Abstract
Covalently circularized and DNA-corralled nanodisc technologies have enabled engineering of large sized-bilayer nanodiscs up to 90 nm. These large nanodiscs have the potential to extend the applicability of nanodisc technology from studying small and medium-sized membrane proteins to acting as surrogate membranes to investigate functional and structural aspects of viral entry. Here, we discuss the recent technical developments leading to construction of large circularized and DNA-corralled nanodiscs and examine their application in viral entry.
Keywords: Covalently circularized Nanodiscs, DNA-corralled Nanodiscs, Viral entry
Introduction
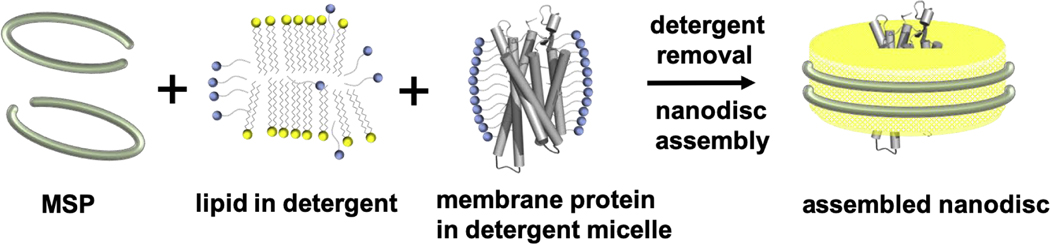
Phospholipid bilayer nanodiscs have attracted great interest from structural biologists as a native-like membrane mimetic for studying membrane proteins. Nanodiscs are detergent-free lipid bilayer models, which enable the study of membrane proteins in a physiologically relevant environment [1–3]. A conventional nanodisc is composed of a phospholipid bilayer patch (~ 8–16 nm in diameter) encircled by two copies of amphipathic helical proteins termed membrane scaffold proteins (MSPs) (Figure 1). MSPs are engineered forms of apolipoprotein A-1, which is the major component of high-density lipoprotein. The hydrophobic residues of MSP interact with the lipid acyl chain; the hydrophilic face is located at the outside surface to allow the nanodisc to stay in solution. Any kind of membrane forming lipid can be used to form nanodiscs and the diameter of the captured lipid patch is determined by the length of the scaffold protein[2,3]. Nanodiscs have been used successfully to stabilize many membrane proteins and render them soluble in aqueous buffers, facilitating high-resolution structural determination in lipid bilayer using cryoEM and NMR. Examples of single-particle cryo-EM structures include the tetrameric TRPV1 ion channel (330 KDa) [4]. The TRPV1 structure revealed the mechanism for channel activation by bioactive lipids. Moreover, it showed how certain phospholipid interactions enhance binding of a spider toxin to the channel. Another remarkable cryo-EM structure is that of the ryanodine receptor RyR1 (2.3 MDa) [5]. The structure identified the calcium binding domain and revealed how the calcium allosterically regulates gating of the channel. There are many more examples of cryo-EM structures of membrane proteins in nanodiscs. These examples include the lipid exporter ABCB4[6], MsbA transporter [7], Transient receptor potential channel subfamily A member 1 (TRPA1)[8], a voltage-activated potassium channel[9], CorA magnesium channel[10]. In addition to facilitating the structure determination of a single membrane protein, nanodiscs have been used to assemble and enable the cryo-EM structure of membrane protein complexes such as the ribosome-SecYE complex[11].
Figure 1.
Traditional nanodisc self-assembly process. Membrane scaffold protein (MSP), lipid and target membrane protein solubilized in detergent assemble nanodisc with incorporated membrane protein upon detergent removal.
Nanodiscs have been also used in both solution and solid-state NMR to reveal critical details of small membrane protein structure and function [12,13]. There are many other applications for nanodisc technology which are covered by several excellent reviews [14–16].
Further engineering efforts are expanding the nanodisc size and stability thus extending the potential applications. Our group and others have developed methods to covalently link the N- and C- termini of membrane scaffold protein variants. As a result of the covalent circularization, we and others have produced nanodiscs with a large range of discrete sizes up to 50 nm [17–19]. The 50 nm nanodisc has been successfully used in studying the dynamics of trans-SNARE complexes and investigate the number of SNAREs needed to drive fusion [20]. Moreover, the 50 nm nanodisc has been used to study poliovirus entry [17]. Other efforts to expand the nanodisc size beyond 50 nm include using a DNA origami scaffold to help construct and stabilize nanodiscs within the DNA cavity. These latter efforts led to the production of nanodiscs up to 90 nm, which were employed to investigate poliovirus interactions with its receptor embedded in bilayer[21]. Here, we give a concise overview of the design and applications of these large nanodiscs in studying viral entry.
