Abstract
Replacement of Escherichia coli’s RecBCD function with phage λ’s Red function generates a strain whose chromosome recombines with short linear DNA fragments at a greatly elevated rate. The rate is at least 70-fold higher than that exhibited by a recBC sbcBC or recD strain. The value of the system is highlighted by gene replacement with a PCR-generated DNA fragment. The ΔrecBCD::Plac-red kan replacement allele can be P1 transduced to other E. coli strains, making the hyper-Rec phenotype easily transferable.
Gene replacement in bacteria can be achieved by a variety of techniques. One of the easiest procedures for gene manipulation in Escherichia coli is to place a drug resistance marker near, within, or in place of a cloned gene of interest. Linear DNA, containing the mutated or deleted gene flanked by homologous regions of the chromosome, is then transformed or electroporated into recombination-proficient strains (i.e., recBC sbcBC or recD). Recombination between the bacterial chromosome and both ends of the linear DNA fragment results in gene replacement (13, 19, 34, 46). The recombinants can be easily selected by the presence of the drug resistance marker. Unfortunately, this replacement procedure is restricted to specific RecBCD nuclease-deficient recombination-proficient genetic backgrounds and thus is not universally applicable. Furthermore, the number of mutations generated is limited by the availability of antibiotic resistance markers.
Another general method involves integration of a plasmid containing selectable markers and the mutant gene of interest into the bacterial chromosome by homologous recombination, followed by resolution of the cointegrate, which, depending on the mechanism of resolution, generates either the wild-type locus or gene replacement. Since plasmid sequences are removed during resolution of the cointegrate, gene replacements can be made free from antibiotic resistance markers. Phenotypic screens and/or Southern analyses are then used to identify the replaced allele. The key to this procedure involves the use of vectors that cannot replicate under conditions used for selection of the cointegrate. Examples of such vectors for E. coli include ColE1-derived plasmids, which do not replicate in polA mutants (10, 36), a temperature-sensitive pSC101 replicon (11), and a phagemid-based vector (38). Thermosensitive, pir-dependent, and repA-dependent broad-host-range plasmids for use in gram-positive bacteria have been described (3, 16, 22).
While the cointegration scheme, and versions of it, has been successfully used in a variety of hosts, it has some drawbacks. The replacement allele is actually a composite gene generated by the cointegrate resolution event. Thus, a particular mutation may not be transferred as expected. Also, resolution of the cointegrate occurs at low frequency and may not always give the replacement desired. Lastly, cloning of the gene prior to replacement is required.
This report identifies a new scheme that can generate gene replacement in nearly any E. coli strain at high frequency, does not depend on a cointegrate, and does not require prior cloning of the gene. Bacteriophage λ recombination functions exo, bet, and gam are expressed from a multicopy plasmid and used to promote gene replacement in a wild-type E. coli host following transformation with linear DNA substrates. λ recombination functions promote gene replacement within the lacZ gene at a rate 15 to 130 times higher than recBC sbcBC or recD strains. Since homologous recombination is elevated by the addition of functions to the host, rather than induced by alteration of host functions, this procedure is widely applicable to any E. coli and possibly other bacteria as well. Furthermore, an E. coli strain has been constructed in which the recBCD genes of E. coli have been replaced with a Plac-bet exo operon, together with a kanamycin resistance determinant. Thus, any strain can be made hyper-Rec by P1 transduction. Compared to plasmid-encoded λ Red, chromosome-encoded λ Red promotes even higher rates of recombination. Finally, by using the λ Red recombination system, the wild-type lacZ allele was replaced with a PCR-generated lacZ fragment containing an insertion of the tetracycline resistance gene. Thus, cloning of the gene of interest before replacement is not necessarily required.
MATERIALS AND METHODS
Chemicals and procedures.
Luria-Bertani (LB) medium has been described elsewhere (37). DNA buffer (DB) is 10 mM Tris-HCl (pH 7.5), 5 mM NaCl, and 1 mM EDTA. Precipitation buffer (PB) is 20 mM Tris-HCl (pH 7.4), 10 mM NaCl, 2 mM EDTA, and 0.5 M ammonium acetate. Isopropyl-β-d-thiogalactopyranoside (IPTG) and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) were purchased from Sigma. DNA transformations, P1 transductions, UV sensitivity assays, and conjugational recombination measurements were performed essentially as described earlier (21, 25, 27). DNA fragments were isolated by electrophoresis on bisacrylylcystamine polyacrylamide gels and purified as described by Hansen (12) except that DNA bands were visualized by staining the gels in 0.025% methylene blue, and phenol was equilibrated with 10 mM Tris-HCl to pH 7.0. Following elution of DNA fragments from DEAE-cellulose minicolumns, tRNA (30 μg) was added as carrier. The samples were precipitated with 2 volumes of ethanol, frozen at −80°C for 30 min, collected by centrifugation, and resuspended in 350 μl of PB. The DNA fragments were extracted with phenol, extracted with ether, precipitated with ethanol, and resuspended in DB. The concentrations of DNA fragments were obtained by running portions of the purified samples on 0.7% agarose gels and comparing them with known amounts of digested plasmids.
Plasmids and bacteria.
The λ Red-Gam-producing plasmid pTP223 (28), control plasmid pMC7 (20), and recBCD-containing plasmids pCDK3 and pKM587 (8, 25) have been previously described. Plasmid pJM22 (from M. Susskind) has an 1,100-bp kanamycin resistance determinant from Tn903 cloned into the filled-in EcoRI site of pUR2 (35), with XbaI linkers inserted at the junctions. Plasmids generated in this study were constructed according to standard techniques (37) and are described in Table 1.
TABLE 1.
Plasmids constructed in this study
| Plasmid | Description |
|---|---|
| pKM116 | The filled-in 6,732-bp BspHI fragment from pKM587, containing ptr, recB, and the proximal portion of recD, was ligated to the filled-in EcoRI site of pBR322, with recB reading clockwise, opposite to bla. The EcoRI sites are reconstructed following ligation. |
| pKM118 | The EcoRI site near the recD sequences in pKM116 was removed by partial digestion of pKM116 with EcoRI, followed by treatment with Klenow fragment and dNTPs and religation. |
| pKM119 | The XmaI (filled-in)-PstI fragment of pKM587, containing sequences upstream of recC, was ligated to the PstI-EcoRI (filled-in) fragment of pBR322. The EcoRI site is reconstructed following ligation. |
| pKM120 | The 7,107-bp EcoRI-BamHI fragment from pKM118, containing sequences downstream of recC, was ligated to the 5,333-bp EcoRI-BamHI fragment from pKM119, containing sequences upstream of recC. The tetracycline resistance gene is reconstructed following ligation. |
| pKM121 | The 1,100 bp PstI fragment from pJM22 containing kan was treated with T4 DNA polymerase and dNTPs and ligated into the filled-in EcoRI site of pKM120. |
| pKM123 | The 1,908-bp BamHI-SspI fragment from pCDK3, containing the distal end of recD and downstream sequences, was ligated to the EcoRV-BamHI fragment of pBR322. |
| pKM124 | The 1,100-bp PstI fragment from pJM22 containing kan was treated with T4 DNA polymerase and dNTPs and ligated to the filled-in HindIII site of pKM123. The kan gene is read counterclockwise, in the same direction as bla. |
| pKM125 | The 5,710-bp EcoRI-BamHI fragment from pKM119, containing sequences upstream of recC and ori, was ligated to the 3,041-bp EcoRI-BamHI fragment from pKM124, containing the kanamycin resistance determinant and sequences downstream of recD. |
| pKM126 | The 1,700-bp fragment containing the Plac-bet exo operon, generated by partial digestion of pTP232 with EcoRI, was ligated to the EcoRI site of pKM125. Both red and kan are transcribed counterclockwise, in the direction of sequences upstream of recC. |
| pKM130 | The 2,500-bp PvuII fragment containing most of the lacZ sequence from pMV201 (M. Volkert) was ligated to the PvuII site of pBR322. |
| pKM131 | The 1,100-bp PstI fragment from pJM22 containing kan was treated with T4 DNA polymerase and dNTPs and ligated to the filled-in BclI site of pKM130. |
| pKM132 | pKM130 was digested with EcoRI and MscI, filled in with Klenow fragment and dNTPs, and religated. |
| pKM133 | The filled-in EcoRI and MscI fragment from pKM130, containing the tetracycline resistance determinant, was ligated to the filled-in BclI site of pKM132. |
E. coli strains used and constructed for this study are listed in Table 2. KM19 was constructed as follows. pKM125 was cut with BamHI and NdeI, and the 5-kb linear fragment containing the kanamycin resistance gene fragment, flanked by sequences upstream of recC and downstream of recD, was isolated by electrophoresis on a 5% bisacrylylcystamine polyacrylamide gel (12). Following purification, the linear fragment was transformed into JC9387 (recBC sbcBC), and kanamycin-resistant transformants were selected. KM20 was constructed in a similar manner except that a 7-kb linear DNA fragment, derived from a BamHI and NdeI digest of pKM126, was used. This fragment contains both the kanamycin resistance gene and the Plac-red operon flanked by sequences upstream and downstream of the recBCD region. KM21 was derived by P1 transduction of AB1157 with a lysate grown from KM19 and selection for kanamycin-resistant transductants. The recBCD-deficient phenotype of this strain was verified by its ability to support the growth of both λ red gam χ+ and χ− phage and a deficiency of conjugational recombination. KM22, KM26, and KM29 were derived by P1 transductions of AB1157 (wild type), JC9239 (recF), and KM354 (recJ), respectively, with a lysate grown from KM20 and selection for kanamycin-resistant transductants. The genotype of KM22 was verified by its ability to promote growth of both λ red gam χ+ and χ− phage and its proficiency at conjugational recombination (20% of wild-type levels). (Conjugation recombination in a recBC host carrying a λ red-expressing plasmid had previously been shown to reach 10% of wild-type levels [32].) The recBCD recF genotype of KM26 and the recBCD recJ genotype of KM29 was verified by their extreme UV sensitivities in the absence of IPTG (data not shown).
