Summary
Obesity, in which the functional importance of small nucleolar RNAs (snoRNAs) remains elusive, correlates with the risk for many cancer types. Here, we identify that the serum copies of adipocyte-expressed SNORD46 correlates with body mass index (BMI) and serum SNORD46 antagonizes interleukin-15 (IL15) signaling. Mechanically, SNORD46 binds IL15 via G11 and G11A (a mutation significantly enhances binding affinity) knockin drives obesity in mice. Functionally, SNORD46 blocks IL15-induced, FER kinase-dependent phosphorylation of platelet glycoprotein 4 (CD36) and monoglyceride lipase (MGLL) in adipocytes, leading to inhibited lipolysis and browning. In nature killer (NK) cells, SNORD46 suppresses the IL15-dependent autophagy, leading to reduced viability of obese NK. SNORD46 power inhibitors exhibit anti-obesity effects concurring with improved viability of obese NK, and anti-tumor immunity of CAR-NK cell therapy. Hence, our findings demonstrate the functional importance of snoRNAs in obesity and the utility of snoRNA power inhibitors for antagonizing obesity-associated immune resistance.
Keywords: Small Nucleolar RNA, Interleukin-15, Obesity, Adipocyte, Nature Killer cells, Lipase, Autophagy
Introduction
The global prevalence of obesity has increased dramatically both in adults and youth in the past few decades1. Obesity has been long considered one of the major risk factors for a myriad of chronic diseases, including type 2 diabetes, heart disease, and cancer2. Furthermore, diet-induced obesity accelerates tumor growth through impairing CD8+ T cell function in the tumor microenvironment (TME), indicating that obesity further impedes the immune checkpoint blockade-based treatment effects in cancer patients3. Current therapeutic options are limited in obesity and effective therapeutic opportunities are urgently needed to dampen the broad prevalence of obesity worldwide.
IL15 is well known as a stimulator of CD8+T cells, natural killer (NK) cells and intraepithelial lymphocytes4, while also acting as a circulating regulator of body composition5. IL15 acts as an anti-obesity cytokine by accelerating the lipid catabolism5. IL15 administration decreases white adipose tissue mass and lipoprotein lipase activity in ob/ob mouse without changing the food intake6. IL15 has shown extensive therapeutic prospects in maintaining lipid homeostasis, which is especially important for the treatment of obesity. However, the molecular mechanism underlying the anti-obesity effect of IL15 remains vague. The introduction of chimeric antigen receptor (CAR) to immune cells, including T-cells (CAR-T) and NK cells (CAR-NK), is under rapid development7,8. IL15 has been shown to stimulate NK cells in promoting an anti-tumor response in a variety of murine tumor models9,10. IL15 agonists or recombinant IL15 have been evaluated in clinical trials in combination with immune checkpoint blockers such as anti-PD-L1 antibodies11. The expression of IL15 significantly increased the cytotoxicity and persistence of NK cells in vivo12. However, obesity-dependent impairment of anti-tumor immunity has yet to be overcome.
Serum small nucleolar RNAs (snoRNAs) have raised attention as potential biomarkers for a variety cancer patients13. Here, we profiled snoRNA copies in obese serum, finding that Small Nucleolar RNA, C/D Box 46 (SNORD46) was increased in human donors with obesity. SNORD46 copies correlate with BMI. Our findings suggested that saturated fatty acid triggered SNORD46 expression in a phosphorylation of C/EBPβ (CCAAT/enhancer-binding protein beta)-dependent manner. The adipocyte-expressed SNORD46 is directly associated with IL15 in serum, by which G11A mutant enhanced the affinity of SNORD46-IL15 interaction. We generated knockin mice harboring the G11→A mutation of Snord46 (referred to as Snord46G11A/G11A). Snord46G11A/G11A mice exhibit obesity, reduced energy expenditure, impaired glucose metabolism, and liver steatosis. Mechanistically, ligand-bound IL15 receptor complexes triggered the phosphorylation of MGLL at Y268, resulting in augmented lipolysis activity; and CD36 at Y370, regulating the transportation of non-esterified fatty acids (NEFA, or free fatty acids, FFA), These events led to the accumulation of intracellular NEFA and adipocyte browning. The elevated SNORD46 level or mutated SNORD46 disrupted IL15-IL15 receptor complex, resulting in inhibited signaling events mentioned forehead and obesity. Further, excessive SNORD46 inhibited the IL15-dependent autophagy in NK cells, leading to impaired antitumor immunity of NK cells under obesity condition. Administration of SNORD46 power inhibitors antagonized the inhibitory role of SNORD46 in vivo, leading to the anti-obesity effect and sensitization of the breast and colorectal cancers to CAR-NK cells under obese conditions. Hence, our findings demonstrated the functional importance of snoRNAs in obesity and immune resistance.
Results
Serum SNORD46 correlates with obese and immune suppressive tumor microenvironment
To study snoRNAs in obesity and cancer, we first utilized snoRNA array to determine snoRNA copies in human serum collected from donors with and without obesity, finding that SNORD46 copies are increased in obese serum (Figures 1A, S1A–S1C; Table S1 and Data S1). We further validated SNORD46 copies in human serum collected from 382 donors with BMIs ranging from 16 to 78. The SNORD46, but not U6 copies were significantly increased in the serum of BMI 30-40 group compared with BMI < 25 group, and further elevated in BMI > 40 group (Figure 1B). SNORD46 copies correlated with BMI of donors with obesity (Figure S1D). We then examined SNORD46 expression in human and mouse tissues, finding that adipose tissue exhibited the highest expression level of this snoRNA (Figure S1E). Northern blot confirmed the correlation between serum SNORD46 level and BMI (Figure 1C). Furthermore, SNORD46 expression was upregulated in omental (OM) and subcutaneous (SubQ) adipose, but not brown adipose tissue (BAT) tissue (Figures 1D and S1F). Serum SNORD46 copies was reduced in donors with regular exercise (> 420 min/week) compared with donors without exercise (0 min/week) (Figure 1E). Consistently, serum SNORD46, but not U6 copies negatively correlated with exercise time of donors with or without obesity (Figures 1F and S1G).
Figure 1. SNORD46 correlates with BMI and immune resistance.
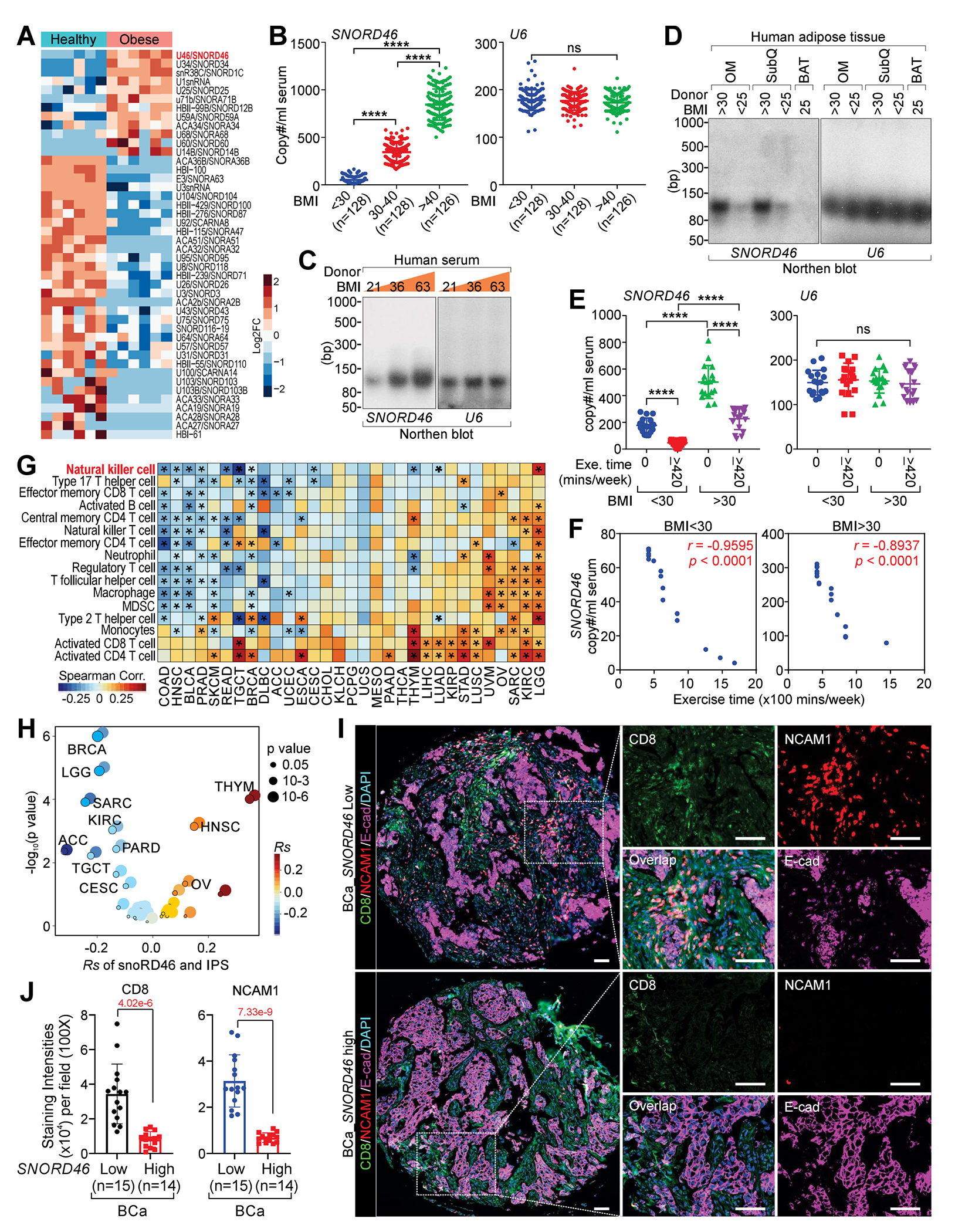
(A) Heatmap of snoRNA expression of indicated human serum samples.
(B) Copy number of SNORD46 (left) or U6 (right) in serum of human donors with BMI < 25, 30-40, or > 40 (n = 128, 128, 126 donors).
(C and D) Northern blotting using indicated probes of indicated human serum (C), or human adipose tissues as indicated (D).
(E) Copy number of SNORD46 (left) or U6 (right) in serum with indicated BMI, without or with exercise (0, ≥ 420 min/week), n = 18 donor per group, one-way ANOVA.
(F) Pearson correlation between SNORD46 copy number and exercise time of donors with BMI < 30 (left) or BMI > 30 (right), n = 18 (BMI < 30), 18 (BMI > 30) donor, Fisher’s exact test.
(G and H) The spearman’s correlation of SNORD46 expression and immune cell type enrichment (G) and immunophenoscore (IPS) (H) in 28 cancer types.
(I and J) Representative multi-IHC images (I) and statistical analysis of CD8 and NCAM1 (J) of breast cancer tissues from serum SNORD46-high or SNORD46-low breast cancer patients. Scale bars, 100 μm. Error bars, SD, n = 15, 14 tissues, unpaired Student’s t-test.
TCGA analysis indicated that tumors harbor elevated SNORD46 level compared with adjacent normal tissues (Figure S1H). SNORD46 level corelated with reduced tumor-resident infiltration of NK cells in colon cancer (COAD), breast cancer (BRCA), and other types of cancer (Figure 1G) and negatively correlated with immunophenotypic score (IPS) (Figure 1H). Tumors from breast cancer patients with high serum SNORD46 copies exhibited reduced tumor-resident CD8+ T cells and NK cells (marked by NCAM1/CD56)14, compared with those with low serum SNORD46 copies (Figures 1I and 1J; Table S2).
Saturated fatty acid induces SNORD46 expression via C/EBPβ
To determine the transcriptional factors that regulate SNORD46 expression, we performed proteomics of isolated chromatin segments (PICh), finding a cohort of transcription factors associated with the promoter region of SNORD46 (Figure 2A; Data S1). We then knocked down individual transcriptional factors in human adipocytes (HAd) validated by immunoblotting and RT-qPCR (Figures S2A and S2B), identifying that C/EBPβ is essential for SNORD46 expression (Figure 2B). Mutagenesis studies confirmed that C/EBPβ T235A abolished SNORD46 expression (Figures 2C and S2C), while C/EBPβ phospho-mimic mutants T235D or T235E, enhanced SNORD46 level in the supernatant of C/EBPβ-deficient human adipocytes (Figure 2C). The recruitment of C/EBPβ to the promoter region of SNORD46 was phospho-T235 dependent (Figure 2D). Cebpb-deficient mouse adipocytes (MAd) exhibited reduced Snord46 expression, which was restored by C/EBPβ T188E mutant (Figures S2D–S2F). We challenged the high-fat diet (HFD)-induced obese mice with the treadmill exercise (TE), finding that HFD induced C/EBPβ T188 phosphorylation, which was reversed upon the TE (Figure 2E). Consistently, HFD-fed mice showed increased serum SNORD46 copies, which was reduced by TE (Figures 2F and 2G). Interestingly, saturated fatty acids induced C/EBPβ (T235) phosphorylation, as well as supernatant SNORD46 copies of human adipocytes (Figures 2H and 2I; Table S3).
Figure 2. C/EBPβ regulates the transcriptional expression of SNORD46.
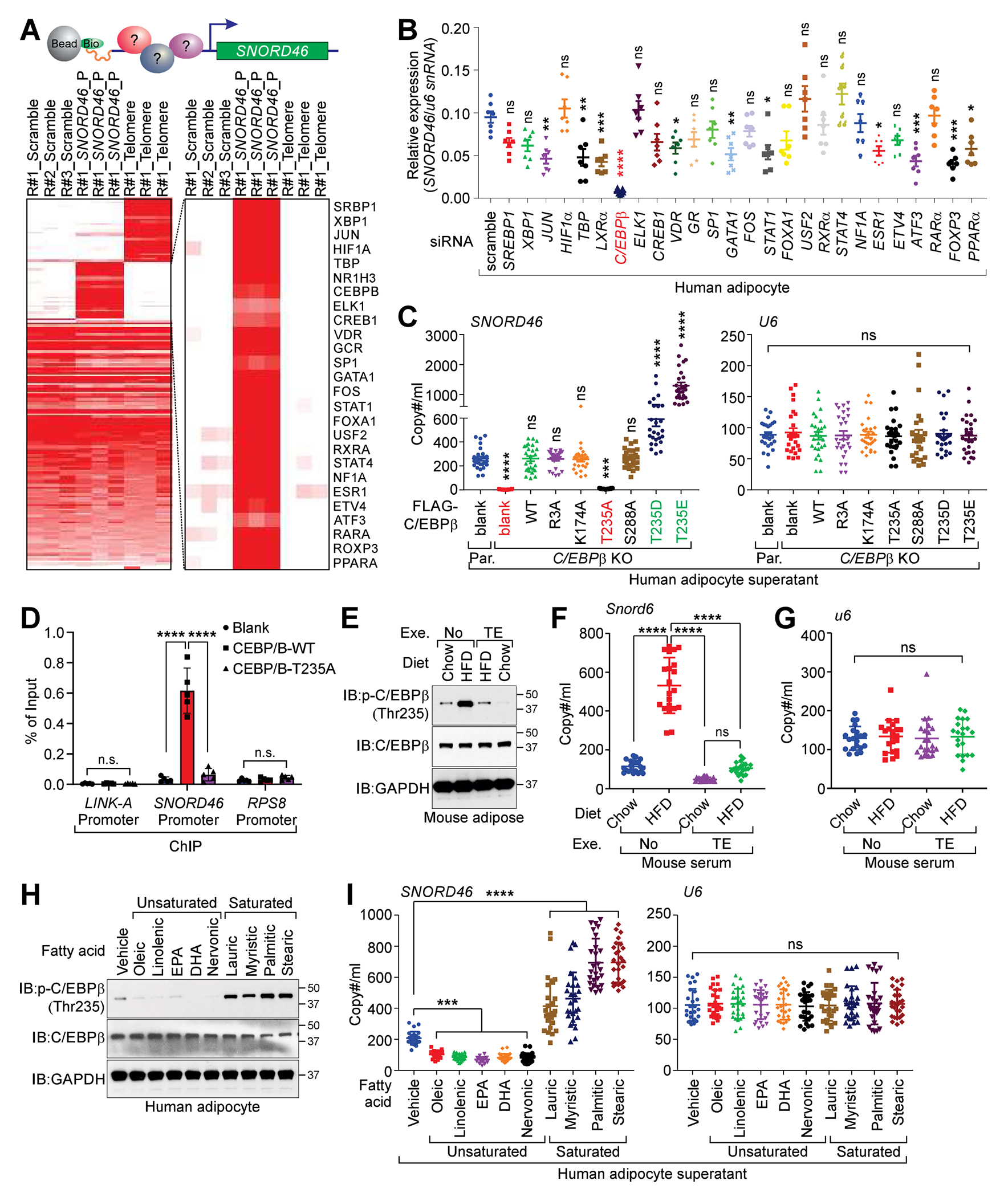
(A) Top: graphic illustration of PICh-MS determination using SNORD46 promoter region. Bottom: heatmap of the PICh-MS protein identification using scramble, SNORD46 promoter or telomere sequence as indicated. R: biological repeat.
