Abstract
The genomes of bacteriophages T4 and RB69 are phylogenetically related but diverge in nucleotide sequence at many loci and are incompatible with each other in vivo. We describe here the biological implications of divergence in a genomic segment that encodes four essential DNA replication proteins: gp45 (sliding clamp), gp44/62 complex (clamp loader), and gp46 (a recombination protein). We have cloned, sequenced, and expressed several overlapping segments of the RB69 gene 46-45.2-(rpbA)-45-44-62 cluster and compared its features to those of the homologous gene cluster from T4. The deduced primary structures of all four RB69 replication proteins and gp45.2 from this cluster are very similar (80 to 95% similarity) to those of their respective T4 homologs. In contrast, the rpbA region (which encodes a nonessential protein in T4) is highly diverged (∼49% similarity) between the two phage genomes and does not encode protein in RB69. Expression studies and patterns of high divergence of intercistronic nucleotide sequences of this cluster suggest that T4 and RB69 evolved similar transcriptional and translational control strategies for the cistrons contained therein, but with different specificities. In plasmid-phage complementation assays, we show that posttranslationally, RB69 and T4 homologs of gp45 and the gp44/62 complex can be effectively exchanged between the two phage replicase assemblies; however, we also show results which suggest that mixed clamp loader complexes consisting of T4 gp62 and RB69 gp44 subunits are not active for phage DNA replication. Thus, specificity of the gp44-gp62 interaction in the clamp loader marks a point of departure between the T4 and RB69 replication systems.
The DNA replicase of bacteriophage T4 is a loosely assembled multiprotein complex that is comparable in composition to the replicases of bacteria and humans (15, 20, 26). Among its components are a DNA polymerase (gp43), product of phage gene 43, a sliding clamp (gp45), a clamp loader (gp44/62 complex), a single-stranded DNA-binding protein (Ssb, gp32), a primase (gp61)-helicase (gp41) complex, a helicase-primase assembly factor (gp59), RNase H (gprnh), DNA ligase (gp30), and probably other proteins. The sliding clamp in the T4 replicase is a gp45 trimer, and the clamp loader is a heteromeric complex consisting of four gp44 subunits and one gp62 subunit that are tightly bound together (11). In addition to their essential roles as accessory proteins to DNA polymerase, gp45 and the gp44/62 complex interact with the T4-modified transcriptional machinery of the Escherichia coli host and are required for the coupling of phage DNA replication to transcription during the late stages of phage development (28, 36).
In previous studies, we observed that gp43 of phage RB69, a distant phylogenetic relative of T4, diverges from its T4 gp43 homolog at several segments of the primary structure (34). We also observed that despite their divergence (74% amino acid similarity overall), RB69 gp43 (when produced by a plasmid) can substitute effectively for the T4 polymerase in a T4 infection, where the replicase complex consists mostly of T4-induced proteins (3, 34). In contrast, T4 gp43 is less effective as a substitute for RB69 gp43 in RB69 DNA replication (34). These observations have suggested that some of the specificity determinants for interactions among phage replication proteins are not identical for the T4 and RB69 DNA replicases. Identification of incompatibilities between T4- and RB69-encoded proteins could help localize sites of specific protein-protein interactions and delineate points of departure between genomic specificities of these two phylogenetically related but distinct phage replication systems.
We describe here an analysis of the RB69 homolog of the T4 gene 46-45.2-rpbA-45-44-62 cluster, which encodes, in addition to the gp45-gp44-gp62 group, another essential replication protein, gp46 (possibly a nuclease [5]), a nonessential RNA polymerase-binding protein (RpbA) (35), and a protein of unknown function, gp45.2 (9). The T4 cluster is also known to harbor several transcriptional and posttranscriptional control signals that regulate biosynthesis of the DNA polymerase accessory proteins (6, 8). Our results here indicate that the primary structures of genes 46, 45, 44, and 62 and their protein products are all highly conserved between T4 and RB69. Gene 45.2 is also highly conserved, which may mean that its protein product is essential in both phage systems. In contrast, the rpbA region is highly diverged between T4 and RB69 and lacks an open reading frame in RB69. Expression studies and examination of nucleotide sequences of untranslated intercistronic regions suggest that T4 and RB69 utilize similar posttranscriptional and transcriptional strategies to control genes 45, 44, and 62; however, the specificities of the control processes are different between the two phage systems. Also, in plasmid-phage complementation assays, we observed that the T4 and RB69 homologs of gp45 and the gp44/62 complex can be functionally exchanged between the two phage replication systems; however, T4 gp62 and RB69 gp44 were not functionally compatible with each other in these assays. Functional incompatibility between clamp loader subunits from the two phylogenetic phage relatives signifies a divergence in the specificities of their DNA replicases.
MATERIALS AND METHODS
Materials.
Vent DNA polymerase (from New England Biolabs Inc., Beverly, Mass.) was used for all PCR amplifications. Other reagents used included restriction endonucleases (Boehringer Mannheim Corp., Indianapolis, Ind., and GIBCO BRL Life Technologies, Gaithersburg, Md.), Sequenase version 2.0 and sequencing kit (Amersham Life Science Inc., Arlington Heights, Ill.), [α-35S]dATP and [35S]methionine (DuPont NEN, Boston, Mass.), and nonradioactive deoxynucleoside triphosphates (Pharmacia Biotech Inc., Piscataway, N.J.). Oligonucleotides used as primers for PCR amplifications and DNA sequencing were made by the Shared Instrumentation Facility at Tulane University School of Medicine, which uses an ABI model 394 DNA/RNA synthesizer and PerSeptive Biosystems Expedite nucleic acid synthesis system.
Bacterial, phage, and plasmid strains.
The E. coli strains and methods used for preparing phage stocks, transforming cells with plasmids, and measuring plasmid-mediated or phage-induced gene expression were the same as those used in other work (32). T4 and RB69 amber mutants and RB69 wild-type phage were obtained from W. B. Wood’s collection (University of Colorado). The T4 strain used as the “wild-type” standard has been maintained in this laboratory (8).
