Abstract
The metabolism of aliphatic epoxides (epoxyalkanes) by the alkene-utilizing actinomycete Nocardia corallina B276 was investigated. Suspensions of N. corallina cells grown with propylene as the carbon source readily degraded propylene and epoxypropane, while suspensions of glucose-grown cells did not. The addition of propylene and epoxypropane to glucose-grown cells resulted in a time-dependent increase in propylene- and epoxypropane-degrading activities that was prevented by the addition of rifampin and chloramphenicol. The expression of alkene- and epoxide-degrading activities was correlated with the high-level expression of several polypeptides not present in extracts of glucose-grown cells. Epoxypropane and epoxybutane degradation by propylene-grown cell suspensions of N. corallina was stimulated by the addition of CO2 and inhibited by the depletion of CO2. Cell extracts catalyzed the carboxylation of epoxypropane to form acetoacetate in a reaction that was dependent on the addition of CO2, NAD+, and a reductant (NADPH or dithiothreitol). In the absence of CO2, epoxypropane was isomerized by cell extracts to form acetone at a rate approximately 10-fold lower than the rate of epoxypropane carboxylation. Methylepoxypropane was found to be a time-dependent, irreversible inactivator of epoxyalkane-degrading activity. These properties demonstrate that epoxyalkane metabolism in N. corallina occurs by a carboxylation reaction forming β-keto acids as products and provide evidence for the involvement in this reaction of an epoxide carboxylase with properties and cofactor requirements similar to those of the four-component epoxide carboxylase enzyme system of the gram-negative bacterium Xanthobacter strain Py2 (J. R. Allen and S. A. Ensign, J. Biol. Chem. 272:32121–32128, 1997). The addition of epoxide carboxylase component I from Xanthobacter strain Py2 to methylepoxypropane-inactivated N. corallina extracts restored epoxide carboxylase activity, and the addition of epoxide carboxylase component II from Xanthobacter Py2 to active N. corallina extracts stimulated epoxide isomerase rates to the same levels observed with the purified Xanthobacter system. Antibodies raised against Xanthobacter strain Py2 epoxide carboxylase component I cross-reacted with a polypeptide in propylene-grown N. corallina extracts with the same molecular weight as component I but did not cross-react with glucose-grown extracts. Together, these results suggest a common pathway of epoxyalkane metabolism for phylogenetically distinct bacteria that involves CO2 fixation and the activity of a multicomponent epoxide carboxylase enzyme system.
Several bacteria, including some strains of Mycobacterium, Xanthobacter, and Nocardia species, are capable of growth with aliphatic alkenes as carbon and energy sources (8, 9, 20). The pathway for alkene metabolism in these organisms involves an initial monooxygenase-catalyzed reaction producing epoxide intermediates, as illustrated for the substrate propylene and the product epoxypropane in equation 1:
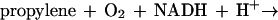 |
1 |
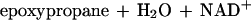 |
While the monooxygenase-catalyzed epoxidation of aliphatic alkenes is a well-characterized reaction (13, 17), less is known about the subsequent metabolism of the epoxide intermediates thus formed. The only characterized pathway for an aliphatic-alkene-utilizing bacterium is that of Xanthobacter strain Py2. In this bacterium, epoxides are further metabolized via a novel ring-opening and carboxylation reaction that requires CO2 as a cosubstrate and forms β-keto acids as products (1, 18). Epoxide carboxylase, the enzyme catalyzing this reaction, requires an oxidant (NAD+) and a reductant (NADPH or a dithiol) (21), which are reduced and oxidized in the course of epoxide carboxylation as illustrated in equation 2 (3):
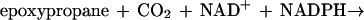 |
2 |
 |
The epoxide carboxylase complex from Xanthobacter strain Py2 has recently been purified and found to consist of four separate proteins (3): component I, a homohexameric protein (41.7-kDa subunits) that is believed to contain the epoxide binding and/or activation site(s); component II, a dimeric flavoprotein (57-kDa subunits) identified as an NADPH:disulfide oxidoreductase which is proposed to generate a reduced thiol(s) essential for catalysis (19); component III, a dimeric protein (26- and 26.2-kDa subunits) with an unidentified catalytic role; and component IV, a homodimeric protein (25.4-kDa subunits) which also has an unidentified catalytic role (3). The mechanistic details involving the interplay of the above-mentioned protein components and cofactors of this complex enzyme have not been elucidated. A preliminary mechanism involving a reductive epoxide ring-opening step, which is followed by an NAD+-dependent oxidation step prior to product release, has been proposed (21). Interestingly, in the absence of CO2, epoxide carboxylase catalyzes the isomerization of aliphatic epoxides to form ketones, although this reaction is apparently of no physiological significance (15, 16). This isomerase activity is, however, relevant to the catalytic mechanism since it demonstrates that a reaction intermediate can alternatively undergo carboxylation to form a β-keto acid or protonation to form a ketone.
Epoxide carboxylation represents a new and novel strategy for biological epoxide activation and transformations. To date, it is the only strategy that has been identified for aliphatic epoxide metabolism by the class of bacteria that grow with short-chain aliphatic alkenes as carbon and energy sources. The question of whether epoxide carboxylation is the predominant strategy used to metabolize epoxides in aliphatic-alkene-utilizing bacteria or whether other characterized transformations (i.e., isomerization [10, 14] or hydration [5, 11]) are utilized as well has been raised. One organism that may aid in addressing this question is the gram-positive bacterium Nocardia corallina B276. This bacterium, isolated with propylene as the source of carbon and energy (8), has been reported to convert 1-alkenes ranging from C2 to C18 in chain length to 1-epoxides, as well as to convert styrene to styrene oxide (7). The alkene monooxygenase which catalyzes these reactions has recently been isolated and shown to be a multiprotein enzyme system (13). However, the pathway(s) by which epoxides are further metabolized in N. corallina B276 has not been studied.