Engineering large nanodiscs
1. Covalent protein circularization
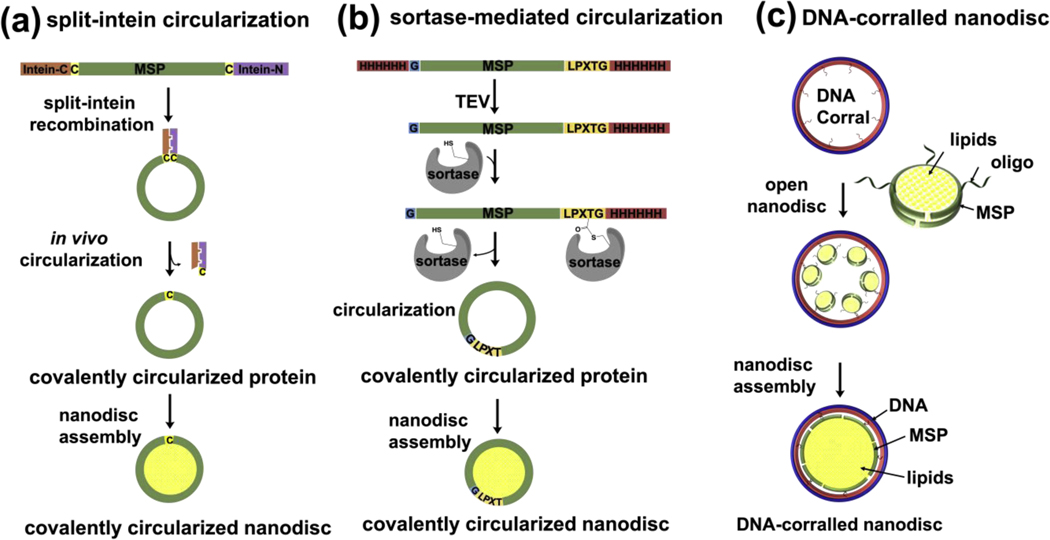
The covalent circularization of proteins results in improved thermal stability [22–24] and proteolytic resistance [24–28]. MSP represents an attractive target for covalent circularization because its N- and C- termini are in close proximity [29,30]. There are several established methods for protein circularization. These methods include the use of intein-fusion proteins [31,32], which permit the covalent circularization through expressed-protein ligation or protein trans-splicing. Alternatively, circularized proteins can be produced by using sortase enzymes [33], butelase enzyme [34] or chemical ligations [35]. Combining protein circularization with MSPs enables the creation of large (> 16 nm) and more stable nanodiscs of well-defined circular and polygonal shapes.
Exploiting the potential advantages of circularized proteins, our group engineered recombinant versions of MSP that can be circularized using sortase and we were able to assemble very stable and homogenous nanodiscs of varying sizes up to 50 nm [17]. We have engineered four different variants of circularizable scaffold proteins: NW9, NW11, NW30, and NW50 which assemble ~ 8.5, 11, 15, 50 nm nanodiscs respectively. The NW constructs contain a TEV protease-cleavable N-terminal His tag followed by a single glycine, and a C-terminal sortase-cleavable His tag (Figure 2b). The presence of these two sites ensures covalent linkage between the N and C termini of NWs while still preserving the function to form nanodiscs. Following the publication of the sortase-mediated circularization of nanodiscs, several other groups worked on optimizing the circularization protocol to improve the yield and minimize byproducts. Yusuf et al [19] used detergent during the circularization reaction in order to improve the yield and minimize the high molecular weight oligomers. Another group aimed at continued improvement of the circularized nanodisc protocol and optimized the original MSP sequence to incorporate extra negatively charged residues. With these modifications , they were able to improve the solubility of MSP and perform the purification and circularization reaction in the absence of any detergents [36]. Furthermore, a recent study demonstrated the production of the circularized MSP by using the in vivo split intein ligation in E. coli (Figure 2a). This split intein method has been shown to yield circularized MSP nanodiscs of varying sizes from 8 nm to 26 nm [18].
Figure 2. Different strategy that have been successfully used for production of large nanodiscs.
(a) Design of the Npu DnaE split-intein MSP fusion construct and mechanism of intein splicing in vivo, leading to the generation of circularized MSP. (b) A general outline of the constructs for making covalently circularized nanodiscs using sortase. Glycine (G) and LPXTG at the N and C termini respectively are necessary for the linkage by sortase. (c) DNA-corralled reconstitution strategy. Addition of detergent and lipids to small nanodisc- decorated barrels followed by dialysis results in the construction of a large bilayer nanodisc within the DNA cavity. Each DNA barrel is decorated with ssDNA overhang as handles to hybridize with ssDNA chemically conjugated small nanodisc.
