Abstract
Pseudomonas sp. strain TW3 is able to oxidatively metabolize 4-nitrotoluene and toluene via a route analogous to the upper pathway of the TOL plasmids. We report the sequence and organization of five genes, ntnWCMAB*, which are very similar to and in the same order as the xyl operon of TOL plasmid pWW0 and present evidence that they encode enzymes which are expressed during growth on both 4-nitrotoluene and toluene and are responsible for their oxidation to 4-nitrobenzoate and benzoate, respectively. These genes encode an alcohol dehydrogenase homolog (ntnW), an NAD+-linked benzaldehyde dehydrogenase (ntnC), a two-gene toluene monooxygenase (ntnMA), and part of a benzyl alcohol dehydrogenase (ntnB*), which have 84 to 99% identity at the nucleotide and amino acid levels with the corresponding xylWCMAB genes. The xylB homolog on the TW3 genome (ntnB*) appears to be a pseudogene and is interrupted by a piece of DNA which destroys its functional open reading frame, implicating an additional and as-yet-unidentified benzyl alcohol dehydrogenase gene in this pathway. This conforms with the observation that the benzyl alcohol dehydrogenase expressed during growth on 4-nitrotoluene and toluene differs significantly from the XylB protein, requiring assay via dye-linked electron transfer rather than through a nicotinamide cofactor. The further catabolism of 4-nitrobenzoate and benzoate diverges in that the former enters the hydroxylaminobenzoate pathway as previously reported, while the latter is further metabolized via the β-ketoadipate pathway.
The pathways of nitroaromatic catabolism are diverse, and removal of the nitro group by both oxidative and reductive reactions has been reported previously (20, 29). Elimination from the aromatic ring as nitrite is mediated by monooxygenases in the catabolism of 2-nitrophenol (39) and 4-nitrophenol (14, 30) and by dioxygenases in the catabolism of 2,4-dinitrotoluene (31), nitrobenzene (24), 2-nitrotoluene (9), and 3-nitrobenzoate (22). In contrast, partial reduction of the nitro group prior to its elimination as ammonia has been described for the catabolism of 4-nitrobenzoate by Comamonas acidovorans (6, 7) and Pseudomonas pickettii (38) and for that of 4-nitrotoluene by Pseudomonas sp. strain TW3 (27) and Pseudomonas sp. strain 4NT (8). In both Pseudomonas sp. strain TW3 (27) and strain 4NT (8), the nitro group of 4-nitrotoluene is retained during the sequential oxidation of the methyl group to form 4-nitrobenzoate in reactions analogous to the first reactions in the TOL plasmid-encoded pathway of toluene catabolism (37). The 4-nitrobenzoate is then assimilated by the reactions first described by Groenewegen et al. (7), in which the nitro group is reduced via 4-hydroxylaminobenzoate to the ring cleavage substrate 3,4-dihydroxybenzoate (protocatechuate) with release of ammonia. The two strains differ in their oxidation of nitrobenzyl alcohol, which is dependent on NAD+ in strain 4NT, but not in TW3, and in the dioxygenase-catalyzed ring cleavage of 3,4-dihydroxybenzoate, which occurs at the 4,5 position in strain 4NT but at the 3,4 position in TW3 (8, 27).
The molecular genetics of nitroaromatic catabolism have not yet been extensively studied. The P. pickettii genes encoding catabolism of 4-nitrobenzoate have been cloned and sequenced previously (38), as have the genes encoding the four-component dioxygenase involved in the catabolism of 2-nitrotoluene (26) and 2,4-dinitrotoluene (32).
In this paper, we extend the list of characterized genes to include the ntn genes involved in the early steps of the pathway between 4-nitrotoluene and 4-nitrobenzoate in Pseudomonas sp. strain TW3.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids.
The bacterial strains and plasmids used in this study are listed in Table 1. Pseudomonas sp. strain TW3 utilizes 4-nitrotoluene as its sole carbon and nitrogen source (27) and was originally isolated from contaminated soil in the United Kingdom. It also grows on toluene.
TABLE 1.
Bacterial strains and plasmids
| Bacterial strain or plasmid | Genotype or phenotypea | Reference or source |
|---|---|---|
| Pseudomonas sp. strain TW3 | 4NT+ 4NBA+ 4NBZate+ | 27 |
| Pseudomonas sp. strain PaW130 | Rifr | 15 |
| E. coli XL1-Blue MRF′ | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac(F′ proAB lacIqZ M15 Tn10 [Tetr]) | Stratagene |
| E. coli TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZM15 ΔlacX74 deoR recA1 araD139 Δ(ara-leu)7679 galU galK rpsL(Strr) endA1 nupG | Invitrogen |
| E. coli BL21(DE3)/pLysS | F−ampT hsdSB (rB− mB−) dcm gal λ(DE3) pLysS(Cmr) | Promega |
| pCK04 | Cmr; derivative of pSC101 containing pWW0 xylUWCMABN on a ∼15-kbp NotI fragment | 17 (GenBank accession no. for operon, D63341) |
| pTW3.1 | 7.5-kbp HindIII clone containing ntnWCMAB* in pUC18 | This study |
| pTW3.1E | 4.5-kbp EcoRI/HindIII subclone of pTW3.1 in pUC18 | This study |
| pTW3.5 | 4.3-kbp XhoI clone containing ntnWCM in pUC18 (cloned into SalI site) | This study |
| pTW3.6 | 6-kbp EcoRI clone containing ntnCMAB* in pUC18 | This study |
| pETntnC | NdeI/EcoRI fragment containing ntnC in pET5a | This study |
4NT, 4-nitrotoluene; 4NBA, 4-nitrobenzyl alcohol; 4NBZate, 4-nitrobenzoate.
Growth media.