In the present paper, evidence that epoxides are metabolized in a CO2-dependent manner in N. corallina, similar to that described for Xanthobacter strain Py2, is presented. The reaction requirements and enzymes involved in epoxide carboxylation in N. corallina B276 are described and compared to those of the epoxide carboxylase reaction of Xanthobacter strain Py2. The similarities and differences between these two epoxide carboxylation systems are of interest in the ongoing effort to gain insight into microbial strategies for the metabolism of epoxides.
MATERIALS AND METHODS
Chemicals.
Tris(hydroxymethyl)aminomethane (Tris) buffer was purchased from Sigma Chemical Co. Horseradish peroxidase color development reagent and a goat anti-rabbit immunoglobulin G-horseradish peroxidase conjugate were purchased from Bio-Rad. Epoxypropane (99% minimum) and 1,2-epoxybutane (99% minimum) were purchased from Aldrich Chemical Co. Methylepoxypropane was purchased from Lancaster Laboratories.
Growth of bacteria and preparation of cell suspensions.
Cultures of N. corallina B276 (ATCC 31338) were grown in sealed 4-liter shake flasks containing 0.8 liter of mineral salts medium (1) with propylene (10% [vol/vol] gas phase) or glucose (10 g/liter) as the carbon source. Cultures were harvested at an A600 of between 1.0 and 3.0 by centrifugation and washed in buffer (50 mM potassium phosphate, pH 7.2) as described previously (1). Cultures of Xanthobacter strain Py2 were grown on propylene (10% [vol/vol] gas phase) or glucose (10 g/liter) as the carbon source and harvested as previously described (1). Cell pastes were stored at −80°C.
Assay of epoxide degradation activity in whole-cell suspensions.
Assays of epoxyalkane degradation by whole-cell suspensions of N. corallina were performed with shaking at 30°C in sealed 9-ml serum vials containing cells, substrate (2 μmol), and buffer (50 mM potassium phosphate, pH 7.2) in a total volume of 1 ml (18). For whole-cell assays performed in the absence of CO2, residual CO2 was removed by sparging buffers and flushing sealed vials with CO2-free nitrogen and including a KOH-saturated filter trap in each of the vials.
Preparation of cell extracts and purified epoxide carboxylase components.
Frozen cell paste (15 to 30 g) of N. corallina or Xanthobacter strain Py2 was thawed and resuspended in 2 volumes of buffer (50 mM Tris-HCl [pH 8.2] containing 1 mM dithiothreitol [DTT], 10% [vol/vol] glycerol, DNase I [0.2 mg/ml], and lysozyme [0.3 mg/ml]). The cell suspension was passed four times through a French pressure cell at 110,000 kDa, and the lysate was clarified by centrifugation at 137,000 × g for 30 min at 4°C. After removal of cell debris, the supernatant was dialyzed for 16 h at 4°C against buffer (50 mM Tris-HCl [pH 8.2] containing 10% [vol/vol] glycerol). The dialysate was stored at −80°C and used as the source of enzyme. Methylepoxypropane-treated cell extracts of N. corallina and Xanthobacter strain Py2 were prepared by a method described previously (2). Purified epoxide carboxylase components I to IV from Xanthobacter strain Py2 were prepared as previously described (2, 3).
Assay of epoxide carboxylase activity in cell extracts.
Epoxide carboxylase activity was measured by monitoring the time-dependent depletion of epoxypropane by gas chromatography as described previously (1). Assays were performed in sealed 9-ml serum vials containing a source of enzyme (cell extract alone or cell extract plus purified epoxide carboxylase components), in 50 mM Tris-HCl (pH 8.2) containing 10% glycerol, using reagents and reaction conditions described previously (1, 2). For assays performed in the absence of CO2, CO2 was removed as described above. Acetoacetate, acetone, and 1,2-epoxybutane were quantified by gas chromatography as previously described (1). Products of epoxyalkane carboxylation were also identified by using 13C-enriched NaHCO3 in the assay mix and analyzing the reaction products by proton-decoupled 13C nuclear magnetic resonance spectrometry (NMR) as described previously (1).
Induction of epoxypropane-degrading activity in batch cultures.
N. corallina cells which had been grown for several generations with either glucose or propylene as the carbon source were used for induction experiments as previously described (6).
SDS-PAGE and immunoblotting procedures.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (12% total gel; 2.7% cross-linker running gel) was performed in a Mini-Protean II apparatus (Bio-Rad) by the Laemmli procedure (12). Electrophoresed proteins were visualized by Coomassie blue staining. Apparent molecular masses of polypeptides were determined by comparison with the Rf values for molecular mass standard proteins. The standards were bovine serum albumin (66.2 kDa), ovalbumin (45 kDa), carbonic anhydrase (31 kDa), and cytochrome c (12.3 kDa). Immunoblot analysis was conducted as described previously, using a polyclonal antiserum raised against Xanthobacter strain Py2 epoxide carboxylase component I or epoxide carboxylase component II (2).
Protein determination.
Protein concentrations were determined by a modified biuret assay with bovine serum albumin as the standard (4). The protein concentration of epoxide carboxylase component II from Xanthobacter strain Py2 was routinely determined by using its reported extinction coefficient (2).
RESULTS
CO2-dependent epoxide degradation in whole-cell suspensions of N. corallina B276.
