‘bottom-up’ mechanism). IBS-C’s microbiome and metabolome resembled HCs, but IBS-C had increased connectivity in the default mode and salience networks compared to IBS-D, which may indicate importance of visceral signals, suggesting a more ‘top-down’ BGM pathophysiology. These BGM characteristics highlight possible mechanistic differences for variations in the IBS bowel habit phenome.
This article is part of the Special Issue on ‘Microbiome & the Brain: Mechanisms & Maladies’.
Keywords: Brain gut microbiome, Irritable bowel syndrome, Bowel subtype, Functional connectivity, Visceral hypersensitivity
Lay summary
Using an integrated systems biology approach, distinct brain-gut-microbiome alterations associated with self-reported predominant stool patterns in IBS patients can be identified.
Graphical Abstract
1. Introduction
Irritable Bowel Syndrome (IBS) is a common, female-predominant disorder of brain-gut interactions, characterized by chronic abdominal pain combined with altered bowel habits (Drossman and Hasler; Heitkemper et al.; Longstreth et al.). The pathophysiology underlying differences in predominant bowel habits and their relationship with specific alterations within the brain-gut-microbiome (BGM) system is incompletely understood (Mayer et al.).
The few studies that have compared brain differences between IBS-C and IBS-D have examined task-states and have shown group-related differences in brain networks involved in integrating emotions (emotional arousal network: EAN), perception (sensorimotor network: SMN, salience network: SAL), visceral functions (central autonomic network: CAN), and pain processing (default mode network: DMN, central executive network: CEN, SMN, SAL, EAN, and others). Prior studies comparing IBS-C, IBS-D, and HCs undergoing aversive rectal stimuli have identified abnormal connectivity in the SAL (Berman et al., 2008; Guleria et al., 2017) and EAN (Berman et al., 2008; Wilder-Smith et al., 2004) in IBS-C, and in the occipital network (OCC) (Guleria et al., 2017) in IBS-D. This may indicate a greater importance of alterations in sensory, emotional, and autonomic responses associated with the perception of visceral pain and discomfort in IBS-C.
Resting state (RS) functional connectivity patterns reported in individuals with functional constipation (physiologically similar to IBS-C) identify altered connections between the SMN and EAN when compared to HCs, furthering the potential aberration of sensory and emotional salience in IBS-C (Zhu et al., 2016). IBS-D patients were found to have enhanced RS brain connectivity compared to HCs involving the CAN, EAN, and DMN (Chen et al., 2021), which may be implicated in autonomic dysfunction, emotional arousal, and perseverative thoughts. When viewed together, these findings have led to the hypothesis that at the brain level IBS-C is characterized by altered sensory processing, whereas IBS-D is characterized primarily by autonomic and emotional dysregulation.
Certain gut microbial metabolites, rather than individual microbes, are associated with clinically meaningful changes in the BGM system (Osadchiy et al., 2018). Higher levels of fecal short chain fatty acid (SCFA) in IBS-D compared to IBS-C have been observed, which may contribute to increased colonic water secretion and diarrhea in IBS-D (Chassard et al., 2012; Mars et al., 2020). Tryptophan (a precursor to serotonin) has been extensively studied in relation to IBS bowel habit subtype (Clarke et al., 2012; Keszthelyi et al., 2015; Vahora et al., 2020). Serotonin production from tryptophan occurs via the rate-limiting enzyme tryptophan hydroxylase located in enterochromaffin cells (ECCs), a conversion which is modulated by SCFAs and secondary bile acids produced by certain gut microbes. Serotonin, acting on various receptor subtypes, stimulates motility, secretion, and visceral sensitivity (Mawe and Hoffman, 2013), and several drugs aimed at the peripheral serotonin system have been used to treat IBS (Labus et al., 2011). Tryptophan’s role in bowel habit subtype is somewhat unclear: higher effective serotonin levels have been found in IBS-D compared to IBS-C (Vahora et al., 2020), and yet tryptophan depletion studies indicate that tryptophan plays a role in IBS-C brain changes in response to noxious stimuli (Labus et al., 2011). While tryptophan metabolites are likely to play a role in BGM alterations in IBS, the mechanisms and contribution to bowel habit alterations remain incompletely understood.
Alterations in the BGM system play a critical role in IBS pathophysiology, including bowel habit subtype. In this study, we used an integrated, systems biology approach to explore the relationship between BGM alterations and bowel habit subtypes of IBS in premenopausal women (summarized in Graphical Abstract). A prior study by our group using the same cohort (Osadchiy et al., 2020) used a systems biology approach to examine associations between brain changes and fecal metabolite changes, but this study did not stratify by bowel habit subtype, nor examine clinical variables. We focused on women because of the increased prevalence and morbidity of IBS in women, as well as known sex-specific differences in clinical presentation, BGM signatures, and treatment responses (Jiang et al., 2013; Kim and Kim, 2018; Labus et al., 2013).
We aimed to test the hypothesis that IBS-C exhibits bowel-habit-specific changes in the brain (e.g., SAL network) that reflect altered sensory and emotional regulation processing of visceral inputs from the ‘top-down.’ We hypothesized that IBS-D would have widespread gut microbiome and metabolome changes (e.g., tryptophan, SCFAs) which may translate to ‘bottom-up’ brain changes (e.g., SMN, DMN). We also investigated common BGM pathways in IBS vs. HCs (e.g., SMN, CAN, and EAN networks).
2. Materials and methods
2.1. Participants
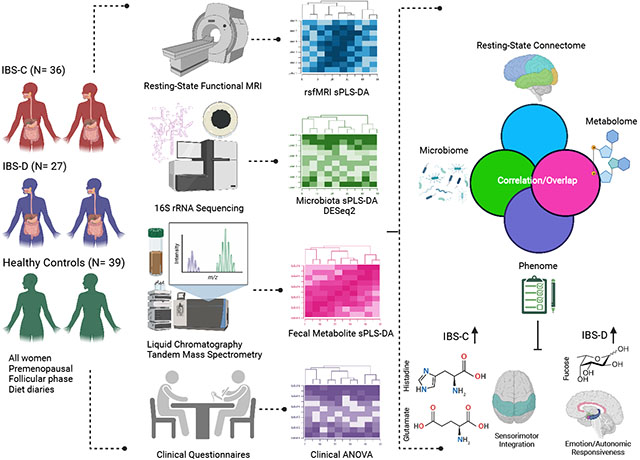
A workflow for the entire methods can be seen in the Graphical Abstract. 102 premenopausal women (36 IBS-C, 27 IBS-D, 39 HCs) were recruited by the Center for Neurobiology of Stress and Resilience at University of California Los Angeles (UCLA) starting in 2016. This cohort was also used in a prior, separate analysis by our group in 2020 (Osadchiy et al., 2020). All IBS patients were evaluated by a gastroenterologist or nurse practitioner with expertise in IBS for presence of Rome IV diagnosis of IBS and bowel habit predominance (Drossman, 2006). The following were exclusion criteria: other gastrointestinal illnesses; eating disorders; rectal prolapse; severe hemorrhoids; gastric, abdominal, or colon surgery; recent steroid use; insulin-dependent diabetes; kidney disease; heart disease; hypertension; cancer; lung disease and/or neurological condition (traumatic brain injury, seizures); major surgery within 6 months of study onset, chronic illness and/or pain conditions; alcohol or drug misuse, psychiatric or developmental disorders impairing self-report, recent clinical trial participation (i.e., within 28 days), MRI contraindications, use of medications that could compromise the interpretation of the findings; ongoing major psychiatric diagnoses or use of psychotropic medications in the past 6 months; use of antibiotics or probiotics in the past 3 months, SSRIs, opioids; and excessive physical exercise (e.g., marathon runners).
Participants provided a stool sample and underwent multimodal brain-imaging studies at UCLA. MRI scans took place during the follicular phase of the menstrual cycle, with stool collection within a week of the scan. All procedures complied with the principles of the UCLA Institutional Review Board and informed consent was obtained from all participants.
2.2. Questionnaires
Clinical and behavioral data taken at baseline are detailed in our “Supplementary Methods: Questionnaires” section. The Bowel Symptom Questionnaire (BSQ) measures the overall symptom severity in IBS and abdominal pain over the past 1 week. It is a reliable, validated tool for patients with chronic gastrointestinal symptoms that assesses self-reported GI symptom severity, bloating, and abdominal pain (Talley et al., 1995). It uses a 0–20 rating scale (ranging from none to most intense imaginable), and a 5-point scale for the severity of IBS symptoms (How bad are your symptoms usually? 1 = none, 2 = mild, 3 = moderate, 4 = severe, 5 = very severe) (Park et al., 2018). The Visceral Sensitivity Index (VSI) is a self-reported scale validated in adult IBS patients that measures GI symptom-specific anxiety (Labus et al., 2004). Other self-report variables used in our analyses included the Diet Questionnaire (Dong et al., 2022), Socioeconomic Status (SES) (Adler et al., 2008), Irritable Bowel Syndrome Symptom Severity Score (IBS-SSS) (Francis et al., 1997), Coping Strategies Questionnaire (CSQ) (Seres et al., 2008), Early Trauma Inventory (ETI) (Bremner et al., 2000), Irritable Bowel Syndrome-Quality of Life (IBS-QOL) (Park et al., 2018), Hospital Anxiety and Depression (HADS) Scale (Zigmond and Snaith, 1983), and Perceived Stress Scale (PSS) (Cohen et al., 1983).
