Abstract
Polyhydroxyalkanoates (PHAs) are a class of carbon and energy storage polymers produced by numerous bacteria in response to environmental limitation. The type of polymer produced depends on the carbon sources available, the flexibility of the organism’s intermediary metabolism, and the substrate specificity of the PHA biosynthetic enzymes. Ralstonia eutropha produces both the homopolymer poly-β-hydroxybutyrate (PHB) and, when provided with the appropriate substrate, the copolymer poly(β-hydroxybutyrate-co-β-hydroxyvalerate) (PHBV). A required step in production of the hydroxyvalerate moiety of PHBV is the condensation of acetyl coenzyme A (acetyl-CoA) and propionyl-CoA to form β-ketovaleryl-CoA. This activity has generally been attributed to the β-ketothiolase encoded by R. eutropha phbA. However, we have determined that PhbA does not significantly contribute to catalyzing this condensation reaction. Here we report the cloning and genetic analysis of bktB, which encodes a β-ketothiolase from R. eutropha that is capable of forming β-ketovaleryl-CoA. Genetic analyses determined that BktB is the primary condensation enzyme leading to production of β-hydroxyvalerate derived from propionyl-CoA. We also report an additional β-ketothiolase, designated BktC, that probably serves as a secondary route toward β-hydroxyvalerate production.
Polyhydroxyalkanoates (PHAs) are a class of naturally occurring polymers which serve as a carbon and energy reserve in numerous bacterial species. Ralstonia eutropha (formerly designated Alcaligenes eutrophus [41]) produces the homopolymer poly(β-hydroxybutyrate) (PHB) and, when provided with propionate in the feedstock, the copolymer poly(β-hydroxybutyrate-co-β-hydroxyvalerate) (PHBV). R. eutropha is used commercially to produce PHBV, which is a biodegradable thermoplastic.
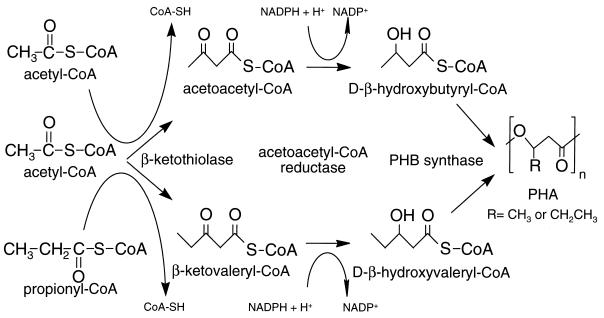
The PHB biosynthetic pathway requires three enzymatic activities: a β-ketothiolase (PhbA), an NADPH-dependent acetoacetyl coenzyme A (acetoacetyl-CoA) reductase (PhbB) and a PHB synthase (PhbC). The first step in production of the homopolymer PHB is catalyzed by β-ketothiolase which condenses two acetyl-CoA molecules to form acetoacetyl-CoA. Formation of the copolymer PHBV requires the additional condensation of acetyl-CoA with propionyl-CoA to form β-ketovaleryl-CoA (Fig. 1). Subsequently, the acetoacetyl-CoA and β-ketovaleryl-CoA are converted into a polymer by the activities of the reductase and synthase. The genes encoding these proteins in R. eutropha reside in an operon which has been well characterized (10, 21, 22, 31, 37).
FIG. 1.
Pathway for production of PHBV from acetyl-CoA and propionyl-CoA. β-Ketothiolase performs the condensation reactions to generate either acetoacetyl-CoA or β-ketovaleryl-CoA. These are reduced by acetoacetyl-CoA reductase (PhbB) and polymerized by PHB synthase (PhbC).
The substrate specificities of these three enzymes are reportedly adequate for production of PHBV copolymer (7–9), but propionate-fed Escherichia coli harboring the R. eutropha phb operon produces essentially PHB homopolymer (35). Moreover, PHBV copolymer can be produced in E. coli after induction of the fatty acid β-oxidation complex, which contains a β-ketothiolase with broad substrate specificity (26, 27, 35). These data suggest that the R. eutropha PHB pathway is capable of producing copolymer, but only in the context of a second β-ketothiolase with broad substrate specificity.
R. eutropha is known to produce at least two β-ketothiolases (7), and at least two distinct plasmid clones which express β-ketothiolase have been isolated from R. eutropha (37). In this work, we analyzed the substrate specificity of the PhbA β-ketothiolase and demonstrated that this enzyme catalyzes thiolysis of β-ketovaleryl-CoA very poorly. We determined that R. eutropha expresses at least two β-ketothiolases in addition to PhbA and that these additional enzymes, which we designate BktB and BktC, efficiently utilize β-ketovaleryl-CoA. We also report the isolation and characterization of bktB (β-ketothiolase B), which encodes the BktB β-ketothiolase required for efficient production of PHBV in R. eutropha.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains utilized in this study are summarized in Table 1. Luria-Bertani (LB) medium contained 10 g of tryptone (Difco), 5 g of yeast extract (Difco), and 5 g of NaCl. All plates contained 1.5% Bacto Agar (Difco). LB-sucrose plates used to select against pJQ200SK and pLO2 derivatives had no NaCl and contained 5% sucrose. M9 minimal medium was prepared according to the method of Miller (18). R. eutropha was grown in Schlegel’s minimal medium (29) containing 0.1% nutrient broth powder (Difco), with the exception that Hoagland’s trace element solution was replaced with SL6 trace element solution (23). Schlegel’s nitrogen-complete (SNC) medium also contained 1 g of NH4Cl per liter, whereas Schlegel’s nitrogen-limited (SNL) medium also contained 0.1 g of NH4Cl per liter. Antibiotics were used at the following levels: E. coli, carbenicillin, 100 μg/ml; chloramphenicol, 25 μg/ml; and kanamycin, 50 μg/ml; R. eutropha, chloramphenicol, 100 μg/ml for broth and 100 to 250 μg/ml for plates; kanamycin, 350 μg/ml; and gentamicin, 10 μg/ml.
TABLE 1.
E. coli and R. eutropha strains used in this study
| Strain | Genotype or description | Source or reference |
|---|---|---|
| E. coli | ||
| DH5α | supE44 Δ(lacZYA-argF)U169 (φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 deoR | 28 |
| LE392 | supE44 supF58 hsdR514 galK2 galT22 metB1 trpR55 lacY1 | 28 |
| S17-1 | C600: recA thi pro hsdR RP-4 2-tet::Mu::kan::Tn7 | 34 |
| EE32 | LE392 harboring pBK6 | D. Dennis |
| EE245 | DH5α harboring pMON25982 | This study |
| EE247 | DH5α harboring pMON25984 | This study |
| R. eutropha | ||
| EE167 | R. eutropha DSM428 (wild type) | Deutsche Sammlung von Mikroorganismen, Göttingen, Germany |
| EE168 | bktB::kan | This study |
| EE169 | bktB::kan | This study |
| EE234 | EE167 harboring pBBR1MCS | This study |
| EE235 | EE167 harboring pMON25901 | This study |
| EE236 | EE168 harboring pBBR1MCS | This study |
| EE237 | EE168 harboring pMON25901 | This study |
| EE238 | EE169 harboring pBBR1MCS | This study |
| EE239 | EE169 harboring pMON25901 | This study |
| EE303 | ΔphbA | This study |
| EE317 | ΔphbA bktB::kan | This study |
Cultures for polymer analysis were grown in 25 ml of the appropriate medium in 250-ml flasks and shaken at 220 rpm for the desired time. Each flask was inoculated with 0.5 ml of a separate 5-ml seed culture containing the appropriate antibiotics. R. eutropha seed cultures were grown in SNC medium containing 0.4% fructose, and E. coli seed cultures were grown in M9 medium containing 0.2% glucose. E. coli was incubated at 37°C, and R. eutropha was incubated at 30°C.