Pseudomonas strains were grown on minimal salts medium (3) supplemented with either solid 4-nitrotoluene (0.5 g/liter) or toluene vapor. 4-Nitrobenzyl alcohol or the sodium salts of 4-nitrobenzoic acid and benzoic acid were added as carbon sources at 5 mM, and sodium succinate was added at 10 mM. Escherichia coli strains were grown on Luria-Bertani medium (28). Where appropriate, ampicillin was added at 100 μg/ml and kanamycin was added at 50 μg/ml.
Chemicals.
Aromatic substrates were obtained from Aldrich Chemical Co.
Enzyme assays.
Cells were harvested by centrifugation, washed with 100 mM Na2HPO4 buffer (pH 7.5), and stored as pellets at −20°C. Cell extracts were prepared by resuspending frozen cell pellets in ice-cold 50 mM Na2HPO4 buffer (pH 7.5) containing 2.5 mM dithiothreitol. Cells were disrupted by passing them through a precooled French pressure cell (SLM Instruments, Inc., Urbana, Ill.), and particulates were removed by centrifugation at 45,000 × g and 4°C for 30 min. Activities of 4-nitrobenzyl alcohol dehydrogenase (4NBADH) and 4-nitrobenzaldehyde dehydrogenase (4NBZDH) were determined spectrophotometrically by dye reduction and NAD+ reduction assays, respectively (27). The presence of any NAD+-dependent 4NBADH activity was tested under the same conditions as those for the dye reduction assay, except that dyes were replaced by NAD+ to a final concentration of 2 mM. The method of Fujisawa (5) was used to assay 3,4-dihydroxybenzoate dioxygenase, and the method of Hegeman (12) was used to assay catechol 1,2-dioxygenase. All assays were carried out at 28°C.
Enzyme purification, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and amino acid sequencing.
A 2-ml bed volume column of Matrex Blue-A (Amicon) was equilibrated with 50 mM Na2HPO4 buffer (pH 7.5) containing 2.5 mM dithiothreitol. The column was loaded with 2 ml of cell extract (containing 10 mg of protein and 12.2 U of 4NBZDH activity) and washed with 10 ml of the same buffer. 4NBZDH was eluted with the same buffer containing 1 mM NAD+, with 2-ml fractions being collected. A total of 1 mg of protein and 9.6 U of 4NBZDH activity were recovered in a volume of 10 ml. Both temperature and time of retention on the column were critical in retaining enzyme activity; loading the column at 4 rather than 20°C or elution after more than 30 min reduced the final yield by half. Fractions containing 4NBZDH activity were pooled, dialyzed overnight against 10 mM (NH4)HCO3 at 4°C, and freeze-dried.
The purified enzyme was subjected to SDS-PAGE by the method of Laemmli (19). The N-terminal amino acid sequences of the intact protein and a tryptic fragment were determined by Mark Wilkinson (University of Liverpool, Liverpool, United Kingdom). Peptides were subjected to N-terminal amino acid sequencing by Edman degradation on an Applied Biosystems 471A pulsed liquid-phase Sequenator.
DNA manipulations.
Unless otherwise stated, standard methods for DNA manipulation were used (28). Total DNA was prepared from Pseudomonas sp. strain TW3 by the method of Ausubel et al. (2), and plasmid DNA was prepared by the sucrose gradient method (33). Plasmid DNA was prepared from E. coli strains with alkaline lysis Miniprep (28) or Qiaprep columns (Qiagen). DNA fragments were recovered from agarose gels with Qiaquick columns (Qiagen). Southern blots and colony lifts were prepared as described by Sambrook et al. (28) with the following modifications: the colony lift lysis solution was 0.5 M NaOH, and the colony filters were neutralized by washing them vigorously in 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Hybridizations were carried out with ECL direct labelling (Amersham) according to the manufacturer’s instructions.
Preparation of xyl probes by PCR.
Gene-specific probes of the xyl genes from TOL plasmid pWW0 were amplified by PCR from plasmid pCK04 carrying the complete xylUWCMABN operon (Table 1). Primers were designed such that each gene was amplified separately, except that xylUW were amplified on the same fragment. Primer sequences were as follows: xylUW (forward), 5′-TTC AGA TTG GTT GCT TTC GCC; xylUW (reverse), 5′-GCT CTT TTG TTT CCC GCA TAA; xylC (forward), 5′-CGT TCG AAA TGG CCC TAA AT; xylC (reverse), 5′-GAT CAG CCC GCA AGC AGT AAC AAC; xylM (forward), 5′-TGG CGG ACC TGC AAG TAT T; xylM (reverse), 5′-AAA AAG AGA CCG TAG AGT TCG TTC; xylA (forward), 5′-AAG CGA AGA GCG AAC GAA C; xylA (reverse), 5′-TTT TGG CCG CAA GAC GAT. PCR amplifications were carried out in a 100-μl reaction volume containing 100 ng of template DNA, 100 pmol of each primer, 200 μM (each) deoxynucleoside triphosphate (dNTP), and 2 U of Taq polymerase (Promega) in the reaction buffer supplied by the manufacturer. An MgCl2 concentration of 2 mM was used for amplification of xylUW, -C, -M, and -A, and 1.5 mM was used for xylB. After a 2-min hot start at 94°C, the reaction mixtures were given 30 cycles of 1 min at 94°C, 1 min at 55°C, and 1.5 min at 72°C.
Expression of ntnC in E. coli.