2.3. Gut microbiome
2.3.1. Collection and storage
Described in detail in published papers (Dong et al., 2020; Osadchiy et al., 2020). Participants were given “at-home kits” with specific instructions regarding the time of stool collection (e.g., time of day and within 2–3 days before the MRI scan). Participants were instructed to immediately freeze the fresh stool after collection. 2–3 consecutive diet diaries were collected from the time of enrollment to the time of the MRI scan and stool collection (1–2 weekdays and 1 weekend). Participants were asked to collect the stool before the first meal of the day. If participants were on antidiarrheal or laxatives, they were asked to refrain from use for 2–3 days before the sample collection. Any deviations from the stool sample collection were documented to account for in the analyses. Fecal samples were stored at −80 °C, then ground into a coarse powder by mortar and pestle under liquid nitrogen and aliquoted for DNA extraction and metabolomic profiling.
2.3.2. Fecal microbial profiling
DNA extraction with bead beating was performed using the QIAGEN Powersoil DNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA), following the manufacturer’s protocol. The V4 hypervariable region of the 16S rRNA gene was then amplified using 515F and 806R primers to generate a sequencing library according to a published protocol (Tong et al., 2014). The PCR products were purified with a commercial kit. The library underwent 2 × 250 sequencing on an Illumina HiSeq 2500 to a mean depth of 250,000 merged sequences per sample. The DADA2 pipeline was used for quality filtering, merging paired-end reads, removing chimera, and assigning taxonomy to each amplicon sequence variant (ASV) using the SILVA 138 reference database (Callahan et al., 2016; Glockner et al., 2017; Quast et al., 2013; Yilmaz et al., 2014).
See Supplementary Material for details on microbiome alpha and beta diversity calculations. To summarize microbial alpha diversity was assessed on datasets using the Chao1 index for species richness and the Shannon index for species evenness. Microbial composition (beta-diversity) was compared across samples by using the DEICODE plugin in QIIME 2 This was then visualized with principal coordinate analysis.
2.3.3. Fecal metabolomics processing
Fecal aliquots were shipped to Metabolon, Inc., and run as a single batch on their global HD4 metabolomics platform (Evans et al., 2009). This involved running methanol-extracted samples through ultrahigh performance liquid chromatography-tandem mass spectroscopy under four separate chromatography and electrospray ionization conditions to separate compounds with a wide range of chemical properties. Missing values in the raw data were completed using median values, and ineffective peaks were removed through the interquartile range denoising method. The internal standard normalization method was employed in the data analysis. The dataset for the multiple classification analysis was compiled from the metabolite profiling results, and a 3-dimensional matrix involving metabolite numbers, sample names, and normalized peak intensities was fed into the MetaboAnlyst web software 3.0 (http://www.metaboanalyst.ca).
2.4. Resting state brain connectivity
2.4.1. Magnetic resonance imaging acquisition
All participants underwent imaging in a 3.0 T S Prisma MRI Scanner (Siemens Healthcare, Erlangen, Germany) for a high-resolution T1 structural scan, and a resting-state functional scan. Acquisition parameters for high resolution T1-weighted images were as follows: echo time/repetition time (TE/TR) = 2.98/2300 ms, inversion time = 900 ms, field of view (FOV) = 240 × 256mm, slice thickness = 1 mm, 176 slices, 240 × 256 acquisition matrix, and isotropic voxel size = 1 mm3. Functional RS MRI scans were acquired with eyes closed using a gradient echo-planar imaging sequence with the following parameters: TE/TR: 28/2000 ms, flip angle = 77°, scan duration = 10 min, FOV = 220 mm, slices = 40, slice thickness = 4.0 mm, voxel resolution: 3.44 × 3.44 × 4mm, and slices were obtained with whole brain coverage.
2.4.2. MRI processing
Preprocessing and quality control of structural images were performed using Statistical Parametric Mapping 12 (Chudler and Dong, 1995). All structural images were skull stripped, segmented, and normalized to the MNI152 T1 template. This created normalized T1 images for every participant, along with segmented images (gray and white matter, and cerebrospinal fluid (CSF)) in normalized space.
2.4.3. Structural image parcellation
T1-image segmentation and cortical and subcortical regional parcellation were conducted using FreeSurfer v.6.0 following the nomenclature described in the Destrieux, Harvard-Oxford subcortical, and Harvard Ascending Arousal Network (AAN) atlases.
2.4.4. Resting-state fMRI preprocessing
Preprocessing and quality control of functional images was done using SPM-12 software (Welcome Department of Cognitive Neurology, London, UK) and involved slice-time correction and motion correction for the six realignment parameters. If any motion above 2 mm translation or 2° rotation was detected, the scan, along with the paired structural scan was discarded. In order to robustly account for motion, root mean squared (RMS) realignment estimates were calculated as robust measures of motion using publicly available MATLAB code from GitHub. The resting state images were then co-registered to their respective anatomical T1 images. Each participant’s T1 normalization parameters were then applied to that participant’s resting state image, resulting in an MNI space normalized resting state image. The resulting images were smoothed with a 5 mm3 Gaussian kernel. For each participant, a sample of the volumes was inspected for any artifacts and anomalies.
2.4.5. Resting-state fMRI network construction
Functional brain networks were constructed using the CONN 17 toolbox in MATLAB (Nieto-Castanon, 2020; Whitfield-Gabrieli and Nieto-Castanon, 2012). Regions from specific atlases were entered as regions of interest (ROIs). All pre-processed, normalized images were corrected for noise using the CompCor method to remove physiological noise, without regressing out the global signal (Chai et al., 2012; Nieto-Castanon, 2020; Whitfield-Gabrieli and Nieto-Castanon, 2012). Confounds for the six motion parameters, along with their first-order temporal derivatives, as well as confounds emerging from white matter and cerebrospinal fluid (CSF), and first-order temporal derivatives of motion, and the RMS were removed using regression. The images were band-passed filtered between 0.008 and 0.09 Hz to reduce low and high frequency noise that are not indicative of intrinsic brain activity (Weissenbacher et al.). Linear measures of ROI-to-ROI functional connectivity were computed using the Fisher-transformed correlation, representing the association between average temporal BOLD time series signals across all voxels within each brain region. The final outputs for each participant consisted of a matrix comprising Fisher-transformed Z correlation values between each ROI.
2.5. Statistical analyses
2.5.1. Data integration for biomarker discovery (DIABLO) analysis
A data integration analysis for biomarker discovery using latent components (DIABLO) was conducted to predict HCs vs IBS-C vs IBS-D. DIABLO is a multi-omics integration method using matrix factorization which has been used in the ‘omics literature (Bhatt et al., 2022; Gavin et al., 2018; Kong et al., 2021; Lee et al., 2019). DIABLO uses a limited number of correlated variables from multiple datasets to predict an outcome (i.e., HC vs IBS-C vs IBS-D). DIABLO is an extension of sparse generalized canonical correlation analysis (Tenenhaus et al., 2014), which generalizes partial least squares analysis for multiple matching datasets (Q), to a supervised learning framework (Rohart et al., 2017; Singh et al., 2019).
Prior to analyses, each individual dataset was preprocessed respective to its datatype (as described above). The identification of near zero variance predictors was determined on the metabolomics and clinical data and then removed with the cutoff being 50% of the values must be distinct with respect to the number of subjects. The MixMC preprocessing framework (Le Cao et al., 2011) as applied to the microbiome data to offset, pre-filter and center-log transform the data. The resting-state functional connectivity, metabolomics and clinical data was scaled and centered.
Individual sPLS models were run between data types first to guide the integration process by obtaining correlations to employ in a data-driven weighted design matrix (Singh et al., 2019). This process is done to create a design matrix for the subsequent DIABLO analysis, not to prefilter any variables. The design matrix is a matrix representing if and by how much each dataset are correlated (i.e., values ranging from 0 to 1) with each other as a hyperparameter in the DIABLO analysis. The design matrix was constructed by taking the correlation values of the first components of each sPLS model (Supplementary Figure 1) (Singh et al., 2019). With the design matrix determined, an initial DIABLO model with 5 components was fit without any variable selection, and global performance was assessed using the 5-fold cross validation repeated 50 times. After determining the number of components to use, the optimal number of variables to be kept per component per block, one component at a time, by defining a grid of values—in this case from 2 to 50—for each component per block, and using 5-fold cross-validation. To ease the interpretability of the model as a smaller number of features can have similar performance, we chose to have 5 features per component per block. The number of components, three, was chosen based on the lowest balanced error rate (BER) and distance metric (maximum distance vs centroids distance vs. mahalanobis distance) across several components. The main output measures for DIABLO are a set of components (i.e., latent variables) chosen in the model, a set of loading vectors (i.e., coefficients assigned to each variable to define each component), and a list of selected variables from each dataset and associated to each component. Loadings are the coefficients assigned to each variable to define each component, and their absolute value represents the importance of each variable in DIABLO. It is important to note that each loading vector is assigned to a particular component, and the loading vectors are obtained so that the covariance between a linear combination of variables and is maximized. Moreover, even if a top feature loading does not have a clear discriminatory effect, it may be highly correlated with other features across ‘omics types. A total of 5 features per each of the three components for each of the 4 data types (60 total features) were used in the final DIABLO model. The final performance of the model was assessed using 5-fold cross validation repeated 50 times and looking at the area under the ROC curve (AUC) to determine how well the model was performing for each group (Table 1). The area under the ROC curve is a way to summarize the overall diagnostic accuracy of the model. The values range from 0 to 1, where 1 represents a perfectly accurate model. A value of 0.5 represents no discriminatory ability. Values between 0.7 and 0.8 are considered acceptable, while between 0.8 and 0.9 is considered excellent, and greater values are considered outstanding. All online tutorials and template scripts to run DIABLO are available at https://github.com/mixOmicsTeam/mixOmics.
Table 1.