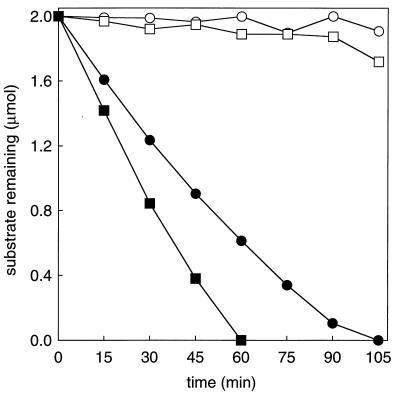
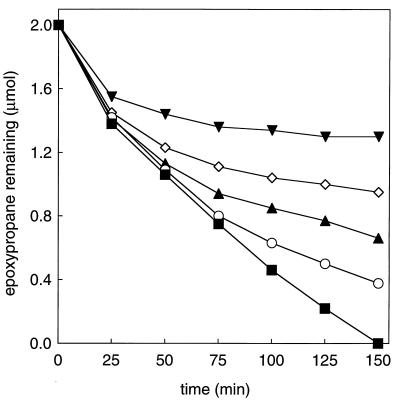
While the oxidation of aliphatic alkenes to epoxides is a well-characterized reaction in N. corallina (8, 13), the pathway by which epoxides are further converted to central metabolites has not been investigated. As an initial step in characterizing the epoxide-converting reaction, the degradation of two aliphatic epoxides, epoxypropane and 1,2-epoxybutane, was investigated using whole-cell suspensions of propylene-grown N. corallina. As shown in Fig. 1, both epoxides were degraded in the presence of CO2 and bicarbonate but not in their absence. These results suggest a role for CO2 as a cosubstrate in epoxide degradation similar to that identified in Xanthobacter strain Py2; i.e., aliphatic epoxides are carboxylated to form the corresponding β-keto acids. Interestingly, in the absence of CO2, cell suspensions and cell extracts of propylene-grown Xanthobacter cells catalyze the isomerization of aliphatic epoxides to form the corresponding ketones (acetone in the case of epoxypropane isomerization; methyl ethyl ketone in the case of 1,2-epoxybutane isomerization) at rates approximately 40% lower than those observed for epoxide carboxylation (1, 18). This epoxide isomerization has been proposed to be a fortuitous reaction of no physiological importance (16). To determine whether a similar CO2-dependent isomerization of epoxides occurs in N. corallina, the rates of epoxypropane and 1,2-epoxybutane degradation were measured in the absence of CO2. As shown in Fig. 1, no significant degradation of either substrate was observed during the time frame of these assays. These results suggest either that the epoxide-converting enzyme of N. corallina does not catalyze epoxide to ketone isomerization or that the rate of isomerization, relative to that of carboxylation, is much lower than that in the corresponding Xanthobacter enzyme system.
FIG. 1.
Requirement of CO2 for epoxyalkane degradation by propylene-grown N. corallina. Assays were performed with whole-cell suspensions (0.5 mg of protein). Closed symbols, assays performed with CO2 and NaHCO3 (50 mM combined concentration); open symbols, assays performed without CO2 and NaHCO3; squares, epoxypropane remaining; circles, 1,2-epoxybutane remaining.
Carboxylation of epoxides to β-keto acids in cell extracts of N. corallina.
As mentioned in the introduction and shown in equation 2, epoxide carboxylase from Xanthobacter strain Py2 couples the carboxylation of epoxyalkanes to the transhydrogenation of NAD+ and NADPH. While NADPH is believed to be the physiological reductant for the reaction, dithiols (e.g., DTT) are capable of substituting for NADPH. With these requirements in mind, the in vitro degradation of epoxypropane and epoxybutane in cell extracts of N. corallina was investigated. As observed for extracts of Xanthobacter strain Py2 (1, 21), a time-dependent consumption of epoxypropane or 1,2-epoxybutane was observed in assay mixtures that contained CO2, NAD+, and either NADPH or DTT. In the absence of both NADPH (or DTT) and NAD+, or in the individual absence of either reductant or oxidant, the rates of epoxyalkane degradation were approximately five- to sevenfold lower at the outset of the assays and decreased during the course of the assays. These results suggest that epoxide degradation in N. corallina is coupled to the transhydrogenation of NAD+ and NADPH, as has been previously shown to occur for the epoxide carboxylase system of Xanthobacter strain Py2 (3). To identify the products of epoxyalkane degradation, assays were performed with cell extract in the presence of NaH13CO3 and the reaction products were analyzed by 13C NMR. The 13C NMR spectrum of the reaction products formed on complete consumption of epoxypropane and 1,2-epoxybutane showed resonance peaks with chemical shifts identical to the C-1 (carboxyl) carbon atoms of acetoacetate and 3-keto-pentanoic acid, respectively (data not shown).
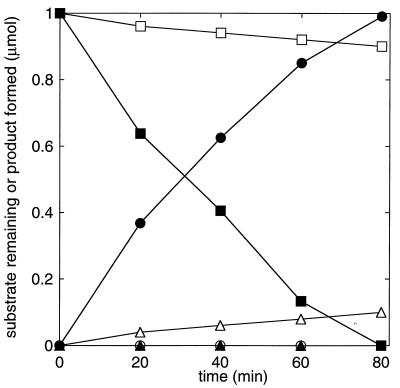
As mentioned above, in the absence of CO2, cell suspensions and extracts of propylene-grown Xanthobacter strain Py2 catalyze the isomerization of epoxides to form ketones at rates approximately 1.5- to 2-fold lower than those observed for carboxylation (1, 18). Epoxide isomerization requires the same cofactors (NAD+ and either NADPH or DTT) as epoxide carboxylation. The effect of CO2 on epoxide degradation was investigated in cell extracts of N. corallina to determine whether any isomerase activity could be detected in vitro and, if so, how the rate of isomerization compared to the rate of carboxylation. As shown in Fig. 2, in the presence of CO2, the time course of epoxypropane consumption was stoichiometric with the time course of acetoacetate formation. In the absence of CO2, a 10-fold-lower rate of epoxypropane degradation was observed, and epoxypropane consumption correlated with the stoichiometric formation of acetone (Fig. 2). Similar results were obtained when NADPH, rather than DTT, was used as the reductant in the assays. These results demonstrate that the epoxide carboxylase of N. corallina is capable of catalyzing epoxide to ketone isomerization, albeit at a much slower rate, relative to epoxide carboxylation, than the epoxide carboxylase from Xanthobacter strain Py2.