The ntnC gene was amplified by PCR from plasmid pTW3.6 with Vent DNA polymerase (New England Biolabs). Primers incorporating NdeI and EcoRI sites in the forward primer and an EcoRI site in the reverse primer were designed. The NdeI site was positioned at the start codon of the ntnC open reading frame. Primer sequences were as follows: ntnC (forward), 5′-TTAAGGAGGAATTCATATGCGGGAA; ntnC (reverse), 5′-GATCAGGAATTCACGAACTGTATC. The EcoRI sites are underlined, and the NdeI site is in bold italics. PCR amplifications were carried out in a 100-μl reaction volume containing 10 ng of template DNA, 100 pmol of each primer, 200 μM (each) dNTP, 6 mM MgSO4, and 1 U of Vent polymerase in the reaction buffer supplied by the manufacturer. After a 2-min hot start at 94°C, the reaction mixtures were given 25 cycles of 1 min at 94°C, 1 min at 56°C, and 1.5 min at 72°C. The PCR product was cloned directly into pCR-blunt (Invitrogen) in E. coli TOP10 without further treatment, with selection on 50 μg of kanamycin per ml. The PCR product was sequenced to confirm that no point mutations had been introduced during the amplification and cloned into the expression vector pET5a (Promega) as an NdeI/EcoRI fragment, placing the ntnC gene in frame with the T7 promoter to create pETntnC. The NtnC protein was expressed in E. coli BL21(DE3)/pLysS (Promega) grown in LB broth to an optical density at 600 nm of 0.3 and induced with 0.2 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h before harvesting. SDS-PAGE of cell extracts showed overexpression of a polypeptide of ∼50 kDa which comigrated with the partially purified wild-type enzyme. A 4NBZDH specific activity of ∼12 U/mg was detected in cell extracts with 4-nitrobenzaldehyde as substrate (compared to ∼4 U/mg from wild-type TW3). No activity was detectable in controls containing the expression vector with no insert.
RNA isolation and RT-PCR.
Cells were grown on minimal medium supplemented with either 4-nitrotoluene, toluene, or succinate until they reached an optical density at 600 nm of 0.3. Total RNA was prepared from 108 cells with RNeasy Mini columns (Qiagen), with elution in 100 μl of water. The RNA was treated with DNase I to remove any genomic DNA contamination by incubation with 1 U of RNase-free DNase (Promega) and 1 U of RNasin (Promega) in 40 mM Tris-HCl (pH 7.9)–10 mM NaCl–10 mM CaCl2–6 mM MgSO4 for 30 min at 37°C. The RNA was cleaned by passage through an RNeasy Mini column prior to use in reverse transcriptase PCR (RT-PCR). RT-PCR was carried out with total RNA from TW3 grown on 4-nitrotoluene, toluene, and succinate with an Access RT-PCR kit (Promega). Amplifications were carried out with the xylM and xylA primer pairs used to prepare the xyl gene probes and the ntnC primer pair used in making the pETntnC expression construct. Comparison of the xylM and xylA primer pairs with their corresponding regions in the ntn operon revealed sufficient identity for these primers to be used to amplify their ntn homologs, and their effectiveness had been demonstrated on TW3 genomic DNA. PCRs were carried out in a 50-μl volume containing 0.5 μg of template RNA, 50 pmol of each primer, 50 μM (each) dNTP, 1 mM MgSO4, 5 U of avian myeloblastosis virus reverse transcriptase, and 5 U of Tfl DNA polymerase in the reaction buffer supplied by the manufacturer. After reverse transcription at 48°C for 45 min, the reaction mixtures were heated to 94°C for 2 min and given 40 cycles of 30 s at 94°C, 1 min at 55°C, and 2 min at 68°C. Negative control reactions to eliminate the possibility of amplifying residual genomic DNA were performed in the same way, except that avian myeloblastosis virus reverse transcriptase was omitted from the reaction mixtures.
DNA sequencing.
DNA sequences were determined by primer-walking fragments cloned in pUC18. Sequencing was carried out by Alta Biosciences (University of Birmingham, Birmingham, United Kingdom) with an Applied Biosystems ABI 373a automated sequencer.
Sequence analysis and alignment methods.
Searches of the GenBank and Swissprot databases were carried out by the BLASTN and BLASTX methods, respectively (1). Pairwise DNA and amino acid alignments were carried out by the method of Needleman and Wunsch (23) with the program GAP contained in the GCG sequence analysis package, version 8.1 (Genetics Computer Group, Inc.).
Nucleotide sequence accession number.
The nucleotide sequence of 6,636 bases is available in GenBank under accession no. AF043544.
RESULTS
Partial purification of 4NBZDH.
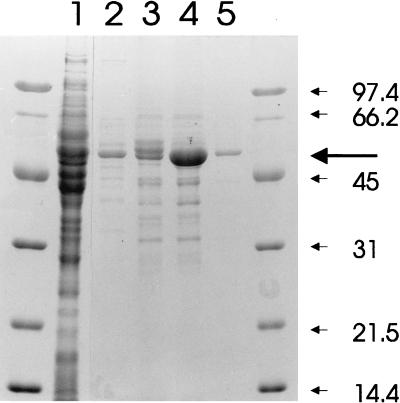
The enzyme was partially purified from extracts of 4-nitrotoluene-grown cells. Passage through a Matrex Blue-A column resulted in a 7.5-fold purification (to a specific activity of 9.1 μmol/min/mg of protein) with an 80% yield (Fig. 1). The amino acid sequences of the N terminus and an internal tryptic fragment were homologous to those of other benzaldehyde dehydrogenases in the data banks but in particular to the benzaldehyde dehydrogenase XylC from the TOL plasmid pWW0 (34), which is part of the similar metabolic sequence for the conversion of toluene to benzoate (Fig. 2).
FIG. 1.
SDS-PAGE of 4NBZDH on a 12% polyacrylamide gel. Lane 1, crude cell extract of TW3 prior to loading onto the dye affinity column; lanes 2 to 5, successive fractions containing 4NBZDH eluted from the column with 1 mM NAD+. Marker sizes are indicated in kilodaltons. The molecular mass of the major band is ∼53 kDa.