2. DNA-Corralled nanodisc
Recently, we reported a method of constructing a large nanodisc inside the cavity of DNA corrals[21]. Each DNA corral recruits a number of ~11 nm diameter nanodiscs that are assembled by noncircularized, oligonucleotide-functionalized MSPs, and directs their reconstitution into a single large nanodisc (Figure 2c). We employed two different sized barrels: 90 and 60 nm outer diameter to reconstitute ~70 or ~45 nm DNA-corralled nanodiscs, respectively. The DNA corral offers a number of useful features including the ability to assemble large nanodiscs while circumventing the need for sortase circularization and production of larger MSP variants. In addition, DNA corral acts as a bumper case to prevent nanodisc aggregation and facilitates control over stoichiometry, geometry, and orientation of inserted proteins through tethering to the corral. The enclosed nanodiscs are relatively stable and tolerant of a broad range of pH levels and divalent ion concentrations. This DNA-corralled nanodisc system was used to reconstitute two membrane-protein clusters and to study poliovirus entry[21]. Because of the unprecedented possibilities of controlling the stoichiometry and orientation of inserted proteins through tethering to the DNA corral, DNA-corralled nanodiscs have great potential in studying the stoichiometry of viral receptor engagement. Moreover, they can be utilized to incorporate multimeric receptor complexes and study the impact of these complexes on fusion and penetration dynamics as well as subsequent viral uncoating. A similar approach to DNA-corralled nanodisc was reported by Iric et al [37] to prepare DNA-encircled bilayer (DEB) structures. The DEB structures are made of multiple copies of an alkylated oligonucleotide hybridized to a single-stranded minicircle. The presence of the alkyl modification enables interactions between negatively charged hydrophilic DNA and lipids.
Virus entry
1. Imaging of membrane interaction
Viruses interact with, modulate and penetrate the cellular membranes during cell entry and exit. In order to ensure successful viral replication, the entry of viruses into host cells requires the disruption of the membrane without compromising cell integrity. For enveloped viruses, transfer of viral genome is achieved by fusion of viral and cellular membranes[38]. This fusion step is facilitated by the glycoproteins on the surface of enveloped viruses. On the other hand, most of the non-enveloped viruses transiently destabilize the host membranes using amphipathic or hydrophobic capsid peptides[39]. Many questions regarding the mechanisms of membrane penetration, genome translocation and disassembly of non-enveloped virus remain unanswered. Thus far, elucidating these mechanisms have been challenging, due in part to technical difficulties relating