FIG. 2.
Effect of CO2 on the time courses of epoxypropane degradation and acetoacetate formation catalyzed by cell extracts of N. corallina. Assays were performed with 7.5 mg of cell extract. Closed symbols, assays performed with CO2 and NaHCO3 (60 mM); open symbols, assays performed without CO2 and NaHCO3; squares, epoxypropane remaining; circles, acetoacetate formed; triangles, acetone formed.
Induction of epoxide-degrading activity in N. corallina and identification of inducible polypeptides.
The alkene monooxygenase and epoxide carboxylase enzymes in Xanthobacter strain Py2 are inducible proteins that are not expressed in cultures grown on other carbon sources, such as glucose or acetone (6, 16, 18). Induction of alkene monooxygenase and epoxide carboxylase activities by the addition of alkenes or epoxides to glucose-grown Xanthobacter strain Py2 results in the de novo synthesis of new polypeptides, several of which are expressed at high levels and are hence readily visible on SDS-PAGE gels of cell extracts (6). Several of these polypeptides represent components of the alkene monooxygenase and epoxide carboxylase enzymes. For example, the polypeptides constituting epoxide carboxylase components I (42 kDa) and II (57 kDa) and two subunits of the three-subunit alkene monooxygenase (the 43- and 53-kDa subunits) are readily visible in gels of cell extracts (2, 6, 17). To obtain more information about similarities and possible differences between the Xanthobacter and N. corallina epoxide carboxylases, we investigated whether the alkene- and epoxide-degrading enzymes of N. corallina were similarly induced and, if so, whether similar patterns of polypeptides could be observed in induced cells.
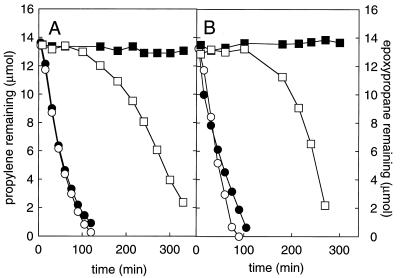
To investigate the inducible nature of the N. corallina system, batch cultures were grown with either glucose or propylene as the carbon source and then transferred to serum bottles to which propylene or epoxypropane was added. The time courses of substrate disappearance in the bottles were used as a measure of the presence and/or induction of alkene- and epoxide-degrading activities. As shown in Fig. 3, the bacterial cells from cultures grown with propylene as the carbon source rapidly degraded propylene (Fig. 3A) or epoxypropane (Fig. 3B) from the outset of the assays. The rates of propylene and epoxypropane degradation by these cells were not affected by the addition of rifampin and chloramphenicol. In contrast, the cells from cultures grown on glucose did not degrade propylene or epoxypropane to any noticeable degree within the first 100 min of incubation, during which time propylene-grown cells had consumed nearly all of the propylene or epoxypropane added to these assay mixtures (Fig. 3). After this initial lag period, the propylene and epoxypropane began to disappear, and the rates of propylene and epoxypropane depletion increased over time. In contrast, no significant propylene or epoxypropane degradation was observed when rifampin and chloramphenicol were added to the glucose-grown cells (Fig. 3). These data demonstrate that the alkene monooxygenase and epoxide carboxylase enzymes in N. corallina are repressed in cultures grown on glucose as the carbon source and induced by the addition of propylene or epoxypropane.
FIG. 3.
Requirement of new protein synthesis for the degradation of propylene and epoxypropane by N. corallina grown with glucose as the carbon source. Assays were conducted in 150-ml serum bottles containing 15 ml of glucose-grown cells (A600 = 1.47) or propylene-grown cells (A600 = 0.93) that had been transferred from 200-ml cultures grown in shake flasks. Closed symbols, assay bottles containing chloramphenicol (10 mg) and rifampin (5 mg); open symbols, assay bottles without chloramphenicol or rifampin present; circles, propylene-grown cells; squares, glucose-grown cells. (A) Propylene remaining; (B) epoxypropane remaining.
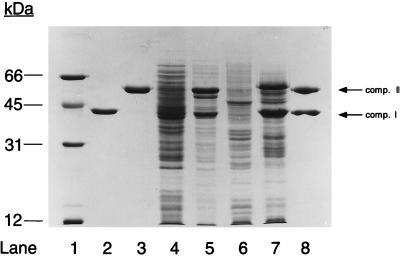
The polypeptide banding patterns of cell extracts prepared from glucose-grown (noninduced) and propylene-grown (induced) cultures of Xanthobacter strain Py2 and N. corallina are presented in Fig. 4. As observed for Xanthobacter strain Py2, unique polypeptides not visible in glucose-grown cells are readily visible in propylene-grown N. corallina cells. Presumably, some or all of these polypeptides represent subunits of the alkene monooxygenase and epoxide carboxylase enzymes. The alkene monooxygenase from N. corallina was recently purified and found to consist of three separate components: a monomeric reductase (40 kDa), a monomeric small effector protein (14 kDa), and a heterodimeric oxygenase (35- and 53-kDa subunits) (13). Based on the purification schemes for the three proteins, it can be concluded that the reductase and small protein are expressed at relatively low levels and would not be easily visualized in extracts run on SDS-PAGE gels (13). In contrast, the oxygenase would be expected to constitute approximately 3% of the soluble cell protein (13). Inducible polypeptides with apparent molecular masses of 35 and 53 kDa are present in the cell extract of propylene-grown N. corallina (Fig. 4); these polypeptides could conceivably represent the two oxygenase subunits. The most highly induced proteins in propylene-grown N. corallina extracts have apparent molecular masses of 42 and 59 kDa on SDS-PAGE (Fig. 4). Clearly, these polypeptides are not associated with the alkene monooxygenase and may be components of the epoxide carboxylase. The relative abundance and apparent molecular masses of these polypeptides are similar to those of components I (42 kDa on SDS-PAGE) and II (57 kDa on SDS-PAGE) of the epoxide carboxylase complex from Xanthobacter strain Py2 (Fig. 4).