Plasmids pSP72 and pSP73 (Promega Corp., Madison, Wis.) carry multicloning sites bracketed by T7 and SP6 promoters. They were used to clone DNA fragments for in vitro transcriptions and preparation of radiolabeled RNA probes (riboprobes) as previously described (34). The pGWM1, pLY900, and pLY965 cloning vectors that were used for plasmid-mediated expression of cloned T4 and RB69 genes were derived from the λcI857pLN-derived expression vector pGW7, which had been constructed and given to us by Geoff Wilson (8, 16, 37). These plasmids allow expression of cloned genes after heat inactivation (at 42°C) of the vector-encoded λcI857 repressor. The pGWM1 vector is identical to pGW7 except that it carries an XhoI site in place of pGW7’s SphI site. The pLY900 vector was derived from pGWM1 by inserting the multicloning site of pSP72 (from the BglII to the XhoI sites) into the BamHI-to-XhoI interval of pGWM1. The pLY965 vector (derived from pLY900) lacks the HpaI-to-EcoRV DNA interval of pLY900; i.e., pLY965 is deleted for much of the λN gene whereas pLY900 possesses this transcription antitermination gene function.
Screening for gene 45, 44, and 62 sequences in an RB69 genomic library.
The methods for constructing and screening the λZAPII RB69 genomic library used here have been described elsewhere (34). Figure 1 shows maps of the T4 and RB69 gene 46-43 clusters and diagrams of the approach that we used to retrieve and subclone RB69 DNA fragments containing genes 45, 44, and 62. Several λZAPII recombinants containing at least portions of these genes were initially identified by screening the library with a 32P-labeled riboprobe made by in vitro transcription of a cloned internal SspI fragment from the neighboring RB69 gene 43 (SP101 probe [Fig. 1]). These clones were sequenced, and those containing RB69 genes 44, 62, and regA in addition to at least portions of genes 45 and 43 were used for further studies. Based on sequence information obtained directly from the λZAPII clones, either specific segments of the λZAPII recombinants were subcloned directly or their counterparts were amplified by PCR from purified phage DNA and then subcloned in T7 expression vectors for the preparation of other riboprobes that were used in additional library screening. The studies reported here used mostly pSP72 recombinants expressing the PBY16 and SPR45-5 DNA segments (diagrammed in Fig. 1) for synthesizing hybridization riboprobes.
FIG. 1.
Diagrammatic representation of λZAPII genomic library screening for RB69 DNA fragments (A) and partial restriction maps of the gene 46-43 regions of T4 and RB69 (B). Endonucleases DraI and SspI were used in the preparation of the λZAPII library (34). The solid horizontal bars designated PBS3K1, SP101, and SPR45-5 (A) represent 32P-labeled riboprobes that were used to identify recombinant plasmids carrying the DNA fragments PBY16, PBS3, and LY6, respectively (see Materials and Methods). The SP101 probe corresponds to an internal SspI fragment of RB69 gene 43, PBS3K1 corresponds to a KpnI deletion of PBS3, and SPR45-5 corresponds to a 3′-terminal gene 45 segment that was generated from purified RB69 phage DNA by PCR amplification. ▿ in panel A denotes a terminal deletion for the respective gene. Restriction site abbreviations in panel B: H, HindIII; Sa, SalI; Sc, SacI; P, PstI.
PCR.
PCR amplifications were carried out in 100 μl of reaction mixtures containing 500 nM each of the two primers used, 0.046 nM RB69 genomic DNA, 0.4 mg of bovine serum albumin (New England Biolabs) per ml, 1.5 mM each dGTP and dCTP, 2 mM each dATP and dTTP, and 4 U of Vent DNA polymerase. The reaction buffer contained 10 mM KCl, 20 mM Tris-Cl (pH 8.8 at 25°C), 10 mM (NH4)2SO4, 6 mM MgSO4, and 0.1% Triton X-100. Reactions were carried out in a model 9600 or 2400 Perkin-Elmer GeneAmp PCR system and begun with incubations at 94°C (5 min) followed by 25 cycles of 94°C for 45 s, 48°C for 45 s, and 72°C for 60 s to 2 min. These conditions were particularly suitable for amplifying desired DNA segments from the AT-rich genomes of T4 and RB69. PCR products were subsequently digested with the appropriate restriction endonucleases and cloned into the desired vectors for sequencing or expression studies.
Isolation of RB69 genomic DNA.
RB69 DNA was isolated from enriched Hershey agar (EHA) plate lysates (2,000 to 5,000 plaques/100-mm-diameter petri dish) of wild-type or mutant RB69 phage. The lysed lawn from each plating was harvested into 4 ml of chloroform-saturated M9 buffer (42 mM Na2HPO4, 22 mM KH2PO4, 19 mM NH4Cl, 9 mM NaCl, 1 μM FeCl3, 1 mM MgCl2, 0.001% gelatin) and stored in a refrigerator for several hours. Cell debris and agar were then removed by centrifugation at 9,600 × g (10 min) in the cold, and the supernatant was treated with DNase I (2 μg/ml) and RNase A (2 μg/ml) for 1 h. The phage was then precipitated in 10% polyethylene glycol 6000 at 1 M NaCl and resuspended in 10 mM Tris (pH 7.4)–0.2 M MgCl2 buffer. Excess polyethylene glycol 6000 was removed by extraction with chloroform. To isolate phage DNA, the purified RB69 preparation was treated with proteinase K (50 μg/ml) and sodium dodecyl sulfate (SDS; 0.5%, final concentration) at 65°C for about 2 h. This mixture was then extracted with phenol and phenol-chloroform (1:1) before the DNA was precipitated by the addition of 0.1 volume of 3 M sodium acetate and 4 volumes of 100% ethanol (−20°C). DNA fibers were spooled out of the ethanol solution, rinsed twice in tubes containing cold 70% ethanol, transferred to a microcentrifuge tube, dried under vacuum, and redissolved at an A260 of ∼10 for use without further purification. Lysates and DNA from wild-type and mutant T4 strains were similarly prepared, except that yields were higher than with RB69.
Burst size measurements.
In the plasmid-phage complementation assays for Fig. 7, the T4 and RB69 genes of interest were cloned under T7 promoter control in pSP72 or pSP73 and their complementing activities were measured as previously described for the polymerase genes of these phages (34). Briefly, E. coli BL21(DE3) cells harboring the plasmid-borne wild-type T4 or RB69 gene(s) under study were grown to 3 × 108/ml in M9S medium and infected at 30°C by using 0.1 volume of the phage mutant to be complemented (multiplicity of infection of 0.1). At 20 min postinfection, samples were removed and plated on the permissive host (E. coli CR63/5) for the phage mutant to measure infective centers (transmission coefficients). The remainder of the culture was aerated for 100 min before being lysed with chloroform and assayed for phage yield on E. coli CR63/5. We define burst size as the phage yield per average infective center titer obtained with a wild-type phage control, which was assumed to yield 100% transmission.