TABLE 2.
E. coli strains used in this studya
| Strain | Genotype | Reference or source |
|---|---|---|
| Wild type | ||
| W3310 | lacIq (F+) | 5 |
| AB1157 | argE3 his-4 leuB6 proA2 thr-1 ara-14 galK2 lacY1 mtl-1 xyl-5 thi-1 rpsL31 tsx-33 supE44 | A. J. Clark |
| Isogenic to AB1157 | ||
| JC10287 | Δ(srlR-recA)304 | A. J. Clark |
| JC9387 | recB21 recC22 sbcB15 sbcC201 | A. J. Clark |
| JC9239 | recF143 | A. J. Clark |
| KM353 | recD | 25 |
| KM354 | recJ | 25 |
| KM19 | Δ(recC ptr recB recD)::kan sbcB15 sbcC201 | This study |
| KM20 | Δ(recC ptr recB recD)::Plac-bet exo kan sbcB15 sbcC201 | This study |
| KM21 | Δ(recC ptr recB recD)::kan | This study |
| KM22 | Δ(recC ptr recB recD)::Plac-bet exo kan | This study |
| KM26 | Δ(recC ptr recB recD)::Plac-bet exo kan recF143 | This study |
| KM29 | Δ(recC ptr recB recD)::Plac-bet exo kan recJ | This study |
Constructions for strains prepared for this study are described in Materials and Methods.
Preparation of electrocompetent cells.
Cultures (20 ml) were started by placing an isolated colony from a selective medium into LB broth, containing tetracycline (15 μg/ml) for hosts carrying pTP223 or kanamycin (25 μg/ml) for cells carrying the ΔrecBCD::Plac-red exo replacement allele. Plasmid-containing cells were grown on a shaker at 37°C for various times before IPTG was added to a final concentration of 1 mM. With cells containing the ΔrecBCD::Plac-red substitution, IPTG was added to the culture at the time of inoculation. When the culture reached a density of 108 cells/ml, the cells were collected by centrifugation, resuspended in an equal volume of 20% glycerol, and recentrifuged at 5,000 × g for 15 min. This step was repeated. The cells were then resuspended in 10 ml of 20% glycerol, collected by centrifugation, and resuspended in 100 μl of 20% glycerol. Cells were either used immediately or frozen in 50-μl aliquots in a dry ice-ethanol bath and stored at −80°C. Cells stored in this way were usable weeks after freezing. Scaled-up versions of this protocol were used to store large numbers of electrocompetent cells.
Electroporation.
Electrocompetent cells were thawed quickly at room temperature and placed on ice. Disposable 0.1-cm electroporation cuvettes (Bio-Rad) were placed in an ice-water bath for 10 min prior to electroporation. DNA samples in DB (5 to 10 μl) were mixed with 50 μl of electrocompetent cells. The mixture was pipetted into the precooled cuvettes (which were dried thoroughly) and electroporated with an in-house instrument (constructed by J. Goguen), consisting of a simple design similar to commercial capacitor-discharge units but using a vacuum relay with tungsten carbide contacts to switch between charge and discharge modes. The peak discharge field used was 20 kV/cm with an RC time constant of 4.2 ms. A 200-ohm resistor was used in series with the cuvette to limit discharge current should arcing occur.
Measurement of recombination frequency.
Immediately following electroporation, 350 μl of LB was added to the cuvette and quickly mixed with the cells by repeated pipetting. The cell suspension was diluted 10-fold in LB and grown by rolling at 37°C for 30 to 45 min. Recombinant titers were determined by plating various dilutions of the cultures on LB plates containing either kanamycin (20 μg/ml) or tetracycline (2.5 μg/ml) (depending on the fragment used), previously spread with 0.2 ml IPTG (0.1 M) and 0.2 ml X-Gal (20 mg/ml in dimethyl formamide). Surviving titers were determined on LB plates containing IPTG; in some experiments, X-Gal was included as well. It was found that having IPTG on the recombinant and surviving titer plates decreased colony counts in some experiments (when plasmid-produced Gam was expressed in recF and recJ mutants) or increased colony counts in others (in strains containing the ΔrecBCD::Plac-red substitution). However, for any one strain, the percent recombination per survivor with or without IPTG on the plates did not vary more than twofold in side-by-side comparisons. Plates were incubated at 37°C for 24 to 36 h, and the fraction of kanamycin (or tetracycline)-resistant Lac− (white) transformants relative to the total number of survivors was calculated. In control samples containing buffer plus electroporated cells (no DNA), no recombinants were detected.
Exonuclease-deficient recombination-proficient recBC sbcBC strains, and to a lesser extent recD strains, do not stably maintain ColE1-related plasmids (1, 2, 33), largely due to the formation of linear plasmid multimers in these strains. As a result, circular plasmid transformation rates (transformants/viable cell) are decreased in these strains (with pBR322, decreased 2-fold in recD and 10-fold in recBC sbcBC strains [unpublished observations]). This decrease in circular plasmid transformation rates is also observed with cells containing the ΔrecBCD::Plac-red allele (decreased 10-fold, with or without IPTG [data not shown]) but not when Red and Gam are supplied from pTP223 (probably as a result of the presence of plasmid-encoded LacI, which keeps expression of red and gam under tight control in the absence of IPTG following transformation). Thus, with JC9387, KM353, and KM22, circular plasmid transformation rates reflect both DNA uptake and circular plasmid stability and cannot be used to normalize linear DNA uptake. As a result, in the electroporation experiments described above, percent recombination is reported as the number of linear DNA transformants per survivor. Consequently, gene replacement frequencies per survivor reported here may be underestimated relative to replacements per competent cell, if after electroporation fewer than 100% of the survivors take up DNA.