FIG. 4.
Gel electrophoretic analysis of propylene-induced polypeptides in N. corallina and comparison to purified epoxide carboxylase components (comp.) I and II from Xanthobacter strain Py2. Lane 1, molecular mass standards (2 μg each); lane 2, epoxide carboxylase component I from Xanthobacter strain Py2 (3 μg); lane 3, epoxide carboxylase component II from Xanthobacter strain Py2 (3 μg); lane 4, glucose-grown cell extract from Xanthobacter strain Py2 (25 μg); lane 5, propylene-grown cell extract from Xanthobacter strain Py2 (25 μg); lane 6, glucose-grown cell extract from N. corallina (25 μg); lane 7, propylene-grown cell extract from N. corallina (25 μg); lane 8, epoxide carboxylase components I (3 μg) and II (3 μg) from Xanthobacter strain Py2.
Immunoblot analysis of glucose- and propylene-grown N. corallina cell extracts.
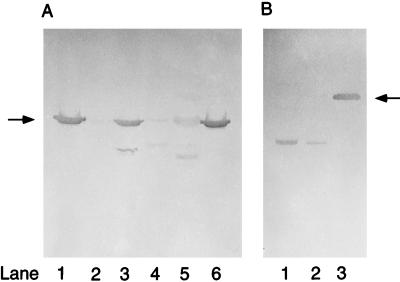
The polypeptides in glucose- and propylene-grown N. corallina cell extracts were analyzed by immunoblotting with polyclonal antibodies raised against purified epoxide carboxylase components I and II from Xanthobacter strain Py2 to determine whether there is any cross-antigenicity between these proteins. Antibodies raised against epoxide carboxylase component I from Xanthobacter strain Py2 cross-reacted weakly with a protein, present in cell extracts of propylene-grown N. corallina B276 cells, that migrated on SDS-PAGE with the same apparent molecular mass as the 42-kDa inducible polypeptide (Fig. 5A). Cross-reaction to a lesser degree was observed at this same position in cell extracts of glucose-grown N. corallina. Antibodies raised against purified epoxide carboxylase component II from Xanthobacter strain Py2 cross-reacted with a protein, present in both glucose- and propylene-grown N. corallina cell extracts, that migrated on SDS-PAGE with an apparent molecular mass of 37 kDa but did not cross-react with the 59-kDa inducible polypeptide (Fig. 5B).
FIG. 5.
Immunoblot analysis of glucose- and propylene-grown cell extracts of N. corallina. (A) Immunoblot prepared using antibodies raised against epoxide carboxylase component I from Xanthobacter strain Py2 as the probe. The arrow indicates the position of component I. Lanes 1 and 6, epoxide carboxylase component I from Xanthobacter strain Py2 (0.5 μg); lane 2, glucose-grown cell extract from Xanthobacter strain Py2 (15 μg); lane 3, propylene-grown cell extract from Xanthobacter strain Py2 (15 μg); lane 4, glucose-grown cell extract from N. corallina (15 μg); lane 5, propylene-grown cell extract from N. corallina (15 μg). (B) Immunoblot prepared using antibodies raised against epoxide carboxylase component II from Xanthobacter strain Py2 as the probe. The arrow indicates the position of component II. Lane 1, glucose-grown cell extract from N. corallina (15 μg); lane 2, propylene-grown cell extract from N. corallina (15 μg); lane 3, epoxide carboxylase component II from Xanthobacter strain Py2 (0.5 μg).
Inhibition of epoxide carboxylase activity in N. corallina by methylepoxypropane.
As mentioned in the introduction, epoxide carboxylase from Xanthobacter Py2 is a multiprotein enzyme complex consisting of four separate proteins that are required for epoxide carboxylation or isomerization (3). Since epoxide carboxylation in cell extracts of N. corallina has cofactor requirements identical to those of the Xanthobacter strain Py2 epoxide carboxylase, it would be reasonable to speculate that N. corallina epoxide carboxylase functions as a multiprotein complex as well. One mechanistic probe that might prove useful in addressing this question is the substrate analog methylepoxypropane, which has previously been characterized as a time-dependent, irreversible inactivator of epoxide carboxylase from Xanthobacter strain Py2 (2). Methylepoxypropane differs from epoxypropane in the presence of a methyl group rather than a hydrogen substituent on the C-2 carbon atom. Epoxide carboxylation (or isomerization) requires the abstraction of hydrogen, possibly as a hydride, from the C-2 carbon and the formation of a carbonyl carbon at this center (21). Methylepoxypropane is proposed to undergo a covalent reaction with the catalytic, active-site-containing component of epoxide carboxylase but, due to the lack of an abstractable hydride, not to react further to form a product (2). The addition of epoxide carboxylase component I, but not that of component II, III, or IV, to methylepoxypropane-inactivated cell extracts of Xanthobacter strain Py2 restored epoxide carboxylase activity, demonstrating that component I is the specific target of inactivation and suggesting that it contains the epoxide binding and/or activation site(s) (3).
