Abstract
The dynamics and assembly of bacterial cell division protein FtsZ were monitored in individual, growing and dividing Escherichia coli cells in real time by microculture of a merodiploid strain expressing green fluorescent protein (GFP)-tagged FtsZ. Cells expressing FtsZ-GFP at levels less than or equivalent to that of wild-type FtsZ were able to grow and divide over multiple generations, with their FtsZ rings visualized by fluorescence. During the late stages of cytokinesis, which constituted the last one-fourth of the cell cycle, the lumen of the FtsZ ring disappeared as the whole structure condensed. At this time, loops of FtsZ-GFP polymers emanated outward from the condensing ring structure and other unstable fluorescent structures elsewhere in the cell were also observed. Assembly of FtsZ rings at new division sites occurred within 1 min, from what appeared to be single points. Interestingly, this nucleation often took place in the predivisional cell at the same time the central FtsZ ring was in its final contraction phase. This demonstrates directly that, at least when FtsZ-GFP is being expressed, new division sites have the capacity to become fully functional for FtsZ targeting and assembly before cell division of the mother cell is completed. The results suggest that the timing of FtsZ assembly may be normally controlled in part by cellular FtsZ concentration. The use of wide-field optical sectioning microscopy to obtain sharp fluorescence images of FtsZ structures is also discussed.
FtsZ is one of several proteins essential for cell division in Escherichia coli (10) and acts at a very early stage of cytokinesis before midcell constriction is visible (1; for three excellent recent reviews, see references 8, 14, and 21). FtsZ is a GTPase (11, 24, 29) that has significant structural similarity to tubulin (19). Like tubulin, FtsZ can assemble into microtubule-like polymers (9, 15, 25). Recently, these polymers have been shown to have dynamic properties in vitro, as they can grow from single nucleation sites and eventually interconnect (34). Probably all prokaryotes, including archaea and chloroplasts, contain FtsZ (5, 23, 27, 33). It is likely that FtsZ evolved into the three types of tubulin proteins that are keystones of the eukaryotic cytoskeleton.
FtsZ localizes to the leading edge of the invaginating inner membrane in E. coli and other prokaryotes and likely defines the cell division plane (6, 20, 31). Based on immunoelectron microscopic data, the active assembled polymer is thought to exist as a continuous ring. Therefore, the behavior of FtsZ seems to be more like actin than like tubulin. FtsZ forms protofilament sheets and bundles in vitro, suggesting that the FtsZ ring in vivo comprises such a superstructure (15, 34). However, the FtsZ polymeric ring has not been directly observed in electron microscopic sections. It is also not known precisely how the FtsZ structure changes during the cell cycle, particularly during invagination, and whether it is truly dynamic in vivo. Recent results suggest that FtsZ has different polymerization states (3, 22) and that the FtsZ structure may be more unstable during the constriction process than before constriction (2). A major question in the field is whether FtsZ is actively involved in generating the cytokinetic force or is simply the scaffold to which septum-synthesizing enzymes are recruited and on which they exert inward pressure (14, 21, 30).
Previously, we showed that a fusion of FtsZ with green fluorescent protein (GFP) was able to localize to the native FtsZ ring in E. coli cells (22). Here, by using three-dimensional image reconstruction of cells containing FtsZ-GFP, we show for the first time that FtsZ dynamics can be visualized directly in individual, growing E. coli cells throughout the entire cell cycle. The advantage of monitoring individual cells is that rapid protein dynamics can be observed in real time.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Strain WM720 was made by transforming E. coli BSP853 (W3110 rnc40::ΔTn10), obtained from R. Britton, with plasmid pZG (22), which contains FtsZ-GFP under lac promoter control, and lacIq in plasmid pBC (Stratagene). Cells were grown in Luria-Bertani medium (10 g of tryptone per liter, 5 g of NaCl per liter, 5 g of yeast extract per liter) at 30°C to early exponential phase (optical density, 0.3) and then induced with 40 μM IPTG (isopropyl-β-d-thiogalactopyranoside) for 40 min before transfer to agar (see next section). Cells induced with this concentration of IPTG exhibited normal FtsZ rings and cell division for several hours after induction, indicating that the extra FtsZ-GFP did not significantly perturb cell physiology during the time used for the experiments.
Preparation of cells for fluorescence microscopy.
For vital membrane staining, FM4-64 vital dye (Molecular Probes, Eugene, Oreg.) was added to the culture at a final concentration of 30 μM. For observation of GFP fluorescence in growing cells, a glass slide was covered with a thin layer of Luria-Bertani medium plus 1.5% agar; then, a drop of culture was added to the top of the agar and allowed to dry at room temperature for 2 min, and a cover glass was placed over the slide. Growth of the microcolonies was then monitored by light microscopy at room temperature. Under the conditions of this assay, the cells grew with an average generation time of about 2 h. Immunofluorescence microscopy (IFM) with anti-FtsZ was performed on fixed cells essentially as described previously (28).
Microscopy and image analysis.