In the experiment following linear transformation into calcium-treated competent cells, recombinant frequencies are reported as transformants per competent cell (comparisons to recBC sbcBC and recD strains were not included in this experiment). Also, X-Gal and IPTG were not included in plates used to determine the number of kanamycin-resistant colonies. In this case, the percentage of lacZ colonies was determined by stabbing 25 to 30 kanamycin-resistant colonies into a plate containing X-Gal and IPTG. In all instances, 100% of tested colonies were white, while the nontransformed parents were blue.
PCRs.
PCR was used to verify strain construction and recombinant formation. Primers (24-mers) used to verify kan or tet insertion into lacZ were as follows: from the lacZ sequence, GTAACCTATCCCATTACGGTCAAT and CGGTTAAATTGCCAACGCTTATTA. Primers used to verify the ΔrecBCD::Plac-red replacement allele were as follows: for the left end, TTTGTTTGCGTTTACTGGCAGATA, TCGTTGACCCACTGGCGTAAATAA, and ACGGCAACGGCCTTGAACTGAAAT (primers 7 to 9, respectively); for the right end, TCGCATCCGGCAATTACGTTTATT, CATCGCATTGCTGATTACGACTAT, and ATCAGGATTATCAATACCATATTT (primers 10 to 12, respectively) (see Fig. 4 for relative positions of these primers). Primers 7 and 10 were used to verify the ΔrecBCD::kan substitution in KM19 and KM21. In PCR analysis of JC9387 (recBC sbcBC), the presumed missense mutations in recB and recC did not alter the size of PCR product expected.
FIG. 4.
Structures of recBCD deletions and Plac-red substitution. Shown are the recBCD regions in wild-type cells (A), KM19 and KM21 (ΔrecBCD::kan) (B), and KM20 and KM22 (ΔrecBCD::Plac-red kan) (C). Included are the relative positions of the PCR primers used to verify the structures of the wild-type and both substitution alleles. In panel C, transcription of red and kan occurs leftward (same direction as argA but opposite the direction of thyA).
PCR mixtures consisted of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 1.9 mM MgCl2, 1 μM primers, 50 μM deoxynucleoside triphosphates (dNTPs), 5% dimethyl sulfoxide, and 2.5 U of AmpliTaq DNA polymerase (Perkin-Elmer) in a volume of 40 μl. After all components (except the polymerase) were added together, a sterile pipette tip was used to pick a colony off an agar plate and thoroughly mix the cells with the PCR reagents. AmpliTaq DNA polymerase was added and mixed; the samples were overlaid with an equal volume of mineral oil (Sigma) and placed in a thermocycler (MJ Research). All programs consisted of an initial step at 95°C for 5 min, followed by 30 cycles of 1 min at 94°C, 1 min at 55°C, and 5 min at 72°C. A final extension step of 7 min at 72°C was included. Samples (10 μl) of the PCR mixtures were run on 0.7% agarose along with a set of standards (1-kb DNA ladder; Gibco-BRL) and stained with ethidium bromide.
RESULTS
λ Red-Gam-promoted gene replacement.
The bacteriophage λ recombination system, known as red, consists of two genes, exo and bet. Exo is a 5′-3′ exonuclease that acts processively on double-stranded (ds) DNA (6, 17). Bet is a single-stranded DNA (ssDNA) binding protein capable of annealing complementary ssDNA strands (15, 23). Bet can stimulate the formation of joint molecules and strand exchange by RecA but cannot promote strand invasion on its own (24). λ Red-mediated recombination is stimulated by the presence of dsDNA ends, generated by cutting of cos by λ terminase prior to packaging or by a restriction endonuclease (29, 41, 44). RecA is not required for Red-promoted recombination unless phage replication is inhibited (43). The Red recombination functions are assisted by the λ Gam function, which inhibits host RecBCD exonuclease (14, 25). While RecBCD is one of the key enzymes in E. coli’s major recombination system, its activation is dependent on the presence of Chi sites (5′GCTGGTGG3′), which are absent in wild-type lambda DNA (39). Thus, Gam’s role is to prevent RecBCD-promoted digestion of λ phage DNA, so that Exo and Bet can gain access to DNA ends to promote recombination.
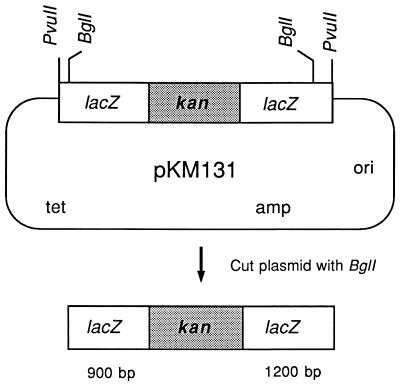
This study was done to examine if expression of λ Red plus Gam, in the absence of other phage functions, might promote gene replacement in E. coli following transformation with linear substrates. A linear DNA substrate was prepared from pKM131, which contains most of the lacZ sequence, interrupted at an internal BclI site by a gene encoding kanamycin resistance (Fig. 1). After digestion of pKM131 with BglI, the 3.2-kb linear lacZ::kan fragment was isolated and purified as described in Materials and Methods. The fragment, which contains the kanamycin resistance determinant flanked by 921 and 1,200 bp of lacZ sequence, was electroporated into various E. coli hosts containing pTP223, a plasmid that expresses gam, bet, and exo under control of Plac (28). Recombinants were identified as white kanamycin-resistant colonies on plates containing X-Gal and IPTG and compared to the total number of survivors.
FIG. 1.
Linear DNA transformation substrate. Plasmid pKM131 contains the 2,557-bp PvuII lacZ fragment inserted into the PvuII site of pBR322, with a 1,100-bp kanamycin resistance-conferring fragment inserted into the BclI site within lacZ. When cut with BglI, pKM131 generates a fragment containing the kan gene, flanked by 921 and 1,200 bp of lacZ sequence. A similar plasmid, pKM133 (not shown), contains the tetracycline resistance determinant from pBR322, flanked by 1,256 and 1,301 bp of lacZ sequence.
Table 3 shows the results of two of these experiments. In experiment A, the purified linear lacZ::kan substrate (described above) was mixed with electrocompetent cells at a concentration of 3 μg/ml and electroporated as described in Materials and Methods. Wild-type cells (AB1157) containing a control plasmid (pMC7) produced no recombinants. When gam, bet, and exo were expressed in vivo in a wild-type host, recombinants were generated at a frequency of 3.6 × 10−5 per survivor, an increase of at least 3 orders of magnitude over cells containing the control plasmid. In a recJ mutant background, the frequency of recombination was further increased twofold. In other experiments (see below), λ Red-promoted recombination was stimulated 2- to 25-fold by deletion of recJ. The lack of dependence on RecJ (a 5′-3′ ssDNA exonuclease) is not surprising in light of the enzymatic activity of λ Exo (a 5′-3′ dsDNA exonuclease). The stimulation of λ Red-promoted recombination by a mutation in recJ suggests that the RecJ exonuclease interferes with the formation or stability of an intermediate generated by the action of λ Red. Interestingly, two strains known for their ability to transform linear DNA substrates (recBCD sbcBC and recD) did so with the lacZ::kan fragment, but at 138- and 620-fold lower frequencies, respectively, than λ Red-Gam-producing strains (Table 3, experiment A). Thus, at least with these substrates, cells expressing λ Red and Gam are more proficient than recBC sbcBC or recD strains in promoting recombination of linear DNA with the E. coli chromosome.
TABLE 3.