FIG. 2.
Amino acid sequences of the N-terminal and internal peptides from the purified TW3 benzaldehyde dehydrogenase. The equivalent regions of the xylC gene and their predicted amino acid sequences are shown for comparison.
Hybridization analysis of TW3 genomic DNA and cloning of the ntnWCMAB* gene cluster.
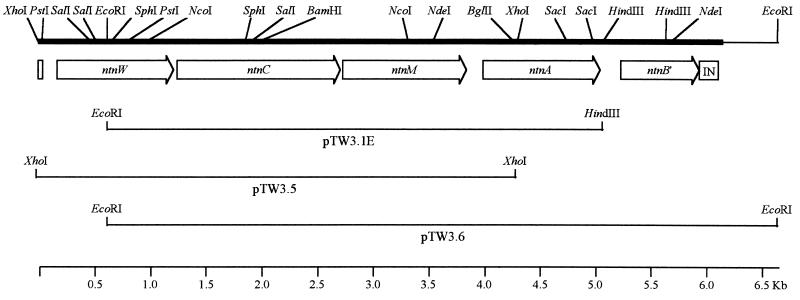
PCR-amplified gene-specific fragments carrying xylC (for benzaldehyde dehydrogenase), xylUW, xylM, and xylA from TOL plasmid pWW0 were used to probe restriction digests of total DNA from TW3. A HindIII fragment (7.5 kb) and an XhoI fragment (4.5 kb) hybridized to xylUW, -C, -M, and -A, but not to xylB, and a 6-kb EcoRI fragment which hybridized to xylC, xylM, xylA, and xylB was also identified. The hybridizing HindIII, XhoI, and EcoRI fragments of the appropriate size ranges were ligated into pUC18 as pTW3.1, pTW3.5, and pTW3.6, respectively (Table 1; Fig. 3). The DNA sequences of the inserts of pTW3.5, pTW3.6, and pTW3.1E, a subclone of pTW3.1, were determined over 6,636 bp.
FIG. 3.
Map of the ntn operon of TW3. The locations of the four open reading frames (ntnW, ntnC, ntnM, and ntnA) and the pseudogene ntnB* are marked by the open arrows, the directions of the arrowheads indicating the directions of transcription. The small, open box to the left of the arrows indicates the 48-bp sequence which shows 100% identity with the terminal 48 bp of xylU, and the open box labelled IN marks the insertion which interrupts the open reading frame of the pseudogene ntnB*. The extent of the nucleotide sequence determined is denoted by the thick solid line. The lines below the open reading frame arrows represent the inserts of recombinant plasmids pTW3.1E, pTW3.5, and pTW3.6, from which the nucleotide sequence was derived. Only restriction sites relevant to the clones and construct depicted in this figure are shown.
Analysis of the nucleotide sequence of ntnWCMAB*.
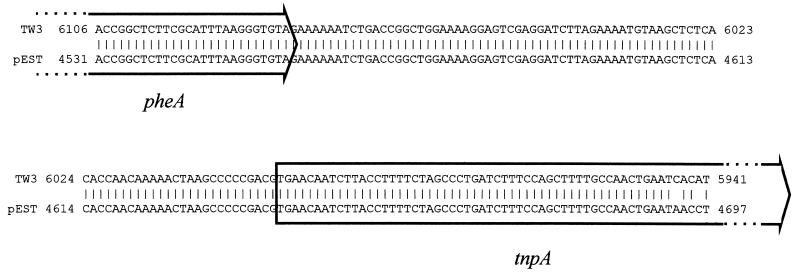
From the nucleotide sequence, the presence of four complete and two partial reading frames which align with the xylUWCMAB genes was deduced, and these have been named ntnUWCMAB* (Fig. 3; Table 2). Nucleotides 1 to 48 are identical to the last 48 bases of pWW0 xylU, implying the presence of a complete ntnU gene immediately upstream of the region sequenced. The complete reading frames are ntnWCMA, encoding an alcohol dehydrogenase of unknown function, a nitrobenzaldehyde dehydrogenase, and the hydroxylase and NADH-acceptor reductase components of nitrotoluene monooxygenase, respectively (Table 2). ntnM is the least homologous to its xyl counterpart, and in contrast, ntnA is 99% identical at the nucleotide level to xylA. Bases 5241 to 5940 show 96% identity with the first 699 bases of the benzyl alcohol dehydrogenase gene xylB but contain a stop codon at position 5535. Beyond position 5940, there is no discernible homology with xylB, suggesting that an insertion-deletion event has disrupted the 3′ end of the xylB homolog which we have designated ntnB*. The sequence between bases 5941 and 6108 is highly homologous to a sequence found in the intergenic region between the pheA (phenol 2-monooxygenase) and tnpA (transposase) genes from Pseudomonas sp. strain EST1001 (16) (Fig. 4), which would suggest that the interruption of the gene might be the residue of a transposition event. Downstream of this putative insertion, the terminal 400 bp of sequence appears to contain the 5′ end of another open reading frame, which we are currently investigating.
TABLE 2.
ntn genes and their gene products
| Gene | No. of bases in comparison | Position in sequence | Nucleotide identity with xyl homolog (%) | Amino acid identity of ORFb (%) | Mr of ORF (thousands) |
|---|---|---|---|---|---|
| ntnUa | 48 | 1–48 | 100 | 100 | |
| ntnW | 1,074 | 174–1220 | 99.6 | 99.1 | 36.9 |
| ntnC | 1,464 | 1252–2715 | 92.4 | 94.9 | 51.8 |
| ntnM | 1,110 | 2740–3849 | 84.1 | 86.2 | 41.6 |
| ntnA | 1,053 | 3999–5051 | 99.7 | 99.1 | 38.5 |
| ntnB*a | 699 | 5241–5940 | 95.6 | 97.4 |
Incomplete open reading frames.