Area under the curve (AUC) values for the integrated DIABLO model by added component. The full integrated model with 3 components give the highest classification accuracy in predicting each group.
| Component 1 | AUC | p-value |
|---|---|---|
| HC vs. Others | 0.7652010 | 0.04067888 |
| IBS-C vs. Others | 0.6769755 | 0.18932110 |
| IBS-D vs. Others | 0.6420480 | 0.15638324 |
| Component 2 | ||
| HC vs. Others | 0.7927955 | 0.01789217 |
| IBS-C vs. Others | 0.7625095 | 0.04736679 |
| IBS-D vs. Others | 0.8006695 | 0.01875788 |
| Component 3 | ||
| HC vs. Others | 0.8151760 | 0.004440287 |
| IBS-C vs. Others | 0.7929240 | 0.020113075 |
| IBS-D vs. Others | 0.8356735 | 0.002993763 |
P-value< 0.05 (significant).
Abbreviations: HC, healthy control; IBS-C, irritable bowel syndrome-constipation; IBS-D, irritable bowel syndrome-diarrhea.
The entire model can be visualized in the circos diagram (Supplementary Figure 2), where the levels of each feature can be visualized on the perimeter of the circle, and correlations between datatypes can be seen inside the circle. Circos diagrams are built on a similarity matrix (Gonzalez et al., 2012) and represent the correlation between variables from different datasets, and a cutoff was chosen as r = 0.7 as this is universally considered a “strong” correlation. The distribution of each variable after standardization and scaling in the DIABLO model across the three groups can be seen in Fig. 1. A relevance network was generated to represent variable associations. Edges between nodes were only drawn if the association was 0.7 or higher (Fig. 2). Nodes/variables extracted from DIABLO that did not have any associations were not represented on the network.
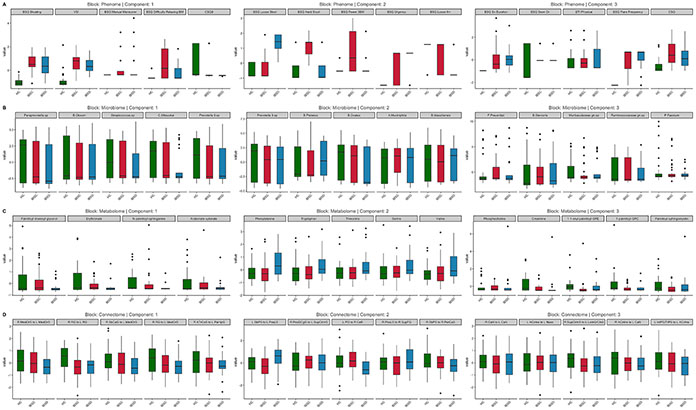
Fig. 1.
Omics Variables Selected by the DIABLO Model Across 3 Components. Standardized values comparing HC, IBS-C, and IBS-D are displayed. Abbreviations: IBSD, Irritable bowel syndrome-diarrhea; IBSC, irritable bowel syndrome-constipation; HC, healthy control; Phenome: BSQ, Bowel Symptom Questionnaire; VSI, Visceral Sensitivity Index; CSQ, The Coping Strategies Questionnaire; ETI, Early Traumatic Inventory. Brain Connectome: MedOrS, Medial Orbital Sulcus; RG, Rectus Gyrus; SbCaG, Subcallosal Gyrus; ATrCoS, Anterior Transverse Collateral Sulcus; PaHipG, Parahippocampal gyrus; SbPS, Subparietal Sulcus; PosLS, Posterior Ramus of the Lateral Sulcus; PosDGgG, Posterior Dorsal Part of the Cingulate Gyrus (dPCC); SupCirIns, Superior Segment of the Circular Sulcus of the Insula; PO, Pontis Oralis; CeB, Cerebellum; SupFG, Superior Frontal Gyrus; PerCaS, Pericallosal Sulcus; CaN, Caudate Nucleus; ACirIns, Anterior Segment of the Circular Sulcus of the Insula; Nacc, Nucleus Accumbens; LoInG/CInS, Long Insular Gyrus and Central Sulcus of the Insula; IntPS/TrPS, Intraparietal Sulcus and Transverse Parietal Sulci.
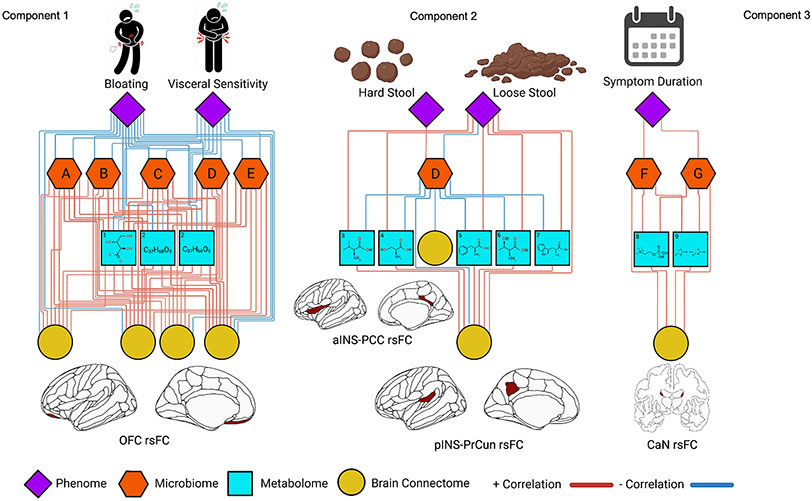
Fig. 2.
Relevance network from the DIABLO analysis depicting the correlation between different ’omics types. Red lines represent positive correlations and blue lines represent negative correlations. Cutoff for the correlations was r = 0.7. Microbiome features include (A) Paraprevotella. sp, (B) Blautia obeum, (C) Streptococcus. sp, (D) Prevotella 9. sp, (E) Catenibacterium mitsuokai (F) Faecalibacterium prausnitzii and (G) Bacteroides stercoris. Metabolome features include (1) erythronate, (2) palmitoyl-linoleoyl-glycerol, (3) valine, (4) serine, (5) phenylalanine, (6) threonine, (7) tryptophan, (8) phosphocholine, and (9) creatinine. Brain connectome features include (1) orbitofrontal cortex (CAN) resting-state functional connectivity (rsFC), (2) rsFC between the anterior insula (SAL) and posterior cingulate (DMN). rsFC between the posterior insula (SMN) and subparietal sulcus (DMN). (3) rsFC between the caudate nuclei (SMN).
3. Results
3.1. Participant demographics and clinical measures (Table 2)
Table 2.
Participant characteristics and clinical data.
| Characteristic | N | HC, N = 38a | IBSC, N = 36a | IBSD, N = 27a | p-valueb | HC vs. IBSC | HC vs. IBSD | IBSC vs. IBSD |
|---|---|---|---|---|---|---|---|---|
| Age | 101 | 27 (8) | 24 (6) | 25 (6) | 0.3 | 0.052 | 0.10 | 0.9 |
| SES | 44 | 5.75 (1.29) | 5.85 (1.60) | 5.50 (1.91) | >0.9 | 0.8 | 0.8 | 0.7 |
| BMI | 101 | 23.99 (2.69) | 23.33 (3.41) | 23.26 (3.80) | 0.2 | 0.4 | 0.4 | >0.9 |
| BSQ - Overall Symptoms | 64 | 3.0 (NA) | 8.0 (4.4) | 10.0 (3.8) | 0.070 | |||
| BSQ - Abdominal Pain | 65 | 4.0 (5.7) | 7.8 (4.5) | 8.2 (4.1) | 0.5 | 0.2 | 0.2 | 0.7 |
| BSQ - Bloating | 94 | 1.5 (1.8) | 11.8 (4.7) | 10.1 (5.4) | <0.001 | <0.001 | <0.001 | 0.11 |
| BSQ - Usual Severity | 65 | 2.00 (1.41) | 3.19 (0.79) | 3.22 (0.58) | 0.3 | 0.026 | 0.024 | 0.9 |
| BSQ - Age Onset | 64 | 19.0 (NA) | 17.7 (5.8) | 17.6 (4.6) | 0.8 | |||
| BSQ - Symptom Duration | 93 | 5.77 (3.11) | 3.61 (1.61) | 2.92 (1.52) | 0.002 | <0.001 | <0.001 | 0.2 |
| BSQ - Symptom Free Duration | 64 | 4.00 (NA) | 3.22 (0.83) | 3.19 (1.08) | 0.6 | |||
| BSQ - Flare Frequency | 64 | 1.00 (NA) | 3.94 (1.37) | 4.33 (1.21) | 0.11 | |||
| BSQ - Flare Now | 64 | 0/1 (0%) | 4/36 (11%) | 6/27 (22%) | 0.4 | |||
| BSQ - Symptom Duration Years | 64 | 1.0 (NA) | 6.7 (6.6) | 7.1 (5.2) | 0.2 | |||
| BSQ - Chest Pain/Pressure | 64 | 1/1 (100%) | 11/36 (31%) | 4/27 (15%) | 0.087 | |||
| BSQ - Fulness, Gas or Bloating | 64 | 1/1 (100%) | 36/36 (100%) | 26/27 (96%) | 0.4 | |||
| BSQ - Distention | 64 | 1/1 (100%) | 28/36 (78%) | 20/27 (74%) | 0.8 | |||
| BSQ - Rectum Fullness | 64 | 0/1 (0%) | 31/36 (86%) | 16/27 (59%) | 0.008 | |||
| BSQ - Urgency | 64 | 0/1 (0%) | 19/36 (53%) | 25/27 (93%) | 0.001 | |||
| BSQ - Nausea | 64 | 0/1 (0%) | 15/36 (42%) | 9/27 (33%) | 0.6 | |||
| BSQ - Belly Pain | 64 | 0/1 (0%) | 32/36 (89%) | 25/27 (93%) | 0.11 | |||
| BSQ - Irregular Bowel Movements | 64 | 1/1 (100%) | 36/36 (100%) | 26/27 (96%) | 0.4 | |||