Electroporationa of a linear DNA fragment (lacZ::kan) into cells containing λ Red-Gam-producing plasmid
| Expt | Strain | Plasmid (functions) | Titer of recombinantsb (104) | Titer of survivors (109) | Recombinants/survivor (10−5) |
|---|---|---|---|---|---|
| A | AB1157 (wild type) | pMC7 (none) | 0 | 5.0 | 0 |
| AB1157 (wild type) | pTP223 (gam, bet, exo) | 7.9 | 2.2 | 3.6 | |
| KM354 (recJ) | pTP223 (gam, bet, exo) | 22.6 | 3.2 | 7.1 | |
| KM353 (recD) | None | 0.0075 | 1.3 | 0.0058 | |
| JC9387 (recBC sbcBC) | None | 0.037 | 1.4 | 0.026 | |
| B | AB1157 (wild type) | pTP223 (gam, bet, exo) | 11.1 | 2.0 | 5.5 |
| JC10287 (recA) | pTP223 (gam, bet, exo) | 0.03 | 0.37 | 0.081 | |
| JC9239 (recF) | pTP223 (gam, bet, exo) | 4.5 | 3.3 | 1.4 | |
| KM354 (recJ) | pTP223 (gam, bet, exo) | 136.0 | 1.95 | 70.0 | |
| JC9387 (recBC sbcBC) | None | 0.85 | 2.3 | 0.37 |
Cells were electroporated with 0.2 μg (experiment A) and 0.9 μg (experiment B) of purified lacZ::kan linear DNA fragment, generated by digestion of pKM131 with BglI.
Titers of electroporation mixture (60 μl); recombinants were scored as kanamycin-resistant white colonies.
In another experiment, cells containing the Red-Gam-producing plasmid pTP223 were electroporated with linear DNA at a final concentration of 15 μg/ml (Table 3, experiment B). Compared to the recBC sbcBC host, the λ Red-Gam-producing strain showed only a 15-fold increase in transformation with the linear substrate. λ Red-promoted recombinant formation was decreased 68-fold by a recA mutation and 4-fold by a recF mutation but increased 12-fold by a recJ mutation.
In addition to electroporation, simple addition of linear DNA to calcium-treated competent cells containing the λ Red-Gam-expressing plasmid was also examined. Table 4 shows the results from one such experiment. The linear lacZ::kan fragment or an equimolar amount of its parent circular plasmid was added to wild-type cells containing pTP223 or a control plasmid. In the absence of λ Red and Gam, no linear DNA transformants were obtained. λ Red and Gam expression, in either AB1157 or W3110 wild-type strains, resulted in a linear transformation rate per competent cell of 0.013 or 0.056%, respectively. As seen with electroporation experiments described above, calcium-mediated linear DNA transformation was stimulated by a mutation in recJ (25-fold in this experiment). For each transformation, between 25 and 30 kanamycin-resistant colonies were tested for activity of lacZ (which is functional in the recipient strains). All tested candidates were lacZ mutants, indicating replacement of the endogenous lacZ gene with the fragment-borne allele. Thus, while similar patterns of linear DNA transformations are seen in λ Red-Gam-producing hosts following calcium-mediated transformation, higher total numbers of transformants are obtainable following electroporation of the linear substrates (presumably because of the higher numbers of cells capable of DNA uptake during electroporation).
TABLE 4.
Transformation of calcium-treated cells by linear DNA fragment(lacZ::kan)a and supercoiled plasmid
| Strain | Resident plasmid (functions) | Transformants
|
Linear transformants/competent cellb | |
|---|---|---|---|---|
| Linear DNA | Circular DNA (105) | |||
| AB1157 (wild type) | pMC7 (control) | 0 | 3.2 | 0 |
| pTP223 (gam, bet, exo) | 24 | 1.8 | 1.3 × 10−4 | |
| KM354 (recJ) | pTP223 (gam, bet, exo) | 522 | 1.6 | 3.3 × 10−3 |
| W3110 (F′ lacIq) | pMC7 (control) | 0 | 1.5 | 0 |
| pTP223 (gam, bet, exo) | 270 | 4.8 | 5.6 × 10−4 | |
Cells were grown in LB containing tetracycline (12 μg/ml); at 108 cells/ml, IPTG was added to the culture (final concentration, 1 mM) for 30 min. The cells were collected by centrifugation, treated with calcium, and transformed with purified linear lacZ::kan DNA fragment (0.12 μg) or an equimolar amount of supercoiled parent plasmid (0.3 μg).
Ratio of transformation titers (linear fragment/circular plasmid).
In other experiments, pTP232, a plasmid that expresses only λ Red (no Gam), did not promote linear DNA transformation of the lacZ::kan substrate following electroporation into wild-type cells (data not shown). Thus, inactivation of RecBCD nuclease by λ Gam (or by mutation) is required to observe λ Red-promoted recombination, as has been observed previously (29, 42). The P22 recombination system consisting of arf, erf, abc1, and abc2, which can substitute for growth and recombination of λ red gam phage (28, 29), was also tested for its ability to promote linear DNA transformation. The P22 system, supplied by pTP178 (9), produced only 5 to 10% of the recombinants per survivor (or competent cell) exhibited by λ Red (data not shown). While the reason for this lower rate of linear DNA transformation by the P22 recombination system is not clear, it may in part have to do with the role of Abc2, a protein which modifies the host’s RecBCD enzyme. A suggested role of Abc2 is to harness the exonuclease activity of RecBCD to promote Chi-independent recombination during a P22 infection (references 26, 27, and 31 and unpublished data). As such, Abc-modified RecBCD becomes part of P22’s recombination system. Overproduction of RecBCD, together with all the components of P22’s recombination system, may be required to attain the levels of linear DNA transformation observed with λ Red and Gam.
Recombinant formation is IPTG dependent.
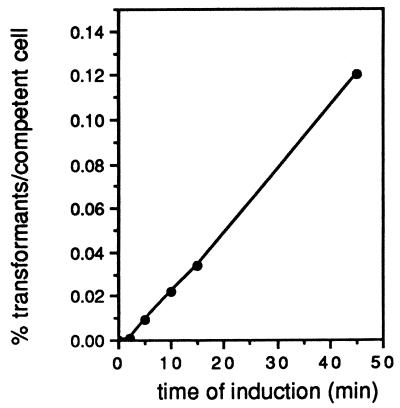
λ Red-Gam-producing cells were grown for various amounts of time in the presence of IPTG, made competent, and transformed with equimolar amounts of linear lacZ::kan fragment and its parent supercoiled plasmid. As shown in Fig. 2, in the absence of IPTG, no recombinants were found. Longer exposure to IPTG resulted in greater amounts of recombinants per competent cell. The highest rate of linear DNA transformation was achieved with a 45-min IPTG induction period, the highest-exposure period tested. Thus, even higher rates of linear DNA transformation may be achievable with longer periods of IPTG induction. Thus, the hyper-Rec phenotype of cells containing the λ Red-Gam-producing plasmid is easily controlled with IPTG.
FIG. 2.
Linear transformation promoted by λ Red is IPTG dependent. Wild-type cells (W3110) containing λ Red-Gam-producing plasmid (pTP223) were incubated for various times in the presence of 1 mM IPTG prior to collection of the cells by centrifugation. Cells were made competent and transformed with equimolar amounts of lacZ::kan fragment (0.12 μg) and parent plasmid pKM131 (0.3 μg). Ratios of transformation titers (fragment/plasmid) were plotted as a function of time of exposure to IPTG.
Characterization of the recombinants.
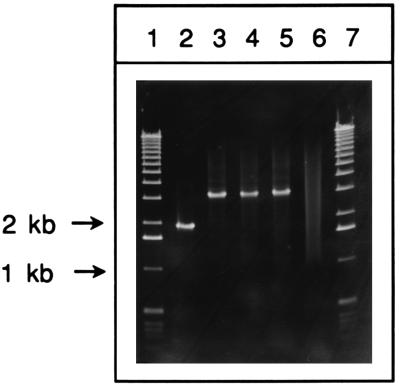
If a double crossover occurred between the lacZ::kan linear DNA substrate and the E. coli chromosome, the recombinant would contain a 1.1-kb insertion of the kan-containing fragment into the chromosomal lacZ locus. The PCR products from the lacZ region in three of the white kanamycin-resistant transformants described in Table 3 were compared to the PCR product of their kanamycin-sensitive blue parent. Figure 3 (lane 2) shows that the PCR product from AB1157 containing pTP223 produces a band of approximately 2 kb (as expected from the predicted PCR product of 1,972 bp), while all three white recombinants produced a 3.1-kb band (lanes 3 to 5) indicative of replacement of the chromosomal lacZ gene by the kan insertion allele. In another analysis, 10 of 10 white kanamycin-resistant transformants all produced the recombinant 3.1-kb band after PCR analysis (data not shown). In addition, sam-ples of recombinants produced with recA and recJ strains containing pTP223 also showed the 3.1-kb recombinant band after PCR analysis (data not shown). While these analyses do not rule out other types of chromosomal insertions into (or deletions of) lacZ, the majority of recombinant products involve simple replacements, likely generated by a double-crossover event.