ORF, open reading frame.
FIG. 4.
Alignment of the reverse complement of part of the ntn gene sequence of Pseudomonas sp. strain TW3 with bases 4531 to 4697 of pEST1226 (16, 25) (GenBank accession no. M57500). The ntn sequence starts at base 5941, the first base in the insert which disrupts the ntnB* pseudogene (see Fig. 3). The pEST1226 sequence spans the 3′ end of the pheA gene (phenol 2-monooxygenase), the intergenic region, and the 5′ end of the tnpA gene as shown.
RT-PCR analysis of transcripts present in TW3.
In order to show whether the ntn genes encode enzymes involved in both 4-nitrotoluene and toluene catabolism, we examined transcripts from cells grown on both these substrates and on succinate as a control. Because Southern hybridizations indicated a single copy of each of the ntnWCMAB* genes, we determined that independent PCR amplification from cDNA made from mRNA of the three structural genes of known functions in the pathway (xylC, xylM, and xylA) would be sufficient to implicate this operon in the catabolism of both substrates (given that ntnW was, like xylW, of unknown function [36] and that ntnB* is apparently a pseudogene). From the primers selected, the expected product sizes for ntnC, ntnM, and ntnA were 1,531, 1,291, and 1,265 bp, respectively.
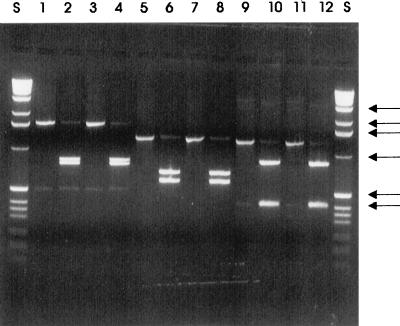
The PCR products obtained together with restriction digests chosen to confirm the presence of expected restriction sites were analyzed by agarose gel electrophoresis. Figure 5 shows that products of the expected sizes were obtained from the total RNA of cells grown on both 4-nitrotoluene and toluene. The presence of restriction sites in the expected positions within the fragments for BamHI and SalI (ntnC), NcoI and NdeI (ntnM), and BglII and SacI (ntnA) was confirmed by digestion. Only the digests of the first of each of these pairs of enzymes are shown in Fig. 5. No products were obtained from total RNA of succinate-grown cells or from reaction mixtures from which the reverse transcriptase had been omitted.
FIG. 5.
Agarose gel electrophoresis of RT-PCR products amplified from TW3 grown on 4-nitrotoluene and toluene. The sizes of molecular size markers in lanes S (1-kb ladder; Gibco BRL) are indicated by arrows (3,054, 2,036, 1,636, 1,018, 506/517, and 396 bp). Lanes: 1, ntnC, nitrotoluene-grown cells (expected size, 1,531 bp); 2, ntnC, nitrotoluene-grown cells cut with BamHI (819 and 714 bp); 3, ntnC, toluene-grown cells (1,531 bp); 4, ntnC, toluene-grown cells cut with BamHI (819 and 714 bp); 5, ntnM, nitrotoluene-grown cells (1,291 bp); 6, ntnM, nitrotoluene-grown cells cut with NcoI (692 and 598 bp); 7, ntnM, toluene-grown cells (1,291 bp); 8, ntnM, toluene-grown cells cut with NcoI (692 and 598 bp); 9, ntnA, nitrotoluene-grown cells (1,265 bp); 10, ntnA, nitrotoluene-grown cells cut with BglII (862 and 402 bp); 11, ntnA, toluene-grown cells (1,265 bp); 12, ntnA, toluene-grown cells cut with BglII (862 and 402 bp). No detectable products were obtained in control reactions, with each pair of primers, from which reverse transcriptase had been omitted or in reactions carried out on succinate-grown cells (data not shown).
Enzyme activities in TW3 and in E. coli expressing ntnC.
The relative activities on different substrates of the wild-type benzaldehyde dehydrogenase expressed during growth on 4-nitrotoluene and toluene and of the ntnC gene product cloned into expression vector pET5a and expressed in E. coli BL21(DE3) were compared. Cell extracts were assayed with benzaldehyde and with its 2-, 3-, and 4-nitro and -methyl analogs. Two independent cultures of each were grown, and triplicate assays were performed with cell extracts against each of the substrates. The activity against 4-nitrobenzaldehyde was arbitrarily taken to be 100% in each case, and activities against the other substrates are expressed relative to this in Table 3. No detectable activity was observed with either of the 2-substituted analogs. Control assays of cell extracts of TW3 grown on succinate and of E. coli BL21(DE3) containing pET5a with no insert gave no detectable activity with any of the substrates.
TABLE 3.
Substrate specificities of 4NBZDH activitiesa
| Substrate | Value for source of cell extract
|
||
|---|---|---|---|
| TW3 grown on 4-nitrotoluene | TW3 grown on toluene | E. coli BL21(DE3) expressing ntnC | |
| 4-Nitrobenzaldehyde | 100 | 100 | 100 |
| 3-Nitrobenzaldehyde | 121 | 126 | 120 |
| 4-Methylbenzaldehyde | 28 | 27 | 25 |
| 3-Methylbenzaldehyde | 22 | 22 | 20 |
| Benzaldehyde | 40 | 43 | 37 |
Activities are expressed as percentages relative to that observed with 4-nitrobenzaldehyde, each value being the mean from two different cultures. No individual value was more than ±3% from the mean.