FIG. 7.
Complementation between phage-encoded and plasmid-encoded T4 and RB69 DNA polymerase accessory proteins. The abilities of polymerase accessory proteins to be functionally exchanged between T4 and RB69 were examined by burst size measurements (Materials and Methods) and qualitative spot tests. (A) Results of plasmid-phage complementation tests involving gene 45 and genes 44 and 62 together; (B and C) results of similar tests involving genes 44 and 62 separately. In the “Spot test” blocks, each pair of spots represents growth responses (cell lysis) from 5 μl of two phage concentrations, ∼104 and ∼107/ml, respectively. Numbers shown in parentheses in the “Relative burst size” blocks are the actual bursts corresponding to the 1.0 reference values for the pairs of infections compared. Note that although the T4 and RB69 counterparts of gp45 and the gp44/62 complex can exchange effectively, with some preferences by the gene functions to support replication of the phage from which they originated (values in panel A), the gp44(RB69)/gp62(T4) combination is largely inactive for phage replication (C). Also note that plasmid-expressed wild-type (wt) RB69 gene 44 is inhibitory to replication of wild-type T4 (T4 wt phage on pRB69g44-bearing host [B]).
Cloning of T4 and RB69 genes in expression vectors.
The DNA regions encompassing genes 45, 44, and 62 of T4 and RB69 were PCR amplified by using either purified phage DNA or plaque suspensions (2) as the source of template. The amplified DNA fragments were purified by elution from agarose gels after electrophoresis and digested with the appropriate restriction enzymes for insertion into the λpL plasmid expression vectors pLY900 (λcI857pLN) and pLY965 (λcI857pL), and the recombinant plasmids were propagated in E. coli CAJ70. Integrity of the T4 and RB69 DNA inserts in the plasmid vectors was confirmed by DNA sequencing and plasmid-phage complementation assays. In measurements of plasmid-mediated expression of cloned genes, cultures of plasmid-bearing cells were grown at 30°C to 5 × 108 cells/ml and then shifted to 42°C and aerated either for 10 min (Fig. 6) or 15 min (Fig. 5) to induce the plasmid-borne λpL before [35S]methionine was added to a final concentration of 5 μCi/ml. Labeling with isotope was for 10 min (Fig. 6) or 15 min (Fig. 5) at 42°C. Cells were subsequently harvested from the cultures and extracts prepared for SDS-polyacrylamide gel electrophoresis (PAGE) analysis as described previously (14).
FIG. 6.
Alignment of the deduced primary structures of the T4 and RB69 counterparts of gp44 and gp62. The boxed region in the gp44 panel marks the putative ATP-binding site of these proteins (25). The graphics below the primary structures depict distribution of amino acid identities between each pair of homologous proteins (Table 1). Also shown are the locations of gp44 and gp62 chain termination mutations that were sequenced (at the nucleotide level) in this work (T4 44amE4408, RB69 44am51, T4 62amE1140 and RB69 62am85) or previously by Hsu (7), i.e., T4 44amN82 and T4 44amB110. The mutant 44amB110, previously classified as tsB110 (4), forms small plaques at 30°C and no plaques at 42°C on sup° hosts.
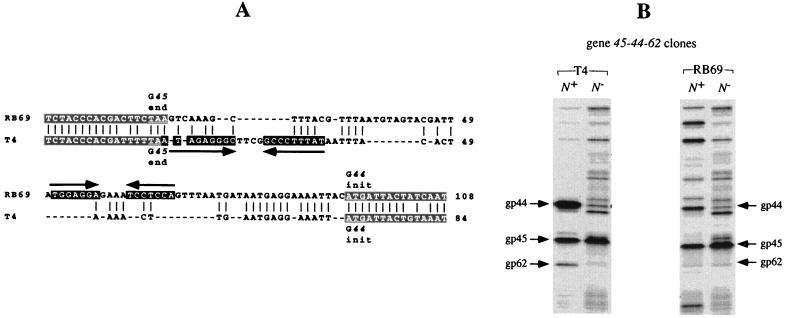
FIG. 5.
Nucleotide sequences of the T4 and RB69 gene 44-62 junctures (A) and expression of cloned gene 62 from RB69 and T4 (B). (A) Open reading frames for genes 44 and 62, which are separated by one base pair at the g44-62 junctures of both T4 and RB69. The nucleotide sequence alignment shown is based on analysis of RB69 DNA clones derived from the λZAPII genomic library as well as PCR-amplified RB69 genomic fragments. The horizontal arrows underline the stem segment of a putative mRNA structure that has been suggested to play a role in translational coupling of gp62 to gp44 synthesis in T4 (29). No such structure can be predicted for RB69. (B) Results of plasmid-mediated expression of comparable genomic segments encompassing the gene 45-62 intervals from T4 mutant 44amN82 and RB69 mutant 44am51. The desired DNA segments were PCR amplified from the respective phage mutants as well as wild-type strains and cloned in λpLN vector, and their plasmid-mediated expression was subsequently analyzed in E. coli CAJ70 as described in Materials and Methods. The short arrows mark the positions of gp45, gp44, and gp62 bands on the SDS-PAGE autoradiogram (10% gel) from the experiment; dots mark the positions of gp44 amber fragments. Note that expression of gene 62 (from either T4 or RB69) is lower when DNA from gene 44 amber mutants is used than with DNA carrying the wild-type gene 44 alleles.
Nucleotide sequence accession number.
The primary structure of RB69 gene 46 as well as the other RB69 genomic sequences reported here may be obtained from GenBank (accession number AF039565) or the corresponding author.
RESULTS
Overview of the RB69 gene 46-43 cluster.
Previous studies had determined the nucleotide sequences of RB69 genes 43 (34) and regA (12), as well as the regA-43 intercistronic region (30, 32). The work described here extended analysis of the RB69 genome to include the ∼5-kb chromosomal segment directly upstream of the regA cistron and established that RB69 and T4 have the same genetic order for homologous cistrons of the gene 46-43 cluster (Fig. 1; see also reference 8). Table 1 provides a summary of the patterns of nucleotide and amino acid sequence conservation and divergence for the RB69 and T4 gene 46-43 clusters and their protein products, and the results to be described below provide a closer view of the nature, distribution, and biological consequences of the divergences.