FIG. 3.
PCR analysis of lacZ::kan recombinants. PCRs were performed as described in Materials and Methods. Lanes: 1 and 7, 1-kb DNA ladder standards (Gibco-BRL); 2, PCR product from AB1157 containing pTP223; 3 to 5, PCR products from three independent linear transformants of AB1157 containing pTP223; 6, control reaction (no cells). The positions of 1- and 2-kb DNA standards are shown by arrows.
Replacement of kan for recC.
A region other than lacZ was also tested for gene replacement promoted by λ Red and Gam. Digestion of pKM121 with PstI generates a 6.3-kb linear fragment containing the kanamycin resistance gene flanked by 2,476 bp upstream and 2,771 bp downstream of recC. The fragment was purified on polyacrylamide gels, mixed with AB1157 containing pTP223 (Red plus Gam) and JC9387 (recBC sbcBC) at a concentration of 2 μg/ml, and electroporated as described in Materials and Methods. Recombinants were scored as kanamycin-resistant transformants. In AB1157 containing a control plasmid, no transformants were found. The recBC sbcBC strain produced recombinants at a frequency of 1.8 × 10−4 per survivor. In AB1157 containing pTP223 (Red plus Gam), gene replacement occurred at a frequency of 0.75 × 10−4 per survivor. (However, gene replacement with this fragment deletes recC, generating a strain with fivefold reduced viability relative to JC9387 [not shown]. For this reason, the true frequency of λ Red-promoted recombination in this strain may be closer to 3.7 × 10−4.) From these data, it is clear that other regions of the chromosome besides lacZ are amenable to λ Red-promoted recombination. The λ Red-Gam-producing strain did not generate recombinants at a higher frequency than the recBC sbcBC strain, as was seen with the lacZ::kan fragment described above. One possible explanation for this result is that the λ Red system is better equipped than the host RecF pathway to recombine linear DNA substrates that contain small regions of homology (∼1,000 bp), a relative advantage that disappears with increasing amounts of homologous DNA. However, I cannot rule out context effects (for example, higher rates of transcription of lacZ relative to recC). Experiments to examine these types of questions are under way.
Moving the Plac-red operon from the plasmid to the chromosome.
Induction of plasmid-encoded gam, bet, and exo results in linear multimerization of the plasmid (30). It is possible that these linear multimers act as competitive inhibitors for λ Red-promoted recombination of electroporated linear DNA substrates. A similar scenario was seen previously when control plasmids in a recD strain, which induces linear multimerization of plasmids, inhibited RecBC-promoted conjugational recombination (25). This hypothesis was tested by placing the Plac-red operon into the chromosome. Since RecBCD must be deactivated to promote λ Red-dependent recombination, the chromosomal region of JC9387 (recBC sbcBC) encoding recC, ptr, recB, and recD was replaced with the Plac-red operon (see Materials and Methods). Since RecBCD is absent in this strain, Gam is predicted not to be required for efficient recombination.
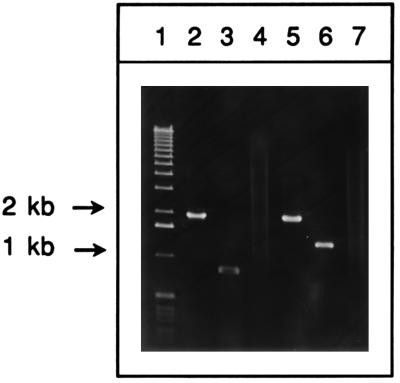
Figure 4 shows the region of the chromosome containing the recBCD genes, the ΔrecBCD::kan and ΔrecBCD::Plac-red kan substitutions, and the PCR primers used to verify the strain constructions. Primers 7, 8, and 9 were included in a PCR with colonies of JC9387 (recBC sbcBC) and KM20 (ΔrecBCD::Plac-red sbcBC). These primers flank the left junction point of the substitution and are predicted to generate a PCR product of 1,982 kb from wild-type E. coli sequence (via primers 7 and 8) and a 738-bp PCR fragment from KM20 (via primers 7 and 9). As seen in Fig. 5 (lanes 2 and 3), the PCR fragments generated in these reactions are consistent with its genotype. A second set of primers (10, 11, and 12) was used to verify the right endpoint of the replacement allele. PCR products of 2,018 and 1,223 bp are predicted from wild-type E. coli sequence and the replacement allele, respectively. In both cases, the expected products were observed (Fig. 5, lanes 5 and 6), verifying the structure of the replacement allele in KM20. This same analysis was performed on KM22, with identical results (data not shown). In another PCR with primers 7 and 10, KM19 gave the 2.0-kb product expected for replacement of the recBCD region with the kan insert (data not shown). The gene for kanamycin resistance flanks the Plac-red operon in KM20, allowing the ΔrecBCD::Plac-red substitution to be easily transferred into other E. coli strains. The ΔrecBCD::Plac-red substitution, as well as a control ΔrecBCD deletion, were transferred into wild-type, recF, and recJ hosts by P1 transduction. The recombination proficiency of these strains was tested with electroporated linear substrates (Table 5).
FIG. 5.
PCR analysis of the ΔrecBCD::Plac-red allele. Lanes: 1, 1-kb DNA ladder standards (Gibco-BRL); 2 to 4, PCR products generated with primers 7, 8, and 9 (Fig. 4) from JC9387 (recBC sbcBC), KM20 (ΔrecBCD::Plac-red sbcBC), and the control reaction (no cells), respectively; 5 to 7, PCR products generated with primers 10, 11, and 12 (Fig. 4) from JC9387 (recBC sbcBC), KM20 (ΔrecBCD::Plac-red sbcBC), and the control reaction (no cells), respectively. The positions of 1- and 2-kb DNA standards are shown by arrows.
TABLE 5.
Electroporation of a linear DNA fragment (lacZ::tet)a into cells containing the ΔrecBCD::Plac-bet exo substitution
| Strain (relevant genotype) | Titer of recombinantsb (105) | Titer of survivorsb (109) | Recombinants/survivor (10−5) |
|---|---|---|---|
| AB1157 (wild type) | 0 | 1.5 | 0 |
| KM19 (ΔrecBCD sbcBC) | 0.13 | 1.7 | 0.76 |
| KM20 (ΔrecBCD::Plac bet exo sbcBC) | 4.2 | 1.8 | 23.3 |
| KM21 (ΔrecBCD) | 0 | 0.35 | 0 |
| KM22 (ΔrecBCD::Plac bet exo)c | 7.0 ± 2.1 | 2.6 ± 1.0 | 27.0 |
| KM26 (ΔrecBCD::Plac bet exo, recF) | 0.03 | 0.19 | 1.6 |
| KM29 (ΔrecBCD::Plac bet exo, recJ) | 1.9 | 0.13 | 146.1 |
| KM353 (recD) | 0.21 | 2.9 | 0.72 |
| JC9387 (recBC sbcBC)c | 0.082 ± 0.027 | 1.9 ± 1.1 | 0.43 |
Purified linear lacZ::tet DNA fragment (0.9 μg), generated by digestion of pKM133 with PvuII, was mixed with 50 μl of electrocompetent cells and electroporated as described in Materials and Methods.