Cell extracts of TW3 grown on 4-nitrotoluene, toluene, and succinate were assayed for NAD+-dependent and the dye-linked NAD+-independent 4NBADH. Both 4-nitrotoluene- and toluene-grown cells exhibited 4NBADH activity, detectable only by the dye reduction assay (specific activities of 139 and 538 mU/mg, respectively, with 4-nitrobenzyl alcohol as substrate). Cells grown on succinate showed no activity detectable by either assay.
Previous work had shown that, when 4-nitrotoluene is used as a growth substrate, the pathway proceeds via 4-nitrobenzoate to 3,4-dihydroxybenzoate (protocatechuate), which is cleaved in the 3,4 position by protocatechuate 3,4-dioxygenase (27). Extracts of toluene- or benzoate-grown cells do not contain significant activity of protocatechuate 3,4-dioxygenase (<5 mU/mg of protein) compared with 4-nitrotoluene-grown cells (100 mU/mg of protein). They do, however, have elevated levels of catechol 1,2-dioxygenase activity (122 and 138 mU/mg of protein, respectively) compared to cells grown on 4-nitrotoluene or succinate (both, <0.5 mU/mg of protein).
DISCUSSION
We have cloned and sequenced five genes from the chromosome of Pseudomonas sp. strain TW3 which bear a striking resemblance, in terms of both sequence identity and gene organization, to the xyl upper pathway operon of plasmid pWW0. In the region analyzed, a sequence of genes, ntn(U)WCMAB*, encoding an alcohol dehydrogenase homolog (ntnW), an NAD+-dependent benzaldehyde dehydrogenase (ntnC), a two-gene toluene monooxygenase (ntnMA), and part of a benzyl alcohol dehydrogenase (ntnB*), occurs. These are in the same order as and have 84 to 99% identity at the nucleotide and amino acid levels with the corresponding xylWCMAB genes from the xyl operon, the roles of which have been demonstrated for the conversion of toluene to benzoate (11).
Southern hybridizations indicated that TW3 contains only one copy of each of these genes, and RT-PCR products indistinguishable in terms of size and restriction sites (Fig. 5) were amplified from mRNA produced during growth on both 4-nitrotoluene and toluene, with primers specific to three key genes, ntnC, ntnM, and ntnA. The identity of the RT-PCR fragments was further confirmed by exposing Southern blots of them to hybridization with a probe of the EcoRI insert of pTW3.6 (bearing ntnCMAB*) which hybridized to all of the products (data not shown). Amplification of these genes during growth on 4-nitrotoluene and toluene, but not on succinate, proves that they play the same role in the metabolism of both substrates. Furthermore, the aldehyde dehydrogenases expressed during growth on both substrates have the same relative activities towards seven different substrates, leading to the conclusion that the same enzyme is present in both. The substrate specificity of the cloned ntnC gene product overexpressed in E. coli is also identical to that determined for the wild-type enzyme present in toluene- and 4-nitrotoluene-grown cells, compounding the evidence that ntnC is involved in the metabolism of both substrates. In addition, the N-terminal and internal amino acid sequences obtained from the aldehyde dehydrogenase which we purified from 4-nitrotoluene-grown TW3 correspond to those predicted from the nucleotide sequence of the cloned ntnC, indicating that this gene is induced during growth on 4-nitrotoluene. It was in fact this amino acid homology which alerted us to the possible relationship between the nitrotoluene genes and the xyl genes when earlier results had suggested that there was no hybridization between the TOL plasmid and DNA from TW3 (27).
One other fact suggests that this gene cluster functions in the catabolism of both toluene and 4-nitrotoluene in TW3. We have confirmed the earlier finding (27) that the benzyl alcohol dehydrogenase expressed during growth on 4-nitrotoluene differs significantly from the TOL plasmid XylB protein by requiring assay via dye-linked electron transfer rather than through a nicotinamide cofactor. We have extended this finding to show that toluene catabolism in TW3 also utilizes this alternative mechanism for benzyl alcohol oxidation, indicating again that the same biochemical route is used for both substrates. The absence of an NAD+-linked alcohol dehydrogenase is explained by the observation that the xylB homolog on the TW3 genome (ntnB*) appears to be a pseudogene and is interrupted by an insert which compromises its role as a functional open reading frame: in fact, even in the region homologous to xylB there is a stop codon which the gene has probably acquired subsequent to its loss of function by the insertion.
The absence of an expressed NAD+-dependent benzyl alcohol dehydrogenase in TW3 growing on toluene shows that NtnW, which, like its counterpart XylW (36), is highly homologous to NAD+-linked alcohol dehydrogenases and especially to benzyl alcohol dehydrogenases, does not function as an alternative enzyme to oxidize benzyl alcohols in this pathway. Similarly, XylW does not appear to be directly involved in the metabolism of toluenes (36).
Although TW3 grows on toluene and contains genes which are highly homologous to the xyl genes of TOL plasmids, it is unable to grow on any of the alkyl-substituted toluenes such as the xylenes that other TOL plasmid-containing strains can utilize (18, 37). This can be attributed to the absence (or lack of expression) of any meta-cleavage pathway in TW3 and the fact that the benzoate formed from toluene is assimilated via the β-ketoadipate pathway, as demonstrated by the induction of its key enzyme catechol 1,2-dioxygenase in both toluene- and benzoate-grown cells. In Pseudomonas, this pathway is specific to benzoate and cannot handle alkyl substituents.