The possibility that methylepoxypropane would serve as an inactivator of epoxide carboxylase activity in N. corallina was investigated by monitoring the degradation of epoxypropane in assay mixtures that contained various concentrations of methylepoxypropane. As shown in Fig. 6, the presence of methylepoxypropane resulted in the concentration- and time-dependent inhibition of epoxide carboxylase activity in N. corallina cell suspensions. No recovery of epoxide carboxylase activity was observed after removal of methylepoxypropane (data not shown), demonstrating that the inhibition is essentially irreversible.
FIG. 6.
Concentration- and time-dependent inactivation of epoxide carboxylase in whole-cell suspensions of N. corallina by methylepoxypropane. Each assay mixture contained cell suspension (0.35 mg of protein) and CO2 and NaHCO3 (50 mM total). Assays were initiated by the addition of 2 μmol of epoxypropane. Symbols: ▪, no methylepoxypropane; ○, 0.5 mM methylepoxypropane; ▴, 1 mM methylepoxypropane; ◊, 2 mM methylepoxypropane; ▾, 4 mM methylepoxypropane.
Restoration of epoxide carboxylase activity in methylepoxypropane-treated N. corallina cell extract by Xanthobacter strain Py2 component I.
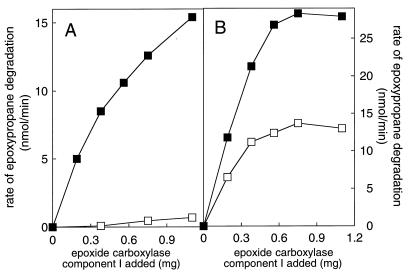
As shown in Fig. 7A, the addition of Xanthobacter epoxide carboxylase component I to methylepoxypropane-inactivated N. corallina cell extract restored epoxide carboxylase activity in a concentration-dependent fashion. The addition of Xanthobacter epoxide carboxylase component II, III, or IV up to the highest amount of component I added (1.1 mg) had no effect on activity (data not shown). Importantly, purified component I did not display epoxide carboxylase activity alone (in the absence of methylepoxypropane-inactivated N. corallina cell extract) or in the presence of N. corallina glucose-grown cell extracts (data not shown). These results demonstrate that the combination of Xanthobacter epoxide carboxylase component I with active N. corallina epoxide carboxylase components provides a heterologous system capable of catalyzing epoxide carboxylation. These results also demonstrate that an N. corallina protein with functions similar to that of Xanthobacter component I is the specific target of methylepoxypropane inactivation.
FIG. 7.
Restoration of epoxide carboxylase activity in methylepoxypropane-treated cell extracts of N. corallina by addition of epoxide carboxylase component I from Xanthobacter strain Py2. Symbols: ▪, assays performed with CO2 and NaHCO3 (60 mM); □, assays performed without CO2 and NaHCO3. (A) Assays of cell extract (4.8 mg) prepared from methylepoxypropane-treated N. corallina; (B) assays of cell extract (5.4 mg) prepared from methylepoxypropane-treated Xanthobacter strain Py2.
As mentioned earlier, there is a significant difference in the relative rates of epoxide carboxylation and isomerization catalyzed by the Xanthobacter (1.5- to 2-fold-lower isomerase rate) and N. corallina (10-fold-lower isomerase rate) epoxide carboxylases. Possibly, component I in Xanthobacter strain Py2 and the analogous methylepoxypropane-sensitive protein in N. corallina are the determinants in this rate difference. To investigate this, the rates of epoxypropane degradation in methylepoxypropane-treated N. corallina cell extracts complemented with Xanthobacter component I were measured in the absence of CO2. As shown in Fig. 7A, the addition of component I did not stimulate the rate of CO2-independent epoxide degradation (indicative of isomerase activity). As a control, the rate of epoxypropane degradation in assay mixtures containing the homologous combination of Xanthobacter component I and methylepoxypropane-inactivated Xanthobacter cell extract was measured in the absence and presence of CO2, and the expected relative ratio of isomerase activity to carboxylase activity was found (Fig. 7B). These results suggest that Xanthobacter component I is not the sole determinant in the ratio of isomerase and carboxylase activities catalyzed by the multiprotein enzyme.
Xanthobacter strain Py2 component II confers epoxide isomerase activity in cell extracts of N. corallina.
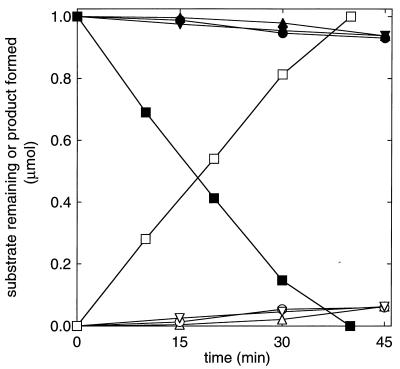
The difference in the relative ratios of epoxide isomerization and carboxylation catalyzed by the Xanthobacter and N. corallina epoxide carboxylase systems was further investigated by individually adding each of the four purified Xanthobacter components to active N. corallina extracts and measuring the resultant rates of epoxide isomerase activity observed in the absence of CO2. As shown in Fig. 8, the addition of component II resulted in a significantly higher rate of isomerase activity in the N. corallina cell extract. In contrast, the addition of component I, III, or IV did not stimulate isomerization to any noticeable degree.
FIG. 8.
Epoxide carboxylase component II from Xanthobacter strain Py2 confers epoxide isomerase activity in cell extracts of N. corallina. Assays were performed with cell extracts of propylene-grown N. corallina (7.5 mg) in the absence of CO2. Closed symbols, epoxypropane remaining; open symbols, acetone produced; inverted triangles, addition of epoxide carboxylase component I (0.8 mg); squares, addition of epoxide carboxylase component II (0.6 mg); circles, addition of epoxide carboxylase component III (0.5 mg); triangles, addition of epoxide carboxylase component IV (0.6 mg).