(B) Relative expression of SNORD46, normalized by U6 snRNA of differentiated human adipocyte harboring indicated siRNAs. Error bars, SD, n = 7 independent experiments, one-way ANOVA.
(C) Copy number of SNORD46 (left) or U6 (right) in supernatant of C/EBPβ parental (Par.) or knockout (KO) adipocytes expressing indicated expression vectors. Error bars, SD, n = 26 donors, one-way ANOVA.
(D) ChIP-qPCR detection of the C/EBPβ occupancy on LINK-A, SNORD46 or RPS8 promoter regions of differentiated human adipocytes expressing indicated expression constructs. Error bars, SD, n = 5 independent experiments, two-way ANOVA.
(E) Immunoblotting (IB) detection of indicated proteins in adipose tissues from chow or HFD-fed mice with or without treadmill exercise (TE).
(F and G) Serum Snord46 (F) or U6 (G) copy number in wild-type mice challenged with chow or HFD followed with or without treadmill exercise. Error bars, SD, n = 21 mice per group, one-way ANOVA.
(H) IB detection of indicated proteins in differentiated human adipocytes supernatant upon indicated stimuli.
(I) Copy number of SNORD46 (left) or U6 (right) in differentiated human adipocytes supernatant upon indicated stimuli. Error bars, SD, n = 26 donors, one-way ANOVA.
SNORD46 is associates with IL15 and modulates IL15-dependent signaling
Liquid chromatography-mass spectrometry (LC-MS) analysis identified that biotinylated SNORD46 was associated with IL15 (Figure 3A; Data S1), while biotinylated miR20-a was associated with AGO2, which is consistent with previous findings15. SNORD46/Snord46-IL15 interaction was validated by Cross-linking immunoprecipitation (CLIP) assay in human and mouse serum respectively (Figures 3B and 3C). The SNORD46/Snord46 motifs responsible for IL15 binding were also identified by CLIP assay (Figure 3C; Data S1). We next determined the single nucleotides of SNORD46 responsible for IL15 binding in a rescue experiment using SNORD46-deficient adipocytes (Figures S3A–S3C), finding that mutation of G11C abolished SNORD46-IL15 interaction (Figure S3D), while G11A enhanced the SNORD46-IL15 interaction, and G11C/U impaired interaction with IL15 (Figure 3D).
Figure 3. SNORD46 physically interact with IL15.
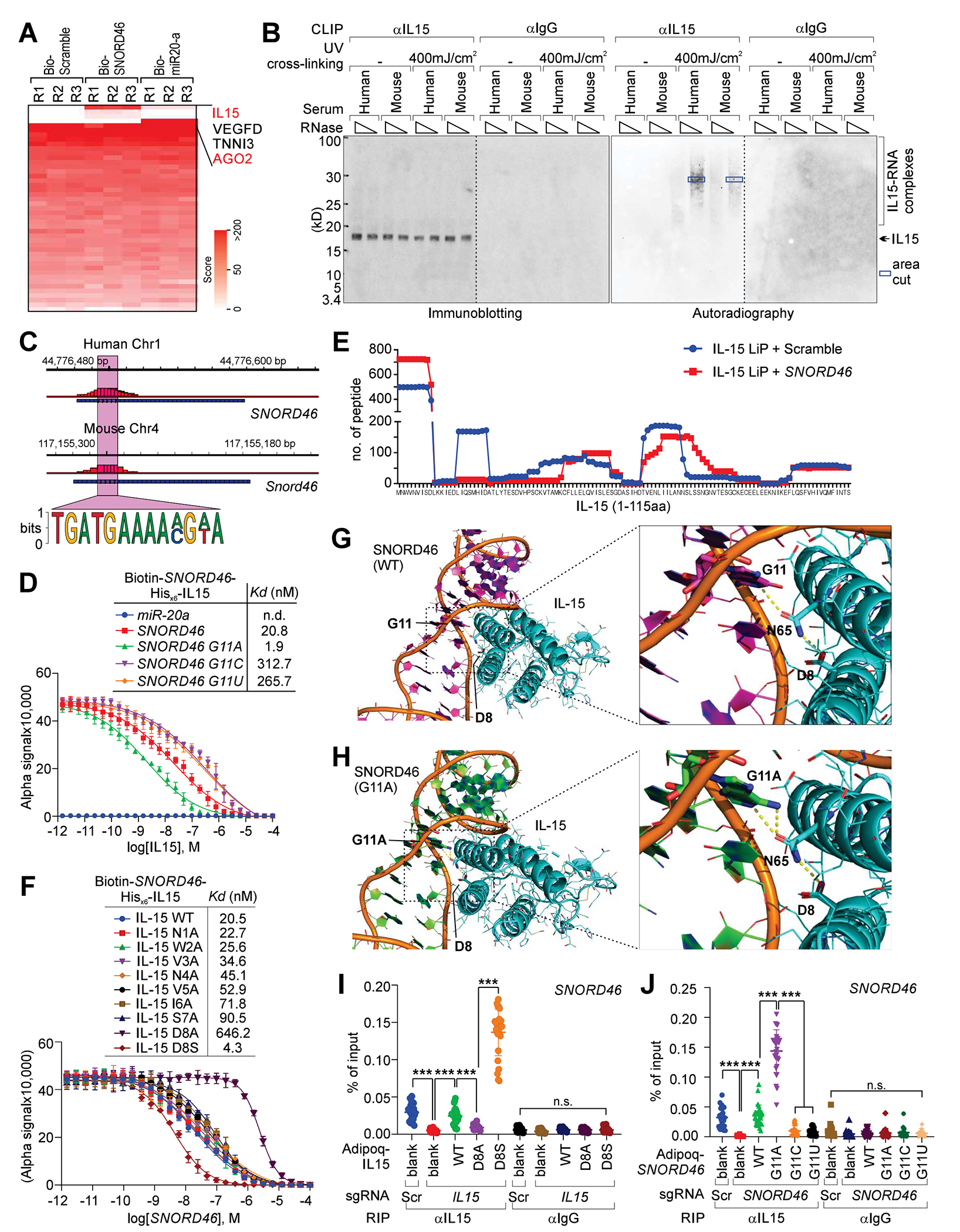
(A) Heatmap of the protein identification score for biotinylated Scramble, SNORD46 or miR-20a pull-down. R: biological replicate.
(B) IB detection (left) and autoradiography (right) of IL15 CLIP assay using human (H) or mice (M) serum. Blue box: RNA-protein complex extracted for the following sanger sequencing.
(C) Summary of SNORD46 sequence responsible for IL15 binding. The chromatin sequences corresponding to RNA (negative-stranded) and RNA motif bound by IL15 are shown.
(D) Competition binding assay to determine Kd of the interaction between His-tagged IL15 and biotinylated-SNORD46 wild-type or mutants. Unlabeled IL15 serve as the competitor. Error bars, SD, n = 3 independent experiments. miR-20a was included as a negative control.
(E) Number of peptides recovered from LiP-MS of IL15 incubated with scramble or SNORD46 RNA. x-axis: amino acid position of full-length IL15.
(F) Competition binding assay to determine Kd of the interaction between His-tagged IL15 wild-type or mutants and biotinylated-SNORD46. Unlabeled SNORD46 serve as competitor. Error bars, SD, n = 3 independent experiments.
(G) Computational modeling of SNORD46–IL15 interaction. Magenta cartoon: SNORD46; Cyan cartoon: IL15; Magenta stick: G11; Cyan stick: D8 and N65.
(H) G11A mutation of SNORD46 strengthens the interaction with IL15. Green cartoon: SNORD46; Cyan cartoon: IL15; Green stick: A11; Cyan stick: D8 and N65.
(I and J) RIP assay using differentiated human adipocytes harboring IL15 sgRNAs (I), or SNORD46 sgRNAs (J) and Adipoq-driven expression of indicated mutants. Error bars, SD, n = 26 donors, one-way ANOVA.
We next performed limited proteolysis (LiP) followed by LC-MS analysis (LiP-MS)16,17 to determine IL15 residues responsible for SNORD46 binding. The LiP-MS analysis confirmed that the peptide recovery of N-termini of IL15 was increased in the presence of SNORD46 RNA oligonucleotides, indicating that a.a. 1-8 of IL15 might be involved in interacting with SNORD46 (Figure 3E; Data S1). Previous structural and mutagenesis studies have indicated that the D8 of IL15 plays important role in mediating IL15 receptor binding18,19. Our mutagenesis screening of IL15 a.a. 1-8 suggested that D8A mutation of IL15 impaired the SNORD46 binding, while the D8S mutant enhanced SNORD46 binding affinity (Figure 3F).
We further validated SNORD46-IL15 interaction by computational modeling using crystal structure of IL15 (PDB ID: 2Z3Q) for RNA-protein docking 20–22(Yan et al., 2017), finding that D8 and N65 of IL15 forms hydrogen bond with SNORD46 G11, which stabilizes the SNORD46-IL15 complex (Figure 3G). The N1 of G11 forms a hydrogen bond with N65, then N65 directly interacts with D8 (Figure 3G). Interestingly, D8-N65 interaction is one of the only two polar interactions between the two alpha helixes (the other one is at the edge of the alpha helix), suggesting that this interaction is critical to maintain the conformation of IL15 to bind SNORD46 (Figure 3G). With the mutation from guanine to adenine at nucleotide 11 of SNORD46, the carbonyl group was replaced with an amine group, which can form an additional hydrogen bond with N65 compared with guanine (Figure 3H). Thus, the binding affinity between SNORD46 and IL15 is increased. With the mutation from aspartic acid to serine at ammino acid 8 of IL15, the carboxylic acid group was replaced with a hydroxyl group (Figure S3E). The hydroxyl group can be both a hydrogen bond acceptor and donor, while carboxylic acid can only accept hydrogens. The hydroxyl group may form a hydrogen bond with the backbone of SNORD46, while maintain the interaction with N65 (Figure S3E). Thus, the interaction between SNORD46 and IL15 is increased. This computational modeling was validated in a RIP rescue experiment using IL15- or SNORD46-depleted human adipocytes with comparable levels of exogenously expressed WT or mutant of IL15 and SNORD46 respectively (Figures 3I, 3J, and S3F–S3I).
IL15 has been suggested to reduce WAT mass in obese mice models23. However, the underlying molecular mechanism is largely unknown. We determined the gene expression profile of human adipocytes (BMI < 30) treated with vehicle or IL15 (GSE210203). Adipocytes isolated from healthy donors exhibited reduced expression of gene signatures related to Adipogenesis and Fatty Acid metabolism upon IL15 stimulation, which is consistent with the previous notion that IL15 exhibits an anti-obesity effect (Figure S3J). Taken together, our findings suggested that SNORD46 physically interact with IL15 and acts as a natural IL15 antagonist in the serum of donors with obesity to hinder the anti-obese effect of IL15.
SNORD46 G11A mutation drives obesity in vivo
Given that SNORD46 is highly conserved between humans and mice, with 92.3% identity, we generated a knockin mouse model harboring a single-nucleotide variant (G11→A) of the mouse Snord46 gene using CRISPR/Cas9-mediated technology (Figure 4A). Male and female homozygous mice harboring the AA mutant (Snord46G11A/G11A) exhibited augmented body weight compared with WT or heterozygous littermates (Figures 4B and 4C). Body composition analysis indicated that Snord46G11A/G11A mice exhibited increased body weight, fat weight, and fat percentage (Figures 4D–4H). Histological analysis indicated that SNORD46G11A/G11A mice exhibited increased adipose tissue, enlarged islet areas, and liver steatosis (Figures 4I and 4J). Comprehensive Lab Animal Monitor System (CLAMP) analysis indicated that Snord46G11A/G11A mice exhibited reduced oxygen consumption (VO2), reduced carbon dioxide production (VCO2), and reduced energy expenditure (Figures 4K, 4L, and S4A–S4D). Female Snord46G11A/G11A mice exhibited increased food intake whereas male Snord46G11A/G11A mice showed unaltered food intake compared with Snord46WT/WT mice (Figure S4E). Male Snord46G11A/G11A mice showed increased water intake in compared with Snord46WT/WT littermates (Figure S4F). Further, Snord46G11A/G11A mice exhibited significantly impaired glucose tolerance (Figures 4M and S4G). Compared with WT littermates, Snord46G11A/G11A mice exhibited reduced body temperature, increased urine volume, blood triglycerides, AST, and ALT concentrations (Figures S4H–S4L). Both male and female Snord46G11A/G11A mice had similar serum leptin level24 compared with WT mice (Figure S4M). The serum Ghrelin24,25 level is unaffected in Snord46G11A/G11A mice compared with WT mice (Figure S4N). Furthermore, both male and female Snord46G11A/G11A mice showed increased serum insulin level compared with Snord46WT/WT mice (Figure S4O). Hence, our findings suggested that SNORD46 contributes to the obesity, which is independent on its neighboring gene RPS8 (Figures S5A–S5D).
Figure 4. SNORD46 G11A drives obesity.
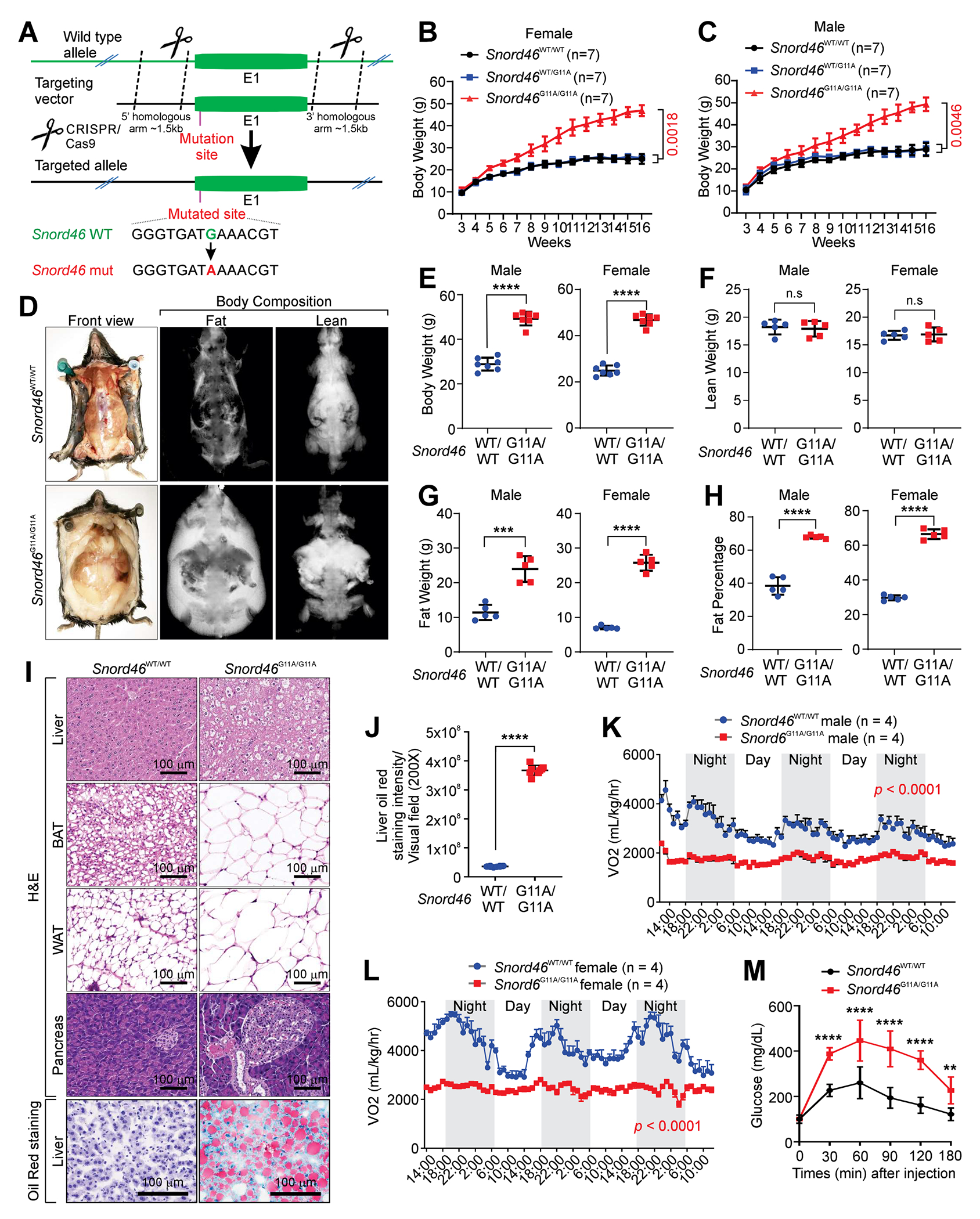
(A) Schematic of CRISPR-Cas9 method used to generate SNORD46G11A/G11A mice.