TABLE 1.
Summary of similarities between homologous segments of the T4 and RB69 gene 46-43 clusters and their protein productsa
| Genetic region | % Nucleotide identity | Protein | % Amino acid identity (similarity) | |
|---|---|---|---|---|
| g46 | 76 | gp46 | 82 (91) | |
| IC46-45.2 | 62 | None | ||
| g45.2 | 73 | gp45.2 | 76 (84) | |
| grpbA(T4)/IC45.2-45(RB69) | 49 | RpbA(T4)/none(RB69) | ||
| g45 | 76 | gp45 | 78 (85) | |
| IC45-44 | 51 | None | ||
| g44 | 74 | gp44 | 78 (88) | |
| g62 | 71 | gp62 | 73 (83) | |
| gregA | 76 | RegA | 79 (82) | |
| ICregA-43 | 50 | None | ||
| g43 | 64 | gp43 | 60 (74) |
Based on the results presented in Fig. 2 to 6 of this report as well as results from other work (see text for references). Note that the rpbA region, which encodes RpbA protein in T4, is presented as an intercistronic region in RB69 (IC46-45.2) since no open reading frame longer than 38 amino acids could be predicted for this region from the nucleotide sequence of this phage (see also Fig. 1 and 3).
The RB69 gene 46-45 region.
RB69 genomic library screening yielded one clone, pLY6 (Fig. 1), that contained most of gene 46 in addition to the ∼3-kb DNA segment directly to its downstream. The LY6 DNA insert and PCR-generated facsimiles amplified from purified RB69 phage DNA were sequenced to obtain information about similarities to the corresponding genomic region from T4. A comparison between the deduced primary structures of RB69 gp46 and its T4 counterpart indicated that the two proteins are identical or similar at ∼91% of amino acid positions (Table 1), with differences being evenly distributed along the linear protein maps (results not shown). No clones were obtained for the region upstream of the assumed fifth codon of RB69 gene 46. This result mirrors similar unsuccessful attempts at cloning an intact T4 gene 46 in the past (unpublished work). Possibly, a functional gene 46 is toxic to host cells even when residually expressed in cloning vehicles.
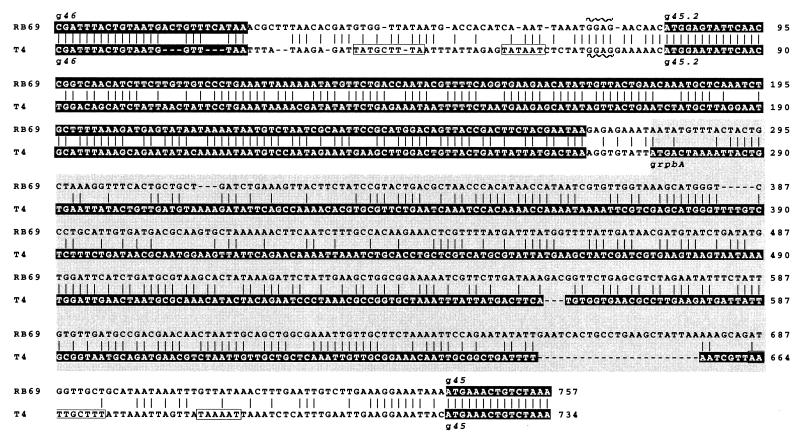
Figure 2 compares the T4 and RB69 nucleotide sequences for the chromosomal region between genes 46 and 45. The RB69 sequence shown is based on examination of both λZAPII library DNA and DNA amplified directly from RB69 phage. In T4, this region encodes the RNA polymerase-binding protein RpbA and is also presumed to encode gp45.2, a protein of unknown function whose existence has been inferred from results of plasmid-mediated expression of the T4 region (9). The nucleotide sequence alignments shown in Fig. 2 suggest that gp45.2 is highly conserved between T4 and RB69 (∼84% amino acid similarity [Table 1]). In contrast, the RpbA protein, which is known to be nonessential (although abundantly produced) in T4, appears to have vanished during the evolution of RB69. Interestingly, the rpbA regions of T4 and RB69 are similar to each other in size, suggesting that spacing of the interval between genes 46 and 45.2 may be important for expression of phage genes in this region of the genome. In Fig. 2, it is also interesting that the nucleotide sequence to the upstream of gene 45 does not harbor any recognizable T4-like MotA promoter sequence elements at positions where the existence of such promoters in T4 has been confirmed (6, 8).
FIG. 2.
Chromosomal region between genes 46 and 45 of T4 and RB69. The nucleotide sequence alignment shown is based on analysis of LY6 DNA in this study (Fig. 1) and analysis of the T4 counterpart in other studies (9, 35). Sequences highlighted in the darkest shading indicate open reading frames. Note the high nucleotide sequence divergence between the RB69 and T4 rpbA regions (lighter shading) compared to other cistrons in the gene 46-62 cluster (see also Fig. 4A); the RB69 rpbA region does not constitute a protein-encoding cistron. Other untranslated intercistronic sequences are unshaded. The boxed regions 5′ terminal to T4 genes 45.2 and 45 indicate the locations of MotA-dependent promoters; note their absence in RB69.
RB69 gene 45 and transcription termination in the gene 45-44 intercistronic region (IC45-44).
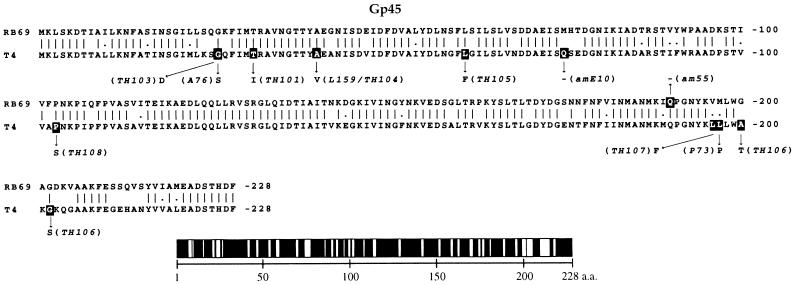
Figure 3 shows an alignment of the deduced primary structures of the T4 and RB69 gene 45 proteins based on determination of nucleotide sequences of λZAPII recombinants and PCR-amplified T4 and RB69 phage DNA segments. The two gp45 phylogenetic variants are ∼85% similar (Table 1), with most of their differences appearing in clusters of amino acid sequence. In contrast, as shown in Fig. 4A, the untranslated region between genes 45 and 44 (IC45-44 [Table 1]) exhibits a high degree of divergence between the genomes of the two phages, both in size and in nucleotide sequence, suggesting that gene 44 expression may be regulated differently in the two phages. In particular, as Fig. 4A depicts, the RB69 IC45-44 region harbors an RNA hairpin structure of different nucleotide sequence from the hairpin that has been implicated in transcription termination in the T4 IC45-44 region (8).