to direct visualization of viral gene delivery and size heterogeneity of liposomes, which are commonly used membrane models for nonenveloped viral entry studies. Previous studies using liposomes have resulted in several low-resolution structural models of cell entry and have shed light on the mechanisms of the early stages of viral entry. Strauss and colleagues have shown that poliovirus 135s particles form a 50 Å elongated extension or “umbilical connector”, attaching the poliovirus to the membrane to allow the passage of the viral genome and initiating the infection[40]. Kumar and colleagues reported that the protein shell of membrane-bound human rhinovirus 2 intimately interacts with the membrane; they did not observe any umbilical connector [41]. Another study reported a cryo-EM structure for a complex of a small nanodisc (10 nm) containing coxsackie-adenovirus receptors with human pathogen coxsackie virus B3 (CVB3)[42]. The structure was resolved to 7.8 Å without or to 3.9 Ǻ with imposed icosahedral symmetry and revealed local dynamics in capsid structures. The authors identified an extension of electron density near the nanodisc membrane and formation of a channel on the surface of the virus capsid. Even though the use of small nanodiscs only minimally interfered with capsid imaging and triggered local structural rearrangement, it failed to trigger RNA release.
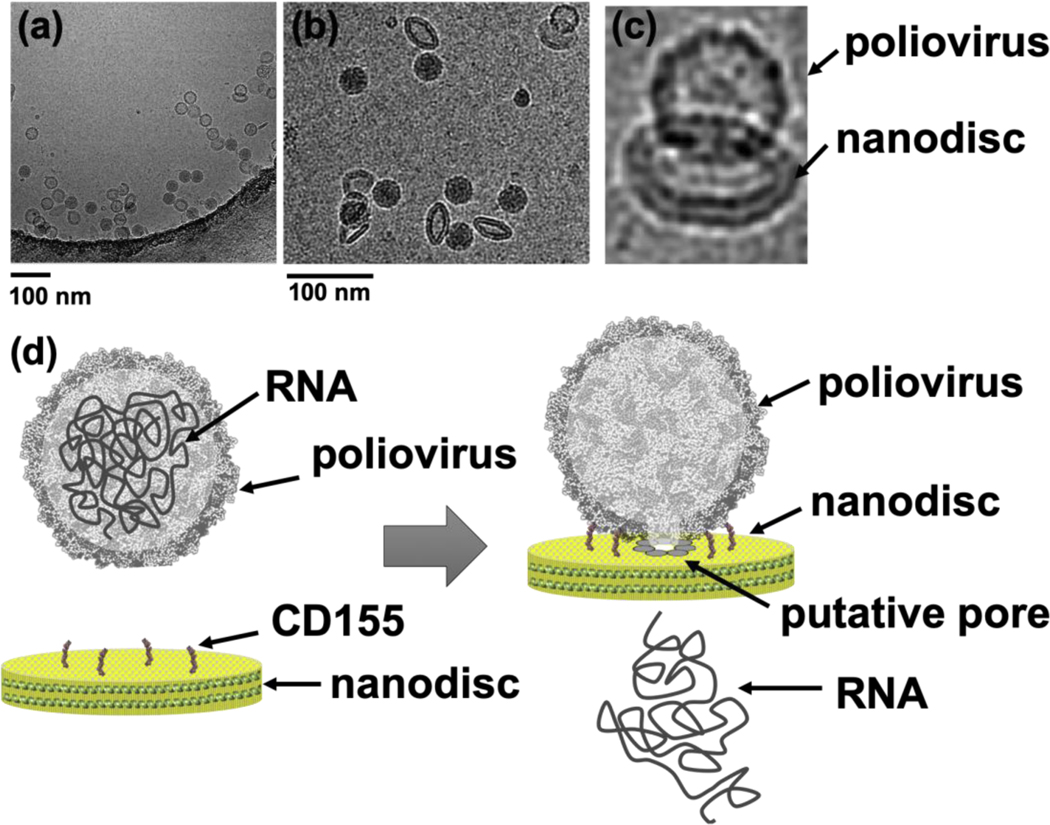
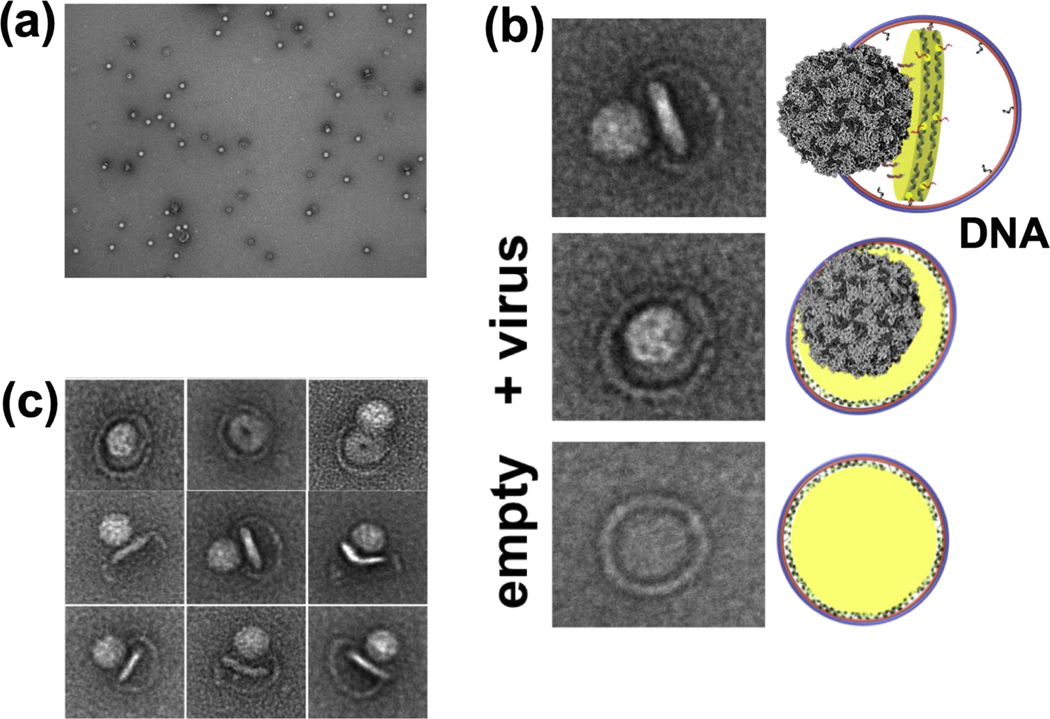
Unlike small nanodiscs, large covalently circularized nanodiscs successfully triggered RNA release when incubated with poliovirus [17]. This could be attributed to the fact that poliovirus can accommodate multiple copies of viral receptors and has enough surface for the RNA-translocation complex during viral uncoating. The quality of the cryo-EM data we collected on the poliovirus–nanodisc complexes (Figure 3) was substantially better than the data collected using the poliovirus-liposome complexes. The cryo-EM images in figure 3a–c show poliovirus bound to 50 nm cNDs decorated with CD155 ectodomain receptors, formation of a putative pore/channel and ejection of RNA across the membrane. Similarly, using 60 nm DNA-corralled nanodiscs functionalized with CD155 ectodomain, we were successful in initiating the receptor mediated uncoating of poliovirus and observed the early steps of virus attachment to the bilayer and pore formations in the nanodisc (Figure 4).
Figure 3.
Poliovirus caught in the act. (a) Cryo-EM image of poliovirus interacting with 50 nm covalently circularized nazanodiscs containing CD155 receptor. (b) Tilted views of the nanodisc were observed in ice. (c) Cryo-EM image of individual viral particle tethered to nanodisc containing CD155. (d) Schematic of pore formation and RNA release from poliovirus capsid across the bilayer nanodisc.