For conventional fluorescence microscopy, images were acquired with an Olympus BX60 microscope equipped with a 100× oil immersion phase-contrast objective, a standard fluorescein isothiocyanate (FITC) filter set for GFP, and an Optronics Engineering DEI-750 24-bit color video camera. Images were captured and digitized with a Scion LG3 framegrabber and manipulated with Adobe Photoshop. Care was always taken to minimize exposure of the bacteria to the blue excitation light to minimize photobleaching. Three-dimensional image reconstruction of fluorescence was performed with a Deltavision wide-field optical sectioning microscope (Applied Precision, Issaquah, Wash.) equipped with a 100× oil-immersion objective and visualized with a cooled charge-coupled device camera, an FITC filter set (GFP), and a rhodamine filter set (FM4-64). z-axis sections were taken at 0.1- to 0.2-μm intervals, for a total of 12 to 20 sections. Since the cell diameters were less than 1 μm, this oversampling ensured that image data would not be omitted. Deconvolution of raw data was performed with five rounds of iteration, and volume views were generated with the maximum-intensity method at best quality.
To obtain fluorescence images of cell cross sections without using deconvolution, it was necessary for bacteria to be simultaneously immobilized and perpendicular to the coverslip plane, which was not possible by placing the cells on agar or by mixing them with agar. This problem was circumvented by the following layering technique. A glass slide was prewarmed at 37°C, and 20 μl of a logarithmic-phase cell culture was dropped on it. Then, 20 μl of 3% low-melting-point agarose was layered above the cells, followed immediately by 4 μl of 0.5% Casamino Acids layered on the top and center of the agarose. The slide was then incubated at 37°C for 1 min. In order to avoid crushing the solidified agarose, two strips of double-sided tape were applied along the edge of the slide, upon which a cover glass was carefully placed.
RESULTS
Normal cell division with fluorescent FtsZ.
To study FtsZ dynamics in growing cells, we used merodiploid E. coli cells expressing native FtsZ from the chromosome and an FtsZ-GFP fusion from the lac promoter on a plasmid, as described previously (22). Although it was reported previously (22) that this fusion failed to complement the ftsZ84 temperature-sensitive mutant, we subsequently found conditions, after finely tuning expression levels with IPTG and glucose, that would allow complementation of the mutant at the restrictive temperature (data not shown). This indicated that the fusion protein was at least partially functional for septation, although the narrow concentration requirement for its activity suggested that it was somewhat less functional than the untagged protein.
To monitor single cells in culture, exponentially growing cells were subjected to a short-term, low-level induction with IPTG and then transferred to thin agar overlays on a microscope slide beneath a coverslip, and the structure and localization of FtsZ-GFP over time in the microculture were monitored by fluorescence microscopy. The cells were grown at room temperature, resulting in a generation time of about 2 h. The results of a typical time course experiment in which cells were monitored through several cell division cycles are shown in Fig. 1. One important conclusion to be drawn from this data is that under these conditions, fluorescent FtsZ/FtsZ-GFP rings were clearly functional through several cycles of cell division, as they constricted and disappeared after cell separation. This is consistent with the ability of FtsZ-GFP, when at low levels in the cell, to colocalize with FtsZ (22) and complement the ftsZ84 mutation. This result is also consistent with the ability of purified FtsZ-GFP to bind and hydrolyze GTP as efficiently as the untagged protein and to coassemble homogeneously with the untagged protein into dynamic protofilament bundles (34). Therefore, it is likely that FtsZ and FtsZ-GFP coassembled to make a mixed ring, although it cannot be ruled out that two adjacent rings, one with FtsZ and one with FtsZ-GFP, were being assembled. Immunoblot analysis with anti-FtsZ showed that under these growth conditions, the levels of FtsZ-GFP were between 50 and 100% of the levels of FtsZ in the cell (data not shown).
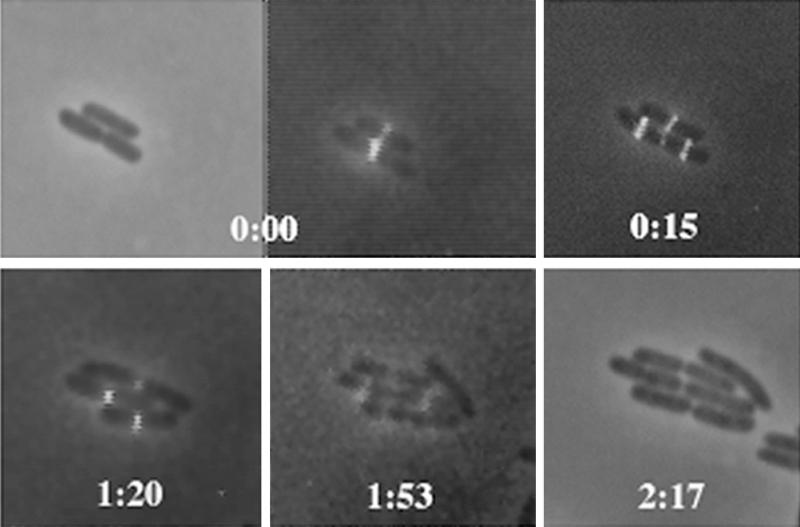
FIG. 1.