FIG. 3.
Comparison between the primary structures of RB69 gp45 and T4 gp45. The amino acid sequence alignment shown is based on determination of the nucleotide sequences for the two respective genes (Materials and Methods; see also references 7 and 24 for the T4 sequence). Also shown are locations of several T4 and RB69 gp45 mutations (darkened sites) that have been sequenced at the nucleotide level (see also reference 7): L159 (a temperature-sensitive mutant) and amE10 are from W. B. Wood’s collection, the TH mutants are temperature-sensitive mutants isolated by Hsu (7), Gly26 is a mutational hotspot (7), and A76 and P73 are tabB mutants (19). The graphic below the primary structures depicts distribution of amino acid (a.a.) identity (black segments) between the two homologous proteins, with unfilled sectors marking nonidentical positions (see also Table 1). Note that the T4 gene 45 sequence shown here is a corrected version of the sequence reported in reference 24 (GenBank accession no. M10160).
FIG. 4.
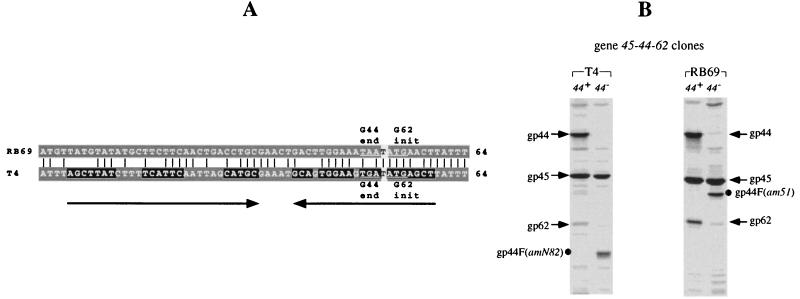
The IC45-44 regions of T4 and RB69 (A) and plasmid-mediated expression of genes 45, 44, and 62 (B). The T4 sequence has been reported previously (23–25; see also reference 8). The RB69 sequence was obtained from four different subclones that were derived from the RB69 λZAPII genomic library and confirmed by determinations on independent clones that were obtained after PCR amplification of genomic DNA. The alignment shown in panel A was purposely configured to depict the highest degree of similarity (51% [Table 1]) between the two sequences. Note that the two intercistronic regions differ in size (50 bp for T4 and 75 bp for RB69). The grey-shaded sequences constitute parts of open reading frames. The black-shaded sequences (underlined by arrows) constitute the stem portions of predicted mRNA secondary structures. The T4 RNA structure functions as part of a transcription termination signal (8). In the experiment for panel B, comparable DNA segments from the T4 and RB69 genomes were inserted in λpLN and λpL vectors and then introduced into E. coli NapIV for measurements of gene expression as described in Materials and Methods. The arrows on the SDS-PAGE autoradiogram indicate positions of the 35S-labeled plasmid-derived phage proteins. A 7.5%-10%-12.5% step gradient gel was used.
The results in Fig. 4B compare expression of the T4 and RB69 gene 45-44-62 subclusters under conditions of transcription termination (λpL plasmid) versus transcription antitermination (λpLN plasmid). We observed that with DNA from either phage, the overall level of expression of gene 44 (measured as gp44 synthesis) by the λpL plasmid was lower than that with the λpLN plasmid, i.e., six- to eightfold with T4 DNA and two- to threefold with RB69 DNA. Measurements based on gp62 synthesis in these experiments were not reliable due to low levels of synthesis of this protein and its poor resolution in some gel assays. Based on these observations, we suspect that the IC45-44 regions of both T4 and RB69 harbor transcription termination signals but that the nucleotide sequences (and perhaps efficiencies) of these signals are different from each other. We should also point out that although the T4 and RB69 gene 44 translation initiation regions appear to be very similar to each other in nucleotide sequence (Fig. 4A), they differ at several nucleotide positions. It is not yet known if these regions have quantitatively different efficiencies of translation initiation.
RB69 genes 44 and 62.
In T4, genes 44 and 62 are known to be translationally coupled (13), and it has been suggested that the coupling involves an RNA hairpin structure at the gene 44-62 juncture (29). An inspection of the RB69 nucleotide sequence at this juncture (Fig. 5A) suggests that no such RNA structure exists in RB69. Yet, as shown in Fig. 5B, amber mutations in gene 44 are polar on gp62 biosynthesis in both RB69 and T4. Thus, one need not invoke the involvement of RNA secondary structure in the mechanism of gp44-gp62 translational coupling.
Species specificity of the polymerase accessory proteins.
As shown in Fig. 6, the deduced primary structures of RB69 gp44 and gp62 are very similar to those of their T4 counterparts, with differences appearing in clusters of amino acid sequence in both cases. In previous studies, we had observed that the more highly diverged DNA polymerases (gp43s) of these two phages (∼74% similar [Table 1]) can partially substitute for each other in phage-plasmid complementation assays (34). Here, we examined whether the more highly conserved polymerase accessory proteins can also be exchanged between the T4 and RB69 replication systems. Figure 7 shows some results of plasmid-phage complementation assays that measured compatibilities between the RB69 and T4 gene products. Although protein amounts could not be quantitated in such assays, the results suggest that gp45 and the gp44/62 complex can indeed be functionally exchanged between T4 and RB69; however, as with gp43, we did observe preferences by the gene functions to support replication of the genomes from which they originated (Fig. 7A). Individually, gp44 and gp62 exhibited even more striking species specificity. That is, a phage-derived T4 gp62 failed to function (0 to 10% biological activity) with plasmid-derived RB69 gp44 (compare T4 44−62+ and RB69 44−62+ infections of pRB69g44-bearing cells in Fig. 7C; also compare RB69 44+62− infections of pRB69g62-bearing and pT4g62-bearing cells in Fig. 7C). The reciprocal combination (i.e., RB69 gene 62 with T4 gene 44) functioned well (compare T4 44−62+ and RB69 44−62+ infections of pT4g44-bearing cells in Fig. 7C; also, note the results with T4 44+62− infection of pRB69g62-bearing cells in Fig. 7C). We should also mention that T4 44amN82, the gene 44 amber mutant used in the experiment for Fig. 7, synthesizes reduced levels of gp62 due to polar effects of the amber mutation (13). Polarity may have caused decreases in burst size in infections of pT4g44-bearing cells with T4 44−62+ phage (i.e., values in Fig. 7C).