Figure 4.
Poliovirus interactions with DNA corralled nanodiscs containing CD155 receptor. (a) Negative-stain TEM images of 60 nm DNA-corralled nanodiscs containing CD155 ectodomain. (b) TEM images of individual Negative-stain TEM images of DCND (60 nm outer diameter) containing CD155 ectodomain interacting with poliovirus. Some of the nanodiscs were partially released from their DNA scaffolds after binding the virus. (c) TEM images of individual viral particles showing the bending of the bilayer and the creation of a pore in nanodisc by the poliovirus. This figure is adapted from reference [21••].
Poliovirus particles have been shown to induce the formation of channels or pores in planar membranes. This was demonstrated by electrophysiology experiments [43] and genetic studies [44]. Danthi at al. have demonstrated that mutations in the VP4 caspid protein alter the ability of poliovirus to form channels and release RNA during infection [31]. This has led to the hypothesis that the insertion of VP4 into membranes enables the formation of pores or channels in host membranes that allow the translocation of the viral genome into the cytoplasm of the cell [45].
2. Role of host lipids in viral entry
Lipids play a critical role in viral entry and replication. Several studies have shown that the alterations of membrane lipid composition can block viral release and entry. For example, a study by Snyder and colleagues shows that the phosphatidylethanolamine lipid cooperates with the reovirus membrane penetration peptide to facilitate viral particle uncoating and enables the structural transition of intermediate subviral particles [46]. Another study demonstrated that the infectivity of bovine rotavirus is directly influenced by lysobisphosphatidic acid during the ESCRT-mediated entry [47]. Similarly, lysobisphosphatidic acid cooperates with the membrane penetration peptide, VP5, of the bluetongue virus to facilitate viral uncoating [48]. On the other hand, Ceramides have been shown to inhibit viral entry, likely due to self segregating into ceramide-rich microdomains[49].
In addition to certain lipids, membrane structures such as lipid rafts have been shown to facilitate the entry of many viruses [50,51]. The diameter of lipid rafts are on the order of 10–50 nm [52] and many viral receptors and/or co-receptors are often localized in the rafts [53].
Host proteins, other than the specific receptor(s) for any given virus, have been shown to play critical roles in membrane penetration by nonenveloped viruses. For example SV40 and human BK viruses, members of the polyomavirus family, have been shown to use EMC1 and SGTA chaperones in addition to the heat shock protein Hsp105 to enable their release from the ER membrane and transportation into the cytosol [54,55]. Another study using a genome-wide haploid genetic screen, has identified the lipid-modifying enzyme PLA2G16 as a host factor that functions early during picornaviruses infection, enabling genome delivery into the cytoplasm [56]. Large nanodiscs can be used to create a functional replication of membrane systems, including duplication of lipid compositions, fluidity and potentially membrane curvature, as well as lipid rafts and asymmetry. Moreover, they can provide a large membrane area to allow the co-incorporation of the specific receptor(s) along with other host protein factors important for the entry and replication of any given virus. Research has only begun to tap the broad potential applications of the large designed nanodiscs.
Conclusions and future directions
This review is an effort to shed light on the recent technical developments leading to the construction of large circularized protein and DNA-corralled nanodiscs and discuss some of the viral entry applications. These nanodiscs are sufficiently large to accommodate multiple copies of any given viral receptor and provide enough surface area to act as a surrogate membrane for the genome-translocation complexes during viral uncoating. They can be used to mimic specific membrane features including lipid compositions and curvature. Although extensive research over the past four decades has resulted in identification of many membrane-penetrating peptides and many models to explain how genome of non-enveloped viruses is released across the bilayer, questions regarding the mechanisms of membrane penetration and the influence of host factors remain unanswered. The receptor-decorated large nanodisc model system represents a very attractive tool, in that it can allow virus structures to be determined in the context of membranes and has already begun to reveal some exciting information about the nature of pore/channel that is triggered by non-enveloped viruses.
Despite rapid progress in advancing nanodisc technology, there are areas that warrant further exploration. Currently it is difficult to create asymmetric nanodiscs whose lipid composition is different on their two faces. Biological membranes are asymmetric with respect to lipid distributions across the bilayer and this asymmetry gives rise to a nonzero intrinsic potential difference between the two sides of the membrane in the absence of ions and has important functional consequences. Therefore, reconstructing this asymmetry in nanodisc system will provide invaluable tool that can be widely used to evaluate the role of this parameter in membrane protein function.
Acknowledgements
The author acknowledges Mike Strauss (McGill University) for his help in imaging poliovirus-nanodisc complex (Figure 3a–c). The author acknowledges the support provided by USA National Institutes of Health (NIH) grant R01GM131401.
Funding
Funding was received for this work.