| BSQ - Most Bothersome Symptom | 64 | >0.9 | ||||||
| Abdominal Distenstion | 0/1 (0%) | 4/36 (11%) | 2/27 (7.4%) | |||||
| Belly Pain | 0/1 (0%) | 5/36 (14%) | 6/27 (22%) | |||||
| Fullness in Rectum | 0/1 (0%) | 1/36 (2.8%) | 0/27 (0%) | |||||
| Fullness/Gas/Bloating | 0/1 (0%) | 12/36 (33%) | 8/27 (30%) | |||||
| Irregular Bowel Habits | 1/1 (100%) | 13/36 (36%) | 9/27 (33%) | |||||
| Urgency | 0/1 (0%) | 1/36 (2.8%) | 2/27 (7.4%) | |||||
| BSQ - Amount of PCP Doctor Visits | 101 | 1.71 (0.69) | 2.58 (1.11) | 2.48 (1.05) | <0.001 | <0.001 | 0.002 | 0.7 |
| BSQ - Consulted Doctor | 81 | 3/18 (17%) | 31/36 (86%) | 23/27 (85%) | <0.001 | |||
| BSQ - Amount of GI Doctor Visits | 71 | 1.21 (0.43) | 1.88 (0.79) | 1.84 (0.75) | 0.009 | 0.006 | 0.011 | 0.9 |
| BSQ - Colicky Baby | 68 | 0.002 | ||||||
| Don’t Know | 13/26 (50%) | 10/23 (43%) | 2/19 (11%) | |||||
| No | 13/26 (50%) | 13/23 (57%) | 12/19 (63%) | |||||
| Yes | 0/26 (0%) | 0/23 (0%) | 5/19 (26%) | |||||
| BSQ - Sensitive Stomach | 68 | 1/26 (3.8%) | 12/23 (52%) | 8/19 (42%) | <0.001 | |||
| BSQ - Breast Fed | 37 | 0.057 | ||||||
| Don’t Know | 3/17 (18%) | 0/10 (0%) | 1/10 (10%) | |||||
| No | 1/17 (5.9%) | 5/10 (50%) | 1/10 (10%) | |||||
| Yes | 13/17 (76%) | 5/10 (50%) | 8/10 (80%) | |||||
| BSQ - Formula Fed | 37 | 0.009 | ||||||
| Don’t Know | 7/17 (41%) | 0/10 (0%) | 1/10 (10%) | |||||
| No | 6/17 (35%) | 1/10 (10%) | 4/10 (40%) | |||||
| Yes | 4/17 (24%) | 9/10 (90%) | 5/10 (50%) | |||||
| BSQ - Mode Delivery | 37 | 0.045 | ||||||
| Don’t Know | 0/17 (0%) | 1/10 (10%) | 0/10 (0%) | |||||
| Elective C-Section | 3/17 (18%) | 1/10 (10%) | 0/10 (0%) | |||||
| Emergency C-Section | 0/17 (0%) | 0/10 (0%) | 3/10 (30%) | |||||
| Vaginal | 14/17 (82%) | 8/10 (80%) | 7/10 (70%) | |||||
| BSQ - Less than 3 BM Per Week | 94 | 1.09 (0.30) | 2.36 (1.44) | 1.15 (0.61) | <0.001 | <0.001 | 0.8 | <0.001 |
| BSQ - Hard Stool | 94 | 1.38 (0.66) | 3.53 (0.84) | 1.42 (0.70) | <0.001 | <0.001 | 0.8 | <0.001 |
| BSQ - Straining | 94 | 1.28 (0.52) | 3.22 (1.10) | 2.04 (1.08) | <0.001 | <0.001 | 0.003 | <0.001 |
| BSQ - Incomplete Evacuation | 94 | 1.22 (0.49) | 3.00 (1.07) | 2.65 (1.06) | <0.001 | <0.001 | <0.001 | 0.14 |
| BSQ - Anorectal Obstruction | 94 | 1.13 (0.34) | 3.03 (1.16) | 1.65 (0.63) | <0.001 | <0.001 | 0.016 | <0.001 |
| BSQ - Manual Maneuver | 94 | 1.00 (0.00) | 1.44 (0.88) | 1.54 (1.14) | 0.011 | 0.026 | 0.013 | 0.7 |
| BSQ - Difficult Relaxing during BM | 92 | 1.10 (0.31) | 2.58 (1.46) | 1.54 (0.81) | <0.001 | <0.001 | 0.11 | <0.001 |
| BSQ - Loose Stool | 94 | 1.28 (0.46) | 1.47 (0.70) | 3.35 (0.75) | <0.001 | 0.2 | <0.001 | <0.001 |
| BSQ - Loose Stool 75% | 50 | 1/9 (11%) | 3/15 (20%) | 15/26 (58%) | 0.011 | |||
| BSQ - Loose Stool > 6 Months | 52 | 2/11 (18%) | 7/15 (47%) | 23/26 (88%) | <0.001 | |||
| BSQ - Bowel Habit and Meds | 94 | <0.001 | ||||||
| Hard stool only with antidiarrheal | 0/32 (0%) | 0/35 (0%) | 10/27 (37%) | |||||
| Loose stool only with laxatives | 2/32 (6.2%) | 23/35 (66%) | 0/27 (0%) | |||||
| Neither | 30/32 (94%) | 12/35 (34%) | 17/27 (63%) | |||||
| BSQ - Feel Constipated | 48 | 1.0 (1.4) | 14.4 (3.7) | 4.7 (5.8) | <0.001 | <0.001 | 0.3 | <0.001 |
| IBS-SSS | 62 | NA (NA) | 231 (100) | 197 (95) | 0.2 | |||
| IBS QoL - Dysphoria | 65 | 98 (2) | 65 (25) | 69 (25) | 0.10 | 0.071 | 0.11 | 0.5 |
| IBS QoL - Interference | 65 | 98 (3) | 69 (24) | 64 (23) | 0.071 | 0.084 | 0.045 | 0.4 |
| IBS QoL - Body Image | 65 | 97 (4) | 58 (24) | 73 (24) | 0.009 | 0.028 | 0.2 | 0.019 |
| IBS QoL - Health Worry | 65 | 100 (0) | 51 (31) | 66 (23) | 0.013 | 0.017 | 0.10 | 0.035 |
| IBS QoL - Food Avoidance | 65 | 100 (0) | 44 (32) | 51 (30) | 0.070 | 0.016 | 0.033 | 0.4 |
| IBS QoL - Social Reaction | 65 | 97 (4) | 69 (24) | 63 (27) | 0.079 | 0.13 | 0.072 | 0.4 |
| IBS QoL - Sexual | 62 | 100 (0) | 69 (30) | −21 (476) | 0.2 | 0.9 | 0.6 | 0.3 |
| IBS QoL - Relationship | 65 | 100 (0) | 78 (20) | 75 (20) | 0.092 | 0.13 | 0.088 | 0.6 |
| IBS QoL - Total | 65 | 99 (2) | 64 (21) | 66 (20) | 0.052 | 0.021 | 0.034 | 0.6 |
| PSS | 96 | 12 (6) | 17 (7) | 15 (6) | 0.016 | 0.007 | 0.2 | 0.3 |
| HAD - Anxiety | 100 | 4.0 (3.0) | 8.7 (4.8) | 6.7 (3.3) | <0.001 | <0.001 | 0.006 | 0.036 |
| HAD - Depression | 100 | 1.26 (1.55) | 3.49 (3.29) | 2.22 (2.19) | <0.001 | <0.001 | 0.12 | 0.047 |
| VSI | 101 | 7 (13) | 43 (17) | 37 (14) | <0.001 | <0.001 | <0.001 | 0.14 |
| CSQ Q7 | 94 | 14 (30) | 9 (23) | 3 (2) | 0.002 | 0.4 | 0.069 | 0.3 |
| CSQ Q8 | 94 | 37 (46) | 9 (23) | 2 (1) | <0.001 | <0.001 | <0.001 | 0.5 |
| CSQ - Catastrophizing Score | 93 | 0.44 (0.70) | 1.57 (1.14) | 1.37 (1.12) | <0.001 | <0.001 | <0.001 | 0.5 |
| ETI - General | 96 | 1.16 (1.41) | 1.68 (1.57) | 1.83 (1.74) | 0.15 | 0.2 | 0.10 | 0.7 |
| ETI - Physical | 96 | 0.68 (1.04) | 0.91 (1.24) | 1.04 (1.43) | 0.7 | 0.4 | 0.3 | 0.7 |
| ETI - Emotional | 96 | 0.50 (1.13) | 1.06 (1.59) | 1.21 (1.47) | 0.043 | 0.093 | 0.055 | 0.7 |
| ETI - Sexual | 96 | 0.42 (0.98) | 0.62 (1.10) | 0.75 (1.19) | 0.4 | 0.4 | 0.2 | 0.6 |
| ETI - Total | 96 | 2.8 (3.2) | 4.3 (3.9) | 4.8 (4.1) | 0.049 | 0.087 | 0.034 | 0.6 |
N = 102 total, IBS-C group N = 36, IBS-D group N = 27, HC group N = 39.
Abbreviations: IBSD, Irritable bowel syndrome-diarrhea; IBSC, irritable bowel syndrome-constipation; HC, healthy control; SES, socioeconomic status; BMI, Body Mass Index; BSQ, Bowel Symptom Questionnaire; IBS-SSS, irritable bowel syndrome symptom severity scale; IBS QoL, Irritable Bowel Syndrome-Quality of Life; PSS, Perceived Stress Scale; HAD; Hospital Anxiety and Depression Scales; VSI, Visceral Sensitivity Index; CSQ, The Coping Strategies Questionnaire; ETI, Early Traumatic Inventory. CSQ Q7: Based on all you do to cope, how much control do you feel you have over it?: 0 = No control - 6 = Complete control. CSQ Q8: Based on all the things you do to cope, how much are you able to decrease it?: 0 = None at all - 6 = Decrease completely.