(B and C) Body weight measurement of female (B) or male (C) Snord46WT/WT, Snord46WT/G11A or Snord46G11A/G11A mice. Error bars, SD, n = 7 mice, one-way ANOVA.
(D-H) Representative images of lean and fat tissues (D), quantification of male (left) or female (right) body weight (E), lean weight (F), fat weight (G), and fat percentage (H) by the dual energy x-ray absorptiometry imaging system from Snord46WT/WT or Snord46G11A/G11A mice. Error bars, SD, n = 5 mice, Student’s t-test.
(I) Representative H&E and Oil Red O staining of indicated tissues from Snord46WT/WT or Snord46G11A/G11A mice. Scale bars, 100 μm.
(J) Statistical analysis of Oil Red O staining intensity of liver sections from Snord46WT/WT or Snord46G11A/G11A mice. Error bars, SD, n = 8 mice, Student’s t-test.
(K and L) CLAMS measurement of oxygen consumption (VO2) in male (K) or female (L) Snord46WT/WT or Snord46G11A/G11A mice. Error bars, SD, n = 4, 4 mice, two-way ANOVA.
(M) Glucose tolerance tests (GTTs) of male Snord46WT/WT or Snord46G11A/G11A mice at the indicated time points. Error bars, SD, n = 8, 5 mice, Student’s t-test for each time point.
IL15 triggers a non-canonical signaling cascades in adipocytes
We aimed to compare the signaling pathways of IL15 in the major organs between Snord46WT/WT and Snord46G11A/G11A mice by identifying IL15-binding proteins. Interestingly, IL15 was associated with IL15Rα and tyrosine-protein kinase JAK3 in brain, heart, liver, skeletal muscle (SKM), spleen, and intestine of Snord46WT/WT mice, which are components of the conventional IL15 signaling pathway11 (Figure 5A; Data S1). In WAT, IL15 was associated with tyrosine-protein kinase FER, Monoglyceride Lipase (MGLL), and Platelet glycoprotein 4 (CD36), triggering a potentially unconventional pathway (Figure 5A). The association between IL15 and conventional/unconventional signaling pathway components were diminished in SNORD46G11A/G11A mice (Figure 5A). The MS also revealed that FER was phosphorylated at Y402; MGLL was phosphorylated at Y268; and CD36 was phosphorylated at Y370 (Figures S5E–S5G). We generated modification specific antibodies targeting human p-MGLL Y268, or p-CD36 Y370, which were validated using peptide-blocking assays (Figures S5H and S5I). Hence, we reason that upon ligand binding, IL15 receptor complex recruits FER, which mediates the downstream signaling cascades. We verified the interactions between mouse IL15 and IL2Rβ, MGLL, and CD36, which were enhanced by IL15 but abolished by SNORD46 G11A mutant in mouse adipocytes (Figure 5B).
Figure 5. IL15 triggers FER-medicated signaling cascades in adipocytes to regulates the enzymatic activities of CD36 and MGLL.
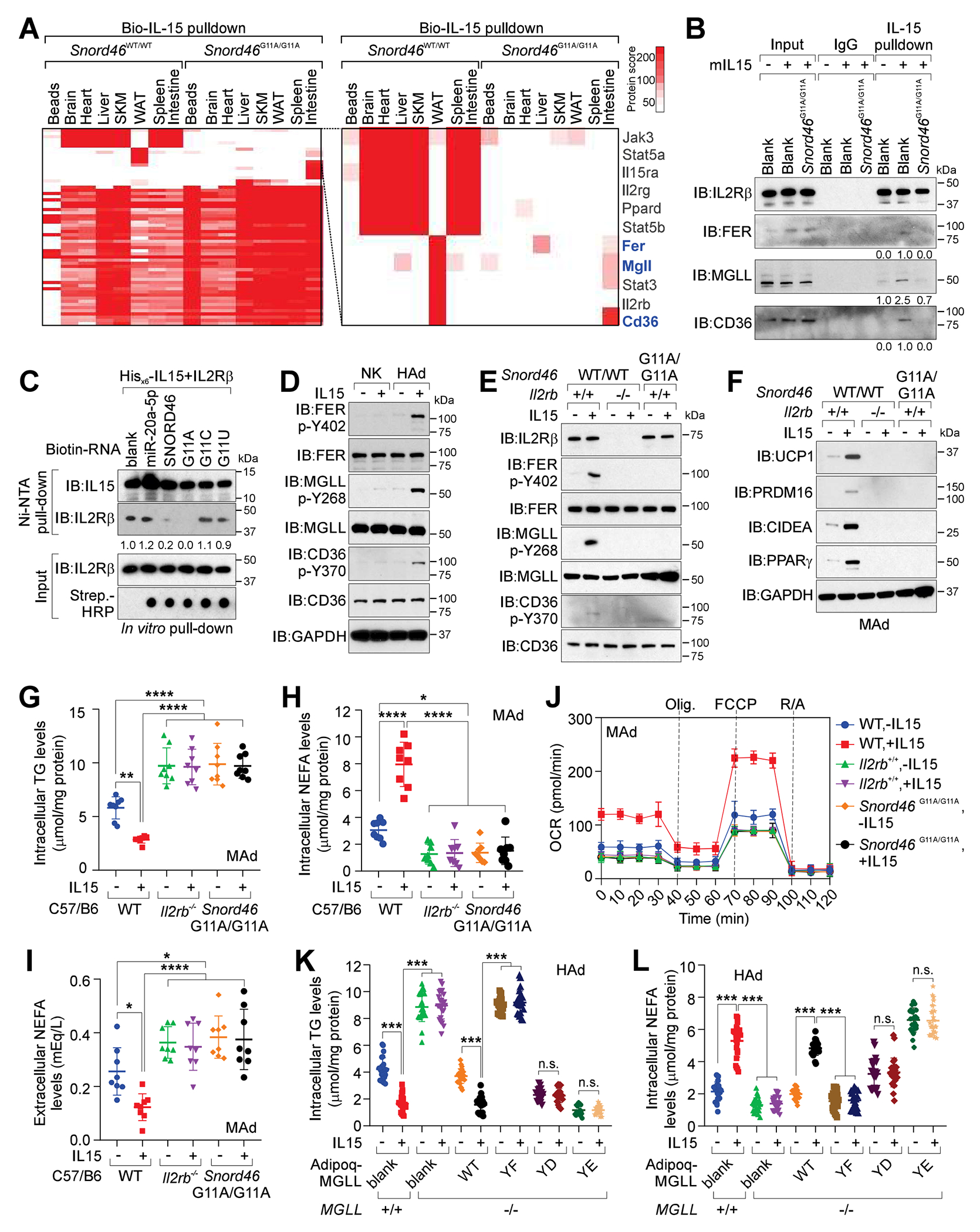
(A) Identification of biotinylated IL15-associated proteins in Snord46WT/WT or Snord46G11A/G11A tissues as indicated. Protein scores are shown.
(B) IL15 pull-down followed by IB detection of indicated proteins in mouse adipocytes with or without IL15 stimulation.
(C) Ni-NTA pull-down using indicated recombinant proteins and biotinylated RNA oligonucleotides, followed by IB detection using indicated antibodies.
(D) IB detection of indicated proteins in NK cells or differentiated human adipocytes (HAd) with or without IL15 stimulation.
(E and F) IB detection of indicated proteins in WT, Il2rb-deficient or Snord46G11A/G11A adipocytes with or without IL15 stimulation.
(G-I) Measurement of intracellular TG (G), intracellular NEFA (H), or extracellular NEFA (I) concentration of differentiated WT, Il2rb−/−or Snord46 G11A/G11A adipocytes with or without IL15 stimulation. Error bars, SD, n = 8 animals per experimental group, one-way ANOVA.
(J) Oxygen consumption rate (OCR) across time for WT, Il2rb−/− and Snord46G11A/G11A mouse adipocytes with or without IL15 stimulation. Dot lines indicate the addition of mitochondrial inhibitors (oligomycin; FCCP; antimycin A/rotenone) (n = 8 wells of adipocytes, error bars: SD).
(K and L) Measurement of intracellular TG (K) and intracellular NEFA (L) levels in MGLL-proficient or -deficient differentiated human adipocytes expressing indicated expression constructs with or without IL15 stimulation. Error bars, SD, n = 24 donors, two-way ANOVA.
To examine whether SNORD46 mutant modulates IL15-IL2Rβ interaction, we performed in vitro pulld-own assay using His-tagged IL15 (Figure 5C). IL15 exhibited adequate interactions with IL2Rβ; in the presence of SNORD46, IL15 WT exhibited reduced interaction with IL2Rβ, which were further abolished in the presence of SNORD46 G11A mutant (Figure 5C). The presence of miR-20a showed minimal effect on the IL15-IL2Rβ interaction (Figure 5C). SNORD46 G11C or G11U mutants exhibited a minimal effect on the interaction between IL15 and IL2Rβ (Figure 5C). Our findings suggested that SNORD46 inhibited the interaction between IL15 and the receptor complex. SNORD46 G11A showed a robust inhibitory effect on the IL15 ligand-receptor binding.
To further characterize IL15-triggered, FER/MGLL/CD36-medicated unconventional pathways in adipocytes, we determined that in adipocytes, IL15 induced the p-FER (Y402), p-MGLL (Y268), p-CD36 (Y370), but not in NK cells (Figure 5D). The IL15 induced-p-FER (Y402), -p-MGLL (Y268), and -p-CD36 (Y370) were abolished by Il2rb depletion or Snord46 G11A mutation (Figure 5E). IL15 also upregulated the protein level of mitochondrial brown fat uncoupling protein 1 (UCP1), PR/SET domain 16 (PRDM16), cell death inducing DFFA like effector A (CIDE-A), and peroxisome proliferator-activated receptor gamma (PPARγ) (Figure 5F), which are essential factors of WAT browning26. The UCP1, PRDM16, CIDEA, or PPARγ levels were reduced by IL2Rb depletion, or SNORD46 G11A mutation (Figure 5F). The reduced UCP1 and PPARγ levels were verified in Snord46G11A/G11A WAT compared with Snord46WT/WT WAT (Figures S5J and S5K).
The obesity phenotype of Snord46G11A/G11A mice inspired us to further validate the concentrations of intracellular/ extracellular non-esterified fatty acids (NEFA) and intracellular triacylglycerol (TG). Our data showed that IL15 triggered reduction of intracellular TG with concurrent increased intracellular NEFA and decreased extracellular NEFA (Figures 5G–5I). Il2rb deficiency or Snord46 G11A mutation in adipocytes abolished IL15’s effect, resulting in elevated intracellular TG, reduced intracellular NEFA, as well as increased extracellular NEFA (Figures 5G–5I). The intracellular NEFA stimulates FA oxidation and oxygen consumption of adipocytes27. We hence determined the oxygen consumption rate (OCR) of mouse adipocytes isolated from Il2rb KO or Snord46G11A/G11A mice, finding that IL15 triggered the oxygen consumption of adipocytes and Il2rb depletion, or Snord46 G11A mutation reduced OCR compared with the adipocytes isolated from WT mice (Figure 5J). Taken together, our findings suggested that IL15 inhibits obesity via an unconventional signaling cascade promoting lipolysis of adipocytes, which can be antagonized by SNORD46.
Phosphorylation of MGLL and CD36 modulates lipolysis and FA transportation
MGLL converts monoacylglycerides to free FA and glycerol and is an essential enzyme in the lipolysis28. Hence, we measured the glycerol production catalyzed by recombinant MGLL, finding that the phosphorylated MGLL, or phospho-mimic mutant Y268E (YE) exhibited increased enzymatic activity compared with unphosphorylated MGLL (Figure S5L). In human adipocytes, IL15 led to enhanced glycerol production, in which the enzymatic activity of MGLL was abolished upon MGLL depletion (Figure S6A). The expression of exogenous MGLL WT, but not YF mutant, rescued the glycerol production in MGLL-deficient cells (Figure S6A). Furthermore, the phospho-mimic mutant YE, but not YD restored enzymatic activities of MGLL (Figure S6A). Consistently, adipocytes with MGLL KO exhibited increased intracellular TG with concurrent reduced intracellular NEFA (Figures 5K and 5L). Expression of MGLL WT restored IL15-induced NEFA catalysis (Figures 5K and 5L). Furthermore, adipocytes with MGLL YE mutant exhibited decreased TG and enhanced NEFA concentration (Figures 5K and 5L). Consequently, adipocytes with MGLL KO exhibited abolished expression of UCP1, PRDM16, CIDEA, and PPARγ, which was restored upon the expression of MGLL WT or YE mutant (Figure S6B). In addition, MGLL WT or YE mutant rescued the OCR of MGLL-deficient adipocytes (Figure S6C).
CD36 is a fatty acid translocase, playing important roles in binding with long chain fatty acids (LCFA) and transportation of LCFAs into cells29. Our findings suggested that CD36-proficient adipocytes exhibited normal FA uptake, which was inhibited by IL15 (Figure S6D). Knockout of CD36 abolished the FA uptake, which was restored by WT CD36 (Figure S6D). The CD36 Y370F (YF) mutant enhanced FA uptake; Y370D (YD) or Y370E (YE) phospho-mimic mutants further reduced FA uptake (Figure S6D). Our findings suggested that the CD36-dependent FA uptake is regulated by the IL15-triggered phosphorylation of CD36 at Y370. It is interesting to observe that the extracellular NEFA concentration of adipocytes was reduced by IL15 (Figure S6E). CD36 depletion resulted in elevated extracellular NEFA, which was restored by exogenous CD36 WT or YD mutant (Figure S6E).
We further strengthen the molecular linkage of SNORD46 to IL15-dependent FER/CD36/MGLL signaling cascade in Il2rb-deficient adipocytes rescued with FER YD, MGLL YE, CD36 YD, or MGLL YE + CD36 YE mutants (Figure S6F). It is noteworthy that FER phospho-mimic mutants (Y420D, YD) restored UCP1, PRDM16, CIDEA, and PPARγ protein levels in Il2rb-deficient adipocytes (Figure S6F). MGLL YE or CD36 YD mutant alone exhibited partial rescue on UCP1, PRDM16, CIDEA, and PPARγ protein levels in Il2rb-deficient adipocytes, however, MGLL YE + CD36 YE mutants robustly rescued the levels of these proteins in Il2rb-deficient adipocytes (Figure S6F). Consistently, Il2rb-deficient adipocytes exhibited elevated intracellular TG and extracellular NEFA concentrations, with concurrent reduced intracellular NEFA concentration, which were restored by FER YD mutant independent on IL15 treatment (Figures S6G–S6I). CD36 YD mutant alone failed to rescue the IL15-induced intracellular TG hydrolysis and concurrent NEFA export in Il2rb-deficient adipocytes (Figures S6G and S6I). However, Il2rb-deficient adipocytes with MGLL YE or MGLL YE + CD36 YE mutants exhibited the restored intracellular TG hydrolysis and concurrent NEFA export (Figures S6G and S6I). Consistent with this notion, MGLL YE or MGLL YE + CD36 YE mutants rescue the OCR of Il2rb-deficient adipocytes (Figure S6J). Taken together, our findings demonstrated that IL15 triggers an unconventional FER/CD36/MGLL-medicated signaling cascades in adipocyte to modulate the cellular FA transportation and lipolysis.