Footnotes
Intellectual Property
We confirm that we have given due consideration to the protection of intellectual property associated with this work and that there are no impediments to publication, including the timing of publication, with respect to intellectual property. In so doing we confirm that we have followed the regulations of our institutions concerning intellectual property.
Research Ethics
We further confirm that any aspect of the work covered in this manuscript that has involved human patients has been conducted with the ethical approval of all relevant bodies and that such approvals are acknowledged within the manuscript.
IRB approval was obtained (required for studies and series of 3 or more cases)
Written consent to publish potentially identifying information, such as details or the case and photographs, was obtained from the patient(s) or their legal guardian(s).
Conflict of interest
The author is a co-founder of NOW Scientific LLC.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Bayburt TH, Carlson JW, Sligar SG: Reconstitution and imaging of a membrane protein in a nanometer-size phospholipid bilayer. J Struct Biol 1998, 123:37–44. [DOI] [PubMed] [Google Scholar]
- 2.Bayburt TH, Grinkova YV, Sligar SG: Self-assembly of discoidal phospholipid bilayer nanoparticles with membrane scaffold proteins. Nano Letters 2002, 2:853–856. [Google Scholar]
- 3.Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG: Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol 2009, 464:211–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gao Y, Cao E, Julius D, Cheng Y: TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 2016, 534:347–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Efremov RG, Leitner A, Aebersold R, Raunser S: Architecture and conformational switch mechanism of the ryanodine receptor. Nature 2015, 517:39–43. [DOI] [PubMed] [Google Scholar]
- 6.Olsen JA, Alam A, Kowal J, Stieger B, Locher KP: Structure of the human lipid exporter ABCB4 in a lipid environment. Nat Struct Mol Biol 2019. [DOI] [PubMed] [Google Scholar]
- 7.Mi W, Li Y, Yoon SH, Ernst RK, Walz T, Liao M: Structural basis of MsbA-mediated lipopolysaccharide transport. Nature 2017, 549:233–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suo Y, Wang Z, Zubcevic L, Hsu AL, He Q, Borgnia MJ, Ji RR, Lee SY: Structural Insights into Electrophile Irritant Sensing by the Human TRPA1 Channel. Neuron 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matthies D, Bae C, Toombes GE, Fox T, Bartesaghi A, Subramaniam S, Swartz KJ: Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs. Elife 2018, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matthies D, Dalmas O, Borgnia MJ, Dominik PK, Merk A, Rao P, Reddy BG, Islam S, Bartesaghi A, Perozo E, et al. : Cryo-EM Structures of the Magnesium Channel CorA Reveal Symmetry Break upon Gating. Cell 2016, 164:747–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frauenfeld J, Gumbart J, Sluis EO, Funes S, Gartmann M, Beatrix B, Mielke T, Berninghausen O, Becker T, Schulten K, et al. : Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat Struct Mol Biol 2011, 18:614–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yoshiura C, Kofuku Y, Ueda T, Mase Y, Yokogawa M, Osawa M, Terashima Y, Matsushima K, Shimada I: NMR analyses of the interaction between CCR5 and its ligand using functional reconstitution of CCR5 in lipid bilayers. J Am Chem Soc 2010, 132:6768–6777. [DOI] [PubMed] [Google Scholar]
- 13.Hagn F, Etzkorn M, Raschle T, Wagner G: Optimized phospholipid bilayer nanodiscs facilitate high-resolution structure determination of membrane proteins. J Am Chem Soc 2013, 135:1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denisov IG, Sligar SG: Nanodiscs in Membrane Biochemistry and Biophysics. Chem Rev 2017, 117:4669–4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rouck JE, Krapf JE, Roy J, Huff HC, Das A: Recent advances in nanodisc technology for membrane protein studies (2012–2017). FEBS Lett 2017, 591:2057–2088. •This review article highlights the different applications of nanodisc technology. The authors reviewed the techniques that have been integrated with nanodisc technology to study membrane proteins.
- 16.Efremov RG, Gatsogiannis C, Raunser S: Lipid Nanodiscs as a Tool for High-Resolution Structure Determination of Membrane Proteins by Single-Particle Cryo-EM. Methods Enzymol 2017, 594:1–30. [DOI] [PubMed] [Google Scholar]
- 17. Nasr ML, Baptista D, Strauss M, Sun ZJ, Grigoriu S, Huser S, Pluckthun A, Hagn F, Walz T, Hogle JM, et al. : Covalently circularized nanodiscs for studying membrane proteins and viral entry. Nat Methods 2017, 14:49–52. ••The authors engineered covalently circularized nanodiscs up to 50 nm in diameter. They used 50-nm nanodisc as a model membrane to study the poliovirus interactions with its receptor , CD155, attached to membrane. EM images show the formation of a putative pore in the nanodisc.