For polymer production, R. eutropha was grown in SNL medium containing 3% fructose and 0.1% sodium propionate. At 24-h intervals, each culture was supplemented with 1 ml of 2.5% sodium propionate, and the cells were harvested 72 h after inoculation.
For polymer production in E. coli, strains were grown in M9 medium containing 1% glucose, the desired amount of sodium propionate, and 100 μg of carbenicillin where appropriate. Cultures were incubated for approximately 40 h prior to harvesting.
DNA manipulations.
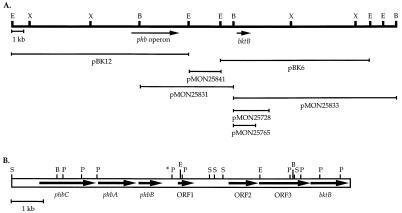
The plasmids utilized in this study are described in Table 2. All cloning was performed by standard procedures (28). The location of bktB was mapped within plasmid pBK6 (37) by sequential deletion of restriction fragments (Fig. 2). Plasmid pMON25728 contains bktB on a 3.9-kb DNA fragment extending from a BglII site about 0.5 kb upstream of bktB to an NcoI site approximately 2 kb downstream of bktB. pMON25765 contains a DNA fragment extending from the aforementioned BglII site to a SalI site 98 bp downstream of bktB, and the R. eutropha DNA is flanked by HindIII sites. Plasmid pMON25901 harbors the bktB-containing HindIII fragment of pMON25765 cloned into the HindIII site of pBBR1MCS (13). Plasmid pMON25831 harbors DNA extending from the BglII site just upstream of bktB to the BglII site within the phb operon.
TABLE 2.
Plasmids used in this study
| Plasmid | Descriptiona | Reference |
|---|---|---|
| pBluescript KS(+) | General-utility E. coli cloning vector, Ampr | Stratagene |
| pBBR1MCS | Broad-host-range cloning vector, Camr | 13 |
| pSE280 | E. coli expression vector | 1 |
| pJQ200SK | Suicide vector for gene replacement, Genr | 25 |
| pLO2 | Suicide vector for gene replacement, Kanr | 15 |
| pAE175 | R. eutropha phbCAB-bktB region harbored in cosmid pVK102 | 37 |
| pBK6 | pUC13 harboring 12-kb EcoRI fragment from pAE175, bktB+ | 37 |
| pPHAΔA | R. eutropha phbCB in pBBR1MCS | D. Dennis |
| pMON25636 | R. eutropha phbA expressed from trc promoter of pSE280 | This study |
| pMON25728 | 3.9-kb BglII-NcoI subclone of pBK6 in pBluescript KS(+), bktB+ | This study |
| pMON25742 | bktB::kan derivative of pMON25728 | This study |
| pMON25765 | 1.8-kb BglII-SalI subclone of pBK6 in pBluescript KS(+), bktB+ | This study |
| pMON25776 | bktB::kan harbored in pJQ200SK | This study |
| pMON25845 | Camr Gens derivative of pMON25776 | This study |
| pMON25901 | 1.8-kb bktB+ fragment from pMON25765 cloned into pBBR1MCS | This study |
| pMON25966 | R. eutropha phbA under control of bktB promoter in pBluescript KS(+) | This study |
| pMON25968 | R. eutropha phbCB and bktB genes in pBBR1MCS | This study |
| pMON25970 | R. eutropha phbCB and phbA genes in pBBR1MCS | This study |
| pMON25982 | R. eutropha phbCB and bktB genes in pBluescript KS(+) | This study |
| pMON25984 | R. eutropha phbCB and phbA genes in pBluescript KS(+) | This study |
| pMON36818 | ΔphbA harbored in pLO2 | This study |
Resistance abbreviations are as follows: Ampr, ampicillin resistant; Camr, chloramphenicol resistant; Genr, gentamicin resistant; Kanr, kanamycin resistant.
FIG. 2.
(A) Restriction map of the region surrounding the phb operon and bktB in pAE175 (37) and the regions of DNA harbored by plasmid vectors pertinent to this work. B, BglII; E, EcoRI; X, XhoI. Beyond the phbCAB-bktB region, the structure of pBK6 does not accurately represent that of the R. eutropha chromosome. (See “DNA sequencing and chromosomal analysis” in Materials and Methods for details.) (B) Restriction map and locations of ORFs within the phbCAB-bktB chromosomal region of R. eutropha. The PstI site marked with an asterisk is the termination site of the previously published sequence (21) and the initiation site of the sequence reported in this work. P, PstI; S, SmaI.
Plasmid pMON25636 harbors a derivative of phbA in which an NcoI site has been engineered via PCR to overlap the initiation ATG codon and a HindIII site has been engineered immediately downstream of phbA. Plasmid pMON25966 contains the phbA NcoI-HindIII fragment of pMON25636 fused to the bktB promoter of pMON25765 at the BspHI site that overlaps the bktB initiation codon in the native gene. The resulting construct contains phbA under bktB promoter control, and the entire gene is flanked by HindIII sites.
Plasmids used for PHA production in E. coli were constructed with the HindIII fragments from pMON25765 and pMON25966 cloned into plasmid pPHAΔA. Plasmid pPHAΔA (obtained from D. Dennis) contains the R. eutropha phb operon from plasmid pJM9238 (11) in pBBR1MCS, but with a deletion of a 995-bp StuI fragment. The deletion inactivates phbA, but both phbC and phbB are expressed. The β-ketothiolase-containing HindIII fragments of pMON25765 and pMON25966 were cloned into the unique HindIII site of pPHAΔA (located upstream of the phb promoter), and the orientation of the insert was determined. Plasmids pMON25968 and pMON25970 harbor the resulting bktB and phbA constructs, respectively, oriented such that the phbCB and bktB promoters are adjacent and divergent. Plasmids pMON25982 and pMON25984 were constructed by excising the entire PHA production cassette from pMON25968 and pMON25970, respectively, with EcoRI and XhoI and cloning the resulting fragments into the EcoRI-XhoI sites of pBluescript KS(+) (Stratagene).
Plasmid pMON25742 (bktB::kan) contains the end-filled 1.7-kb Kanr BamHI fragment of Tn10kan (12) cloned in the end-filled NcoI site within bktB of pMON25728. Plasmid pMON25776 contains an approximately 5-kb ApaI fragment harboring bktB::kan from pMON25742 cloned in the ApaI site of pJQ200SK (25). Plasmid pMON25845 contains a Camr BamHI fragment of pFF589 (provided by R. Maurer) cloned into the BamHI site within the gentamicin resistance gene of pMON25776.