SNORD46 inhibitors antagonize obesity
We next screened SNORD46 power inhibitors that can abolish in vivo SNORD46-IL15 interaction by MS (Figure 6A; Table S10). SNORD46 power inhibitor #4 (referred to as SNORD46 pi) specifically abolished the IL15 protein binding using biotinylated SNORD46 as a bait (Figure 6A; Data S1). In serum of donors with obesity, the specific inhibitory effect of SNORD46 pi and IL15 neutralizing antibody (NAb) on suppressing SNORD46-IL15 interaction was verified by northern blotting and immunoblotting (Figures 6B and S7A–S7C). We further determined the effect of SNORD46 pi on SNORD46-IL15 interaction in human serum (Figure 6C), finding that the presence of SNORD46 pi abolished the interaction between IL15 and SNORD46 G11A mutant with minimal effect on miR-20a-AGO2 interaction (Figure 6C).
Figure 6. SNORD46 power inhibitors antagonize obesity.
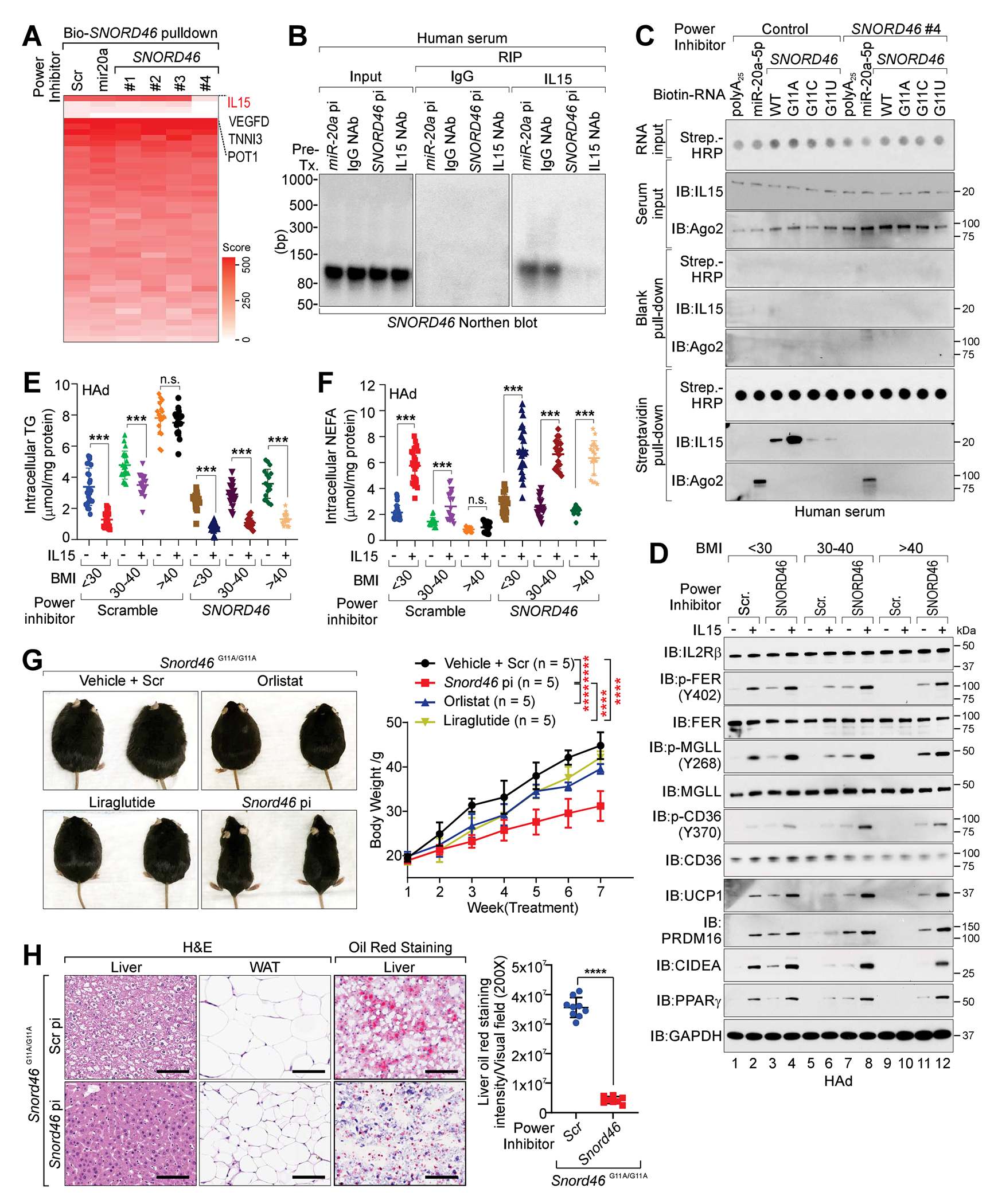
(A) Heatmap of the protein identification score for biotinylated Scramble, SNORD46 or miR-20a pull-down in the presence of indicated power inhibitors. R: biological replicate.
(B) Northernblot of SNORD46 in human serum treated with indicated power inhibitors or antibodies.
(C) IB detection of proteins associated with biotinylated RNA (wild-type and indicated mutants) in human serum treated with indicated power inhibitors.
(D) IB detection of indicated proteins in differentiated human adipocytes collected from donors with BMI < 25, 30-40, or > 40 and treated with IL15 and/or indicated power inhibitors.
(E and F) Measurement of intracellular TG (E) or intracellular NEFA (F) levels in differentiated human adipocytes collected from donors with BMI < 25 (n = 24), 30-40 (n= 19), or > 40 (n = 17) and treated with indicated power inhibitors with or without IL15 stimulation. Error bars, SD, two-way ANOVA.
(G) Left, representative images; right, body weight measurement of Snord46G11A/G11A mice treated with scramble (Scr), Snord46 power inhibitor (pi), Orlistat or Liraglutide as indicated. Error bars, SD, n = 5 mice per group, one-way ANOVA.
(H) Representative H&E and Oil Red O staining of liver or WAT (left) and statistical analysis of staining intensities of liver sections (right) from Snord46G11A/G11A mice treated with scramble or Snord46 power inhibitor. Scale bars, 100 μm. Error bars, SD, n= 9, 8 mice, Student’s t-test.
Compared with adipocytes of BMI < 30 group, BMI > 40 adipocytes exhibited reduced level of p-FER (Y420), p-MGLL (Y268), and p-CD36 (Y370), in the absence of IL15 (Figure 6D, lane 9 vs. lane 1). For BMI < 30 adipocytes, IL15 induced p-FER (Y420), p-MGLL (Y268), and p-CD36 (Y370) respectively (Figure 6D, lane 2 vs. lane 1). However, for BMI > 40 adipocytes, IL15 showed undetected effect on p-FER (Y420), p-MGLL (Y268) and p-CD36 (Y370) (Figure 6D, lane 10 vs. lane 9). SNORD46 pi sensitized BMI < 30 adipocytes to IL15 stimulation, leading to the enhanced p-FER (Y420), p-MGLL (Y268), p-CD36 (Y370) and UCP1, PRDM16, CIDEA and PPARγ protein levels (Figure 6D, lane 4 vs. lane 3). Furthermore, for BMI > 40 adipocytes, SNORD46 pi facilitated the IL15-induced p-FER (Y420), p-MGLL (Y268) and p-CD36 (Y370) respectively, as well as the protein levels of UCP1, PRDM16, CIDEA and PPARα (Figure 6D, lane 12 vs. lane 11).
We next measured the intracellular TG, NEFA and extracellular NEFA concentration of BMI < 30, 30-40, or > 40 adipocytes, finding that IL15 induced reduced intracellular TG, extracellular NEFA with elevated intracellular NEFA concentration of BMI < 30 adipocytes (Figures 6E, 6F, and S7D). However, IL15 showed minimal effect on these for BMI > 40 adipocytes (Figures 6E, 6F, and S7D). SNORD46 pi sensitized BMI > 40 adipocytes to IL15, leading to reduced intracellular TG, elevated intracellular NEFA and decreased extracellular NEFA concentration (Figures 6E, 6F, and S7D). Consistently, BMI > 40 adipocytes exhibited reduced OCR compared with BMI < 30 adipocytes, which can be restored by which SNORD46 pi (Figure S7E).
Furthermore, Snord46 pi decreased the body weight and improved the glucose tolerance of Snord46G11A/G11A mice significantly compared with scramble sequence, Orlistat and Liraglutide (two FDA-approved anti-obesity drugs) (Figures 6G, S7F, and S7G). Snord46 pi treated Snord46G11A/G11A mice exhibited reduced liver steatosis, fatty acid accumulation, and increased protein levels of UCP1 and PPARγ in WAT (Figures 6H and S7H). The HFD-fed mice receiving Snord46 pi treatment also displayed a significant decrease in lipid accumulation in the liver (Figure S7I). Hence, our findings suggested that inhibition of SNORD46 may block SNORD46-IL15 interaction and antagonize obesity in vivo.
Targeting SNORD46 restores anti-tumor immune microenvironment
NK cells in obese patients have been indicated to exhibit reduced cytotoxic capabilities30,31. Hence, we next tested whether the elevated SNORD46 contribute to the reduced cytotoxicity of obese NK cells using NK cells isolated from PBMCs of healthy donors with BMI < 25 (referred to as normal NK) and donors with obesity (BMI > 35) (referred to as obese NK) (Table S4). We first noticed that NK cell number was reduced in donors with obesity compared with normal donors, which was consistent with previous findings30 (Figures S8A and S8B). Similarly, NK cell number was reduced in peripheral blood of Snord46G11A/G11A mice compared with Snord46WT/WT littermates (Figure S8C). Transcription profiling indicated that obese NK cells with high SNORD46 expression exhibited reduced inflammatory response, TNFα signaling, and JAK/STAT3 signaling, which is independent on SNORD46’s neighboring gene RPS8 (Figures 7A and S8D–S8G; GSE213465). Obese NK cells also showed reduced autophagy gene signature (Figure 7B; Data S1), suggesting impaired autophagy response. We validated the reduced expression of LC3A/B in NK cells isolated from BMI > 35 group, compared with normal BMI group (Figure 7C). Consistently, obese human NK cells or mouse Snord46G11A/G11A NK cells showed impaired cell viability in the presence of an autophagy inducer, Tubeimoside I (TBM)32 (Figures 7D and 7E). SNORD46 pi significantly enhanced the cell viability of NK cells isolated from donors with obesity or Snord46G11A/G11A mice (Figures 7F and 7G).
Figure 7. SNORD46 power inhibitors restore the anti-tumor immunity of CAR-NK cells under obesity.
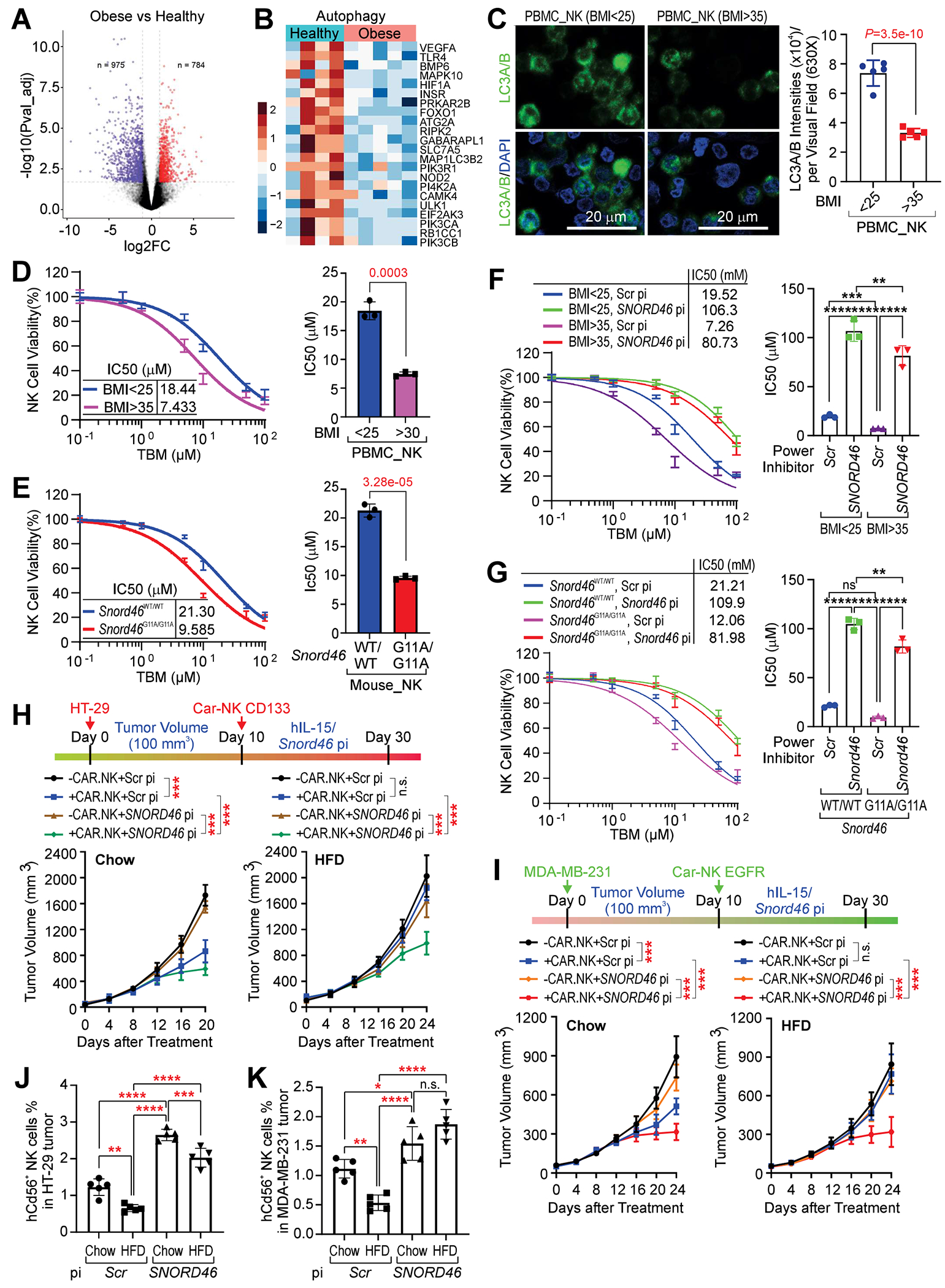
(A) Volcano presentation of the transcription profiling of NK cells isolated from donors with obesity (n = 5) or healthy donors (n = 4).
(B) Heatmap of the expression of Autophagy gene signature of NK cells isolated from donors with obesity (n = 5) or healthy donors (n = 4).
(C) Immunofluorescent labeling using indicated antibodies in NK cells isolated from PBMC of healthy donors (BMI < 25) or donors with obesity (BMI > 35). Scale bars, 20 μm. Error bars, SD, n = 5, 5 donors, Student’s t-test.
(D) Percentage of NK cells viability (left) and IC50 determination of NK cells viability (right) of BMI < 25, or BMI > 35 donors. Error bars, SD, n = 3 independent experiment, Student’s t-test.
(E) Percentage of NK cells viability (left) and IC50 determination of NK cells viability (right) of Snord46 WT/WT or Snord46 G11A/G11A mice. Error bars, SD, n = 3 independent experiment, Student’s t-test.
(F) Percentage of NK cells viability (left) and IC50 determination of NK cells viability (right) of BMI < 25, or BMI > 35 donors treated with scramble or SNORD46 power inhibitor. Error bars, SD, n = 3 independent experiment, one-way ANOVA.
(G) Percentage of NK cells viability (left) and IC50 determination of NK cells viability (right) of Snord46 WT/WT or Snord46 G11A/G11A mice treated with scramble or Snord46 power inhibitor. Error bars, SD, n = 3 independent experiment, one-way ANOVA.