Growth and division of E. coli microcolonies expressing FtsZ-GFP and FtsZ, showing that FtsZ rings containing FtsZ-GFP function normally. Two consecutive cell division cycles are shown, using conventional fluorescence microscopy. The first and last panels are phase-contrast images; note that the last time point lacks a fluorescence image. The other panels were obtained by a digital overlay of phase-contrast and fluorescence images at the times shown. Times are shown in hours and minutes. Note the simultaneous formation of daughter FtsZ rings and contraction of the midcell ring at 0:15 in the bottom cell and at 1:53 in two daughter cells of the original bottom cell.
Assembly of FtsZ rings at daughter division sites before completion of mother cell division.
Interestingly, we found that FtsZ/FtsZ-GFP rings assembled at one-fourth and three-fourths of the cell length (1/4 and 3/4 positions) immediately before the midcell site was fully constricted (Fig. 1, 0:15 time point). This result demonstrates clearly that under our experimental conditions, daughter division sites can become competent for FtsZ assembly while the central septum is being formed, prior to the complete division of the mother cell. The localization of FtsZ-GFP to the 1/4 and 3/4 positions in dividing cells is consistent with the placement of periseptal annuli in the periseptal annulus model (30) as well as a previous proposal of such a possibility based on FtsZ ring formation in cephalexin-treated cells (28).
Our results differ from those of previous IFM experiments, in which FtsZ was not observed to target daughter cell sites before completion of the midcell septum in wild-type cells (1, 18) and newborn cells almost always lacked FtsZ rings (28). To determine whether the expression of the FtsZ-GFP fusion was causing this localization of FtsZ to the quarter sites, we examined a large number of cells with and without the fusion (WM720 and BSP853, respectively) by IFM (Table 1). The high percentage of cells containing FtsZ rings and the presence of FtsZ rings at quarter sites in a small fraction of dividing cells support our GFP data with live cells. In addition, the more frequent quarter site localization in cells expressing FtsZ-GFP suggests that our observations of very early FtsZ targeting are probably due to a small excess of total FtsZ in the cell because of the presence of the fusion protein. Although this excess would be at most twofold, such an increase in FtsZ levels may be sufficient to allow assembly at new division sites as soon as they are ready, whereas normally some time after cell birth may be necessary to build up FtsZ levels to a critical level.
TABLE 1.
FtsZ rings and their positions, as determined by IFMa
| Strain | Total no. of cells counted | % of cells with:
|
|
|---|---|---|---|
| FtsZ rings | FtsZ rings at quarter sitesb | ||
| BSP853 | 795 | 97.8 | 0.5 |
| WM720 | 818 | 95.0 | 4.5 |
Cells of the two strains were grown, induced with IPTG, and processed for IFM as described in Materials and Methods. Digital images of the cells were then analyzed with NIH Image for cell numbers and the presence of FtsZ fluorescence.
Cells were scored as containing FtsZ rings at quarter sites if they had rings at 1/4 and 3/4 of the cell length. Essentially all of these cells contained visible midcell constrictions, as viewed by phase-contrast microscopy; a subset of these cells contained FtsZ fluorescence at the constriction point.
Another possible explanation for our results is that FtsZ-GFP expression slowed down the septation process, thus allowing enough time for the new sites to become active. To test this, we measured the lengths of WM720 and BSP853 cells in the same experiment and found that their length distributions were superimposable (data not shown), with an average cell length of 3.4 μm for both strains. These results rule out a general cell cycle delay induced by expression of the fusion and support the model invoking a threshold level of FtsZ as a prerequisite for assembly at new sites. The low percentage of WM720 cells displaying the quarter site localization is consistent with the transient nature of the localization relative to the generation time. The fact that a few BSP853 cells lacking the fusion also exhibited this localization suggests that this strain has the capacity to target FtsZ to the quarter sites during division on rare occasions, although it is possible that these rare cells would have never divided.
Both the IFM experiments and the live-cell experiments failed to detect FtsZ-GFP assembly at quarter sites prior to the development of a deep midcell constriction (Fig. 1; also see below). This suggests that the localization of FtsZ-GFP to quarter sites occurs simultaneously with the normal disassembly of the medial ring after cytokinesis. Although it appears that this phenomenon occurs mostly when FtsZ levels are elevated, these results have allowed us to detect perhaps the earliest time in the cell cycle at which division sites become competent to assemble FtsZ. It is important to note that in steady-state experiments, ZipA-GFP also seemed to target potential division sites early, perhaps because of its similar ability to recognize the division site as soon as it appears (17).
High-resolution microscopy of the FtsZ ring.