The ΔphbA allele used for disruption of phbA in R. eutropha is the same allele harbored by plasmid pPHAΔA (described above [Table 2]). The particular construct used for deletion of phbA in R. eutropha was derived from pMON25971, which is identical to pMON25970 (described above), except that the HindIII fragment harboring phbA is in the opposite orientation. This orientation places an XbaI site upstream of phbC, with the result that the entire ΔphbA allele is flanked by XbaI sites. The XbaI fragment harboring phbC-ΔphbA-phbB was ligated into the XbaI site of pLO2 (15) to create pMON36818, which was used for disruption of phbA.
DNA sequencing and chromosomal analysis.
Sequencing of the region from the PstI site downstream of phbB through bktB was accomplished by a combination of subcloning and primer walking with both automated and manual sequencing. Automated sequencing utilized the ABI-Prism sequencing kit with AmpliTaq FS DNA polymerase (Perkin-Elmer), and manual sequencing used Sequenase DNA polymerase (U.S. Biochemicals). Sequence analysis was performed with the Wisconsin Package of molecular analysis software (Genetics Computer Group, Madison, Wis.) and the Sequencher DNA sequencing software package (Gene Codes Corporation).
The chromosomal structure of the R. eutropha phbCAB-bktB region was determined by Southern blot hybridization to match the structure identified by DNA sequencing (Fig. 2 and 3). However, these analyses also determined that a region of pAE175 is the product of a cloning artifact. Specifically, the bktB-containing plasmid pBK6 (derived from pAE175) harbors an EcoRI fragment of approximately 12 kb, whereas bktB from the R. eutropha chromosome is harbored on an EcoRI fragment of approximately 4 kb. Two other cosmid clones from R. eutropha, pAE537 and pAE683 (37), also harbor bktB on a 4-kb EcoRI fragment. Additional Southern blots determined that the pBK6 sequence diverges from the wild-type sequence immediately downstream from bktB, at the SalI site at bp 5592 of Fig. 3. Since pAE175 was constructed with a partial SalI digestion of R. eutropha chromosomal DNA, it appears that the phbCAB-bktB region was linked to another portion of the R. eutropha chromosome during the ligation reaction.
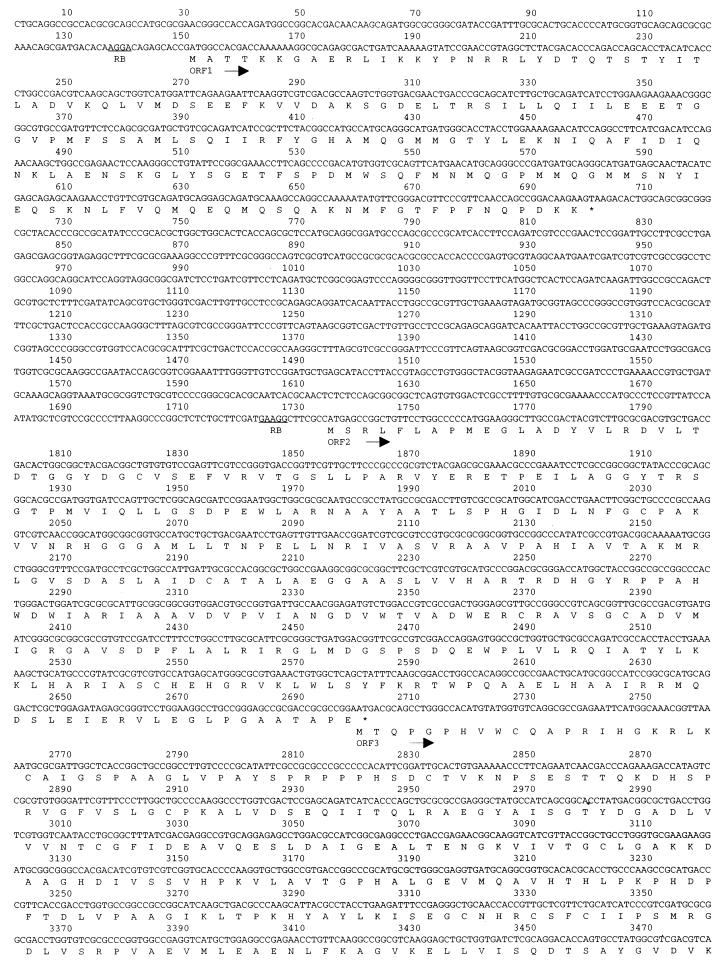
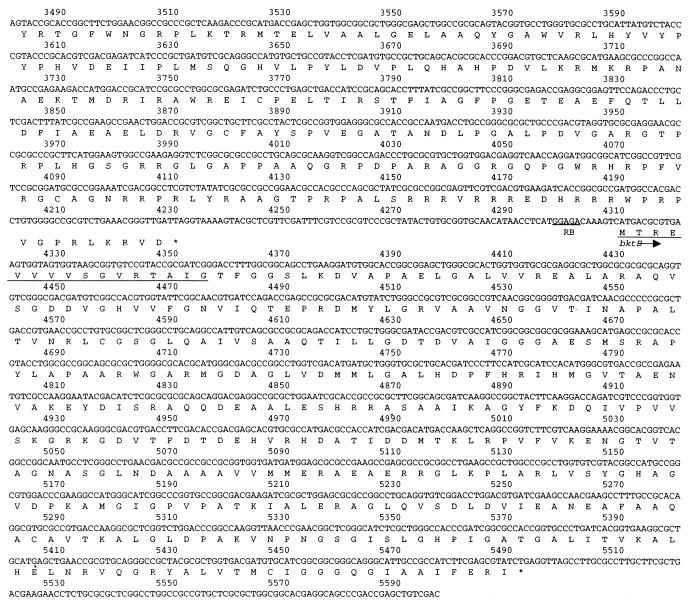
FIG. 3.
Nucleotide sequence and predicted translation products of the R. eutropha chromosomal region downstream of the phb operon through bktB. The sequence shown initiates from the PstI site downstream of phbB in the published phbAB sequence (21). Possible ribosome-binding (RB) sequences upstream of each ORF are underlined. The predicted amino acids underlined within the bktB ORF correspond precisely to those determined by N-terminal sequence analysis of BktB.
Disruption of bktB and phbA.
For disruption of bktB, E. coli S17-1 (34) was transformed with pMON25845 (Table 2), and the resulting strain was mated overnight at 30°C with wild-type R. eutropha H16 (strain EE167) on LB plates containing 0.2% fructose. Exconjugants were purified on NaCl-free LB agar containing 350 μg of kanamycin per ml, 10 μg of gentamicin per ml, 5% sucrose, and 0.2% fructose. Following 3 days of growth at 30°C, several colonies were twice purified and then checked for the absence of BktB by immunoblotting, and the bktB genotype was confirmed by Southern blot hybridization. Strains EE168 and EE169 were isolated from two independent mating events (Table 1).
For disruption of phbA in R. eutropha, E. coli S17-1 (34) was transformed with pMON36818 (Table 2), and the resulting strain was mated overnight at 30°C with wild-type R. eutropha H16 (strain EE167) on LB plates containing 0.2% fructose. Exconjugants were twice purified on LB medium containing 0.2% fructose and 350 μg of kanamycin per ml and then purified on NaCL-free LB medium containing 5% sucrose and 0.2% fructose. Several kanamycin-sensitive strains having reduced β-ketothiolase activity were further purified, and the phbA deletion was verified by Southern blot hybridization. One strain, EE303 (Table 1), was chosen for further analysis.