(H) Top: schematic illustration of CAR-NK cells in HT-29 xenograft tumors. Bottom: tumor volume measurement of HT-29 tumor of chow (left) or HFD-fed animals (right) upon CAR-NK and indicated powder inhibitor treatment. Error bars, SD, n = 5 mice per group, one-way ANOVA.
(I) Top: schematic illustration of CAR-NK cells in MDA-MB-231 xenograft tumors. Bottom: tumor volume measurement of MDA-MB-231 tumor of chow (left) or HFD-fed animals (right) upon CAR-NK and indicated power inhibitor treatment. Error bars, SD, n = 5 mice per group, one-way ANOVA.
(J and K) Percentage of CD56+ NK cells isolated from HT-29 tumor (J) or MDA-MB-231 tumor (K) of chow or HFD-fed animals upon indicated treatment. Error bars, SD, n = 5 mice per group, one-way ANOVA.
To test whether SNORD46 pi augments the cytotoxicity of obesity-associated NK cells and sensitizes tumors to CAR-NK cell therapy under the obese condition, we generated two CAR-iPS cells engineered to express either EGFR-CAR33,34 or CD133-CAR35,36 and were subsequently differentiated into NK cells with high specificity and purity (Figures S8H and S8I). EGFR-CAR-iPS-NK exhibited anti-tumor immunity against the human triple-negative breast cancer (TNBC) cells (MDA-MB-231), while CD133-CAR-iPS-NK cells showed cytotoxicity to the human colorectal adenocarcinoma cells (HT-29) (Figures S8J–S8L). HT-29 tumor growth was significantly inhibited by CD133-CAR-iPS-NK cells under chow condition (Figure 7H, left). However, under the condition of HFD-induced obesity, administration of CD133-CAR-iPS-NK cells showed a marginal effect on the growth of the HT-29 tumor (Figure 7H, right). SNORD46 pi significantly improved the anti-tumor immunity of these CAR-NK cells (Figure 7H). Similarly, EGFR-CAR-iPS-NK cells significantly inhibited the growth of the MDA-MB-231 tumor under the chow-fed condition, but not under HFD-induced obesity (Figure 7I). Animals subjected to SNORD46 pi treatment exhibited reduced MDA-MB-231 tumor burden upon EGFR-CAR-iPS-NK administration under both chow and HFD-fed conditions (Figure 7I). We next determined the status of CAR-iPS-NK cells in tumors of normal or obese mice, finding that both MDA-MB-231 and HT-29 xenograft harbored reduced NK cells upon HFD challenge, which were restored by SNORD46 pi (Figures 7J and 7K). Taken together, these data suggested a precisely targeted combinatorial treatment for obesity and obesity-associated immune resistance through the application of a snoRNA-based approach.
Discussion
Human subject studies suggested that plasma IL15 concentration negatively correlates with BMI, and fat mass37,38. Mice with Il15 knockout (Il15−/−) results in significant increase in weight gain without altering appetite5,23. Transgenic mice with elevated circulating IL15 exhibited reduced weight of body fat and resistant to diet-induced obesity39. Introduction of recombinant IL15 by protein injection40, adenoviral expression vector5 or DNA electrotransfer37, all resulted in reduced fat mass in rodents, but no effect on food intake nor lean body mass. Our research work suggests that SNORD46 inhibits the normal biology of IL15 in lipid metabolism, leading to inhibited lipolysis and adipose browning, which is unlikely indirect effect on modified energy intake. The exact role of IL15 and SNORD46 signaling axis in obesity warrants the future investigation using tissue specific Il15/Snord46 knockout mice.
MGLL plays an important role in converting monoacylglycerides to FFA and glycerol, which is the rate-limiting step of lipolysis41. Mgll knockout mice exhibit resistance to obesity induced by HFD42. Histidine-269 plays an essential role in the catalytic triad of MGLL, forming a hydrogen bond with substrate43. Hence, it is possible that the phosphorylation of Y268 identified by our studies may affect the release of substrate and lead to altered enzymatic activity of MGLL. Genetic evidence indicated that Cd36-deficient mice exhibited significant decrease in binding and uptake of oxidized low-density lipoprotein44, signifying the essential role of CD36 in lipid transportation. Our findings suggested the FER-mediated phosphorylation of CD36 inhibits FA uptake. We reason that the phosphorylation of CD36 is likely to modulate the cycling of CD36. The roles of other lipase, including ATGL, HSL, and lipid transporters in IL15 induced lipolysis warrants further investigation.
Overall, our studies demonstrated the underlying mechanism of obesity in an IL15 and SNORD46-dependent manner. In this study, we focus on targeting SNORD46 by pi as a promising therapeutic strategy for alleviating obesity and augmenting the anti-tumor immunity of CAR-NK cells. Thus, in the future, it is worthwhile to study the snoRNAs as the primary interplaying factors that constitute metabolism, immunological reactions, and cancer with a focus on mechanistic aspects. Finally, our studies reveal the possibility of metabolic interventions at non-coding RNA level to enhance the efficacy of immunotherapeutic and conventional approaches for future anti-obesity and -cancer treatments.
Limitations of the study
In in vitro biochemical studies, we identified that SNORD46-IL15 interaction is significantly enhanced by SNORD46 G11A mutation. Although genetic knockin SNORD46 G11A mutation drives the obesity and spontaneous tumor development in multiple organs of mice, it is not clear if this mutation affects IL15 signaling to regulate obesity or contribute to NK cell pathophysiology and cancer development in human. Technically, since most experiments were conducted with mouse and human adipocytes, we appreciate its intraindividual variability, and genome editing in these adipocytes did not reach complete KO. Moreover, we cannot fully exclude putative off-target effects of the SNORD46 inhibitor used. Our Snord46WT/G11A mice did not show any phenotypes. It is possible that the expression of the knockin allele does not follow a simple gene dosage effect. The wild-type allele may compensate for the effects of the knockin allele, preventing the obese phenotype from manifesting. This compensation could occur at the transcriptional, translational, or post-translational levels.
STAR★Methods
Resource Availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Liuqing Yang (lyang7@mdanderson.org).
Materials availability
All unique/stable reagents generated in this study are available from the lead contact under Material Transfer Agreement.
Data and code availability
The bulk RNA-seq of human adipocytes with or without IL15 stimulation were deposited to GEO (GSE210203). The bulk RNA-seq of NK cells isolated from healthy donors and donors with obesity were deposited to GEO (GSE213465).
Data S1: Unprocessed source data underlying all blots and graphs, related to Figures 1, 2, 3, 4, 5, 6, 7, and S1–S8. This file includes: 1) Uncropped hi-resolution scans of all the blots; 2) Excel file containing the values that were used to create all graphs in the paper; 3) Normalized expression value of snoRNA PCR array; 4) Protein identification data of PICh-MS; 5) Protein identification data of SNORD46-binding proteins; 6) Sanger sequencing results of IL15 CLIP assays; 7) Peptide identification data of IL15 LiP-MS assay; 8) Protein identification data of IL15 binding proteins in the major organs of Snord46WT/WT or Snord46G11A/G11A mice; 9) Mass spectrometry verification data of SNORD46 power inhibitors; 10) Normalized expression value of bulk-RNA seq in NK cells.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Experimental model and subject details
Study approval
Human tissue studies were deemed exempt by the Institutional Review Board (IRB) of MD Anderson Cancer Center due to the fact that only de-identified human samples were utilized, while the human induced pluripotent stem (hiPS) cell studies are approved by HEIP Stem Cell committee of MD Anderson Cancer Center. All animal studies were approved by the Institutional Animal Care and Use Committee (IACUC) of MD Anderson Cancer Center.
Tissue, serum, and primary cells
Human serum samples from donors with BMI and clinical parameters were purchased from the following commercial vendors: Asterand Bioscience, BioIVT, Proteogenex, Reprocell USA and Coriell Biorepository (Table S1). Human breast cancer tumor tissues and paired serum samples with clinical information were obtained from Discovery Life Sciences. Clinical information is listed in Table S2. Primary human pre-adipocytes were purchased from Zen-Bio, Cell Applications and Celprogen. BMI and clinical parameters are listed in Table S3. Human PBMC from healthy donors or donors with obesity were obtained from STEMCELL Technologies. BMI and clinical parameters are listed in Table S4. Human and mouse tissue total RNA were purchased from Zyagen.
Mouse models and in vivo treatment
By using CRISPR/Cas9 Extreme Genome Editing System (EGE™ Biocytogen), we knocked in a one-nucleotide mutant to mouse SNORD46 gene Chr4:117,155,277 C>T in C57BL/6N mice to achieve the mouse Snord46 G11A mutant, which is the residue conserved in human SNORD46 G11A. C57BL/6, Cebpb−/− mice, Il2rb−/− mice used in this study were purchased from The Jackson Laboratory. To establish the high fat diet (HFD) animal model, mice were fed with HFD (Research Diets) starting at 6-weeks age. To obtain unbiased and reliable results, mice were randomly grouped for all experiments and at least five mice were used in each group. For pi treatment, Snord46WT/WT and Snord46G11A/G11A mice were treated with Snord46 pi (1 mg/kg, subQ, every 3 days). For the treatment of Orlistat or Liraglutide, Snord46G11A/G11A mice were treated with orlistat by oral (10 mg/kg/day)45 or Liraglutide via subQ delivering (0.4 mg/kg/day)46. To identify the IL15 binding proteins in WAT, biotinylated IL15 were injected to the Snord46WT/WT or Snord46G11A/G11A mice (10 mg/kg, ip. Once). 24-h post injection, major organs were collected, homogenized for streptavidin pull-down followed by mass spectrometry, which is described below. All animals were housed under a 12-h light/ 12-h dark cycle in the animal facility with free access to water and food. Sample size was indicated in each figure.
Pluripotent Stem Cells
The hiPS cell lines derived from skin fibroblasts of healthy donor were obtained from Human Stem Cell Core (HSCC) (Baylor College of Medicine) and cultured individually on hESC-Qualified Matrigel (Corning) coated plates and maintained in mTeSR™ plus medium (STEMCELL Technologies).
Method details
Plasmid, siRNAs, transfection, lentiviruses production and transduction
SNORD46 and SNORD46 DNA sequence was synthesized by GenScript and cloned into pGEM-3Z vector (Promega) for in vitro transcription and into the pcDNA3.1 (+) vector (Life technologies) for mammalian expression. Mammalian expression vector for C/EBPβ, IL15, SNORD46 and corresponding mutants were constructed by VectorBuilder and packed in lentivirus. IL15, MGLL wild-type, and mutant sequences were synthesized and cloned into pET-28a (+) (GeneScript). The recombinant protein was expressed in E. coli strain BL21-CodonPlus (DE3)-RIPL (Agilent Technologies) and purified using HisPur™ Cobalt Purification Kit (Thermo Scientific). ON-TARGETplus siRNAs SMARTpool (Dharmacon™) used in this study were transfected using SilenceMag siRNA Delivery Reagent based on the Magnetofection™ technology (OZ Biosciences). Lentiviruses were produced using pPACKH1 HIV Lentivector Packaging Kit (System Biosciences). Lentiviral transduction of sgRNAs and expression constructs were conducted using ViroMag CRISPR (OZ Biosciences) and ViroMag Stem Transduction Enhancer respectively.
Human snoRNA PCR array
The total RNA from serum of 6 healthy donors (BMI < 25) and 6 donors with morbid obesity (BMI > 40) were purified with the miRNeasy Serum/Plasma Kit (Qiagen) followed by cDNA synthesis using rtStar™ tRNA Pretreatment & First-Strand cDNA Synthesis Kit (ArrayStar). SnoRNA expression analysis was performed using nrStar™ Human snoRNA PCR Array (ArrayStar), which analyzed 359 H/ACA and C/D box snoRNAs. HK (Housekeeping genes; Internal Controls): 8 human housekeeping genes ACTB, B2M, Gusb, Hsp90ab1, GAPDH, 5S rRNA, 28S rRNA, and 18S rRNA were included as the internal qPCR normalization references.
Human adipocyte culture and treatment
Human preadipocytes were obtained from various vendors and were cultured with DMEM/F12 medium with 5% fetal calf serum (added penicillin/streptomycin) at 37°C at 5% CO2. Adipogenic conversion was then promoted for 14 days by changing medium to DMEM/F12 without serum addition (penicillin/streptomycin), supplemented with 66 nM insulin, 100 nM dexamethasone, 0.5 mM IBMX, 0.1 mg/ml pioglitazone, 1 nM triiodo-L-thyronine, and 10 mg/ml human transferrin. After 5 days of incubation, medium was used as described before but without IBMX and pioglitazone for 9 more days47. Gene knockdown was performed to the preadipocytes by introducing siRNA and were further induced into mature adipocytes after confirming the expression level of individual genes. Gene targets were knocked out using the CRISPR/Cas9 genome editing system (VectorBuilder) followed by expressing the individual indicated expression constructs in preadipocytes. For supplementation assays, 5 days after the start of the induction process, the indicated unsaturated and saturated fatty acid were added directly to the cell culture well at each medium change. Treatment of adipocytes with fatty acids (Nu-Chek-Prep) were performed as previously described48,49.
DNA and RNA isolation, quantitative real-time PCR
snoRNA was extracted from the human/mouse serum and adipocyte supernatant using miRNeasy Serum/Plasma Kit (Qiagen). Total RNA or snoRNA was isolated from human/mouse adipose tissue and adipocytes using RNeasy Lipid Tissue Mini Kit (Qiagen). For snoRNA detection, 1 μl of miRNeasy Serum/Plasma Spike-In Control (Qiagen) was added before RNA extraction as the reference gene for the following quantitative experiments. The cel-miR39-3p RNA spike-in template was used in combination with the UniSp6 RNA spike-in template provided with the miRCURY LNA RT Kit (Qiagen) as the cDNA synthesis control. Quantitative real-time PCR was performed using the miRCURY LNA SYBR Green PCR Kit (Qiagen). For mRNA detection, reverse transcription and real-time PCR were performed using iScript™ cDNA Synthesis Kit and SsoAdvanced Universal SYBR Green Supermix (Bio-Rad). The RNA copy number was performed as previously described50. Briefly, the total RNA of the serum, adipocyte supernatant, and adipocyte samples were extracted and detected by qRT-PCR. A standard curve was generated by a serial dilution of in vitro transcribed SNORD46 RNA, using 728908.2 for human SNORD46 and 710082.8 for mouse Gm26330 as the molecular weight, and the total RNA per cell was estimated to be 20 pg. The primers used in these studies are listed in Table S5.
Northern blot
The RNA samples were separated by polyacrylamide gel electrophoresis with 6 M urea as a denaturing agent. The separated RNA sequences were transferred to the nylon membrane and subjected to blotting using NorthernMax® Kit (Thermo Fisher Scientific). The in vitro-produced biotinylated probes targeting SNORD46/Snord46 or the control RNA were hybridized with RNA sequences on the nylon membrane, separately. After removing the non-hybridized probe, the blot membrane was then subjected to avidin attached to HRP (horseradish peroxidase). The labeled probe was detected by chemiluminescence to evaluate the molecular weight and the abundance of the RNAs in each sample.
Proteomics of isolated chromatin segments (PICh)
PICh was carried out as previously described51 using the following 2’Fluoro-RNA probes for hybridization: Destiobiotin-108 atom tether- AaCgCaCaAcTgTcAcAcGgAa (SNORD46 probe); Destiobiotin-108 atom tether-TtAgGgTtAgGgTtAgGgTtAgGgt (Telomere probe); Destiobiotin-108 atom tether-GaTgTgTgGaTgTggAtGtGgAtgTgg (Scramble probe).
Chromatin immunoprecipitation (ChIP)
Chromatin was prepared from human adipocytes after cross-linking for 10 min at room temperature with 1% formaldehyde-containing medium, using truChIP™ Chromatin Shearing Kit with Formaldehyde (Covaris). Nuclear extracts were sonicated using M220 Focused-ultrasonicator (Covaris) to obtain DNA fragments averaging 200 bp in length followed by immunoprecipitation overnight with 10 μg of C/EBPβ antibody (Table S6). Immunoprecipitation was washed in RIPA buffer [50 mM HEPES-KOH (pH 8), 500 mM LiCl, 1 mM EDTA (pH 8), 1% NP-40, 0.7% sodium deoxycholate followed by a TE/50 mM NaCl wash. Elution was performed at 65°C for 15 min with elution buffer [50 mM Tris-HCl (pH 8), 10 mM EDTA (pH 8), 1% SDS] and reverse crosslinked at 65°C overnight together with input. DNA was recovered by ChIP DNA Clean & Concentrator (Zymo Research). RT-qPCR was performed using SYBR green and CFX96 Real-Time PCR System (Bio-Rad) with primers listed in Table S5.