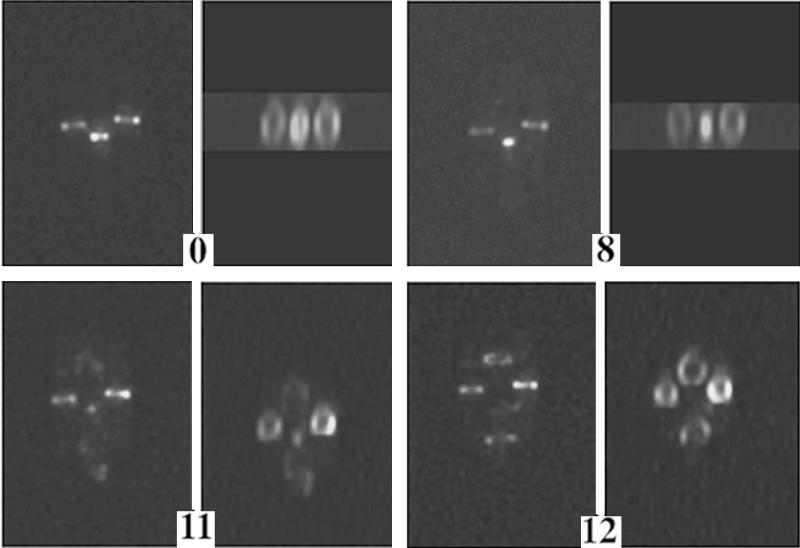
To obtain a more detailed, three-dimensional picture of FtsZ structure throughout the cell division cycle, we monitored FtsZ-GFP fluorescence in individual, growing cells by using a wide-field optical sectioning technique previously applied to cells in steady state (22). This technique uses deconvolution algorithms to sharpen two-dimensional images obtained with a conventional fluorescence microscope (4). The deconvolved images are then rotated to make a three-dimensional reconstruction. Figure 2 shows three cells; only the middle cell is undergoing cytokinesis during the time course. The fluorescent FtsZ structure can clearly be observed to contract to a dot and disappear. This time course suggests three major points. (i) FtsZ initially nucleates assembly at a point corresponding to the potential division sites (the 1/4 and 3/4 positions), polymerizing outward along the cell periphery to form arcs (Fig. 2, middle cell). (ii) The transition between the 11- and 12-min time points for the middle cell is equivalent to that in the 0:15 panel of Fig. 1 but with greater resolution, showing simultaneous assembly of FtsZ at daughter sites and disappearance at midcell. (iii) The assembly of the FtsZ structure is detectably complete within 1 min. A more detailed time course and discussion of the last step of septation are presented below.
FIG. 2.
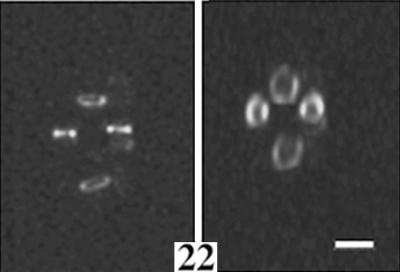
Three-dimensional image reconstruction of a shorter time course showing a fluorescence image of three adjacent growing cells, in vertical orientation, with the central cell undergoing complete septation and cell separation. Cells in the left-hand panels are viewed without rotation; the other panels show cells viewed with rotation to highlight the FtsZ annular structure as it condenses and duplicates. Times are shown in minutes; complete division of the central cell occurred between the 12- and 22-min time points. Note the formation of arcs in the 11-min panel that form nearly complete rings in the 12-min panel. Also note the fluorescence at the bottom pole of the dividing cell in the 11-min panel. Bar, 1.3 μm.
Many of the images of the FtsZ rings obtained by deconvolution and subsequent rotation appeared to exhibit discontinuities in their fluorescence (Fig. 2), suggesting that the rings were not uniform. Even rings from IFM experiments appeared to have segmented fluorescence after deconvolution and rotation (data not shown). To independently confirm whether this observation was true or was an artifact of the optical sectioning, we obtained conventional fluorescence images of cells containing FtsZ-GFP rings that were parallel to the plane of the coverslip so that they could be viewed directly in cross section (see Materials and Methods). It is clear from these images that the FtsZ rings are uniformly fluorescent, with no detectable discontinuities (Fig. 3). Therefore, it is likely that the gaps in fluorescence that we observed with rotated images from deconvolution stem from z-axis resolution problems. Since z-axis resolution is limited by the thickness of the optical sections, when out-of-focus light is subtracted out by the deconvolution algorithm, dropouts can occur. These dropouts are most frequent at the top and bottom of the cell, which usually is captured in only one optical section, whereas the sides of the cell are captured in several optical sections. Therefore, despite the stunningly crisp images delivered by deconvolution techniques, it is important not to overinterpret the z-axis components of these images, particularly in small cells such as bacteria and in cells with fine structures such as FtsZ rings.
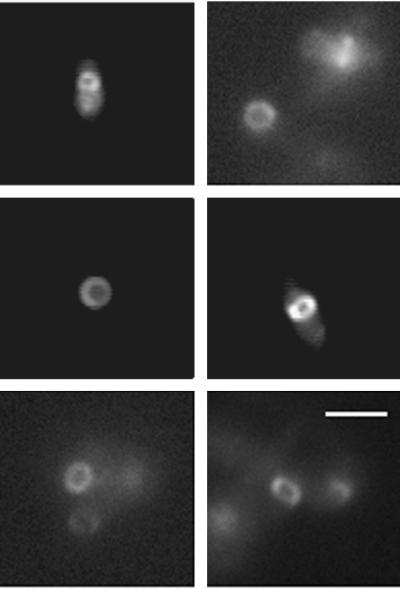
FIG. 3.