We also observed that a plasmid-expressed RB69 gene 44 inhibited growth of an infecting wild-type T4 phage (T4 wt infection of pRB69g44-bearing cells [Fig. 7B]). This observation may mean that performed RB69 gp44 (pRB69g44 expression) can form gp44/62 complexes with phage-derived T4 gp62 and that inhibition results from competition for T4 gp62 between the T4 and RB69 gp44 homologs; however, other explanations are also possible.
DISCUSSION
The T4 genomic region that specifies DNA polymerase and the DNA polymerase accessory proteins (T4 gene 46-43 cluster [Fig. 1]) is known to include three types of genetic elements: structural genes for essential replication proteins (gp46, gp45, gp44/62, and gp43), structural genes for seemingly nonessential regulatory proteins (RpbA and RegA), and untranslated intercistronic nucleotide sequences that harbor signals for transcriptional and posttranscriptional regulation of the structural genes (8). In addition, the T4 gene 46-43 cluster contains the open reading frame for a protein of unknown function, gp45.2, which has been detected in experiments involving plasmid-mediated expression of the cloned putative T4 gene (9). T4 gp45.2 has not been visualized in extracts of phage-infected cells, and no T4 gene 45.2 mutants have yet been described; however, transcripts of the gene 45.2 region can be detected in extracts of T4-infected cells and belong to a subclass of prereplicative RNAs that are subject to cleavage (and probably inactivation) by a nonessential T4-induced RNase (9, 21, 31). The processing enzyme, termed RegB protein, is stimulated by host ribosomal protein S1 and cleaves several T4-induced RNAs at GGAG sequences in untranslated initiation regions as well as some translated sites (22). Considering the high degree of conservation of the gene 45.2 sequence between T4 and RB69 (Table 1), we suspect that gp45.2 plays an essential role in propagation of both phage strains; however, this remains to be shown. In contrast, the T4 rpbA gene product has been shown to be nonessential for T4 growth in E. coli (35), and we are not surprised by its extinction during evolution of phage RB69. Maintenance of an RpbA function in T4 may have been associated with a requirement for this RNA polymerase-binding protein in certain natural hosts for T4 phage that are not shared by RB69.
It is obvious from our results that signals for quantitative regulation of essential phage replication proteins from the gene 46-43 cluster have been more prone to evolutionary divergence than have sites that determine the quality (structure) of these proteins (Table 1). Results of the T4-RB69 comparisons reported here and previously by Wang et al. (32, 34) probably demonstrate a common phenomenon in biology that regulatory sites and substances are designed to allow for wide variations in gene product dosage without causing adverse effects on organismal survival. For example, in the T4 system, differential overproduction of DNA polymerase by translational-operator-constitutive mutations (1, 30) or polymerase accessory proteins by regA mutations (14) is consistent with normal levels of DNA replication and phage production. That is, although the maintenance of regulated gene expression may have been essential for natural selection of the T4 and RB69 genomes, the precise titration of dosage of phage-induced replication proteins does not appear to be critical for assembly of functional phage DNA replication complexes.
Based on nucleotide sequence comparisons and results of expression studies, we can surmise that T4 and RB69 evolved different specificities of transcriptional control for the gp45-gp44-gp62 group and different levels of synthesis of these polymerase accessory proteins. We have noted that the T4 and RB69 IC45-44 regions both harbor RNA hairpin structures but that these hairpins are formed from different nucleotide sequences. In T4, RNA end mapping assays as well as expression studies have implicated the IC45-44 hairpin in transcription termination (8). The results reported here suggest that RB69 also terminates transcription in this region but by utilizing a signal different from that used by T4. Also, RB69 appears to lack T4-like MotA-dependent (middle-mode) transcription initiation sequences (27) upstream of genes 45.2 and 45 (Fig. 2), and so it is unclear if this phage possesses any MotA-like function or has evolved a different type of protein-DNA recognition specificity for initiating middle-mode transcription. In regard to posttranscriptional events, it also remains to be determined if T4 and RB69 have similar RNA processing activities for transcripts of this gene cluster (8) and if the two phage systems utilize such processing as well as translational repression (by their respective RegA proteins) to regulate molar ratios of the clamp loader and sliding-clamp proteins in infected cells. We have observed some nucleotide sequence differences between the known RegA-binding sites (translational operators) for T4 genes 45 and 44 (17) and the putative corresponding sites in RB69 (Fig. 4), and it will be interesting to find out if these differences reflect a divergence in RegA protein specificity between the two phages. A difference in RNA-binding specificities has been observed between the DNA polymerases of these phages (32).
We are particularly intrigued by the RB69-T4 divergence that we see on both sides of the translated gene 44-62 juncture. At the nucleotide sequence level, the differences predict that contiguity of the two cistrons is encompassed in an RNA secondary structure in the T4 but not the RB69 transcript (Fig. 5; see also reference 29). Since translational coupling of gp62 to gp44 synthesis can be demonstrated to occur in both phage systems, we conclude that RNA secondary structure is not a necessary component of the coupling mechanism between these two contiguous cistrons. Rather, a weak translation initiation site for gene 62 and close proximity of, and overlap with, the gene 44 termination region are probably the major determinants of the coupling. In addition, we suggest that translational coupling of these two cistrons was conserved in evolution, despite RNA sequence divergence, because it provided the advantage of smaller diffusion distances between gp44 and gp62 during assembly of gp44/62 complexes. Possibly, gp62 is captured by gp44 tetramers soon after translation is initiated at the gene 62 segment of the polycistronic message. In addition, translational coupling appears to be a determinant of the final gp44/gp62 ratio in the clamp loader (13), which may be another indication of cotranslational control of gp44/62 complex assembly.