In vitro pull-down, immunoprecipitation, and immunoblotting
For in vitro protein-protein interaction studies, the recombinant His-tagged full length IL15, and its mutants were incubated with biotinylated SNORD46 or the control molecule at 4 °C for 16 h, and the Ni-NTA pull-down was performed in the presence of IL2RB using Pierce™ His Protein Interaction Pull-Down Kit (Thermo Fisher Scientific). The pull-downs were resolved by SDS–PAGE and analyzed by immunoblotting with the appropriate antibodies. The immunoprecipitation was performed using Dynabeads™ Co-Immunoprecipitation Kit (Thermo Fisher Scientific). The immunoprecipitates were separated by SDS–PAGE and immunoblotted with listed antibodies (Table S6). Visualization of the immunoblots was performed by Clarity Western ECL Substrate (Bio-Rad) according to the manufacturer’s instructions.
RNA pull-down and mass spectrometry analysis
To identify SNORD46-binding proteins in serum, SNORD46 pull-down was performed as previously described17,52. Briefly, biotin-labeled SNORD46 RNAs were in vitro transcribed with Biotin RNA Labeling Mix (Millipore Sigma) and MEGAscript® Transcription Kit (Thermo Fisher Scientific), further purified by RNA Clean & Concentrator-5 (Zymo Research). Human serum proteins were purified using Aurum Serum Protein Mini Kit (Bio-Rad). The BcMag™ Monomer avidin Magnetic Beads (Bioclone) were prepared according to the manufacturer’s instructions and then immediately subjected to RNA (20 μg) capture in RNA capture buffer [20 mM Tris-HCl (pH 7.5), 1M NaCl, 1mM EDTA] as previously described52. The eluted RNA-protein complexes were denatured, reduced, alkylated, and digested with immobilized trypsin (Promega) for mass spectrometry analysis at MD Anderson Cancer Center Proteomics Facility.
UV-Crosslinking and Immunoprecipitation (CLIP) assay and RIP assay
Human serum was UV crosslinked on ice with three irradiations of 254 nm UV-light at 400 mJ/cm2 in a UV crosslinker (Agilent). CLIP was performed using the IL15 antibody (Table S6) as previously described53. RNA-protein complexes of interest were then partially purified by immunoprecipitation, and non-covalently associated RNAs were removed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). RNA-protein complexes were subjected to SDS-PAGE and autoradiography. The RNA fragments were extracted, ligated, reverse transcribed, and ligated into the vector pCR-Blunt for Sanger sequencing. RIP assay was performed using Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore). Human serum was immunoprecipitated using 2.5 μg of normal rabbit IgG, anti-IL15 antibody or anti-Argonaute-2 antibody (Table S6). Immunoprecipitation of IL15-target and non-target associated RNA was validated by quantitative PCR (qPCR). CFX Manager software v.3.1 (Bio-Rad) was used for data acquisition for qPCR.
Determination of Kd value using Alpha assay
Alpha binding assays were used to quantitatively assess the interaction between SNORD46 and IL15 using biotinylated SNORD46 and His-tagged IL15 as the donor and acceptor pair17. The dissociation constant (Kd) was determined via a competition experiment in which WT SNORD46, or IL15 and their mutants were titrated (two-fold dilution) from 100 μM to 1 pM, separately. Streptavidin donor beads and anti-His6 AlphaLISA acceptor beads were used in these assays (PerkinElmer). The plates were read on the EnSpire Multimode plate reader (PerkinElmer). The competitive inhibition curves were calculated based on alpha signal readings by fitting to a log (inhibitor) versus response-variable slope (four parameters) model (GraphPad Prism 9).
Limited proteolysis (LiP)-coupled liquid chromatography–mass spectrometry (LC-MS)
LiP followed by LC-MS was adapted from LiP-SRM analysis16. Briefly, bacterially expressed IL15 (2 mg/ml) alone, in the presence of Scramble RNA or SNORD46 RNA (2 mM) were incubated in buffer (20 mM HEPES, pH7.5, 150 mM KCl and 10 mM MgCl2) and Protease K at room temperature for 5 min as limited proteolysis. The digestion was stopped by transferring the reaction mixture to a tube containing guanidine hydrochloride crystals to a final concentration of 7.4 M and by boiling for 3 min. The digestion mixtures were then subjected to complete Staphylococcal peptidase I and Arg-C proteinase digestion. The peptides were subjected to LC-MS analysis at Proteomic and Metabolic core facility of MD Anderson Cancer Center. The numbers of peptides recovered from LiP-MS for each sample were normalized using only IL15 wild-type. Number of peptides recovered from LiP-MS is shown.
Computational modeling of SNORD46-IL15 Interaction
The Secondary structures of snoRNAs were predicted using RNAfold webserver54. The minimum free energy structure was selected for further investigation. The 3D structure of snoRNAs were predicted using Rosetta software55. The computational RNA structure models with highest scores will be used for the RNA-Protein docking studies. The crystal structure of IL15 was retrieved from the protein data bank (PDB ID: 2Z3Q). Protein preparation and energy minimization were processed using Schrodinger software suite. Then we performed RNA-Protein docking studies using multiple webservers including HDOCK20, 3dRPC21, and NPDock22. The binding poses with best scores were selected for examination of detailed interactions. Visualization of the RNA-Protein structure complexes and generation of final figures were performed using PyMOL v2.4.
Animal metabolic studies
Whole-body composition parameters were measured in male or female Snord46WT/WT or Snord46G11A/G11A mice at age of 12-weeks old by Faxitron Specimen Radiography System (Faxitron X-Ray Corp., Wheeling, IL) to precisely measure total body fat and lean mass. Mice were housed in the Oxymax/CLAMS (The Columbus Instruments Comprehensive Lab Animal Monitoring System) metabolic cage system from Columbus Instruments for 4 days with ad libitum access to food and water. VCO2, VO2, RER, and activity were measured by the Oxymax system. CLAMS was used to record the food and water intake, O2 consumption, CO2 expenditure, and energy expenditure. Mice (12-weeks old) were randomly grouped, and four mice were included in each group. Data reading and recording proceeded seven times per hour, and each mouse was housed and recorded for 72 h.
Tissue collection and immunohistochemistry (IHC)
Unless otherwise indicated, mice were fasted for 4-6 h, anesthetized with isoflurane, blood collected by heart puncture, and then sacrificed. Tissues were dissected, weighed, and either dipped in liquid nitrogen or fixed in 10% formalin solution. Pectoralis, triceps, quadriceps, gastrocnemius, and tibialis anterior muscles were dissected on one side of the body. The tissues fixed in formalin were processed and embedded in paraffin. The samples were sectioned into 4 μm thickness for the H&E, and IHC staining. Lipid droplet accumulation in the liver was visualized using Oil Red O (Millipore Sigma) staining of frozen liver sections prepared in an optimum cutting temperature (O.C.T.) compound (Fisher Scientific). Histopathology images were acquired with a light microscope (Olympus).
Blood analyses
Whole blood was collected by tail bleeding or cardiac puncture from mice fasted for 4-6 h. Total cholesterol (TC) and triglycerides (TG) were determined by enzymatic assays using commercial kits (BioAssay Systems). Alanine aminotransferase (ALT) and aspartate aminotransferase (AST) levels were measured using Alanine Transaminase Colorimetric Activity Assay Kit (Cayman Chemical) and EnzyChrom™ Aspartate Transaminase Assay Kit (BioAssay Systems). Serum levels of Leptin (R and D Systems), Insulin (Thermo Fisher Scientific), and Ghrelin (Millipore Sigma) were measured by ELISA assay according to the manufacturer’s instructions respectively.
Glucose and insulin tolerance tests
Insulin (ITT) and glucose (GTT) tolerance tests were performed on 6-h fasted male and female mice fed with chow or HFD. Glucose values were measured using the AimStrip® Plus Blood Glucose Meter (VWR, PA) by tail snip. Glucose (1g per kg body weight) and human insulin (0.75 U per kg body weight) were injected intraperitoneally (i.p.) after baseline glucose levels were measured in each mouse, and blood glucose levels were measured 15, 30, 45, 60, 90, and 120 min after injection.
Metabolic assays
Human adipocytes were cultured in vitro, and the cells and the supernatant were collected after the treatment with indicated cytokines. The intracellular NEFA and triglyceride were detected by Lipolysis Assay Kit (Zen-Bio) and Triglyceride Assay Kit (Zen-Bio) respectively. Extracellular/supernatant and serum NEFA were detected by Serum/Plasma Fatty Acid Detection Kit (Zen-Bio). OCR of adipocytes was measured using Seahorse XF Cell Mito Stress Test (Agilent Technologies). Briefly, human or mouse adipocytes were resuspended in XF assay media onto a XF24 cell plate pre-coated with Cell-Tak (Corning) followed by sequentially treatment with 1 μM oligomycin; 1 μM phenylhydrazone (FCCP); and 0.5 μM mixture including rotenone and antimycin A according to the instruction. Seahorse XFe Wave Software (Agilent) was applied to analyze the data.
Measurement of MGLL activity
MGLLL activity was measured as previously reported56. The reaction was set up in 200 ul assay buffer (10 mM Tris-HCl 1 mM EDTA pH 7.4, plus 0.1% fatty acid-free BSA) containing 25 ng of purified MGLL, 10 μM cold 2-arachidonoylglycerol (Cayman Chemical) and [3H]-2-arachidonoylglycerol (American Radiolabeled Chemicals) as substrate at indicated concertation. The reaction mixture was incubated for 15 min at 37°C and then stopped by adding 400 μL of activated acid-washed charcoal). The contents were shaken at 37 °C for 90 seconds at 750 RPM, incubated at room temperature for 30 min, and then centrifuged at 2,000 x g for 10 min at room temperature. The 40 ul of aqueous phase was removed and combined with 160 ul of Ultima Gold scintillation liquid (PerkinElmer). Samples were then counted using a Beckman LS 6500 scintillation counter to measure the amount of [3H]-glycerol in the supernatant cocktail (CPMs). The CPMs were converted to uM/min/mg protein based on a [3H]-glycerol standard curve using MasterPlex® curve fitting software (Hitachi Solutions America). The MGLL activity (kcat)was calculated by Michaelis-Menten model using GraphPad Prism 9 software.
Assay of fatty acid uptake
[3H]-oleic acid metabolic flux assay adapted from previous studies57,58 was used to evaluate the CD36 activity in human adipocytes. The CPMs of cell lysate were counted using a Beckman LS 6500 scintillation counter and converted to uM/min/mg protein based on a [3H]-oleic acid standard curve using MasterPlex® curve fitting software (Hitachi Solutions America). The CD36 activity (kcat) was calculated by Michaelis-Menten model using GraphPad Prism 9 software.
NK cell isolation, cell viability determination, bulk RNA sequencing and analysis
NK cells were isolated form human PBMC. Human PBMC from healthy donors or donors with obesity were obtained from STEMCELL Technologies. PBMC cells were negatively selected using CD3 as marker, the CD3− cells were further selected by using CD56 as the positive selective marker, according to manufacturer’s instruction (STEMCELL Technologies). The CD3-CD56+ NK cells were used in the following experiments. The cell viability of NK cells was determined using Trypan Blue dye, and automated cell counter (Bio-Rad), with or without Tubeimoside I (TBM) at the indicated concentration. The percentage of live cells normalized by the number of total cells were shown. Bulk RNA seq were performed by Novogene. RNAseq raw output fastq files were mapped to human transcriptome (genome version GENCODE v35, hg38) using Salmon (version 0.14.1), STAR2 (version 2.6.0b), and htseq (version 0.11.0) to produce raw gene counts and log2 transformed TPM reads. Differential gene expression was performed using raw counts and R DESeq2 package and results confirmed using TPM and R limma package. Pathway analyses were performed using GSEA and IPA software. Raw data and processed gene expression table were deposited to GEO (GSE213465).
Immunofluorescence and mIHC staining
IF staining was performed using NK cells isolated from human PBMC of healthy donors or donors with obesity. The cells were fixed with 4% paraformaldehyde solution and immune labeled using antibodies listed in the Table S6. The images were visualized with a Zeiss Axioskop2 Plus microscope. All immunostained slides were scanned on the APERIO ScanScope XT (Leica Biosystems) for quantification by digital image analysis. The quantification of IF staining density was performed by Image-Pro Plus 6.0 software (Media Cybernetics) and calculated based on the average staining intensity and the percentage of positively stained cells. For mIHC staining, formalin-fixed, paraffin-embedded (FFPE) tissue sections were cut at a thickness of 4 μm using a HistoCore BIOCUT Manual Rotary Microtome (Leica Biosystems). Staining of the sections with indicated antibodies was conducted using Opal™ Polaris 7 Color Automation IHC Detection Kit (Akoya Biosciences) and imaged with a PE Vectra Polaris Automated Quantitative Pathology Imaging System (PerkinElmer).
Flow cytometry
Cells were stained with various antibodies (Table S6) at room temperature for 15 min, washed with PBS and then re-suspended in PBS containing 1:200 dilution of Zombie Violet fixable viability dyes (BioLegend). Cells were then incubated for 15 min under room temperature and were fixed with 1.5% formaldehyde for 20 min under room temperature, washed one time and resuspended in FACS buffer (PBS with 5% fetal calf serum) before analysis on flow cytometer. Flow cytometry was performed on an LSR II system (BD Biosciences), and data were analyzed using FlowJo software (BD Biosciences).
CAR-NK cell generation and in vitro cytotoxicity
The hiPS cells derived from male donor were transfected with a CD133-CAR and the hiPS cells derived from female donor were transfected with an EGFR CAR. The CAR-iPS cells were then differentiated to NK cells by using the STEMdiff™ NK Cell Kit (STEMCELL Technologies). In brief, the CAR-iPS cells were induced to generate embryoid bodies for downstream lymphoid differentiation, after a total of 12 days culture, then CD34+ cells are enriched by EasySep™ positive selection. Then the hiPS-derived CD34+ cells were cultured in StemSpan™ Lymphoid Progenitor Expansion Medium for 14 days, followed by 14 days of culture using StemSpan™ NK Cell Generation Kit to generate CD56+ NK cells. Cells were harvested and analyzed for CD56, NKp46, NKp44, NKp30, NKG2D, and KIR expression by flow cytometry. MDA-MB-231 or HT-29 were incubated with EGFR-CAR-iPS-NK or CD133-CAR-iPS-NK cells at the 8:1 ratio in triplicate wells of U-bottomed 96-well plates. Target cell death was detected 6 h later by adding YOYO™-1 Iodide (491/509) (Thermo Fisher Scientific). The YOYO staining intensities were scanned on the APERIO ScanScope XT (Leica Biosystems) for quantification by digital image analysis. The quantification of IF staining density was performed by Image-Pro Plus 6.0 software (Media Cybernetics) and calculated based on the average staining intensity and the percentage of positively stained cells.
CAR-NK cell target tumorigenesis studies
6-8 weeks-old female NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, Jackson Laboratory) were raised under specific pathogen-free conditions. Animals were grouped randomly, and 3-5 mice were put into each cage. NSG mice under both chow and HFD-fed conditions were inoculated with 5×106 MDA-MB231 cells orthotopically into the mammary fat pad, or 5×105 HT-29 cells were implanted subcutaneously. Scramble or Snord46 power inhibitors (10 mg/kg/day, IP injection) were administrated when tumor cells inoculation. EGFR-CAR-iPS-NK or CD133-CAR-iPS-NK (1×107) cells were injected intravenously (every 7 days, 3 times in total) to tumor-bearing mice when tumor volume reached approximately 50 mm3. Tumor size was measured every 4 days using a caliper, and tumor volume was calculated using the standard formula: 0.54×L×W2, where L is the longest diameter and W is the shortest diameter.