Titers of electroporation mixture (60 μl); recombinants were scored as tetracyclin-resistant white colonies.
Trials with KM22 and JC9387 were done four times; standard deviations for titers are included. Electroporation of other strains was performed only once in this experiment.
Since these strains are kanamycin resistant, a lacZ::tet-containing plasmid, pKM133 (Table 1), was prepared. After digestion of pKM133 with PvuII, the 3.9-kb linear lacZ::tet fragment was isolated and purified as described in Materials and Methods. The fragment, which contains the tetracycline resistance determinant flanked by 1,256 and 1,301 bp of lacZ sequence, was electroporated into strains containing the ΔrecBCD::Plac-red substitution. As seen in Table 5, no recombinants were obtained with wild-type cells. When the ΔrecBCD::Plac-red substitution was transferred into a recBC sbcBC background (KM20), recombinants appeared at a frequency of 2.3 × 10−4 per survivor, a 53-fold increase relative to JC9387, its recBC sbcBC parent. When a ΔrecBCD allele (no red) was transferred into an sbcBC background (KM19), no increase in recombination relative to JC9387 (recBC sbcBC) was observed. When the ΔrecBCD::Plac-red substitution was moved into a wild-type background (KM22), recombination proficiency with the linear lacZ::tet fragments was retained (63-fold higher relative to recBC sbcBC). Thus, sbcBC is not required for λ Red-promoted recombination, as expected. In constructs where the recBCD region was deleted without inserting the Plac-red operon (KM21), no recombinants were detected. Similar to the pattern seen with plasmid-encoded λ Red and Gam, RecF was required (decreased 17-fold in a recF background) and RecJ was inhibitory (increased 5-fold in a recJ background). A recD strain (KM353) showed a frequency of recombination similar to that for the recBC sbcBC strain (JC9387). As with the plasmid-encoded λ Red-promoted recombinants described above, the structures of these recombinants were tested by PCR analysis. All of 12 white tetracycline-resistant colonies from electroporation of KM22 with the lacZ::tet substrate generated a 3.4-kb fragment following PCR analysis of the chromosomal lacZ locus (data not shown), verifying that most the recombinants were formed by simple replacement of lacZ, an event likely generated by a double-crossover mechanism.
In summary, even though the copy number of the Plac-red operon was reduced significantly when the operon was moved from a multicopy plasmid to the chromosome, the frequency of recombinants per survivor increased fivefold (compare AB1157 containing pTP223 in Table 3, experiment B, with KM22 in Table 5). This comparison may not be fair, however, since the lacZ::tet fragment used for Table 5 has 20% more homologous DNA relative to the lacZ::kan fragment used for Table 3 (see the legend to Fig. 1 for details). A more appropriate comparison is between the ratios of recombination rates promoted by λ Red and RecF pathways, when λ Red is produced from a plasmid rather than the chromosome, a ratio which increases from 15-fold to 63-fold (at linear DNA concentrations of 15 μg/ml; compare Table 3, experiment B, and Table 5). This ratio is even higher (130-fold) in circumstances where lower concentrations of DNA are used (Table 3, experiment A).
Properties of E. coli (ΔrecBCD Plac-red).
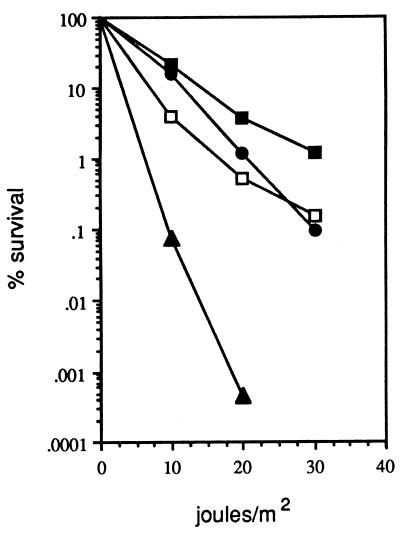
KM22, containing the ΔrecBCD Plac-red allele, was compared to wild-type cells and KM21 (ΔrecBCD) for viability, UV sensitivity, and proficiency at conjugational recombination. By determining the number of visible cells that eventually form CFU, KM22 (in the presence of IPTG) was found to be similar to wild-type cells, displaying nearly 100% viability (data not shown). In the absence of IPTG, KM22 showed about 50% viability, while KM21 (ΔrecBCD) showed only 10% viability (data not shown). A comparison of the UV survival patterns among these strains is shown in Fig. 6. This experiment reveals that λ Red can suppress the UV sensitivity of KM21 (ΔrecBCD). In fact, expression of λ Red in KM22 increased the level of survival (relative to wild-type cells) as much as 10-fold at 30 J/m2. In the absence of IPTG, KM22 showed decreased survival rates, but still higher than that exhibited by KM21, likely caused by low-level expression of λ red. Finally, KM22 was proficient at conjugational recombination (20% of wild-type levels) compared to KM21 (1% of wild-type levels) (data not shown). Thus, single-copy λ red in place of recBCD in the E. coli chromosome suppresses many of the defects exhibited by the ΔrecBCD strain, making hosts containing the ΔrecBCD Plac-red allele easy to work with for strain construction.
FIG. 6.
UV resistance of cells containing the ΔrecBCD::Plac-red allele. AB1157 (wild type; circles), KM21(ΔrecBCD; triangles), and KM22 (ΔrecBCD::Plac-red; squares) were grown to 2 × 108 cells/ml, spun down, resuspended in minimal medium, and kept on ice. Cells were diluted in minimal medium, spread on LB plates, exposed to the indicated doses of UV, and grown overnight in the dark. KM22 was grown and plated in the presence (closed squares) and absence (open squares) of 1 mM IPTG. Unirradiated plates were used to determine the total number of cells plated.
Gene replacement with a PCR product.
The DNA from one of the PCRs used to verify the structure of the lacZ::tet replacement into KM22 described above (containing ∼1 to 2 μg of DNA) was purified by phenol extraction, concentrated by ethanol precipitation, and electroporated into KM22. This fragment has the tetracycline resistance gene from pBR322 (1.4 kb) flanked by 1,054 and 918 bp of lacZ sequence. Following electroporation of a portion of this PCR-generated DNA sample into KM22, white tetracycline-resistant transformants were generated at a frequency of 6.5 × 10−5 per survivor. No transformants were generated when an equal amount of the purified PCR product was electroporated into a recBC sbcBC strain. Thus, the higher efficiency of the ΔrecBCD::Plac-red substitution, relative to recBC sbcBC, allows greater success of linear transformation with DNA fragments generated by PCR, showing that gene replacement can be performed in E. coli without prior cloning of the gene of interest.
DISCUSSION
It is known that the bacteriophage λ recombination system Red, in the absence of E. coli’s RecBCD activity, can act on linear DNA substrates to promote general homologous recombination. Given the break-join mechanism proposed for λ Red-mediated recombination (40), as well as the absence of any hotspot requirements, it seemed likely that λ Red would be well suited for the promotion of gene replacement in E. coli. This report shows that λ Red, together with λ’s anti-RecBCD function Gam, when produced from a plasmid, is capable of promoting high levels of recombination between linear DNA fragment and the E. coli chromosome. What is noteworthy is that wild-type cells expressing λ Red and Gam, or cells containing the ΔrecBCD::Plac-red substitution, consistently showed higher levels of recombinants per survivor than recBC sbcBC or recD strains, hosts typically used for gene replacement in E. coli, when cells were transformed with linear DNA containing ∼2,000 bp of homology. The levels of λ Red-promoted recombination varied between 15- and 130-fold greater than that seen with the recBC sbcBC strain, depending on the amount of DNA electroporated, the extent of flanking homology, and how the λ Red functions were supplied.