These results demonstrate that toluene and 4-nitrotoluene induce a common set of genes in TW3 whose products convert the hydrocarbons to the corresponding carboxylic acids. These are then assimilated by a divergent set of reactions, the β-ketoadipate pathway for benzoate and the hydroxylaminobenzoate pathway for 4-nitrobenzoate described for Comamonas (7). This suggests that the ntn gene cluster has been acquired by either transposition or recombination from a TOL or related plasmid but has undergone insertional inactivation of its xylB homolog. A dehydrogenase encoded by a gene from elsewhere in its genome has been recruited to carry out the conversion of the benzyl alcohols to benzoates, resulting in the dye-linked activity found in both toluene- and 4-nitrotoluene-grown cells. TW3 is a wild-type counterpart to the laboratory strain constructed by Michan and coworkers (21), who recently expanded the substrate range of a 4-nitrobenzoate-utilizing strain of Pseudomonas to degrade 4-nitrotoluene by inserting the xylUWCMABN operon into its chromosome on a constructed mini-Tn5 transposon. We are currently investigating the 4-nitrotoluene-utilizing strain Pseudomonas sp. strain 4NT, isolated by Haigler and Spain (8), which has an NAD+-linked benzyl alcohol dehydrogenase, and preliminary results indicate that this too appears to carry a cluster of xyl-homologous upper pathway genes with no insertion inactivating the xylB homolog.
It is remarkable that the insertion within ntnB* also has a relationship to the TOL plasmid pWW0. It is homologous to part of a sequence which comprises two genes involved in catabolism of phenol, pheAB, derived from Pseudomonas strain EST1001. The two phe genes are expressed only as a result of their recombination within a sequence derived from a 17-kb transposon, Tn4562, which is part of pWW0 and its deletion derivative pWW0-8 (17, 26). Although it is not explicit in the report (25), the tnpA gene, the 5′ end of which is homologous to the disrupting sequence (Fig. 4), is presumed to be the gene for the Tn4562-encoded transposase and must therefore be derived from pWW0 itself. It therefore seems possible that the ntn gene sequence has been acquired by strain TW3 from a pWW0-like plasmid by a recombination or transposition event which at the same time has inactivated the ntnB gene.
This data from TW3 supports the proposition (4, 10, 13, 35) that these aromatic pathways have evolved by the coacquisition by strains of genetic modules containing the operons or parts of operons. The xylUWCMABN-like module can confer the ability to grow on toluene and alkyl toluenes when in the presence of a meta pathway which will assimilate benzoate and alkyl benzoates (as on TOL plasmids) or can confer the ability to grow on toluene when in a host with a β-ketoadipate pathway for benzoate utilization or can confer the ability to grow on 4-nitrotoluene when present with a 4-nitrobenzoate pathway (as found in TW3).
ACKNOWLEDGMENTS
We thank Ruben Kok for the gift of plasmid pCK04 and Mark Wilkinson for advice on peptide sequence analysis.
This work was supported by a grant from the Chemical and Pharmaceutical Directorate of the Biotechnology and Biological Sciences Research Council.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D E, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 3.Bauchop T, Elsden S R. The growth of microorganisms in relation to their energy supply. J Gen Microbiol. 1960;23:457–469. doi: 10.1099/00221287-23-3-457. [DOI] [PubMed] [Google Scholar]
- 4.Cane P, Williams P A. A restriction map of naphthalene catabolic plasmid pWW60-1 and the location of some of its catabolic genes. J Gen Microbiol. 1986;132:2919–2929. [Google Scholar]
- 5.Fujisawa H. Protocatechuate 3,4-dioxygenase (Pseudomonas) Methods Enzymol. 1970;17A:526–529. [Google Scholar]
- 6.Groenewegen P E J, deBont J A M. Degradation of 4-nitrobenzoate via 4-hydroxylaminobenzoate and 3,4-dihydroxybenzoate in Comamonas acidovorans NBA-10. Arch Microbiol. 1992;158:381–386. [Google Scholar]
- 7.Groenewegen P E J, Breeuwer P, Vanhelvoort J M L M, Langenhoff A A M. Novel degradative pathway of 4-nitrobenzoate in Comamonas acidovorans NBA-10. J Gen Microbiol. 1992;138:1599–1605. doi: 10.1099/00221287-138-8-1599. [DOI] [PubMed] [Google Scholar]
- 8.Haigler B E, Spain J C. Biodegradation of 4-nitrotoluene by Pseudomonas sp. strain 4NT. Appl Environ Microbiol. 1993;59:2239–2243. doi: 10.1128/aem.59.7.2239-2243.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haigler B E, Wallace W H, Spain J C. Biodegradation of 2-nitrotoluene by Pseudomonas sp. strain JS42. Appl Environ Microbiol. 1994;60:3466–3469. doi: 10.1128/aem.60.9.3466-3469.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harayama S. Codon usage patterns suggest independent evolution of two catabolic operons on toluene-degradative plasmid TOL pWW0 of Pseudomonas putida. J Mol Evol. 1994;39:328–335. doi: 10.1007/BF00163150. [DOI] [PubMed] [Google Scholar]
- 11.Harayama S, Rekik M, Wubbolts M, Rose K, Leppik R A, Timmis K N. Characterization of five genes in the upper-pathway operon of TOL plasmid pWW0 from Pseudomonas putida and identification of the gene products. J Bacteriol. 1989;171:5048–5055. doi: 10.1128/jb.171.9.5048-5055.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hegeman G D. Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida. I. Synthesis of enzymes by the wild type. J Bacteriol. 1966;91:1140–1154. doi: 10.1128/jb.91.3.1140-1154.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horn J M, Harayama S, Timmis K N. DNA sequence determination of the TOL plasmid (pWW0) xylGFJ genes of Pseudomonas putida: implications for the evolution of aromatic catabolism. Mol Microbiol. 1991;5:2459–2474. doi: 10.1111/j.1365-2958.1991.tb02091.x. [DOI] [PubMed] [Google Scholar]
- 14.Jain R K, Dreisbach J H, Spain J C. Biodegradation of p-nitrophenol via 1,2,4-benzenetriol by an Arthrobacter sp. Appl Environ Microbiol. 1994;60:3030–3032. doi: 10.1128/aem.60.8.3030-3032.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keil H, Keil S, Pickup R W, Williams P A. Evolutionary conservation of genes coding for meta pathway enzymes within TOL plasmids pWW0 and pWW53. J Bacteriol. 1985;164:887–895. doi: 10.1128/jb.164.2.887-895.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kivisaar M, Horak R, Kasak L, Heinaru A, Habicht J. Selection of independent plasmids determining phenol degradation in Pseudomonas putida and the cloning and expression of genes encoding phenol monooxygenase and catechol 1,2-dioxygenase. Plasmid. 1990;24:25–36. doi: 10.1016/0147-619x(90)90022-5. [DOI] [PubMed] [Google Scholar]
- 17.Kok, R. Personal communication.