DISCUSSION
The results of the studies presented in this paper demonstrate that epoxide metabolism in N. corallina B276 proceeds by a CO2-dependent reaction that forms β-keto acids as products. This represents only the second example of biological epoxide carboxylation, the other being that of Xanthobacter strain Py2 (1, 18). These results demonstrate that epoxide carboxylation is not an isolated phenomenon and that it may, in fact, be the sole or predominant strategy for epoxide metabolism by the class of bacteria that grow with aliphatic alkenes as carbon and energy sources. N. corallina and Xanthobacter strain Py2 are phylogenetically distinct bacteria; N. corallina is a gram-positive actinomycete, while Xanthobacter strain Py2 is a gram-negative rod. The fact that two unrelated bacteria utilize the same pathway of aliphatic alkene and epoxide metabolism provides support for the idea that this is a fundamental microbial strategy for the utilization of these compounds.
The initial characterization of epoxide carboxylase from N. corallina suggests that it, like epoxide carboxylase from Xanthobacter strain Py2, functions as a multiprotein complex and requires an oxidant (NAD+) and a reductant (NADPH or DTT) as cofactors. In the purified epoxide carboxylase system from Xanthobacter strain Py2, the carboxylation of epoxides is coupled to the transhydrogenation of NAD+ and NADPH (equation 2). Presumably, epoxide carboxylation in N. corallina has the same reaction stoichiometry. These cofactor requirements, and the associated transhydrogenation, are unprecedented among the known carboxylases. The mechanism of epoxide carboxylation and the roles that the individual components play in the reaction are not well understood. Epoxide carboxylase component I from Xanthobacter strain Py2 has been proposed to play a key role in epoxide binding and activation, based on the specific inactivation of this component by the irreversible inactivator methylepoxypropane (2). Epoxide carboxylase component II has been proposed to function as an NADPH:disulfide oxidoreductase which generates a reduced thiol, possibly on component I, that serves as a nucleophile for attack on and ring opening of the epoxide substrate (19). It is unclear what role(s) epoxide carboxylase components III and IV play in the carboxylation reaction, but they may be involved in the reduction of NAD+ and/or the formation of the protein-protein complexes necessary for catalysis (3).
By analogy, proteins with functions similar to those of Xanthobacter epoxide carboxylase components I to IV are likely to be involved in epoxide carboxylation in N. corallina. Evidence for the involvement of a protein with component I activity was provided by the demonstration that Xanthobacter strain Py2 component I restored epoxide carboxylase activity when added to cell extracts of N. corallina that had been inactivated by methylepoxypropane (Fig. 7). Furthermore, induction of epoxide carboxylase activity in N. corallina led to the high-level expression of a polypeptide that migrated on SDS-PAGE with a molecular mass nearly identical to that of component I from Xanthobacter strain Py2 (Fig. 4) and that cross-reacts with polyclonal antibodies raised against component I. Together, these results suggest that a protein homologous to component I is required for epoxide carboxylation in N. corallina. A polypeptide with a molecular weight and level of expression similar to those of component II is present in cell extracts of propylene-grown N. corallina as well (Fig. 4), suggesting that this polypeptide may function in a role analogous to that of component II, i.e., in the oxidation of NADPH and the reduction of a disulfide. This polypeptide did not, however, cross-react to any detectable degree with polyclonal antibodies raised against Xanthobacter strain Py2 component II. At present, we have not obtained evidence that proteins analogous to components III and IV are present in N. corallina, although we suspect that such proteins are required. In Xanthobacter strain Py2, the levels of expression of components III and IV are significantly lower than those of components I and II, and they cannot be distinguished from the protein background on one-dimensional SDS-PAGE gels of cell extracts (3). Additional experiments, culminating with the purification and characterization of the necessary components from N. corallina, will be required to definitively define the proteins required by this multiprotein enzyme system.
While there are many apparent similarities between the epoxide carboxylase systems of Xanthobacter strain Py2 and N. corallina, there is one striking difference: the lower level of isomerase activity exhibited by N. corallina epoxide carboxylase when CO2 is excluded from the assays. While epoxide isomerase activity was detected in cell extracts of N. corallina, the rate of isomerization was significantly lower (approximately five- to sevenfold) than that observed with cell extracts or purified components from Xanthobacter strain Py2. In Xanthobacter strain Py2, epoxide-to-ketone isomerization is apparently a fortuitous reaction of no physiological importance (16). The observation that isomerization can occur is, however, significant since it demonstrates that a reaction intermediate that can alternatively undergo carboxylation (with CO2 present) or protonation (without CO2 present) is generated. Since component I has been implicated in epoxide binding and activation in the Xanthobacter system (2, 3), and since the addition of purified component I from Xanthobacter strain Py2 restored epoxide carboxylase activity to methylepoxypropane-inactivated N. corallina extracts (Fig. 7A), it seemed reasonable to predict that differences in the respective component I proteins might be the determinant in the observed differences in epoxide carboxylation and isomerization in the two systems. It was therefore surprising to find that the addition of purified component II (the NADPH:disulfide oxidoreductase) from Xanthobacter strain Py2 to N. corallina extracts conferred an increase in epoxide isomerase activity to the level observed for the Xanthobacter system while components I, III, and IV had no stimulatory effect (Fig. 8). At present we have no clear explanation for these observations. Possibly, component II, as well as component I, contains a portion of the epoxide binding and activation sites or accepts an intermediate formed at some point during the reaction from another protein component. Differences in the properties of the respective component II proteins could thus result in increased or decreased rates of protonation when CO2 is not present. The binding of component II to component I may also confer conformational changes that make the active site more or less accessible for proton abstraction from water or a general base. Once purified sources of the N. corallina epoxide carboxylase proteins are available, it will be interesting to compare their molecular properties to those of the Xanthobacter proteins and to investigate the ratios of carboxylase and isomerase activities when heterologous mixtures of the various components are combined. These studies should provide further insight into microbial strategies for the metabolism of xenobiotic compounds generated as intermediates in unsaturated-hydrocarbon metabolism.