With the linear DNA lacZ substrates containing about 1,000 bp of homology on each end, wild-type cells expressing plasmid-encoded λ Red and Gam were more efficient at linear DNA transformation than a recBC sbcBC strain, especially with lower concentrations of DNA (Table 3, experiment A). This result may reflect the ability of λ Red to work more efficiently, possibly because of the higher copy number of Exo and Bet relative to whatever functions act to initiate recombination events via the RecF pathway. Alternatively, the higher efficiency of λ Red functions may reflect a higher affinity for DNA ends or greater catalytic activity relative to the comparable functions of the RecF pathway. The higher efficiency of the λ Red system (and possibly other phage recombination systems) may reflect the circumstances of phage biology, where recombination may be restricted to small regions of homology (e.g., terminal redundancy) and has to take place within a short interval of the life cycle right after infection or just prior to packaging.
Regions other than lacZ can act as a substrate for λ Red-promoted recombination. A ΔrecC::kan fragment containing 2,400 and 2,700 bp of homologous E. coli DNA sequences was electroporated both into wild-type cells expressing plasmid-encoded Red and Gam and into recBC sbcBC cells. In this case, λ Red was as good as, but not better than, the RecF pathway in promoting gene replacement. However, in another experiment, a wild-type strain expressing λ Red was 100 times more efficient than a recD strain in generating linear transformation leading to deletion of the nei gene (endonuclease 8) in E. coli (45). In this case, flanking homology was ∼2,500 bp on one side and ∼3,500 bp on the other. DNA dose-response curves for both the λ Red and RecF-promoted linear DNA transformation should reveal if λ Red’s higher efficiency is dependent on the levels of DNA, the length of homologous DNA, or both.
In an effort to increase the rate of linear transformation, the λ Red functions were moved from the plasmid into the chromosome. It was speculated that since λ Red promotes rolling-circle replication of plasmids, perhaps the linear multimers compete with λ Bet and Exo for electroporated substrates. I found that chromosomally encoded λ Red promoted recombination 63-fold higher than in a recBC sbcBC strain, a 4.2-fold increase in the relative rates of linear transformation compared to plasmid-encoded λ Red. This result is interpreted as follows: the decrease in copy number of λ Red is more than compensated for by the absence of competitive plasmid multimers, resulting in a higher rate of linear DNA transformation when λ Red is encoded by the chromosome rather than a plasmid. Alternatively (or in addition), the increase may reflect the inability of Gam to totally inactivate RecBCD, as has been observed previously during conjugational recombination (27). This latter possibility can be answered by placing both Red and Gam in a chromosomal region other than recBCD.
A recent study taking advantage of the RecBCD pathway of recombination used Chi sites properly situated on each side of a 6.5-kb linear DNA fragment to promote gene replacement in E. coli (7). The advantage of this system is that it uses the RecBCD pathway of recombination, which does not require any modification of (or addition to) the host functions. Thus, Chi-promoted gene replacement may be applicable to other bacteria, if their respective Chi hotspot sequences can be identified, as has been done with Lactococcus lactis (4). The disadvantages of Chi-promoted recombination is the requirement for specific sequences in the linear DNA substrate, as well as the relatively low frequency of gene replacement (2 × 10−5 to 7 × 10−5). It is noteworthy that the frequencies of λ Red-promoted gene replacement in wild-type hosts reported here were 2- to 10-fold higher than those reported for RecBCD, even though the λ Red substrates used here contained shorter regions of homology relative to the substrate used in the RecBCD study (2.1 kb of homology versus 3 kb). The advantage of λ Red-promoted gene replacement was highlighted here by the ability to transform E. coli with a PCR-generated fragment containing only about 1,000 bp of homology on each end; a recBC sbcBC strain was not transformed with this substrate. Thus, by prudent selection of PCR primers and protocols, one is likely to be able to construct any type of gene replacement, insertion, or deletion desirable.
Assuming that the lacZ::kan fragment and its parent plasmid pKM131 are transformed with equal efficiency, the highest replacement frequency per competent cell reported here was 3.3 × 10−3 (transformation of recJ hosts bearing λ Red-Gam-producing plasmid [Table 4]). This replacement frequency is about 100-fold less than that observed with conjugation recombination. It is possible that this frequency can be driven even higher by increased expression of λ Red in vivo and/or by electroporation with greater amounts of linear DNA substrates (unpublished data).
Any E. coli strain transduced with the ΔrecBCD Plac-red substitution should be proficient for gene replacement following electroporation with linear DNA substrates; the exceptions, of course, are those containing mutations in genes involved in assisting λ Red-promoted recombination (e.g., recA). In strains intolerant of a ΔrecBCD allele (e.g., polA and dam strains), induction of λ Red-promoted recombination has the potential to suppress lethalities. KM22 (ΔrecBCD Plac-red) is fully viable and grows well even in the absence of IPTG, though the circular plasmid transformation rate in this strain is 10-fold lower than in wild-type cells (a result likely due to plasmid linear multimerization, as seen in recBC sbcBC strains). If necessary, the replaced allele should be moved to a background of choice. In cases where the host lacks an efficient system of backcrossing, one could use pTP223 to transiently generate proficiency for gene replacement, followed by plating on fusaric acid plates to select for loss of the plasmid following nonselective growth (18). Finally, it is possible that plasmid-encoded λ Red and Gam would stimulate gene replacement in other bacteria as well, if the λ Gam function is active at inhibiting the hosts’ RecBCD enzyme. To this end, anti-RecBCD functions from phages other than those that infect E. coli, such as the abc function from phage P22 (27), may be useful for gene replacement in other bacterial species.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grant AI18234 and a small grant project (105061) from the University of Massachusetts Medical Center.
I thank Anthony Poteete, Anita Fenton, and Milan Jucovic for comments on the manuscript. I thank Jon Goguen for construction and advice on use of the electroporation apparatus.