Quantification and statistical analysis
The experiment was set up to use 3-8 samples/repeats per experiment/group/condition to detect a 2-fold difference with power of 80% and at the significance level of 0.05 by a two-sided test for significant studies. Each of these experiments was independently repeated 3-5 times. Results were reported as mean ± standard deviation (SD) of at least three independent experiments, as indicated by figure legends. Each exact n value was indicated in the corresponding figure legend. Statistical analysis was performed using GraphPad Prism 9 software. Comparisons were analyzed by unpaired Student’s t-test or one-way ANOVA test (n.s., p > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001), as indicated in individual figures. For survival analysis, Kaplan-Meier survival curves were compared using the log rank test.
Supplementary Material
Table S2. Clinical information of tumor tissues and paired serum samples used in this study, related to Figure 1.
Table S3. Clinical information of human primary adipocyte donors, related to Figures 2, 3, 5 and 6.
This table contains 3 tabs:
Tab 1: Clinical parameters of human primary adipocyte donors from Zen-Bio
Tab 2: Clinical parameters of human primary adipocyte donors from Cell Applications
Tab 3: Clinical parameters of human primary adipocyte donors Celprogen
Table S1. Clinical information of human serum used in this study, related to Figures 1, 3, 5 and 6.
This table contains 5 tabs:
Tab 1: Clinical parameters of human serum from Asterand Bioscience
Tab 2: Clinical parameters of human serum from BioIVT
Tab 3: Clinical parameters of human serum from Proteogenex
Tab 4: Clinical parameters of human serum from Reprocell USA
Tab 5: Clinical parameters of human serum from Coriell Biorepository
Table S4. Clinical information of human PBMC used for NK isolation, related to Figure 7.
Table S5. Oligonucleotides used in this study.
Table S6. Antibodies used in this study.
Data S1. Unprocessed source data underlying all blots and graphs, related to Figures 1, 2, 3, 4, 5, 6, 7, and S1–S8.
- Uncropped hi-resolution scans of all the blots.
- Excel file containing the values that were used to create all graphs in the paper.
- Normalized expression value of snoRNA PCR array, related to Figure 1.
- Protein identification of PICh-MS, related to Figure 2. This table contains 9 tabs:
- Tab 1: List of proteins associated with scramble probes, replicate #1
- Tab 2: List of proteins associated with scramble probes, replicate #2
- Tab 3: List of proteins associated with scramble probes, replicate #3
- Tab 4: List of proteins associated with SNORD46 promoter probes, replicate #1
- Tab 5: List of proteins associated with SNORD46 promoter probes, replicate #2
- Tab 6: List of proteins associated with SNORD46 promoter probes, replicate #3
- Tab 7: List of proteins associated with telomere probes, replicate #1
- Tab 8: List of proteins associated with telomere probes, replicate #2
- Tab 9: List of proteins associated with telomere probes, replicate #3
- Protein identification of SNORD46-binding proteins, related to Figure 3. This table contains 9 tabs:
- Tab 1: Biotinylated scramble RNA pull-down in human serum, replicate #1
- Tab 2: Biotinylated scramble RNA pull-down in human serum, replicate #2
- Tab 3: Biotinylated scramble RNA pull-down in human serum, replicate #3
- Tab 4: Biotinylated SNORD46 RNA pull-down in human serum, replicate #1
- Tab 5: Biotinylated SNORD46 RNA pull-down in human serum, replicate #2
- Tab 6: Biotinylated SNORD46 RNA pull-down in human serum, replicate #3
- Tab 7: Biotinylated miR-20a RNA pull-down in human serum, replicate #1
- Tab 8: Biotinylated miR-20a RNA pull-down in human serum, replicate #2
- Tab 9: Biotinylated miR-20a RNA pull-down in human serum, replicate #3
- Sanger sequencing results of IL15 CLIP assays, related to Figure 3. This table contains 2 tabs:
- Tab 1: Sanger sequencing results of IL15 CLIP assay in human serum
- Tab 2: Sanger sequencing results of IL15 CLIP assay in mouse serum
- Peptide identification of IL15 LiP-MS assay, related to Figure 3. This table contains 3 tabs:
- Tab 1: Peptide identified from IL15 LiP-MS assay
- Tab 2: Peptide identified from IL15 LiP-MS assay in the presence of scramble RNA.
- Tab 3: Peptide identified from IL15 LiP-MS assay in the presence of SNORD46 RNA
- Protein identification of IL15 binding proteins in the major organs of Snord46WT/WT or Snord46G11A/G11A mice, related to Figure 5. This table contains 16 tabs:
- Tab 1: List of proteins associated with IgG in Snord46WT/WT mouse tissues
- Tab 2: List of proteins associated with IL15 in brain tissues of Snord46WT/WT mice
- Tab 3: List of proteins associated with IL15 in heart tissues of Snord46WT/WT mice
- Tab 4: List of proteins associated with IL15 in liver tissues of Snord46WT/WT mice
- Tab 5: List of proteins associated with IL15 in SKM tissues of Snord46WT/WT mice
- Tab 6: List of proteins associated with IL15 in WAT of Snord46WT/WT mice
- Tab 7: List of proteins associated with IL15 in spleen tissues of Snord46WT/WT mice
- Tab 8: List of proteins associated with IL15 in intestine tissues of Snord46WT/WT mice
- Tab 9: List of proteins associated with IgG in Snord46WT/WT mouse tissues
- Tab 10: List of proteins associated with IL15 in brain tissues of Snord46G11A/G11A mice
- Tab 11: List of proteins associated with IL15 in heart tissues of Snord46G11A/G11A mice
- Tab 12: List of proteins associated with IL15 in liver tissues of Snord46G11A/G11A mice
- Tab 13: List of proteins associated with IL15 in SKM tissues of Snord46G11A/G11A mice
- Tab 14: List of proteins associated with IL15 in WAT of Snord46G11A/G11A mice
- Tab 15: List of proteins associated with IL15 in spleen tissues of Snord46G11A/G11A mice
- Tab 16: List of proteins associated with IL15 in intestine tissues of Snord46G11A/G11A mice
- Mass spectrometry verification of SNORD46 power inhibitors, related to Figure 6. This table contains 6 tables:
- Tab 1: List of serum SNORD46-binding proteins in the presence of scramble power inhibitors
- Tab 2: List of serum SNORD46-binding proteins in the presence of miR-20a power inhibitors
- Tab 3: List of serum SNORD46-binding proteins in the presence of SNORD46 power inhibitor #1
- Tab 4: List of serum SNORD46-binding proteins in the presence of SNORD46 power inhibitor #2
- Tab 5: List of serum SNORD46-binding proteins in the presence of SNORD46 power inhibitor #3
- Tab 6: List of serum SNORD46-binding proteins in the presence of SNORD46 power inhibitor #4
- Normalized expression value of bulk-RNA seq in NK cells, related to Figure 7.
Acknowledgements
This work was supported by CPRIT grant RR150085 and NIH R01CA262623 (to CPRIT Scholar in Cancer Research L.H.); NIH R01CA231011 and R01CA255080, CPRIT RR180259, and DoD BC180196 grants (to C.L.); and NIH R01CA218036 and R01CA269489, DoD BC181384, CPRIT RP200423, and AACR-The Mark Foundation for Cancer Research 20-60-51 grants (to L.Y.). Z.T. is a CPRIT scholar in cancer research, and Z.T. thanks the CPRIT for research funding support (RR220039). This research is supported in part by The University of Texas MD Anderson Cancer Center SPORE in Hepatocellular Carcinoma grant NIH P50CA217674.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Footnotes
Declaration of interests
M.C. reports grants and personal fees from ImmunoGenesis, Inc., personal fees from Alligator Bioscience, Inc., ImmunOS, Inc., Oncoresponse, Inc., Nurix, Inc., Aptevo, Inc., Kineta, Inc., Xencor, Inc., Agenus, Inc., Adagene, Inc., Astrazeneca, Inc., outside the submitted work; K.R. and The University of Texas MD Anderson Cancer Center have an institutional financial conflict of interest with Takeda Pharmaceutical and Affimed GmbH. K.R. participates on the Scientific Advisory Board for GemoAb, AvengeBio, Virogin Biotech, GSK, Caribou Biosciences, Navan Technologies and Bayer. B.G. is an inventor on patent applications involving targeting ferroptosis in cancer therapy, and reports personal fees from Guidepoint Global, Cambridge Solutions, and NGM Bio.
References
- 1.Hales CM, Carroll MD, Fryar CD, and Ogden CL (2017). Prevalence of Obesity Among Adults and Youth: United States, 2015-2016. NCHS Data Brief, 1–8. [PubMed] [Google Scholar]
- 2.Berrington de Gonzalez A, Hartge P, Cerhan JR, Flint AJ, Hannan L, MacInnis RJ, Moore SC, Tobias GS, Anton-Culver H, Freeman LB, et al. (2010). Body-mass index and mortality among 1.46 million white adults. N Engl J Med 363, 2211–2219. 10.1056/NEJMoa1000367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ringel AE, Drijvers JM, Baker GJ, Catozzi A, Garcia-Canaveras JC, Gassaway BM, Miller BC, Juneja VR, Nguyen TH, Joshi S, et al. (2020). Obesity Shapes Metabolism in the Tumor Microenvironment to Suppress Anti-Tumor Immunity. Cell 183, 1848–1866 e1826. 10.1016/j.cell.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma A, Koka R, and Burkett P (2006). Diverse functions of IL-2, IL-15, and IL-7 in lymphoid homeostasis. Annu Rev Immunol 24, 657–679. 10.1146/annurev.immunol.24.021605.090727. [DOI] [PubMed] [Google Scholar]
- 5.Barra NG, Reid S, MacKenzie R, Werstuck G, Trigatti BL, Richards C, Holloway AC, and Ashkar AA (2010). Interleukin-15 contributes to the regulation of murine adipose tissue and human adipocytes. Obesity (Silver Spring) 18, 1601–1607. 10.1038/oby.2009.445. [DOI] [PubMed] [Google Scholar]
- 6.Alvarez B, Carbo N, Lopez-Soriano J, Drivdahl RH, Busquets S, Lopez-Soriano FJ, Argiles JM, and Quinn LS (2002). Effects of interleukin-15 (IL-15) on adipose tissue mass in rodent obesity models: evidence for direct IL-15 action on adipose tissue. Biochim Biophys Acta 1570, 33–37. 10.1016/s0304-4165(02)00148-4. [DOI] [PubMed] [Google Scholar]
- 7.Ahmad A, Uddin S, and Steinhoff M (2020). CAR-T Cell Therapies: An Overview of Clinical Studies Supporting Their Approved Use against Acute Lymphoblastic Leukemia and Large B-Cell Lymphomas. Int J Mol Sci 21. 10.3390/ijms21113906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xia J, Minamino S, and Kuwabara K (2020). CAR-expressing NK cells for cancer therapy: a new hope. Biosci Trends 14, 354–359. 10.5582/bst.2020.03308. [DOI] [PubMed] [Google Scholar]
- 9.Tang F, Zhao LT, Jiang Y, Ba de N, Cui LX, and He W (2008). Activity of recombinant human interleukin-15 against tumor recurrence and metastasis in mice. Cell Mol Immunol 5, 189–196. 10.1038/cmi.2008.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu P, Steel JC, Zhang M, Morris JC, and Waldmann TA (2010). Simultaneous blockade of multiple immune system inhibitory checkpoints enhances antitumor activity mediated by interleukin-15 in a murine metastatic colon carcinoma model. Clin Cancer Res 16, 6019–6028. 10.1158/1078-0432.CCR-10-1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson TO, and Schluns KS (2017). The potential and promise of IL-15 in immuno-oncogenic therapies. Immunol Lett 190, 159–168. 10.1016/j.imlet.2017.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu E, Tong Y, Dotti G, Shaim H, Savoldo B, Mukherjee M, Orange J, Wan X, Lu X, Reynolds A, et al. (2018). Cord blood NK cells engineered to express IL-15 and a CD19-targeted CAR show long-term persistence and potent antitumor activity. Leukemia 32, 520–531. 10.1038/leu.2017.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steinbusch MM, Fang Y, Milner PI, Clegg PD, Young DA, Welting TJ, and Peffers MJ (2017). Serum snoRNAs as biomarkers for joint ageing and post traumatic osteoarthritis. Sci Rep 7, 43558. 10.1038/srep43558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Acker HH, Capsomidis A, Smits EL, and Van Tendeloo VF (2017). CD56 in the Immune System: More Than a Marker for Cytotoxicity? Front Immunol 8, 892. 10.3389/fimmu.2017.00892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, and Joshua-Tor L (2012). The structure of human argonaute-2 in complex with miR-20a. Cell 150, 100–110. 10.1016/j.cell.2012.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng Y, De Franceschi G, Kahraman A, Soste M, Melnik A, Boersema PJ, de Laureto PP, Nikolaev Y, Oliveira AP, and Picotti P (2014). Global analysis of protein structural changes in complex proteomes. Nat Biotechnol 32, 1036–1044. 10.1038/nbt.2999. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Tan Z, Zhang Y, Zhang Z, Hu Q, Liang K, Jun Y, Ye Y, Li YC, Li C, et al. (2021). A noncoding RNA modulator potentiates phenylalanine metabolism in mice. Science 373, 662–673. 10.1126/science.aba4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Collins L, Tsien WH, Seals C, Hakimi J, Weber D, Bailon P, Hoskings J, Greene WC, Toome V, and Ju G (1988). Identification of specific residues of human interleukin 2 that affect binding to the 70-kDa subunit (p70) of the interleukin 2 receptor. Proc Natl Acad Sci U S A 85, 7709–7713. 10.1073/pnas.85.20.7709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pettit DK, Bonnert TP, Eisenman J, Srinivasan S, Paxton R, Beers C, Lynch D, Miller B, Yost J, Grabstein KH, and Gombotz WR (1997). Structure-function studies of interleukin 15 using site-specific mutagenesis, polyethylene glycol conjugation, and homology modeling. J Biol Chem 272, 2312–2318. 10.1074/jbc.272.4.2312. [DOI] [PubMed] [Google Scholar]
- 20.Yan Y, Zhang D, Zhou P, Li B, and Huang SY (2017). HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res 45, W365–W373. 10.1093/nar/gkx407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang Y, Li H, and Xiao Y (2018). 3dRPC: a web server for 3D RNA-protein structure prediction. Bioinformatics 34, 1238–1240. 10.1093/bioinformatics/btx742. [DOI] [PubMed] [Google Scholar]
- 22.Tuszynska I, Magnus M, Jonak K, Dawson W, and Bujnicki JM (2015). NPDock: a web server for protein-nucleic acid docking. Nucleic Acids Res 43, W425–430. 10.1093/nar/gkv493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lacraz G, Rakotoarivelo V, Labbe SM, Vernier M, Noll C, Mayhue M, Stankova J, Schwertani A, Grenier G, Carpentier A, et al. (2016). Deficiency of Interleukin-15 Confers Resistance to Obesity by Diminishing Inflammation and Enhancing the Thermogenic Function of Adipose Tissues. PLoS One 11, e0162995. 10.1371/journal.pone.0162995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klok MD, Jakobsdottir S, and Drent ML (2007). The role of leptin and ghrelin in the regulation of food intake and body weight in humans: a review. Obes Rev 8, 21–34. 10.1111/j.1467-789X.2006.00270.x. [DOI] [PubMed] [Google Scholar]
- 25.Kojima M, Hosoda H, Date Y, Nakazato M, Matsuo H, and Kangawa K (1999). Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature 402, 656–660. 10.1038/45230. [DOI] [PubMed] [Google Scholar]