Mean (SD); n/N (%), p-value <0.05.
Kruskal-Wallis rank sum test; Fisher’s exact test; Pearson’s Chi-squared test.
IBS-C and IBS-D showed significant differences in frequency and consistency of bowel habits, including fewer than 3 bowel movements in a day (IBS–C > IBS-D, p = 0.0002), hard stool (IBS–C > IBS-D, p < 0.001) and loose stool (IBS-D > IBS-C, p < 0.001). While both IBS-C and IBS-D had significantly increased scores for anxiety and depression compared to HCs, no statistically significant differences between the IBS-bowel subtype groups were identified.
3.2. Microbiome alpha and beta diversity differences by IBS subtype
PCoA analysis showed no significant differences in alpha or beta diversity between IBS-C, IBS-D, and HCs (see Supplementary Figures 3, 4, and 5).
3.3. DIABLO identifies a multi-omic signature able to classify healthy controls from IBS subtypes
A highly correlated multi-omics signature was able to successfully classify HCs vs IBS-C vs IBS-D (Table 1). The achieved area under the curve (AUC) was 82% (p = 0.004) for HC. vs others, 79% (p = 0.02) for IBS-C vs others, and 84% (p = 0.003) for IBS-D vs others.
On component 1, phenome features in order of importance included the BSQ (bloating, manual maneuver, and difficulty relaxing during a bowel movement subscales), VSI, and CSQ (Based on all the things you do to cope, how much are you able to decrease it?). Microbiome features in order of importance included the Paraprevotella spp., Blautia obeum, Streptococcus spp., Mitsuokai spp. and Prevotella 9 spp. Metabolome features in order of importance included palmitoyl-linoleoyl-glycerol, erythronate, N-palmitoyl-sphinganine, and arabonate-xylonate. Resting-state connectome features in order of importance included connectivity between the left and right medial orbital sulcus (CAN), left and right rectus gyrus (CAN), between the right subcentral gyrus (SMN) and left medial orbital sulcus (CAN), between the right rectus gyrus and left medial orbital sulcus (CAN), and between the right anterior transverse collateral sulcus (DMN) and left parahippocampal gyrus (EAN).
On component 2, phenome features in order of importance included the BSQ (loose stool, hard stool, the fewer than 3 bowel movements per week, urgency, loose stool for more than 6 months). Microbiome features in order of importance included the Prevotella 9 spp., Bacteriodes plebeius, Bacteriodes ovatus, Akkermansia muciniphila, and Bacteriodes massiliensis. Metabolome features in order of importance included phenylalinine, tryptophan, threonine, serine, and valine. Resting-state connectome features in order of importance included connectivity between the left subparietal sulcus and left posterior ramus of the lateral sulcus (SMN), between the right posterior-dorsal part of the cingulate gyrus (DMN) and left superior segment of the circular sulcus of the insula (SAL), the left pontis oralis (CAN) and right cerebellum, the right posterior ramus of the lateral sulcus (SMN) and right superior frontal gyrus (CEN), and right subparietal sulcus (SMN) and right pericallosal sulcus (EAN).
On component 3, phenome features in order of importance included the BSQ (symptom duration, if the patient had seen a doctor, flare frequency), ETI (physical subscale), and CSQ. Microbiome features in order of importance included Faecalibacterium prausnitzii, Bacteriodes stercoris, Muribaculaceae family, Ruminococcaceae family, and Phascolarctobacterium faecium. Metabolome features of importance included phosphocholine, creatinine, 1-1-enyl-palmitoyl-GPE, 1-palmitoyl-GPC, and palmitoyl-sphingomyelin. Resting-state connectome features in order of importance included connectivity between the left and right caudate nucleus (SMN), between the left anterior segment of the circular sulcus of the insula and nucleus accumbens (SMN), the right superior segment of the circular sulcus of the insula and left long insular gyrus and central sulcus of the insula, and the right anterior segment of the circular sulcus of the insula and left caudate nucleus and left intraparietal sulcus.
4. Discussion
The current study aimed to test the hypothesis that IBS patients that differ in their reported predominant bowel habit are characterized by distinct biological alterations within the BGM system. Our findings support that in addition to shared features, the highly correlated multi-omic signature across the phenome, microbiome, metabolome, and resting-state brain connectome can differentiate IBS subtypes and from healthy controls.
4.1. IBS bowel habits are associated with alterations in the BGM system
4.1.1. Component 1
A BGM signature involving key IBS hallmarks, altered levels of multiple microbial taxa, and regions of the brain invovled in sensory and autonomic processing differentiated IBS from healthy controls in our study.
Bloating (BSQ-Bloating) and Visceral Sensitivity Index (VSI) were the most important clinical variables to separate IBS from HCs in this study and thus served as clinical proxies for IBS. Compared to healthy controls, both IBS-C and IBS-D had lower abundance of the 5 bacterial taxa most important in differentiating IBS from HCs (Paraprevotella spp., Blautia obeum, Streptococcus spp., Catenibacterium mitsuokai, and Prevotella 9 spp.). This echoes prior work generally showing a dysbiosis in IBS (Casen et al., 2015; Jeffery et al., 2012; Pozuelo et al., 2015), though studies have been mixed. Our study raises the possibility that dysbiosis in IBS may be largely explained by its role in visceral sensitivty and bloating, as supported by their consistently negative relationship with these taxa in our integrated analysis. For example, Blautia spp. and specifically Blautia obeum have been shown to be inversely related to inflammation, obesity, and pathogenic bacteria (Liu et al., 2021). Therefore, our finding that Blautia obeum was negatively associated with visceral pain and bloating may suggest an underlying inflammatory pathway, associating the microbiome to IBS symptomatology. Prior work supports that visceral hypersensitivity specifically may have an etiologic root in dysbiotic intestines (Chichlowski and Rudolph, 2015), where improving the dysbiosis with, for example, a probiotic, has improved visceral hypersensitivity in animal and human models (Parkes et al., 2008).
The lower brain connectivity involving the orbitofrontal cortex (OFC) shown in both IBS-C and IBS-D compared to HC may implicate an impairment in pain regulatory pathways. The OFC encodes associations between sensory stimuli and emotionally relevant internal states, interfaces with the CAN, and regulates descending pain pathways (Mayer et al., 2006, 2015). Lower OFC connectivity may thus translate to less regulatory ability to modulate descending pain pathways (from the ‘top-down’), which is consistent with the negative association between the OFC and bloating/visceral sensitivity in our IBS cohort.
Taken together, the indirect relationship between select gut microbes/metabolites and visceral sensitivity and bloating, coupled with the direct relationship of these microbial/metabolomic factors with OFC connectivity alterations, suggests that interactions in the gut may mediate central autonomic and descending pain pathways to produce symptomatology in IBS.
4.1.2. Component 2
BGM connections between loose stool, tryptophan and phenylalanine, and key default mode and salience regions differentiated IBS-D from IBS-C. Loose Stool (BSQ-Loose Stool) here serves as a proxy for IBS-D, and Hard Stool (BSQ-Hard Stool) for IBS-C, as these were the most important clinical variables to separate IBS-C from IBS-D in the integrated model.
When comparing Loose Stool and Hard Stool, Prevotella spp. emerges as the only highly important microbial variable in the classifier, with lower abundance in both IBS-C and IBS-D, but with a specifically positive association with Hard Stool and a negative association with Loose Stool. Disturbances in Prevotella spp. have been established in IBS literature (Su et al., 2018; Wu et al., 2019; Zhu et al., 2021), hypothesized to produce a deleterious effect via carbohydrate fermentation, inflammation, and visceral hypersensitivity (Ley, 2016; Pandiyan et al., 2019). Given the specific relationships to bowel habit, it is possible that Prevotella spp. may exert differential effects in IBS-C compared to IBS-D.
In the metabolome, IBS-D had higher levels of multiple metabolites compared to IBS-C and HCs, but tryptophan as well as phenylalanine are of particular interest. Tryptophan has a powerful role in the gut, largely through its role in gut serotonin production and modulation. Microbes can produce and degrade tryptophan (the precursor to serotonin) independently of the host, which affects overall intestinal serotonin production. However, they can also modulate serotonin synthesis and activity indirectly through producing metabolites such as SCFAs, which can promote the conversion of tryptophan to serotonin (via tryptophan hydroxylase) and regulate serotonin transporter activity and expression (Layunta et al., 2021). Gut serotonin increases motility, secretion, and visceral sensitivity (Mawe and Hoffman, 2013) – all hallmarks of diarrhea-predominant bowel habits. Postprandial plasma serotonin levels vary significantly between IBS subtypes and HCs (Grasberger et al., 2013), possibly as a function of serotonin transporter (SERT) expression, leading to higher effective serotonin level in IBS-D compared to IBS-C (Vahora et al., 2020). Our study’s positive association between tryptophan and loose stool supports ongoing studies on modulation of gut serotonin for potential therapeutic benefit in patients with IBS-D. Similarly, phenylalanine’s elevated levels in IBS-D compared to other groups is interesting in the context of motility. Phenylalanine is a precursor to a number of key neurotransmitters that directly affect colonic motility, such as dopamine and norepinephrine (Lou, 1994), whose roles are beginning to be recognized as potential pathophysiological and therapeutic factors in IBS (Gros et al., 2021).
Both tryptophan and phenylalanine had a positive association with increased brain connectivity between the SMN (posterior insula) and the DMN (subparietal sulcus) as they related to loose stool. These brain regions are known for processing aversive sensory stimuli from the viscera and interfacing with self-related homeostasis, respectively. Previous work by our group has demonstrated a positive association between gut tryptophan and DMN connectivity (Osadchiy et al., 2020). Individuals with chronic diarrhea may have abnormal tryptophan- and phenylalanine-mediated signaling traveling from the viscera to the posterior insula, influencing self-related thoughts, emotions and pain perception. Together, these may produce the IBS-D phenotype.