- 18. Miehling J, Goricanec D, Hagn F: A Split-Intein-Based Method for the Efficient Production of Circularized Nanodiscs for Structural Studies of Membrane Proteins. Chembiochem 2018, 19:1927–1933. •The authors used in vivo split intein ligation system in E. coli to produce covalently circularized nanodiscs. They assembeled nanodiscs of varying sizes from 8 nm to 26 nm.
- 19.Yusuf Y, Massiot J, Chang YT, Wu PH, Yeh V, Kuo PC, Shiue J, Yu TY: Optimization of the Production of Covalently Circularized Nanodiscs and Their Characterization in Physiological Conditions. Langmuir 2018, 34:3525–3532. [DOI] [PubMed] [Google Scholar]
- 20. Bao H, Das D, Courtney NA, Jiang Y, Briguglio JS, Lou X, Roston D, Cui Q, Chanda B, Chapman ER: Dynamics and number of trans-SNARE complexes determine nascent fusion pore properties. Nature 2018, 554:260–263. The authors employed fusion assays that use nanodiscs to trap pores in their initial open state. They provided direct experimental support for the idea that a certain number of SNAREs is needed to hold fusion pores open. Moreover, they used 50 nm nanodisc in studying the dynamics of trans-SNARE complexes and investigating the number of SNAREs needed to drive fusion.
- 21. Zhao Z, Zhang M, Hogle JM, Shih WM, Wagner G, Nasr ML: DNA-Corralled Nanodiscs for the Structural and Functional Characterization of Membrane Proteins and Viral Entry. J Am Chem Soc 2018, 140:10639–10643. ••This report is about manufacturing large-sized nanodiscs using DNA-origami barrels as scaffolding corrals. The authors produced large nanodiscs by first decorating the inside of DNA barrels with small lipid-bilayer nanodiscs, which open up when adding extra lipid to form large nanodiscs. They assembeled nanodiscs of diameters up to 90 nm. They demonstrated the potential of these nanodiscs as model membranes to study poliovirus entry.
- 22.Stevens CA, Semrau J, Chiriac D, Litschko M, Campbell RL, Langelaan DN, Smith SP, Davies PL, Allingham JS: Peptide backbone circularization enhances antifreeze protein thermostability. Protein Sci 2017, 26:1932–1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trabi M, Craik DJ: Circular proteins--no end in sight. Trends Biochem Sci 2002, 27:132–138. [DOI] [PubMed] [Google Scholar]
- 24.Iwai H, Pluckthun A: Circular beta-lactamase: stability enhancement by cyclizing the backbone. FEBS Lett 1999, 459:166–172. [DOI] [PubMed] [Google Scholar]
- 25.Qi X, Xiong S: Intein-mediated backbone cyclization of VP1 protein enhanced protection of CVB3-induced viral myocarditis. Sci Rep 2017, 7:41485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Popp MW, Ploegh HL: Making and breaking peptide bonds: protein engineering using sortase. Angew Chem Int Ed Engl 2011, 50:5024–5032. [DOI] [PubMed] [Google Scholar]
- 27.Borra R, Camarero JA: Recombinant expression of backbone-cyclized polypeptides. Biopolymers 2013, 100:502–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popp MW, Dougan SK, Chuang TY, Spooner E, Ploegh HL: Sortase-catalyzed transformations that improve the properties of cytokines. Proc Natl Acad Sci U S A 2011, 108:3169–3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bibow S, Polyhach Y, Eichmann C, Chi CN, Kowal J, Albiez S, McLeod RA, Stahlberg H, Jeschke G, Guntert P, et al. : Solution structure of discoidal high-density lipoprotein particles with a shortened apolipoprotein A-I. Nat Struct Mol Biol 2017, 24:187–193. [DOI] [PubMed] [Google Scholar]
- 30.Ajees AA, Anantharamaiah GM, Mishra VK, Hussain MM, Murthy HM: Crystal structure of human apolipoprotein A-I: insights into its protective effect against cardiovascular diseases. Proc Natl Acad Sci U S A 2006, 103:2126–2131. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Elleuche S, Poggeler S: Inteins, valuable genetic elements in molecular biology and biotechnology. Appl Microbiol Biotechnol 2010, 87:479–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Evans TC Jr., Benner J, Xu MQ: The cyclization and polymerization of bacterially expressed proteins using modified self-splicing inteins. J Biol Chem 1999, 274:18359–18363. [DOI] [PubMed] [Google Scholar]
- 33.Antos JM, Popp MW, Ernst R, Chew GL, Spooner E, Ploegh HL: A straight path to circular proteins. J Biol Chem 2009, 284:16028–16036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen GK, Wang S, Qiu Y, Hemu X, Lian Y, Tam JP: Butelase 1 is an Asx-specific ligase enabling peptide macrocyclization and synthesis. Nat Chem Biol 2014, 10:732–738. [DOI] [PubMed] [Google Scholar]