- 26.Calderon-Dominguez M, Mir JF, Fucho R, Weber M, Serra D, and Herrero L (2016). Fatty acid metabolism and the basis of brown adipose tissue function. Adipocyte 5, 98–118. 10.1080/21623945.2015.1122857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosen ED, and Spiegelman BM (2014). What we talk about when we talk about fat. Cell 156, 20–44. 10.1016/j.cell.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nomura DK, Long JZ, Niessen S, Hoover HS, Ng SW, and Cravatt BF (2010). Monoacylglycerol lipase regulates a fatty acid network that promotes cancer pathogenesis. Cell 140, 49–61. 10.1016/j.cell.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tran TT, Poirier H, Clement L, Nassir F, Pelsers MM, Petit V, Degrace P, Monnot MC, Glatz JF, Abumrad NA, et al. (2011). Luminal lipid regulates CD36 levels and downstream signaling to stimulate chylomicron synthesis. J Biol Chem 286, 25201–25210. 10.1074/jbc.M111.233551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elaraby E, Malek AI, Abdullah HW, Elemam NM, Saber-Ayad M, and Talaat IM (2021). Natural Killer Cell Dysfunction in Obese Patients with Breast Cancer: A Review of a Triad and Its Implications. J Immunol Res 2021, 9972927. 10.1155/2021/9972927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michelet X, Dyck L, Hogan A, Loftus RM, Duquette D, Wei K, Beyaz S, Tavakkoli A, Foley C, Donnelly R, et al. (2018). Metabolic reprogramming of natural killer cells in obesity limits antitumor responses. Nat Immunol 19, 1330–1340. 10.1038/s41590-018-0251-7. [DOI] [PubMed] [Google Scholar]
- 32.Feng X, Zhou J, Li J, Hou X, Li L, Chen Y, Fu S, Zhou L, Li C, and Lei Y (2018). Tubeimoside I induces accumulation of impaired autophagolysosome against cervical cancer cells by both initiating autophagy and inhibiting lysosomal function. Cell Death Dis 9, 1117. 10.1038/s41419-018-1151-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xia L, Zheng ZZ, Liu JY, Chen YJ, Ding JC, Xia NS, Luo WX, and Liu W (2020). EGFR-targeted CAR-T cells are potent and specific in suppressing triple-negative breast cancer both in vitro and in vivo. Clin Transl Immunology 9, e01135. 10.1002/cti2.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu S, Li A, Liu Q, Li T, Yuan X, Han X, and Wu K (2017). Chimeric antigen receptor T cells: a novel therapy for solid tumors. J Hematol Oncol 10, 78. 10.1186/s13045-017-0444-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Y, Chen M, Wu Z, Tong C, Dai H, Guo Y, Liu Y, Huang J, Lv H, Luo C, et al. (2018). CD133-directed CAR T cells for advanced metastasis malignancies: A phase I trial. Oncoimmunology 7, e1440169. 10.1080/2162402X.2018.1440169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vora P, Venugopal C, Salim SK, Tatari N, Bakhshinyan D, Singh M, Seyfrid M, Upreti D, Rentas S, Wong N, et al. (2020). The Rational Development of CD133-Targeting Immunotherapies for Glioblastoma. Cell Stem Cell 26, 832–844 e836. 10.1016/j.stem.2020.04.008. [DOI] [PubMed] [Google Scholar]
- 37.Nielsen AR, Hojman P, Erikstrup C, Fischer CP, Plomgaard P, Mounier R, Mortensen OH, Broholm C, Taudorf S, Krogh-Madsen R, et al. (2008). Association between interleukin-15 and obesity: interleukin-15 as a potential regulator of fat mass. J Clin Endocrinol Metab 93, 4486–4493. 10.1210/jc.2007-2561. [DOI] [PubMed] [Google Scholar]
- 38.Febbraio MA (2014). Role of interleukins in obesity: implications for metabolic disease. Trends Endocrinol Metab 25, 312–319. 10.1016/j.tem.2014.02.004. [DOI] [PubMed] [Google Scholar]
- 39.Quinn LS, Anderson BG, Strait-Bodey L, Stroud AM, and Argiles JM (2009). Oversecretion of interleukin-15 from skeletal muscle reduces adiposity. Am J Physiol Endocrinol Metab 296, E191–202. 10.1152/ajpendo.90506.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carbo N, Lopez-Soriano J, Costelli P, Alvarez B, Busquets S, Baccino FM, Quinn LS, Lopez-Soriano FJ, and Argiles JM (2001). Interleukin-15 mediates reciprocal regulation of adipose and muscle mass: a potential role in body weight control. Biochim Biophys Acta 1526, 17–24. 10.1016/s0304-4165(00)00188-4. [DOI] [PubMed] [Google Scholar]
- 41.Taschler U, Radner FP, Heier C, Schreiber R, Schweiger M, Schoiswohl G, Preiss-Landl K, Jaeger D, Reiter B, Koefeler HC, et al. (2011). Monoglyceride lipase deficiency in mice impairs lipolysis and attenuates diet-induced insulin resistance. J Biol Chem 286, 17467–17477. 10.1074/jbc.M110.215434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dione N, Lacroix S, Taschler U, Deschenes T, Abolghasemi A, Leblanc N, Di Marzo V, and Silvestri C (2020). Mgll Knockout Mouse Resistance to Diet-Induced Dysmetabolism Is Associated with Altered Gut Microbiota. Cells 9. 10.3390/cells9122705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Labar G, Bauvois C, Borel F, Ferrer JL, Wouters J, and Lambert DM (2010). Crystal structure of the human monoacylglycerol lipase, a key actor in endocannabinoid signaling. Chembiochem 11, 218–227. 10.1002/cbic.200900621. [DOI] [PubMed] [Google Scholar]
- 44.Febbraio M, Abumrad NA, Hajjar DP, Sharma K, Cheng W, Pearce SF, and Silverstein RL (1999). A null mutation in murine CD36 reveals an important role in fatty acid and lipoprotein metabolism. J Biol Chem 274, 19055–19062. 10.1074/jbc.274.27.19055. [DOI] [PubMed] [Google Scholar]
- 45.Chu J, Xing C, Du Y, Duan T, Liu S, Zhang P, Cheng C, Henley J, Liu X, Qian C, et al. (2021). Pharmacological inhibition of fatty acid synthesis blocks SARS-CoV-2 replication. Nat Metab 3, 1466–1475. 10.1038/s42255-021-00479-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Buganova M, Pelantova H, Holubova M, Sediva B, Maletinska L, Zelezna B, Kunes J, Kacer P, Kuzma M, and Haluzik M (2017). The effects of liraglutide in mice with diet-induced obesity studied by metabolomics. J Endocrinol 233, 93–104. 10.1530/JOE-16-0478. [DOI] [PubMed] [Google Scholar]
- 47.Hemmrich K, von Heimburg D, Cierpka K, Haydarlioglu S, and Pallua N (2005). Optimization of the differentiation of human preadipocytes in vitro. Differentiation 73, 28–35. 10.1111/j.1432-0436.2005.07301003.x. [DOI] [PubMed] [Google Scholar]
- 48.Lee JY, Sohn KH, Rhee SH, and Hwang D (2001). Saturated fatty acids, but not unsaturated fatty acids, induce the expression of cyclooxygenase-2 mediated through Toll-like receptor 4. J Biol Chem 276, 16683–16689. 10.1074/jbc.M011695200. [DOI] [PubMed] [Google Scholar]
- 49.Joshi-Barve S, Barve SS, Amancherla K, Gobejishvili L, Hill D, Cave M, Hote P, and McClain CJ (2007). Palmitic acid induces production of proinflammatory cytokine interleukin-8 from hepatocytes. Hepatology 46, 823–830. 10.1002/hep.21752. [DOI] [PubMed] [Google Scholar]
- 50.Fey A, Eichler S, Flavier S, Christen R, Hofle MG, and Guzman CA (2004). Establishment of a real-time PCR-based approach for accurate quantification of bacterial RNA targets in water, using Salmonella as a model organism. Appl Environ Microbiol 70, 3618–3623. 10.1128/AEM.70.6.3618-3623.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dejardin J, and Kingston RE (2009). Purification of proteins associated with specific genomic Loci. Cell 136, 175–186. 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang Y, Li Y, Hu Q, Xi Y, Xing Z, Zhang Z, Huang L, Wu J, Liang K, Nguyen TK, et al. (2020). The lncRNA H19 alleviates muscular dystrophy by stabilizing dystrophin. Nat Cell Biol 22, 1332–1345. 10.1038/s41556-020-00595-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jensen KB, and Darnell RB (2008). CLIP: crosslinking and immunoprecipitation of in vivo RNA targets of RNA-binding proteins. Methods Mol Biol 488, 85–98. 10.1007/978-1-60327-475-3_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gruber AR, Lorenz R, Bernhart SH, Neubock R, and Hofacker IL (2008). The Vienna RNA websuite. Nucleic Acids Res 36, W70–74. 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watkins AM, Rangan R, and Das R (2020). FARFAR2: Improved De Novo Rosetta Prediction of Complex Global RNA Folds. Structure 28, 963–976 e966. 10.1016/j.str.2020.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Douglass JD, Zhou YX, Wu A, Zadroga JA, Gajda AM, Lackey AI, Lang W, Chevalier KM, Sutton SW, Zhang SP, et al. (2015). Global deletion of MGL in mice delays lipid absorption and alters energy homeostasis and diet-induced obesity. J Lipid Res 56, 1153–1171. 10.1194/jlr.M058586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stremmel W (1988). Uptake of fatty acids by jejunal mucosal cells is mediated by a fatty acid binding membrane protein. J Clin Invest 82, 2001–2010. 10.1172/JCI113820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hao JW, Wang J, Guo H, Zhao YY, Sun HH, Li YF, Lai XY, Zhao N, Wang X, Xie C, et al. (2020). CD36 facilitates fatty acid uptake by dynamic palmitoylation-regulated endocytosis. Nat Commun 11, 4765. 10.1038/s41467-020-18565-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2. Clinical information of tumor tissues and paired serum samples used in this study, related to Figure 1.
Table S3. Clinical information of human primary adipocyte donors, related to Figures 2, 3, 5 and 6.
This table contains 3 tabs:
Tab 1: Clinical parameters of human primary adipocyte donors from Zen-Bio
Tab 2: Clinical parameters of human primary adipocyte donors from Cell Applications
Tab 3: Clinical parameters of human primary adipocyte donors Celprogen
Table S1. Clinical information of human serum used in this study, related to Figures 1, 3, 5 and 6.
This table contains 5 tabs:
Tab 1: Clinical parameters of human serum from Asterand Bioscience
Tab 2: Clinical parameters of human serum from BioIVT
Tab 3: Clinical parameters of human serum from Proteogenex
Tab 4: Clinical parameters of human serum from Reprocell USA
Tab 5: Clinical parameters of human serum from Coriell Biorepository
Table S4. Clinical information of human PBMC used for NK isolation, related to Figure 7.
Table S5. Oligonucleotides used in this study.
Table S6. Antibodies used in this study.
Data S1. Unprocessed source data underlying all blots and graphs, related to Figures 1, 2, 3, 4, 5, 6, 7, and S1–S8.
- Uncropped hi-resolution scans of all the blots.
- Excel file containing the values that were used to create all graphs in the paper.
- Normalized expression value of snoRNA PCR array, related to Figure 1.
- Protein identification of PICh-MS, related to Figure 2. This table contains 9 tabs:
- Tab 1: List of proteins associated with scramble probes, replicate #1
- Tab 2: List of proteins associated with scramble probes, replicate #2
- Tab 3: List of proteins associated with scramble probes, replicate #3
- Tab 4: List of proteins associated with SNORD46 promoter probes, replicate #1
- Tab 5: List of proteins associated with SNORD46 promoter probes, replicate #2
- Tab 6: List of proteins associated with SNORD46 promoter probes, replicate #3
- Tab 7: List of proteins associated with telomere probes, replicate #1
- Tab 8: List of proteins associated with telomere probes, replicate #2
- Tab 9: List of proteins associated with telomere probes, replicate #3
- Protein identification of SNORD46-binding proteins, related to Figure 3. This table contains 9 tabs:
- Tab 1: Biotinylated scramble RNA pull-down in human serum, replicate #1
- Tab 2: Biotinylated scramble RNA pull-down in human serum, replicate #2
- Tab 3: Biotinylated scramble RNA pull-down in human serum, replicate #3
- Tab 4: Biotinylated SNORD46 RNA pull-down in human serum, replicate #1
- Tab 5: Biotinylated SNORD46 RNA pull-down in human serum, replicate #2
- Tab 6: Biotinylated SNORD46 RNA pull-down in human serum, replicate #3
- Tab 7: Biotinylated miR-20a RNA pull-down in human serum, replicate #1
- Tab 8: Biotinylated miR-20a RNA pull-down in human serum, replicate #2
- Tab 9: Biotinylated miR-20a RNA pull-down in human serum, replicate #3
- Sanger sequencing results of IL15 CLIP assays, related to Figure 3. This table contains 2 tabs:
- Tab 1: Sanger sequencing results of IL15 CLIP assay in human serum
- Tab 2: Sanger sequencing results of IL15 CLIP assay in mouse serum
- Peptide identification of IL15 LiP-MS assay, related to Figure 3. This table contains 3 tabs:
- Tab 1: Peptide identified from IL15 LiP-MS assay
- Tab 2: Peptide identified from IL15 LiP-MS assay in the presence of scramble RNA.
- Tab 3: Peptide identified from IL15 LiP-MS assay in the presence of SNORD46 RNA
- Protein identification of IL15 binding proteins in the major organs of Snord46WT/WT or Snord46G11A/G11A mice, related to Figure 5. This table contains 16 tabs:
- Tab 1: List of proteins associated with IgG in Snord46WT/WT mouse tissues
- Tab 2: List of proteins associated with IL15 in brain tissues of Snord46WT/WT mice
- Tab 3: List of proteins associated with IL15 in heart tissues of Snord46WT/WT mice
- Tab 4: List of proteins associated with IL15 in liver tissues of Snord46WT/WT mice
- Tab 5: List of proteins associated with IL15 in SKM tissues of Snord46WT/WT mice
- Tab 6: List of proteins associated with IL15 in WAT of Snord46WT/WT mice
- Tab 7: List of proteins associated with IL15 in spleen tissues of Snord46WT/WT mice
- Tab 8: List of proteins associated with IL15 in intestine tissues of Snord46WT/WT mice
- Tab 9: List of proteins associated with IgG in Snord46WT/WT mouse tissues
- Tab 10: List of proteins associated with IL15 in brain tissues of Snord46G11A/G11A mice
- Tab 11: List of proteins associated with IL15 in heart tissues of Snord46G11A/G11A mice
- Tab 12: List of proteins associated with IL15 in liver tissues of Snord46G11A/G11A mice
- Tab 13: List of proteins associated with IL15 in SKM tissues of Snord46G11A/G11A mice
- Tab 14: List of proteins associated with IL15 in WAT of Snord46G11A/G11A mice
- Tab 15: List of proteins associated with IL15 in spleen tissues of Snord46G11A/G11A mice
- Tab 16: List of proteins associated with IL15 in intestine tissues of Snord46G11A/G11A mice
- Mass spectrometry verification of SNORD46 power inhibitors, related to Figure 6. This table contains 6 tables:
- Tab 1: List of serum SNORD46-binding proteins in the presence of scramble power inhibitors
- Tab 2: List of serum SNORD46-binding proteins in the presence of miR-20a power inhibitors
- Tab 3: List of serum SNORD46-binding proteins in the presence of SNORD46 power inhibitor #1
- Tab 4: List of serum SNORD46-binding proteins in the presence of SNORD46 power inhibitor #2
- Tab 5: List of serum SNORD46-binding proteins in the presence of SNORD46 power inhibitor #3
- Tab 6: List of serum SNORD46-binding proteins in the presence of SNORD46 power inhibitor #4
- Normalized expression value of bulk-RNA seq in NK cells, related to Figure 7.
Data Availability Statement
The bulk RNA-seq of human adipocytes with or without IL15 stimulation were deposited to GEO (GSE210203). The bulk RNA-seq of NK cells isolated from healthy donors and donors with obesity were deposited to GEO (GSE213465).
Data S1: Unprocessed source data underlying all blots and graphs, related to Figures 1, 2, 3, 4, 5, 6, 7, and S1–S8. This file includes: 1) Uncropped hi-resolution scans of all the blots; 2) Excel file containing the values that were used to create all graphs in the paper; 3) Normalized expression value of snoRNA PCR array; 4) Protein identification data of PICh-MS; 5) Protein identification data of SNORD46-binding proteins; 6) Sanger sequencing results of IL15 CLIP assays; 7) Peptide identification data of IL15 LiP-MS assay; 8) Protein identification data of IL15 binding proteins in the major organs of Snord46WT/WT or Snord46G11A/G11A mice; 9) Mass spectrometry verification data of SNORD46 power inhibitors; 10) Normalized expression value of bulk-RNA seq in NK cells.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