More broadly, when focusing on loose stool, associations between brain connectivity and clinical symptoms are almost entirely indirect, as they “pass through” the gut microbiome and metabolome (rather than directly associate). Taken together, our results support a ‘bottom-up’ directionality (gut microbiome and metabolite changes largely influencing brain changes) in the BGM’s role in IBS-D for future diagnostic and therapeutic investigations. By contrast, overall, IBS-C’s microbiome and metabolomic profiles were far more similar to HC than to IBS-D. Patients with IBS-C had greater connectivity between the anterior insula (aINS) and the posterior cingulate cortex (PSS). The aINS is a key hub of the salience network and receives direct sensory input from visceral and sensory afferents (Seeley et al., 2007). The PCC is a key hub of the DMN and typically shows a decrease in activation conversely with activation of the SAL (Menon, 2015). Greater connectivity between these two networks in the current clinical context has been linked to a low intrinsic attention to symptoms and may be a compensatory mechanism to constant constipation (Kucyi et al., 2013). Thus, our results support that IBS-C may function with more of a ‘top-down’ mechanism in the BGM compared to IBS-D.
4.1.3. Component 3
BGM alterations involving symptom duration, select microbial taxa, and the caudate differentiate IBS from HC.
Elevated levels of Bacteroides stercoris in individuals with IBS-C and IBS-D compared to healthy controls supports prior work: disturbances in Bacteroides stercoris gut populations are known to contribute to dysbiosis in IBS patients (Casen et al., 2015). The positive association between Symptom Duration in Years (another clinical variable proxy differentiating IBS-C and IBS-D from HCs) and Bacteroides stercoris may indicate a role for this taxon in the ‘maintenance’ phase, or chronicity, of the syndrome. However, Faecalibacterium prausnitzii is considered a highly beneficial bacteria and its reduction has been linked to many intestinal and inflammatory diseases (Lopez-Siles et al., 2020). F. prausnitzii and its phylogroups’ abundance indices have also been shown to successfully discriminate between IBS and inflammatory bowel disease (Lopez-Siles et al., 2020). In our study, symptom duration was positively associated with Faecalibacterium prausnitzii; whether this is a coping mechanism for the chronicity of IBS symptomatology remains to be elucidated in further research.
The positive association between symptom duration, these microbes, and the caudate nuclei may support an integrated BGM signature for chronicity of symptoms in IBS. The caudate nucleus has been implicated in pain modulation, such that a greater amount of activation is associated with diminished pain reactivity (Borsook et al., 2010; Lineberry and Vierck, 1975; Wunderlich et al., 2011). Diminished reactivity of the basal ganglia, including the caudate has previously been implicated in the chronification of pain. Chronification of pain symptoms, due to its diminishing effect on the capacity of the caudate nuceli, reflects a shift away from acute pain circuitry and more to emotion and reward circuitry (Hashmi et al., 2013). Our current results provide further potential mechanistic insight to this pathway, as symptom duration is related to caudate nuclei connectivity via differing relationships of Faecalibacterium prausnitzii, Bacteroides stercoris, phosphocholine, and creatinine, but further research is needed to uncover their role.
Overall, the chronicity of bowel habit symptoms in IBS may implicate abnormalities in several key gut microbes and functional connections in the brain involving the caudate nuclei.
4.2. Limitations
The cross-sectional nature of our study precludes the ability to make any causal inferences over time and during symptom fluctuations. Our study is an exploratory analysis that needs to be validated with an independent, large cohort in order to confirm our findings. Study results do not allow us to differentiate between the importance of primary central and peripheral alterations playing a role in IBS symptom generation, including bowel habit subtype. As predominant bowel habits were self-reported and not based on physiological alterations in specific gut functions (secretion, motility, visceral sensitivity) correlations with biological microbial and brain alterations are limited. Visceral perception was not assessed in the subjects, making statements about the involvement of certain mechanisms (alterations in certain brain networks or metabolites) in visceral sensitivity or bowel function speculative.
5. Clinical implications and conclusions
Our study serves as a foundation upon which to assess potential pathophysiological and therapeutic targets in mediating bowel habit subtypes of IBS. The association between abnormal connectivity involving central autonomic and descending pain regulatory networks, gut microbiome/metabolite changes, and select IBS symptoms outlines a compelling BGM signature in IBS. Even though the cross-sectional nature of our study does not allow us to identify causative relationships, our findings suggest that aberrant autonomic processing and modulation of descending pain pathways may play a role in visceral hypersensitivity and bloating (which are hallmarks of IBS as compared to healthy controls), as well as influence the gut microbiome and metabolome. These findings highlight the need for future studies exploring whether neuropsychiatric interventions that modulate orbitofrontal cortex connectivity decrease visceral hypersensitivity, bloating, and the changes in the gut microenvironment in IBS patients.
In IBS-D, our findings show a correlation between high levels of gut metabolites tryptophan and phenylalanine and aberrant connectivity in brain regions involved in processing unpleasant visceral stimuli (SMN) and self-related thoughts (DMN). Supporting the extensive work already establishing tryptophan as an important gut metabolite in IBS pathophysiology, these results suggest that increased tryptophan in the gut may lead to loose stool, and tryptophan-related signaling may travel to the posterior insula and increase pain perception and emotional salience in IBS-D. Suggesting a ‘bottom-up’ signaling direction, our study carves a path for future mechanistic, integrated studies exploring the manipulation of gut tryptophan in IBS-D (by way of dietary or microbial interventions) and its effect on diarrhea and SMN and DMN brain connectivity. IBS-C’s microbiome and metabolome resembled HC; however, the increased connectivity in the default mode (DMN) and salience (SAL) networks compared to IBS-D may indicate abnormalities in the emotional physiological processing of visceral signals. IBS-C’s relatively isolated brain changes may indicate a more ‘top-down’ mechanism to produce the constipation-predominant phenome.
The link between the chronicity of IBS symptoms, B. stercoris and F. prausnitzii, and brain connectivity in the caudate nuclei may influence the chronification of IBS symptoms. Interventions targeting the caudate or these microbes and measuring the effect on IBS symptomatology would help elucidate their potential role in the pathophysiology of IBS symptom burden over time.
Supplementary Material
Acknowledgements
The authors would like to acknowledge support from the Neuroimaging Core (NIC; personnel Priten Vora, Cathy Liu) and the Clinical Studies Core (Jean Stains, Suzanne Smith) at the G. Oppenheimer Center for Neurobiology of Stress and Resilience at the University of California Los Angeles, CA, USA for help with the neuroimaging processing, including creation of the necessary datasets utilized in these analyses; and for help with data collection. We would also like to acknowledge Parth R. Bhatt and his company parth.pro (www.parth.pro) in assistance with creating the figures.
Grant support
This research was supported by grants from the National Institutes of Health including K23 DK106528 (AG), ULTR001881/DK041301 (UCLA CURE/CTSI Pilot and Feasibility Study; AG), R01 MD015904 (AG), R03 DK121025 (AG), U54 DK123755 (EAM/LC), P50 DK064539 (EAM), R01 DK048351 (EAM), P30 DK041301 (CURE), R01 CA270027 (BME), and pilot funds provided for brain scanning by the UCLA Ahmanson-Lovelace Brain Mapping Center.
Abbreviations
- BGM
Brain-Gut-Microbiome
- IBS
Irritable Bowel Syndrome
- IBS-C
constipation-predominant Irritable Bowel Syndrome
- IBS-D
diarrhea-predominant Irritable Bowel Syndrome
- fMRI
functional magnetic resonance imaging
- PLS-DA
partial least squares discriminant analysis
- ANOVA
analysis of variance
- CAN
central autonomic network
- EAN
emotional arousal network
- CEN
central executive network
- SMN
sensorimotor network
- OCC
occipital network
- DMN
default mode network
Footnotes
Data transparency statement
Deidentified individual participant data (clinical, brain, and microbiome) can be shared upon request and will be made available through the Center’s pain repository portal (https://www.painrepository.org/). To access the data, participants will fill out a user agreement, upon which access to the data will be made available through a secure password protected portal.
Declaration of competing interest
AG is scientific advisor to Yamaha. EAM is a scientific advisory board member of Danone, Axial Biotherapeutics, Amare, Mahana Therapeutics, Pendulum, Bloom Biosciences, Seed and APC Microbiome Ireland. All other authors have nothing to disclose.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neuropharm.2022.109381.
Data availability
Data will be made available on request.