REFERENCES
- 1.Bassett C L, Kushner S R. Exonucleases I, III, and V are required for stability of ColE1-related plasmids in Escherichia coli. J Bacteriol. 1984;157:661–664. doi: 10.1128/jb.157.2.661-664.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biek D P, Cohen S N. Identification and characterization of recD, a gene affecting plasmid maintenance and recombination in Escherichia coli. J Bacteriol. 1986;167:594–603. doi: 10.1128/jb.167.2.594-603.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biswas I, Gruss A, Ehrlich S D, Maguin E. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J Bacteriol. 1993;175:3628–3635. doi: 10.1128/jb.175.11.3628-3635.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Biswas I, Maguin E, Ehrlich S D, Gruss A. A 7-base-pair sequence protects DNA from exonucleolytic degradation in Lactococcus lactis. Proc Natl Acad Sci USA. 1995;92:2244–2248. doi: 10.1073/pnas.92.6.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brent R, Ptashne M. Mechanism of action of the lexA gene product. Proc Natl Acad Sci USA. 1981;78:4202–4208. doi: 10.1073/pnas.78.7.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carter D M, Radding C M. Role of exonuclease and β protein of phage lambda in genetic recombination. II. Substrate specificity and the mode of action of lambda exonuclease. J Biol Chem. 1971;246:2502–2510. [PubMed] [Google Scholar]
- 7.Dabert P, Smith G R. Gene replacement with linear DNA fragments in wild type Escherichia coli: enhancement by chi sites. Genetics. 1997;145:877–889. doi: 10.1093/genetics/145.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dykstra C C, Prasher D, Kushner S R. Physical and biochemical analysis of the cloned recB and recC genes of Escherichia coli K-12. J Bacteriol. 1984;157:21–27. doi: 10.1128/jb.157.1.21-27.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fenton A C, Poteete A R. Genetic analysis of the erf region of the bacteriophage P22 chromosome. Virology. 1984;134:148–160. doi: 10.1016/0042-6822(84)90280-0. [DOI] [PubMed] [Google Scholar]
- 10.Gutterson N I, Koshland D E. Replacement and amplification of bacterial genes with sequences altered in vitro. Proc Natl Acad Sci USA. 1983;80:4894–4898. doi: 10.1073/pnas.80.16.4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamilton C M, Aldea M, Washburn B K, Babitzke P, Kushner S R. New method for generating deletions and gene replacements in Escherichia coli. J Bacteriol. 1989;171:4617–4622. doi: 10.1128/jb.171.9.4617-4622.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansen J N. Use of solubilized acrylamide gels for the use of DNA fragments suitable for sequence analysis. Anal Biochem. 1981;116:146–151. doi: 10.1016/0003-2697(81)90337-7. [DOI] [PubMed] [Google Scholar]
- 13.Jasin M, Schimmel P. Deletions of an essential gene in Escherichia coli by site-specific recombination with linear DNA fragments. J Bacteriol. 1984;159:783–786. doi: 10.1128/jb.159.2.783-786.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karu A E, Sakaki Y, Echols H, Lynn S. The γ protein specified by bacteriophage λ. J Biol Chem. 1975;250:7377–7387. [PubMed] [Google Scholar]
- 15.Kmiec E, Holloman W K. β protein of bacteriophage λ promotes renaturation of DNA. J Biol Chem. 1981;256:12636–12639. [PubMed] [Google Scholar]
- 16.Leenhouts K, Buist G, Bolhuis A, tenBerge A, Kiel J, Mierau I, Dabrowska M, Venema G, Kok J. A general system for generating unlabelled gene replacements in bacterial chromosomes. Mol Gen Genet. 1996;253:217–224. doi: 10.1007/s004380050315. [DOI] [PubMed] [Google Scholar]
- 17.Little J W. An exonuclease induced by bacteriophage λ. II. Nature of the enzymic reaction. J Biol Chem. 1967;242:679. [PubMed] [Google Scholar]
- 18.Maloy S R, Nunn W D. Selection for loss of tetracycline resistance by Escherichia coli. J Bacteriol. 1981;145:1110–1112. doi: 10.1128/jb.145.2.1110-1111.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marinus M G, Carraway M, Frey A Z, Brown L, Arraj J A. Insertion mutations in the dam gene of Escherichia coli K-12. Mol Gen Genet. 1983;191:288–289. doi: 10.1007/BF00327681. [DOI] [PubMed] [Google Scholar]
- 20.Mauer R, Meyers B J, Ptashne M. Gene regulation at the right operator (OR) of bacteriophage λ. I. OR3 and autogenous control by repressor. J Mol Biol. 1980;139:147–161. doi: 10.1016/0022-2836(80)90302-2. [DOI] [PubMed] [Google Scholar]
- 21.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 22.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muniyappa K, Radding C M. The homologous recombination system of phage λ: pairing activities of β protein. J Biol Chem. 1986;261:7472–7478. [PubMed] [Google Scholar]
- 24.Muniyappa K, Shaner S L, Tang S S, Radding C M. Mechanism of the concerted action of RecA protein and helix destabilizing proteins in homologous recombination. Proc Natl Acad Sci USA. 1984;81:2757–2761. doi: 10.1073/pnas.81.9.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy K C. λ Gam protein inhibits the helicase and χ-stimulated recombination activities of Escherichia coli RecBCD enzyme. J Bacteriol. 1991;173:5808–5821. doi: 10.1128/jb.173.18.5808-5821.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Murphy K C. Biochemical characterization of P22 phage-modified Escherichia coli RecBCD enzyme. J Biol Chem. 1994;269:22507–22516. [PubMed] [Google Scholar]
- 27.Murphy K C, Lewis L J. Properties of Escherichia coli expressing bacteriophage P22 Abc (anti-RecBCD) proteins, including inhibition of Chi activity. J Bacteriol. 1993;175:1756–1766. doi: 10.1128/jb.175.6.1756-1766.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poteete A R, Fenton A C. Lambda red-dependent growth and recombination of phage P22. Virology. 1984;134:161–167. doi: 10.1016/0042-6822(84)90281-2. [DOI] [PubMed] [Google Scholar]
- 29.Poteete A R, Fenton A C. Efficient double-strand break-stimulated recombination promoted by the general recombination systems of phages λ and P22. Genetics. 1993;134:1013–1021. doi: 10.1093/genetics/134.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poteete A R, Fenton A C, Murphy K C. Modulation of Escherichia coli RecBCD activity by the bacteriophage λ Gam and P22 Abc functions. J Bacteriol. 1988;170:2012–2021. doi: 10.1128/jb.170.5.2012-2021.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Poteete A R, Fenton A C, Semerjian A V. Bacteriophage P22 accessory recombination function. Virology. 1991;182:316–323. doi: 10.1016/0042-6822(91)90675-2. [DOI] [PubMed] [Google Scholar]
- 32.Poteete A R, Volkert M R. Activation of recF-dependent recombination in Escherichia coli by bacteriophage λ and P22-encoded functions. J Bacteriol. 1988;170:4379–4381. doi: 10.1128/jb.170.9.4379-4381.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ream L W, Crisona N J, Clark A J. ColE1 plasmid stability in Exol− and ExoV− strains of Escherichia coli K-12. In: Schlessinger D, editor. Microbiology—1978. Washington, D.C: American Society for Microbiology; 1978. pp. 78–80. [Google Scholar]
- 34.Russell C B, Thaler D S, Dahlquist F W. Chromosomal transformation of Escherichia coli recD strains with linearized plasmids. J Bacteriol. 1989;171:2609–2613. doi: 10.1128/jb.171.5.2609-2613.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruther U. Construction and properties of a new cloning vehicle, allowing direct screening for recombinant plasmids. Mol Gen Genet. 1980;178:475–477. doi: 10.1007/BF00270503. [DOI] [PubMed] [Google Scholar]
- 36.Saarilahti H T, Palva E T. In vivo transfer of chromosomal mutations onto multicopy plasmids utilizing polA strains: cloning of an ompR2 mutation in Escherichia coli K-12. FEMS Microbiol Lett. 1985;26:27–33. [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Slater S, Maurer R. Simple phage-based system for generating allele replacements in Escherichia coli. J Bacteriol. 1993;175:4260–4262. doi: 10.1128/jb.175.13.4260-4262.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stahl F W, Crasemann J M, Stahl M M. Rec-mediated recombinational hot spot activity in bacteriophage lambda. III. Chi mutations are site-mutations stimulating rec-mediated recombination. J Mol Biol. 1975;94:203–212. doi: 10.1016/0022-2836(75)90078-9. [DOI] [PubMed] [Google Scholar]
- 40.Stahl F W, Fox M S, Faulds D, Stahl M M. Break-join recombination in phage λ. Genetics. 1990;125:463–474. doi: 10.1093/genetics/125.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stahl F W, Kobayashi I, Stahl M M. In phage λ, cos is a recombinator in the Red pathway. J Mol Biol. 1985;181:199–209. doi: 10.1016/0022-2836(85)90085-3. [DOI] [PubMed] [Google Scholar]
- 42.Stahl F W, Stahl M M. A role for recBC nuclease in the distribution of crossovers along unreplicated chromosomes of phage λ. Mol Gen Genet. 1974;131:27–30. doi: 10.1007/BF00269384. [DOI] [PubMed] [Google Scholar]
- 43.Stahl F W, Stahl M M, Malone R E. Red-mediated recombination of phage lambda in a recA−recB− host. Mol Gen Genet. 1978;159:207–211. [Google Scholar]
- 44.Thaler D S, Stahl M M, Stahl F W. Double-chain-cut sites are recombination hotspots in the Red pathway of phage λ. J Mol Biol. 1987;195:75–87. doi: 10.1016/0022-2836(87)90328-7. [DOI] [PubMed] [Google Scholar]
- 45.Volkert, M. Personal communication.
- 46.Winans S C, Elledge S J, Krueger J H, Walker G C. Site-directed insertion and deletion mutagenesis with cloned fragments in Escherichia coli. J Bacteriol. 1985;161:1219–1221. doi: 10.1128/jb.161.3.1219-1221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]