We have also examined the patterns of primary structure conservation versus divergence between T4 and RB69 homologs of the replication proteins for indicators of structure-function relationships and clues to determinants of specificities of interactions among these proteins. For example, in previous work, we recognized that modular structure of the multifunctional T4 DNA polymerase is reflected in distribution of dissimilar segments of amino acid sequence between the T4 and RB69 gp43 variants (33, 34). Linear segments of gp43 that bear determinants for essential catalytic functions and interactions are very highly conserved (32) and probably underlie the abilities of the T4 and RB69 enzymes to partially substitute for each other in plasmid-phage complementation assays (34). The polymerase accessory proteins are more highly conserved than the polymerase, although here also, primary structure differences between T4 and RB69 homologs appear to occur nonrandomly (Fig. 3 and 6). At least some of the differences between accessory protein homologs are related to a divergence in their biological specificities. In particular, results of our plasmid-phage complementation assays suggest that T4 gp62 and RB69 gp44 can form a gp44/62 complex but that the complex is inactive for phage DNA replication (Fig. 7). It is known that gp62 is indispensable in phage replication and that it stimulates a DNA-dependent ATPase activity that is intrinsic to the gp44 component of the gp44/62 complex (23). Additional stimulation of the ATPase is achieved via interaction of the gp44/62 complex with gp45, i.e., during the clamp-loading event (10, 23). Recently, we constructed chimeric gene 44(RB69)-gene 62(T4) clones and observed that these complement T4 44−62− mutants only very poorly and that isolated gp44(RB69)/gp62(T4) complexes from these clones exhibit very low ATPase activity in vitro (unpublished results). Functional incompatibility between T4 gp62 and RB69 gp44 would suggest that the sites for mutual recognition between the two types of subunits of the clamp loader have coevolved in each phage system to maintain not only specific binding between the two proteins but also formation of the appropriate final quarternary structure required for biological activity of the heteromeric complex. Since most of the differences between the gp44 and gp62 homologs studied here occur in clusters of primary structure, it may be possible to localize the amino acid determinants for specificity of the gp44-gp62 interaction via the construction of RB69-T4 chimeras of each of these two proteins. Such an approach has helped localize some of the RNA-binding specificity determinants of gp43 (32). The identification of specificity differences and incompatibilities between other phylogenetic variants of DNA replication proteins encoded by phages of the T4 morphotype (18) would facilitate analysis of structure and assembly of the DNA replicase, as well as other multiprotein complexes of this group of DNA viruses.
ACKNOWLEDGMENTS
We thank Chien-Chia Wang, whose screening of the RB69 genomic library provided λZAPII clones that initiated these studies. We also thank Sandy Ng for carrying out the experiment for Fig. 4 and helping with manuscript preparation and Bill Konigsberg (Yale University) for stimulating discussions. Gary Latham (University of Oregon) provided confirmatory T4 gene 45 sequence information and helped clarify ambiguities in GenBank files (Fig. 3).
This work was supported by NIGMS grants GM18842 and GM54627. L.-S. Yeh also received predoctoral stipend support from Tulane University Medical Center.
REFERENCES
- 1.Andrake M D, Karam J D. Mutational analysis of the mRNA operator for T4 DNA polymerase. Genetics. 1991;128:203–213. doi: 10.1093/genetics/128.2.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carlson K, Miller E S. General procedures. In: Karam J D, Drake J W, Kreuzer K N, Mosig G, Hall D W, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, Miller E S, editors. Molecular biology of bacteriophage T4. Washington, D.C: American Society for Microbiology; 1994. pp. 427–437. [Google Scholar]
- 3.Dressman H K, Wang C-C, Karam J D, Drake J W. Retention of replication fidelity by a DNA polymerase functioning in a distantly related environment. Proc Natl Acad Sci USA. 1997;94:8042–8046. doi: 10.1073/pnas.94.15.8042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Epstein R H, Bolle A, Steinberg C, Kellenberger E, Boy de la Tour E, Chevalley R, Edgar R, Susman M, Denhardt C, Lielausis I. Physiological studies of conditional lethal mutants of bacteriophage T4D. Cold Spring Harbor Symp Quant Biol. 1963;28:375–392. [Google Scholar]
- 5.Gram H, Rüger W. Genes 55, αgt, 47 and 46 of bacteriophage T4: the genomic organization as deduced by sequence analysis. EMBO J. 1985;4:257–264. doi: 10.1002/j.1460-2075.1985.tb02344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guild N, Gayle M, Sweeney R, Hollingsworth T, Modeer T, Gold L. Transcriptional activation of bacteriophage T4 middle promoters by the motA protein. J Mol Biol. 1988;199:241–258. doi: 10.1016/0022-2836(88)90311-7. [DOI] [PubMed] [Google Scholar]
- 7.Hsu T. Developmental control of bacteriophage T4 gene expression: transcriptional and post-transcriptional mechanics. Ph.D. dissertation. Charleston, S.C: Medical University of South Carolina; 1991. [Google Scholar]
- 8.Hsu T, Karam J D. Transcriptional mapping of a DNA replication gene cluster in bacteriophage T4. Sites for initiation, termination and mRNA processing. J Biol Chem. 1990;265:5303–5316. [PubMed] [Google Scholar]
- 9.Hsu T, Wei R, Dawson M, Karam J. Identification of two new bacteriophage T4 genes that may have roles in transcription and DNA replication. J Virol. 1987;68:366–374. doi: 10.1128/jvi.61.2.366-374.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jarvis T C, Paul L S, Hockensmith J W, von Hippel P H. Structural and enzymatic studies of the T4 DNA replication system. II. ATPase properties of the polymerase accessory protein complex. J Biol Chem. 1989;264:12717–12729. [PubMed] [Google Scholar]
- 11.Jarvis T C, Paul L S, von Hippel P H. Structural and enzymatic studies of the T4 DNA replication system. I. Physical characterization of the polymerase accessory protein complex. J Biol Chem. 1989;264:12709–12716. [PubMed] [Google Scholar]
- 12.Jozwik C E, Miller E S. Regions of bacteriophage T4 and RB69 RegA translational repressor proteins that determine RNA-binding specificity. Proc Natl Acad Sci USA. 1992;89:5053–5057. doi: 10.1073/pnas.89.11.5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karam J, Bowles M, Leach M. Expression of bacteriophage T4 genes 45, 44 and 62. I. Discoordinate synthesis of the T4 45- and 44- proteins. Virology. 1979;94:192–203. doi: 10.1016/0042-6822(79)90449-5. [DOI] [PubMed] [Google Scholar]
- 14.Karam J, McCulley C, Leach M. Genetic control of mRNA decay in T4 phage-induced Escherichia coli. Virology. 1977;76:685–700. doi: 10.1016/0042-6822(77)90251-3. [DOI] [PubMed] [Google Scholar]
- 15.Kelman Z, O’Donnell M. DNA polymerase III holoenzyme: structure and function of a chromosomal replicating machine. Annu Rev Biochem. 1995;64:171–200. doi: 10.1146/annurev.bi.64.070195.001131. [DOI] [PubMed] [Google Scholar]
- 16.Miller E S, Karam J, Dawson M, Trojanowska M, Gauss P, Gold L. Translational repression: biological activity of plasmid-encoded bacteriophage T4 RegA protein. J Mol Biol. 1987;194:397–410. doi: 10.1016/0022-2836(87)90670-x. [DOI] [PubMed] [Google Scholar]
- 17.Miller, E. S., J. D. Karam, and E. Spicer. Control of translation initiation: mRNA structure and protein repressors, p. 193–208. In J. D. Karam, J. W. Drake, K. N. Kruezer, G. Mosig, D. W. Hall, F. A. Eiserling, L. W. Black, E. K. Spicer, E. Kutter, K. Carlson, and E. S. Miller (ed.), Molecular biology of bacteriophage T4. American Society for Microbiology, Washington, D.C.