- 35.Cowper B, Craik DJ, Macmillan D: Making ends meet: chemically mediated circularization of recombinant proteins. Chembiochem 2013, 14:809–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johansen NT, Tidemand FG, Nguyen T, Rand KD, Pedersen MC, Arleth L: Circularized and solubility-enhanced MSPs facilitate simple and high-yield production of stable nanodiscs for studies of membrane proteins in solution. FEBS J 2019, 286:1734–1751. [DOI] [PubMed] [Google Scholar]
- 37.Iric K, Subramanian M, Oertel J, Agarwal NP, Matthies M, Periole X, Sakmar TP, Huber T, Fahmy K, Schmidt TL: DNA-encircled lipid bilayers. Nanoscale 2018, 10:18463–18467. [DOI] [PubMed] [Google Scholar]
- 38.White JM, Whittaker GR: Fusion of Enveloped Viruses in Endosomes. Traffic 2016, 17:593–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banerjee M, Johnson JE: Activation, exposure and penetration of virally encoded, membrane-active polypeptides during non-enveloped virus entry. Curr Protein Pept Sci 2008, 9:16–27. [DOI] [PubMed] [Google Scholar]
- 40.Strauss M, Levy HC, Bostina M, Filman DJ, Hogle JM: RNA transfer from poliovirus 135S particles across membranes is mediated by long umbilical connectors. J Virol 2013, 87:3903–3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kumar M, Blaas D: Human rhinovirus subviral a particle binds to lipid membranes over a twofold axis of icosahedral symmetry. J Virol 2013, 87:11309–11312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee H, Shingler KL, Organtini LJ, Ashley RE, Makhov AM, Conway JF, Hafenstein S: The novel asymmetric entry intermediate of a picornavirus captured with nanodiscs. Sci Adv 2016, 2:e1501929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tosteson MT, Wang H, Naumov A, Chow M: Poliovirus binding to its receptor in lipid bilayers results in particle-specific, temperature-sensitive channels. J Gen Virol 2004, 85:1581–1589. [DOI] [PubMed] [Google Scholar]
- 44.Danthi P, Tosteson M, Li QH, Chow M: Genome delivery and ion channel properties are altered in VP4 mutants of poliovirus. J Virol 2003, 77:5266–5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bubeck D, Filman DJ, Hogle JM: Cryo-electron microscopy reconstruction of a poliovirusreceptor-membrane complex. Nat Struct Mol Biol 2005, 12:615–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Snyder AJ, Danthi P: Lipids Cooperate with the Reovirus Membrane Penetration Peptide to Facilitate Particle Uncoating. J Biol Chem 2016, 291:26773–26785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diaz-Salinas MA, Silva-Ayala D, Lopez S, Arias CF: Rotaviruses reach late endosomes and require the cation-dependent mannose-6-phosphate receptor and the activity of cathepsin proteases to enter the cell. J Virol 2014, 88:4389–4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Patel A, Mohl BP, Roy P: Entry of Bluetongue Virus Capsid Requires the Late Endosomespecific Lipid Lysobisphosphatidic Acid. J Biol Chem 2016, 291:12408–12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cremesti AE, Goni FM, Kolesnick R: Role of sphingomyelinase and ceramide in modulating rafts: do biophysical properties determine biologic outcome? FEBS Lett 2002, 531:47–53. [DOI] [PubMed] [Google Scholar]
- 50.Ewers H, Romer W, Smith AE, Bacia K, Dmitrieff S, Chai W, Mancini R, Kartenbeck J, Chambon V, Berland L, et al. : GM1 structure determines SV40-induced membrane invagination and infection. Nat Cell Biol 2010, 12:11–18; sup pp 11–12. [DOI] [PubMed] [Google Scholar]
- 51.Chan RB, Tanner L, Wenk MR: Implications for lipids during replication of enveloped viruses. Chem Phys Lipids 2010, 163:449–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simons K, Sampaio JL: Membrane organization and lipid rafts. Cold Spring Harb Perspect Biol 2011, 3:a004697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lorizate M, Krausslich HG: Role of lipids in virus replication. Cold Spring Harb Perspect Biol 2011, 3:a004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dupzyk A, Williams JM, Bagchi P, Inoue T, Tsai B: SGTA-Dependent Regulation of Hsc70 Promotes Cytosol Entry of Simian Virus 40 from the Endoplasmic Reticulum. J Virol 2017, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bagchi P, Inoue T, Tsai B: EMC1-dependent stabilization drives membrane penetration of a partially destabilized non-enveloped virus. Elife 2016, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Staring J, von Castelmur E, Blomen VA, van den Hengel LG, Brockmann M, Baggen J, Thibaut HJ, Nieuwenhuis J, Janssen H, van Kuppeveld FJ, et al. : PLA2G16 represents a switch between entry and clearance of Picornaviridae. Nature 2017, 541:412–416. ••In this study, the authors have identified the lipid-modifying enzyme PLA2G16 as a host factor that functions early during picornaviruses infection, enabling their genome delivery into the cytoplasm.