Strain EE317, a phbA bktB strain (Table 1), was produced by integrating pMON25845 in EE303 and then isolating a bktB::kan derivative as described above for construction of strains EE168 and EE169. The genotype of EE317 was confirmed by Southern blotting.
Complementation of bktB in R. eutropha.
Mobilization of pBBR1MCS and pMON25901 (Table 2) into R. eutropha was performed by mating E. coli S17-1 (34) harboring the appropriate plasmid with the appropriate R. eutropha recipient strain on LB plates containing 0.2% fructose. Exconjugants were purified for isolation on LB plates containing 250 μg of chloramphenicol per ml, 10 μg of gentamicin per ml, and 0.2% fructose, and the plates were incubated for 4 days at 30°C. Surviving colonies were twice purified on LB-fructose plates containing 100 μg of chloramphenicol per ml. Plasmids were purified from each strain, and their structures were verified by digestion with a variety of restriction endonucleases.
Purification of BktB and PhbA.
All protein concentrations were determined by the method of Bradford (3) with reagents obtained from Bio-Rad Laboratories. Bovine serum albumin (obtained from Sigma) was used as a protein standard.
For purification of BktB, E. coli EE32 (Table 1) was grown in a 2-liter Bioflow III fermentor (New Brunswick Scientific) at 37°C for 16 h in Terrific Broth (18) containing carbenicillin. Cells were harvested by centrifugation, washed once with phosphate-buffered saline (28), and frozen at −20°C until lysis. This produced approximately 100 ml of concentrated cell paste.
Approximately 30 ml of cell paste was resuspended in 90 ml of lysis buffer [10 mM Tris (pH 8.0), 1 mM dithiothreitol (DTT), 2 mM 4-(2-aminoethyl)benzenesulfonyl fluoride (AEBSF)] and the cells were disrupted by sonication. The lysate was cleared by centrifugation for 10 min at 31,000 × g in a Beckman SA-17 rotor. Protein was precipitated from the cleared lysate (approximately 100 ml) with 80% saturated ammonium sulfate, and the pellet was redissolved in 50 ml of 1 M ammonium sulfate. The solution was passed through a 0.2-μm-pore-diameter filter and then was loaded onto a phenyl-Sepharose fast-performance liquid chromatography (FPLC) column (1 by 10 cm) (Pharmacia). Buffer A was 1 mM DTT–20 mM sodium phosphate (pH 7.0), and buffer B was 1 mM DTT–20 mM sodium phosphate (pH 7.0) containing 1 M ammonium sulfate. The gradient was 100 to 20% buffer B in 40 min and then 20 to 0% buffer B in 20 min. The flow rate was 2 ml per min, and one fraction was collected per minute. Fractions 42 to 51, which contained most of the β-ketothiolase activity and which were pooled, were concentrated to 5 ml with a Centriprep-10 (Amicon, Inc.), desalted with a PD-10 column (Pharmacia), passed through a 0.2-μm-pore-diameter filter, and further separated on a Mono-Q HR5/5 FPLC column (Pharmacia). Buffer C was 1 mM DTT–10 mM Tris (pH 7.8), and buffer D was 1 mM DTT–10 mM Tris (pH 7.8) containing 1 M KCl. The gradient was 0 to 10% buffer D in 10 min, 10 to 35% buffer D in 40 min, and then 35 to 100% buffer D in 20 min. The flow rate was 1 ml per min, and 1 fraction was collected per minute. Most BktB eluted in fractions 18 through 24. These fractions were pooled, and the pooled material was at least 95% pure as estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Pro-Blue staining (Daiichi). The final solution (6 ml containing 1.6 mg of protein/ml) contained BktB at 65 U/mg (assayed with acetoacetyl-CoA). This material was used for kinetic analyses and to generate antisera against BktB.
Material from a separate, smaller-scale purification utilizing the same frozen cell paste was used for N-terminal sequence analysis. In this case, fraction 23 from the final Mono-Q chromatography step was concentrated in a Centricon-10 column and washed several times with 10 mM NaHCO3 buffer to remove Tris buffer. The final material (0.2 ml at 0.68 mg/ml) was directly N-terminally sequenced by the Monsanto protein analysis laboratory.
PhbA was purified from E. coli DH5α harboring pMON25636, a plasmid in which R. eutropha phbA is under control of an IPTG (isopropyl-β-d-thiogalactopyranoside)-inducible promoter (Table 2). A 200-ml shaken-flask culture of this strain was grown to mid-log phase (Klett reading of 45 units) at 37°C in LB medium containing carbenicillin, and then cells were induced by adding IPTG to 0.5 mM. After 2 more h of growth, the cells were harvested by centrifugation and frozen until use. Cells were disrupted by sonication, and the protein was purified as described above, except that the phenyl-Sepharose column was omitted. PhbA elutes from Mono-Q earlier than BktB, with the peak activity occurring in fractions 12 and 13 of this preparation. These two 1-ml fractions were combined, providing a final solution containing 0.6 mg of total protein per ml. The material was at least 95% pure, as estimated by SDS-PAGE and Pro-Blue staining (Daiichi).
Substrate synthesis and β-ketothiolase enzyme assays.
Preparation of β-ketoacyl-CoA substrate precursors trans-2,3-pentenoyl-CoA and trans-2,3-hexenoyl-CoA was accomplished by the procedure of Schulz (32), essentially as described previously, except on a larger scale (100 μmol of starting trans-2,3-pentenoic or trans-2,3-hexenoic acid and 100 μmol of CoA). Another modification to the procedure was the purification of the enoyl-CoA products by semi-prep C8 reverse-phase chromatography.
β-Ketothiolase was assayed in the thiolysis direction by two methods, depending on whether the substrate was acetoacetyl-CoA (method 1) or β-ketovaleryl-CoA or β-ketohexanoyl-CoA (method 2). Method 1 is similar to that described previously (19) and monitors the disappearance of acetoacetyl-CoA (40 μM) spectrophotometrically at 304 nm in a mixture of 150 mM EPPS (N-[2-hydroxyethyl]piperazine-N′-[3-propanesulfonic acid] (pH 8.0) and 50 mM MgCl2. CoA was used at 100 μM, and the total reaction volume was 1.0 ml. The reaction was initiated by the addition of enzyme. Method 2 is principally the same; however, β-ketovaleryl-CoA or β-ketohexanoyl-CoA was generated in situ by quantitative conversion of the respective enoyl-CoA by using crotonase (3 U), β-hydroxyacyl-CoA dehydrogenase (5 U), lactate dehydrogenase (10 U), 1.5 mM pyruvate, and 0.4 mM NAD+ in a reaction similar to that described by Haywood et al. (7). Enoyl-CoA at a concentration of 40 μM, 100 μM CoA, and the buffering system described above were used for this study. The extinction coefficients for acetoacetyl-CoA, β-ketovaleryl-CoA, and β-ketohexanoyl-CoA at 304 nm were 19.5, 12.2, and 14.0 mM−1 cm−1, respectively, in this buffer system (36) and were used for all activity calculations. The two β-ketothiolase assay methods gave equivalent results, as demonstrated by comparing the results obtained with the in situ generation of acetoacetyl-CoA from crotonyl-CoA (method 2) with results obtained with acetoacetyl-CoA directly (method 1 [36]). One unit of enzyme activity is defined as the amount of enzyme required to convert 1 μmol of β-ketoacyl-CoA to product per min at 25°C.