- 18.Monod C, Repoila F, Kutateladze M, Tétart F, Krisch H M. The genome of the pseudo T-even bacteriophages, a diverse group that resembles T4. J Mol Biol. 1997;267:237–249. doi: 10.1006/jmbi.1996.0867. [DOI] [PubMed] [Google Scholar]
- 19.Nelson M A, Ericson M, Gold L, Pulitzer J F. The isolation and characterization of TabR bacteria: host that restrict bacteriophage T4 rII mutants. Mol Gen Genet. 1982;188:60–68. [Google Scholar]
- 20.Nossal N G. The bacteriophage T4 DNA replication fork. In: Karam J D, Drake J W, Kreuzer K N, Mosig G, Hall D W, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, Miller E S, editors. Molecular biology of bacteriophage T4. Washington, D.C: American Society for Microbiology; 1994. pp. 43–53. [Google Scholar]
- 21.Ruckman J, Parma D, Tuerk C, Hall D H, Gold L. Identification of T4 gene required for bacteriophage mRNA processing. New Biol. 1989;1:54–65. [PubMed] [Google Scholar]
- 22.Ruckman J, Ringquist S, Brody E, Gold L. The bacteriophage T4 regB ribonuclease. Stimulation of the purified enzyme by ribosomal protein S1. J Biol Chem. 1994;269:26655–26662. [PubMed] [Google Scholar]
- 23.Rush J, Lin T C, Quinones M, Spicer E K, Douglas I, Williams K R, Konigsberg W H. The 44P subunit of the T4 DNA polymerase accessory protein complex catalyzes ATP hydrolysis. J Biol Chem. 1989;264:10943–10953. [PubMed] [Google Scholar]
- 24.Spicer E K, Noble J A, Nossal N G, Konigsberg W H, Williams K R. Bacteriophage T4 gene 45. J Biol Chem. 1982;257:8972–8979. [PubMed] [Google Scholar]
- 25.Spicer E K, Nossal N G, Williams K R. Bacteriophage T4 gene 44 DNA polymerase accessory protein. Sequences of gene 44 and its protein product. J Biol Chem. 1984;259:15425–15432. [PubMed] [Google Scholar]
- 26.Stillman B. Smart machines at the DNA replication fork. Cell. 1994;78:725–728. doi: 10.1016/s0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- 27.Stitt B, Hinton D. Regulation of middle-mode transcription. In: Karam J D, Drake J W, Kreuzer K N, Mosig G, Hall D W, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, Miller E S, editors. Molecular biology of bacteriophage T4. Washington, D.C: American Society for Microbiology; 1994. pp. 142–160. [Google Scholar]
- 28.Tinker R L, Williams K P, Kassavetis G A, Geiduschek E P. Transcriptional activation by a DNA-tracking protein: structural consequences of enhancement at the T4 late promoter. Cell. 1994;77:225–237. doi: 10.1016/0092-8674(94)90315-8. [DOI] [PubMed] [Google Scholar]
- 29.Trojanowska M, Miller E S, Karam J, Stormo G, Gold L. The bacteriophage T4 regA gene; primary sequence of a translational repressor. Nucleic Acids Res. 1984;12:5979–5993. doi: 10.1093/nar/12.15.5979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tuerk C, Eddy S, Parma D, Gold L. Autogenous translational operator recognized by bacteriophage T4 DNA polymerase. J Mol Biol. 1990;213:749–761. doi: 10.1016/S0022-2836(05)80261-X. [DOI] [PubMed] [Google Scholar]
- 31.Uzan M, Favre R, Brody E. A nuclease that cuts specifically in the ribosome binding site of some T4 mRNAs. Proc Natl Acad Sci USA. 1988;85:8895–8899. doi: 10.1073/pnas.85.23.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang C-C, Pavlov A, Karam J D. Evolution of RNA-binding specificity in T4 DNA polymerase. J Biol Chem. 1997;272:17703–17710. doi: 10.1074/jbc.272.28.17703. [DOI] [PubMed] [Google Scholar]
- 33.Wang J, Sattar A K M, Wang C-C, Karam J D, Konigsberg W H, Steitz T A. Crystal structure of a pol α family replication DNA polymerase from bacteriophage RB69. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- 34.Wang C-C, Yeh L-S, Karam J D. Modular organization of T4 DNA polymerase. Evidence from phylogenetics. J Biol Chem. 1995;270:26558–26564. doi: 10.1074/jbc.270.44.26558. [DOI] [PubMed] [Google Scholar]
- 35.Williams K P, Kassavetis G A, Esch F S, Geiduschek E P. Identification of the gene encoding an RNA polymerase-binding protein of bacteriophage T4. J Virol. 1987;61:597–599. doi: 10.1128/jvi.61.2.597-599.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Williams K P, Kassavetis G A, Herendeen D R, Geiduschek E P. Regulation of late-gene expression. In: Karam J D, Drake J W, Kreuzer K N, Mosig G, Hall D W, Eiserling F A, Black L W, Spicer E K, Kutter E, Carlson K, Miller E S, editors. Molecular biology of bacteriophage T4. Washington, D.C: American Society for Microbiology; 1994. pp. 161–175. [Google Scholar]
- 37.Wilson, G. Personal communication.