References
- Adler N, Singh-Manoux A, Schwartz J, Stewart J, Matthews K, Marmot MG, 2008. Social status and health: a comparison of British civil servants in Whitehall-II with European- and African-Americans in CARDIA. Soc. Sci. Med 66, 1034–1045. [DOI] [PubMed] [Google Scholar]
- Berman SM, Naliboff BD, Suyenobu B, Labus JS, Stains J, Ohning G, Kilpatrick L, Bueller JA, Ruby K, Jarcho J, Mayer EA, 2008. Reduced brainstem inhibition during anticipated pelvic visceral pain correlates with enhanced brain response to the visceral stimulus in women with irritable bowel syndrome. J. Neurosci 28, 349–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt RR, Gupta A, Labus JS, Liu C, Vora PP, Jean S, Naliboff BD, Mayer EA, 2022. A neuropsychosocial signature predicts longitudinal symptom changes in women with irritable bowel syndrome. Mol. Psychiatr 27, 1774–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsook D, Upadhyay J, Chudler EH, Becerra L, 2010. A key role of the basal ganglia in pain and analgesia–insights gained through human functional imaging. Mol. Pain 6, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremner JD, Vermetten E, Mazure CM, 2000. Development and preliminary psychometric properties of an instrument for the measurement of childhood trauma: the Early Trauma Inventory. Depress. Anxiety 12, 1–12. [DOI] [PubMed] [Google Scholar]
- Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP, 2016. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casen C, Vebo HC, Sekelja M, Hegge FT, Karlsson MK, Ciemniejewska E, Dzankovic S, Froyland C, Nestestog R, Engstrand L, Munkholm P, Nielsen OH, Rogler G, Simren M, Ohman L, Vatn MH, Rudi K, 2015. Deviations in human gut microbiota: a novel diagnostic test for determining dysbiosis in patients with IBS or IBD. Aliment. Pharmacol. Ther 42, 71–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai XJ, Castanon AN, Ongur D, Whitfield-Gabrieli S, 2012. Anticorrelations in resting state networks without global signal regression. Neuroimage 59, 1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassard C, Dapoigny M, Scott KP, Crouzet L, Del’homme C, Marquet P, Martin JC, Pickering G, Ardid D, Eschalier A, Dubray C, Flint HJ, Bernalier-Donadille A, 2012. Functional dysbiosis within the gut microbiota of patients with constipated-irritable bowel syndrome. Aliment. Pharmacol. Ther 35, 828–838. [DOI] [PubMed] [Google Scholar]
- Chen XF, Guo Y, Lu XQ, Qi L, Xu KH, Chen Y, Li GX, Ding JP, Li J, 2021. Aberrant intraregional brain activity and functional connectivity in patients with diarrhea-predominant irritable bowel syndrome. Front. Neurosci 15, 721822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichlowski M, Rudolph C, 2015. Visceral pain and gastrointestinal microbiome. J Neurogastroenterol Motil 21, 172–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chudler EH, Dong WK, 1995. The role of the basal ganglia in nociception and pain. Pain 60, 3–38. [DOI] [PubMed] [Google Scholar]
- Clarke G, McKernan DP, Gaszner G, Quigley EM, Cryan JF, Dinan TG, 2012. A distinct profile of tryptophan metabolism along the Kynurenine pathway downstream of toll-like receptor activation in irritable bowel syndrome. Front. Pharmacol 3, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S, Kamarck T, Mermelstein R, 1983. A global measure of perceived stress. J. Health Soc. Behav 24, 385–396. [PubMed] [Google Scholar]
- Dong TS, Guan M, Mayer EA, Stains J, Liu C, Vora P, Jacobs JP, Lagishetty V, Chang L, Barry RL, Gupta A, 2022. Obesity is associated with a distinct brain-gut microbiome signature that connects Prevotella and Bacteroides to the brain’s reward center. Gut Microb. 14, 2051999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong TS, Gupta A, Jacobs JP, Lagishetty V, Gallagher E, Bhatt RR, Vora P, Osadchiy V, Stains J, Balioukova A, Chen Y, Dutson E, Mayer EA, Sanmiguel C, 2020. Improvement in uncontrolled eating behavior after laparoscopic sleeve gastrectomy is associated with alterations in the brain-gut-microbiome Axis in obese women. Nutrients 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drossman DA, 2006. The functional gastrointestinal disorders and the Rome III process. Gastroenterology 130, 1377–1390. [DOI] [PubMed] [Google Scholar]
- Drossman DA, Hasler WL, 2016. Rome IV-functional GI disorders: disorders of gut-brain interaction. Gastroenterology 150, 1257–1261. [DOI] [PubMed] [Google Scholar]
- Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E, 2009. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal. Chem 81, 6656–6667. [DOI] [PubMed] [Google Scholar]
- Francis CY, Morris J, Whorwell PJ, 1997. The irritable bowel severity scoring system: a simple method of monitoring irritable bowel syndrome and its progress. Aliment. Pharmacol. Ther 11, 395–402. [DOI] [PubMed] [Google Scholar]
- Gavin PG, Mullaney JA, Loo D, Cao KL, Gottlieb PA, Hill MM, Zipris D, Hamilton-Williams EE, 2018. Intestinal metaproteomics reveals host-microbiota interactions in subjects at risk for type 1 diabetes. Diabetes Care 41, 2178–2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glockner FO, Yilmaz P, Quast C, Gerken J, Beccati A, Ciuprina A, Bruns G, Yarza P, Peplies J, Westram R, Ludwig W, 2017. 25 years of serving the community with ribosomal RNA gene reference databases and tools. J. Biotechnol 261, 169–176. [DOI] [PubMed] [Google Scholar]
- Gonzalez I, Cao KA, Davis MJ, Dejean S, 2012. Visualising associations between paired ‘omics’ data sets. BioData Min. 5, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasberger H, Chang L, Shih W, Presson AP, Sayuk GS, Newberry RD, Karagiannides I, Pothoulakis C, Mayer E, Merchant JL, 2013. Identification of a functional TPH1 polymorphism associated with irritable bowel syndrome bowel habit subtypes. Am. J. Gastroenterol 108, 1766–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros M, Gros B, Mesonero JE, Latorre E, 2021. Neurotransmitter dysfunction in irritable bowel syndrome: emerging approaches for management. J. Clin. Med 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guleria A, Karyampudi A, Singh R, Khetrapal CL, Verma A, Ghoshal UC, Kumar D, 2017. Mapping of brain activations to rectal balloon distension stimuli in male patients with irritable bowel syndrome using functional magnetic resonance imaging. J Neurogastroenterol Motil 23, 415–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashmi JA, Baliki MN, Huang L, Baria AT, Torbey S, Hermann KM, Schnitzer TJ, Apkarian AV, 2013. Shape shifting pain: chronification of back pain shifts brain representation from nociceptive to emotional circuits. Brain 136, 2751–2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitkemper M, Jarrett M, Bond EF, Chang L, 2003. Impact of sex and gender on irritable bowel syndrome. Biol. Res. Nurs 5, 56–65. [DOI] [PubMed] [Google Scholar]
- Jeffery IB, O’Toole PW, Ohman L, Claesson MJ, Deane J, Quigley EM, Simren M, 2012. An irritable bowel syndrome subtype defined by species-specific alterations in faecal microbiota. Gut 61, 997–1006. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Dinov ID, Labus J, Shi Y, Zamanyan A, Gupta A, Ashe-McNalley C, Hong JY, Tillisch K, Toga AW, Mayer EA, 2013. Sex-related differences of cortical thickness in patients with chronic abdominal pain. PLoS One 8, e73932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keszthelyi D, Troost FJ, Jonkers DM, van Eijk HM, Dekker J, Buurman WA, Masclee AA, 2015. Visceral hypersensitivity in irritable bowel syndrome: evidence for involvement of serotonin metabolism–a preliminary study. Neuro Gastroenterol. Motil 27, 1127–1137. [DOI] [PubMed] [Google Scholar]
- Kim YS, Kim N, 2018. Sex-Gender differences in irritable bowel syndrome. J Neurogastroenterol Motil 24, 544–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong G, Ellul S, Narayana VK, Kanojia K, Ha HTT, Li S, Renoir T, Cao KL, Hannan AJ, 2021. An integrated metagenomics and metabolomics approach implicates the microbiota-gut-brain axis in the pathogenesis of Huntington’s disease. Neurobiol. Dis 148, 105199. [DOI] [PubMed] [Google Scholar]
- Kucyi A, Salomons TV, Davis KD, 2013. Mind wandering away from pain dynamically engages antinociceptive and default mode brain networks. Proc. Natl. Acad. Sci. U. S. A 110, 18692–18697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labus JS, Bolus R, Chang L, Wiklund I, Naesdal J, Mayer EA, Naliboff BD, 2004. The Visceral Sensitivity Index: development and validation of a gastrointestinal symptom-specific anxiety scale. Aliment. Pharmacol. Ther 20, 89–97. [DOI] [PubMed] [Google Scholar]
- Labus JS, Gupta A, Coveleskie K, Tillisch K, Kilpatrick L, Jarcho J, Feier N, Bueller J, Stains J, Smith S, Suyenobu B, Naliboff B, Mayer EA, 2013. Sex differences in emotion-related cognitive processes in irritable bowel syndrome and healthy control subjects. Pain 154, 2088–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labus JS, Mayer EA, Jarcho J, Kilpatrick LA, Kilkens TO, Evers EA, Backes WH, Brummer RJ, van Nieuwenhoven MA, 2011. Acute tryptophan depletion alters the effective connectivity of emotional arousal circuitry during visceral stimuli in healthy women. Gut 60, 1196–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layunta E, Buey B, Mesonero JE, Latorre E, 2021. Crosstalk between intestinal serotonergic system and pattern recognition receptors on the microbiota-gut-brain Axis. Front. Endocrinol 12, 748254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Cao KA, Boitard S, Besse P, 2011. Sparse PLS discriminant analysis: biologically relevant feature selection and graphical displays for multiclass problems. BMC Bioinf. 12, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AH, Shannon CP, Amenyogbe N, Bennike TB, Diray-Arce J, Idoko OT, Gill EE, Ben-Othman R, Pomat WS, van Haren SD, Cao KL, Cox M, Darboe A, Falsafi R, Ferrari D, Harbeson DJ, He D, Bing C, Hinshaw SJ, Ndure J, Njie-Jobe J, Pettengill MA, Richmond PC, Ford R, Saleu G, Masiria G, Matlam JP, Kirarock W, Roberts E, Malek M, Sanchez-Schmitz G, Singh A, Angelidou A, Smolen KK, Consortium E, Brinkman RR, Ozonoff A, Hancock REW, van den Biggelaar AHJ, Steen H, Tebbutt SJ, Kampmann B, Levy O, Kollmann TR, 2019. Dynamic molecular changes during the first week of human life follow a robust developmental trajectory. Nat. Commun 10, 1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, 2016. Gut microbiota in 2015: Prevotella in the gut: choose carefully. Nat. Rev. Gastroenterol. Hepatol 13, 69–70. [DOI] [PubMed] [Google Scholar]