Polymer analysis.
Cells from each culture were harvested by centrifugation, washed once with absolute ethanol, and then dried overnight at 70°C. The dried pellets were weighed, and then polymer was extracted by immersion of the cell powder in 5 ml of chloroform and heating at 100°C for 5 h. After cooling, the chloroform was filtered through glass wool and collected in glass tubes. The chloroform was evaporated, and the polymer was removed and weighed.
Polymer composition was analyzed by methanolysis of the polymer, followed by gas chromatography of the methyl ester residues. In a glass, screw-cap tube, 5 mg of polymer sample was dissolved in 1 ml of chloroform containing 3 μmol of benzoate per ml as an internal standard. One milliliter of 15% sulfuric acid in methanol was added, and then the tubes were tightly capped and incubated at 100°C for 2.5 h. The tubes were cooled to room temperature and then placed on ice, and the solution was extracted with 0.5 ml of water to remove sulfuric acid. The organic layer was transferred to a clean tube containing approximately 200 mg of sodium sulfate as a drying agent, the mixture was mixed with a Vortex for approximately 1 min, and then solids were removed by centrifugation. The supernatant solution was transferred to vials for chromatography.
Gas chromatography was performed according to a variation of the method of Braunegg et al. (4). The gas chromatograph was a Varian 3400 equipped with a Varian 8200 autosampler, a 1077 split/splitless injector, a 1-ml Hamilton gas-tight syringe, a J & W DB-5 capillary column (0.25 mm by 30 m; 0.25-μm film thickness), and a flame ionization detector. The flow rate of helium carrier gas was 2.5 ml per min, with a 50:1 sample/split ratio at injection. The initial column temperature of 60°C was held for 3 min, and then the temperature was increased by 10°C per min up to 240°C. Finally, the column temperature was raised by 30°C per min to 300°C and held for 10 min. Standards were methyl-(d)-hydroxybutyrate and methyl-(d)-hydroxyvalerate (Fluka, Ronkonkoma, N.Y.).
Nucleotide sequence accession number.
The GenBank accession number for the DNA sequence reported herein is AF026544.
RESULTS
Purification and enzymatic characterization of BktB and PhbA.
The R. eutropha genes sufficient for PHB biosynthesis were previously isolated on cosmid pAE175 (37). Subcloning of pAE175 identified two distinct EcoRI fragments, harbored in plasmids pBK12 and pBK6, that express β-ketothiolase activity. Plasmid pBK12 harbors the phb operon, which includes the phbA β-ketothiolase gene. Plasmid pBK6 carries a second, distinct β-ketothiolase gene that we have designated bktB (β-ketothiolase B) of R. eutropha.
To compare the kinetic properties of the β-ketothiolases, recombinant E. coli strains were used to produce BktB and PhbA (Materials and Methods). The proteins were purified, and the thiolysis reaction was performed with various substrates (Table 3). Both enzymes efficiently catalyze the thiolysis of acetoacetyl-CoA. However, the specific activity of PhbA is approximately 150-fold lower when β-ketovaleryl-CoA is the substrate, and no thiolysis by PhbA of β-ketohexanoyl-CoA was detected. In contrast, BktB prefers β-ketovaleryl-CoA and also efficiently catalyzes the thiolysis of β-ketohexanoyl-CoA. We infer from the thiolysis data that PhbA and BktB can also catalyze the condensation reactions, and in a separate series of experiments, we have confirmed the ability of these enzymes to catalyze the expected condensations (36).
TABLE 3.
Thiolytic cleavage by PhbA and BktB
| β-Ketothiolase | β-Ketothiolase sp act (U/mg) for substratea:
|
||
|---|---|---|---|
| C4 | C5 | C6 | |
| PhbA | 162 | 1.2 | NDb |
| BktB | 65 | 177 | 87 |
One unit is equal to 1 μmol of substrate degraded per min. C4, acetoacetyl-CoA; C5, β-ketovaleryl-CoA; C6, β-ketohexanoyl-CoA.
ND, no activity was detected.
Subcloning and sequencing of bktB and surrounding DNA.
Starting with plasmid pBK6, serial subcloning experiments isolated bktB on pMON25765, which harbors a 1.8-kb fragment extending from the BglII site upstream of bktB to a SalI site just downstream of bktB. Additional overlapping clones derived from pAE175 linked bktB to the phb operon (Fig. 2A).
The entire region from bktB to the phb operon was sequenced. A map of this region appears in Fig. 2, and the DNA sequence appears in Fig. 3. This sequence is contiguous with the previously published sequence of phbAB of R. eutropha (10, 21 [GenBank accession no. J04987]). The phb operon lies approximately 4.6 kb upstream of bktB. The 16 N-terminal amino acids predicted by the bktB sequence match those identified by amino acid sequencing of purified BktB protein (Materials and Methods). bktB is approximately 66% identical to phbA at the nucleotide level, and the two encoded proteins are 53% identical and 61% similar (Fig. 4). BktB contains 394 amino acid residues and has a calculated molecular mass of 40,903 Da.
FIG. 4.
Comparison of the predicted amino acid sequences of PhbA (top) and BktB (bottom) β-ketothiolase proteins from R. eutropha. The two proteins are 51% identical and 61% similar, with similarity extending across the entire sequence. The cysteine residues designated by arrows are the absolutely conserved active site cysteines (20, 39). Comparison was performed with the Gap program of the Wisconsin Package.
The phbCAB-bktB intergenic region contains three additional open reading frames (ORFs) that are likely to be expressed (Fig. 2B and 3). All three ORFs show significant homology to putative genes from other organisms, but none of these putative genes has been assigned a function (see Discussion).
bktB can complement phbA for PHA production in E. coli.
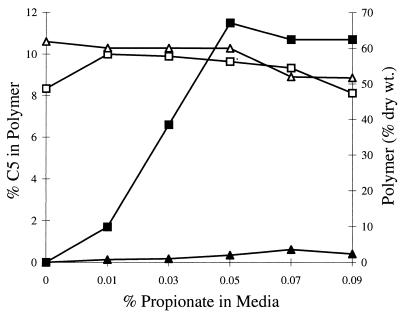
The abilities of PhbA and BktB to produce PHA were compared by using E. coli. A pair of plasmids was constructed, with each plasmid harboring phbCB and with the two differing only in the β-ketothiolase ORF (Materials and Methods). Strains harboring these plasmids were grown in M9 minimal medium containing 1% glucose and various amounts of propionate. After about 40 h of growth, the cells were harvested and the polymer was analyzed (Fig. 5). In these assays, bktB and phbA were essentially equivalent in their ability to produce high levels of polymer. The strain harboring bktB incorporated β-hydroxyvalerate into polymer much more efficiently than a strain harboring phbA. However, the amount of β-hydroxyvalerate incorporated into polymer reached a plateau when the media contained 0.05% or more propionate.
FIG. 5.