Visualization of FtsZ ring cross sections in immobilized, live cells. Shown are different cells, perpendicular to the plane of the coverslip, in which the FtsZ-GFP fluorescence displays a complete, uniform ring. Bar, 2 μm.
Formation of an FtsZ spiral.
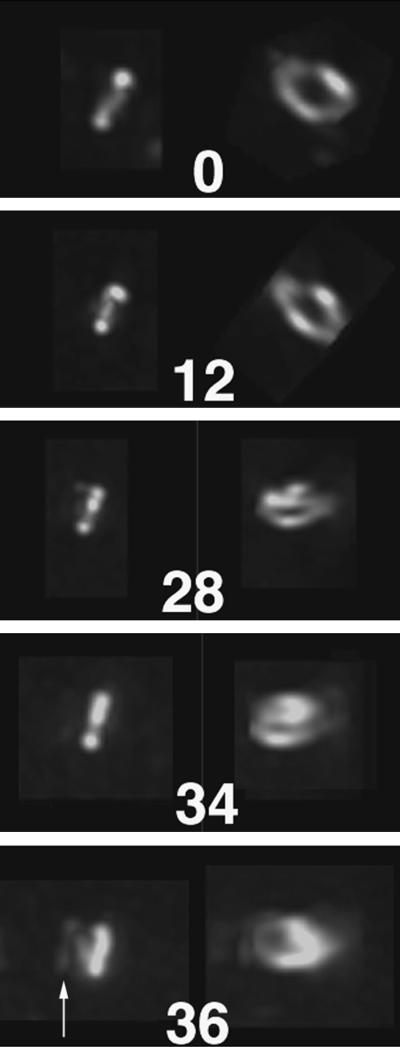
It was shown previously that higher levels of FtsZ-GFP expression correlated with the generation of FtsZ spirals (22). Such spirals appear to be nonfunctional for cell division. Fluorescent spiral structures were also observed when FtsZ was overexpressed in the presence of FtsA-GFP (22) and ZipA-GFP (data not shown). Furthermore, FtsZ spirals were observed by IFM of an ftsZ26 mutant, and these spiral structures can invaginate to form dramatic spiral septa (3). We took advantage of our ability to monitor FtsZ dynamics by examining the formation of a spiral in an atypical cell with deconvolution imaging (Fig. 4). Initially, the cell shown contained a bright fluorescent midcell ring. However, instead of constricting, this ring became distorted, and fluorescent polymers emanated outward from the central structure in a spiral (Fig. 4, 36-min panel). It is likely that the formation of the spiral may have been a result of higher levels of FtsZ-GFP relative to FtsZ in this particular cell, which failed to divide within several hours after the images were taken.
FIG. 4.
Formation of a spiral FtsZ-GFP polymer at midcell. Shown is a time course of a fluorescent FtsZ ring that ultimately fails to participate in septation and instead forms a clear spiral that emanates away from the midcell. The cell, expressing FtsZ-GFP, was grown in microculture and failed to divide over the several hours of growth. Times are shown in minutes. Images on the left for each time point are unrotated, whereas images on the right are rotated to reveal the cross-sectional structure. An arrow in the bottom panel highlights the developing spiral.
FtsZ dynamics during the final stages of cytokinesis.
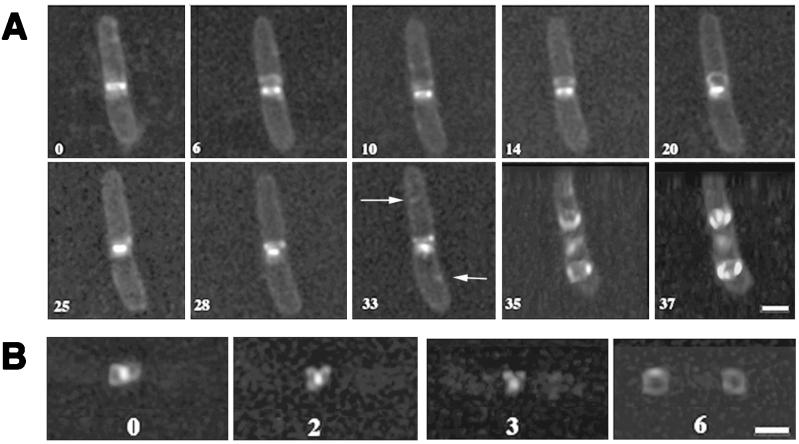
Analysis of the FtsZ-GFP structure in the later stages of cytokinesis, defined as the last quarter or approximately 30 min before cell separation when midcell constriction is easily detectable, revealed several interesting structural changes. As the FtsZ-GFP structure condensed at the leading edge of the cell invagination, the lumen of the ring could no longer be resolved, and finally just a small dot of fluorescence remained (Fig. 2, 8- and 11-min time points; also data not shown). This constriction at the leading edge of the invagination was also observed in the original immunogold studies of FtsZ rings (6). A detailed and high-resolution time course of an entire single cell undergoing late cytokinesis is shown in Fig. 5A.
FIG. 5.