- Lineberry CG, Vierck CJ, 1975. Attenuation of pain reactivity by caudate nucleus stimulation in monkeys. Brain Res. 98, 119–134. [DOI] [PubMed] [Google Scholar]
- Liu X, Mao B, Gu J, Wu J, Cui S, Wang G, Zhao J, Zhang H, Chen W, 2021. Blautia-a new functional genus with potential probiotic properties? Gut Microb. 13, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longstreth GF, Thompson WG, Chey WD, Houghton LA, Mearin F, Spiller RC, 2006. Functional bowel disorders. Gastroenterology 130, 1480–1491. [DOI] [PubMed] [Google Scholar]
- Lopez-Siles M, Aldeguer X, Sabat-Mir M, Serra-Pages M, Duncan SH, Flint HJ, Garcia-Gil LJ, Martinez-Medina M, 2020. Evaluation of bacterial biomarkers to aid in challenging inflammatory bowel diseases diagnostics and subtype classification. World J. Gastrointest. Pathophysiol 11, 64–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou HC, 1994. Dopamine precursors and brain function in phenylalanine hydroxylase deficiency. Acta Paediatr. Suppl 407, 86–88. [DOI] [PubMed] [Google Scholar]
- Mars RAT, Yang Y, Ward T, Houtti M, Priya S, Lekatz HR, Tang X, Sun Z, Kalari KR, Korem T, Bhattarai Y, Zheng T, Bar N, Frost G, Johnson AJ, van Treuren W, Han S, Ordog T, Grover M, Sonnenburg J, D’Amato M, Camilleri M, Elinav E, Segal E, Blekhman R, Farrugia G, Swann JR, Knights D, Kashyap PC, 2020. Longitudinal multi-omics reveals subset-specific mechanisms underlying irritable bowel syndrome. Cell 183, 1137–1140. [DOI] [PubMed] [Google Scholar]
- Mawe GM, Hoffman JM, 2013. Serotonin signalling in the gut-functions, dysfunctions and therapeutic targets. Nat. Rev. Gastroenterol. Hepatol 10, 473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer EA, Labus JS, Tillisch K, Cole SW, Baldi P, 2015. Towards a systems view of IBS. Nat. Rev. Gastroenterol. Hepatol 12, 592–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer EA, Naliboff BD, Craig AD, 2006. Neuroimaging of the brain-gut axis: from basic understanding to treatment of functional GI disorders. Gastroenterology 131, 1925–1942. [DOI] [PubMed] [Google Scholar]
- Mayer EA, Savidge T, Shulman RJ, 2014. Brain-gut microbiome interactions and functional bowel disorders. Gastroenterology 146, 1500–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon V., 2015. Salience network. In: Toga A (Ed.), Brain Mapping: an Encyclopedia Reference. Academic Press: Elsevier, pp. 597–611. [Google Scholar]
- Nieto-Castanon A, 2020. Handbook of Functional Connectivity Magnetic Resonance Imaging Methods in CONN. Hilbert Press, Boston, Massachusetts. [Google Scholar]
- Osadchiy V, Labus JS, Gupta A, Jacobs J, Ashe-McNalley C, Hsiao EY, Mayer EA, 2018. Correlation of tryptophan metabolites with connectivity of extended central reward network in healthy subjects. PLoS One 13, e0201772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osadchiy V, Mayer EA, Gao K, Labus JS, Naliboff B, Tillisch K, Chang L, Jacobs JP, Hsiao EY, Gupta A, 2020. Analysis of brain networks and fecal metabolites reveals brain-gut alterations in premenopausal females with irritable bowel syndrome. Transl. Psychiatry 10, 367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandiyan P, Bhaskaran N, Zou M, Schneider E, Jayaraman S, Huehn J, 2019. Microbiome dependent regulation of tregs and Th17 cells in mucosa. Front. Immunol 10, 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Naliboff BD, Shih W, Presson AP, Videlock EJ, Ju T, Kilpatrick L, Gupta A, Mayer EA, Chang L, 2018. Resilience is decreased in irritable bowel syndrome and associated with symptoms and cortisol response. Neuro Gastroenterol. Motil 30, e13155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkes GC, Brostoff J, Whelan K, Sanderson JD, 2008. Gastrointestinal microbiota in irritable bowel syndrome: their role in its pathogenesis and treatment. Am. J. Gastroenterol 103, 1557–1567. [DOI] [PubMed] [Google Scholar]
- Pozuelo M, Panda S, Santiago A, Mendez S, Accarino A, Santos J, Guarner F, Azpiroz F, Manichanh C, 2015. Reduction of butyrate- and methane-producing microorganisms in patients with Irritable Bowel Syndrome. Sci. Rep 5, 12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glockner FO, 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohart F, Gautier B, Singh A, Le Cao KA, 2017. mixOmics: an R package for ’omics feature selection and multiple data integration. PLoS Comput. Biol 13, e1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeley WW, Menon V, Schatzberg AF, Keller J, Glover GH, Kenna H, Reiss AL, Greicius MD, 2007. Dissociable intrinsic connectivity networks for salience processing and executive control. J. Neurosci 27, 2349–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seres G, Kovacs Z, Kovacs A, Kerekgyarto O, Sardi K, Demeter P, Meszaros E, Tury F, 2008. Different associations of health related quality of life with pain, psychological distress and coping strategies in patients with irritable bowel syndrome and inflammatory bowel disorder. J. Clin. Psychol. Med. Settings 15, 287–295. [DOI] [PubMed] [Google Scholar]
- Singh A, Shannon CP, Gautier B, Rohart F, Vacher M, Tebbutt SJ, Le Cao KA, 2019. DIABLO: an integrative approach for identifying key molecular drivers from multi-omics assays. Bioinformatics 35, 3055–3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T, Liu R, Lee A, Long Y, Du L, Lai S, Chen X, Wang L, Si J, Owyang C, Chen S, 2018. Altered intestinal microbiota with increased abundance of Prevotella is associated with high risk of diarrhea-predominant irritable bowel syndrome. Gastroenterol Res Pract 2018, 6961783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talley NJ, Boyce PM, Owen BK, Newman P, Paterson KJ, 1995. Initial validation of a bowel symptom questionnaire and measurement of chronic gastrointestinal symptoms in Australians. Aust. N. Z. J. Med 25, 302–308. [DOI] [PubMed] [Google Scholar]
- Tenenhaus A, Philippe C, Guillemot V, Le Cao KA, Grill J, Frouin V, 2014. Variable selection for generalized canonical correlation analysis. Biostatistics 15, 569–583. [DOI] [PubMed] [Google Scholar]
- Tong M, Jacobs JP, McHardy IH, Braun J, 2014. Sampling of intestinal microbiota and targeted amplification of bacterial 16S rRNA genes for microbial ecologic analysis. Curr. Protoc. Im 107, 7 41 41–47 41 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahora IS, Tsouklidis N, Kumar R, Soni R, Khan S, 2020. How serotonin level fluctuation affects the effectiveness of treatment in irritable bowel syndrome. Cureus 12, e9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenbacher A, Kasess C, Gerstl F, Lanzenberger R, Moser E, Windischberger C, 2009. Correlations and anticorrelations in resting-state functional connectivity MRI: a quantitative comparison of preprocessing strategies. Neuroimage 47, 1408–1416. [DOI] [PubMed] [Google Scholar]
- Whitfield-Gabrieli S, Nieto-Castanon A, 2012. Conn: a functional connectivity toolbox for correlated and anticorrelated brain networks. Brain Connect. 2, 125–141. [DOI] [PubMed] [Google Scholar]
- Wilder-Smith CH, Schindler D, Lovblad K, Redmond SM, Nirkko A, 2004. Brain functional magnetic resonance imaging of rectal pain and activation of endogenous inhibitory mechanisms in irritable bowel syndrome patient subgroups and healthy controls. Gut 53, 1595–1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KQ, Sun WJ, Li N, Chen YQ, Wei YL, Chen DF, 2019. Small intestinal bacterial overgrowth is associated with Diarrhea-predominant irritable bowel syndrome by increasing mainly Prevotella abundance. Scand. J. Gastroenterol 54, 1419–1425. [DOI] [PubMed] [Google Scholar]
- Wunderlich AP, Klug R, Stuber G, Landwehrmeyer B, Weber F, Freund W, 2011. Caudate nucleus and insular activation during a pain suppression paradigm comparing thermal and electrical stimulation. Open Neuroimaging J. 5, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glockner FO, 2014. The SILVA and "All-species living tree project (LTP)" taxonomic frameworks. Nucleic Acids Res. 42, D643–D648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Cai W, Zheng J, Li G, Meng Q, Liu Q, Zhao J, von Deneen KM, Wang Y, Cui G, Duan S, Han Y, Wang H, Tian J, Zhang Y, Nie Y, 2016. Distinct resting-state brain activity in patients with functional constipation. Neurosci. Lett 632, 141–146. [DOI] [PubMed] [Google Scholar]
- Zhu X, Hong G, Li Y, Yang P, Cheng M, Zhang L, Li Y, Ji L, Li G, Chen C, Zhong C, Jin Y, Yang M, Xiong H, Qian W, Ding Z, Ning K, Hou X, 2021. Understanding of the site-specific microbial patterns towards accurate identification for patients with diarrhea-predominant irritable bowel syndrome. Microbiol. Spectr 9, e0125521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond AS, Snaith RP, 1983. The hospital anxiety and depression scale. Acta Psychiatr. Scand 67, 361–370. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.