Comparison of PhbA and BktB in synthesis of PHBV in E. coli. Both strains harbored a plasmid expressing R. eutropha phbCB plus a β-ketothiolase gene, either bktB or phbA (see Materials and Methods). Polymer accumulation and composition were monitored as a function of the propionate in the medium. Total polymer (□) and the β-hydroxyvalerate fraction of the polymer (▪) in strain EE245, which expresses bktB, are shown, as are total polymer (▵) and the β-hydroxyvalerate fraction of the polymer (▴) in strain EE247, which expresses phbA.
R. eutropha bktB mutants are impaired in their ability to produce PHBV when grown on propionate.
To determine if BktB normally plays a role in PHA synthesis, a bktB::kan allele was used to replace the bktB locus of wild-type R. eutropha. Two independent kanamycin-resistant strains, EE168 and EE169, were characterized. Both strains were determined by Southern blotting to carry the bktB::kan allele, and both are negative for BktB protein as determined by immunoblotting (36).
We compared β-ketothiolase activity and polymer production by wild-type R. eutropha and two independent bktB::kan mutants (Table 4). When β-ketothiolase was assayed with acetoacetyl-CoA as a substrate, essentially no difference was seen between extracts from wild-type R. eutropha and the bktB mutants. However, the specific activity of β-ketothiolase capable of metabolizing β-ketovaleryl-CoA is approximately 80% lower in the bktB mutants than in the wild-type strain. This decreased ability to metabolize β-ketovaleryl-CoA was reflected in the composition of the polymer produced when the various strains were grown in the presence of propionate. The β-hydroxyvalerate component of the polymer in the bktB::kan strains was approximately one-third that found in the wild-type strain. This deficiency is complemented by a plasmid, pMON25901, that carries bktB+. In fact, overproduction of BktB from this plasmid led to slightly increased incorporation of β-hydroxyvalerate into PHA. The overexpression of β-ketothiolase (cumulatively, approximately threefold when assayed on acetoacetyl-CoA) did not significantly affect the total amount of polymer accumulated by R. eutropha in these fermentation end-point assays.
TABLE 4.
β-Ketothiolase activity and polymer composition in R. eutropha bktB::kan mutants and in strains overexpressing bktB
| R. eutropha strain | bktB genotype | Plasmida | β-Ketothiolase activity with substrateb
|
PHA formationc
|
|||
|---|---|---|---|---|---|---|---|
| Sp act (μmol degraded/min/mg of protein)
|
% (dry wt) PHA | % C5 in polymer | |||||
| C5 | C4 | C5/C4 ratio | |||||
| EE234 | Wild type | pBBR1MCS | 0.78 ± 0.1 | 2.5 ± 0.3 | 0.31 | 66 ± 3 | 11.9 ± 2.6 |
| EE236 | bktB::kan | pBBR1MCS | 0.19 ± 0.2 | 2.4 ± 0.6 | 0.08 | 63 ± 7 | 3.9 ± 0.9 |
| EE238 | bktB::kan | pBBR1MCS | 0.12 ± 0.04 | 2.3 ± 0.3 | 0.05 | 64 ± 5 | 3.7 ± 0.7 |
| EE235 | Wild type | pMON25901 | 16.4 ± 1.8 | 6.7 ± 1.2 | 2.45 | 67 ± 6 | 16.9 ± 2.5 |
| EE237 | bktB::kan | pMON25901 | 18.8 ± 5.3 | 7.5 ± 3.8 | 2.50 | 70 ± 6 | 19.3 ± 3.3 |
| EE239 | bktB::kan | pMON25901 | 17.2 ± 4.7 | 8.7 ± 3.9 | 1.97 | 60 ± 11 | 15.9 ± 4.0 |
pMON25901 harbors bktB+ in the broad-host-range plasmid pBBR1MCS.
Specific activities were derived from cleared-cell extracts of at least four independent cultures. C5, β-ketovaleryl-CoA; C4, acetoacetyl-CoA.
Polymer production data were derived from six independent cultures grown in 3% fructose plus 0.1% propionate.
An additional β-ketothiolase is expressed in R. eutropha.
R. eutropha bktB mutants retain significant β-ketothiolase activity capable of degrading β-ketovaleryl-CoA (Table 4). This activity is inconsistent with the substrate specificity of PhbA, so we examined R. eutropha for the presence of additional β-ketothiolases.
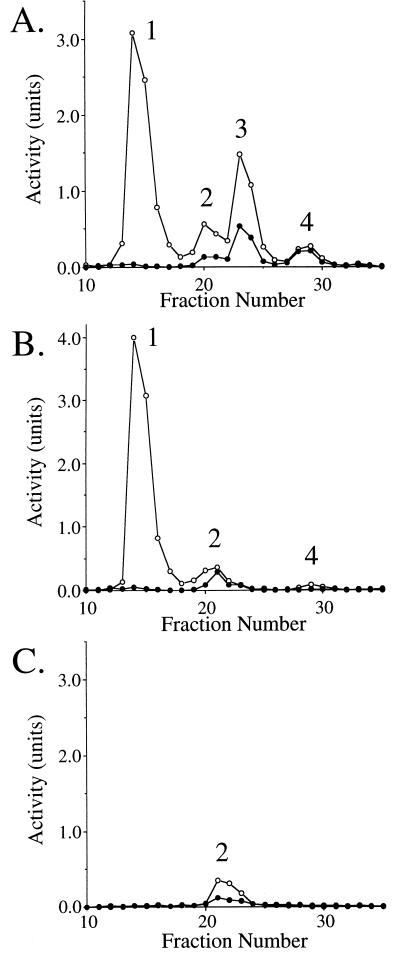
Figure 6 shows anion-exchange chromatography of extracts from wild-type R. eutropha (Fig. 6A), a bktB mutant (Fig. 6B), and a phbA bktB double mutant (Fig. 6C), all grown on fructose. Four peaks of β-ketothiolase activity were distinguishable in extracts of wild-type R. eutropha. PhbA eluted first, as a large peak containing activity primarily restricted to acetoacetyl-CoA utilization. Peak 2 was smaller than peak 1, but more active when assayed on β-ketovaleryl-CoA. We designated this activity as BktC, although we cannot rule out the presence of multiple β-ketothiolases within this activity peak. Peak 3 contains BktB, which provides most of the activity capable of metabolizing β-ketovaleryl-CoA. Peak 4 is a small peak eluting after the primary BktB peak. We have not yet fully characterized the contents of peak 4, but immunoblot analyses suggest that BktB is a component (reference 36 and described below).
FIG. 6.
Mono-Q chromatography of extracts from the R. eutropha wild-type strain EE167 (A), the bktB::kan strain EE168 (B), and the ΔphbA bktB::kan strain EE317 (C), all grown on fructose. Chromatographic conditions were as described in Materials and Methods. The enzymatic activity of β-ketothiolase was assayed in the thiolysis direction with either acetoacetyl-CoA (○) or β-ketovaleryl-CoA (•) as the substrate. The number designations of the individual peaks resolved from each strain are shown within each panel. Note that peak 1 is missing from the phbA mutant, peak 3 is missing from the bktB mutants, and peaks 1, 3, and 4 are missing from the double mutant.
Figure 6B shows anion-exchange chromatography of extract from a bktB::kan mutant. The absence of BktB (peak 3) is clear in this chromatogram, and the relative area of peak 4 is also diminished. Peak 2 (BktC) accounts for almost all of the residual activity capable of degrading β-ketovaleryl-CoA. In Fig. 6C, the presence of BktC is clearly demonstrated in a strain lacking both PhbA and BktB. Peak 4 also disappears in the double mutant, suggesting that peak 4 normally contains BktB and PhbA, apparently in some modified form.