Depolymerization and reorganization of FtsZ during the last stages of septation, as shown by three-dimensional reconstruction. (A) A single growing cell during cytokinesis. To aid visualization of the cell outline, the membrane was stained with FM4-64 vital dye. Note the disappearance of the loop formed at 20 min by the 25-min time point, followed by reappearance at 28 min. Arrows point to putative nucleation sites for FtsZ assembly in daughter cells. Also note that at 35 min, the FtsZ structures were formed at the future daughter cell division sites while the old FtsZ structure was still located as a highly condensed form at midcell. In order to show FtsZ rings in the daughter cell more clearly, the cells at 35 and 37 min are shown at a tilted angle. (B) A more detailed picture of a single growing cell, showing dynamics of the FtsZ loops. The bright dot is the condensed FtsZ ring, and the rings at the 6-min time point are formed in the middles of daughter cells. The numbers throughout the figure represent the time course, in minutes, after the first image was acquired. Bars, 1.3 μm.
One surprising finding was that during the process of contraction, loops of fluorescence often emanated from the central structure. Close examination and rotation of these structures revealed that two FtsZ-GFP loops emerged from the parent structure in a figure eight morphology and were oriented such that they pointed away from the division site. A detailed examination of one such loop is shown in Fig. 5B. Interestingly, the loops were transient. As with the assembly of the FtsZ ring, the dissipation of these loops did not appear to be uniform and took less than 6 min under our experimental conditions (Fig. 5B). In the whole-cell time course (Fig. 5A), the loop appears at the 6-min time point and appears to change direction and intensity until it is most obvious at 20 min. At the 25- to 33-min time points, the loop appears to be collapsed; it then disappears when the medial ring disappears. During the time interval represented in this series, we also observed transient fluorescent segments in other parts of the cell, including the cell poles and other locations not corresponding to the 1/4 and 3/4 positions (Fig. 2, 11-min panel; Fig. 5A, 0- and 10-min panels). These may reflect a normal process of attempted transient assembly by FtsZ-GFP, perhaps due to the appearance of a division site signal. These segments and the loops also may be artifacts of the GFP tag or the presence of excess FtsZ in the cell, although the specific locations of attempted assembly would argue that transient, low-affinity division site signals may still exist. If the polar fluorescence observed only at this time point represents FtsZ-GFP, then it might reflect attempted polar assembly because of the natural affinity of FtsZ for polar sites. Rapid inactivation of FtsZ assembly could then occur via the min system (12).
As was shown above, during the very last stages of cytokinesis the FtsZ loops disappeared at the same time that new FtsZ rings were nucleating assembly at daughter cell division sites. This nucleation was clearly visible as a dot at the 1/4 position and another at the 3/4 position (Fig. 5A, 33-min panel). Interestingly, the dots appeared to be on opposite sides of the dividing cell.
DISCUSSION
In this work, we have monitored the movement of FtsZ-GFP in individual, growing E. coli cells in real time over the entire cell division cycle. We have shown that FtsZ-GFP is capable of rapid, dynamic assembly and disassembly. Our work with live cells complements a previous IFM study of fixed cells with a mutant FtsZ protein (FtsZ84), in which the protein appeared to rapidly reassemble and disassemble after shifts between nonpermissive and permissive temperatures (2).
One major question in bacterial cell division is how assembly and disassembly of FtsZ and the septation complex are regulated to occur only once per cell cycle. The transient FtsZ-GFP loops that we observed during the late stages of cytokinesis may reflect the loss of FtsZ from the condensing, contracting central structure. The forces behind the condensation of the FtsZ structure are not known, but one possibility is that these loops represent FtsZ polymers that are released, springlike, from the constraints of the condensing structure and that the apparently repeated nature of loop formation could reflect the continuous need to remove FtsZ subunits from the structure as it contracts. Another possible explanation is that the loops are involved in providing the cytokinetic force by pushing the daughter cell walls apart. Since FtsZ-GFP is incorporated into the FtsZ structure, it is possible that the loops normally do not exist but are a way to remove some of the GFP-tagged protein. IFM analysis failed to reveal obvious loops (32), but the background fluorescence in IFM is higher and it is difficult to resolve such fine structures, even with deconvolution.
Attempts by FtsZ-GFP to assemble elsewhere in the cell were also apparent in the time-lapse analysis. Although it is possible that these attempts also represent artifacts of the fusion protein, we favor the possibility that these events are aborted assembly events at low-affinity sites in the cell by higher-than-normal levels of FtsZ. Such events might not occur at lower FtsZ levels, but they indicate that assembly of unstable structures can occur at multiple locations. Future IFM studies of cells with slightly elevated levels of FtsZ should provide additional evidence for or against these possibilities.