ACKNOWLEDGMENT
This work was supported by National Institutes of Health grant GM51805.
REFERENCES
- 1.Allen J R, Ensign S A. Carboxylation of epoxides to β-keto acids in cell extracts of Xanthobacter strain Py2. J Bacteriol. 1996;178:1469–1472. doi: 10.1128/jb.178.5.1469-1472.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen J R, Ensign S A. Characterization of three protein components required for functional reconstitution of the epoxide carboxylase multienzyme complex from Xanthobacter strain Py2. J Bacteriol. 1997;179:3110–3115. doi: 10.1128/jb.179.10.3110-3115.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen J R, Ensign S A. Purification to homogeneity and reconstitution of the individual components of the epoxide carboxylase multiprotein enzyme complex from Xanthobacter strain Py2. J Biol Chem. 1997;272:32121–32128. doi: 10.1074/jbc.272.51.32121. [DOI] [PubMed] [Google Scholar]
- 4.Chromy V, Fischer J, Kulhanek V. Re-evaluation of EDTA-chelated biuret reagent. Clin Chem. 1974;20:1362–1363. [PubMed] [Google Scholar]
- 5.de Bont J A M, van Dijken J P, van Ginkel C G. The metabolism of 1,2-propanediol by the propylene oxide utilizing bacterium Nocardia A60. Biochim Biophys Acta. 1982;714:465–470. [Google Scholar]
- 6.Ensign S A. Aliphatic and chlorinated alkenes and epoxides as inducers of alkene monooxygenase and epoxidase activities in Xanthobacter strain Py2. Appl Environ Microbiol. 1996;62:61–66. doi: 10.1128/aem.62.1.61-66.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Furuhashi K, Shintani M, Takagi M. Effects of solvents on the production of epoxides by Nocardia corallina B-276. Appl Microbiol Biotechnol. 1986;23:218–223. [Google Scholar]
- 8.Furuhashi K, Taoka A, Uchida S, Karube I, Suzuki S. Production of 1,2-epoxyalkanes from 1-alkenes by Nocardia corallina B-276. Eur J Appl Microbiol Biotechnol. 1981;12:39–45. [Google Scholar]
- 9.Habets-Crützen A Q H, Brink L E S, van Ginkel C G, de Bont J A M, Tramper J. Production of epoxides from gaseous alkenes by resting-cell suspensions and immobilized cells of alkene-utilizing bacteria. Appl Microbiol Biotechnol. 1984;20:245–250. [Google Scholar]
- 10.Hartmans S, Smits J P, van der Werf M J, Volkering F, de Bont J A M. Metabolism of styrene oxide and 2-phenylethanol in the styrene-degrading Xanthobacter strain 124X. Appl Environ Microbiol. 1989;55:2850–2855. doi: 10.1128/aem.55.11.2850-2855.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacobs M H J, van den Wijngaard A J, Pentenga M, Janssen D B. Characterization of the epoxide hydrolase from an epichlorohydrin-degrading Pseudomonas sp. Eur J Biochem. 1991;202:1217–1222. doi: 10.1111/j.1432-1033.1991.tb16493.x. [DOI] [PubMed] [Google Scholar]
- 12.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 13.Miura A, Dalton H. Purification and characterization of the alkene monooxygenase from Nocardia corallina B-276. Biosci Biotechnol Biochem. 1995;59:853–859. [Google Scholar]
- 14.Nöthe C, Hartmans S. Formation and degradation of styrene oxide stereoisomers by different microorganisms. Biocatalysis. 1994;10:219–225. [Google Scholar]
- 15.Sluis M K, Ensign S A. Purification and characterization of acetone carboxylase from Xanthobacter strain Py2. Proc Natl Acad Sci USA. 1997;94:8456–8461. doi: 10.1073/pnas.94.16.8456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sluis M K, Small F J, Allen J R, Ensign S A. Involvement of an ATP-dependent carboxylase in a CO2-dependent pathway of acetone metabolism by Xanthobacter strain Py2. J Bacteriol. 1996;178:4020–4026. doi: 10.1128/jb.178.14.4020-4026.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Small F J, Ensign S A. Alkene monooxygenase from Xanthobacter strain Py2: purification and characterization of a four-component system central to the bacterial metabolism of aliphatic alkenes. J Biol Chem. 1997;272:24913–24920. doi: 10.1074/jbc.272.40.24913. [DOI] [PubMed] [Google Scholar]
- 18.Small F J, Ensign S A. Carbon dioxide fixation in the metabolism of propylene and propylene oxide by Xanthobacter strain Py2. J Bacteriol. 1995;177:6170–6175. doi: 10.1128/jb.177.21.6170-6175.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swaving J, de Bont J A M, Westphal A, de Kok A. A novel type of pyridine nucleotide-disulfide oxidoreductase is essential for NAD+- and NADPH-dependent degradation of epoxyalkanes by Xanthobacter strain Py2. J Bacteriol. 1996;178:6644–6646. doi: 10.1128/jb.178.22.6644-6646.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Ginkel C G, de Bont J A M. Isolation and characterization of alkene-utilizing Xanthobacter spp. Arch Microbiol. 1986;145:403–407. [Google Scholar]
- 21.Weijers C A G M, Jongejan H, Franssen M C R, de Groot A, de Bont J A M. Dithiol- and NAD-dependent degradation of epoxyalkanes by Xanthobacter Py2. Appl Microbiol Biotechnol. 1995;42:775–781. [Google Scholar]