PhbA, BktB, and BktC were all expressed when R. eutropha was grown on fructose as the sole carbon source and are distinguishable from the β-oxidative β-ketothiolase, which is expressed only during growth on fatty acids and prefers acyl-CoAs of longer chain lengths (36). The gene encoding the β-oxidative enzyme from R. eutropha has been cloned on pAE65 (36, 37).
DISCUSSION
R. eutropha constitutively expresses at least three different β-ketothiolases, designated PhbA, BktB, and BktC. PhbA is catalytically the most active, with BktB and BktC activities present at lower levels. Production of PHBV from acetyl-CoA and propionyl-CoA requires β-ketothiolase to produce both acetoacetyl-CoA and β-ketovaleryl-CoA. However, PhbA, the β-ketothiolase encoded within the phb operon, is essentially limited to production of acetoacetyl-CoA, as inferred from the thiolysis activity results. The genetic and biochemical evidence presented herein establishes BktB as the β-ketothiolase primarily responsible for generating β-ketovaleryl-CoA during growth on fructose and propionate. BktC appears to be a less active contributor to β-ketovaleryl-CoA formation.
The phb operon and bktB map approximately 4.6 kb apart on the R. eutropha chromosome, and the intervening region contains three ORFs that are likely to be expressed. ORF1 is highly homologous to ORFs identified near the pha genes of Rhizobium, Thiocystis, and Chromatium species (16, 17, 40), suggesting that the ORF1 product participates in PHA metabolism. ORF2 and ORF3 apparently form an operon with overlapping termination and initiation codons, respectively. ORF2 is similar to the E. coli yhdG and yohI ORFs, members of an apparently ubiquitous class of genes with homologs in yeast and humans (14). ORF3 is highly similar to E. coli f441 and to other putative genes in both bacteria and archaea. No function has been assigned to any ORF2 or ORF3 homolog.
Complementation studies with E. coli demonstrate that bktB can be used to produce PHBV copolymer in a recombinant system. The β-hydroxyvalerate composition of the polymer increased linearly up to about 0.05% propionate in the media and then reached a plateau (Fig. 5). This plateau is probably due to saturation of propionyl-CoA formation, since E. coli acs (acetyl-CoA synthetase) is repressed under the growth conditions in these experiments (5). Enhancement of propionate import and activation should yield significantly higher incorporation of β-hydroxyvalerate into polymer (26, 27), potentially opening E. coli as a system for copolymer production.
Our findings confirm and extend those of Haywood et al. (7), who partially purified two β-ketothiolases from R. eutropha, designated β-ketothiolases A and B. These two preparations probably correspond to PhbA and BktB, respectively, although the relatively high level of activity on acetoacetyl-CoA reported for β-ketothiolase B suggests that their preparation also contained an enzyme that prefers acetoacetyl-CoA, such as PhbA and/or BktC. We also noted that the BktB-containing peak from wild-type R. eutropha showed a preference for catalysis of acetoacetyl-CoA (Fig. 6), whereas pure BktB preferred β-ketovaleryl-CoA (Table 3). It is possible that PhbA and/or BktC forms mixed oligomers with BktB, resulting in this shift of substrate specificity. Alternatively, other factors may affect the substrate specificity of BktB.
Our data establish a role for BktB in PHA synthesis, but it is likely that BktB and BktC also function during degradation of fatty acids (7). Although we did not assay substrates larger than β-ketohexanoyl-CoA, the substrate flexibility of BktB probably extends at least to β-ketodecanoyl-CoA (7). BktB may, therefore, play an auxiliary role in β-oxidation, including partial degradation of short- and medium-chain fatty acids which do not induce the β-oxidative complex.
Finally, PhbA, BktB, and BktC may all play a critical role as scavengers of carbon, allowing PHA formation when the available carbon cannot be metabolized directly. This situation occurs under anaerobic conditions. R. eutropha is strictly respiratory but can grow anaerobically in the presence of an alternative electron acceptor such as nitrate or nitrite (2). R. eutropha also induces a number of enzymes in response to low O2, indicating that it normally encounters anaerobic conditions (6, 24, 30, 38). Finally, anaerobic conditions induce PHA production in R. eutropha (33). The ability to form PHA is advantageous in anaerobic conditions for at least two reasons: (i) PHA formation serves as an electron sink (33), and (ii) carbon can be scavenged and stored for use when O2 becomes available. In this respect, R. eutropha seems well adapted to a niche at the aerobic-anaerobic boundary. Here, it would encounter acetate and propionate produced by anaerobic fermentation, and both of these compounds could be channeled into polymer by BktB and BktC. Being motile, R. eutropha could potentially migrate between environments, collecting carbon in the anaerobic zone and consuming it in the aerobic zone. In this scenario, PHA production becomes central to cellular survival, providing a means for a respiratory organism to exploit the anaerobic environment.
ACKNOWLEDGMENTS
We thank Damien Rodriguez, Catharine Gunter, Greg Thorne, Jun Wu, and Sunny Gilbert for excellent technical assistance; Bärbel Friedrich, Doug Dennis, and Russ Maurer for providing plasmids; and Barry Goldman for critical reading of the manuscript.
REFERENCES
- 1.Amann E, Brosius J, Ptashne M. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene. 1983;25:167–178. doi: 10.1016/0378-1119(83)90222-6. [DOI] [PubMed] [Google Scholar]
- 2.Bowien B, Schlegel H G. Physiology and biochemistry of aerobic hydrogen-utilizing bacteria. Annu Rev Microbiol. 1981;35:405–452. doi: 10.1146/annurev.mi.35.100181.002201. [DOI] [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 4.Braunegg G, Sonnleitner B, Lafferty R M. A rapid gas chromatographic method for the determination of poly-β-hydroxybutyric acid in microbial biomass. Eur J Appl Microbiol Biotechnol. 1978;6:29–37. [Google Scholar]
- 5.Clark D P, Cronan J E., Jr . Two-carbon compounds and fatty acids as carbon sources. In: Neidhart F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 343–357. [Google Scholar]
- 6.Cramm R, Siddiqui R A, Friedrich B. Primary sequence and evidence for a physiological function of the flavohemoprotein of Alcaligenes eutrophus. J Biol Chem. 1994;269:7349–7354. [PubMed] [Google Scholar]
- 7.Haywood G W, Anderson A J, Chu L, Dawes E A. Characterization of two 3-ketothiolases possessing differing substrate specificities in the polyhydroxyalkanoate synthesizing organisms Alcaligenes eutrophus. FEMS Microbiol Lett. 1988;52:91–96. [Google Scholar]
- 8.Haywood G W, Anderson A J, Chu L, Dawes E A. The role of NADH- and NADPH-linked acetoacetyl-CoA reductases in the poly-3-hydroxybutyrate synthesizing organisms Alcaligenes eutrophus. FEMS Microbiol Lett. 1988;52:259–264. [Google Scholar]
- 9.Haywood G W, Anderson A J, Dawes E A. The importance of PHB synthase substrate specificity in polyhydroxyalkanoate synthesis by Alcaligenes eutrophus. FEMS Microbiol Lett. 1988;57:1–6. [Google Scholar]
- 10.Janes B, Hollar J, Dennis D. Molecular characterization of the poly-β-hydroxybutyrate biosynthetic pathway of Alcaligenes eutrophus H16. In: Dawes E A, editor. New biosynthetic biodegradable polymers of industrial interest from microorganisms. Amsterdam, The Netherlands: Kluwer Publishers; 1990. pp. 175–190. [Google Scholar]
- 11.Kidwell J, Valentin H E, Dennis D. Regulated expression of the Alcaligenes eutrophus pha biosynthesis genes in Escherichia coli. Appl Environ Microbiol. 1995;61:1391–1398. doi: 10.1128/aem.61.4.1391-1398.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- 13.Kovach M E, Elzer P H, Hill D S, Robertson G T, Farris M A, Roop II R M, Peterson K M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 14.Kranz, R. Personal communication.