Another new observation presented here is the appearance, in wild-type cells expressing FtsZ-GFP, of new FtsZ rings precisely when the old FtsZ ring has successfully completed its task and is disassembling. The most likely explanation for the differences between our results and previous findings is the small excess of FtsZ in the form of FtsZ-GFP. Normally, there is only enough FtsZ for one ring per cell (13), so that newborn cells, despite having competent midcell division sites, must wait until FtsZ levels are above a critical concentration. Such a concentration may be related to the 2 μM concentration required for in vitro assembly of FtsZ protofilament bundles (34). Under our experimental conditions, twofold-higher FtsZ/FtsZ-GFP levels would result in FtsZ in excess of what is required at midcell to assemble at new division sites as soon as they become active. Since FtsZ-GFP assembly was never observed at quarter sites earlier than the late midcell constriction stage, one possibility is that this is the earliest that the new division sites become active for FtsZ recruitment. Another possibility is that the division sites are present even earlier but that FtsZ can assemble there only when the central structure is disassembling, releasing large amounts of free FtsZ. Interestingly, much of this released protein may immediately participate in the assembly process at new sites. The important conclusion from our data is that in our system, the elusive signal for forming a new division site occurs not in newborn cells, as would be predicted by existing models, but instead earlier in the cell cycle, in the oldest cells undergoing cytokinesis. The localization of periseptal annuli (30), as well as F plasmid, to the 1/4 and 3/4 sites of predivisional cells (16, 26) implies that structures that may be prerequisites to the division site are already in place and active at these positions. This possibility is supported by recent studies showing that FtsZ or FtsZ84 localizes to the 1/4 and 3/4 sites in cells that fail to divide at midcell (1, 2, 28, 35), which is also true for a GFP fusion to FtsK in nondividing cells (35). However, the important distinction between our present results and these is that our cells with FtsZ-GFP divided normally, without any obvious cell cycle delay. Therefore, the localization of FtsZ-GFP to the quarter sites in our experiments is clearly not due to premature disassembly of the medial FtsZ ring but rather is likely due to excess FtsZ that is recruited to the new division sites. The fact that we never saw FtsZ localize to these sites until septation was nearly complete argues that FtsZ targeting requires sufficient cellular FtsZ levels and perhaps an additional assembly factor that might be released upon segregation of the daughter nucleoids.
FtsZ appears to nucleate at new division sites as a fluorescent dot, followed by what seems to be an arc of fluorescence that quickly becomes a complete ring. This process is very difficult to see, even in our real-time studies with deconvolution. However, these results strongly support recent work by Addinall and Lutkenhaus on a spherical mutant of E. coli in which FtsZ appeared to nucleate at a point and then polymerize as an arc (3). Therefore, it is likely that the model proposed by Addinall and Lutkenhaus can be generalized to normal, rod-shaped E. coli cells.
It will be interesting to find out if the dynamic and structural behavior of FtsZ observed here is also conserved in more distantly related prokaryotic species, such as gram-positive bacteria, mycoplasmas, and archaea. Moreover, it would be interesting to study the dynamics of FtsZ assembly in strains carrying mutations in ftsW, which appears to destabilize the FtsZ ring (7). The future use of in vivo reporters with different emission wavelengths, combined with deconvolution imaging of individual wild-type and mutant cells, should allow a more detailed study of the structure and function of the prokaryotic cell division machine as it functions in real time.
ACKNOWLEDGMENTS
We thank X. Yu, D. Ehrhardt, P. Christie, D. Hereld, P. Zuber, and F. Cabral for valuable discussions and T. Vida for reagents.
This work was supported by grants from the NSF (MCB-9513521) and the Texas Advanced Research Program (011618-016) to W.M.
REFERENCES
- 1.Addinall S G, Bi E, Lutkenhaus J. FtsZ ring formation in fts mutants. J Bacteriol. 1996;178:3877–3884. doi: 10.1128/jb.178.13.3877-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addinall S G, Cao C, Lutkenhaus J. Temperature shift experiments with an ftsZ84(Ts) strain reveal rapid dynamics of FtsZ localization and indicate that the Z ring is required throughout septation and cannot reoccupy division sites once constriction has initiated. J Bacteriol. 1997;179:4277–4284. doi: 10.1128/jb.179.13.4277-4284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Addinall S G, Lutkenhaus J. FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol Microbiol. 1996;22:231–237. doi: 10.1046/j.1365-2958.1996.00100.x. [DOI] [PubMed] [Google Scholar]
- 4.Agard D A, Hiraoka Y, Shaw P, Sedat J W. Fluorescence microscopy in three dimensions. Methods Cell Biol. 1989;30:353–377. doi: 10.1016/s0091-679x(08)60986-3. [DOI] [PubMed] [Google Scholar]