- 18.Kunz D A, Chapman P J. Catabolism of pseudocumene and 3-ethyltoluene by Pseudomonas putida (arvilla) mt-2: evidence for new functions of the TOL (pWWO) plasmid. J Bacteriol. 1981;146:179–191. doi: 10.1128/jb.146.1.179-191.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Marvin-Sikkema F D, de Bont J A M. Degradation of nitroaromatic compounds by microorganisms. Appl Microbiol Biotechnol. 1994;42:499–507. doi: 10.1007/BF00173912. [DOI] [PubMed] [Google Scholar]
- 21.Michan C, Delgado A, Haidour A, Lucchesi G, Ramos J L. In vivo construction of a hybrid pathway for metabolism of 4-nitrotoluene in Pseudomonas fluorescens. J Bacteriol. 1997;179:3036–3038. doi: 10.1128/jb.179.9.3036-3038.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nadeau L J, Spain J C. The bacterial degradation of m-nitrobenzoic acid. Appl Environ Microbiol. 1995;61:840–843. doi: 10.1128/aem.61.2.840-843.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Needleman S B, Wunsch C D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- 24.Nishino S F, Spain J C. Degradation of nitrobenzene by a Pseudomonas pseudoalcaligenes. Appl Environ Microbiol. 1993;59:2520–2525. doi: 10.1128/aem.59.8.2520-2525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nurk A, Kasak L, Kivisaar M. Sequence of the gene (pheA) encoding phenol monooxygenase from Pseudomonas putida sp. EST1001: expression in Escherichia coli and Pseudomonas putida. Gene. 1991;102:13–18. doi: 10.1016/0378-1119(91)90531-f. [DOI] [PubMed] [Google Scholar]
- 26.Parales J V, Kumar A, Parales R E, Gibson D T. Cloning and sequencing of the genes encoding 2-nitrotoluene dioxygenase from Pseudomonas sp. JS42. Gene. 1996;181:57–61. doi: 10.1016/s0378-1119(96)00462-3. [DOI] [PubMed] [Google Scholar]
- 27.Rhys-Williams W, Taylor S C, Williams P A. A novel pathway for the catabolism of 4-nitrotoluene by Pseudomonas. J Gen Microbiol. 1993;139:1967–1972. doi: 10.1099/00221287-139-9-1967. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Spain J C. Biodegradation of nitroaromatic compounds. Annu Rev Microbiol. 1995;49:523–555. doi: 10.1146/annurev.mi.49.100195.002515. [DOI] [PubMed] [Google Scholar]
- 30.Spain J C, Gibson D T. Pathway for biodegradation of para-nitrophenol in a Moraxella sp. Appl Environ Microbiol. 1991;57:812–819. doi: 10.1128/aem.57.3.812-819.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spanggord R J, Spain J C, Nishino S F, Mortelmans K E. Biodegradation of 2,4-dinitrotoluene by a Pseudomonas sp. Appl Environ Microbiol. 1991;57:3200–3205. doi: 10.1128/aem.57.11.3200-3205.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suen W C, Haigler B E, Spain J C. Dinitrotoluene dioxygenase from Burkholderia sp. strain DNT—similarity to naphthalene dioxygenase. J Bacteriol. 1996;178:4926–4934. doi: 10.1128/jb.178.16.4926-4934.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wheatcroft R, Williams P A. Rapid methods for the study of both stable and unstable plasmids in Pseudomonas. J Gen Microbiol. 1981;124:433–437. doi: 10.1099/00221287-124-2-433. [DOI] [PubMed] [Google Scholar]
- 34.Williams P A, Murray K. Metabolism of toluene and xylenes by Pseudomonas putida (arvilla) mt-2: evidence for a TOL plasmid. J Bacteriol. 1974;120:416–423. doi: 10.1128/jb.120.1.416-423.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Williams P A, Sayers J R. The evolution of pathways for aromatic hydrocarbon oxidation in Pseudomonas. Biodegradation. 1994;5:195–217. doi: 10.1007/BF00696460. [DOI] [PubMed] [Google Scholar]
- 36.Williams P A, Shaw L M, Pitt C W, Vrecl M. xylUW, two genes at the start of the upper pathway operon of TOL plasmid pWW0, appear to play no essential part in determining its catabolic phenotype. Microbiology. 1996;143:101–107. doi: 10.1099/00221287-143-1-101. [DOI] [PubMed] [Google Scholar]
- 37.Worsey M J, Williams P A. Metabolism of toluene and xylenes by Pseudomonas putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975;124:7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yabannavar A V, Zylstra G J. Cloning and characterization of the genes for p-nitrobenzoate degradation from Pseudomonas pickettii YH105. Appl Environ Microbiol. 1995;61:4284–4290. doi: 10.1128/aem.61.12.4284-4290.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeyer J, Kocher H P, Timmis K N. Influence of para-substituents on the oxidative metabolism of o-nitrophenols by Pseudomonas putida B2. Appl Environ Microbiol. 1986;52:334–339. doi: 10.1128/aem.52.2.334-339.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]