- 15.Lenz O, Schwartz E, Dernedde J, Eitinger M, Friedrich B. The Alcaligenes eutrophus H16 hoxX gene participates in hydrogenase regulation. J Bacteriol. 1994;176:4385–4393. doi: 10.1128/jb.176.14.4385-4393.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liebergesell M, Steinbüchel A. Cloning and nucleotide sequences of genes relevant for biosynthesis of poly(3-hydroxybutyric acid) in Chromatium vinosum strain D. Eur J Biochem. 1992;209:134–150. doi: 10.1111/j.1432-1033.1992.tb17270.x. [DOI] [PubMed] [Google Scholar]
- 17.Liebergesell M, Steinbüchel A. Cloning and molecular analysis of the poly(3-hydroxybutyric acid) biosynthetic genes of Thiocystis violacea. Appl Microbiol Biotechnol. 1993;38:493–501. doi: 10.1007/BF00242944. [DOI] [PubMed] [Google Scholar]
- 18.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 19.Nishimura T, Saito T, Tomita K. Purification and properties of β-ketothiolase from Zoogloea ramigera. Arch Microbiol. 1978;116:21–27. doi: 10.1007/BF00408729. [DOI] [PubMed] [Google Scholar]
- 20.Palmer M A J, Differding E, Gamboni R, Williams S F, Peoples O P, Walsh C T, Sinskey A J, Masamune S. Biosynthetic thiolase from Zoogloea ramigera: evidence for a mechanism involving CYS-378 as the active site base. J Biol Chem. 1991;266:8369–8375. [PubMed] [Google Scholar]
- 21.Peoples O P, Sinskey A J. Poly-β-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16. Characterization of the genes encoding β-ketothiolase and acetoacetyl-CoA reductase. J Biol Chem. 1989;264:15293–15297. [PubMed] [Google Scholar]
- 22.Peoples O P, Sinskey A J. Poly-β-hydroxybutyrate (PHB) biosynthesis in Alcaligenes eutrophus H16. Identification and characterization of the PHB polymerase gene (phbC) J Biol Chem. 1989;264:15298–15303. [PubMed] [Google Scholar]
- 23.Pfennig N. Rhodopseudomonas globiformis, sp. N., a new species of Rhodospirillaceae. Arch Microbiol. 1974;100:197–206. [Google Scholar]
- 24.Probst I, Wolf G, Schlegel H G. An oxygen-binding flavohemoprotein from Alcaligenes eutrophus. Biochim Biophys Acta. 1979;576:471–478. doi: 10.1016/0005-2795(79)90422-7. [DOI] [PubMed] [Google Scholar]
- 25.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 26.Rhie H G, Dennis D. Role of fadR and atoC(Con) mutations in poly-(3-hydroxybutyrate-co-3-hydroxyvalerate) synthesis in recombinant pha+Escherichia coli. Appl Environ Microbiol. 1995;61:2487–2492. doi: 10.1128/aem.61.7.2487-2492.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rhie H G, Dennis D. The function of ackA and pta genes is necessary for poly-(3-hydroxybutyrate-co-3-hydroxyvalerate) synthesis in recombinant pha+Escherichia coli. Can J Microbiol. 1995;41:200–206. doi: 10.1139/m95-188. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Schlegel H G, Kaltwasser H, Gottschalk G. Ein Submersverfahren zur kultur wasserstoffoxydierender bakterien: Wachstumsphysiologische untersuchungen. Arch Mikrobiol. 1961;38:209–222. [PubMed] [Google Scholar]
- 30.Schlegel H G, Vollbrecht D. Formation of the dehydrogenases for lactate, ethanol and butanediol in the strictly aerobic bacterium Alcaligenes eutrophus. J Gen Microbiol. 1980;117:475–481. [Google Scholar]
- 31.Schubert P A, Steinbüchel A, Schlegel H G. Cloning of the Alcaligenes eutrophus genes for synthesis of poly-β-hydroxybutyrate. J Bacteriol. 1988;170:5837–5847. doi: 10.1128/jb.170.12.5837-5847.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulz H. Long chain enoyl-coenzyme-A hydratase from pig heart. J Biol Chem. 1992;249:2704–2709. [PubMed] [Google Scholar]
- 33.Schuster E, Schlegel H G. Chemolithotrophes Wachstum von Hydrogenomonas H16 im Chemostaten mit elektrolytischer Knallgaserzeugung. Arch Mikrobiol. 1967;58:380–409. [PubMed] [Google Scholar]
- 34.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 35.Slater S, Gallaher T, Dennis D. Production of poly-(3-hydroxybutyrate-co-3-hydroxyvalerate) in a recombinant Escherichia coli strain. Appl Environ Microbiol. 1992;58:1089–1094. doi: 10.1128/aem.58.4.1089-1094.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slater, S., M. Tran, K. Houmiel, and K. Gruys. Unpublished data.
- 37.Slater S C, Voige W H, Dennis D E. Cloning and expression in Escherichia coli of the Alcaligenes eutrophus H16 poly-β-hydroxybutyrate biosynthesis pathway. J Bacteriol. 1988;170:4431–4436. doi: 10.1128/jb.170.10.4431-4436.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steinbüchel A, Kuhn M, Niedrig M, Schlegel H G. Fermentation enzymes in strictly aerobic bacteria: comparative studies on strains of the genus Alcaligenes and on Nocardia opaca and Xanthobacter autotrophicus. J Gen Microbiol. 1983;129:2825–2835. [Google Scholar]
- 39.Thompson S, Mayer F, Peoples O P, Masamune S, Sinskey A J, Walsh C T. Mechanistic studies on β-ketoacyl thiolase from Zoogloea ramigera: identification of the active-site nucleophile as Cys89, its mutation to Ser89, and kinetic and thermodynamic characterization of wild-type and mutant enzymes. Biochemistry. 1989;28:5735–5742. doi: 10.1021/bi00440a006. [DOI] [PubMed] [Google Scholar]
- 40.Tombolini R, Povolo S, Buson A, Squartini A, Nuti M P. Poly-β-hydroxybutyrate (PHB) biosynthetic genes in Rhizobium meliloti 41. Microbiology. 1995;141:2553–2559. doi: 10.1099/13500872-141-10-2553. [DOI] [PubMed] [Google Scholar]
- 41.Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuchi Y. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia pickettii (Ralston, Palleroni and Doudoroff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol Immunol. 1995;39:897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x. [DOI] [PubMed] [Google Scholar]