- 5.Baumann P, Jackson S P. An archaebacterial homologue of the essential eubacterial cell division protein FtsZ. Proc Natl Acad Sci USA. 1996;93:6726–6730. doi: 10.1073/pnas.93.13.6726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature (London) 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 7.Boyle D S, Khattar M M, Addinall S G, Lutkenhaus J, Donachie W D. ftsW is an essential cell-division gene in Escherichia coli. Mol Microbiol. 1997;24:1263–1273. doi: 10.1046/j.1365-2958.1997.4091773.x. [DOI] [PubMed] [Google Scholar]
- 8.Bramhill D. Bacterial cell division. Annu Rev Cell Dev Biol. 1997;13:395–424. doi: 10.1146/annurev.cellbio.13.1.395. [DOI] [PubMed] [Google Scholar]
- 9.Bramhill D, Thompson C M. GTP-dependent polymerization of Escherichia coli FtsZ protein to form tubules. Proc Natl Acad Sci USA. 1994;91:5813–5817. doi: 10.1073/pnas.91.13.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai K, Lutkenhaus J. ftsZ is an essential cell division gene in Escherichia coli. J Bacteriol. 1991;173:3500–3506. doi: 10.1128/jb.173.11.3500-3506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Boer P, Crossley R, Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature (London) 1992;359:254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]
- 12.de Boer P A J, Crossley R E, Rothfield L I. A division inhibitor and a topological specificity factor coded for by the minicell locus determine the proper placement of the division site in Escherichia coli. Cell. 1989;56:641–649. doi: 10.1016/0092-8674(89)90586-2. [DOI] [PubMed] [Google Scholar]
- 13.Donachie W D, Begg K J. “Division potential” in Escherichia coli. J Bacteriol. 1996;178:5971–5976. doi: 10.1128/jb.178.20.5971-5976.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erickson H P. FtsZ, a tubulin homologue in prokaryote division. Trends Cell Biol. 1997;7:362–367. doi: 10.1016/S0962-8924(97)01108-2. [DOI] [PubMed] [Google Scholar]
- 15.Erickson H P, Taylor D W, Taylor K A, Bramhill D. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci USA. 1996;93:519–523. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon G S, Sitnikov D, Webb C D, Teleman A, Straight A, Losick R, Murray A W, Wright A. Chromosome and low copy plasmid segregation in E. coli: visual evidence for distinct mechanisms. Cell. 1997;90:1113–1121. doi: 10.1016/s0092-8674(00)80377-3. [DOI] [PubMed] [Google Scholar]
- 17.Hale C A, de Boer P. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 18.Levin P A, Losick R. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Dev. 1996;10:478–488. doi: 10.1101/gad.10.4.478. [DOI] [PubMed] [Google Scholar]
- 19.Löwe J, Amos L A. Crystal structure of the bacterial cell-division protein FtsZ. Nature (London) 1998;391:203. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- 20.Lutkenhaus J. FtsZ ring in bacterial cytokinesis. Mol Microbiol. 1993;9:403–410. doi: 10.1111/j.1365-2958.1993.tb01701.x. [DOI] [PubMed] [Google Scholar]
- 21.Lutkenhaus J, Addinall S G. Bacterial cell division and the Z ring. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 22.Ma X, Ehrhardt D W, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Margolin W, Wang R, Kumar M. Isolation of an ftsZ homolog from the archaebacterium Halobacterium salinarium: implications for the evolution of FtsZ and tubulin. J Bacteriol. 1996;178:1320–1327. doi: 10.1128/jb.178.5.1320-1327.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukherjee A, Dai K, Lutkenhaus J. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc Natl Acad Sci USA. 1993;90:1053–1057. doi: 10.1073/pnas.90.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukherjee A, Lutkenhaus J. Guanine nucleotide-dependent assembly of FtsZ into filaments. J Bacteriol. 1994;176:2754–2758. doi: 10.1128/jb.176.9.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niki H, Hiraga S. Subcellular distribution of actively partitioning F plasmid during the cell division cycle in E. coli. Cell. 1997;90:951–957. doi: 10.1016/s0092-8674(00)80359-1. [DOI] [PubMed] [Google Scholar]
- 27.Osteryoung K W, Vierling E. Conserved cell and organelle division. Nature (London) 1995;376:473–474. doi: 10.1038/376473b0. [DOI] [PubMed] [Google Scholar]
- 28.Pogliano J, Pogliano K, Weiss D S, Losick R, Beckwith J. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc Natl Acad Sci USA. 1997;94:559–564. doi: 10.1073/pnas.94.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raychaudhuri D, Park J T. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature (London) 1992;359:251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- 30.Rothfield L I, Justice S S. Bacterial cell division: the cycle of the ring. Cell. 1997;88:581–584. doi: 10.1016/s0092-8674(00)81899-1. [DOI] [PubMed] [Google Scholar]
- 31.Rothfield L I, Zhao C-R. How do bacteria decide where to divide? Cell. 1996;84:183–186. doi: 10.1016/s0092-8674(00)80971-x. [DOI] [PubMed] [Google Scholar]
- 32.Sun, Q., and W. Margolin. Unpublished data.
- 33.Wang X, Lutkenhaus J. FtsZ ring: the eubacterial division apparatus conserved in archaebacteria. Mol Microbiol. 1996;21:313–320. doi: 10.1046/j.1365-2958.1996.6421360.x. [DOI] [PubMed] [Google Scholar]
- 34.Yu X-C, Margolin W. Ca2+-mediated GTP-dependent dynamic assembly of bacterial cell division protein FtsZ into asters and polymer networks in vitro. EMBO J. 1997;16:5455–5463. doi: 10.1093/emboj/16.17.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu X-C, Tran A H, Sun Q, Margolin W. Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J Bacteriol. 1998;180:1296–1304. doi: 10.1128/jb.180.5.1296-1304.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]